- 1 Department of Medical Microbiology and Immunology, University of Alberta, Edmonton, AB, Canada
- 2 Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, BC, Canada
Vibrio cholerae, the marine bacterium responsible for the diarrheal disease cholera, utilizes a multitude of virulence factors to cause disease. The importance of two of these factors, the toxin co-regulated pilus (TCP) and cholera toxin (CT), has been well documented for pandemic O1 and epidemic O139 serogroups. In contrast, endemic non-O1 and non-O139 serogroups can cause localized outbreaks of cholera-like illness, often in the absence of TCP and CT. One virulence mechanism used by these strains is the type VI secretion system (T6SS) to export toxins across the cell envelope and confer toxicity toward eukaryotic and prokaryotic organisms. The V. cholerae strain V52 (an O37 serogroup strain) possesses a constitutively active T6SS and was responsible for an outbreak of gastroenteritis in Sudan in 1968. To evaluate a potential role of the T6SS in the disease cholera, we compared the T6SS clusters of V. cholerae strains with sequenced genomes. We found that the majority of V. cholerae strains, including one pandemic strain, contain intact T6SS gene clusters; thus, we propose that the T6SS is a conserved mechanism that allows pandemic and endemic V. cholerae to persist both in the host and in the environment.
Introduction
Diarrheal diseases including cholera are a major cause of morbidity and mortality in developing countries worldwide (Kosek et al. (2003); Bryce et al., 2005). Several million cases and over 120,000 deaths result annually from cholera caused by the marine bacterium Vibrio cholerae (World Health Organization, 1995). Upon ingestion of contaminated water, V. cholerae passes through the gastric acid barrier in the stomach and colonizes the small intestine (Zhu and Mekalanos, 2003) with the help of toxin-coregulated pili (TCP). The bacteria secrete cholera toxin (CT) that, when internalized by epithelial cells of the small intestine, causes major water efflux into the gut lumen (Sack et al., 2004). As a result, V. cholerae leaves the human host during diarrheal purges (Larocque et al., 2005).
Over 200 different V. cholerae serogroups have been classified so far, but only the O1 serogroup strains spread globally and cause pandemics. Bacteria from this serogroup (and others like O139) use TCP to autoaggregate in the small intestine and secrete CT (Sack et al., 2004). Besides CT and TCP, V. cholerae O1 serogroup strains require accessory toxins to cause cholera, as live vaccine strains with deleted CT and TCP genes still cause severe diarrhea (Fullner et al., 2002). These accessory toxins include virulence factors such as repeats-in-toxin (RTX) toxin, the zinc-dependent metalloprotease, hemagglutinin/protease (HA/protease), and HlyA hemolysin (Fullner et al., 2002). In contrast, most other serogroups (so-called non-O1, non-O139) are either avirulent or cause severe gastroenteritis (often in the absence of cholera toxin), but have not yet spread globally (Dziejman et al., 2002). In an attempt to identify virulence factors that function independently of CT and TCP, we studied the genetic basis for the virulent behavior of V. cholerae O37 serogroup strain V52, a highly virulent strain that caused 460 cases and 125 deaths during a local outbreak in Sudan in 1968 (Zinnaka and Carpenter, 1972). Using the social amoeba Dictyostelium discoideum as a host model, we found that the type VI secretion system (T6SS) functions as a potent virulence determinant (Pukatzki et al., 2006).
It is not clear if expression of the T6SS is restricted to certain V. cholerae serogroups or whether it contributes to the human disease cholera. In this Perspective Article, we evaluate the possible role of the T6SS in the disease cholera by analyzing the presence of T6SS gene clusters in the genomes of various V. cholerae strains, including those responsible for the sixth (1899–1923) and the current seventh pandemic (>1961) (Sack et al., 2004).
The V. Cholerae T6SS
Using the social amoeba D. discoideum as a host model, we identified a large conserved cluster of 17 genes (VCA0107–0123) plus two smaller auxiliary clusters (VCA0017–VCA0021 and VC1415–VC1416) in the genome of V. cholerae strain V52 that encode a novel secretion system – the T6SS (Pukatzki et al., 2006).
Although most genes in the V. cholerae type VI clusters (Figure 1A) are conserved hypothetical genes with unknown functions, a few genes have putative functions as suggested by bioinformatic analysis (Pukatzki et al., 2009), and a series of gene deletions linked these genes to virulence toward D. discoideum (Pukatzki et al., 2006, 2007). Based on bioinformatics and experimental data, T6SS genes fall into three functional classes: structural, regulatory, and effector.
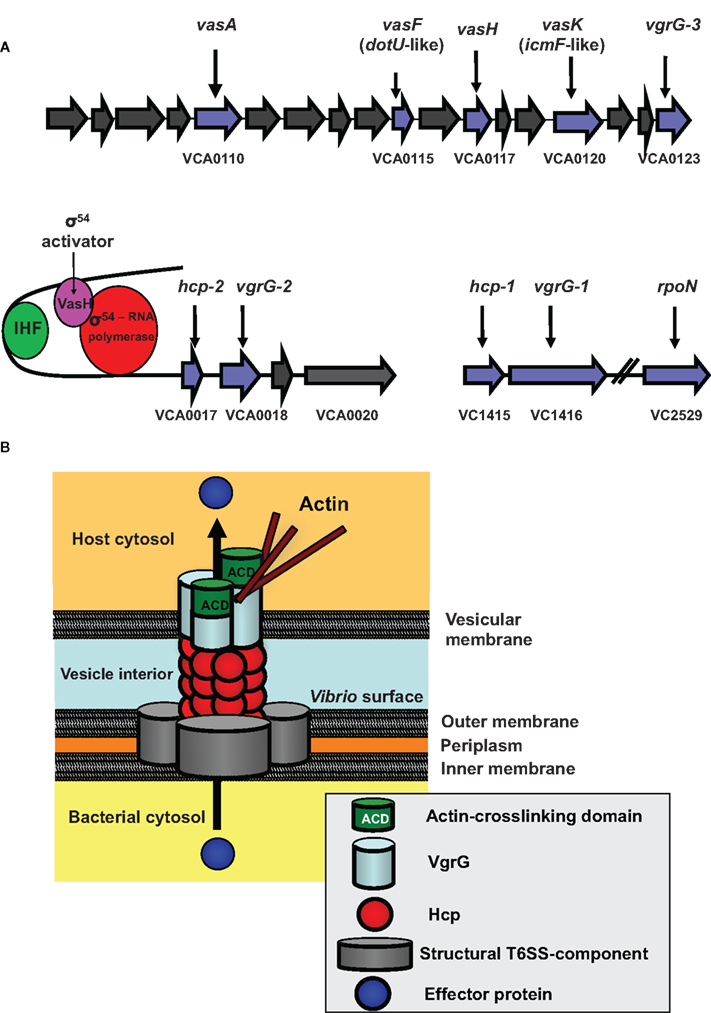
Figure 1. The V. cholerae Type VI Secretion System (T6SS). (A) The T6SS gene clusters. The three gene clusters contain genes of predicted (blue) and unknown (gray) functions. VCA0117 encodes the gene designated vasH. Also shown is the VasH-dependent regulation of the T6SS genes. (IHF, integration host factor). (B) Model for T6SS mechanics. Structural components form the base of the T6SS in the bacterial inner and outer membrane. A surface-exposed Hcp tube decorated with a VgrG trimer punctures a vesicular membrane to translocate effectors into the host cell (see text for details).
Structural Components
We do not know how the T6SS proteins assemble to form a secretion apparatus; however, several type VI proteins are structurally related to components of the T4 bacteriophage tail complex (Kanamaru et al., 2002). This protein complex is located at the distal tip of the syringe-shaped phage tail and allows the phage to puncture the bacterial envelope and inject its DNA into a bacterium. We speculate that bacteria deploy their type VI secretion machinery in a similar fashion to puncture host membranes. We identified four T6SS components that show high structural homology with T4 bacteriophage proteins. These are the hemolysin-coregulated protein (Hcp) and three VgrG (valine-glycine-repeat protein G) proteins (VgrG1–3). Export of these four proteins requires the putative structural gene product VasK, which shows high similarity to the IcmF component of the type IV secretion system (T4SS) of Legionella pneumophila (Segal et al., 1998).
Bioinformatic analyses predict that the T6SS contains a tube-like structure. Monomers that could assemble into such a tube are the small 19-kD Hcp-proteins encoded by the two alleles VCA0017 and VC1415, which share 98% similarity and encode identical proteins. In support of this model, Ballister et al. (2008) found that recombinant Hcp from Pseudomonas aeruginosa readily assembles in vitro into hexameric rings to form nanotubes. Our bioinformatic analyses of VgrGs revealed that these T6SS components are members of a novel protein family (Pukatzki et al., 2007). The recently solved crystal structure (Leiman et al., 2009) of a VgrG molecule from uropathogenic Escherichia coli confirmed our bioinformatic prediction that VgrG proteins form a trimeric cap similar to the cell-puncturing device of the T4 bacteriophage (Pukatzki et al., 2007) (Figure 1B).
The current model (Figure 1B) proposes that after assembly of the VgrG-cap is completed, Hcp molecules form a hexameric ring that may dock underneath the VgrG trimer to build a doublet ring with a continuous central pore. As more hexameric Hcp rings form and attach to the existing structure, the Hcp tube, decorated with a single trimeric VgrG-cap at its tip, may be pushed across the V. cholerae membrane into the extracellular space. Interestingly, two VgrGs (VgrG-1 and VgrG-3) have evolved additional functions, such as actin-crosslinking, and serve as effectors when pushed into the host cytosol (Ma et al., 2009). Because the VgrG-cap lacks a channel, the tube is likely to be a gated complex (Leiman et al., 2009), in which case, the cap would need to dissociate from the complex for soluble substrates to pass through the Hcp channel. Recent findings by Ma et al. suggest that the actin-crosslinking domain of VgrG-1 is translocated in vivo, because infection of infant mice with V. cholerae strain V52 results in a T6SS-dependent inflammatory response and actin-crosslinking in cells of the small intestine (Ma and Mekalanos, 2010).
Regulatory Components
Our transposon mutagenesis screen identified two type VI regulators – VasH, positioned in the center of the large type VI cluster (Figure 1A), and rpoN, encoding the alternative sigma factor-54 (Pukatzki et al., 2006). Both vasH and rpoN mutants were attenuated in their ability to infect and kill D. discoideum. As vasH and rpoN mutant phenotypes are indistinguishable, they may act in the same regulatory pathway (Pukatzki et al., 2006).
VasH encodes a 59-kD protein that has the hallmarks of a sigma-54-dependent regulator with domains that may function as (i) an N-terminal regulatory domain, (ii) an AAA + central core with a putative ATP hydrolysis function, and (iii) a DNA binding domain with a conserved helix-turn-helix motif (Shingler, 1996; Schumacher et al., 2006) (Figure 2B). A vasH-null strain does not produce Hcp and is avirulent toward D. discoideum (Pukatzki et al., 2006). Analyses of both hcp promoters revealed conserved binding sites for integration host factor and sigma-54 (Williams et al., 1996). Integration host factor bends DNA such that a sigma-54 activator bound to a distant upstream activation sequence can directly interact with the promoter-bound sigma-54 of the RNA polymerase-holoenzyme (Stonehouse et al., 2008). We predict that the same mechanism is true for VasH, which is highly expressed during human infections (Lombardo et al., 2007). VasH is polymorphic in V. cholerae (Figures 2B and C) and it will be exciting to determine if this heterogeneity contributes to the strain-specific regulation of T6SS genes.
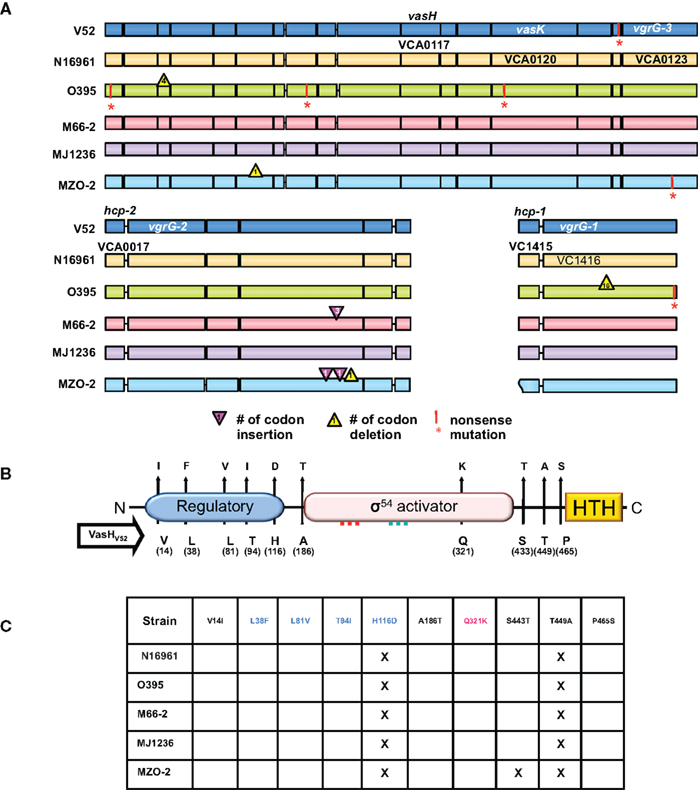
Figure 2. T6SS clusters in V. cholerae strains with sequenced genomes. (A) Genomic alignments. Nucleotide sequences used for this study were obtained from Broad Institute and NCBI databases and compared with the blastn program from the stand-alone NCBI BLAST application (version 2.2.21). In-house Perl scripts were written to parse BLAST output and to compare genomic sequences of gene models for variations. BioPerl (http://www.bioperl.org/) modules were used. Regions containing the T6SS clusters of various V. cholerae strains (names on the left) were aligned. Each gene is indicated by a box in a color distinct for a particular strain. A vertical bar and an asterisk indicate the 3′-end of a truncated gene. Codon insertions are indicated by red triangles, while deletions are indicated by yellow triangles with numbers that indicate the insertion/deletion size (number of codons). Rough ends of a gene indicate that the complete sequence was not available. (B) Polymorphic domains of VasH. Graphical depiction of VasH from the O37 serogroup strain V52 (V52_VasH) with its putative N-terminal regulatory domain (blue), core sigma-54 activator domain (pink), and C-terminal helix-turn-helix (HTH) domain (yellow) (drawn to scale). Conserved amino acid substitutions are indicated by arrows. The residue and its position in VasH of V52 is indicated below the line and the substitutions found in other strains are indicated above the arrowhead. (C) Allelelic VasH variants. Listed are VasH variants of different V. cholerae strains compared to VasH in V52. Amino acid substitutions are listed for each variant. Font color indicates in which of the three domains the substitution is located.
Effector Components
It is not known if molecules commonly called “effectors” are transported through the central channel of the type VI apparatus. Candidates for such soluble effectors are the autoinducer-2 (AI-2) binding protein RbsB from Rhizobium leguminosarum (Bladergroen et al., 2003), EvpP from Edwardsiella tarda (Zheng and Leung, 2007), and AaiC from enteroaggregative E. coli (Dudley et al., 2006); however, V. cholerae does not carry homologs of these putative T6SS substrates.
Distribution of T6SS Clusters Among V. Cholerae Strains
While the gene content differs tremendously between V. cholerae serogroups and strains in respect to virulence genes – including the genes encoding cholera toxin or the genes encoding the type III secretion system (T3SS) (Tam et al., 2007) – all V. cholerae serogroups sequenced thus far contain all three gene clusters involved in the T6SS (Figure 2A) (Pukatzki et al., 2006).
To assess the distribution of T6SS clusters in V. cholerae genomes, we compared V. cholerae strains for which sequenced genomes are available. As illustrated in Figure 2A, we included the O1 classical strain O395 (sixth pandemic), O1 El Tor strains M66-2 (pre-seventh pandemic), and N16961 (seventh pandemic), the El Tor strain MJ-1236 that carries a classical biotype CTX prophage (so-called hybrid strain), and the non-O1/non-O139 strains V52 (O37 serogroup) and MZO-2 (O14 serogroup). Both V52 (Zinnaka and Carpenter, 1972) and MZO-2 (GenBank Taxonomy No. 417398) have been responsible for gastrointestinal illness that failed to become pandemic. The genome of V52 was used as the reference genome as this strain produces a functional T6SS (Pukatzki et al., 2006). The large T6SS cluster of N16961 contains 285 single nucleotide substitutions of which 49 are non-synonymous substitutions that result in amino acid changes in all 17 genes of the V. cholerae type VI cluster.
We found a complete T6SS complement in each strain tested. The classical O1 strain O395 has four non-sense mutations, including a mutation in VCA0120 (vasK) that is essential for T6SS function (Pukatzki et al., 2006); thus the T6SS of the O1 strain O395 may be non-functional. Interestingly, one gene, VCA0122, encodes an 80-amino acid polypeptide in N16961, O395, M66-2, MJ-1236, and MZO-2, while the V52 genome encodes a shorter 70-residue protein with a unique 27 residue C-terminus. It must be noted that truncation of the VCA0122-encoded protein in V52 does not correlate with impaired T6SS function, as V52 readily engages in T6SS-mediated toxicity (Pukatzki et al., 2006). The unique VCA0122-encoded protein may contribute to the constitutive production of the V52 T6SS. The satellite cluster VCA0017-22 appears to be intact in all strains. While hcp-2 (VCA0017) and vgrG-2 (VCA0018) are highly polymorphic genes encoding full-length products, VCA0020 contains three extra codons in M66-2 and two extra codons in MZO-2 plus a one-codon deletion. The genes hcp-1 (VC1415) and vgrG-1 (VC1416) are highly conserved and share identical sequences in V52, N16961, M66-2, and MJ-1236. Hcp-1 of O395 shares an identical sequence with these strains, however its vgrG-1 contains a 16-codon in-frame deletion and a non-sense mutation. Furthermore, VgrG-1 of MZO-2 is highly polymorphic and differs from V52 in 15 residues.
From a genomic perspective there is reason to believe that many V. cholerae strains, including post-sixth pandemic strains, carry functional T6SSs. Even though important T6SS genes are polymorphic, the vast majority of nucleotide substitutions are synonymous and do not alter the sequence of the encoded protein. Substitutions that alter the protein are for the most part conserved. The fact that most mutations are silent and do not lead to truncated gene products suggests to us that V. cholerae is subjected to selective pressure that maintains an operative T6SS. It should be noted that even though V52 belongs to the non-O1/non-O139 group of V. cholerae, it is more closely related to the classical O1 strain O395 than to the El Tor strain N16961 (Chun et al., 2009); yet, O395 accumulated at least four non-sense mutations in the T6SS cluster while N16961 accumulated none. Thus, unlike N16961, O395 might have lost the need for a functional T6SS and has accumulated disabling mutations in the T6SS cluster.
Expression of the T6SS Complement
If some endemic and pandemic V. cholerae strains carry a functional T6SS complement, we need to understand why some strains use their system constitutively under laboratory conditions while others do not (Pukatzki et al., 2006). One explanation is that strains regulate their systems differently: while some clinical and environmental V. cholerae strains express the T6SS constitutively, pandemic strains may be more selective. There is precedence for strain-specific regulation of virulence genes in V. cholerae: the ToxR regulons of classical and El Tor strains require different in vitro growth conditions to express the genes encoding CT and TCP, while the classical O1 strain 569B expresses CT and TCP constitutively (Majumder et al., 1996). We hypothesize that the T6SS behaves similarly such that pandemic strains require yet to be identified signals, while V52 expresses its T6SS in the absence of such signals.
The fact that the T6SS genes identified in V52 are maintained in the pandemic El Tor strain N16961 implies that the T6SS genes are under selection in N16961 and have not drifted. One reason that N16961 does not kill amoebae may be that this strain uses a regulatory T6SS network that is not activated under laboratory conditions. Genomic comparison of vasH (VCA0117), the gene that encodes the T6SS regulator VasH, revealed that VasH is polymorphic and differs between strains (Figures 2B and C). The polymorphisms occur in interesting places: 10 conserved amino acid substitutions give rise to 10 variants. Five substitutions occur in the putative regulatory N-terminal domain, and the only substitution in the activator core domain is located just 27 residues downstream from the Walker B motif. Three substitutions are located between the core domain and the DNA-binding domain. It will be interesting to see if the polymorphic nature of the T6SS cluster, including vasH, accounts for the different utilization of this virulence determinant.
A Working Model
If the T6SS is a universal feature used by V. cholerae, then how does the T6SS contribute to the persistent and virulent behavior of V. cholerae? We recently demonstrated that V. cholerae uses its T6SS to kill eukaryotic phagocytes (Pukatzki et al., 2006). The structural homology of the T6SS and the cell-puncturing device of the T4 bacteriophage suggests that the T6SS can target bacteria, an activity that has been demonstrated experimentally for V. cholerae (MacIntyre et al., 2010), P. aeruginosa and Burkholderia thailandensis (Hood et al., 2010; Schwarz et al., 2010).
We propose that V. cholerae uses its T6SS to outcompete bacterial neighbors as well as eukaryotic predators like mammalian immune cells and amoebae. As shown in Figure 3, V. cholerae must colonize the small intestine to initiate an infection. However, the epithelial lining of the small intestine contains commensal bacteria, some of which secrete products harmful to V. cholerae (step 1). V. cholerae passes the gastric acid barrier can reach the lumen of the small intestine (step 2) where it may turn on its T6SS to kill commensal bacteria to clear competitors from epithelial surfaces required for colonization (step 3). Activation of the T6SS likely involves the putative transcriptional activator VasH, which we hypothesize activates RNA polymerase bound to the sigma-54 binding sites of selective promoters. We do not know how VasH is regulated, but bioinformatics analysis allows us to speculate that VasH may function as the receiver module of a yet to be identified two-component system that is activated in response to unknown external stimuli. Once activated, VasH triggers the expression of genes encoding important structural proteins such as Hcp and VgrG. As V. cholerae multiplies and continues to kill adjacent commensals, it begins to secrete cholera toxin (step 4). The infection is recognized by the host, and phagocytic immune cells invade the area to clear the infection. As V. cholerae is phagocytosed, the T6SS is activated to kill the infected immune cells (step 5), thereby allowing the infection to proceed. If this model is correct, it would be interesting to determine if there are distinct environmental signals that activate the T6SS depending on whether they are challenged by immune cells or other prokaryotic competitors. The release of cholera toxin triggers massive water efflux into the gut lumen, and V. cholerae leaves the small intestine during diarrheal purges (step 6). V. cholerae leaving the human host is primed to ward off bacterial and amoeboid predators in the environment they are about to encounter (step 7).

Figure 3. A model for the role of the T6SS in persistence and virulence of V. cholerae. (1) The small intestine colonized with commensal bacteria is (2) invaded by V. cholerae cells that passed the gastric acid barrier. (3) Upon descent into the ducts between adjacent villi of the small intestine, V. cholerae assembles the T6SS on its surface (indicated by black spikes) in order to kill commensal microorganisms. (4) The sterile patches generated allow V. cholerae to colonize and multiply. (5) Incoming immune cells that ingest V. cholerae are killed by the T6SS-mediated toxicity, allowing the infection to proceed. (6) Secretion of cholera toxin results in massive water efflux into the host lumen, and V. cholerae cells leave the human host during diarrheal purges. (7) V. cholerae exiting the body maintain T6SS activation in preparation for bacterial and eukaryotic predators in the environment.
Conclusions
The predominant presence of T6SS genes in V. cholerae genomes suggests a fundamental function for this pathway. Based on the toxicity displayed by this secretion system in V52, we hypothesize that the T6SS and toxin translocation play key roles in allowing V. cholerae to outcompete other bacteria and phagocytic cells (i.e., amoebae and macrophages) and thus persist in human hosts and in the environment. If this hypothesis is correct, the T6SS would maximize the pathogenicity imposed by strain-specific cholera toxin and other virulence factors. As many environmental and enteric Gram-negative proteobacteria carry T6SS genes (Folkesson et al., 2002; Gray et al., 2002; Das and Chaudhuri, 2003; Nano et al., 2004; Rao et al., 2004; Parsons and Heffron, 2005; Dudley et al., 2006; Mougous et al., 2006; de Bruin et al., 2007; Mougous et al., 2007; Schell et al., 2007; Shalom et al., 2007; Zheng and Leung, 2007; Aubert et al., 2008; Suarez et al., 2008; Yen et al., 2008), future research will determine if use of the T6SS for competition purposes is a common theme among microbes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Daniele Provenzano (University of Texas Brownsville) and members of the Pukatzki laboratory for helpful discussions. We are grateful to Marcia Craig for critically reviewing the manuscript and to Steven McAuley for help with the figures. Stefan Pukatzki is a recipient of the Alberta Heritage Foundation for Medical Research Scholar Award. Lukasz Wieteska is an IAESTE (The International Association for the Exchange of Students for Technical Experience) scholar. Work in Stefan Pukatzki’s laboratory is supported by the Canadian Institute for Health Research Operating Grant MOP-84473, and Alberta Innovates – Health Solutions (funded by the Alberta Heritage Foundation for Medical Research Endowment Fund). Research for this project in Nansheng Chen’s laboratory is supported by a Discovery Grant of the National Sciences and Engineering Research Council (NSERC) of Canada. Nansheng Chen is a Michael Smith Foundation for Health Research (MSFHR) Scholar and a CIHR New Investigator.
References
Aubert, D. F., Flannagan, R. S., and Valvano, M. A. (2008). A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect. Immun. 76, 1979–1991.
Ballister, E. R., Lai, A. H., Zuckermann, R. N., Cheng, Y., and Mougous, J. D. (2008). In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc. Natl. Acad. Sci. U.S.A. 105, 3733–3738.
Bladergroen, M. R., Badelt, K., and Spaink, H. P. (2003). Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant Microbe. Interact. 16, 53–64.
Bryce, J., Boschi-Pinto, C., Shibuya, K., and Black, R. E. (2005). WHO estimates of the causes of death in children. Lancet 365, 1147–1152.
Chun, J., Grim, C. J., Hasan, N. A., Lee, J. H., Choi, S. Y., Haley, B. J., Taviani, E., Jeon, Y. S., Kim, D. W., Lee, J. H., Brettin, T. S., Bruce, D. C., Challacombe, J. F., Detter, J. C., Han, C. S., Munk, A. C., Chertkov, O., Meincke, L., Saunders, E., Walters, R. A., Huq, A., Nair, G. B., and Colwell, R. R. (2009). Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 106, 15442–15447.
Das, S., and Chaudhuri, K. (2003). Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol. 3, 287–300.
de Bruin, O. M., Ludu, J. S., and Nano, F. E. (2007). The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 7, 1.
Dudley, E. G., Thomson, N. R., Parkhill, J., Morin, N. P., and Nataro, J. P. (2006). Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 61, 1267–1282.
Dziejman, M., Balon, E., Boyd, D., Fraser, C. M., Heidelberg, J. F. and Mekalanos, J. J. (2002). Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. U.S.A. 99, 1556–1561.
Folkesson, A., Lofdahl, S., and Normark, S. (2002). The Salmonella enterica subspecies I specific centisome 7 genomic island encodes novel protein families present in bacteria living in close contact with eukaryotic cells. Res. Microbiol. 153, 537–545.
Fullner, K. J., Boucher, J. C., Hanes, M. A., Haines, G. K. III, Meehan, B. M., Walchle, C., Sansonetti, P. J., and Mekalanos, J. J. (2002). The contribution of accessory toxins of Vibrio cholerae O1 El Tor to the proinflammatory response in a murine pulmonary cholera model. J. Exp. Med. 195, 1455–1462.
Gray, C. G., Cowley, S. C., Cheung, K. K., and Nano, F. E. (2002). The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol. Lett. 215, 53–56.
Hood, R. D., Singh, P., Hsu, F., Guvener, T., Carl, M. A., Trinidad, R. R., Silverman, J. M., Ohlson, B. B., Hicks, K. G., Plemel, R. L., Li, M., Schwarz, S., Wang, W. Y., Merz, A. J., Goodlett, D. R., and Mougous, J. D. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37.
Kanamaru, S., Leiman, P. G., Kostyuchenko, V. A., Chipman, P. R., Mesyanzhinov, V. V., Arisaka, F., and Rossmann, M. G. (2002). Structure of the cell- puncturing device of bacteriophage T4. Nature 415, 553–557.
Kosek, M., Bern, C., and Guerrant, R. L. (2003). The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull. World Health Organ 81, 197–204.
Larocque, R. C., Harris, J. B., Dziejman, M., Li, X., Khan, A. I., Faruque, A. S., Faruque, S. M., Nair, G. B., Ryan, E. T., Qadri, F., Mekalanos, J. J., and Calderwood, S. B. (2005). Transcriptional profiling of Vibrio cholerae recovered directly from patient specimens during early and late stages of human infection. Infect. Immun. 73, 4488–4493.
Leiman, P. G., Basler, M., Ramagopal, U. A., Bonanno, J. B., Sauder, J. M., Pukatzki, S., Burley, S. K., Almo, S. C., and Mekalanos, J. J. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A.106, 4154–4159.
Lombardo, M. J., Michalski, J., Martinez-Wilson, H., Morin, C., Hilton, T., Osorio, C. G., Nataro, J. P., Tacket, C. O., Camilli, A., and Kaper, J. B. (2007). An in vivo expression technology screen for Vibrio cholerae genes expressed in human volunteers. Proc. Natl. Acad. Sci. U.S.A. 104, 18229–18234.
Ma, A. T., McAuley, S., Pukatzki, S., and Mekalanos, J. J. (2009). Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host. Microbe 5, 234–243.
Ma, A. T., and Mekalanos, J. J. (2010). In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 4365–4370.
MacIntyre, D., Miyata, S.T., Kitaoka, M., and Pukatzki, S. (2010). The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. USA. October 25 [Epub ahead of print].
Majumder, R., Sengupta, S., Khetawat, G., Bhadra, R. K., Roychoudhury, S., and Das, J. (1996). Physical map of the genome of Vibrio cholerae 569B and localization of genetic markers. J. Bacteriol. 178, 1105–1112.
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., Goodman, A. L., Joachimiak, G., Ordonez, C. L., Lory, S., Walz, T., Joachimiak, A., and Mekalanos, J. J. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530.
Mougous, J. D., Gifford, C. A., Ramsdell, T. L., and Mekalanos, J. J.. (2007). Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell. Biol. 9, 797–803.
Nano, F. E., Zhang, N., Cowley, S. C., Klose, K. E., Cheung, K. K., Roberts, M. J., Ludu, J. S., Letendre, G. W., Meierovics, A. I., Stephens, G., and Elkins, K. L. (2004). A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 186, 6430–6436.
Parsons, D. A., and Heffron, F. (2005). sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect. Immun. 73, 4338–4345.
Pukatzki, S., Ma, A. T., Revel, A. T., Sturtevant, D., and Mekalanos, J. J. (2007). Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513.
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., Heidelberg, J. F., and Mekalanos, J. J. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533.
Pukatzki, S., McAuley, S. B., and Miyata, S. T. (2009). The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12, 11–17.
Rao, P. S., Yamada, Y., Tan, Y. P., and Leung, K. Y. (2004). Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol. Microbiol. 53, 573–586.
Schell, M. A., Ulrich, R. L., Ribot, W. J., Brueggemann, E. E., Hines, H. B., Chen, D., Lipscomb, L., Kim, H. S., Mrazek, J., Nierman, W. C., and Deshazer, D. (2007). Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 64, 1466–1485.
Schumacher, J., Joly, N., Rappas, M., Zhang, X., and Buck, M. (2006). Structures and organisation of AAA + enhancer binding proteins in transcriptional activation. J. Struct. Biol. 156, 190–199.
Schwarz, S., West, T. E., Boyer, F., Chiang, W. C., Carl, M. A., Hood, R. D., Rohmer, L., Tolker-Nielsen, T., Skerrett, S. J., and Mougous, J. D. (2010). Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6, 1–14. doi: 10.1371/journal.ppat.1001068.
Segal, G., Purcell, M., and Shuman, H. A. (1998). Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U.S.A.95, 1669–1674.
Shalom, G., Shaw, J., and Thomas, M. (2007). In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153, 2689–2699.
Shingler, V. (1996). Signal sensing by sigma 54-dependent regulators: derepression as a control mechanism. Mol. Microbiol. 19, 409–416.
Stonehouse, E., Kovacikova, G., Taylor, R. K., and Skorupski, K. (2008). Integration host factor positively regulates virulence gene expression in Vibrio cholerae. J. Bacteriol. 190, 4736–4748.
Suarez, G., Sierra, J. C., Sha, J., Wang, S., Erova, T. E., Fadl, A. A., Foltz, S. M., Horneman, A. J., and Chopra, A. K. (2008). Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44, 344–361.
Tam, V. C., Serruto, D., Dziejman, M., Brieher, W., and Mekalanos, J. J. (2007). A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe 1, 95–107.
Williams, S. G., Varcoe, L. T., Attridge, S. R., and Manning, P. A. (1996). Vibrio cholerae Hcp, a secreted protein coregulated with HlyA. Infect. Immun. 64, 283–289.
World Health Organization. (1995). Meeting on the Potential Role of New Cholera Vaccines in the Prevention and Control of Cholera outbreaks during Acute Emergencies. in Document CDR/GPV/95.1 Geneva, World Health Organization.
Yen, Y. T., Bhattacharya, M., and Stathopoulos, C. (2008). Genome-wide in silico mapping of the secretome in pathogenic Yersinia pestis KIM. FEMS Microbiol. Lett. 279, 56–63.
Zheng, J., and Leung, K. Y. (2007). Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66, 1192–1206.
Keywords: Vibrio cholerae, cholera, type VI secretion system
Citation: Miyata ST, Kitaoka M, Wieteska L, Frech C, Chen N and Pukatzki S (2010) The Vibrio cholerae type VI secretion system: evaluating its role in the human disease cholera. Front. Microbio. 1:117. doi: 10.3389/fmicb.2010.00117
Received: 21 August 2010;
Paper pending published: 27 September 2010;
Accepted: 06 October 2010;
Published online: 15 November 2010
Edited by:
John S. Gunn, Ohio State University, USAReviewed by:
Rita Tamayo, University of North Carolina at Chapel Hill, USA;Guangming Zhong, University of Texas Health Science Center, USA
Copyright: © 2010 Miyata, Kitaoka, Wieteska, Frech, Chen and Pukatzki. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Stefan Pukatzki, Department of Medical Microbiology & Immunology, University of Alberta, 1-63 Medical Sciences Building, Edmonton, AB, T6G 2H7 Canada. e-mail: spukatzki@ualberta.ca
