Community structure and population genetics of Eastern Mediterranean polychaetes
- 1Biology Department, University of Crete, Heraklion, Greece
- 2Hellenic Centre for Marine Research, Institute of Marine Biology, Biotechnology and Aquaculture, Heraklion, Greece
- 3Department of Biology, Chemistry and Pharmacology, Institute of Biology, Free University of Berlin, Berlin, Germany
Species and genetic diversity are often found to co-vary since they are influenced by external factors in similar ways. In this paper, we analyse the genetic differences of the abundant polychaete Hermodice carunculata (Pallas, 1766) during two successive years at two locations in northern Crete (Aegean Sea) and compare them to other populations in the Mediterranean Sea and the Atlantic Ocean. The genetic analysis is combined with an analysis of ecological divergence of the total polychaete community structure (beta diversity) at these locations. The phylogenetic analysis of all included H. carunculata populations revealed two main clades, one exclusively found in the Mediterranean and a second occurring in both the Mediterranean and the Atlantic. Genetic diversity indices reveal unexpectedly high differences between the two Cretan populations, despite the absence of apparent oceanographic barriers. A similarly high divergence, represented by a high beta diversity index, was observed between the polychaete communities at the two locations. This comparatively high divergence of the genetic structure of a dominant species and the total polychaete community might be explained by the strong influence of local environmental factors as well as inter-specific interactions between the dominance of a single species and the members of the community.
Introduction
The marine environment provides many opportunities for dispersal of individuals within and among populations (Cowen and Sponaugle, 2009). Benthic invertebrate taxa usually have very limited dispersal potential as adults, whereas many species have pelagic larval stages that facilitate dispersal (Weersing and Toonen, 2009). The dispersal ability of a taxon is directed by a number of factors such as historical processes, environmental conditions, currents and life history traits such as the duration of the larval stage (Pringle and Wares, 2007; Jolly et al., 2009). These factors can lead to a variety of distribution pattens—from cosmopolitan distributions of species to the existence of populations with significant genetic differences even at close geographic distances (Hohenlohe, 2004; Hart and Marko, 2010; Derycke et al., 2013; Iacchei et al., 2013; Vergara-Chen et al., 2013).
The evolution of genetic diversity patterns depends mostly on the interplay of mutation, random genetic drift, gene flow and natural selection (Hartl and Clark, 2007). These processes can be studied through population genetics and phylogeographic approaches (Derycke et al., 2013). Over the last two decades, the development of molecular techniques has allowed the in-depth study of population structure and dynamics in benthic invertebrates (e.g., Duran et al., 2004). Mitochondrial genes and especially the cytochrome c oxidase subunit I (mtCOI) are widely used and have been proved useful for both DNA barcoding of species (http://www.barcodeoflife.org/) and population genetic analysis (e.g., Avise, 1994; Galtier et al., 2009). For instance, mtDNA data have been widely used to assess the temporal and spatial fluctuations of haplotype diversity in natural populations. Many studies investigate cryptic species along environmental gradients (e.g., Jolly et al., 2005; Barroso et al., 2010), but few focus on the population structure within a specified area (e.g., Schulze et al., 2000; Craft et al., 2010; Chust et al., 2013).
At the community level, a series of mathematical approaches and indices exist to calculate quantitative estimates of divergence. Whittaker (1960) introduced the term “beta diversity” (often referred to as turnover diversity) to describe the diversification between assemblages within a certain geographic area. Since then, 24 different (dis-)similarity coefficients have been proposed to quantify beta diversity (Legendre and Cáceres, 2013).
There is a conceptual analogy between beta diversity of community structure and genetic divergence between populations of a species. Theory predicts that environmental and stochastic processes act at all levels of the biological organization and thus cause species diversity and genetic diversity to co-vary in time and space (Etienne and Olff, 2004; Vellend and Geber, 2005). A number of studies have identified the parallel influence of local processes on species and genetic diversity (e.g., Vellend, 2003; Papadopoulou et al., 2011), as well as the mutual shaping of community diversity and the genetic diversity of its members (e.g., Booth and Grime, 2003; Whitham et al., 2003; Hughes et al., 2008). If these theories hold true, beta diversity and population genetic diversity can be expected to change at similar rates between locations. Indeed, Baselga et al. (2013) have found such correlations in communities of water beetles across Europe, and Papadopoulou et al. (2011) revealed that beta diversity and haplotype diversity of tenebrionid beetle communities on the Aegean Islands declined at similar rates with increasing distances.
Furthermore, there are strong indications that the genetic diversity of a single species can have effects on the diversity of a whole community if that species has a dominant or keystone role in the ecosystem (Treseder and Vitousek, 2001; Whitham et al., 2003). If the genetic diversity of certain species is shown to change at similar rates to the species diversity of a community, these species could be used as a proxy for time-consuming and costly whole-community analyses (e.g., Féral et al., 2003). In this paper, we analyse the genetic differences of the abundant polychaete Hermodice carunculata (Pallas, 1766) during two successive years at two locations in northern Crete (Aegean Sea). The results are combined with an analysis of ecological divergence of the total polychaete community structure between these locations. This allows us to assess to what extent the genetic diversity of a single, dominant species reflects the overall community diversity, and provides a basis for the future exploration of such species as biological indicators.
Materials and Methods
Study Area
The analysis was based on samples collected previously in the framework of the NaGISA project [http://www.coml.org/projects/natural-geography-shore-areas-nagisa; datasets were published by Faulwetter et al. (2011) and Chatzigeorgiou et al. (2014)]. Samples were taken from two locations in northern Crete, Alykes and Elounda (35.41583, 24.98785; 35.25166, 25.75833; Figure 1). Both locations are characterized by a moderate wave exposure and a continuous hard bottom habitat, densely covered by algae, with Cystoseira spp. and Sargassum spp. being the most abundant in terms of surface coverage. The locations are 60 km away from each other and they seem to receive no detectable impact caused by human activities (Chatzigeorgiou et al., 2012).
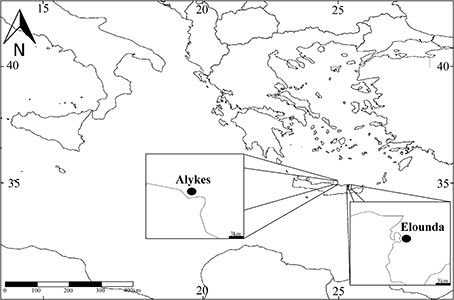
Figure 1. Map of the study sites, located in Crete: Alykes in the central part of the island and Elounda in the eastern part.
Specimen Collection and Processing
Sampling was conducted following the NaGISA protocol (Iken and Konar, 2003); details on the sampling procedure can be found in Chatzigeorgiou et al. (2012). All samples were preserved in 98% ethanol. In total, more than 8000 individuals were sorted and identified as 182 different polychaetes species, using the most recent literature. The species composition varied in terms of sampling depth and sampling site. Very few species were found in all sampling depths and sampling sites. From these, 40 individuals of Hermodice carunculata (Pallas, 1766) were randomly selected from each sampling location (20 per sampling year) for genetic analysis (see also paragraph “Model species” below). Primary morphometric characters (wet weight, length, width and number of chaetigers) were measured on each individual. Subsequently, the internal parts of each individual were removed and the remaining tissues were first cleaned and then processed for the analysis (Vasileiadou et al., 2012).
Ecological Analyses
For the purpose of this study, the original dataset was modified by combining the abundances of each depth, and analyses of beta diversity were based on the total species pool in each location per year. Although many indices have been suggested, the bw (Whittaker, 1960; Magurran, 1988; Southwood and Henderson, 2000) was selected as the most suitable for comparing geographic variation among populations, according to Wilson and Shmida (1984). The following equation was used:
where,
a is the total number of species occurring in both sampling locations, b the total number of species occurring only in the first sampling location and c the total number of species occurring only in second location. The beta diversity between two locations can assume values between zero (complete similarity) and one (complete dissimilarity) (Koleff et al., 2003).
A Mann-Whitney test (Mann and Whitney, 1947) was applied in order to test for differences between the measured morphological characters of the individuals belonging to the two populations.
Model Species
Hermodice carunculata (Polychaeta, Amphinomidae) is an abundant and important predator and scavenger on hard substrates in circumtropical coastal waters. The larvae of amphinomid species are unique among polychaetes, in possessing two elongated feeding tentacles (Mileikovsky, 1961). This allows the larva to cover great distances by staying for a longer period in the water column and probably provides distant populations with a high connectivity potential. This hypothesis was confirmed by Ahrens et al. (2013), who found no substantial genetic differentiation in the H. carunculata populations he examined, from the Mediterranean Sea (Malta and Crete) to the Atlantic Ocean (Caribbean Sea, Gulf of Mexico and Gulf of Guinea), and consequently assigned them to a single species. The species was chosen for this analysis for four reasons: (a) its ecological function, as one of the major predators in rocky and reef environments (e.g., Vreeland and Lasker, 1989), (b) its high frequency and abundance in the above habitats, (c) stable COI protocols for the species exist and, (d) a number of sequence data sets are available from a large geographic scale (e.g., Ahrens et al., 2013).
DNA Purification, Amplification and Sequencing
A small tissue sample (maximally 2 mg) was removed from the abdominal part of the individuals. In cases where individuals were too small, the tissue sample was collected from the widest part of the body. DNA purification was accomplished using a Nucleospin Tisseuekit XS (MACHEREY-NAGEL GmbH and Co. KG, Dueren, Germany). In order to determine the average concentrations of extracted DNA and the contamination with protein and phenol in the samples, a spectrophotometer was used for nucleic acid measurement (Nanodrop -1000). The universal DNA primers COI- LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) were used for the amplification of the COI gene (Folmer et al., 1994). The amplification reaction mix contained 1 μ l 10x buffer (with 1.5 mM Mg2+), 0.6 μ l MgCl2 (25 μ M), 0.2 μ l dNTPs (100 mM), 0.25 μ l of each primer (10 mM) and 0.1 μ l Taq DNA Polymerase (5 U/μ l) for a total volume of 10 μ l per reaction. The DNA template concentration was ~50 ng/μ l. The optimal PCR conditions were found by means of a temperature gradient PCR: 94°C for 5 min; 36 cycles with 94°C for 1 min, 44°C for 1 min, 72°C for 1 min; final extension at 72°C for 3 min. Amplifications were carried out in a BioRad Thermal Cycler T-100 PCR machine and PCR products were purified by using the Ethanol precipitation protocol. Cycle sequencing using the BigDye Terminator (Life Technologies) chemistry was conducted in 10 μ l volumes, contained 2 μ l BigDye Enzyme, 0.6 μ l of primer, 1 μ l of reaction buffer and 2 μ l of PCR product. The sequence reaction were performed with: an initial step at 96°C for 3 min, 35 cycles at 96°C for 10 s, 50°C for 15 s, 60°C for 4 min. Reactions were purified with EDTA (0,5 M, pH 8), NaAc (3 M, pH 4,3) and 98% Ethanol. Sequences were analyzed using an ABI 3730 Genetic Analyzer (Applied Biosystems, in the Laboratory of Genetics in HCMR), and resulting electrochromatograms were edited in BioEdit ver 7.2 by assembling the forward and reverse fragments from each specimen and deleting the primer regions. All sequences were submitted to GenBank with accession numbers KF878397-KF878476.
Sequence Analysis
Edited sequences were aligned in MEGA 5 (Tamura et al., 2011) via ClustalW using default settings (Larkin et al., 2007). The number of distinct haplotypes for COI was calculated using Arlequin 3.5.1.3 (Excoffier and Lischer, 2010). Phylogenetic trees were constructed using three different methods: (a) Bayesian inference (BI), (b) maximum likelihood (ML) and (c) neighbor-joining (NJ). The first method was done with the MrBayes 3.2.2 software (Huelsenbeck et al., 2002), using average branch lengths, while the other two were conducted with the Mega 5 software, using the Tamura-Nei model. For all the above methods a bootstrap option (n = 1,000) was used to calculate branch supporting values. Previously published data from Crete, Malta, Gulf of Mexico, Caribbean Sea and Gulf of Guinea were also included in the analyses (Ahrens et al., 2013, with accession numbers: Crete KC017526-8, 30; Malta KC017553-56; Caribbean Sea KC017521,24-25, KC017552; Gulf of Mexico KC017587-90 and Gulf of Guinea KC0 17480-2, KC017569). Taking into account the high rates (around 30%) of the genetic distances observed among other amphinomid genera (e.g., Borda et al., 2012) no out-group could be selected for the phylogenetic analyses. Therefore, the tree constriction midpoint rooted technique was selected, following Ahrens et al. (2013).
Additionally, the following indices were calculated, for each sampling year and for both years combined: haplotype diversity (h), nucleotide diversity (π) and fixation index (FST) using Arlequin 3.5.1.3. These analyses were run at the scale of (a) sampling location, which means that samples from the same location were grouped together (b) sites within Mediterranean sea and (c) sites between Mediterranean and Atlantic. For (b) and (c) data from Ahrens et al. (2013) were used. Phylogenetic networks were derived from the analyses using Network 4.6.1.1 software (Bandelt et al., 1999). The values of the fixation index range from zero to one, where zero implies complete panmixis, that is, that the two populations are interbreeding freely, and one implies that all genetic variation is explained by the population structure, and that the two populations do not share any genetic diversity at all.
Finally, a Mann-Whitney test was performed order to identify differences in the distribution of the haplotypes between the sampling sites (Alykes and Elounda) and between the sampling years (2007 and 2008).
Results
Morphometric Diversity
The total length of the individuals ranged from 1.54 to 9.97 cm and the maximum width from 0.54 to 1.75 cm. The number of the chaetigers varied from 21 to 78 and the wet weight of the individuals ranged from 0.002 to 2.9 g. The Mann-Whitney test showed no significant differences between the populations sampled from the two locations (Table 1).
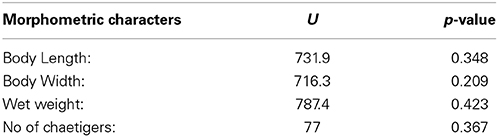
Table 1. Results of the Mann-Whitney test, comparing the values of the primary morphometric characters measured on the individuals from the two sampling locations.
Beta Diversity
Overall, a very high species diversity was observed in the polychaete assemblages sampled in the two Cretan locations with a total number of 182 species, 126 of which occur in both sites. Out of the total species number, 18 species were exclusive to Alykes and 38 to Elounda. The beta diversity (bw) value calculated from the above numbers is 0.181. In addition, bw values were calculated for each sampling year separately. In the first sampling year, the bw value was higher than in the second year (0.213 and 0.167, respectively).
Phylogeny
The COI sequencing analyses resulted in a joined alignment of 607 bp for the 80 individuals, with a total number of 23 mutations. A total of 15 haplotypes was found, of which seven were found exclusively in Elounda, seven exclusively in Alykes and a single one was shared between the two locations. Among the exclusive haplotypes, four were found only once (singletons), three of them in Alykes and one in Elounda. The shared haplotype, as expected, had a high frequency at both locations, but was the most common only in Alykes. In addition, the Mann-Whitney test performed on the frequencies of the above haplotypes at two different levels (sampling site and year), demonstrated that they were independent only in the case of the site (Year, U = 35.5, (N1 = 7, N2 = 9) P = 0.072; Sampling site, U = 38.5, (N1 = 8, N2 = 8) P = 0.023). The above haplotypes from both sampling locations are illustrated on the phylogenetic network of Figure 2.
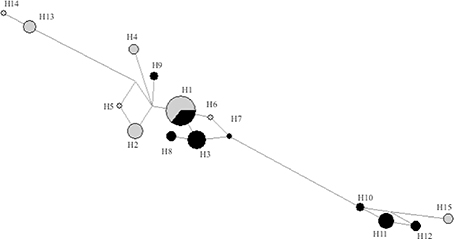
Figure 2. Phylogenetic network for the H. carunculata populations studied. Haplotypes from individuals from Alykes are represented with gray color, those from Elounda with black. Circle size is proportionate to the frequency of the haplotypes and the distances proportional to the number of the evolution steps. H: haplotype.
The phylogenetic analyses of COI sequence data via BI resulted in the consensus tree depicted in Figure 3. Two main clades can be distinguished, each with several subclades. Clades I and II were separated with high branch support values (BI posterior probability = 1.0, NJ Bootstrap = 98% and ML bootstrap = 97%).
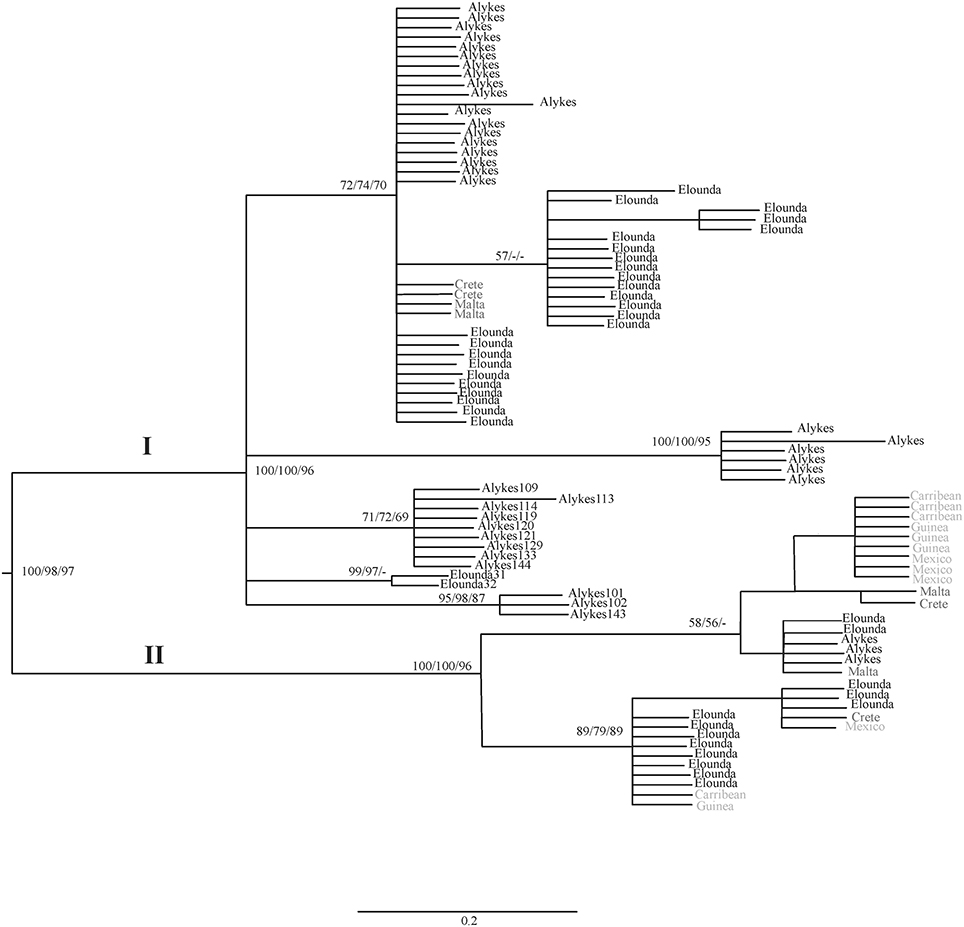
Figure 3. Consensus tree constructed using Bayesian Inference for the COI marker. Branch support values (higher than 50%) for Bayesian Inference, Maximum likelihood and Neighbor-joining are shown in each clade, respectively (_/_/_). Black: Cretan individuals and Mediterranean individuals from GenBank. Light gray: GenBank deposited data from outside the Mediterranean (Ahrens et al., 2013).
Molecular Diversity
Haplotype diversity (h) and nucleotide diversity (π) indices were calculated for each sampling location separately, for each sampling year and for both years combined (Table 2). Both indices showed higher values in the site of Elounda. The fixation index FST calculated from the two H. carunculata populations resulted in a value of 0.192 (P < 0.001) between two NaGISA sites (2007: 0.194, P < 0.001; 2008: 0.189, P < 0.001) while the FST value was 0.204 between the other Mediterranean sites and 0.224 between the Mediterranean and Atlantic sites.
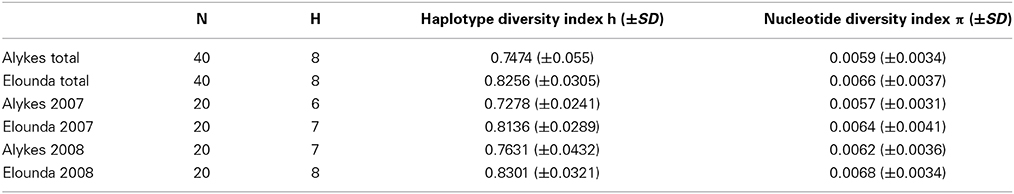
Table 2. Diversity indices values, calculated for the two sampling locations and two sampling years; ±SD: standard deviation.
Discussion
Both the mitochondrial and nuclear markers such as COI, 16S and 18S have been extensively used for the assessment of the genetic structure of populations in many geographic areas (e.g., Reeb and Avise, 1990). The dispersion potential of the species, the presence or absence of clear barriers and the effective population size are important components affecting the genetic population structure (Foll and Gaggiotti, 2006). In the case of the annelids, a rather complex pattern seems to have emerged from these factors. Schulze (2006) found that despite the absence of planktonic larvae in eunicid polychaetes, long distance dispersal is possible in at least some lineages. Within the genera of the family Amphinomidae, both ends of the spectrum have been found: Strongly divergent populations have been detected in the genus Eurythoe (considered to be cryptic species, Barroso et al., 2010), and widely distributed populations representing single species, such as those of Hermodice carunculata (Ahrens et al., 2013). Although the existing references for the reproductive biology and larval stages of amphinomids are scarce (e.g., Allen, 1957; Kudenov, 1974; Giangrande, 1997), teleplanic and rostraria larval forms are typical of the family. Both forms are planktonic and especially that of the unique rostraria with elongated feeding tentacles supports the theory of a long distance larval dispersal potential. However, Glasby (2005), based on a study of local polychaete endemism, postulated that the presence of a pelagic larval form does not necessarily imply a gene flow capable of homogenizing distant populations or preventing speciation.
The phylogenetic analysis of H. carunculata populations has revealed two main clades. These clades are separated having strong branch support values. The sequences included in Clade I reflect the long isolation of Atlantic and Mediterranean populations, having led to divergence and lineage sorting, although selective factors cannot be excluded to have added to such divergence. Clade II includes subclades consisting of individuals from both Mediterranean and from Mediterranean populations. The latter is indicative of the gene flow mechanism through which high connection rates between distant populations can be recorded. Ahrens et al. (2013) proposed that this gene flow mechanism is largely facilitated by factors such as sea water currents and ship transportation (ballast water and hull fouling), and which are capable of establishing connections between the H. carunculata populations in the Mediterranean Sea and the Atlantic Ocean.
Despite the ability of the taxon to maintain gene flow over long distances, the two Cretan populations show an important degree of differentiation according to the FST index, when compared to the populations within the Mediterranean and between the Mediterranean—Atlantic sites. The small geographic distance between the two sampling sites in Crete is reflected by lineage sorting, which in turn leads to fairly distinct branches for each location on the haplotype network. Among the 15 found haplotypes, only one is shared between the two sampling sites. This pattern implies a very limited or no gene flow, which could be by explained by a small effective population size (Slatkin, 1987). Effective population size is a key component affecting genetic population structure, as exemplified by differences in the case of the genetic structure between two nematode species (Caenorhabditis remanei and C. elegans) with differing effective population sizes due to different breeding behavior (Cutter et al., 2006; Charlesworth and Charlesworth, 2010). The effective population size may also be the main factor that accounts for low divergence in benthic nematodes (Bik et al., 2010) which show huge population sizes and a the high number of cryptic species, and the high divergence usually found in polychaetes with much smaller effective population sizes (Ahrens et al., 2013 and references therein).
Considering such a relatively quick lineage sorting, it is difficult to understand the occurrence of Clade II with representative samples in both the Atlantic and the Mediterranean. On the other hand, in other invertebrate species, distinct oceanographic conditions, occurring at spatial scales of just a few kilometers, can be responsible for the detected genetic differentiation in populations (Palumbi, 2004). This has also been reported for many polychaete species (e.g., Jolly et al., 2005; Schulze, 2006; Rockman, 2012). In this study, haplotype and nucleotide diversity indices values present higher values in Elounda than in Alykes. This suggests that the populations of H. carunculata in Elounda may be larger and more stable than in Alykes (Lowe et al., 2004). The less stable a population is, the higher is the genetic drift, which in turn, may cause gene variants to disappear (Scaps, 2002). This is most probably the reason for the increased number of haplotype singletons found in Alykes.
The beta diversity between the two polychaete assemblages was found to be rather high, compared to cases in which larger geographic distances or contrasted ecosystems with geographically isolated populations were analyzed (e.g., Legendre and Cáceres, 2013). Such high values of beta diversity can be considered as the consequence of habitat diversification (Whittaker, 1972). Given the apparent homogeneity of the sublittoral habitats in the region, the absence of apparent oceanographic barriers, the shallow water currents along the coasts of Crete and the relatively small distance between the two locations, they cannot be considered as geographically isolated from each other. Therefore, the past and present environmental conditions have to be considered as the major factor responsible for the rather strong divergence of these assemblages.
The two indices (FST and bw) represent different components of biodiversity in a certain area. Beta diversity reflects the differences between whole communities and thus represent species diversity, whereas FST is a measure of population differentiation due to genetic structure. In the Cretan locations, both the differences between the two populations of H. carunculata (based on molecular diversity) and the divergence of the polychaete taxo-communities (represented by bw) are comparatively high, given the apparent absence of any barriers. This indicates that environmental processes in the two areas must act in a similar way on both the composition of the communities and on the genetic diversity (Vellend and Geber, 2005). Such positive correlations between species and genetic diversity have been reported before (e.g., Vellend, 2003; Papadopoulou et al., 2011; Struebig et al., 2011; Baselga et al., 2013), and our results are in line with these findings. The dominant role of H. carunculata in the studied ecosystem, being an abundant carnivore, might be an additional explanation for the corresponding patterns between species and genetic divergence. Interactions between the species and the remaining community could therefore have led to a co-variation of the two levels of the biological organization (Whitham et al., 2003). If future analyses with additional locations show that these results are constant, the species could be a candidate for an indicator species, allowing rapid assessment of the divergence of two communities without the need for time-consuming identification of thousands of specimens.
Our research has highlighted a region and a taxonomic group worthy of further investigation, and is a promising approach for studies of community structuring and evolution, as well as for the determination of biological indicator species. In the present study, we have combined a standard community ecology analysis with population genetics and have successfully highlighted a scientific field of interest for follow up studies. Although the results of the present study are limited by the fact that only two locations are compared, the indices are rather constant over the two successive years, indicating that the populations and communities are not subjected to strong fluctuations over time. Furthermore, due to the high number of species in the community and the high number of individuals chosen for the genetic analysis the results are not statistically biased by a small sampling size, a common problem in similar analyses (Nazareno and Jump, 2012). Future studies should include additional locations as well as additional species and nuclear markers for the population genetics analysis. To complete this image, life history traits of the studied species as well as environmental factors such as oceanographic currents should be included to understand in detail the factors affecting the co-variation of species and genetic diversity.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors thank Dr. P. Divanac and Mr. G. Skouradakis for assistance in the preparation of the sampling equipment. The two anonymous referees are thanked for their comments which significantly improved this paper. This study is an outcome of the EuroNaGISA Network and forms part of the biodiversity core project of the IMBBC.
References
Ahrens, J. B., Borda, E., Barroso, R., Paiva, P. C., Campbell, A. M., Wolf, A., et al. (2013). The curious case of Hermodice carunculata (Annelida: Amphinomidae): evidence for genetic homogeneity throughout the Atlantic Ocean and adjacent basins. Mol. Ecol. 22, 2280–2291. doi: 10.1111/mec.12263
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Allen, M. J. (1957). Histochemical studies on developmental stages of polychaetous annelids. Anat. Rec. 128, 515–516.
Avise, J. C. (1994). Molecular Markers, Natural History and Evolution. New York, NY;London: Chapman & Hall.
Bandelt, H. J., Forster, P., and Rohl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Barroso, R., Klautau, M., Solé-Cava, A. M., and Paiva, P. C. (2010). Eurythoe complanata (Polychaeta: Amphinomidae), the “cosmopolitan” fireworm, consists of at least three cryptic species. Mar. Biol. 157, 69–80. doi: 10.1007/s00227-009-1296-9
Baselga, A., Fujisawa, T., Crampton-Platt, A., Bergsten, J., Foster, P. G., Monaghan, M. T., et al. (2013). Whole-community DNA barcoding reveals a spatio-temporal continuum of biodiversity at species and genetic levels. Nat. Commun. 4:1892. doi: 10.1038/ncomms2881
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bik, H. M., Thomas, W. K., Lunt, D. H., and Lambshead, P. J. D. (2010). Low endemism, continued deep-shallow interchanges, and evidence for cosmopolitan distributions in free-living marine nematodes (order Enoplida). Evol. Biol. 10:389. doi: 10.1186/1471-2148-10-389
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Booth, R. E., and Grime, J. P. (2003). Effects of genetic impoverishment on plant community diversity. J. Ecol. 91, 721–730. doi: 10.1046/j.1365-2745.2003.00804.x
Borda, E., Kudenov, J. D., Bienhold, C., and Rouse, G. W. (2012). Towards a revised Amphinomidae (Annelida, Amphinomida): description and affinities of a new genus and species from the Nile Deep-sea Fan, Mediterranean Sea. Zool. Scri. 41, 307–325. doi: 10.1111/j.1463-6409.2012.00529.x
Charlesworth, B., and Charlesworth, D. (2010). Elements of Evolutionary Genetics. Colorado, CO: Roberts and Company Publishers.
Chatzigeorgiou, G., Faulwetter, S., and Arvanitidis, C. (2014). Polychaetes From Two Subtidal Rocky Shores of the North Coast of Crete, Collected for the NaGISA Project 2007-2008. 18300 Records. Available online at: http://lifewww-00.her.hcmr.gr:8080/medobis/resource.do?r=nagisa_species_2007_2008
Chatzigeorgiou, G., Faulwetter, S., López, E., Sardá, R., and Arvanitidis, C. (2012). Can coastal biodiversity measured in four Mediterranean sites be representative of the region? a test for the robustness of the NaGISA protocol by using the hard substrate syllid (Annelida, Polychaeta) taxo-communities as a surrogate. Hydrobiologia 691, 147–156. doi: 10.1007/s10750-012-1065-5
Chust, G., Aitor Albaina, A., Aranburu, A., Borja, A., Diekmann, O. E., Estonba, A., et al. (2013). Connectivity, neutral theories and the assessment of species vulnerability to global change in temperate estuaries. Estuar. Coast. Shelf Sci. 131, 52–63. doi: 10.1016/j.ecss.2013.08.005
Cowen, R. K., and Sponaugle, S. (2009). Larval dispersal and marine population connectivity. Ann. Rev. Mar. Sci. 1, 443–466. doi: 10.1146/annurev.marine.010908.163757
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Craft, K. J., Pauls, S. U., Darrow, K., Miller, S. E., Hebert, P. D. N., Helgen, L. E., et al. (2010). Population genetics of ecological communities with DNA barcodes: an example from New Guinea Lepidoptera. Proc. Natl. Acad. Sci. U.S.A. 107, 5041–5046. doi: 10.1073/pnas.0913084107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cutter, A. D., Baird, S. E., and Charlesworth, D. (2006). High nucleotide polymorphism and the decay of linkage disequilibrium in wild populations of Caenorhabditis remanei and C. elegans. Genetics 174, 901–913. doi: 10.1534/genetics.106.061879
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Derycke, S., Backeljau, T., and Moens, T. (2013). Dispersal and gene flow in free-living marine nematodes. Front. Zool. 10, 1–12. doi: 10.1186/1742-9994-10-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Duran, S., Pascual, M., Estoup, A., and Turon, X. (2004). Strong population structure in the marine sponge Crambe crambe (Poecilosclerida) as revealed by microsatellite markers. Mar. Ecol. 13, 511–522. doi: 10.1046/j.1365-294X.2004.02080.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Etienne, R. S., and Olff, H. (2004). A novel genealogical approach to neutral biodiversity theory. Ecol. Lett. 7, 170–175. doi: 10.1111/j.1461-0248.2004.00572.x
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Faulwetter, S., Chatzigeorgiou, G., Galil, B. S., and Arvanitidis, C. (2011). Eastern Mediterranean Syllidae From Three Locations in Crete and Israel. 1123 Records. Available online at: http://lifewww-00.her.hcmr.gr:8080/medobis/resource.do?r=easternmedsyllids
Féral, J. P., Fourt, M., Perez, T., Warwick, R. M., Emblow, C., Heip, C., et al. (2003). European Marine Biodiversity Indicators. Yerseke: NIOO-CEME.
Foll, M., and Gaggiotti, O. (2006). Identifying the environmental factors that determine the genetic structure populations. Genetics 174, 875–891. doi: 10.1534/genetics.106.059451
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 3, 294–299.
Galtier, N., Nabholz, B., Glemin, S., and Hurst, G. D. D. (2009). Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 18, 4541–4550. doi: 10.1111/j.1365-294X.2009.04380.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Giangrande, A. (1997). Polychaetes reproductive patterns, life cycle and life histories: an overview. Oceanograph. Mar. Biol. 35, 323–386.
Glasby, C. J. (2005). Polychaete distribution patterns revisited: an historical explanation. Mar. Ecol. 26, 235–245. doi: 10.1111/j.1439-0485.2005.00059.x
Hart, M. W., and Marko, P. B. (2010). It's about time: divergence, demography, and the evolution of developmental modes in marine invertebrates. Integr. Comp. Biol. 50, 643–661. doi: 10.1093/icb/icq068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hartl, D., and Clark, A. (2007). Principles of Population Genetics. Sunderland, MA: Sinauer Associates.
Hohenlohe, P. A. (2004). Limits to gene flow in marine animals with planktonic larvae: models of Littorina species around Point Conception, California. Biol. J. Lin. Soc. 82, 169–187. doi: 10.1111/j.1095-8312.2004.00318.x
Huelsenbeck, J. P., Larget, B., Miller, R. E., and Ronquist, F. (2002). Potential applications and pitfalls of Bayesian inference of phylogeny. Syst. Biol. 51, 673–688. doi: 10.1080/10635150290102366
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hughes, A. R., Inouye, B. D., Johnson, M. T. J., Underwood, N., and Vellend, M. (2008). Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623. doi: 10.1111/j.1461-0248.2008.01179.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Iacchei, M., Ben-Horin, T., Selkoe, K. A., Bird, C. E., García-Rodríguez, F. J., and Toonen, R. J. (2013). Combined analyses of kinship and FST suggest potential drivers of chaotic genetic patchiness in high gene-flow populations. Mol. Ecol. 22, 3476–3494. doi: 10.1111/mec.12341
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Iken, K., and Konar, B. (2003). Natural Geography in Nearshore Areas (NaGISA): the Nearshore component of the Census of Marine Life. Gayana 67, 153–160. doi: 10.4067/S0717-65382003000200004
Jolly, M. T., Guyard, P., Ellien, C., Gentil, F., Viard, F., Thiébaut, E., et al. (2009). Population genetics and hydrodynamic modeling of larval dispersal dissociate contemporary patterns of connectivity from historical expansion into European shelf seas in the polychaete Pectinaria koreni. Limnol. Oceanogr. 54, 2089–2106. doi: 10.4319/lo.2009.54.6.2089
Jolly, M. T., Jollivet, D., Gentil, F., Thiébaut, E., and Viard, F. (2005). Sharp genetic break between Atlantic and English Channel populations of the polychaete Pectinaria koreni, along the North coast of France. Heredity 94, 23–32. doi: 10.1038/sj.hdy.6800543
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Koleff, P., Gaston, K. J., and Lennon, J. J. (2003). Measuring beta diversity for presence-absence data. J. Anim. Ecol. 72, 367–382. doi: 10.1046/j.1365-2656.2003.00710.x
Kudenov, J. D. (1974). The reproductive biology of Eurythoe complanata (Pallas, 1766) (Polychaeta, Amphinomidae). Ph.D. dissertation
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). ClustalW and ClustalX version 2. Bioinformatics, 23, 2497–2498. doi: 10.1093/bioinformatics/btm404
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Legendre, P., and Cáceres, M. (2013). Beta diversity as the variance of community data: dissimilarity coefficients and partitioning. Ecol. Lett. 16, 951–963. doi: 10.1111/ele.12141
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lowe, A., Harris, S., and Ashton, P. (2004). Ecological Genetics: Design, Analysis and Application. Oxford: Blackwell.
Mann, H. B., and Whitney, D. R. (1947). On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 18, 50–60. doi: 10.1214/aoms/1177730491
Mileikovsky, S. A. (1961). Assignment of two rostraria-type polychaete larvae from the plankton of the northwest Atlantic to species Amphinome pallasi Quatrefages, 1865 and Chloenea atlantica McIntosh, 1885 (Polychaeta, Errantia, Amphinomorpha). Dokl. Biol. Sci. 141, 1109–1112.
Nazareno, A. G., and Jump, A. S. (2012). Species-genetic diversity correlations in habitat fragmentation can be biased by small sample sizes. Mol. Ecol. 21, 2847–2849. doi: 10.1111/j.1365-294X.2012.05611.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pallas, P. S. (1766). Miscellanea Zoologica Quibus Novae Imprimis Atque Obscurae Animalium Species Describuntur Et Observationibus Iconibusque Illustrantur. Hagae Comitum: Apud Petrum van Cleef.
Palumbi, S. R. (2004). Marine reserves and ocean neighborhoods: the spatial scale of marine populations and their management. Ann. Rev. Env. Resour. 29, 31–68. doi: 10.1146/annurev.energy.29.062403.102254
Papadopoulou, A., Anastasiou, I., Spagopoulou, F., Stalimerou, M., Terzopoulou, S., Legakis, A., et al. (2011). Testing the species-genetic diversity correlation in the aegean archipelago: toward a haplotype-based macroecology? Am. Nat. 178, 241–255. doi: 10.1086/660828
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pringle, J. M., and Wares, J. P. (2007). Going against the flow: maintenance of alongshore variation in allele frequency in a coastal ocean. Mar. Ecol. Prog. Ser. 335, 69–84. doi: 10.3354/meps335069
Reeb, C. A., and Avise, J. C. (1990). A genetic discontinuity in a continu- ously distributed species: mitochondrial DNA in the American oyster, Crassostrea virginica. Genetics 124, 397–406.
Rockman, M. V. (2012). Patterns of nuclear genetic variation in the poecilogonous polychaete Streblospio benedicti. Integr. Comp. Biol. 52, 173–180. doi: 10.1093/icb/ics083
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scaps, P. (2002). A review of the biology, ecology and the potential use of the common ragworm Hediste diversicolor (O.F. Muller) (Annelida: Polychaeta). Hydrobiologia 470, 203–218. doi: 10.1023/A:1015681605656
Schulze, A. (2006). Phylogeny and genetic diversity of palolo worms (Palola, Eunicidae) from the Tropical North Pacific and the Caribbean. Biol. Bull. 210, 25–37. doi: 10.2307/4134534
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schulze, S. R., Rice, S. A., Simon, J. L., and Karl, S. A. (2000). Evolution of poecilogony and the biogeography of North American populations of the polychaete Streblospio. Evolution 54, 1247–1259. doi: 10.1111/j.0014-3820.2000.tb00558.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Slatkin, M. (1987). Gene flow and geographic structure of natural populations. Science 236, 787–792. doi: 10.1126/science.3576198
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Struebig, M., Kingston, T., Petit, E. J., Zubaid, A., Mohd-Adnan, A., and Rossiteri, S. J. (2011). Parallel declines in species and genetic diversity in tropical forest fragments. Ecol. Lett. 14, 582–590. doi: 10.1111/j.1461-0248.2011.01623.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tamura, K., Peterson, D., and Peterson, N. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Treseder, K. K., and Vitousek, P. M. (2001). Potential ecosystem-level effects of genetic variation among populations of Metrosideros polymorpha from a soil fertility gradient in Hawaii. Oecologia 126, 266–275. doi: 10.1007/s004420000523
Vasileiadou, K., Sarropoulou, E., Tsigenopoulos, C., Reizopoulou, S., Nikolaidou, A., Orfanidis, S., et al. (2012). Genetic vs community diversity patterns of macrobenthic species : preliminary results from the lagoonal ecosystem. Transitional Waters Bull. 6, 20–33. doi: 10.1285/i1825229Xv6n2p20
Vellend, M. (2003). Island biogeography of genes and species. Am. Nat. 162, 358–365. doi: 10.1086/377189
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vellend, M., and Geber, M. A. (2005). Connections between species diversity and genetic diversity. Ecol. Lett. 8, 767–781. doi: 10.1111/j.1461-0248.2005.00775.x
Vergara-Chen, C., Gonzalez-Wanguemert, M., Marcos, C., and Perez-Ruzafa, A. (2013). Small-scale genetic structure of Cerastoderma glaucum in a lagoonal environment: potential significance of habitat discontinuity and unstable population dynamics. J. Mollus. Stud. 79, 230–240. doi: 10.1093/mollus/eyt015
Vreeland, H. V., and Lasker, H. R. (1989). Selective feeding of the polychaete Hermodice carunculata Pallas on Caribbean gorgonians. J. Exp. Mar. Biol. Ecol. 129, 265–277. doi: 10.1016/0022-0981(89)90108-1
Weersing, K., and Toonen, R. J. (2009). Population genetics, larval dispersal, and connectivity in marine systems. Mar. Ecol. Prog. Ser. 393, 1–12. doi: 10.3354/meps08287
Whitham, T. G., Young, W. P., Martinsen, G. D., Gehring, C. A., Schweitzer, J. A., Shuster, S. M., et al. (2003). Community and ecosystem genetics: a consequence of the extended phenotype. Ecol. 84, 559–573. doi: 10.1890/0012-9658(2003)084[0559:CAEGAC]2.0.CO;2
Whittaker, R. H. (1960). Vegetation of the Siskiyou Mountains, Oregon and California. Ecol. Monogr. 30, 279–338. doi: 10.2307/1943563
Whittaker, R. H. (1972). Evolution and measurement of species diversity. Taxon 21, 213–251. doi: 10.2307/1218190
Keywords: Hermodice carunculata, mtCOI, NaGISA, rocky shore, beta diversity, fixation index
Citation: Chatzigeorgiou G, Sarropoulou E, Vasileiadou K, Brown C, Faulwetter S, Kotoulas G and Arvanitidis CD (2014) Community structure and population genetics of Eastern Mediterranean polychaetes. Front. Mar. Sci. 1:47. doi: 10.3389/fmars.2014.00047
Received: 18 June 2014; Accepted: 15 September 2014;
Published online: 06 October 2014.
Edited by:
Alberto Basset, University of Salento, ItalyReviewed by:
Guillem Chust, AZTI-Tecnalia, SpainKatherine Dafforn, University of New South Wales, Australia
Copyright © 2014 Chatzigeorgiou, Sarropoulou, Vasileiadou, Brown, Faulwetter, Kotoulas and Arvanitidis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giorgos Chatzigeorgiou, Hellenic Centre for Marine Research, Institute of Biology Biotechnology and Aquaculture, Former American Base at Gournes, 71500 Crete, Greece e-mail: chatzigeorgiou@hcmr.gr
 Giorgos Chatzigeorgiou
Giorgos Chatzigeorgiou Elena Sarropoulou2
Elena Sarropoulou2  Katerina Vasileiadou
Katerina Vasileiadou Christina Brown
Christina Brown Sarah Faulwetter
Sarah Faulwetter Christos D. Arvanitidis
Christos D. Arvanitidis