
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 12 December 2014
Sec. Microbial Immunology
Volume 5 - 2014 | https://doi.org/10.3389/fimmu.2014.00581
This article is part of the Research TopicHow Salmonella Infection Can Inform on Mechanisms of Immune Function and HomeostasisView all 16 articles
Salmonella are facultative intracellular pathogens. Salmonella infection occurs mainly by expression of two Salmonella pathogenicity Islands (SPI-1 and SPI-2). SPI-1 encodes transcriptional factors that participate in the expression of virulence factors encoded in the island. However, there are transcriptional factors encoded outside the island that also participate in the expression of SPI-1-encoded genes. Upon infection, bacteria are capable of avoiding the host immune response with several strategies that involve several virulence factors under the control of transcriptional regulators. Interestingly, LeuO a transcriptional global regulator which is encoded outside of any SPI, is proposed to be part of a complex regulatory network that involves expression of several genes that help bacteria to survive stress conditions and, also, induces the expression of porins that have been shown to be immunogens and can thus be considered as antigenic candidates for acellular vaccines. Hence, the understanding of the LeuO regulon implies a role of bacterial genetic regulation in determining the host immune response.
Salmonella enterica are Gram-negative bacterial pathogens capable of infecting human beings and other vertebrates, and causing substantial morbidity and mortality (1, 2). In human beings, most of Salmonella serovars can cause infections in the small intestine and hence gastroenteritis; yet a small percentage of Salmonella serovars can cause a systemic infection, such as typhoid fever by the Typhi serovar (3). Control of Salmonella infection is difficult, in part due to the capacity of the bacterium to tolerate environmental stress, to its widespread distribution, multiple drug resistance, and adaptability (4). They infect human beings and other animals by the fecal–oral route, via contaminated food and water.
After oral acquisition, Salmonella resists low pH in the stomach and colonizes the intestinal tract and some cells can disseminate to cause systemic infection of organs such as liver and spleen (1). Salmonella virulence factors as well as host immune responses are determinant in the infectious process developed in the pathology (5). S. enterica Typhimurium and Typhi serovars interact with host cells through the activities mainly of two type three secretion systems (TTSS), encoded in two pathogenicity islands, 1 and 2 (SPI-1 and SPI-2) (6, 7). While SPI-1 participates in bacterial cell entry into non-phagocytic epithelial cells, SPI-2 is required for intracellular maintenance of the bacteria in a specialized membranous compartment (8). Salmonella internalization is mediated by effectors encoded in SPI-1: SopE, SopE2, and SopB, which activate the Rho family of GTPases Rac1, Cdc42 and RhoG (9, 10). These bacterial effectors promote a transcriptional reprograming in host cells, which in turn leads to the expression of pro-inflammatory cytokines, which could be essential for the initiation of diarrhea, a hallmark of acute Salmonella infection. Recently, it has been observed that the expression of the pro-inflammatory cytokine interleukin 22 (IL-22) can be exploited by pathogens, such as Salmonella, to suppress the growth of their closest competitors thereby enhancing pathogen colonization of mucosal surfaces (11–13).
Upon infection of intestinal epithelial cells, early transcriptional host responses occur characteristically after the stimulation of the innate immune receptors (14). However, the Salmonella-induced responses are unique in that this pathogen is capable of stimulating them independently of innate immune receptors (12), which are largely inactive in the intestinal epithelial cells due to robust negative regulatory mechanisms (15–17). After internalization in epithelial cells, bacteria traverse the intestinal epithelium and can invade M-cells overlying Peyer’s patches, as well as being captured by dendritic cells directly from the intestinal lumen (18).
Systemic infection requires intracellular survival and replication, while Salmonella-macrophage interactions are essential for bacterial virulence, disease, pathology and chronic infection (19–21). Immunity to intra-macrophage pathogens (i.e., Salmonella) requires the infected host to generate a robust and sustained CD4 Th1 response (22). Salmonella infection of inbred mouse strains induces a robust CD4+ T-cell response that is essential toward protective immunity to secondary infection (23–27). Salmonella also induces CD8+ T-cells and antibody responses that can contribute to the resolution of infection (25, 27, 28). The first study to successfully characterize Salmonella-specific CD4+ T-cell clones identified the target antigen of these T-cells as an I-Ak epitope within the central hypervariable portion of bacterial flagellin encoded by the FliC gene (29). Subsequently, additional MHC class II epitopes were identified in the same protein and thus flagellin remains the most thoroughly defined target antigen in the Salmonella infection model (30, 31). Additional studies have shown that immunization with flagellin provides a modest degree of protective immunity to Salmonella infection, usually defined by slightly lower bacterial counts or a delay in time to death after infection. Thus, flagellin is a well-defined target antigen of CD4+ T-cells during Salmonella infection and this response contributes modestly to protective immunity in vivo (32, 33). Among other antigens, the outer membrane proteins (OMPs) are particularly important. In a murine model, the highly abundant OmpC and OmpF porins (34) can induce long-term antibody responses with high bactericidal capacity, and they even confer protection against challenge with Salmonella Typhi (35, 36).
LeuO is part of the LysR-type transcriptional regulators (LTTRs), the largest family of transcriptional regulators in prokaryotes. In consequence, they regulate a wide variety of genes that are involved in a diversity of cellular functions such as biosynthesis of amino acids, catabolism of aromatic compounds, antibiotic resistance, oxidative stress response, nitrogen fixation, quorum sensing and virulence (Figure 1) (37–40). Many structural studies have shown an organization of an N-terminal DNA-binding domain (DBD) with a winged Helix-Turn-Helix (wHTH) motif; and a long linker helix (LH) involved in dimerization that connects the DBD with the C-terminal effector binding domain (EBD) or regulatory domain (RD) (37, 41–43). These regulators are proteins between 300 and 350 residues, mostly acting as transcriptional activators that bind to A–T rich DNA sequences in similar positions.
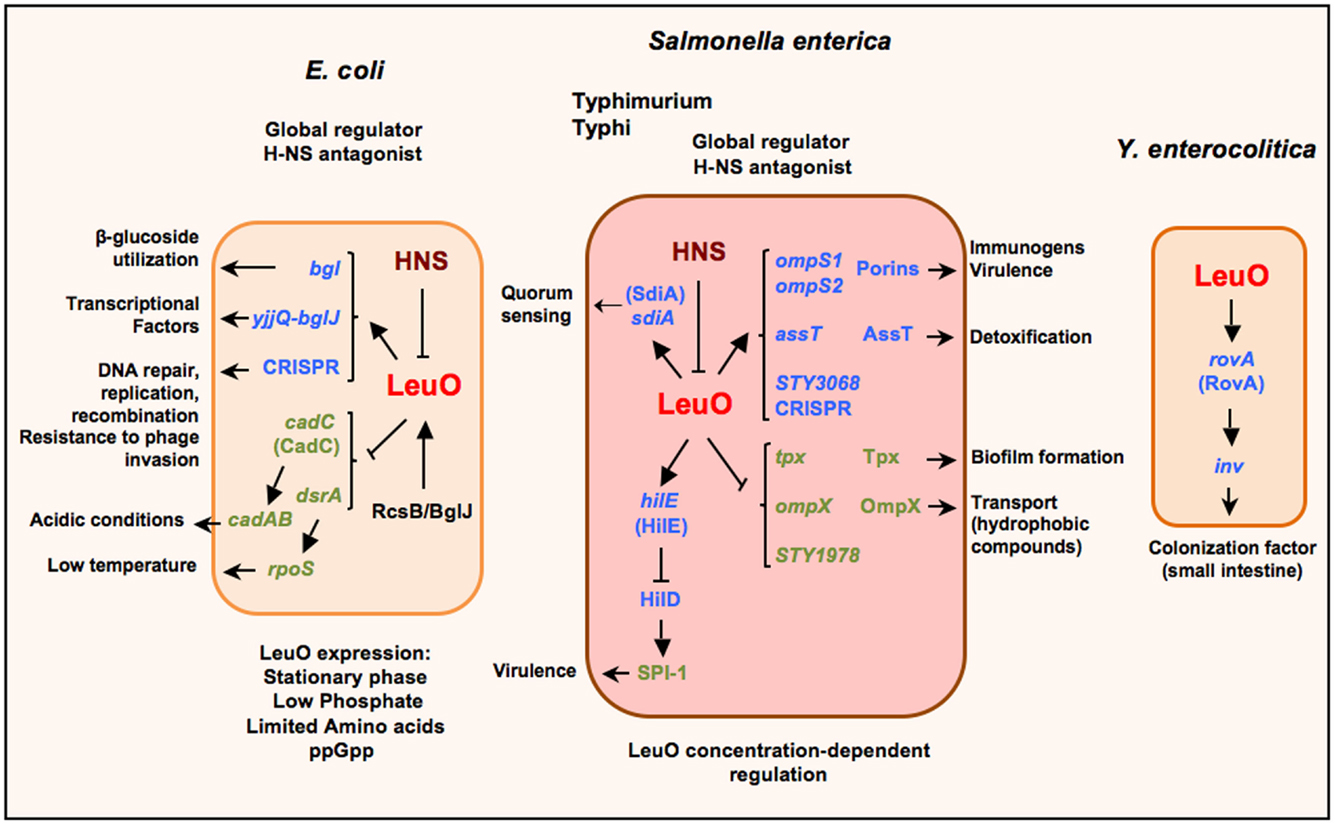
Figure 1. Schematic representation of the LeuO regulon in Escherichia coli, S. enterica serovars Typhimurium and Typhi, and Yersinia enterocolitica. LeuO is a dual regulator that can induce the expression of several genes (arrows) and also is capable of repressing gene expression (lines). When acting as a repressor it has been suggested to function as a backup for H-NS; nevertheless in several cases LeuO acts as a derepressor of gene expression by displacement or prevention of H-NS repression. Recently, LeuO has been denominated as a global antagonist of H-NS in E. coli and in S. enterica serovar Typhimurium. The expression of leuO is repressed by H-NS, although there are some stress conditions when LeuO can be detected in E. coli. Also, in Salmonella it has been described as an interesting case of differential control of transcriptional regulation, which depends on LeuO concentration. Parentheses depict the proteins coded by the indicated genes. Small arrows denote the several functions for the LeuO-regulated genes.
In the classical model of action, LTTRs activate the transcription of a divergent gene and repress their own transcription, independently of the presence of a co-inducer or effector (small signal molecule); although there are exceptions where no co-inducer is required and in most of these cases they act as repressors (37). Therefore, the members of the family have been described as dual regulators (44). Nevertheless, there are examples where the LTTR positively autoregulates its expression; and some LTTRs can have more gene targets that they activate or repress, involved in different cellular process, different from those divergently located with respect to the gene for the regulator (39). Even more, as addressed below, LeuO is an interesting case due to the fact that it can act as derepressor, and has been shown to have complex DNA-binding sites (45, 46).
The first report of the LeuO regulator was by the localization of the leuO gene between the leuABCD and ilvIH operons; upon which it was included in the LysR family due to its amino acid sequence similitude with other members of the family (47, 48). Based on the localization of its gene, LeuO was presumed to be a leuABCD regulator, although Leu auxotrophy was not observed in a leuO mutant strain (49).
Nevertheless, since the first report of LeuO as a transcriptional regulator, it was shown to be involved in the regulation of genes important for bacterial survival in stringent conditions (Figure 1). Thereby, when LeuO was overexpressed in E. coli it was found to repress cadC: this was the result of searching for genes that can complement an H-NS mutant strain, thus providing an insight about a relationship between LeuO and H-NS (50). CadC activates the cadAB operon, an important system expressed under acidic conditions (51). H-NS is a global regulator that acts as a nucleoid protein (52, 53). Later, LeuO was determined to reduce rpoS translation (which encodes S sigma factor) by repression of the small regulatory DsrA-RNA, who positively regulates rpoS translation, mainly at low temperature (54). Both cadC and dsrA are repressed by H-NS (55, 56). Interestingly, in both cases, LeuO indirectly represses the cadAB operon expression and RpoS translation.
According with a LeuO-dual role regulator, it was found to be a positive regulator of bgl and yjjQ-bglJ operons in E. coli. Later, it was demonstrated that LeuO counteracts H-NS repression (49, 57, 58). The bgl operon is involved in the utilization of some β-glucosides as salicin and arbutirin; and the yjjQ-bglJ genes encode for a transcriptional factor belonging to the LuxR family. These operons are repressed by H-NS in a wild type genotype (59) (Figure 1).
In several studies in Salmonella Typhimurium, a model called cis-acting promoter relay mechanism has been described that involves LeuO and DNA local supercoiling in a complex regulatory interplay, in a strain with a mutated promoter of leuABCD (pleuO-500), and a suppressor mutation in topA (60–62). In this complex regulatory mechanism, the Leucine-responsive regulator protein (Lrp) elicits changes in local DNA supercoiling by ilvIH promoter activation, exposing the leuO regulatory region upon which leuO can be transcribed (63–65). Also, there are H-NS binding sites in the regulatory region of leuO: hence the system appears to be repressed by changes in local supercoiling and LeuO prevents a cis-spreading of H-NS enhancing positive autoregulation and permits leuABCD transcription (66–69).
Studies in S. enterica serovar Typhi (Figure 1) have shown that overexpression of LeuO induces the expression of two quiescent genes that encode for the OmpS1 and OmpS2 porins (70, 71). An interesting observation was that the LeuO concentration differentially affects ompS1 and ompS2 expression. The ompS2 gene is expressed at lower concentrations of LeuO, whereas ompS1 is expressed at higher concentrations where ompS2 expression is repressed. Moreover, for the first time, in a detailed study of ompS1 expression, LeuO was shown to exert an antagonist role toward H-NS (71). The relevance of this observation is that such function had not been reported for other LTTRs members until now. Interestingly, members of other transcriptional regulators families such as VirF (AraC/XilS), RovA (SlyA/Hor), and Ler (H-NS/StpA) have been described as antagonists of H-NS mainly on genes involved in virulence (72–74).
In a subsequent study to pursue more targets in Salmonella Typhi, LeuO was found to also positively regulate assT and STY3070; and negatively ompX, tpx and STY1978 (Figure 1). These genes are involved in a variety of cellular functions (75). AssT is a putative arylsulfate sulfotransferase that has been proposed to be involved in detoxification by transforming toxic phenolic derivatives into non-toxic compounds (76). The global regulators H-NS and LeuO regulate the assT-dsbL-dsbI cluster expression negatively and positively, respectively, and this regulation depends on specific growth conditions (77). STY3070 in Salmonella was later determined to be the casC gene of the CRISPR/Cas system; and its repression was found to depend also on Lrp, and its expression induced in minimal media independent of LeuO (78).
The CRISPR/Cas system in Escherichia coli has been involved in DNA repair, replication and recombination and is proposed to confer resistance to phage invasion in bacteria and archaea, thus the suggestion that it is an ancient defense mechanism (79). Interestingly, LeuO was shown to be an antagonist of H-NS in the CRISPR-system in E. coli (80). OmpX is an OMP that is homolog to PagC and Rck and Ail proteins of Salmonella and Yersinia, respectively. When overexpressed, it has been observed to increase sigma E activity; and the lack of ompX increased the tolerance to sodium dodecyl sulfate and antibiotics, thus appearing to affect the transport of hydrophobic compounds across the membrane (81–84). Tpx is a thiol peroxidase that codes for a periplasmic antioxidant enzyme that is induced during the exponential growth phase and during biofilm formation (85). It is important to notice that LeuO down-regulates proteins that are involved in the resistance to different pH conditions (83). Another down-regulated gene was STY1978, which codes for a hypothetical protein without an association to any cellular process until now. In this report, LeuO was denominated as a global regulator and opened the possibility that LeuO could have more targets depending on the growth conditions (75).
In Y. enterocolitica, LeuO was found to positively regulate rovA and, in turn, H-NS also negatively regulates its expression (86) (Figure 1). RovA is a MarA/SlyA type regulator that regulates inv gene expression in response to temperature and growth phase (87).
In E. coli, by SELEX screening, LeuO was found to regulate genes involved in sulfa drug sensitivity and to increase its own expression during transition into stationary phase and after a week of culture, where H-NS concentration decreased (Figure 1). Even more, a global antagonistic interplay between H-NS and LeuO was proposed, acting on some genes involved in stress response, such as cryptic chaperone/usher-type fimbriae. In addition, mutants in leuO and in some fimbrial genes were defective or altered in biofilm formation (88, 89).
In S. enterica serovar Typhimurium, LeuO was reported to increase sdiA expression in low levels (90) (Figure 1). SdiA is proposed to respond to signals produced by other organisms (91, 92) and recently was found to be active in gut in response to AHLs (N-acyl homoserine lactones) a quorum sensing signal produced by other species (93–96).
In a genomic study in S. enterica serovar Typhimurium, using ChIP-chip, the LeuO regulon members were extended to include SPI-1 (Figure 1) and SPI-2 genes. In addition, the differential binding of LeuO and regulation of genes was observed depending on the concentration of LeuO. Another important observation was the intragenic binding; hence opening the possibility that LeuO could act as a negative regulator preventing the progress of transcription or as nucleoid structure protein. The finding of LeuO co-binding at various sites with H-NS and RNA polymerase confirms the notion of the antagonist role of LeuO, although they could likely be acting together to regulate a large number of genes. Moreover, the possible interaction with RNA polymerase and H-NS would suggest another mechanism of LeuO regulation (45, 46).
In this respect, the structural properties of LeuO as an LTTR member have been initially explored: finding that it is active as a tetramer, that the mechanisms for induction and repression of gene expression appear to be different, and that there are relevant interactions between the N- and C-termini (97).
In the Salmonella Typhi and E. coli wild type genomic backgrounds, LeuO expression is silenced by H-NS (unpublished data). Nevertheless, in E. coli and Salmonella Typhimurium, leuO expression has been detected when grown under stress conditions, especially in the stationary phase under nutrient limitation. Nevertheless, leuO is not under the control of rpoS; although its expression requires the presence of ppGpp in stationary phase (54, 63, 98, 99). Interestingly, LeuO was shown to be essential to restore cellular growth, after a 2-h delay in a media lacking isoleucine, valine, and leucine (100).
Also, LeuO expression was detected in a phosphate-restricted media (98); and recently it was shown that the expression of the leuO gene can be activated by the RcsB and BglJ regulators (58, 101)
Even though LeuO is expressed at very low level in standard laboratory conditions, it seems that in vivo it has a role in bacterial survival. In this manner, in a mouse and in a Caenorhabditis elegans model of infection, a S. enterica serovar Typhimurium leuO mutant showed to be attenuated in virulence. Also, in Vibrio cholera, biofilm formation was reduced in a deleted leuO strain (102–104).
Virulence attenuation in a murine model was reported for the ompC ompF double mutant (105). In addition, it has been observed that the OmpC and OmpF porins induced long-term antibody response with bactericidal capacity and conferred protection against challenge with Salmonella Typhi (35, 36). Nevertheless, these major porins are expressed at very high levels in standard laboratory conditions. In addition, strains lacking ompS1 and ompS2 are attenuated for virulence, suggesting that besides lacking the LeuO regulator the absence of OmpS1 and OmpS2 porins affected bacterial survival (103). Virulence attenuation of mutated strains in leuO and ompS1 and ompS2 quiescent genes offers evidence that they are expressed in vivo. Even though the specific role of these porins in Salmonella virulence is not clear, it has been shown that the major porins are passive diffusion channels of solutes, nutrients and toxins through the outer bacterial membrane that might allow bacteria to grow in different environments and to be resistant to drugs (106).
Recently it was found that OmpS1 and OmpS2 induced a strong immune response in the mouse, and a single dose conferred a significant protection against Salmonella Typhi. The immunostimulatory properties of OmpS1 and OmpS2 porins further reinforce the notion that they could be expressed following host infection. These studies are relevant because they open the possibility of using these porins as antigens for the development of vaccines against typhoid fever and other non-typhoidal salmonellosis (107).
Moreover, in a recent report it was shown that the activation of leuO transcription in S. enterica serovar Typhimurium represses expression of pathogenicity island 1 (SPI-1) and inhibits invasion of epithelial cells (108). Two different modes of action were found: the major one that involves the induction of hilE transcription by LeuO (Figure 1) and another one that was HilE-independent. HilE is a regulator encoded outside SPI-1 that represses hilD expression. HilD is one of the transcriptional factors encoded in SPI-1 that positively controls the expression of other genes in the island (109, 110). It has been suggested that LeuO repression of SPI-1 genes may occur under growth conditions where H-NS, for unknown reasons, has failed to perform such repression.
The possibility of LeuO acting as a backup for H-NS has two implications: one is that it could allow Salmonella to confront the hostile free-living conditions where SPI-1 gene expression has a high cost in bacterial growth; and two, it might ensure the specific, sequential, and appropriate level of SPI-1 gene expression in the intestine (111, 112). Due to the fact that H-NS in Salmonella is considered as a genome sentinel that silences horizontally acquired genes (113–115), LeuO could be acting as a backup regulator for H-NS, highlighting the subtleties and contrasts of the LeuO mode of action. Thus, the proposed role of LeuO as an activator or as a repressor depending on its concentration could explain this differential gene regulation.
LeuO is an example of a global regulator whose level of expression is an important issue, since this has an effect on its many regulated genes that are involved in a variety of cellular functions, such as virulence and bacterial survival. The levels of expression could thus have spatial and temporal consequences as well. In addition, knowledge of LeuO-regulated genes has been important in the study of the immune response induced by Salmonella, such as that elicited by the quiescent porins, which are protein components of the outer membrane. This has opened the possibility for the development of typhoid fever vaccines and perhaps as adjuvants for others vaccines.
It is intriguing that conditions known at present for LeuO expression are extreme and that in many studies it has to be overexpressed to analyze its function. Furthermore, no co-inducer of LeuO is known until now. These are some of the subjects that pose challenges for the future.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Edmundo Calva was supported by grants from CONACyT, Mexico (No. 82383 and 179946).
1. Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review. Immunol Cell Biol (2007) 85(2):112–8. doi:10.1038/sj.icb.7100007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Yue M, Schifferli DM. Allelic variation in Salmonella: an underappreciated driver of adaptation and virulence. Front Microbiol (2014) 4:419. doi:10.3389/fmicb.2013.00419
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. Mittrucker HW, Kaufmann SH. Immune response to infection with Salmonella typhimurium in mice. J Leukoc Biol (2000) 67(4):457–63.
4. Chen HM, Wang Y, Su LH, Chiu CH. Nontyphoid Salmonella infection: microbiology, clinical features, and antimicrobial therapy. Pediatr Neonatol (2013) 54(3):147–52. doi:10.1016/j.pedneo.2013.01.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. de Jong HK, Parry CM, van der Poll T, Wiersinga WJ. Host-pathogen interaction in invasive salmonellosis. PLoS Pathog (2012) 8(10):e1002933. doi:10.1371/journal.ppat.1002933
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Galan JE. Salmonella interactions with host cells: type III secretion at work. Annu Rev Cell Dev Biol (2001) 17:53–86. doi:10.1146/annurev.cellbio.17.1.53
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Waterman SR, Holden DW. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol (2003) 5(8):501–11. doi:10.1046/j.1462-5822.2003.00294.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Hannemann S, Gao B, Galán JE. Salmonella modulation of host cell gene expression promotes its intracellular growth. PLoS Pathog (2013) 9(10):e1003668. doi:10.1371/journal.ppat.1003668
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Patel JC, Galán JE. Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J Cell Biol (2006) 175(3):453–63. doi:10.1083/jcb.200605144
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Patel JC, Galán JE. Manipulation of the host actin cytoskeleton by Salmonella – all in the name of entry. Curr Opin Microbiol (2005) 8(1):10–5. doi:10.1016/j.mib.2004.09.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Hobbie S, Chen LM, Davis RJ, Galán JE. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J Immunol (1997) 159(11):5550–9.
12. Bruno VM, Hannemann S, Lara-Tejero M, Flavell RA, Kleinstein SH, Galán JE. Salmonella typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog (2009) 5(8):e1000538. doi:10.1371/journal.ppat.1000538
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, et al. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity (2014) 40(2):262–73. doi:10.1016/j.immuni.2014.01.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol (2005) 3(4):281–94. doi:10.1038/nrmicro1126
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Lee J, Mo JH, Shen C, Rucker AN, Raz E. Toll-like receptor signaling in intestinal epithelial cells contributes to colonic homoeostasis. Curr Opin Gastroenterol (2007) 23(1):27–31. doi:10.1097/MOG.0b013e3280118272
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Shibolet O, Podolsky DK. TLRs in the Gut. IV. Negative regulation of toll-like receptors and intestinal homeostasis: addition by subtraction. Am J Physiol Gastrointest Liver Physiol (2007) 292(6):G1469–73. doi:10.1152/ajpgi.00531.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Lang T, Mansell A. The negative regulation of toll-like receptor and associated pathways. Immunol Cell Biol (2007) 85(6):425–34. doi:10.1038/sj.icb.7100094
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Fink SL, Cookson BT. Pyroptosis and host cell death responses during Salmonella infection. Cell Microbiol (2007) 9(11):2562–70. doi:10.1111/j.1462-5822.2007.01036.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Lindgren SW, Stojiljkovic I, Heffron F. Macrophage killing is an essential virulence mechanism of Salmonella typhimurium. Proc Natl Acad Sci U S A (1996) 93(9):4197–201. doi:10.1073/pnas.93.9.4197
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Wijburg OL, Simmons CP, van Rooijen N, Strugnell RA. Dual role for macrophages in vivo in pathogenesis and control of murine Salmonella enterica serovar Typhimurium infections. Eur J Immunol (2000) 30(3):944–53. doi:10.1002/1521-4141(200003)30:3<944::AID-IMMU944>3.3.CO;2-T
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Monack DM, Bouley DM, Falkow S. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J Exp Med (2004) 199(2):231–41. doi:10.1084/jem.20031319
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. McSorley SJ. Immunity to intestinal pathogens: lessons learned from Salmonella. Immunol Rev (2014) 260(1):168–82. doi:10.1111/imr.12184
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Mittrucker HW, Kohler A, Kaufmann SH. Characterization of the murine T-lymphocyte response to Salmonella enterica serovar Typhimurium infection. Infect Immun (2002) 70(1):199–203. doi:10.1128/IAI.70.1.199-203.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Srinivasan A, Foley J, McSorley SJ. Massive number of antigen-specific CD4 T cells during vaccination with live attenuated Salmonella causes interclonal competition. J Immunol (2004) 172(11):6884–93. doi:10.4049/jimmunol.172.11.6884
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Nauciel C. Role of CD4+ T cells and T-independent mechanisms in acquired resistance to Salmonella typhimurium infection. J Immunol (1990) 145(4):1265–9.
26. Mastroeni P, Villarreal-Ramos B, Hormaeche CE. Role of T cells, TNF alpha and IFN gamma in recall of immunity to oral challenge with virulent Salmonellae in mice vaccinated with live attenuated aro-Salmonella vaccines. Microb Pathog (1992) 13(6):477–91. doi:10.1016/0882-4010(92)90014-F
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. O’Donnell H, Pham OH, Li LX, Atif SM, Lee SJ, Ravesloot MM, et al. Toll-like receptor and inflammasome signals converge to amplify the innate bactericidal capacity of T helper 1 cells. Immunity (2014) 40(2):213–24. doi:10.1016/j.immuni.2013.12.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Mastroeni P, Villarreal-Ramos B, Hormaeche CE. Adoptive transfer of immunity to oral challenge with virulent Salmonellae in innately susceptible BALB/c mice requires both immune serum and T cells. Infect Immun (1993) 61(9):3981–4.
29. Cookson BT, Bevan MJ. Identification of a natural T-cell epitope presented by Salmonella-infected macrophages and recognized by T cells from orally immunized mice. J Immunol (1997) 158(9):4310–9.
30. Bergman MA, Cummings LA, Alaniz RC, Mayeda L, Fellnerova I, Cookson BT. CD4+-T-cell responses generated during murine Salmonella enterica serovar Typhimurium infection are directed towards multiple epitopes within the natural antigen FliC. Infect Immun (2005) 73(11):7226–35. doi:10.1128/IAI.73.11.7226-7235.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Salazar-Gonzalez RM, McSorley SJ. Salmonella flagellin, a microbial target of the innate and adaptive immune system. Immunol Lett (2005) 101(2):117–22. doi:10.1016/j.imlet.2005.05.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. McSorley SJ, Cookson BT, Jenkins MK. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J Immunol (2000) 164(2):986–93. doi:10.4049/jimmunol.164.2.986
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Strindelius L, Degling Wikingsson L, Sjoholm I. Extracellular antigens from Salmonella enteritidis induce effective immune response in mice after oral vaccination. Infect Immun (2002) 70(3):1434–42. doi:10.1128/IAI.70.3.1434-1442.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Blanco F, Isibasi A, Raul Gonzalez C, Ortiz V, Paniagua J, Arreguin C, et al. Human cell mediated immunity to porins from Salmonella typhi. Scand J Infect Dis (1993) 25(1):73–80. doi:10.1080/00365549309169673
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Isibasi A, Ortiz-Navarrete V, Paniagua J, Pelayo R, Gonzalez CR, García JA, et al. Active protection of mice against Salmonella typhi by immunization with strain-specific porins. Vaccine (1992) 10(12):811–3. doi:10.1016/0264-410X(92)90041-H
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Secundino I, López-Macias C, Cervantes-Barragán L, Gil-Cruz C, Ríos-Sarabia N, Pastelín-Palacios R, et al. Salmonella porins induce a sustained, lifelong specific bactericidal antibody memory response. Immunology (2006) 117(1):59–70. doi:10.1111/j.1365-2567.2005.02263.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Schell MA. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol (1993) 47:597–626. doi:10.1146/annurev.mi.47.100193.003121
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Tropel D, van der Meer JR. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol Mol Biol Rev (2004) 68(3):474–500. doi:10.1128/MMBR.68.3.474-500.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Maddocks SE, Oyston PC. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology (2008) 154(Pt 12):3609–23. doi:10.1099/mic.0.2008/022772-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Pareja E, Pareja-Tobes P, Manrique M, Pareja-Tobes E, Bonal J, Tobes R. Extratrain: a database of extragenic regions and transcriptional information in prokaryotic organisms. BMC Microbiol (2006) 6:29. doi:10.1186/1471-2180-6-29
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Muraoka S, Okumura R, Ogawa N, Nonaka T, Miyashita K, Senda T. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J Mol Biol (2003) 328(3):555–66. doi:10.1016/S0022-2836(03)00312-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Sainsbury S, Lane LA, Ren J, Gilbert RJ, Saunders NJ, Robinson CV, et al. The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators. Nucleic Acids Res (2009) 37(14):4545–58. doi:10.1093/nar/gkp445
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Zhou X, Lou Z, Fu S, Yang A, Shen H, Li Z, et al. Crystal structure of ArgP from Mycobacterium tuberculosis confirms two distinct conformations of full-length LysR transcriptional regulators and reveals its function in DNA binding and transcriptional regulation. J Mol Biol (2010) 396(4):1012–24. doi:10.1016/j.jmb.2009.12.033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Perez-Rueda E, Collado-Vides J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res (2000) 28(8):1838–47. doi:10.1093/nar/28.8.1838
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Dillon SC, Espinosa E, Hokamp K, Ussery DW, Casadesús J, Dorman CJ. LeuO is a global regulator of gene expression in Salmonella enterica serovar Typhimurium. Mol Microbiol (2012) 85(6):1072–89. doi:10.1111/j.1365-2958.2012.08162.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Hernández-Lucas I, Calva E. The coming of age of the LeuO regulator. Mol Microbiol (2012) 85(6):1026–8. doi:10.1111/j.1365-2958.2012.08175.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Hertzberg KM, Gemmill R, Jones J, Calvo JM. Cloning of an EcoRI-generated fragment of the leucine operon of Salmonella typhimurium. Gene (1980) 8(2):135–52. doi:10.1016/0378-1119(80)90033-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
48. Henikoff S, Haughn GW, Calvo JM, Wallace JC. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A (1988) 85(18):6602–6. doi:10.1073/pnas.85.18.6602
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Ueguchi C, Ohta T, Seto C, Suzuki T, Mizuno T. The LeuO gene product has a latent ability to relieve bgl silencing in Escherichia coli. J Bacteriol (1998) 180(1):190–3.
50. Shi X, Bennett GN. Effects of multicopy LeuO on the expression of the acid-inducible lysine decarboxylase gene in Escherichia coli. J Bacteriol (1995) 177(3):810–4.
51. Soksawatmaekhin W, Kuraishi A, Sakata K, Kashiwagi K, Igarashi K. Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol Microbiol (2004) 51(5):1401–12. doi:10.1046/j.1365-2958.2003.03913.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Rimsky S, Spassky A. Sequence determinants for H1 binding on Escherichia coli lac and gal promoters. Biochemistry (1990) 29(15):3765–71. doi:10.1021/bi00467a024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Durrenberger M, La Teana A, Citro G, Venanzi F, Gualerzi CO, Pon CL. Escherichia coli DNA-binding protein H-NS is localized in the nucleoid. Res Microbiol (1991) 142(4):373–80. doi:10.1016/0923-2508(91)90106-K
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Klauck E, Bohringer J, Hengge-Aronis R. The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol Microbiol (1997) 25(3):559–69. doi:10.1046/j.1365-2958.1997.4911852.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Shi X, Waasdorp BC, Bennett GN. Modulation of acid-induced amino acid decarboxylase gene expression by HNS in Escherichia coli. J Bacteriol (1993) 175(4):1182–6.
56. Yamashino T, Ueguchi C, Mizuno T. Quantitative control of the stationary phase-specific sigma factor, sigma S, in Escherichia coli: involvement of the nucleoid protein H-NS. EMBO J (1995) 14(3):594–602.
57. Bartowsky E, Normark S. Purification and mutant analysis of Citrobacter freundii AmpR, the regulator for chromosomal AmpC beta-lactamase. Mol Microbiol (1991) 5(7):1715–25. doi:10.1111/j.1365-2958.1991.tb01920.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Stratmann T, Madhusudan S, Schnetz K. Regulation of the yjjQ-bglJ operon, encoding LuxR-type transcription factors, and the divergent yjjP gene by H-NS and LeuO. J Bacteriol (2008) 190(3):926–35. doi:10.1128/JB.01447-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Bertin P, Lejeune P, Laurent-Winter C, Danchin A. Mutations in bglY, the structural gene for the DNA-binding protein H1, affect expression of several Escherichia coli genes. Biochimie (1990) 72(12):889–91. doi:10.1016/0300-9084(90)90008-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
60. Lilley DM, Higgins CF. Local DNA topology and gene expression: the case of the leu-500 promoter. Mol Microbiol (1991) 5(4):779–83. doi:10.1111/j.1365-2958.1991.tb00749.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Chen D, Bowater R, Dorman CJ, Lilley DM. Activity of a plasmid-borne leu-500 promoter depends on the transcription and translation of an adjacent gene. Proc Natl Acad Sci U S A (1992) 89(18):8784–8. doi:10.1073/pnas.89.18.8784
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Wu HY, Tan J, Fang M. Long-range interaction between two promoters: activation of the leu-500 promoter by a distant upstream promoter. Cell (1995) 82(3):445–51. doi:10.1016/0092-8674(95)90433-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Fang M, Wu HY. Suppression of leu-500 mutation in topA+ Salmonella typhimurium strains. The promoter relay at work. J Biol Chem (1998) 273(45):29929–34. doi:10.1074/jbc.273.45.29929
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Fang M, Wu HY. A promoter relay mechanism for sequential gene activation. J Bacteriol (1998) 180(3):626–33.
65. Wu HY, Fang M. DNA supercoiling and transcription control: a model from the study of suppression of the leu-500 mutation in Salmonella typhimurium topA-strains. Prog Nucleic Acid Res Mol Biol (2003) 73:43–68. doi:10.1016/S0079-6603(03)01002-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Chen CC, Fang M, Majumder A, Wu HY. A 72-base pair AT-rich DNA sequence element functions as a bacterial gene silencer. J Biol Chem (2001) 276(12):9478–85. doi:10.1074/jbc.M010501200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
67. Chen CC, Ghole M, Majumder A, Wang Z, Chandana S, Wu HY. LeuO-mediated transcriptional derepression. J Biol Chem (2003) 278(39):38094–103. doi:10.1074/jbc.M300461200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Chen CC, Wu HY. LeuO protein delimits the transcriptionally active and repressive domains on the bacterial chromosome. J Biol Chem (2005) 280(15):15111–21. doi:10.1074/jbc.M414544200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Chen CC, Chou MY, Huang CH, Majumder A, Wu HY. A cis-spreading nucleoprotein filament is responsible for the gene silencing activity found in the promoter relay mechanism. J Biol Chem (2005) 280(6):5101–12. doi:10.1074/jbc.M411840200 Epub 2004/12/08.,
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
70. Fernández-Mora M, Puente JL, Calva E. OmpR and LeuO positively regulate the Salmonella enterica serovar Typhi ompS2 porin gene. J Bacteriol (2004) 186(10):2909–20. doi:10.1128/JB.186.10.2909-2920.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
71. De la Cruz MA, Fernández-Mora M, Guadarrama C, Flores-Valdez MA, Bustamante VH, Vázquez A, et al. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol Microbiol (2007) 66(3):727–43. doi:10.1111/j.1365-2958.2007.05958.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by VirF and repression by H-NS. J Bacteriol (1993) 175(19):6142–9.
73. Bustamante VH, Santana FJ, Calva E, Puente JL. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol Microbiol (2001) 39(3):664–78. doi:10.1046/j.1365-2958.2001.02209.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. Heroven AK, Nagel G, Tran HJ, Parr S, Dersch P. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol Microbiol (2004) 53(3):871–88. doi:10.1111/j.1365-2958.2004.04162.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
75. Hernández-Lucas I, Gallego-Hernández AL, Encarnación S, Fernández-Mora M, Martínez-Batallar AG, Salgado H, et al. The LysR-type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar Typhi. J Bacteriol (2008) 190(5):1658–70. doi:10.1128/JB.01649-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
76. Kang JW, Kwon AR, Kim DH, Choi EC. Cloning and sequencing of the astA gene encoding arylsulfate sulfotransferase from Salmonella typhimurium. Biol Pharm Bull (2001) 24(5):570–4. doi:10.1248/bpb.24.570
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Gallego-Hernández AL, Hernández-Lucas I, De la Cruz MA, Olvera L, Morett E, Medina-Aparicio L, et al. Transcriptional regulation of the assT-dsbL-dsbI gene cluster in Salmonella enterica serovar Typhi IMSS-1 depends on LeuO, H-NS, and specific growth conditions. J Bacteriol (2012) 194(9):2254–64. doi:10.1128/JB.06164-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
78. Medina-Aparicio L, Rebollar-Flores JE, Gallego-Hernández AL, Vázquez A, Olvera L, Gutiérrez-Rios RM, et al. The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J Bacteriol (2011) 193(10):2396–407. doi:10.1128/JB.01480-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
79. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science (2007) 315(5819):1709–12. doi:10.1126/science.1138140
80. Westra ER, Pul U, Heidrich N, Jore MM, Lundgren M, Stratmann T, et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol Microbiol (2010) 77(6):1380–93. doi:10.1111/j.1365-2958.2010.07315.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
81. Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev (1993) 7(12B):2618–28. doi:10.1101/gad.7.12b.2618
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
82. Mecsas J, Welch R, Erickson JW, Gross CA. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J Bacteriol (1995) 177(3):799–804.
83. Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J Bacteriol (2002) 184(15):4246–58. doi:10.1128/JB.184.15.4246-4258.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
84. Otto K, Hermansson M. Inactivation of ompX causes increased interactions of type 1 fimbriated Escherichia coli with abiotic surfaces. J Bacteriol (2004) 186(1):226–34. doi:10.1128/JB.186.1.226-234.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Kim YH, Lee Y, Kim S, Yeom J, Yeom S, Seok Kim B, et al. The role of periplasmic antioxidant enzymes (superoxide dismutase and thiol peroxidase) of the Shiga toxin-producing Escherichia coli O157:H7 in the formation of biofilms. Proteomics (2006) 6(23):6181–93. doi:10.1002/pmic.200600320
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Lawrenz MB, Miller VL. Comparative analysis of the regulation of rovA from the pathogenic yersiniae. J Bacteriol (2007) 189(16):5963–75. doi:10.1128/JB.00528-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
87. Revell PA, Miller VL. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol Microbiol (2000) 35(3):677–85. doi:10.1046/j.1365-2958.2000.01740.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Shimada T, Yamamoto K, Ishihama A. Involvement of the leucine response transcription factor LeuO in regulation of the genes for sulfa drug efflux. J Bacteriol (2009) 191(14):4562–71. doi:10.1128/JB.00108-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
89. Shimada T, Bridier A, Briandet R, Ishihama A. Novel roles of LeuO in transcription regulation of E. coli genome: antagonistic interplay with the universal silencer H-NS. Mol Microbiol (2011) 82(2):378–97. doi:10.1111/j.1365-2958.2011.07818.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
90. Turnbull AL, Kim W, Surette MG. Transcriptional regulation of sdiA by cAMP-receptor protein, LeuO, and environmental signals in Salmonella enterica serovar Typhimurium. Can J Microbiol (2012) 58(1):10–22. doi:10.1139/W11-101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J Bacteriol (2001) 183(19):5733–42. doi:10.1128/JB.183.19.5733-5742.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
92. Smith JN, Ahmer BM. Detection of other microbial species by Salmonella: expression of the SdiA regulon. J Bacteriol (2003) 185(4):1357–66. doi:10.1128/JB.185.4.1357-1366.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. Ahmer BM, Smith JN, Dyszel JL, Lindsay A. Methods in cell-to-cell signaling in Salmonella. Methods Mol Biol (2007) 394:307–22. doi:10.1007/978-1-59745-512-1_15
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
94. Gorshkov V, Petrova O, Gogoleva N, Gogolev Y. Cell-to-cell communication in the populations of enterobacterium Erwinia carotovora ssp. atroseptica SCRI1043 during adaptation to stress conditions. FEMS Immunol Med Microbiol (2010) 59(3):378–85. doi:10.1111/j.1574-695X.2010.00684.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
95. Mei GY, Yan XX, Turak A, Luo ZQ, Zhang LQ. AidH, an alpha/beta-hydrolase fold family member from an Ochrobactrum sp. strain, is a novel N-acylhomoserine lactonase. Appl Environ Microbiol (2010) 76(15):4933–42. doi:10.1128/AEM.00477-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. Weeks JN, Galindo CL, Drake KL, Adams GL, Garner HR, Ficht TA. Brucella melitensis VjbR and C12-HSL regulons: contributions of the N-dodecanoyl homoserine lactone signaling molecule and LuxR homologue VjbR to gene expression. BMC Microbiol (2010) 10:167. doi:10.1186/1471-2180-10-167
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
97. Guadarrama C, Medrano-López A, Oropeza R, Hernández-Lucas I, Calva E. The Salmonella enterica Serovar Typhi LeuO global regulator forms tetramers: residues involved in oligomerization, DNA binding and transcriptional regulation. J Bacteriol (2014) 196(12):2143–54. doi:10.1128/JB.01484-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
98. VanBogelen RA, Olson ER, Wanner BL, Neidhardt FC. Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli. J Bacteriol (1996) 178(15):4344–66.
99. Fang M, Majumder A, Tsai KJ, Wu HY. ppGpp-dependent leuO expression in bacteria under stress. Biochem Biophys Res Commun (2000) 276(1):64–70. doi:10.1006/bbrc.2000.3440
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
100. Majumder A, Fang M, Tsai KJ, Ueguchi C, Mizuno T, Wu HY. LeuO expression in response to starvation for branched-chain amino acids. J Biol Chem (2001) 276(22):19046–51. doi:10.1074/jbc.M100945200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
101. Stratmann T, Pul U, Wurm R, Wagner R, Schnetz K. RcsB-BglJ activates the Escherichia coli leuO gene, encoding an H-NS antagonist and pleiotropic regulator of virulence determinants. Mol Microbiol (2012) 83(6):1109–23. doi:10.1111/j.1365-2958.2012.07993.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
102. Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog (2006) 2(2):e11. doi:10.1371/journal.ppat.0020011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
103. Rodríguez-Morales O, Fernández-Mora M, Hernández-Lucas I, Vázquez A, Puente JL, Calva E. Salmonella enterica serovar Typhimurium ompS1 and ompS2 mutants are attenuated for virulence in mice. Infect Immun (2006) 74(2):1398–402. doi:10.1128/IAI.74.2.1398-1402.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
104. Moorthy S, Watnick PI. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol Microbiol (2005) 57(6):1623–35. doi:10.1111/j.1365-2958.2005.04797.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
105. Chatfield SN, Dorman CJ, Hayward C, Dougan G. Role of OmpR-dependent genes in Salmonella typhimurium virulence: mutants deficient in both ompC and ompF are attenuated in vivo. Infect Immun (1991) 59(1):449–52.
106. Gil F, Ipinza F, Fuentes J, Fumeron R, Villarreal JM, Aspee A, et al. The ompW (porin) gene mediates methyl viologen (paraquat) efflux in Salmonella enterica serovar Typhimurium. Res Microbiol (2007) 158(6):529–36. doi:10.1016/j.resmic.2007.05.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
107. Moreno-Eutimio MA, Tenorio-Calvo A, Pastelín-Palacios R, Pérez-Shibayama C, Gil-Cruz C, López-Santiago R, et al. Salmonella Typhi OmpS1 and OmpS2 porins are potent protective immunogens with adjuvant properties. Immunology (2013) 139(4):459–71. doi:10.1111/imm.12093
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
108. Espinosa E, Casadesús J. Regulation of Salmonella enterica pathogenicity island 1 (SPI-1) by the LysR-type regulator LeuO. Mol Microbiol (2014) 91(6):1057–69. doi:10.1111/mmi.12500
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
109. Fahlen TF, Mathur N, Jones BD. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol Med Microbiol (2000) 28(1):25–35. doi:10.1111/j.1574-695X.2000.tb01453.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
110. Lostroh CP, Lee CA. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect (2001) 3(14–15):1281–91. doi:10.1016/S1286-4579(01)01488-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
111. Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, et al. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog (2011) 7(7):e1002143. doi:10.1371/journal.ppat.1002143
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
112. Bustamante VH, Calva E. LeuO, a dormant sentinel for SPI-1? Mol Microbiol (2014) 91(6):1054–6. doi:10.1111/mmi.12514
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
113. Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog (2006) 2(8):e81. doi:10.1371/journal.ppat.0020081
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
114. Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science (2006) 313(5784):236–8. doi:10.1126/science.1128794
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: LeuO, Typhi, OmpS1, OmpS2, H-NS, porins
Citation: Guadarrama C, Villaseñor T and Calva E (2014) The subtleties and contrasts of the LeuO regulator in Salmonella Typhi: implications in the immune response. Front. Immunol. 5:581. doi: 10.3389/fimmu.2014.00581
Received: 17 July 2014; Accepted: 30 October 2014;
Published online: 12 December 2014.
Edited by:
Adam F. Cunningham, University of Birmingham, UKReviewed by:
Xun Suo, China Agricultural University, ChinaCopyright: © 2014 Guadarrama, Villaseñor and Calva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edmundo Calva, Instituto de Biotecnología, UNAM, Av. Universidad 2001, Cuernavaca, Morelos 62210, Mexico e-mail:ZWNhbHZhQGlidC51bmFtLm14
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.