- Viral Immunobiology, Institute of Experimental Immunology, University of Zürich, Zurich, Switzerland
Macroautophagy delivers cytoplasmic constituents for lysosomal degradation. Because MHC class II molecules are loaded with lysosomal products for CD4+ T-cell stimulation, macroautophagy supports intracellular antigen processing onto MHC class II molecules. The molecular machinery of macroautophagy, however, does not only support this autophagic antigen processing, but seems to also modify extracellular antigen uptake for MHC class II presentation, antigen exocytosis, and packaging for improved cross-presentation onto MHC class I molecules. The different membrane trafficking pathways with LC3-associated phagocytosis, compartment for unconventional protein secretion, and DRibbles as well as the role that autophagic proteins play in them will be discussed in this review.
Introduction
Autophagy describes several pathways, by which cytoplasmic constituents gain access to lysosomal degradation (1). During one of these autophagic pathways, called macroautophagy, a dedicated vesicle, the autophagosome, is formed, which engulfs with two membranes its cytoplasmic cargo and then fuses with lysosomes for degradation of its inner autophagosomal membrane and its contents.
The Molecular Machinery of Macroautophagy
The extensive membrane remodeling events that first generate the cup-shaped isolation membrane, which then closes to autophagosomes and gets directed to fusion with late endosomes or lysosomes, require >30 essential proteins, which were termed autophagy-related gene (Atg) products. They are grouped in distinct complexes (ULK1 complex, Atg9L, the class III PI3K complex, the WIPI–Atg2 complex, the Atg12 conjugation complex, and the Atg8 conjugation complex). The Atg1/ULK1 complex (ULK1/2, Atg13, Atg101, and FIP200) is under metabolic control and can be activated by AMP activated protein kinase (AMPK) and inhibited by target of rapamycin (TOR), both through phosphorylation. The ULK1 complex induces macroautophagy during nutrient depletion to recycle cellular constituents for survival. This constitutes the canonical function of macroautophagy. The ULK1 complex in turn phosphorylates members of the class III PI3K complex (Vps34, Atg14L, Beclin-1, and Vps15), including Atg6/Beclin-1, which activates phosphatidylinositol 3-phosphate (PI3P) deposition at membrane sites like the ER, where autophagosomes are generated. Atg9L most likely recruits membranes to these sites after phosphorylation by the ULK1 complex. The PI3P marks recruit WIPI–Atg2 complexes to these membrane sites, which function as landing platforms for the Atg12–Atg5/Atg16L1 complexes via direct binding of Atg16L1 to WIPI2. These interactions lead to autophagosome initiation.
For autophagosome elongation, the Atg12–Atg5/Atg16L1 complexes assemble after ubiquitin-like coupling of Atg12–Atg5 via the E1- and E2-like enzymes, Atg7 and Atg10, and binding of Atg16L1 to the resulting conjugate. The Atg12–Atg5/Atg16L1 complex fulfills the E3-like enzymatic activity for the other ubiquitin-like system that couples Atg8 to phosphatidylethanolamine (PE) in the assembling autophagosome membrane called the phagophore or isolation membrane. Prior to conjugation five amino acids are removed from the C-terminus of Atg8 by the protease Atg4, and Atg8 is then activated and conjugated by Atg7 and Atg3. Atg8 has eight mammalian orthologs (two splice variants of LC3A, LC3B, LC3C, GABARAP, and GABARAPL1-3). The PE coupled forms of these Atg8 orthologs can be distinguished from their cytosolic form by SDS gel electrophoresis. LC3B-PE is assessed most often for this purpose and migrates faster (LC3B-II) than its cytosolic form (LC3B-I). These autophagic protein tags are conjugated to the outer and inner membrane of the developing autophagosome and seem to mediate membrane fusion events during phagophore elongation and recruit substrates into autophagosomes via LC3 interacting regions (LIRs). Once the autophagosome is completed the Atg12–Atg5/Atg16L1 complex dissociates and Atg4 also cleaves Atg8 from the outer membrane. These molecular events lead to autophagosome completion.
For autophagosome maturation, however, residual Atg8 at the outer autophagosome membrane might facilitate migration along microtubules and fusion with lysosomes via recruitment of FYVE and coiled coil domain containing 1 (FYCO1) (2) and pleckstrin homology domain containing protein family member 1 (PLEKHM1) proteins (3). Alternatively, FYCO1 and PLEKHM1 could also be recruited by the lipid composition of the outer autophagosomal membrane, including PI3P. PLEKHM1 in turn seems to recruit the homotypic fusion and vacuole protein sorting (HOPS) complex, the GTPase Rab7 that is involved in late endosome and autophagosome fusion with lysosomes, and even the SNARE syntaxin 17 (STX17), which executes outer autophagosome membrane fusion with lysosomes. Autophagosome fusion with lysosomes is again regulated by type III PI3K complexes containing Vps34, Beclin-1, and VPS15. PI3K complexes containing Rubicon inhibit, whereas UVRAG containing complexes accelerate autophagosome fusion with lysosomes. These findings indicate that some components of the fusion machinery are also recruited by PI3P to the autophagosomal membrane. Thus, macroautophagy utilizes a sophisticated membrane remodeling and fusion machinery, which recently has been found to support other membrane trafficking events. This involvement of Atg proteins in endocytosis and exocytosis for antigen processing toward MHC restricted antigen presentation will be discussed in this review.
LC3-Associated Phagocytosis
MHC class I and II molecules present peptide products of proteasomal and lysosomal proteolysis to CD8+ cytotoxic and CD4+ helper T-cells, respectively (4). Classically, endocytosed material is delivered to lysosomes and its fragments are then presented on MHC class II molecules. One prominent endocytic pathway is phagocytosis and Atg proteins have been found to regulate it during LC3-associated phagocytosis (LAP) (5) (Figure 1). During LAP, its cargo needs to engage a subset of phagocytic and/or pathogen-associated molecular pattern (PAMP) receptors to recruit NADPH oxidase 2 (NOX2) to the phagosomal membrane (6, 7). These receptors can be members of the toll-like receptor (TLR) family, primarily TLR2, or the C-type lectin Dectin-1, the T-cell immunoglobulin mucin protein 4 (TIM4) or Fc receptors for immunoglobulins (5, 7–10). NOX2 either allows coupling of LC3 to phagosomes or prevents its uncoupling by Atg4, and this LC3 lipidation is independent of the Atg1/ULK1 complex (8). Irrespective of the exact mechanism, LC3 is coupled to the cytosolic side of the limiting phagosomal membrane (7) and influences the fate of these LAP phagosomes. In some cell types, primarily mouse macrophages, LAP phagosomes seem to fuse more rapidly than LC3 negative phagosomes with lysosomes (5, 10, 11), whereas in human macrophages, conventional and plasmacytoid dendritic cells (DCs), LAP vesicles seem to be stabilized and can fuse with TLR containing endosomes (7, 12). Acceleration of LAP phagosome fusion with lysosomes might be because of more efficient transport along microtubules via LC3 binding to FYVE and coiled-coil domain containing 1 (FYCO1) protein (11). Stabilized LAP phagosomes do not recruit Rab7 and do not seem to contain classical autophagy cargo like the LIR-positive sequestosome 1/p62 protein (7). Prior to delayed fusion of LAP vesicles with lysosomes, LC3 is uncoupled from the phagosomal membrane (7). This delayed phagosomal processing might be beneficial for continuous MHC class II antigen presentation of phagocytosed antigens. Indeed, LAP formation was required for prolonged antigen processing of Candida antigens for MHC class II presentation to specific human CD4+ T-cell clones in vitro (7). Similarly, Dectin-1 bound antigens were also more efficiently presented to mouse CD4+ T-cells (9). Finally, CD4+ T-cell priming after antigen injection into mice with Atg5-deficient DCs was compromised as compared to wild-type mice, suggesting that LAP improves MHC class II presentation of exogenous antigen in vivo, in addition to the role of Atg5 in endogenous antigen processing for MHC class II presentation (13). Indeed, retention of phagocytosed antigens by LAP might account in part for more efficient antigen presentation after attenuated lysosomal degradation (14). Therefore, regulation of phagocytosis by autophagic proteins seems to enhance MHC class II antigen presentation of extracellular antigens.
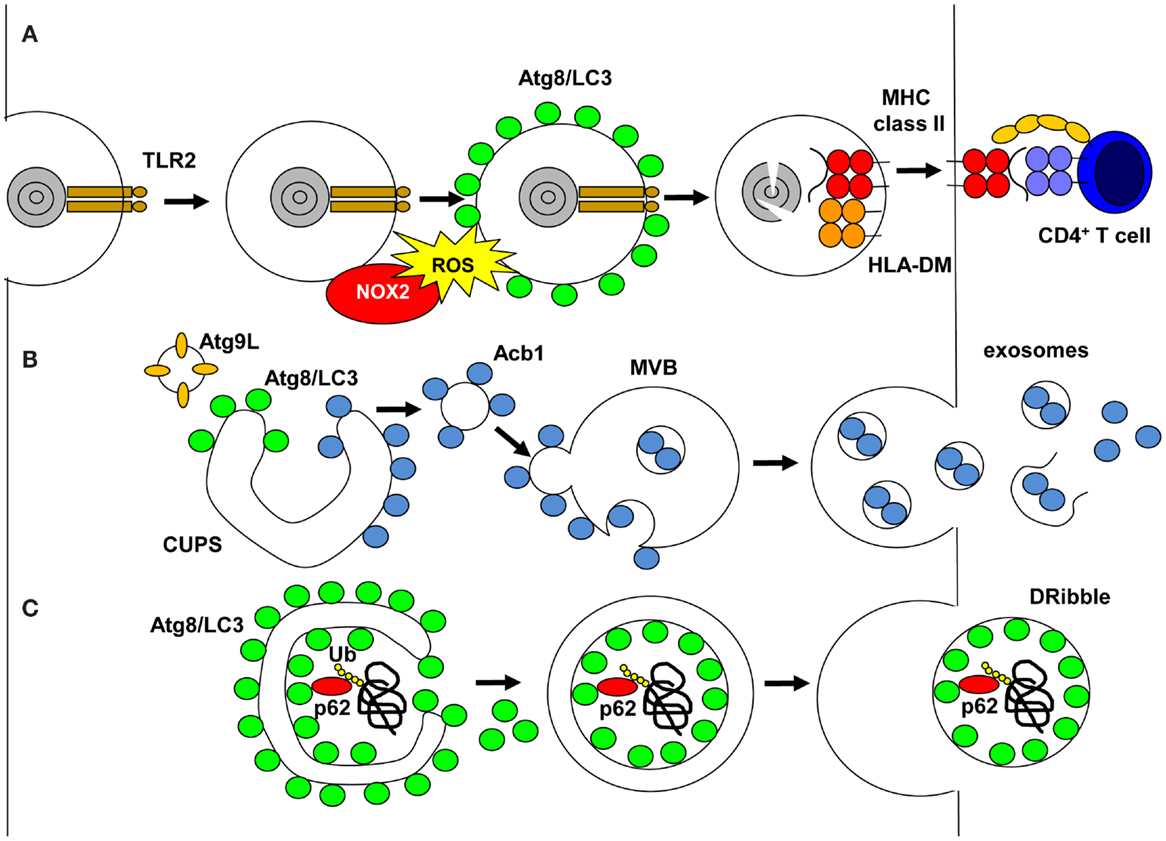
Figure 1. Unconventional pathways that use autophagic proteins. (A) LC3-associated phagocytosis (LAP) is engaged on TLR2 ligand phagocytosis, which recruits NADPH oxidase (NOX2). NOX2 produce reactive oxygen species (ROS), which are required to recruit or maintain Atg8/LC3 at the phagosomal membrane. Atg8/LC3 needs to be cleaved from the phagosomal membrane for phagosomes to fuse with MHC class II loading compartments, and their cargo is degraded by lysosomal hydrolysis and fragments loaded onto MHC class II molecules for stimulation of CD4+ T-cells. (B) During unconventional secretion of proteins without signal peptides for co-translational insertion into the ER, Atg8/LC3 and Atg9L cooperate to expand the compartment for unconventional protein secretion (CUPS). Proteins to be secreted, like acyl-CoA binding protein 1 (Acb1), bind on the cytosolic side to these membranes, then vesicles from the CUPS fuse with multivesicular bodies (MVBs). Acb1 is internalized into intravesicular membranes by invagination and then released in exosomes after MVB fusion with the cell membrane. (C) Proteasomal inhibition diverts ubiquitinated proteins, including defective ribosomal products, into autophagosomes via sequestosome 1/p62 binding to polyubiquitin and Atg8/LC3. The inner autophagosomal membrane with this cargo is released as defective ribosomal products-containing autophagosome-rich blebs (DRibbles) if lysosomal degradation is inhibited.
Compartment for Unconventional Protein Secretion
Autophagy-related gene proteins are also involved in unconventional membrane trafficking in the opposite direction of phagocytosis, namely signal peptide independent secretion (15). A number of proteins leave eukaryotes not via the classical ER to Golgi pathway and lack signal peptides for co-translational insertion into the ER. These include acyl-CoA binding protein 1 (Acb1), interleukin-1β, interleukin-33, tissue transglutaminase, galectin-3, macrophage migration inhibitory factor, and insulin-degrading enzyme (15). Recently, it was found that Acb1 and IL-1β require autophagic proteins for their secretion, namely Atg1, 5, 6, 7, 8, 9, 11, 12, and 17 (16–18). Moreover, release of microbial peptides and secretory lysosomes was also found to be dependent on this autophagic core machinery (19–21). Interestingly, the secretion of Acb1 during yeast and ameba starvation requires the formation of a membrane structure close to the ER exit site that seems to be composed of cis- and trans-Golgi membranes and is called the compartment for unconventional protein secretion (CUPS) (22, 23) (Figure 1). It contains PI3P, Atg9, Atg8, the Golgi-associated protein GRASP65 and Vps23 of the ESCRT-1 complex for multivesicular body (MVB) formation. Moreover, after starvation the CUPS is absorbed into the ER (23). Therefore, the CUPS might be a continuous membrane, which is elongated like the isolation membrane with the help of Atg proteins, but might never form a double-membrane surrounded autophagosome. Instead, the cargo for unconventional secretion seems to get attached to the cytosolic side of the membrane (15). Small Acb1-coated vesicles might then get released from CUPS and fuse with an endosomal compartment, on the surface of which they require the ESCRT-1 complex for invagination of the endosomal membrane to generate MVBs. These MVBs fuse then with the cell membrane to release exosomes that contain the unconventionally secreted substrate. Indeed, the surface membrane SNARE Sso1 has been found to be required for unconventional Acb1 secretion (17). Moreover, part of the IL-1β released from activated macrophages is membrane engulfed and therefore protease resistant (24). This unconventional secretion pathway could also give rise to immunostimulatory exosomes, which have been proposed for efficient antigen delivery vesicles that already contain antigenic peptide loaded MHC molecules (25).
Defective Ribosomal Products-Containing Autophagosome-Rich Blebs
Such exosomal fractions might be further modified by autophagosomes, which are not degraded in lysosomes. Indeed, loss of Atg5, Atg6/Beclin-1, or Atg12 compromised tumor and influenza A virus-infected cells to donate antigens for efficient cross-presentation (26, 27). Ubiquitinated proteins and defective ribosomal products, including viral and tumor antigens, were enriched in vesicular fractions of cell supernatants after blocking proteasomal and lysosomal degradation (28) (Figure 1). In the presence of proteasomal inhibition, ubiquitinated substrates of the proteasome seem to aggregate and get incorporated into autophagosomes via sequestosome1/p62, a protein that anchors ubiquitinated proteins to LC3 for autophagic substrate recruitment (29). Blocking lysosomal degradation might then generate autophagolysosomes with MVB characteristics, from which the inner autophagosomal membranes and their cargo might be released into the vesicular supernatant fraction. Similar pathways of arrested macroautophagy might be used by viruses to exit cells (30–33). These vesicular fractions were coined defective ribosomal products-containing autophagosome-rich blebs (DRibbles) and can be endocytosed in a CLEC9A-dependent fashion by antigen presenting cells (28). DRibbles seem to be more efficient for cross-presentation on MHC class I molecules to CD8+ T-cells than cell lysates and soluble antigens (34). DRibbles were similarly processed for MHC class II presentation to CD4+ T-cells like whole cell lysates, but better than soluble viral antigens. The more efficient MHC class I cross-presentation could be further enhanced in human peripheral blood mononuclear cells by GM-CSF and TLR3 agonist addition (34). Furthermore, DRibbles immunization of mice protected from challenge with melanoma, lung carcinoma, breast carcinoma, and sarcoma cell lines (29, 35). Thus, frustrated macroautophagy of proteasomal substrates might lead to secretion of DRibbles, which could be a superior antigen format for cross-presentation on MHC class I molecules.
Conclusion
LAP, CUPS, and DRibbles bear witness to non-canonical functions of Atg proteins. They have in common with macroautophagy that they regulate membrane trafficking and cargo delivery to topologically extracellular compartments (endosomes or cell supernatants during endocytosis and exocytosis, respectively). Atg proteins might facilitate membrane fusion events and recruit substrates and adaptor proteins during the trafficking of LAP- and CUPS-derived vesicles as well as during DRibbles secretion. A better understanding of these non-canonical Atg pathways should provide insights into how groups of Atg proteins can be used by different cellular pathways in a modular fashion to achieve vesicle elongation, fusion with other vesicles and substrate recruitment. This knowledge would then also provide a better understanding of macroautophagy itself, the pathway that defined Atgs. Because MHC-restricted antigen presentation relies on cellular trafficking events to produce antigenic fragments, load them into MHC peptide binding grooves, and then transport the products to the cell surface for T-cell recognition, interference with the various Atg-assisted pathways undoubtedly modulates T-cell recognition of antigens and these pathways should be harnessed during vaccination.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported grants from Cancer Research Switzerland (KFS-3234-08-2013), Worldwide Cancer Research (14-1033), KFSPMS and KFSPHHLD of the University of Zurich, the Baugarten Foundation, the Sobek Foundation, Fondation Acteria, the Swiss Vaccine Research Institute, and the Swiss National Science Foundation (310030_143979 and CRSII3_136241).
References
1. Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol (2011) 27:107–32. doi: 10.1146/annurev-cellbio-092910-154005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, et al. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol (2010) 188:253–69. doi:10.1083/jcb.200907015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell (2015) 57:39–54. doi:10.1016/j.molcel.2014.11.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol (2013) 31:443–73. doi:10.1146/annurev-immunol-032712-095910
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature (2007) 450:1253–7. doi:10.1038/nature06421
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, et al. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A (2009) 106:6226–31. doi:10.1073/pnas.0811045106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Romao S, Gasser N, Becker AC, Guhl B, Bagajic M, Vanoaica LD, et al. Essential autophagy proteins stabilize pathogen containing phagosomes for prolonged MHC class II antigen processing. J Cell Biol (2013) 203:757–66. doi:10.1083/jcb.201308173
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol (2011) 13:1335–43. doi:10.1038/ncb2363
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Ma J, Becker C, Lowell CA, Underhill DM. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J Biol Chem (2012) 287:34149–56. doi:10.1074/jbc.M112.382812
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A (2011) 108:17396–401. doi:10.1073/pnas.1113421108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Ma J, Becker C, Reyes C, Underhill DM. Cutting edge: FYCO1 recruitment to dectin-1 phagosomes is accelerated by light chain 3 protein and regulates phagosome maturation and reactive oxygen production. J Immunol (2014) 192:1356–60. doi:10.4049/jimmunol.1302835
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity (2012) 37:986–97. doi:10.1016/j.immuni.2012.09.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity (2010) 32:227–39. doi:10.1016/j.immuni.2009.12.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Delamarre L, Pack M, Chang H, Mellman I, Trombetta ES. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science (2005) 307:1630–4. doi:10.1126/science.1108003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Malhotra V. Unconventional protein secretion: an evolving mechanism. EMBO J (2013) 32:1660–4. doi:10.1038/emboj.2013.104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J (2011) 30:4701–11. doi:10.1038/emboj.2011.398
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol (2010) 188:527–36. doi:10.1083/jcb.200911154
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Manjithaya R, Anjard C, Loomis WF, Subramani S. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol (2010) 188:537–46. doi:10.1083/jcb.200911149
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature (2008) 456(7219):259–63. doi:10.1038/nature07416
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell (2010) 141:1135–45. doi:10.1016/j.cell.2010.05.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. DeSelm CJ, Miller BC, Zou W, Beatty WL, van Meel E, Takahata Y, et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell (2011) 21:966–74. doi:10.1016/j.devcel.2011.08.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Bruns C, McCaffery JM, Curwin AJ, Duran JM, Malhotra V. Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion. J Cell Biol (2011) 195:979–92. doi:10.1083/jcb.201106098
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Cruz-Garcia D, Curwin AJ, Popoff JF, Bruns C, Duran JM, Malhotra V. Remodeling of secretory compartments creates CUPS during nutrient starvation. J Cell Biol (2014) 207:695–703. doi:10.1083/jcb.201407119
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Rubartelli A, Cozzolino F, Talio M, Sitia R. A novel secretory pathway for interleukin-1 beta, a protein lacking a signal sequence. EMBO J (1990) 9:1503–10.
25. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol (2014) 30:255–89. doi:10.1146/annurev-cellbio-101512-122326
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Li Y, Wang LX, Yang G, Hao F, Urba WJ, Hu HM. Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res (2008) 68:6889–95. doi:10.1158/0008-5472.CAN-08-0161
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Uhl M, Kepp O, Jusforgues-Saklani H, Vicencio JM, Kroemer G, Albert ML. Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell Death Differ (2009) 16:991–1005. doi:10.1038/cdd.2009.8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Yi Y, Zhou Z, Shu S, Fang Y, Twitty C, Hilton TL, et al. Autophagy-assisted antigen cross-presentation: autophagosome as the argo of shared tumor-specific antigens and DAMPs. Oncoimmunology (2012) 1:976–8. doi:10.4161/onci.20059
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Twitty CG, Jensen SM, Hu HM, Fox BA. Tumor-derived autophagosome vaccine: induction of cross-protective immune responses against short-lived proteins through a p62-dependent mechanism. Clin Cancer Res (2011) 17:6467–81. doi:10.1158/1078-0432.CCR-11-0812
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Jackson WT, Giddings TH Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol (2005) 3:e156. doi:10.1371/journal.pbio.0030156
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol (2009) 186:255–68. doi:10.1083/jcb.200903070
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Nowag H, Guhl B, Thriene K, Romao S, Ziegler U, Dengjel J, et al. Macroautopphagy proteins assist Epstein Barr virus production and get incorporated into the virus particles. EBio Medicine (2014) 1:116–25. doi:10.1016/j.ebiom.2014.11.007
33. Nowag H, Münz C. Diverting autophagic membranes for exocytosis. Autophagy (2015) 11:425–7. doi:10.1080/15548627.2015.1009793
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Ye W, Xing Y, Paustian C, van de Ven R, Moudgil T, Hilton TL, et al. Cross-presentation of viral antigens in dribbles leads to efficient activation of virus-specific human memory T cells. J Transl Med (2014) 12:100. doi:10.1186/1479-5876-12-100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Li Y, Wang LX, Pang P, Cui Z, Aung S, Haley D, et al. Tumor-derived autophagosome vaccine: mechanism of cross-presentation and therapeutic efficacy. Clin Cancer Res (2011) 17:7047–57. doi:10.1158/1078-0432.CCR-11-0951
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Atg, exocytosis, endocytosis, unconventional secretion, TLRs
Citation: Münz C (2015) Of LAP, CUPS, and DRibbles – unconventional use of autophagy proteins for MHC restricted antigen presentation. Front. Immunol. 6:200. doi: 10.3389/fimmu.2015.00200
Received: 12 March 2015; Accepted: 13 April 2015;
Published: 29 April 2015
Edited by:
Monique Gannagé, University of Geneva, SwitzerlandReviewed by:
Paul Roche, National Institutes of Health, USAVeronika Lukacs-Kornek, University of Saarland, Germany
Copyright: © 2015 Münz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian Münz, Viral Immunobiology, Institute of Experimental Immunology, University of Zürich, Winterthurerstrasse 190, Zürich CH-8057, Switzerland, christian.muenz@uzh.ch
 Christian Münz
Christian Münz