Absence of genetic differentiation in the coral Pocillopora verrucosa along environmental gradients of the Saudi Arabian Red Sea
- 1Red Sea Research Center, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
- 2Helmholtz Center for Ocean Research, GEOMAR, Kiel, Germany
- 3Department of Marine Biology, Faculty of Marine Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
The Red Sea is the world's northernmost tropical sea. The 2000 km long, but narrow basin creates distinct environmental conditions along its latitudinal spread. The Red Sea displays a pronounced salinity gradient from 41 to 37 PSU (north to south) with an opposing temperature gradient from 21 to 27°C in the north to 27–33.8°C in the south. The Red Sea further displays a decreasing nutrient gradient from south to north that can also influence underwater light fields due to higher phytoplankton content and turbidity. Despite this strong variation in temperature, salinity, nutrients, and light conditions, the Red Sea supports large and diverse coral reef ecosystems along its nearly entire coastline. Only few studies have targeted whether these prevailing gradients affect genetic connectivity of reef organisms in the Red Sea. In this study, we sampled the abundant reef-building coral Pocillopora verrucosa from 10 reefs along a latitudinal gradient in the Red Sea covering an area of more than 850 km. We used nine Pocillopora microsatellite markers to assess the underlying population genetic structure and effective population size. To assure the exclusion of cryptic species, all analyzed specimens were chosen from a single mitochondrial lineage. Despite large distances between sampled regions covering pronounced, but smooth temperature and salinity gradients, no significant genetic population structure was found. Rather, our data indicate panmixia and considerable gene flow among regions. The absence of population subdivision driven by environmental factors and over large geographic distances suggests efficient larval dispersal and successful settlement of recruits from a wide range of reef sites. It also advocates, broadcast spawning as the main reproductive strategy of Pocillopora verrucosa in the Red Sea as reflected by the absence of clones in sampled colonies. These factors might explain the success of Pocillopora species throughout the Indo-Pacific and Arabian Seas.
Introduction
The Red Sea features an environment that is characterized by both, strong latitudinal temperature and salinity gradients. From north to south, sea surface temperature (SST) increases from 21 to up to 33.8°C and salinity decreases from 41 PSU to 37 PSU (Edwards, 1987; Sofianos and Johns, 2007; Raitsos et al., 2013; Kürten et al., 2014). The Red Sea thereby displays an antagonistic salinity-temperature profile, which is in contrast to other seas (e.g., the Mediterranean or the Gulf of Mexico). The Red Sea is further characterized by a lack of fresh water river inlets and low precipitation rates, being surrounded by hot arid land masses, and by high evaporation rates (~2 m year−1) due to strong hot winds (Morcos, 1970; Sofianos et al., 2002; Siddall et al., 2003; Sirocko, 2003). In the south, however, water exchange occurs between the Indian Ocean and the Red Sea through the very narrow and shallow connection at Bab el Mandeb. This connection allows cooler and less saline oceanic waters to enter the Red Sea. The water is quickly warmed up in the rather shallow southern Red Sea and gradually decreases in temperature further north, while salinity increases toward the north due to high evaporation (Murray and Johns, 1997; Sofianos et al., 2002). Due to monsoon-driven seasonal upwelling off of Jemen's coast in the Gulf of Aden, waters entering the Red Sea in the south are also higher in nutrients (Kürten et al., 2014). The nutrients are gradually depleted toward the north. Thus, in the southern Red Sea productivity is higher leading to higher phytoplankton concentrations and consequently to higher turbidity (Raitsos et al., 2013; Kürten et al., 2014).
Habitat delimitations of aquatic species are considerably driven by temperature, light (Schwalbach et al., 2005; Zinser et al., 2007), salinity, and nutrients (Hewson et al., 2006; Foster et al., 2007), and the prevailing strong environmental and physicochemical gradients in the Red Sea can potentially shape genetic divergence of populations among marine species. This was shown by a recent study in the Red Sea analyzing the population structure of a coral reef fish, where the environmental gradients were found to predict the genetic population structure (Nanninga et al., 2014). Whether this applies for Red Sea corals as well, remains to be investigated. Yet, studies have repeatedly shown that corals are sensitive to environmental conditions, including salinity, turbidity, nutrients and temperature (Hoegh-Guldberg, 1999; Brainard et al., 2013).
The reef-building coral Pocillopora verrucosa is commonly found throughout the Red Sea and the Indo-Pacific, mostly in shallow waters (Veron and Pichon, 1976). It is a hermaphrodite broadcast-spawner, which releases its gametes during daytime in the Red Sea (Bouwmeester et al., 2011). Despite its wide distribution and success in various habitats, P. verrucosa, as well as other pocilloporids, have been recognized to be highly susceptible to environmental change (Marshall and Baird, 2000; McClanahan, 2004; Ziegler et al., 2014). A recent study by Pinzón et al. (2013) identified between five and eight genetically distinct lineages (i.e., species) of Pocillopora in the Indo-Pacific with minimal or no hybridization between them. The same study identified two distinct mitochondrial Open Reading Frame (mORF) haplotype clusters in the Red Sea that may represent two distinct species. These were not resolved by microsatellite genotyping, and appeared “damicornis-like” in some regions and “verrucosa-like” in other regions. Accordingly, for concordance of data, any study on Pocillopora in the Red Sea should discriminate between these two taxonomic units.
In this study, we analyzed specimens of Pocillopora verrucosa along the coast of Saudi Arabia employing nine microsatellite markers (Magalon et al., 2004; Starger et al., 2008) to reveal the underlying population genetic structure. To assure the exclusion of cryptic species, all specimens were chosen from a single mitochondrial lineage (“type 3” sensu Pinzón et al., 2013). Our further aim was to assess whether population differentiation correlates to geographic region and/or prevailing environmental conditions along a latitudinal environmental gradient. Our analysis provides a point of reference for the genetic structure of Pocillopora verrucosa in the Red Sea as well as hypotheses regarding the genetic diversity of coral populations and recruits.
Materials and Methods
Coral Sampling and Ethics Statement
The reefs sampled in this study do not fall under any legislative protection or special designation as a marine/environmental protected area. The Saudi Coastguard Authority issued sailing permits to the sites that include coral collection. Pocillopora verrucosa is listed on the ICUN Red List (http://www.iucnredlist.org/details/133197/0). Corals were sampled in accordance with ethical standards for the care and use of invertebrate animals. Coral branches of 1 to 10 cm in length were collected from colonies of the morphotype Pocillopora verrucosa from 2011 to 2013 at 10 sites from 3 geographical regions along the Saudi Arabian Red Sea coast. The regions included Al Wajh (WAJ) in the north, Thuwal (THU) in the center, and Doga (DOG) in the south. Each region was represented by 3–4 reefs, which were separated by 2–5 km. Samples were fixed in salt saturated DMSO solution and stored at −20°C until further processing. We sampled a total of 361 colonies from multiple offshore reefs (Table 1). Colonies were at least 5 m distant to each other in order to avoid collecting clonal corals. We sampled four reefs around Al Wajh (WAJ) and collected 44 samples from reef number one (WR1: 26°11′15.05″N, 36°20′57.21″E) as well as 29 samples from reef number two (WR2: 26°09′59.80″N, 36°23′32.20″E) and reef number three (WR3: 26°14′28.30″N, 36°26′25.30″E), and 30 samples from reef number four (WR4: 26°11′06.20″N, E36°22′58.90″E). In the central region, i.e., around Thuwal (THU), we collected 48 samples from each of the following reefs: Shib Nizar (TR1: 22°19′09.00″N, 38°51′16.00″E), Abu Madafi (TR2: 22°04′02.00″N, 38° 46′09.00″E) and Abu Roma (TR3: 22°30′4.008″N, 83°55′17.00″E). In the southern Red Sea, i.e., around Doga (DOG), we collected 29 samples from reef one (DR1: 19°38′06.40″N, 40°34′31.30″E) and reef three (DR3: 19°39′56.49″N, 40°37′21.59″E) as well as 27 samples from reef two (DR2: 19°36′50.50″N, 40°38′17.50″E). Prevailing environmental conditions of sampled reefs are detailed in Sawall et al. (2014b) and Kürten et al. (2014) (Table 1).
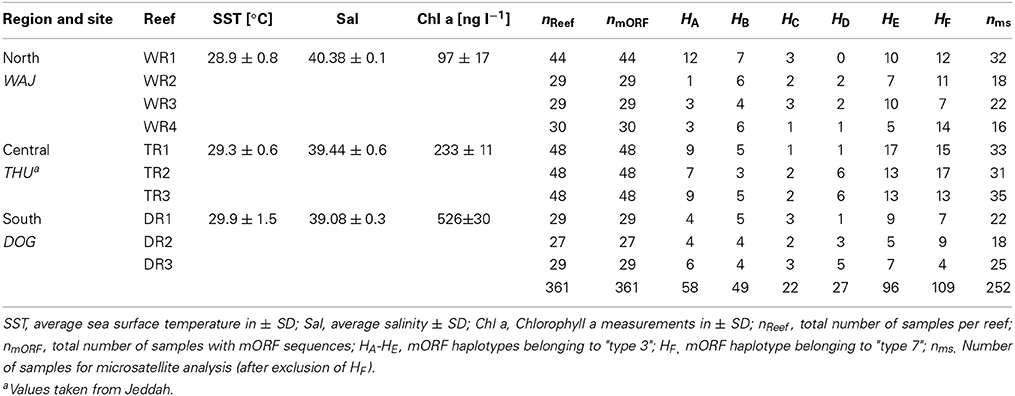
Table 1. Sampling sites, environmental conditions, and number of samples for mitochondrial (mORF) and nuclear (ms) marker analyses.
DNA Extraction
Coral DNA was extracted using the Qiagen DNeasy Plant Mini Kit following the manufacturer's protocol with minor modifications. Briefly, coral pieces of ~5 mm diameter were cut with sterile bone clippers and placed in a sterile 1.5 ml Eppendorf tube filled with ~0.5 ml of 0.5 mm glass beads (Biospecs) and 400 μl AP1 Qiagen Lysis Buffer. Tubes were bead-beaten with the Qiagen Tissue Lyser II at highest frequency (30 Hz) for 90 s, and DNA was eluted once with 80 μl Qiagen AE elution buffer. Subsequently, the DNA concentration was measured using a NanoDrop 2000 (ThermoScientific) and concentrations were adjusted between 10 and 40 ng/μl for PCR amplifications. The same diluted aliquots were used for mitochondrial (mORF) and nuclear DNA (microsatellite) amplifications.
Mitochondrial Open Reading Frame Amplification and Analysis
A mitochondrial Open Reading Frame (mORF) of unknown function was amplified using the primers FATP6.1 5′-TTTGGGSATTCGTTTAGCAG-3′ and RORF 5′-SCCAATATGTTAAACASCATGTCA-3′ as described and developed by Flot and Tillier (2007) for the genus Pocillopora. PCR reactions were carried out in 12 μl reactions containing 6 μl Qiagen Multiplex PCR Kit mix, 1.25 μmol of each primer, 3.75 μl water and 1 μl DNA (10–40 ng) as template. Cycle conditions were: 94°C for 15 min, followed by 35 cycles of 94°C for 30 s, 53°C for 45 s, 72°C for 90 s, and a final extension at 72°C for 10 min. PCR products were cleaned with Illustra ExoStar 1-step (Biotech) following the manufacturer's protocol. Fragments were bi-directionally sequenced on a Sanger ABI 3730XL at the Bioscience Core lab of KAUST. For sequence analysis, forward and reverse sequences were aligned, assembled, and trimmed in CodonCodeAligner version 3.7.1 (Codon-Code Corporation, Centerville, MA, USA). Only high quality unambiguous sequences were used to build a contig of bi-directionally sequenced mORFs. Resulting contigs were exported to MEGA 5.1 (Tamura et al., 2011) to check for gaps or indels and subsequently imported into DNAsp 5.10.1 (Librado and Rozas, 2009) to determine distinct haplotypes. A Maximun Likelihood phylogenetic tree (1000 bootstrap replicates) was then built using the program MEGA 5.1 for all distinct haplotypes belonging to type 3 specimens to exclude the possibility of skewed results originating from the analysis of distinct Pocillopora lineages. Type 3 was also the most abundant mitochondrial lineage among our samples.
Microsatellite Amplification and Analysis
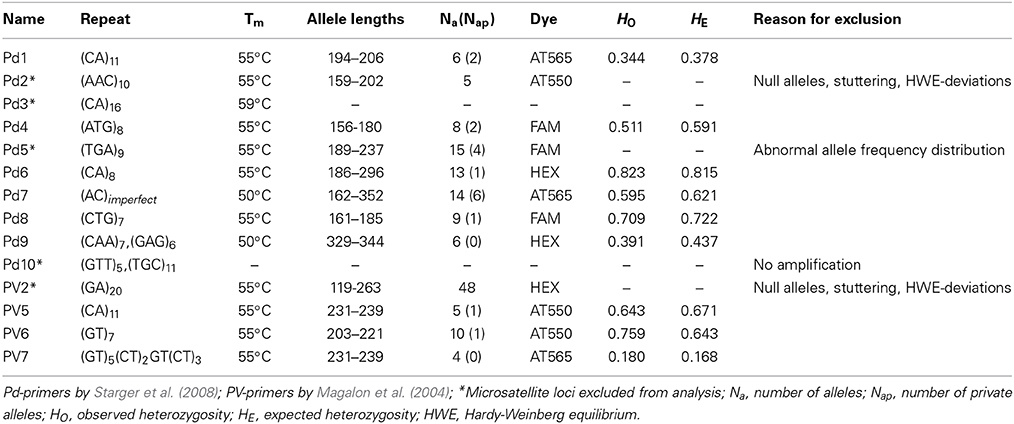
A total of 14 microsatellite loci developed by Magalon et al. (2004) and Starger et al. (2008) were tested (Table 2). For each primer pair, the forward primers were labeled with red (AT565), yellow (AT550), green (HEX), or blue (FAM) fluorescent dyes. PCR fragment lengths, quantity, and quality were checked and estimated with the QIAxcel DNA High Resolution Kit. Thirteen primer pairs gave amplifications and were successfully multiplexed (MPLX) with touchdown PCR conditions detailed in the following. MPLX No. 1, 2, 3: 95°C for 15 min, 10 cycles at 94°C for 60 s, 57°C for 90 s (−0.2°C/cycle), 72°C for 60 s, followed by 13 cycles of 94°C for 60 s, 55°C for 90 s, 72°C for 60 s, followed by 72°C for 30 min; MPLX No. 4: 95°C for 15 min, 10 cycles at 94°C for 60 s, 52°C for 90 s (−0.2°C/per cycle), 72°C for 60 s, followed by 13 cycles of 94°C for 60 s, 51°C for 90 s, 72°C for 60 s, followed by 72°C for 30 min. A final elongation time of 30 min was performed to assure 3′ adenylation of PCR products yielding equal lengths for identical alleles in the fragment analysis. Multiplexed PCR products were diluted 1:25 with MilliQ water (EMD Millipore Corporation) prior to fragment analysis. Fragment lengths were detected on a Sanger ABI 3730XL.
For analysis of microsatellites, ABI files were imported to GeneMapper 4.0 (Applied Biosystems, Inc.) for genotyping. Alleles were manually and independently scored at least four times. Alleles that yielded ambiguous results were re-amplified with more template DNA and/or lower annealing temperature, and alleles of different PCR setups were subsequently compared for accurate and consistent allele scoring. The Microsatellite Toolkit 3.1.1 (Park, 2001) was used to detect missing alleles (diploid) and erroneous data (e.g., alleles, subpopulation names, data format, characters). Further, the software was used to identify clonal colonies (matching multi-locus-samples with up to 4 mismatches) within the data set. MicroChecker 2.2.3 (Van Oosterhout et al., 2004) was used to detect the presence of null alleles, errors in allele sizes due to stuttering, and large allele dropouts by comparing observed and expected homozygote and heterozygote frequencies. We employed CONVERT 1.3.1 (Glaubitz, 2004) and PDG SPIDER 2.0 (Lischer and Excoffier, 2012) to convert the data into different file formats.
Population differentiation was calculated over all populations with GENEPOP 4.1 (Raymond and Rousset, 1995) using the Markov Chain (MC) algorithm by Guo and Thompson (1992) with 10,000 burn-in, 100 batches, and 5000 MC Monte Carlo (MCMC) steps. Bonferroni adjusted P-values were used to assign the significance of differentiation according to the FST-values, also calculated with GENEPOP 4.1. Other applicable measurements of differentiation were calculated with GenAlEx 6.5 (Peakall and Smouse, 2006, 2012), i.e., differentiation coefficient D, G'ST, and pairwise fixation indices FST. STRUCTURE 2.2.3 (Pritchard et al., 2000; Falush et al., 2003, 2007; Hubisz et al., 2009) was used to assess population structure based on Bayesian multi-locus clustering. The number of putative subpopulations (K) based on the STRUCTURE analysis was calculated with STRUCTURE HARVESTER's Evanno Method (Earl and Vonholdt, 2012). Prior runs of STRUCTURE were performed using 100,000 MCMC steps and 50,000 burn-in for K = 1 to K = 10 (maximum number of sampled reefs) in order to firstly infer the range of possible populations. For the final iterations, K was set from 1 to 4 with 1,000,000 MCMC and a burn-in period of 500,000, with 5 runs per parameter set and model without providing the sample's location as a prior. Results from all models were compiled before using STRUCTURE HARVESTER to give the least model-biased number of putative genetic clusters, i.e., subpopulations. A graphical representation of the final STRUCTURE output based on the output of K = 3 and 5 iterations with the latter parameter set was generated using output files from STRUCTURE HARVESTER and processed in CLUMPP 1.1.2 (Jakobsson and Rosenberg, 2007) and edited with DISTRUCT 1.1 (Rosenberg, 2004).
Estimates on the effective population size (Ne) were calculated using approximate Bayesian computation from the set of microsatellite genotypes with ONeSAMP 1.2 (Tallmon et al., 2008) using the following parameters: #num samples 252, #num polymorphic loci 9, #min Ne size 4, #max generation size 10,000, #iterations 50,000 (Supplemental File S1).
Results
Pocillopora Lineage Selection via mORF
Based on results from a study by Pinzón et al. (2013), we used the mORF marker to identify distinct mitochondrial lineages for P. verrucosa and to rule out analyzing data from several (cryptic) species. Sequences of ~850 bp were generated and single base pair mutations could be clearly identified due to exclusion of low quality sequences from the alignments and the absence of ambiguous peaks. We identified 6 distinct haplotypes (A to F) in our data, which were about equally represented among all sampled reefs (Table 1). Haplotypes B, C, E, and F were previously identified by Pinzón et al. (2013) studying P. verrucosa form the Red Sea (GenBank accession numbers: haplotype B JX994083; haplotype C JX994080, haplotype E JX994075; haplotype F: JX994084). Haplotypes A and D represented novel haplotypes in our study (GenBank accession numbers: haplotype A KP238127; haplotype D KP238128). Analysis of these haplotypic data in MEGA suggested clustering of haplotypes A-E corresponding to “type 3” sensu Pinzón et al. (2013), and a second cluster constituted by haplotype F corresponding to “type 7” sensu (Pinzón et al., 2013). We only considered samples for “type 3” for the population genetics analysis in order to confidently exclude possible biases arising from the presence of genotypic data from different lineages. From a total of 361 coral colonies, 252 samples belonged to “type 3.” The set of 252 samples is henceforth referred to as P. verrucosa and was used for the remainder of the analyses. It is worth mentioning that we could not detect morphological differences between the putative different mitochondrial lineages (types 3 and 7) based on photographical vouchers of the sampled colonies (data not shown) emphasizing the usefulness of genetic data in differentiating mitochondrial lineages. This is also in accordance with Pinzón et al. (2013) who reported on morphologically scattered “verrucosa-” and “damicornis-like” phenotypes among discrete genetic lineages. However, using more sophisticated methods [e.g., scanning electron microscopy (SEM)], Schmidt-Roach et al. (2014) demonstrated that morphological groupings are congruent with mitochondrial molecular phylogenies within species of the Pocillopora damicornis species complex.
Population Structure and Effective Population Size of Pocillopora verrucosa in the Red Sea
We assessed suitability of 14 microsatellites on P. verrucosa colonies collected from northern, central, and southern reefs in the Red Sea (Table 2). From the 14 microsatellite loci tested, a total of 9 microsatellites successfully amplified and were selected based on overall data quality and the absence of null alleles. None of these 9 microsatellites showed deviation from the Hardy-Weinberg (HW) Equilibrium (Table 2). The number of alleles per microsatellite locus ranged from 4 to 10 (Table 2).
We did not detect significant linkage disequilibrium (LD) among microsatellite loci pairs (MC parameters: 10,000 de-memorization, 1000 batches, 10,000 iteration steps per batch) except between Pd1 and Pd8 (LRT = 0.0021 ± 0.0003 SE, P = 0.0019 ± 0.0003 SE). These loci were not excluded because there was no LD between Pd1 and Pd8 when inferred within our three sampling regions (i.e., WAJ, THU, DOG) and since none of the previous studies on Pocillopora that used these two markers found significant LD between them (Starger et al., 2008, 2010; Souter, 2010; Combosch and Vollmer, 2011). Tests on heterozygote deficiency (MC parameters as former) were significant for locus Pd7 (P = 0.0000 ± 0.0000 SE; FIS = 0.0463 (Weir and Cockerham, 1984). However, when looking within the three sampling regions, significant disequilibrium from HW was only found for the WAJ samples (P = 0.000 ± 0.0019 SE; FIS = 0.0116) and not overall samples when using Fisher's method (P = 0.0551; χ2 = 12.3251). Hence, we kept Pd7, as it was a highly variable marker in our data set providing a potentially higher resolution on more recent population structure (ecological timescale). Further, other studies used this marker without finding heterozygote deficiency in P. verrucosa populations (Yeoh and Dai, 2009; Pinzón et al., 2012; Schmidt-Roach et al., 2012).
Inference of fixation indices (i.e., FST) based on ANOVA by GENEPOP for our three sampling regions shows that FIS (individual to subpopulation = 0.0193; P = 0.087) values were lower than FIT values (individual to total population = 0.0198, P = 0.079), but FST values (subpopulations to total population = 0.0005, P = 0.340) values were the lowest among all three. Consequently, allele sizes of subpopulations had no explanatory value for the variance of the total data and there was no correlation between the genetic variance and the geographic location of the sample. Taken together, genetic differentiation between reefs/sites (n = 10) and regions (n = 3) was absent indicating that the variance between individuals from a sampling region was not significantly different from the variance between individuals of the total population.
FST values have a high comparative value between studies as they are commonly used as a proxy of genetic differentiation within a population and among its constituting subpopulations. However, the interpretation of FST from highly variable loci is debatable as a high number of alleles can lead to an underestimation of differentiation when using FST (e.g., Hedrick, 1999; Balloux et al., 2000; Hedrick, 2005; Jost, 2008; Meirmans and Hedrick, 2011). For this reason, we chose to derive additional parameters for estimating the magnitude of genetic divergence among populations of P. verrucosa. We calculated pairwise DEST (Jost, 2008) and G'ST (Hedrick, 1999) between sampling regions, which represent more recently developed parameters of genetic differentiation, and subsequently compared DEST and G'ST to pairwise FST values (Table 3). DEST and G'ST estimators could not capture or resolve differentiation between our sampling regions either. Rather, pairwise FST values as well as DEST and G'STestimators all indicate absence of genetic differentiation and extensive gene flow between reefs and regions (Table 3). To further confirm this, we applied STRUCTURE analysis to find the optimal number of populations. Independent from the implemented type of model, K = 1 (i.e., one single population) was the most likely number of genetic clusters implemented during preliminary (with K = 1–10) and final (K = 1–4) STRUCTURE runs. This was further confirmed in a subsequent analysis with STRUCTURE HARVESTER (Figure 1).
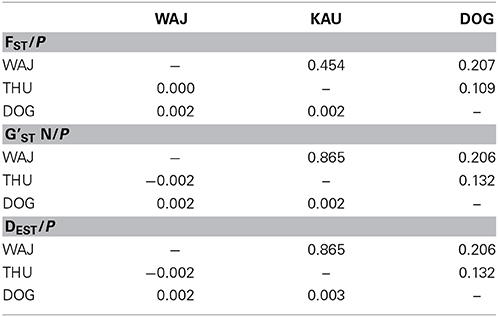
Table 3. Coefficients for genetic differentiation assessment between populations pairs (below the diagonal) and associated P-values (above the diagonal).
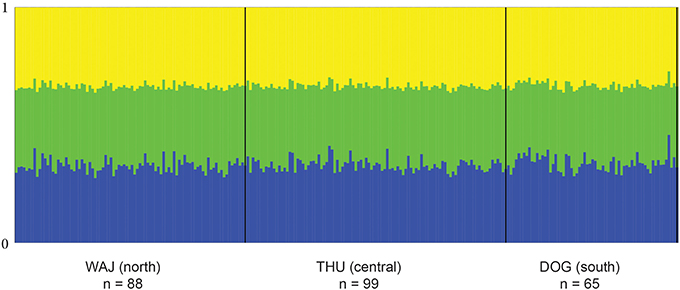
Figure 1. Averaged bar plot graph of STRUCTURE results for K = 3. Each vertical line of the x-axis represents a single individual. The two bold vertical lines divide the sampling sites WAJ (north), THU (central), and DOG (south) with numbers of sampled colonies indicated (n). Each color represents one of the 3 inferred clusters (K). Individual lines are broken into color segments, with lengths proportional to the probability of each inferred cluster assignment.
We calculated effective population size (Ne) from the distribution of microsatellite genotypes to assess genetic diversity, and consequently quantitative trait heritability (“evolutionary potential”). Decreased evolvability is often considered the most relevant evolutionary consequence of small population sizes (Wang, 2005; Palstra and Ruzzante, 2008). The average Ne estimate from our set of microsatellite data from P. verrucosa colonies was calculated to be 116.1598 (median: 115.9605; lower 95% CL 66.6535; upper 95% CL 215.9047).
Discussion
Population Structure of P. verrucosa in the Red Sea
In this study, the population structure of P. verrucosa in the Saudi Arabian Red Sea was investigated over a distance of 850 km along a unique latitudinal environmental gradient. Our sampled sites covered a wide range of the Saudi Arabian Red Sea, but even between the most distant reefs investigated, i.e., 850 km between WAJ and DOG, no genetic differentiation was found at neutral loci. Rather, P. verrucosa in the Red Sea seems to belong to a single, panmictic population (FST = 0.0005, P = 0.340) with an estimated effective population size of (Ne) 116.1598. Palstra and Ruzzante (2008) empirically estimated a median unbiased Ne of 260 (among 83 studies) for wild populations including amphibians, birds, crustaceans, fish, insects, mammals, molluscs, reptiles, and plants. In this regard, the estimated Ne from our data seems within the order of magnitude of other wild populations. Further, Richards et al. (2008) estimated global Ne's for a number of Acropora species that were drastically different depending on the species investigated (e.g., A. spathulata had a mean effective global population size of 3611 ± 1805 and A. rongelapensis of 25 ± 13) and argued that this might be a result of unidirectional hybrids. For the majority of common coral species though, effective Ne's are unknown, but it is likely that local Ne sizes are substantially smaller than the global estimates from Richards et al. (2008).
Despite the reasonably large area investigated here, future studies should incorporate sampling from the northernmost region of the Gulf of Aqaba as well as from the southernmost region of the Red Sea, and possibly from outside the Red Sea (Gulf of Aden). This is to test whether the absence of population differentiation further holds true for regions featuring more extreme environmental conditions for coral growth. Results from Nanninga et al. (2014) have found genetically distinct populations of sponges and clownfish in the Red Sea below 17°N (i.e., ~250 km south of DOG, our southernmost sampling site) where physical changes in the reef ecosystems and reef-scape were also visually apparent. The observed pattern in these studies might likely arise not only from prevailing higher temperatures, but also from highly nutrient enriched waters and incoming genetic diversity introduced from the Gulf of Aden.
Absence of genetic differentiation has also been observed for P. verrucosa around East Africa over a distance of 697 km (Souter, 2010). As in the Red Sea, the P. verrucosa population reproduces via broadcast spawning suggesting that the reproductive mode might play an important role in geographic dispersal. Earlier studies on the connectivity of other Pocillopora species also came to similar conclusions regarding the link between reproductive mode and geographic dispersal (Benzie et al., 1995; Ayre et al., 1997; Ridgway et al., 2001; Miller and Ayre, 2008a,b). However, the markers employed (e.g., allozymes) had lower resolution in comparison to the faster evolving microsatellite markers used in this study. Ayre et al. (1997) described gene flow even between sites that were up to 1200 km apart based on data from allozymes. The absence of population differentiation over northern, central, and southern reefs in the Red Sea spanning an area of more than 850 km indicates that larvae of P. verrucosa are able to disperse over long distances from their reef of origin and successfully settle in new reefs. Three main features are responsible for successful larval dispersal and recruitment (and consequently gene flow after reaching reproductive maturity) over large geographic regions: (1) the reproductive mode of the species should allow long distance dispersal, (2) water currents should support the dispersive pathways, and (3) recruits have to be (genetically) equipped to survive and succeed in a wide range of physicochemical conditions. These will be discussed in the following.
Clonal Structure and Reproductive Mode
In order to avoid sampling of clonal colonies, we collected corals that were at least 5 m apart from each other following a design employed in other studies (Baums et al., 2006; Combosch and Vollmer, 2011). However, identical multi-locus genets were identified within samples of many studies [e.g., Souter et al., 2009 (P. verrucosa; 5 m distant colonies); Combosch and Vollmer, 2011 (P. damicornis; samples from 10 m distant colonies)]. In most cases this was interpreted as partial presence of asexual reproduction among colonies from the species under study. Interestingly, in our data we did not detect a single identical multi-locus genotype. This indicates that either P. verrucosa in the Red Sea reproduces exclusively sexually, or we did not sample clonal colonies due to their generally low presence, or simply by chance. Given that this study was based on nine highly resolving microsatellite loci (previous studies were mainly based on five or six loci), we advocate the first explanation. In contrast to our results, Maier et al. (2005) found significant genetic structure and isolation by distance in a coral of the Pocilloporidae family, Seriatopora hystrix, in the Red Sea, although the study was based on a comparatively smaller geographic range (northern Red Sea only, ~600 km). S. hysterix, in comparison to P. verrucosa, is a viviparous brooding species (Ayre and Resing, 1986) that also showed significant population structure in the Great Barrier Reef (Ayre and Dufty, 1994). A potential high level of genetic heterozygosity in P. verrucosa from the Red Sea can have important implications for the species ability to adapt to changing environments and warrants further studies. Additionally, even though previous studies report on asexual (clones) and sexual coral recruits among Pocillopora species, this might be the result of analyzing samples from distinct mitochondrial lineages which can lead to the inclusion of cryptic species (e.g., one species reproducing exclusively asexually and the other species sexually and thus, analyzing colonies from both reproductive forms). Taken together, the broadcast spawning P. verrucosa from the Red Sea seems to reproduce exclusively sexually, which allows for long distance dispersal of gametes and a high level of heterozygosity.
Currents and Larval Transport
While the mode of reproduction is an important factor affecting connectivity among coral reefs, currents also play a crucial role in genetic dispersal (i.e., gene flow) in marine environments. Currents can produce physical barriers (Baums et al., 2005; Thornhill et al., 2008) as well as powerful transport routes for aquatic larvae (e.g., Pineda et al., 2007). There is only little information on current flow patterns in the Red Sea, but the main driver of surface currents in the Red Sea is presumably a dominant North to South wind (Sofianos et al., 2002; Sofianos and Johns, 2007; Raitsos et al., 2013). Furthermore, permanent clockwise (i.e., anticyclonic) and counterclockwise (i.e., cyclonic) eddies can be found in the Red Sea (Sofianos and Johns, 2007; Raitsos et al., 2013) (Figure 2). Studies have shown that eddies in some cases can work like baskets transporting larvae and gametes through water bodies, but at the same time keeping their dispersal restricted to the range of the eddy (Lobel and Robinson, 1986; Sammarco and Andrews, 1989; Wolanski et al., 1989). In addition, given that the Red Sea is comparatively narrow (notably ~200 km width near THU), cyclonic water currents might become significant vehicles for the transport of larvae from one shore to the other (from East to West and vice versa), but further studies are needed to confirm this. Taken together, the wind patterns and permanent eddies in the Red Sea are potentially promoting larval dispersal along a latitudinal and longitudinal gradient of the Red Sea, but further studies are needed to confirm this. The reproductive and larval features of a coral (here: Pocillopora verrucosa) combined with powerful oceanic water transport would increase gene flow over long distances, and thereby assist panmixia within most of the Red Sea.
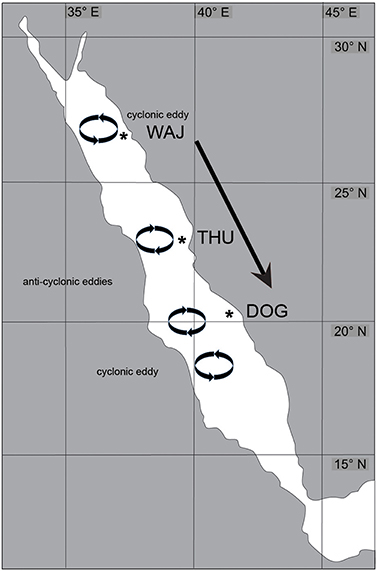
Figure 2. Overview of Pocillopora verrucosa sampling sites and wind and current patterns in the Red Sea. Sampling sites WAJ (north), THU (central), and DOG (south) are indicated by an asterisk. The predominant wind direction among the study sites is from North to South (arrow). A permanent cyclonic eddy influences current direction in the northernmost part (circle) and is followed by two anti-cyclonic eddies (circles). The southernmost circle visualizes the presence of another cyclonic eddy in this area. The flow directions created by these eddies potentially influence coral larval transport across the Red Sea.
Strategies to Success Across the Environmental Gradient
Apart from a suitable mode of reproduction and effective means of larval dispersal, the genetic makeup of dispersed larval populations need to provide genotypes that are able to survive and succeed in prevailing environmental conditions. Our study yielded no data that indicate significant genetic differentiation between coral colonies from P. verrucosa across a latitudinal gradient in the Red Sea despite pronounced differences in prevailing environmental conditions (i.e., temperature, salinity, and nutrients). It is important to note that lack of genetic divergence at neutral loci does not constitute evidence of lack of genetic divergence elsewhere in the genome. Rather, selection can result in genetic divergence at specific, adaptation-related loci without any detectable divergence at neural loci between populations as reviewed in Nosil et al. (2009 and references therein). For this reason, further studies are needed to determine the role of phenotypic plasticity on the one hand or loci-specific adaptation on the other hand.
Pocilloporidae are reproductively most successful in the Red Sea in comparison to other scleractinian families (Glassom et al., 2004), although they are commonly regarded as a coral family that is vulnerable to environmental change (Glynn, 1993, 1996; Marshall and Baird, 2000). A deeper understanding into the success of corals in the Red Sea might come from hologenome-targeted analyses that uncover how member species (i.e., coral animal, dinoflagellate symbionts, bacterial community) of the coral metaorganism interact with each other to select for the most advantageous coral holobiont composition for prevailing environmental conditions (Rosenberg et al., 2007). In particular, the microbial community structure of the coral might be subject to change given the short generation time and greater versatility of microorganisms (Bayer et al., 2013). This is further supported by coral disease studies that usually show more diverse and less structured (i.e., less selected) microbial communities in compromised corals (Sunagawa et al., 2009; Kellogg et al., 2012; Roder et al., 2014a,b). With regard to the dinoflagellate symbionts associated with P. verrucosa, Sawall et al. (2014a) found that symbiont clade compositions differed in specimens from the most northern and the most southern ends of the Red Sea. Such differentiation has also been observed in other coral reef ecosystems (Baums et al., 2010; Lien et al., 2013). Differences in algal symbiont assemblage might feature different physiological capacities and thereby contribute to the success of P. verrucosa at areas of the Red Sea with more extreme conditions (Sawall et al., 2014a). Consequently, complementary analyses of the corals' algal symbiont and bacterial communities along a latitudinal gradient in the Red Sea is highly warranted and promises to gain further insights into the adaptability and resilience of corals to extreme environments and environmental change.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the KAUST BioScience Core Lab for sequence data generation. We would also like to thank Shobhit Agrawal for assistance with data analysis generation. Research reported in this publication was supported by the King Abdullah University of Science and Technology (KAUST).
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmars.2015.00005/abstract
References
Ayre, D. J., and Dufty, S. (1994). Evidence for restricted gene flow in the viviparous coral Seriatopora hystrix on Australia's Great Barrier Reef. Evolution 48, 1183–1201. doi: 10.2307/2410377
Ayre, D. J., Hughes, T. P., and Standish, R. J. (1997). Genetic differentiation, reproductive mode, and gene flow in the brooding coral Pocillopora damicornis along the Great Barrier Reef, Australia. Mar. Ecol. Prog. Ser. 159, 175–187. doi: 10.3354/meps159175
Ayre, D. J., and Resing, J. M. (1986). Sexual and asexual production of planulae in reef corals. Mar. Biol. 90, 187–190. doi: 10.1007/BF00569126
Balloux, F., Brünner, H., Lugon-Moulin, N., Hausser, J., and Goudet, J. (2000). Microsatellites can be misleading: an empirical and simulation study. Evolution 54, 1414–1422. doi: 10.1111/j.0014-3820.2000.tb00573.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baums, I. B., Johnson, M. E., Devlin-Durante, M. K., and Miller, M. W. (2010). Host population genetic structure and zooxanthellae diversity of two reef-building coral species along the Florida Reef Tract and wider Caribbean. Coral Reefs 29, 835–842. doi: 10.1007/s00338-010-0645-y
Baums, I. B., Miller, M. W., and Hellberg, M. E. (2005). Regionally isolated populations of an imperiled Caribbean coral, Acropora palmata. Mol. Ecol. 14, 1377–1390. doi: 10.1111/j.1365-294X.2005.02489.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baums, I. B., Paris, C. B., and Cherubin, L. M. (2006). A bio-oceanographic filter to larval dispersal in a reef-building coral. Limnol. Oceanogr. 51, 1969–1981. doi: 10.4319/lo.2006.51.5.1969
Bayer, T., Neave, M. J., Alsheikh-Hussain, A., Aranda, M., Yum, L. K., Mincer, T., et al. (2013). The Microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl. Environ. Microbiol. 79, 4759–4762. doi: 10.1128/AEM.00695-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Benzie, J. A. H., Haskell, A., and Lehman, H. (1995). Variation in the genetic composition of coral (Pocillopora damicornis and Acropora palifera) populations from different reef habitats. Mar. Biol. 121, 731–739. doi: 10.1007/BF00349309
Bouwmeester, J., Berumen, M. L., and Baird, A. H. (2011). Daytime broadcast spawning of Pocillopora verrucosa on coral reefs of the central Red Sea. Galaxea J. Coral Reef Stud. 13, 23–24. doi: 10.3755/galaxea.13.23
Brainard, R. E., Weijerman, M., Eakin, C. M., McElhany, P., Miller, M. W., Patterson, M., et al. (2013). Incorporating climate and ocean change into extinction risk assessments for 82 coral species. Conserv. Biol. 27, 1169–1178. doi: 10.1111/cobi.12171
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Combosch, D. J., and Vollmer, S. V. (2011). Population genetics of an ecosystem-defining reef coral Pocillopora damicornis in the Tropical Eastern Pacific. PLoS ONE 6:e21200. doi: 10.1371/journal.pone.0021200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Earl, D. A., and Vonholdt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Edwards, A. J. (1987). Key Environments: Red Sea. International Union for Conservation of Nature and Natural Resources. IV Series. Oxford, New York, NY: Pergamon Press.
Falush, D., Stephens, M., and Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587. doi: 10.1111/j.1471-8286.2007.01758.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Falush, D., Stephens, M., and Pritchard, J. K. (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7, 574–578. doi: 10.1111/j.1471-8286.2007.01758.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Flot, J.-F., and Tillier, S. (2007). The mitochondrial genome of Pocillopora (Cnidaria: Scleractinia) contains two variable regions: the putative D-loop and a novel ORF of unknown function. Gene 401, 80–87. doi: 10.1016/j.gene.2007.07.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Foster, R. A., Subramaniam, A., Mahaffey, C., Carpenter, E. J., Capone, D. G., and Zehr, J. P. (2007). Influence of the Amazon River plume on distributions of free-living and symbiotic cyanobacteria in the western tropical north Atlantic Ocean. Limnol. Oceanogr. 52, 517–532. doi: 10.4319/lo.2007.52.2.0517
Glassom, D., Zakai, D., and Chadwick-Furman, N. E. (2004). Coral recruitment: a spatio-temporal analysis along the coastline of Eilat, northern Red Sea. Mar. Biol. 144, 641–651. doi: 10.1007/s00227-003-1243-0
Glaubitz, J. C. (2004). convert: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol. Ecol. Notes 4, 309–310. doi: 10.1111/j.1471-8286.2004.00597.x
Glynn, P. W. (1993). Coral reef bleaching: ecological perspectives. Coral Reefs 12, 1–17. doi: 10.1007/BF00303779
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Glynn, P. W. (1996). Coral reef bleaching: facts, hypotheses and implications. Glob. Change Biol. 2, 495–509. doi: 10.1111/j.1365-2486.1996.tb00063.x
Guo, S. W., and Thompson, E. A. (1992). Proportion performing the exact test of hardy-weinberg proportion for multiple alleles. Biometrics 48, 361–372. doi: 10.2307/2532296
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hedrick, P. W. (1999). Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution 53, 313–318. doi: 10.2307/2640768
Hedrick, P. W. (2005). A standardized genetic differentiation measure. Evolution 59, 1633–1638. doi: 10.1111/j.0014-3820.2005.tb01814.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hewson, I., Capone, D. G., Steele, J. A., and Fuhrman, J. A. (2006). Influence of Amazon and Orinoco offshore surface water plumes on oligotrophic bacterioplankton diversity in the west tropical Atlantic. Aquat. Microb. Ecol. 43, 11–22. doi: 10.3354/ame043011
Hoegh-Guldberg, O. (1999). Climate change, coral bleaching and the future of the worldís coral reefs. Mar. Freshw. Res. 50, 839–866. doi: 10.1071/MF99078
Hubisz, M. J., Falush, D., Stephens, M., and Pritchard, J. K. (2009). Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 9, 1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jakobsson, M., and Rosenberg, N. A. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. doi: 10.1093/bioinformatics/btm233
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jost, L. (2008). G ST and its relatives do not measure differentiation. Mol. Ecol. 17, 4015–4026. doi: 10.1111/j.1365-294X.2008.03887.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kellogg, C. A., Piceno, Y. M., Tom, L. M., Desantis, T. Z., Zawada, D. G., and Andersen, G. L. (2012). PhyloChip microarray comparison of sampling methods used for coral microbial ecology. J. Microbiol. Methods 88, 103–109. doi: 10.1016/j.mimet.2011.10.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kürten, B., Al-Aidaroos, A. M., Struck, U., Khomayis, H. S., Gharbawi, W. Y., and Sommer, U. (2014). Influence of environmental gradients on C and N stable isotope ratios in coral reef biota of the Red Sea, Saudi Arabia. J. Sea Res. 85, 379–394. doi: 10.1016/j.seares.2013.07.008
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lien, Y., Keshavmurthy, S., Nakano, Y., Plathong, S., Huang, H., Hsu, C., et al. (2013). Host genetics and Symbiodinium D diversity in a stress-tolerant scleractinian coral, Oulastrea crispata, in the West Pacific. Mar. Ecol. Prog. Ser. 473, 163–177. doi: 10.3354/meps10041
Lischer, H. E. L., and Excoffier, L. (2012). PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28, 298–299. doi: 10.1093/bioinformatics/btr642
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lobel, P. S. S., and Robinson, A. R. (1986). Transport and entrapment of fish larvae by ocean mesoscale eddies and currents in Hawaiian waters. Deep Sea Res. A. Oceanogr. Res. Pap. 33, 483–500. doi: 10.1016/0198-0149(86)90127-5
Magalon, H., Samadi, S., Richard, M., Adjeroud, M., and Veuille, M. (2004). Development of coral and zooxanthella-specific microsatellites in three species of Pocillopora (Cnidaria, Scleractinia) from French Polynesia. Mol. Ecol. Notes 4, 206–208. doi: 10.1111/j.1471-8286.2004.00618.x
Maier, E., Tollrian, R., Rinkevich, B., and Nürnberger, B. (2005). Isolation by distance in the scleractinian coral Seriatopora hystrix from the Red Sea. Mar. Biol. 147, 1109–1120. doi: 10.1007/s00227-005-0013-6
Marshall, P. A., and Baird, A. H. (2000). Bleaching of corals on the Great Barrier Reef: differential susceptibilities among taxa. Coral Reefs 19, 155–163. doi: 10.1007/s003380000086
McClanahan, T. R. (2004). The relationship between bleaching and mortality of common corals. Mar. Biol. 144, 1239–1245. doi: 10.1007/s00227-003-1271-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Meirmans, P. G., and Hedrick, P. W. (2011). Assessing population structure: F(ST) and related measures. Mol. Ecol. Resour. 11, 5–18. doi: 10.1111/j.1755-0998.2010.02927.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miller, K. J., and Ayre, D. J. (2008a). Population structure is not a simple function of reproductive mode and larval type: insights from tropical corals. J. Anim. Ecol. 77, 713–724. doi: 10.1111/j.1365-2656.2008.01387.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miller, K. J., and Ayre, D. J. (2008b). Protection of genetic diversity and maintenance of connectivity among reef corals within marine protected areas. Conserv. Biol. 22, 1245–1254. doi: 10.1111/j.1523-1739.2008.00985.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morcos, S. A. (1970). Physical and chemical oceanography of the Red Sea. Oceanogr. Mar. Biol. 8, 73–202.
Murray, S. P., and Johns, W. (1997). Direct observations of seasonal exchange through the Bab el Mandab Strait. Geophys. Res. Lett. 24, 2557–2560. doi: 10.1029/97GL02741
Nanninga, G. B., Saenz-Agudelo, P., Manica, A., and Berumen, M. L. (2014). Environmental gradients predict the genetic population structure of a coral reef fish in the Red Sea. Mol. Ecol. 23, 591–602. doi: 10.1111/mec.12623
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nosil, P., Funk, D. J., and Ortiz-Barrientos, D. (2009). Divergent selection and heterogeneous genomic divergence. Mol. Ecol. 18, 375–402. doi: 10.1111/j.1365-294X.2008.03946.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palstra, F. P., and Ruzzante, D. E. (2008). Genetic estimates of contemporary effective population size: what can they tell us about the importance of genetic stochasticity for wild population persistence? Mol. Ecol. 17, 3428–3447. doi: 10.1111/j.1365-294X.2008.03842.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Park, S. D. E. (2001). The Excel Microsatellite Toolkit. Available online at: ucd.ie/sdepark/ms-toolkit/ [Accessed August 2007]
Peakall, R., and Smouse, P. E. (2006). genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecolo. Notes 6, 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pineda, J., Hare, J., and Sponaugle, S. (2007). Larval transport and dispersal in the coastal ocean and consequences for population connectivity. Oceanography 20, 22–39. doi: 10.5670/oceanog.2007.27
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pinzón, J. H., Reyes-Bonilla, H., Baums, I. B., and Lajeunesse, T. C. (2012). Contrasting clonal structure among Pocillopora (Scleractinia) communities at two environmentally distinct sites in the Gulf of California. Coral Reefs 31, 765–777. doi: 10.1007/s00338-012-0887-y
Pinzón, J. H., Sampayo, E., Cox, E., Chauka, L. J., Chen, C. A., Voolstra, C. R., et al. (2013). Blind to morphology: genetics identifies several widespread ecologically common species and few endemics among Indo-Pacific cauliflower corals (Pocillopora, Scleractinia). J. Biogeogr. 40, 1595–1608. doi: 10.1111/jbi.12110
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Raitsos, D. E., Pradhan, Y., Brewin, R. J. W., Stenchikov, G., and Hoteit, I. (2013). Remote sensing the phytoplankton seasonal succession of the Red Sea. PLoS ONE 8:e64909. doi: 10.1371/journal.pone.0064909
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Raymond, M., and Rousset, F. (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249.
Richards, Z. T., Van Oppen, M. J. H., Wallace, C. C., Willis, B. L., and Miller, D. J. (2008). Some rare Indo-Pacific coral species are probable hybrids. PLoS ONE 3:e3240. doi: 10.1371/journal.pone.0003240
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ridgway, T., Hoegh-Guldberg, O., and Ayre, D. J. (2001). Panmixia in Pocillopora verrucosa from South Africa. Mar. Biol. 139, 175–181. doi: 10.1007/S002270100573
Roder, C., Arif, C., Bayer, T., Aranda, M., Daniels, C., Shibl, A., et al. (2014a). Bacterial profiling of White Plague Disease in a comparative coral species framework. ISME J. 8, 31–39. doi: 10.1038/ismej.2013.127
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roder, C., Arif, C., Daniels, C., Weil, E., and Voolstra, C. R. (2014b). Bacterial profiling of White Plague Disease across corals and oceans indicates a conserved and distinct disease microbiome. Mol. Ecol. 23, 965–974. doi: 10.1111/mec.12638
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rosenberg, N. A. (2004). DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Notes 4, 137–138. doi: 10.1046/j.1471-8286.2003.00566.x
Sammarco, P. W., and Andrews, J. C. (1989). The Helix experiment: differential localized dispersal and recruitment patterns in Great Barrier Reef corals. Limnol. Oceanogr. 34, 896–912. doi: 10.4319/lo.1989.34.5.0896
Sawall, Y., Al-Sofyani, A., Banguera-Hinestroza, E., Voolstra, R. C., and Voolstra, C. R. (2014a). Spatio-temporal analyses of Symbiodinium physiology of the coral Pocillopora verrucosa along large-scale nutrient and temperature gradients in the Red Sea. PLoS ONE 9:e103179. doi: 10.1371/journal.pone.0103179
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sawall, Y., Al-Sofyani, A., Kuerten, B., Al-Aidaroos, A. M., Hoang, B. X., Marimuthu, N., et al. (2014b). Coral communities, in contrast to fish communities, maintain a high assembly similarity along the large latitudinal gradient along the Saudi Red Sea Coast. J. Ecosyst. Ecogr. S4:003. doi: 10.4172/2157-7625.S4-003
Schmidt-Roach, S., Lundgren, P., Miller, K. J., Gerlach, G., Noreen, A. M. E., and Andreakis, N. (2012). Assessing hidden species diversity in the coral Pocillopora damicornis from Eastern Australia. Coral Reefs 32, 161–172. doi: 10.1007/s00338-012-0959-z
Schmidt-Roach, S., Miller, K. J., Lundgren, P., and Andreakis, N. (2014). With eyes wide open: a revision of species within and closely related to the Pocillopora damicornis species complex (Scleractinia; Pocilloporidae) using morphology and genetics. Zool. J. Linn. Soc. 170, 1–33. doi: 10.1111/zoj.12092
Schwalbach, M. S., Brown, M., and Fuhrman, J. A. (2005). Impact of light on marine bacterioplankton community structure. Aquat. Microb. Ecol. 39, 235–245. doi: 10.3354/ame039235
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Siddall, M., Rohling, E. J., Almogi-Labin, A., Hemleben, C., Meischner, D., Schmeltzer, I., et al. (2003). Sea-level fluctuations during the last glacial cycle. Nature 423, 853–858. doi: 10.1038/nature01690
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sirocko, F. (2003). Ups and downs in the Red Sea. Nature 423, 813–814. doi: 10.1038/423813a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sofianos, S. S., and Johns, W. E. (2007). Observations of the summer Red Sea circulation. J. Geophys. Res. 112, 1978–2012. doi: 10.1029/2006JC003886
Sofianos, S. S., Johns, W. E., and Murray, S. P. (2002). Heat and freshwater budgets in the Red Sea from direct observations at Bab el Mandeb. Deep Sea Res. 49, 1323–1340. doi: 10.1016/S0967-0645(01)00164-3
Souter, P. (2010). Hidden genetic diversity in a key model species of coral. Mar. Biol. 157, 875–885. doi: 10.1007/s00227-009-1370-3
Souter, P., Henriksson, O., Olsson, N., and Grahn, M. (2009). Patterns of genetic structuring in the coral Pocillopora damicornis on reefs in East Africa. BMC Ecol. 9:19. doi: 10.1186/1472-6785-9-19
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Starger, C. J., Barber, P. H., and Baker, A. C. (2010). The recovery of coral genetic diversity in the Sunda Strait following the 1883 eruption of Krakatau. Coral Reefs 29, 547–565. doi: 10.1007/s00338-010-0609-2
Starger, C. J., Yeoh, S. S. R., Dai, C.-F., Baker, A. C., and Desalle, R. (2008). Ten polymorphic STR loci in the cosmopolitan reef coral, Pocillopora damicornis. Mol. Ecol. Resour. 8, 619–621. doi: 10.1111/j.1471-8286.2007.02017.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sunagawa, S., Desantis, T. Z., Piceno, Y. M., Brodie, E. L., Desalvo, M. K., Voolstra, C. R., et al. (2009). Bacterial diversity and White Plague Disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 3, 512–521. doi: 10.1038/ismej.2008.131
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tallmon, D. A., Koyuk, A., Luikart, G., and Beaumont, M. A. (2008). COMPUTER PROGRAMS: onesamp: a program to estimate effective population size using approximate Bayesian computation. Mol. Ecol. Resour. 8, 299–301. doi: 10.1111/j.1471-8286.2007.01997.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thornhill, D. J., Mahon, A. R., Norenburg, J. L., and Halanych, K. M. (2008). Open-ocean barriers to dispersal: a test case with the Antarctic Polar Front and the ribbon worm Parborlasia corrugatus (Nemertea: Lineidae). Mol. Ecol. 17, 5104–5117. doi: 10.1111/j.1365-294X.2008.03970.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Veron, J. E. N., and Pichon, M. (1976). Scleractinia of Eastern Australia. Part I: Families Thamnasteriidae, Astrocoeniidae, Pocilloporidae. Canberra: Australian Institute of Marine Science (AIMS); Australian Government Publishing Service.
Wang, J. (2005). Estimation of effective population sizes from data on genetic markers. Philos. Trans. R. Sos. Lond. B Biol. Sci. 360, 1395–1409. doi: 10.1098/rstb.2005.1682
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weir, B. S., and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. doi: 10.2307/2408641
Wolanski, E., Burrage, D., and King, B. (1989). Trapping and dispersion of coral eggs around Bowden Reef, Great Barrier Reef, following mass coral spawning. Cont. Shelf Res. 9, 479–496. doi: 10.1016/0278-4343(89)90011-3
Yeoh, S.-R., and Dai, C.-F. (2009). The production of sexual and asexual larvae within single broods of the scleractinian coral, Pocillopora damicornis. Mar. Biol. 157, 351–359. doi: 10.1007/s00227-009-1322-y
Ziegler, M., Roder, C., Büchel, C., and Voolstra, C. (2014). Limits to physiological plasticity of the coral Pocillopora verrucosa from the central Red Sea. Coral Reefs 33, 1115–1129. doi: 10.1007/s00338-014-1192-8
Keywords: coral reef, Red Sea, Pocillopora verrucosa, population genetics, connectivity, biogeography
Citation: Robitzch V, Banguera-Hinestroza E, Sawall Y, Al-Sofyani A and Voolstra CR (2015) Absence of genetic differentiation in the coral Pocillopora verrucosa along environmental gradients of the Saudi Arabian Red Sea. Front. Mar. Sci. 2:5. doi: 10.3389/fmars.2015.00005
Received: 11 September 2014; Accepted: 19 January 2015;
Published online: 11 February 2015.
Edited by:
Andrew Stanley Mount, Clemson University, USAReviewed by:
Mikhail V. Matz, The University of Texas at Austin, USAChaolun Allen Chen, Academia Sinica, Taiwan
Copyright © 2015 Robitzch, Banguera-Hinestroza, Sawall, Al-Sofyani and Voolstra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian R. Voolstra, Red Sea Research Center, King Abdullah University of Science and Technology, Building 2, Room 2226, 23955-6900 Thuwal, Saudi Arabia e-mail: christian.voolstra@kaust.edu.sa
 Vanessa Robitzch
Vanessa Robitzch Eulalia Banguera-Hinestroza
Eulalia Banguera-Hinestroza Yvonne Sawall
Yvonne Sawall Abdulmohsin Al-Sofyani
Abdulmohsin Al-Sofyani Christian R. Voolstra
Christian R. Voolstra