- Department of Immunology, University of Toronto, Sunnybrook Research Institute, Toronto, ON, Canada
The NKR-P1 receptors were identified as prototypical natural killer (NK) cell surface antigens and later shown to be conserved from rodents to humans on NK cells and subsets of T cells. C-type lectin-like in nature, they were originally shown to be capable of activating NK cell function and to recognize ligands on tumor cells. However, certain family members have subsequently been shown to be capable of inhibiting NK cell activity, and to recognize proteins encoded by a family of genetically linked C-type lectin-related ligands. Some of these ligands are expressed by normal, healthy cells, and modulated during transformation, infection, and cellular stress, while other ligands are upregulated during the immune response and during pathological circumstances. Here, we discuss historical and recent developments in NKR-P1 biology that demonstrate this NK receptor–ligand system to be far more complex and diverse than originally anticipated.
Introduction
Natural killer (NK) cells are innate lymphocytes that recognize and respond to a variety of different pathological target cells via cytotoxicity and secretion of type I helper cytokines, most notably IFN-γ. However, they can also secrete TNF-α, GM-CSF, and chemokines, to initiate crosstalk with the adaptive immune system. Target cell recognition is mediated by a variety of receptors on NK cells that detect specific cognate ligands, which in turn are differentially expressed on pathological versus normal cells (1). Healthy cells broadly express a number of inhibitory ligands (including MHC-I molecules), which are frequently lost during pathological transformation, infection, or cell stress; this has been termed “missing-self” recognition (1, 2). On the other hand, most stimulatory ligands are minimally expressed by healthy cells, but are strongly upregulated during cellular pathologies; this phenomenon has been termed “induced-self” recognition (3). Typically, both missing-self and induced-self recognition events operate simultaneously to shift the balance from NK cell tolerance to induction of NK cell activation, via an integration of target cell recognition signals (3, 4).
Many NK cell receptors are encoded by genes linked to the NK gene complex (NKC), located on chromosome 6 in mice, chromosome 4 in rats, and chromosome 12 in humans (5–7) (Figure 1). The NKC is also conserved among several other species, including dogs (7), cattle (where it is split between two chromosomes) (7), and chickens (where it is genetically linked to the MHC region) (8). However, a number of NK cell receptor gene families are linked to the leukocyte receptor cluster (LRC), located on mouse chromosome 7, rat chromosome 1, and human chromosome 19 (9–12). There are also numerous other NK cell receptors encoded in various regions in the genome, including the SAP/SLAM family of receptors found on mouse and human chromosome 1 (13, 14), the natural cytotoxicity receptors (NCR1, 2, 3) (15, 16), and others. Within these regions, most NK cell receptors can be broadly classified into two structurally divergent categories: immunoglobulin-like receptors (e.g., KIR, NCR) and C-type lectin-like receptors (e.g., KLR, Ly49).
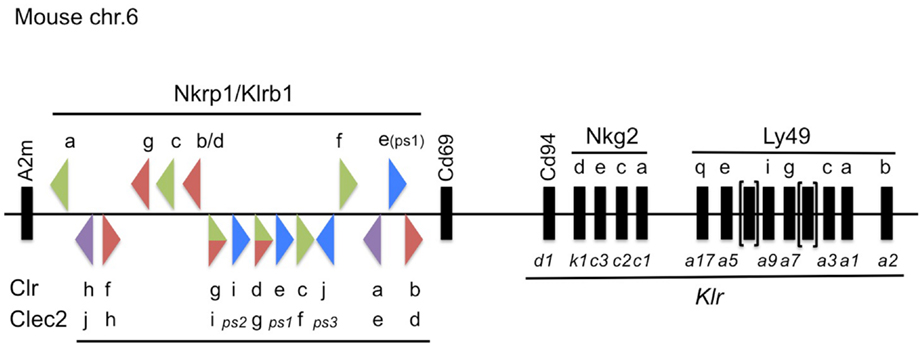
Figure 1. Updated map of the NKC on mouse chromosome 6, highlighting the Nkrp1/Klrb1 and Clr/Clec2 receptor–ligand system. The mouse Nkrp1 receptor genes (official gene nomenclature, Klrb1) are highlighted at the top, with the Clr ligand genes (official gene nomenclature, Clec2) denoted below, located between the A2m and Cd69 genes (17). The triangles depict the gene orientation (plus or minus strand) and their color signifies the known or suspected function of the gene products in regulating NK cell activity, as follows: co-stimulatory (green); co-inhibitory (red); bifunctional (green/red); pseudogene (ps, blue); or unknown (purple). Other select NK receptor genes located telomeric to Cd69 are also depicted with common and official (Klr) gene nomenclatures. Not to scale.
Among the earliest identified group of NK cell receptors is the NKR-P1 family (encoded by the Klrb1 genes; centromeric to Cd69 in the NKC) [Reviewed in Ref. (17)] (Figure 1). This family of receptors is somewhat unique within the NKC, because their ligands are other C-type lectin-related proteins (Clr; encoded by the Clec2 genes), and the Clr/Clec2 loci are genetically interspersed amongst the Nkrp1/Klrb1 receptor genes themselves (18–20) (Figure 1). Although a number of functional interactions have been demonstrated to date between different NKR-P1 receptors and Clr ligands in mice (21), rats (22), and humans (23), many of the receptor–ligand interactions remain unknown, and most have unknown function. This review will outline the historical and recent discoveries surrounding the NKR-P1:Clr systems in rodents and humans, and provide an update on their nomenclature, as well as known expression, structure, and function.
Historical Perspective
Discovery of the NKR-P1 (Klrb1/CD161/Ly-55/Ly-59/Clec5b) Receptors
The first receptor identified to be selectively expressed by NK cells was the mouse NK1 alloantigen, which was discovered by Glimcher et al. in 1977 and found to be differentially expressed across mouse strains (24) [reviewed in Ref. (17)]. The development of a specific monoclonal antibody (PK136 mAb) facilitated its designation as the NK1.1 antigen, permitting the detection and purification of NK cells in select inbred mouse strains (25, 26). Subsequently, the NK1.1 antigen was shown to possess activating function (27, 28), providing direct evidence that NK cells express receptors that may be capable of recognizing cognate ligands on target cells (29). However, the identity of the NK1.1 antigen would remain unknown for several years (30).
In 1989, Chambers et al. generated a mAb (3.2.3) against a cell surface antigen present at high density on rat NK cells and purified rat lymphokine-activated killer (LAK) cells (31). The 3.2.3 antibody was shown to induce redirected NK cell cytotoxicity against FcR+ targets, as well as exocytosis of NK cell cytolytic granules, classifying it as an activating receptor. They called the antigen NKR-P1A (32). Since ligation of mouse NK1.1 and rat NKR-P1A both induced NK cell-mediated cytotoxicity, the hypothesis arose that they could represent homologous structures (28).
Consequently, Giorda et al. screened a B6-strain mouse LAK cDNA library using the rat NKR-P1A cDNA, and identified three mouse NKR-P1 homologs, which they called NKR-P1A (clone 2), NKR-P1B (clone 34), and NKR-P1C (clone 40) (33). The cloned sequences corresponding to the mouse NKR-P1 cDNA were found to exist in different sizes, suggestive of alternative splicing. Overall, these cDNA shared between 61–87% identity at the amino acid level with rat NKR-P1A, with high similarity existing in the extracellular lectin-like region, including several C residues and N-linked glycosylation sites. The discovery and designation of the NKC in mice in 1991 showed that the NKR-P1 receptor loci were distinct from the Ly49 receptor loci, despite their common expression and structural similarity (34). Importantly, however, their genetic linkage demonstrated that a specific location on mouse chromosome 6 was dedicated to NK cell function (5). With the physical mapping of the NKR-P1 genes and the locus encoding the NK1.1 antigen to the same region of mouse chromosome 6, along with their similar expression, structure, and function, it became increasingly likely that the NK1.1 antigen belonged to the NKR-P1 family, and this was formally demonstrated by Ryan et al. in 1992, via expression cloning of the mouse Nkrp1cB6 cDNA using PK136 mAb (35).
However, it later became clear that the strain-dependent expression of the NK1.1 antigen was not only due to allelic expression of the Nkrp1cB6 gene product. In 1999, two groups demonstrated that Nkrp1b gene products from the Swiss-NIH and SJL strains also reacted with the NK1.1 mAb, PK136 (36, 37). Furthermore, the NKR-P1BSw/SJL receptors inhibited NK cell function rather than activating NK cells like NKR-P1C. In these studies, the cloned NKR-P1BB6 (NKR-P1D) gene products did not react with NK1.1 mAb, nor did the NKR-P1CSw/SJL gene products, making it unclear whether they were alleles of existing genetic loci or new genes. In any case, these results demonstrated that polymorphisms exist at both the mouse Nkrp1b and Nkrp1c loci (see below).
In 2001, a BAC contig of the Nkrp1 gene cluster in the B6 mouse strain allowed for the identification of several new genomic sequences, including Nkrp1d (Nkrp1bB6), Nkrp1e, and Nkrp1f (38). The Nkrp1d sequence is 90% similar to that of Nkrp1b, and likely represents an allele of the Nkrp1b locus, rather than a new gene, since the coding sequence resembles that of the cloned NKR-P1BB6 cDNA reported above (see also below) (37, 39). Nkrp1e contains an early stop codon in exon-3, and splicing of intron-5 is predicted to create a frame-shift in the open reading frame (ORF), suggesting it may represent a pseudogene, at least in the B6-strain (38). The Nkrp1f gene appears to be intact and is predicted to code for a functional protein. Work in our lab has identified the latest mouse family member, Nkrp1g (21, 39) (Figure 1).
Subsequent investigation into the nature of strain-dependent NK1.1 reactivity showed that it was due, at least in part, to a single amino acid substitution in the NKR-P1B gene products (and presumably the NKR-P1C gene products, although this remains to be shown) (39). The more recent development of NKR-P1BB6 (NKR-P1D) mAb (18, 40) has shown that, unlike NKR-P1C, which is expressed uniformly by all NK cells in B6 mice, NKR-P1B is variegated and only expressed at high levels on a subset of mature NK cells (36, 37, 41). Previous Southern blot analyses and more recent aCGH analyses, paired with phenotypic NK subset comparisons, have shown this trend to be a generalized phenomenon across many strains (34, 42).
A recent comprehensive study by Hao et al. provided an in-depth computational analysis of the NKC regions from many species, including the rat Nkrp1 genes (7). The rat NKC has at least four Nkrp1 genes, predicted to represent orthologs of the Nkrp1a/c, Nkrp1b/d, Nkrp1f, and Nkrp1g gene sets (Figure 2). Like Nkrp1bB6, the rat Nkrp1c gene likely represents a divergent Nkrp1b allele present in the PVG rat strain (Nkrp1bPVG; see below) (43). While the activating function of the rat NKR-P1A receptor was known since the late 1980s, the inhibitory function of the rat NKR-P1B isoform was not shown until much later. Here, the Miller lab transfected human YTS cells with a rat cDNA library and sorted cells using the 10/78 mAb (similar NKR-P1A reactivity to 3.2.3 mAb) (44). Sequence analysis of positive clones fell into two groups corresponding to either rat NKR-P1A or NKR-P1B. Using chromium release assays, they showed that the rat NKR-P1B clones inhibited NK cytotoxicity. It is now clear that both the NKR-P1A/B gene products from some rat strains react with the 10/78 and 3.2.3 mAb, demonstrating that polymorphisms also exist in selected rat Nkrp1 loci (45).
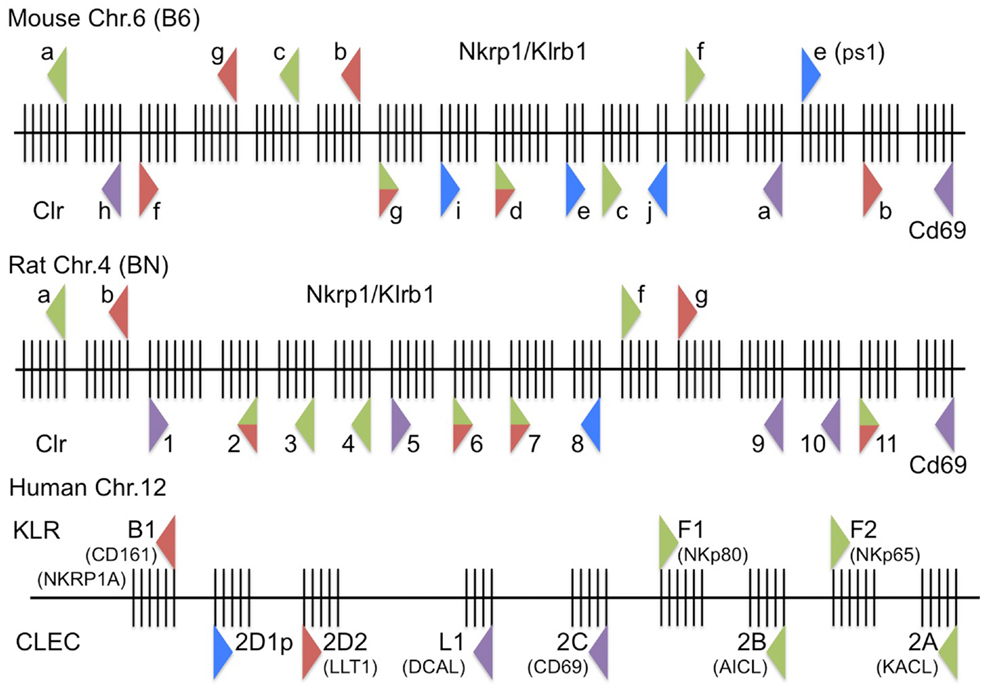
Figure 2. Updated high-resolution maps of the rodent and human Nkrp1/KLRB/F and Clr/CLEC2 receptor–ligand systems. (Top) The mouse Nkrp1/Klrb1 receptor genes and Clr/Clec2 ligand genes on mouse chromosome 6 (B6-strain reference sequence, genome build: December 2011, GRCm38/mm10) are depicted, along with their orientation (triangle direction), known or suspected function (triangle color), and intron–exon structure (vertical exon lines); triangle representations are as in Figure 1. (Middle) The rat Nkrp1/Klrb1 receptor genes and Clr/Clec2 ligand genes on rat chromosome 4 (BN-strain reference sequence, genome build: November 2004, Baylor 3.4/rn4) are depicted as above. (Bottom) The human KLRB/F receptor genes and CLEC ligand genes on human chromosome 12 (genome build: February 2009, GRCh37/hg19) are depicted as above. Not to scale.
The closest human homolog, NKR-P1A (CD161/KLRB1), was identified in 1994 through the development of the DX1 mAb against human LAK cells (46). Human NKR-P1A shares 46% homology at the amino acid level with rat NKR-P1A and 46–47% homology with the mouse NKR-P1 proteins, but significantly lower homology with other C-type lectin-like NK cell receptors. Ligation of human NKR-P1A with DX1 mAb did not induce cytotoxicity against FcR-bearing P815 cells or have an effect on lysis of K562 target cells. However, when added to NK clones with the spontaneous ability to lyse P815 cells, cytotoxicity was inhibited, while F(ab′)2 of anti-NKR-P1A mAb did not inhibit, suggesting FcR cross-linking was important. Paradoxically, NK cell clones that did not spontaneously kill P815 targets could have a small proportion induced to kill P815 in the presence of NKR-P1A mAb. In 2005, two groups showed that the NKR-P1A receptor was indeed inhibitory on human NK cells, demonstrating it to be a likely functional homolog of the mouse and rat NKR-P1B receptors (47, 48). Also similar to the rodent NKR-P1B, only a subset of human NK cells express NKR-P1A (46). However, the existence of another putative inhibitory receptor in rodents, NKR-P1G, calls into question orthology of human NKR-P1A with rodent NKR-P1B (7). Whereas several different NKR-P1 transcripts could be detected by Northern blot in mouse NK cells (33, 49), only one band was detected in human NK cells (46). Nonetheless, an apparent lack of stimulatory NKR-P1 homologs in the human genome was recently re-investigated by the Steinle group, who suggested that the human KLRF1/2 gene products, located telomeric to CD69, may actually represent divergent activating NKR-P1 homologs (see below) (23) (Figure 2).
Discovery of the Clr (Clec2d/Ocil/LLT1) Ligands
While investigating bone morphogenesis, Zhou et al. discovered a gene product they designated osteoclast inhibitory lectin (Ocil), after its ability to inhibit osteoclast formation when expressed on osteoblast cells (50). They went on to show that Ocil was a prototypical member of a group of similar gene products in mice (Ocil, Ocilrp1, Ocilrp2) (51). The extracellular domains of all three gene products are similar to each other at the amino acid (>70%) and nucleotide (~90%) levels. Around the same time, Plougastel et al. identified a novel set of Clr genes by sequencing B6-strain BACs of the NKC region centromeric to Cd69 (52). The Clr gene products were shown to possess 40% amino acid identity to the lectin-like domain of CD69, and 80–90% homology at the nucleotide level to each other. They named the genes Clr-a, -b, -c, -d, -e, -f, -g, where Clr-b was identical to Ocil, Clr-g was identical to a splice variant of Ocilrp2, and Clr-d (and perhaps Clr-c) were similar to splice variants of Ocilrp1 (51, 52). Since then, new family members, namely Clr-h, Clr-i, and Clr-j, were discovered by hybridizing partial Clr PCR products to mouse BALB/c and 129-strain BACs (21, 39) (Figure 1). While the majority of the Clr genes are predicted to code for functional proteins, Clr-e likely represents a pseudogene, due to a frame-shift (52, 53); Clr-i is a predicted pseudogene, due to multiple stop codons in all reading frames (21, 53); and Clr-j is only a gene fragment (53). Unfortunately, several nomenclature issues complicate the Clr literature, including synonyms and the existence of splice isoforms (see below) (Figure 1).
Subsequent to the identification of the Ocil/Clr gene family, several screens aimed at identifying NKR-P1 ligands demonstrated interactions between gene products of the two families [reviewed in Ref. (17)]. Briefly, interactions between the NKR-P1B/D:Clr-b as well as NKR-P1F:Clr-g receptor–ligand pairs were shown using cellular assays (involving BWZ.36 reporter cells bearing CD3ζ/NKR-P1 fusion receptors), NKR-P1 tetramers, and blocking mAbs for NKR-P1BB6 (a.k.a., NKR-P1D; 2D12 mAb) or Clr-b (4A6 mAb) (18, 20). These results were significant in that they showed an interaction between the NKR-P1 receptors, which were previously thought to recognize oligosaccharides (54, 55), and protein ligands. Thus, the NKR-P1 receptors are notable in that they recognize other Clr proteins encoded by the Ocil/Clr/Clec2 ligand genes linked to the Nkrp1/Klrb1 receptor genes. More recently, additional interactions have been shown in the mouse system, including strain-dependent conservation of NKR-P1B/D:Clr-b, NKR-P1F:Clr-c, -d, -g, and NKR-P1G:Clr-d, -f, -g (21) (Figure 1).
The first ligand identified in the rat system was a viral immunoevasin, RCTL, a spliced ORF with C-type lectin-like sequence homology identified in 2001 (56), derived from the rat cytomegalovirus-English isolate [RCMV-E; Mhv8 (57)]. rctl is expressed as an early gene upon infection of rat embryonic fibroblast (REF) cells (45, 56). The RCTL gene product appears to functionally replace the endogenous Clr-b-like ligand (rat Clec2d11), which is rapidly lost during RCMV-E infection; notably, both the host and viral ligands are recognized by specific allele(s) of the NKR-P1B receptor in certain rat strains (45). Prior to this work, no information was available regarding the rat Ocil/Clr gene family, until a breakthrough publication in 2006 by Hao et al. described the complete repertoire of NKC-associated Clec-like genes in several species (7). This in-depth analysis outlined at least 11 rat Clec2d-like genes or pseudogenes, making the rat system the most complex family identified to date. Due to close sequence homology and an inability to determine strict orthology with the mouse family members, the rat Clr genes were simply designated Clec2d1-11 (centromeric to telomeric), with a close relative of mouse Clr-b (mouse Clec2d8) predicted to be rat Clec2d11, based upon sequence, structure, and genomic position (7). Indeed, rat Clec2d11 was shown to be recognized by two distinct host NKR-P1B alleles (45). In 2009, the promoter region of a rat Ocil homolog was first described (58); however, the gene annotated rat Ocil is another Clr family member, rat Clec2d5 (7).
The first in-depth analysis of rat NKR-P1 ligands, which adopted a rat Clr-1–11 nomenclature based upon the corresponding Clec2d1–11 assignments, used an NFAT-driven GFP-reporter cell assay (BWN3G, similar to the BWZ.36 LacZ-reporter cell assay), to show that rat NKR-P1A and NKR-P1B (cluster 1 receptors) recognize Clr-11, while NKR-P1F and NKR-P1G (cluster 2 receptors) recognize an overlapping set of ligands, namely Clr-2, -3, -4, -6, -7 and Clr-2, -6, -7, respectively (59) (Figure 2). Follow-up work by this group showed that the mouse and rat NKR-P1F/G receptors are highly xenoreactive, cross-recognizing corresponding ligands from the other species and allowing some degree of assignment of functional orthology (22).
In the human system, a Clr sequence was discovered in the NKC in 1999 by Boles et al. by searching the EST database with a consensus sequence of known human C-type lectin-like receptors (60). They screened an NK cell cDNA library and identified an 850-bp cDNA, designated lectin-like transcript-1 (LLT1), and later generated a mouse anti-LLT1 mAb (L9.7) by immunizing AKR/J2 mice with LLT1 fusion proteins (61). LLT1 is similar in structure and function to the mouse Clr gene products, and indeed has been given the synonymous gene designation, CLEC2D, as the mouse Clr-b locus (Clec2d). Actually, Hao et al. identified two homologs, CLEC2D1p (a predicted pseudogene) and CLEC2D2 (which encodes LLT1) (7) (Figure 2). However, LLT1 also shares significant homology with other rodent Clr gene products, as well as other human gene products, such as KACL (CLEC2A), AICL (CLEC2B), and CD69 (CLEC2C) (17, 23, 60, 62, 63). As with mouse Ocil/Clr-b, human OCIL was also discovered separately from LLT1 in 2004 (64), but the cDNA encodes an identical protein to LLT1; human LLT1/OCIL is 42% identical to mouse Ocil/Clr-b at the protein level.
The above features of LLT1 made it an attractive candidate for interaction with the human NKR-P1A receptor (61). Indeed, in 2005, simultaneous publications from two groups formally demonstrated an interaction between the human NKR-P1A (KLRB1) receptor and the LLT1 (CLEC2D) ligand, using LLT1 multimers and liposomes, reciprocal NKR-P1A/LLT1 reporter cell assays, as well as functional cytotoxicity and IFN-γ production assays (47, 48). More recent studies have identified several splice variants of the human CLEC2D gene, only one of which (LLT1) appears to functionally bind the human NKR-P1A receptor (65) (Oscar Aguilar; see GenBank KF958454-9, KF971856-9). As mentioned above, the Steinle lab has also reported significant CLEC2 group homology between KACL (CLEC2A, linked to its receptor, KLRF2), AICL (CLEC2B, linked to its receptor, KLRF1), CD69 (CLEC2C, unknown receptor; between CLEC2D and CLEC2A/B), and LLT1 (CLEC2D, linked to its receptor, KLRB1) (23) (Figure 2).
Nomenclature Issues and Functional Homology
The NKR-P1:Clr receptor–ligand system has historically suffered from considerable nomenclature issues, across different species as well as among distinct strains within a given species. In the mouse system, the receptors have been variably known as NK1/NK1.1 (an alloantigen between mouse strains), Ly-55(a–c)/Ly-59 (lymphocyte antigens), NKR-P1 (A–G, the common names), Cd161 (after the human receptor designation), Klrb1 (the official gene nomenclature, with members a–g), and Clec5b (according to the C-type lectin-like nomenclature). While the official gene nomenclature could have been designated killer-cell lectin-like receptor group-B, with numerical family numbers (i.e., Klrb1-7; following the similar Klra1-n designations for Ly49a–x), it has instead adopted a hybrid lettered gene nomenclature, Klrb1a–g, after their common names. Thus, NKR-P1A is encoded by Klrb1a, etc. However, this is complicated by the fact that NKR-P1B and NKR-P1D are likely highly divergent alleles of the same or a similarly evolved single genetic locus; thus, NKR-P1B is encoded by Klrb1b, which is also known as Klrb1d in the B6 mouse strain (i.e., NKR-P1BB6 is synonymous with NKR-P1D). Following this, the remaining family members are NKR-P1C (encoded by Klrb1c), NKR-P1F (Klrb1f), and NKR-P1G (Klrb1g), plus one predicted pseudogene, NKR-P1E (Klrb1e or Klrb1-ps1) (53). While NKR-P1CB6 (Klrb1cB6) is widely thought to represent “the” NK1.1 antigen, it is actually only crossreactive as “one” NK1.1 antigen in the B6 and related mouse strains, as NKR-P1B alleles from several strains (e.g., NKR-P1BSw/SJL) also encode NK1.1 antigens, and the NK1.1 mAb was originally derived by immunization of (C3H × BALB)F1 mice with CE-strain splenocytes and bone marrow (BM) cells (not B6 cells) (26). In addition, the Nkrp1g gene has also been referred to as a second Nkrp1e-like sequence (Nkrp1e2) (17), and most recently just Klrb1, perhaps signifying the closest predicted human KLRB1 ortholog or functional homolog.
Furthering this complexity, as with any gene family, the discovery of the Ocil/Clr gene products by multiple groups has complicated the ligand nomenclature, which has been officially designated the Clec2 gene family. Thus, the known mouse Clr ligands include Clr-a (encoded by Clec2e), Clr-b (Clec2d), Clr-c (Clec2f), Clr-d (Clec2g), Clr-f (Clec2h), and Clr-g (Clec2i). There are also a number of predicted pseudogenes, including Clr-e (Clec2-ps1; Clec2d5), three recently discovered members, Clr-h (Clec2j), Clr-i (Clec2k/Clec2-ps2), Clr-j (Clec2-ps3) (53), and an unlinked, more distantly related family member recently annotated Bacl (Clec2l), although Bacl is not encoded in the NKC (66). However, as mentioned above, under alternative nomenclature, other names have been assigned historically to the gene products: Clr-a (Clec2e, Clec2d7), Clr-b (Clec2d, Ocil, Clec2d8), Clr-c (Clec2f, Clr-z, Clec2d6), Clr-d (Clec2g, Ocilrp1, Ddv10, Clr-x, Clec2d4), Clr-e (Clec2-ps1, Clec2d5), Clr-f (Clec2h, Clec2d2), Clr-g (Clec2i, Ocilrp2, Dcl1, LCL-1(a–d), Clec2d3), Clr-h (Clec2j; Clec2d1), Clr-i (Clec2k, Clec2-ps2), and Clr-j (Clec2-ps3) (7, 53). Note that there are no annotated Clec2a-c genes in the mouse genome database, but these have been assigned to the human KACL (CLEC2A), AICL (CLEC2B), and CD69 (CLEC2C) loci. Also, note that an alternative myeloid receptor named Clec2 exists, which is distinct from the Clec2 gene family and is encoded by the Clec1b gene (67). To date, many of the Clr have unknown receptors and most have unknown physiological function. However, the known mouse receptor:ligand pairs to date include NKR-P1B/D:Clr-b, NKR-P1F:Clr-c, -d, -g, and NKR-P1G:Clr-d, -f, -g in the B6, 129, and BALB/c strains (18, 20–22, 40) (Figure 1).
Adding to this diversity, the rat NKC contains 4 Nkrp1/Klrb1 and 11 Clr/Clec2 genes (7), and has had its share of nomenclature issues. The rat NKR-P1 receptors are fairly straightforward, and have been classified into two distinct structural and functional clusters, NKR-P1A/B (cluster 1), and NKR-P1F/G (cluster 2) (59). However, as in the mouse system, a highly divergent allele of the NKR-P1B receptor in the PVG strain (NKR-P1BPVG) has been alternately referred to as NKR-P1C (43). In addition, early references to NKR-P1F-like sequences were also called Klrb1c/NKR-P1C (68), and NKR-P1G-like sequences were also called Klrb1d/NKR-P1E. Notwithstanding this, the receptor designations have largely been resolved in recent literature. As above, human KLRB1 may be more closely related to rat Klrb1g than Klrb1b (7), but this remains to be demonstrated.
Nonetheless, there are significant deviations in the ligand nomenclature, and due to the complexity and homology of the rat Clec2 family, gene annotations are still being worked out in the genome browsers1, with many being given RGD or LOC designations (e.g., RGD1564464/LOC500331 for Clec2d1/Clr1; see below). To nucleate the gene family, Hao et al. presented the earliest complete characterization of the rat Clr genes, and designated them numerically, centromere to telomere, as Clec2d1–11 (7). There have been few updated attempts to rename these genes according to the mouse nomenclature, or to fully deduce speculated orthologies [if indeed possible (7, 69)], so they have simply been referred to as Clr-1–11 (22, 43, 59). However, the annotated rat Ocil gene is Clec2d5 (Clr-5), distinct from a likely functional homolog of mouse Ocil/Clr-b, rat Clr-11 (Clec2d11), which is also the demonstrated rat NKR-P1B ligand (22, 45, 59). To confuse matters further, there is an annotated rat Clec2d-like-1 sequence (abbreviated Clec2dl1; not Clec2d11, or Clr-11) that is actually rat Clec2d6, or Clr-6. Notwithstanding these discrepancies, the known interactions in the rat system include: NKR-P1A/B:Clr-11, NKR-P1F:Clr-2, -3, -4, -6, -7, and NKR-P1G:Clr-2, -6, -7 in various rat strains (22, 45, 59). Interestingly, the mouse and rat NKR-P1F/G proteins have been labeled “promiscuous” because they share overlapping ligand specificity, cross-react in a xenogeneic manner with their respective ligands, and react with many rat tumor target cells (22). Indeed, the mouse NKR-P1F/G receptors can respond equivalently or better to their rat Clr ligand counterparts using functional reporter assays (e.g., mouse NKR-P1G also weakly binds rat Clr-4) (22) (Figure 2).
In humans, the system is relatively simple, apart from some synonymous designations and recent developments. The lone human NKR-P1A receptor (KLRB1/CD161/CLEC5B) possesses inhibitory function upon binding to the closest Clr-related ligand, LLT1 (CLEC2D/OCIL/CLAX) (47, 48). This interaction is functionally similar to the rodent NKR-P1B:Clr-b interaction; however, the receptor and ligand also share strong similarity with another rodent (putative) inhibitory interaction, NKR-P1G:Clr-d, -f, -g. Hao et al. originally identified two human Clec2d homologs, where CLEC2D1p is a predicted pseudogene (a.k.a., LOC374443), and CLEC2D2 encodes LLT1 (7). However, recent work has speculated that the missing human functional equivalents of the rodent stimulatory NKR-P1 receptors may actually be located telomeric to CD69 (CLEC2C) as part of a greater KLRB/F family intermingled amongst CLEC2 family ligands (7). Here, Nkp80 (KLRF1; CLEC5C) and NKp65 (KLRF2) are genetically linked with their respective ligands, AICL (CLEC2B) (62, 70) and KACL (CLEC2A) (63, 71). Of note, CD69 (CLEC2C) itself and a newly characterized non-NKC-encoded BACL (CLEC2L) have no known binding partners, but notably the latter is genetically linked with another lectin-like receptor, KLRG2 (CLEC15B) (66). There is also another more distantly related gene encoding DCAL (CLECL1), located between LLT1 (CLEC2D) and CD69 (CLEC2C) (Figure 2).
Expression
Mouse
Transcripts for all mouse Nkrp1 genes have been detected in the spleen, thymus, lymph nodes, and other hematopoietic tissues, essentially wherever NK cells are found (53). Additionally, the lung and intestine have shown expression of almost all Nkrp1 transcripts (53). Nkrp1b/d is unique in showing expression within the tongue and bladder (53). Within these tissues, Nkrp1a, Nkrp1b/d, Nkrp1c, and Nkrp1f transcripts (Nkrp1g has not yet been examined in detail) are present in NK cells, while Nkrp1c is also found in NKT cells (53). Subsets of cells classified by various sort markers as tissue DCs and macrophages have also been shown to express Nkrp1b/d (53). Nkrp1f transcripts are also present in BM DC/monocyte precursors and lymph node endothelial cells (53), and expression has also been reported in DC/APC (72, 73). Nkrp1b/d transcripts are also highly expressed in innate lymphoid cells (ILC), including ILC1, ILC2, ILC3, and LTi-like cells (David Allan, manuscript submitted). They are also enriched in a CD8αα+ subset of IEL; notably, these latter IEL also express greatly enriched levels of transcripts for Nkrp1a, upregulated almost 30-fold in CD8αα+ versus CD8αβ+ IEL (74). While Nkrp1c is found in almost all NK and NKT cells, expression of Nkrp1b/d is found at high levels in only a subset of NK cells (50–70%), but few NKT cells (~10%) (40) (see also Peter Chen et al., Munir Rahim et al., manuscripts submitted).
Consistent with the transcript data, most NKR-P1 receptors are expressed on mouse NK cells; however, few NKT cells express surface NKR-P1 receptors, with the exception of NKR-P1C, which is expressed at intermediate levels by NKT cells (40). Mouse NKR-P1B/D is only detected at significant levels on a subset of NK cells (~50–70%, depending on the strain), whereas the remainder expresses only low levels (18, 40). Strain-dependent NK1.1 expression is detectable on NK cells from a number of strains (CE, B6, BTBR, KK, NZB, NZW, C57L, C58, Ma/My, ST, SJL, FVB, and NIH-Swiss mice, but not 129, BALB/c, A/J, AKR, CBA, C3H, DBA, LG, PL, or SM mice); however, expression of the particular NK1.1 antigen involved (NKR-P1B or NKR-P1C) seems to be dictated by whether the majority of NK cells are NK1.1+ (NKR-P1C), or whether the NK1.1 expression is variegated (NKR-P1B), as determined by gating on DX5+ or NKp46+ NK cells (39, 40, 42). Surface expression of mouse NKR-P1G has not been documented yet, but it is predicted to be variegated, like NKR-P1B/D. Interestingly, both activating and inhibitory NKR-P1 isoforms are rapidly downregulated from the cell surface following ligation by plate-bound antibody or cognate ligands (40). On the other hand, surface NK1.1 (NKR-P1C) expression is induced on conventional CD8+ T cells following viral infection and activation by other stimuli (75–78). Expression of NK1.1 isoforms (NKR-P1B > NKR-P1C) is also found on immature thymocytes, thymic NK cells, fetal blood progenitors, subsets of mucosal T cells, and some NKp46+ gut ILC (36, 79–86).
Expression of the mouse Clr ligand genes is variable and highly regulated. Mouse Ocil (Clr-b) was reported to be expressed broadly in most tissues (52, 53), but is also prevalent during bone development and upregulated in response to osteotropic factors like retinoic acid, 1,25-dihydroxyvitamin D3, parathyroid hormone, IL-1α, and IL-11 (50). This regulation of Ocil appears to be unique, as Ocilrp1 (Clr-d) and Ocilrp2 (Clr-g) were not regulated by these treatments, despite Ocilrp2 being co-localized in the same tissues as Ocil (51). At the transcript level, Clr-a is only expressed in the intestines, and even then only at low levels (53). Clr-b is expressed in all nucleated hematopoietic cells, some non-hematopoietic cells, and within most tissues, with the exception of the brain, which appears to be devoid of Clr and Nkrp1 transcript expression (20, 50, 53). In addition, Clr-b transcript expression is tightly regulated on healthy versus pathological target cells. While expressed highly in normal cells, the transcripts are lost following transformation (20), viral infection (45), and genotoxic and cellular stress (87). Clr-c is expressed at low levels in the tongue, spleen, thymus, ovaries, testes, and lymph node tissues; Clr-d transcripts are uniquely present in the eye; and Clr-f is expressed in the liver and very highly in the kidney and intestine, specifically in intestinal epithelial cells and kidney tubular epithelial cells (52, 53). Clr-g is expressed in the spleen, thymus, and lymph node, particularly in intestinal epithelial cells, as well as LAK cells (52, 53); notably, distinct Clr-g splice isoforms exist (a.k.a., LCL-1a, -1b, -1c, -1d) that may have distinct expression patterns (72, 73). Expression of the remaining family members requires further investigation.
In terms of cell surface and protein expression, Clr-b has been the most widely studied, as detected using 4A6 mAb (20, 87). It appears to be a constitutive surface marker of healthy cells, as nearly all nucleated hematopoietic cells express it, as do normal embryonic and adult skin fibroblasts (20, 87). In line with this, Clr-b is frequently downregulated on pathological target cells, including most hematopoietic tumor cell lines (20), virally infected cells (e.g., MCMV, RCMV) (45), and cells undergoing genotoxic or cellular stress (87). Thus, like MHC-I molecules, Clr-b demonstrates a “missing-self” recognition pattern, expressed by healthy cells but lost on target cells undergoing pathological changes, making them more susceptible to NK cell effector function. Interestingly, the downregulation of Clr-b occurs at both the transcript and protein levels, suggesting that transcriptional, as well as post-transcriptional and post-translational mechanisms, play important roles in regulating surface Clr-b expression, and therefore regulating NK cell activity (45, 87). Notably, loss of cell surface Clr-b expression on stressed cells is abrogated by inhibitors of the ubiquitin–proteasome and endolysosomal pathways (e.g., MG132, lactacystin, chloroquine), many of which also affect autophagy (87). Expression of the other mouse Clr family members at the cell surface has been less documented due to a paucity of specific mAb.
Rat
Where studied, expression in the rat system appears to be largely similar to that of the mouse. Rat NKR-P1A transcripts are found in NK cells and NKT cells, and within tissues that contain these cells (32). As in the mouse, the mAb that define rat NKR-P1 expression (3.2.3, 10/78 mAb) identify all NK cells from most strains, where they react with rat NKR-P1A, but these mAb also crossreact with the NKR-P1B inhibitory receptor on certain strains (44, 88). In PVG rats, two major subsets of NK cells can be phenotypically and functionally classified based upon their variegated expression of Nkrp1b: NKR-P1B+ NK cells (largely CD94/NKG2A+ but Ly49−); and Ly49s3+ NK cells (mostly NKR-P1B− Ly49+ and responsive to MHC-I− targets) (43, 89). This pattern is suggestive of subsets differentially regulated by MHC-I-dependent and MHC-I-independent recognition mechanisms. In addition, further characterization has revealed Nkrp1b to be highly expressed by unique subset(s) of circulating and liver/gut-resident lymphocytes, some of which may include mature/activated NK cells and/or ILC-like cells; these cells are enriched for CD8α, CD25, CD93, CX3CR1, but depleted for CD62L, CD27, and CD11b (90). Interestingly, Nkrp1g transcripts are exclusively expressed in the Ly49s3+ subset, while Nkrp1f is expressed in both subsets (22). This may be unique to the rat system, since mouse Nkrp1(a, -c, -f, -g) appear to be more equally expressed by the variegated Nkrp1b+/− NK cell subsets (22). Little is known regarding the surface expression of the rat NKR-P1F and NKR-P1G receptors, although they are presumably expressed on the surface of NK cells. Expression on ILC subsets remains to be determined.
To date, little is known about the expression of the rat Clr molecules, in part due to a paucity of specific mAb. As seen with mouse Clr-b, rat Clr-11 (Clec2d11) is expressed broadly on ex vivo hematopoietic cells (as assessed using R3A8 mAb) (45). Moreover, expression is rapidly lost in REF cells following infection with RCMV-E (45). Whether or not this applies to other Clr family members needs to be tested; notably, expression of another Clr family member, Clec2d5, remained relatively unaffected or increased. General expression profiles of some of the rat Clrs have been inferred using qRT-PCR (69) or NKR-P1-bearing reporter cells (BWN3G) mixed with various types of stimulator cells (59). Ligand(s) for NKR-P1A/BF344 (most likely Clr-11) are expressed by ex vivo splenocytes, peritoneal exudate cells, and to a lesser degree on thymocytes, lymph node cells, and BM cells (59). This overlaps the expression visualized using reporter cells bearing the NKR-P1BPVG and NKR-P1BWAG alleles, which appear to respond more strongly and uniformly to most or all of the above cell types (especially BM cells) (45, 59). Similarly, NKR-P1F ligand(s) (most likely some combination of Clr-2, -3, -4, -6, -7) are expressed on peritoneal exudate cells, splenocytes, BM cells, thymocytes, and lymph node cells (59). NKR-P1G ligands (likely Clr-2, -6, -7) seem to be more restricted to peritoneal exudate cells, splenocytes, and BM cells (59). However, it should be kept in mind that an apparently restricted pattern may be partly due to non-uniform expression or a differential reactivity of rodent reporter cells expressing low or high-affinity receptors (e.g., NKR-P1BF344 versus NKR-P1BPVG/WAG) (45, 59). Interestingly, the RNK-16 rat NK cell line appears to express ligands for all the rat NKR-P1 receptors, while some tumor cells also express certain Clr ligands (22, 59). In one promoter study, binding sites for Sp1 family transcription factors such as Sp1 and Sp7 were shown to have an important role in the regulation of rat Ocil expression; however, the annotated rat Ocil gene is Clec2d5 (Clr-5) not rat Clec2d11 (Clr-11) (58).
Human
Human NKR-P1A is expressed on the majority of (but not all) CD56bright CD16− and CD56dim CD16+ NK cells, as well as subsets of CD4+ and CD8+ TCRαβ+ T cells, NKT cells, and TCRγδ+ T cells (48, 91, 92). It appears to be expressed on a higher proportion of memory versus naïve T cells (46, 93). Induction of NKR-P1A occurs in the fetal liver early in developmental ontogeny (46, 91), and it is one of the earliest markers of human NK cell development (94–97). It is also expressed de novo on CD34− and CD34+ immature thymocytes, and is induced on thymocytes upon culture in rIL-2 (98). Surface NKR-P1A is upregulated on immature and mature NK cells upon exposure to IL-12 (94, 99, 100). NKR-P1A has also been reported to be a marker of all Th17 cell subsets [where it is induced by RORC (101)], human ILC and LTi subsets (102, 103), as well as a novel subset of FoxP3+ “Treg” found in both healthy and arthritic humans that secrete pro-inflammatory cytokines (104). CD4+ NKR-P1A+ T cells are capable of migration in transwell assays and transendothelial migration in vitro (105). Acquisition of NKR-P1A has also been reported to be an early event in monocyte differentiation and on DCs (106).
In contrast to mouse Clr-b and rat Clr-11, and perhaps more akin to other rodent Clr, human LLT1 transcripts are only expressed at low levels in NK cells, T cells, B cells, and osteoblasts, but are undetectable in monocytes (65, 91, 107). In addition, LLT1 is expressed in some tumor lines, and its expression is greatly increased by mitogen stimulation or activation of lymphocytes, including LAK cells, NK cells, T cells, B cells, and DC (65, 91, 107). Induction in osteoblasts has also been documented using IL-1α and PGE2 (64). While freshly isolated B cells and DCs express little LLT1 mRNA, transcript levels are augmented following infection with viral pathogens (e.g., influenza, HSV-1, EBV, HIV) or TLR agonists (i.e., TLR-3, -4, -7, -8, -9), in particular CpG DNA treatment (91, 107). Again unlike rodent Clr-b/Clr-11, LLT1 protein is not detected at the surface under naïve, resting conditions; however, mirroring transcript expression, surface LLT1 is highly upregulated upon activation with the stimuli noted above. Induction on B cells also occurs following IgM cross-linking or CD40 ligation. LLT1 expression is also inducible on T lymphocytes upon stimulation via CD3 cross-linking, PMA, PHA, and IL-2 (61, 91, 107). On DC, the most potent inducers of LLT1 expression are CpG DNA, polyI:C, LPS, TLR-7/8 agonists, and IFN-γ (107). LLT1 is also induced on NK cells upon activation, following incubation with certain NK-sensitive target cells (91, 107). It remains to be determined whether NKR-P1A:LLT1 interactions regulate NK and T cell functions in cis, in addition to recognition of target cells in trans (108, 109).
Expression of the other human CLEC2 ligands is more restricted. As the names suggest, KACL (CLEC2A) is expressed in keratinocytes, AICL (CLEC2B) is induced upon activation of hematopoietic cells, CD69 (a.k.a., EA-1/AIM; CLEC2C) is induced early after activation of T and NK lymphocytes and other hematopoietic cells, BACL (CLEC2L) is expressed in brain tissue, and the more distantly related DCAL (CLECL1) is expressed in dendritic cells (23, 110).
Structure
Genomic
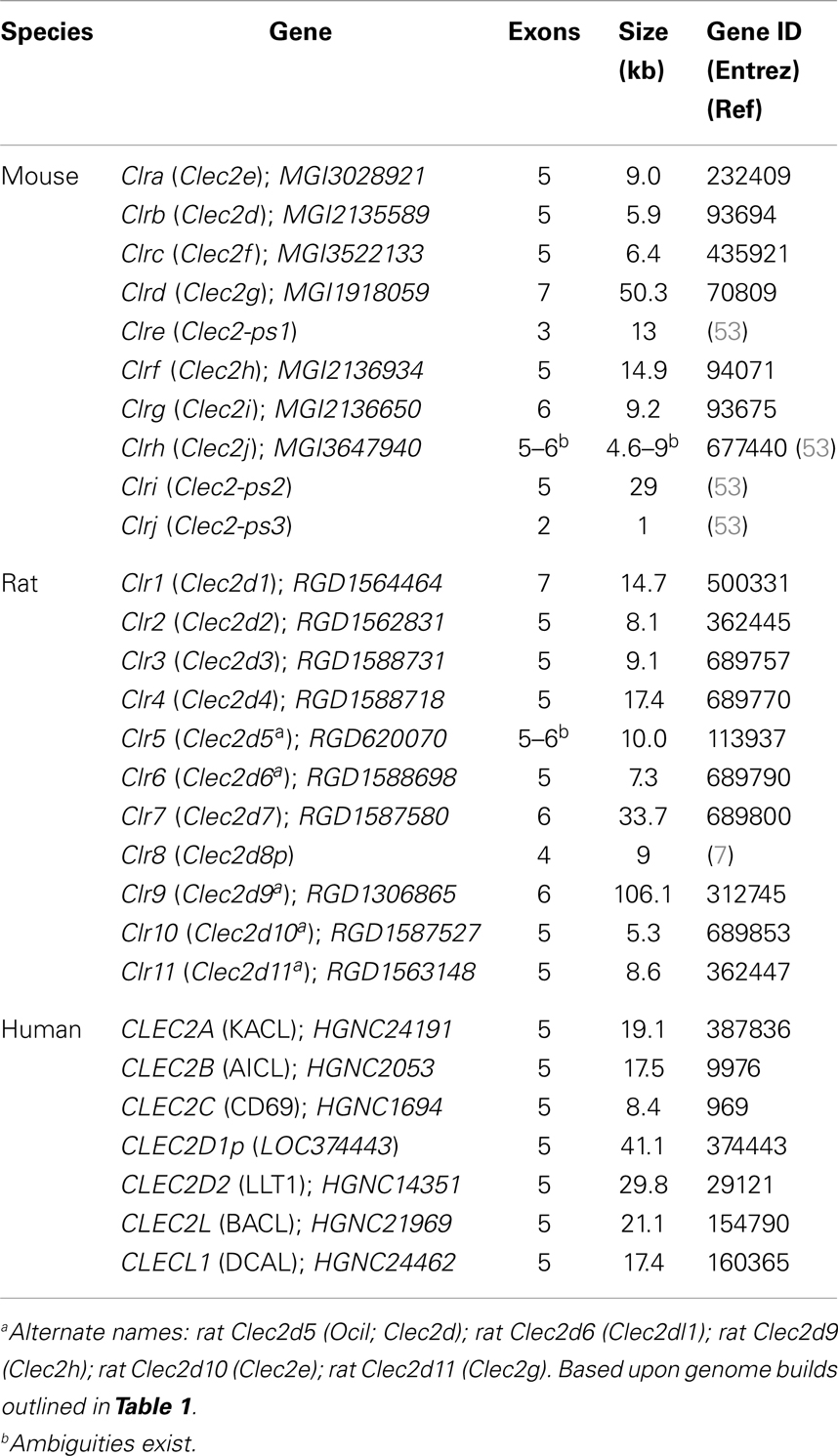
In the mouse B6-strain reference genome, all of the Klrb1 genes have six coding exons: exon-1 codes for the cytoplasmic domain, exon-2 for the transmembrane domain, exon-3 for the stalk region, and exons-4, -5, -6 for the extracellular lectin-like domain (21, 38). This gene structure appears to be conserved for the Klrb1 genes in the BALB/c and 129 strains (53), human KLRB1 (46), as well as the Klrb1 genes in the rat BN-strain reference genome, with the exception of rat Nkrp1f (Klrb1c/f), which may have five to six exons, depending upon reference genome build (7) (Table 1). Notably, the human KLRF1 and KLRF2 genes also have a six-exon gene structure (Figure 2; Table 1).
The Clec2 genes in the mouse and rat also have a similar genomic structure. All the genes have five exons, with the following exceptions: mouse Clrg (Clec2i, six exons); mouse Clrd (Clec2g, seven exons); mouse Clre (Clec2-ps1, three exons); mouse Clrj (Clec2-ps3, two exons); rat Clr1 (Clec2d1, seven exons); rat Clr5 (Clec2d5, five to six exons); rat Clr7 (Clec2d7, six exons); rat Clr8 (Clec2d8p, four exons); and rat Clr9 (Clec2d9, six exons) (Table 2). The human CLEC2D gene that encodes LLT1 (a.k.a., CLEC2D2) also has five exons, as does the upstream CLEC2D1p pseudogene (7). The human CLEC2A and CLEC2B genes also possess a five-exon gene structure, and the same is true for the CLEC2C gene that encodes CD69 (23, 110). The recently described CLEC2L (BACL) gene, which is not linked to the NKC, also possesses five exons, but the less related CLECL1 (DCAL) gene linked to the NKC only has four exons (Figure 2; Table 2).
The promoter regions of the rodent Nkrp1 and Clec2 genes remain poorly characterized. There were early attempts to analyze the upstream regions of the mouse Nkrp1 genes in different strains (49, 111). The proximal promoters appear to be TATA-less, relying on initiator (Inr) and downstream promoter elements (DPE) (111). The mouse Nkrp1 genes also appear to possess a three-promoter organization similar to that seen for the mouse Ly49 genes (112). Several predicted transcription factor-binding sites in the distal and proximal promoter regions are speculated to be important in regulating Nkrp1 expression, including consensus Ets-1, Ikaros, GATA, TCF-1, Sp1, NFAT, and Oct-1 sites (111). In addition, a number of upstream regulatory elements have been mapped, including transcriptional start sites (TSS), alternative exons, and a distal DNase hypersensitive site that may function as an enhancer element. Luciferase-based reporter assays have demonstrated that a 600-bp core promoter drives expression in both T/NK lineage cells, while a 9.76-kb upstream region enhances expression in NK-like cells but represses expression in pre-T cells, suggesting lineage-specific regulation (111). Northern blot analysis has revealed differential expression of Nkrp1a-c transcripts in select strains, but culture of LAK cells in IL-2 could induce Nkrp1 expression where low (49).
In the mouse Clr system, Ocil/Clrb and Ocilrp2/Clrg have been reported to possess an inverted TATA box upstream of their TSS, while Ocilrp1/Clrd has a more traditional TATA box (51). Both Ocilrp1/Clrd and Ocilrp2/Clrg have been reported to generate alternative splice variants (51), and an independent characterization of LCL-1 (Ocilrp2/Clrg) transcripts suggests that up to four different splice variants exist (a–d) (72, 73). The human OCIL (CLEC2D/LLT1) gene lacks an inverted TATA sequence, instead containing a GAATCA sequence upstream of the TSS (64). The human CLEC2D gene has also been reported to generate several alternative splice variants (other than LLT1), some of which may be functional (e.g., some code for proteins retained in the ER as heterodimers with LLT1, while others lack a transmembrane domain) (65) (Oscar Aguilar, see GenBank KF958454-9, KF971856-9). As mentioned above, Sp1 transcription factor-binding sites (Sp1, Sp7) were shown to regulate expression of the rat Ocil gene (Clec2d5; Clr-5) (58).
Protein
The NKR-P1 and Clr proteins belong to group-V of the 14 C-type lectin superfamily groups (7, 113). C-type lectins are a class of glycoprotein characterized by their Ca2+ dependence, conserved disulfide-linked cysteine residues, and functional carbohydrate recognition domains (113). The NKR-P1 and Clr proteins are designated as “lectin-like” because they primarily bind other proteins, and they possess somewhat atypical conservation of their cysteine residues, such that they may have lost the residues and loop structures required to coordinate divalent calcium ions, and thus may lack high-affinity Ca2+-dependent binding to carbohydrates. For example, the mouse Clr and human LLT1 are all missing the C5 residue in their lectin-like domain, and the C4 residue is also absent from mouse Clr-b and Clr-g (52). The implications of these changes require further investigation. However, it should be noted that the NKR-P1 receptors were first reported to possess functional carbohydrate recognition domains (54, 55). In addition, the human LLT1/OCIL and mouse Clr-b/Ocil proteins have been reported to bind high-molecular weight carbohydrates (including sulfated forms) in a Ca2+-independent manner, blockable using soluble carbohydrates (including heparin and chondroitin sulfate) (64).
Initial work on the structure of the mouse NKR-P1 proteins demonstrated that they likely exist as disulfide-linked homodimers. For example, NKR-P1D (NKR-P1BB6) is ~90 kDa under non-reducing conditions, but ~47 kDa under reducing conditions, when assessed using 2D12 mAb (18). The Clr proteins also likely exist as homodimers, but heterodimers between different Clr have also been speculated to exist (18). Crystal structures for the mouse NKR-P1 proteins are just beginning to be published and assessed. Although early co-crystal structures were first reported in 2003 for the mouse NKR-P1F:Clr-g and NKR-P1D:Clr-b proteins (the latter reported under conditions requiring mutagenesis of certain residues), these have not been published (18). Published work suggests that mouse NKR-P1AB6 and NKR-P1CB6 have a similar structure, with slight differences explainable by their amino acid sequences (e.g., NKR-P1AB6 has a higher Y residue content than NKR-P1CB6) (114). Mouse NKR-P1CB6 also has a better-defined conformation of disulfide bridges than NKR-P1AB6, suggesting that NKR-P1C is more rigid and stable; indeed, this is supported in the literature regarding their individual stable expression at the cell surface on NK cells (40). However, it should be noted that the NKR-P1CBALB receptor lacks a potentially crucial conserved C4 residue (position C122S) that may have implications for its expression and function, in addition to its lack of NK1.1 reactivity due to a S191T substitution (which is also found in the functional NKR-P1BBALB receptor) (39, 115).
Rat NKR-P1A and NKR-P1B protein structures have also been compared. Interestingly, rat NKR-P1A is more similar to mouse NKR-P1A/C than to rat NKR-P1B (114). The rat NKR-P1B protein has more α-helical content (including an additional α-helix) than rat NKR-P1A. The human NKR-P1A protein has been modeled on these rodent NKR-P1 structures, where it more closely resembles NKR-P1B than other isoforms, in that it is predicted to retain an additional α-helix (114).
In general, all of the NKR-P1 proteins seem to possess a fold similar to other C-type lectin-like domains, including at least two α-helices and two anti-parallel β-sheets. They have a β-core composed of long anti-parallel β-strands that form a central pillar, flanked at one end by short β-strands and at the other end by a β-sheet (114). This core is surrounded by α-helices (two to three, depending on the isoform). All appear to have three disulfide bonds, except mouse NKR-P1CBALB, which has only two (114). Comparison of all known NKR-P1 ectodomains revealed the highest conservation in the β-core, less conservation in the loop regions, with the longest loop suggested to play a role in ligand specificity (114). A mouse NKR-P1A crystal structure reported recently suggests that a claw-like structure may be important for ligand interaction (116).
A recent biochemical study of the human NKR-P1A:LLT1 proteins have shown that they interact with a Kd ~ 50 μM (117). Reciprocal mutagenesis of the proteins also provided a partially validated structural model of the interaction, supporting a dimeric interaction with several important contact residues. Even more recent co-crystal structures of the high-affinity (Kd ~ 2 nM) human NKp65:KACL interaction have suggested a conserved docking topology for the interaction of genetically linked C-type lectin-like receptor–ligand pairs in the NKC (118). Here, two NKp65 receptor monomers, with limited dimeric interface between them, contact a single dimeric KACL ligand bivalently at two distinct but symmetrical sites in a butterfly-shaped complex. A similarity in this study was noted to the compact Ly49C dimer interacting independently with two monomeric H-2Kb MHC-I ligands (119). Whether or not this applies to other NKC-encoded receptor–ligand pairs remains to be elucidated, but the remaining genetically linked receptor–ligand interactions have all been shown to interact with much lower affinities (micromolar versus nanomolar) (118).
Much less is known about the structure of the rodent Clr or human LLT1 proteins. Human LLT1 has been modeled on a CD69 structural backbone (65), and models have also been proposed based upon mutagenesis studies (117), as well as the human NKp65:KACL crystal structure (118). Recently, the structure and biophysical properties of mouse Clr-g were determined by expressing soluble portions of the extracellular lectin-like domain (120). In this structure, Clr-g exists as a monomer or dimer, depending on the size of the ectodomain used. In terms of secondary structure, it has 2 α-helices, 2 anti-parallel β-sheets composed of 3 β-strands each, and 2 disulfide bonds, with a dimeric interface involving 10 hydrogen bonds. Mouse Clr-g has mostly positive electrostatic potential, while its cognate receptor, NKR-P1F, has negative potential, suggesting electrostatic potential may be a driving force in their contact (120). Notably, models of the other Clr, and other C-type lectin-like proteins, are predicted to possess distinct surface electrostatic potential in this work. Mouse Clr-g also shares almost ~47% sequence identity with human KACL, and predictive models of most of the human and mouse NKR-P1:Clr receptor–ligand interactions have been outlined based upon the human NKp65:KACL interaction (118).
Function
Signaling
Signaling via NK cell receptors is hypothesized to deliver either an activating signal or to exert inhibitory effects on downstream signaling cascades, and the NKR-P1 family is no exception to this notion of paired or balanced recognition systems. Early experiments demonstrated that Syk acts as a common signaling element important for both FcR-initiated and natural cytotoxicity in NK cells (121). However, additional pathways involving Src-family and PI3K kinases are also involved in triggering natural cytotoxicity (122–124).
In this light, it was shown that the mouse NKR-P1C receptor activates both redirected cytotoxicity and IFN-γ production in NK and NKT cells, and that association with the FcRγ adaptor protein was crucial for these functions (28, 125). Furthermore, while FcRγ homodimers were shown to be required for both optimal NKR-P1C expression and signaling in both NK cells and NKT cells (125), FcRγ/CD3ζ heterodimers may be sufficient for residual NKR-P1C cell surface expression, in contrast to CD16 expression, which is dependent upon FcRγ homodimers and is negatively regulated by CD3ζ (via FcRγ/CD3ζ heterodimers) (126). In addition, the rat NKR-P1A receptor was shown to induce Ca2+ flux, phosphatidylinositol turnover, PLA2 activation, arachidonic acid release, heterotrimeric G-protein activation, redirected cytotoxicity, and granule exocytosis by rat NK cells and the RNK-16 cell line (127–129). Subsequent analysis of a number of NKR-P1 protein sequences revealed the presence of potential PLC-γ1/2 motifs (YxxL), SH3-binding proline-rich domains, Lck-recruitment motifs (CxCPR/H), and variably present consensus ITIM motifs (ΦxYxxΦ) in the rodent but not human NKR-P1 receptors (68). The activating rodent NKR-P1 receptors were later shown to contain a conserved positively charged amino acid residue (R) in their transmembrane domain, likely for FcRγ adaptor association (based upon a distinct transmembrane R amino acid position versus K for DAP12-associated receptors), while the inhibitory rodent NKR-P1 receptors contain consensus ITIM motifs (36, 68, 130).
Notably, the rodent NKR-P1 CxCPR/H motifs, similar to those found in the CD4 and CD8 T cell co-receptors, were shown to functionally recruit Lck by immunoprecipitation, yeast two-hybrid analysis, and functional mutagenesis (131, 132). In addition, Lck was also shown to be necessary for efficient NKR-P1C-mediated redirected lysis, as cytotoxicity was diminished in BM-derived LAK cells from Lck−/− mice (whereas splenic LAK cells may use another Src-family member to compensate) (132). The ITIM motif in the mouse NKR-P1B receptor was also shown to recruit the SHP-1 phosphatase upon pervanadate stimulation (and to a lesser extent SHP-2) (36, 132). A consensus ITIM motif (133) is also present in the mouse and rat NKR-P1G receptors, which are presumably inhibitory; however, mouse NKR-P1G lacks the consensus Lck-recruitment motif (21). The functional consequences of the loss of the di-cysteine motif remain unknown, but may suggest a more direct inhibitory receptor function versus a co-inhibitory receptor function, if the CD4/8 T cell co-receptors provide any reference. The mouse and rat NKR-P1F receptors may either be stimulatory, co-stimulatory, or function as co-receptors in terms of signaling (40), as they retain a charged transmembrane residue, yet mouse NKR-P1F lacks the conserved YxxL motif, instead containing a Y residue in a different position and context (21).
Like the rat NKR-P1A receptor (127), the mouse NKR-P1C receptor (NK1.1 in B6 mice) has also been shown to signal IFN-γ production in part via PI3K (p110γ > δ) activity (134), suggestive of some co-stimulatory function, akin to NKG2D isoforms (135–138). Given the specific expression of NKR-P1C on NKT cells [but not other mouse NKR-P1 receptors (40)], its ability to recruit Lck (akin to a T cell co-receptor), and its partial signaling via the PI3K pathway (similar to CD28, NKG2D), it could serve as a co-receptor or costimulatory receptor (signal 2) for TCR activation (signal 1) on NKT cells in their recognition of CD1d-restricted ligands. Since the mouse NKR-P1C ligand(s) remain unknown, it is possible that endogenous or foreign glycolipids (and/or glycoproteins) could be recognized simultaneously by the NKT cell TCR and this lectin-like receptor to facilitate dual recognition (co-receptor or accessory function) and/or co-stimulation. Unlike conventional MHC-restricted CD4+ and CD8+ T cells, NKT cells are not yet known to express a co-receptor for CD1, although they are tightly regulated by SAP/SLAM family interactions (13, 14). On the other hand, conventional NK cells express most NKR-P1 receptor isoforms [whereas NKT cells more uniquely express NKR-P1C (40)], and CD1d over-expression on target cells has been reported to inhibit NK cell function via an as yet unidentified receptor (139, 140).
Interestingly, the cytoplasmic tail of the human NKR-P1A receptor contains neither a strong consensus ITIM motif (ΦxYxxΦ) (133), nor an intact Lck-binding motif (CxCPR/H) (68). Nonetheless, the human NKR-P1A cytoplasmic tail does contain a tyrosine residue in an atypical motif (AxYxxL) that may function as a weak ITIM (48, 68), and the receptor has been shown by immunoprecipitation and Western blotting to form large complexes containing the Src-family kinases, Lck, Fyn, and Lyn in Brij-58-solubilized NK cells (141). However, association with SHP-1 has not been demonstrated (47, 48), and Lck association was not observed in CD161+ NKT cells (142). The receptor has been shown by yeast two-hybrid and co-immunoprecipitation studies to associate with acid sphingomyelinase (aSM) in primary human cell lines and NK cells (143). Ligation using CD161 mAb results in association of CD161 with aSM in detergent-resistant membranes, activation of aSM activity, production of ceramide, PI3K-dependent activation of the Akt and ERK pathways, and proliferation (143). Ligation of human NKR-P1A on thymocytes has been reported to inhibit cytotoxicity but enhance proliferation (98), expression on T cells enhances transendothelial migration (105), and ligation on macrophages and DC fluxes intracellular calcium and leads to IL-1α and IL-12 production (106). More recently, human NKR-P1A has also been shown to be expressed specifically by Th17 cells (101, 144), MR1-restricted MAIT cells (145), and a unique “Treg” subset (104), where its function(s) are still being evaluated (92).
Autoimmunity and Disease Association
Human NKR-P1A was shown to be preferentially expressed on γδ T cells expressing the Vδ2 chain (93). Interestingly, the proportion of γδ T cells and IL-17+ T cells expressing NKR-P1A is higher in MS patients than in healthy donors (93, 146). Upregulation of human NKR-P1A on γδ T cells also occurs following exposure to IL-12, as it does for NK cells, but only for the Vδ2+ population, because Vδ1+ cells express lower levels of the IL-12Rβ2 subunit (93). These NKR-P1A+Vδ2+ T cells also undergo transendothelial migration in an NKR-P1A-dependent manner in vitro. Since MS patients have more of these T cells, it has been proposed that they use NKR-P1A to migrate to local lymph nodes, recirculate, and extravasate into the brain, leading to exacerbated MS symptoms (93).
NKR-P1A+ T cells can also be detected in inflammatory infiltrates in patients suffering from psoriasis (144) and Crohn’s disease (147), with 20-fold more NKR-P1A+CD4+ T cells present in patients with Crohn’s disease than healthy patients (147). Whether this is due to upregulation of NKR-P1A on reactive T cells, or exacerbated production or expansion of pre-existing NKR-P1A+ Th17 cells, requires further investigation (92). Importantly, however, an inflammatory bowel disease (IBD) susceptibility SNP in the human LLT1/OCIL (CLEC2D) gene was recently found to be associated with Crohn’s disease, but not ulcerative colitis (148).
Interestingly, an N19K substitution in the human LLT1/OCIL (CLEC2D) gene has been found to be associated with reduced bone mineral density in postmenopausal women (149). Genome-wide association studies should reveal new interactions between this receptor–ligand system and human disease.
Viral Infection
Viral evasion of host innate and adaptive immunity is a well-documented evolutionary occurrence. In 2001, a spliced C-type lectin-like ORF was identified in the RCMV-E genome (56). The gene product was designated RCTL, and it was later shown to closely resemble the coding sequence of an endogenous rat Clec2d-like sequence, Clec2d11, a predicted functional homolog of mouse Clr-b (Clec2d) (45). Indeed, an mAb specific for the RCTL protein (R3A8 mAb) was also shown to crossreact with the host rat Clr-11 (Clec2d11) protein. Expression of rctl was detected as early as 3 h post-infection (characterizing it as an early gene), while expression of host rat Clec2d11 was reciprocally lost from the cell surface, replaced by RCTL (although heterodimers of host Clec2d11 and RCTL may exist). BWZ.36 reporter cells expressing a CD3ζ/RCTL fusion receptor responded strongly to 293T transfectants expressing the rat NKR-P1B receptor (indeed, only the NKR-P1BWAG allele), suggesting a direct interaction between RCTL and the inhibitory NK cell receptor. RCTL expression on infected cells moderately inhibited WAG-strain LAK cell activity, an effect that was abrogated with either R3A8 mAb blockade or infection using an RCMV ΔRCTL-mutant virus (45). The ΔRCTL-mutant virus exhibited lower splenic and liver titers in vivo, but only in rat strains where the RCTL protein interacted strongly with the NKR-P1B allele. Notably, weak recognition of RCTL was also shown by the activating rat NKR-P1A receptor, suggesting that the host and pathogen were evolving and counter-evolving this innate recognition mechanism. Recently, downregulation of Clr-b has also been shown to occur upon infection with poxviruses, including vaccinia virus and ectromelia virus, where loss of the surface protein was dependent upon active virus infection but not late viral gene synthesis (150). It will be interesting to investigate the roles of similar CMV-encoded genes with lectin-like homology [e.g., r153 in RCMV-Maastricht (151)] in NK cell recognition, as well as whether these CMV immunoevasins are capable of heterodimerizing with host proteins (e.g., rat Clec2d1–11).
Induction of LLT1 surface expression has been shown to occur upon infection with several viruses (e.g., EBV, HIV, influenza, HSV) (91, 107). In addition, LLT1 induction by TLR agonists (e.g., TLR3, 4, 7, 8, 9) as well as other immune stimuli (such as TCR/BCR cross-linking, mitogens, CD40, etc.) suggests that LLT1 interaction with human NKR-P1A also regulates immune responses during infection, particularly viral and bacterial infections (91, 107). In this light, several lectin-like viral ORF (including those from vaccinia and other poxviruses) have been speculated to play roles in the regulation of host immune responses, but none of these have been evaluated to date (56).
T Cell Co-Stimulation and Adaptive Immunity
Few reports to date have examined the effects of the rodent NKR-P1:Clr receptor–ligand system on the adaptive immune response. Ligation of an Ocilrp2 splice isoform (LCL-1; Clr-g) expressed on activated T cells by NKR-P1F on B cells and DC has been reported in mice, leading to enhanced TCR/CD28-mediated T cell proliferation and IL-2 production (73). However, the topology of this interaction would not be predicted by the somewhat restricted expression of NKR-P1F mainly within NK and T cells, with splice variants of Clr-g being more broadly expressed (53). Thus, in light of the recently identified “promiscuous” NKR-P1F:Clr-c, -d, -g and NKR-P1G:Clr-d, -f, -g interactions (21), the reagents used in these studies may possess multiple specificities (e.g., polyclonal anti-Clr-g Ab could cross-react with other Clr, Ocilrp2–Ig fusion protein could recognize both NKR-P1F/G, NKR-P1F tetramer could recognize Clr-c/d/g). Nonetheless, Clr-g was reported to be induced more rapidly after CD3 cross-linking than other co-stimulatory molecules (e.g., OX40, 4-1BB) (73). Also, the co-stimulatory effect of Clr-g was shown to require and synergize with CD28, while OX40 and 4-1BB can act independently of CD28 (73). In a follow-up study, silencing of Clr-g resulted in impaired T cell proliferation and IL-2 production, due to an inability of the responding T cells to phosphorylate Lck, reorganize the actin cytoskeleton, form TCR caps, and transduce NFκB signals (72). These studies highlight an important issue regarding the perceived roles of “receptor” versus “ligand” (or vice versa) in NKR-P1:Clr interactions (51). Notably, the mouse Clr-b protein has been suggested to possess a TRAF2-interaction motif (130), supporting the possibility of “reverse signaling” in regulation of NFκB signals by the Clr proteins on target cells and APC, but the function of this motif has not been investigated yet.
In the human system, CD161 on T cells may function as a costimulatory receptor or accessory molecule for recognition of CD1d (142). Ligation of CD161 on NKT cells did not directly activate cytokine secretion (IFN-γ, IL-4), nor cause proliferation. However, co-ligation of CD3 and CD161 could augment cytokine production and proliferation, and this could be diminished using soluble CD161 mAb. Similarly, while LLT1 inhibits cytotoxicity and IFN-γ production via NKR-P1A on NK cells, it appears to costimulate IFN-γ production by NKT cells (47, 107). Reciprocal studies have suggested that LLT1 itself may signal on NK and NKT cells to augment IFN-γ production via a Src-family–ERK signaling pathway (61, 152).
Ribozyme
A recent publication examined the presence of discontinuous hammerhead ribozymes in mammalian genomes and identified several such self-cleaving RNA motifs within the 3′-UTRs of mouse Clec2e (Clr-a), mouse Clec2d (Clr-b), and rat Clec2d11 (Clr-b-like), along with others in the 3′-UTRs of predicted Clec2-like genes in other species (153). Discontinuous hammerhead ribozymes differ from other ribozymes in that they exist as two fragments (enzyme, substrate) in cis, separated by an insertion of variable length sequence into the stem-1 loop, rather than being contiguous [reviewed in Ref. (154–156)]. In the mouse Clec2d, mouse Clec2e, and rat Clec2d11 gene products, this insertion varies from ~250 to ~690 to ~790 nucleotides, respectively, and serves to separate the substrate sequence (upstream) from the enzyme sequence (downstream) (153). In addition, these ribozyme sequences were shown to be catalytically active (with the mouse Clec2d ribozyme being more active than the Clec2e ribozyme), reducing luciferase reporter expression by ~80% due to autocatalytic reporter RNA degradation (153). The net effect of these cleavage events would be predicted to separate the mRNA 5′-cap and coding sequence from the poly-A tail of the transcript, leading to altered RNA stability. This suggests another possible level of regulation (post-transcriptional) of the rodent Clec2d and Clec2e gene products, either enhancing mRNA turnover to provide more efficient “missing-self” recognition, or providing a molecular switch to turn on and off expression of the gene products during immune responses.
Interestingly, other closely related Clec2 genes contain substrate-like motifs in their 3′-UTRs (in the absence of corresponding enzyme motifs), each with similar sequence identity to the substrates in Clec2d and Clec2e (153). It would be interesting to see whether these sequences can act in trans as substrates for either the Clec2d or Clec2e enzyme components, akin to micro-RNA-mediated regulation.
Evolution
It is clear that the NKR-P1:Clr (Klrb:Clec2) systems, although genetically linked, have undergone significant evolution between species and between strains within a given species. It has been proposed that the creation and deletion of Ly49 receptor genes can occur by unequal crossing over during meiosis, generating new inhibitory or stimulatory Ly49 by swapping the cytosolic and transmembrane exons (which encode function) with the ectodomain exons (which dictate specificity), even incorporating pseudogene exons (17). The NKR-P1:Clr system is no exception to this form of evolution, and the head-to-tail orientation of some genes suggests how this may occur. Notably, the Nkrp1b and Nkrp1c genes are arranged such that even a continuous transcript may be possible, splicing together the inhibitory region of NKR-P1B and the NK1.1+ ectodomain of NKR-P1C, generating a novel inhibitory receptor that is NK1.1+ with new affinity and/or specificity in certain mouse strains (Figure 2). This is also possible for other Nkrp1 receptor and Clr ligand genes in mice, rats, and perhaps humans. This arrangement would be ideal for counter-evolving stimulatory receptors to directly recognize viral decoys that inhibit NK cell function by co-opting self ligands, or for modulating the specificity of self-specific inhibitory receptors to counter–evade interaction with such decoys (45). In addition, the promiscuity of the NKR-P1F/G receptors for overlapping ligands may have evolved under selection pressure from viral evasion strategies. Whether or not additional novel viral immunoevasins exist that may target the NKR-P1:Clr systems is currently under investigation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Dr. David S. J. Allan, Oscar A. Aguilar, and William Kuan Lun Chu for contributions and critical reading of the manuscript. This work was supported in part by an Operating Grant from the Canadian Institutes of Health Research (CIHR 106491 to James R. Carlyle). Christina L. Kirkham was supported by a Natural Science and Engineering Research Council Canada Graduate Scholarship-Masters (NSERC CGS-M) Award and an Ontario Graduate Scholarship (OGS) Award. James R. Carlyle holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund, USA.
Footnote
References
1. Lanier LL. NK cell recognition. Annu Rev Immunol (2005) 23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526
2. Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature (1986) 319:675–8. doi:10.1038/319675a0
3. Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nat Rev Immunol (2006) 6:520–31. doi:10.1038/nri1863
4. Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell (2010) 142:847–56. doi:10.1016/j.cell.2010.08.031
5. Yokoyama WM, Seaman WE. The Ly-49 and NKR-P1 gene families encoding lectin-like receptors on natural killer cells: the NK gene complex. Annu Rev Immunol (1993) 11:613–35. doi:10.1146/annurev.immunol.11.1.613
6. Dissen E, Ryan JC, Seaman WE, Fossum S. An autosomal dominant locus, Nka, mapping to the Ly-49 region of a rat natural killer (NK) gene complex, controls NK cell lysis of allogeneic lymphocytes. J Exp Med (1996) 183:2197–207. doi:10.1084/jem.183.5.2197
7. Hao L, Klein J, Nei M. Heterogeneous but conserved natural killer receptor gene complexes in four major orders of mammals. Proc Natl Acad Sci U S A (2006) 103:3192–7. doi:10.1073/pnas.0511280103
8. Rogers SL, Gobel TW, Viertlboeck BC, Milne S, Beck S, Kaufman J. Characterization of the chicken C-type lectin-like receptors B-NK and B-lec suggests that the NK complex and the MHC share a common ancestral region. J Immunol (2005) 174:3475–83. doi:10.4049/jimmunol.174.6.3475
9. Wagtmann N, Rojo S, Eichler E, Mohrenweiser H, Long EO. A new human gene complex encoding the killer cell inhibitory receptors and related monocyte/macrophage receptors. Curr Biol (1997) 7:615–8. doi:10.1016/S0960-9822(06)00263-6
10. Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome (1999) 10:154–60. doi:10.1007/s003359900961
11. Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity (2001) 15:363–74. doi:10.1016/S1074-7613(01)00197-2
12. Martin AM, Kulski JK, Witt C, Pontarotti P, Christiansen FT. Leukocyte Ig-like receptor complex (LRC) in mice and men. Trends Immunol (2002) 23:81–8. doi:10.1016/S1471-4906(01)02155-X
13. Dong Z, Veillette A. How do SAP family deficiencies compromise immunity? Trends Immunol (2010) 31:295–302. doi:10.1016/j.it.2010.05.008
14. Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol (2011) 29:665–705. doi:10.1146/annurev-immunol-030409-101302
15. Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol (2001) 19:197–223. doi:10.1146/annurev.immunol.19.1.197
16. Biassoni R, Bottino C, Cantoni C, Moretta A. Human natural killer receptors and their ligands. Curr Protoc Immunol (2002) Chapter 14(Unit 14):10. doi:10.1002/0471142735.im1410s46
17. Carlyle JR, Mesci A, Fine JH, Chen P, Belanger S, Tai LH, et al. Evolution of the Ly49 and Nkrp1 recognition systems. Semin Immunol (2008) 20:321–30. doi:10.1016/j.smim.2008.05.004
18. Iizuka K, Naidenko OV, Plougastel BF, Fremont DH, Yokoyama WM. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol (2003) 4:801–7. doi:10.1038/ni954
19. Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol (2003) 3:304–16. doi:10.1038/nri1055
20. Carlyle JR, Jamieson AM, Gasser S, Clingan CS, Arase H, Raulet DH. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc Natl Acad Sci U S A (2004) 101:3527–32. doi:10.1073/pnas.0308304101
21. Chen P, Belanger S, Aguilar OA, Zhang Q, St-Laurent A, Rahim MM, et al. Analysis of the mouse 129-strain Nkrp1-Clr gene cluster reveals conservation of genomic organization and functional receptor-ligand interactions despite significant allelic polymorphism. Immunogenetics (2011) 63:627–40. doi:10.1007/s00251-011-0542-8
22. Kveberg L, Dai KZ, Inngjerdingen M, Brooks CG, Fossum S, Vaage JT. Phylogenetic and functional conservation of the NKR-P1F and NKR-P1G receptors in rat and mouse. Immunogenetics (2011) 63:429–36. doi:10.1007/s00251-011-0520-1
23. Vogler I, Steinle A. Vis-a-vis in the NKC: genetically linked natural killer cell receptor/ligand pairs in the natural killer gene complex (NKC). J Innate Immun (2011) 3:227–35. doi:10.1159/000324112
24. Glimcher L, Shen FW, Cantor H. Identification of a cell-surface antigen selectively expressed on the natural killer cell. J Exp Med (1977) 145:1–9. doi:10.1084/jem.145.1.1
25. Brooks CG, Burton RC, Pollack SB, Henney CS. The presence of NK alloantigens on cloned cytotoxic T lymphocytes. J Immunol (1983) 131:1391–5.
26. Koo GC, Peppard JR. Establishment of monoclonal anti-Nk-1.1 antibody. Hybridoma (1984) 3:301–3. doi:10.1089/hyb.1984.3.301
27. Hackett J Jr, Tutt M, Lipscomb M, Bennett M, Koo G, Kumar V. Origin and differentiation of natural killer cells. II. Functional and morphologic studies of purified NK-1.1+ cells. J Immunol (1986) 136:3124–31.
28. Karlhofer FM, Yokoyama WM. Stimulation of murine natural killer (NK) cells by a monoclonal antibody specific for the NK1.1 antigen. IL-2-activated NK cells possess additional specific stimulation pathways. J Immunol (1991) 146:3662–73.
29. Ryan JC, Niemi EC, Nakamura MC, Seaman WE. NKR-P1A is a target-specific receptor that activates natural killer cell cytotoxicity. J Exp Med (1995) 181:1911–5. doi:10.1084/jem.181.5.1911
30. Sentman CL, Hackett J Jr, Moore TA, Tutt MM, Bennett M, Kumar V. Pan natural killer cell monoclonal antibodies and their relationship to the NK1.1 antigen. Hybridoma (1989) 8:605–14. doi:10.1089/hyb.1989.8.605
31. Chambers WH, Vujanovic NL, Deleo AB, Olszowy MW, Herberman RB, Hiserodt JC. Monoclonal antibody to a triggering structure expressed on rat natural killer cells and adherent lymphokine-activated killer cells. J Exp Med (1989) 169:1373–89. doi:10.1084/jem.169.4.1373
32. Giorda R, Rudert WA, Vavassori C, Chambers WH, Hiserodt JC, Trucco M. NKR-P1, a signal transduction molecule on natural killer cells. Science (1990) 249:1298–300. doi:10.1126/science.2399464
33. Giorda R, Trucco M. Mouse NKR-P1. A family of genes selectively coexpressed in adherent lymphokine-activated killer cells. J Immunol (1991) 147:1701–8.
34. Yokoyama WM, Ryan JC, Hunter JJ, Smith HR, Stark M, Seaman WE. cDNA cloning of mouse NKR-P1 and genetic linkage with Ly-49. Identification of a natural killer cell gene complex on mouse chromosome 6. J Immunol (1991) 147:3229–36.
35. Ryan JC, Turck J, Niemi EC, Yokoyama WM, Seaman WE. Molecular cloning of the NK1.1 antigen, a member of the NKR-P1 family of natural killer cell activation molecules. J Immunol (1992) 149:1631–5.
36. Carlyle JR, Martin A, Mehra A, Attisano L, Tsui FW, Zuniga-Pflucker JC. Mouse NKR-P1B, a novel NK1.1 antigen with inhibitory function. J Immunol (1999) 162:5917–23.
37. Kung SK, Su RC, Shannon J, Miller RG. The NKR-P1B gene product is an inhibitory receptor on SJL/J NK cells. J Immunol (1999) 162:5876–87.
38. Plougastel B, Matsumoto K, Dubbelde C, Yokoyama WM. Analysis of a 1-Mb BAC contig overlapping the mouse Nkrp1 cluster of genes: cloning of three new Nkrp1 members, Nkrp1d, Nkrp1e, and Nkrp1f. Immunogenetics (2001) 53:592–8. doi:10.1007/s002510100367
39. Carlyle JR, Mesci A, Ljutic B, Belanger S, Tai LH, Rousselle E, et al. Molecular and genetic basis for strain-dependent NK1.1 alloreactivity of mouse NK cells. J Immunol (2006) 176:7511–24. doi:10.4049/jimmunol.176.12.7511
40. Aust JG, Gays F, Mickiewicz KM, Buchanan E, Brooks CG. The expression and function of the NKRP1 receptor family in C57BL/6 mice. J Immunol (2009) 183:106–16. doi:10.4049/jimmunol.0804281
41. Liu J, Morris MA, Nguyen P, George TC, Koulich E, Lai WC, et al. Ly49I NK cell receptor transgene inhibition of rejection of H2b mouse bone marrow transplants. J Immunol (2000) 164:1793–9. doi:10.4049/jimmunol.164.4.1793
42. Higuchi DA, Cahan P, Gao J, Ferris ST, Poursine-Laurent J, Graubert TA, et al. Structural variation of the mouse natural killer gene complex. Genes Immun (2010) 11:637–48. doi:10.1038/gene.2010.48
43. Kveberg L, Back CJ, Dai KZ, Inngjerdingen M, Rolstad B, Ryan JC, et al. The novel inhibitory NKR-P1C receptor and Ly49s3 identify two complementary, functionally distinct NK cell subsets in rats. J Immunol (2006) 176:4133–40. doi:10.4049/jimmunol.176.7.4133
44. Li J, Rabinovich BA, Hurren R, Shannon J, Miller RG. Expression cloning and function of the rat NK activating and inhibitory receptors NKR-P1A and -P1B. Int Immunol (2003) 15:411–6. doi:10.1093/intimm/dxg046
45. Voigt S, Mesci A, Ettinger J, Fine JH, Chen P, Chou W, et al. Cytomegalovirus evasion of innate immunity by subversion of the NKR-P1B:Clr-b missing-self axis. Immunity (2007) 26:617–27. doi:10.1016/j.immuni.2007.03.013
46. Lanier LL, Chang C, Phillips JH. Human NKR-P1A. A disulfide-linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J Immunol (1994) 153:2417–28.
47. Aldemir H, Prod’homme V, Dumaurier MJ, Retiere C, Poupon G, Cazareth J, et al. Cutting edge: lectin-like transcript 1 is a ligand for the CD161 receptor. J Immunol (2005) 175:7791–5. doi:10.4049/jimmunol.175.12.7791
48. Rosen DB, Bettadapura J, Alsharifi M, Mathew PA, Warren HS, Lanier LL. Cutting edge: lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J Immunol (2005) 175:7796–9. doi:10.4049/jimmunol.175.12.7796
49. Giorda R, Weisberg EP, Ip TK, Trucco M. Genomic structure and strain-specific expression of the natural killer cell receptor NKR-P1. J Immunol (1992) 149:1957–63.
50. Zhou H, Kartsogiannis V, Hu YS, Elliott J, Quinn JM, McKinstry WJ, et al. A novel osteoblast-derived C-type lectin that inhibits osteoclast formation. J Biol Chem (2001) 276:14916–23. doi:10.1074/jbc.M011554200
51. Zhou H, Kartsogiannis V, Quinn JM, Ly C, Gange C, Elliott J, et al. Osteoclast inhibitory lectin, a family of new osteoclast inhibitors. J Biol Chem (2002) 277:48808–15. doi:10.1074/jbc.M209059200
52. Plougastel B, Dubbelde C, Yokoyama WM. Cloning of Clr, a new family of lectin-like genes localized between mouse Nkrp1a and Cd69. Immunogenetics (2001) 53:209–14. doi:10.1007/s002510100319
53. Zhang Q, Rahim MM, Allan DS, Tu MM, Belanger S, Abou-Samra E, et al. Mouse Nkrp1-Clr gene cluster sequence and expression analyses reveal conservation of tissue-specific MHC-independent immunosurveillance. PLoS One (2012) 7:e50561. doi:10.1371/journal.pone.0050561
54. Bezouska K, Vlahas G, Horvath O, Jinochova G, Fiserova A, Giorda R, et al. Rat natural killer cell antigen, NKR-P1, related to C-type animal lectins is a carbohydrate-binding protein. J Biol Chem (1994) 269:16945–52.
55. Bezouska K, Yuen CT, O’Brien J, Childs RA, Chai W, Lawson AM, et al. Oligosaccharide ligands for NKR-P1 protein activate NK cells and cytotoxicity. Nature (1994) 372:150–7. doi:10.1038/372150a0
56. Voigt S, Sandford GR, Ding L, Burns WH. Identification and characterization of a spliced C-type lectin-like gene encoded by rat cytomegalovirus. J Virol (2001) 75:603–11. doi:10.1128/JVI.75.2.603-611.2001
57. Ettinger J, Geyer H, Nitsche A, Zimmermann A, Brune W, Sandford GR, et al. Complete genome sequence of the English isolate of rat cytomegalovirus (Murid herpesvirus 8). J Virol (2012) 86:13838. doi:10.1128/JVI.02614-12
58. Quan JX, Zheng F, Li XX, Hu LL, Sun ZY, Jiao YL, et al. Cloning and analysis of rat osteoclast inhibitory lectin gene promoter. J Cell Biochem (2009) 106:599–607. doi:10.1002/jcb.22036
59. Kveberg L, Dai KZ, Westgaard IH, Daws MR, Fossum S, Naper C, et al. Two major groups of rat NKR-P1 receptors can be distinguished based on chromosomal localization, phylogenetic analysis and Clr ligand binding. Eur J Immunol (2009) 39:541–51. doi:10.1002/eji.200838891
60. Boles KS, Barten R, Kumaresan PR, Trowsdale J, Mathew PA. Cloning of a new lectin-like receptor expressed on human NK cells. Immunogenetics (1999) 50:1–7. doi:10.1007/s002510050679
61. Mathew PA, Chuang SS, Vaidya SV, Kumaresan PR, Boles KS, Pham HT. The LLT1 receptor induces IFN-gamma production by human natural killer cells. Mol Immunol (2004) 40:1157–63. doi:10.1016/j.molimm.2003.11.024
62. Hamann J, Montgomery KT, Lau S, Kucherlapati R, Van Lier RA. AICL: a new activation-induced antigen encoded by the human NK gene complex. Immunogenetics (1997) 45:295–300. doi:10.1007/s002510050208
63. Spreu J, Kienle EC, Schrage B, Steinle A. CLEC2A: a novel, alternatively spliced and skin-associated member of the NKC-encoded AICL-CD69-LLT1 family. Immunogenetics (2007) 59:903–12. doi:10.1007/s00251-007-0263-1
64. Hu YS, Zhou H, Myers D, Quinn JM, Atkins GJ, Ly C, et al. Isolation of a human homolog of osteoclast inhibitory lectin that inhibits the formation and function of osteoclasts. J Bone Miner Res (2004) 19:89–99. doi:10.1359/jbmr.0301215
65. Germain C, Bihl F, Zahn S, Poupon G, Dumaurier MJ, Rampanarivo HH, et al. Characterization of alternatively spliced transcript variants of CLEC2D gene. J Biol Chem (2010) 285:36207–15. doi:10.1074/jbc.M110.179622
66. Lysenko O, Schulte D, Mittelbronn M, Steinle A. BACL is a novel brain-associated, non-NKC-encoded mammalian C-type lectin-like receptor of the CLEC2 family. PLoS One (2013) 8:e65345. doi:10.1371/journal.pone.0065345
67. Colonna M, Samaridis J, Angman L. Molecular characterization of two novel C-type lectin-like receptors, one of which is selectively expressed in human dendritic cells. Eur J Immunol (2000) 30:697–704. doi:10.1002/1521-4141(200002)30:2<697::AID-IMMU697>3.0.CO;2-M
68. Ryan JC, Seaman WE. Divergent functions of lectin-like receptors on NK cells. Immunol Rev (1997) 155:79–89. doi:10.1111/j.1600-065X.1997.tb00941.x
69. Flornes LM, Nylenna O, Saether PC, Daws MR, Dissen E, Fossum S. The complete inventory of receptors encoded by the rat natural killer cell gene complex. Immunogenetics (2010) 62:521–30. doi:10.1007/s00251-010-0455-y
70. Welte S, Kuttruff S, Waldhauer I, Steinle A. Mutual activation of natural killer cells and monocytes mediated by NKp80-AICL interaction. Nat Immunol (2006) 7:1334–42. doi:10.1038/ni1402
71. Spreu J, Kuttruff S, Stejfova V, Dennehy KM, Schittek B, Steinle A. Interaction of C-type lectin-like receptors NKp65 and KACL facilitates dedicated immune recognition of human keratinocytes. Proc Natl Acad Sci U S A (2010) 107:5100–5. doi:10.1073/pnas.0913108107
72. Tian W, Feng B, Liou HC. Silencing OCILRP2 leads to intrinsic defects in T cells in response to antigenic stimulation. Cell Immunol (2005) 235:72–84. doi:10.1016/j.cellimm.2005.07.004
73. Tian W, Nunez R, Cheng S, Ding Y, Tumang J, Lyddane C, et al. C-type lectin OCILRP2/Clr-g and its ligand NKRP1f costimulate T cell proliferation and IL-2 production. Cell Immunol (2005) 234:39–53. doi:10.1016/j.cellimm.2005.04.021
74. Denning TL, Granger SW, Mucida D, Graddy R, Leclercq G, Zhang W, et al. Mouse TCRalphabeta+CD8alphaalpha intraepithelial lymphocytes express genes that down-regulate their antigen reactivity and suppress immune responses. J Immunol (2007) 178:4230–9. doi:10.4049/jimmunol.178.10.6654-a
75. Assarsson E, Kambayashi T, Sandberg JK, Hong S, Taniguchi M, Van Kaer L, et al. CD8+ T cells rapidly acquire NK1.1 and NK cell-associated molecules upon stimulation in vitro and in vivo. J Immunol (2000) 165:3673–9. doi:10.4049/jimmunol.165.7.3673
76. Kambayashi T, Assarsson E, Michaelsson J, Berglund P, Diehl AD, Chambers BJ, et al. Emergence of CD8+ T cells expressing NK cell receptors in influenza A virus-infected mice. J Immunol (2000) 165:4964–9. doi:10.4049/jimmunol.165.9.4964
77. Slifka MK, Whitton JL. Activated and memory CD8+ T cells can be distinguished by their cytokine profiles and phenotypic markers. J Immunol (2000) 164:208–16. doi:10.4049/jimmunol.164.1.208
78. McMahon CW, Zajac AJ, Jamieson AM, Corral L, Hammer GE, Ahmed R, et al. Viral and bacterial infections induce expression of multiple NK cell receptors in responding CD8(+) T cells. J Immunol (2002) 169:1444–52. doi:10.4049/jimmunol.169.3.1444
79. Ballas ZK, Rasmussen W. NK1.1+ thymocytes. Adult murine CD4-, CD8- thymocytes contain an NK1.1+, CD3+, CD5hi, CD44hi, TCR-V beta 8+ subset. J Immunol (1990) 145:1039–45.
80. Carlyle JR, Michie AM, Furlonger C, Nakano T, Lenardo MJ, Paige CJ, et al. Identification of a novel developmental stage marking lineage commitment of progenitor thymocytes. J Exp Med (1997) 186:173–82. doi:10.1084/jem.186.2.173
81. Carlyle JR, Michie AM, Cho SK, Zuniga-Pflucker JC. Natural killer cell development and function precede alpha beta T cell differentiation in mouse fetal thymic ontogeny. J Immunol (1998) 160:744–53.
82. Carlyle JR, Zuniga-Pflucker JC. Requirement for the thymus in alphabeta T lymphocyte lineage commitment. Immunity (1998) 9:187–97. doi:10.1016/S1074-7613(00)80601-9
83. Carlyle JR, Zuniga-Pflucker JC. Lineage commitment and differentiation of T and natural killer lymphocytes in the fetal mouse. Immunol Rev (1998) 165:63–74. doi:10.1111/j.1600-065X.1998.tb01230.x
84. Carlyle JR, Zuniga-Pflucker JC. Regulation of NK1.1 expression during lineage commitment of progenitor thymocytes. J Immunol (1998) 161:6544–51.
85. Guy-Grand D, Azogui O, Celli S, Darche S, Nussenzweig MC, Kourilsky P, et al. Extrathymic T cell lymphopoiesis: ontogeny and contribution to gut intraepithelial lymphocytes in athymic and euthymic mice. J Exp Med (2003) 197:333–41. doi:10.1084/jem.20021639
86. Vivier E, Spits H, Cupedo T. Interleukin-22-producing innate immune cells: new players in mucosal immunity and tissue repair? Nat Rev Immunol (2009) 9:229–34. doi:10.1038/nri2522
87. Fine JH, Chen P, Mesci A, Allan DS, Gasser S, Raulet DH, et al. Chemotherapy-induced genotoxic stress promotes sensitivity to natural killer cell cytotoxicity by enabling missing-self recognition. Cancer Res (2010) 70:7102–13. doi:10.1158/0008-5472.CAN-10-1316
88. Kveberg L, Dai KZ, Dissen E, Ryan JC, Rolstad B, Vaage JT, et al. Strain-dependent expression of four structurally related rat Ly49 receptors; correlation with NK gene complex haplotype and NK alloreactivity. Immunogenetics (2006) 58:905–16. doi:10.1007/s00251-006-0154-x
89. Kveberg L, Jimenez-Royo P, Naper C, Rolstad B, Butcher GW, Vaage JT, et al. Two complementary rat NK cell subsets, Ly49s3+ and NKR-P1B+, differ in phenotypic characteristics and responsiveness to cytokines. J Leukoc Biol (2010) 88:87–93. doi:10.1189/jlb.0110039
90. Inngjerdingen M, Kveberg L, Vaage JT. A novel NKR-P1B(bright) NK cell subset expresses an activated CD25(+)CX(3)CR1(+)CD62L(-)CD11b(-) CD27(-) phenotype and is prevalent in blood, liver, and gut-associated lymphoid organs of rats. J Immunol (2012) 188:2499–508. doi:10.4049/jimmunol.1003939
91. Rosen DB, Cao W, Avery DT, Tangye SG, Liu YJ, Houchins JP, et al. Functional consequences of interactions between human NKR-P1A and its ligand LLT1 expressed on activated dendritic cells and B cells. J Immunol (2008) 180:6508–17. doi:10.4049/jimmunol.180.10.6508
92. Fergusson JR, Fleming VM, Klenerman P. CD161-expressing human T cells. Front Immunol (2011) 2:36. doi:10.3389/fimmu.2011.00036
93. Poggi A, Zocchi MR, Costa P, Ferrero E, Borsellino G, Placido R, et al. IL-12-mediated NKRP1A up-regulation and consequent enhancement of endothelial transmigration of V delta 2+ TCR gamma delta+ T lymphocytes from healthy donors and multiple sclerosis patients. J Immunol (1999) 162:4349–54.
94. Bennett IM, Zatsepina O, Zamai L, Azzoni L, Mikheeva T, Perussia B. Definition of a natural killer NKR-P1A+/CD56-/CD16- functionally immature human NK cell subset that differentiates in vitro in the presence of interleukin 12. J Exp Med (1996) 184:1845–56. doi:10.1084/jem.184.5.1845
95. Loza MJ, Zamai L, Azzoni L, Rosati E, Perussia B. Expression of type 1 (interferon gamma) and type 2 (interleukin-13, interleukin-5) cytokines at distinct stages of natural killer cell differentiation from progenitor cells. Blood (2002) 99:1273–81. doi:10.1182/blood.V99.4.1273
96. Farag SS, Caligiuri MA. Human natural killer cell development and biology. Blood Rev (2006) 20:123–37. doi:10.1016/j.blre.2005.10.001
97. Freud AG, Caligiuri MA. Human natural killer cell development. Immunol Rev (2006) 214:56–72. doi:10.1111/j.1600-065X.2006.00451.x
98. Poggi A, Costa P, Morelli L, Cantoni C, Pella N, Spada F, et al. Expression of human NKRP1A by CD34+ immature thymocytes: NKRP1A-mediated regulation of proliferation and cytolytic activity. Eur J Immunol (1996) 26:1266–72. doi:10.1002/eji.1830260613
99. Azzoni L, Zatsepina O, Abebe B, Bennett IM, Kanakaraj P, Perussia B. Differential transcriptional regulation of CD161 and a novel gene, 197/15a, by IL-2, IL-15, and IL-12 in NK and T cells. J Immunol (1998) 161:3493–500.
100. Poggi A, Costa P, Tomasello E, Moretta L. IL-12-induced up-regulation of NKRP1A expression in human NK cells and consequent NKRP1A-mediated down-regulation of NK cell activation. Eur J Immunol (1998) 28:1611–6. doi:10.1002/(SICI)1521-4141(199805)28:05<1611::AID-IMMU1611>3.0.CO;2-6
101. Maggi L, Santarlasci V, Capone M, Peired A, Frosali F, Crome SQ, et al. CD161 is a marker of all human IL-17-producing T-cell subsets and is induced by RORC. Eur J Immunol (2010) 40:2174–81. doi:10.1002/eji.200940257
102. Mjosberg JM, Trifari S, Crellin NK, Peters CP, Van Drunen CM, Piet B, et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol (2011) 12:1055–62. doi:10.1038/ni.2104
103. Crellin NK, Trifari S, Kaplan CD, Cupedo T, Spits H. Human NKp44+IL-22+ cells and LTi-like cells constitute a stable RORC+ lineage distinct from conventional natural killer cells. J Exp Med (2010) 207:281–90. doi:10.1084/jem.20091509
104. Pesenacker AM, Bending D, Ursu S, Wu Q, Nistala K, Wedderburn LR. CD161 defines the subset of FoxP3+ T cells capable of producing proinflammatory cytokines. Blood (2013) 121:2647–58. doi:10.1182/blood-2012-08-443473
105. Poggi A, Costa P, Zocchi MR, Moretta L. Phenotypic and functional analysis of CD4+ NKRP1A+ human T lymphocytes. Direct evidence that the NKRP1A molecule is involved in transendothelial migration. Eur J Immunol (1997) 27:2345–50. doi:10.1002/eji.1830270932
106. Poggi A, Rubartelli A, Moretta L, Zocchi MR. Expression and function of NKRP1A molecule on human monocytes and dendritic cells. Eur J Immunol (1997) 27:2965–70. doi:10.1002/eji.1830271132
107. Germain C, Meier A, Jensen T, Knapnougel P, Poupon G, Lazzari A, et al. Induction of lectin-like transcript 1 (LLT1) protein cell surface expression by pathogens and interferon-gamma contributes to modulate immune responses. J Biol Chem (2011) 286:37964–75. doi:10.1074/jbc.M111.285312
108. Back J, Malchiodi EL, Cho S, Scarpellino L, Schneider P, Kerzic MC, et al. Distinct conformations of Ly49 natural killer cell receptors mediate MHC class I recognition in trans and cis. Immunity (2009) 31:598–608. doi:10.1016/j.immuni.2009.07.007
109. Ito D, Iizuka YM, Katepalli MP, Iizuka K. Essential role of the Ly49A stalk region for immunological synapse formation and signaling. Proc Natl Acad Sci U S A (2009) 106:11264–9. doi:10.1073/pnas.0900664106
110. Bartel Y, Bauer B, Steinle A. Modulation of NK cell function by genetically coupled C-type lectin-like receptor/ligand pairs encoded in the human natural killer gene complex. Front Immunol (2013) 4:362. doi:10.3389/fimmu.2013.00362
111. Ljutic B, Carlyle JR, Zuniga-Pflucker JC. Identification of upstream cis-acting regulatory elements controlling lineage-specific expression of the mouse NK cell activation receptor, NKR-P1C. J Biol Chem (2003) 278:31909–17. doi:10.1074/jbc.M212869200
112. Anderson SK. Transcriptional regulation of NK cell receptors. Curr Top Microbiol Immunol (2006) 298:59–75.
113. Drickamer K. Two distinct classes of carbohydrate-recognition domains in animal lectins. J Biol Chem (1988) 263:9557–60.
114. Sovova Z, Kopecky V Jr, Pazderka T, Hofbauerova K, Rozbesky D, Vanek O, et al. Structural analysis of natural killer cell receptor protein 1 (NKR-P1) extracellular domains suggests a conserved long loop region involved in ligand specificity. J Mol Model (2011) 17:1353–70. doi:10.1007/s00894-010-0837-y
115. Rozbesky D, Kavan D, Chmelik J, Novak P, Vanek O, Bezouska K. High-level expression of soluble form of mouse natural killer cell receptor NKR-P1C(B6) in Escherichia coli. Protein Expr Purif (2011) 77:178–84. doi:10.1016/j.pep.2011.01.013
116. Kolenko P, Rozbesky D, Vanek O, Bezouska K, Hasek J, Dohnalek J. Structure of the H107R variant of the extracellular domain of mouse NKR-P1A at 2.3 A resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun (2011) 67:1519–23. doi:10.1107/S1744309111046203
117. Kamishikiryo J, Fukuhara H, Okabe Y, Kuroki K, Maenaka K. Molecular basis for LLT1 protein recognition by human CD161 protein (NKRP1A/KLRB1). J Biol Chem (2011) 286:23823–30. doi:10.1074/jbc.M110.214254
118. Li Y, Wang Q, Chen S, Brown PH, Mariuzza RA. Structure of NKp65 bound to its keratinocyte ligand reveals basis for genetically linked recognition in natural killer gene complex. Proc Natl Acad Sci U S A (2013) 110:11505–10. doi:10.1073/pnas.1303300110
119. Dam J, Guan R, Natarajan K, Dimasi N, Chlewicki LK, Kranz DM, et al. Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2K(b). Nat Immunol (2003) 4:1213–22. doi:10.1038/ni1006
120. Skalova T, Kotynkova K, Duskova J, Hasek J, Koval T, Kolenko P, et al. Mouse Clr-g, a ligand for NK cell activation receptor NKR-P1F: crystal structure and biophysical properties. J Immunol (2012) 189:4881–9. doi:10.4049/jimmunol.1200880
121. Brumbaugh KM, Binstadt BA, Billadeau DD, Schoon RA, Dick CJ, Ten RM, et al. Functional role for Syk tyrosine kinase in natural killer cell-mediated natural cytotoxicity. J Exp Med (1997) 186:1965–74. doi:10.1084/jem.186.12.1965
122. Colucci F, Di Santo JP, Leibson PJ. Natural killer cell activation in mice and men: different triggers for similar weapons? Nat Immunol (2002) 3:807–13. doi:10.1038/ni0902-807
123. Colucci F, Schweighoffer E, Tomasello E, Turner M, Ortaldo JR, Vivier E, et al. Natural cytotoxicity uncoupled from the Syk and ZAP-70 intracellular kinases. Nat Immunol (2002) 3:288–94. doi:10.1038/ni764
124. Zompi S, Hamerman JA, Ogasawara K, Schweighoffer E, Tybulewicz VL, Di Santo JP, et al. NKG2D triggers cytotoxicity in mouse NK cells lacking DAP12 or Syk family kinases. Nat Immunol (2003) 4:565–72. doi:10.1038/ni930
125. Arase N, Arase H, Park SY, Ohno H, Ra C, Saito T. Association with FcRgamma is essential for activation signal through NKR-P1 (CD161) in natural killer (NK) cells and NK1.1+ T cells. J Exp Med (1997) 186:1957–63. doi:10.1084/jem.186.12.1957
126. Arase H, Suenaga T, Arase N, Kimura Y, Ito K, Shiina R, et al. Negative regulation of expression and function of Fc gamma RIII by CD3 zeta in murine NK cells. J Immunol (2001) 166:21–5. doi:10.4049/jimmunol.166.1.21
127. Ryan JC, Niemi EC, Goldfien RD, Hiserodt JC, Seaman WE. NKR-P1, an activating molecule on rat natural killer cells, stimulates phosphoinositide turnover and a rise in intracellular calcium. J Immunol (1991) 147:3244–50.
128. Al-Aoukaty A, Rolstad B, Maghazachi AA. Functional coupling of NKR-P1 receptors to various heterotrimeric G proteins in rat interleukin-2-activated natural killer cells. J Biol Chem (1997) 272:31604–8. doi:10.1074/jbc.272.50.31604
129. Grazia Cifone M, Roncaioli P, Cironi L, Festuccia C, Meccia A, D’Alo S, et al. NKR-P1A stimulation of arachidonate-generating enzymes in rat NK cells is associated with granule release and cytotoxic activity. J Immunol (1997) 159:309–17.
130. Mesci A, Ljutic B, Makrigiannis AP, Carlyle JR. NKR-P1 biology: from prototype to missing self. Immunol Res (2006) 35:13–26. doi:10.1385/IR:35:1:13
131. Campbell KS, Giorda R. The cytoplasmic domain of rat NKR-P1 receptor interacts with the N-terminal domain of p56(lck) via cysteine residues. Eur J Immunol (1997) 27:72–7. doi:10.1002/eji.1830270111
132. Ljutic B, Carlyle JR, Filipp D, Nakagawa R, Julius M, Zuniga-Pflucker JC. Functional requirements for signaling through the stimulatory and inhibitory mouse NKR-P1 (CD161) NK cell receptors. J Immunol (2005) 174:4789–96. doi:10.4049/jimmunol.174.8.4789
133. Burshtyn DN, Yang W, Yi T, Long EO. A novel phosphotyrosine motif with a critical amino acid at position -2 for the SH2 domain-mediated activation of the tyrosine phosphatase SHP-1. J Biol Chem (1997) 272:13066–72. doi:10.1074/jbc.272.20.13066
134. Tassi I, Cella M, Gilfillan S, Turnbull I, Diacovo TG, Penninger JM, et al. p110gamma and p110delta phosphoinositide 3-kinase signaling pathways synergize to control development and functions of murine NK cells. Immunity (2007) 27:214–27. doi:10.1016/j.immuni.2007.07.014
135. Gilfillan S, Ho EL, Cella M, Yokoyama WM, Colonna M. NKG2D recruits two distinct adapters to trigger NK cell activation and costimulation. Nat Immunol (2002) 3:1150–5. doi:10.1038/ni857
136. Diefenbach A, Tomasello E, Lucas M, Jamieson AM, Hsia JK, Vivier E, et al. Selective associations with signaling proteins determine stimulatory versus costimulatory activity of NKG2D. Nat Immunol (2002) 3:1142–9. doi:10.1038/ni858
137. Karimi M, Cao TM, Baker JA, Verneris MR, Soares L, Negrin RS. Silencing human NKG2D, DAP10, and DAP12 reduces cytotoxicity of activated CD8+ T cells and NK cells. J Immunol (2005) 175:7819–28. doi:10.4049/jimmunol.175.12.7819
138. Rabinovich B, Li J, Wolfson M, Lawrence W, Beers C, Chalupny J, et al. NKG2D splice variants: a reexamination of adaptor molecule associations. Immunogenetics (2006) 58:81–8. doi:10.1007/s00251-005-0078-x
139. Chang CS, Brossay L, Kronenberg M, Kane KP. The murine nonclassical class I major histocompatibility complex-like CD1.1 molecule protects target cells from lymphokine-activated killer cell cytolysis. J Exp Med (1999) 189:483–91. doi:10.1084/jem.189.3.483
140. Huang MM, Borszcz P, Sidobre S, Kronenberg M, Kane KP. CD1d1 displayed on cell size beads identifies and enriches an NK cell population negatively regulated by CD1d1. J Immunol (2004) 172:5304–12. doi:10.4049/jimmunol.172.9.5304
141. Cerny J, Fiserova A, Horvath O, Bezouska K, Pospisil M, Horejsi V. Association of human NK cell surface receptors NKR-P1 and CD94 with Src-family protein kinases. Immunogenetics (1997) 46:231–6. doi:10.1007/s002510050267
142. Exley M, Porcelli S, Furman M, Garcia J, Balk S. CD161 (NKR-P1A) costimulation of CD1d-dependent activation of human T cells expressing invariant V alpha 24 J alpha Q T cell receptor alpha chains. J Exp Med (1998) 188:867–76. doi:10.1084/jem.188.5.867
143. Pozo D, Vales-Gomez M, Mavaddat N, Williamson SC, Chisholm SE, Reyburn H. CD161 (human NKR-P1A) signaling in NK cells involves the activation of acid sphingomyelinase. J Immunol (2006) 176:2397–406. doi:10.4049/jimmunol.176.4.2397
144. Cosmi L, De Palma R, Santarlasci V, Maggi L, Capone M, Frosali F, et al. Human interleukin 17-producing cells originate from a CD161+CD4+ T cell precursor. J Exp Med (2008) 205:1903–16. doi:10.1084/jem.20080397
145. Dusseaux M, Martin E, Serriari N, Peguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood (2011) 117:1250–9. doi:10.1182/blood-2010-08-303339
146. Lock C, Hermans G, Pedotti R, Brendolan A, Schadt E, Garren H, et al. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat Med (2002) 8:500–8. doi:10.1038/nm0502-500
147. Kleinschek MA, Boniface K, Sadekova S, Grein J, Murphy EE, Turner SP, et al. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J Exp Med (2009) 206:525–34. doi:10.1084/jem.20081712
148. Wolfkamp SC, Verstege MI, Vogels EW, Meisner S, Verseijden C, Stokkers PC, et al. Single nucleotide polymorphisms in C-type lectin genes, clustered in the IBD2 and IBD6 susceptibility loci, may play a role in the pathogenesis of inflammatory bowel diseases. Eur J Gastroenterol Hepatol (2012) 24:965–70. doi:10.1097/MEG.0b013e328354f3d5
149. Pineda B, Laporta P, Cano A, Garcia-Perez MA. The Asn19Lys substitution in the osteoclast inhibitory lectin (OCIL) gene is associated with a reduction of bone mineral density in postmenopausal women. Calcif Tissue Int (2008) 82:348–53. doi:10.1007/s00223-008-9135-4
150. Williams KJ, Wilson E, Davidson CL, Aguilar OA, Fu L, Carlyle JR, et al. Poxvirus infection-associated downregulation of C-type lectin-related-b prevents NK cell inhibition by NK receptor protein-1B. J Immunol (2012) 188:4980–91. doi:10.4049/jimmunol.1103425
151. Brocchieri L, Kledal TN, Karlin S, Mocarski ES. Predicting coding potential from genome sequence: application to betaherpesviruses infecting rats and mice. J Virol (2005) 79:7570–96. doi:10.1128/JVI.79.12.7570-7596.2005
152. Bambard ND, Mathew SO, Mathew PA. LLT1-mediated activation of IFN-gamma production in human natural killer cells involves ERK signalling pathway. Scand J Immunol (2010) 71:210–9. doi:10.1111/j.1365-3083.2009.02367.x
153. Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature (2008) 454:899–902. doi:10.1038/nature07117
154. Birikh KR, Heaton PA, Eckstein F. The structure, function and application of the hammerhead ribozyme. Eur J Biochem (1997) 245:1–16. doi:10.1111/j.1432-1033.1997.t01-3-00001.x
155. Hammann C, Luptak A, Perreault J, De La Pena M. The ubiquitous hammerhead ribozyme. RNA (2012) 18:871–85. doi:10.1261/rna.031401.111
Keywords: natural killer cell, innate immunity, C-type lectin-like, Nkrp1, CD161, Clr, Ocil, LLT1
Citation: Kirkham CL and Carlyle JR (2014) Complexity and diversity of the NKR-P1:Clr (Klrb1:Clec2) recognition systems. Front. Immunol. 5:214. doi: 10.3389/fimmu.2014.00214
Received: 15 January 2014; Paper pending published: 24 March 2014;
Accepted: 28 April 2014; Published online: 02 June 2014.
Edited by:
Andrew P. Makrigiannis, University of Ottawa, CanadaReviewed by:
Martin Herrmann, Universitätsklinikum Erlangen, GermanyEric O. Long, National Institute of Allergy and Infectious Diseases, USA
Copyright: © 2014 Kirkham and Carlyle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James R. Carlyle, Department of Immunology, University of Toronto, Sunnybrook Research Institute, 2075 Bayview Avenue, Toronto, ON, M4N 3M5, Canada e-mail: james.carlyle@utoronto.ca
 Christina L. Kirkham
Christina L. Kirkham James R. Carlyle
James R. Carlyle