A Deg-protease family protein in marine Synechococcus is involved in outer membrane protein organization
- Scripps Institution of Oceanography, University of California at San Diego, La Jolla, CA, USA
Deg-family proteases are a periplasm-associated group of proteins that are known to be involved in envelope stress responses and are found in most microorganisms. Orthologous genes SYNW2176 (in strain WH8102) and sync_2523 (strain CC9311) are predicted members of the Deg-protease family and are among the few genes induced by copper stress in both open ocean and coastal marine Synechococcus strains. In contrast to the lack of a phenotype in a similar knockout in Synechocystis PCC6803, a SYNW2176 knockout mutant in strain WH8102 was much more resistant to copper than the wild-type. The mutant also exhibited a significantly altered outer membrane protein composition which may contribute to copper resistance, longer lag phase after transfer, low-level consistent alkaline phosphatase activity, and an inability to induce high alkaline phosphatase activity in response to phosphate stress. This phenotype suggests a protein-quality-control role for SYNW2176, the absence of which leads to a constitutively activated stress response. Deg-protease family proteins in this ecologically important cyanobacterial group thus help to determine outer membrane responses to both nutrients and toxins.
Introduction
Marine picocyanobacteria including Synechococcus and its sister taxon Prochlorococcus are found throughout the world oceans and contribute significantly to primary production and carbon cycling (Li, 1994; Richardson and Jackson, 2007). Marine Synechococcus is a highly diverse group consisting of multiple clades/species with different strategies for environmental niche adaptation that contribute to their global success (Ferris and Palenik, 1998; Rocap et al., 2002; Scanlan et al., 2009). For example, in an earlier paper comparing an open ocean strain, Synechococcus sp. strain WH8102 (WH8102), and a coastal strain, Synechococcus sp. strain CC9311 (CC9311), we found striking differences in sensitivity to copper and in the transcriptional response to elevated copper (Stuart et al., 2009). Despite the diversity in response within the group, marine Synechococcus species in general are quite sensitive to elevated copper levels, when compared to other phytoplankton groups (Brand et al., 1986). Understanding the similarities in the responses of Synechococcus strains to copper is key to understanding the nature of copper sensitivity in this globally important group.
In our previous work we found a few similarities in the transcriptional response to elevated copper, including a shared osmoregulatory-like response (Stuart et al., 2009). One gene that was up-regulated in both Synechococcus strains was a predicted serine protease (SYNW2176 in WH8102 and sync_2523 in CC9311), which is a member of the Deg-family of serine proteases. This family is characterized by a protease domain and one or more C-terminal PDZ domains. It is found in most organisms from bacteria to humans, and in bacteria is often involved in tolerance to protein folding stress (Clausen et al., 2002). Most bacteria have at least one of these proteins, but many have multiple paralogs, which can have variable functions. The family has been well-characterized in Escherichia coli (E. coli) which has three Deg proteases, DegS, DegP, and DegQ, that are either periplasmic or embedded in the membrane (reviewed in Meltzer et al., 2009). All three appear to be involved in cell envelope stress, and regulation of bacterial envelope perturbations in E. coli have been well characterized (Raivio, 2005). DegS serves a regulatory role by sensing damaged outer membrane proteins and subsequently degrading an anti-sigma factor (RseA) initiating an envelope-stress regulatory cascade. DegS is the only one of the three Deg-proteases that is essential in E. coli (Alba et al., 2001). DegP can act as both a protease and a chaperone depending on temperature, and is thought to be a protective factor regulated by both the Cpx and σE envelope stress pathways (Raffa and Raivio, 2002; Meltzer et al., 2009). DegQ is perhaps the least well-characterized of the three in E. coli, but overexpression has been shown to partially rescue a DegP null phenotype and DegQ exhibits higher chaperone and lower protease activity in vitro than DegP, indicating overlapping functions for these two (Waller and Sauer, 1996; Bai et al., 2011).
In E. coli, the majority of redox active copper is in the periplasm (Macomber et al., 2007) and many copper response mechanisms involve periplasmic copper detoxification so copper stress and cell envelope stress can be related. Indeed, in E. coli, CopA, a cytoplasmic copper efflux protein, is regulated by CpxR, part of an envelope stress response pathway. CutC, a copper chaperone, is regulated by RpoE, which is part of the σE envelope stress response pathway (Rensing and Grass, 2003). Although DegP is not induced by copper stress in E. coli, which has much higher tolerance to elevated copper levels than marine Synechococcus, DegP is induced by zinc stress and is also regulated by RpoE (Egler et al., 2005).
In a fresh water cyanobacterium, Synechocystis PCC6803 (Synechocystis), the three Deg proteases (slr1204/htrA/degP, sll1679/hhoA/degQ, and sl1427/hhoB/degS) are all localized in the inner membrane and periplasm (Roberts et al., 2012) and have transcriptionally distinct but overlapping responses: all respond to light to dark transitions; htrA and hhoB respond to salt stress; only hhoB responds to high light; only htrA responds to cold shock (Jansen et al., 2005). In Synechocystis, mutants in which only two of the three are inactivated exhibit no phenotype; all three need to be knocked out in order to observe a phenotype which then includes both light and heat sensitivity as well as a hyperpiliated outer membrane (Barker et al., 2006). Thus, in contrast to E. coli where DegS is an essential protein, all three Deg proteins in Synechocystis are nonessential and appear to have overlapping functions, despite differing in transcript abundance and induction patterns. Additionally, recombinantly expressed versions of all these Synechocystis proteins degrade unfolded substrates and some phycobiliproteins in vitro (Diao et al., 2011; Huesgen et al., 2011). This suggests a role in protein quality control through sensing and processing of improperly folded proteins.
In marine Synechococcus WH8102, SYNW2176 has a varied transcriptional response to stressors. Along with strong upregulation in response to copper stress, SYNW2176 is slightly upregulated in response to early phosphate starvation and in a phosphate regulatory gene mutant (ptrA/SYNW1019). It is downregulated in response to late phosphate starvation, in a phosphate regulatory gene knockout mutant (SYNW0947) and in co-culture with Vibrio parahaemolyticus (which can induce phosphate stress) (Tai et al., 2009; Tetu et al., 2009; Thomas et al., 2009; Ostrowski et al., 2010). Transcript abundance of a SYNW2176 homolog in Synechococcus sp. strain WH7803 (SynWH7803_2187) is induced by methyl viologen and H2O2, but not high light (Blot et al., 2011). The two other Deg-family protease paralogs (SYNW0608, SYNW1537) do not appear to be differentially regulated by the conditions tested so far including copper toxicity, nickel-deprivation, phosphate limitation, co-culturing with Vibrio parahemolyticus, variable nitrogen source and DNA damaging agents (Su et al., 2006; Stuart et al., 2009; Tai et al., 2009; Tetu et al., 2009, 2013; Dupont et al., 2012). Given the varied nature of stressors that affect transcript abundance of this gene, we predicted that SYNW2176 is involved in envelope stress response in marine Synechococcus.
In this study we inactivated SYNW2176 and characterized the stress response and outer membrane protein composition of the mutant in order to better understand the overlap between envelope stress and copper stress in marine Synechococcus.
Materials and Methods
Phylogenetic Analyses of Cyanobacterial Deg-Family Proteases
Deg-family protease homologs were identified from sequenced genomes of 11 marine Synechococcus strains (WH8102, CC9605, BL107, CC9902, WH78903, WH7805, RS9917, CC9311, RS9916, WH5701, and RCC307) and 12 Prochlorococcus strains (MIT9303, MIT9313 CCMP1375, MIT9211, MIT9812, MIT9301, AS960, MIT9215, MIT9515, CCMP1986, NATL2A, and NATL1A). Deg-family proteases are characterized by the presence of both a C-terminal PDZ domain and a trypsin domain and identified in NCBI as COG0265. Homologs were first identified using the MicrobesOnline database (Price et al., 2009) to search for paralogs and homologs of SYNW2176 and COG0265. These results were then verified using published data of genome-wide comparisons of the 11 Synechococcus strains conducted by Dufresne et al. (2008) where the three groups were separately clustered for Synechococcus (Clusters 8096, 8095, and 13). The genes were also searched against the ProPortal database (Kelly et al., 2012) where the genes are grouped in one cluster (CyCog4045). Finally, a reciprocal BLAST was done for each gene found against its own respective genome to identify any paralogs that might have been missed. From this search a total of 59 Deg-family proteases were identified in the 23 genomes searched. The amino acid sequences were then aligned using MUSCLE (Edgar, 2004), trimmed manually and curated using Gblocks 0.91 (Talavera and Castresana, 2007). A maximum likelihood tree was then built using PhyML (WAG substitution model, 4 substitution rate categories) and branch support values were calculated with an approximate likelihood ratio test (aLRT SH-like) (Anisimova and Gascuel, 2006; Dereeper et al., 2008; Guindon et al., 2010). The alignment was later run through ProtTest (Abascal et al., 2005) which determined that an LG substitution model was best. The LG-based tree and the WAG-based tree were very similar, with one difference that included a change in position for the two group 3 Prochlorococcus genes within the group 3 branch. Since this was of uncertain significance and the support values for the WAG-based tree were better, the WAG-based tree is shown. TreeDyn was used for initial tree rendering. Conserved neighborhoods were assessed using MicrobesOnline (Price et al., 2009).
QRTPCR of WH8102 Following Copper Addition
Three-liter batch cultures of WH8102 were grown to mid-exponential phase in chelexed synthetic ocean water (SOW) (Dupont et al., 2008; Stuart et al., 2009) and 10 μM CuEDTA (pCu 10.1),1 μM CuEDTA (pCu 11.1) or water (control) was added. For the 10 μM addition, 500 ml of culture was harvested by centrifugation at 1, 2, 4, 6, and 8 h after addition in three biological replicates. For the 1 μM addition samples were harvested at 2 and 24 h after addition in two biological replicates. RNA was extracted from the cell pellet, cDNA synthesized and QRTPCR performed as described in Stuart et al. (2009), with the exception that QRTPCR was normalized to total RNA, 250 ng of total RNA was used in triplicate cDNA reactions for each sample. DNA contamination controls with no reverse transcription were performed with every reaction. Primers for genes assayed are listed in Table S2. Three technical replicates were ran for each of the three biological replicates. A genomic DNA standard curve was used to calculate average transcript abundance for each sample, with subtraction of DNA-contaminant control. Biological triplicates were then averaged and log2 fold changes between no-Cu controls and Cu-treated samples were calculated for each gene and time point. Statistical significance of the fold change was calculated by a one-sample t-test and a p-value less than 0.05 was considered significant.
Insertional Inactivation of SYNW2176
Gene inactivations were conducted as described in Brahamsha (1996b) with several modifications in vector construction for the selection of double recombinants. Synechococcus sp. strain SYNW2176- in which the gene SYNW2176 is interrupted by an antibiotic resistance cassette, kanamycin (kanR) was constructed as follows. A 1997-bp region surrounding the 1125-bp gene was PCR-amplified (Figure S2, Table S2, 2176 dbl primers) and cloned into pCR®2.1-TOPO® (Life Technologies, Carlsbad CA) according to the manufacturer's specifications. The vector was then digested with EcoRI and the 1997 bp + 10 bp insert was ligated into the EcoRI site of pBR322 (New England Biolabs, Ipswich, MA). Next the vector was digested with SmaI and the kanamycin resistance cassette (kanR) from pUC4K (Taylor and Rose, 1988) was cut out of the pUC4K (using BamHI digestion, gel purification and blunting) and ligated into the SmaI cut site. The construct was then digested with EcoRI and the insert was ligated into pRL271, a vector for sacB-mediated positive selection for double recombinants in gram-negative bacteria (Cai and Wolk, 1990). The new vector, pRL271_kanR_2176 was then transformed into E. coli MC1061 (pRK24, pRL528) and introduced via conjugation into Synechococcus sp. strain WH8102 as described in Brahamsha (1996b). The pour plates contained 5% sucrose to select for double recombinants, and 25 μg/ml kanamycin. Mutants were verified using PCR (Figure S2, Table S2).
Cell Culture and Growth Assays
One liter batch cultures of Synechococcus sp. strain WH8102 (WH8102), and SYNW2176- were grown in chelexed SOW with stirring as previously described in Stuart et al. (2009). For mutant cultures, kanamycin was added at a final concentration of 20 μg/ml to 50 ml stock cultures that were transferred weekly, but antibiotics were not added to 1 L growth assay cultures, because kanamycin may bind copper. For growth assays, cultures were grown to early log phase and 50 ml were aliquoted into acid-washed glass flasks and placed on a shaker plate at 23°C. Cultures were given 1 day in these conditions to adjust to the transfer, after which CuEDTA was added in triplicate at 10, 4, 1, or 0.4 μM and sterile milliQ water was added to the controls. Using the MINEQL algorithm (Westall et al., 1976) this approximately corresponds to equilibrium free copper levels equivalent to pCu 10.1, pCu 10.5, pCu 11.1, and pCu 11.5. Flow cytometry samples were collected daily for up to 7 days and were fixed and analyzed as described in Stuart et al. (2009).
Outer Membrane Protein Preparations and Band Excision
Outer membrane protein preparation was done as described in Brahamsha (1996a) and McCarren and Brahamsha (2007) with the exception that SOW medium was used for culturing and washing instead of SN. Briefly, 1 L of WH8102 or 2176- was grown up to the mid-exponential phase. For copper stress conditions, CuEDTA was added during mid-exponential at pCu 10.5 (4 μM) and samples were incubated for 6 h before harvesting. 10 ml of culture was filtered through a 0.2 μm polycarbonate filter and the spent medium was frozen at −80°C. Cells were harvested by centrifugation, washed with sterile medium, and resuspended in 50 mM Tris pH 8.0, 50 mM EDTA and 15% sucrose and incubated on ice for 30 min. Samples were subsequently spun down at 10,000 × g for 10 min at 4°C. The cell pellet from which the outer membrane had been stripped was frozen and the supernatant containing the outer membrane fraction was spun at 28,000 × g for 90 min at 4°C. The resulting high speed pellet (HSP), which consists of the outer membrane, was resuspended in sterile water. The high-speed supernatant (HSS), which contains periplasmic proteins as well as cell-surface proteins solubilized by EDTA, and the spent medium fraction were concentrated with an Amicon Ultra 4 30,000 MWCO filter (Millipore, Billerica, MA). Protein fractions were run on NuPAGE Novex Tris-Acetate 3–8% gels (Life Technologies, Carlsbad, CA) under denaturing conditions according to the manufacturer's specifications. For imaging, gels were stained with SYPRO Ruby stain (Life Technologies, Carlsbad, CA) according to the manufacturer's specifications. For band excision, Coomassie Blue R-250 (Thermo Scientific) was used to stain according to the manufacturer's specifications. Excised bands were sequenced by the UCSD Biomolecular/Proteomics Mass Spectrometry Facility.
OMP Fraction LC-MS-MS
Sample preparation (Guttman et al., 2009)
Protein samples were diluted in TNE (50 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA) buffer. RapiGest SF reagent (Waters Corp.) was added to the mix to a final concentration of 0.1% and samples were boiled for 5 min. TCEP (Tris (2-carboxyethyl) phosphine) was added to 1 mM (final concentration) and the samples were incubated at 37°C for 30 min. Subsequently, the samples were carboxymethylated with 0.5 mg/ml of iodoacetamide for 30 min at 37°C followed by neutralization with 2 mM TCEP (final concentration). Proteins samples prepared as above were digested with trypsin (trypsin:protein ratio—1:50) overnight at 37°C. RapiGest was degraded and removed by treating the samples with 250 mM HCl at 37°C for 1 h followed by centrifugation at 14,000 rpm for 30 min at 4°C. The soluble fraction was then added to a new tube and the peptides were extracted and desalted using Aspire RP30 desalting columns (Thermo Scientific).
LC-MS-MS
Trypsin-digested peptides were analyzed by high pressure liquid chromatography (HPLC) coupled with tandem mass spectroscopy (LC-MS/MS) using nano-spray ionization (McCormack et al., 1997). The nanospray ionization experiments were performed using a TripleTof 5600 hybrid mass spectrometer (ABSCIEX) interfaced with nano-scale reversed-phase HPLC (Tempo) using a 10 cm–100 micron ID glass capillary packed with 5-μm C18 Zorbax™ beads (Agilent Technologies, Santa Clara, CA). Peptides were eluted from the C18 column into the mass spectrometer using a linear gradient (5–60%) of ACN (Acetonitrile) at a flow rate of 250 μl/min for 1 h. The buffers used to create the ACN gradient were: Buffer A (98% H2O, 2% ACN, 0.2% formic acid, and 0.005% TFA) and Buffer B (100% ACN, 0.2% formic acid, and 0.005% TFA). MS/MS data were acquired in a data-dependent manner in which the MS1 data was acquired for 250 ms at m/z of 400–1250 Da and the MS/MS data was acquired from m/z of 50–2000 Da. For Independent data acquisition (IDA) parameters MS1-TOF 250 ms, followed by 50 MS2 events of 25 ms each. The IDA criteria, over 200 counts threshold, charge state +2–4 with 4 s exclusion. Finally, the collected data were analyzed using MASCOT® (Matrix Sciences) and Protein Pilot 4.0 (ABSCIEX) for peptide identifications.
Spectral count analysis
Analysis of spectra was done using a spectral counting statistical method. Briefly, ProteinPilot software v.4.0 (Applied Biosystems, MDS-Sciex) was used to identify and quantify proteins and generate confidence numbers for each spectra with the Paragon search algorithm (Shilov et al., 2007). All protein spectra with less than 95% confidence to a Synechococcus WH8102 protein were discarded. All the remaining hits in each sample were summed for a total number of spectral counts per sample. Raw spectral counts ranged between 397 and 760 in the insoluble fraction (HSP) and 869 and 1655 in the soluble fraction (HSS). Then the number of hits per protein were summed and divided through by the total to give a percentage of total spectral counts for each protein in order to compare protein levels between samples. Duplicates were averaged and a standard deviation calculated. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (Vizcaino et al., 2014) via the PRIDE partner repository with the dataset identifier PXD000989.
Alkaline Phosphatase Activity Assay and P Analysis
Alkaline phosphatase activity assays and phosphate analysis were carried out using colorimetric assays as described in Tai et al. (2009). Briefly, for alkaline phosphatase activity 180 μl of sample of standard were put in a microplate and 20 μl 135 mM p-nitrophenyl phosphate in 1 M Tris (pH 8.0) added. Plates were read at 405 nM absorbance at 0, 30, 60, and 90 min. Phosphate concentrations were measured using a standard ammonium molybdate assay, samples were diluted 20-fold and 200 μl was added to a microplate with 20 μl reagent, incubated for 30 min and the absorbance at 880 nM was measured. Duplicate 50 ml cultures were inoculated with either wild-type WH8102 or SYNW2176- into SOW media and assayed for 14 days. Cell counts were determined by flow cytometry using a BD FACSort as described in Stuart et al. (2009).
Results
Phylogenetic Grouping of Deg Proteases
Eleven sequenced genomes of marine Synechococcus all contain three Deg-family proteases, which can be separated phylogenetically into three main groups. The picocyanobacterial deg-family proteases have only one PDZ domain, compared with the E. coli family, which has two. Of the 12 Prochlorococcus genomes included in the analysis only two (strains MIT9313 and MIT9303, underlined) had a group 3 Deg protease (Figure 1A). MIT9313 and MIT9303 have much larger genomes than the other 10 Prochlorococcus strains analyzed (2.4 and 2.6 Mbp vs. 1.6–1.8 Mbp). In addition, the group 1 proteins from MIT9313 and MIT9303 grouped with the Synechococcus sequences in contrast with a more typical phylogenetic placement at the base of Prochlorococcus gene trees as seen in their group 2 proteins. The three Deg-family proteases previously studied from the freshwater cyanobacterium Synechocystis PCC6803 were most similar to each other. Based on pairwise BLASTp scores group 1 member SYNW2176 was more similar to HhoA (DegQ) and group 2 member SYNW0608 was more similar to HtrA (DegP). However, the alignment based trees using all members of the group did not support this. It is thus difficult to predict whether group 1 is homologous to HtrA/DegP or HhoA/DegQ. Although there was no clear predicted operon for any of the three Deg protease groups, the surrounding genes for each of these groups in Synechococcus were also well conserved (Figure 1B).
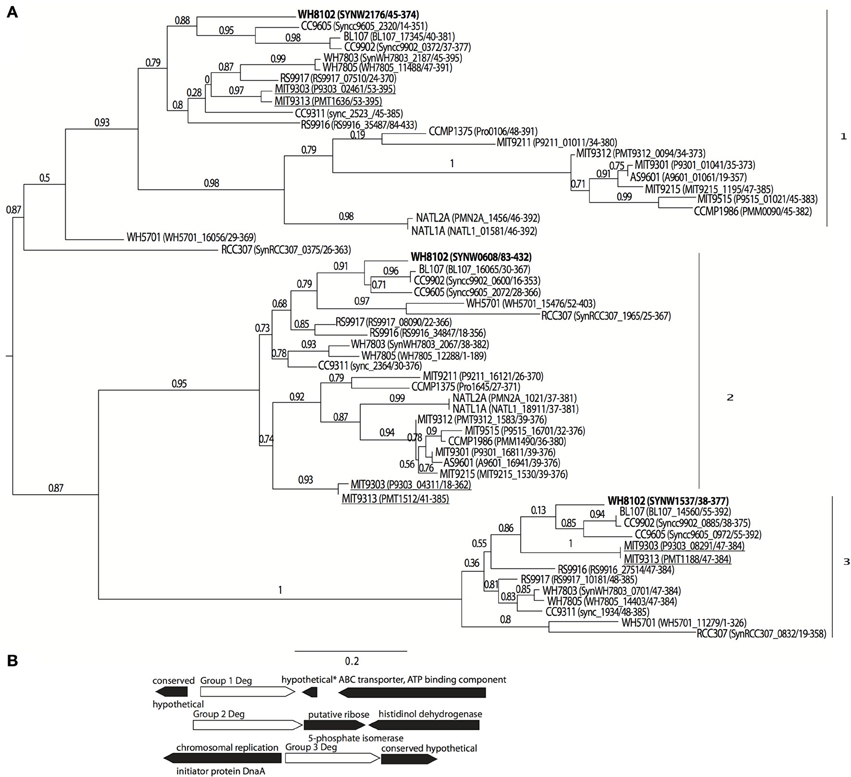
Figure 1. Phylogenetic grouping of Deg-family proteases in picocyanobacteria. (A) Maximum likelihood tree based on amino acid sequence alignment in 11 marine Synechococcus, and 12 Prochlorococcus strains. Protein sequences were aligned with MUSCLE (Edgar, 2004) and the tree was built using PhyML (WAG substitution model) and TreeDyn (Dereeper et al., 2008; Guindon et al., 2010). Branch support values calculated using approximate likelihood ratio test (aLRT SH-like). Labels are strain (locus number/amino acid positions aligned). WH8102 versions are bolded and Prochlorococcus strains with a group 3 member are underlined (B) Schematic of conserved gene neighborhood surrounding the aligned proteins from Synechococcus members in tree. “*” indicates gene not conserved in all strain gene neighborhoods.
SYNW2176 Shows Sustained Induction of Expression in Response to High and Moderate Copper
We used QRTPCR to investigate induction over time in response to copper stress (pCu 10.1 addition). In previous microarray expression work on copper stress, we found that SYNW2176 was induced after 2 h of copper stress at both pCu 10.1 (Cu2+ 10−10 M; 10 μM CuEDTA) and pCu 11.1 (Cu2+ 10−11 M; 1 μM CuEDTA). The other two paralog Deg-proteases (SYNW0608, SYNW1537) were not differentially regulated by copper stress in either strain WH8102 or CC9311 (Stuart et al., 2009). We looked at expression of three genes including SYNW2176, SYNW0807 (a predicted sensor histidine kinase thought to be involved in osmoregulatory response) and SYNW0514 (predicted GroEL chaperonin). The putative specialized stress response genes, SYNW2176 and SYNW0807, both sustained induction over at least 6 h in response to copper stress whereas for GroEL, the general stress response gene, induction was only detected at 2 h after copper stress (Figure 2). We also looked at expression following a lower copper addition (pCu 11.1) over 24 h and found that SYNW2176 was induced at 2 h and maintained induction over 24 h (Figure S1). In contrast, GroEL was downregulated and SYNW0807 was not induced at 2 h by this lower level of copper, but by 24 h had increased (Figure S1). Based on expression data which showed sustained induction to both high and moderate copper additions, we expected that a SYNW2176 mutant would be more sensitive to copper.
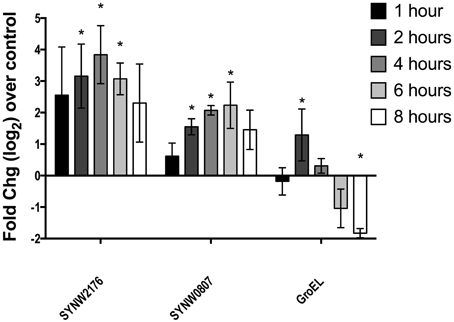
Figure 2. Gene expression following copper addition. QRTPCR of SYNW2176 and two other genes induced in WH8102 2 h after pCu10.1 addition. All values are calculated as log2fold changes over the no-copper control. Error bars represent one standard deviation between three biological replicates. Stars represent statistical significance (p < 0.05) of one sample t-test (mean = 0).
SYNW2176- is Tolerant to Higher Levels of Cu than Wild Type
We inactivated Deg-family protease SYNW2176 and investigated growth in response to copper. Insertional inactivation of SYNW2176 through double recombination (designated strain “SYNW2176-”) was validated using PCR (Figure S2, Table S2). Surprisingly, growth assays with copper additions showed that SYNW2176- exhibited a clear increased tolerance to copper additions relative to the wild-type (Figure 3). The wild-type strain showed decreased growth rates relative to the no-Cu control in response to copper levels as low as pCu 11.5 (0.4 μM CuEDTA, Cu2+ at 10−11.5 M) and failed to grow at all at pCu 11.1 (1 μM CuEDTA), whereas SYNW2176- only started to show a significantly decreased growth rate at pCu10.5 (4 μM CuEDTA).
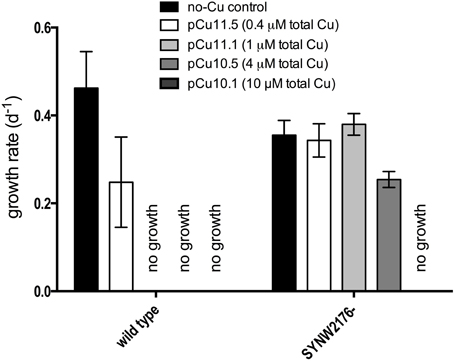
Figure 3. Response to copper stress in mutant and wild type cultures. Growth rates calculated from slope of linear regression on ln-transformed cells/ml from three biological replicates over at least 3 days of exponential growth after treatment. Error bars represent one standard deviation between three biological replicates.
Cu-Responsive Outer Membrane Proteins in Wild-Type Cells
Since envelope stress may affect the outer membrane composition we looked at copper induced changes in wild-type outer membrane fractions (Brahamsha, 1996a; McCarren and Brahamsha, 2007). The technique generates two outer membrane fractions, an insoluble HSP consisting of the outer membrane and a soluble high speed supernatant (HSS) fraction largely consisting of cell surface proteins solubilized by the EDTA treatment. Using HPLC tandem mass spectrometry (“shotgun proteomics”) we sequenced single samples of insoluble outer membrane protein preparations (HSP) from both a control and 6-h copper stress (pCu 10.5) in WH8102. A total of 49 proteins were detected (with at least two unique peptide hits above 95% confidence) with 47 proteins detected in the control condition and 26 under copper stress (Table S1). The two proteins found only under copper stress were SYNW0418, a hypothetical protein of unknown function, and SYNW2077, a 50S ribosomal protein (Table S1).
WH8102 has four genes annotated as possible porins, of which only two were detected in our insoluble outer membrane fractions, SYNW2224, and SYNW2227. Under copper shock these two porins (SYNW2224 and SYNW2227) appeared to be downregulated because peptide spectral count percentages (proportion of total spectral counts) from both porins combined were lower under copper (7.4%) than the control condition (10.4%). Also notable under copper shock were higher levels of the periplasmic phosphate binding ABC transporter protein (SYNW1018) and undetectable levels of SwmB, a cell surface motility protein (Table S1). SYNW2176 itself was not detected in either sample and given that it has transmembrane domains this may indicate that it is located in the inner membrane or conversely its abundance is below the detection limit. Copper treatment clearly caused changes in relative protein abundance in the cell surface/outer membrane compartment of the cells.
SYNW2176- has an Altered Outer Membrane Composition Under Exponential Growth
We examined the outer membrane protein composition of SYNW2176- in two ways, first using gel electrophoresis and then shotgun proteomics. We compared the outer membrane profiles of the wild type and the SYNW2176- mutant under exponential growth. The gels showed a distinctly different pattern of bands in SYNW2176- relative to the wild-type in the insoluble HSP fraction (Figure 4). Most notably, there was an additional band in the mutant at around 120 kDA. This band was also detectable in the soluble HSS fraction (Figure S3A). These differences remained unchanged 6 h after copper additions (Figure S3B). There was also a distinct difference in the ~80 kDA region in the mutant in the soluble and insoluble fractions. In the mutant the band was sharper and darker relative to the wild-type.
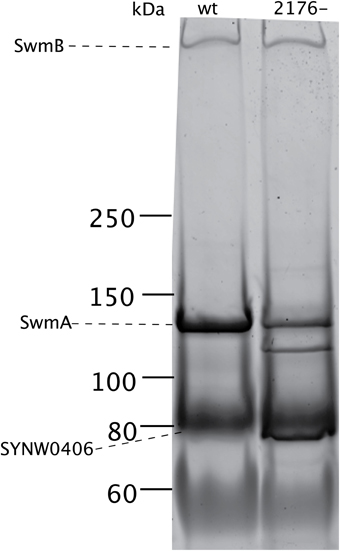
Figure 4. Outer membrane protein differences between wild type and mutant cultures. SDS-PAGE protein gel (3–8% Tris-Acetate) of insoluble outer membrane fraction (HSP). Dashed lines indicate previously identified abundant outer membrane proteins (Brahamsha, 1996a; McCarren and Brahamsha, 2007).
Duplicate biological replicates of the soluble (HSS) and insoluble (HSP) fractions of outer membrane samples from SYNW2176- and wild-type in exponential growth were also analyzed with shotgun proteomics. In the insoluble fraction a total of 11 proteins were detected in both SYNW2176- biological replicates (with at least two unique peptide hits above 95% confidence) and 12 proteins were detected in both wild-type replicates. In the soluble fraction 13 proteins were detected in SYNW2176- and 12 in wild-type. There were a total of 22 proteins detected in at least one set of biological replicates (Table 1).
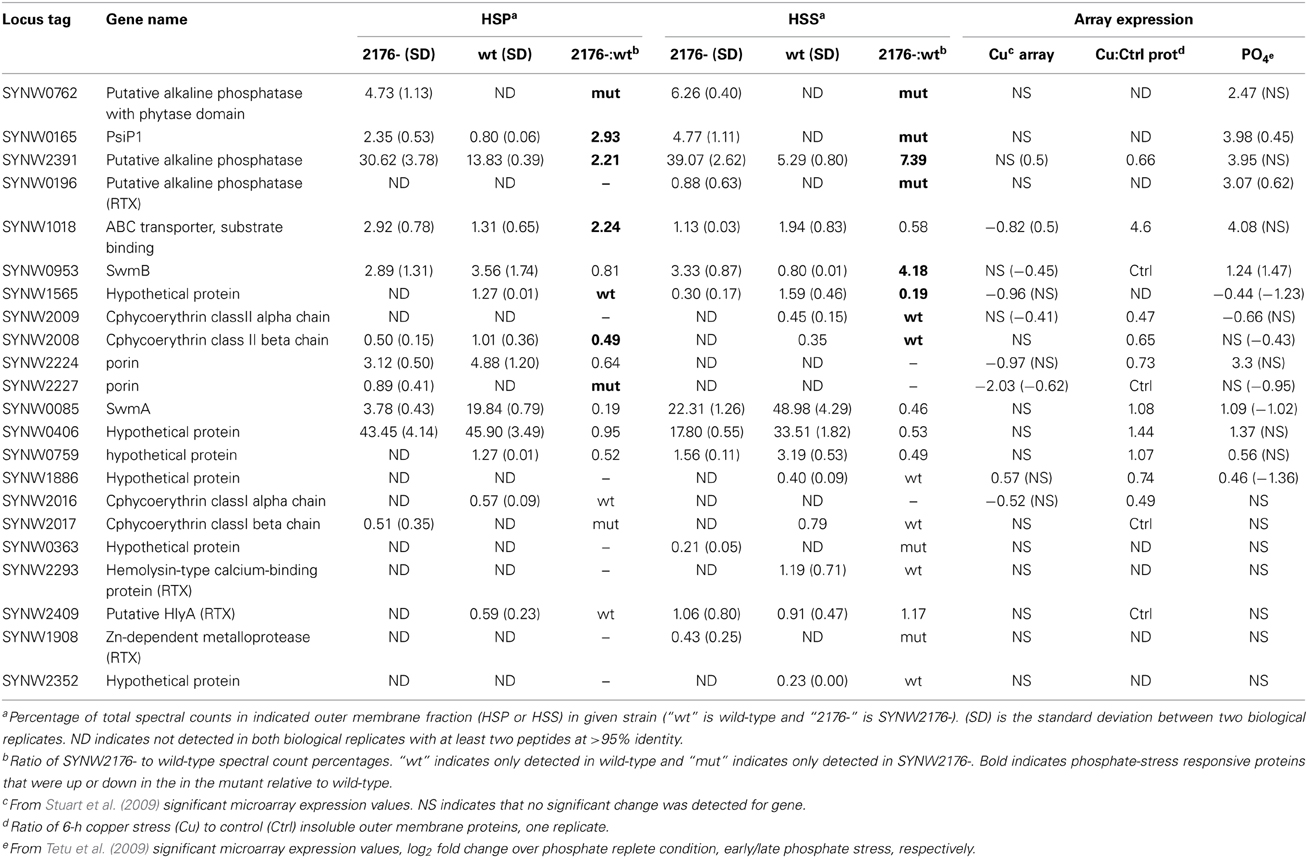
Table 1. Comparison of outer membrane proteins detected in wild-type and SYNW2176- (Average % spectral counts).
Several differences in protein levels were detected between the wild-type and mutant, a few of which were also induced in wild-type under copper stress (Table 1). One difference was in the abundant porin, SYNW2224, which was lower in both the mutant and under copper stress compared to wild-type. Also, higher levels of SYNW1018 (an ABC transporter phosphate-binding protein) were found in both the mutant and under copper stress compared to wild-type.
The less abundant porin SYNW2227 was only detected in the mutant, and it was not detected following 6 h of copper stress to the wild-type. Comparing the two porins revealed that predicted isoelectric points of SYNW2227 and SYNW2224 were drastically different. Modeling of the outer membrane loops using published methods (Bagos et al., 2004) revealed far fewer positive residues in the outer loops of SYNW2227 (Figure S4, Table S4). SYNW2227 is also missing two cysteine residues on the predicted outer loop portion of the protein that are present in SYNW2224 and one of the other undetected porins, SYNW2223. These differences between the porins suggest that SYNW2227 may have reduced permeability to copper or reduced sensitivity to copper due to the fewer cysteines potentially sensitive to copper binding.
Insertional Inactivation of SYNW2176 Results in the Expression of Proteins Normally Induced Under Phosphate Limitation
There was little overlap between proteins upregulated in the SYNW2176- mutant and genes identified from our previous transcriptional work on Cu-stress that we predicted would be upregulated in the mutant given its copper tolerance (Table 1). Instead, there was a strong correlation with phosphate-stress inducible genes (Table 1). Several studies have looked at the phosphate limitation response in Synechococcus WH8102, including two microarray expression studies by Tetu et al. (2009) and Thomas et al. (2009) and a study by Mazard et al. (2012) that compared the phosphorus stress response in several Synechococcus strains. Of the 22 identified proteins in our study, 10 are differentially transcriptionally regulated under phosphate-starvation and similarly differ in protein abundance in the SYNW2176- mutant relative to wild-type (Table 1).
The most apparent difference was the presence of an additional protein in the mutant, SYNW0762, which is encoded by a phosphate-stress inducible gene. SYNW0762 is annotated as a hypothetical protein whose closest BLAST hit is to an atypical alkaline phosphatase with a phytase domain in Synechococcus WH7803 (Table 1). The protein has two large repeat regions that are identified as WD40/YVTN repeat-like domains. When the additional band seen in the 120 kDa region of mutant outer membrane preparations was excised and sequenced, this protein was the most abundant hit (Figure S3B, labeled “1,” Table S3). Also, SYNW0165, a phosphate-inducible protein of unknown function, was almost 3-fold higher in the mutant insoluble fraction and in the soluble fraction it was only detected in the mutant. Two putative alkaline phosphatases, SYNW2391, and SYNW0196, both of which are induced by phosphate-stress (Tetu et al., 2009) were also detected at higher levels in the mutant.
Two of the proteins identified were both phosphate inducible and copper stress responsive—the phosphate ABC transporter SYNW1018 and SwmB were both more abundant in the mutant. SYNW1018 was 2.2-fold more abundant in the mutant insoluble fraction and SwmB was 4-fold higher in the mutant soluble fraction. Additionally, two proteins that are down-regulated transcriptionally by phosphate stress in the wild-type (Tetu et al., 2009), SYNW2009 and SYNW1565, were less abundant in the mutant. It should be noted that five phosphate-stress inducible proteins did not follow this pattern: the aforementioned porin SYNW2224, SwmA, and SYNW0759, were more abundant in wild-type; SYNW0406 for which no difference in protein abundance was detected; and SYNW1886 which was only detectable in the wild-type. SYNW2224, the porin that is down-regulated in the mutant, is an interesting exception since it is one of only two proteins detected that was upregulated under phosphate limitation but downregulated under copper stress. All outer membrane preparations were obtained from exponentially growing cultures (in media with 86 μM PO4) that were not expected to be phosphate-limited at this stage of growth.
Alkaline Phosphatase Activity is not Induced in the SYNW2176- Mutant in Response to Phosphate Depletion
Considering the abundance of phosphate-stress inducible proteins expressed in SYNW2176- we compared alkaline phosphatase activity, a hallmark of the onset of phosphate limitation, between SYNW2176- and the wild-type. Mutant and wild-type cultures were inoculated in phosphate-replete media, and alkaline phosphatase activity and phosphate levels were measured during growth. The mutant exhibited alkaline phosphatase activity that did not change significantly over the growth period (Figure 5). By contrast, in wild-type, as expected, alkaline phosphatase activity was very low until day 11, when phosphate levels began to decrease, and then increased dramatically. In the mutant, low activity was detected by day 6, when phosphate levels were still high, and activity stayed low but detectable throughout the rest of the experiment. The mutant grew slightly more slowly than the wild-type under these conditions, but by day 6 after inoculation both strains were in exponential phase (Figure 5B). Since the alkaline phosphatase assay has a low sensitivity threshold, to confirm the early time points when cell numbers were low we replicated the experiment and concentrated the samples 50-fold before measurement. From this we were able to confirm that while in exponential growth (days 6 and 7), the mutant does have slightly higher activity (8.3 × 10−10 μmoles/min/106 cells/ml ± 4.0 × 10−10) than the wild-type (3.58 × 10−10 μmoles/min/106 cells/ml ± 4.6*10−11) while both had high phosphate levels (wild-type 50 μM ± 1.4 and mutant 74 μM ± 8.3). The mutant maintains alkaline phosphatase activity around 1.0 × 10−9 μmoles/min/106 cells/ml never having levels above 2.0 × 10−9 μmoles/min/106 cells/ml, whereas the wild-type has very low activity until phosphate levels decrease after which it induces activity sharply to around 6.0 × 10−9 μmoles/min/106 cells/ml. From both these experiments it is clear that the mutant has constitutive, low alkaline phosphatase activity that is not induced by phosphate levels. This suggests either a constitutive stress response or an inability to sense phosphate levels.
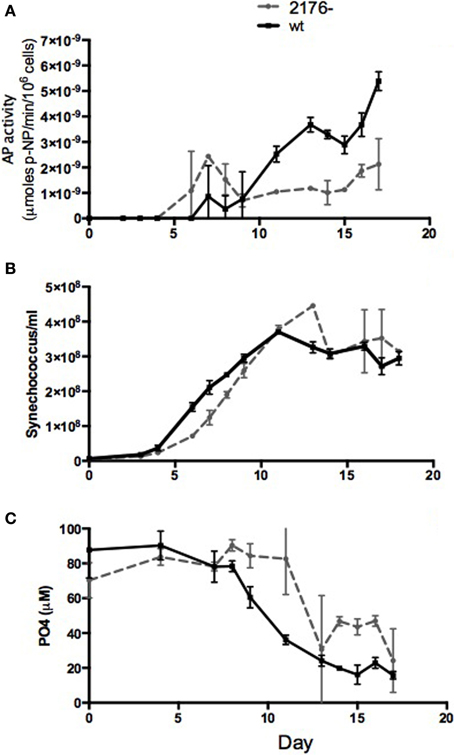
Figure 5. Comparison of alkaline phosphatase activity in wild type and mutant cultures over 16 days. (A) alkaline phosphatase activity. AP activity is reported as μmol of para-nitrophenol (p-NP) produced per minute per 106 cells, (B) cell counts, and (C) phosphate levels.
Discussion
Stress-Induced Expression of Group 1 Deg-Family Proteases in Marine Synechococcus
The phylogenetic grouping of the Deg-family proteases in marine Synechococcus shows three distinct groups. The group 1 Deg-proteases appear to have an inducible function distinct from the other 2 groups. Our transcriptional data show that SYNW2176 exhibits a sustained high level of induction following copper stress for at least 24 h and is quite sensitive to copper as it was induced by 2 h at the lower copper level as well (Figures 2, S1). This is in contrast to GroEL chaperonin, which is induced only at 2 h following the high copper addition and suggests a more specialized stress response for SYNW2176, perhaps to envelope stress, as in E. coli (Meltzer et al., 2009). Published expression data also shows inducible activity in the group 1 members with SYNW2176 and sync_2523 both upregulated in response to copper addition and SYNW2176 slightly upregulated in response to early phosphate stress and downregulated in response to late phosphate starvation (Stuart et al., 2009; Tetu et al., 2009). Additionally, the group 1 member SynWH7803_2187 is up in response to methyl viologen and H2O2, also oxidative stressors (Blot et al., 2011). In contrast, neither group 2 nor 3 members in CC9311 or WH8102 are upregulated in response to copper, and in WH7803, the group 3 member is only slightly upregulated in response to methyl viologen and H2O2 (Blot et al., 2011).
Most of the Prochlorococcus strains do not have a group 3 Deg-protease suggesting that group 3 may serve a redundant function that can be compensated for by the other two in the more streamlined Prochlorococcus genomes. Prochlorococcus strains that have been tested are more sensitive to copper than Synechococcus. Strain MED4 (which lacks a group 3 member) is in fact more tolerant to copper than MIT9313 and SS120 (Mann et al., 2002). There currently are not equivalent transcriptional stress response experiments on Prochlorococcus. However, from data available on high light stress in Prochlorococcus strain MED4, there is upregulation of the group 2 member in response to high light and blue light, but no regulation of the group 1 member in response to light, nitrogen, phosphate, or iron stress (Martiny et al., 2006; Steglich et al., 2006; Tolonen et al., 2006; Thompson et al., 2011). In a manner more similar to Synechococcus, Prochlorococcus MIT9313, which has three Deg proteases, upregulates its group 1 member in response to nitrogen limitation but does not appear to upregulate either of its other Deg proteases in response to nitrogen, phosphate, or iron stress (Martiny et al., 2006; Tolonen et al., 2006; Thompson et al., 2011). From these limited data it appears that the Prochlorococcus strains lacking a group 3 member may have different induction responses to stress for the remaining two Deg proteases.
SYNW2176- Copper-Resistant Mutant Phenotype
Our data on the SYNW2176- mutant reveals a complex phenotype. This includes increased copper tolerance and a significantly altered outer membrane composition with a putative alkaline phosphatase/phytase (SYNW0762) not detected under in wild type. The mutant outer membrane also has an extra porin and lower levels of the main porin (which is downregulated by copper in wild type cells.). The phenotype also includes higher abundance of a copper-inducible outer membrane protein (SYNW1018); higher levels of many phosphate-limitation-induced proteins; a longer lag phase after transfer; and a lack of phosphate limitation-inducible alkaline phosphatase activity. While diverse in nature, all of these could point to a constitutive stress response.
Several hypotheses can be put forward to explain this complex phenotype. SYNW2176 could be part of a regulatory cascade that responds to envelope stresses and regulates a certain set of outer membrane proteins in response to these stresses. In its absence some of these proteins are constitutively expressed whereas others are not induced. One complication for this hypothesis is that it does not correlate directly with previous transcriptional microarray data. If SYNW2176 is a regulatory protein, we would expect to see other genes responding when it is up-or down-regulated. However, although SYNW2176 is up in response to copper and early phosphate stress and down in response to late phosphate stress, we do not see a correlation with other genes, like SYNW0762 (the atypical alkaline phosphatase/phytase protein only detectable in the mutant) between these datasets.
A simpler hypothesis is that SYNW2176 functions in protein quality control, as does DegP in E. coli. If this is the case, the mutant phenotype could be the result of a buildup of damaged membrane and periplasmic proteins, that would otherwise be degraded. This buildup could be sensed by other cellular systems, perhaps other Deg-family proteins, leading to a specific stress response regulating porins and other membrane proteins. Indeed, in E. coli ΔdegP mutants have activated σE and Cpx pathways, likely a result of envelope stress from the buildup of damaged proteins usually degraded by DegP (Leiser et al., 2012). Additionally, activation of the σE pathway can result in a reduction of the major OMP protein levels (Johansen et al., 2006). This buildup of damaged proteins and active envelope stress response could lead to a decreased permeability or transport of copper.
How copper crosses the outer membrane is still not entirely clear but porins can be regulated by copper, can bind copper, and may allow passive import of unchelated copper (Yoneyama and Nakae, 1996; Caille et al., 2007; Balasubramanian et al., 2011). Moreover, a porin-deficient (most likely OmpC and OmpF) mutant in E. coli is resistant to elevated copper levels, indicating such changes can be sufficient to lead to increased copper resistance (Lutkenhaus, 1977). However, this is complicated by the fact that a single OmpC mutant is copper sensitive (Egler et al., 2005), indicating that porins may vary in their ability to let copper into the periplasm. There is some evidence that the charged residues in the outer loops of porins contribute to voltage gating in Haemophilus influenzae (Arbing et al., 2000). Our comparison of the outer loop charged residues of the two porins suggest they may allow diffusion of very different sets of substrates. In addition, copper could potentially react with cysteines on SYNW2224 so one could speculate that its decrease in SYNW2176- and the new presence of the SYNW2227 porin lacking cysteines could be a mechanism increasing copper resistance. Given this, our data suggests that the two porins may have different permeability to copper ions and changing the levels of these porins may change copper permeability and copper resistance.
There could also be a role for SYNW0762, SYNW1018 or SwmB, all of which were much higher in the mutant than wild type although it is not clear what these would do to increase Cu resistance. Phosphatases and phytases have been linked to uranium resistance in bacteria (Martinez et al., 2007; Nilgiriwala et al., 2008; Yung et al., 2014) and copper resistance in fungi (Tsekova and Galabova, 2003), likely through biomineralization of the metals with phosphate. While the marine chemical environment is quite different it is possible that these extra alkaline phosphatases like SYNW0762 may play a role in copper detoxification directly. SYNW1018 is part of a well-conserved group of phosphate-binding proteins, PstS, found in bacteria and in E. coli is part of the high-affinity phosphate transport system (Makino et al., 1989). Oligotrophic Synechococcus strains including WH8102 tend to have multiple paralogs of this protein and it is unclear what different functions the four present in WH8102 may have (Scanlan et al., 2009).
The high number of phosphate-stress inducible proteins in the mutant indicates constitutive phosphate stress. High affinity phosphate transport in Synechococcus involves both a membrane-bound ABC transporter and a periplasmic phosphate-binding protein as well as a phosphate-sensing histidine kinase, PhoR, that may be membrane bound (Scanlan et al., 2009). Thus, a buildup of damaged periplasmic and membrane proteins in the mutant could disproportionately affect phosphate transport as compared to nitrate transport, for example, which is not transported by ABC family transporters but by a permease (Sakamoto et al., 1999). Reduced transport of phosphate could in turn lead to phosphate limitation in the presence of high phosphate levels and induction of a phosphate limitation response. Indeed, Tetu et al. (2009) found changes in expression of many outer membrane proteins in response to phosphate starvation as well as regulation of SYNW2176 (up slightly under early phosphate stress and down under late phosphate stress), so the mutant's distinct phenotype regarding phosphate acquisition is not surprising. During exponential growth SYNW2176- had a higher level of alkaline phosphatase activity than wild-type, which had virtually undetectable levels until phosphate levels decreased (Figure 5). It also has higher levels of many phosphate-inducible proteins than wild-type in exponential growth as well as in low-phosphate growth (Table 1). Despite higher levels of proteins involved in phosphate stress response, or perhaps because of it, the mutant had a longer lag time (Figure 5), all of which may indicate the costs of constitutively activated stress.
While quantification of damaged proteins in the periplasm is beyond the scope of this work, our data nevertheless shows the major role that SYNW2176 plays in determining outer membrane composition, perhaps through protease activity on specific membrane proteins. Our data clearly show that the presence/absence of SYNW2176 affects expression of many outer membrane proteins potentially playing a role in outer membrane permeability to a number of substrates. The Deg-proteases are moreover well-conserved in marine Synechococcus and their phylogeny appears to inform function. Our data also reveal the potential importance of porins in copper tolerance in marine Synechococcus. As is clear from the dramatic increase in copper tolerance in the mutant and the significantly altered outer membrane protein composition, Synechococcus outer membrane composition is key to understanding the copper sensitivity of this group. The outer membrane functions as both a barrier to toxins and a gateway for nutrients and copper is at the intersection of these functions as it is both a toxin and a micronutrient. Identifying proteins that contribute to copper permeability can provide insight into outer membrane function and how marine Synechococcus maintains a balance between importing limiting nutrients and keeping out toxins.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
National Science Foundation Grants (MCB0744334, DEB-1233085) supported this project. Many thanks to M. Ghassemian from the UCSD Biomolecular/Proteomics Mass Spectrometry Facility, the PRIDE team, J. Golden for sharing plasmid pRL271, C. Dupont for QRTPCR assistance and trace metal culturing and B. Pedler for advice on PO4 measurements.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmars.2014.00016/abstract
References
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. doi: 10.1093/bioinformatics/bti263
Alba, B. M., Zhong, H. J., Pelayo, J. C., and Gross, C. A. (2001). degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide sigma(E) activity. Mol. Microbiol. 40, 1323–1333. doi: 10.1046/j.1365-2958.2001.02475.x
Anisimova, M., and Gascuel, O. (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552. doi: 10.1080/10635150600755453
Arbing, M. A., Dahan, D., Boismenu, D., Mamer, O. A., Hanrahan, J. W., and Coulton, J. W. (2000). Charged residues in surface-located loops influence voltage gating of porin from Haemophilus influenzae type b. J. Membr. Biol. 178, 185–193. doi: 10.1007/s002320010026
Bagos, P. G., Liakopoulos, T. D., Spyropoulos, I. C., and Hamodrakas, S. J. (2004). PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 32, W400–W404. doi: 10.1093/nar/gkh417
Bai, X. C., Pan, X. J., Wang, X. J., Ye, Y. Y., Chang, L. F., Leng, D., et al. (2011). Characterization of the structure and function of Escherichia coli DegQ as a representative of the DegQ-like proteases of bacterial HtrA family proteins. Structure 19, 1328–1337. doi: 10.1016/j.str.2011.06.013
Balasubramanian, R., Kenney, G. E., and Rosenzweig, A. C. (2011). Dual pathways for copper uptake by methanotrophic bacteria. J. Biol. Chem. 286, 37313–37319. doi: 10.1074/jbc.M111.284984
Barker, M., De Vries, R., Nield, J., Komenda, J., and Nixon, P. J. (2006). The Deg proteases protect Synechocystis sp PCC 6803 during heat and light stresses but are not essential for removal of damaged D1 protein during the photosystem two repair cycle. J. Biol. Chem. 281, 30347–30355. doi: 10.1074/jbc.M601064200
Blot, N., Mella-Flores, D., Six, C., Le Corguille, G., Boutte, C., Peyrat, A., et al. (2011). Light history influences the response of the marine cyanobacterium Synechococcus sp WH7803 to oxidative stress. Plant Physiol. 156, 1934–1954. doi: 10.1104/pp.111.174714
Brahamsha, B. (1996a). An abundant cell-surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 93, 6504–6509. doi: 10.1073/pnas.93.13.6504
Brahamsha, B. (1996b). A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 62, 1747–1751.
Brand, L. E., Sunda, W. G., and Guillard, R. R. L. (1986). Reduction of marine-phytoplankton reproduction rates by copper and cadmium. J. Exp. Mar. Biol. Ecol. 96, 225–250. doi: 10.1016/0022-0981(86)90205-4
Cai, Y. P., and Wolk, C. P. (1990). Use of a conditionally lethal gene in Anabaena sp-strain PCC-7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172, 3138–3145.
Caille, O., Rossier, C., and Perron, K. (2007). A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J. Bacteriol. 189, 4561–4568. doi: 10.1128/jb.00095-07
Clausen, T., Southan, C., and Ehrmann, M. (2002). The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10, 443–455. doi: 10.1016/s1097-2765(02)00658-5
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Diao, H., Zhou, T., Zhang, J., Zhao, K., Zhou, M., and Yang, C. (2011). Heterogenous expression of Synechocystis sp. PCC 6803 deg proteases and their possible roles on phycobiliprotein degradation in vitro. J. Wuhan Univ. Technol. Mater. Sci. Ed. 26, 1049–1058. doi: 10.1007/s11595-011-0361-7
Dufresne, A., Ostrowski, M., Scanlan, D., Garczarek, L., Mazard, S., Palenik, B., et al. (2008). Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9:R90. doi: 10.1186/gb-2008-9-5-r90
Dupont, C. L., Barbeau, K., and Palenik, B. (2008). Ni uptake and limitation in marine Synechococcus strains. Appl. Environ. Microbiol. 74, 23–31. doi: 10.1128/aem.01007-07
Dupont, C. L., Johnson, D. A., Phillippy, K., Paulsen, I. T., Brahamsha, B., and Palenik, B. (2012). Genetic identification of a high-affinity Ni transporter and the transcriptional response to Ni deprivation in Synechococcus sp. strain WH8102. Appl. Environ. Microbiol. 78, 7822–7832. doi: 10.1128/AEM.01739-12
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Egler, M., Grosse, C., Grass, G., and Nies, D. H. (2005). Role of the extracytoplasmic function protein family sigma factor RpoE in metal resistance of Escherichia coli. J. Bacteriol. 187, 2297–2307. doi: 10.1128/JB.187.7.2297-2307.2005
Ferris, M. J., and Palenik, B. (1998). Niche adaptation in ocean cyanobacteria. Nature 396, 226–228. doi: 10.1038/24297
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Guttman, M., Betts, G. N., Barnes, H., Ghassemian, M., Van Der Geer, P., and Komives, E. A. (2009). Interactions of the NPXY microdomains of the low density lipoprotein receptor-related protein 1. Proteomics 9, 5016–5028. doi: 10.1002/pmic.200900457
Huesgen, P. F., Miranda, H., Lam, X., Perthold, M., Schuhmann, H., Adamska, I., et al. (2011). Recombinant Deg/HtrA proteases from Synechocystis sp. PCC 6803 differ in substrate specificity, biochemical characteristics and mechanism. Biochem. J. 435, 733–742. doi: 10.1042/BJ20102131
Jansen, T., Kidron, H., Taipaleenmaki, H., Salminen, T., and Maenpaa, P. (2005). Transcriptional profiles and structural models of the Synechocystis sp PCC 6803 Deg proteases. Photosyn. Res. 84, 57–63. doi: 10.1007/s11120-005-0475-x
Johansen, J., Rasmussen, A. A., Overgaard, M., and Valentin-Hansen, P. (2006). Conserved small non-coding RNAs that belong to the sigma(E) regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 364, 1–8. doi: 10.1016/j.jmb.2006.09.004
Kelly, L., Huang, K. H., Ding, H., and Chisholm, S. W. (2012). ProPortal: a resource for integrated systems biology of Prochlorococcus and its phage. Nucleic Acids Res. 40, D632–D640. doi: 10.1093/nar/gkr1022
Leiser, O. P., Charlson, E. S., Gerken, H., and Misra, R. (2012). Reversal of the Delta degP Phenotypes by a Novel rpoE Allele of Escherichia coli. PLoS ONE 7:e33979. doi: 10.1371/journal.pone.0033979
Li, W. K. W. (1994). Primary production of prochlorophytes, cyanobacteria, and eukaryotic ultraphytoplankton - measurements from flow cytometric sorting. Limnol. Oceanogr. 39, 169–175. doi: 10.4319/lo.1994.39.1.0169
Lutkenhaus, J. F. (1977). Role of a major outer membrane-protein in Escherichia-coli. J. Bacteriol. 131, 631–637.
Macomber, L., Rensing, C., and Imlay, J. A. (2007). Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 189, 1616–1626. doi: 10.1128/jb.01357-06
Makino, K., Shinagawa, H., Amemura, M., Kawamoto, T., Yamada, M., and Nakata, A. (1989). Signal transduction in the phosphate regulon of Escherichia-coli involves phosphotransfer between PhoR and PhoB proteins. J. Mol. Biol. 210, 551–559. doi: 10.1016/0022-2836(89)90131-9
Mann, E. L., Ahlgren, N., Moffett, J. W., and Chisholm, S. W. (2002). Copper toxicity and cyanobacteria ecology in the Sargasso Sea. Limnol. Oceanogr. 47, 976–988. doi: 10.4319/lo.2002.47.4.0976
Martinez, R. J., Beazley, M. J., Taillefert, M., Arakaki, A. K., Skolnick, J., and Sobecky, P. A. (2007). Aerobic uranium (VI) bioprecipitation by metal-resistant bacteria isolated from radionuclide- and metal-contaminated subsurface soils. Environ. Microbiol. 9, 3122–3133. doi: 10.1111/j.1462-2920.2007.01422.x
Martiny, A. C., Coleman, M. L., and Chisholm, S. W. (2006). Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc. Natl. Acad. Sci. U.S.A. 103, 12552–12557. doi: 10.1073/pnas.0601301103
Mazard, S., Wilson, W. H., and Scanlan, D. J. (2012). Dissecting the physiological response to phosphorus stress in marine Synechococcus isolates (Cyanophyceae). J. Phycol. 48, 94–105. doi: 10.1111/j.1529-8817.2011.01089.x
McCarren, J., and Brahamsha, B. (2007). SwmB, a 1.12-megadalton protein that is required for nonflagellar swimming motility in Synechococcus. J. Bacteriol. 189, 1158–1162. doi: 10.1128/jb.01500-06
McCormack, A. L., Schieltz, D. M., Goode, B., Yang, S., Barnes, G., Drubin, D., et al. (1997). Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal. Chem. 69, 767–776. doi: 10.1021/ac960799q
Meltzer, M., Hasenbein, S., Mamant, N., Merdanovic, M., Poepsel, S., Hauske, P., et al. (2009). Structure, function and regulation of the conserved serine proteases DegP and DegS of Escherichia coli. Res. Microbiol. 160, 660–666. doi: 10.1016/j.resmic.2009.07.012
Nilgiriwala, K. S., Alahari, A., Rao, A. S., and Apte, S. K. (2008). Cloning and overexpression of alkaline phosphatase PhoK from Sphingomonas sp. strain BSAR-1 for bioprecipitation of uranium from alkaline solutions. Appl. Environ. Microbiol. 74, 5516–5523. doi: 10.1128/AEM.00107-08
Ostrowski, M., Mazard, S., Tetu, S. G., Phillippy, K., Johnson, A., Palenik, B., et al. (2010). PtrA is required for coordinate regulation of gene expression during phosphate stress in a marine Synechococcus. ISME J. 4, 908–921. doi: 10.1038/ismej.2010.24
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Raffa, R. G., and Raivio, T. L. (2002). A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 45, 1599–1611. doi: 10.1046/j.1365-2958.2002.03112.x
Raivio, T. L. (2005). Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 56, 1119–1128. doi: 10.1111/j.1365-2958.2005.04625.x
Rensing, C., and Grass, G. (2003). Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27, 197–213. doi: 10.1016/s0168-6445(03)00049-4
Richardson, T. L., and Jackson, G. A. (2007). Small phytoplankton and carbon export from the surface ocean. Science 315, 838–840. doi: 10.1126/science.1133471
Roberts, I. N., Lam, X. T., Miranda, H., Kieselbach, T., and Funk, C. (2012). Degradation of PsbO by the Deg protease HhoA is thioredoxin dependent. PLoS ONE 7:e45713. doi: 10.1371/journal.pone.0045713
Rocap, G., Distel, D. L., Waterbury, J. B., and Chisholm, S. W. (2002). Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68, 1180–1191. doi: 10.1128/aem.68.3.1180-1191.2002
Sakamoto, T., Inoue-Sakamoto, K., and Bryant, D. A. (1999). A novel nitrate/nitrite permease in the marine cyanobacterium Synechococcus sp strain PCC 7002. J. Bacteriol. 181, 7363–7372.
Scanlan, D. J., Ostrowski, M., Mazard, S., Dufresne, A., Garczarek, L., Hess, W. R., et al. (2009). Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73, 249–299. doi: 10.1128/mmbr.00035-08
Shilov, I. V., Seymour, S. L., Patel, A. A., Loboda, A., Tang, W. H., Keating, S. P., et al. (2007). The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655. doi: 10.1074/mcp.T600050-MCP200
Steglich, C., Futschik, M., Rector, T., Steen, R., and Chisholm, S. W. (2006). Genome-wide analysis of light sensing in Prochlorococcus. J. Bacteriol. 188, 7796–7806. doi: 10.1128/jb.01097-06
Stuart, R. K., Dupont, C. L., Johnson, D. A., Paulsen, I. T., and Palenik, B. (2009). Coastal strains of marine Synechococcus species exhibit increased tolerance to copper shock and a distinctive transcriptional response relative to those of open-ocean strains. Appl. Environ. Microbiol. 75, 5047–5057. doi: 10.1128/aem.00271-09
Su, Z. C., Mao, F. L., Dam, P., Wu, H. W., Olman, V., Paulsen, I. T., et al. (2006). Computational inference and experimental validation of the nitrogen assimilation regulatory network in cyanobacterium Synechococcus sp WH 8102. Nucleic Acids Res. 34, 1050–1065. doi: 10.1093/nar/gkj496
Tai, V., Paulsen, I. T., Phillippy, K., Johnson, D. A., and Palenik, B. (2009). Whole-genome microarray analyses of Synechococcus-Vibrio interactions. Environ. Microbiol. 11, 2698–2709. doi: 10.1111/j.1462-2920.2009.01997.x
Talavera, G., and Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
Taylor, L. A., and Rose, R. E. (1988). A correction in the nucleotide-sequence of the Tn903 kanamycin resistance determinant in Puc4K. Nucleic Acids Res. 16, 358. doi: 10.1093/nar/16.1.358
Tetu, S. G., Brahamsha, B., Johnson, D. A., Tai, V., Phillippy, K., Palenik, B., et al. (2009). Microarray analysis of phosphate regulation in the marine cyanobacterium Synechococcus sp WH8102. ISME J. 3, 835–849. doi: 10.1038/ismej.2009.31
Tetu, S. G., Johnson, D. A., Varkey, D. R., Phillippy, K., Stuart, R. K., Dupont, C., et al. (2013). Impact of DNA damaging agents on genome-wide transcriptional profiles in two marine Synechococcus species. Front. Microbiol. 4:232. doi: 10.3389/fmicb.2013.00232
Thomas, E. V., Phillippy, K. H., Brahamsha, B., Haaland, D. M., Timlin, J. A., Elbourne, L. D., et al. (2009). Statistical analysis of microarray data with replicated spots: a case study with Synechococcus WH8102. Comp. Funct. Genomics 2009:950171. doi: 10.1155/2009/950171
Thompson, A. W., Huang, K., Saito, M. A., and Chisholm, S. W. (2011). Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability. ISME J. 5, 1580–1594. doi: 10.1038/ismej.2011.49
Tolonen, A. C., Aach, J., Lindell, D., Johnson, Z. I., Rector, T., Steen, R., et al. (2006). Global gene expression of Prochlorococcus ecotypes in response to changes in nitrogen availability. Mol. Syst. Biol. 2:53. doi: 10.1038/msb4100087
Tsekova, K., and Galabova, D. (2003). Phosphatase production and activity in copper (II) accumulating Rhizopus delemar. Enzyme Microb. Technol. 33, 926–931. doi: 10.1016/j.enzmictec.2003.06.001
Vizcaino, J. A., Deutsch, E. W., Wang, R., Csordas, A., Reisinger, F., Rios, D., et al. (2014). ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226. doi: 10.1038/nbt.2839
Waller, P. R. H., and Sauer, R. T. (1996). Characterization of degQ and degS, Escherichia coli genes encoding homologs of the DegP protease. J. Bacteriol. 178, 1146–1153.
Westall, J. C., Zachary, J. L., and Morel, F. M. M. (1976). MINEQL - general algorithm for computation of chemical-equilibrium in aqueous systems. Abstr. Pap. Am. Chem. Soc. 172, 8.
Yoneyama, H., and Nakae, T. (1996). Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiology 142, 2137–2144. doi: 10.1099/13500872-142-8-2137
Keywords: marine Synechococcus, copper, Deg protease, envelope stress, phosphate stress
Citation: Stuart RK, Brahamsha B and Palenik B (2014) A Deg-protease family protein in marine Synechococcus is involved in outer membrane protein organization. Front. Mar. Sci. 1:16. doi: 10.3389/fmars.2014.00016
Received: 24 April 2014; Paper pending published: 15 May 2014;
Accepted: 13 June 2014; Published online: 30 June 2014.
Edited by:
Irene Wagner-Doebler, Helmholtz-Centre for Infection Research (HZI), GermanyReviewed by:
Adam Michael Reitzel, University of North Carolina at Charlotte, USAManuel Ignacio Aranda Lastra, King Abdullah University of Science and Technology, Saudi Arabia
Copyright © 2014 Stuart, Brahamsha and Palenik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian Palenik, Scripps Institution of Oceanography, University of California at San Diego, 3110 Hubbs Hall, La Jolla, CA 92093-0202, USA e-mail: bpalenik@ucsd.edu
†Present address: Rhona K. Stuart, Lawrence Livermore National Laboratory, Livermore, USA
 Rhona K. Stuart
Rhona K. Stuart Bianca Brahamsha
Bianca Brahamsha  Brian Palenik
Brian Palenik