Detection of bacteria bearing resistant biofilm forms, by using the universal and specific PCR is still unhelpful in the diagnosis of periprosthetic joint infections
- 1Department of Biopathology and Clinical Microbiology, Aeginition Hospital, Athens Medical School, Athens, Greece
- 2Department of Nursing, Faculty of Human Movement and Quality of Life Sciences, University of Peloponnese, Sparta, Greece
- 32nd Orthopaedic Department, Konstantopouleio General Hospital, University of Athens, Athens, Greece
- 4National Influenza Reference Laboratory of Southern Greece, Institut Pasteur Hellénique, Athens, Greece
- 5Department of Statistics and Insurance Science, University of Piraeus, Piraeus, Greece
- 6Department of Microbiology, Medical School, National and Kapodistrian University of Athens, Athens, Greece
- 7Department of Microbiology, Medical School, University of Thessaly, Larissa, Greece
Intraoperative conventional bacteriological cultures were compared with different polymerase chain reaction (PCR) methods in patients with total joint arthroplasties. The isolated bacteria were investigated for biofilm formation, and the biofilm forming strains, in their planktonic and biofilm forms, were further tested for their antimicrobial resistance against several clinically important antimicrobials. Forty four bone and joint samples were included and classified as infected or non-infected according to standard criteria for periprosthetic hip and knee infections. For the bacteriological diagnosis, conventional culture, two types of universal PCR and species specific PCR for three selected pathogens (Staphylococcus aureus, Staphylococcus epidermidis, and Pseudomonas aeruginosa) were applied. Biofilm formation determination was performed by the tissue culture plate method. Antimicrobial susceptibility of the planktonic bacteria was performed by the minimal inhibitory concentration determination and, of the biofilm forms, by the minimal inhibitory concentration for bacterial regrowth from the biofilm. Twenty samples were culture positive, with S. epidermidis, S. aureus, or P. aeruginosa. All PCR methods were very ineffective in detecting only one pathogen. All isolates were biofilm positive and their biofilm forms, were highly resistant. In this study, compared to PCR, culture remains the “gold standard.” The biofilm formation by the causative bacteria and the concomitant manifold increased antimicrobial resistance may explain the clinical failure of treatment in some cases and should be considered in the future for therapeutic planning.
Introduction
Periprosthetic joint infection (PJI) is one of the most serious complications causing a high grade of morbidity in patients with total joint arthroplasty. Actually, the rate of infection after total hip or knee arthroplasty ranges from below 1 up to 5% and it rises slightly in case of revision procedures (1, 2).
The accurate diagnosis of bone and joint infections, has for long been confounded by the difficulty of retrieval and detection of microorganisms, and is still a challenge to the treating physician (1, 3).
The clinical diagnosis can be assisted by laboratory tests like white cell count and differentiation, erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), and others, but there is no gold standard, having a perfect sensitivity and specificity for diagnosing the infection, apart from identifying the infective bacteria (4).
The distinction between septic and non-septic loosening is difficult in most cases, impeding clinical evaluation (5). The risk of a PJI increases greatly due to the long stay, in the body, of the implant, which can be colonized by microbes from a distant infectious focus through the hematogenous route at any time following implantation (1). Therefore, an increase in the number of PJIs is expected in the coming years.
Currently, periprosthetic infection is frequently diagnosed by isolation of one or more organisms from the periprosthetic tissue or fluid with use of conventional culture techniques, and the culture’s results are usually considered the standard, which are compared with other diagnostic tests (6).
Among the various bacterial virulence factors, a very important one, is the ability to form biofilms (7). In a biofilm, the bacteria are attached to each other and adhering to the artificial materials and medical devices such as contact lenses, artificial heart valves, joint replacements, etc., they produce an extracellular polymeric substance (ESP) consisting of carbohydrate (exopolysaccharide) (8). The production of biofilms protects the bacteria from the effect of the antibiotic action, in addition to the inhibition of the phagocytic cells, and impedes the function of the T- and B-lymphocytes (9–11).
The detection and isolation of causative microorganisms is the first important step for the successful treatment of PJI (12). Many diagnostic methods are applied in order to isolate the causative microorganisms in patients with symptoms of a failed arthroplasty. Some conventional methods, like culture of aspirated joint fluid, can be performed preoperatively. However, the preoperative aspiration does not always precede and the surgeons often do not become suspicious of an infection until the revision arthroplasty operation is underway (4).
Molecular techniques overcome some of the limitations of conventional microbiological diagnostic procedures. More than 10 years have passed since the introduction of molecular methods into the diagnosis of orthopedic infections; these methods are still a matter of research and discussion. Gallo et al. (13) as well as Spangehl et al. (14) investigated how the polymerase chain reaction (PCR) might play an essential role in PJI diagnosis. They pointed out the advantages of accuracy and speed, as well as the possible disadvantages, like the false-positive results, misinterpretation, and the expensive equipment requirements (13, 14).
Despite the numerous studies already published, the role of molecular techniques, like PCR, in the diagnosis of PJI, still remains vague and unclear. Therefore, the effectiveness of these techniques has not yet been verified in the routine of the clinical microbiology laboratory diagnosis for PJI (5).
The aim of the present study is to compare the conventional cultures with two PCR methods (universal and specific), for the fast and accurate diagnosis of PJI in 44 patients who had a primary or failed total joint arthroplasty. In addition, the isolated bacteria were investigated with respect to biofilm formation. The biofilm forming strains, in their planktonic and biofilm forms, were further tested for their antimicrobial resistance against several clinically important antimicrobials, used for the treatment of PJI.
Materials and Methods
Patients
The study included 44 bone and joint samples collected intraoperatively from an equal number of patients. Thirteen from patients with a preoperative diagnosis of infected, and 31 from patients with a preoperative diagnosis of non-infected be based on a clinician’s independent medical judgment (depending on the history of the patient, the clinical examination, such as constant pain, warmth and effusion, erythema, delayed healing of the wound, plain x-ray, and other patient individualized criteria). All the patients were treated in the 1st Department of Orthopedic Surgery “ATTIKON” University Hospital of the Athens Medical School. The age of the patients ranged from 31 to 85 years. Patients were classified in two groups, with respect to the final clinical diagnosis, as infected or non-infected cases.
The clinical determination of deep infection (deep around the artificial implants) was according to the criteria of international standard of PJI (15).
All the patients underwent preoperative general blood examination including white blood cell (WBC) count, ESR, and CRP determination.
Sample Collection and Culture
The patient samples used in the study were bone, tissue, or aspiration fluid, which had been taken intraoperatively from the patients with total hip or total knee arthroplasty (more than one sample had been collected from each patient), as routine diagnostic procedure. Bacteriological examination included conventional culture on growth media for aerobic and anaerobic bacteria and direct sample microscopy of Gram stained smears. The identification of the isolated bacteria was performed by conventional bacteriological methods: API-E and API-NE (Biomerieux, Marcy-I’ Etoile/France) for gram-negative bacteria and coagulase and DNAase for Staphylococcus spp.
Biofilm Production Determination
All isolated bacterial strains were investigated for their ability to form biofilms by the tissue culture plate (TCP) method as described by Christensen et al. (16) and Baldassarri et al. (17), with a modification in duration of incubation, which was extended to 24 h (18).
Briefly, a bacterial suspension was prepared from a blood agar plate culture in trypticase soy broth at opacity of 0.5 McFarland standards and cultured overnight at 37°C. The next day, 100 μl of the overnight culture were added to 200 μl tryptose broth and placed in a micro titer tray well, mixed and incubated overnight at 37°C. The next day, the wells were carefully emptied and washed three times with phosphate buffered saline (PBS). The plate was allowed to dry at 60°C for 1 h and then stained with Hucker’s crystal violet (2 g crystal violet, 20 ml 95% alcohol, 0.8 g ammonium oxalate, and 80 ml distilled water). The excess stain was washed off with distilled water, excess water was removed, and the plates were read with an ELISA reader at 570 nm (19). The cut-off value was calculated as mean ± 2SD of the values of 10 wells processed the same way but without bacteria. Values above the cut-off were considered positive for biofilm formation. Each strain was tested in quadruplicate.
MIC Determination
Antimicrobial susceptibility of the planktonic bacterial forms was performed and interpreted by determination of the minimal inhibitory concentration (MIC) using the standard broth dilution method according to the guidelines of the Clinical Laboratory Standards Institute (20, 21). The antimicrobials included, were those of importance in the clinical practice for treating the isolated bacterial species: ciprofloxacin, moxifloxacin, erythromycin, linezolid, daptomycin, teicoplanin, vancomycin tigecyclin, and cotrimoxazole for Staphylococcus aureus and Staphylococcus epidermidis and imipenem, meropenem, ceftazidime, aztreonam, tobramycin, ciprofloxacin, amikacin, cefepime, and cotrimoxazole for Pseudomonas aeruginosa.
Minimal Inhibitory Concentration for Bacterial Regrowth from the Biofilm Determination
The strains producing biofilms were further tested for their antimicrobial susceptibility by determination of the minimal inhibitory concentration for bacterial regrowth from the biofilm (MICBR) using a modified broth dilution method as described previously (22).
Serial dilutions of the antimicrobial in Mueller Hinton broth, corresponding to the concentrations used for the MIC determination of the planktonic forms, were prepared and poured into the micro titer plates, which contained the bacterial biofilm and incubated at 35°C for 48 h. The growth of planktonic bacteria was visualized by the development of turbidity in the medium. The MICBR was defined as the lowest concentration showing no growth in the medium as observed by a complete clarity. Each strain was tested in quadruplicate.
The results were assessed using the breakpoints given by the guidelines of the CLSI (20, 21).
Polymerase Chain Reaction
Bacterial DNA extraction was performed by means of the protocol of the “Insta Gene Matrix” method (Bio Rad Laboratories, CA, USA) The extracted DNA was stored at −20°C until the time of use.
The types of PCR that were performed for the detection of the causative pathogens were: a) two types of universal PCR (Nr.1 and 2) detecting the 16S rRNA gene by different protocols (23, 24), followed by sequencing of the product for the identification of the species, and b) the species specific PCR, for three selected pathogens: S. aureus (25), S. epidermidis (26), and P. aeruginosa (27). All the PCR primers and annealing temperatures are depicted in Table 1. For all PCR methods, controls were run in parallel with extracted DNA from the following reference strains: Escherichia coli ATCC 25922, P. aeruginosa ATCC 27853, S. epidermidis ATCC 35984, S. aureus ATCC 29213, and Streptococcus pneumoniae ATCC 49619.

Table 1. PCR primers and annealing temperatures for the detection of bacteria causing periprosthetic joint infections.
Each reaction of PCR consisted 0.4 μM of forward primer, 0.4 μM of reverse primer, 2.5 units/reaction HotStar Taq DNA Polymerase (Qiagen), 1X PCR Buffer provides a final concentration of 1.5 mM MgCl2, 200 μM of each deoxynucleotide triphosphates (dNTP), 5 μl DNA sample, and PCR-grade water until they completed 50 μl of reaction volume. The conditions we used are: 1 cycle (94°C for 5 min), 35 cycles (94°C for 1 min, annealing temperature as described in Table 1 for each pair of primers for 1 min, 72°C for 1 min), and 1 cycle (72°C for 10 min), storing at 4°C.
Statistical Analysis
The statistical analysis was performed using the statistical package SPSS for Windows (version 20.0) in order to disclose any significant differences between the percentages of antimicrobial susceptibility of the planktonic and the biofilm bacterial forms. The analysis was done by applying an appropriate hypothesis test concerning the difference between the proportions of two samples. The normal approximation to the binomial distribution was used. Additionally, appropriate parametric and non-parametric tests were used for comparing quantitative variables such as CRP, ESR, and WBC.
Results
Sample Origin, Sample Species, Culture, and Inflammation Markers
A total of 44 samples were analyzed in this study: 20 were culture positive, with the following organisms: S. epidermidis (15 isolates), S. aureus (4 isolates), P. aeruginosa (1 isolate), and 24 were culture negative results (Tables 2A,B).
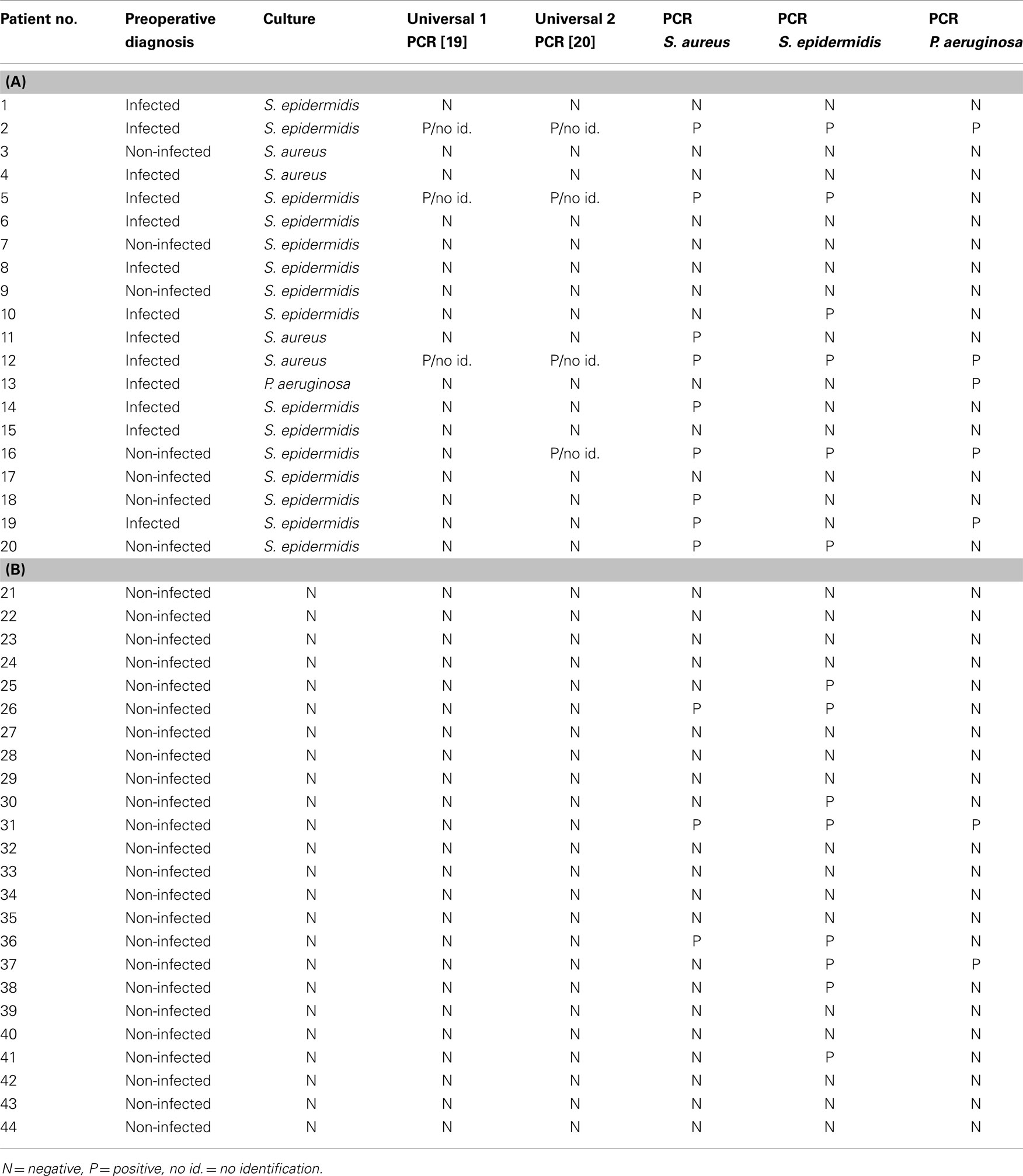
Table 2. The results of universal and specific PCR technique for the culture (A) positive samples and (B) negative samples.
There was no statistically significant relation, either between culture results and patients’ gender or sample origin (total hip or total knee). However, the sample species proved to be critical for the culture outcome: the tissue and bone samples gave significantly more positive culture results than the aspiration fluid (p < 0.001, results not shown).
Sensitivity and specificity of the culture in relation to the presumed preoperative evaluation were 100 and 77.4%, respectively with a Receiver operating characteristic (ROC) area of 0.812–0.962 (ROC value = 0.887).
All three inflammation markers, CRP, ESR, and WBC were significantly higher in patients with a positive culture (p < 0.001 for all, Table 3).
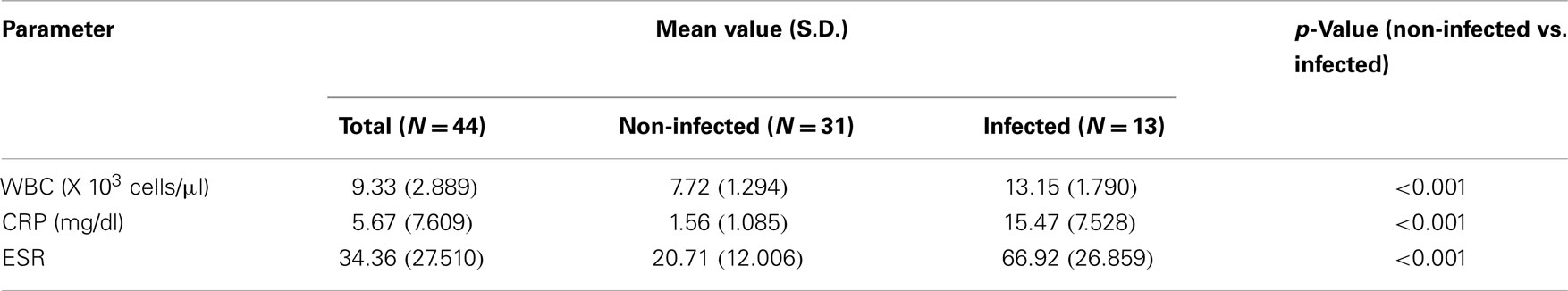
Table 3. White blood cell (WBC), C-reactive protein (CRP), and erythrocyte sedimentation rate (ESR) preoperative determination in the blood of the patients subject to total joint arthroplasty.
Polymerase Chain Reaction
Both universal PCR methods showed very contradictory results in relation to culture. Although there was an agreement between negative culture and negative PCR for both universal PCR tests, they both failed to detect bacterial 16S rDNA in most culture positive samples. When 16S rDNA was detected, identification on species level through sequencing was impossible; as in all 16S rDNA positive samples, the sequencing reaction was blocked after approximately 60 bp, probably due to the presence of more than one bacterial species (Tables 2A,B). All three specific PCR tests were equally ambiguous, being very ineffective in detecting only one pathogen and giving very discrepant results in relation to culture (Tables 2A,B; Figure 1).
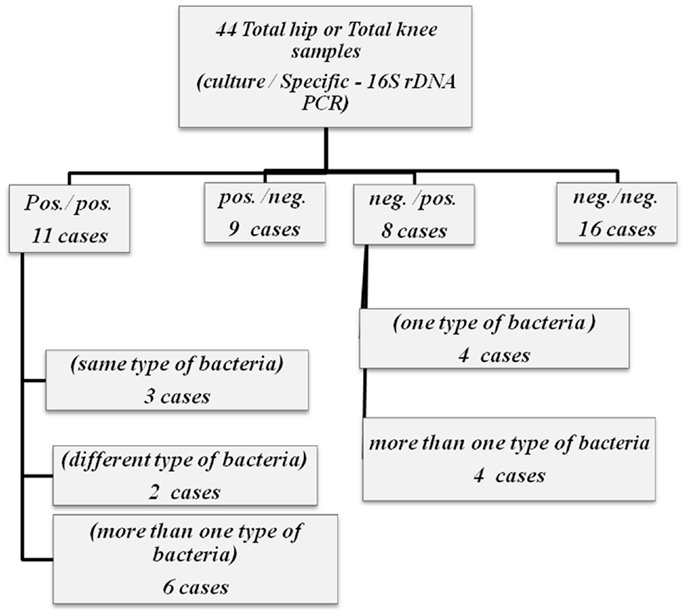
Figure 1. Flipchart summarizing the results of both culture and specific – 16S rDNA for 44 total hip and total knee samples.
In reference to the preoperative clinical evaluation, sensitivities and specificities of the PCR methods were as follows: universal PCR 1 23.1 and 100% (24), universal PCR 2 23.1 and 96.8% (23), PCR specific for S. aureus 46.2 and 80.6%, PCR specific for S. epidermidis 30.8 and 67.7%, and P. aeruginosa 30.8 and 90.3%.
Biofilm formation, MIC, and MICBR
All the strains were positive for biofilm production. The great majority of the biofilm forms were resistant to all antimicrobials. In antimicrobial concentrations far higher than the breakpoints, bacterial regrowth from the biofilms was still possible. The results for S. aureus and S. epidermidis are shown in Table 4. Regarding the MIC (value in microgram per milliliter in parenthesis) of the one isolated strain of P. aeruginosa, the strain was susceptible to meropenem ( ≤1), ceftazidime (2), aztreonam (4), tobramycin ( ≤2), ciprofloxacin (0.25), amikacin ( ≤8), and cefepime (2) and intermediate to imipenem (4). The respective MICBR values were at least the fourfold of the MIC values for imipenem, meropenem, ceftazidime, aztreonam, tobramycin, and cefepime, showing significant resistance of the biofilm forms with respect to bacterial regrowth (p < 0.001). For ciprofloxacin and amikacin, the MICBR values were identical to the MIC values; thus, these two antibiotics seemed to suppress in vitro P. aeruginosa regrowth from the biofilm under corresponding clinical therapeutic dosages.
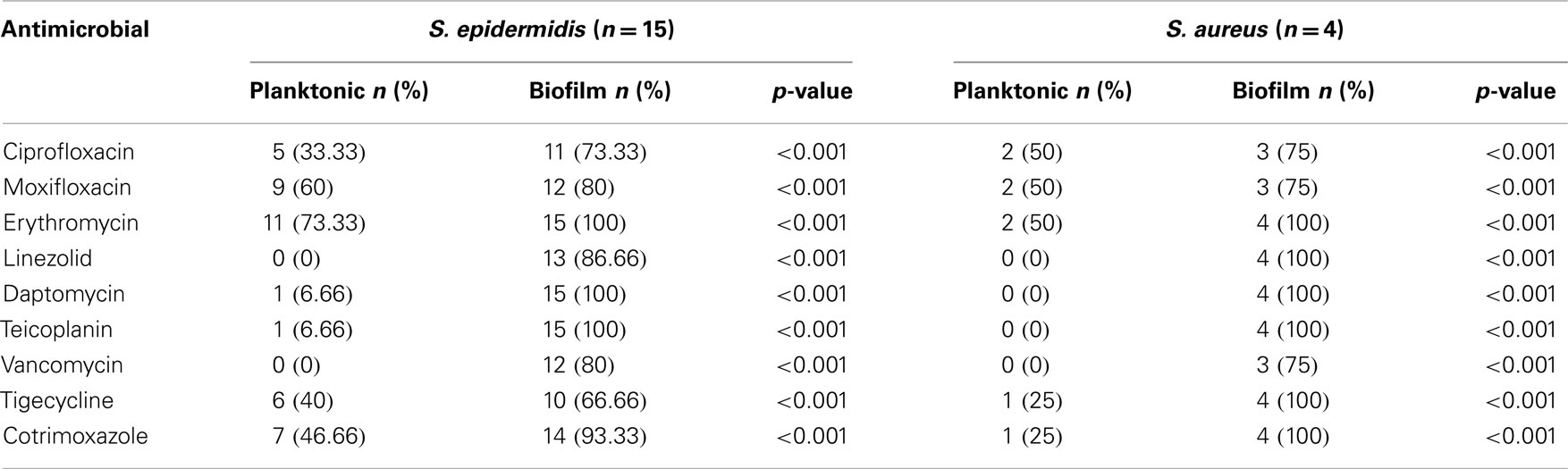
Table 4. Antimicrobial resistance rates of planktonic and biofilm forms of S. epidermidis and S. aureus strains isolated from the samples of the patients with periprosthetic joint infection.
Discussion
The diagnosis of the prosthetic joint infection includes a set of laboratory tests. In the present study, like in previous reports (6, 28, 29), the traditional first line laboratory tests including the inflammation markers CRP, ESR, and WBC proved to have a very good sensitivity, when correlated with the clinical evaluation (Table 3).
Culture is still the gold standard for diagnosis of prosthetic joint infection, offering the possibility of the antimicrobial susceptibility testing, but it is time-consuming in identifying the causative microorganisms. However, the drawbacks of the cultural procedures are the limited sensitivity, in addition to the false negative results in patients receiving antimicrobials (6, 30). Our study comprised 44 cases, 20 cultures-positive, and 24 negative (more than one sample had been taken from each patient). In seven cases with a presumed preoperative absence of infection (non-infected), culture was positive, which may have been through sample contamination during handling (collection, transport, and processing).
In a prospective study involving revision of arthroplasty in 297 patients with a total of 41 infections, Atkins et al. (31) pointed out that only 65% of all samples collected from the infected joints were culture positive. They recommended collecting five or six culture samples from each patient and suggested that the accurate diagnosis of infection should be considered as the growth of the identical microorganism on culture in three or more samples (6). Practically, this procedure is difficult, but at the same time, it could increase the possibility of detecting the causative microorganism.
All isolated bacterial strains were positive for biofilm formation in vitro, by the TCP performed as previously described (16, 17). The in vivo biofilm synthesis gives bacteria the ability to cause infection and impedes their cultural isolation from the samples because the biofilms adhere strongly to the colonized biological surface. The use of ultrasound to expel bacterial cells from the biofilms adherent to the surface of removed implants (sonication) increases the effectiveness and the sensitivity of microbiological studies to determine the underlying microorganisms, but, at the same time, increases the risk of contamination through the more complicated sample handling (32, 33).
The antimicrobial resistance rates of the planktonic and the biofilm forms of the isolated bacteria, tested by the MIC and MICBR determination, showed increased antimicrobial resistance of the biofilm forms to the vast majority of the antimicrobials, with MICs far above those of the planktonic forms and above the breakpoints corresponding to the therapeutic clinical implementation (Table 4). This may explain the failure of treatment in some cases, where despite the antimicrobial susceptibility to a certain antimicrobial in vitro, the infection still exists after the appropriate treatment (clinical resistance) (34–37). Although there are no standard procedures for the determination of MICBR, our results are in agreement with previously reported data, with respect to the role of biofilms in the increase of the bacterial antimicrobial resistance (38). Furthermore, they confirm that the experimental conditions used, led to biofilm formation. Biofilm production in S. epidermidis and P. aeruginosa belongs to the most significant virulence factors for the expression of pathogenicity in infectious diseases (16, 38, 39).
The most important part in this study was the application and evaluation of PCR. Based on the literature, PCR can be considered one of the very helpful diagnostic tools, used in cases of arthritis, especially when culture is negative. Fenollar et al. (3) reported remarkable results for cloning and sequencing of 16S rDNA amplicons. They found a perfect compatibility between culture and PCR results in 475 of 525 samples (90.5%). Kordelle et al. (40) sequenced the 16S rDNA amplicons and found 100% agreement between PCR and culture, but the study included only seven cases of PJI. In the present study, the universal 16S rDNA PCR technique was totally unhelpful for the bacterial identification. Although it detected bacterial DNA in many cases, with a significant agreement with culture (p-value 0.031), there was no identification of the DNA product after sequencing. This problem might be due to the presence of more than one bacterial species (including the causative microorganism), caused through sample handling and processing. To be effectual, the universal PCR requires specimens containing a single bacterial strain; otherwise, the identification after sequencing is impossible.
The specific PCR showed better results but still very discrepant compared to culture, the p-values were 0.014, 0.583, and 0.226 for PCR S. aureus, S. epidermidis, and P. aeruginosa, respectively in comparison with culture. Published results from different studies on the use of PCR to detect prosthetic joint infection have made PCR a technique not yet widely accepted in routine examination. The sensitivity and specificity of broad-range PCR from synovial fluid and/or tissue for the diagnosis of prosthetic joint infection have been reported to be between 50–92% and 65–94%, respectively (41–44). In the present report, sensitivity and specificity of all the PCR methods applied were very diverse and in none of the cases did they fulfill the criteria of a reliable diagnostic method for the bacteriological diagnosis of PJI (45).
In the present report, the molecular methods proved to be inappropriate for a reliable bacterial diagnosis, the culture remaining still “the gold standard.” When compared to conventional microbiological procedures, PCR analysis is still hindered by higher costs, false-positive results, and interpretative problems. Currently, under these circumstances it is not justifiable to introduce molecular methods into the schemes used to diagnose prosthetic joint infection (13).
The detection and identification of bacterial RNA, rather than DNA, can be a new approach to the laboratory diagnosis of prosthetic joint infection, by reverse transcription. RNA is present only in viable bacteria and, thus, it could be more reliable in disclosing active infections. On the other hand, the much shorter half-life of RNA would make its role as a contaminant less likely (46). Rasouli et al. have been evaluating the new approach of multiplex PCR (Ibis T5000 universal Biosensor), which depend on pan-genomic amplification, and mass spectrometry for culture negative cases in patients with suspicion of PJI and the results were promising (15).
In conclusion, our results showed that the molecular diagnostic methods, like PCR, did not increase the detection rate of prosthetic joint infection, compared to culture. Improved PCR methods may be considered in the near future and play an important role in the diagnosis of bone infections as a complement to culture, in cases where a small amount of samples are available for examination, or when culture is negative after 24 h of incubation for patients with suspected prosthetic joint infection, as well as in the diagnosis of samples taken from patients receiving antimicrobial therapy.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
AAOS, American Academy of Orthopaedic Surgeons; APIE, APINE, microorganism identification kits for gram-negative bacteria (biomerieux API, 69280 Marcy-I’ Etoile/France); CLSI, Clinical Laboratory Standards Institute; CRP, C-reactive protein; dNTP, deoxynucleotide triphosphates; ESR, erythrocyte sedimentation rate; MIC, minimal inhibitory concentration; MICBR, minimal inhibitory concentration for bacterial regrowth from the biofilm; PBS, phosphate buffered saline; PJI, periprosthetic joint infections; ROC, receiver operating characteristic; TCP, tissue culture plate; WBC, white blood cell.
References
1. Zimmerli W, Trampuz A, Ochsner PE. Prosthetic-joint infections. N Engl J Med (2004) 351:1645–54. doi: 10.1056/NEJMra040181
2. Sauer P, Gallo J, Kesselova M, Kolar M, Koukalova D. Universal primers for detection of common bacterial pathogens causing prosthetic joint infection. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub (2005) 149:285–8. doi:10.5507/bp.2005.043
3. Fenollar F, Roux V, Stein A, Drancourt M, Raoult D. Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections. J Clin Microbiol (2006) 44:1018–28. doi:10.1128/JCM.44.3.1018-1028.2006
4. Oethinger M, Warner DK, Schindler SA, Kobayashi H, Bauer TW. Diagnosing periprosthetic infection: false-positive intraoperative Gram stains. Clin Orthop Relat Res (2011) 469:954–60. doi:10.1007/s11999-010-1589-9
5. Marin M, Garcia-Lechuz JM, Alonso P, Villanueva M, Alcala L, Gimeno M, et al. Role of universal 16S rRNA gene PCR and sequencing in diagnosis of prosthetic joint infection. J Clin Microbiol (2012) 50:583–9. doi:10.1128/JCM.00170-11
6. Bauer TW, Parvizi J, Kobayashi N, Krebs V. Diagnosis of periprosthetic infection. J Bone Joint Surg Am (2006) 88:869–82. doi:10.2106/JBJS.E.01149
7. Gristina AG, Oga M, Webb LX, Hobgood CD. Adherent bacterial colonization in the pathogenesis of osteomyelitis. Science (1985) 228:990–3. doi:10.1126/science.4001933
8. Sawhney R, Berry V. Bacterial biofilm formation, pathogenicity, diagnostics and control: an overview. Indian J Med Sci (2009) 63:313–21. doi:10.4103/0019-5359.55113
9. Gray ED, Peters G, Verstegen M, Regelmann WE. Effect of extracellular slime substance from Staphylococcus epidermidis on the human cellular immune response. Lancet (1984) 1:365–7. doi:10.1016/S0140-6736(84)90413-6
10. Johnson GM, Lee DA, Regelmann WE, Gray ED, Peters G, Quie PG. Interference with granulocyte function by Staphylococcus epidermidis slime. Infect Immun (1986) 54:13–20.
11. Naylor PT, Myrvik QN, Gristina A. Antibiotic resistance of biomaterial-adherent coagulase-negative and coagulase-positive Staphylococci. Clin Orthop Relat Res (1990) 261:126–33.
12. Achermann Y, Vogt M, Leunig M, Wust J, Trampuz A. Improved diagnosis of periprosthetic joint infection by multiplex PCR of sonication fluid from removed implants. J Clin Microbiol (2010) 48:1208–14. doi:10.1128/JCM.00006-10
13. Gallo J, Raska M, Dendis M, Florschutz AV, Kolar M. Molecular diagnosis of prosthetic joint infection. A review of evidence. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub (2004) 148:123–9. doi:10.5507/bp.2004.022
14. Spangehl MJ, Younger AS, Masri BA, Duncan CP. Diagnosis of infection following total hip arthroplasty. Instr Course Lect (1998) 47:285–95.
15. Rasouli MR, Harandi AA, Adeli B, Purtill JJ, Parvizi J. Revision total knee arthroplasty: infection should be ruled out in all cases. J Arthroplasty (2012) 27(1239–1243):e1231–2. doi:10.1016/j.arth.2011.01.019
16. Christensen GD, Simpson WA, Bisno AL, Beachey EH. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun (1982) 37:318–26.
17. Baldassarri L, Simpson WA, Donelli G, Christensen GD. Variable fixation of staphylococcal slime by different histochemical fixatives. Eur J Clin Microbiol Infect Dis (1993) 12:866–8. doi:10.1007/BF02000411
18. Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A. Detection of biofilm formation among the clinical isolates of Staphylococci: an evaluation of three different screening methods. Indian J Med Microbiol (2006) 24:25–9. doi:10.4103/0255-0857.19890
19. Garcia P, Benitez R, Lam M, Salinas AM, Wirth H, Espinoza C, et al. Coagulase-negative Staphylococci: clinical, microbiological and molecular features to predict true bacteraemia. J Med Microbiol (2004) 53:67–72. doi:10.1099/jmm.0.04994-0
20. CLSI. M07-A9: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard – Ninth Edition. Wayne, PA: Clinical and Laboratory Standard Institute (2013).
21. CLSI. M100-S23: Performance Standards for Antimicrobial Susceptibility Testing; Twenty Third Informational Supplement. Wayne, PA: Clinical and Laboratory Standard Institute (2013).
22. Papavasileiou K, Papavasileiou E, Tseleni-Kotsovili A, Bersimis S, Nicolaou C, Ioannidis A, et al. Comparative antimicrobial susceptibility of biofilm versus planktonic forms of Salmonella enterica strains isolated from children with gastroenteritis. Eur J Clin Microbiol Infect Dis (2010) 29:1401–5. doi:10.1007/s10096-010-1015-y
23. Woo PC, Ng KH, Lau SK, Yip KT, Fung AM, Leung KW, et al. Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles. J Clin Microbiol (2003) 41:1996–2001. doi:10.1128/JCM.41.5.1996-2001.2003
24. Siala M, Jaulhac B, Gdoura R, Sibilia J, Fourati H, Younes M, et al. Analysis of bacterial DNA in synovial tissue of Tunisian patients with reactive and undifferentiated arthritis by broad-range PCR, cloning and sequencing. Arthritis Res Ther (2008) 10:R40. doi:10.1186/ar2398
25. Martineau F, Picard FJ, Roy PH, Ouellette M, Bergeron MG. Species-specific and ubiquitous-DNA-based assays for rapid identification of Staphylococcus aureus. J Clin Microbiol (1998) 36:618–23.
26. Martineau F, Picard FJ, Roy PH, Ouellette M, Bergeron MG. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J Clin Microbiol (1996) 34:2888–93.
27. Spilker T, Coenye T, Vandamme P, Lipuma JJ. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol (2004) 42:2074–9. doi:10.1128/JCM.42.5.2074-2079.2004
28. Spangehl MJ, Masri BA, O’Connell JX, Duncan CP. Prospective analysis of preoperative and intraoperative investigations for the diagnosis of infection at the sites of two hundred and two revision total hip arthroplasties. J Bone Joint Surg Am (1999) 81:672–83.
29. Moyad TF, Thornhill T, Estok D. Evaluation and management of the infected total hip and knee. Orthopedics (2008) 31:581–8; quiz 589–90. doi:10.3928/01477447-20080601-22
30. Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, et al. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev (2006) 19:165–256. doi:10.1128/CMR.19.1.165-256.2006
31. Atkins BL, Athanasou N, Deeks JJ, Crook DW, Simpson H, Peto TE, et al. Prospective evaluation of criteria for microbiological diagnosis of prosthetic-joint infection at revision arthroplasty. The OSIRIS Collaborative Study Group. J Clin Microbiol (1998) 36:2932–9.
32. Trampuz A, Piper KE, Jacobson MJ, Hanssen AD, Unni KK, Osmon DR, et al. Sonication of removed hip and knee prostheses for diagnosis of infection. N Engl J Med (2007) 357:654–63. doi:10.1056/NEJMoa061588
33. Evangelopoulos DS, Stathopoulos IP, Morassi GP, Koufos S, Albarni A, Karampinas PK, et al. Sonication: a valuable technique for diagnosis and treatment of periprosthetic joint infections. ScientificWorldJournal (2013) 2013:5. doi:10.1155/2013/375140
34. Kunin CM, Steele C. Culture of the surfaces of urinary catheters to sample urethral flora and study the effect of antimicrobial therapy. J Clin Microbiol (1985) 21:902–8.
35. Nickel JC, Ruseska I, Wright JB, Costerton JW. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother (1985) 27:619–24. doi:10.1128/AAC.27.4.619
36. Gristina AG, Hobgood CD, Webb LX, Myrvik QN. Adhesive colonization of biomaterials and antibiotic resistance. Biomaterials (1987) 8:423–6. doi:10.1016/0142-9612(87)90077-9
37. Costerton JW, Khoury AE, Ward KH, Anwar H. Practical measures to control device-related bacterial infections. Int J Artif Organs (1993) 16:765–70.
38. Dunne WM Jr. Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev (2002) 15:155–66. doi:10.1128/CMR.15.2.155-166.2002
39. Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev (2002) 15:167–93. doi:10.1128/CMR.15.2.167-193.2002
40. Kordelle J, Hossain H, Stahl U, Schleicher I, Haas H. Usefulness of 16S rDNA polymerase-chain-reaction (PCR) in the intraoperative detection of infection in revision of failed arthroplasties. Z Orthop Ihre Grenzgeb (2004) 142:571–6. doi:10.1055/s-2004-832343
41. Panousis K, Grigoris P, Butcher I, Rana B, Reilly JH, Hamblen DL. Poor predictive value of broad-range PCR for the detection of arthroplasty infection in 92 cases. Acta Orthop (2005) 76:341–6. doi:10.1080/00016470510030805
42. Vandercam B, Jeumont S, Cornu O, Yombi JC, Lecouvet F, Lefevre P, et al. Amplification-based DNA analysis in the diagnosis of prosthetic joint infection. J Mol Diagn (2008) 10:537–43. doi:10.2353/jmoldx.2008.070137
43. De Man FH, Graber P, Luem M, Zimmerli W, Ochsner PE, Sendi P. Broad-range PCR in selected episodes of prosthetic joint infection. Infection (2009) 37:292–4. doi:10.1007/s15010-008-8246-1
44. Gomez E, Cazanave C, Cunningham SA, Greenwood-Quaintance KE, Steckelberg JM, Uhl JR, et al. Prosthetic joint infection diagnosis using broad-range PCR of biofilms dislodged from knee and hip arthroplasty surfaces using sonication. J Clin Microbiol (2012) 50:3501–8. doi:10.1128/JCM.00834-12
45. Pfeiffer D, editor. Interpreting diagnostic test. 1st ed. Veterinary Epidemiology: An Introduction. Oxford: Wiley-Blackwell (2010). 83 p.
Keywords: periprosthetic joint infection, culture, PCR, biofilms, antimicrobial resistance
Citation: Zegaer BH, Ioannidis A, Babis GC, Ioannidou V, Kossyvakis A, Bersimis S, Papaparaskevas J, Petinaki E, Pliatsika P and Chatzipanagiotou S (2014) Detection of bacteria bearing resistant biofilm forms, by using the universal and specific PCR is still unhelpful in the diagnosis of periprosthetic joint infections. Front. Med. 1:30. doi: 10.3389/fmed.2014.00030
Received: 23 July 2014; Paper pending published: 13 August 2014;
Accepted: 31 August 2014; Published online: 16 September 2014.
Edited by:
Evangelos Giamarellos-Bourboulis, University of Athens Medical School, GreeceReviewed by:
Magdalena Chirilă, Iuliu Haţieganu University of Medicine and Pharmacy, RomaniaIris Spiliopoulou, University of Patras, Greece
Copyright: © 2014 Zegaer, Ioannidis, Babis, Ioannidou, Kossyvakis, Bersimis, Papaparaskevas, Petinaki, Pliatsika and Chatzipanagiotou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stylianos Chatzipanagiotou, Department of Biopathology and Clinical Microbiology, Athens Medical School, Aeginition Hospital, Vass. Sophias av. 72, Athens 115 28, Greece e-mail: schatzipa@gmail.com; schatzi@med.uoa.gr
 Batool H. Zegaer
Batool H. Zegaer Anastasios Ioannidis2
Anastasios Ioannidis2
 Athanassios Kossyvakis
Athanassios Kossyvakis Joseph Papaparaskevas
Joseph Papaparaskevas Paraskevi Pliatsika
Paraskevi Pliatsika Stylianos Chatzipanagiotou
Stylianos Chatzipanagiotou