- Fungal Pathogenesis Laboratory, Centre for Infectious Diseases and Microbiology, Sydney Medical School-Western, Westmead Millennium Institute, University of Sydney at Westmead Hospital, Westmead, NSW, Australia
Many pathogenic microbes, including many fungi, produce phospholipases which facilitate survival of the pathogen in vivo, invasion and dissemination throughout the host, expression of virulence traits and evasion of host immune defense mechanisms. These phospholipases are either secreted or produced intracellularly and act by physically disrupting host membranes, and/or by affecting fungal cell signaling and production of immunomodulatory effectors. Many of the secreted phospholipases acquire a glycosylphosphatidylinositol sorting motif to facilitate membrane and/or cell wall association and secretion. This review focuses primarily on the role of two members of the phospholipase enzyme family, phospholipase B (Plb) and phosphatidylinositol (PI)-specific phospholipase C (PI-C/Plc), in fungal pathogenesis and in particular, what has been learnt about their function from studies performed in the model pathogenic yeast, Cryptococcus neoformans. These studies have revealed how Plb has adapted to become an important part of the virulence repertoire of pathogenic fungi and how its secretion is regulated. They have also provided valuable insight into how the intracellular enzyme, Plc1, contributes to fungal fitness and pathogenicity – via a putative role in signal transduction pathways that regulate the production of stress-protecting pigments, polysaccharide capsule, cell wall integrity, and adaptation to growth at host temperature. Finally, this review will address the role fungal phospholipases have played in the development of a new class of antifungal drugs, which mimic their phospholipid substrates.
The Phospholipase Enzyme Family and the Activities of Fungal PLB and PLC
Phospholipases are a heterogeneous group of enzymes that hydrolyse one or more ester linkages in glycerophospholipids (Figure 1), which are the major components of cell membranes and lung surfactant. Each glycerophospholipid consists of a glycerol backbone containing one (lysophospholipid) or two (phospholipid) ester linkages to hydrophobic fatty acid chains, and a third ester linkage to a phosphate head group. The phosphate head group is also linked by an ester bond to a variety of polar moieties, such as choline, ethanolamine, serine, and inositol. Phospholipases are categorized into five groups depending on which ester bond is hydrolysed: A1, A2 B, C, and D (Figure 1). Two major phospholipases associated with pathogenicity in fungi are phospholipase B (Plb) and phosphatidylinositol (PI)-preferring phospholipase C (PI-PLC/Plc). Plb enzymes remove both fatty acyl chains from glycerophospholipids. They may also have lysophospholipase (LPL) and lysophospholipase-transacylase (LPTA) activities, both of which can contribute to the detoxification of potentially cellulolytic lysophospholipids. Specifically, the LPL activity removes the single fatty acyl chain on a lysophospholipid to produce glycerophosphocholine (GPC) and the LPTA activity re-esterifies an acyl chain onto a lysophospholipid, thus regenerating a glycerophospholipid (Figure 2). It has also been proposed that PLB activity is, in fact, a combination of phospholipase A2 (PLA2) and LPL activities. PI-PLCs (or Plcs) remove the polar head group from PI, which can be present as either mono- or diphosphorylated inositol or as the glycosylphosphatydylinositol (GPI) anchor on some membrane and cell wall proteins, including fungal Plbs (Figure 1). Both Plb and PI-PLC can be secreted or remain intracellular. Many Plb enzymes have a GPI anchor which provides an additional level of secretory regulation and appears to serve as a sorting signal for efficient cellular export of fungal Plbs. The lipid derived products of Plb and PI-PLC may act as secondary messengers in signal transduction, or exert immunomodulatory effects.
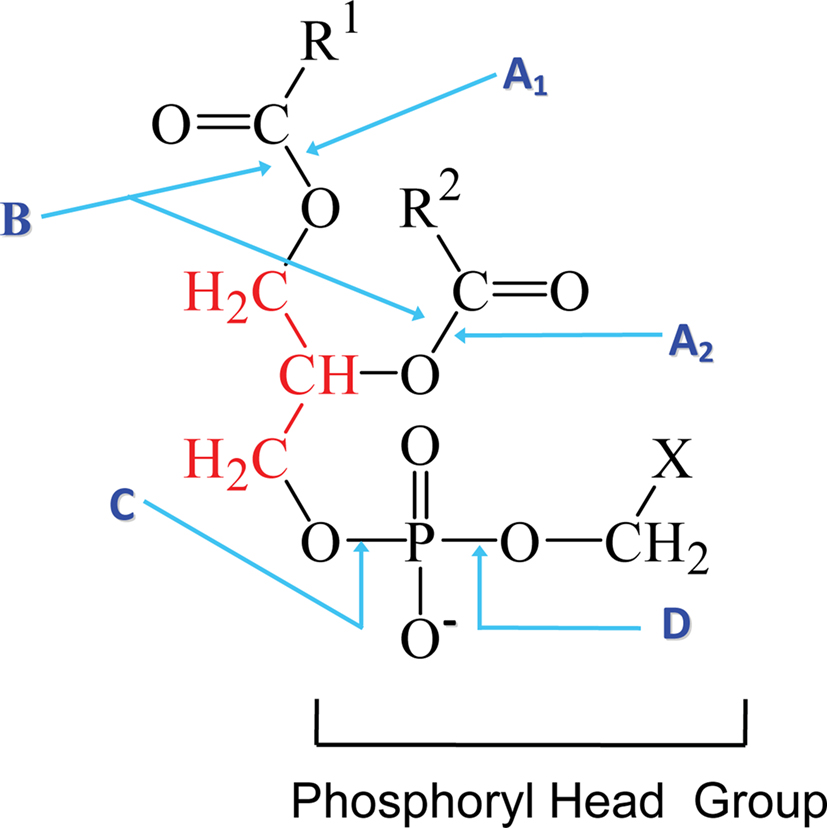
Figure 1. The phospholipid cleavage sites of phospholipase enzymes. The phospholipid glycerol backbone is shown in red. Phospholipase A1 (A1) cleaves a fatty acyl ester at the sn1 position while phospholipase A2 (A2) removes the fatty acid at the sn2 position to yield a lysophospholipid and a fatty acid. Phospholipase B (B) removes both acyl chains from the sn1 and sn2 positions on the glycerol backbone. The phosphodiester bond in the head group proximal to the glycerol backbone is cleaved by phospholipase C (C) to yield diacylglycerol and the phosphoryl head group. PI-PLC enzymes perform this cleavage when X is inositol, which may, or may not, be linked to phosphates or a sugar chain composed predominantly of mannosyl sugars in the case of a GPI anchor. Cleavage by phospholipase D (D) at the phosphodiester bond proximal to X, yields phosphatidic acid and X, which can be either ethanolamine, serine, inositol, or choline. Adapted from the PhD thesis of Dr Rosemary A. Siafakas.
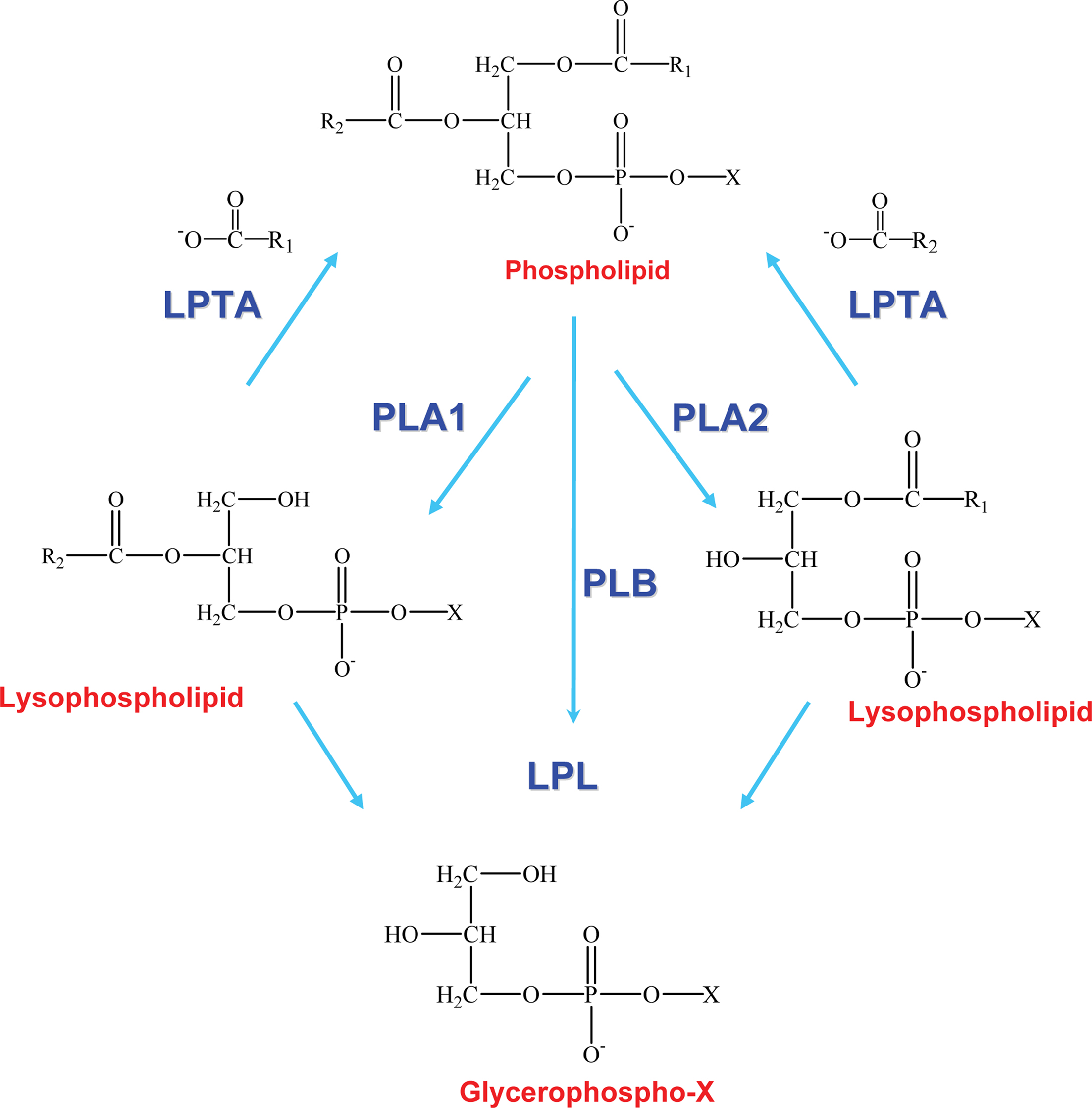
Figure 2. Enzymatic activities of phospholipase B enzymes. Phospholipase B (PLB) activity is a combination of PLA1 and PLA2 activities (or PLA and LPL activity) and generates glycerophospho compounds. Lysophospholipid, a short-lived intermediate, is also a substrate for lysophospholipase (LPL) and lysophospholipase-transacylase (LPTA) activities, which rapidly hydrolyse or re-esterify lysophospholipid, respectively. R1 and R2 represent fatty acid chains in the sn1 and sn2 position of the phospholipid, respectively. X refers to the head group (e.g., serine, ethanolamine, choline, or inositol). Adapted from the PhD thesis of Dr Rosemary A. Siafakas.
Cryptococcus Neoformans – An Effective Model Fungal Pathogen for Characterizing Phospholipase Virulence Determinants
The basidiomycetous fungal pathogen, Cryptococcus neoformans, has a predilection to cause infection in patients with AIDS and is responsible for an estimated one million cases of meningitis and 675,000 deaths per year in HIV-infected patients alone (Park et al., 2009). Infections also occur in immunocompetent individuals (Chau et al., 2010). The other pathogenic Cryptococcus species, C. gattii, mainly infects immunocompetent individuals and was responsible for an outbreak of cryptococcosis that was first noted in 1999 on Vancouver Island, British Columbia (BC), Canada (Ma et al., 2007). There are two varieties of C. neoformans, C. neoformans var. grubii (Serotype A) and C. neoformans var. neoformans (Serotype D), with serotype A being the more clinically prevalent and virulent serotype (Martinez et al., 2001). The predominant natural habitat of C. neoformans is nitrogen-rich avian guano, soil and possibly decaying material from trees. It has been suggested that its pathogenicity for mammals evolved as a consequence of phagocytic interactions with soil ameba (Alvarez et al., 2009). In immunocompetent individuals, phagocytosis of inhaled spores or small dessicated yeast cells by macrophages may result either in clearing of the infection or establishment of a latent infection in the lung. However, in the immunocompromised individual there is evidence that macrophages may also transport C. neoformans to the central nervous system where it establishes severe life-threatening meningoencephalitis (Charlier et al., 2009). Not only is C. neoformans an important pathogen in its own right, it is also an excellent model for studying fungal pathogenesis, as the recently sequenced haploid genomes of both serotypes A and D are amenable to a range of genetic manipulation techniques, including targeted gene disruption and RNAi and robust animal models of infection are available to assess the role of genes in virulence. Much of our knowledge about fungal phospholipase enzymatic activities and their contribution to pathogenesis has been derived from studies performed on cryptococcal enzymes. In fact, C. neoformans is the only fungal pathogen in which the effect of absence of the Plc1-encoding gene on fungal fitness and pathogenicity has been determined (Chayakulkeeree et al., 2008).
Phospholipase B Enzymes Implicated in Fungal Virulence
It is well-established through gene deletion and reconstitution experiments that phospholipases contribute to the virulence repertoire of pathogenic fungi, despite their presence in the mainly non-pathogenic and industrial yeasts. Pathogenic fungi that secrete phospholipase B include C. neoformans (Chen et al., 1997b; Santangelo et al., 1999), Candida albicans (Ibrahim et al., 1995; Mirbod et al., 1995; Theiss et al., 2006), and Aspergillus fumigatus (Shen et al., 2004). The most well characterized phospholipase B enzymes produced by fungal pathogens are phospholipase B 1, 2, and 5 from C. albicans (Sugiyama et al., 1999; Mukherjee et al., 2001; Theiss et al., 2006) and phospholipase B1 from C. neoformans (Chen et al., 2000) and Cryptococcus gattii (Wright et al., 2004) (CaPlb1/2/5, CnPlb1, CgPlb1, respectively). Only CaPlb2 does not play a significant role in virulence while the role of CgPlb1 in virulence has not been assessed. CnPlb1, CaPlb1, and CgPlb1 all possess PLB, LPL and LPTA enzymatic activities, with properties that differ to each other and to Plb1 homologs produced by industrial yeasts (Takahashi et al., 1991; Mirbod et al., 1995; Chen et al., 2000).
The C. neoformans genome contains a single PLB gene, PLB1, with an established role in virulence (Cox et al., 2001). In addition to CnPlb1, C. neoformans produces a Plb1-independent phospholipase A1, which degrades 1-palmitoyl-2-arachidonoyl phosphatidylcholine. Neither the gene encoding this enzyme, nor the protein it specifies, have been isolated (Wright et al., 2007) and its role in virulence has not been determined. C. neoformans also produces two additional LPL/LPTA enzymes, CnLyso1 and CnLpl1, which were characterized in serotype D (Coe et al., 2003) and C. gattii (Wright et al., 2004), respectively. The CnLYSO1 found in serotype D itself is not predicted to be secreted as it has no secretory leader peptide, but appears to affect the secretion of CnPlb1 (Coe et al., 2003). Its role in virulence is unknown. Interestingly, although CnLpl1 found in serotype A does contain a putative secretory leader peptide, it does not contribute to the pool of secreted PLB activity, at least in cultures maintained on agar; this pool was attributed solely to CnPlb1 activity (Cox et al., 2001). Disruption of the CnLPL1 homolog in serotype A altered neither growth nor virulence of C. neoformans in a mouse model (M. Chayakulkeeree, and J. T. Djordjevic, unpublished observation). It may function in association with CnLyso1 to remove toxic lysophospholipids (Wright et al., 2007). These three enzymes may provide functional redundancy by maintaining cryptococcal growth in the absence of CnPlb1.
The genome of C. albicans encodes a Plb multigene family with five putative members. The best studied genes in this family are CaPLB1, CaPLB2, and CaPLB5 which encode putative secreted proteins (Leidich et al., 1998; Sugiyama et al., 1999; Mukherjee et al., 2001; Theiss et al., 2006). Only CaPLB1 and CaPLB5 have an established role in virulence (Leidich et al., 1998; Hoover et al., 1998; Mukherjee et al., 2001; Theiss et al., 2006). Abrogation of CaPlb1 and CaPlb5 enzymatic activity by inactivation of CaPLB1 and CaPLB5, respectively, rendered the mutant strains less virulent in systemic mouse models of infection as measured by host tissue colonization (Leidich et al., 1998; Mukherjee et al., 2001; Theiss et al., 2006). Loss of these genes coincided with reduced LPTA/PLB and PLA2 activity, respectively. Neither the role of CnPlb1 and CaPlb5 in virulence nor their enzymatic properties have been determined. The genome of A. fumigatus encodes three PLB genes (Shen et al., 2004). However, their role in virulence has not yet been established. It is important to note that not all pathogenic fungi secrete significant amounts of Plb. One such example is Candida parapsilosis where, due to contradictory findings among various investigators, the role of secreted Plb in pathogenesis is unclear (Trofa et al., 2008).
PLB Homologs in Pathogenic Fungi Differ from those in Non-Pathogenic Fungi
PLB genes are present in non-pathogenic yeasts including Saccharomyces cerevisiae, Debaryomyces (Torulaspora) hansenii, Penicillium notatum, Schizosaccharomyces pombe, Torulaspora delbrueckii, Neurospora crassa, and Kluyveromyces lactis (reviewed in Ghannoum, 2000). Extensive enzymatic characterization of the Plb enzymes secreted by the pathogenic yeasts, C. neoformans and C. albicans, and a comparison of their properties to the Plb homologs produced by non-pathogenic yeasts, has revealed why these enzymes are more adapted to a role in virulence (Chen et al., 2000). Firstly, the specific activities of purified cryptococcal and candidal Plb1 enzymes are 10- to 200-fold higher than the corresponding homologs from non-pathogenic fungi (Lee et al., 1994; Mirbod et al., 1995; Merkel et al., 1999; Chen et al., 2000). Secondly, the major membrane phospholipids, dipalmityolphosphatidylcholine (DPPC) and phosphatidylglycerol, which are also the most abundant components of lung surfactant, and lysophosphatidylcholine, are preferred substrates of CnPlb1, whereas the less accessible, acidic inner-leaflet phospholipids, phosphatidylserine, phosphatidylinositol, and phosphatidic acid are preferred substrates of Plbs from non-pathogenic fungi (Witt et al., 1984; Saito et al., 1991; Oishi et al., 1996; Merkel et al., 1999). This observation suggests that Plb1 secreted by pathogenic fungi is more likely to digest host cell membranes and surfactant than Plb1 secreted by non-fungal pathogens. In support of this, the extent of cell damage caused by culture supernatants prepared from C. albicans (62%) was less than that caused by the CaPLB1-deficient strain and S. cerevisiae (35 and 4%, respectively) as assessed using a thiazolium blue-based assay (Ghannoum, 2000). Thirdly, a range of protein modifying agents differentially affect the activities of CnPlb1, Plb homologs from non-pathogenic fungi and mammalian secretory PLA2 (Chen et al., 2000). The millimolar quantities of acetic acid secreted by C. neoformans in vitro (Bubb et al., 1999) and in experimental lung and brain cryptococcomas (Himmelreich et al., 2003) may create a microenvironment favorable for CnPlb1, which has an acidic pH optimum (Chen et al., 2000). As C. neoformans is well-adapted to intracellular survival in the acidified phagolysosome (pH 5.0) of human macrophages (Levitz et al., 1999), secreted CnPlb1 may be a contributing survival factor.
Evidence for Production of Fungal PLB in vivo
There is evidence to suggest that Plb1 produced by pathogenic fungi is also produced during infection in vivo. CaPlb1 was identified immunologically in host tissue (Ghannoum, 2000; Mukherjee et al., 2001) and the GPC product of CnPlb1 is present in lung and brain cryptococcomas produced in a rat model (Himmelreich et al., 2003; Sorrell et al., 2006). Furthermore, antibodies to CaPlb1 and CnPlb1 have been identified in the serum of infected patients (Ghannoum, 2000; Santangelo et al., 2005). In addition, blood isolates of C. albicans generally secrete much higher levels of CaPlb1 than isolates from the oral cavity of healthy individuals (Ibrahim et al., 1995) or isolates from wounds or urine (Price et al., 1982).
Factors Regulating CNPLB1 Cellular Export to the Periphery and Secretion
Cryptococcal and candidal Plbs with an established role in virulence (CnPlb1, CaPlb1, CaPlb2, and CaPlb5) contain a secretory leader peptide at the N-terminus and we have confirmed that deletion of this motif in CnPlb1 abolishes its secretion within a heterologous expression system (Djordjevic et al., 2005). This motif permits entry of nascent Plb into the endoplasmic reticulum (ER) where it is folded and acquires asparagines (N)-linked glycosylation which in the case of CnPlb1 is essential for activity (Chen et al., 2000) and export and secretion (Turner et al., 2006). The Plbs are then transported to the surface and subsequently secreted. Both CnPlb1 and CaPlb1 have been detected in fungal secretions and within host tissue. In the case of CnPlb1, secretion still occurs despite the acquisition a glycosylphosphatidylinositol (GPI) anchor which permits CnPlb1 association with membranes and the cell wall, suggesting that CnPlb1 must be released from the anchor prior to secretion. In contrast to CnPlb1, CaPlb1, and CaPlb2 do not acquire a GPI anchor, while CaPlb5 (and CaPlb3, CaPlb4) are predicted to acquire one (Theiss et al., 2006). Of the Plbs produced by A. fumigatus, AfPlb2, is the only Plb not predicted to be secreted or GPI anchored. It is not known why some Plbs involved in virulence acquire a GPI anchor and some do not. However, in addition to permitting membrane and cell wall localization, the anchor in CnPlb1 appears to play a role in regulating CnPlb1 export and secretion.
GPI anchors and rafts Following export through the secretory pathway to the cell periphery, some fungal GPI-anchored proteins are retained at the plasma membrane while others are released from it by an undefined cleavage mechanism within the GPI anchor (Lu et al., 1994; Kollar et al., 1997; Fujii et al., 1999; Gagnon-Arsenault et al., 2008). Cleavage allows the GPI-anchored protein to be transported to the cell wall and covalently cross-linked to β-1,6-glucans via the GPI anchor remnant (Kapteyn et al., 1994; Lu et al., 1994) or secreted. The means by which fungal GPI-anchored proteins are differentially targeted to the membrane or cell wall are poorly understood. However, the signals reside in the C-terminus (Mao et al., 2008). We have found CnPlb1 to be associated with both membranes and the cell wall fraction (Siafakas et al., 2006; Siafakas et al., 2007).
In fungi, GPI-anchored proteins cluster within sphingolipid/sterol-rich microdomains, or rafts, within the plasma membrane (Muniz and Riezman, 2000; Insenser et al., 2006). We also demonstrated that treatment of cryptococci with the raft disrupting agent, methyl-β-cyclodextrin, which binds to sterols, releases CnPlb1 protein and enzyme activity into the external milieu, consistent with CnPlb1 also localizing within plasma membrane rafts following its transport to the cell periphery (Siafakas et al., 2006). We also found that CnPlb1-containing membranes are enriched in sterols, insoluble in Triton X-100 at 4°C and of a higher buoyant density than Triton X-100-soluble membranes (Siafakas et al., 2006), consistent with the properties of rafts (Bagnat et al., 2000). We proposed that raft membranes serve to cluster CnPlb1 at the cell surface to allow efficient access to enzyme substrate and/or to provide a mechanism for rapid release to the external environment (Siafakas et al., 2006). However, since both raft assembly and the sorting of GPI anchored proteins into raft-enriched secretory vesicles occurs within the ER (Sutterlin et al., 1997; Bagnat et al., 2000; Muniz and Riezman, 2000), the association of CnPlb1 with cryptococcal rafts may also be an early secretory event.
GPI anchors are also essential for export of CnPlb1 to the periphery as CnPlb1 export is blocked when GPI anchor synthesis is blocked (Siafakas et al., 2007). In addition, deletion of the GPI anchor consensus motif, which serves as a temporary ER retention signal until fully synthesized GPI anchors are substituted for it, resulted in a higher ratio of secreted to cell-associated CnPlb1 in a heterologous S. cerevisiae expression system due to the lack of CnPlb1 retention in the membrane and cell wall (Djordjevic et al., 2005). However despite this, absence of a GPI anchor resulted in a greater than 50% reduction in the rate of CnPlb1 export and secretion, consistent with export of truncated protein being less efficient than that of native CnPlb1 (Djordjevic et al., 2005). Thus, the acquisition of the GPI anchor in CnPlb1 serves to expedite CnPlb1 transport through the secretory pathway, and to localize CnPlb1 at the fungal cell periphery.
N-linked Glycosylation. Using site-directed mutagenesis, we also showed that altering just a single N-linked glycosylation site out of the potential 17 that exist in CnPlb1, caused a retardation in cellular export and secretion of CnPlb1 within the heterologous host, S. cerevisiae, irrespective of GPI anchor status, consistent with N-linked glycosylation being more crucial for CnPlb1 traffic than acquisition of the GPI anchor. Specifically, CnPlb1 export and secretion was compromised when three individual glycosylation sites, Asn56, Asn430, and Asn550, were changed to Ala (Turner et al., 2006).
Surface-release mechanisms Following CnPlb1 export to raft membranes, some CnPlb1 is relocated to the cell wall and/or is secreted, though the mechanism of release is not known (Siafakas et al., 2006, 2007). The membrane GPI anchor is a potential substrate for (G)PI-PLC, (G)PI-PLD (de Almeida and Heise, 1993; Kristiansen and Richter, 2002) and/or a number of different proteases (Muller et al., 1996; Parkin et al., 2004; Gagnon-Arsenault et al., 2008) and/or α-mannosidase (Fujii et al., 1999; Kitagaki et al., 2002). We recently showed that secretion of CnPlb1 is dependent on PLC1 which encodes a PI-PLC (Chayakulkeeree et al., 2008). However the mechanism by which PLC1 achieves this is unclear (discussed in more detail below). Cell wall CnPlb1 associates with β-1,6-glucan (Siafakas et al., 2007) which, in C. neoformans, is much more abundant than β-1,3-glucan relative to other yeasts (Doering, 2009). β-1,6-glucans are also associated with secreted CnPlb1, suggesting that secreted CnPlb1 is derived from the cell wall reservoir (Siafakas et al., 2007). Disruption of specific KRE family genes in C. neoformans (KRE5 and KRE6/SKN1), which in S. cerevisiae are required for synthesis of β-1,6-glucan, caused a reduction in the β-1,6-glucan content of the cell wall (Gilbert et al., 2010). This reduction in β-1,6-glucan coincided with compromised cell wall integrity and reduced retention of CnPlb1 in the cell wall (Gilbert et al., 2010) supporting the previous finding that CnPlb1 is retained in the cell wall via a linkage to β-1,6-glucan (Siafakas et al., 2007). That β-1,6-glucans are retained on secreted CnPlb1 suggests that extracellular endo β-glucanases may be involved in the release of CnPlb1 from the cell wall, in addition to fulfilling a traditional role in cell wall remodeling.
Extracellular glucanase activity, including α-mannosidase, α- and β-glucosidase, α- and β-galactosidase, β-xylosidase, β-glucuronidase, and endo β-1,3-glucanase is associated with the capsule and/or the cell wall of C. neoformans and some of the substrates of these enzymes can support yeast growth (Macekova et al., 2006). Synthesis of the majority of these enzymes is regulated by glucose repression (Macekova et al., 2006). It has been demonstrated that many cells die as they enter stationary phase due to erosion of their cell walls by autolysis, and that this coincides with an increase in externalization of β-1,3-glucanase activity with a pH optimum of between 3.0 and 3.5 (Farkas et al., 2009). As stationary phase is also the time when CnPlb1 secretion is optimal, it is tempting to speculate that extracellular β-glucanases can release cell wall proteins, such as CnPlb1, into the external environment, while simultaneously making cell wall sugar available to nutrient-depleted cells in stationary phase. Six endo β-1,3-glucanase isoforms have been extracted from cryptococcal cell walls (Macekova et al., 2006). The C. neoformans genome encodes a family of four putative β-1-3-glucanases which could be involved in the release of CnPlb1. Cleavage of β-1,3-glucans, which in yeast are attached to β-1,6-glucans (Aimanianda et al., 2009), would result in the release of β-1-6-glucans and their attached proteins. Using gene deletion analysis, our laboratory is investigating the role of β-1-3-glucanases in CnPlb1 cell wall retention and secretion.
In addition to CnPlb1, the externalization of two other virulence determinants, laccase (Lac 1) and capsular glucuronoxylomannan (GXM), is dependent on fungal secretion pathways. Recent studies in C. neoformans defined separate pathways for export of Lac1 and GXM, which utilize SEC6-dependent exosomes of the lysosomal trafficking pathway and SAV1(SEC4)-dependent vesicles, respectively (Yoneda and Doering, 2006; Panepinto et al., 2009). Recently we determined that CnPlb1 cellular export and secretion in C. neoformans, but not that of Lac1 and GXM, is dependent on a gene homologs to SEC14 from S. cerevisiae, which encodes a PI-transfer protein (PITP). This finding provides further support for the existence of distinct cellular pathways for export of virulence determinants in C. neoformans (unpublished data). Recently, several cryptococcal virulence determinants including laccase, urease, and soluble GXM polysaccharide, but not CnPlb1 to a significant extent, were shown to be transported across the cryptococcal cell wall to the external milieu via SEC6-dependent exosomes (Rodrigues et al., 2007, 2008; Casadevall et al., 2009; Panepinto et al., 2009). The mechanism of release of these virulence determinants from exosomes is unknown, but might involve the membrane hydrolytic action of cell wall-associated phospholipases such as CnPlb1.
Role of PLB1 in Pathogenesis
The relative importance of the three phospholipase B activities (PLB, LPL, and LPTA) in fungal pathogenesis may vary in the different host microenvironments. It has been shown that the LPL and LPTA activities of CnPlb1 can reacylate lysophospholipids and reverse the damage caused to neutrophils by other secreted cryptococcal components (Santangelo et al., 1999). The roles of the various activities in pathogenesis is discussed in more detail below.
Host Adhesion/Penetration and Dissemination
Plb enzymes are proven fungal invasions in C. albicans and C. neoformans. The first barriers encountered by infectious C. neoformans propagules are lung surfactant and lung epithelial cells. DPPC and PG, which are abundant in lung surfactant and dioleoyl PC (DOPC), which is present in the outer leaflet of mammalian cell membranes, are preferred substrates of CnPlb1 (Chen et al., 1997a, 2000). Hydrolysis of these phospholipids by CnPlb1 would potentially facilitate penetration of host tissue and establishment of cryptococcal infection. In support of this, partially purified CnPlb1 hydrolyses lung surfactant (Santangelo et al., 1999) and facilitates adherence of cryptococci to lung epithelial cells (Ganendren et al., 2006). CnPlb1 also facilitated the entry of endotracheally administered cryptococci into lung interstitial macrophages although the mechanism of traversal across the lung epithelial barrier remains unknown. The reduced lung burdens of cryptococci observed after infection with the CnPlb1 deletion mutant (Δplb1) may be explained by decreased invasive potential; it is also possible that multiplication within the host is inhibited. CnPlb1 is also essential for dissemination of C. neoformans from the lung to the brain via the blood, and possibly, the lymphatic system (Cox et al., 2001; Noverr et al., 2003; Santangelo et al., 2004). However, CnPlb1 is not essential for the establishment of neurological infections (Santangelo et al., 2004).
Cryptococcus neoformans is a facultative intracellular pathogen that survives and replicates within the intracellular phagolysosome of macrophages. Yeast–macrophage interactions result in several outcomes which potentially drive the establishment, progression, and dissemination of infection. Using a non-activated murine alveolar macrophage (AM) cell line, MHS-1, Noverr et al. (2003) showed that CnPlb1 is required for C. neoformans survival in the presence of these macrophages. Cox et al. (2001) showed that CnPlb1 promotes cryptococcal budding at early time points (1–3.5 h) post-phagocytosis by an activated J774 macrophage cell line. In contrast, phagocytosis of strain H99 (7.3% ± 1.2) by the phorbol-12-myristate 13-acetate-activated human macrophage-like cell line, THP-1, was significantly greater than that of CnΔplb1 (4.3% ± 0.8) (Wright et al., 2007). Not only does CnPlb1 facilitate entry of cryptococci into interstitial macrophages in the lung, but as demonstrated by Santangelo et al. (2004), it is also required for dissemination from the lungs to regional lymph nodes where cryptococci were observed to multiply within Langerhans-type giant cells (Santangelo et al., 2004). Higher brain burdens were observed following intravenous injection of lung macrophages infected with cryptococci compared with free cryptococci. Since the brain burden was independent of CnPlb1 it was concluded by the authors that macrophages provide a vehicle for dissemination of cryptococci to the brain (Santangelo et al., 2004). Further support for this was provided by Charlier et al. (2009).
CnPlb1 may also be required for cryptococci to exit macrophages and thereby avoid killing by macrophage immune defense mechanisms. Escape can occur either by macrophage lysis and cell death, or by non-lytic extrusion via a process that does not affect the viability of either C. neoformans or the host macrophage (vomocytosis) (Alvarez and Casadevall, 2006, 2007; Ma et al., 2006, 2007). The acidic environment of macrophage phagolysosomes provides an optimal environment for CnPlb1 catalytic activity. Feldmesser et al. (2000) reported disruptions in the phagolysosomal membrane of engulfed cryptococci which they attributed to secreted CnPlb1 and/or proteases. Such membrane disruptions may allow lysis-associated egress of C. neoformans from macrophages. Secreted CnPlb1 was recently found to facilitate non-lytic extrusion of C. neoformans from macrophages; two CnPlb1 strains defective in CnPlb1 secretion, due to CnPLB1 deletion or deletion of a secretory factor required for normal secretion of CnPlb1, exhibited reduced rates of vomocytosis, and failure to disseminate to the brain in a mouse model (manuscript submitted).
Although the role of secreted CaPlb1 in adhesion of C. albicans to host epithelium is not well-established, there is evidence that CaPlb1 does play a role in damage to, and penetration of, host epithelium, as culture supernatants from wild-type C. albicans caused twice as much damage to epithelial cells as those from a CaPLB1-deficient strain (Ghannoum, 2000). Furthermore, CaPlb1 activity is concentrated at the growing tip of the invading hyphae (Mukherjee et al., 2001).
Immunomodulation
CnPlb1 is essential for the synthesis of bioactive anti-inflammatory eicosanoids by C. neoformans, both in vitro and in vivo (Noverr et al., 2002, 2003), and therefore has the potential to suppress the host immune system, promoting the survival, and dissemination of C. neoformans within the host. Arachidonic acid (AA) is the precursor of eicosanoids. C. neoformans lacks arachidonic acid (AA), but readily takes it up in vitro (Wright et al., 2007). Furthermore during phagocytosis, macrophage-derived AA can be incorporate into cryptococcal lipids by a process that is dependent on the LPTA activity of CnPlb1 (Wright et al., 2007). CnPlb1 is also essential for the synthesis of eicosanoids derived from diarachidonyl PC (Noverr et al., 2003). However, Wright et al. (2007) found evidence to suggest that free AA generated by the host, presumably by cPLA2, is the substrate for CnPlb1-dependent eicosanoid production in C. neoformans, as neither wild type nor CnΔplb1 could release AA from the sn-2 position of the more physiologically relevant phospholipid, palmitoylarachidyl PC (PAPC). In C. albicans, the PLA2 activity of CaPlb5 can release AA from PAPC (Theiss et al., 2006). This is consistent with one role for CaPlb5 in pathogenesis, being to generate bioactive lipid mediators, such as AA, from host membranes. This AA potentially provides precursors for pro- or anti-inflammatory compounds including eicosanoids (Theiss et al., 2006). This process may also coincide with the breakdown of host cell membranes leading to cell lysis.
Fungal Nutrition, Membrane Remodeling, Signaling and Cell Wall Integrity
Cryptococcus neoformans can utilize both phospholipids and fatty acids as a sole carbon source, and CnPlb1 is required for the uptake and esterification of fatty acids during stress (Wright et al., 2007). Thus during infection, CnPlb1-dependent hydrolysis of phospholipids in host membranes could provide an energy source and confer a survival advantage on C. neoformans within the low glucose environments of the macrophage phagolysosome and cryptococcomas, particularly as CnPlb1 enzymatic activity is energy-independent (Wright et al., 2007). Consistent with this hypothesis, Cn(Δplb1 has a slower rate of intracellular growth than the wild-type strain at early time points post-phagocytosis (Cox et al., 2001). Wright et al. (2007) also demonstrated CnPlb1-dependence of metabolism of the physiologically relevant lipids, DPPC and DOPC, and the cellulolytic compound lysoPC under stress conditions, and of detoxification of lysoPC by LPTA.
The CnPlb1-derived product of phosophatidylcholine, GPC, which is produced within cryptococcomas, in vivo, could be derived from either the yeast or the host. However we have reported that GPC is present in supernatants of cryptococci cultured in vitro (Bubb et al., 1999). As the secretion medium lacked phospholipid and CnPlb1 is the only enzyme with PLB activity secreted under these conditions, the GPC must have been derived from cryptococcal lipid by the hydrolytic action of CnPlb1. Localization of CnPlb1 in the outer leaflet of membranes with lipid raft-like properties, provides circumstantial evidence for a role within membranes (Siafakas et al., 2006). Interestingly however, PLB activity is, in fact, suppressed in the raft environment (Siafakas et al., 2006) suggesting that sequestration of CnPlb1 in lipid rafts may be a mechanism to prevent fungal membrane damage and/or regulate lipid remodeling within the outer membrane leaflet. Presumably any membrane homeostatic effect of CnPlb1 would occur only after release from the GPI anchor. Although lipid rafts generally facilitate signal activation, there is precedent for suppression of enzyme activity within the raft environment, in this case for a metalloproteinase (Kim and Chung, 2008).
It is also plausible that CnPlb1 is involved in fungal cell signaling following hydrolysis of the GPI anchor, which simultaneously releases CnPlb1 from raft membranes and generates the signaling molecule, diacylglycerol (DAG). In support of this, GPI anchored proteins are thought to be involved in signal transduction in mammalian cells (Stulnig et al., 1997). CnPlb1 could also regulate lysophosphatidic acid (LPA) levels which, in mammalian cells, are associated with regulation of PKC (Holmstrom et al., 2010). CnPlb1 also contributes to cell wall integrity (Chayakulkeeree et al., 2008), either by the physical nature of its GPI-anchored status in the cell wall, by maintaining the integrity of the membrane to which the cell wall is fixed or via a role in cell wall-related signal transduction involving PKC. In fungi, PKC is an enzyme within the PKC/Mpk1 cell wall integrity pathway, but is also a central regulator of signal transduction pathways that impacts on host cell physiology (Gerik et al., 2008).
Molecular Modeling and Site-Directed Mutagenesis Reveal a Single Active Site in CNPLB1
Previous studies demonstrated that several agents, including a CnPlb1 peptide-directed antibody, differentially inhibit either the LPL/LPTA, or the PLB activities of secreted CnPlb1 (Chen et al., 2000; Ganendren et al., 2004), suggesting that CnPlb1 has more than one active site. However, site-directed mutagenesis and molecular modeling in CnPlb1 (Jones et al., 2007) based on the X-ray crystal structure of the catalytic domain of mammalian cytosolic phospholipase A2 (cPLA2) (Dessen, 2000), revealed that CnPlb1 has only a single active site consisting of the same catalytic residues (S146, D392, and R108) as those involved in catalysis of nearly all fungal Plbs (Pickard et al., 1996) and human cPLA2 (S228, D549, and R200) (Jones et al., 2007). Comparison of the CnPlb1 molecular model and the cPLA2 crystal structure revealed the presence of a hydrophobic binding pocket, which accommodated the catalytic residues in identical positions, with S/D forming a catalytic dyad (Jones et al., 2007). In both structures, R108/R200 is sufficiently distanced from the S/D catalytic dyad to stabilize binding of the phospholipid head group and to allow cleavage of the acyl chain of the substrate, by the catalytic dyad (Jones et al., 2007). In silico substrate docking studies revealed that each phospholipid acyl chain is accommodated by an upper and a lower hydrophobic binding tract and that fatty acyl cleavage at the sn-2 ester bond occurs within the upper binding tract. Sequence hypervariability was identified in the CnPlb1 and cPLA2 upper binding tracts, providing a potential explanation for why cPLA2 has a preference for AA in the sn-2 position and removes only this fatty acid (Dessen, 2000), whereas CnPlb1 prefers either palmitoyl or oleoyl fatty acid in both the sn-1 and sn-2 positions and can remove both via the PLB activity (Chen et al., 2000) but cannot remove AA from the sn-2 position (Wright et al., 2007).
Sequence hypervariability was also identified in the binding tracts of the CnPlb1 active site providing a potential explanation for why various inhibitors differentially affect the three CnPlb1 enzyme activities. This could occur if the different inhibitors exhibited a preference for interaction with one binding tract over another. Alternatively, the initial point of contact of the substrate with the enzyme may not be the active site and may differ for phospholipids and lysophospholipids. Molecular modeling also revealed the absence of a “lid” within the CnPlb1 cap domain which, in cPLA2, occludes the active site and is involved in interfacial activation. However, since CnPlb1 also displays interfacial activation in the absence of a lid, an alternative explanation is that this region is involved in substrate specificity, consistent with hypervariability in this region among cPLA2 isoforms and between cPLA2 and CnPlb1. Hence the lid region may function to recognize the appropriate lipid substrate in a membrane.
Fungal PLBS as Antifungal Drug Targets
In light of the paucity of antifungal drug classes, the increasing incidence of relapsing invasive fungal infections and primary drug resistance, and unfavorable toxicity/bioavailability profiles of currently marketed antifungals, investigation of alternative compounds as stand-alone or synergistic therapies, is urgently needed. Fungal Plb and/or its biosynthetic pathway are novel targets for drug development and compounds with structural similarities to LysoPC, which is a preferred substrate of CnPlb1, have already been assessed for their antifungal effect. For many, there was a correlation between antifungal and anti-CnPlb1 activity but not all demonstrated selectivity for CnPlb1 compared with mammalian phospholipase, and some were toxic to mammalian cells.
Early studies using four commercially available compounds [alexidine dihydrochloride (compound A), dioctadecyldimethylammonium bromide (compound O), 1,12 bis-(tributylphosphonium)-dodecane dibromide (compound P), and decamethonium dibromide (compound D)], showed both selectivity for CnPlb1 and antifungal activity. The best CnPlb1 inhibitors (compounds A and P) displayed the most potent antifungal activity. Compound A was highly selective for secreted and cell-associated PLB activities and showed no inhibition of mammalian PLA2. Compound O, which was specific for secretory and cytosolic LPL and LPTA activities and membrane-associated PLB activity, was not antifungal (Ganendren et al., 2004). Non-specific detergent-like action of the compounds was ruled out by the fact that they exhibited a selective inhibitory effect on the different enzyme activities.
These initial observations prompted us to synthesize and test three classes of compound with structural similarities to lysoPC, as potential antifungal agents. Alkyl-bis(phosphonium) analogs of these were active against yeasts and filamentous fungi including A. fumigatus, A. terreus, and Scedosporium apiospermum, with MICs similar to those of Amphotericin B (Obando et al., 2007). Although promising, the selectivity of these prototype compounds for CnPlb1 was unsatisfactory In contrast, the most promising compound of a novel structural class of bis-pyridinium compounds, was selectively active against CnPlb1, non-toxic to human erythrocytes and nucleated mammalian cells and exhibited broad-spectrum antifungal activity (Obando et al., 2009). This compound is currently being tested for toxicity in animals.
Our search for compounds with substrate similarities to phospholipids yielded an additional candidate drug, miltefosine (hexadecylphosphocholine), which has been marketed as an oral formulation for the treatment of leishmaniasis. Miltefosine is a metabolically stable lysophospholipid analog which we reported to have broad-spectrum fungicidal activity in vitro against yeasts and molds, including resistant species such as Candida krusei, C. glabrata, S. prolificans, Fusarium species, and Zygomycetes (Widmer et al., 2006). MICs are well below serum levels achieved by oral dosing with miltefosine and there is very preliminary evidence of an effect in a mouse model of cryptococcosis and as combination salvage therapy in a child with severe, non-responsive, S. prolificans infection (Kesson et al., 2009). Because of its undesirable gastrointestinal side-effects, the antifungal mode(s) of action of miltefosine are under investigation as a prelude to the development of more potent, less toxic analogs. To date, we have established that miltefosine does inhibit CnPlb1, but only at high concentrations, suggesting that this is not its major mode of action (Widmer et al., 2006). Analogs of miltefosine have been synthesized and are under investigation.
In conclusion, although the mature CnPlb1 enzyme is a potential target for antifungal drug development, inhibitor compounds investigated so far appear to exert their antifungal effect through additional pathways. An alternative approach under active investigation is to seek to inhibit cellular synthesis and/or secretion of CnPlb1 (Siafakas et al., 2007).
Role of Fungal Phospholipase C Enzymes in Fungal Fitness and Pathogenicity
How Fungal PLCS Differ from the Mammalian Homologs
The C. neoformans genome contains two genes, PLC1 and PLC2, encoding putative phospholipase C enzymes (Chayakulkeeree et al., 2008). CnPLC1 encodes a protein that is predicted to be 609 amino acids (CnPlc1), with X and Y catalytic domains that are characteristic of eukaryotic Plcs, including the homologs from C. albicans (CaP1c1) (Kunze et al., 2005) and S. cerevisiae (ScPlc1) (Flick and Thorner, 1993; Yoko-o et al., 1993). Like CaP1c1 and ScPlc1, CnPlc1 contains a C2 domain which is predicted to bind Ca2+, and is 26% similar at the amino acid level to both proteins (Chayakulkeeree et al., 2008). In contrast to ScPlc1 and the mammalian Plc-Δ homolog which is most similar (32%) to the fungal homologs, CnPlc1 and CaPlc1 lack the EF hand regulatory domain suggesting differences in their mechanism of action. Furthermore, all of the fungal Plcs, lack the pleckstrin homology (PH) membrane binding domain which is present in mammalian Plc-Δ. Other potentially significant differences between the fungal and mammalian enzymes include the presence of an unusually long N-terminal extension in the fungal enzymes (Flick and Thorner, 1993; Bennett et al., 1998), and observation that fungal enzymes are predicted to be basic while the rat and bovine enzymes are acidic (Flick and Thorner, 1993). These differences, combined with the fact that deletion of fungal PLC1 genes leads to severely compromised growth phenotypes (Flick and Thorner, 1993; Payne and Fitzgerald-Hayes, 1993; Yoko-o et al., 1993; Chayakulkeeree et al., 2008), or lethality in the case of C. albicans (Kunze et al., 2005), highlights the potential of Plc enzymes as antifungal drug targets. In contrast to C. neoformans and C. albicans, ScPLC1 is the sole Plc in S. cerevisiae.
CnPLC2 encodes a 430 amino acid protein which contains an X catalytic domain but no C2 domain. It is therefore more similar to prokaryotic PI-PLCs, many of which are proven pathogenicity determinants (Poussin and Goldfine, 2005; Poussin et al., 2009). Although not essential for pathogenicity in C. neoformans, CnPlc2 may provide some functional redundancy as deletion of CnPLC1 is not lethal (Chayakulkeeree et al., 2008). In contrast, deletion of CaPLC1 is lethal in C. albicans, despite the presence of two prokaryote-like Plc-encoding genes, CaPLC2 and CaPLC3, which therefore cannot functionally compensate for loss of CaPLC1 (Kunze et al., 2005). CaPlc2 and CaPlc3 both share homology to PlcA (a PI-PLC), which is a known virulence factor of the human pathogen, Listeria monocytogenes (Wei et al., 2005). However in contrast to L. monocytogenes, CaPlc2 and CaPlc3 are dispensable for virulence and yeast–macrophage interactions. Additionally, they are not required for yeast–hyphal switching, despite augmented mRNA expression under hypha-inducing conditions (Knechtle et al., 2005). The function of CnPlc2, CaPlc2, and CaPlc3 remains to be elucidated.
PLC – Role in Fungal Pathogenicity
Plcs hydrolyse the membrane-proximal phosphoester bond of the phosphoryl head group of PI phospholipids and/or their phosphorylated and glycosylated derivatives [PI(4)P/PI(4,5)P/PIP2 and (G)PI, respectively]. The eukaryote-type enzymes prefer PIP2 as a substrate, over PI, while the prokaryote types prefer PI. Plcs from bacteria and some protozoan parasites are more efficient at cleaving GPI anchors and are therefore sometimes referred to as (G)PI-PLCs (Heinz et al., 1998). The release of CnPlb1 from membranes in C. neoformans, which allows its translocation to the cell wall and/or the extracellular environment, may be instigated by (G)PI-PLC activity. Surprisingly though, deletion of the more prokaryote-like CnPLC2 in C. neoformans, did not alter CnPlb1 secretion and was dispensable for phenotypic expression of pathogenic traits and virulence in an animal model (Chayakulkeeree et al., 2008), consistent with any potential CnPlc2 functionality being fully compensated for by CnPlc1. In contrast, deletion of the more eukaryote-like CnPLC1 (CnΔplc1), abrogated Plb1 secretion in broth cultures of C. neoformans (Chayakulkeeree et al., 2008). As CnPlc1 is not predicted to be secreted, and would therefore not have access to extracellular GPI anchors, the CnPlb1 secretion block in CnΔplc1 should not be due to compromised GPI anchor cleavage in CnΔplc1. Attempts to make a CnPLC1/PLC2 double deletion mutant were unsuccessful suggesting that loss of these two genes is lethal and that CnPlc2 is functional even though it cannot fully compensate for the loss of CnPlc1 (Chayakulkeeree et al., 2008).
CnPLC1 is essential for a number of other key survival and pathogenic determinants, including the ability of C. neoformans to grow at physiological temperature; which predictably leads to avirulence of CnΔplc1 in a mouse infection model (Chayakulkeeree et al., 2008). Even in a Caenorhabditis elegans killing assay performed at the permissible growth temperature of 28°C, CnΔplc1 was less virulent than WT, demonstrating an effect not solely explained by the 37°C growth defect of this mutant (Chayakulkeeree et al., 2008). Furthermore, CnΔplc1 could not make the stress-protecting pigment, melanin, due to suppression of transcription of the laccase-encoding gene, LAC1, and displayed a cell morphology defect, compromised cell wall integrity and hypersusceptibility to clinically available antifungal drugs (Chayakulkeeree et al., 2008). Recently, we also demonstrated that capsules cannot be induced in CnΔplc1 (Unpublished observation). Thus in contrast to C. albicans, C. neoformans can compensate for loss of PLC1 at temperatures of ≤30°C. This is most likely achieved through functional adaptation by CnPlc2 which may account for the residual Plc activity in CnΔplc1 cell lysates (Chayakulkeeree et al., 2008).
PLC – Role in Other Fungi
PLC1 orthologs in other fungi appear to have a similar function to CnPLC1. However, unlike in C. neoformans and S. cerevisae, CaPLC1 is an essential gene, as attempts to disrupt both CaPLC1 alleles failed and disruption of the second allele was only possible in mutants with CaPLC1 gene duplication (Kunze et al., 2005). Furthermore, growth of C. albicans was blocked in the presence of the Plc-specific inhibitor, ET-18 (Kunze et al., 2005). A conditional CaPLC1 mutant had phenotypes similar to the PLC1 deletion mutant in S. cerevisiae (Flick and Thorner, 1993; Payne and Fitzgerald-Hayes, 1993) and C. neoformans (Chayakulkeeree et al., 2008), including sensitivity to osmotic stress, elevated temperature and growth on carbon sources other than glucose (Kunze et al., 2005). Additionally, this conditional mutant had defects in hyphal formation (Kunze et al., 2005). Other defects apparent in the ScPLC1 deletion mutant are in morphology/budding and chromosome segregation (Payne and Fitzgerald-Hayes, 1993).
Phospholipases – A Role in Non-Fungal Pathogens
Phospholipases are virulence determinants of bacteria and protozoa, as well as fungi, and some examples are given below. Rickettsia rickettsii, the causative agent of life-threatening Rocky Mountain spotted fever, is an obligate intracellular bacterium. Phospholipase A2 is involved in rickettsial entry into host cells, escape from macrophage phagosomes and host cell lysis (Silverman et al., 1992). Phospholipase A also facilitates dissemination from the host cell by two protozoan species, Toxoplasma gondii and Entamoeba histolytica (Ghannoum, 2000). Recently, a protein with phospholipase A activity was identified from R. typhi (Rahman et al., 2010).
PI-PLC from L. monocytogenes promotes escape of the bacterium from macrophage phagolysosmes via a mechanism that is dependent on host Pkcβ, which may be activated by PI-PLC-generated DAG (Poussin and Goldfine, 2005; Poussin et al., 2009). The broad-range phospholipase C, PC-PLC, is also involved in vacuolar lysis by L. monocytogenes, allowing bacterial growth within the host cell cytosol, and spread from cell to cell via actin-based motility. Recently, it was shown that slowing the kinetics of vacuolar maturation in L. moncyotogenes creates an environment conducive for PC-PLC-assisted vacuolar escape (Burrack et al., 2009). In another bacterium, Clostridium perfringens, Plc causes gas gangrene (Kennedy et al., 2009).
CNPLC1 – A Potential Branch Point Enzyme for Integrating Virulence-Related Signal Transduction
There are at least six signaling pathways in C. neoformans that regulate virulence (reviewed in Kozubowski et al., 2009). The CnPlc1-dependent virulence phenotypes (melanin, capsule, high temperature growth, and cell wall integrity) are regulated by three of these: the cAMP/protein kinase A(Pka) pathway, Ca2+/Calcineurin pathway and the protein kinase C1 (Pkc1)/mitogen activated protein kinase 1 (Mpk1) pathway. High temperature growth and cell wall integrity are regulated through the Ca2+/Calcineurin and Pkc1/Mpk1 pathway. There is also evidence of cross-talk between the Ca2+/calcineurin and the Pkc1/Mpk1 pathways to regulate cell wall integrity in C. neoformans. Firstly, the mRNA of CnFKS1, which is a gene target of the Pkc1/Mpk1 pathway, is upregulated by loss of calcineurin function (Kraus et al., 2003). Secondly, inhibition of calcineurin by FK506 induces CnFKS1 mRNA expression and activates CnMpk1 (Kraus et al., 2003). Finally, melanin, capsule, and mating are regulated through the cAMP/Pka pathway (Alspaugh et al., 1997; Alspaugh et al., 2002). Whether production and/or secretion of the invasin, CnPlb1, is also regulated by one of these signaling pathways, is yet to be determined.
Hydrolysis of PIP2 and, to a lesser extent of PI, was reduced in CnΔplc1 (Chayakulkeeree et al., 2008), providing evidence that CnPlc1 is a functional enzyme involved in signaling. Preferential hydrolysis of PIP2 over PI generates two intracellular secondary messengers, 1,2-diacylglycerol (DAG) (the sole hydrolysis product of PI) and inositol 1,4,5-trisphosphate (IP3) which can activate the Pkc1/Mpk1 and Ca2+/Calcineurin pathways, respectively. It has been shown that DAG generated by the sphingolipid biosynthetic pathway activates CnPkc1 to regulate cell wall integrity and melanin production (Heung et al., 2005). Thus, CnPlc1-generated DAG could play a similar role. In support of this, CnΔplc1 has a cell wall integrity defect and Mpk1 is not activated in CnΔplc1 following cell wall perturbation (Chayakulkeeree et al., 2008). Thus, CnPlc1 can potentially integrate signaling through both the Pkc1/Mpk1 and Ca2+/Calcineurin pathways to regulate high temperature growth and cell wall integrity, and potentially, melanin production (see Figure 3). CnPlc1 signal transduction may be influenced by PI-3 kinase, which is a potential competitor for PIP2 substrate and regulator of melanization and autophagy virulence traits in C. neoformans (Hu et al., 2008), and the phospholipid-binding protein, Cts1, which functions coordinately with calcineurin in C. neoformans to regulate growth and cell separation (Fox et al., 2003).
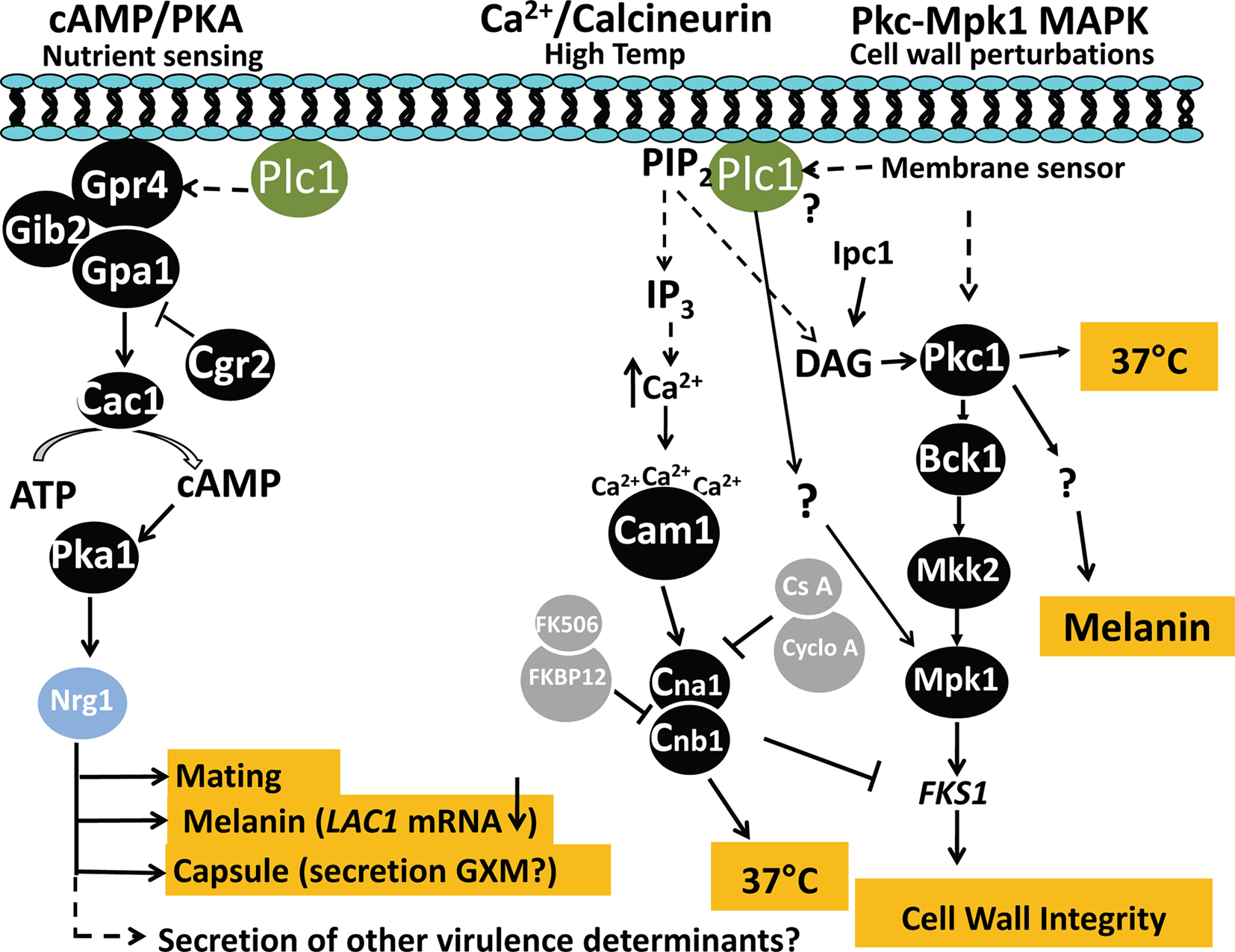
Figure 3. Proposed role of CnPlc1 in pathogenicity-related signaling in C. neoformans. 1. Cyclic AMP (cAMP)/Protein kinase A (Pka) signaling pathway. A change in extracellular nutrient levels trigger activation of the integral membrane-associated G-protein coupled receptor, GPR4, causing its associated Gα subunit (Gpa1) to activate adenylyl cyclase (Cac1), resulting in production of cAMP. cAMP binds to regulatory subunits (Pkr1) of the Pka complex (not shown) to release an active form of the catalytic subunit (Pka1), which phosphorylates the transcription factor, Nrg1. A GTPase-activating protein (Crg2) negatively regulates Gpa1. Gib2, Gβ subunit. Based on studies in S. cerevisiae and our own published and preliminary data in C. neoformans demonstrating the requirement for CnPlc1 in melanin production (via LAC1 transcription), capsule production and secretion of virulence determinants (Chayakulkeeree et al., 2008), CnPlc1 may be linked to cAMP signaling possibly by regulating the activation of Gpa1 as indicated by the dashed line. Lac1, Laccase 1; GXM, glucuronic acid-xylose-mannan. 2. Ca2+/calcineurin signaling pathway. This pathway primarily regulates high temperature growth in C. neoformans, but not in S. cerevisiae. High temperatures may trigger CnPlc1 to hydrolyse PIP2, producing IP3, which potentially raises intracellular Ca2+ (see dashed line) by regulating Ca2+ pumps/channels in the plasma membrane and ER. Cytosolic Ca2+ is sensed by calmodulin (Cam1), which activates calmodulin kinases (CaMKs) (not shown) and the calcineurin complex (Cna1 and Cnb1) to support growth at 37°C. Cyclosporine A (CsA) and FK506, bind cyclophilin A and FKBP12, respectively, to inhibit calcineurin; PIP2, phosphatidylinositol 4,5-bisphosphate. Calcineurin is also essential for hyphal elongation during mating and monokaryotic fruiting (not shown). 3. Pkc1/Mpk1 MAPK signaling pathway. This pathway conveys cell wall perturbation signals detected by unknown membrane sensors, to the nucleus. CnPlc1 is essential for activation of Mpk1 in response to cell wall perturbation in a process that may, or may not be, mediated by activation of Pkc1 via the CnPlc1 hydrolysis product, DAG (see dashed line). Activated Pkc1 phosphorylates MAPKKK (Bck1) and subsequent phosphorylations of MAPKK (Mkk2) by Bck1 and MAPK (Mpk1) by Mkk2 to upregulate transcription of FKS1, which encodes β-1,3-glucan synthase. Expression of FKS1 is also negatively regulated by calcineurin. DAG produced by the sphingolipid synthetic pathway enzyme inositolphosphoceramide 1 synthase (Ipc1), and possibly by CnPlc1, also activates CnPkc1 to regulate melanin production independently of Mpk1 activation (Heung et al., 2005). Pkc1 is also required for host temperature growth (Gerik et al., 2008).
In S. cerevisiae, there is evidence that ScPlc1 signals through cAMP-regulated pathways. Firstly, ScPlc1 modulates interaction of the putative nutrient sensing G-protein coupled receptor, Gpr1p, with the Gα protein, Gpa2p, effecting cAMP-regulated filamentation (Ansari et al., 1999). Secondly, cAMP synthesis is decreased in ScΔplc1 (Demczuk et al., 2008). In C. neoformans, a protein complex of Gpr4p, Gpa1p, and Crg2p, functions to control cAMP signaling via Pka (Kozubowski et al., 2009) to regulate melanin and capsule production, and mating. Support for CnPlc1 being linked to this signaling pathway is that CnΔplc1 has a melanin defect, as a result of reduced LAC1 mRNA transcription, and a capsule defect (Chayakulkeeree et al., 2008) and unpublished data, respectively (see Figure 3). A comparative transcriptomic study of a C. neoformans Pka catalytic subunit deletion mutant, and the WT, identified a role for the cAMP/Pka pathway in secretion and, in particular, secretion of capsular polysaccharide (Hu et al., 2007). However, secretion of protein-based virulence factors was not assessed. Further support for CnPlc1 being linked to the cAMP/Pka pathway in C. neoformans, is that CnPlb1 secretion is defective in CnΔplc1 (Chayakulkeeree et al., 2008). A comparison of secreted CnPlb1 in Pka mutants and WT is currently under investigation to determine whether this pathway does, in fact, play a role in controlling the secretion of pathogenicity determinants other than capsular polysaccharide.
In summary, CnPlc1 may represent an important branchpoint signaling enzyme and its relative contribution to individual signaling pathways will be determined through epistasis studies. These studies will confirm whether or not three individual signaling pathways constitute a regulatory network in which CnPlc1 plays a central role. The elucidation of CnPlc1 function within the model fungal pathogen, C. neoformans, will therefore fill some essential gaps within our understanding of signaling networks of pathogenic fungi, which can differ substantially from those of S. cerevisiae, and invites investigation of similar pathways in other pathogenic fungi.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author would like to thank Mr Stephen Schibeci and Professor Tania Sorrell for proof-reading this review.
References
Aimanianda, V., Clavaud, C., Simenel, C., Fontaine, T., Delepierre, M., and Latge, J. P. (2009). Cell wall beta-(1,6)-glucan of Saccharomyces cerevisiae: structural characterization and in situ synthesis. J. Biol. Chem. 284, 13401–13412.
Alspaugh, J. A., Perfect, J. R., and Heitman, J. (1997). Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11, 3206–3217.
Alspaugh, J. A., Pukkila-Worley, R., Harashima, T., Cavallo, L. M., Funnell, D., Cox, G. M., Perfect, J. R., Kronstad, J. W., and Heitman, J. (2002). Adenylyl cyclase functions downstream of the Galpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryotic Cell 1, 75–84.
Alvarez, M., Burn, T., Luo, Y., Pirofski, L. A., and Casadevall, A. (2009). The outcome of Cryptococcus neoformans intracellular pathogenesis in human monocytes. BMC Microbiol. 9, 51.
Alvarez, M., and Casadevall, A. (2006). Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr. Biol. 16, 2161–2165.
Alvarez, M., and Casadevall, A. (2007). Cell-to-cell spread and massive vacuole formation after Cryptococcus neoformans infection of murine macrophages. BMC Immunol. 8, 16.
Ansari, K., Martin, S., Farkasovsky, M., Ehbrecht, I. M., and Kuntzel, H. (1999). Phospholipase C binds to the receptor-like GPR1 protein and controls pseudohyphal differentiation in Saccharomyces cerevisiae. J. Biol. Chem. 274, 30052–30058.
Bagnat, M., Keranen, S., Shevchenko, A., and Simons, K. (2000). Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. U.S.A. 97, 3254–3259.
Bennett, D. E., McCreary, C. E., and Coleman, D. C. (1998). Genetic characterization of a phospholipase C gene from Candida albicans: presence of homologous sequences in Candida species other than Candida albicans. Microbiology 144 (Pt 1), 55–72.
Bubb, W. A., Wright, L. C., Cagney, M., Santangelo, R. T., Sorrell, T. C., and Kuchel, P. W. (1999). Heteronuclear NMR studies of metabolites produced by Cryptococcus neoformans in culture media: identification of possible virulence factors. Magn. Reson. Med. 42, 442–453.
Burrack, L. S., Harper, J. W., and Higgins, D. E. (2009). Perturbation of vacuolar maturation promotes listeriolysin O-independent vacuolar escape during Listeria monocytogenes infection of human cells. Cell. Microbiol. 11, 1382–1398.
Casadevall, A., Nosanchuk, J. D., Williamson, P., and Rodrigues, M. L. (2009). Vesicular transport across the fungal cell wall. Trends Microbiol. 17, 158–162.
Charlier, C., Nielsen, K., Daou, S., Brigitte, M., Chretien, F., and Dromer, F. (2009). Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect. Immun. 77, 120–127.
Chau, T. T., Mai, N. H., Phu, N. H., Nghia, H. D., Chuong, L. V., Sinh, D. X., Duong, V. A., Diep, P. T., Campbell, J. I., Baker, S., Hien, T. T., Lalloo, D. G., Farrar, J. J., and Day, J. N. (2010). A prospective descriptive study of cryptococcal meningitis in HIV uninfected patients in Vietnam – high prevalence of Cryptococcus neoformans var grubii in the absence of underlying disease. BMC Infect. Dis. 10, 199.
Chayakulkeeree, M., Sorrell, T. C., Siafakas, A. R., Wilson, C. F., Pantarat, N., Gerik, K. J., Boadle, R., and Djordjevic, J. T. (2008). Role and mechanism of phosphatidylinositol-specific phospholipase C in survival and virulence of Cryptococcus neoformans. Mol. Microbiol. 69, 809–826.
Chen, S. C., Muller, M., Zhou, J. Z., Wright, L. C., and Sorrell, T. C. (1997a). Phospholipase activity in Cryptococcus neoformans: a new virulence factor? J. Infect. Dis. 175, 414–420.
Chen, S. C., Wright, L. C., Santangelo, R. T., Muller, M., Moran, V. R., Kuchel, P. W., and Sorrell, T. C. (1997b). Identification of extracellular phospholipase B, lysophospholipase, and acyltransferase produced by Cryptococcus neoformans. Infect. Immun. 65, 405–411.
Chen, S. C., Wright, L. C., Golding, J. C., and Sorrell, T. C. (2000). Purification and characterization of secretory phospholipase B, lysophospholipase and lysophospholipase/transacylase from a virulent strain of the pathogenic fungus Cryptococcus neoformans. Biochem. J. 347, 431–439.
Coe, J. G., Wilson, C. F., Sorrell, T. C., Latouche, N. G., and Wright, L. C. (2003). Cloning of CnLYSO1, a novel extracellular lysophospholipase of the pathogenic fungus Cryptococcus neoformans. Gene 316, 67–78.
Cox, G. M., McDade, H. C., Chen, S. C., Tucker, S. C., Gottfredsson, M., Wright, L. C., Sorrell, T. C., Leidich, S. D., Casadevall, A., Ghannoum, M. A., and Perfect, J. R. (2001). Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 39, 166–175.
de Almeida, M. L., and Heise, N. (1993). Proteins anchored via glycosylphosphatidylinositol and solubilizing phospholipases in Trypanosoma cruzi. Biol. Res. 26, 285–312.
Demczuk, A., Guha, N., Nguyen, P. H., Desai, P., Chang, J., Guzinska, K., Rollins, J., Ghosh, C. C., Goodwin, L., and Vancura, A. (2008). Saccharomyces cerevisiae phospholipase C regulates transcription of Msn2p-dependent stress-responsive genes. Eukaryotic Cell 7, 967–979.
Dessen, A. (2000). Structure and mechanism of human cytosolic phospholipase A(2). Biochim. Biophys. Acta 1488, 40–47.
Djordjevic, J. T., Del Poeta, M., Sorrell, T. C., Turner, K. M., and Wright, L. C. (2005). Secretion of cryptococcal phospholipase B1 (PLB1) is regulated by a glycosylphosphatidylinositol (GPI) anchor. Biochem. J. 389, 803–812.
Doering, T. L. (2009). How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Annu. Rev. Microbiol. 63, 223–247.
Farkas, V., Takeo, K., Macekova, D., Ohkusu, M., Yoshida, S., and Sipiczki, M. (2009). Secondary cell wall formation in Cryptococcus neoformans as a rescue mechanism against acid-induced autolysis. FEMS Yeast Res. 9, 311–320.
Feldmesser, M., Kress, Y., Novikoff, P., and Casadevall, A. (2000). Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 68, 4225–4237.
Flick, J. S., and Thorner, J. (1993). Genetic and biochemical characterization of a phosphatidylinositol-specific phospholipase C in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 5861–5876.
Fox, D. S., Cox, G. M., and Heitman, J. (2003). Phospholipid-binding protein Cts1 controls septation and functions coordinately with calcineurin in Cryptococcus neoformans. Eukaryotic Cell 2, 1025–1035.
Fujii, T., Shimoi, H., and Iimura, Y. (1999). Structure of the glucan-binding sugar chain of Tip1p, a cell wall protein of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1427, 133–144.
Gagnon-Arsenault, I., Parise, L., Tremblay, J., and Bourbonnais, Y. (2008). Activation mechanism, functional role and shedding of glycosylphosphatidylinositol-anchored Yps1p at the Saccharomyces cerevisiae cell surface. Mol. Microbiol. 69, 982–993.
Ganendren, R., Carter, E., Sorrell, T., Widmer, F., and Wright, L. (2006). Phospholipase B activity enhances adhesion of Cryptococcus neoformans to a human lung epithelial cell line. Microbes Infect. 8, 1006–1015.
Ganendren, R., Widmer, F., Singhal, V., Wilson, C., Sorrell, T., and Wright, L. (2004). In vitro antifungal activities of inhibitors of phospholipases from the fungal pathogen Cryptococcus neoformans. Antimicrob. Agents Chemother. 48, 1561–1569.
Gerik, K. J., Bhimireddy, S. R., Ryerse, J. S., Specht, C. A., and Lodge, J. K. (2008). PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryotic Cell 7, 1685–1698.
Ghannoum, M. A. (2000). Potential role of phospholipases in virulence and fungal pathogenesis. Clin. Microbiol. Rev. 13, 122–143, table of contents.
Gilbert, N. M., Donlin, M. J., Gerik, K. J., Specht, C. A., Djordjevic, J. T., Wilson, C. F., Sorrell, T. C., and Lodge, J. K. (2010). KRE genes are required for beta-1,6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans. Mol. Microbiol. 76, 517–534.
Heinz, D. W., Essen, L. O., and Williams, R. L. (1998). Structural and mechanistic comparison of prokaryotic and eukaryotic phosphoinositide-specific phospholipases C. J. Mol. Biol. 275, 635–650.
Heung, L. J., Kaiser, A. E., Luberto, C., and Del Poeta, M. (2005). The role and mechanism of diacylglycerol-protein kinase C1 signaling in melanogenesis by Cryptococcus neoformans. J. Biol. Chem. 280, 28547–28555.
Himmelreich, U., Allen, C., Dowd, S., Malik, R., Shehan, B. P., Mountford, C., and Sorrell, T. C. (2003). Identification of metabolites of importance in the pathogenesis of pulmonary cryptococcoma using nuclear magnetic resonance spectroscopy. Microbes Infect. 5, 285–290.
Holmstrom, T. E., Mattsson, C. L., Wang, Y., Iakovleva, I., Petrovic, N., and Nedergaard, J. (2010). Non-transactivational, dual pathways for LPA-induced Erk1/2 activation in primary cultures of brown pre-adipocytes. Exp. Cell Res. 316, 2664–2675.
Hoover, C. I., Jantapour, M. J., Newport, G., Agabian, N., and Fisher, S. J. (1998). Cloning and regulated expression of the Candida albicans phospholipase B (PLB1) gene. FEMS Microbiol. Lett. 167, 163–169.
Hu, G., Hacham, M., Waterman, S. R., Panepinto, J., Shin, S., Liu, X., Gibbons, J., Valyi-Nagy, T., Obara, K., Jaffe, H. A., Ohsumi, Y., and Williamson, P. R. (2008). PI3K signaling of autophagy is required for starvation tolerance and virulenceof Cryptococcus neoformans. J. Clin. Invest. 118, 1186–1197.
Hu, G., Steen, B. R., Lian, T., Sham, A. P., Tam, N., Tangen, K. L., and Kronstad, J. W. (2007). Transcriptional regulation by protein kinase A in Cryptococcus neoformans. PLoS Pathog. 3:e42.
Ibrahim, A. S., Mirbod, F., Filler, S. G., Banno, Y., Cole, G. T., Kitajima, Y., Edwards, J. E., Nozawa, Y., and Ghannoum, M. A. (1995). Evidence implicating phospholipase as a virulence factor of Candida albicans. Infect. Immun. 63, 1993–1998.
Insenser, M., Nombela, C., Molero, G., and Gil, C. (2006). Proteomic analysis of detergent-resistant membranes from Candida albicans. Proteomics 6 (Suppl. 1), S74–S81.
Jones, P. M., Turner, K. M., Djordjevic, J. T., Sorrell, T. C., Wright, L. C., and George, A. M. (2007). Role of conserved active site residues in catalysis by phospholipase B1 from Cryptococcus neoformans. Biochemistry 46, 10024–10032.
Kapteyn, J. C., Montijn, R. C., Dijkgraaf, G. J., and Klis, F. M. (1994). Identification of beta-1,6-glucosylated cell wall proteins in yeast and hyphal forms of Candida albicans. Eur. J. Cell Biol. 65, 402–407.
Kennedy, C. L., Lyras, D., Cheung, J. K., Hiscox, T. J., Emmins, J. J., and Rood, J. I. (2009). Cross-complementation of Clostridium perfringens PLC and Clostridium septicum alpha-toxin mutants reveals PLC is sufficient to mediate gas gangrene. Microbes Infect. 11, 413–418.
Kesson, A. M., Bellemore, M. C., O'Mara, T. J., Ellis, D. H., and Sorrell, T. C. (2009). Scedosporium prolificans osteomyelitis in an immunocompetent child treated with a novel agent, hexadecylphospocholine (miltefosine), in combination with terbinafine and voriconazole: a case report. Clin. Infect. Dis. 48, 1257–1261.
Kim, H. N., and Chung, H. S. (2008). Caveolin-1 inhibits membrane-type 1 matrix metalloproteinase activity. BMB Rep. 41, 858–862.
Kitagaki, H., Wu, H., Shimoi, H., and Ito, K. (2002). Two homologous genes, DCW1 (YKL046c) and DFG5, are essential for cell growth and encode glycosylphosphatidylinositol (GPI)-anchored membrane proteins required for cell wall biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 46, 1011–1022.
Knechtle, P., Goyard, S., Brachat, S., Ibrahim-Granet, O., and d'Enfert, C. (2005). Phosphatidylinositol-dependent phospholipases C Plc2 and Plc3 of Candida albicans are dispensable for morphogenesis and host-pathogen interaction. Res. Microbiol. 156, 822–829.
Kollar, R., Reinhold, B. B., Petrakova, E., Yeh, H. J., Ashwell, G., Drgonova, J., Kapteyn, J. C., Klis, F. M., and Cabib, E. (1997). Architecture of the yeast cell wall. Beta(1→6)-glucan interconnects mannoprotein, beta(1→3-glucan, and chitin. J. Biol. Chem. 272, 17762–17775.
Kozubowski, L., Lee, S. C., and Heitman, J. (2009). Signalling pathways in the pathogenesis of Cryptococcus. Cell. Microbiol. 11, 370–380.
Kraus, P. R., Fox, D. S., Cox, G. M., and Heitman, J. (2003). The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 48, 1377–1387.
Kristiansen, S., and Richter, E. A. (2002). GLUT4-containing vesicles are released from membranes by phospholipase D cleavage of a GPI anchor. Am. J. Physiol. Endocrinol. Metab. 283, E374–E382.
Kunze, D., Melzer, I., Bennett, D., Sanglard, D., MacCallum, D., Norskau, J., Coleman, D. C., Odds, F. C., Schafer, W., and Hube, B. (2005). Functional analysis of the phospholipase C gene CaPLC1 and two unusual phospholipase C genes, CaPLC2 and CaPLC3, of Candida albicans. Microbiology 151, 3381–3394.
Lee, K. S., Patton, J. L., Fido, M., Hines, L. K., Kohlwein, S. D., Paltauf, F., Henry, S. A., and Levin, D. E. (1994). The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J. Biol. Chem. 269, 19725–19730.
Leidich, S. D., Ibrahim, A. S., Fu, Y., Koul, A., Jessup, C., Vitullo, J., Fonzi, W., Mirbod, F., Nakashima, S., Nozawa, Y., and Ghannoum, M. A. (1998). Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. J. Biol. Chem. 273, 26078–26086.
Levitz, S. M., Nong, S. H., Seetoo, K. F., Harrison, T. S., Speizer, R. A., and Simons, E. R. (1999). Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect. Immun. 67, 885–890.
Lu, C. F., Kurjan, J., and Lipke, P. N. (1994). A pathway for cell wall anchorage of Saccharomyces cerevisiae alpha-agglutinin. Mol. Cell. Biol. 14, 4825–4833.
Ma, H., Croudace, J. E., Lammas, D. A., and May, R. C. (2006). Expulsion of live pathogenic yeast by macrophages. Curr. Biol. 16, 2156–2160.
Ma, H., Croudace, J. E., Lammas, D. A., and May, R. C. (2007). Direct cell-to-cell spread of a pathogenic yeast. BMC Immunol. 8, 15.
Macekova, D., Farkas, V., Kishida, E., and Takeo, K. (2006). Ecto-glycanases and metabolic stability of the capsule in Cryptococcus neoformans. J. Basic Microbiol. 46, 470–479.
Mao, Y., Zhang, Z., Gast, C., and Wong, B. (2008). C-terminal signals regulate targeting of glycosylphosphatidylinositol-anchored proteins to the cell wall or plasma membrane in Candida albicans. Eukaryotic Cell 7, 1906–1915.
Martinez, L. R., Garcia-Rivera, J., and Casadevall, A. (2001). Cryptococcus neoformans var. neoformans (serotype D) strains are more susceptible to heat than C. neoformans var. grubii (serotype A) strains. J. Clin. Microbiol. 39, 3365–3367.
Merkel, O., Fido, M., Mayr, J. A., Pruger, H., Raab, F., Zandonella, G., Kohlwein, S. D., and Paltauf, F. (1999). Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J. Biol. Chem. 274, 28121–28127.
Mirbod, F., Banno, Y., Ghannoum, M. A., Ibrahim, A. S., Nakashima, S., Kitajima, Y., Cole, G. T., and Nozawa, Y. (1995). Purification and characterization of lysophospholipase-transacylase (h-LPTA) from a highly virulent strain of Candida albicans. Biochim. Biophys. Acta 1257, 181–188.
Mukherjee, P. K., Seshan, K. R., Leidich, S. D., Chandra, J., Cole, G. T., and Ghannoum, M. A. (2001). Reintroduction of the PLB1 gene into Candida albicans restores virulence in vivo. Microbiology 147, 2585–2597.
Muller, G., Gross, E., Wied, S., and Bandlow, W. (1996). Glucose-induced sequential processing of a glycosyl-phosphatidylinositol-anchored ectoprotein in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 442–456.
Muniz, M., and Riezman, H. (2000). Intracellular transport of GPI-anchored proteins. EMBO J. 19, 10–15.
Noverr, M. C., Cox, G. M., Perfect, J. R., and Huffnagle, G. B. (2003). Role of PLB1 in pulmonary inflammation and cryptococcal eicosanoid production. Infect. Immun. 71, 1538–1547.
Noverr, M. C., Toews, G. B., and Huffnagle, G. B. (2002). Production of prostaglandins and leukotrienes by pathogenic fungi. Infect. Immun. 70, 400–402.
Obando, D., Pantarat, N., Handke, R., Koda, Y., Widmer, F., Djordjevic, J. T., Ellis, D. H., Sorrell, T. C., and Jolliffe, K. A. (2009). Synthesis, antifungal, haemolytic and cytotoxic activities of a series of bis(alkylpyridinium)alkanes. Bioorg. Med. Chem. 17, 6329–6339.
Obando, D., Widmer, F., Wright, L. C., Sorrell, T. C., and Jolliffe, K. A. (2007). Synthesis, antifungal and antimicrobial activity of alkylphospholipids. Bioorg. Med. Chem. 15, 5158–5165.
Oishi, H., Tsuda, S., Watanabe, Y., and Tamai, Y. (1996). Purification and some properties of phospholipase B from Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 60, 1087–1092.
Panepinto, J., Komperda, K., Frases, S., Park, Y. D., Djordjevic, J. T., Casadevall, A., and Williamson, P. R. (2009). Sec6-dependent sorting of fungal extracellular exosomes and laccase of Cryptococcus neoformans. Mol. Microbiol. 71, 1165–1176.
Park, B. J., Wannemuehler, K. A., Marston, B. J., Govender, N., Pappas, P. G., and Chiller, T. M. (2009). Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23, 525–530.
Parkin, E. T., Watt, N. T., Turner, A. J., and Hooper, N. M. (2004). Dual mechanisms for shedding of the cellular prion protein. J. Biol. Chem. 279, 11170–11178.
Payne, W. E., and Fitzgerald-Hayes, M. (1993). A mutation in PLC1, a candidate phosphoinositide-specific phospholipase C gene from Saccharomyces cerevisiae, causes aberrant mitotic chromosome segregation. Mol. Cell. Biol. 13, 4351–4364.
Pickard, R. T., Chiou, X. G., Strifler, B. A., DeFelippis, M. R., Hyslop, P. A., Tebbe, A. L., Yee, Y. K., Reynolds, L. J., Dennis, E. A., Kramer, R. M., and Sharp, J. D. (1996). Identification of essential residues for the catalytic function of 85-kDa cytosolic phospholipase A2. Probing the role of histidine, aspartic acid, cysteine, and arginine. J. Biol. Chem. 271, 19225–19231.
Poussin, M. A., and Goldfine, H. (2005). Involvement of Listeria monocytogenes phosphatidylinositol-specific phospholipase C and host protein kinase C in permeabilization of the macrophage phagosome. Infect. Immun. 73, 4410–4413.
Poussin, M. A., Leitges, M., and Goldfine, H. (2009). The ability of Listeria monocytogenes PI-PLC to facilitate escape from the macrophage phagosome is dependent on host PKCbeta. Microb. Pathog. 46, 1–5.
Price, M. F., Wilkinson, I. D., and Gentry, L. O. (1982). Plate method for detection of phospholipase activity in Candida albicans. Sabouraudia 20, 7–14.
Rahman, M. S., Ammerman, N. C., Sears, K. T., Ceraul, S. M., and Azad, A. F. (2010). Functional characterization of a phospholipase A(2) homolog from Rickettsia typhi. J. Bacteriol. 192, 3294–3303.
Rodrigues, M. L., Nakayasu, E. S., Oliveira, D. L., Nimrichter, L., Nosanchuk, J. D., Almeida, I. C., and Casadevall, A. (2008). Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryotic Cell 7, 58–67.
Rodrigues, M. L., Nimrichter, L., Oliveira, D. L., Frases, S., Miranda, K., Zaragoza, O., Alvarez, M., Nakouzi, A., Feldmesser, M., and Casadevall, A. (2007). Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryotic Cell 6, 48–59.
Saito, K., Sugatani, J., and Okumura, T. (1991). Phospholipase B from Penicillium notatum. Meth. Enzymol. 197, 446–456.
Santangelo, R., Zoellner, H., Sorrell, T., Wilson, C., Donald, C., Djordjevic, J., Shounan, Y., and Wright, L. (2004). Role of extracellular phospholipases and mononuclear phagocytes in dissemination of cryptococcosis in a murine model. Infect. Immun. 72, 2229–2239.
Santangelo, R. T., Chen, S. C., Sorrell, T. C., and Wright, L. C. (2005). Detection of antibodies to phospholipase B in patients infected with Cryptococcus neoformans by enzyme-linked immunosorbent assay (ELISA). Med. Mycol. 43, 335–341.
Santangelo, R. T., Nouri-Sorkhabi, M. H., Sorrell, T. C., Cagney, M., Chen, S. C., Kuchel, P. W., and Wright, L. C. (1999). Biochemical and functional characterisation of secreted phospholipase activities from Cryptococcus neoformans in their naturally occurring state. J. Med. Microbiol. 48, 731–740.
Shen, D. K., Noodeh, A. D., Kazemi, A., Grillot, R., Robson, G., and Brugere, J. F. (2004). Characterisation and expression of phospholipases B from the opportunistic fungus Aspergillus fumigatus. FEMS Microbiol. Lett. 239, 87–93.
Siafakas, A. R., Sorrell, T. C., Wright, L. C., Wilson, C., Larsen, M., Boadle, R., Williamson, P. R., and Djordjevic, J. T. (2007). Cell wall-linked cryptococcal phospholipase B1 is a source of secreted enzyme and a determinant of cell wall integrity. J. Biol. Chem. 282, 37508–37514.
Siafakas, A. R., Wright, L. C., Sorrell, T. C., and Djordjevic, J. T. (2006). Lipid rafts in Cryptococcus neoformans concentrate the virulence determinants phospholipase B1 and Cu/Zn superoxide dismutase. Eukaryotic Cell 5, 488–498.
Silverman, D. J., Santucci, L. A., Meyers, N., and Sekeyova, Z. (1992). Penetration of host cells by Rickettsia rickettsii appears to be mediated by a phospholipase of rickettsial origin. Infect. Immun. 60, 2733–2740.
Sorrell, T. C., Wright, L. C., Malik, R., and Himmelreich, U. (2006). Application of proton nuclear magnetic resonance spectroscopy to the study of Cryptococcus and cryptococcosis. FEMS Yeast Res. 6, 558–566.
Stulnig, T. M., Berger, M., Sigmund, T., Stockinger, H., Horejsi, V., and Waldhausl, W. (1997). Signal transduction via glycosyl phosphatidylinositol-anchored proteins in T cells is inhibited by lowering cellular cholesterol. J. Biol. Chem. 272, 19242–19247.
Sugiyama, Y., Nakashima, S., Mirbod, F., Kanoh, H., Kitajima, Y., Ghannoum, M. A., and Nozawa, Y. (1999). Molecular cloning of a second phospholipase B gene, caPLB2 from Candida albicans. Med. Mycol. 37, 61–67.
Sutterlin, C., Doering, T. L., Schimmoller, F., Schroder, S., and Riezman, H. (1997). Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J. Cell. Sci. 110 (Pt 21), 2703–2714.
Takahashi, M., Banno, Y., Shikano, Y., Mori, S., and Nozawa, Y. (1991). Purification and characterization of lysophospholipase-transacylase of pathogenic fungus Candida albicans. Biochim. Biophys. Acta 1082, 161–169.
Theiss, S., Ishdorj, G., Brenot, A., Kretschmar, M., Lan, C. Y., Nichterlein, T., Hacker, J., Nigam, S., Agabian, N., and Kohler, G. A. (2006). Inactivation of the phospholipase B gene PLB5 in wild-type Candida albicans reduces cell-associated phospholipase A2 activity and attenuates virulence. Int. J. Med. Microbiol. 296, 405–420.
Trofa, D., Gacser, A., and Nosanchuk, J. D. (2008). Candida parapsilosis, an emerging fungal pathogen. Clin. Microbiol. Rev. 21, 606–625.
Turner, K. M., Wright, L. C., Sorrell, T. C., and Djordjevic, J. T. (2006). N-linked glycosylation sites affect secretion of cryptococcal phospholipase B1, irrespective of glycosylphosphatidylinositol anchoring. Biochim. Biophys. Acta 1760, 1569–1579.
Wei, Z., Zenewicz, L. A., and Goldfine, H. (2005). Listeria monocytogenes phosphatidylinositol-specific phospholipase C has evolved for virulence by greatly reduced activity on GPI anchors. Proc. Natl. Acad. Sci. U.S.A. 102, 12927–12931.
Widmer, F., Wright, L. C., Obando, D., Handke, R., Ganendren, R., Ellis, D. H., and Sorrell, T. C. (2006). Hexadecylphosphocholine (miltefosine) has broad-spectrum fungicidal activity and is efficacious in a mouse model of cryptococcosis. Antimicrob. Agents Chemother. 50, 414–421.
Witt, W., Schweingruber, M. E., and Mertsching, A. (1984). Phospholipase B from the plasma membrane of Saccharomyces cerevisiae. Separation of two forms with different carbohydrate content. Biochim. Biophys. Acta 795, 108–116.
Wright, L. C., Payne, J., Santangelo, R. T., Simpanya, M. F., Chen, S. C., Widmer, F., and Sorrell, T. C. (2004). Cryptococcal phospholipases: a novel lysophospholipase discovered in the pathogenic fungus Cryptococcus gattii. Biochem. J. 384, 377–384.
Wright, L. C., Santangelo, R. M., Ganendren, R., Payne, J., Djordjevic, J. T., and Sorrell, T. C. (2007). Cryptococcal lipid metabolism: phospholipase B1 is implicated in transcellular metabolism of macrophage-derived lipids. Eukaryotic Cell 6, 37–47.
Yoko-o, T., Matsui, Y., Yagisawa, H., Nojima, H., Uno, I., and Toh-e, A. (1993). The putative phosphoinositide-specific phospholipase C gene, PLC1, of the yeast Saccharomyces cerevisiae is important for cell growth. Proc. Natl. Acad. Sci. U.S.A. 90, 1804–1808.
Keywords: Cryptococcus neoformans, phospholipase B, phosphatidylinositol-specific phospholipase C, PI-PLC/Plc, pathogenicity, drug development, secretion, signaling
Citation: Djordjevic JT (2010) Role of phospholipases in fungal fitness, pathogenicity, and drug development – lessons from Cryptococcus neoformans. Front. Microbio. 1:125. doi:10.3389/fmicb.2010.00125
Received: 23 August 2010; Accepted: 25 October 2010;
Published online: 11 November 2010.
Edited by:
Ana Traven, Monash University, AustraliaReviewed by:
Joshua D. Nosanchuk, Albert Einstein College of Medicine, USA;Andrew Alspaugh, Duke University School of Medicine, USA;
Bernhard Hube, International Leibniz Research School, Germany
Copyright: © 2010 Djordjevic. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Julianne Teresa Djordjevic, Fungal Pathogenesis Laboratory, Centre for Infectious Diseases and Microbiology, Sydney, Medical School-Western, Westmead Millennium Institute, University of Sydney at Westmead, Hospital, Westmead, NSW 2145, Australia. e-mail: julie_djordjevic@wmi.usyd.edu.au
