Resistance to OsHV-1 Infection in Crassostrea gigas Larvae
- 1Ifremer, RBE-SG2M-LGPMM, La Tremblade, France
- 2Ifremer, ODE-Littoral-LERPC, La Tremblade, France
The ostreid herpesvirus (OsHV-1) is one of the major diseases that affect the Pacific oyster Crassostrea gigas. Selective breeding programs were recently shown to improve resistance easily to OsHV-1 infections in spat, juvenile, and adult oysters. Nevertheless, this resistance has never been investigated in larvae, whereas this developmental stage has crucial importance for the production of commercial hatcheries, as well as explaining the abundance of spatfall. A first trial tested several viral suspensions at several concentrations using contaminated water with OsHV-1 in 4- and 10-day-old larvae that were produced from an unselected broodstock. In follow up on the results, one viral suspension at a final concentration of 10+6 OsHV-1 DNA copies per L was used to assess resistance to OsHV-1 infection in C. gigas larvae that were produced from selected and unselected broodstock. A second trial evaluated OsHV-1 resistance in larvae from both broodstocks in trials 2a, 2b, and 2c with 4, 10, and 16-day-old larvae for 7 days, which corresponded to post D larvae, umbo larvae, and eyed larvae, respectively. The mortality of unchallenged larvae for both stocks were low (< 15%) at day 7 in trials 2a and 2b, whereas it ranged from 48 to 56% in trial 2c. More interestingly, selected larvae had significantly lower mortality than unselected larvae when exposed to OsHV-1 in all of the trials. Thus, the mortality was 11 and 49% for the selected larvae at day 7 post-exposure in trials 2a and 2c, respectively, in comparison with 84 and 97% for the unselected larvae. Although this difference in mortality was observed at day 5 in trial 2b, it was reduced at day 7, to 86 and 98% for the selected and unselected larvae, respectively. For the first time in the literature, the difference in mortality or the delayed onset of mortality between selected and unselected larvae have indicated a genetic resistance to OsHV-1 infection at the larval stage. Such finding should facilitate the selective breeding programs focusing on resistance to OsHV-1 infection by reducing the span of the genetic evaluation, and thus decreasing its cost.
Introduction
Global oyster production is largely driven by the Pacific oyster Crassostrea gigas, which represented 97% of production, totaling 4.7 million tons in 2012 and 4 billion US dollars (FAO, 2014). Disease resistance has been investigated in numerous oyster species, with significant responses to selection in improving resistance to protozoan, bacteria and viruses such as Haplosporidium nelsoni and Perkinsus marinus in Crassostrea virginica (Ford and Haskin, 1987; Ragone Calvo et al., 2003), Marteilia sydneyi and Bonamia roughleyi in Saccostrea glomerata (Dove et al., 2013), Bonamia ostreae in Ostrea edulis (Naciri-Graven et al., 1998), Roseovarius crassostrea in C. virginica (Barber et al., 1998), and Ostreid herpesvirus type 1 (OsHV-1) in C. gigas (Dégremont et al., 2015c). Most of these studies have focused on the spat and adult stages, which could be explained by the cultivation of these stages by oyster farmers (Dégremont et al., 2015a). However, disease resistance is poorly investigated in larvae, although disease could have a significant impact on spatfall in the wild. Similarly, hatchery production has increased tremendously in recent times, in particular in C. gigas in France, and larval mortality related to disease remains a threat to oyster farmers in terms of the regularity of spat production throughout the whole year.
Pathogens reportedly induce larval mortality in several oyster species, such as Vibrio coralliilyticus, V. tubiashii, or Vibrio spp in C. gigas and C. virginica (Estes et al., 2004; Mersni-Achour et al., 2015; Richards et al., 2015) or V. neptunius and V. ostreicida in O. edulis (Prado et al., 2005, 2014). With respect to the ostreid herpes virus, OsHV-1 has been detected since 1991 during mortality-inducing outbreaks in C. gigas larvae, which were produced in hatcheries in France (Nicolas et al., 1992) and New Zealand (Hine et al., 1992). More recently, OsHV-1 was also detected in larvae that were produced by hatcheries in South Korea, but there is no information on the health status of the larvae (Hwang et al., 2013). The experimental horizontal transmission of OsHV-1 within C. gigas larvae was successfully developed under controlled laboratory conditions in 2-day-old larvae (Le Deuff et al., 1994), 3-day-old larvae (Arzul et al., 2001), and 7-day-old larvae (Burge and Friedman, 2012). Nevertheless, all these studies employed larvae that were produced from unselected broodstock.
In France, higher mortality is now routinely reported in C. gigas spat and juveniles in relation to OsHV-1, which has been the case since 2008 (EFSA, 2010, 2015). A specific genotype called OsHV-1 μVar (GenBank accession no. HQ842610) was ascribed to this mortality (Segarra et al., 2010), which was also reported in New Zealand (Keeling et al., 2014), Australia (Jenkins et al., 2013), and several European countries (Peeler et al., 2012; Roque et al., 2012). Regardless of the OsHV-1 genotype, the selective breeding program was shown to easily enhance OsHV-1 resistance in C. gigas spat, juveniles, and adults (Dégremont et al., 2013, 2015b), and this genetic resistance is higher for older and larger animals (Dégremont, 2013; Dégremont et al., 2015c). Nevertheless, this resistance was never previously investigated in larvae.
This study describes the first investigation of resistance to OsHV-1 infection in C. gigas larvae from unselected broodstock and broodstock that was selected for its higher OsHV-1 resistance. Two trials were designed to track the mortality rate in larvae under experimental infection conditions. The first trial was set up to test different viral concentrations (based on the number of OsHV-1 DNA copies) with 4-day-old (trial 1a) and 10-day-old (trial 1b) unselected larvae. The second trial investigated OsHV-1 resistance in larvae with selected and unselected broodstocks. This trial was performed in 4-day-old (trial 2a), 10-day-old (trial 2b), and 16-day-old (trial 2c) larvae corresponding to D or post D veliger larvae (80–100 μm), umbo larvae (110–200 μm), and eyed larvae (250–350 μm), respectively. Their mortality was recorded at days 3, 5, and 7 post-exposure, and the detection of OsHV-1 DNA was performed by real-time quantitative PCR in larvae at each date, as well as in seawater at days 0 and 7.
Materials and Methods
Broodstocks
The first broodstock consisted of wild adult oysters that were sampled from the dock at La Tremblade in 2010, and it was used to produce the unselected larvae. The second broodstock was the eighth generation of a bi-parental family that had been produced and selected for its higher resistance to the summer mortality phenomenon in C. gigas in France (Dégremont et al., 2007, 2010). This family was then reproduced over eight generations from 2002 to 2010, with ~30 parents per generation. The family was subsequently found to have higher resistance to OsHV-1 from 2001 to 2003 (Dégremont, 2003) and to OsHV-1 μVar in the context of the massive mortality that has been occurring in France since 2008 (Dégremont, 2011; Dégremont et al., 2016). The selected broodstock was used to produce selected larvae. Both broodstocks were held until spawning in the conditioning room of the Ifremer hatchery in La Tremblade, by maintaining the oysters in a seawater temperature at 20°C. This water was UV-treated and enriched with the alga Skeletonema costatum.
Spawn and Larvae
Unselected larvae were produced in April of 2011 for trials 1a and b, and unselected and selected larvae were produced in June 2011 for trials 2 a, b, and c (Table 1). For each spawn, ripe oysters were opened and tissue was sampled from the gonad; this sample was spread on a slide and used to identify the sexes of the oysters under a microscope. The numbers of parents used for each cross ranged from 6 to 14 (Table 1). For each parent, the gametes were collected by stripping the gonad, which were successively sieved to remove large (>60 μm) and small (< 20 μm) tissue debris for the eggs and to only remove the large (>60 μm) ones for the sperm. Within the broodstock, eggs from all the females were mixed and sperm from all the males were added. After fertilization, embryos were transferred into 150-L tanks containing filtered and UV-treated seawater at 25°C, which was changed three times per week. The larvae were fed daily with the algae Isochrysis galbana and S. costatum, and they remained in the hatchery until their evaluation during experimental infection trials. Density was adjusted from 50 larvae mL−1 at early-stage larvae to three larvae mL−1 for late-stage larvae, which it is commonly used by commercial hatcheries. Such densities were then used for the experimental infections.
Viral Suspensions
Viral suspensions were prepared according to Schikorski et al. (2011b). In brief, gills and mantles from OsHV-1 μVar-infected oyster spat were dissected and pooled together in a 50 mL sterile tube. All subsequent dilutions were made with 0.22 μm-filtered artificial seawater (ASW). The total mass of the tissues was weighed, and 10 volumes of 0.22 μm ASW were added to the tube (9 mL of seawater per g of tissue). The tissues were then crushed on ice with an Ultra-Turrax® mixer (3 × 5 s). Following centrifugation (1000 g, 5 min, 4°C), the supernatant was placed in a new tube and diluted by adding four volumes of ASW. Finally, the clarified tissue homogenate was filtered consecutively through syringe filters at 5, 2, 0.45, and 0.22 μm pore sizes under sterile conditions. Filtered tissue homogenates were stored at 4°C until use.
Trial 1: Experimental Infection Trials with OsHV-1 μVar on Unselected Larvae
Before comparing the resistance of the selected and unselected larvae under OsHV-1 infection in a second trial, a first trial was set up by using unselected larvae from wild oysters to test several viral suspensions at several concentrations. The objective of this trial was to choose one viral suspension at one concentration that will induce mortality during a span of 1 week in C. gigas larvae.
Trial 1a on 4 Day-Old Larvae at Sizes Ranging from 80 to 100 μm
Four-day-old larvae with sizes ranging from 80 to 100 μm were taken from the hatchery and then transferred into a quarantine room to perform the experimental infection with OsHV-1 μvar. Three viral suspensions (Vir1, Vir2, and Vir3) were tested, each of which were tested with three final concentrations (10+7, 10+8, and 10+9 OsHV-1 DNA copies L−1), which corresponded to a total of nine conditions (Table 1). For each condition, the larvae (n = 50 larvae mL−1) were placed in two 5 L tanks in 3 L of 1.0 μm-filtered and UV-treated seawater containing 60 μL of Flumisol® at 32% (a quinolone antibiotic) and the viral suspension. According to the density and the concentration, it corresponded to 2 × 10+2, 2 × 10+3, and 2 × 10+4 OsHV-1 DNA copies per larva for the final concentrations 10+7, 10+8, and 10+9 OsHV-1 DNA copies L−1, respectively. Additionally, two control tanks were tested using the same protocol, but without adding the viral suspension. Each tank was aerated, and the seawater temperature was maintained at a 23°C room temperature. The larvae were not fed. After 5 days, the seawater in each tank was transferred into a 5 L beaker and then homogenized, and 260 mL was sampled for the mortality estimation. This volume was then filtered through a 60 μm screen to catch dead and live larvae, which were then transferred into a 15 mL tube. Finally, the tube was gently mixed, and its mortality counts were conducted under a binocular magnifier using three drops of 200 μL each, which contained the larvae. This step was included to increase the larval density and better estimate the mortality rates in a small drop. Larvae that were swimming or showing any movements (without any visible protists inside the shell) were considered alive, and the others were dead (empty shells, degradation of the tissues with brownish and undistinguishable tissues). OsHV-1 detection was performed on larvae at day 0, before their transfer in the quarantine room, and on larvae from each tank at day 5. Similarly, seawater from each tank was also screened at day 0 before adding the larvae and after adding the viral suspension as well as at day 5 at the final counting. All samples were stored at −20°C until disease screening.
Trial 1b on 10 Day-Old Larvae with Sizes Ranging from 110 to 200 μm
Similarly, from the unselected larvae that were maintained in the hatchery, 10-day-old larvae at sizes ranging from 110 to 200 μm were placed in the quarantine room for experimental infection by OsHV-1 μvar. The same protocol was used with the following modifications. One viral suspension (Vir2) was tested at four concentrations, namely 10+5, 10+6, 10+7, and 10+8 OsHV-1 DNA copies L−1, corresponding to 2, 20, 200, and 2000 OsHV-1 DNA copies per larva, respectively. Mortality was estimated 5 and 7 days post-exposure, and the volume that remained after taking a 260 mL sample for mortality counts was returned to the 5 L beaker at day 5. OsHV-1 DNA detection was performed on the larvae at days 0, 5, and 7 and in the water at days 0 and 7 as described in trial 1a. All samples were stored at −20°C until disease screening.
Trial 2: Experimental Infection Trials with OsHV-1 μVar on Unselected and Selected Larvae
Trial 2a on 4 Day-Old Larvae with Sizes Ranging from 80 to 100 μm
Four-day-old unselected larvae at sizes ranging from 80 to 100 μm were transferred from the hatchery to the quarantine room for the experimental infections by OsHV-1 μvar as well as 4-day-old selected larvae. For each group of larvae, six 5 L tanks containing 3 L each of 1.0 μm filtered and UV-treated seawater containing 60 μL of Flumisol® at 32% were used. Two treatments were tested with three of the tanks that were used as control treatments, which is hereafter named the unchallenged condition, and for the second treatment, the Vir2 viral suspension was added to the three other tanks to obtain a final concentration of 10+6 OsHV-1 DNA copies L−1, which is hereafter named the challenged condition. Each tank was aerated, the seawater temperature was maintained at a 23°C room temperature, and the larvae were not fed. The density per tank was ~33 larvae mL−1, corresponding to a final concentration of 30.3 OsHV-1 DNA copies per larva. The mortality was estimated by following the same protocol used in trial 1b on days 3, 5, and 7 post-exposure. OsHV-1 DNA detection was performed on larvae at days 0, 3, 5, and 7 as well as in the water at days 0 and 7 as described in trial 1a. All samples were stored at −20°C until disease screening.
Trial 2b on 10-Day-Old Larvae with Sizes Ranging from 110 to 200 μm
Similarly, 10-day-old larvae with sizes ranging from 110 to 200 μm from the same spawns used in trial 2a (but kept in the hatchery) were transferred in the quarantine room. The same protocol used in trial 2a was used, except that the density per tank was ~27 larvae mL−1, corresponding to a final concentration of 37.0 OsHV-1 DNA copies per larva. OsHV-1 DNA detection was performed in the larvae at days 0, 3, 5, and 7 as well as in the water at days 0 and 7. All samples were stored at −20°C until disease screening.
Trial 2c on 16 Day-Old Larvae with Sizes Ranging from 250 to 350 μm
Sixteen-day-old larvae with sizes ranging from 250 to 350 μm, which came from the same spawns used in trial 2a but were kept in the hatchery, were transferred in the quarantine room. The same protocol used in trial 2a was used here, except that the density per tank was ~3 larvae mL−1 corresponding to a final concentration of 333.3 OsHV-1 DNA copies per larva. OsHV-1 DNA detection was performed on the larvae at days 0, 3, 5, and 7 as well as on the water at days 0 and 7. All samples were stored at −20°C until disease screening.
OsHV-1 DNA Quantification
Total DNA was extracted from the collected larvae as described in the mortality estimation step (see trial 1a) by using ~50–200 larvae per sample. Those larvae were crushed with a piston pellet and then DNA was extracted with a QIAamp tissue mini kit according to the manufacturer's protocol as described in Schikorski et al. (2011a). Similarly, total DNA was extracted from the seawater sampled after adding the viral suspension and before adding the larvae at day 0, as well as at day 5 or 7 post-exposure, depending of the trial. For each tank, the total DNA was extracted from 100 μL of seawater. A final elution of the DNA that was extracted from larval samples was performed with 100 μL of double-distilled water and 50 μL for seawater samples. The DNA concentration was analyzed spectrophotometrically (NANODROP®). A dilution of the larval DNA samples was performed to reach a final DNA concentration of 4 ng μl−1. DNA that was extracted from seawater was used without dilution. Each DNA sample was tested in duplicate qPCR tests.
The detection and quantification of OsHV-1 DNA were performed for larvae and seawater with the SYBR® green real-time PCR protocol described by Pépin et al. (2008) as adapted for use with DPFor/DPRev primers to target the OsHV-1 DNA polymerase sequence (ORF 100) (Pépin, 2013). The results were expressed as the viral DNA copy number per ng of total DNA and the viral DNA copy number per μL for larvae and seawater, respectively.
Statistical Analysis
For each count, the mortality was analyzed by binomial logistic regression throughout the Genmod procedure in SAS® 9.4 software.
In using 4-day-old unselected larvae for trial 1a, the following model was used:
where Yij is the probability of larval death when exposed to the ith viral suspension (Vir1, Vir2, and Vir3) at the jth concentration (10+7, 10+8, and 10+9 OsHV-1 DNA copies L−1), and μ is the intercept. When a significant interaction was observed, the SLICE option was used. This approach allowed for a more powerful analysis than when the model was rerun for each effect because the degrees of freedom are not reduced (Littell et al., 2002).
For 10-day-old unselected larvae in trial 1b, the model was reduced to test the OsHV-1 concentration, and statistical analyses were performed on days 5 and 7 post-exposure.
In trial 2a, 2b, and 2c, mortality was analyzed 7 days post-exposure by using the following model:
where Yij is the probability of larval death in the ith stock (unselected or selected larvae) for the jth treatment (unchallenged or challenged by OsHV-1), and μ is the intercept.
For trials 2a, 2b, and 2c, and only for the challenged treatment, the quantification of OsHV-1 DNA was log (Y + 1) transformed. For larvae, we analyzed the data through an ANOVA using the GLM procedure and by using the following model:
where Yi is the DNA copy number per mg of fresh tissue detected for the ith stock (unselected or selected larvae) at the jth sampling date (days 3, 5, and 7 post-exposure).
For seawater samples, data were analyzed using the following model:
where Yij is the DNA copy number per μl of seawater detected for the ith stock (unselected or selected larvae) at the jth sampling date (day 0 and day 7 post-exposure).
Results
TRIAL 1: Experimental Infection Trials with OsHV-1 μVar using Unselected Larvae
Trial 1a on 4-Day-Old Larvae with Sizes Ranging from 80 to 100 μm
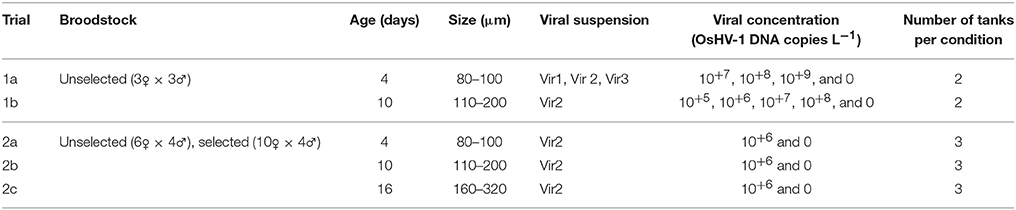
Mortality was low in unselected 4-day-old control larvae (2.1%), and ranged from 3 to 99.4% in larvae exposed to OsHV-1 at day 5 post-exposure (Figure 1). A significant interaction was found between the viral suspensions (Vir 1, Vir 2, and Vir 3) and concentrations (10+7, 10+8, and 10+9) at day 5 post exposure (χ2 = 121.75; p < 0.0001). For Vir 1, the larvae did not exhibit mortality at 10+7 and 10+8 OsHV-1 DNA copies L−1 but they had significantly higher mortality at 10+9 OsHV-1 DNA copies L−1 (χ2 = 279.60; p < 0.0001). For Vir 2 and 3, high mortality was observed regardless of the Vir2 concentration (χ2 = 0.33; p = 0.85) and Vir3 concentration (χ2 = 4.93; p = 0.09).
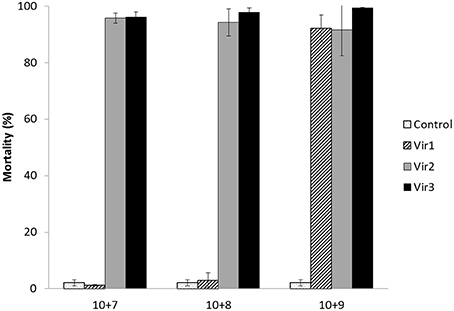
Figure 1. Mortality (% ± SD) of 4 days old unselected oyster larvae at day 5 post-exposure in trial 1a. Three viral suspensions (Vir1, Vir2, and Vir3) and three concentrations were used (10+7, 10+8, and 10+9 DNA copies L−1).
Trial 1b on 10 Day-Old Larvae with Sizes Ranging from 110 to 200 μm
For 10-day-old unselected larvae, the mortality at day 5 post-exposure remained lower than 3% for the control and those exposed to a final concentration of 10+5 and 10+6 OsHV-1 DNA copies L−1. Significantly higher mortality (18.1 and 18.6%) was found for larvae that were exposed to the two higher concentrations of OsHV-1 DNA (10+7 and 10+8; χ2 = 83.68; p < 0.0001; Figure 2). Two days later, the control larvae still did not exhibit mortality (< 3%), and the mortality increased from 63.1% at 10+5 to 98.3% at 10+8 (Figure 2) and was significantly different among the concentrations (χ2 = 119.19; p < 0.0001).
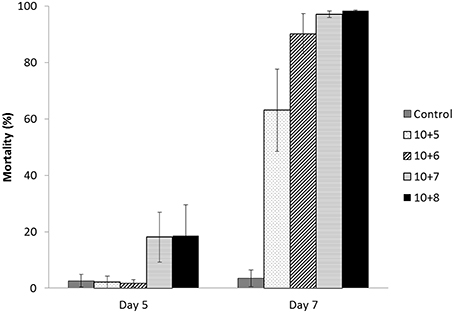
Figure 2. Mortality (%± SD) of 10 days old unselected oyster larvae at days 5 and 7 post-exposure in trial 1b. One viral suspension (Vir2) and four concentrations were used (10+5, 10+6, 10+7, and 10+8 OsHV-1 DNA copies L−1).
Trial 2: Experimental Infection Trials by OsHV-1 μVar using Unselected and Selected Larvae
Trial 2a on 4 Day-Old Larvae at Sizes Ranging from 80 to 100 μm
For 4-day-old larvae at sizes ranging from 80 to 100 μm, the mortality of the unchallenged larvae was low (< 20%) at the end of the trial on day 7 for both the unselected and selected larvae (Figure 3A). A similar mortality pattern was also observed for the selected larvae that were exposed to OsHV-1. By contrast, the unselected larvae that were challenged by OsHV-1 did not exhibit mortality on day 3, but moderate mortality was observed for one of the three tanks (36.3%) on day 5 and remained low for the two others (5.4 and 6.9%). On day 7, mortality was high (83.8%) for the unselected larvae that were challenged by OsHV-1, ranging from 63.8 to 94.3%, depending on the tank (Figure 3A).
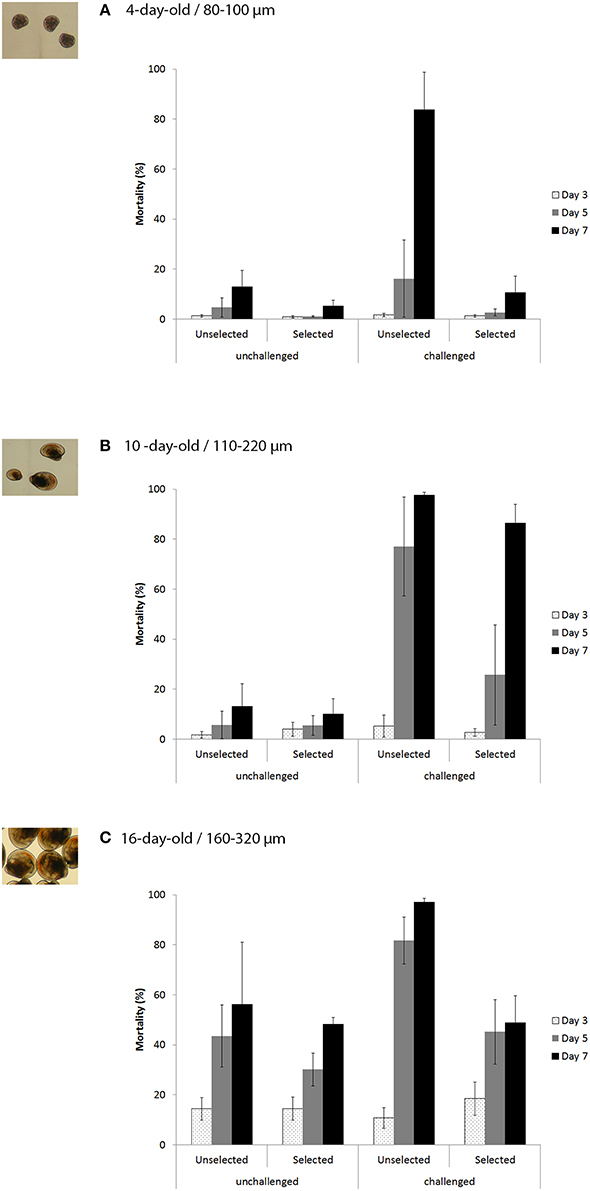
Figure 3. Mortality (% ± SD) of the unselected and selected larvae on days 3, 5, and 7 post-exposure for the unchallenged and challenged conditions (10+6 OsHV-1 DNA copies L−1) in trial 2a (A), trial 2b (B), and trial 2c (C) corresponding to 4, 10, and 17-day-old larvae, respectively and 80–100, 110–200, and 160–320 μm larvae, respectively.
Statistical analyses revealed a significant interaction between stocks and treatments (χ2 = 22.73; p < 0.0001; Table 2). At the stock level, unselected larvae had significantly higher mortality when exposed to OsHV-1 than those that were not exposed (χ2 = 103.75; p < 0.0001), whereas selected larvae had similar and low mortality under both conditions (χ2 = 2.82; p < 0.0001). At the treatment level, unselected larvae had significantly higher mortality than selected larvae under both treatments, although the level of significance was much higher under the challenge condition (Table 2).
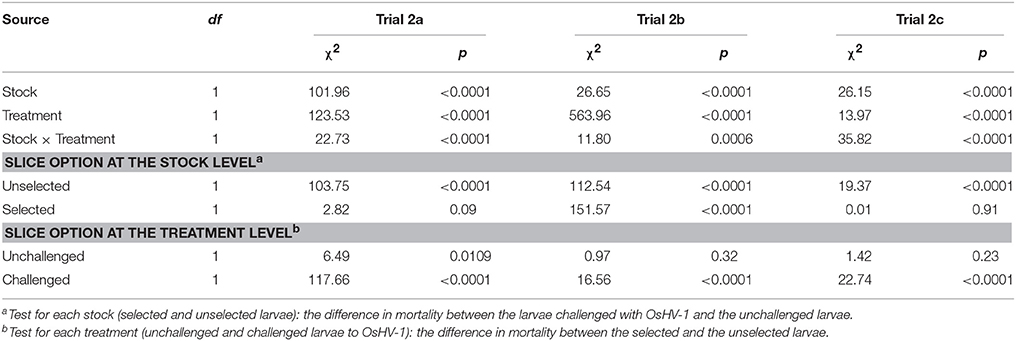
Table 2. Logit analysis of the mortality at the end of the trial on day 7 for unchallenged and challenged larvae when using selected OsHV-1-resistant or unselected broodstocks for 4, 10, and 16 day-old larvae in trials 2a, 2b, and 2c, respectively.
Trial 2b on 10-Day-Old Larvae with Sizes Ranging from 110 to 200 μm
Similar to the results obtained for the unchallenged condition in trial 2a, the 10-day-old unchallenged larvae had low mortality for both stocks on day 7 (< 20%) (Figure 3B). Larvae that were exposed to OsHV-1 did not exhibit mortality on day 3, and their mortality was moderate for the selected larvae (25.7%) and high for the unselected larvae (77.1%) on day 5 (Figure 3B). On day 7, the mortality was 86.4 and 97.7% for the selected and unselected larvae, respectively (Figure 3B).
Significant interactions between the stocks and treatments were evidenced on day 7 (χ2 = 11.80; p = 0.0006; Table 2). At the stock level, larvae that were exposed to OsHV-1 had significantly higher mortality than those that were not exposed in both stocks on day 7 (Table 2). At the treatment level, the unselected and selected larvae had similar mortality for the unchallenged condition (χ2 = 0.97; p = 0.32), and the selected larvae had significantly lower mortality than the unselected larvae when exposed to OsHV-1 on day 7 (χ2 = 16.56; p < 0.0001; Table 2).
Trial 2c on 16 Day-Old Larvae with Sizes Ranging from 250 to 350 μm
The 16-day-old larvae exhibited a mortality trend that regularly increased from day 3 to day 7 for both stocks under the unchallenged condition, reaching 48.2 and 56.1% for the selected and unselected larvae, respectively (Figure 3C). A similar mortality pattern was also observed for the selected larvae that were exposed to OsHV-1, with a final mortality of 48.9% on day 7. The unselected larvae experienced low mortality (10.7%) on day 3, which rapidly increased to 81.7% on day 5 and 97.1% on day 7 (Figure 3C).
Interactions between stocks and treatments in terms of mortality were again significant at day 7 (χ2 = 35.82; p < 0.0001; Table 2). At the stock level, selected larvae that were exposed to OsHV-1 had similar mortality to those that were unexposed (χ2 = 0.01; p = 0.91). In contrast, unselected larvae exposed to OsHV-1 had significantly higher mortality than those not exposed to the virus (χ2 = 19.37; p < 0.0001; Table 2). At the treatment level, no significant difference in mortality was found between the unselected larvae and the selected larvae under the unchallenged condition (χ2 = 1.42; p = 0.23). Conversely, unselected larvae that were exposed to OsHV-1 had significantly higher mortality than selected larvae (χ2 = 22.74; p < 0.0001; Table 2).
OsHV-1 Quantification
Before the experimental infections, OsHV-1 DNA was not detected in the larvae from all trials.
OsHV-1 Quantification in the Seawater and Larvae in Trial 1a
For 4-day-old unselected larvae in trial 1a, OsHV-1 DNA was detected in the seawater at day 0 (< 10+1 μL−1) for the control. Higher amounts were found in seawater and larvae at day 5, but they were still at a low level (10+3 copies per μL or ng of DNA) for the control (Table 3). For the other conditions, OsHV-1 DNA was detected at a low level in the seawater at day 0 (< 10+3 μL−1) and at a much higher level at day 5, ranging from 10+4 to 10+5 DNA copies per μL (Table 3). Similarly, a high amount of OsHV-1 DNA was detected in the larvae at day 5 for all conditions, ranging from 10+5 to 10+7 copies per ng of total DNA except for Vir1 at the lowest concentration tested, for which it was only 10+2 (Table 3).
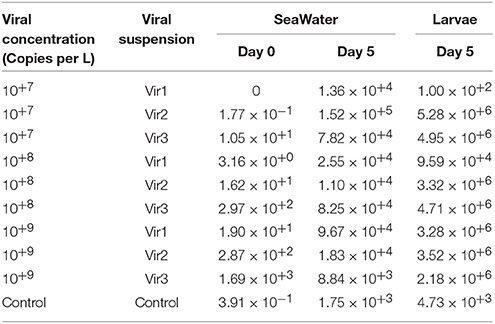
Table 3. Quantification of OsHV-1 DNA in seawater (copies per μL) at days 0 and 5 post-exposure, and in larvae (copies per ng of DNA) at day 5 in trial 1a.
OsHV-1 Quantification in Seawater and Larvae from Trial 1b
In assessing 10-day-old larvae for trial 1b, OsHV-1 DNA was detected in the seawater at day 0 at a low level (< 10+2) and at a higher level at day 7 post-exposure, with concentrations ranging from 10+2 for the control to 10+4 for the infected conditions (Table 4). For the larvae, the amount of OsHV-1 DNA remained low in the control at days 5 and 7 (< 10+1), and it reached 10+6 DNA copies per ng of DNA at days 5 and 7 (Table 4).
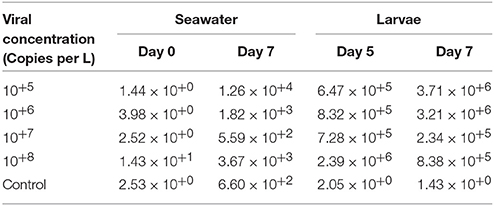
Table 4. Quantification of OsHV-1 DNA in seawater (copies per μL) at days 0 and 5 post-exposure, and in larvae (copies per ng of DNA) at days 5 and 7 in trial 1b.
OsHV-1 Quantification in the Seawater from Trial 2abc
For 4, 10, and 16-day-old selected larvae and unselected larvae in trial 2abc, OsHV-1 was not detected in the seawater from the unchallenged condition at day 0, but it was detected for all challenged conditions after adding the viral suspension (Vir 2) containing in average 15 copies of OsHV-1 DNA per μL, and ranging from 2.94 10−1 to 4.38 10+1 copies of OsHV-1 DNA per μL, depending of the trial (Table 5).
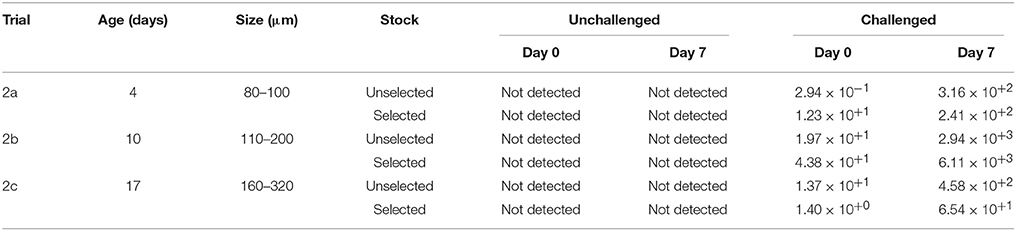
Table 5. Quantification of OsHV-1 DNA in seawater (copies per μL) for the unchallenged and challenged conditions in trial 2a, trial 2b, and trial 2c.
At day 7, similar copy numbers were observed for trial 2abc, except that a higher amount of DNA copies were detected for both stocks under the challenge condition ranging from 6.54 × 10+1 copies DNA copies per μL for the selected larvae in trial 2c to 6.11 × 10+3 copies per μL for the selected larvae in trial 2b (Table 5).
None of the factors (stocks, date of sampling and the interaction) was significant in trial 2a (p > 0.05). In trial 2b, only the date of sampling factor was found significant with higher amount of OsHV-1 DNA at day 7 post-exposure than at day 0 (p < 0.0001). Concerning the trial 2c, only the interaction was not significant (p > 0.05). Thus, the seawater sampled in tanks containing the selected larvae had a significant lower amount of OsHV-1 DNA than the seawater sampled in tanks containing the unselected larvae (p = 0.0010). Significant higher amount of OsHV-1 DNA was detected at day 7 than at day 0 (p < 0.0001).
OsHV-1 Quantification in Larvae from Trial 2abc
For 4, 10, and 16-day-old selected larvae and unselected larvae in trial 2abc, OsHV-1 DNA was detected in the larvae from the unchallenged condition, but it remained low, at 10+3 DNA copies per ng of total DNA, throughout the duration of all the experiments (Figures 4A–C). By contrast, a high level of OsHV-1 DNA was detected in unselected and selected larvae for the challenge condition.
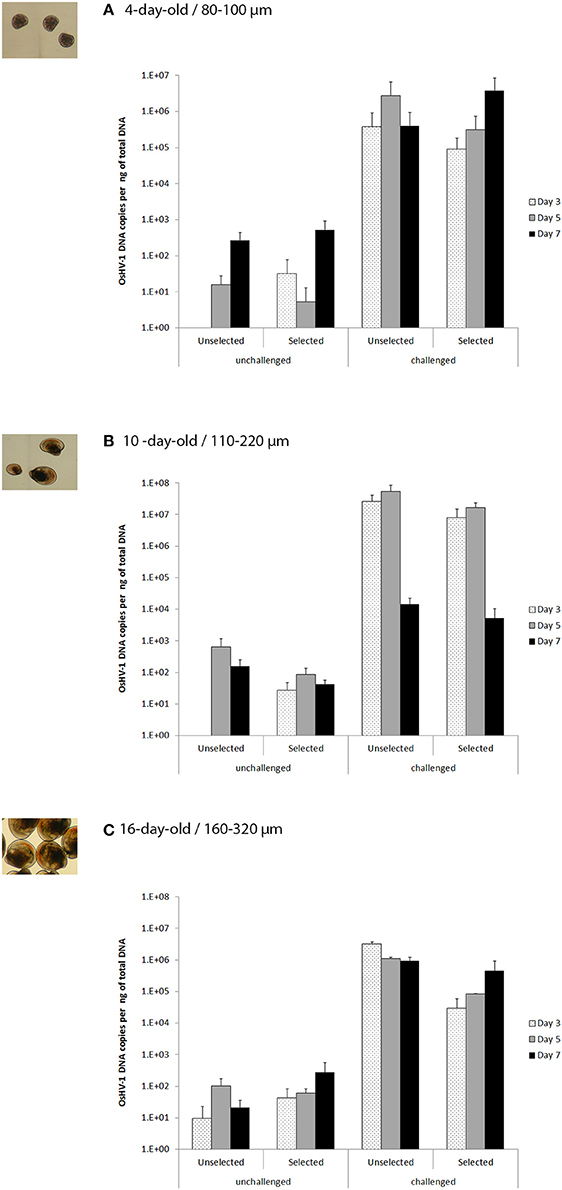
Figure 4. OsHV-1 DNA copies per ng of total DNA (% + SD) in unselected and selected larvae on days 3, 5, and 7 post-exposure for the unchallenged and challenged conditions (0 and 10+6 OsHV-1 DNA copies L−1, respectively) in trial 2a (A), trial 2b (B), and trial 2c (C) corresponding to 4, 10, and 17 day-old larvae, respectively and 80–100, 110–200, and 160–320 μm larvae, respectively.
Thus, in trial 2a, the OsHV-1 DNA copies per ng of total DNA ranged from 10+5 to 10+6 for both unselected and selected larvae at days 3, 5, and 7 (Figure 4A). In a closer look, the OsHV-1 amount reached a peak at day 5 post-exposure for the unselected larvae, while it was at day 7 for the selected larvae. None of the factors (stocks, date of sampling and the interaction) was significant in trial 2a (p > 0.05).
For trial 2b, a high amount of OsHV-1 (10+7) was observed for both stocks at days 3 and 5, and it dropped significantly to 10+4 at day 7 (p < 0.0001; Figure 4B). The interaction between stocks and date of sampling was not significant (p = 0.66), and the amount of OsHV-1 did not significantly differ between the selected and the unselected larvae (p = 0.06).
For trial 2c, the OsHV-1 amount was again high for the unselected larvae reaching a peak at day 3 (3.31 × 10+6), and remained at this level, although it slightly decreased at days 5 and 7 (1 × 10+6 and 9.32 × 10+5, respectively). By contrast, the amount of OsHV-1 DNA increased regularly from day 3 (10+4) to day 7 (10+5). Interaction and date of sampling were not significant (p > 0.05) while the amount of OsHV-1 DNA for the unselected larvae was significantly higher than for selected larvae (p = 0.0067).
Discussion
The major finding of this study revealed lower mortality from OsHV-1 in larvae through day 7 post-exposure that were produced from disease-resistant broodstock in comparison with larvae that were produced from unselected broodstock (Figures 3A–C). Consequently, OsHV-1 infection resistance in C. gigas could be observed at all stages, as previously demonstrated in spat, juveniles and adults (Dégremont, 2013; Dégremont et al., 2013). To our knowledge, this is the first report on a positive response to selection in oyster larvae. A previous study used an experimental challenge with bacterial pathogens in larvae from three disease-resistant lines of the eastern oysters C. virginica (Gomez-Leon et al., 2008). Unfortunately, the line that was selected for its higher resistance to Roseovarius oyster disease at the juvenile stage was not tested at the larval stage, and the two other oyster lines, which were selected for their higher resistance to P. marinus and/or H. nelsoni, were evaluated against three bacterial pathogens, namely two Vibrio spp strains and Roseovarius crassostrea. Nevertheless, the difference in mortality between lines at the larval stage was observed at 20°C, suggesting that resistance to pathogens could be present in the oyster larvae (Gomez-Leon et al., 2008). Our study strengthens their findings.
We observed high mortality in all larval stages exposed to OsHV-1 μvar from D hinge to eyed larvae. The previous studies on the horizontal transmission of OsHV-1 in larvae primarily focused on 2–3-day-old larvae (Le Deuff et al., 1994; Arzul et al., 2001) or the 7-day-old larvae without any indication about the sizes of the larvae (Burge and Friedman, 2012). At these ages, we assumed that the authors had primarily studied D larvae and small umbo larvae. Consequently, C. gigas larvae are susceptible to OsHV-1 during their whole planktonic existence. This result suggests that OsHV-1 is a permanent threat for hatcheries, and OsHV-1 could be an important regulator of spatfall in C. gigas in the open environment. Spawning usually occurs during the summer when the seawater temperature favors the OsHV-1 disease, which is commonly higher than 16°C in France (Pernet et al., 2012; Dégremont, 2013). Thus, the absence of significant C. gigas spatfall from 2009 to 2011 in Arcachon Bay could be explained by the high mortality caused by OsHV-1 in larvae. The pathogen hypothesis was unfortunately not investigated, but others were tested, such as the spatial distribution of broodstocks, their trophic competition, the presence of pollutants or a decrease or change in trophic resources (Bernard et al., 2014). However, some assays for OsHV-1 detection were performed on frozen samples from an oyster larval abundance survey in Marennes Oléron Bay. These samples were collected during the spawning season in 2009, 2010, and 2011, and they revealed a positive signal for viral DNA in the larvae (unpublished data). It would be interesting to extend studies to determine whether exposed larvae are able to settle and become spat.
For the larvae that were experimentally infected by OsHV-1, the quantity of the virus found in seawater increased significantly in all trials from days 0 to days 5 and 7 (Tables 3–5). These results suggest that OsHV-1 replicated in the larvae, which were then released in the seawater. This finding is consistent with the experimental OsHV-1 challenge in C. gigas spat, for which viral DNA was not detected at time 0, but it increased to 10+3 copies per μL by 5 days later (Schikorski et al., 2011a). In contrast to the results of Schikorski's study, OsHV-1 DNA was also detected in seawater at day 0 and days 5 or 7, and in larvae at days 5 or 7 for the unchallenged condition, but the viral copy level was low (< 10+3). Despite the presence of viral DNA, no mortality was reported in unchallenged larvae in all trials, excepted for in trial 2c, which will be explained below, suggesting that this is just a trace of viral DNA and not virulent particles. Its detection could be explained by the seawater that was pumped into the Marennes-Oléron Bay, where mortality related to OsHV-1 was reported during this period (Dégremont et al., 2016). Thus, filtration at 1.0 μm and UV-treated seawater likely removed most viral particles, and the OsHV-1 DNA we detected was likely inactive viral particles or degraded DNA. This finding is consistent with a hypothesis suggesting that OsHV-1 may be carried on abioseston particles and organic matter and that the removal of the putative particulate vector of OsHV-1 from seawater using filtration at 5 μm enabled C. gigas spat to survive in spite of the presence of OsHV-1 μVar in the water supply (Whittington et al., 2015).
For 4-day-old unselected larvae (trial 2a), the unselected and selected larvae did not suffer from mortality at day 3 (Figure 3A). In fact, the amount of OsHV-1 detected in the larvae increased from none at day 0 to 10+5 copies per ng of DNA at day 3(Figure 4A), suggesting the active multiplication of the virus in both stocks. This fast proliferation in the larvae was already reported during the experimental infection of larvae by OsHV-1 (Le Deuff et al., 1994; Burge and Friedman, 2012). Although the amount of OsHV-1 was slightly higher at day 5 for both stocks, unselected larvae had 8-fold times more OsHV-1 DNA detected than selected larvae, and only unselected larvae started dying. This is in agreement with finding obtained in OsHV-1-resistant and unselected in C. gigas spat tested in field condition (Dégremont, 2011). A high-mortality outbreak related to OsHV-1 was observed between days 5 and 7 post-exposure, again only for the unselected larvae. Although the OsHV-1 amount slightly decreased in unselected larvae from days 5 to 7, it was still increasing in selected larvae during this period, which still did not exhibit significant mortality (Figures 3A, 4A). Dégremont (2011) reported that the mortality peak immediately followed the viral load peak in C. gigas spat, suggesting that selected larvae could have experienced mortality if the trial had lasted longer. At this age, selected larvae seemed to be resistant to OsHV-1 infection, or the onset of mortality related to OsHV-1 took longer to appear, underlying the genetic resistance of C. gigas in small larvae.
For 10-day-old larvae (trial 2b), mortality came earlier, between days 3 and 5 post-exposure for unselected larvae, which was concomitant with the large amount of OsHV-1 DNA on both dates (Figures 3B, 4B). Though mortality was high for the unselected larvae at day 5 (77.1%), this mortality level was only observed 2 days later for the selected larvae. Thus, for the first time, we demonstrated that selected larvae can suffer high mortality from OsHV-1. This finding also revealed that the onset of mortality was delayed for the selected larvae, confirming their higher genetic resistance in comparison with unselected larvae. Even if the selected larvae experienced mortality from OsHV-1, this finding is consistent with the results found under field conditions. Indeed, resistant stocks always experienced mortality related to OsHV-1 infection in spat and juveniles, sometimes reaching 50%, whereas unselected stocks experienced higher mortality, usually reaching 90% (Dégremont, 2013; Dégremont et al., 2015c). One hypothesis to explain the higher mortality in larvae than in spat for the selected stock could be related to their younger age and smaller size. OsHV-1 resistance was shown to increase with age and size, and older animals are generally less affected (Pernet et al., 2012; Dégremont, 2013). Another hypothesis could be related to the development of the immune system. Tirape et al. (2007) showed that the immune system develops gradually throughout the life of the animal, and the animal is then able to respond to stress. In addition, the expression of certain genes related to the immune profile was variable depending on the developmental stage of the larvae. The last hypothesis relies on the experimental protocol itself. Our experimental infection was performed in flasks without renewing the seawater and without adding food contrasting with the study conducted by Burge and Friedman (2012) where those were renewed or fed daily. These conditions could have favored the disease, given that OsHV-1 mortality decreased with water renewal in C. gigas spat (Petton et al., 2015) and that energy reserves were diminished in infected oysters (Tamayo et al., 2014). To conclude with trial 2b, the significant decrease in the amount of OsHV-1 from 10+7 at day 5 to 10+4 at day 7 may be the consequence of the decreased replication of OsHV-1 because of the lack of live larvae between days 5 and 7 for both stocks (Figure 4C). This is in agreement with the findings obtained by Burge and Friedman (2012).
For the first time, 16-day-old larvae were evaluated for OsHV-1 resistance during 2c. These larvae exhibited moderate mortality, reaching 50% at day 7 for selected or unselected larvae under unchallenged conditions as well as for selected larvae under the challenged condition (Figure 3C). According to the quantification of OsHV-1, those findings were not related to OsHV-1. To explain this mortality, we hypothesized that in addition to the absence of feeding, the larvae were at a sensitive step of their lives, given that they were eyed larvae, and near metamorphosis at that. Nevertheless, when exposed to OsHV-1, unselected larvae exhibited higher mortality than selected larvae, which was related to OsHV-1 according to the OsHV-1 DNA detected at days 3, 5, and 7. At this stage, unselected larvae had a significant larger amount of OsHV-1 DNA than selected larvae (Figure 4C), strongly supporting the idea that genetic resistance was present at this stage of development.
In following up with our findings, the next step is to study the molecular mechanisms and protein expression in unselected and selected larvae before, during and after an experimental infection with the OsHV-1 virus. It also appears that the larval model would be a powerful approach to understanding genetic resistance to OsHV-1 better as well as facilitating a selective breeding program. Indeed, the family that showed higher resistance to OsHV-1 infection could be identified a couple days after spawning, allowing it to reduce the span of the genetic evaluation, and thus decreasing the cost of the breeding program. Finally, monitoring oyster larvae through settlement to juveniles when exposed to OsHV-1 should also useful to better understand the abundance of spatfall, which is of great importance for oyster farmers.
Author Contributions
LD and JP designed the experiment. LD and ST produced the larvae. ST performed the infection experiments. ST, JP, and BM conducted the disease analyses. LD analyzed the data. LD, JP, and BM wrote and edited the manuscript.
Funding
This work was supported by the Institut Français de Recherche pour l'Exploitation de la Mer through the project “Amélioration par la sélection.”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Pascal Phélipot and Jean-Christophe Billy for their help in the hatchery (algae and seawater supply). We thank Delphine Tourbiez and Nicole Faury for help with the preparation of the viral suspensions.
References
Arzul, I., Renault, T., and Lipart, C. (2001). Experimental herpes-like viral infections in marine bivalves: demonstration of interspecies transmission. Dis. Aquat. Org. 46, 1–6. doi: 10.3354/dao046001
Barber, B. J., Davis, C. V., and Crosby, M. A. (1998). Cultured oysters, Crassostrea virginica, genetically selected for fast growth in the Damariscotta River, Maine, are resistant to mortality caused by Juvenile Oyster Disease (JOD). J. Shellfish Res. 17, 1171–1175.
Bernard, I., Allain, G., Auby, I., Plus, M., Drean, T., Gasmi, S., et al. (2014). Etude de la Fécondité de L'huître Creuse Dans le Bassin d'Arcachon en 2013 en Lien Avec la Qualité de L'eau. Compte-Rendu Final du Programme FeLiBA.
Burge, C. A., and Friedman, C. S. (2012). Quantifying Ostreid Herpesvirus (OsHV-1) genome copies and expression during transmission. Microb. Ecol. 63, 596–604. doi: 10.1007/s00248-011-9937-1
Dégremont, L. (2003). Etude des Bases Génétiques de la Mortalité Estivale et des Relations Avec la Croissance Chez Les Juvéniles de Crassostrea gigas. Ph.D., Université de Caen Basse Normandie.
Dégremont, L. (2011). Evidence of herpesvirus (OsHV-1) resistance in juvenile Crassostrea gigas selected for high resistance to the summer mortality phenomenon. Aquaculture 317, 94–98. doi: 10.1016/j.aquaculture.2011.04.029
Dégremont, L. (2013). Size and genotype affect resistance to mortality caused by OsHV-1 in Crassostrea gigas. Aquaculture 416–417, 129–134. doi: 10.1016/j.aquaculture.2013.09.011
Dégremont, L., Bédier, E., and Boudry, P. (2010). Summer mortality of hatchery-produced Pacific oyster spat (Crassostrea gigas). II. Response to selection for survival and its influence on growth and yield. Aquaculture 299, 21–29. doi: 10.1016/j.aquaculture.2009.11.017
Dégremont, L., Ernande, B., Bedier, E., and Boudry, P. (2007). Summer mortality of hatchery-produced Pacific oyster spat (Crassostrea gigas). I. Estimation of genetic parameters for survival and growth. Aquaculture 262, 41–53. doi: 10.1016/j.aquaculture.2006.10.025
Dégremont, L., Garcia, C., and Allen, S. K. Jr. (2015a). Genetic improvement for disease resistance in oysters: a review. J. Invertebr. Pathol. 131, 226–241. doi: 10.1016/j.jip.2015.05.010
Dégremont, L., Guyader, T., Tourbiez, D., and Pépin, J.-F. (2013). Is horizontal transmission of the Ostreid herpesvirus OsHV-1 in Crassostrea gigas affected by unselected or selected survival status in adults to juveniles? Aquaculture 408–409, 51–57. doi: 10.1016/j.aquaculture.2013.05.025
Dégremont, L., Lamy, J.-B., Pépin, J.-F., Travers, M.-A., and Renault, T. (2015b). New insight for the genetic evaluation of resistance to ostreid herpesvirus infection, a worldwide disease, in Crassostrea gigas. PLoS ONE 10:e0127917. doi: 10.1371/journal.pone.0127917
Dégremont, L., Ledu, C., Maurouard, E., Nourry, M., and Benabdelmouna, A. (2016). Effect of ploidy on the mortality of Crassostrea gigas spat caused by OsHV-1 in France using unselected and selected OsHV-1 resistant oysters. Aquacult. Res. 47, 777–786. doi: 10.1111/are.12536
Dégremont, L., Nourry, M., and Maurouard, E. (2015c). Mass selection for survival and resistance to OsHV-1 infection in Crassostrea gigas spat in field conditions: response to selection after four generations. Aquaculture 446, 111–121. doi: 10.1016/j.aquaculture.2015.04.029
Dove, M. C., Nell, J. A., and O'Connor, W. A. (2013). Evaluation of the progeny of the fourth-generation Sydney rock oyster Saccostrea glomerata (Gould, 1850) breeding lines for resistance to QX disease (Marteilia sydneyi) and winter mortality (Bonamia roughleyi). Aquac. Res. 44, 1791–1800. doi: 10.1111/are.12012
EFSA (2010). Scientific opinion on the increased mortality events in Pacific oysters, Crassostrea gigas. EFSA J. 8, 1–60. doi: 10.2903/j.efsa.2010.1894
Estes, R. M., Friedman, C. S., Elston, R. A., and Herwig, R. P. (2004). Pathogenicity testing of shellfish hatchery bacterial isolates on Pacific oyster Crassostrea gigas larvae. Dis. Aquat. Org. 58, 223–230. doi: 10.3354/dao058223
FAO (2014). Fisheries and Aquaculture Software. FishStatJ - Software for Fishery Statistical Time Series. Rome: FAO Fisheries and Aquaculture Department. Available online at: http://www.fao.org/fishery/statistics/software/fishstatj/en (Accessed).
Ford, S. E., and Haskin, H. H. (1987). Infection and mortality patterns in strains of oysters Crassostrea virginica selected for resistance to the parasite Haplosporidium nelsoni (MSX). J. Parasitol. 73, 368–376. doi: 10.2307/3282092
Gomez-Leon, J., Villamil, L., Salger, S. A., Sallum, R. H., Remacha-Trivino, A., Leavitt, D. F., et al. (2008). Survival of eastern oysters Crassostrea virginica from three lines following experimental challenge with bacterial pathogens. Dis. Aquat. Org. 79, 95–105. doi: 10.3354/dao01902
Hine, P. M., Wesney, B., and Hay, B. E. (1992). Herpesviruses associated with mortalities among hatchery-reared larval Pacific oysters Crassostrea gigas. Dis. Aquat. Org. 12, 135–142. doi: 10.3354/dao012135
Hwang, J. Y., Park, J. J., Yu, H. J., Hur, Y. B., Arzul, I., Couraleau, Y., et al. (2013). Ostreid herpesvirus 1 infection in farmed Pacific oyster larvae Crassostrea gigas (Thunberg) in Korea. J. Fish Dis. 36, 969–972. doi: 10.1111/jfd.12093
Jenkins, C., Hick, P., Gabor, M., Spiers, Z., Fell, S. A., Gu, X. N., et al. (2013). Identification and characterisation of an ostreid herpesvirus-1 microvariant (OsHV-1 mu-var) in Crassostrea gigas (Pacific oysters) in Australia. Dis. Aquat. Org. 105, 109–126. doi: 10.3354/dao02623
Keeling, S. E., Brosnahan, C. L., Williams, R., Gias, E., Hannah, M., Bueno, R., et al. (2014). New Zealand juvenile oyster mortality associated with ostreid herpesvirus 1-an opportunistic longitudinal study. Dis. Aquat. Org. 109, 231–239. doi: 10.3354/dao02735
Le Deuff, R.-M., Nicolas, J.-L., Renault, T., and Cochennec, N. (1994). Experimental transmission of a Herpes-like virus to axenic larvae of Pacific oyster, Crassostrea gigas. Bull. Eur. Assoc. Fish Pathol. 14, 69–72.
Littell, R. C., Stroup, W. W., and Freund, R. J. (2002). SAS® for Linear Models. Cary, NC: SAS Institute Inc.
Mersni-Achour, R., Cheikh, Y. B., Pichereau, V., Doghri, I., Etien, C., Dégremont, L., et al. (2015). Factors other than metalloprotease are required for full virulence of French Vibrio tubiashii isolates in oyster larvae. Microbiology 161, 997–1007. doi: 10.1099/mic.0.000058
Naciri-Graven, Y., Martin, A. G., Baud, J. P., Renault, T., and Gérard, A. (1998). Selecting the flat oyster Ostrea edulis (L.) for survival when infected with the parasite Bonamia ostreae. J. Exp. Mar. Biol. Ecol. 224, 91–107.
Nicolas, J.-L., Comps, M., and Cochennec, N. (1992). Herpes-like virus infecting Pacific oyster larvae, Crassostrea gigas. Bull. Eur. Assoc. Fish Pathol. 12, 11–13.
Peeler, E. J., Allan Reese, R., Cheslett, D. L., Geoghegan, F., Power, A., and Thrush, M. A. (2012). Investigation of mortality in Pacific oysters associated with Ostreid herpesvirus-1 μVar in the Republic of Ireland in 2009. Prev. Vet. Med. 105, 136–143. doi: 10.1016/j.prevetmed.2012.02.001
Pépin, J. F. (2013). Short Technical Report for OsHV-1 Detection and Quantification by Real Time Polynerase Chain Reaction using OsHV-1 DNA Polymerase Sequence. Available online at: http://archimer.ifremer.fr/doc/00137/24814/
Pépin, J. F., Riou, A., and Renault, T. (2008). Rapid and sensitive detection of ostreid herpesvirus 1 in oyster samples by real-time PCR. J. Virol. Methods 149, 269–276. doi: 10.1016/j.jviromet.2008.01.022
Pernet, F., Barret, J., Le Gall, P., Corporeau, C., Dégremont, L., Lagarde, F., et al. (2012). Mass mortalities of Pacific oysters Crassostrea gigas reflect infectious diseases and vary with farming practices in the Mediterranean Thau lagoon, France. Aquacult. Environ. Interact. 2, 215–237. doi: 10.3354/aei00041
Petton, B., Boudry, P., Alunno-Bruscia, M., and Pernet, F. (2015). Factors influencing disease-induced mortality of Pacific oysters Crassostrea gigas. Aquacult. Environ. Interact. 6, 205–222. doi: 10.3354/aei00125
Prado, S., Dubert, J., Romalde, J. L., Toranzo, A. E., and Barja, J. L. (2014). Vibrio ostreicida sp nov., a new pathogen of bivalve larvae. Int. J. Syst. Evol. Microbiol. 64, 1641–1646. doi: 10.1099/ijs.0.051417-0
Prado, S., Romalde, J. L., Montes, J., and Barja, J. L. (2005). Pathogenic bacteria isolated from disease outbreaks in shellfish hatcheries. First description of Vibrio neptunius as an oyster pathogen. Dis. Aquat. Organ. 67, 209–215. doi: 10.3354/dao067209
Ragone Calvo, L. M. R., Calvo, G. W., and Burreson, E. M. (2003). Dual disease resistance in a selectively bred eastern oyster, Crassostrea virginica, strain tested in Chesapeake Bay. Aquaculture 220, 69–87. doi: 10.1016/S0044-8486(02)00399-X
Richards, G. P., Watson, M. A., Needleman, D. S., Church, K. M., and Häse, C. C. (2015). Mortalities of Eastern and Pacific Oyster Larvae caused by the pathogens Vibrio coralliilyticus and Vibrio tubiashii. Appl. Environ. Microbiol. 81, 292–297. doi: 10.1128/AEM.02930-14
Roque, A., Carrasco, N., Andree, K. B., Lacuesta, B., Elandaloussi, L., Gairin, I., et al. (2012). First report of OsHV-1 microvar in Pacific oyster (Crassostrea gigas) cultured in Spain. Aquaculture 324, 303–306. doi: 10.1016/j.aquaculture.2011.10.018
Schikorski, D., Faury, N., Pepin, J. F., Saulnier, D., Tourbiez, D., and Renault, T. (2011a). Experimental ostreid herpesvirus 1 infection of the Pacific oyster Crassostrea gigas: kinetics of virus DNA detection by q-PCR in seawater and in oyster samples. Virus Res. 155, 28–34. doi: 10.1016/j.virusres.2010.07.031
Schikorski, D., Renault, T., Saulnier, D., Faury, N., Moreau, P., and Pepin, J.-F. (2011b). Experimental infection of Pacific oyster Crassostrea gigas spat by ostreid herpesvirus 1: demonstration of oyster spat susceptibility. Vet. Res. 42, 27. doi: 10.1186/1297-9716-42-27
Segarra, A., Pépin, J. F., Arzul, I., Morga, B., Faury, N., and Renault, T. (2010). Detection and description of a particular Ostreid herpesvirus 1 genotype associated with massive mortality outbreaks of Pacific oysters, Crassostrea gigas, in France in 2008. Virus Res. 153, 92–99. doi: 10.1016/j.virusres.2010.07.011
Tamayo, D., Corporeau, C., Petton, B., Quéré, C., and Pernet, F. (2014). Physiological changes in Pacific oyster Crassostrea gigas exposed to the herpesvirus OsHV-1 μvar. Aquaculture 432, 304–310. doi: 10.1016/j.aquaculture.2014.05.023
Tirape, A., Bacque, C., Brizard, R., Vandenbulcke, F., and Boulo, V. (2007). Expression of immune-related genes in the oyster Crassostrea gigas during ontogenesis. Dev. Comp. Immunol. 31, 859–873. doi: 10.1016/j.dci.2007.01.005
Whittington, R. J., Hick, P. M., Evans, O., Rubio, A., Alford, B., Dhand, N., et al. (2015). Protection of Pacific oyster (Crassostrea gigas) spat from mortality due to ostreid herpesvirus 1 (OsHV-1 μVar) using simple treatments of incoming seawater in land-based upwellers. Aquaculture 437, 10–20. doi: 10.1016/j.aquaculture.2014.11.016
Keywords: larvae, Crassostrea gigas, OsHV-1, disease resistance, mortality
Citation: Dégremont L, Morga B, Trancart S and Pépin J-F (2016) Resistance to OsHV-1 Infection in Crassostrea gigas Larvae. Front. Mar. Sci. 3:15. doi: 10.3389/fmars.2016.00015
Received: 17 November 2015; Accepted: 28 January 2016;
Published: 15 February 2016.
Edited by:
Francesca Carella, University of Naples Federico II, ItalyReviewed by:
Colleen A. Burge, University of Maryland, Baltimore County, USAGionata De Vico, University of Naples Federico II, Italy
Noèlia Carrasco, Institut de Recerca i Tecnologia Agroalimentàries, Spain
Copyright © 2016 Dégremont, Morga, Trancart and Pépin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lionel Dégremont, lionel.degremont@ifremer.fr
†Present Address: Suzanne Trancart, Labéo Frank Duncombe, Caen, France
 Lionel Dégremont
Lionel Dégremont Benjamin Morga
Benjamin Morga Suzanne Trancart
Suzanne Trancart Jean-François Pépin
Jean-François Pépin