- School of Kinesiology and Health Science, Faculty of Health, York University, Toronto, ON, Canada
Gene conversion (GCV), a mechanism mediated by activation-induced cytidine deaminase (AID) is well established as a mechanism of immunoglobulin diversification in a few species. However, definitive evidence of GCV-like events in human immunoglobulin genes is scarce. The lack of evidence of GCV in human rearranged immunoglobulin gene sequences is puzzling given the presence of highly similar germline donors and the presence of all the enzymatic machinery required for GCV. In this study, we undertook a computational analysis of rearranged IGHV3-23*01 gene sequences from common variable immunodeficiency (CVID) patients, AID-deficient patients, and healthy individuals to survey “GCV-like” activities. We analyzed rearranged IGHV3-23*01 gene sequences obtained from total PBMC RNA and single-cell polymerase chain reaction of individual B cell lysates. Our search identified strong evidence of GCV-like activity. We observed that GCV-like tracts are flanked by AID hotspot motifs. Structural modeling of IGHV3-23*01 gene sequence revealed that hypermutable bases flanking GCV-like tracts are in the single stranded DNA (ssDNA) of stable stem-loop structures (SLSs). ssDNA is inherently fragile and also an optimal target for AID. We speculate that GCV could have been initiated by the targeting of hypermutable bases in ssDNA state in stable SLSs, plausibly by AID. We have observed that the frequency of GCV-like events is significantly higher in rearranged IGHV3-23-*01 sequences from healthy individuals compared to that of CVID patients. We did not observe GCV-like events in rearranged IGHV3-23-*01 sequences from AID-deficient patients. GCV, unlike somatic hypermutation (SHM), can result in multiple base substitutions that can alter many amino acids. The extensive changes in antibody affinity by GCV-like events would be instrumental in protecting humans against pathogens that diversify their genome by antigenic shift.
Introduction
Diverse mechanisms form the primary immune repertoire. In many species, the presence of multiple functional variable domain encoding gene segments allows a large and significant diversity in the primary antibody repertoire by the mechanism of V(D)J recombination alone (Tonegawa, 1983; Alt et al., 1987; Pascual and Capra, 1991). In contrast, chicken has a sole functional VH and a sole functional VL gene segment, and hence, the primary diversity generated through V(D)J recombination is minimal. In chicken, the predominant and powerful mechanism of antibody diversification is “Gene Conversion” (GCV; Reynaud et al., 1987, 1989; Sayegh et al., 1999; Arakawa and Buerstedde, 2004; Ratcliffe, 2006). GCV is a “copy and paste” type of non-reciprocal transfer of genetic information between homologous DNA sequences (Wysocki and Gefter, 1989). Recombination events in mammals facilitating non-reciprocal transfer of sequences are called “GCV-like events” because the unidirectional transfer of sequence is inferred from the sequence homology between the donor and recipient sequences (indirectly) rather than in organisms like fungi where non-reciprocity of events can be seen directly, in single cells (Radding, 1978; Hastings, 1988). In chicken, over 25 pseudo variable gene segments lie upstream of the sole functional variable gene segment. These upstream pseudo genes are (or can be) used as templates to replace homologous sequences of functionally rearranged VJ and VDJ gene segments (McCormack et al., 1991; Reynaud et al., 1994). Multiple rounds of GCV events, thereby, diversify the primary antibody repertoire. In rabbit, although there are multiple functional and non-functional VH gene segments, 80–90% of rearrangements utilize only one VH gene segment, VH1, with the other VH gene segments being utilized as templates for GCV (Becker and Knight, 1990; Weinstein et al., 1994; Winstead et al., 1999; Sehgal et al., 2000). Although the combinatorial diversity at the light chain loci in rabbit is much higher than that at the heavy chain loci, GCV is visibly used to diversify the light chain gene segments (Sehgal et al., 2000). There is also evidence of GCV for the diversification of Vλ repertoire of cattle (Parng et al., 1996; Lucier et al., 1998). Some good evidence of GCV has been found in mice (Dildrop et al., 1982; Krawinkel et al., 1983; David et al., 1992; Selsing et al., 1996; D’Avirro et al., 2002, 2005; Tsai et al., 2002). GCV in human rearranged immunoglobulin genes have not been identified except for one recent study (Darlow and Stott, 2006).
It appears that species that employ GCV as predominant diversification mechanism have limited functional gene segments or restricted usage of them. Additionally, in these species it appears that most V(D)J recombination directed gene rearrangements occur only early in their life cycle and cease to occur in adult life (McCormack and Thompson, 1990; Reynaud et al., 1991; Griebel and Ferrari, 1994; Parng et al., 1996; Meyer et al., 1997). Immunoglobulin diversification in chickens, rabbits, and cattle occurs by additional mechanisms upon antigenic challenge but GCV is a key mechanism. Infrequent GCV events in humans could be due to the species’ high dependence on V(D)J recombination and hence there is less need for GCV to generate the vast, diverse primary antibody repertoire. Moreover, in humans, gene rearrangements occur throughout life facilitating extensive and continuing diversity. In species where vast diversity can be generated by combinatorial joining of multiple functional gene segments or in species such as sheep where diversity seems to be generated in most part by somatic hypermutation (SHM; Reynaud et al., 1991; Reynaud et al., 1995), it is not necessary to conclude that GCV is an unlikely mechanism of diversification. Immunoglobulin VH genes belong to multigene families (Hood et al., 1975). Members of a multigene family share extensive sequence homology and thus can participate in interactions such as recombination and GCV that can multiply their diversity. Species such as chickens and rabbits provide a unique opportunity to analyze GCV since the limited usage of functional V gene segments makes the identification of germline counterparts quite straight forward. Humans can utilize several hundred functional V gene segments making it difficult to assign exact germline counterparts for their diversified genes. The dominant diversifying mechanisms could well mask GCV tracts. Multiple rounds of GCV events may appear to be SHMs since not only is it difficult to identify the germline segments that participated in GCV but also SHMs may be superimposed on a GCV event further masking the process.
Activation-induced cytidine deaminase (AID) is an enzyme required for immunoglobulin diversity (Muramatsu et al., 2000; Revy et al., 2000). The absence of AID in B cells results in the absence of three immunoglobulin diversification mechanisms: SHM, class-switch recombination (CSR), and GCV (Muramatsu et al., 2000; Revy et al., 2000; Arakawa et al., 2002; Harris et al., 2002). An AID-induced lesion is the initiating lesion in all three processes. The downstream consequences where the AID-induced lesion is resolved, however, are different. AID converts cytidine to uracils, a process that results in high levels of AID-dependent resected-end DSBs’s in V-genes (Bross et al., 2002; Papavasiliou and Schatz, 2002; Wu et al., 2003; Zan et al., 2003). Resected ends (3′) are involved in homologous recombination (HR) repair. HR is considered to the mechanism of GCV in immunoglobulin gene segments (Bezzubova and Buerstedde, 1994; Papavasiliou and Schatz, 2000). Further DSB’s in V-genes have been shown to preferentially target RGYW motifs, with embedded AID hotspot motifs (WRC/GYW; Bross et al., 2000; Papavasiliou and Schatz, 2000). Data also suggest that unidirectional GCV requires chromatin to be in an open state (Roth and Roth, 2000; Schlissel, 2000; Seo et al., 2005; Kurosawa et al., 2010), which, during transcription, generates the single stranded regions required for AID action (Dickerson et al., 2000; Bransteitter et al., 2003; Pham et al., 2003; Ramiro et al., 2003; Odegard and Schatz, 2006; Larijani et al., 2007). It is puzzling to envision the absence or lack of GCV in human rearranged immunoglobulin gene segments when there is presence of required machinery for GCV, i.e., from AID to enzymes required for HR and their resolution by mismatch repair (MMR) and error-prone polymerases (Seki et al., 2005; Liu and Schatz, 2009).
In this study, we made use of the development of genomic databases and bioinformatics software to analyze the rearranged IGHV3-23*01 gene from common variable immunodeficiency (CVID) patients, AID-deficient patients, and healthy individuals to explore “GCV-like” activities. We chose IGHV3-23*01 rearrangements because they comprise approximately 25% of all IGHV3 family rearrangements in normal adult peripheral B cells and the germline number of the IGHV3 family gene segments allowed the analyses to be significant. We analyzed mutated IGHV3-23*01 sequences obtained by two methods (1) RT-PCR of total PBMC RNA (2) Single-cell polymerase chain reaction (PCR) of individual B cell lysates.
During our search for germline donors for mutations in rearranged IGHV3-23*01, we found examples of GCV-like events that could have contributed to the diversity of IGHV3-23*01 post VDJ rearrangement. In our previous studies (Duvvuri et al., 2011, 2012) we provided evidence for defective DNA repair mechanisms in a subgroup of CVID patients. These patients had functional AID as evidenced by the presence of SHM and by a switch in immunoglobulin class of their immunoglobulin genes. In the current analyses rearranged IGHV3-23*01 from these CVID patients showed a significantly reduced frequency of GCV-like events compared to that of healthy individuals. We did not find any evidence of GCV-like events from our analysis of sequences from AID-deficient patients in the data bases. Using modeling software and analysis, we demonstrate the presence of sequences and structures associated with site-specific recombination in the regions flanking GCV-like events. Finally we discuss the contribution of GCV in affinity maturation and receptor editing.
Materials and Methods
DNA Sequence Data Collection
For the CVID patients, IGHV3-23 (also called VH26) cDNAs were obtained by Dr. C. A. Reynaud at the Necker Institute, Paris as described (Levy et al., 1998). Briefly, the sequences were amplified by RT-PCR using total PBMC RNA as template. A “downstream” CΓ primer was used to ensure that the sequences arose from class-switched B cells. The CΓA primer (GTCCTTGACCAGGCAGCCCAG) was used for single strand cDNA synthesis using an RT-PCR kit (Stratagene). PCR was then performed with 0.5 unit of Pfu polymerase on cDNA. The following primers were used for amplification: V3–23 leader exon (GGCTGAGCTGGCTTTTTCTTGTGG-3′) and CΓB (AAGACCGATGGGCCCTTGGTGG). The PCR conditions were 35 cycles (940°C, 45 s; 650°C, 1.5 min; 720°C, 2 min).
Healthy control samples (C1–C5 series) were obtained as described (Pan-Hammarström et al., 2006). Briefly, total RNA was extracted from PBMC using RNeasy RNA purification kits (QIAGEN) and first-strand cDNA synthesis was performed with a CΓA primer (GTCCTTGACCAGGCAGCCCAG) using a cDNA synthesis kit (GE Healthcare). The primers used for amplification of VH3-CΓ and VH3–23-CΓ transcripts were VH3-consensus (aatctagaGGTGCAGCTGGTGGAGTC), VH3–23 (tctagaGGCTGAGCTGGCTTTTTCTTGTGG), and CΓB (cagtcgacAAGACCGATGGGCCCTTGGTGG). The oligonucleotides contained a restriction site, (underlined, a XbaI site in the VH3-consensus or VH3–23 primer and a SalI site in the CΓB primer) for directional cloning of the PCR products.
Detailed analysis of IGHV3-23*01 sequence data from CVID patients and healthy individuals is in Duvvuri et al. (2011, 2012).
Database of IGHV3-23*01 Mutated Sequences from Human Peripheral B Cells using Single-Cell Polymerase Chain Eaction
In total we have screened 636 VH productive rearrangements from healthy controls for IGHV3-23*01 rearranged sequences. The final analysis included 81, IGHV3-23*01 sequences from healthy controls from different subsets of B cells and details are as followed: 15, IGHV3-23*01 sequences from CD19+IgM+ B cells, 52, IGHV3-23*01 sequences from CD19+IgM+CD5+ and CD19+IgM+CD5− B cells (Brezinschek et al., 1995, 1997; Dörner et al., 1998); 11, IGHV3-23*01 sequences from IgD+CD27+ B cells; and 3, IGHV3-23*01 sequences from IgD−CD27+ B cells (Longo et al., 2008). From AID-deficient patients, in total we have screened 1150 VH productive rearrangements for IGHV3-23*01 rearranged sequences. The final analysis included 117, IGHV3-23*01 sequences from CD19+IgM+CD5+ and D19+IgM+CD5- B cells (Longo et al., 2008). All the sequences have been obtained from NCBI.
Computational Approaches used for the Identification of GCV-Like Tracts in IGHV3-23*01 Sequences
Gene conversion-like events in IGHV3-23*01 gene were sought employing two different computational approaches. The first approach employed the tool GENECONV1 (Sawyer, 1989). GENECONV is the highly ranked recombination analysis software and is included in the RDP2 analysis suite. Since we are analyzing hypermutated IGHV3-23*01 gene sequences, it is possible to have GCV tracts masked by point mutations (as a result of SHM) and thus GENECONV would underestimate the occurrence of GCV events.
All possible germline sequences of rearranged (hypermutated) IGHV3-23*01 sequences were identified in JoinSolver alignment program available at http://joinsolver.niaid.nih.gov/.
As a second approach, we visually checked the alignments between rearranged and germline sequences in BioEdit, a biological sequence alignment editor (using default parameters), to identify possible GCV tracts.
GENECONV Detection Methodology
GENCONV is a program specifically designed to identify GCV events. GENECONV searches for potential past GCV events by searching for highly similar sequences in an entire alignment. It determines the statistical significance of observed fragment lengths by computing global and pairwise P-values. Sequence pairs with low P-value, (by default P-value < 0.05) are considered statistically significant. Pairwise P-values are assigned based on the comparison of each fragment with the maximum fragment length that might have been expected for that sequence pair by chance. Pairwise P-values are approximate P-values based on Karlin and Altschul P-value (Karlin and Altschul, 1990), which is the basis of the highly used BLAST method for finding sequence matches in DNA or protein databases. Global P-values, on the other hand, are more conservative because they are based on a comparison of each fragment for all possible pairs reported from an alignment. Global P-values are assigned based on permutations and GENECONV by default considers 10,000 permutations for each tested sequence alignment. GENECONV default settings for the analysis of alignments of three or more sequences were used for the entire analysis. Additionally, we also used – Dumpjseq option, which predicts the bases immediately adjacent to the endpoints of significant GCV fragments displayed as: before <Frag…..ment> After. Input for GENECONV is an alignment of IGHV3-23*01 DNA sequences with germline VH sequences. Possible donor germline sequences were identified by screening each of the rearranged (hypermutated) IGHV3-23*01 sequences (n = 161 from CVID patients by RT-PCR of total PBMC RNA; n = 117 from AID-deficient patients by single-cell PCR of individual B cell lysates; n = 40 from healthy controls by RT-PCR of total PBMC RNA and n = 81 from healthy controls by single-cell PCR of individual B cell lysates).
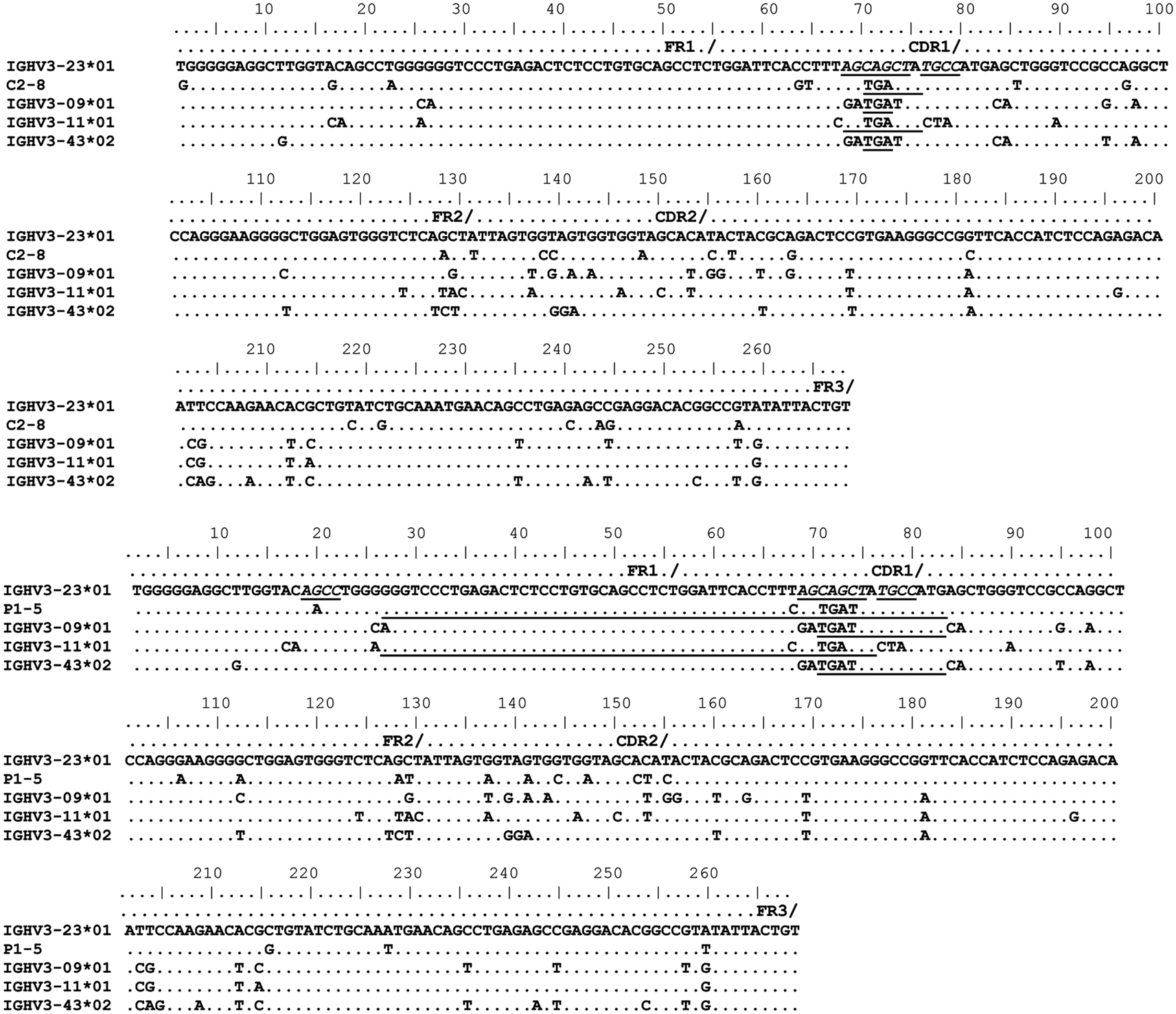
Figure 1. Gene conversion-like events between rearranged IGHV3-23*01 gene sequences and germline VH sequences identified by GENECONV. Alignments of rearranged IGHV3-23*01 gene sequences and germline donor VH sequences demonstrating the minimum and maximum homology at GCV-like events. Topmost sequence is the germline of rearranged IGHV3-23*01 gene sequences from a healthy individual (C2–8) and a CVID patient (P1–5). Potential donor germline IGHV3 family gene segments are shown below the rearranged IGHV3-23*01 sequences. GCV-like tract is underlined. Flanking SHM/AID hotspot motifs are underlined and italicized.
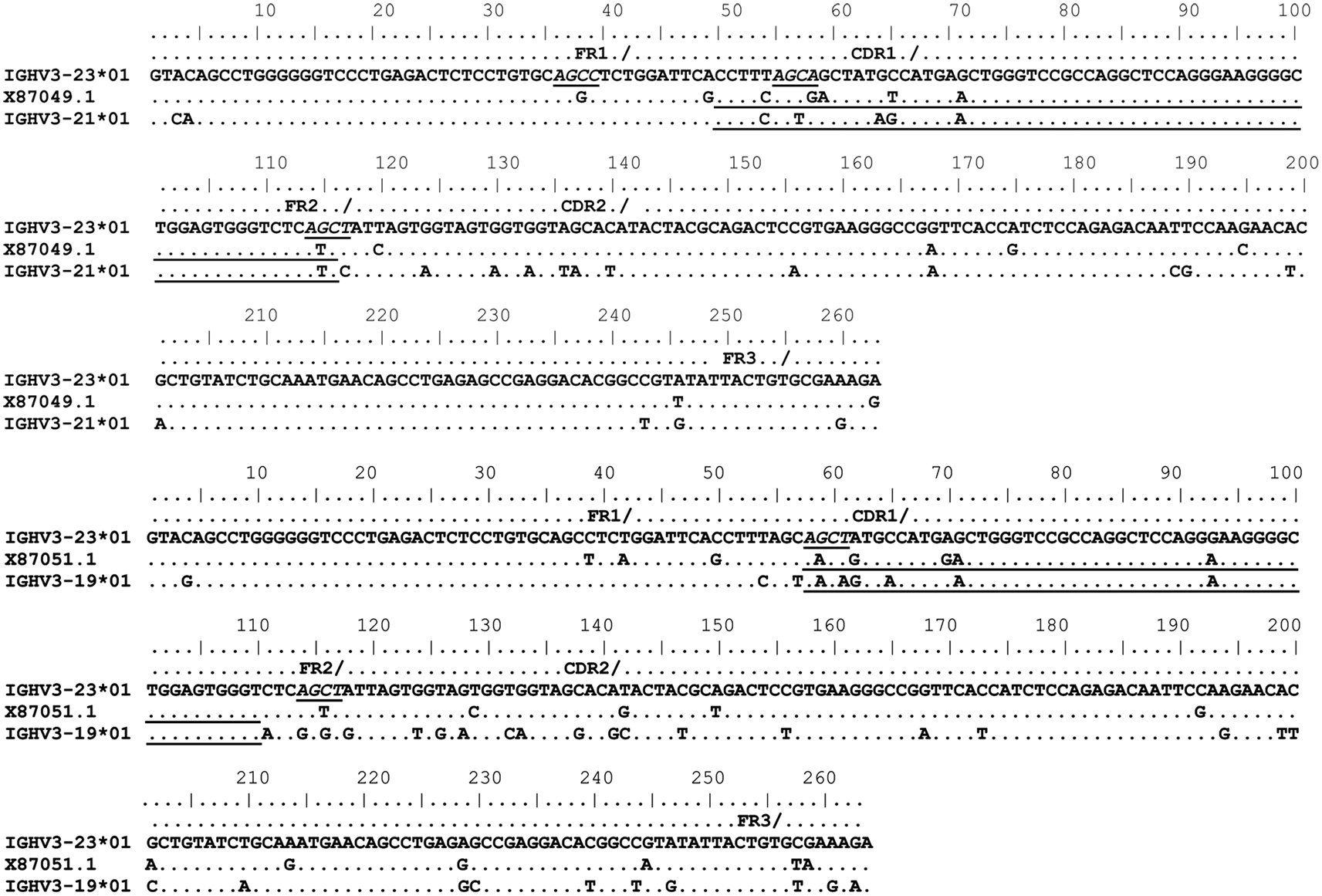
Figure 2. Gene conversion-like events between rearranged IGHV3-23*01 gene sequences and germline VH sequences superimposed by somatic hypermutation (SHM). IGHV3-23*01 mutated sequences obtained by single-cell PCR from CD19+IgM+B cells (Brezinschek et al., 1995, 1997; Dörner et al., 1998; Longo et al., 2008). Topmost sequence is the germline of rearranged IGHV3-23*01 gene sequences from healthy individuals (X87050.1, X87049.1, and X87051.1) Potential donor germline IGHV3 family gene segments are shown below the rearranged IGHV3-23*01 sequences. GCV-like tract is underlined. Flanking SHM/AID hotspot motifs are underlined and italicized.
DNA Secondary Structural Analysis
Mfg is a tool to analyze DNA secondary structures. It was used to analyze the IGHV3-3*01 germline gene during simulated transcription. In a given window size “mfg” folds all nucleotides successively, beginning with each base and predicts the most stable (−ΔG) stem-loop structures (SLSs), in which that base is unpaired and therefore assumed to have longest period of exposure time to AID. −ΔG values document the negative free energy. The more negative the −ΔG value, the more likely is the stable is the SLS. Mfg calculates the frequency with which a specific base is unpaired in the most stable SLS during simulated transcription giving the result as a “percent unpaired.” More detailed information on the software is available in Wright et al. (2003, 2004, 2008a,b, 2011) and on the web site www.dbs.umt.edu/research_labs/wrightlab/upload/mfg.html. A base is called unpaired or paired when present in the loop or in the stem respectively.
Results
Identification of Homology between IGHV3-23*01 and Potential Germline GCV Donor’s VH Regions
For evidence of GCV-like diversification we first searched for homologies with the germline VH genes that could serve as donors. Templated mutations in GCV require some (∼>80%) degree of homology between donor and recipient target sequences. Table 1 shows the global homology between IGHV3-23*01 gene sequence and all potential GCV-like donors identified. IGHV3-23*01 is the germline sequence of the rearranged IGHV3-23*01 sequences being analyzed.
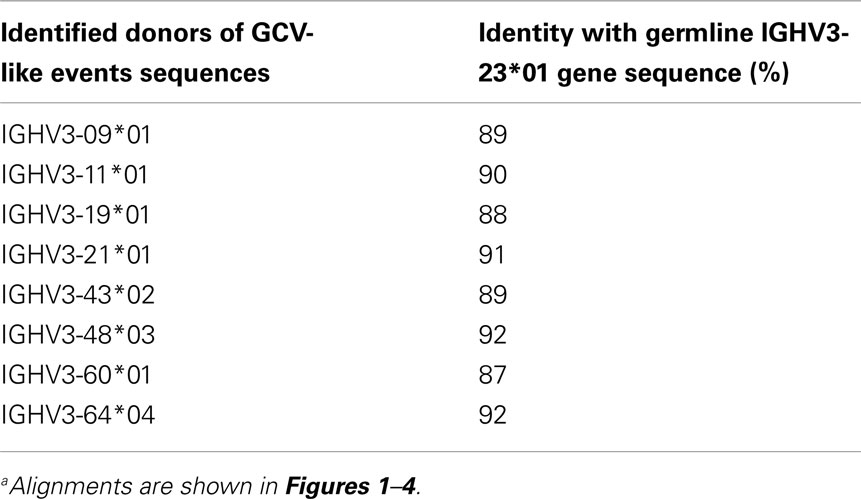
Table 1. Identities between germline IGHV3-23*01 gene sequence and potential germline VH donors for GCV-like eventsa.
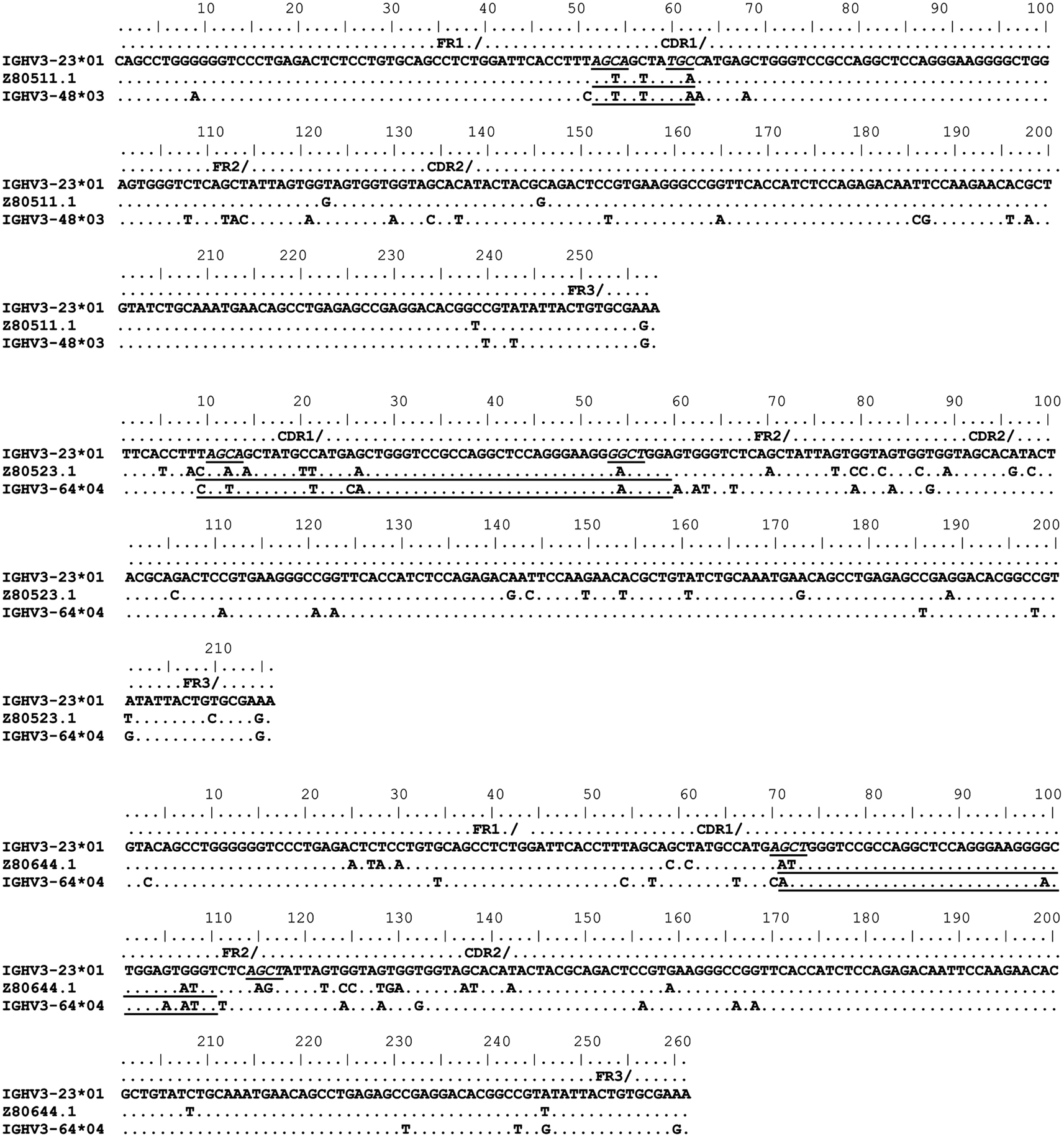
Figure 3. Gene conversion-like events between rearranged IGHV3-23*01 gene sequences and germline VH sequences superimposed by somatic hypermutation (SHM). IGHV3-23*01 mutated sequences obtained by single-cell PCR from CD19+IgM+CD5+ B cells (Brezinschek et al., 1995, 1997; Dörner et al., 1998; Longo et al., 2008). Topmost sequence is the germline of rearranged IGHV3-23*01 gene sequences from healthy individuals (Z80511.1, Z80523.1, and Z80644.1). Potential donor germline IGHV3 family gene segments are shown below the rearranged IGHV3-23*01 sequences. GCV-like tract is underlined. Flanking SHM/AID hotspot motifs are underlined and italicized.
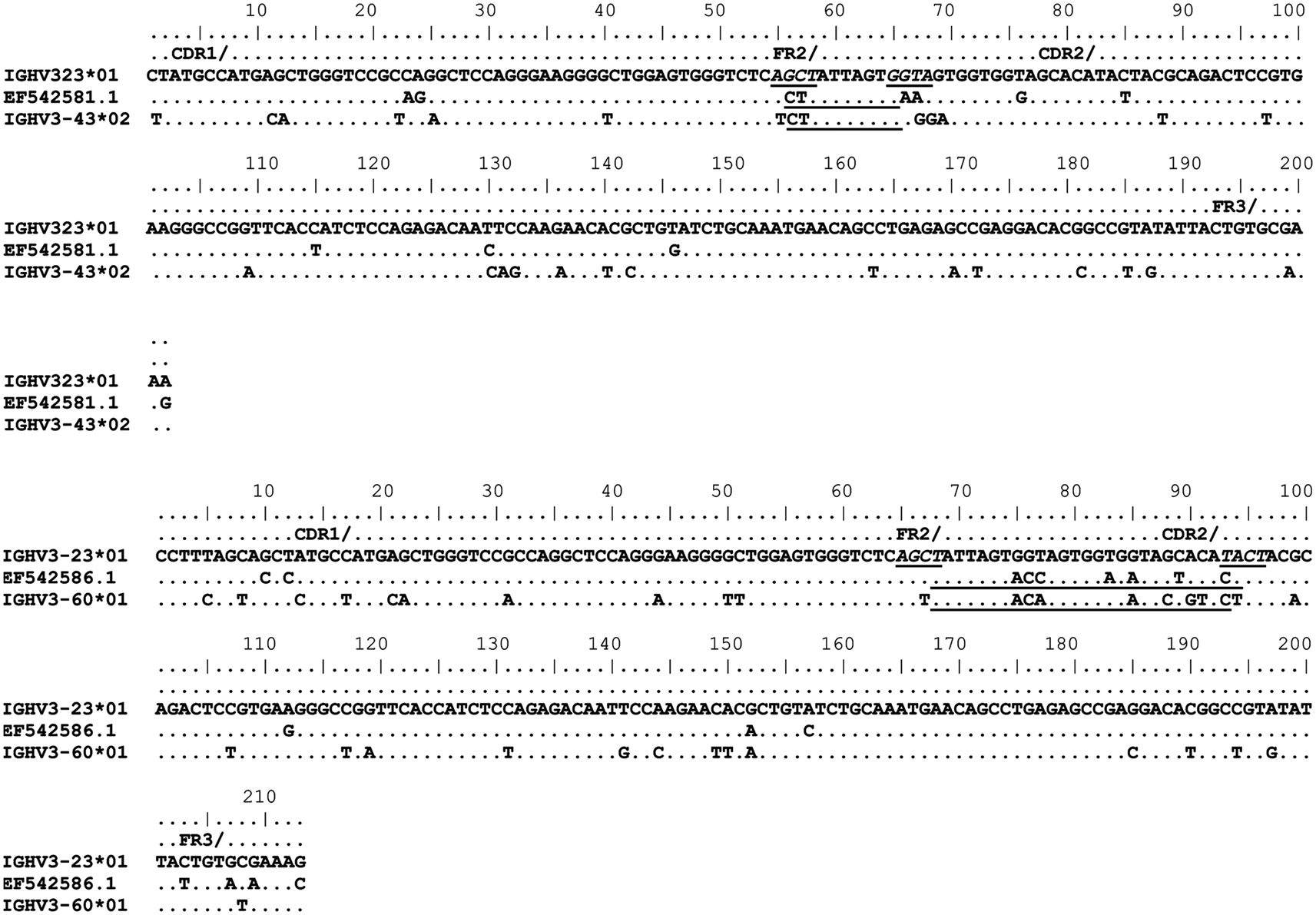
Figure 4. Gene conversion-like events between rearranged IGHV3-23*01 gene sequences and germline VH sequences superimposed by somatic hypermutation (SHM). IGHV3-23*01 mutated sequences obtained by single-cell PCR from CD19+IgD+CD27+ B cells (Brezinschek et al., 1995, 1997; Dörner et al., 1998; Longo et al., 2008). Topmost sequence is the germline of rearranged IGHV3-23*01 gene sequences from healthy individuals (EF542581.1 and EF542586.1). Potential donor germline IGHV3 family gene segments are shown below the rearranged IGHV3-23*01 sequences. GCV-like tract is underlined. Flanking SHM/AID hotspot motifs are underlined and italicized.
Characterization of GCV-Like Events in Rearranged IGHV3-23*01 Sequences
To identify GCV-like events in rearranged IGHV3-23*01 gene sequences, we searched the germline human IGVH locus data bases for matches to the mutated IGHV3-23*01 gene sequences. The IGHV3-23*01 gene sequences were collected from data bases of sequences from healthy individuals, CVID, and AID-deficient patients, Figure 1 shows statistically significant regions of GCV-like events identified by GENECONV in rearranged IGHV3-23*01 gene sequences, that had undergone affinity maturation as evidenced by the presence of SHM (henceforth referred to as hypermutated IGHV3-23*01 gene sequences). Figure 1 contains the analysis of IGHV3-23*01 gene sequences extracted from a healthy individual and a CVID patient, total PBMC RNA; Figures 2–4 contains analysis of IGHV3-23*01 gene sequences extracted from single-cell PCR of B cells from healthy individuals (details in Materials and Methods). No evidence of GCV-like events was identified in from IGHV3-23*01 sequences extracted from single-cell PCR of B cells from AID-deficient patients. Germline VH donors, tract length, and P-values from GENECONV are shown in Tables 2 and 3. The observed base substitutions in GCV-like events show clustered nucleotide differences, an unvarying characteristic of GCV, and thus striking evidence for GCV. Inspection of the IGVH locus2 revealed that both upstream and downstream germline VH genes were involved in the diversity of rearranged IGHV3-23*01 gene sequences through GCV-like mechanisms. Germline VH gene donors downstream of IGHV3-23*01 are: IGHV3-9*01, IGHV3-11*01, IGHV3-19*01, and IGHV3-21*01. Germline VH gene donors upstream of IGHV3-23*01 are: IGHV3-43*02, IGHV3-48*03, IGHV3-60*01 (a pseudogene), and IGHV3-64*04. All germline donor VH genes are functional unless so noted. The implication of these results is discussed below.
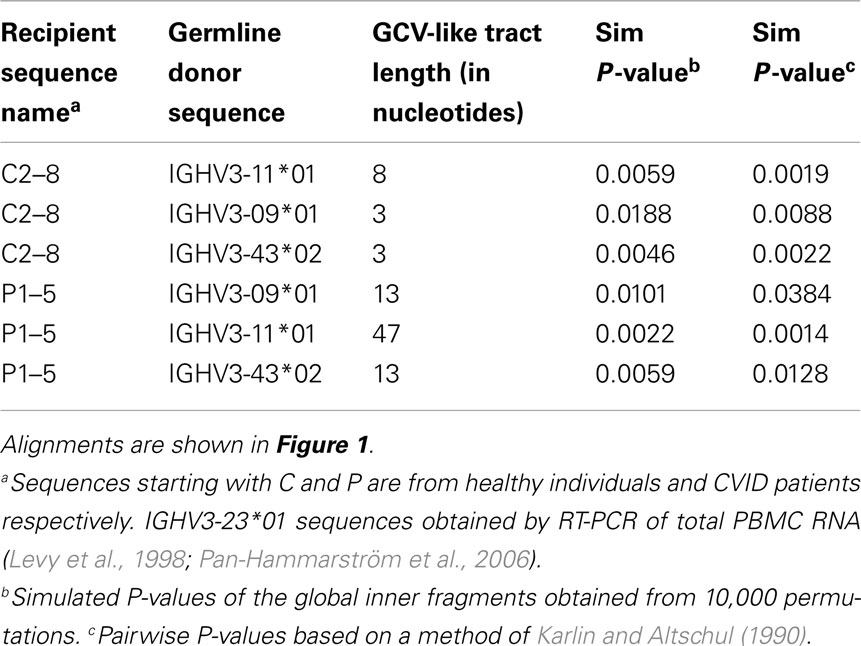
Table 2. Characteristics of GCV-like tracts predicted by GENECONV in rearranged IGHV3-23*01 gene sequences.
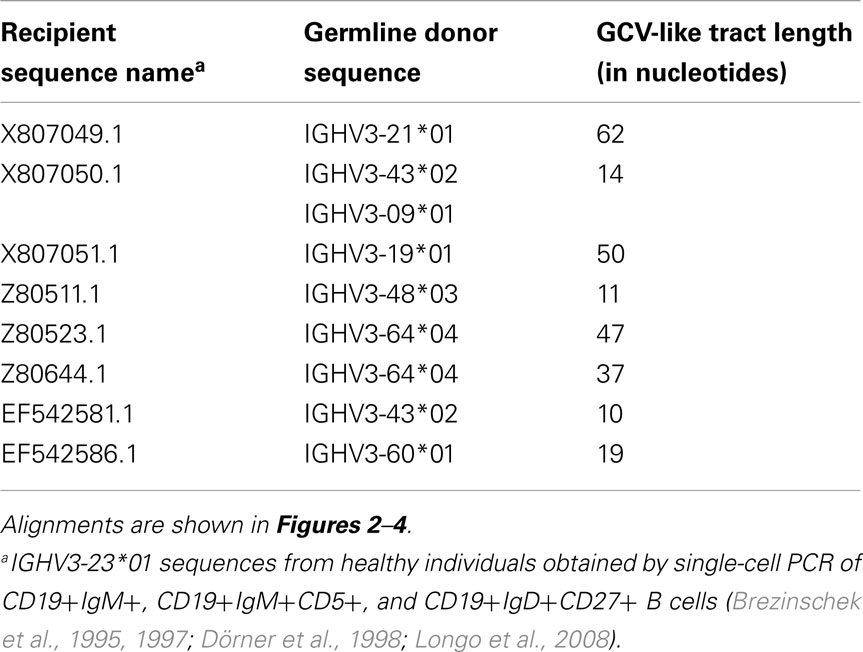
Table 3. Characteristics of GCV-like tracts superimposed by SHM (identified by visual inspection of alignments).
Germline VH Donors for Silent Mutations
As shown in Figures 1–4, GCV-like events were found to be spread in all regions of the VH sequence (CDR1, FR2, CDR2, and FR3). All base substitutions are shown in Tables 4 and 5. Replacement mutations are the result of both mutagenic mechanisms and the intense selection pressure that replacement mutations have undergone during affinity maturation. In order to distinguish between changes due to mutations from changes due to antigenic selection, we analyzed silent mutations for germline matches. Silent mutations as a result of GCV-like events represented more than 25% of nucleotide changes (Tables 4 and 5).
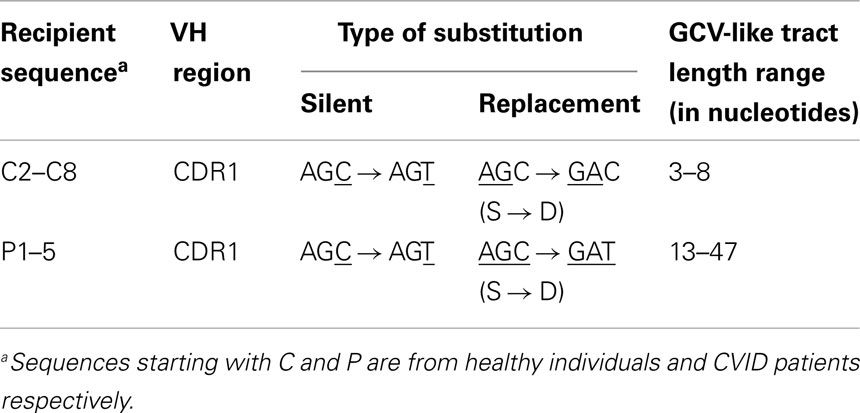
Table 4. Characteristics of base substitutions in GCV-like tracts shown in Figure 1.
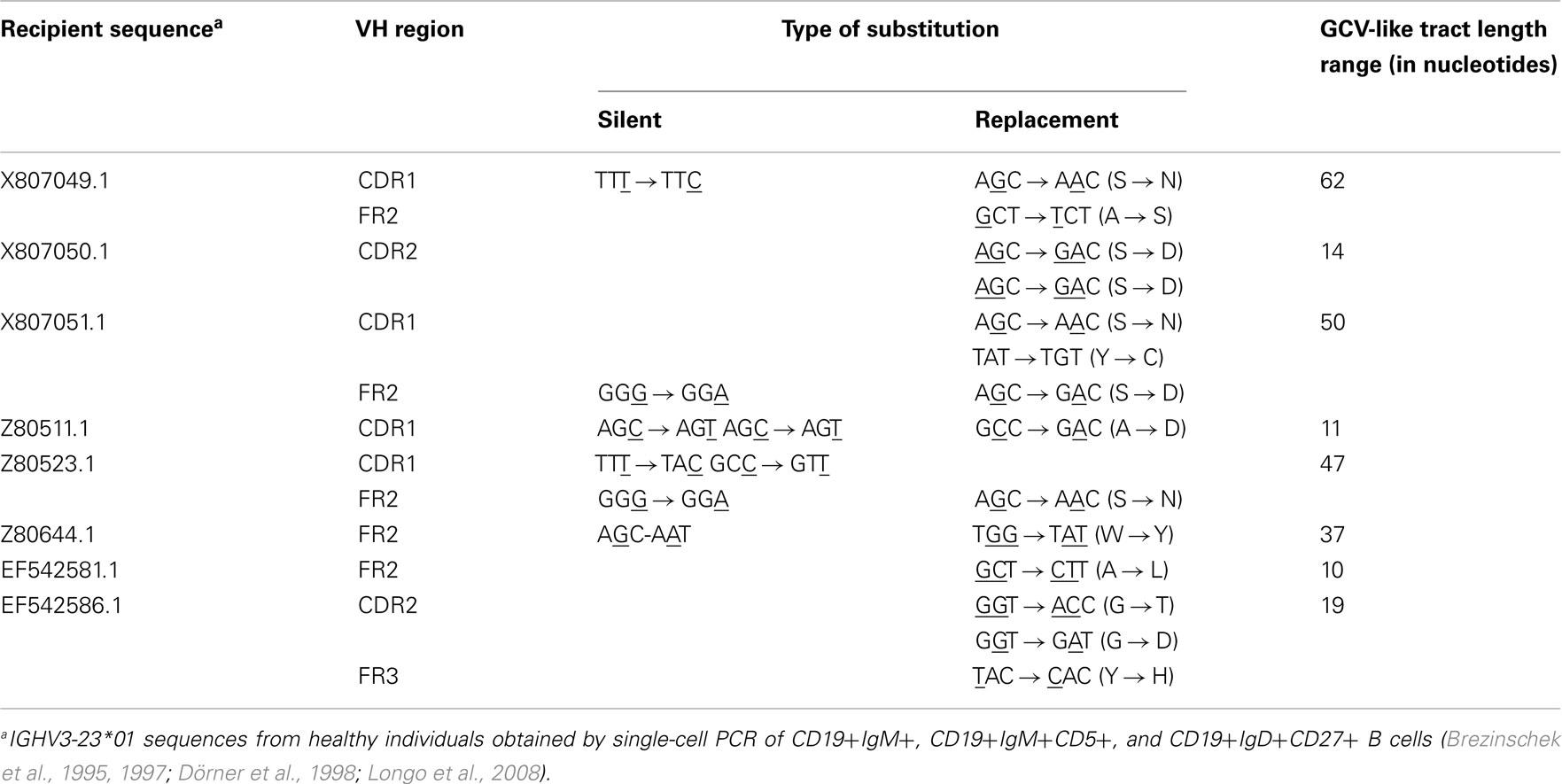
Table 5. Characteristics of base substitutions in GCV-like tracts superimposed by SHM shown in Figures 2–4.
Presence of Sequence-Specific Motifs Potentially Involved in Site-Specific Recombination
Several specific sequences or motifs are recognized by specialized enzymes that can cause DSBs and thus initiate HR through GCV or reciprocal exchanges (Flanagan et al., 1984; Kilpatrick et al., 1984; López-Correa et al., 2001; Lee et al., 2002; Chuzhanova et al., 2009). SHM, GCV, and CSR have all been shown to be initiated by DSBs (Brenner and Milstein, 1966; Wuerffel et al., 1997; Papavasiliou and Schatz, 2000; Chen et al., 2001; Petersen et al., 2001; Catalan et al., 2003; Rush et al., 2004; Tang and Martin, 2006). Like SHM, DSBs also preferentially target SHM hotspot motifs, RGYW/WRCY, in rearranged immunoglobulin genes (Papavasiliou and Schatz, 2000, 2002; Bross et al., 2002; Wu et al., 2003; Zan et al., 2003). Figures 1–4, show the flanking regions of GCV-like tracts with SHM/AID hotspot motifs italicized and underlined in the germline IGHV3-23*01 sequence. We speculate, based on strong evidence in the literature that these SHM motifs could have played a role in initiating the DSB’s and/or could be the targets of AID for resection of 5′ ends.
Presence of Structures that Promote GCV
Regions of the genome that tend to form non-B DNA conformations, such as stable SLSs, are susceptible to DSB’s, which in turn could initiate GCV through HR (Bacolla and Wells, 2004; Bacolla et al., 2004; Wang and Vasquez, 2006; Chen et al., 2007; Wells, 2007). The generation of SLS’s in immunoglobulin genes is coupled to transcription. Transcription can alter the DNA conformation which can fold into secondary structures exposing DNA largely in single stranded state. Single stranded DNA (ssDNA) is inherently fragile and can be the target of enzymes generating DSBs.
With substantial evidence showing that RGYW/WRCY motifs are preferential targets of DSB’s and resected ends during SHM, we undertook structural analysis of bases that are present at or nearest to the break points of GCV-like tracts (5′ and 3′). The hypermutable bases in the flanking regions of GCV-like tracts for which structures are drawn are italicized in Figures 1–4. Since SHM (and AID) targeted bases are predominantly C’s and G’s, we analyzed the paired or unpaired state of C’s or G’s at or nearest to GCV-break points. We used the germline non-transcribed strand (NTS) of IGHV3-23*01 for the structural analysis through simulated transcription using the DNA secondary structural analysis tool, “mfg.” “Mfg” predicts the most stable SLS in which a particular base is unpaired. The propensity of a base to be in the single stranded condition is quantified as the percent unpaired (%UP) and the stabilities of the SLSs as −ΔGs’ are shown in Figure 5. Figure 5 represents the most stable SLSs in which hypermutable bases that are present at GCV-break points are exposed in unpaired or in ssDNA state (C’s or G’s, shown with arrows). This structural analysis allows the speculation that the observed GCV-like tracts in IGHV3-23*01 sequences are initiated by the DSB’s in the sequence-specific motifs owing to their localization in the ssDNA state in stable SLSs.
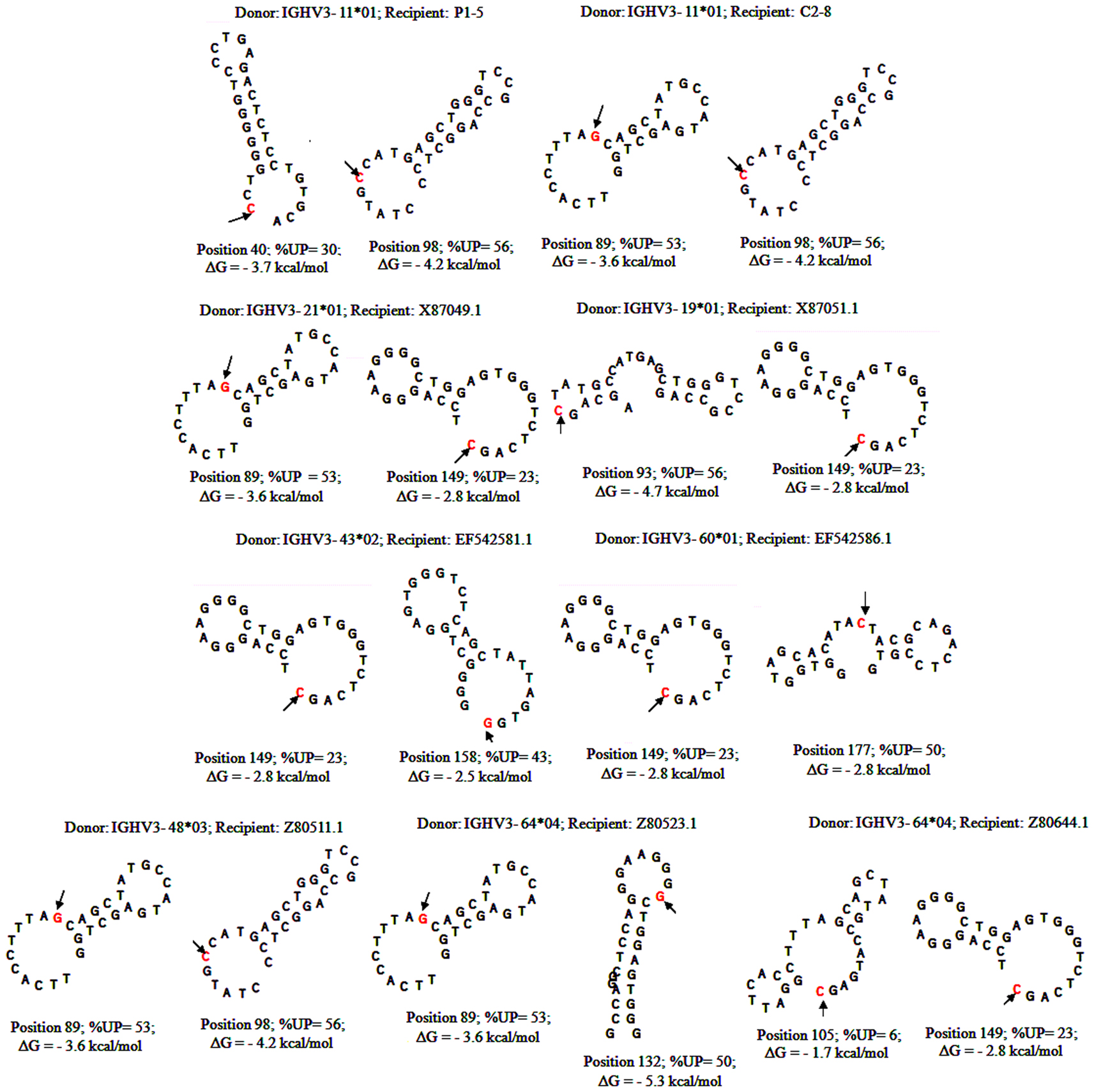
Figure 5. Stem-loop structures of hypermutable bases in SHM/AID hotspot motifs flanking 5′ and 3′of GCV-like tracts. DNA secondary structures formed by folding germline IGHV3-23 sequence in program “mfg” during simulated transcription (Wright et al., 2003, 2004, 2008a,b). Each structure flanking GCV-like tracts are named as the rearranged IGHV3-23*01 sequences shown in Figures 1–4. The hypermutable bases are shown with arrows. DNA secondary structural parameters, −ΔG, % Unpaired (%UP) correspond to hypermutable base (shown with arrows).
Comparison of GCV-Like Events between Healthy Individuals, CVID Patients, and AID-Deficient Patients
Our previous studies (Duvvuri et al., 2011, 2012) revealed SHM patterns suggestive of defects in DNA repair mechanisms in some CVID patients. Given the role of DNA repair processes in GCV mechanisms, we compared the occurrence of GCV-like events between two cohorts, CVID pts and healthy individuals. Rearranged IGHV3-23*01 gene sequences from CVID patients showed significantly lower levels of GCV-like events compared to those of healthy individuals [1 event/161 sequences (0.621%) vs. 8 events/121 sequences (6.61%) respectively; Fisher’s two-tailed P-value = 0.0057].
No GCV-like events were observed from any rearranged IGHV3-23*01 sequences of AID-deficient patients.
In the analysis, although germline donors could be identified for single base changes, evidence of GCV-like events required clustered templated mutations. Hence, only clustered mutations were considered as evidence of GCV-like events. This criterion very likely will underestimate the actual number of GCV-like events.
Discussion
Our computational analysis of rearranged human IGHV3-23*01 sequences led to the identification of GCV-like events. GCV as a means of generating antibody diversity is extensive in some species especially in chicken (Reynaud et al., 1987, 1989, 1994; McCormack et al., 1991; Sayegh et al., 1999; Arakawa and Buerstedde, 2004; Ratcliffe, 2006). Definitive evidence of GCV-like events in human rearranged immunoglobulin genes has been scarce (Darlow and Stott, 2006).
Excluding Potential PCR Artifacts
VDJ chimeric molecules resembling somatic GCV can be produced during PCR in vitro by a process referred to as “template jumping” (Pääbo et al., 1990; Brakenhoff et al., 1991). In order to identify whether PCR artifacts were the likely cause for the GCV-like events, we analyzed rearranged mutated IGHV3-23*01 sequences obtained from single-cell PCR from individual B cell lysates from healthy individuals and AID-deficient patients (shown in Figures 2–4). AID is absolutely required for GCV in immunoglobulin genes (Arakawa et al., 2002). The presence of any GCV-like event in rearranged IGHV3-23*01 sequences in AID-deficient patients would provide evidence of PCR artifacts.
In Figure 2, the rearranged sequences from single-cell PCR, X87049.1, and X87051, had evidence of templated mutations from germline VH3 donors downstream of the IGHV3-23*01 gene. During V(D)J recombination at the human IGVH locus downstream VH gene segments are deleted. Since these GCV-like events are from single-cell PCR, the possible source of VH donors for these GCV-like events could be from either the sister chromatid or the homolog, that is, trans germline VH donors. Our analysis on rearranged IGHV3-23*01 sequences from AID-deficient patients did not reveal any GCV-like events, including no sequences that could have been chimeric PCR artifacts. This result strongly implicates the source of our identified GCV-like events as being from an in vivo mechanism that requires AID.
For the GCV-like events identified in IGHV3-23*01 mutated sequences in Figure 1, were obtained from total PBMC RNA, we explored whether these events could be due to artifactual PCR “jumping events.” Mutated IGHV3-23*01 sequences shown in Figure 1, are from two independent studies. C2–8 is from healthy individual (Pan-Hammarström et al., 2006) and P-5 is from a CVID patient (Levy et al., 1998). C2–8 and P1–5 sequences have GCV-like transfer in the same region. It is unlikely that two similar PCR-chimeras could be isolated from independent PCR reactions. In addition, sequences C2–8 and P1–5 resemble IGHV3-23*01 downstream of the GCV-like event (underlined region), and not the donor IGVH sequence. Thus for these sequences to be produced by a PCR-artifact, two “jump events” during PCR, would be required: One “jump event” from the IGHV3-23*01 template to a donor sequences and the second from a donor sequence to IGHV3-23*01. And identical “jump events” would have had to occur in two different labs. We consider these scenarios to be unlikely and thus we reason that the observed GCV-like transfer is most likely the result of an in vivo GCV-like mechanism.
The mechanism for the templated mutations shown in Figure 6 remains questionable. These mutated IGHV3-23*01 sequences were obtained from total PBMC RNA (Levy et al., 1998; Pan-Hammarström et al., 2006). Sinkora et al. (2000) demonstrated that PCR generated VDJ chimeras occur predominantly in FR3 regions followed by FR2 (2000). Hence we considered whether the GCV-like events, detailed in Figure 6, could be the result of PCR-chimerism (“jump events”). Given that the examples shown in Figure 6 have templated and un-templated mutations, a characteristic expected of extensive in vivo antibody diversification, the possibility of them being true GCV-like events remains.
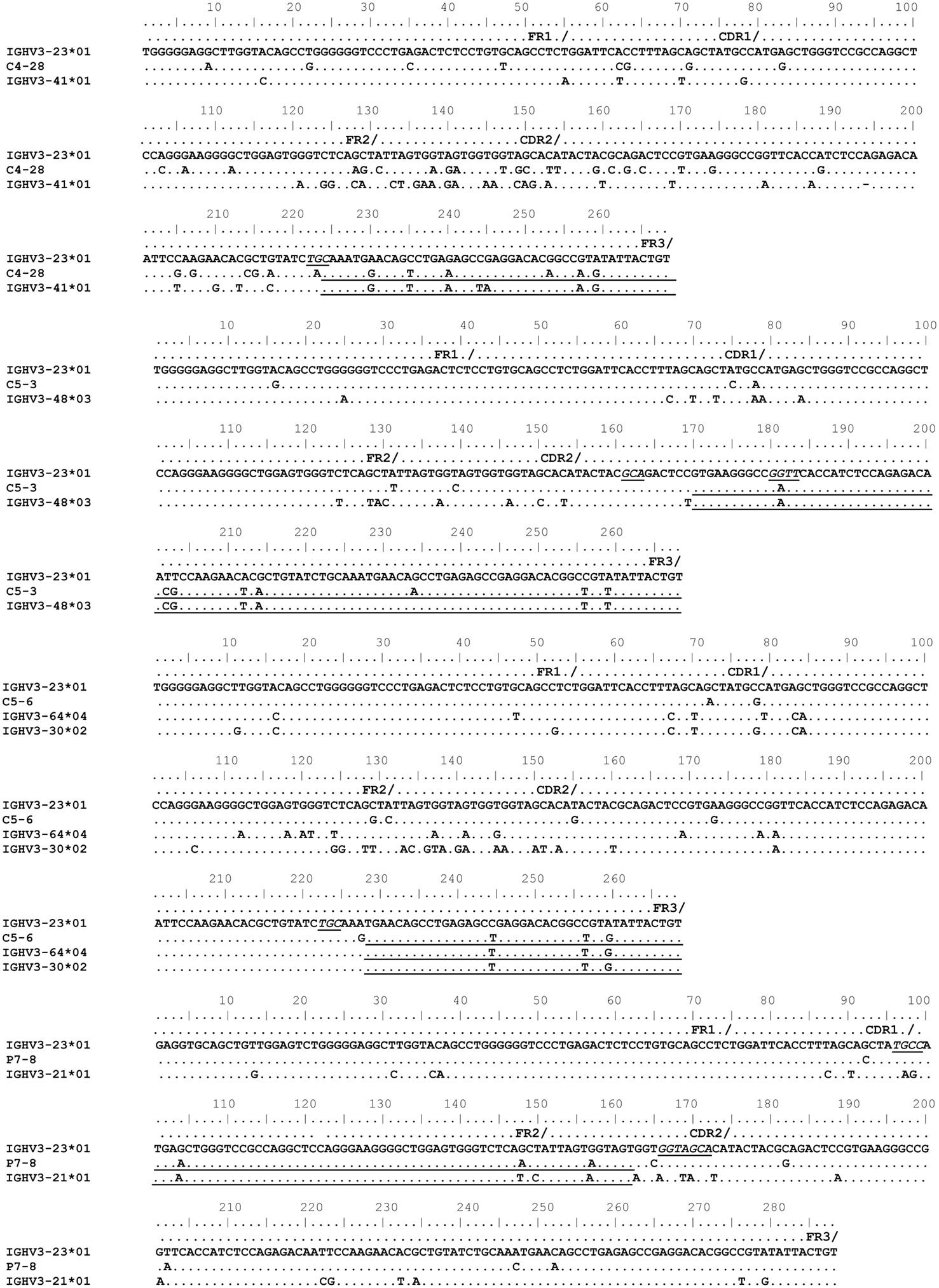
Figure 6. Gene conversion-like events between rearranged IGHV3-23*01 gene sequences and germline VH sequences superimposed by somatic hypermutation (SHM). Topmost sequence is the germline of rearranged IGHV3-23*01 gene sequences from healthy individuals (C4–28, C5–3, C5–6) and a CVID patient (P7–8). Potential donor germline IGHV3 family gene segments are shown below the rearranged IGHV3-23*01 sequences. GCV-like tracts are underlined. Flanking SHM/AID hotspot motifs are underlined and italicized.
Critical Observations of GCV-Like Events in Human Rearranged IGHV3-23*01 Gene Sequences
As the name implies, HR takes place between sequences that have considerable similarity which is critical to stabilize DNA pairing. All the identified germline GCV-like VH donors in this study belong to the VH3 family, a highly homologous multigene family. We found high similarity (>87%) between the potential germline VH donors and the IGHV3-23*01 gene sequence. These findings are consistent with the observations that GCV is favored between highly similar genes usually with identity >80% (Shen and Huang, 1986; David et al., 1992; Xu et al., 2008). The length of identical region also significantly affects the probability of GCV. It has been shown that GCV can occur with regions of identity as small as ∼10 bp (David et al., 1992). We have identified minimum (3 nts) and maximum (62 nts) sequence identities between donor and recipient sequences that can serve as targets for GCV and our findings are consistent with those reported in literature (David et al., 1992). All of the identified templated mutations are clustered nucleotide differences. Clustering is strong evidence for GCV. It is very unlikely that clustered templated diversity observed in the rearranged IGHV3-23*01 gene sequence is the result of independent somatic hypermutational events.
The germline donor VH genes identified were upstream and downstream of the rearranged IGHV3-23*01 gene. The sources of donor DNA could be from sequences upstream of the rearranged gene or from homologous DNA on the homolog or another chromosome or from episomal remnants of VH to DHJH rearrangements. Results of several studies over the years have suggested that CSR can occur between the chromosomes, although the predominant mode of recombination is intrachromosomal. Trans-chromosomal recombination between IGVH and Cα allotypes through GCV has been documented in rabbits (Tosi et al., 1970; Pernis et al., 1973). Kingzette et al. (1998) documented trans-chromosomal recombination at a high frequency (7%) within the IGVH locus of rabbit B lymphocytes. Recombination between transgenic VHDJH gene and the endogenous IGVH locus has also been shown to occur by interchromosomal GCV in mice (Giusti and Manser, 1994). These studies demonstrated that GCV can occur in trans. Given this evidence, it seemed reasonable to us to expect GCV-like events could occur in trans in humans.
Sequence and Structural Consideration of GCV-Like Events
By modeling the dynamics of IGHV3-23*01 DNA folding during simulated transcription, we demonstrated the presence of hypermutable bases, flanking GCV-like tracts, in ssDNA of stable SLSs.
There are two models to explain the generation of DSB’s in DNA that initiate HR which can lead to GCV. In the first model, the excision of an AID-induced G:U lesion can result in an abasic site, which is then cleaved by apurinic/apyrimidinic endonucleases to generate DSB’s that can initiate HR (Casali and Zan, 2004). In the second model, AID is dispensable for the generation of blunt-ended DSB’s that preferentially occur in RGYW/WRCY motifs (Unniraman et al., 2004). However, AID is critical for the generation of resected DSB’s that initiate strand invasion and the subsequent repair of resected ends by HR in error-prone fashion (Papavasiliou and Schatz, 2002). Similarly to blunt end DSB’s, resected ends also target RGYW/WRCY motifs (Papavasiliou and Schatz, 2002; Zan et al., 2003). Whichever is the mechanism for the generation of DSB’s that initiate HR, hypermutable bases of the SHM hotspot motifs in ssDNA of stable SLSs can provide the substrates for the initial or resected cleavage.
A model could be proposed for the role of transcription-induced DNA conformation in initiating HR: studies have shown a correlation between transcription strength and the degree of negative supercoiling. High rates of transcription as in VH genes can induce supercoiling which in turn can promote the formation of large DNA secondary structures (Liu and Wang, 1987; Staudt and Lenardo, 1991; Wright, 2000; Schmidt et al., 2006). Supercoiled DNA and/or the large DNA secondary structures can promote HR by bringing homologous DNA sequences closer together for strand exchange. The hypermutable bases in ssDNA state in SLSs can be targeted for DSB’s to initiate HR.
Common Mutations are Products of GCV-Like Events
One replacement mutation (Ser → Asp) at codon 31 that was contained in clustered mutations is prominent in both healthy individuals and CVID patients. This codon (AGC) at position 31 in CDR1 region was found to be highly mutable in other independent mutational studies with IGHV3-23*01 gene. Repeated codon 31 mutations are apparently selected mutations following affinity maturation. The question is, is this selected mutation from GCV or from independent SHM events. There have been instances where independently generated antibodies shared four consecutive identical amino acids in CDR2 and interestingly recognized the same antigen. In that study, the authors could not identify any obvious germline donors for the observed clustered mutations. The absence of identified donors led to the conclusion that the shared identical mutations were not products of GCV but likely were acquired in independent SHM events under intense selection pressure for such mutated regions (Berek et al., 1987; Allen et al., 1988; Claflin et al., 1989; Clarke et al., 1990; Wysocki et al., 1990). A high incidence of shared mutations in independent products of SHM has been observed in humans (van der Stoep et al., 1993).
Although independent SHM events cannot be ruled out, GCV-like events could be responsible for these shared mutations at codon 31 of IGHV3-23*01 gene for the following reasons: the reoccurrence of identical mutations in mutated versions of the same germline gene (IGHV3-23*01), the identification of germline donors, and the length of matches (3–47 nts). These reasons, consistent with those reported in literature for GCV provide evidence that the observed mutations are templated and the products of single recombinational events.
GCV-like Events in Healthy Individuals, CVID, and AID-Deficient Patients
We have observed that the frequency of GCV-like events is significantly higher in rearranged IGHV3-23*01 sequences from healthy individuals compared to that of CVID patients. CVID is a heterogeneous disorder of chiefly unknown cause (Sneller, 2001). As the mechanism of GCV in human rearranged immunoglobulin gene sequences is unknown, we are unable to conclude that CVID patients have an enzymatic/mechanistic defect in GCV. However, in the view of our previous study that some CVID patients have defective MMR resolution of AID-induced mutations, it remains that DNA repair defects in CVID patients could that be responsible for the lower frequency of GCV-like events. Additional support to our proposition comes from genetic association studies in CVID patients that identified Mre11-Rad50-Nbs1 (MRN) complex defects in their screening of 27 DNA repair genes (Offer et al., 2010). MRN complex plays a crucial role in DSB damage response in eukaryotic damage response via both the HR and NHEJ pathways (Lamarche et al., 2010). CVID patients demonstrate increased propensity to cancer and increased sensitivity to ionizing radiation, both plausible outcomes of defective/inappropriate DNA repair mechanisms (Hermaszewski and Webster, 1993; Vorechovský et al., 1993; Filipovich et al., 1994; Palanduz et al., 1998; Gompels et al., 2003).
Our sequence analysis of IGHV3-23*01 mutated sequences from AID-deficient patients (Longo et al., 2008) did not reveal any evidence of GCV-like events. Our findings corroborates the findings of other studies which have demonstrated that AID is absolutely required for GCV in immunoglobulin genes (Arakawa et al., 2002).
Supposition for Rarity of GCV-Like Events in Human Rearranged Immunoglobulin Genes
Although GCV can generate rapid, extensive changes in immunoglobulin genes in response to certain antigens, it is not frequently observed in humans.
Gene conversion may not be required since considerable diversity is present in primary immune repertoire and fine tuning through SHM is enough to generate potent immune response. The identification of templated mutations in human rearranged IGHV3-23*01 sequences steered us to search for alternate explanations.
The complexity of the mammalian immunoglobulin loci might impose some unknown limitations for GCV in mammals. It could also be due to the lack of homologous donor VH in accessible chromatin (Xu and Selsing, 1994; Weill and Reynaud, 1996; Sale et al., 2001). There could be some mechanistic differences between species that use GCV as a predominant mechanism of antibody diversification compared to the species that do not. For example, humans have low frequency of HR activity compared to chickens (Miyagawa et al., 2002). Low recombination frequency might be the reason for the lack of GCV in human rearranged immunoglobulin genes, despite the presence of highly homologous germline donors. Still another explanation is the presence of multiple numbers of homologous sequences that might result in more than one GCV event in a rearranged sequence and make it difficult to the identify donor sequences.
The use of identical sequences from sister chromatid rather than homologous (non-identical) sequences predominantly for HR and the subsequent repair in error-prone manner can result in mutations that will appear as un-templated mutations, even though they are products of GCV-like events. Alternatively, high fidelity repair by HR using sister chromatid as template would make GCV-like tracts invisible. Support comes from studies that demonstrated that in hypermutating B cells, DSB’s occur predominantly in S/G2 phases of cell cycle (Papavasiliou and Schatz, 2000). HR is preferentially used in S/G2 phases of cell cycle for DNA lesion repair (Rothkamm et al., 2003). The cell cycle requirement for HR likely reflects the necessity of sister chromatids as template for DNA repair through HR. In such a scenario, GCV-like mechanism through HR would result in indiscernible GCV-like events, thus leading to assumption that GCV-like events are absent or rare, as in human rearranged immunoglobulin diversification.
Diversification by GCV-Like Events
The purpose GCV affinity maturation in humans remains unclear. The contribution to affinity maturation by GCV can be quite different from that of SHM. GCV, unlike SHM, can result in multiple base substitutions that can alter many amino acids. In chicken and rabbits a single round of GCV event has been shown to affect 4–19 codons (Reynaud et al., 1987, 1989; Becker and Knight, 1990). Thus the changes in affinity generated by GCV collectively are greater than the changes in affinity made by individual base substitutions. Such extensive changes in affinity as a result of GCV could be critical for clearance of highly mutating pathogens. Germline VH donors are evolutionarily selected. It is advantageous to the species to encode the critical diversity needed at birth in the germline configuration (Benedict and Kearney, 1999). Through mechanisms, like GCV, evolutionarily fixed germline diversity can be transferred to rearranged immunoglobulin genes. On similar lines of receptor editing, the GCV base substitutions in CDRs can altogether change the specificity of antigenic receptor rather than just altering the affinity of receptor toward the antigen (characteristic of SHM). Such type of receptor editing based on multiple templated mutations can prevent self-reactivity of antigen receptors without too much wastage of the primary immune repertoire.
In this study we have identified GCV-like events in human rearranged IGHV3-23*01 gene sequences, that account for 6.61% in the healthy individuals. Given the potential for extensive changes in antibody affinity and/or specificity, this frequency of GCV-like events would be instrumental in protecting humans against pathogens that diversify their genome by antigenic shift.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Bhargavi Duvvuri is supported by the Ontario Graduate Scholarship and The President Susan Mann Dissertation Scholarship. Gillian E. Wu is funded by The Arthritis Society. The authors would like to thank the reviewers for their valuable comments and suggestions to improve the quality of the paper.
Footnotes
References
Allen, D., Simon, T., Sablitzky, F., Rajewsky, K., and Cumano, A. (1988). Antibody engineering for the analysis of affinity maturation of an anti-hapten response. EMBO J. 7, 1995–2001.
Alt, F. W., Blackwell, T. K., and Yancopoulos, G. D. (1987). Development of the primary antibody repertoire. Science 238, 1079–1087.
Arakawa, H., and Buerstedde, J. M. (2004). Immunoglobulin gene conversion: insights from bursal B cells and the DT40 cell line. Dev. Dyn. 229, 458–464.
Arakawa, H., Hauschild, J., and Buerstedde, J. M. (2002). Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 295, 1301–1306.
Bacolla, A., Jaworski, A., Larson, J. E., Jakupciak, J. P., Chuzhanova, N., Abeysinghe, S. S., O’Connell, C. D., Cooper, D. N., and Wells, R. D. (2004). Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl. Acad. Sci. U.S.A. 101, 14162–14167.
Bacolla, A., and Wells, R. D. (2004). Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 279, 47411–47414.
Becker, R. S., and Knight, K. L. (1990). Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbits. Cell 63, 987–997.
Benedict, C. L., and Kearney, J. F. (1999). Increased junctional diversity in fetal B cells results in a loss of protective anti-phosphorylcholine antibodies in adult mice. Immunity 10, 607–617.
Berek, C., Jarvis, J. M., and Milstein, C. (1987). Activation of memory and virgin B cell clones in hyperimmune animals. Eur. J. Immunol. 17, 1121–1129.
Bezzubova, O. Y., and Buerstedde, J. M. (1994). Gene conversion in the chicken immunoglobulin locus: a paradigm of homologous recombination in higher eukaryotes. Experientia 50, 270–276.
Brakenhoff, R. H., Schoenmakers, J. G., and Lubsen, N. H. (1991). Chimeric cDNA clones: a novel PCR artifact. Nucleic Acids Res. 19, 1949.
Bransteitter, R., Pham, P., Scharff, M., and Goodman, M. (2003). Activation induced cytidine deaminase deaminates deoxycytidine on single stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. U.S.A. 100, 4102–4107.
Brezinschek, H. P., Brezinschek, R. I., and Lipsky, P. E. (1995). Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J. Immunol. 155, 190–202.
Brezinschek, H. P., Foster, S. J., Brezinschek, R. I., D�r, T., Domiati-Saad, R., and Lipsky, P. E. (1997). Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(−)/IgM+ B cells. J. Clin. Invest. 99, 2488–2501.
Bross, L., Fukita, Y., McBlane, F., Demolliere, C., Rajewsky, K., and Jacobs, H. (2000). DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity 13, 589–597.
Bross, L., Muramatsu, M., Kinoshita, K., Honjo, T., and Jacobs, H. (2002). DNA double-strand breaks: prior to but not sufficient in targeting hypermutation. J. Exp. Med. 195, 1187–1192.
Casali, P., and Zan, H. (2004). Class switching and Myc translocation: how does DNA break? Nat. Immunol. 5, 1101–1103.
Catalan, N., Selz, F., Imai, K., Revy, P., Fischer, A., and Durandy, A. (2003). The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch mu region. J. Immunol. 171, 2504–2509.
Chen, J. M., Cooper, D. N., Chuzhanova, N., Férec, C., and Patrinos, G. P. (2007). Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 8, 762–775.
Chen, X., Kinoshita, K., and Honjo, T. (2001). Variable deletion and duplication at recombination junction ends: implication for staggered double-strand cleavage in class-switch recombination. Proc. Natl. Acad. Sci. U.S.A. 98, 13860–13865.
Chuzhanova, N., Chen, J. M., Bacolla, A., Patrinos, G. P., Férec, C., Wells, R. D., and Cooper, D. N. (2009). Gene conversion causing human inherited disease: evidence for involvement of non-B-DNA-forming sequences and recombination-promoting motifs in DNA breakage and repair. Hum. Mutat. 30, 1189–1198.
Claflin, J. L., George, J., Dell, C., and Berry, J. (1989). Patterns of mutations and selection in antibodies to the phosphocholine-specific determinant in Proteus morganii. J. Immunol. 143, 3154–3063.
Clarke, S., Rickert, R., Wloch, M. K., Staudt, L., Gerhard, W., and Weigert, M. (1990). The BALB/c secondary response to the Sb site of influenza virus hemagglutinin. Nonrandom silent mutation and unequal numbers of VH and Vk mutations. J. Immunol. 145, 2286–2296.
Darlow, J. M., and Stott, D. I. (2006). Gene conversion in human rearranged immunoglobulin genes. Immunogenetics 58, 511–522.
David, V., Folk, N. L., and Maizels, N. (1992). Germ line variable regions that match hypermutated sequences in genes encoding murine anti-hapten antibodies. Genetics 132, 799–811.
D’Avirro, N., Truong, D., Luong, M., Kanaar, R., and Selsing, E. (2002). Gene conversion-like sequence transfers between transgenic antibody V genes are independent of RAD54. J. Immunol. 169, 3069–3075.
D’Avirro, N., Truong, D., Xu, B., and Selsing, E. (2005). Sequence transfers between variable regions in a mouse antibody transgene can occur by gene conversion. J. Immunol. 175, 8133–8137.
Dickerson, S., Market, E., Besmer, E., and Papavasiliou, F. N. (2000). AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197, 1291–1296.
Dildrop, R., Bruggemann, M., Radbruch, A., Rajewsky, K., and Beyreuther, K. (1982). Immunoglobulin V region variants in hybridoma cells. II. Recombination between V genes. EMBO J. 1, 635–640.
Dörner, T., Brezinschek, H. P., Foster, S. J., Brezinschek, R. I., Farner, N. L., and Lipsky, P. E. (1998). Comparable impact of mutational and selective influences in shaping the expressed repertoire of peripheral IgM+/CD5− and IgM+/CD5+ B cells. Eur. J. Immunol. 28, 657–668.
Duvvuri, B., Duvvuri, V. R., Grigull, J., Martin, A., Pan-Hammarström, Q., Wu, G. E., and Larijani, M. (2011). Altered spectrum of somatic hypermutation in common variable immunodeficiency disease characteristic of defective repair of mutations. Immunogenetics 63, 1–11.
Duvvuri, B., Duvvuri, V. R., Wu, J., and Wu, G. E. (2012). Stabilised DNA secondary structures with increasing transcription localise hypermutable bases for somatic hypermutation in IGHV3-23. Immunogenetics. [Epub ahead of print].
Filipovich, A. H., Mathur, A., Kamat, D., Kersey, J. H., and Shapiro, R. S. (1994). Lymphoproliferative disorders and other tumors complicating immunodeficiencies. Immunodeficiency 5, 91–112.
Flanagan, J. G., Lefranc, M. P., and Rabbitts, T. H. (1984). Mechanisms of divergence and convergence of the human immunoglobulin alpha 1 and alpha 2 constant region gene sequences. Cell 36, 681–688.
Giusti, A. M., and Manser, T. (1994). Somatic generation of hybrid antibody H chain genes in transgenic mice via interchromosomal gene conversion. J. Exp. Med. 179, 235–248.
Gompels, M. M., Hodges, E., Lock, R. J., Angus, B., White, H., Larkin, A., Chapel, H. M., Spickett, G. P., Misbah, S. A., and Smith, J. L. (2003). Lymphoproliferative disease in antibody deficiency: a multi-centre study. Clin. Exp. Immunol. 134, 314–320.
Griebel, P. J., and Ferrari, G. (1994). Evidence for a stromal cell-dependent, self renewing B cell population in lymphoid follicles of the ileal Peyer’s patch of sheep. Eur. J. Immunol. 24, 401–409.
Harris, R. S., Sale, J. E., Petersen-Mahrt, S. K., and Neuberger, M. S. (2002). AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 12, 435–38.
Hastings, P. J. (1988). “Conversion events in fungi,” in Genetic Recombination, eds R. Kucherlapati, and G. Smith (Washington, DC: American Society for Microbiology), 397–428.
Hermaszewski, R. A., and Webster, A. D. (1993). Primary hypogammaglobulinaemia: a survey of clinical manifestations and complications. Q. J. Med. 86, 31–42.
Hood, L., Campbell, J. H., and Elgin, S. C. R. (1975). The organization, expression, and evolution of antibody genes and other multigene families. Annu. Rev. Genet. 9, 305–353.
Karlin, S., and Altschul, S. F. (1990). Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl. Acad. Sci. U.S.A. 87, 2264–2268.
Kilpatrick, M. W., Klysik, J., Singleton, C. K., Zarling, D. A., Jovin, T. M., Hanau, L. H., Erlanger, B. F., and Wells, R. D. (1984). Intervening sequences in human fetal globin genes adopt left-handed Z helices. J. Biol. Chem. 259, 7268–7274.
Kingzette, M., Spieker-Polet, H., Yam, P. C., Zhai, S. K., and Knight, K. L. (1998). Trans-chromosomal recombination within the Ig heavy chain switch region in B lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 95, 11840–11845.
Krawinkel, U., Zoebelein, G., Bruggemann, M., Radbruch, A., and Rajewsky, K. (1983). Recombination between antibody heavy chain variable-region genes: evidence for gene conversion. Proc. Natl. Acad. Sci. U.S.A. 80, 4997–5001.
Kurosawa, K., Lin, W., and Ohta, K. (2010). Distinct roles of HDAC1 and HDAC2 in transcription and recombination at the immunoglobulin loci in the chicken B cell line DT40. J. Biochem. 148, 201–207.
Lamarche, B. J., Orazio, N. I., and Weitzman, M. D. (2010). The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 584, 3682–3695.
Larijani, M., Petrov, A. P., Kolenchenko, O., Berru, M., Krylov, S. N., and Martin, A. (2007). AID associates with single-stranded DNA with high affinity and a long complex half-life in a sequence-independent manner. Mol. Cell. Biol. 27, 20–30.
Lee, H. H., Niu, D. M., Lin, R. W., Chan, P., and Lin, C. Y. (2002). Structural analysis of the chimeric CYP21P/CYP21 gene in steroid 21-hydroxylase deficiency. J. Hum. Genet. 47, 517–522.
Levy, Y., Gupta, N., Le Deist, F., Garcia, C., Fischer, A., Weill, J. C., and Reynaud, C. A. (1998). Defect in IgV gene somatic hypermutation in common variable immuno-deficiency syndrome. Proc. Natl. Acad. Sci. U.S.A. 95, 13135–13140.
Liu, L. F., and Wang, J. C. (1987). Super coiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U.S.A. 84, 7024–7027.
Liu, M., and Schatz, D. G. (2009). Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 30, 173–181.
Longo, N. S., Satorius, C. L., Plebani, A., Durandy, A., and Lipsky, P. E. (2008). Characterization of Ig gene somatic hypermutation in the absence of activation-induced cytidine deaminase. J. Immunol. 181, 1299–1306.
López-Correa, C., Dorschne, M., Brems, H., Lázaro, C., Clementi, M., Upadhyaya, M., Dooijes, D., Moog, U., Kehrer-Sawatzki, H., Rutkowski, J. L., Fryns, J. P., Marynen, P., Stephens, K., and Legius, E. (2001). Recombination hotspot in NF1 microdeletion patients. Hum. Mol. Genet. 10, 1387–1392.
Lucier, M. R., Thompson, R. E., Waire, J., Lin, A. W., Osborne, B. A., and Goldsby, R. A. (1998). Multiple sites of V lambda diversification in cattle. J. Immunol. 161, 5438–5444.
McCormack, W. T., and Thompson, C. B. (1990). Chicken Ig L variable region gene conversion display pseudogene donor preference and 5′ to 3′ polarity. Genes Dev. 4, 548–558.
McCormack, W. T., Tjoelker, L. W., and Thompson, C. B. (1991). Avian B-cell development: generation of an immunoglobulin repertoire by gene conversion. Annu. Rev. Immunol. 9, 219.
Meyer, A., Parng, C. L., Hansal, S. A., Osborne, B. A., and Goldsby, R. A. (1997). Immunoglobulin gene diversification in cattle. Int. Rev. Immunol. 15, 165–183.
Miyagawa, K., Tsuruga, T., Kinomura, A., Usui, K., Katsura, M., Tashiro, S., Mishima, H., and Tanaka, K. (2002). A role for RAD54B in homologous recombination in human cells. EMBO J. 21, 175–180.
Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y., and Honjo, T. (2000). Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563.
Odegard, V. H., and Schatz, D. G. (2006). Targeting of somatic hypermutation. Nat. Rev. Immunol. 6, 573–583.
Offer, S. M., Pan-Hammarström, Q., Hammarström, L., and Harris, R. S. (2010). Unique DNA repair gene variations and potential associations with the primary antibody deficiency syndromes IgAD and CVID. PLoS ONE 5, e12260. doi:10.1371/journal.pone.0012260
Pääbo, S., Irwin, D. M., and Wilson, A. C. (1990). DNA damage promotes jumping between templates during enzymatic amplification. J. Biol. Chem. 265, 4718–4721.
Palanduz, S., Palanduz, A., Yalcin, I., Somer, A., Ones, U., Ustek, D., Ozturk, S., Salman, N., Guler, N., and Bilge, H. (1998). In vitro chromosomal radiosensitivity in common variable immune deficiency. Clin. Immunol. Immunopathol. 86, 180–182.
Pan-Hammarström, Q., Lähdesmäki, A., Zhao, Y., Du, L., Zhao, Z., Wen, S., Ruiz-Perez, V. L., Dunn-Walters, D. K., Goodship, J. A., and Hammarström, L. (2006). Disparate roles of ATR and ATM in immunoglobulin class switch recombination and somatic hypermutation. J. Exp. Med. 203, 99–110.
Papavasiliou, F. N., and Schatz, D. G. (2000). Cell-cycle-regulated DNA double stranded breaks in somatic hypermutation of immunoglobulin genes. Nature 408, 216–221.
Papavasiliou, F. N., and Schatz, D. G. (2002). The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J. Exp. Med. 195, 1193–1198.
Parng, C.-L., Hansal, S., Goldsby, R. A., and Osborne, B. A. (1996). Mechanisms of immunoglobulin gene diversification in cattle. J. Immunol. 157, 5478–5486.
Pascual, V., and Capra, J. D. (1991). Human immunoglobulin heavy-chain variable region genes: organization, polymorphism, and expression. Adv. Immunol. 49, 1.
Pernis, B., Forni, L., Dubiski, S., Kelus, A. S., Mandy, W. J., and Todd, C. W. (1973). Heavy chain variable and constant region allotypes in single rabbit plasma cells. Immunochemistry 10, 281–285.
Petersen, S., Casellas, R., Reina-San-Martin, B., Chen, H. T., Difilippantonio, M. J., Wilson, P. C, Hanitsch, L., Celeste, A., Muramatsu, M., Pilch, D. R., Redon, C., Ried, T., Bonner, W. M., Honjo, T., Nussenzweig, M. C., and Nussenzweig, A. (2001). AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature 414, 660–665.
Pham, P., Bransteitter, R., Petruska, J., and Goodman, M. (2003). Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424, 103–107.
Radding, C. M. (1978). Genetic recombination: strand transfer and mismatch repair. Annu. Rev. Biochem. 47, 847–880.
Ramiro, A. R., Stavropoulos, P., Jankovic, M., and Nussenzweig, M. C. (2003). Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 14, 14.
Ratcliffe, M. J. (2006). Antibodies, immunoglobulin genes, and the bursa of fabricius in chicken b cell development. Dev. Comp. Immunol. 30, 101–118.
Revy, P., Muto, T., Levy, Y., Geissmann, F., Plebani, A., Sanal, O., Catalan, N., Forveille, M., Dufourcq-Labelouse, R., Gennery, A., Tezcan, I., Ersoy, F., Kayserili, H., Ugazio, A. G., Brousse, N., Muramatsu, M., Notarangelo, L. D., Kinoshita, K., Honjo, T., Fischer, A., and Durandy, A. (2000). Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 102, 565–575.
Reynaud, C.-A., Anquez, V., Grimal, H., and Weill, J.-C. (1987). A hyperconversionm echanismg eneratesth e chicken light chain preimmune repertoire. Cell 48, 379–388.
Reynaud, C. A., Bertocci, B., Dahan, A., and Weill, J. C. (1994). Formation of the chicken B-cell repertoire: ontogenesis, regulation of Ig gene rearrangement, and diversification by gene conversion. Adv. Immunol. 57, 353.
Reynaud, C.-A., Dahan, A., Anquez, V., and Weill, J.-C. (1989). Somatic hyperconversion diversifies the single V, gene of the chicken with a high incidence in the D region. Cell 59, 171–183.
Reynaud, C. A., Garcia, C., Hein, W. R., and Weill, J. C. (1995). Hypermutation generating the sheep immunoglobulin repertoire is an antigen-independent process. Cell 80, 115.
Reynaud, C. A., Mackay, C. R., Muller, R. G., and Weill, J. C. (1991). Somatic generation of diversity in a mammalian primary lymphoid organ: the sheep ileal Peyer’s patches. Cell 64, 995.
Roth, D. B., and Roth, S. Y. (2000). Unequal access: regulating V(D)J recombination through chromatin remodeling. Cell 103, 699–702.
Rothkamm, K., Krüger, I., Thompson, L. H., and Löbrich, M. (2003). Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 23, 5706–5715.
Rush, J. S., Fugmann, S. D., and Schatz, D. G. (2004). Staggered AID-dependent DNA double strand breaks are the predominant DNA lesions targeted to S mu in Ig class switch recombination. Int. Immunol. 16, 549–557.
Sale, J. E., Calandrini, D. M., Takata, M., Takeda, S., and Neuberger, M. S. (2001). Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature 412, 921–926.
Sayegh, C. E., Drury, G., and Ratcliffe, M. J. (1999). Efficient antibody diversification by gene conversion in vivo in the absence of selection for V(D)J-encoded determinants. EMBO J. 18, 6319–6328.
Schlissel, M. S. (2000). Perspectives: transcription. A tail of histone acetylation and DNA recombination. Science 287, 438–440.
Schmidt, K. H., Reimers, J. M., and Wright, B. E. (2006). The effect of promoter strength, supercoiling and secondary structure on mutation rates in Escherichia coli. Mol. Microbiol. 60, 1251–1261.
Sehgal, D., Schiaffella, E., Anderson, A. O., and Mage, R. G. (2000). Generation of heterogeneous rabbit anti-DNP antibodies by gene conversion and hypermutation of rearranged VL and VH genes during clonal expansion of B cells in splenic germinal centers. Eur. J. Immunol. 30, 3634–3364.
Seki, M., Gearhart, P. J., and Wood, R. D. (2005). DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 6, 1143–1148.
Selsing, E., Xu, B., and Sigurdardottir, D. (1996). Gene conversion and homologous recombination in murine B cells. Semin. Immunol. 8, 151–158.
Seo, H., Masuoka, M., Murofushi, H., Takeda, S., Shibata, T., and Ohta, K. (2005). Rapid generation of specific antibodies by enhanced homologous recombination. Nat. Biotechnol. 23, 731–735.
Shen, P., and Huang, H. V. (1986). Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112, 441–457.
Sinkora, M., Sun, J., and Butler, J. E. (2000). Antibody repertoire development in fetal and neonatal piglets. V. VDJ gene chimeras resembling gene conversion products are generated at high frequency by PCR in vitro. Mol. Immunol. 37, 1025–1034.
Staudt, L. M., and Lenardo, M. J. (1991). Immunoglobulin gene transcription. Annu. Rev. Immunol. 9, 373–398.
Tang, E. S., and Martin, A. (2006). NHEJ-deficient DT40 cells have increased levels of immunoglobulin gene conversion: evidence for a double strand break intermediate. Nucleic Acids Res. 34, 6345–6351.
Tosi, S. L., Dubiski, S., and Mage, R. G. (1970). Distribution of allotypic specificities A1, A2, A14, and A15 among immunoglobulin G molecules. J. Immunol. 104, 641–647.
Tsai, H. F., D’Avirro, N., and Selsing, E. (2002). Gene conversion-like sequence transfers in a mouse antibody transgene: antigen selection allows sensitive detection of V region interactions based on homology. Int. Immunol. 14, 55–64.
Unniraman, S., Zhou, S., and Schatz, D. G. (2004). Identification of an AID-independent pathway for chromosomal translocations between the Igh switch region and Myc. Nat. Immunol. 5, 1117–1123.
van der Stoep, N., van der Linden, J., and Logtenberg, T. (1993). Molecular evolution of the human immunoglobulin E response: high incidence of shared mutations and clonal relatedness among epsilon VH5 transcripts from three unrelated patients with atopic dermatitis. J. Exp. Med. 177, 99–107.
Vorechovský, I., Scott, D., Haene, M. R., and Webster, D. A. (1993). Chromosomal radiosensitivity in common variable immune deficiency. Mutat. Res. 290, 255–264.
Wang, G., and Vasquez, K. M. (2006). Non-B DNA structure-induced genetic instability. Mutat. Res. 598, 103–119.
Weill, J. C., and Reynaud, C. A. (1996). Rearrangement/hypermutation/gene conversion: when, where and why? Immunol. Today 17, 92–97.
Weinstein, P. D., Anderson, A. O., and Mage, R. G. (1994). Rabbit IgH sequences in appendix germinal centers: VH diversification by gene conversion-like and hypermutation mechanisms. Immunity 1, 647–659.
Wells, R. D. (2007). Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 32, 271–278. [Erratum in Trends Biochem. Sci. 32, 399].
Winstead, C. R., Zhai, S. K., Sethupathi, P., and Knight, K. L. (1999). Antigen induced somatic diversification of rabbit IgH genes: gene conversion and point mutation. J. Immunol. 162, 6602–6612.
Wright, B. E. (2000). A biochemical mechanism for nonrandom mutations and evolution. J. Bacteriol. 182, 2993–3001.
Wright, B. E., Reschke, D. K., Schmidt, K. H., Reimers, J. M., and Knight, W. (2003). Predicting mutation frequencies in stem-loop structures of derepressed genes: implications for evolution. Mol. Microbiol. 48, 429–441.
Wright, B. E., Schmidt, K. H., Davis, N., Hunt, A. T., and Minnick, M. F. (2008a). II. Correlations between secondary structure stability and mutation frequency during somatic hypermutation. Mol. Immunol. 45, 3600–3608.
Wright, B. E., Schmidt, K. H., Minnick, M. F., and Davis, N. (2008b). I VH gene transcription creates stabilized secondary structures for coordinated mutagenesis during somatic hypermutation. Mol. Immunol. 45, 3589–3599.
Wright, B. E., Schmidt, K. H., Hunt, A. T., Reschke, D. K., and Minnick, M. F. (2011). Evolution of coordinated mutagenesis and somatic hypermutation in VH5. Mol. Immunol. 49, 537–548.
Wright, B. E., Schmidt, K. H., and Minnick, M. F. (2004). Mechanisms by which transcription can regulate somatic hypermutation. Genes Immun. 5, 176–182.
Wu, X., Feng, J., Komori, A., Kim, E. C., Zan, H., and Casali, P. (2003). Immunoglobulin somatic hypermutation: double-strand DNA breaks, AID and error-prone DNA repair. J. Clin. Immunol. 23, 235–246.
Wuerffel, R. A., Du, J., Thompson, R. J., and Kenter, A. L. (1997). Ig Sgamma3 DNA-specific double strand breaks are induced in mitogen-activated B cells and are implicated in switch recombination. J. Immunol. 159, 4139–4144.
Wysocki, L. J., and Gefter, M. L. (1989). Gene conversion and the generation of antibody diversity. Annu. Rev. Biochem. 58, 509–531.
Wysocki, L. J., Gefter, M. L., and Margolies, M. N. (1990). Parallel evolution of antibody variable regions by somatic processes: consecutive shared somatic alterations in VH genes expressed by independently generated hybridomas apparently acquired by point mutation and selection rather than by gene conversion. J. Exp. Med. 172, 315–323.
Xu, B., and Selsing, E. (1994). Analysis of sequence transfers resembling gene conversion in a mouse antibody transgene. Science 265, 1590–1593.
Xu, S., Clark, T., Zheng, H., Vang, S., Li, R., Wong, G. K., Wang, J., and Zheng, X. (2008). Gene conversion in the rice genome. BMC Genomics 9, 93. doi:10.1186/1471-2164-9-93
Keywords: human immunoglobulin gene, gene conversion, homologous recombination, activation-induced cytidine deaminase, DNA secondary structure, affinity maturation
Citation: Duvvuri B and Wu GE (2012) Gene conversion-like events in the diversification of human rearranged IGHV3-23*01 gene sequences. Front. Immun. 3:158. doi: 10.3389/fimmu.2012.00158
Received: 05 April 2012; Accepted: 25 May 2012;
Published online: 15 June 2012.
Edited by:
Zhixin (Jason) Zhang, University of Nebraska Medical Center, USAReviewed by:
Patrick C. Wilson, University of Chicago, USAMiles D. Lange, University of Nebraska Medical Center, USA
Copyright: © 2012 Duvvuri and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Bhargavi Duvvuri, School of Kinesiology and Health Science, York University, 4700 Keele Street, 124 Farquharson Building, Toronto, ON, Canada M3J 1P3. e-mail: bhargavi@yorku.ca
