- 1UM2, Centre d’Immunologie de Marseille-Luminy (CIML), Aix-Marseille University, Marseille, France
- 2U1104, Institut National de la Santé et de la Recherche Médicale (INSERM), Marseille, France
- 3UMR7280, Centre National de la Recherche Scientifique (CNRS), Marseille, France
- 4Virologie et Immunologie Moléculaires UR892, Institut National de la Recherche Agronomique, Jouy-en-Josas, France
- 5INSERM U1016, Institut Cochin, Paris, France
- 6CNRS UMR8104, Paris, France
- 7Université Paris Descartes, Paris, France
- 8Assistance Publique-Hôpitaux de Paris (AP-HP), Hôpital Cochin, Paris, France
Dendritic cells (DCs) were initially defined as mononuclear phagocytes with a dendritic morphology and an exquisite efficiency for naïve T-cell activation. DC encompass several subsets initially identified by their expression of specific cell surface molecules and later shown to excel in distinct functions and to develop under the instruction of different transcription factors or cytokines. Very few cell surface molecules are expressed in a specific manner on any immune cell type. Hence, to identify cell types, the sole use of a small number of cell surface markers in classical flow cytometry can be deceiving. Moreover, the markers currently used to define mononuclear phagocyte subsets vary depending on the tissue and animal species studied and even between laboratories. This has led to confusion in the definition of DC subset identity and in their attribution of specific functions. There is a strong need to identify a rigorous and consensus way to define mononuclear phagocyte subsets, with precise guidelines potentially applicable throughout tissues and species. We will discuss the advantages, drawbacks, and complementarities of different methodologies: cell surface phenotyping, ontogeny, functional characterization, and molecular profiling. We will advocate that gene expression profiling is a very rigorous, largely unbiased and accessible method to define the identity of mononuclear phagocyte subsets, which strengthens and refines surface phenotyping. It is uniquely powerful to yield new, experimentally testable, hypotheses on the ontogeny or functions of mononuclear phagocyte subsets, their molecular regulation, and their evolutionary conservation. We propose defining cell populations based on a combination of cell surface phenotyping, expression analysis of hallmark genes, and robust functional assays, in order to reach a consensus and integrate faster the huge but scattered knowledge accumulated by different laboratories on different cell types, organs, and species.
Introduction
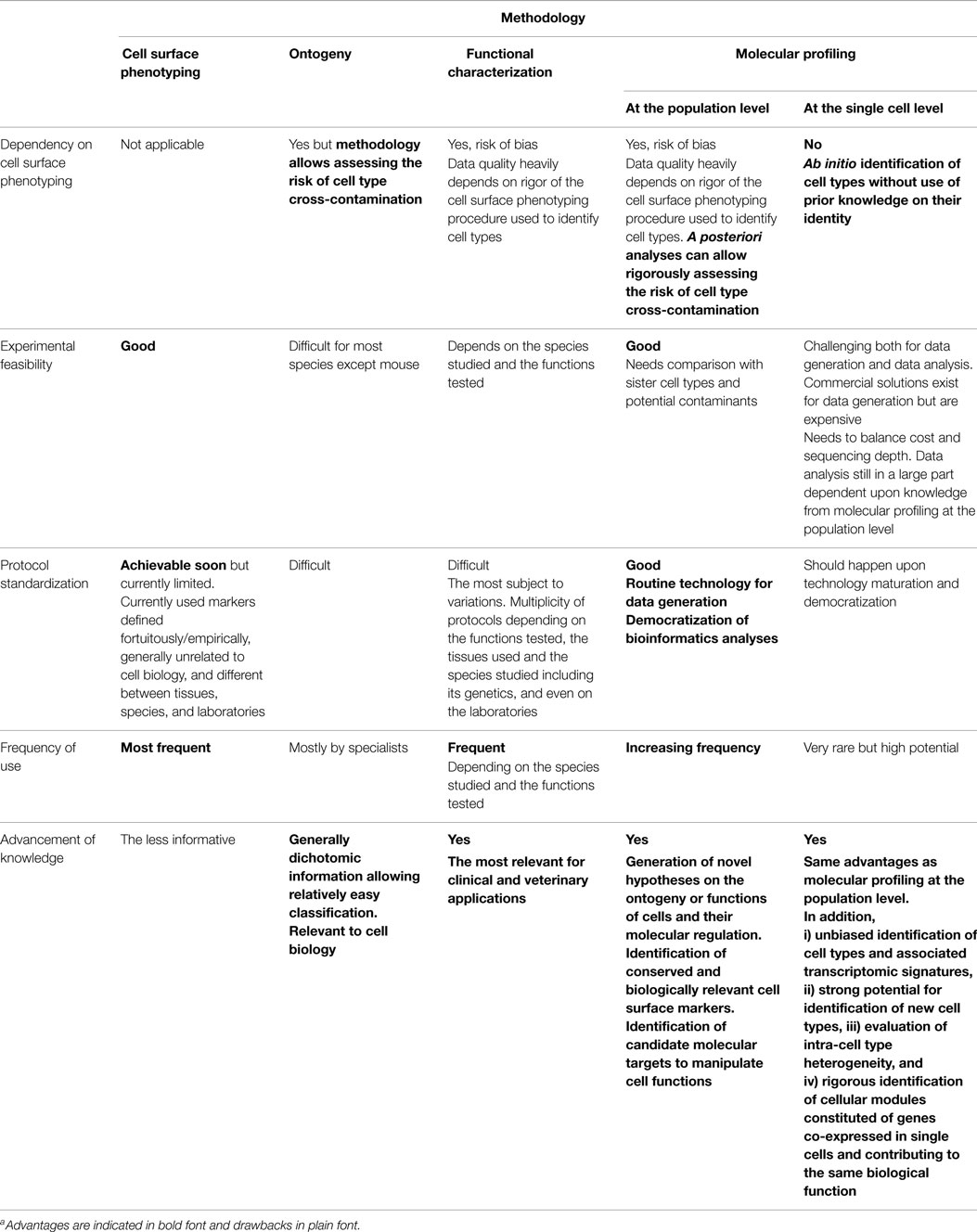
The immune system includes a large variety of myeloid and lymphoid cell types which develop through distinct ontogenic pathways, express specific phenotypes, and exert specialized functions. The mononuclear phagocytes form a complex group of myeloid cells that encompass three major cell types, i.e., monocytes, macrophages, and dendritic cells (DC), together with their proximal progenitors. These three cell types contribute to maintain host integrity by shaping the innate and adaptive immune defense, a generic function related to their common phagocytic properties and their capacity to present antigen to T cells. These functions are also shared by other types of professional antigen-presenting cells (APCs), in particular B lymphocytes. However, different types of APCs are primarily devoted to distinct functions (Figure 1). B cells produce antibodies. Monocytes patrol the organism for the detection of pathogens and dominantly display inflammatory and oxidative stress response. Macrophages mainly perform microbicidal, scavenging, and tissue trophic/maintenance functions. DC are uniquely efficient for antigen-specific activation of naïve T lymphocytes, a process called T-cell priming. Indeed, DC were initially defined by their dendritic morphology and their exquisite capacity for T-cell priming. DC include two main cell types, the plasmacytoid DC (pDC) that are expert in type I interferon synthesis upon viral stimulation and the conventional DC (cDC) that are specialized in antigen capture, processing, and presentation for T-cell priming. Two cDC subsets can be distinguished based on a further segregation of functions. XCR1+ cDC1 are particularly efficient in CD8+ T-cell activation and cross-presentation, at least in mice. XCR1− cDC2 are most efficient for T helper cell priming, in particular polarization toward Th2 or Th17, and for the promotion of humoral immunity. Importantly, an additional layer of complexity is generated by the plasticity of the different mononuclear cell types, which display modified phenotypes and functions contingent to the anatomical microenvironment where they reside or when exposed to pathogens or inflammation. For instance, monocytes adopt a dendritic morphology and antigen-presentation functions in inflammatory settings (1–3) as well as when located in the dermis (4–6), leading to their designation as monocyte-derived DC (MoDC). Langerhans cells, long considered to be DC due to their morphology and antigen-presentation function, are now known as a type of tissue macrophages (7–13). More generally, the gene expression programs, phenotypes, and functional properties of macrophages are strongly influenced by their tissue of residence. Finally, not only XCR1+ cDC but also other DC subsets including pDC and XCR1− cDC can also efficiently cross-present antigens to CD8+ T cells when appropriately stimulated (14–22). Thus, the plasticity of the mononuclear phagocyte responses superimposes onto the segregation of phenotypes and functions attributed to subsets (Figure 2), which can lead to confusion in the definition of the different cell types if only based on functional assays. Hence, morphologic, phenotypic, and functional criteria are not sufficient to rigorously define mononuclear phagocyte subsets, and to properly discriminate what are distinct cell types as opposed to different developmental or activation states of a given cell type. Complementary or robust alternative criteria are needed to rigorously define the identity of the mononuclear phagocyte subsets.
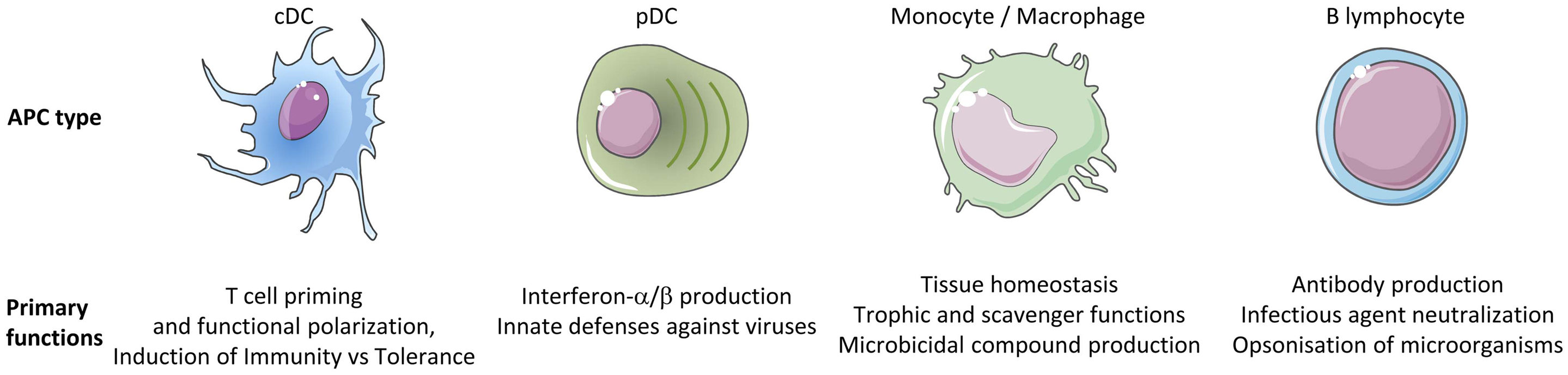
Figure 1. Different types of APCs are specialized in distinct primary functions. cDC are uniquely efficient for the priming and functional polarization of T cells. Although other APCs also contribute to this process, this does not represent their primary functions. Hence, cDC play a central and non-redundant role in the orchestration of adaptive immunity.
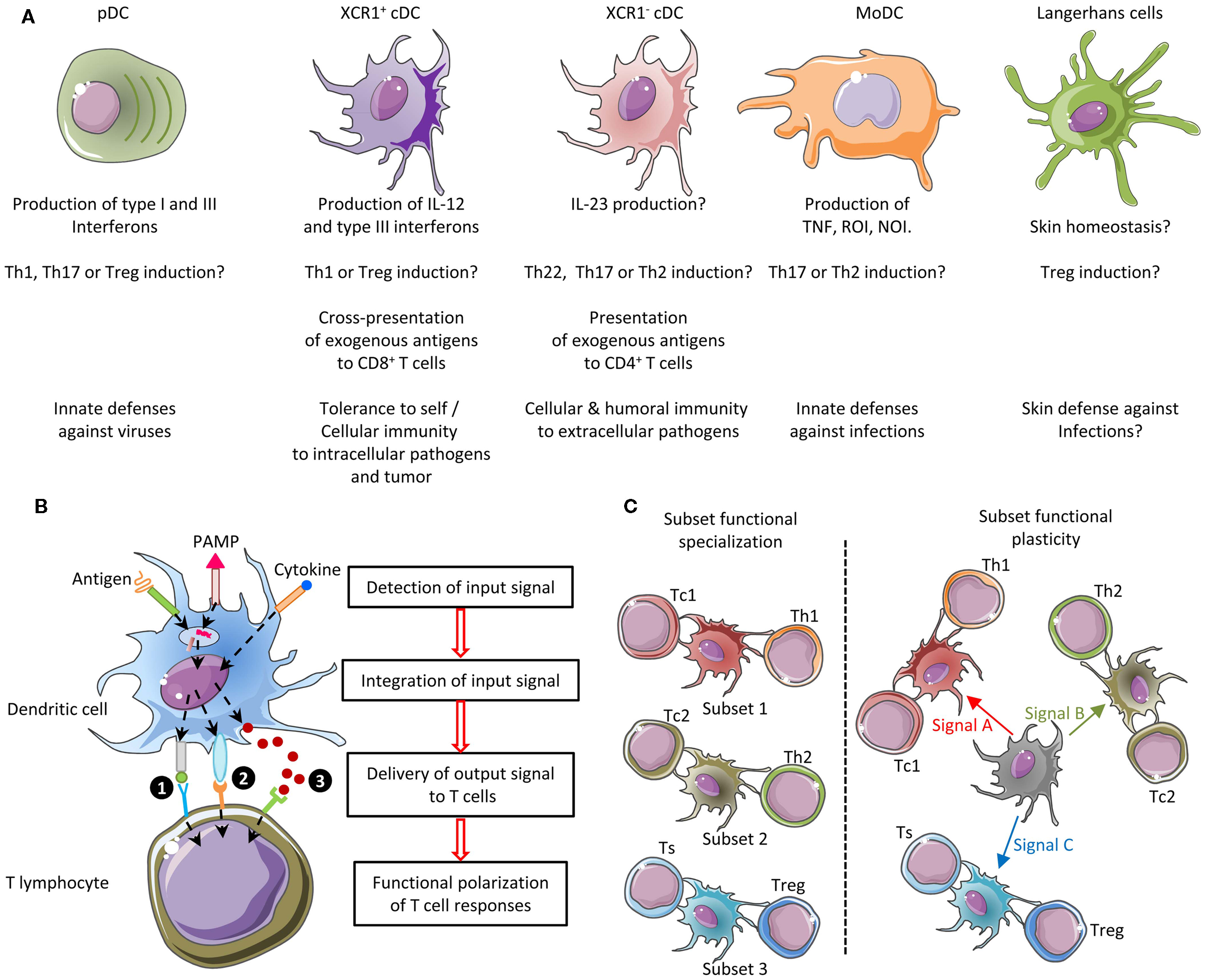
Figure 2. Combined functional specialization and plasticity of DC subsets allows mounting different types of adaptive immune responses adapted to the various natures of the threats to be faced. (A) Five DC subsets can be defined in mice based in part on their functional specialization: pDC, XCR1+ cDC, XCR1− cDC, MoDC, and Langerhans cells. Certain DC subsets are more efficient than others to exert a specific function, because they are intrinsically genetically built to activate this function faster and in more diverse settings. (B) The function of each DC subset is relatively plastic. Three types of output signals are delivered by DC to T cells and instruct their functional polarization: (1) ligands for the T-cell receptor (antigenic peptides presented in association with MHC molecules), (2) co-stimulation, and (3) cytokines. Co-stimulation and cytokine signals can be either activating (e.g., CD86 and IL-12, respectively) or inhibitory (e.g., PD-L1 and IL-10, respectively). Different cytokines induce distinct types of helper T-cell responses. For example, IL-12 primarily promotes Th1, IL-4 promotes Th2, and IL-23 promotes Th17. Each DC subset can sense a specific array of microbial or danger signals. Integration of the particular combination of input signals received by the DC in a given pathophysiological context determines the precise type of maturation ensuing and hence the combination of output signals delivered to T cells. As a result, different DC subsets can exert similar or complementary functions depending on the physiopathological context. (C) The combination of functional specialization and plasticity of subsets allows DC responses to be highly flexible and thus to react rapidly to different threats by coupling the type of danger sensed to the most appropriate type of immune response to induce for protection. However, this flexibility can lead to confusion if attempting to define DC subsets only on functional specialization. NOI, nitric oxide intermediates; ROI, radical oxygen intermediates; Th, T helper cell; Tc, cytotoxic T cell; Treg, regulatory T cell; Ts, T suppressor cell.
Mononuclear phagocyte subsets were recently shown to develop from distinct progenitors and/or under the instruction of different transcription factors or cytokines. cDC and pDC derive from a dedicated bone marrow precursor, the common DC progenitor, with a differentiation potential strictly restricted to this hematopoietic lineage. pDC and cDC homeostasis exquisitely depends on the growth factor FLT3-L. pDC development strictly depends on the transcription factors TCF4 (E2-2) and SPIB both in mouse and human, XCR1+ cDC development on the master transcription factor IRF8 at least in mice, and XCR1− cDC development on IRF4. Macrophages derive from a monocytic precursor, either of embryonic origin as in the case of Langerhans cells and microglia, or at least in part from circulating blood monocytes as in the case of gut macrophages. Egress of classical monocytes from the bone marrow into the blood strictly depends on the chemokine receptor CCR2. As a consequence, in competitive mixed bone marrow reconstitution experiments in mice, all cell types derived from circulating blood monocytes are primarily reconstituted from wild-type cells and not from CCR2-deficient cells. Hence, it has been proposed that the study of their developmental pathway, in other words ontogeny, was the best way to classify mononuclear phagocyte cell types, at least in the mouse model where the knowledge in DC subset properties is also the most advanced. Indeed, in this model, genetically modified animals unambiguously permit to track the development of cell types and to dissect their phenotypes and functions, in different contexts in vivo. However, the identity and functions of the different mononuclear phagocyte subsets need to be established outside of the mouse model, in animal species where ontogenic studies cannot be easily conducted, in order to accelerate translation of our advanced knowledge on the functioning of the mouse immune system toward clinical and/or economical applications to sustain global human health. Very promising vaccine and immunomodulatory strategies have been developed in mouse models based on DC subset targeting (23–35). The translation of these strategies to human and other species has not yet reached the expected success, likely due to insufficient knowledge in the identity and function of homologous DC subsets across species. This knowledge is needed in biomedical model species, primarily in non-human primates, and also in alternative models such as pigs that share physiological and anatomical similarities with humans – for instance skin and lung structural properties – and that present sensitivity to human pathogens of great importance for public health such as influenza. In addition, this knowledge is needed for companion and sport animals, and for animals of the agro-economy, such as ruminants, pigs, poultry, and fishes, with the goal to improve vaccination strategies against pathogens responsible for major economic losses, to decrease antibiotic use and to ameliorate animal welfare. These species, as well as wild animals, are also targets or reservoirs for major zoonotic pathogens whose control could thus benefit from new vaccine strategies targeting DC subsets in these animal species. This raises the question how to best define DC subset identity and functions in a way that can be extrapolated from mouse to human and other species, for clinical applications as well as for a better understanding of the evolution of the immune system.
Different Methodologies to Define the Identity of Immune Cell Types, with Their Advantages and Drawbacks
Several methodologies have been proposed to define cell types. They include cell surface phenotyping and morphology, ontogeny, functional characterization, molecular profiling at population level, and molecular profiling at single cell level. We will discuss the specific drawbacks and advantages of each of these approaches (Table 1).
Cell Surface Phenotyping and Morphology
Cell surface phenotyping generally is a mandatory first step for all other proposed methodologies aiming at defining DC subsets. It may be skipped only for particular experiments of molecular profiling at single cell level and perhaps for functional tests based on validated protocols for specific depletion of the targeted cell subset in vivo. Indeed, phenotypic characterization/identification of DC subsets is necessary either to purify them for morphological analysis, functional assays, or molecular profiling, or to compare their characteristics in tissues or bulk cell suspensions (expression of lineage reporters in cell fate mapping experiments, anatomical location, maturation status, cytokine production, interactions with T cells…). Phenotypic characterization through cell surface phenotyping by flow cytometry is the method of DC subset identification the easiest to perform and the most frequently used. No single cell surface marker has been found to be sufficient for identification of a given DC subset, except for XCR1 expression on mouse and human XCR1+ cDC (18, 36–42) and maybe BDCA2 or LILRA4 expression on human pDC (43–46). Thus, to rigorously identify any given DC subset in any species with a limited risk of contamination by another cell type, most of the time complex combinations of multiple markers are required, often including the use of exclusion marker to ensure lack of contamination of the cell population targeted by other cell types sharing with it many positive markers. For example, the CD8α+ subset of mouse pDC can heavily contaminate mouse lymphoid organ-resident XCR1+ cDC when defined phenotypically as Lineage− CD11c+CD8α+ (47–49). This problem can be solved by exclusion of SiglecH+ or CCR9+ cells or by using XCR1 as a positive marker. Similarly, other cells including MoDC or activated CD1c (BDCA1)+ XCR1− cDC can heavily contaminate human XCR1+ cDC when defined phenotypically as Lineage− HLA-DR+CD141 (BDCA3)+ (41, 50, 51). This problem can be solved by using CADM1 or XCR1 as additional positive markers (41, 52). Rigorous phenotypic identification of XCR1− cDC (mouse CD11b+ cDC and human CD1c+ cDC) can be much more challenging, since these cells can be difficult to discriminate from MoDC, in particular under inflammation settings (53, 54). Identification of DC based on oligoparameter phenotyping is even more at risk of inaccuracy in other species, due to the limited panel of available antibodies directed to surface markers and to the poor knowledge in surface marker expression selectivity in non-DC cell types. However, major advances have recently been made to refine strategies for DC subset identification by cell surface phenotyping, in part based on novel knowledge gained through ontogeny and molecular profiling studies as will be discussed below. Hence, protocols for DC subset identification by cell surface phenotyping might soon become standardized, at least in mouse and human. This would allow better comparison of data across laboratories and limit the risk of use of inappropriate protocols leading to improper data interpretation. Special attention should be given to enzymatic dissociation that can strongly modify cell surface marker detection. Ideally, universal phenotyping protocols could be designed, allowing to considerably simplify the current nomenclatures for DC subsets by using the same name and similar marker combinations to identify homologous cell types irrespective of their tissues and species of origin (55–57). Moreover, the markers used to define and name DC subsets could be chosen based on their relevance to the biology of these cells, contrary to the current situation where the markers used were discovered fortuitously/empirically and may not be linked to the biology of the eponymous cells, as is the case for CD8α and CD141 for mouse and human XCR1+ cDC, respectively. However, when identifying a potentially new subset of DC or studying in a novel context a potentially known DC subset, a number of precautions need to be taken for data interpretation, including confirmation of conclusions by complementary methods such as ontogeny, functional, or molecular profiling studies.
Ontogeny
Ontogeny studies in mice, in particular studies on the dependence of DC subset development on transcription factors, have been instrumental in identifying the homologies between lymphoid tissue-resident CD8α+ cDC and the CD103+CD11b− cDC present in non-lymphoid tissues and migrating into the draining lymph nodes once activated (58). These studies, together with gene expression profiling analyses (9, 40), ultimately allowed grouping mouse CD8α+ cDC and CD103+CD11b− cDC together under the umbrella of the XCR1+ cDC subset (38, 40, 59, 60). The recent discrimination of mouse CD11b+ cDC from MoDC has also been largely based on the analysis of the role of specific chemokine or growth factor receptors on cell type development in vivo, namely CCR2 dependence as a characteristic of monocytic origin and FLT3 dependence as a proof of cDC identity (2, 3, 6, 61). In addition, mouse CD11b+ cDC development was shown to selectively depend on the IRF4 transcription factor (62, 63). Moreover, the establishment of the concept that mouse bona fide DC constitute a separate hematopoietic lineage, and the discrimination between mouse CD11b+ cDC and MoDC, were confirmed using mutant animals allowing to track natural precursor–progeny relationships in vivo through irreversible fluorescent tagging of all daughter cells of a given type of hematopoietic progenitor, based on Cre-mediated conditional activation of a floxed reporter gene under the control of the constitutive Rosa26 promoter, an experimental strategy-coined fate mapping (64). Based on the important contribution of ontogenic studies for rigorous delineation of the identity of mouse DC subsets and of their lineage relationships, it has been proposed to use ontogeny as a primary methodology for the classification of mononuclear cell subsets in all species (57). Recent methodological progress has now made rigorous ontogenic studies applicable to human DC subsets, by using surrogate models of DC development from human CD34+ hematopoietic progenitors, either in vitro (41, 65, 66) or in vivo in alymphoid mice (66–68). Such approaches have allowed demonstrating remarkable similarities in the ontogeny of mouse and human DC subsets. For example, knock-down experiments performed by transducing human CD34+ hematopoietic progenitors with shRNA-expressing lentiviral vectors allowed to show that human pDC development critically depends on the transcription factor SPIB including in vivo in humanized mice (67), and that human XCR1+ cDC development depends on the transcription factor BATF3 in vitro but not in vivo in humanized mice (68). Moreover, the pathway for the development of human pDC, XCR1+ cDC, and XCR1− cDC was very recently demonstrated to be similar to that described for mouse DC subsets, with the identification of the human homologs to the mouse common DC progenitor and pre-cDC (66, 69). The role of candidate genes susceptible to affect DC development can even be assessed in vivo in humans in the rare cases where patients have been identified with primary immune deficiencies resulting from natural mutations in such genes (70). Strategies are being developed to actively search for human primary immunodeficiencies affecting DC development as experiments of nature allowing deciphering the molecular mechanisms regulating this biological process (71). However, ontogenic studies will often not be applicable in human for rigorous assessment of the identity of DC subsets, for example when studying a potentially known DC subset in a novel physiopathological context, including characterization of the DC subsets present in steady-state non-lymphoid tissues (50) or infiltrating tumors and their draining lymph nodes (72, 73) or isolated from infected/inflamed tissues. In addition, rigorous ontogenic studies will be very difficult to perform in many species, because (i) precursor/progeny relationships remain very difficult to evaluate in vivo through cell fate mapping or cell transfer experiments, (ii) in vivo analysis of cell subset development dependence on growth factors or transcription factors cannot be reasonably done due to operational and/or financial reasons, and (iii) in vitro models of bona fide DC development are currently lacking (74). Hence, the use of other methodologies will be necessary to prove DC subset identity in these various conditions.
Functional Characterization
Ideally, cell types should be defined based on the array of functions they can exert, because this definition links identity to function and is hence the most relevant to understand the functioning of the immune system and to harness the biology of DC subsets for improving health care of humans and of other species. In addition, cell type definitions based on their functional specialization could be the most universal across tissues and species. However, functional assays are often the hardest to perform experimentally and can be the most subject to variations depending on assays and experimental conditions. This is especially the case for assays aiming at comparing the ability of different DC subsets to activate T cells. If one aims at precisely comparing the cell-intrinsic ability of different DC subsets to process and present antigens, a number of potentially confounding factors must be taken into account to design the experiment in order to reduce the risk of inappropriate interpretation of results. Adequate steps must be taken to preserve the viability of DC subsets and control for it. This implies adding to each isolated DC subset the appropriate cytokines or growth factors necessary for their survival, for example GM-CSF for cDC and IL-3 for human pDC. For instance, sorted XCR1+ cDC show a lower ex vivo survival as compared to XCR1− cDC in mice and sheep (75, 76). Sorting of DC subset by positive selections may affect DC subset responses due to antibody-mediated receptor stimulation (43, 77–79). This also implies including a positive control consisting in DC subsets pulsed with optimal epitopic peptides, to assess on antigen-specific T-cell priming by DC the impact of other factors than DC subset-intrinsic differences in antigen processing and presentation, not only differences in DC subset viability but also in delivery of co-stimulation or cytokine signals. In this regard, for a fair comparison between DC subsets, they should each be matured by stimulation with an appropriate adjuvant. PolyI:C is much more efficient than LPS for the activation of human XCR1+ DC while it is the reverse for the activation of human MoDC. TLR7 or TLR9 ligands, but not TLR3 or TLR8 ligands, are potent activators of human pDC. Another layer of complexity is due to fundamental differences in the design of experiments in different species. While the gold standard for antigen processing and presentation assays in mice is the measurement of the activation of TcR-transgenic naïve T cells, this is not possible in other species where various surrogate readouts are used including antigen-specific re-activation of antigen-experienced T-cell clones or polyclonal T-cell lines or even proliferation of allogeneic T cells. It is known that significant differences exist in mice in the signals required for naïve T-cell priming, antigen-experienced T-cell re-activation, or allogeneic T-cell proliferation induction. Therefore, the same exact function is not fairly tested in different species. Furthermore, in species outside mice and humans, the use of epitopic peptide control requires to have accurate MHC typing and knowledge of the corresponding optimal peptides, which are generally unavailable. In addition, for accessibility reasons, the DC subsets used generally derived from different anatomic compartments depending on the species. For example, spleen DC subsets are often used in mice, blood, or tonsil DC in humans and lymph DC in sheep, which can further confound rigorous interpretation of the results when differences are observed between species. Finally, while inbred mice with defined sanitary status are generally used to limit the variability of the responses between individuals, this is not the case for other species including humans where the considerable heterogeneity in the genotypes, environments, and immune histories of individuals contribute to the strong variability of their responses (80). Hence, even for mouse experiments, there is a strong need for standardization of functional assays assessing the ability of DC subsets to process and present antigens and to functionally polarize T cells. Moreover, when attempting to compare DC subset functional specialization across two species, efforts should be made to use comparable experimental designs in both species. Thus, while functional characterization is highly desirable when identifying a potentially new subset of DC or studying in a novel context a potentially known DC subset, the identity of DC subsets must first be studied through alternative approaches measuring cell type-specific parameters that are less strongly influenced by the tissue microenvironment and the genetic or immune history of populations, and for which experimental protocols are relatively well standardized.
Molecular Profiling at the Population Level
As the ontogeny and functions of cell types are instructed by specific gene expression modules, cell type identity can be defined by its molecular fingerprinting, including through gene expression profiling (81, 82). Homologous cell types between species can be defined as “those cells that evolved from the same precursor cell type in the last common ancestor” (82). This implies that homologous cell types must exhibit closer molecular fingerprints and gene expression programs than non-homologous cell types. Thus, it should be possible to decipher the identity of immune cell types of virtually all vertebrate species, by establishing their gene signatures and comparing them to the transcriptomic fingerprints of the well-characterized immune cell types of the mouse referent species. This is indeed an approach we pioneered to compare mouse spleen and human blood DC subsets (39) and later extended to comparison with sheep lymph cDC subsets (76), mouse DC subsets across tissues (40), as well as chicken spleen and pig skin mononuclear phagocyte subsets (83, 84). This approach allowed us to rigorously demonstrate for the first time to the best of our knowledge that human CD1c+ cDC and CD141+ cDC were homologous to mouse CD11b+ cDC and CD8α+ cDC, respectively (39, 85). This was later confirmed by us and others based on phenotypic, functional, and ontogeny studies (18, 37, 50, 65, 86). In addition, this approach permitted to show that cDC split into XCR1+ and XCR1− subsets in migrating skin lymph DC in sheep, a species belonging to the Laurasiatherians, which is a mammalian order distant from the mouse and human Euarchontoglires (76). This approach also provided the first compelling evidence for existence of bona fide cDC and macrophages in chicken, showing that diversification in mononuclear phagocyte cell types appeared in a common ancestor to mammals and reptiles (83). Comparative transcriptomics also led to recognize CADM1 and SIRPα as surface molecules whose conserved expression throughout distant species can be used as a first phenotyping step to identify XCR1+ and XCR1− cDC subsets in any mammal (76). Notably, CADM1 is a highly conserved molecule, presenting about 90% identity across mammalian orthologs, thus allowing using commercial anti-human CADM1 antibodies for cellular staining in distant species (76, 84). We found the Xcr1 gene among genes specifically expressed in mouse spleen CD8α+ DC when compared to a number of other immune cell types [see Supplementary Material “Additional file 5; gb-2008-9-1-r17-s5.xls” from Robbins et al. (39), specifically in the “CD8a_DC_gene_signature” established from our microarray data and confirmed from our own re-analysis of the microarray dataset independently generated by Dudziak et al. (87)]. Specific expression of the Xcr1 protein on mouse lymphoid tissue-resident CD8α+ DC and its functions were first unveiled in the pioneering report from the group of Kroczek (36), who showed that CD8+ T-cell cross-priming depends on their ability to secrete the Xcr1 ligand Xcl1 in experimental models where either the OVA coupled to an anti-CD205 Ab or OVA-expressing allogeneic pre-B cells are administrated in vivo. Xcr1 expression on CD8α+ DCs was also found to be critical for the optimal induction of CD8+ T-cell responses upon Listeria monocytogenes infection (18). Importantly, comparative transcriptomics revealed XCR1 as a specific and universal marker for XCR1+ cDC across tissues and species. This was initially shown in human, mice, and sheep (18, 37, 76) and subsequently in non-human primates and pigs (18, 37, 38, 40, 52, 59, 60). Altogether, these studies were critical for the current proposal of cDC subset classification into XCR1+ and XCR1− cDC (38, 40). Many other recent studies have demonstrated the power of gene expression profiling to determine with a high degree of certainty the identity of mononuclear phagocyte subsets in a tissue where they had not been rigorously studied before or to identify homologous subsets of mononuclear phagocytes across species (5, 6, 8, 9, 50, 88–90). Importantly, standardized protocols for generation and analysis of gene expression data are routinely performed in many laboratories, platforms, or commercial companies in many countries. The corresponding costs have strongly decreased over the last decade and continue to go down. Hence, gene expression profiling at the population level is a very robust and reproducible methodology that is feasible in virtually all species where tools are available or can be developed to phenotypically identify and purify candidate cell subsets. However, potentially confounding factors must be taken into account to design experiments in order to reduce the risk of inappropriate interpretation of results (Figure 3). First and foremost, great care and rigor must be exerted in designing the experimental sampling protocol for cell subset purification, inasmuch as minor contamination by another cell type can dramatically impact the gene expression profile obtained. Hence, it is critical to carefully design the marker combination used to purify the different cell populations to be studied, and to control cell purity prior to the generation of the gene expression data. Second, to allow proper analysis of the gene expression profiles of the targeted cell type, appropriate cell type controls must be included, encompassing sister cell types as well as cell types that could be potential contaminants due to their expression of several of the markers used for positive selection of the targeted cell type. These controls are critical to allow assessing the risk of contamination by another cell type (49).
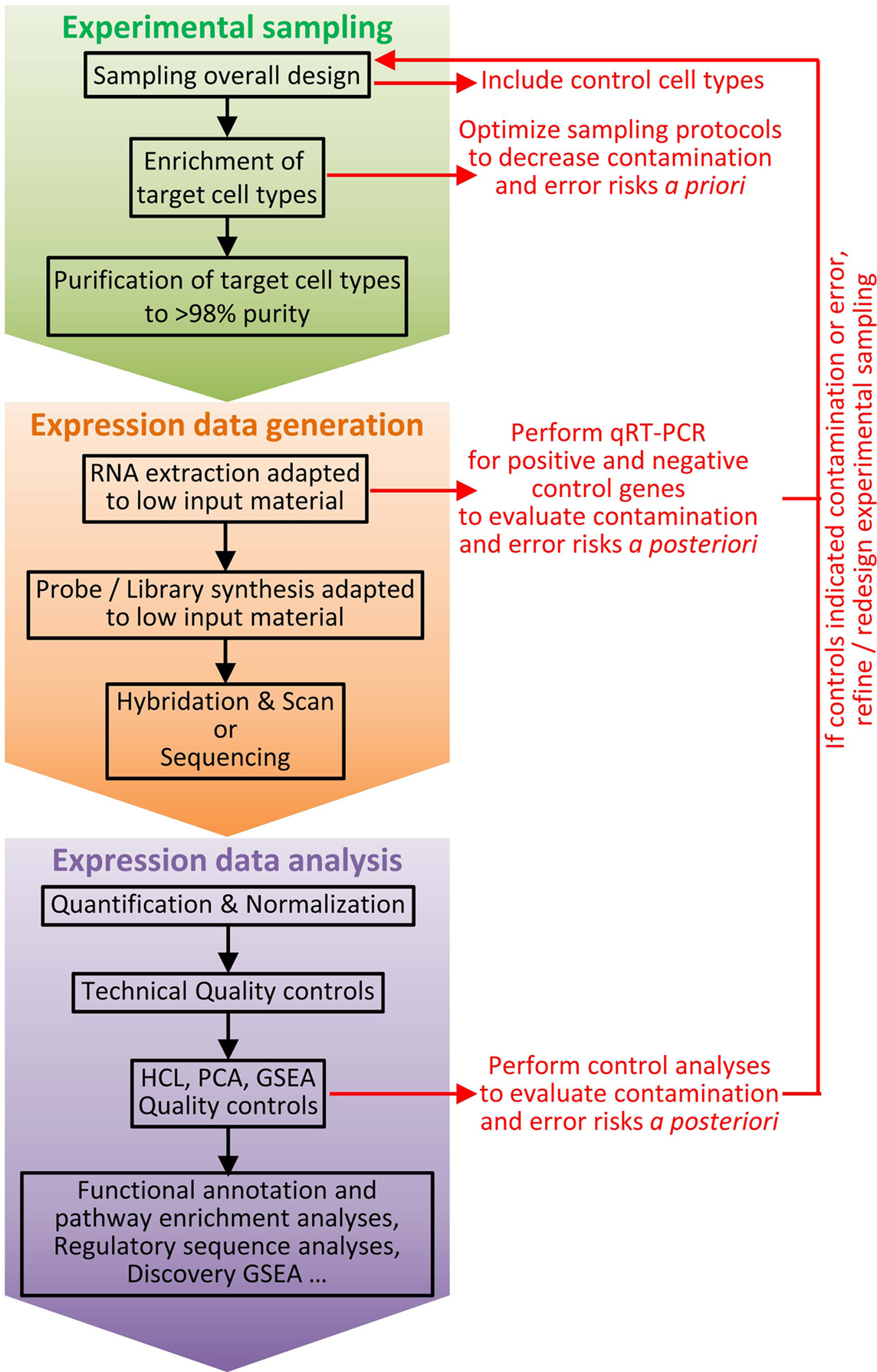
Figure 3. Workflow for cell type identification by molecular profiling at the population level. Molecular profiling at the population level can be very informative for cell type identification. However, inappropriate interpretation can occur if confounding factors are not taken into account. Hence, it is critical to carefully design experiments and to establish a rigorous workflow, including a number of key control samples and quality check procedures. The experimental sampling protocol must be optimized to decrease a priori the risk of cross-contamination between cell types or of error resulting in selection of another cell type than the one wanted. Purity of cell types must be assessed immediately after sampling (e.g., by flow cytometry). Positive and negative cell type controls must be included, such as sister cell types and potential contaminant populations. Once molecular expression data have been obtained, after technical quality has been validated by classical controls, additional specific quality controls must be performed to a posteriori ensure of lack of cross-contamination between cell subsets or to evaluate the risk of misinterpretation of the results. HCL, hierarchical clustering; PCA, principal component analysis; GSEA, Gene Set Enrichment Analysis.
Molecular Profiling at the Single Cell Level
Recent technological advances now allow performing high throughput RNA sequencing at single cell levels with high sensitivity and processivity. Transcriptomic analyses at the single cell level could solve most of the issues raised in the previous section for molecular profiling at the population level. Indeed, because it alleviates the necessity to purify cells on imperfect and potentially confounding phenotypic marker combinations, analysis at the single cell level should allow unbiased identification of potentially all cell types and their associated transcriptomic signatures. It also solves the issue of cross-contamination between cell types, since the identity of each single cell is established a posteriori based on the analysis of its gene expression program. In addition, the generation of gene expression data for many individual cells of the same type should increase statistical power to define genes co-expressed at the single cell level and to define cell type-specific transcriptomic modules (81). As a proof-of-principle, single cell gene expression profiling recently allowed the unbiased and de novo identification of the different cell types of spleen (91) and central nervous system (92, 93) via the description of their molecular identity, starting from the bulk population of all the cells that could be extracted from the organ without any prior enrichment procedure. However, molecular profiling at the single cell level cannot be used without prior phenotype-based enrichment for very rare cell types, and it is difficult to apply to species in which genome has not yet been completely assembled. To obtain complete information, including on functionally important genes for which few mRNA are expressed per cell, it is necessary to sequence at a sufficient depth of about one million reads per cell, which today still represents a very high cost when multiplied by the number of individual cells and conditions. This is all the more the case since, likewise for molecular profiling at population level, correct interpretation of the data requires that sister cell types as well as cell types that could be potential contaminants are included in the experimental design. Moreover, the technology for single cell RNA sequencing is not yet democratized, since it is challenging both for sample preparation and for data analysis. For standardization of high quality sample preparation, commercial solutions exist but are very expensive. For data analysis, there is no consensus yet on how the data should be mathematically modeled for adequate removal of background signal and for discrimination of false negative signal due to sampling bias in the pool of the cell mRNA as opposed to true lack of gene expression. In addition, the interpretation of the RNA-seq data on single cells is still largely based on the transcriptomic/molecular identity of cell types that are deduced from microarray analysis of purified cell pools (91). Hence, molecular profiling at the population level currently represents a more sustainable strategy for most laboratories.
Recent Advances Brought by Comparative Transcriptomics at the Population Level for Defining the Identity and Functions of Mononuclear Phagocyte Subsets and Their Molecular Regulation
In this section, we will review major advances brought forward by comparative transcriptomics at the population level for defining the identity and functions of mononuclear phagocyte subsets and their molecular regulation.
Gene expression profiling of cell types with apparent ambiguous phenotype or functions allowed to rigorously establish their identity, which could be achieved properly strictly contingent to their comparison with all candidate sister cell subsets as well as more distantly related cell types. Hence, we and others showed that human blood Lineage-CD16+ cells are non-classical monocytes (39, 88) and not DC as was sometimes claimed (94–96). Similarly, analysis of human skin CD14+ cell expression of the transcriptomic fingerprints independently established for cDC, monocytes, and macrophages provided critical evidence that these cells are monocyte-derived macrophages (5) while they were previously designated as DC (4). Transcriptomic analyses were also instrumental to demonstrate the homology of this human dermal cell type with the murine CD11b+Ly6C−CD64lo–hi (6) and pig CD163+ (84) skin subsets. We were also able to show that cell populations claimed to correspond to novel cell types actually corresponded to a distinct differentiation or activation state of an already known cell type, for example establishing that the so-called interferon killer DC correspond to a particular activation state of NK cells (39). Furthermore, we showed that, upon many types of in vivo or in vitro stimulation, human and murine pDC and cDC undergo a remodeling of their gene expression program related to their plasticity, including induction of NFκB and IFN target genes, but still keep the canonical gene expression associated to their subset identity (41, 97). In particular, contrary to what other researchers hypothesized (98), gene expression profiling showed that activated pDC are not undergoing a cell fate conversion into a novel type of cDC (97).
Gene expression profiling also allowed aligning subsets of mononuclear cells across tissues (6, 8, 9, 40, 55, 99), establishing cell type homologies across species (5, 39, 50, 76, 83–85, 88, 89, 100), and rigorously examining the proximity of in vitro-derived subsets of mononuclear cells with those naturally existing in vivo (39, 41, 66, 101). These studies allowed significantly advancing the ontogeny and functional characterization of mononuclear phagocyte subsets based on the novel hypotheses that can be inferred from the analysis of the gene expression programs of the cells and from their comparison with other well-characterized cell types.
The study of the functional specialization of human DC subsets was strongly boosted by the demonstration of their transcriptomic homologies with mouse DC subsets (39, 85) which was recognized as a major breakthrough in the field (37, 53, 102–104) and acknowledged to have been impossible to draw from studies based on a limited set of molecular markers (105). In particular, this led to test whether human XCR1+ cDC could be more efficient for cross-presentation than other human DC subsets. Even though the extent to which human XCR1+ cDC are more efficient for cross-presentation than other human DC subsets is debated, the results from the functional studies performed independently by many teams concurrently demonstrate that these cells excel at cross-presentation of cell-associated antigens (18, 19, 37, 41, 86, 106) and of particulate antigens delivered through FcγR, through late endosomal targeting (21, 107) or upon polyI:C stimulation (18, 41, 86, 108). In addition, in sheep, the skin lymph migrating XCR1+ cDC spontaneously displayed a higher efficiency of soluble antigen-presentation to specific CD8+ T cells, as compared to XCR1− cDC (76).
Based on the demonstration of the striking transcriptomic similarities between mouse and human subsets of mononuclear cells, and on knowledge on the ontogeny of these cells in the mouse (109, 110), we proposed that, similar to their mouse counterparts, human pDC and cDC constitute a specific family of cells within the hematopoietic tree, should derive from a common progenitor with a DC-restricted differentiation potential, and could be derived in vitro from human CD34+ progenitor cells in part under the instruction of the FLT3-L growth factor (39, 85), all of which was later confirmed experimentally (41, 65, 66, 69, 111, 112).
Very importantly, comparative genomics of immune cell subsets yielded conserved transcriptomic fingerprints for each of these cell types (39), a novel knowledge which considerably accelerated the deciphering of the molecular mechanisms regulating the development and functions of leukocytes as reviewed in Table 2 (18, 36, 59, 100, 113–127). Finally, this approach uniquely allowed identifying conserved and biologically relevant cell surface markers for each subset of mononuclear cells which could enable considerably simplifying the nomenclature for DC subsets by using the same name and similar marker combinations to identify homologous cell types irrespective of their tissues and species of origin (55–57).
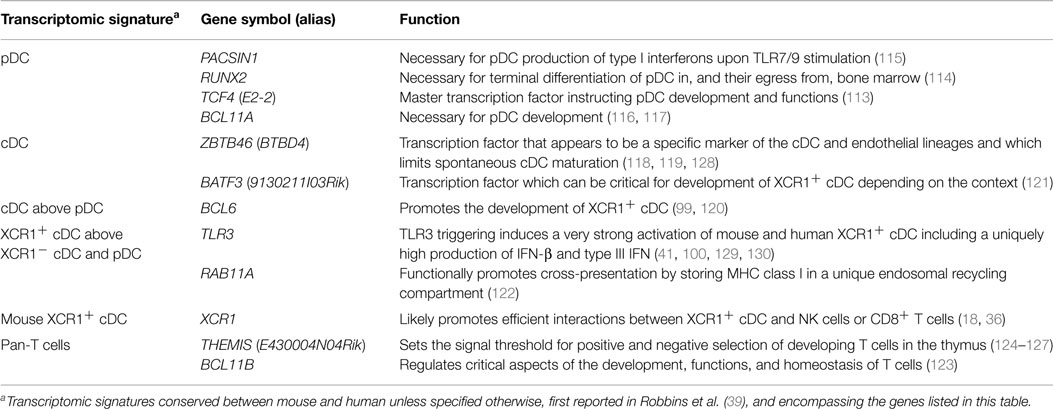
Table 2. Genes which selective expression pattern in immune cell types was uncovered through comparative genomics and which functions in these cells were deciphered later.
Conclusion and Perspectives
While it might be the case in the future for single cell RNA-seq, currently no single method is sufficient to allow the best possible classification of DC. Hence, ideally, all available methods (cell surface phenotyping, gene expression profiling, functional analyses, and ontogeny) should be combined together to define DC subset identity. However, such a combination of approaches cannot be used to define cell subsets in many instances due to technical, financial, or ethical limitations. Taking these limitations into consideration, the data reviewed here show that comparative transcriptomics at the population level is currently the most robust and feasible way to define the identity of cell types. Indeed, because the ontogeny and functions of cell types are instructed by specific gene expression modules, cell type identity can be defined in a universal and unbiased way by its molecular fingerprinting, including through gene expression profiling (81). However, due to its dependency on pre-selection of cell populations based on their expression patterns of a few cell surface molecules, gene expression profiling at the cell population level is imperfect and may require iterative steps of refined cell type isolation and gene expression profiling as illustrated in Figure 3. Hence, it is all the more important that each step of the procedure is performed and rigorously quality controlled according to the best standards in the field.
Cell purity is fundamental. It is important to design a sampling method specific for each study, through identification of the most robust criteria available in the current state of the art for purification of the target cell type based on phenotypic, morphologic, or anatomical characteristics. Cell enrichment is necessary for rare cell types among bulk populations. It relies on the depletion of other populations (MACS or EasySep™ for instance). The marker combination for negative selection must not unwillingly remove a population of interest. For instance, some antibody cocktails for human DC enrichment use anti-CD16 monoclonal antibodies, so as to deplete NK cells, but this should be proscribed for the study of non-classical, CD16+ monocytes. Positive selection by magnetic or flow cytometry sorting is most often required after cell enrichment. Antibody labeling must be clear-cut, with separate peaks and/or selection of the events with the highest labeling and the lowest potential contamination by other populations. This selection implies the use of marker combinations specific for the population of interest, since specific markers are rarely available. XCR1 is a rare instance of a conserved marker so far only expressed on a discrete DC population. To the best of our knowledge, reliable commercial reagent are available for XCR1 staining only for mouse and rat, but XCR1 staining can also be achieved with fluorescently labeled recombinant XCL1 (40, 41, 52), a strategy that is amenable to many species in which XCL1 sequence is known. CLEC4C alias BDCA2 and LILRA4 alias ILT7 are specific markers for human pDC, but their engagement induces inhibitory signals which for instance reduce pDC production of type I interferons after stimulation (43, 77–79, 131). Although selectively expressed at high levels on human pDC in the blood or lymphoid organs under steady-state conditions, NRP1 alias BDCA4 can be induced on activated cDC and is also expressed on other cell types including neurons, endothelial cells, and tumor cells (132, 133). CD123 is a good marker to help identifying pDC in non-human primates, but it also labels mastocytes which are present in the blood or in lymphoid organs (134). Cell purity must be controlled in each experiment, by flow cytometry re-analysis just after sorting, and as one of the first step of transcriptomic analysis by examining the expression of negative and positive control genes (expression of genes that should be expressed only on other populations including potential contaminants, and expression of genes characteristic for the population of interest including but not restricted to genes coding for the molecules used for positive selection) (Figure 3).
The quality and quantity of mRNA must be adequate, even when cell numbers are low. RNA extraction kits adapted to low cell number samples may be required. mRNA quality must be controlled by electrophoresis. A linear amplification protocol must be used, that has been validated for yielding results from low input RNA showing a strong correlation with the results obtained with higher RNA input and a classical amplification procedure.
For bioinformatics analyses, the dataset must include sister cell types as well as the cell types the most likely to contaminate the cell type of interest, or at least be compatible for integrative analysis with a reference dataset including these control populations. Several independent methods for data analysis should be used, to ensure robustness of interpretation. Beyond relative classification of the cell types of the dataset by classical approaches computing the overall distance between their gene expression programs as performed by hierarchical clustering or principal component analysis, the identity of cell types can also be reliably inferred from the analysis of their relative expression of robust cell type-specific gene signatures established from re-analysis of public gene chip databases and/or from published articles.
Novel advances are being brought through molecular profiling of subsets of mononuclear cells. In addition to steady-state conditions, populations can be analyzed after stimulation to identify the specific activation pathways elicited in pure cell populations or upon interaction between different cell types (41, 97, 135–137). In addition to unbiased analysis of the cellular composition of different organs (91, 93), transcriptomic profiling at the single cell level will allow studying heterogeneity in gene expression within one cell type with the hope to link it to functional heterogeneity (138) and eventually with the former history/epigenetic imprinting of each cell. Comparative transcriptomic studies allowed us and others to identify in humans, non-human primates, pig, sheep, and chicken cDC subsets homologous to those well described in mice (5, 18, 39, 50, 52, 62, 76, 83–85). These studies suggest that similar cDC subsets already existed in the last common ancestor of birds and mammals. Conserved gene modules appear during evolution to elicit new functions (81, 82). For instance, regarding T helper lineage diversification during evolution, contrary to bony fishes, the elephant shark, a cartilaginous fish, has been reported to lack genes encoding for critical transcription factors or cytokines instructing the development or involved in the functions of Th2, Th17, and Treg cells, such as RORC and FOXP3, IL-4, IL-21, IL-23, and IL-2 (139). This suggests that the genes required for the development of the different T helper lineages might have appeared progressively as modules during evolution starting in bony fishes and with late development of the Treg and Th17 lineages (81). Comparative genomics of mononuclear phagocyte subsets and single cell gene expression profiling will critically help identifying novel gene modules and their associated immune functions. In pDC, evolutionarily conserved co-expression of TCF4, RUNX2, TLR7, TLR9, UNC93B1, MYD88, IRAK4, IRF7, and PACSIN1 might represent part of a gene module instructing the functional specialization of this cell type in high level production of type I interferon in response to sensing of oligonucleotide sequences of viral or autologous origin. In XCR1+ cDC, evolutionarily conserved co-expression of CLEC9A, SYK, RAB11A, RAB7B, SEPT3, SNX22, TLR3, CADM1, and XCR1 might represent part of a gene module instructing the functional specialization of this cell type in CD8+ T-cell activation and specifically in cross-presentation of cell-associated antigens. In any case, the discovery of the sets of genes that are tightly co-expressed in DC subsets across various tissues and species, not only at the population level but also at the single cell level, should allow identifying the gene modules instructing DC subset functions. Characterization of the members of these gene modules which role in DC is unknown yet should strongly contribute to increase our knowledge on DC subset functional specialization and their molecular regulation. Of note, not all of these gene modules might harbor the same differential pattern of expression between DC subsets in different animal species. Some functions have gained or lost expression in specific cell subsets in some species which should correlate with similar changes in the expression patterns of the corresponding gene modules. For instance, IL-12 is produced both by pDC and cDC in mice, but only by cDC in humans, while antigen cross-presentation appears to be more strongly associated with XCR1+ cDC in mice than in humans (18, 19, 22). Isolation and comparison of mononuclear phagocyte subsets from homologous organs in different species may help understand how the anatomical compartmentalization of these cells is established and affects their functions, including local interaction with specific cell types and chemokines. Dating when during evolution pDC as well as classical and non-classical monocyte subsets appeared, and in which anatomical compartments they reside in the species the most distant to humans and mice, may give novel insights into the core functions of these populations.
In vivo manipulation of DC can promote and orient immune responses based on the intrinsic functional properties of the DC subset targeted and can be advantageously used for prophylactic vaccination or immunotherapy against cancer or infections. This strategy can benefit from the knowledge gained from the expression profiling of DC subsets and their alignment across species. Notably, based on their homology with mouse XCR1+ cDC, human XCR1+ cDC can be considered as a promising target when cross-presentation is desirable, in particular for fighting cancer or infections by intracellular pathogens (23, 24, 29, 72, 73, 140–143). Moreover, because it is specifically expressed in XCR1+ cDC in a conserved manner in evolution, and it has been successfully used for in vivo delivery of antigens specifically to XCR1+ cDC to vaccinate mice (23, 24), XCR1 can be considered for a universal DC targeting strategy in potentially all vertebrate species. Interestingly, the targeting of XCR1 can be achieved with targeting units composed of recombinant XCL1 fused to protective antigens in the form of vaccibodies (24), a strategy that is amenable to many species in which the XCL1 sequence is known. Although more broadly expressed in the DC lineage at least in mice, CLEC9A is also an interesting target since it directly promotes cross-presentation of the material it binds, probably by delivering it into appropriate endosomes (144, 145), and because it is selectively expressed to high levels on XCR1+ cDC in humans, sheep, and mice (25, 32, 76, 146) although it may not be the case in some other species such as pig. Arguments in favor or against the targeting of XCR1+ cDC in the clinic are summarized in Table 3. The identification of XCR1+ cDC in companion and sport animals, and in animals of the agro-economy, such as ruminants, pigs, poultry, and fishes, will allow designing better vaccines to protect them against infections in order to ameliorate animal welfare and to prevent pandemics causing severe economic losses. It will also contribute to a global public health strategy because some of these animal species as well as wild animals are targets or reservoirs for major zoonotic pathogens. The identification of XCR1+ cDC in rhesus macaques and in pigs opens the way to preclinical vaccination studies in these species which are close to humans. Vaccibodies based on XCL1 dimers coupled to influenza or SIV proteins are planned to be used for vaccination of pigs or rhesus macaques, respectively, and induction of immune responses and protection against infection. pDC targeting could also be considered as an interesting alternative for vaccination against viruses or tumors (20, 147, 148), or for the induction of cross-tolerance to treat autoimmune diseases or food allergies (149, 150).
A synthetic list of phenotypic, transcriptomic, and functional hallmarks which have already allowed conserved identification of different DC and monocyte subsets in humans and mice is presented in Table 4. The present Special Issue and future workshop on DC nomenclature will help reach a consensus panel for practical definition of the populations, in order to integrate faster the huge, but scattered knowledge accumulated by different laboratories in different cell types, species, and organs.
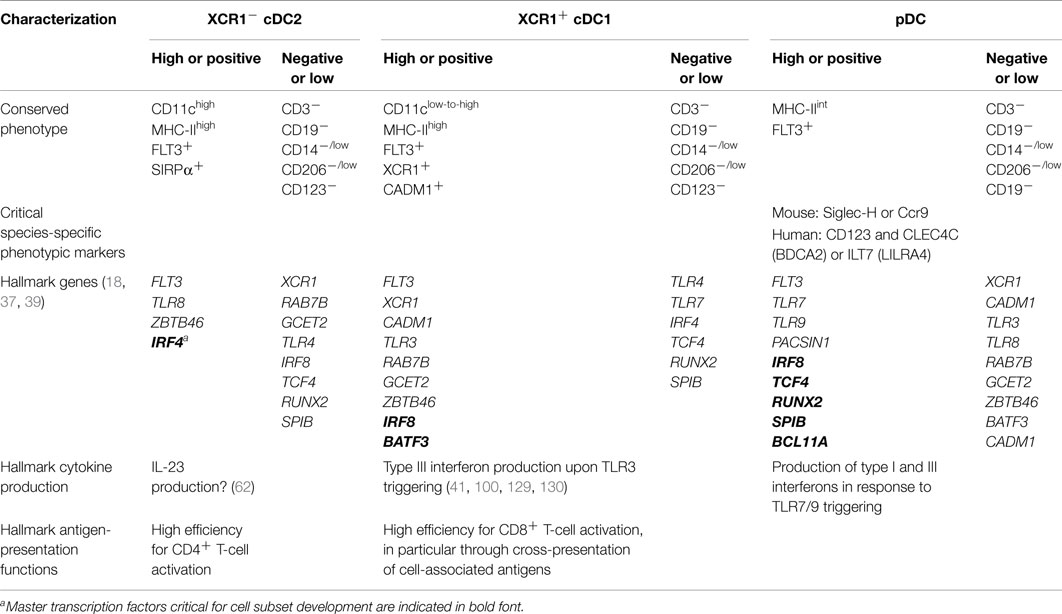
Table 4. Practical guidelines for consistent definition of DC subsets across mouse and human tissues with potential applicability to other mammals.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The studies performed in the laboratories are supported by funding from INSERM, CNRS, INRA, a CNRS-AP-HP collaboration, Aix-Marseille University, Université Paris Descartes, Université Paris Diderot, ANRS, the Agence Nationale de la Recherche (ANR) (PhyloGenDC, ANR-09-BLAN-0073-02 and DCskin-VacFlu, ANR-11-ISV3-0001), the “Integrative Biology of Emerging Infectious Diseases” Labex (ANR-10-LABX-62-IBEID), the DCBIOL Labex (ANR-11-LABEX-0043, grant ANR-10-IDEX-0001-02 PSL*), and the A*MIDEX project (ANR-11-IDEX-0001-02) funded by the French Government’s “Investissements d’Avenir” program managed by the ANR, as well as the European Community’s Seventh Framework Programme FP7/2007–2013 (European Research Council Starting Grant Agreement number 281225 for MD including salary support to TPVM). We thank past and present members of our laboratories and Dr. Rémi Cheynier for their contribution to studies on DC and for discussions. Schematic representations of cells are adapted from Servier Medical Art Powerpoint image bank (smart.servier.fr/servier-medical-art). We apologize for not quoting certain studies because of space limitations.
References
1. Cheong C, Matos I, Choi JH, Dandamudi DB, Shrestha E, Longhi MP, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell (2010) 143(3):416–29. doi:10.1016/j.cell.2010.09.039
2. Plantinga M, Guilliams M, Vanheerswynghels M, Deswarte K, Branco-Madeira F, Toussaint W, et al. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity (2013) 38(2):322–35. doi:10.1016/j.immuni.2012.10.016
3. Langlet C, Tamoutounour S, Henri S, Luche H, Ardouin L, Gregoire C, et al. CD64 expression distinguishes monocyte-derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J Immunol (2012) 188(4):1751–60. doi:10.4049/jimmunol.1102744
4. Klechevsky E, Morita R, Liu M, Cao Y, Coquery S, Thompson-Snipes L, et al. Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity (2008) 29(3):497–510. doi:10.1016/j.immuni.2008.07.013
5. McGovern N, Schlitzer A, Gunawan M, Jardine L, Shin A, Poyner E, et al. Human dermal CD14(+) cells are a transient population of monocyte-derived macrophages. Immunity (2014) 41(3):465–77. doi:10.1016/j.immuni.2014.08.006
6. Tamoutounour S, Guilliams M, Montanana Sanchis F, Liu H, Terhorst D, Malosse C, et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity (2013) 39(5):925–38. doi:10.1016/j.immuni.2013.10.004
7. Hoeffel G, Wang Y, Greter M, See P, Teo P, Malleret B, et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J Exp Med (2012) 209(6):1167–81. doi:10.1084/jem.20120340
8. Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol (2012) 13(11):1118–28. doi:10.1038/ni.2419
9. Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol (2012) 13(9):888–99. doi:10.1038/ni.2370
10. Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, et al. Stroma-derived interleukin-34 controls the development and maintenance of Langerhans cells and the maintenance of microglia. Immunity (2012) 37(6):1050–60. doi:10.1016/j.immuni.2012.11.001
11. Perdiguero EG, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature (2014) 518(7540):547–51. doi:10.1038/nature13989
12. Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science (2012) 336(6077):86–90. doi:10.1126/science.1219179
13. Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol (2012) 13(8):753–60. doi:10.1038/ni.2360
14. Dadaglio G, Fayolle C, Zhang X, Ryffel B, Oberkampf M, Felix T, et al. Antigen targeting to CD11b+ dendritic cells in association with TLR4/TRIF signaling promotes strong CD8+ T cell responses. J Immunol (2014) 193(4):1787–98. doi:10.4049/jimmunol.1302974
15. Desch AN, Gibbings SL, Clambey ET, Janssen WJ, Slansky JE, Kedl RM, et al. Dendritic cell subsets require cis-activation for cytotoxic CD8 T-cell induction. Nat Commun (2014) 5:4674. doi:10.1038/ncomms5674
16. Mouries J, Moron G, Schlecht G, Escriou N, Dadaglio G, Leclerc C. Plasmacytoid dendritic cells efficiently cross-prime naive T cells in vivo after TLR activation. Blood (2008) 112(9):3713–22. doi:10.1182/blood-2008-03-146290
17. Ballesteros-Tato A, Leon B, Lund FE, Randall TD. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8(+) T cell responses to influenza. Nat Immunol (2010) 11(3):216–24. doi:10.1038/ni.1838
18. Crozat K, Guiton R, Contreras V, Feuillet V, Dutertre CA, Ventre E, et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J Exp Med (2010) 207(6):1283–92. doi:10.1084/jem.20100223
19. Segura E, Durand M, Amigorena S. Similar antigen cross-presentation capacity and phagocytic functions in all freshly isolated human lymphoid organ-resident dendritic cells. J Exp Med (2013) 210(5):1035–47. doi:10.1084/jem.20121103
20. Hoeffel G, Ripoche AC, Matheoud D, Nascimbeni M, Escriou N, Lebon P, et al. Antigen crosspresentation by human plasmacytoid dendritic cells. Immunity (2007) 27(3):481–92. doi:10.1016/j.immuni.2007.07.021
21. Cohn L, Chatterjee B, Esselborn F, Smed-Sorensen A, Nakamura N, Chalouni C, et al. Antigen delivery to early endosomes eliminates the superiority of human blood BDCA3+ dendritic cells at cross presentation. J Exp Med (2013) 210(5):1049–63. doi:10.1084/jem.20121251
22. Mittag D, Proietto AI, Loudovaris T, Mannering SI, Vremec D, Shortman K, et al. Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J Immunol (2011) 186(11):6207–17. doi:10.4049/jimmunol.1002632
23. Hartung E, Becker M, Bachem A, Reeg N, Jakel A, Hutloff A, et al. Induction of potent CD8 T cell cytotoxicity by specific targeting of antigen to cross-presenting dendritic cells in vivo via murine or human XCR1. J Immunol (2015) 194(3):1069–79. doi:10.4049/jimmunol.1401903
24. Fossum E, Grodeland G, Terhorst D, Tveita AA, Vikse E, Mjaaland S, et al. Vaccine molecules targeting Xcr1 on cross-presenting DCs induce protective CD8(+) T-cell responses against influenza virus. Eur J Immunol (2015) 45(2):624–35. doi:10.1002/eji.201445080
25. Caminschi I, Proietto AI, Ahmet F, Kitsoulis S, Shin Teh J, Lo JC, et al. The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood (2008) 112(8):3264–73. doi:10.1182/blood-2008-05-155176
26. Caminschi I, Vremec D, Ahmet F, Lahoud MH, Villadangos JA, Murphy KM, et al. Antibody responses initiated by Clec9A-bearing dendritic cells in normal and Batf3(-/-) mice. Mol Immunol (2012) 50(1–2):9–17. doi:10.1016/j.molimm.2011.11.008
27. Idoyaga J, Lubkin A, Fiorese C, Lahoud MH, Caminschi I, Huang Y, et al. Comparable T helper 1 (Th1) and CD8 T-cell immunity by targeting HIV gag p24 to CD8 dendritic cells within antibodies to Langerin, DEC205, and Clec9A. Proc Natl Acad Sci U S A (2011) 108(6):2384–9. doi:10.1073/pnas.1019547108
28. Lahoud MH, Ahmet F, Kitsoulis S, Wan SS, Vremec D, Lee CN, et al. Targeting antigen to mouse dendritic cells via Clec9A induces potent CD4 T cell responses biased toward a follicular helper phenotype. J Immunol (2011) 187(2):842–50. doi:10.4049/jimmunol.1101176
29. Li J, Ahmet F, Sullivan LC, Brooks AG, Kent SJ, De Rose R, et al. Antibodies targeting Clec9A promote strong humoral immunity without adjuvant in mice and non-human primates. Eur J Immunol (2014) 45(3):854–64. doi:10.1002/eji.201445127
30. Park HY, Light A, Lahoud MH, Caminschi I, Tarlinton DM, Shortman K. Evolution of B cell responses to Clec9A-targeted antigen. J Immunol (2013) 191(10):4919–25. doi:10.4049/jimmunol.1301947
31. Joffre OP, Sancho D, Zelenay S, Keller AM, Reis e Sousa C. Efficient and versatile manipulation of the peripheral CD4+ T-cell compartment by antigen targeting to DNGR-1/CLEC9A. Eur J Immunol (2010) 40(5):1255–65. doi:10.1002/eji.201040419
32. Sancho D, Mourao-Sa D, Joffre OP, Schulz O, Rogers NC, Pennington DJ, et al. Tumor therapy in mice via antigen targeting to a novel, DC-restricted C-type lectin. J Clin Invest (2008) 118(6):2098–110. doi:10.1172/JCI34584
33. Bonifaz LC, Bonnyay DP, Charalambous A, Darguste DI, Fujii S, Soares H, et al. In vivo targeting of antigens to maturing dendritic cells via the DEC-205 receptor improves T cell vaccination. J Exp Med (2004) 199(6):815–24. doi:10.1084/jem.20032220
34. Nchinda G, Kuroiwa J, Oks M, Trumpfheller C, Park CG, Huang Y, et al. The efficacy of DNA vaccination is enhanced in mice by targeting the encoded protein to dendritic cells. J Clin Invest (2008) 118(4):1427–36. doi:10.1172/JCI34224
35. Do Y, Didierlaurent AM, Ryu S, Koh H, Park CG, Park S, et al. Induction of pulmonary mucosal immune responses with a protein vaccine targeted to the DEC-205/CD205 receptor. Vaccine (2012) 30(45):6359–67. doi:10.1016/j.vaccine.2012.08.051
36. Dorner BG, Dorner MB, Zhou X, Opitz C, Mora A, Guttler S, et al. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity (2009) 31(5):823–33. doi:10.1016/j.immuni.2009.08.027
37. Bachem A, Guttler S, Hartung E, Ebstein F, Schaefer M, Tannert A, et al. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J Exp Med (2010) 207(6):1273–81. doi:10.1084/jem.20100348
38. Gurka S, Hartung E, Becker M, Kroczek RA. Mouse conventional dendritic cells can be universally classified based on the mutually exclusive expression of XCR1 and SIRPα. Front Immunol (2015) 6:35. doi:10.3389/fimmu.2015.00035
39. Robbins SH, Walzer T, Dembele D, Thibault C, Defays A, Bessou G, et al. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol (2008) 9(1):R17. doi:10.1186/gb-2008-9-1-r17
40. Crozat K, Tamoutounour S, Vu Manh TP, Fossum E, Luche H, Ardouin L, et al. Cutting edge: expression of XCR1 defines mouse lymphoid-tissue resident and migratory dendritic cells of the CD8alpha+ type. J Immunol (2011) 187(9):4411–5. doi:10.4049/jimmunol.1101717
41. Balan S, Ollion V, Colletti N, Chelbi R, Montanana-Sanchis F, Liu H, et al. Human XCR1+ dendritic cells derived in vitro from CD34+ progenitors closely resemble blood dendritic cells, including their adjuvant responsiveness, contrary to monocyte-derived dendritic cells. J Immunol (2014) 193(4):1622–35. doi:10.4049/jimmunol.1401243
42. Yamazaki C, Miyamoto R, Hoshino K, Fukuda Y, Sasaki I, Saito M, et al. Conservation of a chemokine system, XCR1 and its ligand, XCL1, between human and mice. Biochem Biophys Res Commun (2010) 397(4):756–61. doi:10.1016/j.bbrc.2010.06.029
43. Dzionek A, Sohma Y, Nagafune J, Cella M, Colonna M, Facchetti F, et al. BDCA-2, a novel plasmacytoid dendritic cell-specific type II C-type lectin, mediates antigen capture and is a potent inhibitor of interferon alpha/beta induction. J Exp Med (2001) 194(12):1823–34. doi:10.1084/jem.194.12.1823
44. Rissoan MC, Duhen T, Bridon JM, Bendriss-Vermare N, Peronne C, de Saint Vis B, et al. Subtractive hybridization reveals the expression of immunoglobulin-like transcript 7, Eph-B1, granzyme B, and 3 novel transcripts in human plasmacytoid dendritic cells. Blood (2002) 100(9):3295–303. doi:10.1182/blood-2002-02-0638
45. Cao W, Rosen DB, Ito T, Bover L, Bao M, Watanabe G, et al. Plasmacytoid dendritic cell-specific receptor ILT7-Fc epsilonRI gamma inhibits toll-like receptor-induced interferon production. J Exp Med (2006) 203(6):1399–405. doi:10.1084/jem.20052454
46. Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S, et al. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol (2000) 165(11):6037–46. doi:10.4049/jimmunol.165.11.6037
47. Dalod M, Salazar-Mather TP, Malmgaard L, Lewis C, Asselin-Paturel C, Briere F, et al. Interferon alpha/beta and interleukin 12 responses to viral infections: pathways regulating dendritic cell cytokine expression in vivo. J Exp Med (2002) 195(4):517–28. doi:10.1084/jem.20011672
48. Bar-On L, Birnberg T, Lewis KL, Edelson BT, Bruder D, Hildner K, et al. CX3CR1+ CD8alpha+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc Natl Acad Sci U S A (2010) 107(33):14745–50. doi:10.1073/pnas.1001562107
49. Vu Manh T-P, Dalod M. Characterization of dendritic cell subsets through gene expression analysis. Methods Mol Biol (2015) (in press).
50. Haniffa M, Shin A, Bigley V, McGovern N, Teo P, See P, et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity (2012) 37(1):60–73. doi:10.1016/j.immuni.2012.04.012
51. Chu CC, Ali N, Karagiannis P, Di Meglio P, Skowera A, Napolitano L, et al. Resident CD141 (BDCA3)+ dendritic cells in human skin produce IL-10 and induce regulatory T cells that suppress skin inflammation. J Exp Med (2012) 209(5):935–45. doi:10.1084/jem.20112583
52. Dutertre CA, Jourdain JP, Rancez M, Amraoui S, Fossum E, Bogen B, et al. TLR3-responsive, XCR1+, CD141(BDCA-3)+/CD8alpha+-equivalent dendritic cells uncovered in healthy and simian immunodeficiency virus-infected rhesus macaques. J Immunol (2014) 192(10):4697–708. doi:10.4049/jimmunol.1302448
53. Dutertre CA, Wang LF, Ginhoux F. Aligning bona fide dendritic cell populations across species. Cell Immunol (2014) 291(1–2):3–10. doi:10.1016/j.cellimm.2014.08.006
54. Mildner A, Jung S. Development and function of dendritic cell subsets. Immunity (2014) 40(5):642–56. doi:10.1016/j.immuni.2014.04.016
55. Guilliams M, Henri S, Tamoutounour S, Ardouin L, Schwartz-Cornil I, Dalod M, et al. From skin dendritic cells to a simplified classification of human and mouse dendritic cell subsets. Eur J Immunol (2010) 40(8):2089–94. doi:10.1002/eji.201040498
56. Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, et al. Nomenclature of monocytes and dendritic cells in blood. Blood (2010) 116(16):e74–80. doi:10.1182/blood-2010-02-258558
57. Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol (2014) 14(8):571–8. doi:10.1038/nri3712
58. Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol (2013) 31:563–604. doi:10.1146/annurev-immunol-020711-074950
59. Bachem A, Hartung E, Guttler S, Mora A, Zhou X, Hegemann A, et al. Expression of XCR1 characterizes the Batf3-dependent lineage of dendritic cells capable of antigen cross-presentation. Front Immunol (2012) 3:214. doi:10.3389/fimmu.2012.00214
60. Becker M, Guttler S, Bachem A, Hartung E, Mora A, Jakel A, et al. Ontogenic, phenotypic, and functional characterization of XCR1(+) dendritic cells leads to a consistent classification of intestinal dendritic cells based on the expression of XCR1 and SIRPalpha. Front Immunol (2014) 5:326. doi:10.3389/fimmu.2014.00326
61. Tamoutounour S, Henri S, Lelouard H, de Bovis B, de Haar C, van der Woude CJ, et al. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol (2012) 42(12):3150–66. doi:10.1002/eji.201242847
62. Schlitzer A, McGovern N, Teo P, Zelante T, Atarashi K, Low D, et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity (2013) 38(5):970–83. doi:10.1016/j.immuni.2013.04.011
63. Persson EK, Uronen-Hansson H, Semmrich M, Rivollier A, Hagerbrand K, Marsal J, et al. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity (2013) 38(5):958–69. doi:10.1016/j.immuni.2013.03.009
64. Schraml BU, van Blijswijk J, Zelenay S, Whitney PG, Filby A, Acton SE, et al. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell (2013) 154(4):843–58. doi:10.1016/j.cell.2013.07.014
65. Poulin LF, Salio M, Griessinger E, Anjos-Afonso F, Craciun L, Chen JL, et al. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J Exp Med (2010) 207(6):1261–71. doi:10.1084/jem.20092618
66. Lee J, Breton G, Oliveira TY, Zhou YJ, Aljoufi A, Puhr S, et al. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J Exp Med (2015) 212(3):385–99. doi:10.1084/jem.20141442
67. Schotte R, Nagasawa M, Weijer K, Spits H, Blom B. The ETS transcription factor Spi-B is required for human plasmacytoid dendritic cell development. J Exp Med (2004) 200(11):1503–9. doi:10.1084/jem.20041231
68. Poulin LF, Reyal Y, Uronen-Hansson H, Schraml BU, Sancho D, Murphy KM, et al. DNGR-1 is a specific and universal marker of mouse and human Batf3-dependent dendritic cells in lymphoid and nonlymphoid tissues. Blood (2012) 119(25):6052–62. doi:10.1182/blood-2012-01-406967
69. Breton G, Lee J, Zhou YJ, Schreiber JJ, Keler T, Puhr S, et al. Circulating precursors of human CD1c+ and CD141+ dendritic cells. J Exp Med (2015) 212(3):401–13. doi:10.1084/jem.20141441
70. Hambleton S, Salem S, Bustamante J, Bigley V, Boisson-Dupuis S, Azevedo J, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med (2011) 365(2):127–38. doi:10.1056/NEJMoa1100066
71. Jardine L, Barge D, Ames-Draycott A, Pagan S, Cookson S, Spickett G, et al. Rapid detection of dendritic cell and monocyte disorders using CD4 as a lineage marker of the human peripheral blood antigen-presenting cell compartment. Front Immunol (2013) 4:495. doi:10.3389/fimmu.2013.00495
72. Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell (2014) 26(5):638–52. doi:10.1016/j.ccell.2014.09.007
73. Woo SR, Corrales L, Gajewski TF. Innate immune recognition of cancer. Annu Rev Immunol (2015) 33:445–74. doi:10.1146/annurev-immunol-032414-112043
74. Summerfield A, Auray G, Ricklin M. Comparative dendritic cell biology of veterinary mammals. Annu Rev Anim Biosci (2015) 3:533–57. doi:10.1146/annurev-animal-022114-111009
75. Vremec D, Hansen J, Strasser A, Acha-Orbea H, Zhan Y, O’Keeffe M, et al. Maintaining dendritic cell viability in culture. Mol Immunol (2015) 63(2):264–7. doi:10.1016/j.molimm.2014.07.011
76. Contreras V, Urien C, Guiton R, Alexandre Y, Vu Manh TP, Andrieu T, et al. Existence of CD8alpha-like dendritic cells with a conserved functional specialization and a common molecular signature in distant mammalian species. J Immunol (2010) 185(6):3313–25. doi:10.4049/jimmunol.1000824
77. Fanning SL, George TC, Feng D, Feldman SB, Megjugorac NJ, Izaguirre AG, et al. Receptor cross-linking on human plasmacytoid dendritic cells leads to the regulation of IFN-alpha production. J Immunol (2006) 177(9):5829–39. doi:10.4049/jimmunol.177.9.5829
78. Jahn PS, Zanker KS, Schmitz J, Dzionek A. BDCA-2 signaling inhibits TLR-9-agonist-induced plasmacytoid dendritic cell activation and antigen presentation. Cell Immunol (2010) 265(1):15–22. doi:10.1016/j.cellimm.2010.06.005
79. Tavano B, Boasso A. Effect of immunoglobin-like transcript 7 cross-linking on plasmacytoid dendritic cells differentiation into antigen-presenting cells. PLoS One (2014) 9(2):e89414. doi:10.1371/journal.pone.0089414
80. Duffy D, Rouilly V, Libri V, Hasan M, Beitz B, David M, et al. Functional analysis via standardized whole-blood stimulation systems defines the boundaries of a healthy immune response to complex stimuli. Immunity (2014) 40(3):436–50. doi:10.1016/j.immuni.2014.03.002
81. Achim K, Arendt D. Structural evolution of cell types by step-wise assembly of cellular modules. Curr Opin Genet Dev (2014) 27:102–8. doi:10.1016/j.gde.2014.05.001
82. Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet (2008) 9(11):868–82. doi:10.1038/nrg2416
83. Vu Manh TP, Marty H, Sibille P, Le Vern Y, Kaspers B, Dalod M, et al. Existence of conventional dendritic cells in Gallus gallus revealed by comparative gene expression profiling. J Immunol (2014) 192(10):4510–7. doi:10.4049/jimmunol.1303405
84. Marquet F, Vu Manh TP, Maisonnasse P, Elhmouzi-Younes J, Urien C, Bouguyon E, et al. Pig skin includes dendritic cell subsets transcriptomically related to human CD1a and CD14 dendritic cells presenting different migrating behaviors and T cell activation capacities. J Immunol (2014) 193(12):5883–93. doi:10.4049/jimmunol.1303150
85. Crozat K, Guiton R, Guilliams M, Henri S, Baranek T, Schwartz-Cornil I, et al. Comparative genomics as a tool to reveal functional equivalences between human and mouse dendritic cell subsets. Immunol Rev (2010) 234(1):177–98. doi:10.1111/j.0105-2896.2009.00868.x
86. Jongbloed SL, Kassianos AJ, McDonald KJ, Clark GJ, Ju X, Angel CE, et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med (2010) 207(6):1247–60. doi:10.1084/jem.20092140
87. Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, Yamazaki S, et al. Differential antigen processing by dendritic cell subsets in vivo. Science (2007) 315(5808):107–11. doi:10.1126/science.1136080
88. Cros J, Cagnard N, Woollard K, Patey N, Zhang SY, Senechal B, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity (2010) 33(3):375–86. doi:10.1016/j.immuni.2010.08.012
89. Ingersoll MA, Spanbroek R, Lottaz C, Gautier EL, Frankenberger M, Hoffmann R, et al. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood (2010) 115(3):e10–9. doi:10.1182/blood-2009-07-235028
90. Segura E, Touzot M, Bohineust A, Cappuccio A, Chiocchia G, Hosmalin A, et al. Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity (2013) 38(2):336–48. doi:10.1016/j.immuni.2012.10.018
91. Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science (2014) 343(6172):776–9. doi:10.1126/science.1247651
92. Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci (2015) 18(1):145–53. doi:10.1038/nn.3881
93. Zeisel A, Manchado ABM, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science (2015) 347(6226):1138–42. doi:10.1126/science.aaa1934
94. Schakel K, Kannagi R, Kniep B, Goto Y, Mitsuoka C, Zwirner J, et al. 6-Sulfo LacNAc, a novel carbohydrate modification of PSGL-1, defines an inflammatory type of human dendritic cells. Immunity (2002) 17(3):289–301. doi:10.1016/S1074-7613(02)00393-X
95. Lindstedt M, Lundberg K, Borrebaeck CA. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J Immunol (2005) 175(8):4839–46. doi:10.4049/jimmunol.175.8.4839
96. Dobel T, Kunze A, Babatz J, Trankner K, Ludwig A, Schmitz M, et al. FcgammaRIII (CD16) equips immature 6-sulfo LacNAc-expressing dendritic cells (slanDCs) with a unique capacity to handle IgG-complexed antigens. Blood (2013) 121(18):3609–18. doi:10.1182/blood-2012-08-447045
97. Vu Manh TP, Alexandre Y, Baranek T, Crozat K, Dalod M. Plasmacytoid, conventional, and monocyte-derived dendritic cells undergo a profound and convergent genetic reprogramming during their maturation. Eur J Immunol (2013) 43(7):1706–15. doi:10.1002/eji.201243106
98. Reizis B, Bunin A, Ghosh HS, Lewis KL, Sisirak V. Plasmacytoid dendritic cells: recent progress and open questions. Annu Rev Immunol (2011) 29:163–83. doi:10.1146/annurev-immunol-031210-101345
99. Watchmaker PB, Lahl K, Lee M, Baumjohann D, Morton J, Kim SJ, et al. Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat Immunol (2014) 15(1):98–108. doi:10.1038/ni.2768
100. Lauterbach H, Bathke B, Gilles S, Traidl-Hoffmann C, Luber CA, Fejer G, et al. Mouse CD8alpha+ DCs and human BDCA3+ DCs are major producers of IFN-lambda in response to poly IC. J Exp Med (2010) 207(12):2703–17. doi:10.1084/jem.20092720
101. Lundberg K, Albrekt AS, Nelissen I, Santegoets S, de Gruijl TD, Gibbs S, et al. Transcriptional profiling of human dendritic cell populations and models – unique profiles of in vitro dendritic cells and implications on functionality and applicability. PLoS One (2013) 8(1):e52875. doi:10.1371/journal.pone.0052875
102. Reizis B. Regulation of plasmacytoid dendritic cell development. Curr Opin Immunol (2010) 22(2):206–11. doi:10.1016/j.coi.2010.01.005
103. Villadangos JA, Shortman K. Found in translation: the human equivalent of mouse CD8+ dendritic cells. J Exp Med (2010) 207(6):1131–4. doi:10.1084/jem.20100985
104. Naik SH. Demystifying the development of dendritic cell subtypes, a little. Immunol Cell Biol (2008) 86(5):439–52. doi:10.1038/icb.2008.28
105. Soumelis V, Pattarini L, Michea P, Cappuccio A. Systems approaches to unravel innate immune cell diversity, environmental plasticity and functional specialization. Curr Opin Immunol (2015) 32C:42–7. doi:10.1016/j.coi.2014.12.007
106. Deauvieau F, Ollion V, Doffin AC, Achard C, Fonteneau JF, Verronese E, et al. Human natural killer cells promote cross-presentation of tumor cell-derived antigens by dendritic cells. Int J Cancer (2015) 136(5):1085–94. doi:10.1002/ijc.29087
107. Flinsenberg TW, Compeer EB, Koning D, Klein M, Amelung FJ, van Baarle D, et al. Fcgamma receptor antigen targeting potentiates cross-presentation by human blood and lymphoid tissue BDCA-3+ dendritic cells. Blood (2012) 120(26):5163–72. doi:10.1182/blood-2012-06-434498
108. Nizzoli G, Krietsch J, Weick A, Steinfelder S, Facciotti F, Gruarin P, et al. Human CD1c+ dendritic cells secrete high levels of IL-12 and potently prime cytotoxic T-cell responses. Blood (2013) 122(6):932–42. doi:10.1182/blood-2013-04-495424
109. Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol (2007) 8(11):1207–16. doi:10.1038/ni1518
110. Naik SH, Sathe P, Park HY, Metcalf D, Proietto AI, Dakic A, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol (2007) 8(11):1217–26. doi:10.1038/ni1522
111. Thordardottir S, Hangalapura BN, Hutten T, Cossu M, Spanholtz J, Schaap N, et al. The aryl hydrocarbon receptor antagonist StemRegenin 1 promotes human plasmacytoid and myeloid dendritic cell development from CD34+ hematopoietic progenitor cells. Stem Cells Dev (2014) 23(9):955–67. doi:10.1089/scd.2013.0521
112. Proietto AI, Mittag D, Roberts AW, Sprigg N, Wu L. The equivalents of human blood and spleen dendritic cell subtypes can be generated in vitro from human CD34(+) stem cells in the presence of fms-like tyrosine kinase 3 ligand and thrombopoietin. Cell Mol Immunol (2012) 9(6):446–54. doi:10.1038/cmi.2012.48
113. Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, Locksley R, et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell (2008) 135(1):37–48. doi:10.1016/j.cell.2008.09.016
114. Sawai CM, Sisirak V, Ghosh HS, Hou EZ, Ceribelli M, Staudt LM, et al. Transcription factor Runx2 controls the development and migration of plasmacytoid dendritic cells. J Exp Med (2013) 210(11):2151–9. doi:10.1084/jem.20130443
115. Esashi E, Bao M, Wang YH, Cao W, Liu YJ. PACSIN1 regulates the TLR7/9-mediated type I interferon response in plasmacytoid dendritic cells. Eur J Immunol (2012) 42(3):573–9. doi:10.1002/eji.201142045
116. Ippolito GC, Dekker JD, Wang YH, Lee BK, Shaffer AL III, Lin J, et al. Dendritic cell fate is determined by BCL11A. Proc Natl Acad Sci U S A (2014) 111(11):E998–1006. doi:10.1073/pnas.1319228111
117. Wu X, Satpathy AT, Kc W, Liu P, Murphy TL, Murphy KM. Bcl11a controls Flt3 expression in early hematopoietic progenitors and is required for pDC development in vivo. PLoS One (2013) 8(5):e64800. doi:10.1371/journal.pone.0064800
118. Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med (2012) 209(6):1153–65. doi:10.1084/jem.20112675
119. Satpathy AT, Kc W, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med (2012) 209(6):1135–52. doi:10.1084/jem.20120030
120. Ohtsuka H, Sakamoto A, Pan J, Inage S, Horigome S, Ichii H, et al. Bcl6 is required for the development of mouse CD4+ and CD8alpha+ dendritic cells. J Immunol (2011) 186(1):255–63. doi:10.4049/jimmunol.0903714
121. Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science (2008) 322(5904):1097–100. doi:10.1126/science.1164206
122. Nair-Gupta P, Baccarini A, Tung N, Seyffer F, Florey O, Huang Y, et al. TLR signals induce phagosomal MHC-I delivery from the endosomal recycling compartment to allow cross-presentation. Cell (2014) 158(3):506–21. doi:10.1016/j.cell.2014.04.054
123. Avram D, Califano D. The multifaceted roles of Bcl11b in thymic and peripheral T cells: impact on immune diseases. J Immunol (2014) 193(5):2059–65. doi:10.4049/jimmunol.1400930
124. Fu G, Vallee S, Rybakin V, McGuire MV, Ampudia J, Brockmeyer C, et al. Themis controls thymocyte selection through regulation of T cell antigen receptor-mediated signaling. Nat Immunol (2009) 10(8):848–56. doi:10.1038/ni.1766
125. Johnson AL, Aravind L, Shulzhenko N, Morgun A, Choi SY, Crockford TL, et al. Themis is a member of a new metazoan gene family and is required for the completion of thymocyte positive selection. Nat Immunol (2009) 10(8):831–9. doi:10.1038/ni.1769
126. Kakugawa K, Yasuda T, Miura I, Kobayashi A, Fukiage H, Satoh R, et al. A novel gene essential for the development of single positive thymocytes. Mol Cell Biol (2009) 29(18):5128–35. doi:10.1128/MCB.00793-09
127. Lesourne R, Uehara S, Lee J, Song KD, Li L, Pinkhasov J, et al. Themis, a T cell-specific protein important for late thymocyte development. Nat Immunol (2009) 10(8):840–7. doi:10.1038/ni.1768
128. Meredith MM, Liu K, Kamphorst AO, Idoyaga J, Yamane A, Guermonprez P, et al. Zinc finger transcription factor zDC is a negative regulator required to prevent activation of classical dendritic cells in the steady state. J Exp Med (2012) 209(9):1583–93. doi:10.1084/jem.20121003
129. Yoshio S, Kanto T, Kuroda S, Matsubara T, Higashitani K, Kakita N, et al. Human blood dendritic cell antigen 3 (BDCA3)(+) dendritic cells are a potent producer of interferon-lambda in response to hepatitis C virus. Hepatology (2013) 57(5):1705–15. doi:10.1002/hep.26182
130. Zhang S, Kodys K, Li K, Szabo G. Human type 2 myeloid dendritic cells produce interferon-lambda and amplify interferon-alpha in response to hepatitis C virus infection. Gastroenterology (2013) 144(2):414–25e7. doi:10.1053/j.gastro.2012.10.034
131. Cao W, Bover L, Cho M, Wen X, Hanabuchi S, Bao M, et al. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J Exp Med (2009) 206(7):1603–14. doi:10.1084/jem.20090547
132. Kolodkin AL, Levengood DV, Rowe EG, Tai YT, Giger RJ, Ginty DD. Neuropilin is a semaphorin III receptor. Cell (1997) 90(4):753–62. doi:10.1016/S0092-8674(00)80535-8
133. Soker S, Takashima S, Miao HQ, Neufeld G, Klagsbrun M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell (1998) 92(6):735–45. doi:10.1016/S0092-8674(00)81402-6
134. Nascimbeni M, Perie L, Chorro L, Diocou S, Kreitmann L, Louis S, et al. Plasmacytoid dendritic cells accumulate in spleens from chronically HIV-infected patients but barely participate in interferon-alpha expression. Blood (2009) 113(24):6112–9. doi:10.1182/blood-2008-07-170803
135. Baranek T, Manh TP, Alexandre Y, Maqbool MA, Cabeza JZ, Tomasello E, et al. Differential responses of immune cells to type I interferon contribute to host resistance to viral infection. Cell Host Microbe (2012) 12(4):571–84. doi:10.1016/j.chom.2012.09.002
136. Ruscanu S, Jouneau L, Urien C, Bourge M, Lecardonnel J, Moroldo M, et al. Dendritic cell subtypes from lymph nodes and blood show contrasted gene expression programs upon Bluetongue virus infection. J Virol (2013) 87(16):9333–43. doi:10.1128/JVI.00631-13
137. Banchereau R, Baldwin N, Cepika AM, Athale S, Xue Y, Yu CI, et al. Transcriptional specialization of human dendritic cell subsets in response to microbial vaccines. Nat Commun (2014) 5:5283. doi:10.1038/ncomms6283
138. Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature (2013) 498(7453):236–40. doi:10.1038/nature12172
139. Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature (2014) 505(7482):174–9. doi:10.1038/nature12826
140. van der Aa E, van Montfoort N, Woltman AM. BDCA3CLEC9A human dendritic cell function and development. Semin Cell Dev Biol (2014) (in press). doi:10.1016/j.semcdb.2014.05.016
141. Gallois A, Bhardwaj N. A needle in the ‘cancer vaccine’ haystack. Nat Med (2010) 16(8):854–6. doi:10.1038/nm0810-854
142. Tullett KM, Lahoud MH, Radford KJ. Harnessing human cross-presenting CLEC9A(+)XCR1(+) dendritic cells for immunotherapy. Front Immunol (2014) 5:239. doi:10.3389/fimmu.2014.00239
143. Meixlsperger S, Leung CS, Ramer PC, Pack M, Vanoaica LD, Breton G, et al. CD141+ dendritic cells produce prominent amounts of IFN-alpha after dsRNA recognition and can be targeted via DEC-205 in humanized mice. Blood (2013) 121(25):5034–44. doi:10.1182/blood-2012-12-473413
144. Sancho D, Joffre OP, Keller AM, Rogers NC, Martinez D, Hernanz-Falcon P, et al. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature (2009) 458(7240):899–903. doi:10.1038/nature07750
145. Zelenay S, Keller AM, Whitney PG, Schraml BU, Deddouche S, Rogers NC, et al. The dendritic cell receptor DNGR-1 controls endocytic handling of necrotic cell antigens to favor cross-priming of CTLs in virus-infected mice. J Clin Invest (2012) 122(5):1615–27. doi:10.1172/JCI60644
146. Huysamen C, Willment JA, Dennehy KM, Brown GD. CLEC9A is a novel activation C-type lectin-like receptor expressed on BDCA3+ dendritic cells and a subset of monocytes. J Biol Chem (2008) 283(24):16693–701. doi:10.1074/jbc.M709923200
147. Tel J, Aarntzen EH, Baba T, Schreibelt G, Schulte BM, Benitez-Ribas D, et al. Natural human plasmacytoid dendritic cells induce antigen-specific T-cell responses in melanoma patients. Cancer Res (2013) 73(3):1063–75. doi:10.1158/0008-5472.CAN-12-2583
148. Tel J, Sittig SP, Blom RA, Cruz LJ, Schreibelt G, Figdor CG, et al. Targeting uptake receptors on human plasmacytoid dendritic cells triggers antigen cross-presentation and robust type I IFN secretion. J Immunol (2013) 191(10):5005–12. doi:10.4049/jimmunol.1300787
149. Guery L, Hugues S. Tolerogenic and activatory plasmacytoid dendritic cells in autoimmunity. Front Immunol (2013) 4:59. doi:10.3389/fimmu.2013.00059
150. Goubier A, Dubois B, Gheit H, Joubert G, Villard-Truc F, Asselin-Paturel C, et al. Plasmacytoid dendritic cells mediate oral tolerance. Immunity (2008) 29(3):464–75. doi:10.1016/j.immuni.2008.06.017
151. Yu CI, Becker C, Wang Y, Marches F, Helft J, Leboeuf M, et al. Human CD1c+ dendritic cells drive the differentiation of CD103+ CD8+ mucosal effector T cells via the cytokine TGF-beta. Immunity (2013) 38(4):818–30. doi:10.1016/j.immuni.2013.03.004
152. Fuertes MB, Kacha AK, Kline J, Woo SR, Kranz DM, Murphy KM, et al. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med (2011) 208(10):2005–16. doi:10.1084/jem.20101159
Keywords: mononuclear phagocytes, comparative genomics, human, non-human primates, mouse, pig, sheep, chicken
Citation: Vu Manh T-P, Bertho N, Hosmalin A, Schwartz-Cornil I and Dalod M (2015) Investigating evolutionary conservation of dendritic cell subset identity and functions. Front. Immunol. 6:260. doi: 10.3389/fimmu.2015.00260
Received: 27 February 2015; Accepted: 11 May 2015;
Published: 02 June 2015
Edited by:
Florent Ginhoux, Singapore Immunology Network, SingaporeReviewed by:
Richard A. Kroczek, Robert Koch-Institute, GermanyMuzlifah Aisha Haniffa, Newcastle University, UK
Copyright: © 2015 Vu Manh, Bertho, Hosmalin, Schwartz-Cornil and Dalod. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marc Dalod, Centre d’Immunologie de Marseille-Luminy (CIML), Parc Scientifique et Technologique de Luminy, Case 906, Marseille Cedex 09 F-13288, France, dalod@ciml.univ-mrs.fr
 Thien-Phong Vu Manh
Thien-Phong Vu Manh Nicolas Bertho
Nicolas Bertho Anne Hosmalin
Anne Hosmalin Isabelle Schwartz-Cornil
Isabelle Schwartz-Cornil Marc Dalod
Marc Dalod