- Department of Microbiology, Carver College of Medicine, University of Iowa, Iowa City, IA, USA
The genes encoding the heavy and light chains of swine antibodies are organized in the same manner as in other eutherian mammals. There are ∼30 VH genes, two functional DH genes and one functional JH gene, 14–60 Vκ genes, 5 Jκ segments, 12–13 functional Vλ genes, and two functional Jλ genes. The heavy chain constant regions encode the same repertoire of isotypes common to other eutherian mammals. The piglet models offers advantage over rodent models since the fetal repertoire develops without maternal influences and the precocial nature of their multiple offspring allows the experimenter to control the influences of environmental and maternal factors on repertoire development postnatally. B cell lymphogenesis in swine begins in the fetal yolk sac at 20 days of gestation (DG), moves to the fetal liver at 30 DG and eventually to the bone marrow which dominates until birth (114 DG) and to at least 5 weeks postpartum. There is no evidence that the ileal Peyers patches are a site of B cell lymphogenesis or are required for B cell maintenance. Unlike rodents and humans, light chain rearrangement begins first in the lambda locus; kappa rearrangements are not seen until late gestation. Dissimilar to lab rodents and more in the direction of the rabbit, swine utilize a small number of VH genes to form >90% of their pre-immune repertoire. Diversification in response to environmental antigen does not alter this pattern and is achieved by somatic hypermutation (SHM) of the same small number of VH genes. The situation for light chains is less well studied, but certain Vκ and Jκ and Vλ and Jλ are dominant in transcripts and in contrast to rearranged heavy chains, there is little junctional diversity, less SHM, and mutations are not concentrated in CDR regions. The transcribed and secreted pre-immune antibodies of the fetus include mainly IgM, IgA, and IgG3; this last isotype may provide a type of first responder mucosal immunity. Development of functional adaptive immunity is dependent on bacterial MAMPs or MAMPs provided by viral infections, indicating the importance of innate immunity for development of adaptive immunity. The structural analysis of Ig genes of this species indicate that especially the VH and Cγ gene are the result of tandem gene duplication in the context of genomic gene conversion. Since only a few of these duplicated VH genes substantially contribute to the antibody repertoire, polygeny may be a vestige from a time before somatic processes became prominently evolved to generate the antibody repertoire. In swine we believe such duplications within the genome have very limited functional significance and their occurrence is therefore overrated.
The Genomic Potential for the Antibody Repertoire in Swine
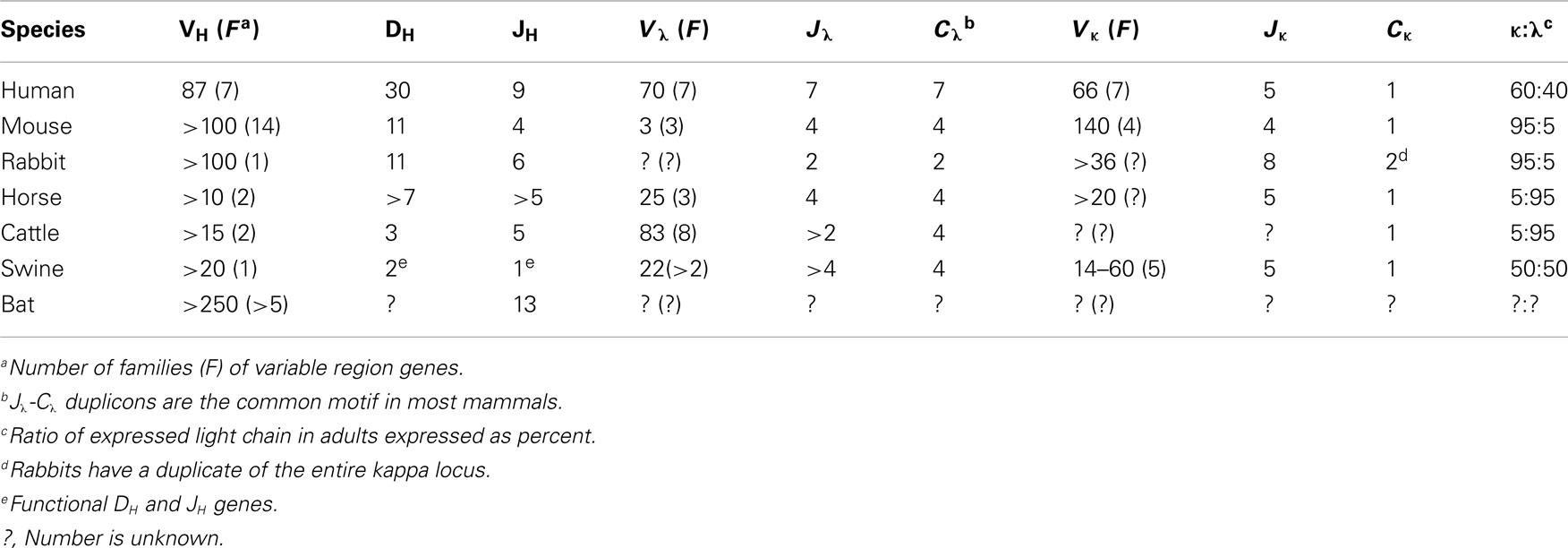
The heavy and light chain genome of swine is organized in the familiar translocon fashion of other eutherian mammals, i.e., placental mammals versus egg-layers. The heavy chain locus contains ∼30 VH genes, all members of the same VH3 family (Sun et al., 1994). There are five DH segments and five JH segments. However only two DH segments and a single JH are functional (Butler et al., 1996; Eguchi-Ogawa et al., 2010; Table 1). Downstream, there are genes encoding Cμ, Cδ, six subclasses of Cγ, Cε, and Cα. Like cattle, Cδ is also associated with a small switch region which may or may not be regularly functional (Zhao et al., 2003; Figure 1A). Putative enhancer and promoter elements and switch regions similar to those described in other mammals have been reported (Sun and Butler, 1997; Eguchi-Ogawa et al., 2010).
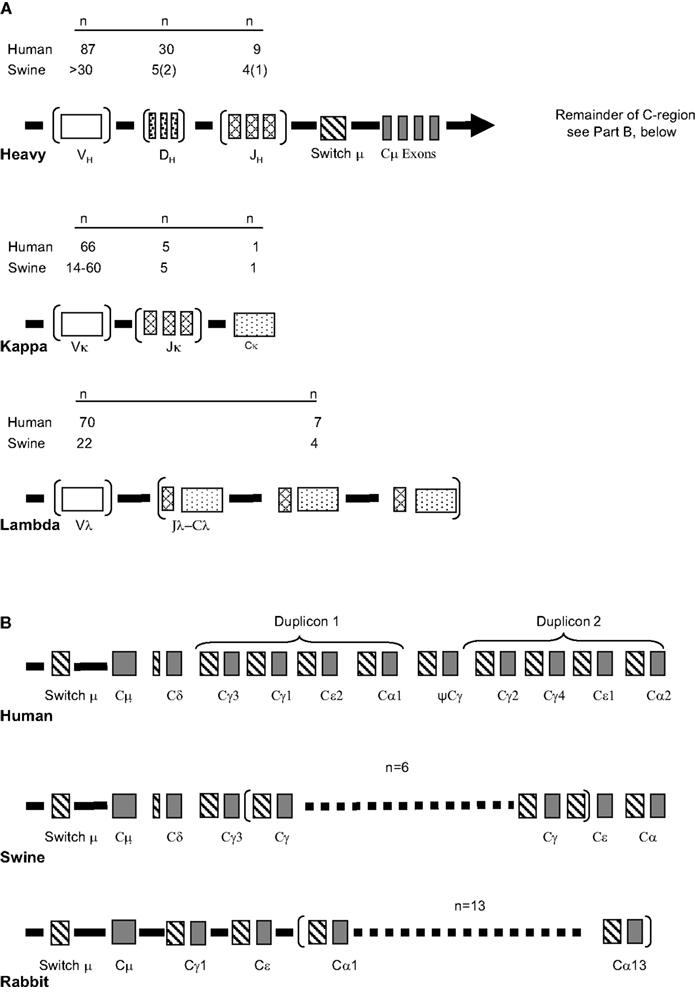
Figure 1. The genomic repertoire of heavy and light chain genes of swine. (A) The light chain and heavy chain variable gene loci compared to humans. Duplicons are illustrated in parentheses with the actual number given in the mini-table above designated as “n.” This includes duplicons of cassettes as in the lambda locus. In the swine only two DH and one JH gene segments are functional (given in parenthesis). (B) Variation in the organization of the heavy chain constant region among human, swine, and rabbit. Modified from Butler et al. (2011c).
The kappa locus is comprised of 14–60 Vκ genes in two families, five Jκ segments and a single Cκ (Figure 1B; Butler et al., 2004; Schwartz et al., 2012a; Table 1). The lambda locus is comprised of 22 Vλ genes, 13 that appear potentially functional and four Jλ genes (Figure 1B; Schwartz et al., 2012b). Usage of kappa and lambda gene elements is discussed in Section “The Pre-Immune Antibody Repertoire of Swine.” The organization of these loci is generally conserved among eutherian mammals although the kappa locus is duplicated in rabbits (Table 1). It is not the purpose of this article to provide a phylogenetic review of the Ig loci of mammals and other vertebrates. We mention only selected species as a reference to help readers less familiar with comparative immunology.
The Piglet Model for Studies of Antibody Repertoire Development
The piglet provides an ideal model for studies on antibody repertoire development for a number of important reasons. First, swine are members of the hoofed mammal group, i.e., Ungulates, which have a form of placentation that, unlike that in rodents, primates, and rabbits, does not allow transport of maternal antibodies and other proteins in utero to the developing fetus (Brambell, 1970; Butler, 1974). Gestation is 114 days which allows 84 days from the time that VDJ rearrangements first appear to study the development of B cells and the antibody repertoire during fetal life in their multiple large fetuses. Because of the placentation described, development during this period is considered intrinsic and not regulated by maternal factors transmitted in utero. Second, the offspring of swine and all Ungulates are Precocial. In the context of development, this refers to the ability of newborn Ungulates to be born totally mobile, with fully functional eyes and protective fur/hair and the ability to immediately forage. Thus piglets can be recovered by Caesarian surgery and reared separately from their mothers in germfree isolators or SPF autosows (Butler et al., 2009a). This allows the experimenter to control the exposure of the postnatal offspring to maternal factors, commensal flora, certain dietary regimes, and infectious agents.
The swine has another major advantage; seven major VH genes, all of the same family, two DH segments and a single JH account for >90% of the VDJ repertoire (Butler et al., 1996; Sun et al., 1998). This is true in the yolk sac (YS) at 20 days of gestation (DG) and continues into adulthood (Butler et al., 2006a, 2011a). Thus all VDJ rearrangements can be recovered from DNA or from transcripts using a single PCR primer set. Cloning these rearrangements and using probes that recognize the CDR regions of each major gene as well as sequence analysis allows vertical studies on the developing repertoire and for its quantification according to a repertoire diversification index (RDI; Butler et al., 2006a, 2011a).
These features collectively allow factors that act during the “critical window” of immune development to be addressed (Figure 2). The critical window in the swine system is the period when innate immunity, “natural antibodies” and passive immunity gain the help of the developing adaptive immune response system to allow offspring survival. Survival of the newborn through this critical period when adaptive responses are poorly developed depends on passive immunity in which the systemic humoral experience of the mother is transmitted via IgG and the mucosal experience by IgA. This is also the time when neonatal tolerance to non-threatening dietary antigens and commensal gut flora become established. These topics are discussed in detail in other reviews (Butler, 1983; Butler and Kehrle, 2005). Relevant to the theme of this chapter is the role played by colonizing gut flora on the development of adaptive immunity and on the diversification of the antibody repertoire.
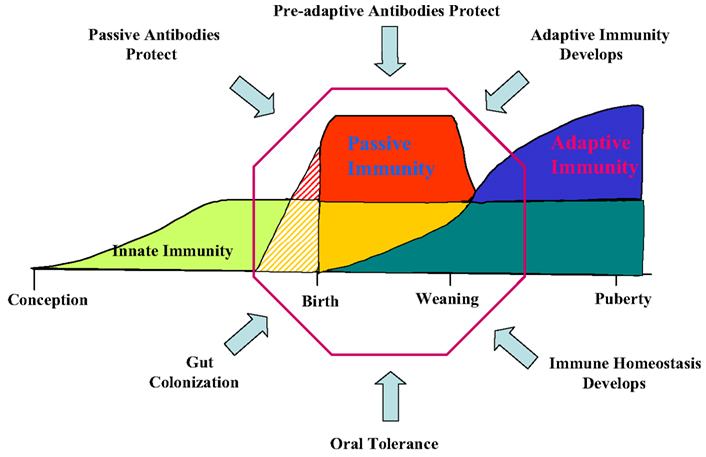
Figure 2. The critical window of immunological development. The factor affecting events in the “window” are indicated. There are a number of cases of superimposing events; e.g., Passive immunity (red) superimposed on innate immunity (green) produces yellow. The “lined section” prepartum applies only to species, e.g., mice and humans, in which passive immunity also takes place in utero. In Ungulates like swine there is no transfer of passive antibodies in utero. Modified from Butler et al. (2006b).
B Cell Development in Swine
Any discussion of antibody repertoire development requires some mention of the B cell lineage from which antibodies are derived. In the fetus, VDJ rearrangements are first seen in YS at 20 DG (Sinkora et al., 2003; Sun et al., 2012a). However signal joint circles (SJC) are difficult to recover at this time, probably because of the slow rate of B cell lymphogenesis at this early time and the rapid rate of their degradation (Figure 3). TdT is expressed and active at this time but N region additions and CDR3 diversity is low compared to older fetuses and young piglets (Sinkora et al., 2003; Butler et al., 2007; Sun et al., 2012a). B cell lymphogenesis moves to the fetal liver at 30 DG and remains active in this organ until the bone marrow (BM) develops at ∼65 DG. SJC are readily detectable at all these sites indicating that it unlikely the B cells found in these sites are immigrants. An interesting feature of heavy chain rearrangements in early B cell lymphogenesis is the unexpected high frequency (>85%) of in-frame rearrangement (Sinkora et al., 2003), suggesting that the rearrangement and selection processes differs between early fetal stages of B cell lymphogenesis and those which occurs later in fetal and adult BM. However, evidence for B-1 and B-2 subpopulations as reported in mice (Herzenberg et al., 1986) has so far not been obtained. We also found no evidence that the ileal Peyer patches (IPP) are a site of B cell lymphogenesis or that they are needed for B cell maintenance (Butler et al., 2011b; Sinkora et al., 2011; Sun et al., 2012a).
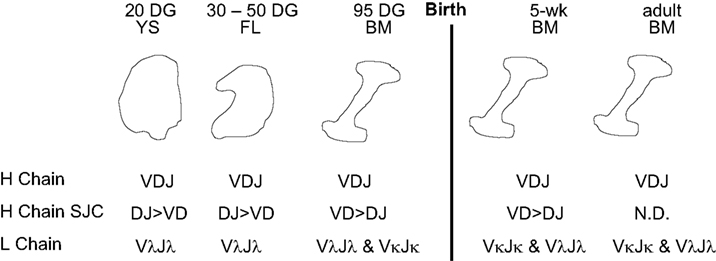
Figure 3. B cell lymphogenesis in swine. Heavy and light chain rearrangements and signal joint circles (SJC) recovered from. YS, yolk sac; FL, fetal liver, BM, bone marrow; DG, day of gestation; N.D., not detected.
A major departure from the pattern seen in lab rodents and humans is the order of light chain rearrangement. Rearrangement begins in the lambda locus at 20 DG in YS and is dominant until in late gestation when kappa rearrangements first appear (Figure 3; Sun et al., 2012a). This may not be unusual given that in Ungulates, lambda usage dominates the repertoire; in cattle, sheep, and horse and it can exceed >90% (Butler, 1997; Table 1). However usage of light chain isotypes in young pigs and adults is roughly equal, similar to humans (Hood et al., 1967; Skvaril et al., 1976; Butler et al., 2005a; Sun et al., 2012a; Table 1). The order of light chain gene segment usage during development has been poorly studied in any Ungulate. However, the IGLV8 (Vλ8) family is exclusively used at early sites of B cell lymphogenesis (Vazquez et al., 2012).
It has been proposed that B cell development among higher vertebrates places species into groups (Lanning et al., 2004). In rodents and primates the BM is the major site of B cell development and diversification whereas in rabbit and sheep, gut associated lymphoid tissues (GALT) are believed to be critical (Lanning et al., 2004; Mage et al., 2006). However, recent findings indicate that swine do not belong to the GALT group (Butler et al., 2011b; Sinkora et al., 2011). This is based on surgical resection of IPP, which did not affect B cell lymphogenesis, repertoire diversification and maintenance of either B or T cell levels. The swine IPP appears to be merely a type of mucosal immune tissue.
Some level of class-switch recombination (CSR) occurs during fetal life, so that IgM, IgG3, and IgA are transcribed and secreted into serum. However, no environmental antigen is present during this time and it is unknown whether this CSR involves germinal center formation. There is also low level somatic hypermutation (SHM) in rearranged VDJs resulting in <10 mutation per kilobase (Butler et al., 2011a). In heavy chain rearrangement these mutations accumulate in CDR regions. In lambda rearrangement mutations are equally distributed in CDR and FR regions, although the total mutation frequency is 10-fold lower than in heavy chain rearrangements (Vazquez et al., 2012). Tests for expression of AID during fetal life have not been undertaken.
The Pre-Immune Antibody Repertoire of Swine
We define the pre-immune repertoire as the one present in fetal and newborn piglets. Conventionally reared swine have been previously exposed to both environmental antigen and regulatory influences from maternal passive antibodies, but isolator piglets are free of such influences. In their circulation, newborns have ∼ 30 μg/ml of IgG, 1 μg/ml of IgM, and 2 μg/ml of IgA (Butler et al., 2001, 2009b). If maintained for 5 weeks in germfree conditions in which dietary protein is the sole source of environmental antigen, IgM and IgA levels increase 3- to 5-fold to ∼ 6 μg/ml, but there is only a 20% increase in IgG to 40–45 μg/ml. Since these animals are immuno-unresponsive (see below) we suspect this small increase is an intrinsic developmental effect much as is the CSR and SHM that occur during fetal life. We base this on the fact that isolator piglets that become immunoresponsive have a 20- to 35-fold increase in serum Ig across all major isotypes. Of the six subclasses of IgG, IgG3 accounts for >60% of the Cγ transcripts in mucosal tissues during the period prior to environmental exposure (Butler and Wertz, 2006).
A notable variation on the theme of repertoire development and diversification from what is presented in textbooks and reviews concerns VH gene usage. Swine use 7 VH genes to form nearly their entire VDJ repertoire starting first in YS and continuing throughout gestation and beyond into postnatal life (Figure 4; Butler et al., 2011a). With minor exceptions, proportional usage remains constant and the usage does not depend on the position of the VH gene in the genome (Eguchi-Ogawa et al., 2010; Butler et al., 2011a). VHG (IGHV2) which is the most 3″ functional gene is seldom used whereas VHN (IGHV15) can account for 13% of the repertoire in YS at 20 DG (Butler et al., 2011a; see Antibody Repertoire Development and the Origin of the Genomic Repertoire). Exceptions to the constancy of VH usage involves decreased usage of VHN in older fetuses and a reciprocal increase in usage of VHC. As will be discussed in more detail in the next section (see Development of Adaptive Immunity Depends on an Encounter with MAMPs) this constancy of VH usage continues after birth in antigenized animals including those reared conventionally and consequently exposed to a plethora of environmental antigens. There is no consistent change in the frequency of usage between the two DH segments and swine have only one functional JH (Butler et al., 1996; Eguchi-Ogawa et al., 2010). The small number of VH, DH, and JH segments used means that combinatorial diversity is very small, i.e., 14 possibilities compared to ∼9000 in humans. Thus, we estimated that junctional diversity in CDR3 accounts for >95% of the swine pre-immune repertoire (Butler et al., 2000a). As indicated above, the frequency of SHM remains low during this period but the complexity of CDR3 is high and the Gaussian spectratype pattern of CDR3 lengths suggests an unselected repertoire (Navarro et al., 2000; Butler et al., 2007).
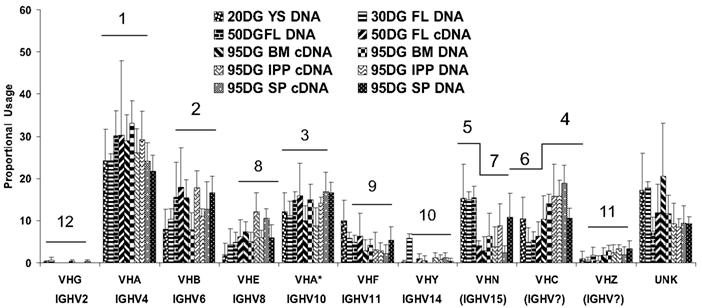
Figure 4. The proportional usage of 11 porcine VH genes during fetal life. VHB and VHB* (IGHV6 and IGHV12) are considered as a group in this analysis. Both the familiar and IMGT nomenclature (if available) are given. Data are based on >5500 VDJ clones and were analyzed using a computer modeling program. The horizontal bars and numbers above each VH gene correspond to the frequency of usage; 1 = highest frequency. For VHN and VHC, the order changes during development. UNK includes VH genes other than those shown and mutated versions of those shown. From Butler et al. (2011a).
As indicated in Section “The Genomic Potential for the Antibody Repertoire in Swine,” the overall genomic structure of the kappa and lambda loci of swine is similar to that in other well-studied mammals. Genomic gene annotation may not totally predict the expressed repertoire. In the case of kappa, 11 functional Vκ genes and 5 Jκ genes are present in the genome, yet 2 Vκ families and 1 Jκ segment account for most of the repertoire (Butler et al., 2004; Schwartz et al., 2012a). In the case of lambda, Vλ usage appears to closely agree with the genomic potential but only two of the four Jλ genes are used (Schwartz et al., 2012b; Vazquez et al., 2012).
Certain features of the pre-immune light chain repertoire differ substantially from those in the heavy chain but these features are not unique to swine. First, SHM is 10-fold lower in light chain rearrangement than in heavy chain rearrangement of piglets from the same population. Furthermore there is little junctional diversity in kappa or lambda rearrangement and CDR3 length is ∼30 ± 2 (Butler et al., 2004; Vazquez et al., 2012). These observations are nearly identical to studies in humans (Victor et al., 1994; Bridges et al., 1995; Girschick and Lipsky, 2001; Richl et al., 2008). Of some notice is that in the pre-immune repertoire, SHM is concentrated in the CDR region of heavy chain rearrangements but both are lower and widely distributed in rearranged light chains (Butler et al., 2006b, 2011a; Vazquez et al., 2012). The significance of this difference is unclear. However, one might speculate that because the antibody binding site is primarily determined by the heavy chain and its CDR3 (Padlan, 1994) light chain may provide only a supporting role and their presence primarily affects the conformation of the heavy chain binding site. Their complete absence in the camelids partially supports this view (Nguyen et al., 2002).
Development of Adaptive Immunity Depends on an Encounter with MAMPs
Colonization and MAMPs
Piglets maintained germfree in isolators for 5–6 weeks show only minor changes in serum Ig levels until after colonized (Butler et al., 2000b, 2009b; see The Pre-Immune Antibody Repertoire of Swine), do not have antibodies to dietary proteins (J.E. Butler and Patrick Weber, unpublished observations) and are unable to respond to T cell dependent (TD) and T cell independent (TI-2) antigens. However, colonization with benign Escherichia coli or a probiotic cocktail, allows responses to both types of antigens (Butler et al., 2002). In lieu of living bacteria, purified MAMPs (bacterial DNA as CpG-ODN, muramyl dipeptide or LPS) have the same affect (Butler et al., 2005b). Thus, bacterial MAMPs provide the adjuvant necessary for innate immune receptors to stimulate the development of adaptive immunity. The impact of such exposure results in 100- to 1000-fold increase in serum Igs (Butler et al., 2009b), CSR to downstream Cγ genes, (Butler et al., 2012a) a 3- to 5-fold increase in the frequency of SHM and a 1–2 log increase in the RDI (Butler et al., 2011a). CpG-ODN and LPS are polyclonal B cell activators and can also expand the existing B cell populations to secrete IgM, IgA, and IgG3 antibodies. However, such expansion cannot be considered a “somatically adapted” repertoire.
Repertoire Diversification Following Infection with RNA Viruses
Viruses have a broad range of effects on adaptive immunity. Some are polyclonal activators while others suppress immune responses by interfering with antigen presentation by a variety of mechanisms (Coutelier et al., 1990; Ehrlich, 1995; Hahn et al., 1998; Acha-Orbea et al., 1999; Hunziker et al., 2003). However some, such as influenza (FLU), stimulate robust antibody responses, the apparent basis of generally high efficacy FLU vaccines. Such viruses generate dsRNA during replication, a known adjuvant (Cunnington and Naysmith, 1975). In piglets, we have studied three pandemic viruses including swine influenza (S-FLU) and another RNA virus called porcine respiratory and reproductive syndrome virus (PRRSV) which acts as a polyclonal activator of B cells in both germfree and colonized piglets and fetuses inoculated in utero. Polyclonal activation by PRRSV results in lymphoid adenopathy, hypergammaglobulinemia, the appearance of autoantibodies and the deposition of immune complexes in kidney (Lemke et al., 2004). Infection with PRRSV expands certain B cells clones that display hydrophobic CDR3s, a feature common to antibodies that comprise the pre-immune repertoire (Butler et al., 2007, 2008; Schelonka et al., 2007). In addition to polyclonal activation there is also diversification of the repertoire, not dissimilar from that seen in piglets infected with S-FLU or colonized with gut flora (Figure 5). However, the degree of repertoire diversification does not parallel the increase in serum Igs (Sun et al., 2012b). This results in only a very small proportion of virus-specific Igs (Lemke et al., 2004).
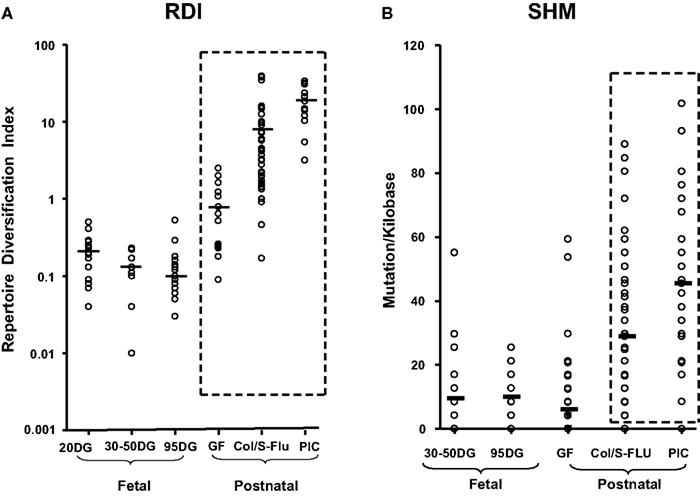
Figure 5. Diversification of the porcine antibody repertoire during fetal and postnatal life expressed as a repertoire diversification index [RDI; (A)] or as the frequency of somatic hypermutation [SHM; (B)]. DG, days of gestation. Values for 95 DG are pooled from clone frequencies recovered from spleen, IPP, and MLN since there were no tissue differences. GF, germfree isolator piglets; Col S-FLU, colonized or S-FLU infected isolator piglets; PIC, parasite-infected young adults reared conventionally. From Butler et al. (2011a).
Infection of isolator piglets with S-FLU results in a robust IgG response to the virus but a much weaker response to irrelevant model TI-2 and TD antigen than does gut colonization (Butler et al., 2012a). Thus, S-FLU does not have the same robust adjuvant impact as does bacterial colonization. This may be due to the fact that S-FLU offers primarily one TLR ligand, double-stranded RNA or that the gut innate immune system is more responsive than that of the respiratory tract. In any case, the result of S-FLU infection supports the concept that MAMPs awakens the adaptive immune system.
A third pandemic disease of swine is porcine circo virus Type 2, a small DNA virus (PCV2; Allan and Ellis, 2000; Merial, 2004). In the piglet model, PCV2 has little or no adjuvant effect in terms of stimulating the production of antibodies to irrelevant model TI-2 and TD antigens. However infection with PCV2 nevertheless results in a generally robust response to a recombinant ORF2 antigen of the virus (Sun et al., 2012c). This IgG response in serum occurs in the context of elevated IgA levels in serum and bronchial-alveolar lavage (BAL). PCV2 infection does not result in polyclonal B cell activation, rather it targets IgA-producing cells. These findings further emphasize that the humoral immune response to any one particular viral infection, should not be projected to other viral infections.
Diversification of the Heavy Variable Region Repertoire is by SHM of VH Genes that Comprise the Pre-Immune Repertoire
The user friendly and simple VDJ system of swine combined with the other advantages of the piglet model (see The Piglet Model for Studies of Antibody Repertoire Development) make it possible to follow the developmental history of the seven major VH genes that comprise >90% of the pre-immune repertoire in postnatal animals under various environmental conditions. The surprising result is that exposure to environmental antigen does not result in the recruitment of other VH genes from the genomic repertoire that are not present in the pre-immune repertoire. Rather, somatic mutants of the “magnificent seven” VH genes that comprise the pre-immune repertoire comprise the adaptive repertoire (Figure 6). In piglets infected with S-FLU, colonized with bacteria or helminth parasites, the same VH genes comprise the repertoire but ∼90% of them are somatically mutated (Butler et al., 2011a; Figure 6). The use of somatic gene conversion, prominent in the rabbit (Knight, 1992; Schiaffella et al., 1999; Winstead et al., 1999) has not been observed in swine.
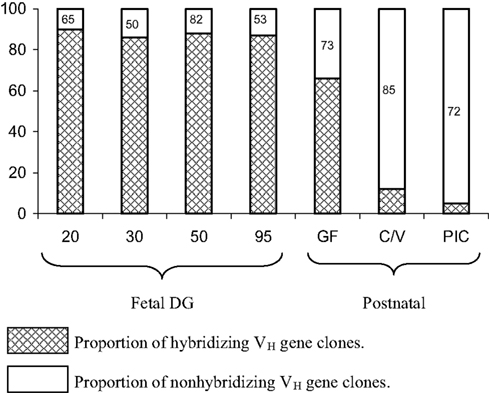
Figure 6. The proportion of VDJ clones from fetal and postnatal piglets that hybridize with cocktails that contain probes for the CDR1 and CDR2 regions of the major seven VH genes used to form the pre-immune repertoire (see Figure 4). Legend for hybridization/non-hybridization is on the figure. Failure to hybridize means that either other VH genes are used or the major seven VH have been mutated to the extent that they no long hybridize with the CDR-specific probes. The number within the bar representing non-hybridizing VH genes indicates the proportion that are the major VH genes as determined by sequence analysis. This means, e.g., that in C/V piglets, 85% are a mutated version of the seven major genes. GF, germfree; C/V, colonized and/or virus infected; PIC, parasite-infected conventional pigs.
Environmental Exposure Results in CSR to Downstream Cγ Genes
We described above that in naïve newborns and fetal piglets, >60% of IgG transcripts encode IgG3. However, fetal infection or postnatal exposure to virus, normal gut flora or parasitic infection reduces IgG3 transcription to ∼5% (Butler et al., 2012a). The resultant IgG is now encoded by downstream Cγ genes of which IgG1 is a major player (Butler and Wertz, 2006). The switch to downstream Cγ genes (Figure 7A) parallels the diversification of their VH genes while the repertoire associated with IgM and IgG3 does not diversify (Figure 7B). These observations are further evidence that environmental exposure of fetal and newborn piglets turns on the machinery of the adaptive immune system. However, since IgA and IgG3 are already transcribed and secreted by the fetus, environmental exposure is not obligatory for CSR.
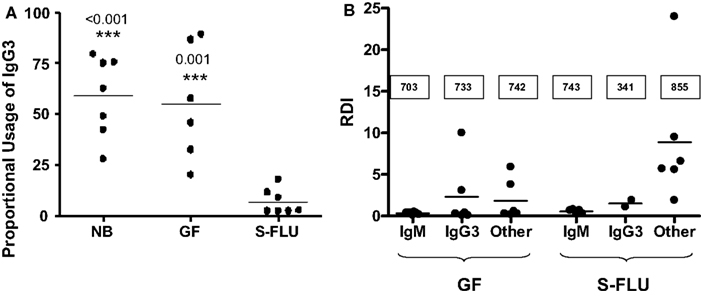
Figure 7. Adaptive changes to the antibody repertoire as a result of postnatal infection with S-FLU. (A) Effect on IgG3 transcription in the tracheal-bronchial lymph node (TBLN). NB, newborn; GF, 5-week germfree isolator piglets; S-FLU, 5-week S-FLU infected isolator piglets. (B) The repertoire diversification index (RDI) for VH genes transcribed with IgM, IgG3 and downstream Cγ transcripts (called: “other”) in GF and S-FLU infected piglets. The boxed values are the number of clones examined. Data indicate that the IgM and IgG3 repertoire does not diversify in S-FLU infection.
The Swine Versus Established Paradigms of Antibody Repertoire Development
The paradigms for development of the mammalian antibody repertoire decorate the pages of immunology textbooks and are embodied in a number of classic reviews (Rajewsky et al., 1987; Cohn and Langman, 1990). Discussed in this section is how well the swine system fits these paradigms. In the swine system, some level of CSR and SHM occurs in the absence of environmental exposure. This may also be the case in lab rodents and in human, but is ambiguous in these species because such changes could result from the regulatory effects of maternal antibodies (Wikler et al., 1980; Rodkey and Adler, 1983; Yamaguchi et al., 1983; Wang and Shlomchik, 1998) or may be the result of antigen trafficking across the maternal-fetal barrier (Tristram, 2005). In any case, newborn piglets enter the world with a “natural antibody repertoire” of IgM, IgA, and IgG3 antibodies; a phenomenon that is probably universal among all vertebrates (Ochsenbein and Zinkernagel, 2000).
In swine, the evolution of antibody repertoire development appears to have followed a somewhat different path than in mice and humans. However, we have been unable to identify vertical studies on VH usage in rodents or humans, equivalent to those done in swine to thoroughly confirm these differences (Sun et al., 1998; Butler et al., 2011a). In swine, proportional VH gene usage appears to be constant from the time of the initial B cell development in the YS to adulthood (Butler et al., 2011a). Adaptive diversification of the repertoire depends on SHM of the same major VH genes that comprise the pre-immune repertoire. In humans and mice, adaptive responses are often ascribed to the selective use of certain VH genes (Sheehan et al., 1993; Glas et al., 2000). Despite these differences between mice and swine, there are many similarities. For example, repertoire diversification is by SHM with mutations accumulating in the CDR regions (Berek and Milstein, 1987; Butler et al., 2006a, 2011a) with no evidence of somatic gene conversion as has been reported in the rabbit and chicken (Reynaud et al., 1987; Becker and Knight, 1990; Schiaffella et al., 1999; Winstead et al., 1999; Ratcliffe, 2006). Consistent with studies in humans, light chains provide limited diversity (see The Pre-Immune Antibody Repertoire of Swine). Also similar to mice and human is the duplication of the Cγ genes and the changes in their expression upon antigenic stimulation (Mossman and Coffman, 1989). This of course differs from lagomorphs that have a single Cγ gene but 13 genes for IgA that comprise their repertoire (Burnett et al., 1989; Figure 1). Like humans and rodents, there is no apparent equivalent to the specialized IgG1 of ruminant artiodactyls that is believed to be essential for passive immunity from mother to young, and which also appears to function as a mucosal antibody (Butler, 1983; Butler and Kehrle, 2005). In regard to mucosal immunity, swine have essentially the same IgA-dependent system as rodents and humans including a well-developed gut-mammary gland axis. Both swine and rodents lack the specialized long-hinged IgA1 of humans and primates, which is especially susceptible to bacterial proteases (Plaut et al., 1974) although H. suis produces an unrelated protease that cleaves both porcine IgA allotypic variants and may well cleave the IgA of most mammals (Mullens et al., 2011).
At this point, information on diversification of the light chain repertoire of swine may be inadequate. At this time there is little to suggest that only a small number of Vλ or Vκ genes comprise the diversified repertoire of the light chains in the manner we have described for the VH genes in this species. Rather, a much larger array of Vk and Vλ genes are used (Butler et al., 2004; Vazquez et al., 2012). However, like mice and humans, length junctional diversity in the light chain repertoire is restricted (Victor et al., 1994; Bridges et al., 1995; Girschick and Lipsky, 2001; Richl et al., 2008). But unlike the camelids, another Ungulate, light chains have not become obsolete (Hamers-Casterman et al., 1993; Nguyen et al., 2002). There is much to favor the idea that light chains help stabilize the heavy chain binding site and allow specificity modification and therefore the rescue of autoreactive B cells through receptor editing (Tiegs et al., 1993).
As regards the adjuvant effect of bacterial and viral MAMPs that act on innate immune receptors, this is most likely a universal phenomenon among higher vertebrates that go on to develop an effective adaptive immune system. In any case, this topic falls outside the main theme of this review.
Antibody Repertoire Development and the Origin of the Genomic Repertoire
Origin and Importance of the Genomic Repertoire
Discussion of the expressed antibody repertoire should also consider its genomic origin. The presence of an annotated gene is not alone evidence for its function; the many breeds of dogs are a poignant example. Insight into why the genomic repertoire of Ig genes greatly exceeds the functional repertoire may help to explain their phylogeny. This can help to explain the redundancy that is a feature of the genomic repertoire which can assure an effective adaptive immune system among higher vertebrates.
Duplication and Genomic Gene Conversion Explains the Porcine VH and Cγ Genomic Repertoire
Figure 8 aligns the sequences of a number of porcine VH genes to show that with minor exceptions, they only differ in their CDR1 and CDR2 regions. Thus, the genomic repertoire is a “mix and match” CDR potpourri, with CDR regions shared among different VH genes. Certain VH genes like VHA and VHA* (IGHV4 and IGHV10) and VHB and VHB* (IGHV6 and IGHV12) are duplications with several mutation; one in the CDR1 region of VHB* and two in FR3 of VHA*. In fact there is evidence that the genes segments encoding these duplicated VH genes and several others were duplicated as a block (Eguchi-Ogawa et al., 2010) similar to the duplicons in the human heavy chain constant region and the JλCλ loci of all studied mammals (Figure 1B). These features of duplicated genes are also seen among the VH genes of bats (Bratsch et al., 2011; Butler et al., 2011d). These comparisons support the hypothesis that the germline VH repertoire in swine results from gene duplication that occurred simultaneously with genomic gene conversion. The same mechanism appears evident when the sequences of the six expressed porcine Cγ genes and their allotypic variants are compared (Figure 9). For example, the allotypic variants of IgG1 and IgG4 have the same hinge exons and the hinge of IgG5a is shared with the allelic variants of IgG6. With exception of IgG5b and IgG3, the Cγ1 domain of all Cγ subclass genes and their alleles are identical.
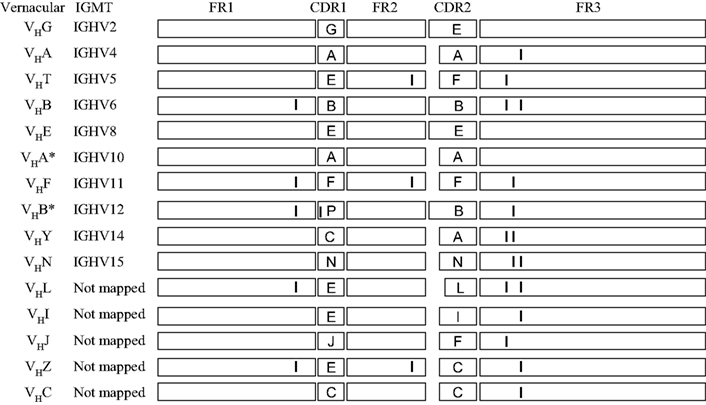
Figure 8. Evidence in support of duplication of VH genes in swine. Diagrammatic comparison of 16 of 32 VH3 family genes described for swine. The sequences are drawn to approximate scale. CDR sequences are identified by capital letter. Only the first fifteen 3′ VH genes have been mapped; pseudogenes are not shown. Those VH genes with the same A,B, etc. CDR designations share the same CDR1 or CDR2 regions. In the case where small differences (genomic point mutations) in FR and CDR regions are involved, vertical lines are drawn.
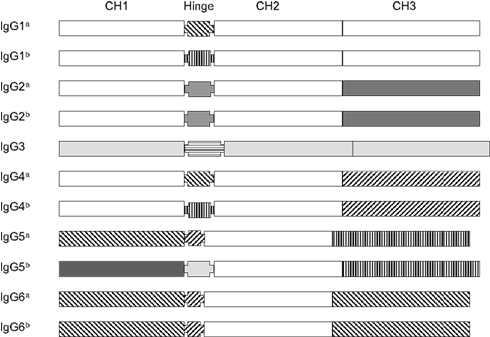
Figure 9. The gene structure of 11 porcine Cγ genes representing the six expressed IgG subclasses and allotypes of swine. Different marking patterns are used to identify domain sequences. Domains with the same pattern indicate they are shared among Cγ genes. Note that no domains are shared with IgG3.
Dendogram analyses of sequence data that are often called phylogenetic studies, indicate that IgG3 is the ancestral IgG for swine. This and other evidence indicates that subclass diversification occurred after speciation (Kehoe and Capra, 1974; Nguyen, 2001; Butler et al., 2009c). We believe the other five Cγ genes then diversified from IgG3. Convincing evidence could be best obtained by studying the Cγ subclasses of other swine related species that are related to the ancestors of the domesticated pig. Consistent with studies on human Cγ (LeFranc et al., 1991), there is also evidence for deletion of some Cγ genes in some pigs (Butler and Wertz, 2006; Eguchi-Ogawa et al., 2012).
The Restricted Use of VH Genes in Swine Questions the Importance of Combinatorial Diversity
As shown in Table 1, lab rodents, rabbits, humans, and bats have and/or express large numbers of VH genes; the same is true for the more primitive Zebrafish (Weinstein et al., 2009). In swine and ruminant artiodactyls, the number is surprisingly low. In swine with ∼30 VH genes, only seven appear to be used/needed for a healthy adaptive immune system. Many of the others may represent only allelic variants, so the actual number of germline VH genes may be <20. This raises a question about the textbook paradigms of the importance of multiple VH genes and combinatorial diversity in generating a protective antibody repertoire. This was tested by Xu and Davis (2000) who used a mouse with a single functional VH gene, but with an intact DH and JH region. They showed this transgenic mouse could make antibodies to nearly all environmental antigens. This can be interpreted to mean that extensive duplication of VH genes is not required for the ability to make antibodies to many specificities. This may explain why ruminant artiodactyls and swine have a small genomic repertoire yet are among the most successful mammals on the planet (Table 1). However the Xu and Davis work also suggests that the critical feature is DH and JH polygeny, which invokes the next question of how swine generate diversity with only two functional diversity segments and a single JH. This brings the focus to junctional diversity in the formation of CDR3 and away from the emphasis on the number of functional VH, DH, and JH segments. CDR3 is considered most important for the specificity of antibodies (Padlan, 1994), whether generated from a genome with many available DH and JH segments, or from a genome with just one or several DH and JH segments. Perhaps this is compensated by light chain diversity, yet the camelids do it without the assistance of light chains (Hamers-Casterman et al., 1993; Nguyen et al., 2002).
Is the Diversification of the IgG Subclasses in Mammals Really Necessary for Species Survival?
Textbooks and reviews emphasize the importance of the division of labor among different antibody isotypes; each constant region permitting some special biological function, i.e., following the accepted concept of structure function relationship. While this is clear for IgM, IgE, IgA, and IgG, applying this to the subclasses of IgG is less convincing, especially in species like the horse, swine, and bats that have large numbers of Cγ variants while the highly successful rabbit lacks subclass variants. We also know that IgD knockout mice behave normally (Nitschke et al., 1993), rabbits lack IgD altogether (Lanning et al., 2003) and even mammals with a gene for IgD apparently do not express it. In humans, deficiencies of individual IgG subclasses are well known but these have not translated to an effect on human health, even in environmentally stress underdeveloped countries (LeFranc et al., 1983; Olsson et al., 1993; Rabbani et al., 1995). In mice, differential IgG subclass expression is related to the balance between inflammatory and regulatory cytokine (Mossman and Coffman, 1989) but there is little evidence that the resulting subclasses make a difference in protective immunity; i.e., does the absence either of IgG1 or IgG2b antibodies really affect protective immunity? Four decades of phenomenological studies in the veterinary world would suggest that the IgG1 subclass is indispensable for passive immunity in ruminant artiodactyls. However, an IgG1 knockout cow or ewe has not been produced to experimentally test this assumption. The same is true in horses in which antibodies of different IgG subclasses have long been described, seven IgG subclasses are known from gene sequences but evidence in support of a unique role for each of these subclass antibodies in the horse immune response is lacking. The study of the many IgG subclasses in swine is only in its infancy because until recently, the subclasses had not been defined (Butler et al., 2009c; Kloep et al., 2012). This lack of progress has been largely due to the lack of IgG subclass reagents for use in immunoassays and, of course, the lack of subclass knockout animals. A solution to the first problem is now underway (Butler et al., 2012a,b)
The point to be made is that with the possible exception of ruminant IgG1, evidence is not strong that subclass diversification of IgG is really necessary for survival of the host especially since rabbits accomplish this with one gene for IgG. Perhaps the IgA polygeny of rabbits compensates for this “deficiency,” but there are no data that address this point and evidence for a unique role for each of the 13 different IgA subclasses in rabbits is missing.
The Origin of VH and Cγ Polygeny
As early as 1932, gene duplication was discussed as an essential feature of the evolutionary process (Haldane, 1932; Bridges, 1936; Ohno, 1970). Various mechanisms are known or proposed including RNA/DNA transposition, non-homologous crossing over, and even entire gene duplication (Woody and McConkey, 2011). The first two are often referred to as genomic gene conversion, although this term seems most appropriate to explain non-homologous crossing over (Meselson and Radding, 1975; Szostak et al., 1983). In humans, mouse and rat genomes, ∼15% of all genes represent tandemly arrayed genes (Shoja and Zhang, 2006). Tandem duplication also creates redundancy (Li et al., 2005) and this is what is seen among Ig genes, especially those encoding the variable heavy and light chain loci and in some species the Cγ genes. Tandem arrayed duplicons are quite often conserved among species (Zhang, 2003); a phenomenon that also appears true for the Ig variable region genes of swine (Eguchi-Ogawa et al., 2010). Since tandem duplicates are mainly attributed to non-homologous cross-overs, many or most emerge as functional genes (Woody and McConkey, 2011). During evolution these duplicons appear to be retained in the genome despite their apparent redundancy (Xue and Fu, 2009). Duplication reduced selective pressure on single genes since one or several of the duplicated “offspring” genes can more rapidly accumulate mutations and therefore assume new functions while not comprising the function of the parent gene (Ohno, 1970; Lynch and Conery, 2000; Kondrashov et al., 2002).
While the exact mechanism remains unknown, evidence points to non-homologous recombination (gene conversion), acting together with gene duplication, as the mechanism for VH and Cγ polygeny in mammals. This is consistent with studies that some proportion of these duplicated genes will be non-functional, i.e., pseudogenes (Wolfe and Shields, 1997). This is specifically seen among the tandem duplicons in the VH, Vκ, Vλ loci, and among especially the Cγ genes of the constant heavy chain sublocus in all higher vertebrates. Among the duplicated VH and Cγ gene in swine, the 5′ region that encodes FR1 or CH1 respectively, is most conserved among duplicons and among species. While this might suggest progressive 5′ to 3′ mutation of the duplicated genes during evolution, this does not seem to be the case for the for VH and Cγ genes of swine. For example, the hinge regions of Cγ genes are most variable in swine and other mammals (Figure 9; Butler et al., 2009c). While the FR regions of porcine VH genes are nearly identical, the CDR1 and CDR2 segments differ and are shared. Thus the CDR regions of VH genes and the hinge segment of Cγ genes are perhaps the best candidates for genomic gene conversion rather than point mutation during evolution.
SHM Reduces the Value of Variable Region Gene Polygeny
Most evolutionary genetic studies of protocaryotic and eucaryotic species focus on changes in the genome that are transmitted to the offspring in Mendelian fashion. Studies on adaptive immunity, a system most developed in mammals, introduced a new mechanism for variability, namely somatic generation of antibody specificity. This process involves somatic rearrangement of gene segments, additions/subtractions of nucleotide at the boundaries of rearranged segments plus SHM of these rearrangements. The rate of the latter is several logs greater than for the mutations that accumulate in the genome.
Well-developed adaptive immune systems that follow this pattern are primarily restricted to mammals and the chicken and are associated with the expression of AID (see below). Such somatic processes are difficult to identify in cartilaginous and bony fishes and are present at reduced frequency in amphibians (Du Pasquier et al., 1998, 2000). At least SHM and CSR in mammals are associated with the formation of germinal centers that involves the expression of a member of the APOBEC family called antigen-activated cytidine deaminase (AID; Honjo et al., 2002). In lieu of this feature, elasmobranches have ∼200 cassettes of fused V-D-J-C genes in their genome although there are some variations on this theme (Dooley and Flajnik, 2006). The genome of Xenopus tropicalis contains 11 VH gene families and 37 Vλ genes encoded at three loci (Qin et al., 2008). Among eutherian mammals, some bats have >250 VH genes, lab rodents and human ∼100, and rabbits ∼200 (Bratsch et al., 2011; Table 1). Many of these VH genes are known to be pseudogenes, consistent with data on tandem gene duplication (Wolfe and Shields, 1997). In general, bats (Chiroptera) and rodents which are considered primitive eutherian mammals, have more VH genes whereas Ungulates, that emerged later, have few VH genes (Table 1). This is superficial support for the hypothesis that the generation of antibody diversity by SHM or somatic gene conversion has minimized the need for large numbers of variable region gene segments and the importance of combinatorial diversity in more recently evolved mammals.
We believe that vertebrates initially evolved the need for multiple VH genes to create a repertoire of specific antibodies but with the subsequent development of somatic rearrangement and especially SHM, the needs for such polygeny became increasingly redundant. Thus, tandemly duplicated VH genes remain in the genome as evolutionary relics.
Lessons from Studies on Antibody Repertoire Development in Piglets
The use of the piglet model to study antibody repertoire development has provided useful information on the B cell system of this species that often differs from the textbook models describing the process. Thus, the basis of the title of the review. It is also inconsistent with proposals that place hoofed mammals in the GALT category in which development of the B cell repertoire depends on hindgut lymphoid tissue (Lanning et al., 2004). Collectively considered, our studies indicate that:
(1) Diversity of the pre-immune repertoire in swine is almost exclusively dependent on junctional diversity in CDR3 of the heavy chain followed by SHM, since only seven VH genes with shared CDRs, two DH genes and one functional JH comprise the functional repertoire and such diversity is greatly restricted in light chain rearrangements.
(2) Repertoire diversification after antigen encounter is by SHM primarily in the CDR regions of the rearranged heavy chain variable regions. Evidence for somatic gene conversion, junctional diversity or SHM in light chains that might contribute to extensive antibody repertoire diversification is lacking.
(3) Light chain rearrangement occurs first in the lambda locus and there is no evidence of a special Vλ gene like λ5 that is used in the earliest phases of B cell lymphogenesis. This raises the question of whether the conventional pre-BCR is present in swine or any artiodactyls.
(4) The prominent hindgut lymphoid tissue of swine, the IPP, is not required for B cell lymphogenesis, maintenance of B cell levels or repertoire development. This opens the question regarding the true function of this lymphoid organ. Here we propose it represents “first responder” mucosal lymphoid tissue. A key factor in this scenario is that IgG3, which is encoded by the most 5′ Cγ gene, shares this same genomic feature with mice, cattle, and humans. Perhaps this primordial IgG provides the “natural antibodies” that target the polysaccharide antigens of intestinal bacteria.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Acha-Orbea, H., Finke, D., Attinger, A., Schmid, S., Wehrli, N., Vacheron, S., Xenarios, I., Scarpellino, L., Toellner, K. M., MacLennan, I. C., and Lutter, S. A. (1999). Interplay between mouse mammary tumor virus and the cellular and humoral immune response. Immunol. Rev. 168, 287–303.
Allan, G. M., and Ellis, J. A. (2000). Porcine circoviruses: a review. J. Vet. Diagn. Invest. 12, 3–14.
Becker, R. S., and Knight, K. L. (1990). Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbit. Cell 63, 987–997.
Berek, C., and Milstein, C. (1987). Mutation drift and repertoire shift in the maturation of the immune response. Immunol. Rev. 96, 23–41.
Brambell, F. W. R. (1970). The Transmission of Passive immunity from Mother to Young. Amsterdam: North-Holland Publishing Company.
Bratsch, S., Wertz, N., Chaloner, K., Kunz, T. H., and Butler, J. E. (2011). The little brown bat displays a highly diverse VH, DH and JH repertoire but little evidence of somatic hypermutation. Dev. Comp. Immunol. 35, 421–430.
Bridges, S. L. Jr., Lee, S. K., Johnson, M. L., Lavelle, J. C., Fowler, P. G., Koopman, W. J., and Schroeder, H. W. Jr. (1995). Somatic mutation and CDR3 lengths of immunoglobulin kappa light chains expressed in patients with rheumatoid arthritis and in normal individuals. J. Clin. Invest. 96, 831–841.
Burnett, R. C., Hanly, W. C., Zhai, S. K., and Knight, K. L. (1989). The IgA heavy chain gene family in rabbits: cloning and sequence analysis of 13 Ca genes. EMBO J. 8, 40410–44047.
Butler, J. E. (1974). “Immunoglobulins of the mammary secretions” in Lactation, a Comprehensive Treatise, Vol. III, Chap. V, eds B. L. Larson and V. Smith (New York: Academic Press), 217–255.
Butler, J. E. (1983). Bovine immunoglobulins: an augmented review. Vet. Immunol. Immunopathol. 4, 43–152.
Butler, J. E. (1997). Immunoglobulin gene organization and the mechanism of repertoire development. Scand. J. Immunol. 45, 455–462.
Butler, J. E., and Kehrle, M. E. Jr. (2005). “Immunocytes and immunoglobulins in milk,” in Mucosal Immunology, 3rd Edn, eds J. Mestecky, M. E. Lamm, W. Strober, J. R. McGhee, L. Mayer and J. Bienenstock (New York: Academic Press), 1763–1793.
Butler, J. E., Lager, K. M., Splichal, I., Francis, D., Kacskovics, I., Sinkora, M., Wertz, N., Sun, J., Zhao, Y., Brown, W. R., DeWald, R., Dierks, S., Muyldermanns, S., Lunney, J. K., McCray, P. B., Rogers, C. S., Welsh, M. J., Navarro, P., Klobasa, F., Habe, F., and Ramsoondar, J. (2009a). The piglet as a model for B cell and immune system development. Vet. Immunol. Immunopathol. 128, 147–170.
Butler, J. E., Sinkora, M., Wertz, N., and Kacskovics, I. (2009b). Immunoglobulins, B cells and repertoire development. Dev. Comp. Immunol. 33, 321–333.
Butler, J. E., Wertz, N., Deschacht, N., and Kacskovics, I. (2009c). Porcine IgG: structure, genetics and evolution. Immunogenetics 61, 209–230.
Butler, J. E., Lemke, C. D., Weber, P., Sinkora, M., and Lager, K. D. (2007). Antibody repertoire development in fetal and neonatal piglets. XIX. Undiversified B cells with hydrophobic HCDR3s preferentially proliferate in PRRS. J. Immunol. 178, 6320–6331.
Butler, J. E., Sun, J., and Navarro, P. (1996). The swine immunoglobulin heavy chain locus has a single JH and no identifiable IgD. Int. Immunol. 8, 1897–1904.
Butler, J. E., Sun, J., Weber, P., Ford, S. P., Rehakova, Z., Sinkora, J., and Lager, K. (2001). Antibody repertoire development in fetal and neonatal piglets. IV. Switch recombination, primarily in fetal thymus occurs independent of environmental antigen and is only weakly associated with repertoire diversification. J. Immunol. 167, 3239–3249.
Butler, J. E., Sun, X.-Z., Wertz, N., Lager, K. M., Urban, J. Jr., Nara, P., and Tobin, G. (2011a). Antibody repertoire development in fetal and neonatal piglets. XXI. VH usage remains constant in fetal piglets and postnatally development. Mol. Immunol. 49, 483–494.
Butler, J. E., Mateo, K., Sun, X.-Z., Wertz, N., Sinkora, M., Harvey, R., and Francis, D. L. (2011b). Antibody repertoire development in fetal and neonatal piglets XX: the ileal Peyers patches are not a site of B cell lymphogenesis and are not required for systemic B cell proliferation and Ig synthesis. J. Immunol. 187, 5141–5149.
Butler, J. E., Sun, X.-Z., and Wertz, N. (2011c). “Immunoglobulin polygeny: an evolutionary perspective” in Gene Duplication, ed. F. Friedberg (Rijeka: InTech), 113–140.
Butler, J. E., Wertz, N., Zhao, Y., Kunz, T. H., Bratsch, S., Whitaker, J., and Schountz, T. (2011d). Two suborders of bats have the canonical isotypes repertoire of other eutherian mammals. Dev. Commun. Immunol. 35, 272–284.
Butler, J. E., Sun, X.-Z., Wertz, N., Vincent, A. L., Zanella, E. L., and Lager, K. M. (2012a). Antibody repertoire development in fetal and neonatal piglets. XVI. Influenza stimulates adaptive immunity, class switch and diversification of the IgG repertoire encoded by downstream Cγ genes. Immunology (in press).
Butler, J. E., Wertz, N., Sun, X.-Z., Lunney, J. K., and Muyldermanns, J. (2012b). Resolution of an immunodiagnostic dilemma: heavy chain chimeric antibodies for species in which plasmocytomas are unknown. Mol. Immunol. (pending).
Butler, J. E., Weber, P., Sinkora, M., Baker, D., Schoenherr, A., Mayer, B., and Francis, D. (2002). Antibody repertoire development in fetal and neonatal piglets. VIII. Colonization is required for newborn piglets to make serum antibodies to T-dependent and type 2 T-independent antigens. J. Immunol. 169, 6822–6830.
Butler, J. E., Weber, P., Sinkora, M., Sun, J., Ford, S. J., and Christenson, R. (2000a). Antibody repertoire development in fetal and neonatal piglets. II. Characterization of heavy chain CDR3 diversity in the developing fetus. J. Immunol. 165, 6999–7011.
Butler, J. E., Sun, J. Weber, P., and Francis, D. (2000b). Antibody repertoire development in fetal and neonatal piglets. III. Colonization of the gastrointestinal tracts results in preferential diversification of the pre-immune mucosal B-cell repertoire. Immunology 100, 119–130.
Butler, J. E., Weber, P., and Wertz, N. (2006a). Antibody repertoire development in fetal and neonatal pigs. XIII. “Hybrid VH genes” and the pre-immune repertoire revisited. J. Immunol. 177, 5459–5470.
Butler, J. E., Sinkora, M., Wertz, N., Holtmeier, W., and Lemke, C. D. (2006b). Development of the neonatal B- and T-cell repertoire in swine: implications for comparative and veterinary immunology. Vet. Res. 37, 417–441.
Butler, J. E., Weber, P., Wertz, N., and Lager, K. M. (2008). Porcine reproductive and respiratory syndrome virus (PRRSV) subverts development of adaptive immunity by proliferation of germline-encoded B cells with hydrophobic HCDR3s. J. Immunol. 180, 2347–2356.
Butler, J. E., and Wertz, N. (2006). Antibody repertoire development in fetal and neonatal piglets. XVII. IgG subclass transcription revisited with emphasis on new IgG3. J. Immunol. 177, 5480–5489.
Butler, J. E., Wertz, N., Sun, J., Wang, H., Lemke, C., Chardon, P., Puimi, F., and Wells, K. (2005a). The pre-immune variable kappa repertoire of swine is selectively generated from certain subfamilies of Vκ2 and one Jκ gene. Vet. Immunol. Immunopathol. 108, 127–137.
Butler, J. E., Francis, D., Freeling, J., Weber, P., Sun, J., and Krieg, A. M. (2005b). Antibody repertoire development in fetal and neonatal piglets. IX. Three PAMPs act synergistically to allow germfree piglets to respond to TI-2 and TD antigens. J. Immunol. 175, 6772–6785.
Butler, J. E., Wertz, N., Wang, H., Sun, J., Chardon, P., Piumi, F., and Wells, K. (2004). Antibody repertoire in fetal and neonatal pigs. VII. Characterization of the pre-immune kappa light chain repertoire. J. Immunol. 173, 6794–6805.
Cohn, M., and Langman, R. E. (1990). The protecton: the unit of humoral immunity selected by evolution. Immunol. Rev. 115, 11–147.
Coutelier, J.-P., Coulie, G., Wauters, P., Heremans, H., and der Logt, J. T. (1990). In vivo polyclonal B-lymphocyte activation elicited by murine viruses. J. Virol. 64, 5383–5388.
Cunnington, P. G., and Naysmith, J. D. (1975). Naturally occurring double-stranded RNA and immune responses. III. Immunogenicity and antigenicity in animals. Immunology 29, 1001–1017.
Dooley, H., and Flajnik, M. F. (2006). Antibody repertoire development in cartilaginous fish. Dev. Comp. Immunol. 30, 43–56.
Du Pasquier, L., Robert, J., Courtet, M., and Mussmann, R. (2000). B cell development in the amphibian Xenopus. Immunol. Rev. 175, 201–213.
Du Pasquier, L., Wilson, M., Greenberg, A. S., and Flajnik, M. F. (1998). Somatic mutation in ectothermic vertebrates: musings on selection and origins. Curr. Top. Microbiol. Immunol. 229, 199–216.
Eguchi-Ogawa, T., Sun, X.-Z., Wertz, N., Uenishi, H., Puimi, F., Chardon, P., Wells, K., Tobin, G. J., and Butler, J. E. (2010). Antibody repertoire development in fetal and neonatal piglets. XI. The relationship of VDJ usage and the genomic organization of the variable heavy chain locus. J. Immunol. 184, 3734–3742.
Eguchi-Ogawa, T., Toki, D., Wertz, N., Butler, J. E., and Uenishchi, H. (2012). Complete structure of the genomic sequence encoding the constant region of the porcine immunoglobulin heavy chain. Mol. Immunol. (in press).
Ehrlich, R. (1995). Selective mechanisms utilized by persistent and oncogenic viruses to interfere with antigen producing and presentation. Immunol. Res. 14, 77–97.
Girschick, H. J., and Lipsky, P. E. (2001). The kappa repertoire of human neonatal B cells. Mol. Immunol. 38, 1113–1127.
Glas, A. M., van Monfort, E. H. N., and Milner, E. C. B. (2000). “The human antibody repertoire: old notions, current realities and VH gene-dependent biases,” in The Antibodies, Vol. 6, eds M. Zanetti and J. D. Capra (Amsterdam: Harwood Academic Publishers), 63–79.
Hahn, G., Jores, R., and Mocarski, E. S. (1998). Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. U.S.A. 95, 3937–3942.
Hamers-Casterman, C., Atarhouch, T., Muylermans, S., Robinson, G., Hamers, C., Bajyana Songa, E., Bendahman, N., and Hamers, R. (1993). Naturally occurring antibodies devoid of light chains. Nature 363, 446–448.
Herzenberg, L. A., Stall, A. M., Lalor, P. A., Sidman, C., Moore, W. A., Parks, D., and Herzenberg, L. A. (1986). The Ly-1 B cell lineage. Immunol. Rev. 93, 81–102.
Honjo, T., Kinoshita, K., and Muranmatsu, M. (2002). Molecular mechanisms of lass switch recombination: linkage with somatic hypermutation. Annu. Rev. Immunol. 20, 165–196.
Hood, L., Gray, W. R., Saunders, B. G., and Dreyer, W. J. (1967). Light chain evolution. Cold Spring Harb. Symp. Quant. Biol. 32, 133–146.
Hunziker, L., Recher, M., Macpherson, A. J., Ciurea, A., Freigang, S., Hengartner, H., and Zinkernagel, R. M. (2003). Hypergammaglobulinemia and autoantibody induction mechanisms in viral infection. Nat. Immunol. 4, 343–349.
Kehoe, J. M., and Capra, J.D. (1974). Nature and significance of immunoglobulin subclasses. NY State J. Med. 74, 489–491.
Kloep, A., Wertz, N., Mendicino, M., and Butler, J. E. (2012). Linkage haplotype for IgG and IgA subclass genes. Immunogenetics 64, 469–473.
Knight, K. L. (1992). Restrictive VH gene usage and generation of antibody diversity in rabbit. Annu. Rev. Immunol. 10, 593–616.
Kondrashov, F. A., Rogozin, I. B., Wolf, Y. I., and Koonin, E. V. (2002). Selection in the evolution of gene duplications. Genome Biol. 3, RESEARCH0008.
Lanning, D., Osbourne, B. A., and Knight, K. L. (2004). “Immunoglobulin genes and generation of antibody repertoires in higher vertebrates: a key role for GALT,” in Molecular Biology of B Cells, eds F. W. Alt, T. Honjo and M. S. Neuberger (London: Elsevier Science Ltd.), 433–448.
Lanning, D. K., Zhao, S. K., and Knight, K. L. (2003). Analysis of the 3′ Cmu region of the rabbit Ig heavy chain locus. Gene 309, 135–144.
LeFranc, G., Chaabani, H., Van Loghem, E., Lefranc, M. P., De Lange, G., and Helal, A. N. (1983). Simultaneous absence of the human IgG1, IgG2, IgG4 and IgA1 subclasses: immunological and immunogenetic considerations. Eur. J. Immunol. 13, 240–244.
LeFranc, M. P., Hammarstrom, L., Smith, C. I., and Lefranc, G. (1991). Gene deletion in the human immunogolubulin heavy chain constant region locus: molecular and immunological analysis. Immunol. Rev. 2, 265–281.
Lemke, C. D., Haynes, J. S., Spaete, R., Adolphson, D., Vorwald, A., Lager, K., and Butler, J. E. (2004). Lymphoid hyperplasia resulting in immune dysregulation is caused by PRRSV infection in pigs. J. Immunol. 172, 1916–1925.
Li, W. H., Yang, J., and Guo, X. (2005). Expression divergence between duplicate genes. Trends Genet. 21, 602–607.
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155.
Mage, R. G., Lanning, D., and Knight, K. L. (2006). B cell and antibody repertoire development in rabbits: the requirement of gut-associated lymphoid tissues. Dev. Comp. Immunol. 30, 137–153.
Merial, Inc. (2004). PCV2 diseases: from research back to the field again, Vol. 5. Kansas City, MO: Corporate Publication by Merial Inc.
Meselson, M. S., and Radding, C. M. (1975). A general model for genetic recombination. Proc. Natl. Acad. Sci. U.S.A. 72, 358–361.
Mossman, T. R., and Coffman, R. L. (1989). TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu. Rev. Immunol. 7, 143–173.
Mullens, M. A., Register, K. B., Bayles, D. O., and Butler, J. E. (2011). Haemophilus parasuis exhibits IgA protease activity but lacks homologs of the IgA protease genes of Haemophilus influenzae. Vet. Microbiol. 153, 407–412.
Navarro, P., Christenson, R., Weber, P., Rothschild, M., Ekhard, G., Lemky, J., and Butler, J. E. (2000). Porcine IgA allotypes are not equally transcribed or expressed in heterozygous swine. Mol. Immunol. 37, 653–664.
Nguyen, V. K. (2001). Generation of Heavy Chain Antibodies in Camelids. Ph.D. thesis, Free University of Brussels, Brussels, 109–111.
Nguyen, V. K., Su, C., Muyldermans, S., and van der Loo, W. (2002). Heavy chain antibodies in Camelids; a case of evolutionary innovation. Immunogenetics 54, 39–47.
Nitschke, L., Kosco, M. L., Kohler, G., and Lamers, M. C. (1993). Immunoglobulin D deficient mice can mount normal immune responses to thymus-independent and -dependent antigens. Proc. Natl. Acad. Sci. U.S.A. 90, 1887–1891.
Ochsenbein, A. F., and Zinkernagel, R. (2000). Natural antibodies and complement link innate and acquired immunity. Immunol. Today 1, 624–630.
Olsson, P. G., Rabbani, H., Hammarstrom, L., and Smith, C. I. (1993). Novel human immunoglobulin heavy chain constant region gene deletion haplotypes characterized by pulsed-field electrophoresis. Clin. Exp. Immunol. 94, 84–90.
Plaut, A. G., Wustar, R. Jr., and Capra, J. D. (1974). Differential susceptibility of human IgA immunoglobulins to streptocococcal IgA proteases. J. Clin. Invest. 54, 1295–1300.
Qin, T., Ren, L., Hu, X., Guo, Y., Fei, J., Pan-Hammarstrom, Q., Butler, J. E., Wu, C., Li, L., Hammarstrom, L., and Zhao, Y. (2008). Genomic organization of the immunoglobulin gene loci in Xenopus tropcallis: evolutionary implications. Dev. Comp. Immunol. 32, 156–165.
Rabbani, H., Kondo, N. Smith, C. I., and Hammarstrom, L. (1995). The influence of gene deletion and duplication within the IGHC locus on serum immunoglobulin subclass levels. Clin. Immunol. Immunopathol. 76, 214–218.
Rajewsky, K., Forster, I., and Cumano, A. (1987). Evolutionary and somatic selection of the antibody repertoire in the mouse. Science 238, 1088–1094.
Ratcliffe, M. J. H. (2006). Antibodies, immunoglobulin genes and the bursa of Fabricius in chicken. B cell development. Dev. Comp. Immunol. 30, 101–118.
Reynaud, C. A., Anquez, V., Daher, A., and Weill, J.-C. (1987). A hyperconversion mechanism generates the chicken pre-immune light chain repertoire. Cell 48, 379–388.
Richl, P., Stern, U., Lipsky, P. E., and Girschick, H. J. (2008). The lambda gene immunoglobulin repertoire of human neonatal B cells. Mol. Immunol. 45, 320–327.
Rodkey, L. S., and Adler, F. L. (1983). Regulation of natural anti-allotypic antibody responses by network induced auto-anti-idiotypic responsiveness of their offspring. J. Exp. Med. 152, 1024–1035.
Schelonka, R. L., Tanner, J., Zhang, Y., Gartland, G. L., Zemlin, M., and Schroeder, H. W. Jr. (2007). Categorized selection of the antibody repertoire in splenic B cells. Eur. J. Immunol. 4, 1010–1021.
Schiaffella, E., D. Sehgal, A.O., Anderson, and Mage, R. G. (1999). Gene conversion and hypermutation during diversification of VH sequences in developing germinal centers of immunized rabbits. J. Immunol. 162, 3984–3995.
Schwartz, J. C., Lefranc, M., and Murtaugh, M. P. (2012a). Evolution of the porcine kappa locus through germline gene conversion. Immunogenetics 64, 303–311.
Schwartz, J. C., Lefranc, M., and Murtaugh, M. P. (2012b). Organization, complexity and allelic diversity of the porcine immunoglobulin lambda locus. Immunogenetics 64, 399–407.
Sheehan, K. M., Mainville, C. V. A., Willert, S., and Brodeur, P. H. (1993). The utilization of individual VH exons in the primary repertoire of adult BALB/c mice. J. Immunol. 151, 5363–5375.
Shoja, V., and Zhang, L. (2006). A roadmap of tandemly arrayed genes in the genomes of human, mouse and rat. Mol. Biol. Evol. 23, 2134–2141.
Sinkora, M., Sun, J., Sinkorova, J., Christenson, R. K., Ford, S. P., and Butler, J. E. (2003). Antibody repertoire development in fetal and neonatal piglets. VI. B cell lymphogenesis occurs in multiple sites with differences in the frequency of in-frame rearrangements. J. Immunol. 170, 1781–1788.
Sinkora, M., Zelena, K., Butler, J. E., Francis, D., Santiago-Mateo, K., Potockova, H., and Sinkorova, J. (2011). Ileal Peyers patches (IPP) are not necessary for B cell development and maintenance and do not contribute significantly to the overall B cell pool in swine. J. Immunol. 187, 5150–5161.
Skvaril, F., Baranden, S., Kuffer, F., and Probst, M. (1976). Changes of kappa/lambda ratio of human serum immunoglobulins in the course of development. Blut 33, 281–284.
Sun, J., and Butler, J. E. (1997). Sequence analysis of swine switch μ, Cμ and Cμm. Immunogenetics 46, 452–460.
Sun, J., Hayward, C., Shinde, R., Christenson, R., Ford, S. P., and Butler, J. E. (1998). Antibody repertoire development in fetal and neonatal piglets. I. Four VH genes account for 80% of VH usage during 84 days of fetal life. J. Immunol. 161, 5070–5078.
Sun, J., Kacskovics, I., Brown, W. R., and Butler, J. E. (1994). Expressed swine VH genes belong to a small VH gene family homologous to human VH III. J. Immunol. 153, 5618–5627.
Sun, X.-Z., Wertz, N., Lager, K., Sinkora, M., Stepanova, K., Tobin, G., and Butler, J. E. (2012a). Antibody repertoire development in fetal and neonatal piglets. XXII. Lambda rearrangement precedes kappa rearrangement during B cell lymphogenesis in swine. Immunol. (British) (in press).
Sun, X.-Z., Wertz, N., Lager, K. L., Tobin, G., and Butler, J. E. (2012b). Antibody repertoire development in fetal and neonatal piglets XXIII: fetal piglets infected with a vaccine strain of PRRS Virus display the same immune dysregulation seen in isolator piglets. Vaccine 30, 3646–3652.
Sun, X.-Z., Wertz, N., Lager, K. L., and Butler, J. E. (2012c). Antibody repertoire development in fetal and neonatal piglets. XV. Porcine circovirus type 2 infection results in serum IgG antibodies to ORF 2, elevated IgA levels but little evidence for immune suppression. Vaccine (pending).
Szostak, J. W., Orr-Weaver, T. L., and Rothstein, R. J. (1983). The double-strand break repair model for recombination. Cell 33, 25–35.
Tiegs, S. L., Russell, D. M., and Nemazee, D. (1993). Receptor editing in self-reactive bone marrow B cells. J. Exp. Med. 177, 1009–1020.
Tristram, D. A. (2005). “Maternal genital tract infection and the neonates,” in Mucosal Immunology, 3rd Edn, Vol II, eds J. Mestecky, M. E. Lamm, W. Strober, J. Bienestock, J. R. McGhee and L. Mayer (Elsevier/Academic Press), 1721–173l.
Vazquez, J., Wertz, N., Sun, J., Wells, K., Sun, X.-Z., and Butler, J. E. (2012). Antibody repertoire development in fetal and neonatal piglets. XI. Characterization of the expressed lambda repertoire. Mol. Immunol. (pending).
Victor, K. D., Vu, K., and Feeney, A. J. (1994). Limited junctional diversity in kappa light chains. Junctional sequences from CD431B2201 early B cell progenitors resemble those from peripheral B cells. J. Immunol. 152, 3467–3475.
Wang, H., and Shlomchik, M. J. (1998). Maternal Ig mediates neonatal tolerance in rheumatoid factor transgenic mice but tolerance breaks down in adult mice. J. Immunol. 160, 2263–2271.
Weinstein, J. A., Jiang, N., White, R. A. III, Fisher, D. S., and Quake, S. R. (2009). High throughput sequencing of the Zebrafish antibody repertoire. Science 324, 807–810.
Wikler, M., Demeur, C., Dewasne, G., and Urbain, J. (1980). Immunoregulatory role of maternal idiotypes. Ontogeny of immune networks. J. Exp. Med. 152, 1024–1035.
Winstead, C. R, Zhai, S. K., Sethupathi, P., and Knight, K. L. (1999). Antigen-induced somatic diversification of rabbit IgA genes: gene conversion and point mutation. J. Immunol. 162, 6602–6612.
Wolfe, K. H., and Shields, D. C. (1997). Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713.
Woody, O. Z., and McConkey, B. J. (2011). “Detection and analysis of functional specialization in duplicated genes,” in Gene Duplication, ed. F. Friedberg (Croatia: InTeck Rijeka), 37–58.
Xu, J. L., and Davis, M. M. (2000). Diversity in the CDR3 region of VH is sufficient for most antibody specificities. Immunity 13, 37–45.
Xue, C., and Fu, Y. (2009). Preservation of duplicate genes by originalization. Genetica 136, 69–78.
Yamaguchi, N., Shimizu, S., Hara, A., and Saito, T. (1983). The effector maternal antigenic stimulation upon the active immune responsiveness of their offspring. Immunology 50, 229–238.
Keywords: antibody repertoire, swine, gene duplication, development
Citation: Butler JE and Wertz N (2012) The porcine antibody repertoire: variations on the textbook theme. Front. Immun. 3:153. doi: 10.3389/fimmu.2012.00153
Received: 13 February 2012; Paper pending published: 21 March 2012;
Accepted: 24 May 2012; Published online: 27 June 2012.
Edited by:
Harry W. Schroeder, University of Alabama at Birmingham, USACopyright: © 2012 Butler and Wertz. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: John E. Butler, Department of Microbiology, Carver College of Medicine, University of Iowa, 3-501 BSB, 51 Newton Road, Iowa City, IA 52242, USA. e-mail: john-butler@uiowa.edu