- 1Division of Immunology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, USA
- 2Department of Microbiology and Immunology, Harvard Medical School, Boston, MA, USA
Whereas the SLAMF-associated protein (SAP) is involved in differentiation of T follicular helper (Tfh) cells and antibody responses, the precise requirements of SLAMF receptors in humoral immune responses are incompletely understood. By analyzing mice with targeted disruptions of the Slamf1, Slamf5, and Slamf6 genes, we found that both T-dependent and T-independent antibody responses were twofold higher compared to those in single knockout mice. These data suggest a suppressive synergy of SLAMF1, SLAMF5, and SLAMF6 in humoral immunity, which contrasts the decreased antibody responses resulting from a defective GC reaction in the absence of the adapter SAP. In adoptive co-transfer assays, both [Slamf1 + 5 + 6]−/− B and T cells were capable of inducing enhanced antibody responses, but more pronounced enhancement was observed after adoptive transfer of [Slamf1 + 5 + 6]−/− B cells compared to that of [Slamf1 + 5 + 6]−/− T cells. In support of [Slamf1 + 5 + 6]−/− B cell intrinsic activity, [Slamf1 + 5 + 6]−/− mice also mounted significantly higher antibody responses to T-independent type 2 antigen. Furthermore, treatment of mice with anti-SLAMF6 monoclonal antibody results in severe inhibition of the development of Tfh cells and GC B cells, confirming a suppressive effect of SLAMF6. Taken together, these results establish SLAMF1, SLAMF5, and SLAMF6 as important negative regulators of humoral immune response, consistent with the notion that SLAM family receptors have dual functions in immune responses.
Introduction
The humoral immune response is crucial for protecting individuals from many infections and eliminating foreign substances. Antibody responses can be induced in a T cell dependent or T cell independent manner. In T-independent immunity, the antibody response occurs directly after B cell activation in T cell deficient mice. In contrast with T-independent responses, T-dependent responses require T cell help for B cell activation and maturation. In the presence of T cell help, B cells undergo robust proliferation and somatic hypermutation of their variable region genes and differentiate into high affinity memory B cells and long-lived plasma cells in the germinal centers (1, 2). Although several subsets of CD4+ T helper cells may be implicated in T-dependent humoral responses, it becomes clear that the follicular helper CD4+ T cell subset [T follicular helper (Tfh)] is a major B cell help provider (3–6). Tfh cells exhibit a phenotype distinct from that of other effector CD4+ T helper cells, as they express the transcription factor B cell lymphoma 6 (Bcl6) that is necessary for the development of Tfh cells and inhibits expression of genes critical for development of other T helper cells (4, 7–9). Although Bcl6 expression by pre-Tfh cells is required, it is not sufficient in vivo for full polarization of Tfh cells. In fact, multiple molecules have been shown to be involved in the differentiation of Tfh cells (3, 4, 6). In addition, Tfh development is highly dependent on B cell responses, as Tfh cells are not found in B cell deficient mice (7, 10, 11). These findings indicate that, through their interaction, GC B cells and Tfh cells reciprocally provide each other with signaling for survival, proliferation, and differentiation.
The signaling lymphocytic activation molecule family (SLAMF) includes nine structurally related Ig-like proteins that are differentially expressed on the surface of hematopoietic cells (12). SLAMF receptors have been shown to function as co-stimulatory molecules and to modulate the activation and differentiation of a wide array of immune cell types involved in both innate and adaptive immune responses (12–14). While most SLAMF receptors serve as self-ligands, SLAMF2 and SLAMF4 interact with each other. Six SLAMF receptors (SLAMF1, SLAMF3, SLAMF4, SLAMF5, SLAMF6, and SLAMF7) carry one or more copies of an immunoreceptor tyrosine-based switch motif (ITSM) in their cytoplasmic tails. This signaling switch motif can recruit SH2 domain-containing signaling molecules such as SLAM-associated protein (SAP) (15). SAP is a cytoplasmic adapter molecule with a single Src homology 2 domain and a small carboxy-terminal region. The SAP family consists of three members: SAP expressing T, NK, and NKT cells, and EAT-2A and EAT-2B (murine) expressing NK cells and APC (12, 16). There is accumulating evidence that SAP and EAT-2 can function as signaling adaptors that link SLAMF receptors to active signaling molecules such as the Src family protein tyrosine kinases Fyn and PI3K (15, 17–21). SAP and EAT-2 have also been shown to act as blockers to outcompete SH2 domain-containing inhibitory molecules SHP1, SHP2, and SHIP1 (22–28).
Deficiencies in the gene that encodes SAP (SH2D1A) result in a primary immunodeficiency called X-linked lymphoproliferative disease (XLP) (29–31). Patients with XLP suffer from fatal infectious mononucleosis, malignant B cell lymphomas, and dysgammaglobulinemia. Defects in humoral responses and lack of germinal center formation are observed in XLP patients and in virally infected or immunized SAP-deficient mice (32–36). Considerable evidence indicates that the humoral immune response defect in XLP patients and SAP−/− mice stems from a defect in CD4+ helper cells because T-dependent antigen responses are defective and are restored after reconstitution with WT CD4+ T cells, but not WT B cells (32, 34, 35). However, the role of SAP in T-dependent humoral responses remains unclear. The SLAM/SAP/FynT axis regulates IL-4 producing Th2 differentiation, as demonstrated by the observation that Slamf1−/− and SAP−/− mice have defective Th2 cytokine secretions (37–39). IL-4 is known to stimulate B cell antibody responses and Ig class switching, but the R78A mutant SAP mice can mount normal T-dependent antibody responses even though this mutant SAP molecule loses Fyn binding motif R78 (40). Recently, Qi and co-workers elegantly showed that SAP-deficient CD4+ T cells cannot form lasting mobile conjugate pairs with cognate B cells in the germinal center while the interaction between SAP-deficient T cells and DC is not affected (41). Since a sustained T-B conjugate allows their comprehensive activation and subsequent differentiation to Tfh cells and GC B cells, unstable T-B cell conjugates may contribute to humoral immune deficiency in SAP-deficient mice and XLP patients.
Compared to severe immunodeficiencies in SAP−/− mice, single ablation of SLAMF receptors causes a mild phenotype (40, 42–44). When mice deficient in SLAMF1 (the prototypic member of the SLAMF receptors) were infected with LCMV, Tfh cell differentiation, germinal center development, and the acute or long-term anti-viral antibody responses were comparable between LCMV-infected WT and Slamf1−/− mice (40). Similar to Slamf1−/− mice, Slamf3−/−, Slamf5−/−, and Slamf6−/− mice showed no defects in response to LCMV (28, 42). This suggests that functional redundancy exists among the SLAMF receptors, which has been confirmed in NKT cell development in pseudo Slamf[1 + 6]-deficient bone marrow reconstituting mice (45) and recent Slamf[1 + 6]−/− mice (46). For more than a decade, investigating the role of SLAMF receptors in SAP-mediated signaling has been difficult due to an inability to generate double or multiple SLAMF receptor deficient mice, as the receptors are closely located on the same chromosome (12). To define roles of SLAMF receptors as well as how they interact with one another in humoral immune responses, we generated Slamf[1 + 6] double knockout and Slamf[1 + 5 + 6] triple knockout mice using a two-time gene targeting technique and Cre/LoxP system. Surprisingly, we found that the combined absence of SLAMF1, SLAMF5, and SLAMF6 results in higher antibody production in response to both T-dependent and T-independent antigens. In addition, the administration of anti-SLAMF6 monoclonal antibody also impairs humoral immune responses in vivo. These observations suggest that SLAMF1, SLAMF5, and SLAMF6 function as negative regulators in T-dependent and T-independent antibody responses.
Materials and Methods
Mice
To generate Slamf[1 + 6]−/− and Slamf[1 + 5 + 6]−/− mice, a B6 bacterial artificial chromosome clone (B6 BAC clone #RP23-77A8) containing the Slamf1 and Slamf6 genes was used to construct a targeting vector with a neomycin resistant cassette flanked by two LoxP sites. SLAMF6 ES cell clones heterozygous for the mutation were generated by standard methods. To generate Slamf1 and Slamf6 double-deficient mice, we used a SLAMF1 targeting vector to retarget the previously generated SLAMF6 mutant ES cell clone that was known to give germline transmission with extremely high frequency. Co-integration of the two targeting vectors on the same chromosome was assessed by in vitro transfection-targeted ES cell clones with a Cre recombinase expression vector. Deletion of the whole Slamf1, Slamf5, and Slamf6 locus was confirmed by PCR (Figures 1A,B). B6 background Slamf5−/− mice have been reported previously (46). Wild-type C57BL/6 (B6) mice were obtained from the Jackson Laboratory. Animal studies were conducted in accordance with the National Research Council’s Guide for the Care and Use of Laboratory Animals and were approved by the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee.
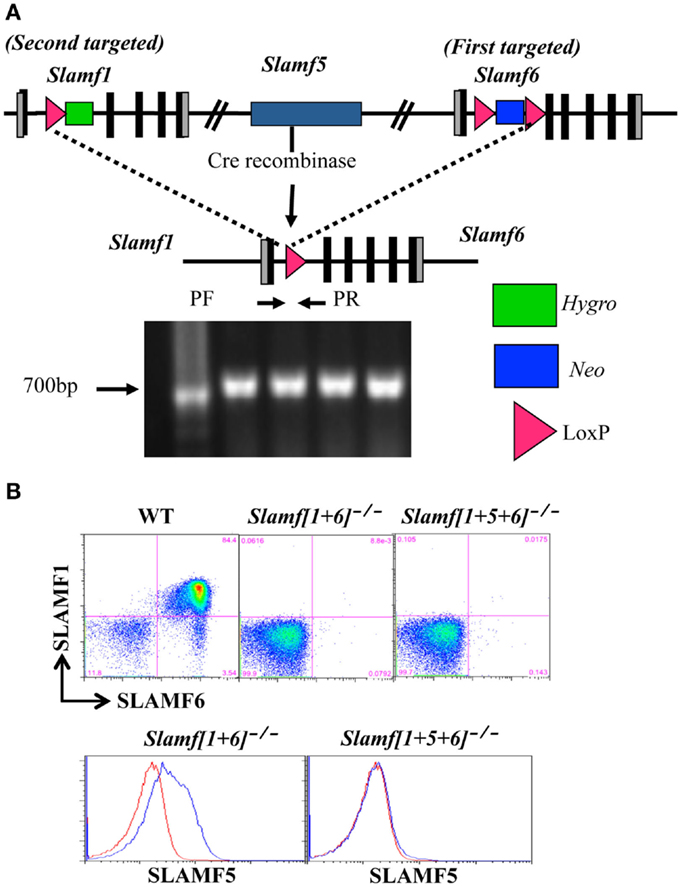
Figure 1. Generation of Slamf[1 + 6]−/− and Slamf[1 + 5 + 6]−/− mice. (A) Schematic representation of the Slamf[1 + 6] and Slamf[1 + 5 + 6] targeting strategy. Top: illustration of the genomic mouse SLAMF1-5-6 locus after targeted replacement of exon 2 and 3 of both Slamf1 and Slamf6 genes. Middle: The Slamf1-5-6 locus after Cre-mediated recombination leading to the deletion of the LoxP site-flanked genomic fragment. Bottom: PCR genotyping primers (PF and PR) used to confirm the junction of Cre-mediated deletion from mouse-tail DNA. (B) Thymocytes from WT, Slamf[1 + 5]−/−, and Slamf[1 + 5 + 6]−/− mice were stained with anti-SLAMF1, anti-SLAMF5, and anti-SLAMF6 antibodies, and the expression of SLAMF1, SLAMF5, and SLAMF6 was evaluated by flow cytometry.
Anti-SLAMF Receptor Antibodies
Rat anti-mouse SLAMF1 mAb (9D1) is specific for the extracellular region of mouse SLAMF1 (25). The anti-mouse SLAMF5 mAb was generated by fusing NS1 myeloma cells with spleen cells obtained from Armenian hamsters, which were immunized three times with mouse SLAMF5-Fc fusion protein (47). Mouse anti-mouse SLAMF6 mAbs (13G3 and 330) are specific for the extracellular region of mouse SLAMF6. Anti-SLAMF1, anti-SLAMF5, and anti-SLAMF6 mAbs used in our studies were purified by affinity chromatography (Harlan Bioproducts for Science). Anti-SLAMF6 F(ab’)2 fragments were generated from whole SLAMF6 mAb, using the Thermo Scientific Pierce F(ab’)2 Preparation Kit (PI-44988) according to manufacturer instructions.
Immunization
Mice were immunized intraperitoneally (i.p.) with 40 μg of NP-ovalbumin (NP-OVA, Biosearch Technology) precipitated with Complete Freund’s Adjuvant (Difco) or Alum. For eliciting TI antigen responses, mice were i.p. immunized with 20 μg of NP28-Ficoll (Biosearch Technologies) or with 10 μg NP-LPS. Mice were bled on day 9 (for T-dependent antigen) or day 7 (for T-independent antigen) post-immunization. NP-specific IgG and IgM titers were determined by ELISA after serial dilutions of the serum.
Adoptive Cell Transfer
Naïve CD4+ and B220+ B cells were purified from the spleens of WT and Slamf[1 + 5 + 6]−/− mice using a magnetic cell sorting kit (Miltenyi Biotec). Rag-1−/− recipient mice were injected with 5 × 106 CD4+ T cells and 10 × 106 B220+ B cells in 200 μl PBS. Mice were immunized with NP-OVA/CFA 7 days after adoptive cell transfer.
ELISA
Serum was collected from mice on day 7 or 9. High binding plates (Costar) were coated overnight at 4°C with [NP(4)-BSA] or [NP(25)-BSA] (50 μg/ml, Biosearch Technologies). Horseradish peroxidase-conjugated sheep anti-mouse IgG antibody (Amersham) was used for detection. Relative affinity of the NP-specific IgG antibodies was calculated from the ratio of antibody binding to low-density hapten [NP(4)-BSA] versus high-density hapten [NP(25)-BSA] coated plates.
Flow Cytometry
Single-cell suspensions of splenocytes, thymocytes, and inguinal lymph nodes were stained with the following antibodies and reagents after blocking non-specific binding with CD16/32 and 15% rabbit-serum: αCD4 (RM4-5), αPD-1 (RMP1-30), αCD44 (IM7), αCD138 (281-2), αB220 (RA3-6B2), αFas (Jo2), αT- and B-cell activation antigen (GL-7), and αIgD (11-26) purchased from eBioscience, BD Pharmingen, or Biolegend. NP32-phycoerythrin was purchased from Biosearch Technologies (N-5070-1). For staining of CXCR5, biotinylated-αCXCR5 (2G8, BD Biosciences) was used, followed by PE-labeled streptavidin (eBioscience). For staining of NP-PE, splenocytes were fixed for 10 min with 4% paraformaldehyde, then were washed with flow cytometry buffer with 0.2% saponin, and stained in the presence of 0.2% saponin. Data were acquired using an LSRII flow cytometer (BD Pharmingen) and analyzed using FlowJo software, Version 8.8.6 (TreeStar Inc.).
Statistical Analysis
Statistical significance was determined by unpaired t-test (two-tailed with equal SD) using Prism software (GraphPad, San Diego, CA, USA). The p-value <0.05 was considered statistically significant.
Results
Generation of Slamf[1 + 6]−/− and Slamf[1 + 5 + 6]−/− Mice
Since the murine SLAMF1, SLAMF5, and SLAMF6 genes are closely linked on mouse chromosome 1, a mouse strain lacking Slamf[1 + 6] or Slamf[1 + 5 + 6] cannot be generated by interbreeding individual Slamf1−/−, Slamf5−/−, and Slamf6−/− mice. To generate Slamf[1 + 6]−/− and Slamf[1 + 5 + 6]−/− mice, we first replaced exons 2 and 3 of the Slamf6 gene with a LoxP-flanked PGK-NeoR cassette in the first targeting event in B6 ES cells (Figure 1A). We next transfected one of the SLAMF6-targeted ES cell clones with a vector that replaced exons 2 and 3 of the Slamf1 gene with a hygromycin resistant gene containing a LoxP site, thus generating Slamf[1 + 6]−/+ ES cells. To identify ES cell clones in which both insertions had taken place on the same chromosome, we removed the LoxP-flanked chromosome fragment of 200 Kb, which includes the Slamf1, Slamf5, and Slamf6 genes. The confirmed Slamf[1 + 6]−/+ ES cell clones were used to generate Slamf[1 + 6]−/− mice. Subsequently, Slamf[1 + 6]−/− mice were bred to CreTg mice to obtain Slamf[1 + 5 + 6]−/− mice (Figure 1A). The absence of Slamf[1 + 6] and Slamf[1 + 5 + 6] expression was confirmed by flow cytometric analyses using SLAMF1, SLAMF5, and SLAMF6 specific antibodies (Figure 1B).
The Number of Marginal Zone B Cells is Significantly Increased in Slamf[1 + 5 + 6]−/− Mice
Thymic development and the number of splenic T cells were not altered in Slamf[1 + 6]−/− or Slamf[1 + 5 + 6]−/− mice (Figures S1A,C in Supplementary Material). However, a close examination of the B cell compartment by flow cytometric analysis revealed that the number and percentage of marginal zone (MZ) B cells (sIgMhiAA4.1-CD19+CD21hiCD23lo-neg) was significantly increased in Slamf[1 + 5 + 6]−/− mice as compared to WT and Slamf[1 + 6]−/− mice (Figures 2A–C). By contrast, the percentage and number of T and B cells and the number of splenocytes in Slamf[1 + 5 + 6]−/− mice were identical to those in WT mice (Figure 2D; Figures S1A–C in Supplementary Material). The population of follicular B cells (AA4.1− CD21+CD23+sIgMint) in the spleens of Slamf[1 + 6]−/− and Slamf[1 + 5 + 6]−/− mice was similar to that in WT mice (Figure 2E). Similarly, the frequencies of CD5+ B1a and CD5− B1b cells were comparable in the peritoneal cavity of mutant and WT mice (Data not shown). Immature peripheral B cells can be divided into three transitional populations based on their surface marker expression, designated as transitional type 1 (T1) (AA4.1+sIgMintCD23-), transitional type 2 (T2) (AA4.1+sIgMhiCD23-), and transitional type 3 (T3) (AA4.1+sIgMintCD23+). Similarly, the frequencies of T1, T2, and T3 immature B cell populations in Slamf[1 + 6]−/− and Slamf[1 + 5 + 6]−/− mice were not significantly different from those of wild-type mice (Figure 2F). Collectively, these findings show that the ablation of SLAMF1, SLAMF5, and SLAMF6 affects the development of MZ B cells, but other B cell subsets and T cell development occur normally in Slamf[1 + 6]−/− and Slamf[1 + 5 + 6]−/− mice.
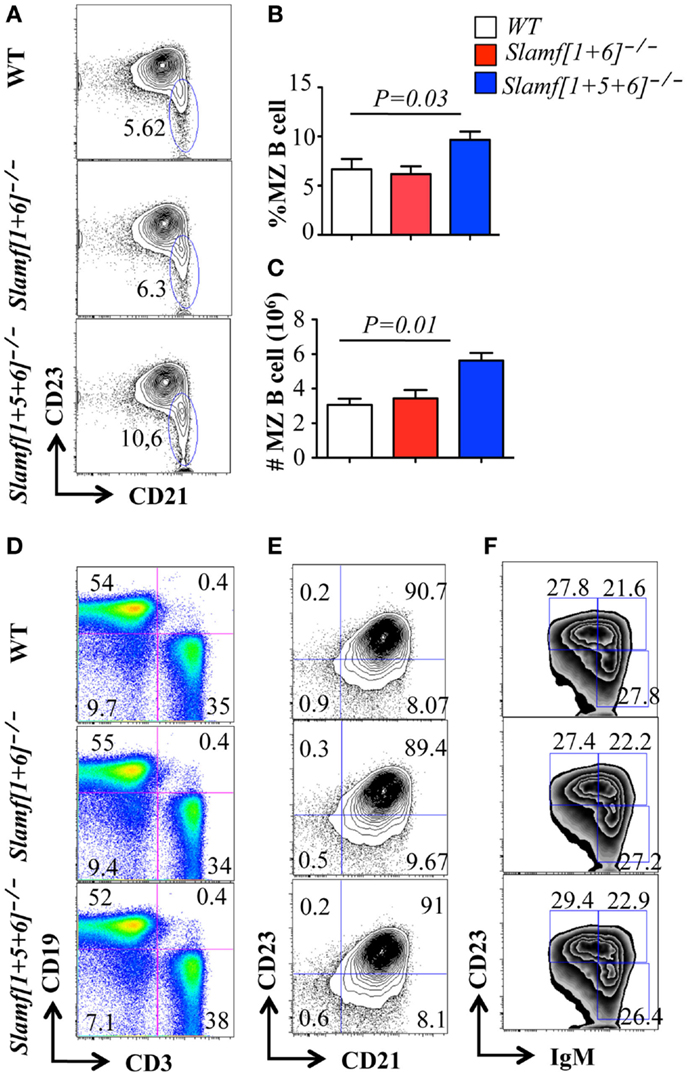
Figure 2. Comparison of B cell and T cell populations in Slamf[1 + 5]−/−, Slamf[1 + 5 + 6]−/−, and WT mice. Flow cytometric analyses of B cell and T cell subsets in the spleens of Slamf[1 + 6]−/−, Slamf[1 + 5 + 6]−/−, and WT mice: (A) CD19+AA4-IgMhi cells in spleens are gated for the expression of CD23 and CD21 to delineate CD21+CD23- marginal zone (MZ) B cells. (B) Percentage of CD19+AA4− IgMhiCD21+ CD23− MZ B cells. (C) The number of CD19+AA4− IgMhiCD21+CD23- MZ B cells. (D) Splenocytes from Slamf[1 + 6]−/−, Slamf[1 + 5 + 6]−/−, and WT mice are stained for surface expression of CD3 and CD19. (E) CD19+ AA4− IgMhiCD21 cells in spleens are gated for the expression of CD23 and CD21 to delineate CD21+CD23+ follicular B cells in the spleens. (F) Transitional B cell subsets in spleens are stained for the expression of CD19, AA4.1, CD23, and IgM: T1 (CD19+AA4+IgMhiCD23-), T2 (CD19+AA4+IgMhiCD23+), and T3 (CD19+AA4+IgMloCD23-).
Enhanced T Cell Dependent Antibody Production in Slamf[1 + 5 + 6]−/− Mice
Although most SLAMF receptors are expressed on the surface of T and B cells, ablation of single SLAMF genes does not lead to significant defects in germinal center formation after protein immunization or viral infection (27, 42, 44). In contrast, the absence of SAP, the SLAMF specific adaptor, leads to a severe defect in humoral response (14, 32, 34), which suggests functional redundancies in the control of antibody responses by SLAMF receptors. To test this hypothesis, we compared NP-specific antibody production by Slamf[1 + 6]−/−, Slamf[1 + 5 + 6]−/−, and WT mice. Slamf[1 + 6]−/− mice, which had been immunized with NP-OVA in CFA, produced similar amounts of NP-specific serum IgM as WT mice (data not shown). However, the level of anti-NP IgG in the serum of Slamf[1 + 6]−/− mice was consistently higher, although statistical analysis did not reach significance (Figure 3A). Surprisingly, the additional disruption of the Slamf5 gene significantly augmented the level of anti-NP IgG in Slamf[1 + 5 + 6]−/− mice (Figure 3A) even though NP-specific IgM production was not altered (data not shown). Affinity maturation of NP-specific IgG was comparable between the mutant and WT mice (data not shown). Because Slamf[1 + 5 + 6]−/− mice produced higher NP-specific IgG compared to Slamf[1 + 6]−/− mice, we reasoned that SLAMF5 signaling might suppress antibody responses. To test this, we then immunized Slamf5−/− mice with NP-OVA. In contrast to a previous report (41), Slamf5 deficiency had no effect on NP-specific antibody production or the development of Tfh cells or GC B cells (Figures 3B–F). Taken together, the data support the notion that SLAMF1, SLAMF5, and SLAMF6 cooperate in the negative regulation of T-dependent antibody responses.
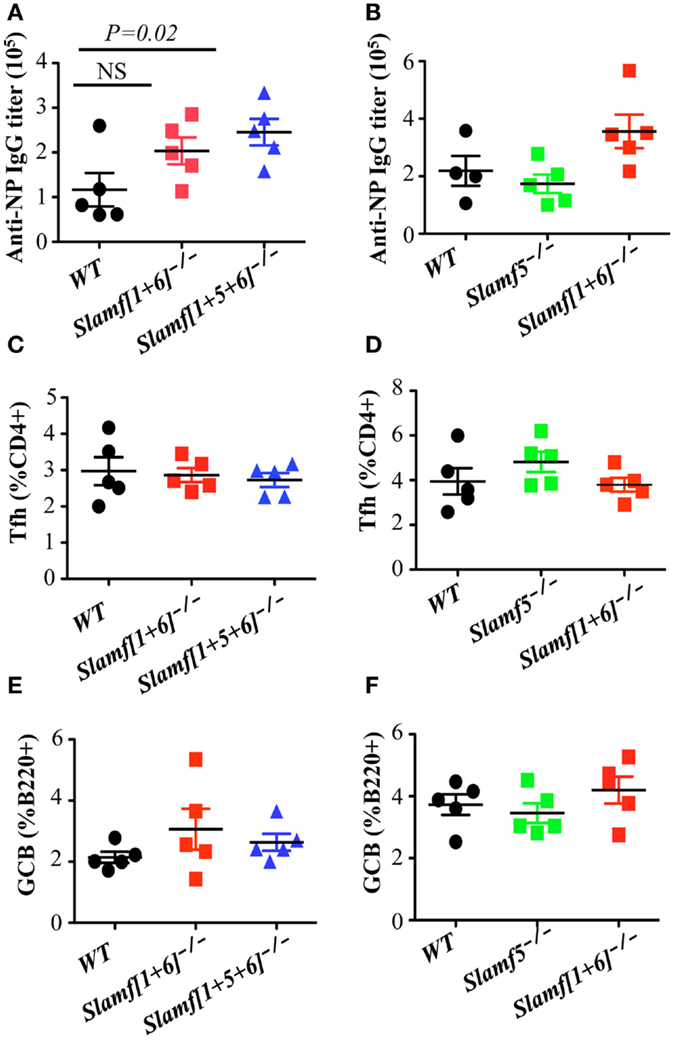
Figure 3. A combination of SLAMF1, SLAMF5, and SLAMF6 negatively regulates T cell dependent antibody responses, but normal Tfh and GCB development is observed in Slamf[1 + 5 + 6]−/− mice. WT, Slamf5−/−, Slamf[1 + 6]−/−, and Slamf[1 + 5 + 6]−/− mice were immunized with 40 μg of NP-OVA and serum was collected on day 9. (A) NP-specific IgG titers for Slamf[1 + 6]−/−, Slamf[1 + 5 + 6]−/−, and WT mice immunized with NP-OVA in CFA were determined by ELISA using NP(4)-BSA coated plates. (B) NP-specific IgG titers for Slamf5−/−, Slamf[1 + 6]−/−, and WT mice immunized with NP-OVA in Alum were determined by ELISA using NP(4)-BSA coated plates. (C) Percentage of Tfh cells (CD4+PD-1+CXCR5+) in the spleens of Slamf[1 + 6]−/−, Slamf[1 + 5 + 6]−/−, and WT mice. (D) Percentage of Tfh cells (CD4+PD-1+CXCR5+) in the spleens of Slamf5−/−, Slamf[1 + 6]−/−, and WT mice. (E) Percentage of Germinal Center B cells (B220+GL7+Fas+) in the spleens of Slamf[1 + 6]−/−, Slamf[1 + 5 + 6]−/−, and WT mice. (F) Percentage of Germinal Center B cells (B220+GL7+Fas+) in the spleens of Slamf5−/−, Slamf[1 + 6]−/−, and WT mice. Data represent at least three independent experiments.
The Combined Absence of SLAMF1, SLAMF5, and SLAMF6 Enhances Antigen Specific Plasma Cell Expansion, but has No Effect on the Development of GC B Cells, Tfh Cells, or T Follicular Regulatory (Tfr) Cells
As strong humoral immune responses, characterized by GC formation and long-lived plasma and memory B cells, are dependent on help provided by CD4+ Tfh cells (4, 5, 48), we next examined whether enhanced T-cell dependent antibody responses in Slamf[1 + 5 + 6]−/− mice are correlated with an increase in Tfh cell differentiation and higher germinal center responses after immunization with NP-OVA. Contrary to our prediction, the percentage and number of Tfh cells was comparable between Slamf[1 + 5 + 6]−/− and WT mice (Figure 3C; Figure S2A in Supplementary Material). Percentages of GC B cells (FAS+ GL-7+) were also unaffected by the combined absence of SLAMF1, SLAMF5, and SLAMF6 (Figure 3E; Figure S2B in Supplementary Material).
Given the increased antibody responses observed in Slamf[1 + 5 + 6]−/− mice, we hypothesized that the absence of SLAMF, SLAMF5, and SLAMF6 may enhance either plasma cell differentiation or their capacity to produce antibody. To test this hypothesis, we evaluated NP-specific plasma cells in NP-OVA immunized mice. Consistent with high antibody responses, the frequency of plasma cells (B220+IgD
-
CD138+) was significantly increased in Slamf[1 + 5 + 6]−/− mice (Figures 4A,B). Furthermore, flow cytometric analysis confirmed an increase in NP-specific plasma cells in immunized Slamf[1 + 5 + 6]−/− mice as compared to wild-type mice (Figures 4C,D). Together, the data indicate that the absence of SLAMF1, SLAMF5, and SLAMF6 has no effect on Tfh and GC B cell development, but that it appears to regulate development of antigen specific plasma cells.
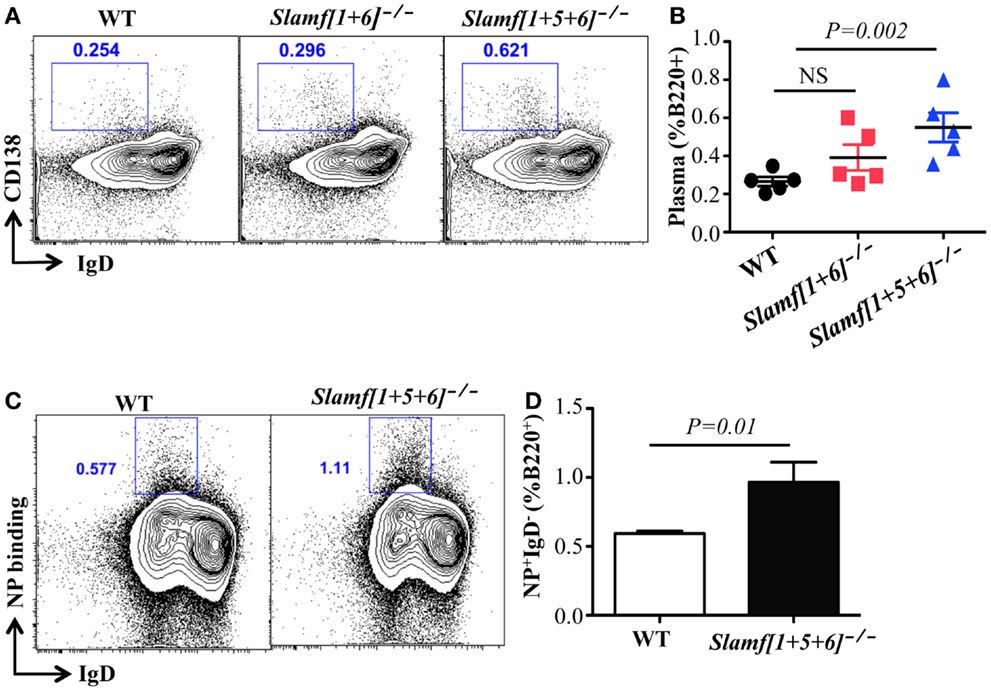
Figure 4. A combined absence of SLAMF1, SLAMF5, and SLAMF6 enhances antigen specific plasma cell expansion. Slamf[1 + 6]−/−, Slamf[1 + 5 + 6]−/−, and WT mice were immunized with NP-OVA in CFA. After 9 days, spleens were isolated and subjected to staining with the indicated antibodies and analyzed by flow cytometry. (A) Representative FACS plots showing B220+IgD-CD138+ plasma cells from the spleens of Slamf[1 + 6]−/−, Slamf[1 + 5 + 6]−/−, and WT mice. (B) Percentage of Plasma cells (B220+IgD-CD138+) from the spleens of Slamf[1 + 6]−/−, Slamf[1 + 5 + 6]−/−, and WT mice. (C) Representative FACS plots showing B220+NP+IgD- NP-specific plasma cells from the spleens of Slamf[1 + 5 + 6]−/− and WT mice. (D) Percentage of NP-staining plasma cells (B220+NP+IgD-) from the spleens of Slamf[1 + 5 + 6]−/− and WT mice. Data represent at least three independent experiments.
A new Treg cell subset termed Tfr cells has recently been identified (49, 50). These cells, which express not only CXCR5 and PD-1 but also the transcription factors Bcl6 and FoxP3, suppress both Tfh cells and GC B cells. As Tfr cell differentiation requires SAP expression (49), it is possible that the absence of SLAMF1, SLAMF5, and SLAMF6 might also cause a defect in Tfr cell development, thereby contributing to the enhanced antibody responses in Slamf[1 + 5 + 6]−/− mice. To assess the impact of the combined absence of SLAMF1, SLAMF5, and SLAMF6 on Tfr cell differentiation, we immunized mice with NP-OVA and analyzed Tfr cells 7 days later. The frequency of Tfr cells (CD4+CXCR5highPD-1highFoxP3+) was not significantly affected in Slamf[1 + 6]−/− or Slamf[1 + 5 + 6]−/− mice (Figure S3A in Supplementary Material). Although the expression of Ki67, a marker used to identify proliferating cells, is slightly decreased in Slamf[1 + 5 + 6]−/− Tfr cells, it is not statistically significant (Figure S3B in Supplementary Material). Thus, while the enhanced antibody production may not result from a defect in Tfr differentiation, its functional inability would not be excluded in Slamf[1 + 5 + 6]−/− mice.
Slamf[1 + 5 + 6]−/− B or T Cells Adoptively Transferred to RAG-1 Deficient Mice Can Induce Enhanced Antibody Responses
As SLAMF1, SLAMF5, and SLAMF6 are expressed on both B cells and T cells, it was not clear on which cell type ablation of their expression was critical for the altered T-dependent antibody responses observed in Slamf[1 + 5 + 6]−/− mice. This prompted us to evaluate potential contributions of Slamf[1 + 5 + 6]−/− T and B cells to the enhanced humoral responses by using the adoptive transfer of naïve T and B cells. To this end, four combinations of CD4+ cells and B cells were transferred into Rag-1−/− recipient mice: WT CD4+ T and WT B cells, Slamf[1 + 5 + 6]−/− CD4+ T and Slamf[1 + 5 + 6]−/− B cells, WT CD4+ T and Slamf[1 + 5 + 6]−/− B cells, and Slamf[1 + 5 + 6]−/− CD4+ T and WT B cells. Seven days post-transfer, the recipient Rag-1−/− mice were immunized with NP-OVA in CFA. Rag-1−/− mice reconstituted with CD4+ T cells and B cells from Slamf[1 + 5 + 6]−/− mice had significantly higher NP-specific antibody production than recipient mice that had been reconstituted with WT CD4+ T cells and B cells (Figure 5). Interestingly, the transfer of Slamf[1 + 5 + 6]−/− B cells together with WT CD4+ T cells was sufficient to induce a stronger antibody response as compared to the transfer of WT CD4+ T cells and B cells. Although the transfer of Slamf[1 + 5 + 6]−/− CD4+ T cells and WT B cells also led to increased titers of NP-specific IgG, the magnitude of the antibody responses was less pronounced (Figure 5). Thus, we concluded that Slamf[1 + 5 + 6]−/− T and B cells both have intrinsic activity, but that the enhanced T-dependent humoral response in Slamf[1 + 5 + 6]−/− mice mainly results from Slamf[1 + 5 + 6]−/− B cells.
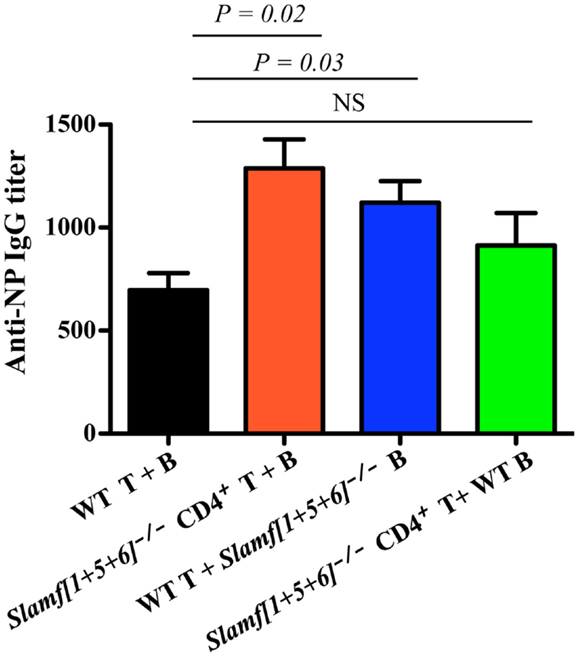
Figure 5. The adoptive transfer of naïve Slamf[1 + 5 + 6]−/− T or B cells enhanced NP-specific antibody responses after co-transfer of WT B or T cells into Rag-1−/− mice. CD4+ T cells (5 × 106) together with 10 × 106 B cells are isolated from WT and Slamf[1 + 5 + 6]−/− mice and transferred into Rag-1−/− recipients in the following four combinations of T and B cells: WT CD4+ T and WT B cells, Slamf[1 + 5 + 6]−/− CD4+ T and Slamf[1 + 5 + 6]−/− B cells, WT CD4+ T and Slamf[1 + 5 + 6]−/− B cells, and Slamf[1 + 5 + 6]−/− CD4+ T and WT B cells. The Rag-1−/− recipients were immunized with 40 μg of NP-OVA in CFA 7 days after the transfer. Mice were sacrificed and NP-specific IgG titers were determined by ELISA day 9 post-immunization. Data are representative of three independent experiments.
Enhanced T-Independent Antibody Responses are Observed in Slamf[1 + 5 + 6]−/− Mice
Because Slamf[1 + 5 + 6]−/− mice have a high frequency of MZ B cells (Figures 2A,B) that are known to participate in responses to T-independent antigens, we questioned whether T-independent antibody responses are affected in Slamf[1 + 5 + 6]−/− mice. Consequently, we examined the SLAMF mutant mice in response to NP-Ficoll, a classical synthetic TI-2 antigen that induces murine Ag-specific B cells to expand, differentiate, and produce NP-specific antibodies. As anticipated, the serum concentrations of NP-specific IgM and IgG3 in Slamf[1 + 5 + 6]−/− mice were increased on day 7 after immunization compared to those in WT mice (Figures 6A,B). In contrast, NP-specific IgM and IgG3 titers were comparable in WT and Slamf[1 + 6]−/− mice (Figures 6A,B). As responses to NP-LPS were not altered (data not shown), the enhanced TI-2 responses are specifically linked to the combined absence of SLAMF1, SLAMF5, and SLAMF6.
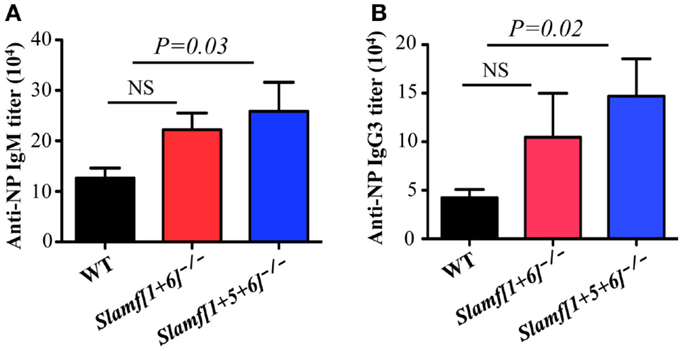
Figure 6. A combined ablation of SLAMF1, SLAMF5, and SLAMF6 shows a selective increase in MZ B cells and enhanced TI-2 antibody responses. WT, Slamf[1 + 6]−/−, and Slamf[1 + 5 + 6]−/− mice were immunized with 20 μg of NP-Ficoll. NP-specific IgM (A) and IgG3 (B) titers were determined at day 7 by ELISA after serial dilutions of the serum. Results are representative of three independent experiments.
Anti-SLAMF6 Antibody Inhibits Humoral Immune Responses
The observation that the ablation of SLAMF1, SLAMF5, and SLAMF6 enhances T-dependent and T-independent antibody responses suggests that they function as potential inhibitory molecules in humoral immune responses. In addition, a recent report indicates that SLAMF6 transmits inhibitory signaling in Tfh differentiation and NKT development in the context of the absence of SAP (27). To directly evaluate whether triggering of a single SLAMF receptor would initiate inhibitory signaling in humoral responses, we first sought to trigger SLAMF6 by using anti-SLAMF6 mAb in NP-OVA-immunized mice. As shown in Figure 7, the treatment of WT mice with anti-SLAMF6 (330) dramatically impaired NP-specific IgG production 9 days post-immunization of NP-OVA (Figure 7A). As negative control, anti-SLAMF6 had no detectable effects on antibody production in Slamf[1 + 6]−/− mice (Figure S4A in Supplementary Material). Noticeably, anti-SLAMF6 injected mice had a significant reduction in IgG high affinity antibody (Figure 7B). Furthermore, NP-specific IgM production and its affinity maturation also were impaired in anti-SLAMF6 injected mice (Figures 7C,D). In correlation with the impaired antibody response, the frequencies and absolute numbers of splenic GC B cells (GL7+Fas+) (Figures 8A–C) and Tfh T cells (CXCR5+PD-1+) (Figures 9A–C) were significantly reduced in anti-SLAMF6 injected mice. A reduced number of plasma cells was also observed in anti-SLAMF6 injected mice (Figures 8D,E), but effector B cell and T cell populations were equivalent in anti-SLAMF6 injected and non-injected mice (Figure 8F and Figures 9D,E). In order to confirm the inhibitory effects of anti-SLAMF6 (330), another anti-SLAMF6 mAb (13G3) was also tested. As expected, a similar inhibitory effect was seen in 13G3-injected mice (Figures S4A,B in Supplementary Material).
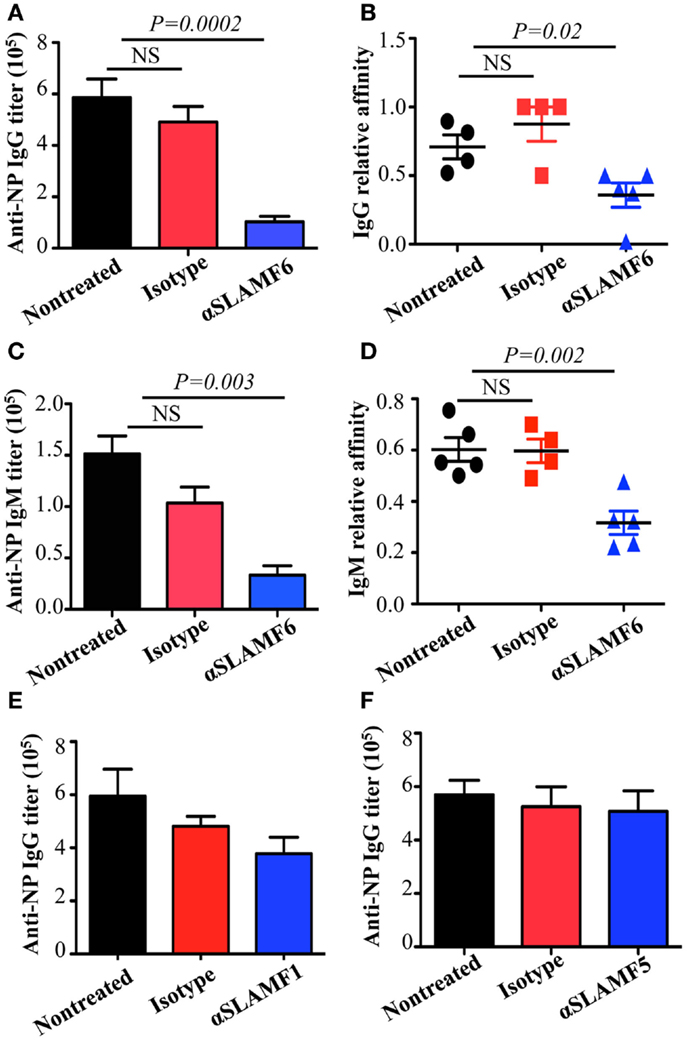
Figure 7. Administration of anti-SLAMF6 antibody has a negative effect on antibody production in protein-immunized WT mice. Mice were immunized with 40 μg NP-OVA in CFA and some were injected with either 250 μg of anti-SLAMF6 (330), anti-SLAMF1 (9D1), anti-SLAMF5 (M5), or mouse Ig isotype control. The mice were sacrificed on day 9 and serum was collected to measure Ig production. (A) NP-specific IgG titers from sera of anti-SLAMF6 injected mice were determined by ELISA. (B) Affinity of NP-specific IgG in immune-sera collected as in (A). (C) NP-specific IgM titers from sera of anti-SLAMF6 injected mice were determined by ELISA. (D) Affinity of NP-specific IgM in immune-sera collected as in (C). (E) NP-specific IgG titers from sera of anti-SLAMF1 injected mice were determined by ELISA. (F) NP-specific IgG titers from sera of anti-SLAMF5 injected or non-injected immunized mice were determined by ELISA. Results are representative of three independent experiments.
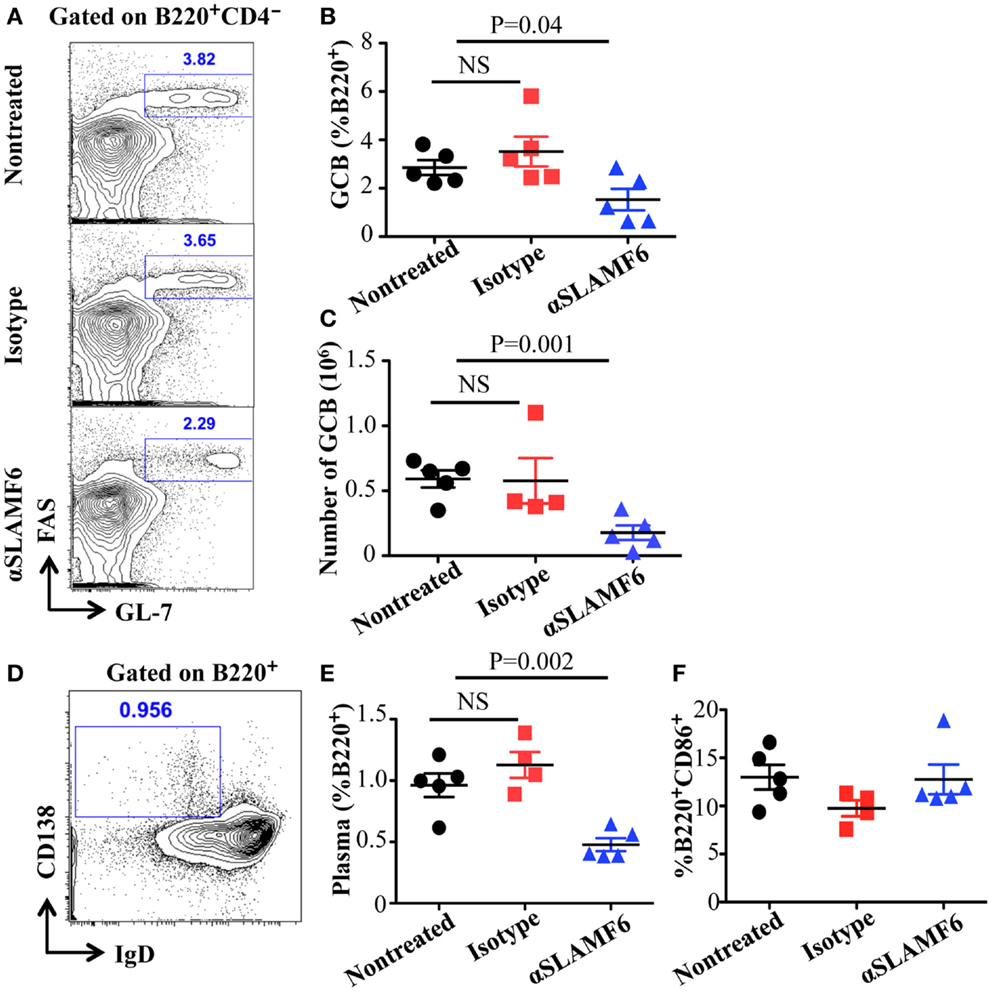
Figure 8. Administration of anti-SLAMF6 (330) antibody has a negative effect on GC B cell differentiation in protein-immunized WT mice. Mice were immunized with 40 μg of NP-OVA in CFA and some mice were injected with either 250 μg anti-SLAMF6 (330) or Ig isotype control. The mice were sacrificed on day 9. (A) Representative flow cytometry staining of B220+GL7+Fas+ Germinal Center B cells in the spleens of anti-SLAMF6, isotype, and non-injected immunized mice. (B) Percentage of Germinal Center B cells (B220+GL7+Fas+) in the spleens of anti-SLAMF6, isotype, and non-injected immunized mice was determined by flow cytometry. (C) The numbers of Germinal Center B cells (B220+GL7+Fas+) in the spleens of anti-SLAMF6, isotype, and non-injected immunized mice were determined by flow cytometry. (D) Representative of flow cytometry staining of B220+IgD-CD138+ plasma cells in the spleens of anti-SLAMF6, isotype, and non-injected immunized mice. (E) Percentage of plasma cells (B220+IgD-CD138+) in the spleens of anti-SLAMF6, isotype, and non-injected immunized mice. (F) Percentage of B220+CD86+ activated B cells in the spleens of anti-SLAMF6, isotype, and non-injected immunized mice. Results are representative of three independent experiments.
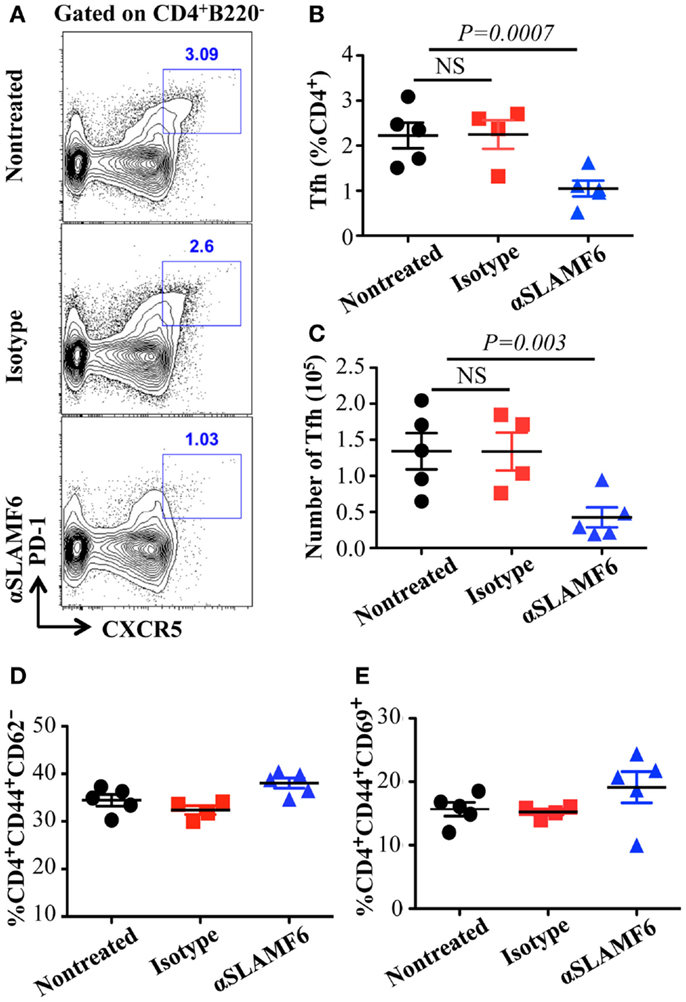
Figure 9. Administration of anti-SLAMF6 (330) antibody has a negative effect on Tfh cell differentiation in protein-immunized WT mice. Mice were immunized with 40 μg of NP-OVA in CFA and some mice were injected with either 250 μg anti-SLAMF6 (330) or Ig isotype control. The mice were sacrificed on day 9. (A) Representative flow cytometry staining of CD4+PD-1+CXCR5+ Tfh cells in the spleens of anti-SLAMF6, isotype, and non-injected immunized mice. (B) Percentage of Tfh cells (CD4+PD-1+CXCR5+) was determined by flow cytometry in the spleens of anti-SLAMF6, isotype, and non-injected immunized mice. (C) The number of Tfh cells (CD4+PD-1+CXCR5+) was determined by flow cytometry in the spleens of anti-SLAMF6, isotype, and non-injected immunized mice. (D) Percentage of CD4+CD44hiCD62lo memory T cells in the spleens of anti-SLAMF6, isotype, and non-injected immunized mice. (E) Percentage of CD4+CD44hiCD69hi activated T cells in the spleens of anti-SLAMF6, isotype, and non-injected immunized mice. Results are representative of three independent experiments.
To address whether the administration of anti-SLAMF6 affects early commitment to antigen specific Tfh cells and GC B cells or late stages of humoral responses, immunized mice were injected with anti-SLAMF6 four days post-immunization, at which point T and B cells are already committed to becoming Tfh and GC B cells (51). Interestingly, the late injection of mice with anti-SLAMF6 did not significantly reduce GC response and antibody production (data not shown). This suggests that the signal initiated by SLAMF6 has efficient inhibition in early Tfh and GC B cell differentiations, but has little effect on late Tfh and GC B cell expansion and antibody production.
To evaluate whether the Fc portion of anti-SLAMF6 influences immune function, anti-SLAMF6 F(ab’)2 fragments were injected into mice along with NP-OVA immunization. Similar to intact anti-SLAMF6, anti-SLAMF6 F(ab’)2 caused a significant decrease in the percentage and number of GC B cells (Figures 10B,C) and Tfh cells (Figures 10D,E). In contrast, an impaired NP-specific antibody production was not observed in the anti-SLAMF6 F(ab’)2 injected mice. To exclude the possibility that NK cells mediate natural cytotoxicity against a variety of immune cells, we examined splenocyte phenotype and levels of NP-specific antibody in NK-depleted WT mice following NP-OVA immunization and anti-SLAMF6 injection. Notably, the depletion of NK cells did not impact the capacity of anti-SLAMF6 to suppress antibody production or the development of Tfh cells and GC B cells (data not shown). Thus, severely impaired antibody production by the injection of anti-SLAMF6 is not due to NK cell-mediated ADCC.
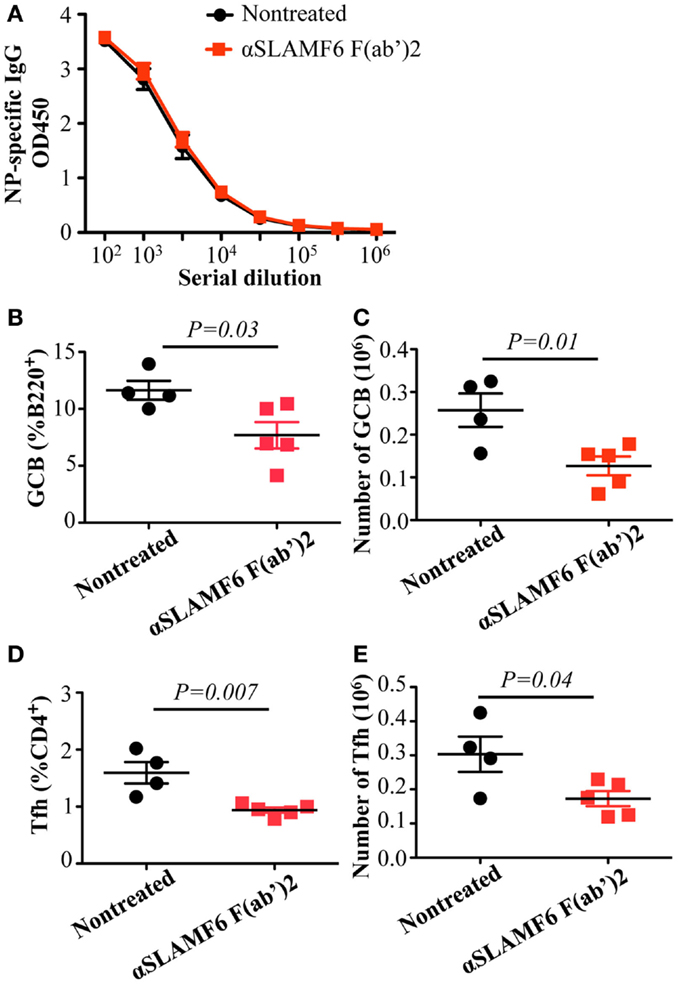
Figure 10. Administration of anti-SLAMF6 (330) F(ab’)2 antibody has a similar negative effect on GC B cell and Tfh cell development in protein-immunized WT mice. Mice were immunized with 40 μg of NP-OVA in CFA and some were injected with 250 μg of anti-SLAMF6 F(ab’)2 on day 0 and day 4. The mice were sacrificed on day 9 and serum was collected to measure IgG production. (A) NP-specific IgG titers were determined by ELISA. (B–C) The percentage and number of Germinal Center B cells (B220+GL7+Fas+) were determined by flow cytometry. (D–E) The percentage and number of Tfh cells (CD4+PD-1+CXCR5+) were determined by flow cytometry. Results are representative of three independent experiments.
Based on the studies using anti-SLAMF6, we next examined the ability of anti-SLAMF1 mAb and anti-SLAMF5 mAb to suppress humoral immune responses in NP-OVA immunized mice. We found that NP-specific antibody production was moderately, but consistently, reduced in anti-SLAMF1 injected mice (Figure 7E). However, anti-SLAMF5 did not suppress antibody production (Figure 7F). In addition, the frequencies of GC B cells and Tfh cells were comparable among the non-injected, anti-SLAMF1, and anti-SLAMF5 injected mice (data not shown). Thus, the results suggest that neither anti-SLAMF1 nor anti-SLAMF5 is alone sufficient to have a significant impact on humoral responses.
Discussion
An important role of SAP in T cell help to B cells has been highlighted in defects in follicular T helper cell differentiation and lack of germinal center development in XLP patients and in virally infected or immunized SAP−/− mice (4, 12, 13, 32, 48, 52). Although SAP has been proven to bind to ITSMs in most SLAMF receptors upon ligand stimulation, how these receptors function in the presence and absence of SAP is poorly understood, particularly in B cells. In this study, we provide in vivo evidence that SLAMF1, SLAMF5, and SLAMF6 synergistically suppress T-dependent and T-independent antibody responses, as Slamf[1 + 5 + 6]−/− mice, but not Slamf[1 + 6]−/− mice or single SLAMF mutant mice, exhibit a significant enhancement of antibody production. Strikingly, an adoptive transfer assay shows that the enhanced antibody responses depend largely on Slamf[1 + 5 + 6]−/− B cells, which indicates that the absence of SLAMF1, SLAMF5, and SLAMF6 induces intrinsic B cell activity. Furthermore, injection of mice with anti-SLAMF6 mAb dramatically reduced antibody responses accompanied by impairing Tfh cell and GC B cell development in spite of a less suppressive effect of anti-SLAMF1 mAb on antibody responses. Our results therefore point to a new and important mechanism by which SLAMF1, SLAMF5, and SLAMF6 regulate humoral responses in B cells.
Maintaining sustained CD4+ T cell adhesion to B cells is required for Tfh differentiation and germinal center development, which allows for important signal transfer between T and B cells. SAP-deficient T cells fail to form a stable T-B cell conjugate (41) and thereby severely impaired development of Tfh cells and GC B cells becomes a hallmark in XLP patients and SAP−/− mice (14, 32, 34, 41). Although most SAP-binding SLAMF receptors are highly expressed on resting and activated T and B cells and implicate a diverse array of lymphocyte functions, including sustained T-B cell conjugates (43, 53), the deficiencies in single SLAMF receptors actually exhibit mild phenotypes. However, in contrast to our current study, Slamf5−/− mice have previously been shown to have a defect in germinal center development and T-dependent antibody production in protein immunization (43). The reason that the same B6 background Slamf5−/− mice strains have different phenotypes is not clear. One explanation for this discrepancy is that the presence of selection marker (Neo) could influence neighboring genes in targeted loci (54). Alternatively, it remains possible that the discordant findings could reflect the different environmental conditions of animal facilities.
Because the functional redundancies in SLAMF-mediated signaling have been demonstrated in NKT cell development (45, 46), we speculate that multiple deficiencies in SLAMF receptors may have a strong influence on humoral immune responses that are able to recapitulate most of the phenotypic alterations observed in SAP−/− mice. Surprisingly, in our in vivo studies comparing T-dependent antibody responses, loss of expression of SLAMF1, SLAMF5, and SLAMF6 receptors actually removed inhibitory signaling and resulted in higher antibody responses (Figure 3A). When the antibody responses in mice lacking Slamf[1 + 6] were compared to those in WT mice, there was also a consistent increase in NP-specific antibody titer, but the effect was less pronounced than that observed in the combined ablation of SLAMF1, SLAMF5, and SLAMF6. Although some variability in antibody responses between mutant and WT mice occurs, statistical analysis always reached significance. These findings indicate that the homophilic binding of SLAMF1, SLAMF5, and SLAMF6 synergistically transmits inhibitory signaling during humoral immune responses. The dual function of SLAMF receptors was initially reported in NK cell studies. In human NK cells, SLAMF4 predominantly functions as an activating receptor because engagement of SLAMF4 with SLAMF2 mediates NK cell cytotoxicity, cell proliferation, and cytokine secretion. However, in NK cells from XLP patients, the SLAMF2–SLAMF4 interaction fails to activate NK cells, but rather inhibits NK-medicated cytolysis (23). Besides SLAMF4, other SLAMF receptors such as SLAMF3, SLAMF5, and SLAMF6 become inhibitory molecules instead of activating receptors in mouse NK cells lacking SAP, EAT-2A, and EAT-2B (55). Consistent with these findings, a similar inhibitory effect of SLAMF6 on humoral responses and NKT cell development was reported in the context of the absence of SAP (27). Collectively, these observations suggest that most SLAMF receptors can mediate either positive or negative signaling, depending on the expression of SLAMF adaptors, SAP and EAT-2. Given that SAP and EAT-2A/B are not expressed in B cells, the homophilic interactions of SLAMF1, SLAMF5, and SLAMF6 between T and B cells or B and B cells would result in preferential binding to inhibitory signaling molecules (e.g., SHP1) in B cells because of the lack of competition of SAP and EAT-2 for ITSMs of SLAMF receptors. This idea is supported by our adoptive transfer assays, in which B cells from Slamf[1 + 5 + 6]−/− mice led to a further enhancement in antibody responses compared to the transfer of Slamf[1 + 5 + 6]−/− T cells. Furthermore, Slamf[1 + 5 + 6]−/− mice display higher antibody responses in the absence of T cell help when immunized with T-independent antigen NP-Ficoll, which directly indicates that an intrinsic B cell hyperactivation exists in Slamf[1 + 5 + 6]−/− mice.
Although the higher antigen specific antibody production does not accompany enhanced Tfh and GC B cell responses in the absence of SLAMF1, SLAMF5, and SLAMF6, increased development of plasma cells is consistently observed in NP-OVA immunized Slamf[1 + 5 + 6]−/− mice (Figures 4A–D). The mechanism regulating plasma cell differentiation is only partly understood. Two transcription factors Bcl-6 and Blimp-1 reciprocally modulate differentiation of GC B cells and plasma cells (52). Cytokines and chemokines also provide crucial survival signals to plasma cells (56). Far less is known about SLAMF receptor-mediated signals for plasma cell differentiation and function. However, in our experiments, we provide interesting evidence that SLAMF1, SLAMF5, and SLAMF6 negatively regulate either plasma cell differentiation and/or expansion in humoral immune responses. Further work will assess how SLAMF receptors are involved in plasma cell development at a molecular level.
The interesting finding in this study is that Slamf[1 + 5 + 6]−/− mice exhibit an increased frequency of MZ B cells. A cell-fate decision between follicular B cells and MZ B cells occurs in the transitional (T2) B cell stage, when T2 B cells differentiate into follicular B cells or MZ B cells after integration of BCR signal strength and signaling via other essential signaling molecules (57–59). Like in GC B cells, SLAMF1, SLAMF5, and SLAMF6 are highly expressed in transitional B cells and MZ B cells (ImmGen.org), and therefore, signaling resulting from their homophilic interaction may implicate differentiation, migration, or survival of MZ B cells. Since differentiation of transitional B cells was not altered in Slamf[1 + 5 + 6]−/− mice, signaling initiated by SLAMF1, SLAMF5, and SLAMF6 seems to play a critical role in controlling the development and/or survival of MZ B cells. It has been described that MZ B cells and B1 cells are prime B cell subpopulations responding to T-independent antigens (60, 61). Interestingly, in spite of increased pools of MZ B cells, Slamf[1 + 5 + 6]−/− mice showed enhanced immune responses to the TI-2 Ag NP-Ficoll, but not to the TI-1 Ag NP-LPS. The fact that enhanced TI-antigen response is only limited to TI-2 antigens suggests that Slamf[1 + 5 + 6]−/− MZ B cell intrinsic activity, not number, seems to be more critical in determining the extent of humoral immunity. Thus, these results provide evidence that synergistic activity of SLAMF1, SLAMF5, and SLAMF6 may be implicated in functional activity of MZ B cells.
Complementary approaches with SLAMF receptor-deficient mice and SLAMF-specific antibodies are important for understanding the functions of their immunoregulatory pathways. Recently, SLAMF6 was found not only to constitutively associate with SHP1 in SAP-sufficient cells, but also to co-distribute with the CD3 complex. The ligation of SLAMF6 can reduce CD3ζ phosphorylation (53). Based on these findings, we hypothesize that crosslinking of SLAMF6 by anti-SLAMF6 mAb may cause high phosphorylation in its ITSM. Subsequently, protein tyrosine phosphatases and lipid phosphatases are preferentially recruited to SLAMF6, particularly in SAP and EAT-2 negative B cells. Surprisingly, we found that treatment of WT mice with anti-SLAMF6 almost recapitulates the phenotype observed in SAP−/− mice, which showed a marked defect in Tfh cell and GC B cell formation and reduced antibody production and affinity maturation. Interestingly, a significant defect in humoral response was not observed when mice were treated with anti-SLAMF6 four days after antigen exposure. This further indicates that SLAMF6-mediated inhibitory signals have distinct roles in the early differentiation of Tfh cells and GC B cells. However, the administration of anti-SLAMF6 F(ab’)2 fragments did not fully suppress NP-specific antibody production even though the development of Tfh cells and GC B cells was significantly impaired (Figure 10). This difference may be due to a shorter half-life of anti-SLAMF6 F(ab’)2, which could prevent it from maintaining sustainable triggering of SLAMF6 receptors during in vivo immune responses. Alternatively, the suppressive effect of anti-SLAMF6 mAb on humoral response might depend on the ability of anti-SLAMF6 Fc to bind to other accessory cells and crosslink the SLAMF6 receptors on Tfh cells and GC B cells. Such crosslinking is necessary for many surface molecules to initiate signaling events. Clearly, further studies are required to determine the contribution of other mechanisms such as complement and non-NK cell-mediated cytotoxicity in suppressing the activity of anti-SLAMF6 mAb. Compared to anti-SLAMF6 mAb, anti-SLAMF1 mAb has a milder, yet consistent, negative effect on antibody production, but anti-SLAMF5 mAb does not show any impact on humoral response. It seems contradictory to the functional redundancy we observed in Slamf[1 + 5 + 6]−/− mice. In fact, SLAMF6 has been shown to initiate dominant signaling in SAP−/− mice because ablation of the Slamf6 gene, but not the Slamf1 gene or Slamf5 gene, can rescue Tfh cell differentiation and antibody responses in SAP−/− mice (27, 44). This indicates that SLAMF6 can complement deficiency of either SLAMF1 or SLAMF5 to facilitate inhibitory signaling in the absence of SAP. However, if Slamf1 and Slamf5 double mutations can be introduced into SAP−/− mice, impaired humoral responses may be partially restored. Similarly, the injection of mice with both anti-SLAMF1 mAb and anti-SLAMF5 mAb may also cause some reduction in antibody production.
By comparing T-dependent and T-independent antigen responses in Slamf5−/−, Slamf[1 + 6]−/−, and Slamf[1 + 5 + 6]−/− mice, we demonstrated for first time that SLAMF1, SLAMF5, and SLAMF6 synergistically regulate humoral immune responses. Genetic interruption of SLAMF1, SLAMF5, and SLAMF6 results in enhanced antibody responses to T-dependent and T-independent antigens. In complementary studies, the administration of anti-SLAMF6 mAb further implicates SLAMF6 as a primary inhibitory member of the SLAMF receptors in antibody responses. The studies suggest that the ligation of SLAMF receptors in SAP-negative B cells (Figure 11) may preferentially recruit inhibitory signaling molecules to immunological synapses and control B cell responses during cognate interaction between T and B cells.
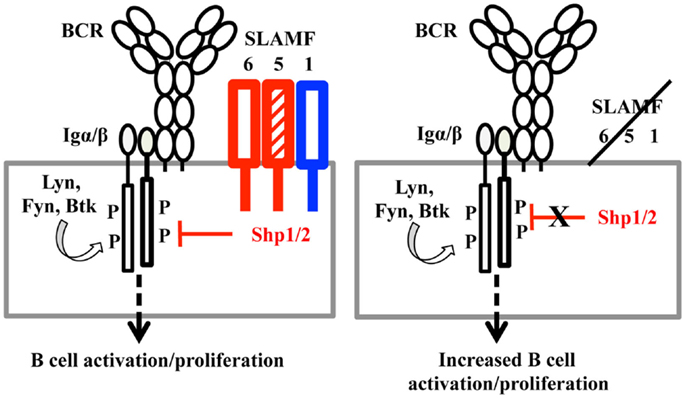
Figure 11. A model of SLAMF receptors: SHP1/2 action on the BCR during B cell activation. When B cells are activated, the ITSMs of SLAMF1, SLAMF5, and SLAMF6 recruit SHP1/2 and translocate these phosphatases to the vicinity of the B cell antigen receptor. Signaling from the BCR is thus down-regulated, maintaining proper response to antigens in humoral responses. When SLAMF1, SLAMF5, and SLAMF6 are deleted from B cells, inhibitory signaling mediated by SLAMF and SHP1/2 is dampened, which induces enhanced humoral responses.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Terhorst Lab for helpful discussions. This work was supported by the National Institutes of Health (PO1 AI-065687).
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2015.00158
References
1. Rajewsky K. Clonal selection and learning in the antibody system. Nature (1996) 381:751–8. doi: 10.1038/381751a0
2. Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell (2010) 143:592–605. doi:10.1016/j.cell.2010.10.032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. Vinuesa CG, Linterman MA, Goodnow CC, Randall KL. T cells and follicular dendritic cells in germinal center B-cell formation and selection. Immunol Rev (2010) 237:72–89. doi:10.1111/j.1600-065X.2010.00937.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Crotty S. Follicular helper CD4 T cells (TFH). Annu Rev Immunol (2011) 29:621–63. doi:10.1146/annurev-immunol-031210-101400
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol (2012) 30:429–57. doi:10.1146/annurev-immunol-020711-075032
6. Ramiscal RR, Vinuesa CG. T-cell subsets in the germinal center. Immunol Rev (2013) 252:146–55. doi:10.1111/imr.12031
7. Johnston RJ, Poholek AC, Ditoro D, Yusuf I, Eto D, Barnett B, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science (2009) 325:1006–10. doi:10.1126/science.1175870
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, et al. Bcl6 mediates the development of T follicular helper cells. Science (2009) 325:1001–5. doi:10.1126/science.1176676
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity (2009) 31:457–68. doi:10.1016/j.immuni.2009.07.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Haynes NM, Allen CD, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol (2007) 179:5099–108. doi:10.4049/jimmunol.179.8.5099
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Glatman Zaretsky A, Taylor JJ, King IL, Marshall FA, Mohrs M, Pearce EJ. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. J Exp Med (2009) 206:991–9. doi:10.1084/jem.20090303
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Calpe S, Wang N, Romero X, Berger SB, Lanyi A, Engel P, et al. The SLAM and SAP gene families control innate and adaptive immune responses. Adv Immunol (2008) 97:177–250. doi:10.1016/S0065-2776(08)00004-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol (2011) 29:665–705. doi:10.1146/annurev-immunol-030409-101302
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Detre C, Keszei M, Garrido-Mesa N, Kis-Toth K, Castro W, Agyemang AF, et al. SAP expression in invariant NKT cells is required for cognate help to support B-cell responses. Blood (2012) 120:122–9. doi:10.1182/blood-2011-11-395913
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, et al. SAP couples Fyn to SLAM immune receptors. Nat Cell Biol (2003) 5:155–60. doi:10.1038/ncb920
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Veillette A. SLAM-family receptors: immune regulators with or without SAP-family adaptors. Cold Spring Harb Perspect Biol (2010) 2:a002469. doi:10.1101/cshperspect.a002469
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Aoukaty A, Tan R. Association of the X-linked lymphoproliferative disease gene product SAP/SH2D1A with 2B4, a natural killer cell-activating molecule, is dependent on phosphoinositide 3-kinase. J Biol Chem (2002) 277:13331–7. doi:10.1074/jbc.M112029200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Cruz-Munoz ME, Dong Z, Shi X, Zhang S, Veillette A. Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function. Nat Immunol (2009) 10:297–305. doi:10.1038/ni.1693
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Wang N, Calpe S, Westcott J, Castro W, Ma C, Engel P, et al. Cutting edge: the adapters EAT-2A and -2B are positive regulators of CD244- and CD84-dependent NK cell functions in the C57BL/6 mouse. J Immunol (2010) 185:5683–7. doi:10.4049/jimmunol.1001974
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Perez-Quintero LA, Roncagalli R, Guo H, Latour S, Davidson D, Veillette A. EAT-2, a SAP-like adaptor, controls NK cell activation through phospholipase Cgamma, Ca++, and Erk, leading to granule polarization. J Exp Med (2014) 211:727–42. doi:10.1084/jem.20132038
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Wilson TJ, Garner LI, Metcalfe C, King E, Margraf S, Brown MH. Fine specificity and molecular competition in SLAM family receptor signalling. PLoS One (2014) 9:e92184. doi:10.1371/journal.pone.0092184
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Tangye SG, Lazetic S, Woollatt E, Sutherland GR, Lanier LL, Phillips JH. Cutting edge: human 2B4, an activating NK cell receptor, recruits the protein tyrosine phosphatase SHP-2 and the adaptor signaling protein SAP. J Immunol (1999) 162:6981–5.
23. Parolini S, Bottino C, Falco M, Augugliaro R, Giliani S, Franceschini R, et al. X-linked lymphoproliferative disease. 2B4 molecules displaying inhibitory rather than activating function are responsible for the inability of natural killer cells to kill Epstein-Barr virus-infected cells. J Exp Med (2000) 192:337–46. doi:10.1084/jem.192.3.337
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Morra M, Lu J, Poy F, Martin M, Sayos J, Calpe S, et al. Structural basis for the interaction of the free SH2 domain EAT-2 with SLAM receptors in hematopoietic cells. EMBO J (2001) 20:5840–52. doi:10.1093/emboj/20.21.5840
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Howie D, Simarro M, Sayos J, Guirado M, Sancho J, Terhorst C. Molecular dissection of the signaling and costimulatory functions of CD150 (SLAM): CD150/SAP binding and CD150-mediated costimulation. Blood (2002) 99:957–65. doi:10.1182/blood.V99.3.957
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Dong Z, Cruz-Munoz ME, Zhong MC, Chen R, Latour S, Veillette A. Essential function for SAP family adaptors in the surveillance of hematopoietic cells by natural killer cells. Nat Immunol (2009) 10:973–80. doi:10.1038/ni.1763
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, et al. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity (2012) 36:986–1002. doi:10.1016/j.immuni.2012.05.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Zhao F, Cannons JL, Dutta M, Griffiths GM, Schwartzberg PL. Positive and negative signaling through SLAM receptors regulate synapse organization and thresholds of cytolysis. Immunity (2012) 36:1003–16. doi:10.1016/j.immuni.2012.05.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Coffey AJ, Brooksbank RA, Brandau O, Oohashi T, Howell GR, Bye JM, et al. Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nat Genet (1998) 20:129–35. doi:10.1038/2424
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Nichols KE, Harkin DP, Levitz S, Krainer M, Kolquist KA, Genovese C, et al. Inactivating mutations in an SH2 domain-encoding gene in X-linked lymphoproliferative syndrome. Proc Natl Acad Sci U S A (1998) 95:13765–70. doi:10.1073/pnas.95.23.13765
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, et al. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature (1998) 395:462–9. doi:10.1038/26683
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Crotty S, Kersh EN, Cannons J, Schwartzberg PL, Ahmed R. SAP is required for generating long-term humoral immunity. Nature (2003) 421:282–7. doi:10.1038/nature01318
33. Hron JD, Caplan L, Gerth AJ, Schwartzberg PL, Peng SL. SH2D1A regulates T-dependent humoral autoimmunity. J Exp Med (2004) 200:261–6. doi:10.1084/jem.20040526
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Morra M, Barrington RA, Abadia-Molina AC, Okamoto S, Julien A, Gullo C, et al. Defective B cell responses in the absence of SH2D1A. Proc Natl Acad Sci U S A (2005) 102:4819–23. doi:10.1073/pnas.0408681102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Cannons JL, Yu LJ, Jankovic D, Crotty S, Horai R, Kirby M, et al. SAP regulates T cell-mediated help for humoral immunity by a mechanism distinct from cytokine regulation. J Exp Med (2006) 203:1551–65. doi:10.1084/jem.20052097
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Kamperschroer C, Roberts DM, Zhang Y, Weng NP, Swain SL. SAP enables T cells to help B cells by a mechanism distinct from Th cell programming or CD40 ligand regulation. J Immunol (2008) 181:3994–4003. doi:10.4049/jimmunol.181.6.3994
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Wu C, Nguyen KB, Pien GC, Wang N, Gullo C, Howie D, et al. SAP controls T cell responses to virus and terminal differentiation of TH2 cells. Nat Immunol (2001) 2:410–4. doi:10.1038/ni0901-823
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Davidson D, Shi X, Zhang S, Wang H, Nemer M, Ono N, et al. Genetic evidence linking SAP, the X-linked lymphoproliferative gene product, to Src-related kinase FynT in T(H)2 cytokine regulation. Immunity (2004) 21:707–17. doi:10.1016/j.immuni.2004.10.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Wang N, Satoskar A, Faubion W, Howie D, Okamoto S, Feske S, et al. The cell surface receptor SLAM controls T cell and macrophage functions. J Exp Med (2004) 199:1255–64. doi:10.1084/jem.20031835
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. McCausland MM, Yusuf I, Tran H, Ono N, Yanagi Y, Crotty S. SAP regulation of follicular helper CD4 T cell development and humoral immunity is independent of SLAM and Fyn kinase. J Immunol (2007) 178:817–28. doi:10.4049/jimmunol.178.2.817
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature (2008) 455:764–9. doi:10.1038/nature07345
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Graham DB, Bell MP, Mccausland MM, Huntoon CJ, Van Deursen J, Faubion WA, et al. Ly9 (CD229)-deficient mice exhibit T cell defects yet do not share several phenotypic characteristics associated with SLAM- and SAP-deficient mice. J Immunol (2006) 176:291–300. doi:10.4049/jimmunol.176.1.291
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Cannons JL, Qi H, Lu KT, Dutta M, Gomez-Rodriguez J, Cheng J, et al. Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity (2010) 32:253–65. doi:10.1016/j.immuni.2010.01.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, et al. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J Immunol (2010) 185:190–202. doi:10.4049/jimmunol.0903505
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity (2007) 27:751–62. doi:10.1016/j.immuni.2007.08.020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. De Calisto J, Wang N, Wang G, Yigit B, Engel P, Terhorst C. SAP-dependent and -independent regulation of innate T cell development involving SLAMF receptors. Front Immunol (2014) 5:186. doi:10.3389/fimmu.2014.00186
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Sintes J, Romero X, De Salort J, Terhorst C, Engel P. Mouse CD84 is a pan-leukocyte cell-surface molecule that modulates LPS-induced cytokine secretion by macrophages. J Leukoc Biol (2010) 88:687–97. doi:10.1189/jlb.1109756
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
48. Qi H. From SAP-less T cells to helpless B cells and back: dynamic T-B cell interactions underlie germinal center development and function. Immunol Rev (2012) 247:24–35. doi:10.1111/j.1600-065X.2012.01119.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med (2011) 17:975–82. doi:10.1038/nm.2425
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Sage PT, Francisco LM, Carman CV, Sharpe AH. The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat Immunol (2013) 14:152–61. doi:10.1038/ni.2496
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nat Immunol (2010) 11:681–8. doi:10.1038/ni.1900
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Ma CS, Nichols KE, Tangye SG. Regulation of cellular and humoral immune responses by the SLAM and SAP families of molecules. Annu Rev Immunol (2007) 25:337–79. doi:10.1146/annurev.immunol.25.022106.141651
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Chu C, Wang Y, Zhang X, Ni X, Cao J, Xu W, et al. SAP-regulated T Cell-APC adhesion and ligation-dependent and -independent Ly108-CD3zeta interactions. J Immunol (2014) 193:3860–71. doi:10.4049/jimmunol.1401660
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Wang N, Wang B, Salio M, Allen D, She J, Terhorst C. Expression of a CD3 epsilon transgene in CD3 epsilon(null) mice does not restore CD3 gamma and delta expression but efficiently rescues T cell development from a subpopulation of prothymocytes. Int Immunol (1998) 10:1777–88. doi:10.1093/intimm/10.12.1777
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Veillette A, Dong Z, Perez-Quintero LA, Zhong MC, Cruz-Munoz ME. Importance and mechanism of “switch” function of SAP family adapters. Immunol Rev (2009) 232:229–39. doi:10.1111/j.1600-065X.2009.00824.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Minges Wols HA, Underhill GH, Kansas GS, Witte PL. The role of bone marrow-derived stromal cells in the maintenance of plasma cell longevity. J Immunol (2002) 169:4213–21. doi:10.4049/jimmunol.169.8.4213
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Cariappa A, Tang M, Parng C, Nebelitskiy E, Carroll M, Georgopoulos K, et al. The follicular versus marginal zone B lymphocyte cell fate decision is regulated by Aiolos, Btk, and CD21. Immunity (2001) 14:603–15. doi:10.1016/S1074-7613(01)00135-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Casola S, Otipoby KL, Alimzhanov M, Humme S, Uyttersprot N, Kutok JL, et al. B cell receptor signal strength determines B cell fate. Nat Immunol (2004) 5:317–27. doi:10.1038/ni1036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Pillai S, Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nat Rev Immunol (2009) 9:767–77. doi:10.1038/nri2656
60. Martin F, Oliver AM, Kearney JF. Marginal zone and B1 B cells unite in the early response against T-independent blood-borne particulate antigens. Immunity (2001) 14:617–29. doi:10.1016/S1074-7613(01)00129-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Vinuesa CG, Chang PP. Innate B cell helpers reveal novel types of antibody responses. Nat Immunol (2013) 14:119–26. doi:10.1038/ni.2511
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: SLAM family receptors, SLAM-associated protein, follicular T helper cells, germinal center B cells, marginal zone B cells, anti-SLAMF6 mAb
Citation: Wang N, Halibozek PJ, Yigit B, Zhao H, O’Keeffe MS, Sage P, Sharpe A and Terhorst C (2015) Negative regulation of humoral immunity due to interplay between the SLAMF1, SLAMF5, and SLAMF6 receptors. Front. Immunol. 6:158. doi: 10.3389/fimmu.2015.00158
Received: 12 February 2015; Paper pending published: 11 March 2015;
Accepted: 23 March 2015; Published online: 14 April 2015.
Edited by:
Michael Sitkovsky, Northeastern University, USAReviewed by:
Christopher E. Rudd, University of Cambridge, UKAntónio Gil Castro, University of Minho, Portugal
Fabio Malavasi, University of Turin Medical School, Italy
Copyright: © 2015 Wang, Halibozek, Yigit, Zhao, O’Keeffe, Sage, Sharpe and Terhorst. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ninghai Wang and Cox Terhorst, Division of Immunology, Beth Israel Deaconess Medical Center, Harvard Medical School, 3 Blackfan Circle, Boston, MA 02115, USA e-mail: nwang@bidmc.harvard.edu; cterhors@bidmc.harvard.edu
 Ninghai Wang
Ninghai Wang Peter J. Halibozek
Peter J. Halibozek Burcu Yigit
Burcu Yigit Hui Zhao1
Hui Zhao1 Michael S. O’Keeffe
Michael S. O’Keeffe Arlene Sharpe
Arlene Sharpe Cox Terhorst
Cox Terhorst