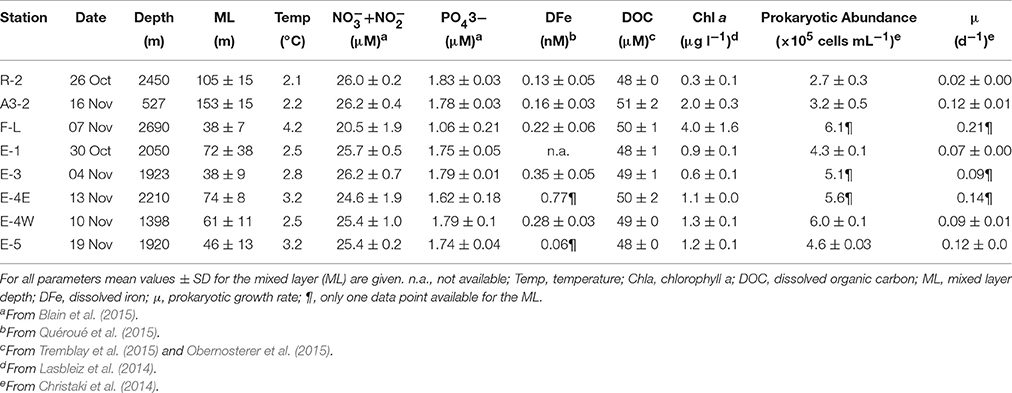
Uptake of Leucine, Chitin, and Iron by Prokaryotic Groups during Spring Phytoplankton Blooms Induced by Natural Iron Fertilization off Kerguelen Island (Southern Ocean)
- 1Antarctic Climate & Ecosystems Cooperative Research Centre, University of Tasmania, Hobart, TAS, Australia
- 2CNRS, Sorbonne Universités, UPMC Univ Paris 06, Laboratoire d'Océanographie Microbienne (LOMIC), Observatoire Océanologique, Banyuls/mer, France
- 3Department of Biological Oceanography, Leibniz Institute for Baltic Sea Research Warnemünde (IOW), Rostock, Germany
Iron and carbon are essential for microbial heterotrophic activity, but the bioavailability of these elements is low in surface waters of the Southern Ocean. Whether the access to iron and carbon differs among phylogenetic groups of prokaryotes is barely known. To address this question we used iron (55FeCl3), and the carbon compounds chitin (3H-Diacetylchitobiose) and leucine (3H-leucine) as model substrates in combination with MICRO-CARD-FISH during spring phytoplankton blooms induced by natural iron fertilization off Kerguelen Island (KErguelen Ocean and Plateau compared Study 2—KEOPS2; October-November 2011). The application of probes at broad phylogenetic levels indicated an overall similar community composition in surface waters at the 8 investigated sites. The relative contributions of the prokaryotic groups to abundance revealed a strong positive relationship with their respective contributions to the leucine-active community (p < 0.0001; r = 0.93). This relationship was much weaker for chitin (p < 0.001; r = 0.51) and absent for iron (p > 0.05; r = 0.26). These results suggest preferential uptake of iron and chitin by some prokaryotic groups. SAR11 and Cytophaga-Flavobacterium-Bacteroides (CFB) were the dominant contributors to the leucine-active community, while CFB and Archaea had the highest contributions to the chitin-active community. By contrast, Gammaproteobacteria, including SAR86, and CFB revealed the highest contributions to the iron-active community. We found several correlations between the group-specific fractions of active cells for a given substrate and most of them included CFB, pointing to the potential importance of microbial interactions for iron and carbon cycling in the Southern Ocean.
Introduction
The Southern Ocean is the largest High Nutrient Low Chlorophyll (HNLC) area where major nutrients are perennially present at high concentrations yet phytoplankton biomass remains low. Surface depletion in iron (Fe) was demonstrated to be the cause of these paradoxical HNLC conditions (Martin et al., 1990). The increase in phytoplankton biomass in response to Fe input and the consequent enhancement of the CO2 uptake were reported in several mesoscale Fe fertilization studies (reviewed in Boyd et al., 2007) and from naturally fertilized regions (Blain et al., 2007; Pollard et al., 2009). These previous studies have reached the same conclusion as to the importance of Fe for the biological pump of carbon in the Southern Ocean.
Despite the numerous studies conducted up to date, it is not well understood how Fe affects heterotrophic prokaryotic metabolism. Fe and carbon are tightly coupled in a suite of metabolic processes crucial for growth. Fe plays a pivotal role in the carbon metabolism because pathways such as glycolysis, the citric acid cycle and processes related to respiration rely on multiple Fe-containing enzymes. A deficiency in Fe ultimately results in a reduction in the metabolic activity with consequences on the energetic status of the organism and its ability to proliferate (Kirchman et al., 2000; Fourquez et al., 2014). Experimental studies using bacterial strains revealed a positive effect of Fe on bacterial growth (Tortell et al., 1996; Fourquez et al., 2014). The response of natural bacterial communities to Fe addition varies among studies (see Obernosterer et al., 2015 for an overview). A common observation is the positive effect of Fe addition on heterotrophic bacteria when incubated in the light and in the presence of phytoplankton. Such effect has been discussed to be due to labile organic matter released by phytoplankton and renders a firm conclusion on Fe or carbon limitation difficult. The picture is further complicated by the potential competition between heterotrophic bacteria and the small representatives of phytoplankton (Fourquez et al., 2015). The close interrelatedness of Fe and carbon renders it challenging to decipher the cycling of these elements within Southern Ocean microbial communities.
In this context, an interesting feature that characterizes surface waters of the Southern Ocean is the low concentration of dissolved organic carbon (DOC, roughly 50 μM; Hansell et al., 2009). This results from the low primary production in Fe-limited surface waters and the large-scale upwelling of low DOC circumpolar Antarctic deep waters. As a consequence, surface water dissolved organic matter (DOM) is characterized by overall low bioavailability. Fe and carbon represent therefore both potentially limiting elements for heterotrophic prokaryotes in Southern Ocean surface waters.
The objective of the present study was to provide a novel perspective on this issue, by investigating the incorporation of Fe and two distinct sources of carbon, leucine and chitin, by major prokaryotic groups in the Southern Ocean using the single-cell approach MICRO-CARD-FISH. In contrast to leucine, a monomeric amino acid, the utilization of the polymer chitin is more complex involving several regulated enzymes as for instance chitinase or β-N-acetyl-hexosaminidases: chitinases cleave the polymer extracellularly into dimers or monomers which are then transported into the cell and further processed (Beier and Bertilsson, 2013). We performed our study during the KEOPS2 project (KErguelen Ocean and Plateau compared Study 2) from 8 October to 30 November 2011 over and downstream of the Kerguelen Plateau in the Southern Ocean. KEOPS2 was performed during the early stage of the phytoplankton bloom that forms annually in the vicinity of the Kerguelen Plateau in response to natural Fe fertilization of the surface waters. During this study we had access to a patchwork of spring phytoplankton blooms induced by large-scale natural Fe fertilization of the Southern Ocean and HNLC-waters, reflecting a range of conditions with respect to the availability of Fe and organic carbon.
Materials and Methods
Sample Sites
Samples were collected in surface waters (20 m depth) at 8 sites in the naturally Fe-fertilized region east of Kerguelen Island and at one site in HNLC waters during Austral spring (Figure 1). The hydrodynamic properties of the study region are presented in detail in Park et al. (2014) and basic biogeochemical parameters are presented in Table 1. The region around Kerguelen Island is characterized by the passage of the Antarctic Circumpolar Current (ACC) that splits into two branches: a main branch circulates to the south of Kerguelen Islands to further join a branch of the Fawn Trough Current (FTC), and a branch that circulates north of Kerguelen Island (Park et al., 2008). A narrow jet of ACC water also flows across the Kerguelen Plateau. This feature corresponds with the northernmost branch of the Polar Front (Park et al., 2014). These hydrographic features result in mesoscale activity that generate contrasted environmental conditions with respect to Fe availability. Station R-2 located south west of Kerguelen Island served as a HNLC station reference. Station F-L represents a bloom site located north of the Polar Front and Station A3-2 is located above the Plateau. Low horizontal advection characterized Station A3-2, Station R-2 and the E-sites (Park et al., 2014), as discussed below. Stations E (E-1, E-3, E-4, and E-5) were located in a recirculation feature south of the Polar Front. Multiple visits were conducted in this feature. The E stations were sampled in a quasi Lagrangian manner (D'Ovidio et al., 2015) and can be considered as a time-series. Briefly, the quasi Lagrangian strategy was based on the deployment of two drifters equipped with GPS and iridium transmission at Station E-1. Their positions were monitored in real time and the subsequent E stations were placed along the trajectory of these drifters. A broader context of the horizontal circulation in the region of interest was provided by two other approaches. Firstly, the trajectories of water parcels were determined by geostrophic currents derived from satellite altimetry (D'Ovidio et al., 2015). This approach computes backward trajectories thereby providing an estimate for the rate at which water parcels are transported by horizontal stirring. Secondly, this altimetry-based approach was validated by the trajectories of 200 World Ocean Experiment-Surface Velocity Program drifters, 48 drifters deployed in an adaptive strategy during the KEOPS2 cruise and 24 drifters during a cruise in January 2014 in the study region (D'Ovidio et al., 2015). Further, 120 trajectories measured between 2011 and 2014 in the study region were collected from the Global Drifter Program. Taken together, this information suggests that the water within the recirculation feature south of the Polar Front is retained over a relatively long time period (roughly 60 days, D'Ovidio et al., 2015).
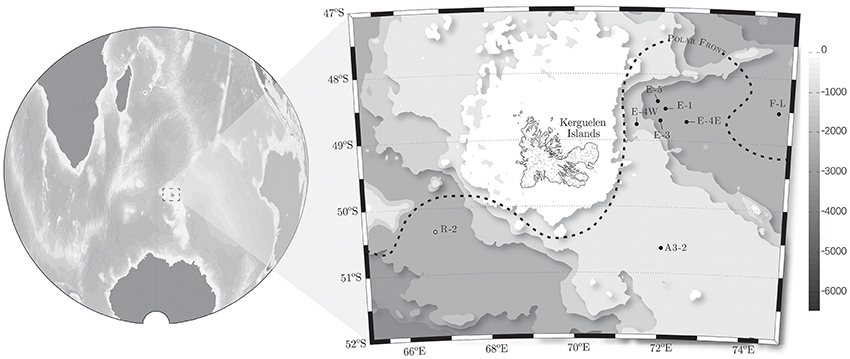
Figure 1. Location of the study area, with a zoom on the Kerguelen Plateau in the Southern Ocean. The positions of the stations located inside the blooms are shown in black dots and the reference station (R-2) outside the bloom is shown as a white dot. The base map represents the bathymetry in meters.
Incubations with Radiolabeled Substrates for Microautoradiography
To determine the single-cell activities of leucine and chitin, raw seawater samples (10 mL) were incubated with either [4,5-3H]-leucine (Perkin Elmer, specific activity 144 Ci mmol−1; 10 nM final concentration) or [3H]-Diacetylchitobiose (Amersham, specific activity 14 Ci mmol−1; 10 nM final concentration) in the dark at in situ temperature for 6–8 h. Controls were fixed with paraformaldehyde (PFA, 2% final concentration) 10 min prior to incubation with the radiolabeled substrates. The incubations were terminated by adding PFA to the samples (2% final concentration). Finally, samples were filtered onto 0.2 μm PC filters (25 mm filter diameter, Nuclepore) and subsequently rinsed with 0.2 μm filtered Milli-Q water. Filters were stored at −20°C until treated.
Incubations with 55Fe are described in detail in Fourquez et al. (2015). Briefly, samples were collected using trace metal clean 10-L modified Niskin bottles set up on an autonomous model 1018 trace metal rosette especially adapted for trace metal work (General Oceanics Inc., USA; Bowie et al., 2015). The seawater collected was further processed in a clean container under a laminar flow hood (ISO class 5). Seawater was filtered at low pressure (<200 mm Hg) through 0.8 μm acid-washed PC filters (47 mm; Millipore) and incubations were performed on the <0.8 μm fraction. Subsamples (100 mL) of filtered seawater were spiked with 55Fe (as 55FeCl3, specific activity 1.83 × 103 Ci mol−1, Perkin Elmer) at final concentrations of 0.2, 1, 5, or 15 nM of 55Fe and they were incubated in the dark at in situ temperature for 1 and up to 7 days. Subsamples (10 mL) were collected on 0.2-μm PC filters (25 mm diameter; Millipore) and rinsed with the washing solution Ti-citrate-EDTA for 2 min. This step allows eliminating 55Fe non-incorporated by cells from the filters. This was followed by 10 rinses of 0.2 μm-filtered seawater of 1 min each (Fourquez et al., 2012). Control treatments of the seawater samples were killed with formaldehyde (2% final concentration) and kept for 1 h at 4°C before the addition of 55Fe. All the filters were dried and kept at −20°C until processed.
The samples for microautoradiography were chosen to (1) limit isotope dilution as determined by saturation curves and (2) reach a quota in 55Fe per cell sufficient for silver grain production during microautoradiography. Fourquez et al. (2012) showed previously that the fraction of cells associated with silver grains was linked to the duration of the 55Fe incubation. For the present data set, we chose the samples at the time point when these two conditions were met. We found that 5 nM (St. A3-2 and E-4E) and 15 nM (St. E-4W and E-5) of 55Fe final concentration and a minimal incubation time of 3 days were necessary to meet these requirements.
Catalyzed Reporter Deposition–Fluorescence In situ Hybridization and Microautoradiography (MICRO-CARD-FISH)
We used catalyzed reporter deposition–fluorescence in situ hybridization (CARD-FISH) on filter sections from seawater samples following the incubation with the radiolabeled compounds. The relative abundances of the bacterial groups and Archaea were determined using the CARD-FISH protocols described in Sekar et al. (2003) and Alonso-Sáez et al. (2007), respectively. We followed the protocol of Cottrell and Kirchman (2003) for the microautoradiographic development of leucine- and chitin-active cells and the protocol of Fourquez et al. (2012) for the development of Fe-active cells. Briefly, for CARD-FISH, filters were embedded in low-melting-point agarose (0.2% final concentration), dried and dehydrated (96% Ethanol, 1 min). To target bacterial groups, the filters were treated with lysozyme (Fluka, 10 mg mL−1, 100 mmol L−1 Tris [pH 8], 50 mM EDTA) for 1 h at 37°C to allow cell wall permeabilization. To target Archaea, or more specifically Crenarchaeota, cell wall permeabilization was performed as described previously (Tischer et al., 2012). The filters were subsequently washed in Milli-Q water and dehydrated in ethanol (96%) for 1 min. Probe working solution was added at a final concentration of 2.5 ng μL−1 and the hybridization was performed at 35°C for 2 h. Bacterial probes and archaeal probes were labeled with FITC and Alexa Fluor® dyes (dyes 488 or 546, Thermo Fisher Scientific, Waltham, MA, USA), respectively.
The probes Eub338-I, -II, and -III were used for the identification of Bacteria and the Eub338-I antisense probe Non338 was used to determine non-specific binding (Glöckner et al., 1999). The probes SAR11-152R, SAR11-441R, SAR11-542R, SAR11-732R were used to target the SAR11 cluster (Morris et al., 2002) and the probe Ros537 to target members of the Roseobacter-Sulfitobacter-Silicibacter group (Eilers et al., 2000a). To target Gammaproteobacteria, we used the probe Gam42a (Manz et al., 1992) and the probes SAR86 (Eilers et al., 2000b) and SAR92 (Obernosterer et al., 2011) were used to target the SAR86 and SAR92 clusters, respectively. The probe CF319a (Manz et al., 1996) was used to identify Bacteroidetes. To target Archaea, we used the probe Arc915 (Auguet and Casamayor, 2008) and to target Crenarchaeota Marine Group 1 we used the probe Cren554 (Alonso-Sáez et al., 2007). Earlier experiments with probe Arc915 in freshwater samples revealed that this probe hybridized in some cases also with bacterial cells (data not shown). Therefore, in order to test the specificity of probe Arc915 in the current sample set, a few filters were hybridized consecutively with the probes EUBI/II/III (Alexa Fluor® 488 dye) and Arch915 (Alexa Fluor® 546 dye). Inactivation of the horseradish peroxidase between the hybridization steps was performed as described elsewhere (Pernthaler and Pernthaler, 2007). We could not detect any unspecific hybridisation via multi-color CARD-FISH with the probes Arc915 and EubI/II/III.
Following the CARD-FISH step, filters were treated for microautoradiography as detailed in Cottrell and Kirchman (2003). Briefly, after hybridization the filters were deposited with cells facing down onto a glass slide previously dipped into a molten (43°C) solution of NTB2 photographic emulsion (Kodak, diluted 50:50 with milli-Q water). The photographic emulsion was left to solidify on ice for a minimum of 10 min before the slides were stored for autoradiographic exposure times that ranged from 2 to 3 days for leucine and chitin to 7 weeks (Fe). All manipulations were conducted in the darkroom under a safe light. Slides were developed for 2 min in Kodak developer as detailed in Cottrell and Kirchman (2003).
All cells were stained before observation by adding one drop of 4′,6-diamidino-2-phenylindole (DAPI, 2 μg mL−1 final concentration) directly prepared in an antifade mountant (1:4 mixture of Vectashield® and Citifluor®) and subsequently detected in images captured with epifluorescent microscopy. The total number of cells (DAPI-cells), cells affiliated with a specific prokaryotic group and cells that assimilated the radiolabeled compound (with silver grains) were counted using a semi-automated microscope (Olympus BX61) and image analysis software (Microbe Counter software) as described previously (Cottrell and Kirchman, 2003). Data were collected from 10 fields of view with at least 500 cells per sample. The proportions of DAPI-cells that were substrate-active were determined as the fraction of total DAPI-cells with silver grains. The composition of the substrate active community was determined as the fraction of cells with silver grains that are probe-positive. The proportion of a given prokaryotic group that was substrate-active was determined as the fraction of probe-positive cells with silver grains.
For a given sample, multiple hybridizations were done with all probes: CARD-FISH only, and CARD-FISH on samples previously incubated with leucine, chitin or iron. The number of replicates varied between 3 and 10 (all probes included) and the variability (coefficient of variation) was on average 24 ± 14% (n = 44) for “high abundant probes” (>5% FISH) and was on average 57 ± 30% (n = 15) for “low abundant probes” (≤5% FISH) such as SAR92 and Cren554.
Bacterial and archaeal group relative abundance was calculated as percentage of probe positive cells vs. total DAPI-cells counts. The relative abundances of bacterial groups (EUB probes) accounted for 81 ± 4% DAPI-cells (mean ± SD, n = 8) and nearly reached a total cover at the sites where Archaea were investigated 98 ± 6% DAPI-cells (mean ± SD, n = 4). The sum of the relative contribution of SAR86 and SAR92 accounted for 101 ± 26% (mean ± SD, n = 8) of the cells identified as Gammaproteobacteria.
Uptake Experiments
Leucine uptake rates were determined as detailed in Christaki et al. (2014). Briefly, 20 ml triplicate samples plus one trichloroacetic acid (TCA)-killed control were incubated with 3H-leucine for 3–8 h at in situ temperature. Incubations were terminated by adding 5% ice-cold TCA and cells were subsequently collected by filtration on 0.22 μm cellulose acetate filters (Millipore) and washed with ice-cold 5% TCA. Filters were dissolved in 1 mL of ethyl acetate and 10 mL of Ultima Gold scintillation cocktail (Packard) before being counted by liquid scintillation.
Chitin uptake rates were estimated at 4 stations (A3-2, F-L, E-3, and E-4W) using a concentration series bioassay (Wright and Hobbie, 1966; Zubkov and Tarran, 2005). Raw seawater samples were incubated with [3H]-Diacetylchitobiose (Amersham, specific activity 14 Ci mmol−1) at final concentrations of 0.5, 1, 2, 3, and 6 nM. Ten mL of each addition were incubated at in situ temperature in the dark for 5, 7, and 10 h. Controls were fixed with PFA (2% final concentration) 10 min prior to the addition of chitin. Incubations were terminated by the addition of PFA (2% final concentration). Samples were filtered onto 0.2 μm nitrocellulose membranes (25 mm diameter; Nuclepore), and the filters rinsed with 0.2 μm filtered Milli-Q water (Beier and Bertilsson, 2011) before determining the radioactivity by liquid scintillation. Chitin uptake rates were estimated from the linear regression of radioactivity against incubation time. The calculated chitin turnover times were plotted against the added concentrations of chitin. The slope of the linear regression provides an estimate of the chitin turnover rate (r2 = 0.918 for E-3; r2 = 0.944 for E-4W; r2 = 0.532 for F-L; r2 = 0.912 for A3-2).
Fe uptake rates were determined in triplicates plus one formol-killed control at all stations following 24 h incubations with 55Fe, when a steady state was achieved (Fourquez et al., 2015). To measure intracellular Fe uptake, cells were collected on nitrocellulose filters (Nuclepore) and immediately rinsed first with the Ti–citrate–EDTA washing solution and then with 0.22 μm filtered seawater as detailed in Fourquez et al. (2015). The radioactivity on filters was counted by liquid scintillation after they dissolved for 24 h in 10 mL of Filter-Count scintillation cocktail (PerkinElmer). Fe uptake rates were calculated considering in situ Fe concentrations and corrected for background radioactivity (see Fourquez et al., 2015).
For each uptake experiment, the radioactivity (in disintegration per minute, DPM) was determined using the Tricarb® scintillation counter and the DPMs of the controls were subtracted from the live incubations.
Statistical Analyses
We used Pearson correlation to determine the relationships between (1) the relative contributions of prokaryotic groups to total abundance and to total substrate active-cells and (2) the percentages of substrate-active cells among different groups. To test whether the percentages of substrate-active cells and their contributions to the substrate-active community were significantly different among prokaryotic groups we applied the Kruskall-Wallis one-way analysis of variance on ranks and the post-hoc Dunn's test if a significant difference was detected by the former. We used this non-parametric test because of the unequal number of observations among prokaryotic groups. We used Pearson correlation to determine the relationships between the environmental parameters measured at each sampling location and the percent of total and group-specific active cells assessed by MICRO-CARD-FISH. This analysis was done for the three substrates of the study. Values for bacterial abundance, Chl a, inorganic nutrients, and bacterial heterotrophic production were log transformed and the percentage of active cells was arcsine transformed for this analysis. Transformed data were according to the Shapiro-Wilk test still not significantly normally distributed, but visual examination of the data revealed that their distribution after transformation was closer to the normal distribution. We performed our test on transformed data using Pearson correlation. We also tested for correlation on both, raw and transformed data using Spearman-rank correlation analysis. No difference in the result was obtained by these statistical approaches.
Results
Environmental Context
The reference Station R-2, located west of the Kerguelen Plateau, revealed low dissolved Fe (DFe, 0.13 ± 0.05 nM) and chlorophyll a concentrations (Chl a, 0.3 ± 0.1 μg L−1) that are typical for surface HNLC waters (Table 1). At this station, prokaryotic abundance, prokaryotic biomass production and respiration (Christaki et al., 2014) and prokaryotic growth rates were lower than those in surface waters of the Fe-fertilized stations (Table 1). A patchwork of phytoplankton blooms induced by natural Fe-fertilization developed east of Kerguelen Island. These blooms varied in terms of phytoplankton biomass and community composition (Lasbleiz et al., 2016) reflecting different stages of development. Above the plateau, at Station A3-2, concentrations of DFe in the surface mixed layer (0.16 ± 0.03 nM) were slightly higher than in HNLC waters, and concentrations of Chl a exceeded those in HNLC waters by 6-fold (2.0 ± 0.03 μg L−1), reflecting a well-developed spring bloom at this site, sustained by recent and persistent Fe-fertilization (Trull et al., 2015). The response of heterotrophic microbes was reflected by 6-fold higher bacterial growth rates (0.12 d−1) than in HNLC surface waters (0.02 d−1; Table 1). At Station E4-W, DFe concentrations (0.17 ± 0.03 nM) were similar to those at Station A3-2, but Chl a concentrations (1.3 ± 0.1 μg L−1) and prokaryotic growth rates (0.09 d−1) remained lower than at the Plateau station. The highest Chl a concentration (4.0 ± 1.6 μg L−1) was observed at Station F-L, located north of the Polar Front. The concurrent relatively low concentrations of nitrate (20 μM as compared to 25 μM at all other sites) indicate an intense bloom event, induced by recent and brief Fe-fertilization (D'Ovidio et al., 2015). DFe concentrations at Station F-L (0.22 ± 0.06 nM) were among the highest determined in the study region (Table 1), and prokaryotic abundance and growth rates (0.21 d−1) exceeded those observed at all other stations in the Fe-fertilized region. Fe-fertilization appears to be less recent but more persistent within the recirculation feature south of the Polar Front (D'Ovidio et al., 2015). In this regard, the Stations E-1, E-3, E-4E and E-5 were sampled in a quasi Lagrangian manner and can be considered to some extent as a succession of sampling in the same water mass over time (D'Ovidio et al., 2015). Concentrations of Chl a increased from 0.6 to 1.2 μg L−1, accompanied by increases in prokaryotic growth rate from 0.07 to 0.12 d−1. The unchanged low concentrations of dissolved organic carbon (mean 49 ± 1 μM; Obernosterer et al., 2015; Tremblay et al., 2015) across all sampling sites are likely a result of the rapid consumption of phytoplankton-derived DOM.
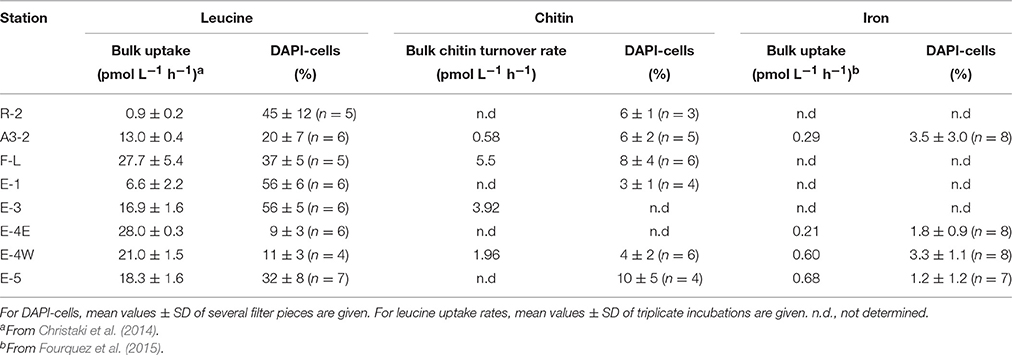
Bulk uptake rates of leucine ranged between 0.9 and 28 pmol L−1 h−1 (n = 8; Christaki et al., 2014) and those of chitin varied between 0.6 and 5.5 pmol L−1 h−1 (n = 4, Table 2). Prokaryotic Fe uptake rates were substantially lower (0.21–0.68 pmol L−1 h−1, n = 4; Fourquez et al., 2015, Table 2). Similarly, the percent DAPI-cells that were substrate-active was highest for leucine (range 11–56%) and varied between 3 and 10% for chitin and between 1 and 4 % for Fe.
Abundance of Prokaryotic Groups
Our CARD-FISH results, using probes at varying phylogenetic levels, reveal an overall similar community composition at all sites (Figure 2A). The dominant bacterial groups were SAR11 (range 34–61% of total DAPI-stained cells) and CFB (range 42–48%, except Station E-1: 25% of total DAPI-cells) (Figure 2B). Roseobacter (3–10% of total DAPI-cells) and the Gammaproteobacterial groups SAR86 (10–13%, except E-4E: 19% of total DAPI-cells) and SAR92 (0.4–6% of total DAPI-cells) revealed lower relative abundances. SAR86 accounted for a large fraction of Gammaproteobacteria (55–95%) at all sites. The relative abundance of Archaea varied between 2 and 19% of total DAPI-cells and Crenarchaeota accounted for 1–63% of Archaeal abundance at the 4 sites considered (Figure 2B). Interestingly our analyses revealed higher abundances of SAR92 and Archaea at the Fe-fertilized sites as compared to the HNCL-site R-2.
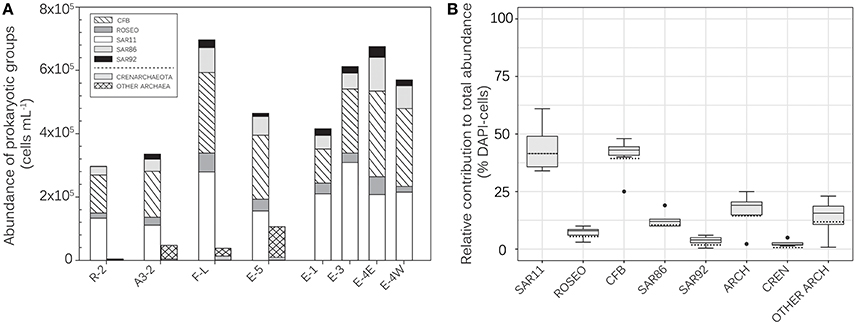
Figure 2. Absolute (A) and relative (B) abundances of prokaryotic groups in surface waters of the study region. Except for Station E-5, no Archaea data are available for the E-sites. The black and the dashed lines across the box represent respectively the median and the mean. The ends of the boxes define the 25th and 75th percentiles and error bars define the 10th and 90th percentiles (data pooled from all stations shown in A). Outliers are plotted in black dots. Values for “other Archaea” were calculated as the difference between Archaea and Crenarchaeota. ARCH, Archaea; CREN, Crenarchaeota; CFB, Cytophaga/Flexibacter/Bacteroides; ROSEO, Roseobacter clade; SAR11, SAR11 cluster; SAR 86, SAR86 cluster; SAR92, SAR92 cluster.
Contribution of Prokaryotic Groups to the Substrate Active Community
The single-cell approach MICRO-CARD-FISH provides two lines of informations: First, the contribution of a prokaryotic group to the bulk substrate active community and second, the fraction of substrate active cells within a prokaryotic group. We observed a pronounced positive relationship between the contribution to abundance and to the leucine-active community (p < 0.0001; r = 0.93; n = 47) across all sites and including all prokaryotic groups (Figure 3A). By contrast, different patterns emerged for chitin and Fe. We observed both more (as indicated by data points above the 1:1 line in Figure 3) and less (as indicated by data points below the 1:1 line Figure 3A) single-cell use of these substrates by the prokaryotic groups than their abundance would suggest. A less pronounced, but still significant correlation was found between these two variables for chitin (p < 0.001; r = 0.51; n = 38), while no significant correlation was found for Fe (p > 0.05; r = 0.32; n = 18).
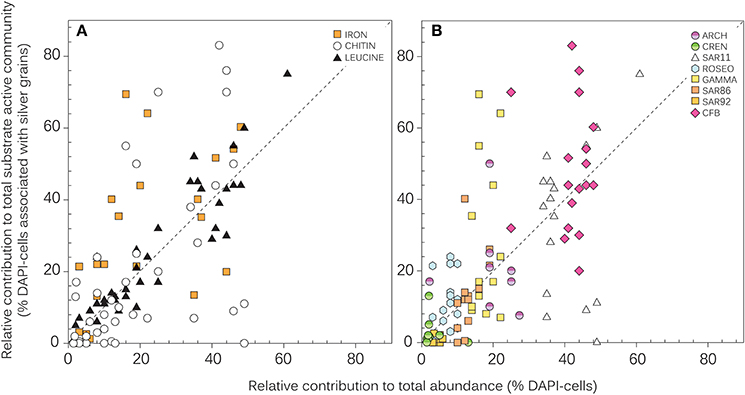
Figure 3. Relationship between the relative contributions of prokaryotic groups to total abundance (% DAPI-cells) and to the total substrate active community (% DAPI-cells associated with silver grains) for the three substrates (A) and for different bacterial groups (B). All stations are pooled. Pearson correlations between the relative contributions to abundance and to the total substrate active community are respectively r = 0.93 (p < 0.00001), r = 0.51 (p < 0.001), and r = 0.26 (p > 0.05) for leucine, chitin, and iron. The dotted line indicates the 1:1 ratio. ARCH, Archaea; CREN, Crenarchaeota; CFB, Cytophaga/Flexibacter/Bacteroides; ROSEO, Roseobacter clade; SAR11, SAR11 cluster; SAR86, SAR86 cluster; SAR92, SAR92 cluster.
To better understand this variability among the three substrates, we present the same data set by highlighting the phylogenetic identity of the bacterial and archaeal groups (Figure 3B). Despite the overall similar composition of the community among stations (Figure 2A), the contribution of distinct bacterial groups and Archaea to the uptake of chitin and Fe varied considerably. As indicated by the positive correlation, the contribution of specific prokaryotic groups to leucine uptake (Figure 4A) was overall reflected by their respective contributions to bulk abundance: in both, the total and the leucine active community, SAR11 represented the highest fraction of community members followed by CFB (Figure 2B). The contribution of SAR11 to the leucine active community (mean 54 ± 11%, n = 8) was significantly higher than that of all the other prokaryotic groups (mean 2–34%, n = 4–8; p < 0.001), except that of CFB (mean 37 ± 7%; n = 8). The contribution of CFB was significantly higher than the contributions of Roseobacter and Crenarcheaota (p < 0.001).
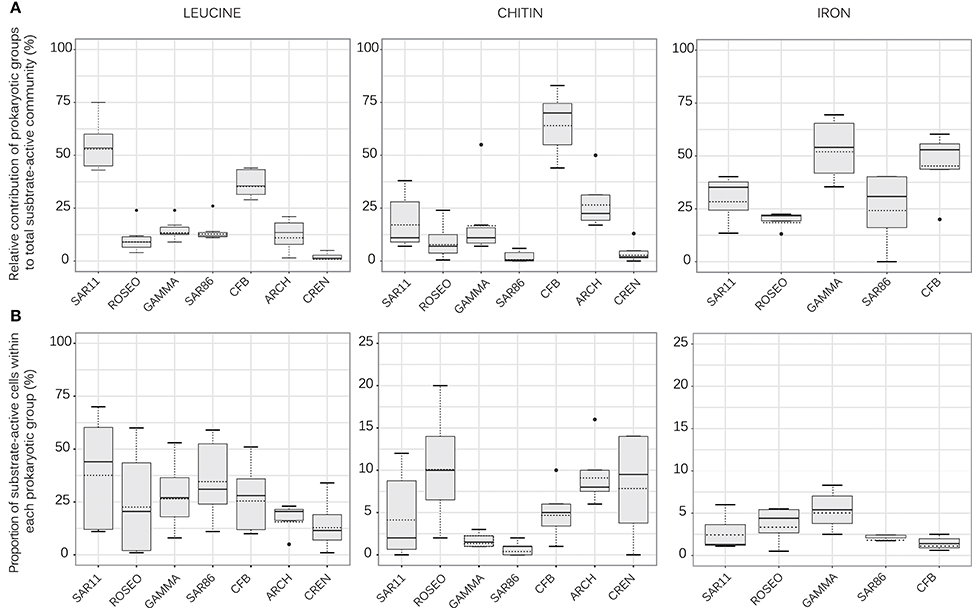
Figure 4. Box plots displaying (A) the relative contribution of prokaryotic groups to the total leucine-, chitin-, and iron- active community (%) and (B) the proportion of leucine-, chitin-, and iron- active cells within each prokaryotic group (%)- note that scale is different for chitin and iron as compared to leucine). The black and the dashed lines across the box represent respectively the median and the mean (all stations included). The ends of the boxes define the 25th and 75th percentiles and error bars define the 10th and 90th percentiles. Outliers are plotted in black dots. ARCH, Archaea; CREN, Crenarchaeota; CFB, Cytophaga/Flexibacter/Bacteroides; ROSEO, Roseobacter clade; SAR11, SAR11 cluster; SAR86, SAR86 cluster; SAR92, SAR92 cluster.
The chitin and Fe-active communities revealed, however, different patterns than the overall community composition. The chitin active community was dominated by CFB and Archaea, but not by SAR11 (Figure 4A). The contribution of CFB (mean 66 ± 15, n = 6) was significantly higher than that of Roseobacter (mean 9 ± 9, n = 6), SAR86 (mean 2 ± 3, n = 5) and Crenarchaeota (mean 4 ± 6, n = 4) (p < 0.001). In contrast, Gammaproteobacteria (mean 53 ± 16, n = 4) and CFB (mean 47 ± 18, n = 4) accounted for the largest fractions of the Fe-active community.
Fraction of Cells That Are Substrate Active
The fraction of substrate-active cells within a prokaryotic group revealed considerable variations among the sampling sites, and this was most pronounced for leucine (Figure 4B). This could be due to the overall higher percentage of DAPI-cells incorporating this substrate (Table 2). The mean proportion of leucine-active cells varied between 15% (Crenarchaeota) and 44% (SAR11). The fractions of cells taking up chitin revealed more pronounced differences among groups. It was interesting to note that the fractions of chitin-active Roseobacter, Archaea and Crenarchaeota (means of 8–10%) were higher than those of Gammaproteobacteria, more specifically SAR86, and SAR11 (means of 1–2%). With respect to Fe, Gammaproteobacteria and Roseobacter had slightly higher fractions of active cells (means of 4%) than SAR11 and CFB (means of 1–2%). However, due to the large variability across sites, none of these differences were significantly different.
We further explored what factors could explain the variability in the fraction of substrate-active cells across sites. We did not obtain any significant correlation between environmental variables (bacterial abundance, Chl a, inorganic nutrients, and bacterial heterotrophic production) and the substrate-specific single-cell activities of the different groups. However, we did obtain several significant correlations between the percent of active cells among different groups for a given substrate (Figure 5). Interestingly, the majority of the correlations were associated with CFB. The number CFB chitin-active cells was positively correlated with that of Gammaproteobacteria and of SAR11, and the number of CFB leucine-active cells was positively correlated with that of Gammaproteobacteria. The number of CFB Fe-active cells was positively correlated with that of Roseobacter.
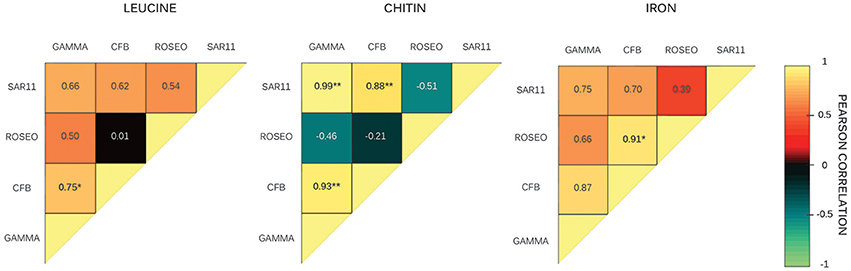
Figure 5. Pearson correlations between the fractions of substrate active cells among bacterial groups for the three different compounds at 4 stations. GAMMA, Gammaproteobacteria; CFB, Cytophaga/Flexibacter/Bacteroides; ROSEO, Roseobacter clade; SAR11, SAR11 cluster. Pearson correlation value is indicated inside each box. Correlations among groups that are statistically significant at 95 or 99% of confidence level are marked with one or two asterisks, respectively.
Discussion
By investigating the single-cell activities of several prokaryotic groups, our study provides a new perspective on the microbial cycling of carbon and Fe in the Southern Ocean. We observed that all investigated prokaryotic groups assimilated Fe and the two distinct carbon substrates leucine and chitin. While the contribution of the prokaryotic groups to the leucine-active community was strongly driven by their relative abundances, their contributions to chitin and Fe uptake appear to be uncoupled from abundance. Our results illustrate pronounced differences in the composition of the leucine, chitin and Fe-active communities and also suggest a preferential use of chitin and Fe. We discuss that interactions among groups in the access of these substrates could explain this pattern.
Our observation of the assimilation of the amino acid leucine by all major prokaryotic groups confirms a large number of studies in temperate and polar environments (see for overviews del Giorgio and Gasol, 2008; Kirchman, 2016). The strong correlation between the contribution to abundance and to the bulk leucine-active community could indicate a non-preferential uptake of this substrate and bottom-up control of heterotrophic prokaryotic activity (Cottrell and Kirchman, 2003). The present study took place in early spring when prokaryotic heterotrophic activity is greatly stimulated by the release of phytoplankton-derived DOM in the study region (Christaki et al., 2014), which in turn affects community composition (Landa et al., 2015). Both C and Fe were identified as growth-limiting factors (Obernosterer et al., 2015). Interestingly, the contributions of several bacterial groups at a fine phylogenetic level (>99% identity of the 16S rRNA gene) to abundance and leucine incorporation were also strongly positively correlated during the declining phase of the bloom above the Kerguelen Plateau (Obernosterer et al., 2011), concomitant with high virus-mediated prokaryotic mortality (Malits et al., 2014). These results could suggest that the access to the limiting resource has a stronger shaping effect on the prokaryotic community composition than top-down control in these perennially cold waters.
In contrast to readily bioavailable low molecular weight substrates, the assimilation of polymers requires more specialized pathways, and the lack of these in some prokaryotic taxa could lead to the use of these substrates by a smaller range of groups (Zimmerman et al., 2013). In the present study, we used the dimer N-Diacteylglucosamine, composed of the two amino sugars N-acetylglucosamines (NAG), as a model for an abundant polymeric compound in the ocean (Souza et al., 2011). This substrate is taken up and subsequently cleaved intracellularly in the periplasmatic space or in the cytoplasm by β-N-acetyl-hexosaminidases into NAG (Beier and Bertilsson, 2013). Even though the uptake of the soluble chitin dimer N-Diacteylglucosamine does not prove the active involvement of cells into chitin polymer degradation, an earlier study could demonstrate that members of bacterial groups that incorporated this substrate at high frequencies were typically also highly abundant in the biofilm attached to chitin particles (Beier and Bertilsson, 2011).
Our observation that CFB are key players during the degradation of chitin confirms previous MICRO-CARD-FISH studies in marine (Cottrell and Kirchman, 2003) and freshwater systems (Beier and Bertilsson, 2011). In chitin-amended microcosms performed in the Southern Ocean the growth of Bacteroidetes, more specifically the genus Reichenbachiella, was stimulated (Wietz et al., 2015). Whole genome sequencing of several strains belonging to this phylum provide the metabolic underpinning of these observations (Bauer et al., 2006; Tang et al., 2012; Kabisch et al., 2014). Interestingly, CFB were the only group that had high contributions to both the chitin- and Fe-active communities in our study. This feature could represent an ecological advantage of CFB that could in part explain their high abundances in surface waters of the Southern Ocean (present study; Simon et al., 1999; Obernosterer et al., 2011) characterized by low concentrations in bioavailable Fe and C.
A more surprising result is the relatively high fraction of chitin-active Archaea, and thus their potential role in the degradation of this substrate. Chitinase genes were detected in the genomes of several crenarchaeotal and euryarchaeotal strains (Staufenberger et al., 2012; García-Fraga et al., 2014, and references therein), but to the best of our knowledge not yet in the abundant marine group I Thaumarchaeota or marine group II (belonging to Euryarchaeota) that are likely abundant among the detected cells in the current study. Crenarchaeota and Euryarchaeota were both shown to assimilate a range of organic substrates and CO2 (Teira et al., 2004; Herndl et al., 2005; Kirchman et al., 2007). Marine group I Thaumarchaeota are considered to be chemolithotrophs and more abundant in deeper layers (Qin et al., 2014; Zhang et al., 2015). In contrast, marine group II Euryarchaeota are mostly heterotrophs and abundant in ocean surface water. Marine group II were suggested to live as particle-attached polymer degraders (Iverson et al., 2012). In line with this current knowledge concerning the ecology of Archaea it seems reasonable to believe that, even if so far no chitinase genes within the three sequenced genomes of marine group II were discovered, some members of this group could be active chitin degraders. Our results demonstrate that members of both Euryarchaeota and Crenarchaeota, and most likely Thaumarchaeota, are involved at least in the final steps of the complex process of chitin degradation.
Copepods are considered the most important source of chitin in the environment (Souza et al., 2011). Exoskeletons, including molts from copepods and marine invertebrate larvae contain chitin (Gooday, 1990) and they could represent one source of this polymer in our study. Mesozooplankton responded to the early stage of the phytoplankton bloom with the production of larvae of copepods and euphausiids (Carlotti et al., 2015). The overall biomasses remained, however, low as compared to those observed during the declining phase of the Kerguelen bloom in late summer (Carlotti et al., 2008). Another potential source that has been given less attention are diatoms, the dominant phytoplankton in the study region (Lasbleiz et al., 2014). In surface waters, heterotrophic prokaryotic activity was strongly driven by phytoplankton-derived organic matter (Christaki et al., 2014; Landa et al., 2015). Whether chitin is provided as growth substrate by diatoms is a particularly interesting question in this context. The genus Thalassiosira was reported to contain chitin synthetase genes, implying chitin production (Durkin et al., 2009). In the present study, small centric diatoms (<25 μm) principally composed of several Thalassiosira species were present at all sites, but accounted for about 60% of total diatom biomass at the Polar Front Station F-L (Laurenceau-Cornec et al., 2015; Lasbleiz et al., 2016). At this site, we observed the highest bulk chitin turnover rates concomitantly with high archaeal and crenarchaeotal abundances (19 and 5% of total DAPI-cells, respectively) and cell-specific chitin activities (14 and 16% of chitin active archaeal and crenarchaeotal cells, respectively) that were higher than at any other site. Whether chitin production and consumption favors associations between specific groups of phytoplankton and Archaea as observed for diverse bacterial taxa (Amin et al., 2012; Durham et al., 2015) remains to be explored. Chitin was found to support between 5% to up to 30% of the bacterial production in the Delaware Estuary (Kirchman and White, 1999). These quantitative estimates together with the observation that all investigated prokaryotic groups contributed to chitin uptake further confirm that this compound is a widespread bioavailable source of organic carbon and nitrogen for prokaryotic growth in the ocean.
Whether Fe uptake strategies and demand differ among microbial taxa and how community composition might in turn affect Fe cycling in the Southern Ocean is an essential, but largely unresolved question. A survey of representative marine bacteria revealed the presence of several pathways for Fe acquisition in most genomes, while siderophore biosynthesis genes were reported to be rare (Hopkinson and Barbeau, 2012). Global metagenomic surveys point to spatial differences in Fe metabolism strategies, including uptake and storage and these studies established indirect links to the distribution of microbial taxa (Desai et al., 2012; Toulza et al., 2012). These genomic based observations complement earlier laboratory experiments demonstrating the capability of marine bacterial strains to consume Fe in different forms, such as inorganic Fe, organic ligand-bound Fe (Granger and Price, 1999; Guan et al., 2001; Weaver et al., 2003) or as heme (Hopkinson et al., 2008), and to produce siderophores (Yamamoto et al., 1994; Martinez et al., 2000). Interestingly, Pelagibacter, a member of the most abundant bacterial group in the study region, lacks entirely most Fe transport and siderophore biosynthesis genes, and thus depend on inorganic Fe uptake (Hopkinson and Barbeau, 2012). According to metagenomic and transcriptomic analyses this genera has non-specialized Fe uptake systems, largely lacks the ability to take up Fe chelates directly and would rather use inorganic Fe (Smith et al., 2010; Thompson et al., 2011). In Candidatus Pelagibacter ubique, the presence of Fe3+ transporters and the lack of TonB-dependent transporters is suspected to be a niche specific adaptation (Smith et al., 2010).
Our study is the first to investigate Fe uptake by different bacterial groups. Because of the low specific activity of 55Fe, the detection of silver grains is dependent on the cellular 55Fe quota (Fourquez et al., 2012). This methodological constraint probably explains the overall low fraction of cells associated with silver grains. The Fe-positive fraction could represent cells with highest Fe uptake activities or requirements, or those cells that have potential Fe-storage capacities. We observed in the present study that all investigated bacterial groups assimilated the micronutrient Fe in a range of environmental conditions, but our study highlights Gammaproteobacteria, in particular SAR86, and CFB to be important players. TonB-dependent transporters were particularly abundant in members belonging to Gammaproteobacteria and CFB across the GOS data set (Tang et al., 2012). These transporters promote the uptake of diverse substrates present at low concentrations, and they have high affinity with Fe complexes (Moeck and Coulton, 1998). In the region of the present study where Fe and carbon are limiting nutrients (Obernosterer et al., 2015), the presence of TonB-dependent transporters could provide an advantage to members of these bacterial groups. Previous studies report strain-specific differences in the cellular Fe quota under both Fe replete and Fe limited conditions (Granger and Price, 1999; Fourquez et al., 2014). The question, however, of whether Fe quota differ among phylogenetically diverse bacterial groups were not addressed in these studies. How variable this basic physiological characteristic is among taxa and how this affects microbial Fe and carbon cycling needs further investigation.
For the three compounds used in the present study we observed high variability in the fraction of substrate-active cells across sites. None of the environmental variables could explain this observation. Possibly small changes in the concentrations of bioavailable DOM and Fe, not measurable with present chemical techniques, could in part account for the observed variability. Changes in within group community composition could also play a role. Using 454 pyrosequencing, the bacterial communities revealed pronounced differences among sites (Landa et al., 2015), suggesting the metabolic capacities for the uptake of the three substrates to vary at finer phylogenetic levels than those resolved via the probes used in this study. An interesting hypothesis in this context is the possible interaction among bacterial groups as suggested by the positive relationships between substrate-active cells belonging to different bacterial groups. The temporal succession of prokaryotic communities driven by specific metabolic capabilities and substrate preferences could represent synergistic interactions in the degradation of DOM (McCarren et al., 2010; Teeling et al., 2012).
Interactions among taxa might be of particular relevance if “leaky” components are involved that are available extracellularly and provided by the activity of a restricted group of community members (Morris et al., 2012). In contrast to leucine, for both, Fe via the active release of siderophores (Granger and Price, 1999; Martinez et al., 2003; Hopkinson and Barbeau, 2012; Morris et al., 2012) and chitin via the extracellular production of hydrolyses products of the chitin polymer from released chitinases (Beier and Bertilsson, 2011), species interactions may therefore be specifically favored. How these and possibly other types of interactions might facilitate the utilization of the limiting resources carbon and Fe in the Southern Ocean and thereby affect the cycling of these elements is a challenging question for future studies.
Author Contributions
Contributed to conception, design, and acquisition of data: MF, EJ, RH and IO. Contributed to analysis and interpretation of data: MF, IO, SB, EJ, and RH. Drafted and revised the article: MF, IO, and SB. Approved the submitted version for publication: MF, IO, SB, EJ, and RH.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We a grateful to S. Blain, the PI of the KEOPS2 project for providing us the opportunity to participate to this cruise. We thank the chief scientist B. Quéguiner, the captain Bernard Lassiette and the crew of the R/V Marion Dufresne for their enthusiasm and help aboard. S. Bertilsson kindly provided radiolabeled N-Diacteylglucosamine. Thanks to P. Catala and S. Rachik for their help with MICRO-CARD-FISH and microscopic analyses. Two reviewers provided constructive comments on a previous version of this manuscript. We also thank Gustave Chiffard for insightful discussions during the development of this project. This work was supported by the French Research program of the INSU-CNRS LEFE−CYBER (Les enveloppes fluides et l'environnement–Cycles biogéochimiques, environnement et ressources), the French ANR (Agence Nationale de la Recherche, SIMI-6 program), the French CNES (Centre National d'Etudes Spatiales) and the French Polar Institute IPEV (Institut Polaire Paul−Emile Victor).
References
Alonso-Sáez, L., Arístegui, J., Pinhassi, J., Gómez-consarnau, L., González, J. M., Vaqué, D., et al. (2007). Bacterial assemblage structure and carbon metabolism along a productivity gradient in the NE Atlantic Ocean. Aquat. Microb. Ecol. 46, 43–53. doi: 10.3354/ame046043
Amin, S. A., Parker, M. S., and Armbrust, E. V. (2012). Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684. doi: 10.1128/MMBR.00007-12
Auguet, J.-C., and Casamayor, E. O. (2008). A hotspot for cold crenarchaeota in the neuston of high mountain lakes. Environ. Microbiol. 10, 1080–1086. doi: 10.1111/j.1462-2920.2007.01498.x
Bauer, M., Kube, M., Teeling, H., Richter, M., Lombardot, T., Allers, E., et al. (2006). Whole genome analysis of the marine Bacteroidetes‘Gramella forsetii’ reveals adaptations to degradation of polymeric organic matter. Environ. Microbiol. 8, 2201–2213. doi: 10.1111/j.1462-2920.2006.01152.x
Beier, S., and Bertilsson, S. (2011). Uncoupling of chitinase activity and uptake of hydrolysis products in freshwater bacterioplankton. Limnol. Oceanogr. 56, 1179–1188. doi: 10.4319/lo.2011.56.4.1179
Beier, S., and Bertilsson, S. (2013). Bacterial chitin degradation-mechanisms and ecophysiological strategies. Front. Microbiol. 4:149. doi: 10.3389/fmicb.2013.00149
Blain, S., Capparos, J., Guéneuguès, A., Obernosterer, I., and Oriol, L. (2015). Distributions and stoichiometry of dissolved nitrogen and phosphorus in the iron-fertilized region near Kerguelen (Southern Ocean). Biogeosciences 12, 623–635. doi: 10.5194/bg-12-623-2015
Blain, S., Quéguiner, B., Armand, L., Belviso, S., Bombled, B., Bopp, L., et al. (2007). Effect of natural iron fertilization on carbon sequestration in the Southern Ocean. Nature 446, 1070–1074. doi: 10.1038/nature05700
Bowie, A. R., van der Merwe, P., Quéroué, F., Trull, T., Fourquez, M., Planchon, F., et al. (2015). Iron budgets for three distinct biogeochemical sites around the Kerguelen archipelago (Southern Ocean) during the natural fertilisation experiment KEOPS-2. Biogeosciences 12, 4421–4445. doi: 10.5194/bg-12-4421-2015
Boyd, P. W., Jickells, T., Law, C. S., Blain, S., Boyle, E. A., Buesseler, K. O., et al. (2007). Mesoscale iron enrichment experiments 1993–2005: synthesis and future directions. Science 315, 612–617. doi: 10.1126/science.1131669
Carlotti, F., Jouandet, M.-P., Nowaczyk, A., Harmelin-Vivien, M., Lefèvre, D., Richard, P., et al. (2015). Mesozooplankton structure and functioning during the onset of the Kerguelen phytoplankton bloom during the KEOPS2 survey. Biogeosciences 12, 4543–4563. doi: 10.5194/bg-12-4543-2015
Carlotti, F., Thibault-Botha, D., Nowaczyk, A., and Lefèvre, D. (2008). Zooplankton community structure, biomass and role in carbon fluxes during the second half of a phytoplankton bloom in the eastern sector of the Kerguelen Shelf (January–February 2005). Deep Sea Res. Part II Top. Stud. Oceanogr. 55, 720–733. doi: 10.1016/j.dsr2.2007.12.010
Christaki, U., Lefèvre, D., Georges, C., Colombet, J., Catala, P., Courties, C., et al. (2014). Microbial food web dynamics during spring phytoplankton blooms in the naturally iron-fertilized Kerguelen area (Southern Ocean). Biogeosciences 11, 6739–6753. doi: 10.5194/bg-11-6739-2014
Cottrell, M. T., and Kirchman, D. L. (2003). Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48, 168–178. doi: 10.4319/lo.2003.48.1.0168
D'Ovidio, F., Della Penna, A., Trull, T. W., Nencioli, F., Pujol, I., Rio, M. H., et al. (2015). The biogeochemical structuring role of horizontal stirring: Lagrangian perspectives on iron delivery downstream of the Kerguelen plateau. Biogeosciences 12, 5567–5581. doi: 10.5194/bgd-12-779-2015
del Giorgio, P. A., and Gasol, J. M. (2008). “Physiological structure and single-cell activity in marine bacterioplankton,” in Microbial Ecology of the Oceans, 2nd Edn., ed D. L. Kirchman (Hoboken, NJ: John Wiley & Sons, Inc.), 243–298. doi: 10.1002/9780470281840.ch8
Desai, D. K., Desai, F. D., and Laroche, J. (2012). Factors influencing the diversity of iron uptake systems in aquatic microorganisms. Front. Microbiol. 3:362. doi: 10.3389/fmicb.2012.00362
Durham, B. P., Sharma, S., Luo, H., Smith, C. B., Amin, S. A., Bender, S. J., et al. (2015). Cryptic carbon and sulfur cycling between surface ocean plankton. Proc. Natl. Acad. Sci. U.S.A. 112, 453–457. doi: 10.1073/pnas.1413137112
Durkin, C. A., Mock, T., and Armbrust, E. V. (2009). Chitin in diatoms and its association with the cell wall. Eukaryot. Cell 8, 1038–1050. doi: 10.1128/EC.00079-09
Eilers, H., Pernthaler, J., and Amann, R. (2000a). Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl. Environ. Microbiol. 66, 4634–4640. doi: 10.1128/AEM.66.11.4634-4640.2000
Eilers, H., Pernthaler, J., Glöckner, F. O., and Amann, R. (2000b). Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66, 3044–3051. doi: 10.1128/AEM.66.7.3044-3051.2000
Fourquez, M., Obernosterer, I., and Blain, S. (2012). A method for the use of the radiotracer 55Fe for microautoradiography and CARD-FISH of natural bacterial communities. FEMS Microbiol. Lett. 337, 132–139. doi: 10.1111/1574-6968.12022
Fourquez, M., Obernosterer, I., Davies, D. M., Trull, T. W., and Blain, S. (2015). Microbial iron uptake in the naturally fertilized waters in the vicinity of the Kerguelen Islands: phytoplankton–bacteria interactions. Biogeosciences 12, 1983–1992. doi: 10.5194/bg-12-1893-2015
Fourquez, M., Devez, A., Schaumann, A., Guéneuguès, A., Jouenne, T., Obernosterer, I., et al. (2014). Effects of iron limitation on growth and carbon metabolism in oceanic and coastal heterotrophic bacteria. Limnol. Oceanogr. 59, 349–360. doi: 10.4319/lo.2014.59.2.0349
García-Fraga, B., da Silva, A. F., López-Seijas, J., and Sieiro, C. (2014). Functional expression and characterization of a chitinase from the marine archaeon Halobacterium salinarum CECT 395 in Escherichia coli. Appl. Microbiol. Biotechnol. 98, 2133–2143. doi: 10.1007/s00253-013-5124-2
Glöckner, F. O., Fuchs, B. M., and Amann, R. (1999). Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65, 3721–3726.
Gooday, G. W. (1990). “The ecology of chitin degradation,” in Advances in Microbial Ecology, ed K. C. Marshall (New York, NY: Springer), 387–430.
Granger, J., and Price, N. M. (1999). The importance of siderophores in iron nutrition of heterotrophic marine bacteria. Limnol. Oceanogr. 44, 541–555. doi: 10.4319/lo.1999.44.3.0541
Guan, L. L., Kanoh, K., and Kamino, K. (2001). Effect of exogenous siderophores on iron uptake activity of marine bacteria under iron-limited conditions. Appl. Environ. Microbiol. 67, 1710–1717. doi: 10.1128/AEM.67.4.1710-1717.2001
Hansell, D., Carlson, C., Repeta, D., and Schlitzer, R. (2009). Dissolved organic matter in the ocean. Oceanography 22, 202–211. doi: 10.5670/oceanog.2009.109
Herndl, G. J., Reinthaler, T., Teira, E., van Aken, H., Veth, C., Pernthaler, A., et al. (2005). Contribution of archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71, 2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005
Hopkinson, B. M., and Barbeau, K. A. (2012). Iron transporters in marine prokaryotic genomes and metagenomes. Environ. Microbiol. 14, 114–128. doi: 10.1111/j.1462-2920.2011.02539.x
Hopkinson, B. M., Roe, K. L., and Barbeau, K. A. (2008). Heme uptake by Microscilla marina and evidence for heme uptake systems in the genomes of diverse marine bacteria. Appl. Environ. Microbiol. 74, 6263–6270. doi: 10.1128/AEM.00964-08
Iverson, V., Morris, R. M., Frazar, C. D., Berthiaume, C. T., Morales, R. L., and Armbrust, E. V. (2012). Untangling Genomes from Metagenomes: revealing an uncultured class of Marine Euryarchaeota. Science 335, 587–590. doi: 10.1126/science.1212665
Kabisch, A., Otto, A., König, S., Becher, D., Albrecht, D., Schüler, M., et al. (2014). Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes “Gramella forsetii” KT0803. ISME J. 8, 1492–1502. doi: 10.1038/ismej.2014.4
Kirchman, D. L. (2016). Growth rates of microbes in the oceans. Ann. Rev. Mar. Sci. 8, 285–309. doi: 10.1146/annurev-marine-122414-033938
Kirchman, D. L., Elifantz, H., Dittel, A. I., Malmstrom, R. R., and Cottrell, M. T. (2007). Standing stocks and activity of Archaea and Bacteria in the western Arctic Ocean. Limnol. Oceanogr. 52, 495–507. doi: 10.4319/lo.2007.52.2.0495
Kirchman, D. L., Meon, B., Cottrell, M. T., Hutchins, D. A., Weeks, D., and Bruland, K. W. (2000). Carbon vs. iron limitation of bacterial growth in the California upwelling regime. Limnol. Oceanogr. 45, 1681–1688. doi: 10.4319/lo.2000.45.8.1681
Kirchman, D. L., and White, J. (1999). Hydrolysis and mineralization of chitin in the Delaware Estuary. Aquat. Microb. Ecol. 18, 187–196. doi: 10.3354/ame018187
Landa, M., Blain, S., Christaki, U., Monchy, S., and Obernosterer, I. (2015). Shifts in bacterial community composition associated with increased carbon cycling in a mosaic of phytoplankton blooms. ISME J. 10, 1–12. doi: 10.1038/ismej.2015.105
Lasbleiz, M., Leblanc, K., Armand, L. K., Christaki, U., Georges, C., Obernosterer, I., et al. (2016). Composition of diatom communities and their contribution to plankton biomass in the naturally iron fertilized region of Kerguelen in the Southern Ocean. FEMS Microbiol. Ecol. 92:fiw171. doi: 10.1093/femsec/fiw171
Lasbleiz, M., Leblanc, K., Blain, S., Ras, J., Cornet-Barthaux, V., Hélias Nunige, S., et al. (2014). Pigments, elemental composition (C, N, P, Si) and stoichiometry of particulate matter, in the naturally iron fertilized region of Kerguelen in the Southern Ocean. Biogeosciences 11, 5931–5955. doi: 10.5194/bg-11-5931-2014
Laurenceau-Cornec, E. C., Trull, T. W., Davies, D. M., Bray, S. G., Doran, J., Planchon, F., et al. (2015). The relative importance of phytoplankton aggregates and zooplankton fecal pellets to carbon export: insights from free-drifting sediment trap deployments in naturally iron-fertilised waters near the Kerguelen Plateau. Biogeosciences 12, 1007–1027. doi: 10.5194/bg-12-1007-2015
Malits, A., Christaki, U., Obernosterer, I., and Weinbauer, M. G. (2014). Enhanced viral production and virus-mediated mortality of bacterioplankton in a natural iron-fertilized bloom event above the Kerguelen Plateau. Biogeosciences 11, 6841–6853. doi: 10.5194/bg-11-6841-2014
Manz, W., Amann, R., Ludwig, W., Vancanneyt, M., and Schleifer, K. H. (1996). Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 142, 1097–1106. doi: 10.1099/13500872-142-5-1097
Manz, W., Amann, R., Ludwig, W., Wagner, M., and Schleifer, K. H. (1992). Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15, 593–600. doi: 10.1016/S0723-2020(11)80121-9
Martin, J. H., Gordon, M. R., and Fitzwater, S. E. (1990). Iron in Antarctic waters. Nature 345, 156–158. doi: 10.1038/345156a0
Martinez, J. S., Carter-Franklin, J. N., Mann, E. L., Martin, J. D., Haygood, M. G., and Butler, A. (2003). Structure and membrane affinity of a suite of amphiphilic siderophores produced by a marine bacterium. Proc. Natl. Acad. Sci. 100, 3754–3759. doi: 10.1073/pnas.0637444100
Martinez, J. S., Zhang, G. P., Holt, P. D., Jung, H.-T., Carrano, C. J., Haygood, M. G., et al. (2000). Self-assembling amphiphilic siderophores from marine bacteria. Science 287, 1245–1247. doi: 10.1126/science.287.5456.1245
McCarren, J., Becker, J. W., Repeta, D. J., Shi, Y., Young, C. R., and Malmstrom, R. R. (2010). Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc. Natl. Acad. Sci. U.S.A. 107, 16420–16427. doi: 10.1073/pnas.1010732107
Moeck, G. S., and Coulton, J. W. (1998). TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28, 675–681. doi: 10.1046/j.1365-2958.1998.00817.x
Morris, J. J., Lenski, R. E., and Zinser, E. R. (2012). The Black Queen Hypothesis : evolution of dependencies through adaptive gene loss. Am. Soc. Microbiol. 3, 1–7. doi: 10.1128/mBio.00036-12
Morris, R. M., Rappé, M. S., Connon, S. A., Vergin, K. L., Siebold, W. A., Carlson, C. A., et al. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806–810. doi: 10.1038/nature01240
Obernosterer, I., Catala, P., Lebaron, P., and West, N. J. (2011). Distinct bacterial groups contribute to carbon cycling during a naturally iron fertilized phytoplankton bloom in the Southern Ocean. Limnol. Oceanogr. 56, 2391–2401. doi: 10.4319/lo.2011.56.6.2391
Obernosterer, I., Fourquez, M., and Blain, S. (2015). Fe and C co-limitation of heterotrophic bacteria in the naturally fertilized region off Kerguelen Islands. Biogeosciences 12, 1983–1992. doi: 10.5194/bg-12-1983-2015
Park, Y.-H., Durand, I., Kestenare, E., Rougier, G., Zhou, M., D'Ovidio, F., et al. (2014). Polar Front around the Kerguelen Isalnds: an up-to-date determination and associated circulation of surface/subsurface waters. J. Geophys. Res. 119, 6575–6592. doi: 10.1002/2014JC010061
Park, Y. H., Roquet, F., Durand, I., and Fuda, J. L. (2008). Large-scale circulation over and around the Northern Kerguelen Plateau. Deep. Res. Part II Top. Stud. Oceanogr. 55, 566–581. doi: 10.1016/j.dsr2.2007.12.030
Pernthaler, A., and Pernthaler, J. (2007). Fluorescence in situ hybridization for the identification of environmental microbes. Methods Mol. Biol. 353, 153–164. doi: 10.1385/1-59745-229-7:153
Pollard, R. T., Salter, I., Sanders, R. J., Lucas, M. I., Moore, C. M., Mills, R. A., et al. (2009). Southern Ocean deep-water carbon export enhanced by natural iron fertilization. Nature 457, 577–580. doi: 10.1038/nature07716
Qin, W., Amin, S. A., Martens-Habbena, W., Walker, C. B., Urakawa, H., Devol, A. H., et al. (2014). Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc. Natl. Acad. Sci. U.S.A. 111, 12504–12509. doi: 10.1073/pnas.1324115111
Quéroué, F., Sarthou, G., Planquette, H. F., Bucciarelli, E., Chever, F., van der Merwe, P., et al. (2015). High variability of dissolved iron concentrations in the vicinity of Kerguelen Island (Southern Ocean). Biogeosciences 12, 3869–3883. doi: 10.5194/bg-12-3869-2015
Sekar, R., Pernthaler, A., Pernthaler, J., Warnecke, F., Posch, T., and Amann, R. (2003). An improved protocol for quantification of freshwater Actinobacteria by Fluorescence in situ Hybridization. Appl. Environ. Microbiol. 69, 2928–2935. doi: 10.1128/AEM.69.5.2928-2935.2003
Simon, M., Glöckner, F. O., and Amann, R. (1999). Different community structure and temperature optima of heterotrophic picoplankton in various regions of the Southern Ocean. Aquat. Microb. Ecol. 18, 275–284. doi: 10.3354/ame018275
Smith, D. P., Kitner, J. B., Norbeck, A. D., Clauss, T. R., Lipton, M. S., Schwalbach, M. S., et al. (2010). Transcriptional and translational regulatory responses to iron limitation in the globally distributed marine bacterium Candidatus pelagibacter ubique. PLoS ONE 5:e10487. doi: 10.1371/journal.pone.0010487
Souza, C. P., Almeida, B. C., Colwell, R. R., and Rivera, I. N. G. (2011). The importance of chitin in the marine environment. Mar. Biotechnol. 13, 823–830. doi: 10.1007/s10126-011-9388-1
Staufenberger, T., Imhoff, J. F., and Labes, A. (2012). First crenarchaeal chitinase found in Sulfolobus tokodaii. Microbiol. Res. 167, 262–269. doi: 10.1016/j.micres.2011.11.001
Tang, K., Jiao, N., Liu, K., Zhang, Y., and Li, S. (2012). Distribution and functions of TonB-dependent transporters in marine bacteria and environments: implications for dissolved organic matter utilization. PLoS ONE 7:e41204. doi: 10.1371/journal.pone.0041204
Teeling, H., Fuchs, B. M., Becher, D., Klockow, C., Gardebrecht, A., Bennke, C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Teira, E., Reinthaler, T., Pernthaler, A., Pernthaler, J., and Herndl, G. J. (2004). Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by Bacteria and Archaea in the deep Ocean. Appl. Environ. Microbiol. 70, 4411–4414. doi: 10.1128/AEM.70.7.4411
Thompson, A. W., Huang, K., Saito, M. A., and Chisholm, S. W. (2011). Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability. ISME J. 5, 1580–1594. doi: 10.1038/ismej.2011.49
Tischer, K., Zeder, M., Klug, R., Pernthaler, J., Schattenhofer, M., Harms, H., et al. (2012). Fluorescence in situ hybridization (CARD-FISH) of microorganisms in hydrocarbon contaminated aquifer sediment samples. Syst. Appl. Microbiol. 35, 526–532. doi: 10.1016/j.syapm.2012.01.004
Tortell, P. D., Maldonado, M. T., and Price, N. M. (1996). The role of heterotrophic bacteria in iron-limited ocean ecosystems. Nature 383, 330–332. doi: 10.1038/383330a0
Toulza, E., Tagliabue, A., Blain, S., and Piganeau, G. (2012). Analysis of the global ocean sampling (GOS) project for trends in iron uptake by surface ocean microbes. PLoS ONE 7:e30931. doi: 10.1371/journal.pone.0030931
Tremblay, L., Caparros, J., Leblanc, K., and Obernosterer, I. (2015). Origin and fate of particulate and dissolved organic matter in a naturally iron-fertilized region of the Southern Ocean. Biogeosciences 12, 607–621. doi: 10.5194/bg-12-607-2015
Trull, T. W., Davies, D. M., Dehairs, F., Cavagna, A.-J., Lasbleiz, M., Laurenceau, E. C., et al. (2015). Chemometric perspectives on plankton community responses to natural iron fertilization over and downstream of the Kerguelen Plateau in the Southern Ocean. Biogeosciences 12, 1029–1056. doi: 10.5194/bg-12-1029-2015
Weaver, R. S., Kirchman, D. L., and Hutchins, D. (2003). Utilization of iron/organic ligand complexes by marine bacterioplankton. Aquat. Microb. Ecol. 31, 227–239. doi: 10.3354/ame031227
Wietz, M., Wemheuer, B., Simon, H., Giebel, H. A., Seibt, M. A., Daniel, R., et al. (2015). Bacterial community dynamics during polysaccharide degradation at contrasting sites in the Southern and Atlantic Oceans. Environ. Microbiol. 17, 3822–3831. doi: 10.1111/1462-2920.12842
Wright, R. R., and Hobbie, E. J. (1966). Use of Glucose and Acetate by Bacteria and Algae in aquatic ecosystems on JSTOR. Ecology 47, 447–464. doi: 10.2307/1932984
Yamamoto, S., Okujo, N., Yoshida, T., Matsuura, S., and Shinoda, S. (1994). Structure and iron transport activity of vibrioferrin, a new siderophore of Vibrio parahaemolyticus. J. Biochem. 115, 868–874.
Zhang, Z., Lo, I. M. C., Zheng, G., Woon, K. S., and Rao, P. (2015). Effect of autotrophic denitrification on nitrate migration in sulfide-rich marine sediments. J. Soils Sediments 15, 1019–1028. doi: 10.1007/s11368-015-1078-6
Zimmerman, A. E., Martiny, A. C., and Allison, S. D. (2013). Microdiversity of extracellular enzyme genes among sequenced prokaryotic genomes. ISME J. 7, 1187–1199. doi: 10.1038/ismej.2012.176
Keywords: iron, chitin, leucine, prokaryotic community composition, MICRO-CARD-FISH, Southern Ocean
Citation: Fourquez M, Beier S, Jongmans E, Hunter R and Obernosterer I (2016) Uptake of Leucine, Chitin, and Iron by Prokaryotic Groups during Spring Phytoplankton Blooms Induced by Natural Iron Fertilization off Kerguelen Island (Southern Ocean). Front. Mar. Sci. 3:256. doi: 10.3389/fmars.2016.00256
Received: 30 May 2016; Accepted: 24 November 2016;
Published: 15 December 2016.
Edited by:
Anton F. Post, University of Rhode Island, USAReviewed by:
Hugh Ducklow, Columbia University, USAPia Moisander, University of Massachusetts Dartmouth, USA
Copyright © 2016 Fourquez, Beier, Jongmans, Hunter and Obernosterer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marion Fourquez, marion.fourquez@gmail.com
 Marion Fourquez
Marion Fourquez Sara Beier
Sara Beier Elanor Jongmans2
Elanor Jongmans2  Robert Hunter
Robert Hunter Ingrid Obernosterer
Ingrid Obernosterer