- 1 Department of Microbiology and Immunology, College of Medicine, University of Louisville, Louisville, KY, USA
- 2 Department of Biology, University of Louisville, Louisville, KY, USA
- 3 Department of Microbiology, Al-Quds University Medical School, Abu Dies, Jerusalem, Palestinian Territory
- 4 Department of Microbiology and Molecular Genetics, The Hebrew University Medical School, Jerusalem, Israel
Microbial pathogens have evolved exquisite mechanisms to interfere and intercept host biological processes, often through molecular mimicry of specific host proteins. Ubiquitination is a highly conserved eukaryotic post-translational modification essential in determining protein fate, and is often hijacked by pathogenic bacteria. The conserved SKP1/CUL1/F-box (SCF) E3 ubiquitin ligase complex plays a key role in ubiquitination of proteins in eukaryotic cells. The F-box protein component of the SCF complex provides specificity to ubiquitination by binding to specific cellular proteins, targeting them to be ubiquitinated by the SCF complex. The bacterial pathogens. Legionella pneumophila, Agrobacterium tumefaciens, and Ralstonia solanacearum utilize type III or IV translocation systems to inject into the host cell eukaryotic-like F-box effectors that interact with the host SKP1 component of the SCF complex to trigger ubiquitination of specific host cells targets, which is essential to promote proliferation of these pathogens. Our bioinformatic analyses have identified at least 74 genes encoding putative F-box proteins belonging to 22 other bacterial species, including human pathogens, plant pathogens, and amebal endosymbionts. Therefore, subversion of the host ubiquitination machinery by bacterial F-box proteins may be a widespread strategy amongst pathogenic bacteria. The findings that bacterial F-box proteins harbor Ankyrin repeats as protein–protein interaction domains, which are present in F-box proteins of primitive but not higher eukaryotes, suggest acquisition of many bacterial F-box proteins from primitive eukaryotic hosts rather than the mammalian host.
Introduction
Bacterial pathogens exhibit an intimate relationship with host cells that involves direct subversion of host pathways by a myriad of microbial derived effectors that are injected into the host cell by specialized type III–VII translocation systems. Pathogens actively evade and manipulate vesicular trafficking to avoid lysosomal degradation, modulate the actin cytoskeleton and interfere with numerous cellular signaling cascades to help establish their replicative niche (Franco et al., 2009; Galan, 2009; Ibarra and Steele-Mortimer, 2009; Isberg et al., 2009). A particularly elegant manipulation of host cell processes is the ability of bacteria to hijack the host ubiquitination system, often through molecular mimicry of host proteins.
Ubiquitination is an essential post-translational protein modification conserved among all eukaryotic organisms. It involves the covalent addition of a 76-amino acid ubiquitin polypeptide to a lysine residue of a target protein. Ubiquitination of proteins regulates a wide range of important cellular processes such as DNA repair, signaling, endocytosis, vesicular trafficking, and cell cycle progression and thus plays a key role in cellular homeostasis (Welchman et al., 2005; Kerscher et al., 2006). The process of ubiquitination involves the addition of a single or several ubiquitin moieties to a target protein. The conjugation of a single ubiquitin, monoubiquitination, often directs protein localization and activity of the targeted protein (Welchman et al., 2005; Kerscher et al., 2006; Raiborg et al., 2006). In contrast, the addition of further ubiquitin moieties, building a complex polyubiquitin chain, often targets a protein for 26S proteasome-mediated degradation or can modulate its biological activity (Welchman et al., 2005; Kerscher et al., 2006). The fate of polyubiquitinated proteins is dependent on the linkage pattern between individual ubiquitin moieties making up the chain (Welchman et al., 2005; Kerscher et al., 2006). Generally, if the ubiquitin moieties are covalently linked via their lysine 48 residues, they target the protein for 26S proteasome-mediated degradation, or if they are linked by the lysine 63 residue this alters protein activity and trafficking in the cell (Pickart and Fushman, 2004). However, the functional implications of other lysine linkages in polyubiquitin chains such as K6, K11, K27, K29, and K33 are not well understood (Xu et al., 2009).
Ubiquitination involves the sequential activity of three classes of enzymes (Kerscher et al., 2006). Initially, ubiquitin is activated by an E1 ubiquitin activating enzyme that binds to the C-terminus of ubiquitin, via a thioester linkage to a cysteine residue in the enzyme (Kirkpatrick et al., 2005). This linked ubiquitin is then transferred to another cysteine residue present within an E2 ubiquitin-conjugating enzyme (Zhang et al., 2005). Humans harbor at least 35 different E2 conjugating enzymes and these are chiefly responsible for directing different polyubiquitin chain patterns (van Wijk and Timmers, 2010). This is followed by a substrate-specific E3 ubiquitin ligase that transfers ubiquitin to the target protein (Ardley and Robinson, 2005). Specificity in this process is provided by the large family of >1000 estimated E3 human enzymes, each of which recognizes a specific substrate (Petroski and Deshaies, 2005). E3 ubiquitin ligases are classified into two major groups, the HECT-type E3 ubiquitin ligases and the RING-type ubiquitin ligases (Ardley and Robinson, 2005). RING-type E3 ubiquitin ligases consist of either a single protein (U-box domain proteins) or a multi-subunit complex such as the SKP1-CUL1-F-box (SCF)-complex (Ardley and Robinson, 2005).
The SCF Complex
The SCF complex (Figure 1) contains the RING-domain protein RING-box 1 (RBX1), cullin 1 (CUL1), S-phase-kinase associated protein 1 (SKP1), and a protein harboring an F-box domain, that directly binds SKP1 (Schulman et al., 2000). These F-box-containing proteins also typically have a leucine-rich repeat (LRR) or WD40 protein-binding domains, which bind specific substrates to be ubiquitinated and target them to the E3 ligase multimeric complex that is bound to the F-box domain (Smith et al., 1999; Kobe and Kajava, 2001) (Figure 1). In humans there are at least ∼70 F-box proteins (Jin et al., 2004), while the plant Arabidopsis has over 700 F-box proteins (Gagne et al., 2002), indicating that regulation of protein ubiquitination by the SCF ligase machinery is a key mechanism controlling protein fate in eukaryotic cells. F-box proteins in both invertebrates and vertebrates have been extensively reviewed (Lechner et al., 2006; Ho et al., 2006, 2008). Given the broad range of cellular processes controlled by SCF-mediated ubiquitination and the large number of F-box proteins giving specificity to the reaction, this mechanism is particularly sensitive to hijacking by bacterial pathogens, allowing these organisms to manipulate the host ubiquitination pathway to their advantage.
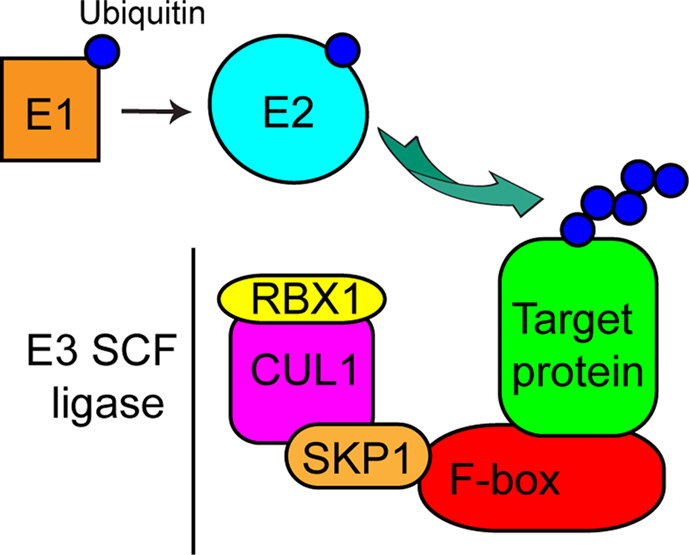
Figure 1. Model of the eukaryotic SCF complex. Cartoon representation of the eukaryotic SCF complex and the E1 activating and E2 conjugating enzymes. E1 transfers ubiquitin to E2, which then conjugates ubiquitin to a target protein associated with the F-box protein within the E3 SCF multi-protein complex.
Several species of pathogenic Gram-negative bacteria harbor elaborate type III–VII secretion systems that inject a myriad of virulence proteins, termed effectors, into the host cell, which modulate various cellular processes to promote proliferation of the invading pathogen (Christie and Cascales, 2005; Cornelis, 2006). Many of these translocated effectors exhibit structure and function mimicry of eukaryotic proteins and this enables them to interfere with various cellular processes including vesicular trafficking, cytoskeletal rearrangement and intracellular trafficking (Franco et al., 2009; Galan, 2009; Hauser, 2009; Ibarra and Steele-Mortimer, 2009; Isberg et al., 2009). A growing number of type III–VII effectors have been shown to manipulate the host cell ubiquitin system through molecular mimicry of the F-box protein component of the SCF E3 ubiquitin ligases (Schrammeijer et al., 1998, 2001; Angot et al., 2006; Kajava et al., 2008; Price et al., 2009). Bacteria do not possess the other enzymatic components of the SCF complex nor the ubiquitination machinery. Therefore, genes encoding bacterial F-box proteins have likely evolved through inter-kingdom horizontal gene transfer from eukaryotes to bacteria during close bacterial/host interaction. This review will discuss recent advances in the understanding of bacterial F-box proteins and their role in manifestation of disease caused by Agrobacterium tumefaciens, Ralstonia solanacearum, and Legionella pneumophila. In addition, bioinformatic analyses of whole bacterial genome sequences have revealed the presence of putative eukaryotic-like F-box proteins in many other bacterial species, including the human pathogen Coxiella burnetii, various plant pathogens, and amebal endosymbionts. It is likely that the strategy to subvert host ubiquitination through molecular mimicry of eukaryotic F-box proteins is a widespread phenomenon among prokaryotes that have a close interaction with eukaryotic cells.
F-Box Proteins of Legionella
The Legionnaires’ disease causing bacterium, L. pneumophila, replicates within alveolar macrophages causing pneumonia (Isberg et al., 2009). The organism is transmitted to humans from the aquatic environment where L. pneumophila replicates within ameba and ciliates (Molmeret et al., 2005; Franco et al., 2009). Co-evolution and adaptation of L. pneumophila to the intracellular lifestyle within ameba in the aquatic environment is believed to have played a major role in its ability to exploit evolutionarily conserved eukaryotic processes that enables its proliferation within human alveolar macrophages (Molmeret et al., 2005; Franco et al., 2009). Within both evolutionarily distant host cells, L. pneumophila evades endocytic fusion and intercepts ER-to-Golgi vesicular traffic to remodel its phagosome into an ER-derived vacuole that is decorated with polyubiquitinated proteins (Kagan and Roy, 2002; Molmeret et al., 2005; Dorer et al., 2006; Shin and Roy, 2008; Isberg et al., 2009). The Dot/Icm type IV secretion system (Segal et al., 1998; Vogel et al., 1998) is triggered upon intimate attachment to the host cell membrane (Franco et al., 2009) to inject into the host cell a cadre of ∼200 effectors to modulate a myriad of cellular processes to re-program the host cell into a proliferation niche (de Felipe et al., 2008; Shin and Roy, 2008; Isberg et al., 2009) and many of these effectors harbor eukaryotic-like domains. Among these are a number of proteins harboring eukaryotic like F-box domains (Figure 2).
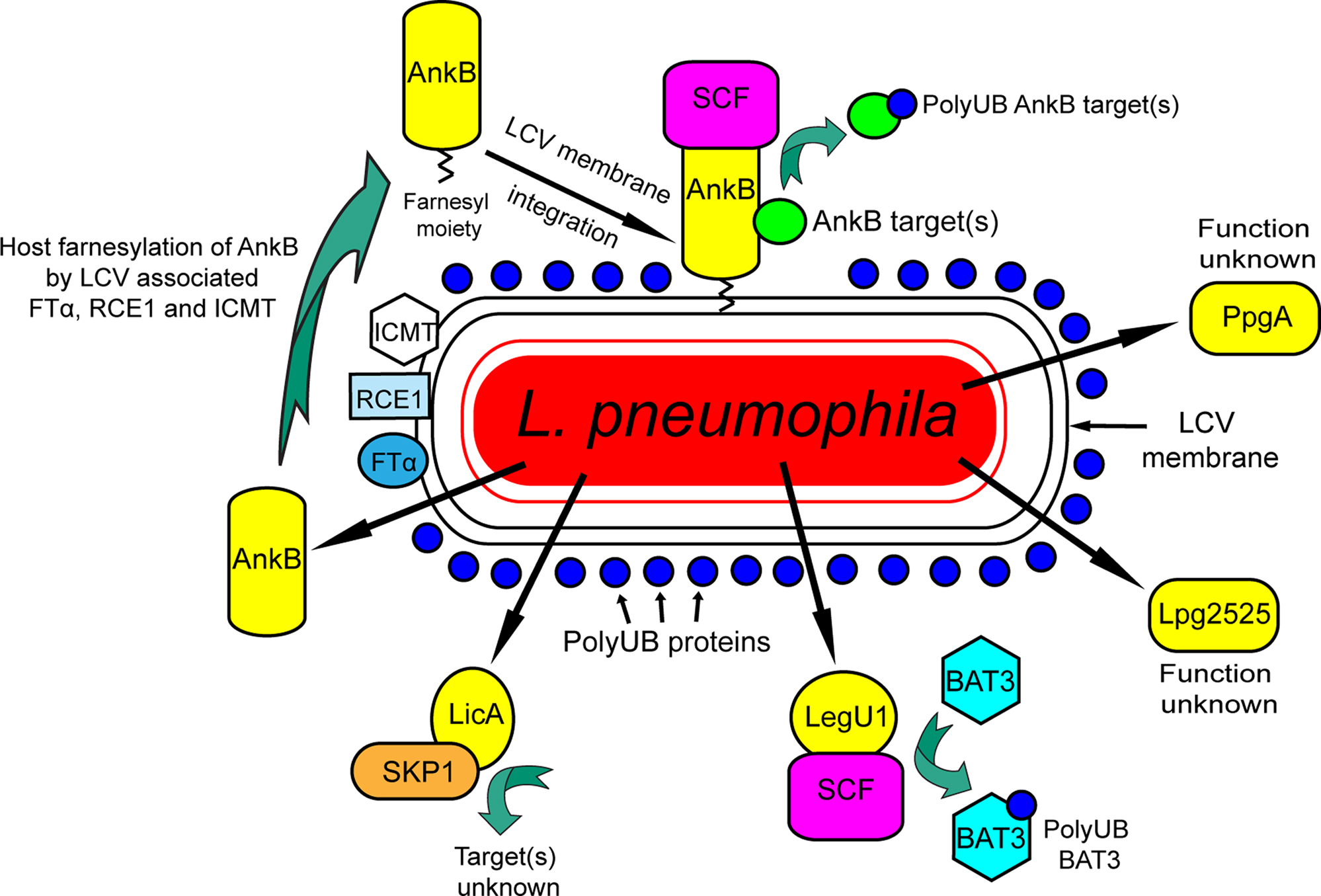
Figure 2. The F-box proteins of Legionella pneumophila. Five F-box proteins of L. pneumophila are translocated into the host cell. Two of these, PpgA and Lpg2525 do not interact with host SKP1 and their function is unknown. LicA interacts with SKP1 but its function is unknown. LegU1 interacts with the SCF complex to mediate ubiquitination of the host protein BAT3. Following translocation, AnkB is farnesylated by the three host enzymes FT, RCE1, and ICMT and is anchored into the cytosolic face of the LCV membrane, where it interacts with the SCF complex and acts as a platform for the docking of polyubiquitinated proteins to the LCV.
The Legionella-containing vacuole (LCV) is decorated with polyubiquitinated proteins, and the Dot/Icm transport system and the F-box effector AnkB are essential for this process (Dorer et al., 2006; Price et al., 2009, 2010a). The Dot/Icm-translocated AnkB effector is a non-canonical F-box protein that harbors two eukaryotic-like ankyrin domains (ANK) that are thought to be involved in protein–protein interaction, and is highly regulated by the growth phase, and is found in all sequenced L. pneumophila strains (Al-Khodor et al., 2008, 2010; Habyarimana et al., 2008; Price et al., 2010a). To date, only confirmed prokaryotic F-box proteins harbor ANK protein–protein interaction domains, instead of the LRR or WD40 domains present in higher eukaryotes (Sonnberg et al., 2008; Blanie et al., 2010). Our examination of the InterPro database indicates a proliferation of proteins harboring F-box and ANK domains in fungi and other lower unicellular eukaryotes including Dictyostelium discoideum, and bacteria, suggesting this domain arrangement is more widespread in primitive eukaryotes and prokaryotes.
The F-box domain of AnkB interacts with the SKP1 component of the SCF1 ubiquitin ligase complex and functions as a platform for the docking of polyubiquitinated proteins to the LCV membrane within primary human macrophages, U937 human macrophage cell line, HEK293 cells, Acanthamoeba, and D. discoideum (Dorer et al., 2006; Price et al., 2009, 2010a,b) (Figure 2). The AnkB effector is essential for proliferation of L. pneumophila within the two evolutionarily distant hosts, mammalian, and protozoan cells, and for intrapulmonary bacterial proliferation and manifestation of pulmonary disease in the mouse model (Al-Khodor et al., 2008; Price et al., 2009). Importantly, the F-box domain as well as the two ANK protein–protein interaction domains, are all essential for the biological function of AnkB (Al-Khodor et al., 2008; Price et al., 2009, 2010a). Currently, the polyubiquitinated targets of AnkB are unknown and they may be of host and/or bacterial origin. The dense accumulation of polyubiquitinated proteins surrounding the LCV indicates that the modulation of activity and turnover of ubiquitinated bacterial effectors or host proteins at the LCV/cytosol interface is essential for the development of a favorable L. pneumophila replicative niche. AnkB reduces ubiquitination of Parvin B (Lomma et al., 2010), a protein that localizes to focal adhesions and lamellipodia. This may appear to be contrary to the action of F-box proteins in promoting ubiquitination, but the reason for this conundrum is not known yet. It is also unclear how reducing Parvin B ubiquitination relates to the recruitment of polyubiquitinated proteins to the LCV (Dorer et al., 2006; Price et al., 2009; Lomma et al., 2010).
Injection of AnkB into the host cell by the by the Dot/Icm type IV secretion system results in exclusive targeting of the effector into the LCV membrane (Price et al., 2010b). Recent data have shown that AnkB is anchored into the cytosolic face of the LCV membrane by host-mediated farnesylation of its C-terminal eukaryotic-like CaaX motif (Price et al., 2010b) (Figure 2). Farnesylation is a highly conserved post-translation lipid modification of eukaryotic proteins that confers hydrophobicity on the modified protein, and its targeting to membranes (Wright and Philips, 2006). Anchoring of AnkB to the LCV membrane via host-mediated farnesylation is essential for its biological function as a platform for the docking of polyubiquitinated proteins to the LCV membrane within macrophages and ameba. The host polyubiquitination and farnesylation machineries are essential for intracellular replication of L. pneumophila within the two evolutionarily distant host cells, and for intrapulmonary proliferation of L. pneumophila in the mouse model of Legionnaires’ disease (Price et al., 2010b) (Figure 2). Therefore, host polyubiquitination and farnesylation are two evolutionarily conserved eukaryotic machineries exploited by the same effector to enable intra-vacuolar proliferation of L. pneumophila within evolutionarily distant hosts (Molmeret et al., 2005; Franco et al., 2009; Price et al., 2010b). These are the first two documented examples of the role of exploitation of highly conserved eukaryotic processes by Legionella within ameba and other primitive eukaryotes in pathogenic evolution of this bacterium to infect humans. Host-mediated farnesylation and anchoring of AnkB to the pathogen-containing vacuolar membrane is novel among the large family of eukaryotic F-box proteins that are cytosolic and not membrane-anchored.
Legionella pneumophila has at least four other proteins that have putative F-box domains and each of these are injected into host cells by the Dot/Icm system (Ensminger and Isberg, 2010) (Figure 2). Interestingly, these other F-box proteins are not found in all the sequenced strains of L. pneumophila. Of these four, Lpg2224 (PpgA) and Lpg2525, which are only found in the Philadelphia strain, do not associate with components of the host ubiquitination machinery, and may therefore play a different role during infection (Ensminger and Isberg, 2010) (Figure 2). Lpg1408 (LicA), which is found in all the sequenced L. pneumophila strains, interacts with SKP1, but not CUL1. This suggests that this protein may form a non-canonical SCF complex containing a different cullin (Ensminger and Isberg, 2010) (Figure 2). Lpg0171 (LegU1) interacts with both SKP1 and CUL1, forming an active E3 ubiquitin ligase complex, and is found in three of the four sequenced L. pneumophila genomes (Ensminger and Isberg, 2010). The LegU1 effector directs the ubiquitination of the host protein BAT3 (Ensminger and Isberg, 2010), which plays key roles in modulating apoptosis, ER stress, p53-regulated expression, and Hsp70 stability (Sasaki et al., 2007; Desmots et al., 2008; Nguyen et al., 2008; Tsukahara et al., 2009) (Figure 2). It is not currently clear what the exact role of LegU1-mediated BAT3 ubiquitination is during the course of infection by L. pneumophila. However, the presence of multiple translocated F-box proteins in L. pneumophila, which hijack the host ubiquitination system, strongly suggests that bacterial molecular mimicry of eukaryotic F-box proteins is very important for the infection by Legionella. Interestingly, both Lpg2525 and LegU1 have putative CaaX farnesylation motifs, similar to AnkB (Price et al., 2010b), and it will be interesting to determine if farnesylation is crucial for their membrane anchoring and biological activity following their injection into the host cell.
Legionella longbeachae is another species of Legionella that cause Legionnaires’ disease (McKinney et al., 1981). Unlike, L. pneumophila which primarily inhabits aquatic environments, L. longbeachae is found predominantly in moist potting soil (Steele et al., 1990). Similar to L. pneumophila, L. longbeachae invades and proliferates within macrophages, and translocates effectors via the Dot/Icm system, although its trafficking following phagocytosis differs from L. pneumophila (Feldman and Segal, 2004; Asare and Abu Kwaik, 2007). The L. longbeachae genome harbors over 30 genes that encode putative proteins with eukaryotic domains, including ANK repeats, U-box, Sel-1, LRRs, and serine–threonine kinase domains (Cazalet et al., 2010; Kozak et al., 2010). In addition, the L. longbeachae genome has three putative genes that encode F-box proteins (LLB0157, LLB3234, LLB3296), and these three all have homologues in L. pneumophila (Kozak et al., 2010) (Figure 4). Interestingly, only LLB0157 was identified by the InterPro F-box motif, again suggesting bacterial F-box proteins are under-represented in databases. The L. pneumophila homologue of LLB0157, LicA is translocated into host cells, and shown to interact with SKP1 but not the other components of the SCF complex (Ensminger and Isberg, 2010). The functions of the L. longbeachae F-box proteins are currently unknown.
Agrobacterium Tumefaciens VirF
The plant pathogen, A. tumefaciens, causes neoplastic growth on several plants species, giving rise to crown gall disease. This bacterium has a type IV secretion system that translocates both protein effectors and a single-stranded DNA molecule (T-DNA) from the tumor-inducing plasmid, which is essential for genetic transformation of the host cell (Gelvin, 2000). The T-DNA is coated after translocation by the bacterial ssDNA binding protein VirE2, packaging the DNA into a nucleoprotein complex called the T-complex (Citovsky et al., 2007). The T-complex then associates with the plant protein VIP1, which interacts with karyopherin-α to enable nuclear import of the T-complex (Ziemienowicz et al., 2001; Citovsky et al., 2004; Li et al., 2005; Lacroix et al., 2008). Before the T-DNA can integrate into the plant chromatin, it is essential that the associated VirE2 and VIP1 proteins be removed. This is achieved by the action of another translocated bacterial protein, VirF (Schrammeijer et al., 2001) (Figure 3), which harbors an F-box domain and determines host tropism for A. tumefaciens (Hirooka et al., 1987). ASK1 and ASK2, the Arabidopsis homologues of the yeast SKP1 protein interact with VirF (Schrammeijer et al., 2001). Furthermore, the F-box domain of VirF is essential for ASK1/2 interaction in vitro and for full virulence of A. tumefaciens during infection of plants (Schrammeijer et al., 2001). VirF directly interacts with VIP1 and this interaction leads to targeting of VIP1 to the SCF complex, whereby it becomes ubiquitinated and subsequently tagged for proteolysis by the proteasome (Tzfira et al., 2004). This process also indirectly targets VirE2 to the proteasome, which allows the T-DNA to integrate into the plant chromatin, and in turn allows successful infection by A. tumefaciens.
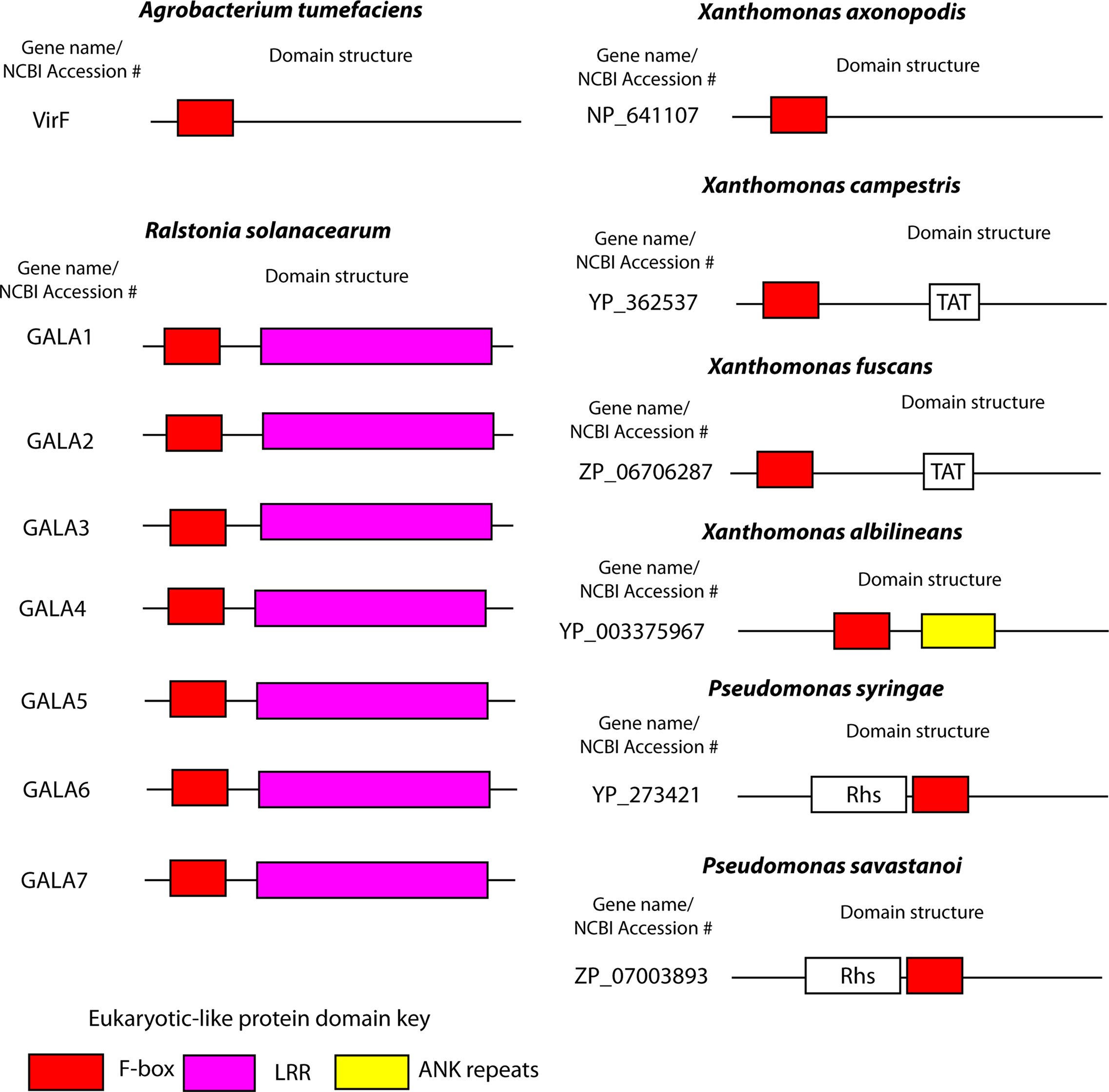
Figure 3. F-box proteins in plant pathogens. Cartoon representations of the domain structures of bacterial F-box proteins found in plant pathogens. Proteins and domain predictions were found at the EMBL-EBI website InterPro website (http://www.ebi.ac.uk/interpro), using the F-box domain motif IPR001810. Genes are identified by their name or NCBI accession number.
Interestingly, for some plant species VirF is not required for genetic transformation by A. tumefaciens (Hirooka et al., 1987). It has been recently shown that A. tumefaciens induces expression of the plant F-box protein VBF, which can functionally replace VirF (Zaltsman et al., 2010), but VBF regulates the level of VIP1 and indirectly VirE2 by targeting VIP1 to the SCF complex, allowing even a virF mutant strain of A. tumefaciens to cause disease (Zaltsman et al., 2010). This indicates that VirF is the direct molecular mimic of the plant VBF protein, but VBF is not found in all plant species. Therefore, it appears that A. tumefaciens has acquired a functional homologue of VBF allowing this bacterium to extend its host range and in turn increase its chance for survival. It will be interesting to determine if other bacterial F-box proteins have direct functional homologues in the host cells.
Ralstonia Solanacearum F-Box Proteins
The Gram-negative plant pathogen, R. solanacearum, has a type III secretion system that translocate over 80 different effectors into plant cells, resulting in bacterial wilt in a number of plant species (Salanoubat et al., 2002; Cunnac et al., 2004). Among these effectors are seven proteins that harbor the F-box domain (Angot et al., 2006) (Figure 3). The R. solanacearum F-box proteins also harbor LRRs, which specify protein–protein interaction (Gagne et al., 2002; Angot et al., 2006), similar to many eukaryotic F-box proteins (Ho et al., 2006). The R. solanacearum F-box proteins have been designated “GALA” proteins due to a GAxALA motif within the LRR (Angot et al., 2006). Interestingly, the domain architecture of the R. solanacearum GALA proteins is similar to plant F-box proteins, and there is evidence to suggest that GALA proteins have been acquired by lateral inter-kingdom gene transfer from plants (Angot et al., 2006; Kajava et al., 2008). The seven GALA proteins of R. solanacearum interact with several of the Arabidopsis SKP1 like proteins (ASKs) (Angot et al., 2006). Interestingly, individual mutations in each of the seven GALA proteins had no effect on the ability of R. solanacearum to cause disease on Arabidopsis and tomato plants (Angot et al., 2006). However a R. solanacearum strain harboring mutations in all seven GALA proteins is essentially avirulent when tested on Arabidopsis, and is significantly less virulent when tested on tomato plants (Angot et al., 2006). Interestingly, infection of another R. solanacearum host plant, Medicago truncatula with a GALA7 mutant strain results in no disease, in contrast to that observed in Arabidopsis and tomato infections (Angot et al., 2006). Furthermore, the F-box domain of GALA7 is crucial for infection of M. truncatula, since complementation of a R. solanacearum GALA7 mutant with a GALA7 gene lacking the F-box domain does not restore virulence (Angot et al., 2006). The essential role of the GALA7 F-box domain for biological function of the protein is analogous to the key role the F-box domain of AnkB plays in L. pneumophila virulence. Furthermore, the essential role of GALA7 in one plant species over another, suggests that GALA7 plays a role in host tropism. To date, the plant proteins targeted by the R. solanacearum GALA proteins are unknown.
Putative F-Box Proteins are Abundant in Other Bacterial Species
To date, bacterial F-box proteins have been characterized in L. pneumophila, A. tumefaciens, and R. solanacearum. Interestingly, in each of these bacterial species, F-box proteins play a key role in virulence through subverting the host ubiquitination system. To explore whether other bacterial species harbor F-box proteins, we have searched the InterPro database using the F-box motif ID IPR001810 and found 74 bacterial proteins that contain the F-box domain (Figures 3–5). These 74 F-box proteins belong to 22 different bacterial species, including human and plant pathogens, and ameba endosymbionts. Interestingly, only three F-box proteins have been found in bacteria that are not known to be pathogenic or do not infect ameba, Lentisphaera araneosa, Aquifex aeolicus, and Eubacterium siraeum. This strongly suggests that acquisition of F-box proteins encoding genes by bacterial pathogens and amebal endosymbionts has been favored by evolution to enable these bacteria to subvert the host ubiquitination machinery. Surprisingly, the IPR001810 motif does not recognize VirF from A. tumefaciens, and only four out of the seven GALA proteins from R. solanacearum, suggesting this motif does not adequately identify bacterial F-box proteins. Therefore, it is quite possible that F-box proteins are more abundant, and are found in a wider range of bacterial species than is currently predicted.
Other Human Pathogens
Coxiella burnetii, the causative agent of Q fever, is a Gram-negative, intracellular pathogen that invades mononuclear phagocytes, where the bacterium replicates within a lysosomal-like vacuole (Voth and Heinzen, 2007). C. burnetii harbors a type IV secretion system, similar to L. pneumophila, which is believed to translocate effector molecules into the host to modulate cellular processes (Zamboni et al., 2003; Zusman et al., 2003; Voth and Heinzen, 2009). This organism harbors a number of potential T4SS effectors that have eukaryotic-like protein domains (Voth and Heinzen, 2009). The InterPro database identified a single C. burnetii gene (AnkD) encoding a putative F-box protein (Uniprot ID B6J1V7, CBU0355) (Figure 4). This protein also harbors ANK repeats, analogous to AnkB of L. pneumophila (Figure 4). Perhaps this C. burnetii protein targets a host protein for ubiquitination via an interaction with the ANK domains and interaction of the host SKP1 with the F-box domain. When AnkD was expressed as an adenylate cyclase fusion in L. pneumophila it was not translocated into mammalian cells (Pan et al., 2008; Voth et al., 2009). However, this may be due to a lack of recognition of this foreign fusion protein by the L. pneumophila type IV translocation apparatus. At least two other C. burnetii proteins have been identified that harbor the F-box domain, CBU0814 and CBUA0014 (Voth and Heinzen, 2009) (Figure 4). These two proteins failed to be identified by the InterPro F-box motif, further suggesting that many bacterial F-box proteins have gone un-annotated. CBU0814 also harbors a regulator of chromosomal condensation (RCC) domain, while CBUA0014 does not have any other recognizable functional domains (Voth and Heinzen, 2009) (Figure 4). The role of these proteins in C. burnetii infection remains unknown, but it is an exciting avenue of research that may enhance the understanding of this obligate intracellular pathogen.
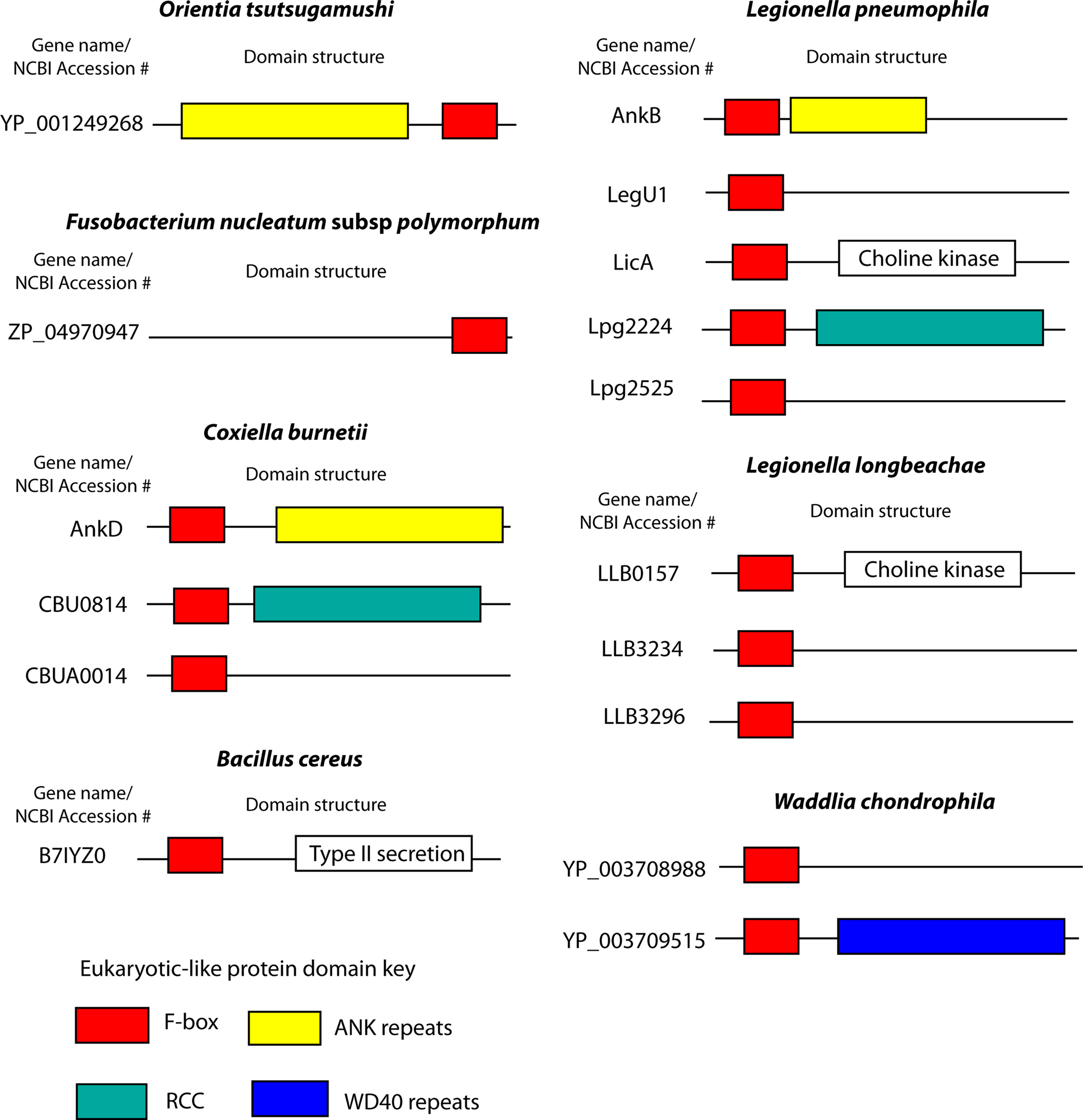
Figure 4. F-box proteins in human pathogens. Cartoon representations of the domain structures of bacterial F-box proteins found in human pathogens. Proteins and domain predictions were found at the EMBL-EBI website InterPro website (http://www.ebi.ac.uk/interpro), using the F-box domain motif IPR001810. Genes are identified by their name or NCBI accession number.
Five other human bacterial pathogens harbor genes encoding putative F-box proteins (Figure 4). Orientia tsutsugamushi is an obligate intracellular Rickettsia and is the causative agent of the zoonosis scrub typhus (Seong et al., 2001). The O. tsutsugamushi genome has an expansive set of genes encoding components of type IV secretion systems and has around 200 genes that encode proteins with eukaryotic-like domains, including histidine kinases, ANK, and tetratricopeptide repeats (Cho et al., 2007). O. tsutsugamushi has a single putative protein (Ank1u9) harboring an F-box domain (Cho et al., 2007) (Figure 4). Interestingly, Ank1u9 has eight ANK repeats at the N-terminus while the F-box domain of this protein is found at the C-terminus (Figure 4). This unusual domain architecture has only been observed in viral F-box proteins (Sonnberg et al., 2008; Blanie et al., 2010). The biological function of Ank1u9 is unknown, however the massive proliferation of proteins with eukaryotic-like domains in O. tsutsugamushi (Cho et al., 2007), and the obligate intracellular lifestyle of this organisms suggests that manipulation of host pathways is essential for successful replication.
Fusobacterium nucleatum is a Gram-negative, anaerobic bacterium that plays a role in the complex polymicrobial infection of periodontal diseases (Moore and Moore, 1994). F. nucleatum can constitute a large proportion of plaque biofilm and amongst the first Gram-negative species to become established in the plaque (Ximenez-Fyvie et al., 2000). F. nucleatum has been shown to attach to and invade gingival epithelia and oral fibroblast cells, and this ability may be crucial for development of periodontitis (Han et al., 2000; Dabija-Wolter et al., 2009). This organism harbors a type IV secretion system, which translocates effector proteins into host cells (Karpathy et al., 2007). A single putative F-box protein is predicted to be encoded by the F. nucleatum genome (Figure 4).
Waddlia chondrophila is an emerging pathogen belonging to the Chlamydiae family and has been associated with miscarriage of fetuses in cows and humans (Rurangirwa et al., 1999; Henning et al., 2002; Baud et al., 2007). This organism may also be associated with respiratory illness in humans (Goy et al., 2009). W. chrondrophila is an obligate intracellular bacterium that replicates within eukaryotic cells and exhibits a typical Chlamydiae biphasic developmental cycle, comprising infectious elementary bodies and non-infectious, replicating reticulate bodies (Goy et al., 2008). In human macrophages, W. chrondrophila evades the endocytic pathway, residing within a vacuole that is localized with endoplasmic reticulum proteins and mitochondria (Croxatto and Greub, 2010). The genome of this organism encodes two putative F-box proteins, and one of these also has WD40 repeats, similar to that of F-box proteins of higher eukaryotes (Figure 4).
The Gram-positive soil dwelling bacterium, Bacillus cereus, which can cause food-borne illness in humans (Guinebretiere et al., 2010), harbors a gene (BCEA0052) encoding a type II secretion protein that also has an F-box domain (Figure 4). This protein has weak BLAST homology to ComG, a protein involved in DNA competence, from Bacillus anthracis. To date no F-box proteins have been identified in B. anthracis. Taken together, diverse human pathogens harbor putative F-box proteins that may affect host ubiquitination, and opens up exciting new paths of research to decipher molecular and biochemical bases of bacterial pathogenesis.
Plant Pathogens
The two bacterial plant pathogens, A. tumefaciens and R. solanacearum utilize F-box proteins to subvert the host ubiquitination system to cause disease. At least six other bacterial plant pathogens encode putative proteins that harbor the F-box domain (Figure 3). Xanthomonas campestris, X. axonopodis, and X. fuscans cause disease on a wide variety of plants (da Silva et al., 2002; Graham et al., 2004; Darsonval et al., 2008). These plant pathogens each harbor Hrp type III secretion systems that are essential for virulence through translocation of effector proteins into host cells (Bogdanove et al., 1996; Kim et al., 2003; Roden et al., 2004; Darsonval et al., 2008). X. campestris, X. axonopodis, and X. fuscans encode a single putative F-box protein with unknown function (Figure 3). X. albilineans is a devastating pathogen of sugar cane crops. Although it does not have genes encoding the Hrp type III secretion, it has a type III secretion system similar to that of Burkholderia pseudomallei that suggests translocation of effector proteins is important for this organism. The genome of X. albilineans encodes a single putative F-box protein (XALc_1472) which also harbors two ANK repeats, similar to AnkB of L. pneumophila (Figure 3).
Pseudomonas syringae pv phaseolicola causes disease on bean plants, and like the Xanthomonads, virulence is dependent on translocation of effector molecules via its type III secretion system (Bender et al., 1999; Joardar et al., 2005). This organism possesses a single putative F-box protein (Figure 3). Indeed there is mounting evidence implicating P. syringae subversion of host ubiquitination through the translocation of the effector AvrPtoB, which has intrinsic E3 ubiquitin ligase activity (Rosebrock et al., 2007). To date, the opportunistic human pathogen, Pseudomonas aeruginosa, does not harbor F-box proteins. Finally P. savastanoi, another devastating plant pathogen, encodes a single putative F-box protein of unknown function (Rodriguez-Palenzuela et al., 2010) (Figure 3). It will be interesting to determine if the F-box proteins in these plant pathogens are crucial for virulence, similar to that already seen for A. tumefaciens and R. solanacearum.
Amebal Endosymbionts
Free-living ameba in the environment often play host to stable obligate intracellular bacterial parasites or endosymbionts belonging to the Chlamydiae, Bacteriodetes, alpha- and beta-Proteobacteria. Protochlamydia amoebophila is a relative of pathogenic Chlamydia species and exhibits a Chlamydia-like developmental cycle within ameba (Collingro et al., 2005). The genome (Horn et al., 2004) encodes a type IV secretion system and at least 11 genes encoding putative F-box proteins (Figure 5). Seven of these F-box proteins harbor WD40 protein interaction domains, similar to F-box proteins of higher eukaryotes (Ho et al., 2006) and one F-box protein harbors ANK repeats, similar to that observed in other bacterial F-box proteins like AnkB of L. pneumophila and lower eukaryotes including ameba.

Figure 5. F-box proteins in amebal endosymbionts. Cartoon representations of the domain structures of bacterial F-box proteins found in amebal symbionts. Proteins and domain predictions were found at the EMBL-EBI website InterPro website (http://www.ebi.ac.uk/interpro), using the F-box domain motif IPR001810. Genes are identified by their name or NCBI accession number.
Protochlamydia acanthamoeba, another ameba endosymbiont, harbors 13 putative F-box proteins, many of which also contain WD40 protein interaction domains (Figure 5). UnlikeP. amoebophila, P. acanthamoeba has been implicated in human disease, causing pneumonia, and can survive and replicate in human macrophages (Casson et al., 2008; Greub, 2009). It will be interesting to determine if P. acanthamoeba F-box proteins target conserved pathways shared between ameba and humans, enabling this organism to survive and replicate in otherwise genetically distant host cells.
Amoebophilus asiaticus is an amebal endosymbiont belonging to the Bacteroidetes group (Schmitz-Esser et al., 2008). Its 1.89 Mbp genome (Schmitz-Esser et al., 2010) devotes up to 8% (129 proteins) of its coding capacity to genes that harbor eukaryotic domains, including proteins with ANK repeats, tetratricopeptide domains, SEL1 domains, LRR and proteins with F-box and U-box domains (Schmitz-Esser et al., 2010) suggesting that subversion of host pathways is crucial for A. asiaticus survival. A total of 15 putative F-box proteins were identified to be encoded by the genome of A. asiaticus (Schmitz-Esser et al., 2010). However, searching with the InterPro F-box motif identified only 13 of the 15 annotated proteins. Most of these F-box proteins also harbor eukaryotic protein interaction domains such as LRR, ANK repeats or the tetratricopeptide repeat (Figure 5). The abundance of F-box proteins in A. asiaticus suggests that subversion of the host ubiquitin system may be important in interaction of this bacterium with the host cell. Legionella drancourtii is a strictly intracellular amebal pathogen, growing in cytoplasmic vacuoles of infected ameba (La Scola et al., 2004). L. drancourtii encodes six putative F-box proteins, and three of these proteins also harbor ANK repeats (Figure 5), analogous to AnkB of L. pneumophila.
The role of F-box proteins in the interaction of ameba with their endosymbionts is unknown. Compared to other bacterial species with known and predicted F-box proteins, amebal endosymbionts have a larger cadre of proteins that have the potential to subvert the host SCF E3 ubiquitination system. This may reflect their obligate and intimate intracellular lifestyle within ameba. Furthermore, the proliferation of F-box proteins in these organisms may allow greater amebal host tropism, increasing chances of survival in an ever changing environment.
Conclusions
Host ubiquitination by the SCF E3 ligases plays an essential role in numerous eukaryotic cell processes making this system particularly sensitive to interference by invading bacteria. Many of the bacterial F-box proteins studied to date in human and plant pathogens play a key role in virulence, indicating that subversion of host ubiquitination through molecular mimicry is an important strategy in unrelated plant and mammalian bacterial pathogens. We show that at least 74 putative F-box proteins are encoded by 22 bacterial species. Many of these bacterial species cause disease in humans or plants by injecting effectors through type III or IV secretion systems, or live in symbiotic relationships with ameba. The abundance of predicted bacterial F-box proteins suggests that molecular mimicry of host F-box proteins involved in the ubiquitination pathway is an emerging and widespread theme among pathogenic bacteria and endosymbionts. It will be crucial to determine if the F-box proteins found in these other bacterial species are translocated into host cells and if they act in a similar fashion to the known bacterial and eukaryotic F-box proteins.
Bacteria do not possess the other enzymatic components of the SCF E3 ubiquitin ligase machinery and do not possess the ubiquitination pathways of eukaryotes. It is most likely that bacterial F-box proteins have been acquired through inter-kingdom horizontal gene transfer from eukaryotic hosts. Interestingly the distribution of eukaryotic protein–protein interaction domains in bacterial F-box proteins reflects the natural hosts of the bacteria. The ANK repeat has not yet been found in F-box proteins of higher eukaryotes, but is prevalent in F-box proteins found in fungi, ameba and other lower eukaryotes. Many of the bacteria described in this review, including the human pathogens L. pneumophila and C. burnetii and amebal endosymbionts have F-box proteins with ANK repeats. The natural hosts of these intracellular bacteria are ameba and other primitive eukaryotes, organisms which also possess F-box proteins with ANK repeats, which are not present in F-box proteins of mammals. This reflects the exquisite co-evolution of bacteria with their primitive host cells and the potential of this interaction in inter-kingdom horizontal gene transfer that allow bacteria to exploit conserved eukaryotic processes. This is clearly illustrated by the AnkB F-box effector of L. pneumophila that exploits the two evolutionarily conserved polyubiquitination and farnesylation eukaryotic machineries to proliferate within primitive and highly evolved eukaryotic cells and hosts.
Numerous questions remain unanswered regarding bacterial F-box proteins. The only confirmed target for a bacterial F-box protein known to date is VirF directed ubiquitination of VIP1 in plants. Identifying the targets of bacterial F-box proteins will be crucial for future research, as they will provide insight into the host mechanisms exploited by bacterial pathogens and the exquisite lengths bacteria have reached through co-evolution with their hosts to proliferate.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Yousef Abu Kwaik is supported by Public Health Service Awards R01AI43965 and R01AI069321 from NIAID and by the commonwealth of Kentucky Research Challenge Trust Fund. The authors declare no competing financial interests.
References
Al-Khodor, S., Al-Quadan, T., and Abu Kwaik, Y. (2010). Temporal and differential regulation of expression of the eukaryotic-like ankyrin effectors of L. pneumophila. Environ. Microbiol. 2, 677–684.
Al-Khodor, S., Price, C. T., Habyarimana, F., Kalia, A., and Abu Kwaik, Y. (2008). A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol. Microbiol. 70, 908–923.
Angot, A., Peeters, N., Lechner, E., Vailleau, F., Baud, C., Gentzbittel, L., Sartorel, E., Genschik, P., Boucher, C., and Genin, S. (2006). Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. U.S.A. 103, 14620–14625.
Asare, R., and Abu Kwaik, Y. (2007). Early trafficking and intracellular replication of Legionella longbeachaea within an ER-derived late endosome-like phagosome. Cell Microbiol. 9, 1571–1587.
Baud, D., Thomas, V., Arafa, A., Regan, L., and Greub, G. (2007). Waddlia chondrophila, a potential agent of human fetal death. Emerg. Infect. Dis. 13, 1239–1243.
Bender, C. L., Alarcon-Chaidez, F., and Gross, D. C. (1999). Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63, 266–292.
Blanie, S., Gelfi, J., Bertagnoli, S., and Camus-Bouclainville, C. (2010). MNF, an ankyrin repeat protein of myxoma virus, is part of a native cellular SCF complex during viral infection. Virol. J. 7, 56.
Bogdanove, A. J., Beer, S. V., Bonas, U., Boucher, C. A., Collmer, A., Coplin, D. L., Cornelis, G. R., Huang, H. C., Hutcheson, S. W., Panopoulos, N. J., and Van Gijsegem, F. (1996). Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol. Microbiol. 20, 681–683.
Casson, N., Michel, R., Muller, K. D., Aubert, J. D., and Greub, G. (2008). Protochlamydia naegleriophila as etiologic agent of pneumonia. Emerg. Infect. Dis. 14, 168–172.
Cazalet, C., Gomez-Valero, L., Rusniok, C., Lomma, M., Dervins-Ravault, D., Newton, H. J., Sansom, F. M., Jarraud, S., Zidane, N., Ma, L., Bouchier, C., Etienne, J., Hartland, E. L., and Buchrieser, C. (2010). Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires’ disease. PLoS Genet. 6, e1000851. doi: 10.1371/journal.pgen.1000851.
Cho, N. H., Kim, H. R., Lee, J. H., Kim, S. Y., Kim, J., Cha, S., Darby, A. C., Fuxelius, H. H., Yin, J., Kim, J. H., Lee, S. J., Koh, Y. S., Jang, W. J., Park, K. H., Andersson, S. G., Choi, M. S., and Kim, I. S. (2007). The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc. Natl. Acad. Sci. U.S.A. 104, 7981–7986.
Christie, P. J., and Cascales, E. (2005). Structural and dynamic properties of bacterial type IV secretion systems (review). Mol. Membr. Biol. 22, 51–61.
Citovsky, V., Kapelnikov, A., Oliel, S., Zakai, N., Rojas, M. R., Gilbertson, R. L., Tzfira, T., and Loyter, A. (2004). Protein interactions involved in nuclear import of the Agrobacterium VirE2 protein in vivo and in vitro. J. Biol. Chem. 279, 29528–29533.
Citovsky, V., Kozlovsky, S. V., Lacroix, B., Zaltsman, A., Dafny-Yelin, M., Vyas, S., Tovkach, A., and Tzfira, T. (2007). Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol. 9, 9–20.
Collingro, A., Toenshoff, E. R., Taylor, M. W., Fritsche, T. R., Wagner, M., and Horn, M. (2005). ‘Candidatus Protochlamydia amoebophila’, an endosymbiont of Acanthamoeba spp. Int. J. Syst. Evol. Microbiol. 55, 1863–1866.
Croxatto, A., and Greub, G. (2010). Early intracellular trafficking of Waddlia chondrophila in human macrophages. Microbiology 156, 340–355.
Cunnac, S., Occhialini, A., Barberis, P., Boucher, C., and Genin, S. (2004). Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53, 115–128.
da Silva, A. C., Ferro, J. A., Reinach, F. C., Farah, C. S., Furlan, L. R., Quaggio, R. B., Monteiro-Vitorello, C. B., Van Sluys, M. A., Almeida, N. F., Alves, L. M., do Amaral, A. M., Bertolini, M. C., Camargo, L. E., Camarotte, G., Cannavan, F., Cardozo, J., Chambergo, F., Ciapina, L. P., Cicarelli, R. M., Coutinho, L. L., Cursino-Santos, J. R., El-Dorry, H., Faria, J. B., Ferreira, A. J., Ferreira, R. C., Ferro, M. I., Formighieri, E. F., Franco, M. C., Greggio, C. C., Gruber, A., Katsuyama, A. M., Kishi, L. T., Leite, R. P., Lemos, E. G., Lemos, M. V., Locali, E. C., Machado, M. A., Madeira, A. M., Martinez-Rossi, N. M., Martins, E. C., Meidanis, J., Menck, C. F., Miyaki, C. Y., Moon, D. H., Moreira, L. M., Novo, M. T., Okura, V. K., Oliveira, M. C., Oliveira, V. R., Pereira, H. A., Rossi, A., Sena, J. A., Silva, C., de Souza, R. F., Spinola, L. A., Takita, M. A., Tamura, R. E., Teixeira, E. C., Tezza, R. I., Trindade dos Santos, M., Truffi, D., Tsai, S. M., White, F. F., Setubal, J. C., and Kitajima, J. P. (2002). Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417, 459–463.
Dabija-Wolter, G., Cimpan, M. R., Costea, D. E., Johannessen, A. C., Sornes, S., Neppelberg, E., Al-Haroni, M., Skaug, N., and Bakken, V. (2009). Fusobacterium nucleatum enters normal human oral fibroblasts in vitro. J. Periodontol. 80, 1174–1183.
Darsonval, A., Darrasse, A., Meyer, D., Demarty, M., Durand, K., Bureau, C., Manceau, C., and Jacques, M. A. (2008). The Type III secretion system of Xanthomonas fuscans subsp. fuscans is involved in the phyllosphere colonization process and in transmission to seeds of susceptible beans. Appl. Environ. Microbiol. 74, 2669–2678.
de Felipe, K. S., Glover, R. T., Charpentier, X., Anderson, O. R., Reyes, M., Pericone, C. D., and Shuman, H. A. (2008). Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 4, e1000117. doi: 10.1371/journal.ppat.1000117.
Desmots, F., Russell, H. R., Michel, D., and McKinnon, P. J. (2008). Scythe regulates apoptosis-inducing factor stability during endoplasmic reticulum stress-induced apoptosis. J. Biol. Chem. 283, 3264–3271.
Dorer, M. S., Kirton, D., Bader, J. S., and Isberg, R. R. (2006). RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog. 2, e34. doi: 10.1371/journal.ppat.0020034.
Ensminger, A. W., and Isberg, R. R. (2010). E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates. Infect. Immun. 78, 3905–3919.
Feldman, M., and Segal, G. (2004). A specific genomic location within the icm/dot pathogenesis region of different Legionella species encodes functionally similar but nonhomologous virulence proteins. Infect. Immun. 72, 4503–4511.
Franco, I. S., Shuman, H. A., and Charpentier, X. (2009). The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell Microbiol. 11, 1435–1443.
Gagne, J. M., Downes, B. P., Shiu, S. H., Durski, A. M., and Vierstra, R. D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99, 11519–11524.
Galan, J. E. (2009). Common themes in the design and function of bacterial effectors. Cell Host Microbe 5, 571–579.
Gelvin, S. B. (2000). Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 223–256.
Goy, G., Croxatto, A., and Greub, G. (2008). Waddlia chondrophila enters and multiplies within human macrophages. Microbes Infect. 10, 556–562.
Goy, G., Croxatto, A., Posfay-Barbe, K. M., Gervaix, A., and Greub, G. (2009). Development of a real-time PCR for the specific detection of Waddlia chondrophila in clinical samples. Eur. J. Clin. Microbiol. Infect. Dis. 28, 1483–1486.
Graham, J. H., Gottwald, T. R., Cubero, J., and Achor, D. S. (2004). Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 5, 1–15.
Greub, G. (2009). Parachlamydia acanthamoebae, an emerging agent of pneumonia. Clin. Microbiol. Infect. 15, 18–28.
Guinebretiere, M. H., Velge, P., Couvert, O., Carlin, F., Debuyser, M. L., and Nguyen-The, C. (2010). The food poisoning power of Bacillus cereus Group strains varies according to phylogenetic affiliation (groups I-VII), not to species affiliation. J. Clin. Microbiol. 48, 3388–3391.
Habyarimana, F., Al-Khodor, S., Kalia, A., Graham, J. E., Price, C. T., Garcia, M. T., and Kwaik, Y. A. (2008). Role for the Ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ. Microbiol. 10, 1460–1474.
Han, Y. W., Shi, W., Huang, G. T., Kinder Haake, S., Park, N. H., Kuramitsu, H., and Genco, R. J. (2000). Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect. Immun. 68, 3140–3146.
Hauser, A. R. (2009). The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7, 654–665.
Henning, K., Schares, G., Granzow, H., Polster, U., Hartmann, M., Hotzel, H., Sachse, K., Peters, M., and Rauser, M. (2002). Neospora caninum and Waddlia chondrophila strain 2032/99 in a septic stillborn calf. Vet. Microbiol. 85, 285–292.
Hirooka, T., Rogowsky, P. M., and Kado, C. I. (1987). Characterization of the virE locus of Agrobacterium tumefaciens plasmid pTiC58. J. Bacteriol. 169, 1529–1536.
Ho, M. S., Ou, C., Chan, Y. R., Chien, C. T., and Pi, H. (2008). The utility F-box for protein destruction. Cell Mol. Life Sci. 65, 1977–2000.
Ho, M. S., Tsai, P. I., and Chien, C. T. (2006). F-box proteins: the key to protein degradation. J. Biomed. Sci. 13, 181–191.
Horn, M., Collingro, A., Schmitz-Esser, S., Beier, C. L., Purkhold, U., Fartmann, B., Brandt, P., Nyakatura, G. J., Droege, M., Frishman, D., Rattei, T., Mewes, H. W., and Wagner, M. (2004). Illuminating the evolutionary history of chlamydiae. Science 304, 728–730.
Ibarra, J. A., and Steele-Mortimer, O. (2009). Salmonella – the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol. 11, 1579–1586.
Isberg, R. R., O'Connor, T. J., and Heidtman, M. (2009). The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24.
Jin, J., Cardozo, T., Lovering, R. C., Elledge, S. J., Pagano, M., and Harper, J. W. (2004). Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18, 2573–2580.
Joardar, V., Lindeberg, M., Jackson, R. W., Selengut, J., Dodson, R., Brinkac, L. M., Daugherty, S. C., Deboy, R., Durkin, A. S., Giglio, M. G., Madupu, R., Nelson, W. C., Rosovitz, M. J., Sullivan, S., Crabtree, J., Creasy, T., Davidsen, T., Haft, D. H., Zafar, N., Zhou, L., Halpin, R., Holley, T., Khouri, H., Feldblyum, T., White, O., Fraser, C. M., Chatterjee, A. K., Cartinhour, S., Schneider, D. J., Mansfield, J., Collmer, A., and Buell, C. R. (2005). Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 187, 6488–6498.
Kagan, J. C., and Roy, C. R. (2002). Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 4, 945–954.
Kajava, A. V., Anisimova, M., and Peeters, N. (2008). Origin and evolution of GALA-LRR, a new member of the CC-LRR subfamily: from plants to bacteria? PLoS One 3, e1694. doi: 10.1371/journal.pone.0001694.
Karpathy, S. E., Qin, X., Gioia, J., Jiang, H., Liu, Y., Petrosino, J. F., Yerrapragada, S., Fox, G. E., Haake, S. K., Weinstock, G. M., and Highlander, S. K. (2007). Genome sequence of Fusobacterium nucleatum subspecies polymorphum – a genetically tractable fusobacterium. PLoS One 2, e659. doi: 10.1371/journal.pone.0000659.
Kerscher, O., Felberbaum, R., and Hochstrasser, M. (2006). Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180.
Kim, J. G., Park, B. K., Yoo, C. H., Jeon, E., Oh, J., and Hwang, I. (2003). Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185, 3155–3166.
Kirkpatrick, D. S., Denison, C., and Gygi, S. P. (2005). Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat. Cell Biol. 7, 750–757.
Kobe, B., and Kajava, A. V. (2001). The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732.
Kozak, N. A., Buss, M., Lucas, C. E., Frace, M., Govil, D., Travis, T., Olsen-Rasmussen, M., Benson, R. F., and Fields, B. S. (2010). Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968. J. Bacteriol. 192, 1030–1044.
La Scola, B., Birtles, R. J., Greub, G., Harrison, T. J., Ratcliff, R. M., and Raoult, D. (2004). Legionella drancourtii sp. nov., a strictly intracellular amoebal pathogen. Int. J. Syst. Evol. Microbiol. 54, 699–703.
Lacroix, B., Loyter, A., and Citovsky, V. (2008). Association of the Agrobacterium T-DNA-protein complex with plant nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 105, 15429–15434.
Lechner, E., Achard, P., Vansiri, A., Potuschak, T., and Genschik, P. (2006). F-box proteins everywhere. Curr. Opin. Plant Biol. 9, 631–638.
Li, J., Krichevsky, A., Vaidya, M., Tzfira, T., and Citovsky, V. (2005). Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 102, 5733–5738.
Lomma, M., Dervins-Ravault, D., Rolando, M., Nora, T., Newton, H. J., Samson, F. M., Sahr, T., Gomez-Valero, L., Jules, M., Hartland, E. L., and Buchrieser, C. (2010). The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol. 12, 1272–1291.
McKinney, R. M., Porschen, R. K., Edelstein, P. H., Bissett, M. L., Harris, P. P., Bondell, S. P., Steigerwalt, A. G., Weaver, R. E., Ein, M. E., Lindquist, D. S., Kops, R. S., and Brenner, D. J. (1981). Legionella longbeachae species nova, another etiologic agent of human pneumonia. Ann. Intern. Med. 94, 739–743.
Molmeret, M., Horn, M., Wagner, M., Santic, M., and Abu Kwaik, Y. (2005). Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71, 20–28.
Moore, W. E., and Moore, L. V. (1994). The bacteria of periodontal diseases. Periodontology 5, 66–77.
Nguyen, P., Bar-Sela, G., Sun, L., Bisht, K. S., Cui, H., Kohn, E., Feinberg, A. P., and Gius, D. (2008). BAT3 and SET1A form a complex with CTCFL/BORIS to modulate H3K4 histone dimethylation and gene expression. Mol. Cell Biol. 28, 6720–6729.
Pan, X., Luhrmann, A., Satoh, A., Laskowski-Arce, M. A., and Roy, C. R. (2008). Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320, 1651–1654.
Petroski, M. D., and Deshaies, R. J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20.
Pickart, C. M., and Fushman, D. (2004). Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616.
Price, C. T., Al-Khodor, S., Al-Quadan, T., and Abu Kwaik, Y. (2010a). Indispensable role for the eukaryotic-like ankyrin domains of the ankyrin B effector of Legionella pneumophila within macrophages and amoebae. Infect. Immun. 78, 2079–2088.
Price, C. T., Al-Quadan, T., Santic, M., Jones, S. C., and Abu Kwaik, Y. (2010b). Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J. Exp. Med. 207, 1713–1726.
Price, C. T., Al-Khodor, S., Al-Quadan, T., Santic, M., Habyarimana, F., Kalia, A., and Kwaik, Y. A. (2009). Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 5, e1000704. doi: 10.1371/journal.ppat.1000704.
Raiborg, C., Slagsvold, T., and Stenmark, H. (2006). A new side to ubiquitin. Trends Biochem. Sci. 31, 541–544.
Roden, J. A., Belt, B., Ross, J. B., Tachibana, T., Vargas, J., and Mudgett, M. B. (2004). A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc. Natl. Acad. Sci. U.S.A. 101, 16624–16629.
Rodriguez-Palenzuela, P., Matas, I. M., Murillo, J., Lopez-Solanilla, E., Bardaji, L., Perez-Martinez, I., Rodriguez-Moskera, M. E., Penyalver, R., Lopez, M. M., Quesada, J. M., Biehl, B. S., Perna, N. T., Glasner, J. D., Cabot, E. L., Neeno-Eckwall, E., and Ramos, C. (2010). Annotation and overview of the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 draft genome reveals the virulence gene complement of a tumour-inducing pathogen of woody hosts. Environ. Microbiol. 12, 1604–1620.
Rosebrock, T. R., Zeng, L., Brady, J. J., Abramovitch, R. B., Xiao, F., and Martin, G. B. (2007). A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448, 370–374.
Rurangirwa, F. R., Dilbeck, P. M., Crawford, T. B., McGuire, T. C., and McElwain, T. F. (1999). Analysis of the 16S rRNA gene of micro-organism WSU 86-1044 from an aborted bovine foetus reveals that it is a member of the order Chlamydiales: proposal of Waddliaceae fam. nov., Waddlia chondrophila gen. nov., sp. nov. Int. J. Syst. Bacteriol. 49 (Pt 2), 577–581.
Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., Billault, A., Brottier, P., Camus, J. C., Cattolico, L., Chandler, M., Choisne, N., Claudel-Renard, C., Cunnac, S., Demange, N., Gaspin, C., Lavie, M., Moisan, A., Robert, C., Saurin, W., Schiex, T., Siguier, P., Thebault, P., Whalen, M., Wincker, P., Levy, M., Weissenbach, J., and Boucher, C. A. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502.
Sasaki, T., Gan, E. C., Wakeham, A., Kornbluth, S., Mak, T. W., and Okada, H. (2007). HLA-B-associated transcript 3 (Bat3)/Scythe is essential for p300-mediated acetylation of p53. Genes Dev. 21, 848–861.
Schmitz-Esser, S., Tischler, P., Arnold, R., Montanaro, J., Wagner, M., Rattei, T., and Horn, M. (2010). The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J. Bacteriol. 192, 1045–1057.
Schmitz-Esser, S., Toenshoff, E. R., Haider, S., Heinz, E., Hoenninger, V. M., Wagner, M., and Horn, M. (2008). Diversity of bacterial endosymbionts of environmental acanthamoeba isolates. Appl. Environ. Microbiol. 74, 5822–5831.
Schrammeijer, B., Hemelaar, J., and Hooykaas, P. J. (1998). The presence and characterization of a virF gene on Agrobacterium vitis Ti plasmids. Mol. Plant Microbe Interact. 11, 429–433.
Schrammeijer, B., Risseeuw, E., Pansegrau, W., Regensburg-Tuink, T. J., Crosby, W. L., and Hooykaas, P. J. (2001). Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr. Biol. 11, 258–262.
Schulman, B. A., Carrano, A. C., Jeffrey, P. D., Bowen, Z., Kinnucan, E. R., Finnin, M. S., Elledge, S. J., Harper, J. W., Pagano, M., and Pavletich, N. P. (2000). Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386.
Segal, G., Purcell, M., and Shuman, H. A. (1998). Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U.S.A. 95, 1669–1674.
Seong, S. Y., Choi, M. S., and Kim, I. S. (2001). Orientia tsutsugamushi infection: overview and immune responses. Microbes Infect. 3, 11–21.
Shin, S., and Roy, C. R. (2008). Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell Microbiol. 10, 1209–1220.
Smith, T. F., Gaitatzes, C., Saxena, K., and Neer, E. J. (1999). The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181–185.
Sonnberg, S., Seet, B. T., Pawson, T., Fleming, S. B., and Mercer, A. A. (2008). Poxvirus ankyrin repeat proteins are a unique class of F-box proteins that associate with cellular SCF1 ubiquitin ligase complexes. Proc. Natl. Acad. Sci. U.S.A. 105, 10955–10960.
Steele, T. W., Lanser, J., and Sangster, N. (1990). Isolation of Legionella longbeachae serogroup 1 from potting mixes. Appl. Environ. Microbiol. 56, 49–53.
Tsukahara, T., Kimura, S., Ichimiya, S., Torigoe, T., Kawaguchi, S., Wada, T., Yamashita, T., and Sato, N. (2009). Scythe/BAT3 regulates apoptotic cell death induced by papillomavirus binding factor in human osteosarcoma. Cancer Sci. 100, 47–53.
Tzfira, T., Vaidya, M., and Citovsky, V. (2004). Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 431, 87–92.
van Wijk, S. J., and Timmers, H. T. (2010). The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 24, 981–993.
Vogel, J. P., Andrews, H. L., Wong, S. K., and Isberg, R. R. (1998). Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876.
Voth, D. E., and Heinzen, R. A. (2007). Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol. 9, 829–840.
Voth, D. E., and Heinzen, R. A. (2009). Coxiella type IV secretion and cellular microbiology. Curr. Opin. Microbiol. 12, 74–80.
Voth, D. E., Howe, D., Beare, P. A., Vogel, J. P., Unsworth, N., Samuel, J. E., and Heinzen, R. A. (2009). The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J. Bacteriol. 191, 4232–4242.
Welchman, R. L., Gordon, C., and Mayer, R. J. (2005). Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 6, 599–609.
Wright, L. P., and Philips, M. R. (2006). Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 47, 883–891.
Ximenez-Fyvie, L. A., Haffajee, A. D., and Socransky, S. S. (2000). Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 27, 648–657.
Xu, P., Duong, D. M., Seyfried, N. T., Cheng, D., Xie, Y., Robert, J., Rush, J., Hochstrasser, M., Finley, D., and Peng, J. (2009). Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145.
Zaltsman, A., Krichevsky, A., Loyter, A., and Citovsky, V. (2010). Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe. 7, 197–209.
Zamboni, D. S., McGrath, S., Rabinovitch, M., and Roy, C. R. (2003). Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 49, 965–976.
Zhang, M., Windheim, M., Roe, S. M., Peggie, M., Cohen, P., Prodromou, C., and Pearl, L. H. (2005). Chaperoned ubiquitylation – crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell. 20, 525–538.
Ziemienowicz, A., Merkle, T., Schoumacher, F., Hohn, B., and Rossi, L. (2001). Import of Agrobacterium T-DNA into plant nuclei: two distinct functions of VirD2 and VirE2 proteins. Plant Cell 13, 369–383.
Keywords: farnesylation, SKP1, AnkB, VirF, GALA, Legionella. Legionnaires’, Agrobacterium, Ralstonia
Citation: Price CTD and Abu Kwaik Y (2010) Exploitation of host polyubiquitination machinery through molecular mimicry by eukaryotic-like bacterial F-box effectors. Front. Microbio. 1:122. doi:10.3389/fmicb.2010.00122
Received: 15 September 2010; Paper pending published: 04 October 2010;
Accepted: 12 October 2010; Published online: 11 November 2010.
Edited by:
Amal Amer, The Ohio State University, USAReviewed by:
Amal Amer, The Ohio State University, USAJohn S. Gunn, The Ohio State University, USA
Dario S. Zamboni, Universidade de São Paulo, Brazil
Copyright: © 2010 Price and Abu Kwaik. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Yousef Abu Kwaik, Department of Microbiology and Immunology, College of Medicine, University of Louisville, Louisville, KY 40202, USA. e-mail: abukwaik@louisville.edu
