Nitric oxide is not a negative regulator of metamorphic induction in the abalone Haliotis asinina
- School of Biological Sciences, University of Queensland, Brisbane, QLD, Australia
Nitric oxide (NO) is a second messenger molecule synthesized by the enzyme nitric oxide synthase (NOS) that requires the molecular chaperone heat shock protein 90 (HSP90) for normal enzymatic activity. Past studies have revealed that both NO and HSP90 act as negative regulators (repressors) of metamorphosis in a diverse range of marine invertebrates, including several molluscan species. Here, we test the role of NO in the metamorphic induction of a Vetigastropod mollusc, the tropical abalone Haliotis asinina. Specifically, we (1) test the effects of NO-manipulating pharmacological agents, (2) measure the temporal expression of NOS and HSP90 genes through metamorphosis, and (3) assess the spatial expression of NOS and HSP90 in larvae. We find that inhibition of NOS reduces rates of metamorphosis, indicating that NO facilitates, rather than represses, induction of metamorphosis in H. asinina. The marked increase in NOS expression in putative sensory cells localized to the anterior foot of competent larvae is consistent with NO as an inductive molecule for metamorphosis. In contrast to NOS, HSP90 transcript abundance decreases at competence and there is no evidence of NOS and HSP90 transcript co-localization. This study provides the first evidence of NO as an inductive facilitator of molluscan metamorphosis. Our experimental data suggest that NO modulates signals derived from live inductive substrates via the larval foot to regulate metamorphosis. Inter-specific comparisons of spatial NOS expression in molluscs suggest that the localized pattern of NOS or its protein product is related to the regulatory action of NO in metamorphosis.
Introduction
The ecological success and stability of marine benthic communities depends upon the recruitment of larvae. These planktonic larvae represent the dispersive phase of a biphasic life cycle that is common among phylogenetically diverse marine invertebrates (Pechenik, 2004; Heyland et al., 2011). To complete the life cycle, free-swimming larvae usually must acquire a state of ontogenic maturation, known as competency (Hadfield et al., 2001), at which time they are able to settle onto an appropriate benthic substrate and undergo metamorphosis into the benthic reproductive form (Hadfield, 1998; Pechenik, 2004). In most species, larval settlement is dependent on contact with specific environmental cues (Hadfield, 1998) that include chemical ligands released from conspecifics, microbial films, and prey species, all of which indicate the suitability of a habitat for post-metamorphic life (Pawlik, 1992).
The ability to discriminate and respond to the appropriate cue is crucial, because settling in the right place is necessary for survival, growth, and reproductive success of post-metamorphic individuals (Rodriguez et al., 1993; Underwood and Keough, 2001). To do so, competent larvae have sensory organs that use cell surface receptors to bind to the appropriate environment-derived ligands. The binding of ligands to their receptors activates conserved biochemical signaling pathways that ultimately coordinate the global morphogenetic events that constitute metamorphosis (Hadfield, 2000; Leise and Hadfield, 2000). The application of pharmacological agents that activate or inhibit parts of these conserved signaling pathways can induce settlement and metamorphosis of many species in vitro (Baxter and Morse, 1987; Degnan and Morse, 1995; Biggers and Laufer, 1999; Eri et al., 1999; Amador-Cano et al., 2006).
One pathway of particular interest in marine invertebrate biology is the nitric oxide (NO) signaling pathway. NO is a highly conserved second messenger molecule that regulates diverse physiological responses in all organism (Colasanti and Venturini, 1998; Palumbo, 2005), including the timing of life cycle transitions in bacteria, fungi, slime mold, plants, and animals (Tao et al., 1997; Kashiwagi et al., 1999; Wilken and Huchzermeyer, 1999; He et al., 2004; Schmidt et al., 2004; Cáceres et al., 2011). The main endogenous source of NO is an enzyme, nitric oxide synthase (NOS), whose normal enzymatic activity requires a molecular chaperone, heat shock protein 90 (HSP90). Under stress-free cellular conditions, HSP90 constitutively stabilizes the functional conformation of NOS, and thus facilitates NO synthesis (García-Cardeña et al., 1998; Song et al., 2001; Yoshida and Xia, 2003).
Endogenous NO has been shown to negatively regulate (repress) the initiation of larval settlement and metamorphosis in multiple species of marine invertebrate representing diverse phyla, including chordates, echinoderms, molluscs, annelids and crustaceans (Froggett and Leise, 1999; Bishop and Brandhorst, 2001; Bishop et al., 2001, 2008; Comes et al., 2007; Pechenik et al., 2007; Biggers et al., 2011; Zhang et al., 2012; Romero et al., 2013). In 2003, Bishop and Brandhorst (2003) first hypothesized that a negative regulatory role of NO is widely conserved in bilaterian life cycle transitions, and results from interactions between NOS and HSP90. This initial hypothesis has since been refined to account for differing specificities of larval responses to inductive cues (Bishop et al., 2006; Hodin, 2006). In generalist larvae—those that can metamorphose spontaneously or in response to a wide variety of cues—a reduction in NO is sufficient to induce metamorphosis; that is, NO acts as a direct regulator of metamorphosis in generalist species (Bishop et al., 2006; Hodin, 2006; Romero et al., 2013). On the other hand, in specialist larvae—those that require very specific inductive cues—a reduction in NO alone is not sufficient to induce metamorphosis, but it does enhance larval sensitivity to the specific cue; that is, NO functions as a modulator of metamorphosis in specialist species (Bishop et al., 2006; Hodin, 2006; Romero et al., 2013). To date, almost all empirical studies have supported both the hypothesis of a negative regulatory role for NO and that of the direct regulatory vs. modulatory NO function in generalist vs. specialist larvae (Froggett and Leise, 1999; Bishop and Brandhorst, 2001; Bishop et al., 2001, 2008; Comes et al., 2007; Pechenik et al., 2007; Biggers et al., 2011; Zhang et al., 2012; Romero et al., 2013). The sole exception so far is a recent report demonstrating a positive regulatory role for NO in metamorphosis of the solitary ascidian Herdmania momus (Ueda and Degnan, 2013); whether this represents a species-specific anomaly or a more widespread phenomenon awaits further data from a diversity of marine taxa.
Four molluscan taxa have been investigated to date for the role of NO in metamorphosis. The application of a NOS inhibitor is sufficient to induce metamorphosis of the generalist larvae of the mud snail Ilyanassa obsoleta and the slipper shell snail Crepidula fornicata (Froggett and Leise, 1999; Pechenik et al., 2007). In the specialist larvae of the nudibranch, Phestilla sibogae, the presence of a NOS inhibitor enhances the metamorphic response to a natural inductive cue (Bishop et al., 2008). Recently, Romero et al. (2013) reported that the direct regulatory vs. modulatory repressive role of NO in generalist vs. specialist larvae is evident even within a single species, namely the sea slug Alderia willowi, which produces both generalist and specialist larvae from a single spawning (Krug, 2001). Consistent with these results, application of an NO donor antagonizes the effect of known inductive cues in I. obsolete, P. sibogae, and A. willowi (Froggett and Leise, 1999; Bishop et al., 2008; Romero et al., 2013).
Immunohistochemistry assays in molluscs have detected NOS protein activity localized to the apical sensory organ (ASO) of I. obsoleta (Thavaradhara and Leise, 2001) and C. fornicata (Pechenik et al., 2007) veliger larvae, and in putative sensory neurons in the edge of the mantle and foot in I. obsolete veligers (Thavaradhara and Leise, 2001). The ASO is a larval-specific sensory structure that is thought to function as a sensory structure to detect inductive cues (Hadfield et al., 2000); it is lost during or immediately after metamorphosis (Croll and Dickinson, 2004; Gifondorwa and Leise, 2006; Croll, 2009). In P. sibogae veligers, NOS expression is detectable in both cerebral and pedal ganglia (Bishop et al., 2008). At time of writing, complete nucleotide sequences of NOS genes have been isolated from five mollusc species and, in all cases, the domain organization of the predicted protein products resembles the neuronal isoform of mammalian NOS (nNOS), further implicating a neuro-active role of NO in molluscs (Korneev et al., 1998; Scheinker et al., 2005; Moroz et al., 2006; Matsuo et al., 2008; Cioni et al., 2011). These results together strongly suggest that, in molluscan larval settlement, NO may process signals from environmentally-derived inductive cues to regulate the activation of morphogenetic signal transduction pathways via the sensory nervous system.
Here, we examine the hypothesis of NO as a phylogenetically conserved negative regulator during the pelagobenthic transition by using the tropical abalone, Haliotis asinina (Mollusca: Vetigastropoda: Haliotidae). Abalone have a pelagobenthic life cycle typical of broadcast spawners; externally fertilized eggs hatch as trochophore larvae and subsequently develop into lecithotrophic (non-feeding) veligers (Sawatpeera et al., 2001). Veligers of H. asinina become competent to settle by 96 h post fertilization (hpf) at 25°C (Jackson et al., 2005). Haliotis asinina produces specialist larvae that require the presence of particular natural substrates for successful settlement and metamorphosis, which can be induced very effectively (>90%) within 48 h by articulated coralline algae of the genus Amphiroa (Williams et al., 2008).
To investigate the role of NO in regulating the H. asinina pelagobenthic transition, we first examine the effects of pharmacological NOS inhibitors and NO donors on the induction of settlement and metamorphosis. We complement these behavioral experiments with NOS and HSP90 gene expression analyses using quantitative reverse transcriptase-PCR by examining NOS and HSP90 expression temporal profiles through the pelagobenthic transition. To address the hypothesis that NO regulates larval settlement in response to exogenous stimulation of larval sensory cells, we use whole mount in situ hybridization to assess spatial expression of both NOS and HSP90. A schematic showing the temporal development of H. asinina and our experimental sampling points is presented in Figure 1.
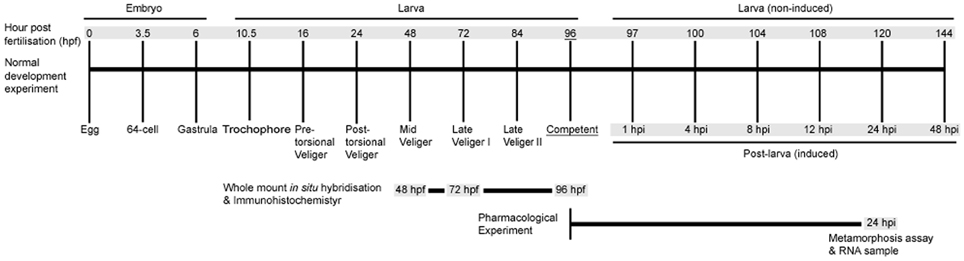
Figure 1. A time course of Haliotis asinina development indicating experimental strategies employed in this study. Developmental stages are indicated by hours post fertilization (hpf) for embryonic and larval development. Post-larval development is indicated by hours post induction (hpi). All metamorphosis assays were initiated at competency (96 hpf). Gray shading indicates times at which RNA was sampled.
Materials and Methods
H. asinina Larval Culture
Reproductively active adult specimens of H. asinina were collected from Heron Island Reef, Great Barrier Reef, Australia (23°27′S; 151°55′E). Detailed protocols for maintenance of collected specimens, spawning, fertilizations, and larval culture were followed in accordance with Williams et al. (2009a). In brief, eggs and sperm from at least three males and three females were collected and fertilized on the night of the natural spawning cycle (Counihan et al., 2001). Embryos and larvae were cultured in 300 mm diameter larval culture chambers with flow-through 10 μm filtered sea water (FSW) at 25°C ± 0.5 until the larvae matured to the competent state (96 hpf) (Jackson et al., 2005). The culture chambers were treated with 20 μg/L rifampicin for 2 h daily to prevent bacterial outbreaks.
Metamorphosis Assays Using Pharmacological Treatments
Because variable metamorphic responses to specific NOS inhibitors have been reported in C. fornicata (Pechenik et al., 2007) and Capitella teleta (Annelida) (Biggers et al., 2011), we elected to test several different NOS inhibitors and NO donors to overcome a potential incompatibility between the pharmacological agents and H. asinina veligers (Table 1). As NOS inhibitors, we used L-nitroarginine-methyl-ester (L-NAME) (Sigma), aminoguanidine hemisulfate (AGH) (Sapphire Bioscience), and S-methylisothiourea sulfate (SMIS) (Sapphire Bioscience). To examine the effects of increased NO on metamorphosis, we used S-nitroso-N-acetyl-penicillamine (SNAP) (Sapphire Bioscience) as a direct NO donor. L-Arginine (Sigma), which is the essential substrate of NOS enzymatic activity, was also applied as an NO donor, since it theoretically increases the internal availability of NO (Stuehr, 2004). All chemicals used here have been shown to affect the rate of metamorphosis in other marine invertebrates (Froggett and Leise, 1999; Bishop and Brandhorst, 2001; Bishop et al., 2001, 2008; Comes et al., 2007; Pechenik et al., 2007; Biggers et al., 2011; Zhang et al., 2012; Romero et al., 2013).
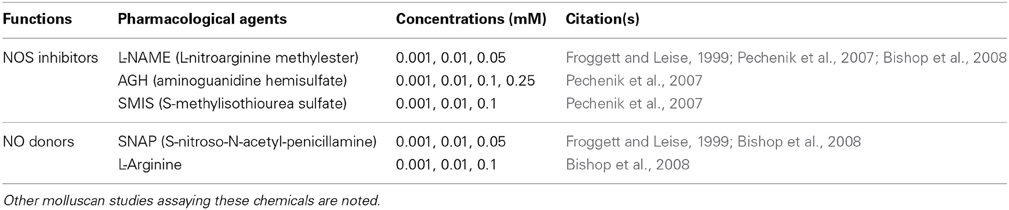
Table 1. Summary of chemicals and their concentrations used in metamorphosis assay of Haliotis asinina.
Stock solutions of 0.5 M L-NAME, 0.5 M AGH, 0.5 M SMIS, and 0.5 M L-Arginine were prepared in 0.22 μm FSW, stored at 4°C, and diluted to final experimental concentrations just prior to the experiments. For SNAP, a stock solution of 0.1 M was prepared in dimethyl sulfoxide (DMSO) immediately before the experiment and diluted to final concentrations just prior to use. To maintain a steady concentration of NO delivered by SNAP during the metamorphosis assay, the solution was renewed every 6 h (Froggett and Leise, 1999). The final concentrations of each chemical used in the experiments are listed in Table 1.
Metamorphosis assays were initiated at competency (96 hpf) (Figure 1) and performed in 6-well 35-mm diameter sterile polycarbonate tissue culture dishes with 10 ml of 0.22 μm FSW per well. The following controls and treatments were included in experiments with each pharmacological agent: (1) FSW only (negative control), (2) FSW with living articulated red coralline algae (CA), Amphiroa ephedraea shards covering approximately 25% of the bottom of the well (positive control), and (3) FSW containing a pharmacological agent either with or without live CA shards (treatments). The live CA used for the experiment were collected from Heron Island reef flat just prior to experimentation, washed in FSW, cleaned of any epiphytes growing on the surface and placed in the wells for immediate use (Williams et al., 2008). Initiation of juvenile shell growth at 24 hpi was used as a definitive indicator of metamorphosis (Williams et al., 2008).
RNA Sample Collection and Preparation for Quantitative Reverse-Transcriptase PCR (qRT-PCR)
To investigate temporal expressional patterns of NOS and HSP90 genes in H. asinina, total ribonucleic acid (RNA) samples were collected during embryonic, larval, and post-larval development (Figure 1). Embryonic and larval samples were collected from the culture chambers described above. To collect the post-larval samples of H. asinina, competent veligers (96 hpf) were exposed to live CA in 12 cm petri dishes with live CA shards. We only collected those individuals crawling on live CA to ensure a clear discrimination between still-swimming larvae and post-larvae that had initiated settlement and metamorphosis.
For all RNA samples, ~1000 individuals were collected for each sampling point, and were preserved in TRI reagent (Sigma) at −80°C prior to extraction. RNA extraction and complementary DNA (cDNA) synthesis followed methods described in Williams et al. (2009a). For the assessment of genomic DNA contamination, no-RT control samples were prepared from the 0.5 μg DNase-treated RNA for the all RNA samples and tested by quantitative RT-PCR (qRT-PCR). All synthesized cDNA and no-RT control samples were stored at −20°C.
Isolation of HasNOS
We isolated a single NOS gene from H. asinina (HasNOS) by degenerate PCR. The degenerate primers were designed from the alignment of NOS derived amino acid sequence from the following species: Lehmannia valentiana (BAF73722), Aplysia californica 1 and 2 (AAK83069 and AAK92211, respectively), Branchiostoma floridae (AAQ02989), Nematostella vectensis (XP001631503), Sepia officinalis A and B (AAS93626 and AAS93627, respectively), and Lottia gigantia (JGI223312) (Figure 2). The degenerate forward (DegF1) and reverse (DegR1) primers were designed from conserved amino acid sequences of CNHIKY and CPADWVW, respectively (Figure 2). The latter amino acid sequence contains a part of the tetrahydrobiopterin (BH4) binding site, which is a highly conserved region of metazoan NOS (Andreakis et al., 2011). The DegF1 and DegR1 sequences were 5′-TGYAAYCAYATHARTAY-3′ and 5′-CCANACCCARTCNSCNGGRCA-3′, respectively. A touch-down PCR profile was used: 94°C for 2 min, 5 cycles at 94°C for 30 s, 55 to 49°C (2°C increment for every 5 cycles) for 30 s, and 70°C for 3 min, 25 cycles at 94°C for 30 s, 47°C for 30 s, and 70°C for 3 min, with a final extension of 72°C for 10 min. Each PCR reaction comprised 1x reaction buffer (Promega), 0.2 mM dNTP, 1 U Taq polymerase (New England Biolab), and 1 μM of each primer in a total volume of 20 μL.
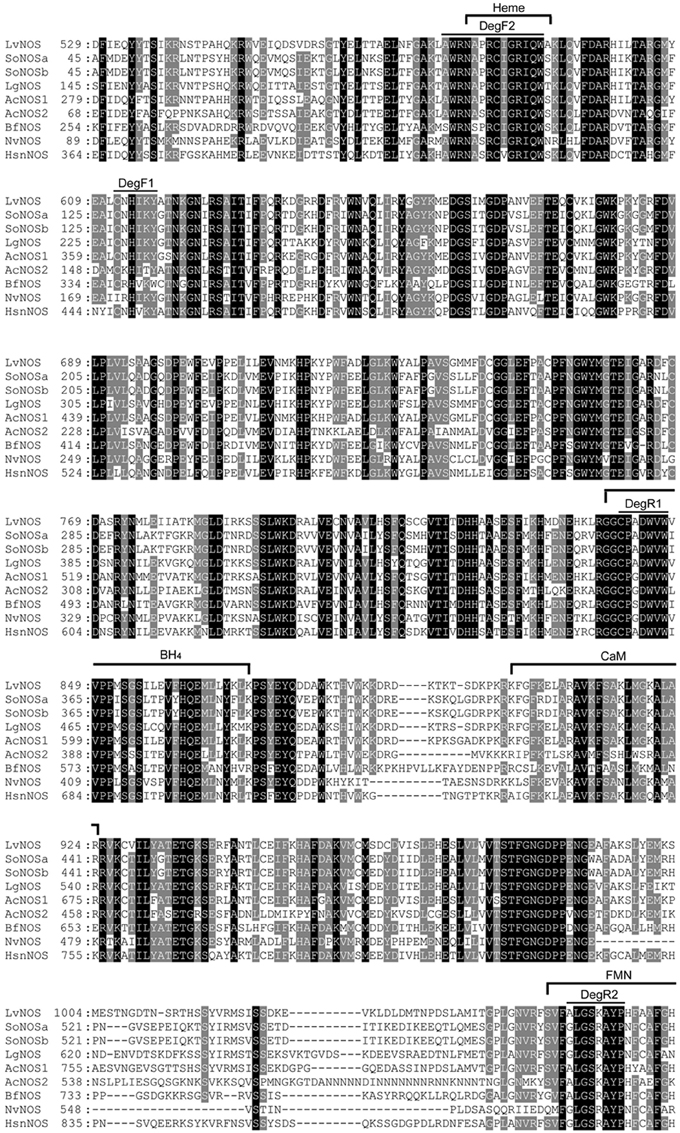
Figure 2. Multiple sequence alignment of NOS derived amino acid sequences. Black shading indicate completely conserved residues; gray shading indicates semi-conserved residues. The locations of amino acid sequence used to design degenerate primers are indicated by DegF1, DegF2, DegR1, and DegR2. Homo sapiens neuronal nitric oxide synthase (HsnNOS) was used for a reference to indicate conserved functional sites: Heme, BH4 (tetrahydrobiopterin), CaM (calmodulin), and FMN (flavin mononucleotide). Lv Lehmannia valentiana, So Sepia officinalis, Lg, Lottia gigantia, Ac Aplysia californica, Bf Branchiostoma floridae, Nv Nematostella vectensis, and Hs Homo sapiens.
The degenerate PCR products were separated and visualized by 1.5 % agarose TAE gel electrophoresis. Products of the expected size (approximately 500 bp) were excised and gel-purified by a silica suspension method (Boyle and Lew, 1995). The purified PCR products were then cloned using the pGEM-T Easy Vector System 1, following the manufacturer's recommendations (Promega). Successfully transformed recombinant colonies were picked and directly added to a second PCR mixture with a final concentration of 1x reaction buffer (Promega), 0.5 mM MgCl2, 0.2 mM dNTP, 1.5 U Taq polymerase (New England Biolab), and 0.25 μM of both M13 forward and reverse primers to amplify inserts. This second PCR reaction used a profile of 94°C for 5 min, 35 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min, with a final extension of 72°C for 10 min. Products were then separated by 1.5 % agarose TAE gel electrophoresis and those of correct size were purified using the silica suspension method (Boyle and Lew, 1995) as described above. The purified PCR products were Sanger-sequenced using reactions containing 1 μL of Big Dye Terminator mix v 3.1 (Applied Biosystems), 1x Big Dye Terminator reaction buffer, 3.2 pmol of primer, and 6–10 ng in a final volume of 10 μL, as recommended by the Australian Genome Research Facility (AGRF). The sequenced products were purified by magnesium sulfate precipitation in accordance with the AGRF protocol and submitted to the AGRF Brisbane node (Queensland, Australia). The resulting sequences were compared to previously-deposited orthologs in the National Center for Biotechnology Information (NCBI) by tBLASTx queries.
We first isolated a HasNOS fragment of 210 bp, sufficient to design a primer set for quantitative real-time PCR (qRT-PCR). To extend the length of the HasNOS sequence, as necessary to produce an adequate length RNA probe for whole mount in situ hybridization analysis, we then designed a set of degenerate primers from AA sequences of AWRNAPRCIGRIQW (DegF2) and ALGSKAYP (DegR2) in aforementioned species, except Lottia gigantia (Figure 2). These amino acid sequences are located in the functional heme and flavin mononucleotide (FMN) binding sites, respectively. Andreakis et al. (2011) found both binding sites in all metazoan NOS orthologs analyzed, showing the highly conserved nature of these two functional sites. The sequences of DegF2 and DegR2 were 5′-CCNMGNTGYATHGGNMGNATHCARTG-3′ and 5′-GRTANGCNCKNSWNCCNARNCC-3′, respectively. This target region is located in the NOS oxygenase domain and encompasses the previously identified HasNOS nucleotide sequence. A touch-down PCR cycle was run as above with modified annealing temperatures from 54 to 46°C (2°C increment for every 5 cycles) and 44°C for the following cycle. The obtained PCR product was used to run a nested-PCR with the degenerate forward primer and a gene-specific reverse primer (5′-CCCGAAGACGCTGCTCGTTCTCC-3′) designed from the fragment of HasNOS nucleotide sequence isolated by the first attempt of isolation. This overlap in primer sets ensured that the two sequenced regions were contiguous. Nested PCRs used a profile of 94°C for 5 min, 35 cycles at 94°C for 30 s, 48°C for 30 s, and 72°C for 1 min, and 72°C for 10 min. Resulting products of the expected size were purified and directly submitted to AGRF for Sanger sequencing. The obtained sequence was again confirmed as a fragment of NOS gene by tBLASTx.
The full sequence of the H. asinina HSP90 gene was previously obtained and named as HasHSP90A (Gunter and Degnan, 2007) [GenBank: EF621884].
qRT-PCR to Assay HasNOS and HasHSP90A Temporal Expression
To analyze the transcriptional profiles of HasNOS and HasHSP90A genes during H. asinina development (embryonic, larval, and post-larval stages) (Figure 1), we performed quantitative real-time PCR (qRT-PCR) as described in Williams and Degnan (2009) with following modifications. About 3.75 ng cDNA template of each sample was mixed in a 15 μL reaction mix comprising cDNA, SYBR Green Master mix (Roche), and 0.17–0.34 μM primer on a Light Cycler 480 (Roche). The following gene-specific primers were designed manually to meet criteria of >45% GC content and >60°C primer melting temperature: HasNOS forward 5′-TGGGTTTGGACGTCCGGAAGAGC-3′ and reverse 5′-CCCGAAGACGCTGCTCGTTCTCC-3′ and HasHSP90A forward 5′-GTTACCTCCCCTTGCTGTATTGTCAC-3′ and reverse 5′-TTGTCAGCATCTGCCTTCTCCTTC-3′.
To normalize the level of transcription for obtaining relative gene expression values, we used the geometric mean of two reference genes - Has-nascent polypeptide- associated complex alpha polypeptide (NACA) (forward 5′-TGTCGCAAGCCAACGTTTCA-3′ and reverse 5′- GACAGCATGTTCAGCACTGGT-3′) and Has-ubiquitin (forward 5′-TGGCAAGCAGTTGGAAGATGGT-3′ and reverse 5′-CAGTTGTACTTGGAGGCCAGGAT-3′). These reference genes were previously chosen by Williams et al. (2009a) from microarray expression data by selecting for transcripts showing the least variable expression across larval and post-larval developmental stages. Their stability within our experimental sample set was confirmed using Genorm software (Vandesompele et al., 2002).
The following qRT-PCR reaction parameters were used: initial denaturation 95°C for 10 min (ramp rate 4.4°C/s), and 40–50 cycles of 95°C for 5 s (ramp rate 4.4°C/s), 58°C for 10 s (ramp rate 2.2°C/s), and 72°C for 20 s (ramp rate 4.4°C/s). Melt curve data acquisition was from 55–95°C with continuous measurement (acquisition/°C = 5). All samples were run in triplicate. The purity of PCR product was confirmed by the presence of only a single peak in the resultant temperature melt curve. For each primer pair, a standard curve was generated to calculate the efficiency of qRT-PCR using a dilution series from the calibrator sample, which was a mixture of 4 μl of all undiluted cDNA samples. In addition to the developmental stages cDNAs, a no-template (H2O) control and the calibrator sample were included for each qRT-PCR run and for each primer pair. The efficiencies of each primer pair and the cycle threshold of each sample were calculated by the second derivative method using Roche Light Cycler 480 software program. Relative expression ratios were calculated as described in Williams and Degnan (2009).
Whole Mount In Situ Hybridization (WMISH) to Assay HasNOS and HasHSP90A Spatial Expression
To determine spatial localization of HasNOS and HasHSP90A during larval development, H. asinina veligers were sampled at 48, 72, and 96 hpf (Figure 1); 48 and 72 hpf are precompetent larvae, and 96 hpf are competent larvae. Fixation and storage of H. asinina larvae for WMISH were carried out as described in Williams and Degnan (2009). Dioxygenin (DIG)-labeled antisense RNA probes for HasNOS and HasHSP90A were synthesized from purified PCR products that were amplified from a mixture of embryonic and larval cDNA collected as described above, using 10× DIG labeled mix (Roche) following the manufacturer's instructions. The HasNOS probe was transcribed from a 702 bp fragment of nucleotide sequence identified above. The probe for HasHSP90A was transcribed exactly as described in Gunter and Degnan (2007). The procedure of WMISH to examine the spatial gene expression was performed as described in Williams and Degnan (2009).
Statistical Analysis
Data collected from metamorphosis assays were analyzed by One-Way analysis of variance (ANOVA) with treatment as a factor. Significant differences among treatments were detected by Tukey's HSD post-hoc testing. Prior to ANOVA, all data were arcsine-transformed to improve the normal distribution of samples. Levene's test was performed to ensure homogeneity of variance among treatments. All statistical analyses were performed in R (R Foundation for Statistical Computing). An alpha value of 0.05 was used to determine a significant difference (Zar, 1984).
Results
NOS Inhibitors Suppress Metamorphosis in Haliotis asinina, but No Donors Have No Effect
Pharmacological experiments to test the role of NO in regulating settlement and metamorphosis in the tropical abalone Haliotis asinina were performed using both NOS inhibitors and NO donors (Table 1). Because competent veligers of H. asinina require the presence of coralline algae (CA) to induce metamorphosis (Williams et al., 2008), pharmacological experiments were conducted both with and without live CA. This allowed us to examine whether application of a pharmacological agent alone is sufficient to induce metamorphosis or sufficient to inhibit the inductive capability of live CA. In the absence of live CA that is already known to be an effective environmental inductive cue for metamorphosis in H. asinina, none of the pharmacological treatments—neither inhibitors nor donors—were able to induce metamorphosis on their own (Figures 3, 4).
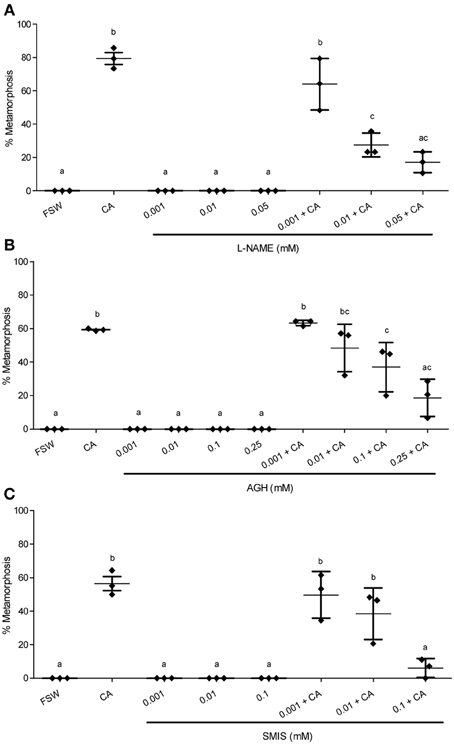
Figure 3. Effect of NOS inhibitors on metamorphosis of Haliotis asinina. (A) L-nitroarginine methyl ester (L-NAME), (B) aminoguanidine hemisulfate (AGH), and (C) S-methylisothiourea sulfate (SMIS) were applied at various concentrations. Filtered sea water (FSW) and Amphiroa ephedraea (CA) were used as negative and positive controls, respectively. The application of NO inhibitors alone without CA completely failed to induce metamorphosis. Data are presented as the mean percentage of larval metamorphosis ± s.e.m. (n = 3, 30 larvae per replicate). Diamonds show the actual percentage of larval metamorphosis in each replicate. Letters above error bars indicate statistically significant differences (P < 0.05), as determined by one-way analysis of variance and Tukey's HSD post-hoc testing.
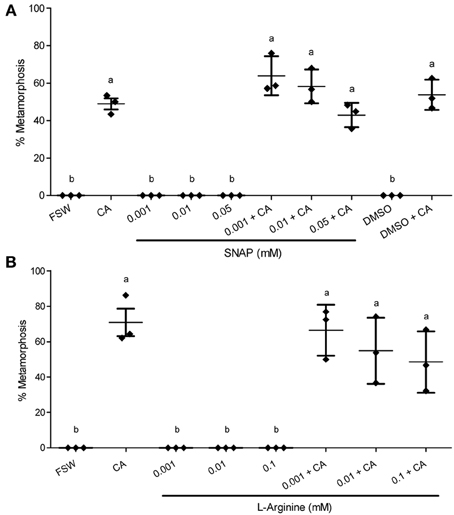
Figure 4. Effect of NO donors on metamorphosis of Haliotis asinina. (A) S-nitroso-N-acetyl-penicillamine (SNAP) and (B) L-Arginine were applied at various concentrations. Filtered sea water (FSW) and Amphiroa ephedraea (CA) were used as negative and positive controls, respectively. Data are presented as the mean percentage of larval metamorphosis ± s.e.m. (n = 3, 30 larvae per replicate). Diamonds show the actual percentage of larval metamorphosis in each replicate. Letters above error bars indicate statistically significant differences (P < 0.05), as determined by one-way analysis of variance and Tukey's HSD post-hoc testing.
In the presence of live CA, however, the application of NOS inhibitors gave results that are strikingly discordant with any molluscan studies published so far. All of the three NOS inhibitors examined—L-NAME, AGH, and SMIS (Table 1)—suppressed the mean percentage of larval metamorphosis in the presence of live CA, and did so in a concentration-dependent manner (Figure 3). The mean percent metamorphosis was significantly less than the positive control in the 0.01 and 0.05 mM L-NAME (Figure 2A), 0.25 mM AGH (Figure 2B), and 0.1 mM SMIS (Figure 2C) treatments in the presence of the live CA. In contrast, in the presence of live CA, neither of the NO donors—we tried both SNAP and L-arginine (Table 1)—significantly affected the mean percentage of larval metamorphosis in comparison with the positive controls (Figure 4). We do not believe that our ability to detect an enhanced rate of metamorphosis in the presence of NO donors was constrained by a ceiling effect, because our positive CA controls achieved only 50–70% metamorphosis in these experiments. In the SNAP experiment, the mean percentage of larval metamorphosis in the solvent control, DMSO, also did not significantly differ from the positive control (Figure 4A).
HasNOS and HasHSP90A Genes have Similar Expression Profiles Through the Onset of Metamorphosis
As an initial step to analyze the expression of NOS and HSP90 in H. asinina, we used degenerate PCR to isolate the partial NOS gene sequence, which was used to design specific primers for qRT-PCR and to synthesize specific probes for in situ hybridization analyses (see below).
Degenerate PCR yielded a putative 702 bp HasNOS product, which was confirmed to be a partial NOS gene sequence by tBLASTx against NCBI databases. The tBLASTx analysis showed very high sequence similarity to other molluscan orthologs, including Stramonita haemastoma (FR667655, E-value: 1e–115), Lehmannia valentiana (AB333805, E-value: 3e–110), and Crassostrea virginica (GQ844865, E-value: 7e–110). The Conserved Domain Database (CDD) confirmed that the isolated HasNOS was part of the NOS oxygenase domain, which was the intended target region of the degenerate primers. This partial Haliotis asinina NOS sequence has been named HasNOS and is available at NCBI [GenBank: KC571824].
Temporal expression profiles of HasNOS and HasHSP90A through embryonic, larval, and post-larval development (Figure 1) were measured by qRT-PCR (Figure 5). Both genes are expressed in all developmental stages examined. HasNOS is low from the egg until 24 hpf (Figure 5A). At 48 hpf, expression increases and then is maintained relatively stable, except for a sharp decrease at 84 hpf. The highest expression level among larvae is found at 144 hpf. Expression of HasNOS in post-larvae (that have initiated metamorphosis) shows more fluctuation in comparison with larvae of the same age that have not initiated metamorphosis. During the early post-larval stages (1–8 hpi), the expression level gradually declines, but sharply increases by 12 hpi, at which time it shows the highest expression level observed among samples induced to metamorphose. The expression pattern for 24 and 48 hpi is similar to that for the corresponding larval samples of 120 and 144 hpf.
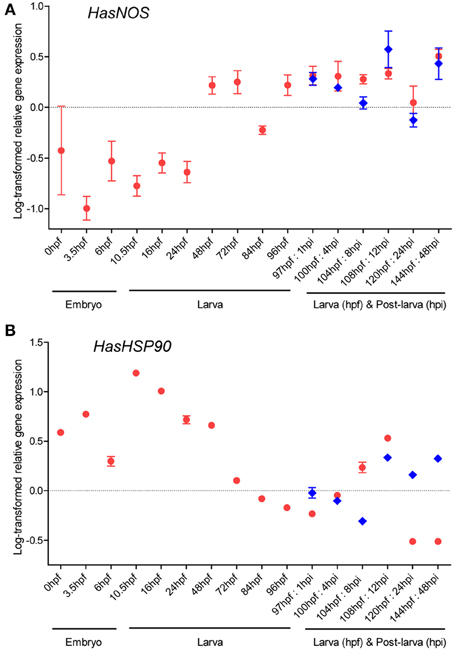
Figure 5. HasNOS (A) and HasHSP90A (B) gene expression through H. asinina normal development. Transcript abundance was assessed by qRT-PCR using mRNA purified from a pool of ~1000 embryos or larvae for each developmental stage (red circles). Transcript abundance in pooled samples of post-larvae induced by coralline algae Amphiroa ephedraea is denoted by blue diamonds. Data are presented as log-transformed mean ± s.e.m. of three technical replicates.
For HasHSP90A, the highest expression is seen at 10.5 hpf (Figure 5B). Expression then consistently declines through larval development until 97 hpf, followed by gradual increases from 100 to 108 hpf. After 108 hpf, the expression level decreases quite dramatically by 120 hpf, at which time it shows the lowest expression level observed among larval samples. Interestingly, the fluctuation of HasHSP90A expression pattern during post-larval development is comparable to that of HasNOS expression. The abundance of HasHSP90A transcript gradually decreases during the early post-larval stages and then increases sharply by 12 hpi, followed by fluctuations around 24–48 hpi.
Whole Mount in situ Hybridization Reveals that HasNOS—but not HSP90A—Transcripts are Localized to the Larval Foot
Whole mount in situ hybridization (WMISH) was performed to examine spatial expression of HasNOS and HasHSP90A in precompetent (48 and 72 hpf) and competent (96 hpf) veligers (Figure 1). At 48 hpf, HasNOS is localized to the larval retractor muscle and to a tight cluster of unidentified cells in the right lateral side of the mantle tissue (Figures 6A,B). This unidentified right mantle expression is maintained in 72 and 96 hpf larvae (Figures 6C–F). Additionally at this time, HasNOS expression appears in the cells aligned along the lateral edge of the foot (Figures 6C–F). By 96 hpf, expression in the foot has expanded across the pedal sole (Figure 6F). Unlike previous studies of spatial expression in other molluscan species (Thavaradhara and Leise, 2001; Pechenik et al., 2007; Bishop et al., 2008), no HasNOS expression is detectable in the larval central nervous system (CNS). Schematic representation of HasNOS localizations is shown in Figure 6G.
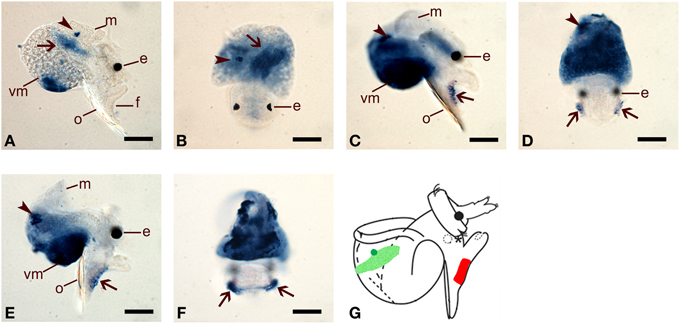
Figure 6. Spatial expression of HasNOS assessed by whole mount in situ hybridisation. Right lateral and dorsal views are shown for each developmental stage. (A,B) 48 hpf pre-competent larvae. Probe concentration of 1 ng/μl was used. Arrows indicate the larval retractor muscle and arrowheads are pointing to a cluster of unidentified cells in the right lateral side of the mantle tissue. (C,D) 72 hpf pre-competent larvae. Probe concentration of 2 ng/μl was used. (E,F) 96 hpf competent larvae. Probe concentration of 2 ng/μl was used. In (C–F), arrows indicate the cells aligned along the bottom of the lateral edge of the foot and arrowheads are pointing a cluster of unidentified cells on the right lateral side of veligers. Expansion of HasNOS expression across the sole is visible in (F). (G) A schematic representation of HasNOS localizations. The localization of a cluster of unidentified cells in the right lateral side of the mantle tissue in 48, 72, and 96 hpf larvae is indicated by green. The localization in the larval retractor muscle in 48 hpf larvae is indicated by sanded green. The localization in the lateral edge of the foot in 72 and 96 hpf larvae is indicated by red. e, eyespot; f, foot; m, mantle; o, operculum; vm, visceral mass. Scale bars: (A–F), 50 μm.
HasHSP90A is clearly localized to mantle tissue in 48 hpf larvae (Figures 7A,B), but by 72 and 96 hpf, transcripts are difficult to detect above background (Figures 7C,D). These results are concordant with the transcriptional abundance indicated by the qRT-PCR assay, in that HasHSP90A continues to decline through larval development (Figure 4B). Schematic representation of HasHSP90A localizations is shown in Figure 7E.
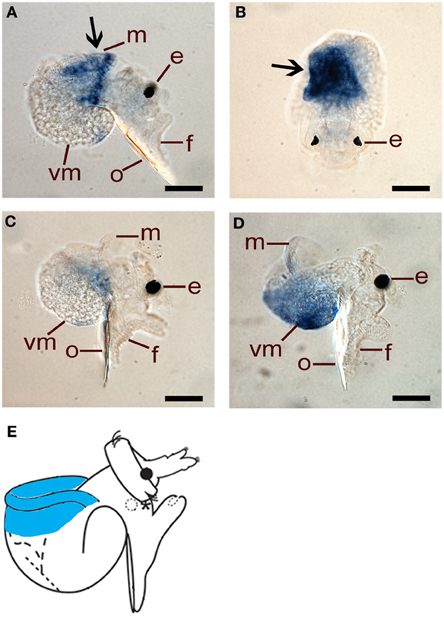
Figure 7. Spatial expression of HasHSP90A assessed by whole mount in situ hybridisation. (A,B) Right lateral and dorsal views of 48 hpf pre-competent larvae. Probe concentration of 1 ng/μl was used. Arrows indicate HasHSP90A localisation in mantle tissue. (C) Right lateral view of 72 hpf pre-competent larvae. Probe concentration of 1 ng/μl was used. (D) Right lateral view of 96 hpf competent larvae. Probe concentration of 1 ng/μl was used. In (C,D) HasHSP90A localisation becomes hardly detectable. (E) A schematic representation of HasHSP90A localization. The localization in mantle tissue is indicated by blue. e, eyespot; f, foot; m, mantle; o, operculum; vm, visceral mass. Scale bars: (A–D), 50 μm.
Discussion
Nitric Oxide Facilitates the Metamorphic Induction of H. asinina
Past studies so far have reported that NO functions as a negative (repressive) regulator of metamorphosis in several molluscs, namely I. obsoleta, C. fornicata, P. sibogae, and A. willowi (Froggett and Leise, 1999; Pechenik et al., 2007; Bishop et al., 2008; Romero et al., 2013). Here we reveal a first time contrasting situation, in which NO does not have a repressive function, but rather appears to assist in the initiation of larval metamorphosis in the abalone Haliotis asinina. In the context of the generalist-specialist larvae hypothesis (Bishop et al., 2006; Hodin, 2006; Romero et al., 2013), our results support the hypothesis for the modulatory role of NO at the initiation of metamorphosis in specialist larvae. However, in contrast to larvae of P. sibogae or A. willowi, in which NO repressively modulates the initiation of metamorphosis, NO in H. asinina appears to inductively modulate the initiation of metamorphosis.
Specifically, all NOS inhibitors examined in the current study—L-NAME, AGH, and SMIS—significantly suppressed metamorphosis in H. asinina in the presence of live CA (Figure 3), indicating that NO is a facilitator of metamorphic induction in this species. Williams et al. (2009b) showed that live CA induces a significantly higher mean percentage of H. asinina larval metamorphosis than does dead (bleached) CA, implying that the biomolecules derived from live CA play a crucial role in the induction of metamorphosis. We propose therefore that the application of NOS inhibitors impedes the signaling required to modulate the biomolecules derived from live CA, resulting in significant suppression of larval metamorphosis.
Importantly, the failure of NO donors examined in this study—SNAP and L-arginine—to induce metamorphosis in the absence of live CA indicates that elevated NO alone cannot act as an inductive agent (Figure 4). This suggests that the activation of at least one other morphogenetic pathway by inductive live CA is required for successful metamorphosis of H. asinina. More than 25 years ago, Baxter and Morse (1987) proposed the existence of two independent metamorphic pathways in the red abalone, Haliotis rufescens. One of these, which constitutes adenylate cyclase cascades coupled with gamma-aminobutyric acid (GABA) receptors, they named the morphogenetic pathway. The other, which constitutes chemosensory lysine receptors regulated by G protein signaling complexes, they named the regulatory pathway. While the sole activation of the morphogenetic pathway by externally-applied GABA can directly induce metamorphosis of H. rufescens larvae, stimulation of the regulatory pathway alone fails to do so (Baxter and Morse, 1987). Remarkably, however, the regulatory pathway can significantly amplify the effect of metamorphic induction by sub-optimal concentrations of GABA, suggesting that the actual role of the regulatory pathway is to increase the sensitivity of the morphogenetic pathway (Baxter and Morse, 1987). The fact that the application of NO donors alone is unable to induce metamorphosis in H. asinina suggests that a similar system of dual metamorphic pathways may exist in H. asinina to that reported in H. rufescens.
An Up-Regulation of HasNOS at Competency is Consistent with NO Facilitating the Initiation of Metamorphosis
Temporal gene expression profiles of HasNOS and HasHSP90A in post-larvae exposed to live CA are similar to each other (Figure 5), tentatively supporting the Bishop and Brandhorst (2003) hypothesis of NOS-HSP90 interaction at the initiation of settlement and metamorphosis. However, this support is not compelling and does not extend to expression profiles during attainment of competence, nor to spatial localization of transcripts. During larval development, the HasNOS expression has a sharp decrease to 84 hpf, but then increases again to 96 hpf, by which time all H. asinina larvae have achieved competency (Figure 5). As discussed above, NO may be playing a critical role in processing signals from inductive biomolecules to regulate the initiation of metamorphosis in H. asinina; therefore, we hypothesize that this increased expression around the time of acquiring competency is a molecular indication that larvae are developmentally mature enough to be able to respond to environmental cues that trigger the critical pelagic-benthic transition. The localization of HasNOS transcripts to the larval foot by 72 hpf (Figures 6C,D) and their expansion across the pedal sole by 96 hpf (Figures 6E,F) also supports this hypothesis (discussed further below). Intriguingly, both HasNOS and HasHSP90A expression show a second spike of up-regulation at 12 hpi (Figure 4), suggesting perhaps a different—and currently unknown—role of NO during post-larval development.
In other molluscs, in which NO acts as a negative regulator of settlement and metamorphosis, NOS expression data is reported only from the mud snail I. obsoleta (Hens et al., 2006) and the slipper shell snail C. fornicata (Taris et al., 2009). In I. obsoleta, the down-regulation of NOS soon after initiation of metamorphosis (Hens et al., 2006) was considered to be consistent with the hypothesis that endogenous NO is necessary for retention of the larval state (Froggett and Leise, 1999). In C. fornicata, NOS expression was reported to gradually increase through larval development, again consistent with a negative regulatory role of NO (Taris et al., 2009). However, NOS expression also then steadily increased during the first 6 h post metamorphosis, and this was attributed to handling stress (Taris et al., 2009). In H. asinina, our data also show some fluctuation in HasNOS expression soon after induction (Figure 5A), and we conclude that proper interpretation of these fluctuations will require additional expression analyses in conjunction with detailed descriptions of the cellular and morphological reorganization during metamorphosis of each species.
H. asinina Larval Foot as a Putative Sensory Structure for the Induction of Metamorphosis
Past studies have provided evidence that the foot of gastropod veliger larvae plays a significant role in the induction of metamorphosis, as it is a potential source of both chemo- and mechano-receptors (Chia and Koss, 1988, 1989; Arkett et al., 1989; Jackson et al., 2005; Stewart et al., 2011). In H. asinina, the localization of Has-tft1, which is thought to be involved in the recognition of inductive cues, was detected in the anterior part of the larval foot (and the cephalic tentacle) in competent larvae (Jackson et al., 2005). More recently, Has-GABAAR-β transcripts were reported localized to the lateral edge of the larval foot, and the application of an anti-GABA receptor protein antibody significantly reduced percent settlement induced by GABA, providing further evidence for the larval foot as a sensory structure to modulate the induction of metamorphosis in competent larvae of H. asinina (Stewart et al., 2011).
Spatial analysis of HasNOS gene expression (Figure 6) also provides evidence for a sensory role of the larval foot in H. asinina metamorphosis. HasNOS transcripts are not localized to the foot of 48 hpf non-competent veligers; however, localization is apparent in the lateral edge of the foot by 72 hpf and expands across the pedal sole by 96 hpf, by which time all larvae are competent to settle (Jackson et al., 2005). At the same times, there is no indication of HasNOS expression in any components of the larval CNS (Figure 6), regardless of whether these components are larval-specific or are retained in the adult CNS post-metamorphosis. Together, these data on NOS gene expression, combined with larval metamorphosis assays, strongly suggest that NO modulates the metamorphic induction of H. asinina larvae via the larval foot that is putatively functioning as a chemosensory structure to receive and process external settlement cues. This result differs from previous reports of NOS localization in the larval CNS (Thavaradhara and Leise, 2001; Pechenik et al., 2007; Bishop et al., 2008), and we propose below a relationship between these contrasting spatial expression patterns and contrasting regulatory roles for NO in molluscan metamorphosis.
A Hypothesis to Explain the Contrasting Regulatory Roles of NO in the Initiation of Molluscan Metamorphosis
A particularly significant outcome of this study is the unequivocal demonstration of NO as an essential molecule for the induction of H. asinina metamorphosis. This is a contrasting situation to all previously published studies on NO in molluscs. Based upon the limited comparative data available for NOS transcript and/or protein localization in gastropod larvae of multiple species, we present here a hypothesis to explain this contrasting function of NO in molluscan metamorphosis. Including the current study, the role of NO in molluscan metamorphosis has demonstrated three discrete patterns so far: (a) a negative regulatory role, whereby metamorphosis is induced solely by application of a NOS inhibitor, as reported in I. obsoleta, C. fornicata, and A. willowi (Froggett and Leise, 1999; Pechenik et al., 2007; Romero et al., 2013); (b) a negative modulatory role, whereby induction of metamorphosis is enhanced by the application of a NOS inhibitor in combination with an inductive substrate, as reported in P. sibogae and A. willowi (Bishop et al., 2008; Romero et al., 2013); and (c) an inductive role, whereby metamorphosis is suppressed by the application of a NOS inhibitor, as reported here for H. asinina. Very interestingly, these patterns are in accordance with variations in spatial NOS or NOS expression in these species.
Spatial expression data for the NOS protein or gene are available for I. obsoleta (Thavaradhara and Leise, 2001), C. fornicata (Pechenik et al., 2007), P. sibogae (Bishop et al., 2008), and H. asinina (this study). Both I. obsoleta and C. fornicata express NOS in the ASO (Thavaradhara and Leise, 2001; Pechenik et al., 2007), which as previously mentioned is a larval-specific sensory structure thought to function mainly to detect settlement cues (Croll and Dickinson, 2004; Croll, 2009). In I. obsoleta, the ASO is completely degraded by programmed cell death by 72 hpi (Gifondorwa and Leise, 2006). Although the fate of the ASO during and immediately after the metamorphosis of C. fornicata has not yet been reported, NOS expression is no longer detected in older veligers (9 days pre-competent and 23 days competent veligers), suggesting a mitigated function of the ASO as the veligers age (Pechenik et al., 2007). In P. sibogae, Bishop et al. (2008) found NOS localized to the cerebral ganglia, pedal ganglia, peripheral nervous systems around pedal ganglia, and the putative sensory cells of the foot. Unlike the ASO, the cerebral and pedal ganglia of P. sibogae become a part of the adult CNS after metamorphosis (Croll and Dickinson, 2004). Although the ASO of P. sibogae is a sensory structure that seems to receive inductive cues and modulate the initiation of settlement and metamorphosis (Hadfield et al., 2000), NOS localization is undetectable in the ASO in this species (Bishop et al., 2008).
In contrast to I. obsoleta, C. fornicata, and P. sibogae, NOS is localized to neither the larval nervous structure nor the CNS in H. asinina competent veligers, but instead to the putative peripheral sensory system of the foot. Therefore, we postulate that the means by which NO exerts its effect on metamorphosis of molluscan larvae may be directly related to the localization of NOS gene or protein with following patterns: (1) when NOS expression is mainly localized to larval-specific nervous structures that have a sensory role in the CNS, NO acts as a negative regulator; (2) when NOS expression is localized to parts of the CNS that are not larval-specific structures, NO acts as a negative modulator; and (3) when NOS expression is localized to peripheral sensory systems, NO acts as a facilitator of metamorphic induction. This hypothesis will be explicitly testable as more, taxonomically diverse studies on NOS regulation and expression in molluscan larval metamorphosis become available.
Conclusion
By integrating metamorphosis assays with pharmacological treatments and HasNOS and HasHSP90A gene expression analyses in the tropical abalone H. asinina, we provide the first evidence for NO acting as an inductive, rather than repressive, agent in the initiation of molluscan metamorphosis. Elevated NO levels alone are not sufficient to induce H. asinina metamorphosis, but NO does appear to modulate substrate-derived inductive signals. Specifically, we propose that the foot of H. asinina veligers acts as a site of chemoreception that modulates—via NO signaling—inductive signals derived from live coralline algae substrates. Inter-specific comparisons suggest that the variable function of NO in molluscan metamorphosis as a negative regulator, a negative modulator, or a facilitator pertains to spatial localization of NOS in the larval-specific CNS, the non-larval specific CNS, or the larval peripheral sensory system, respectively.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Elizabeth Williams, Carmel McDougall, Tim Wollesen, Andrew Calcino, Alexandrea Kranz, and Felipe Aguilera for help with abalone collection and spawning and the staff of the Heron Island Research Station (HIRS) for kindly assisting with animal and laboratory maintenance in the field. We are also grateful to the Degnan lab and to Bernie Degnan for critical suggestions and insights to improve the quality of this manuscript. This research was conducted while Nobuo Ueda held an Endeavour International Postgraduate Research Scholarship provided by Australian Government at the University of Queensland, and was supported by Australian Research Council grants to Sandie M. Degnan.
References
Amador-Cano, G., Carpizo-Ituarte, E., and Cristino-Jorge, D. (2006). Role of protein kinase C, G-protein coupled receptors and calcium flux during metamorphosis of the sea urchin Strongylocentrotus purpuratus. Biol. Bull. 210, 121–131. doi: 10.2307/4134601
Andreakis, N., D'Aniello, S., Albalat, R., Patti, F. P., Garcia-Fernàndez, J., Procaccini, G., et al. (2011). Evolution of the nitric oxide synthase family in metazoans. Mol. Biol. Evol. 28, 163–179. doi: 10.1093/molbev/msq179
Arkett, S. A., Chia, F.-S., Goldberg, J. I., and Koss, R. (1989). Identified settlement receptor cells in a nudibranch veliger respond to specific cue. Biol. Bull. 176, 155–160. doi: 10.2307/1541583
Baxter, G., and Morse, D. E. (1987). G protein and diacylglycerol regulate metamorphosis of planktonic molluscan larvae. Proc. Nat. Acad. Sci. U.S.A. 84, 1867–1870. doi: 10.1073/pnas.84.7.1867
Biggers, W. J., and Laufer, H. (1999). Settlement and metamorphosis of Capitella larvae induced by juvenile hormone-active compounds is mediated by protein kinase C and ion channels. Biol. Bull. 196, 187–198. doi: 10.2307/1542564
Biggers, W. J., Pires, A., Pechenik, J. A., Johns, E., Patel, P., Polson, T., et al. (2011). Inhibitors of nitric oxide synthase induce larval settlement and metamorphosis of the polychaete annelid Capitella teleta. Invertebr. Reprod. Dev. 56, 1–13. doi: 10.1080/07924259.2011.588006
Bishop, C. D., Bates, W. R., and Brandhorst, B. P. (2001). Regulation of metamorphosis in ascidians involves NO/cGMP signaling and HSP90. J. Exp. Zool. 289, 374–384. doi: 10.1002/jez.1019
Bishop, C. D., and Brandhorst, B. P. (2001). NO/cGMP signaling and HSP90 activity represses metamorphosis in the sea urchin Lytechinus pictus. Biol. Bull. 201, 394–404. doi: 10.2307/1543617
Bishop, C. D., and Brandhorst, B. P. (2003). On nitric oxide signaling, metamorphosis, and the evolution of biphasic life cycles. Evol. Dev. 5, 542–550. doi: 10.1046/j.1525-142X.2003.03059.x
Bishop, C. D., Huggett, M. J., Heyland, A., Hodin, J., and Brandhorst, B. P. (2006). Interspecific variation in metamorphic competence in marine invertebrates: the significance for comparative investigations into the timing of metamorphosis. Integr. Comp. Biol. 46, 662–682. doi: 10.1093/icb/icl043
Bishop, C. D., Pires, A., Norby, S.-W., Boudko, D., Moroz, L. L., and Hadfield, M. G. (2008). Analysis of nitric oxide-cyclic guanosine monophosphate signaling during metamorphosis of the nudibranch Phestilla sibogae Bergh (Gastropoda: Opisthobranchia). Evol. Dev. 10, 288–299. doi: 10.1111/j.1525-142X.2008.00238.x
Boyle, J. S., and Lew, A. M. (1995). An inexpensive alternative to glassmilk for DNA purification. Trends Genet. 11:8. doi: 10.1016/S0168-9525(00)88977-5
Cáceres, L., Necakov, A. S., Schwartz, C., Kimber, S., Roberts, I. J. H., and Krause, H. M. (2011). Nitric oxide coordinates metabolism, growth, and development via the nuclear receptor E75. Gene. Dev. 25, 1476–1485. doi: 10.1101/gad.2064111
Chia, F.-S., and Koss, R. (1988). Induction of settlement and metamorphosis of the veliger larvae of the nudibranch, Onchidoris bilamellata. Int. J. Inver. Rep. Dev. 14, 53–70. doi: 10.1080/01688170.1988.10510365
Chia, F.-S., and Koss, R. (1989). The fine structure of the newly discovered propodial ganglia of the veliger larva of the nudibranch Onchidoris bilamellata. Cell Tissue Res. 256, 17–26. doi: 10.1007/BF00224714
Cioni, C., Di Patti, M. C. B., Venturini, G., Modica, M. V., Scarpa, F., Oliverio, M., et al. (2011). Cellular, biochemical, and molecular characterization of nitric oxide synthase expressed in the nervous system of the prosobranch Stramonita haemastoma (Gastropoda, Neogastropoda). J. Comp. Neurol. 520, 364–383. doi: 10.1002/cne.22729
Colasanti, M., and Venturini, G. (1998). Nitric oxide in invertebrates. Mol. Neurobiol. 17, 157–174. doi: 10.1007/BF02802029
Comes, S., Locascio, A., Silvestre, F., D'Ischia, M., Russo, G. L., Tosti, E., et al. (2007). Regulatory roles of nitric oxide during larval development and metamorphosis in Ciona intestinalis. Dev. Biol. 306, 772–784. doi: 10.1016/j.ydbio.2007.04.016
Counihan, R. T., McNamara, D. C., Souter, D. C., Jebreen, E. J., Preston, N. P., Johnson, C. R., et al. (2001). Pattern, synchrony and predictability of spawning of the tropical abalone Haliotis asinina from Heron Reef, Australia. Mar. Ecol. Progr. Ser. 213, 193–202. doi: 10.3354/meps213193
Croll, R. P. (2009). Developing nervous systems in molluscs: navigating the twists and turns of a complex life cycle. Brain Behav. Evolut. 74, 164–176. doi: 10.1159/000258664
Croll, R. P., and Dickinson, A. J. G. (2004). Form and function of the larval nervous system in molluscs. Invertebr. Reprod. Dev. 46, 173–187. doi: 10.1080/07924259.2004.9652620
Degnan, B. M., and Morse, D. E. (1995). Developmental and morphogenetic gene regulation in Haliotis rufescens larvae at metamorphosis. Am. Zool. 35, 391–398.
Eri, R., Arnold, J. M., Hinman, V. F., Green, K. M., Jones, M. K., Degnan, B. M., et al. (1999). Hemps, a novel EGF-like protein, plays a central role in ascidian metamorphosis. Development 126, 5809–5818.
Froggett, S. J., and Leise, E. M. (1999). Metamorphosis in the marine snail Ilyanassa obsoleta, yes or NO? Biol. Bull. 196, 57–62. doi: 10.2307/1543167
García-Cardeña, G., Fan, R., Shah, V., Sorrentino, R., Cirino, G., Papapetropoulos, A., et al. (1998). Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature 392, 821–824.
Gifondorwa, D. J., and Leise, E. M. (2006). Programmed cell death in the apical ganglion during larval metamorphosis of the marine mollusc Ilyanassa obsoleta. Biol. Bull. 210, 109–120. doi: 10.2307/4134600
Gunter, H. M., and Degnan, B. M. (2007). Developmental expression of Hsp90, Hsp70 and HSF during morphogenesis in the vetigastropod Haliotis asinina. Dev. Gene. Evol. 217, 603–612. doi: 10.1007/s00427-007-0171-2
Hadfield, M. G. (1998). The D P Wilson Lecture. Research on settlement and metamorphosis of marine invertebrate larvae: past, present and future. Biofouling 12, 9–29. doi: 10.1080/08927019809378343
Hadfield, M. G. (2000). Why and how marine-invertebrate larvae metamorphose so fast. Semin. Cell. Dev. Biol. 11, 437–443. doi: 10.1006/scdb.2000.0197
Hadfield, M. G., Carpizo-Ituarte, E. J., del Carmen, K., and Nedved, B. T. (2001). Metamorphic competence, a major adaptive convergence in marine invertebrate larvae. Am. Zool. 41, 1123–1131. doi: 10.1668/0003-1569(2001)041[1123:MCAMAC]2.0.CO;2
Hadfield, M. G., Meleshkevitch, E. A., and Boudko, D. Y. (2000). The apical sensory organ of a gastropod veliger is a receptor for settlement cues. Biol. Bull. 198, 67–76. doi: 10.2307/1542804
He, Y., Tang, R., Hao, Y., Stevens, R. D., Cook, C. W., Ahn, S. M., et al. (2004). Nitric oxide represses the Arabidopsis floral transition. Science 305, 1968–1971. doi: 10.1126/science.1098837
Hens, M. D., Fowler, K. A., and Leise, E. M. (2006). Induction of metamorphosis decreases nitric oxide synthase gene expression in larvae of the marine mollusc Ilyanassa obsoleta (Say). Biol. Bull. 211, 208–211. doi: 10.2307/4134543
Heyland, A., Degnan, S. M., and Reitzel, A. M. (2011). “Emerging patterns in the regulation and evolution of marine invertebrate settlement and metamorphosis,” in Mechanisms of Life History Evolution, eds T. Flatt and A. Heyland (New York, NY: Oxford University Press), 29–42.
Hodin, J. (2006). Expanding networks: signaling components in and a hypothesis for the evolution of metamorphosis. Integr. Comp. Biol. 46, 719–742. doi: 10.1093/icb/icl038
Jackson, D. J., Ellemor, N., and Degnan, B. M. (2005). Correlating gene expression with larval competence, and the effect of age and parentage on metamorphosis in the tropical abalone Haliotis asinina. Mar. Biol. 147, 681–697. doi: 10.1007/s00227-005-1603-z
Kashiwagi, A., Hanada, H., Yabuki, M., Kanno, T., Ishikawa, R., Sasaki, J., et al. (1999). Thyroxine enhancement and the role of reactive oxygen species in tadpole tail apoptosis. Free Radical Bio. Med. 26, 1001–1009. doi: 10.1016/S0891-5849(98)00296-2
Korneev, S. A., Piper, M. R., Picot, J., Phillips, R., Korneeva, E. I., and O'Shea, M. (1998). Molecular characterization of NOS in a mollusc: expression in a giant modulatory neuron. J. Neurobiol. 35, 65–76.
Krug, P. J. (2001). Bet-hedging dispersal strategy of a specialist marine herbivore: a settlement dimorphism among sibling larvae of Alderia modesta. Mar. Ecol. Progr. Ser. 213, 177–192. doi: 10.3354/meps213177
Leise, E. M., and Hadfield, M. G. (2000). An inducer of molluscan metamorphosis transforms activity patterns in a larval nervous system. Biol. Bull. 199, 241–250. doi: 10.2307/1543180
Matsuo, R., Misawa, K., and Ito, E. (2008). Genomic structure of nitric oxide synthase in the terrestrial slug is highly conserved. Gene 415, 74–81. doi: 10.1016/j.gene.2008.02.021
Moroz, L. L., Edwards, J., Puthanveettil, S., Kohn, A., Ha, T., Heyland, A., et al. (2006). Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell 127, 1453–1467. doi: 10.1016/j.cell.2006.09.052
Palumbo, A. (2005). Nitric oxide in marine invertebrates: a comparative perspective. Comp. Biochem. Phys. A. 142, 241–248. doi: 10.1016/j.cbpb.2005.05.043
Pawlik, J. R. (1992). Chemical ecology of the settlement of benthic marine invertebrates. Oceanogr. Mar. Biol. Annu. Rev. 30, 273–335.
Pechenik, J. A., Cochrane, D. E., Li, W., West, E. T., Pires, A., and Leppo, M. (2007). Nitric oxide inhibits metamorphosis in larvae of Crepidula fornicata, the slippershell snail. Biol. Bull. 213, 160–171. doi: 10.2307/25066632
Rodriguez, S. R., Ojedal, F. P., and Inestrosa, N. C. (1993). Settlement of benthic marine invertebrates. Mar. Ecol. Progr. Ser. 97, 193–207. doi: 10.3354/meps097193
Romero, M. R., Phuong, M. A., Bishop, C. D., and Krug, P. J. (2013). Nitric oxide signaling differentially affects habitat choice by two larval morphs of the sea slug Alderia willowi: mechanistic insight into evolutionary transitions in dispersal strategies. J. Exp. Biol. 216, 1114–1125. doi: 10.1242/jeb.080747
Sawatpeera, S., Upatham, E., Kruatrachue, M., Chitramvong, Y., Sonchaeng, P., Pumthong, T., et al. (2001). Larval development in Haliotis asinina Linnaeus. J. Shell. Res. 20, 593–601. doi: 10.1111/j.1365-2109.2008.01982.x
Scheinker, V., Fiore, G., Di Cristo, C., Di Cosmo, A., D'Ischia, M., Enikolopov, G., et al. (2005). Nitric oxide synthase in the nervous system and ink gland of the cuttlefish Sepia officinalis: molecular cloning and expression. Biochem. Bioph. Res. Co. 338, 1204–1215. doi: 10.1016/j.bbrc.2005.10.069
Schmidt, I., Steenbakkers, P. J. M., op den Camp, H. J. M., Schmidt, K., and Jetten, M. S. M. (2004). Physiologic and proteomic evidence for a role of nitric oxide in biofilm formation by Nitrosomonas europaea and other ammonia oxidizers. J. Bacteriol. 186, 2781–2788. doi: 10.1128/JB.186.9.2781-2788.2004
Song, Y., Zweier, J., and Xia, Y. (2001). Heat-shock protein 90 augments neuronal nitric oxide synthase activity by enhancing Ca2+/calmodulin binding. Biochem. J. 360, 357–360. doi: 10.1042/0264-6021:3550357
Stewart, P., Williams, E. A., Stewart, M. J., Soonklang, N., Degnan, S. M., Cummins, S. F., et al. (2011). Characterization of a GABA A receptor β subunit in the abalone Haliotis asinina that is upregulated during larval development. J. Exp. Mar. Biol. Ecol. 410, 53–60. doi: 10.1016/j.jembe.2011.10.005
Tao, Y. P., Misko, T. P., Howlett, A. C., and Klein, C. (1997). Nitric oxide, an endogenous regulator of Dictyostelium discoideum differentiation. Development 124, 3587–3595.
Taris, N., Comtet, T., and Viard, F. (2009). Inhibitory function of nitric oxide on the onset of metamorphosis in competent larvae of Crepidula fornicata: a transcriptional perspective. Mar. Genom. 2, 161–167. doi: 10.1016/j.margen.2009.08.002
Thavaradhara, K., and Leise, E. M. (2001). Localization of nitric oxide synthase-like immunoreactivity in the developing nervous system of the snail Ilyanassa obsoleta. J. Neurocytol. 30, 449–456. doi: 10.1023/A:1015669112986
Ueda, N., and Degnan, S. M. (2013). Nitric oxide acts as a positive regulator to indcue metamorphosis of the ascidian Herdmania momus. PLoS ONE. 8:e72797. doi: 10.1371/journal.pone.0072797
Underwood, A. J., and Keough, M. J. (2001). “Supply-side ecology: the nature and consequences of variations in recruitment of intertidal organisms,” in Marine Community Ecology, eds M. D. Bertness, S. D. Gaines, and M. E. Hay (Sunderland: Sinauer Associates), 183–200.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034–research0034.11. doi: 10.1186/gb-2002-3-7-research0034
Wilken, M., and Huchzermeyer, B. (1999). Suppression of mycelia formation by NO produced endogenously in Candida tropicalis. Eur. J. Cell Biol. 78, 209–213. doi: 10.1016/S0171-9335(99)80100-9
Williams, E. A., Craigie, A., Yeates, A., and Degnan, S. M. (2008). Articulated coralline algae of the genus Amphiroa are highly effective natural inducers of settlement in the tropical abalone Haliotis asinina. Biol. Bull. 215, 98–107. doi: 10.2307/25470687
Williams, E. A., Cummins, S., and Degnan, S. M. (2009b). Settlement specifics: effective induction of abalone settlement and metamorphosis corresponds to biomolecular compositon of natural cues. Commun. Integr. Biol. 2, 347–349. doi: 10.4161/cib.2.4.8553
Williams, E. A., Degnan, B. M., Gunter, H., Jackson, D. J., Woodcroft, B. J., and Degnan, S. M. (2009a). Widespread transcriptional changes pre-empt the critical pelagic-benthic transition in the vetigastropod Haliotis asinina. Mol. Ecol. 18, 1006–1025. doi: 10.1111/j.1365-294X.2008.04078.x
Williams, E. A., and Degnan, S. M. (2009). Carry-over effect of larval settlement cue on postlarval gene expression in the marine gastropod Haliotis asinina. Mol. Ecol. 18, 4434–4449. doi: 10.1111/j.1365-294X.2009.04371.x
Yoshida, M., and Xia, Y. (2003). Heat shock protein 90 as an endogenous protein enhancer of inducible nitric-oxide synthase. J. Biol. Chem. 278, 36953–36958. doi: 10.1074/jbc.M305214200
Keywords: metamorphosis, nitric oxide synthase, marine invertebrate, mollusc, larval settlement, heat shock protein 90
Citation: Ueda N and Degnan SM (2014) Nitric oxide is not a negative regulator of metamorphic induction in the abalone Haliotis asinina. Front. Mar. Sci. 1:21. doi: 10.3389/fmars.2014.00021
Received: 20 May 2014; Paper pending published: 16 June 2014;
Accepted: 23 June 2014; Published online: 10 July 2014.
Edited by:
Iliana B. Baums, The Pennsylvania State University, USAReviewed by:
Andrew R. Mahon, Central Michigan University, USAAdam Michael Reitzel, University of North Carolina at Charlotte, USA
William Jarrett Biggers, Wilkes University, USA
Copyright © 2014 Ueda and Degnan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sandie M. Degnan, School of Biological Sciences, University of Queensland, St Lucia, Brisbane, QLD 4072, Australia e-mail: s.degnan@uq.edu.au
†Present Address: Nobuo Ueda, Max Planck Institute for Developmental Biology, Tuebingen, Germany
 Nobuo Ueda
Nobuo Ueda Sandie M. Degnan
Sandie M. Degnan