- 1 Department of Biological Sciences, Bowling Green State University, Bowling Green, OH, USA
- 2 Marine Sciences, Bermuda Institute for Ocean Sciences, St. George’s, Bermuda
In phosphorus-limited marine environments, picocyanobacteria (Synechococcus and Prochlorococcus spp.) can hydrolyze naturally occurring phosphonates as a P source. Utilization of 2-aminoethylphosphonate (2-AEP) is dependent on expression of the phn genes, encoding functions required for uptake, and C–P bond cleavage. Prior work has indicated that expression of picocyanobacterial phnD, encoding the phosphonate binding protein of the phosphonate ABC transporter, is a proxy for the assimilation of phosphonates in natural assemblages of Synechococcus spp. and Prochlorococcus spp (Ilikchyan et al., 2009). In this study, we expand this work to assess seasonal phnD expression in the Sargasso Sea. By RT-PCR, our data confirm that phnD expression is constitutive for the Prochlorococcus spp. detected, but in Synechococcus spp. phnD transcription follows patterns of phosphorus availability in the mixed layer. Specifically, our data suggest that phnD is repressed in the spring when P is bioavailable following deep winter mixing. In the fall, phnD expression follows a depth-dependent pattern reflecting depleted P at the surface following summertime drawdown, and elevated P at depth.
Introduction
Phosphonates, previously thought to be refractory sources of P, are utilized by microbial communities in aquatic environments deprived of inorganic phosphates (Clark et al., 1998; Benitez-Nelson, 2004; Dyhrman et al., 2006; Karl et al., 2008). Phosphonates represent upto 25% of the marine DOP pool (Clark et al., 1998; Kolowith et al., 2001), and sources of naturally occurring phosphonates in the oligotrophic open sea include the nitrogen-fixing cyanobacterium, Trichodesmium erythraeum (Dyhrman et al., 2009). Recent work also indicates that at least three distinct mechanisms are widespread in nature that function to cleave the phosphonate C–P bond (Dyhrman et al., 2006; Gilbert et al., 2009; Kulakova et al., 2009; Martinez et al., 2010; Thomas et al., 2010). For example, Trichodesmium spp. possess a C–P lyase pathway that is distributed widely among bacterial taxa, and exhibits broad substrate specificity (Dyhrman et al., 2006). Marine and freshwater picocyanobacteria typically contain a phosphonatase pathway that exclusively hydrolyzes 2-aminoethyl phosphonate (2-AEP; Quinn et al., 2007; Ilikchyan et al., 2009). Last, a novel third mechanism for marine 2-AEP utilization was recently unmasked by a functional assay (Martinez et al., 2010), indicating that additional phosphonate utilization pathways may await future discovery. Nonetheless, these pathways appear to share common ABC transporters for phosphonates, encoded by the genes phnDCE. Earlier, we have demonstrated that picocyanobacteria of the genera Synechococcus and Prochlorococcus possess and express phnD, encoding the phosphonate binding protein, in a variety of aquatic environments, and that phnD expression can be used as a proxy for phosphonate utilization. Moreover, preliminary studies suggested that expression of Synechococcus spp. phnD reflected the bioavailability of dissolved inorganic phosphorus (DIP) in the Sargasso Sea, whereas constitutive expression was observed for Prochlorococcus spp. phnD (Ilikchyan et al., 2009).
Here, we extended our survey of seasonal environmental expression of phnD along a N–S transect through the Sargasso Sea. The seasonal physical, chemical, and biological characteristics of the Sargasso Sea are well studied (Schroeder and Stommel, 1969). In winter, deep (>80 m) mixing transports cold, nutrient rich waters to the surface, whereas throughout summer and fall, surface waters in the Sargasso Sea are stratified with a shallow (∼50 m), nutrient poor mixed layer. The physical and chemical changes of the surface waters are reflected in the abundance and productivity of phytoplankton (e.g., Menzel and Ryther, 1960, 1961). Picocyanobacteria constitute the majority of photoautotrophic organisms in the Sargasso Sea. Synechococcus spp. reach maximum abundance in April–May shortly after winter mixing, and Prochlorococcus spp. populations peak in summer and fall (DuRand et al., 2001; Casey et al., 2007). The Sargasso Sea is characterized as a phosphorus depleted region with DIP concentrations typically less than 10 nM (e.g., Cotner et al., 1997). Under these conditions, utilization of phosphonates from the DOP pool can be an important survival strategy for picocyanobacteria. Indeed, the expression of phosphonate transporter genes was observed by endemic picocyanobacteria in the Sargasso Sea in October (Ilikchyan et al., 2009). In this study, we have assessed seasonal picocyanobacterial phnD expression at multiple depths and locations in May and October.
Materials and Methods
Environmental Samples
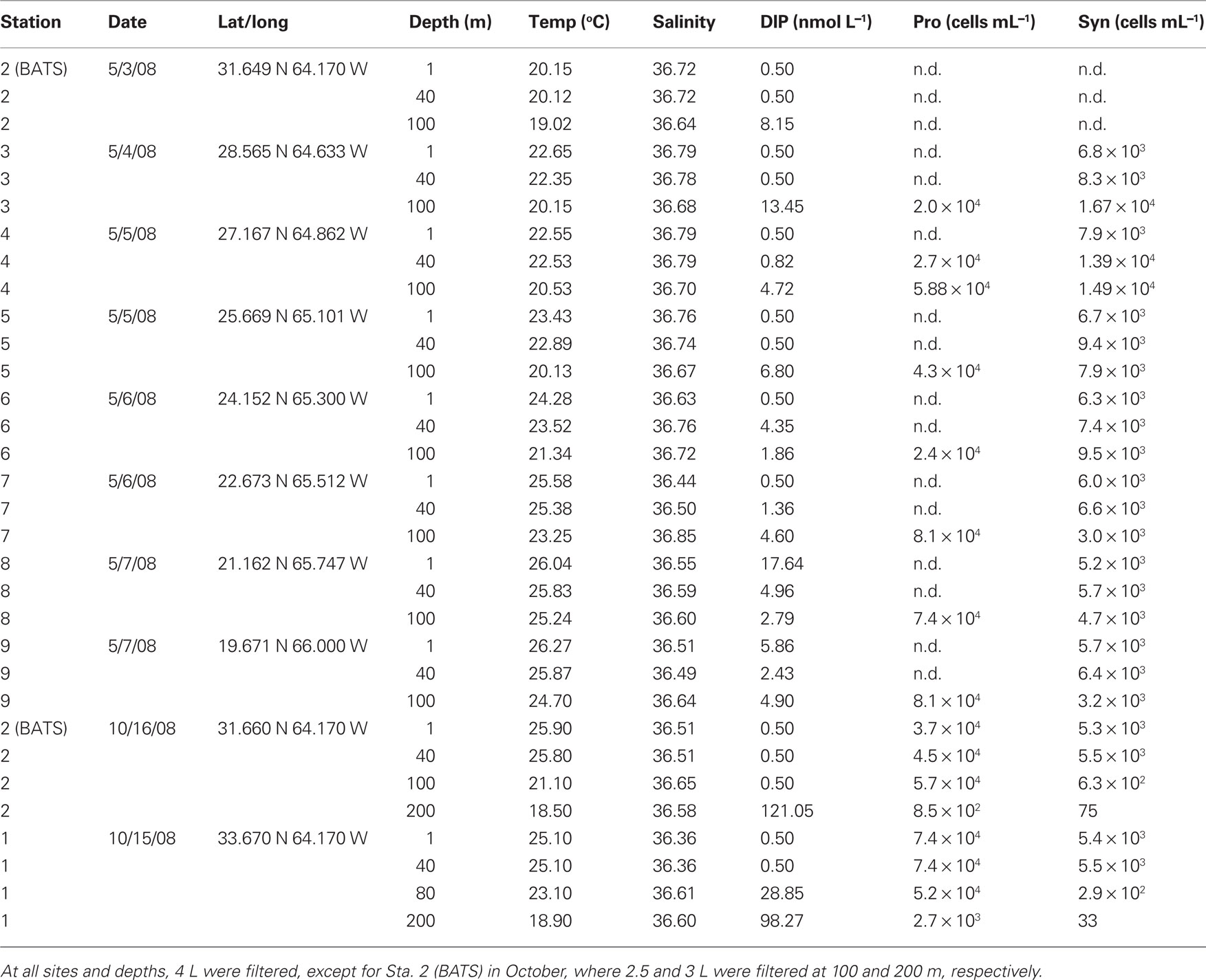
Table 1 and Figure 1 provides details on sampling and station locations for cruises aboard the R/V Bank of Bermuda Atlantic Explorer during May and October 2008. The samples for RNA and DNA extraction were processed as described in Ilikchyan et al. (2009). Briefly, suspended particles from environmental samples of 3–4 L was collected onto 0.22 μm Sterivex cartridge filters by using a peristaltic pump, and cartridges were frozen immediately in liquid nitrogen. Flow rates were maintained at 125 mL min−1. The cartridges were transferred to storage at −80°C prior to RNA and DNA extraction. Chlorophyll a was measured fluorometrically after extraction of filtered seston with 90% acetone (Welschmeyer, 1994).
Nucleic Acid Extraction PCR and RT-PCR Conditions
Extraction of environmental DNA and RNA from a Sterivex filter was performed as described previously (Ilikchyan et al., 2009). All PCR and RT-PCR amplifications with Synechococcus and Prochlorococcus spp. phnD primers were done as described earlier (Ilikchyan et al., 2009). Specifically, the primers employed were: phnD_syn119F: 5′-TCGGNGCMATYCCSGATCAGAACCCSG-3′; phnD_syn734R1: 5′-TTGGGCTGSGCGASCCAGTGGTARTC-3′; phnD_syn731R2: 5′-GGNCGNGCCACCCAGTGGTARTC-3′. Both reverse primers were used in a single reaction. For amplification of the phnD sequence from Prochlorococcus spp. the following primers were used: phnD_pro307F: 5′-GTNATWGCTCAAAGAGATATWGAT-3′; phnD_pro551R: 5′-GTTGCATCATGACTNCCRCTATANCC-3′. rnpB is a single copy gene and encodes for RNase P RNA. The rnpB primers were designed to amplify a 118 bp region specific for picocyanobacteria, RNPB-F: CCGTGAGGAGAGTGCCACAG; RNPB-R: CAGCACCTCTCGATGCTGCTGG. Specificity of the primers was confirmed by the absence of amplification from DNA extracted from Synechococcus sp. PCC7942, Synechocystis sp. PCC6803 and Microcystis sp. M300. All available marine and freshwater picocyanobacterial DNA yielded an amplicon of the correct size. The PCR conditions for amplification with the rnpB primers were as follows: 95°C for 5 min, 30 cycles of 95°C for 30 s, 65°C for 30 s, and 72°C for 30 s, followed by 72°C for 10 min.
For phnD amplification, each PCR (25 μL) contained 1× PCR buffer (Promega), 0.2 mM of each deoxynucleotide (Promega), 0.5 μM of each primer, and 1.0 unit of GoTaq DNA polymerase (Promega), and ca. 10 ng template DNA. For Synechococcus spp. phnD amplification, the temperature profile was 95°C for 5 min, 40 cycles of 95°C for 1 min, an initial annealing temperature of 65°C for 1 min decreasing by 0.5°C each cycle until 55°C was reached, 72°C for 1 min, followed by extension at 72°C for 20 min. For Prochlorococcus spp. phnD primers, the annealing temperature was 57°C decreasing by 0.5°C each cycle until 50°C was reached, and the remaining PCR profile was the same as for Synechococcus spp. phnD. RT-PCR was performed using a OneStep-RT-PCR kit (QIAGEN) according to the manufacturer’s instructions with a PCR profile as stated above. Amounts of RNA used per reaction were ca. 10 ng. Additionally, each reaction was performed without RT to ensure the absence of genomic DNA in the RNA samples.
2-AEP Amendment Experiment
Surface water from station BATS was drawn during the October 2008 BVAL 42 cruise (Table 1). Water was prescreened through a 5 μm polycarbonate membrane, and 2 L volumes were distributed into triplicate acid-cleaned bottles: three control, three phosphate amended (500 nM K2HPO4) and three amended with 500 nM 2-AEP. Bottles were incubated for 48 h in a flow-through deck incubator screened to yield a midday irradiance of 70 μmol quanta m−2 s−1. After 48 h, bottles 1 and 2 for each treatment were filtered to measure extractive chlorophyll (2 × 500 mL each bottle) and particulate organic phosphorus (POP). Bottle 3 from each treatment was filtered onto a Sterivex cartridge for RT-PCR profiling with phnD primer sets.
Alkaline Phosphatase and Particulate Phosphorus Assays
Alkaline phosphatase activity (APA) was measured fluorometrically with methylumbelliferyl phosphate (Sigma) as described elsewhere (Hoppe, 1993; Sterner et al., 2004). The assay was done in triplicate and on the same day when the water samples were collected. Particulate phosphorus was measured as described previously (Hood et al., 2006). Briefly, samples collected on GF/F filters were digested by adding 5% potassium persulfate and autoclaving for 30 min. Liberated soluble reactive phosphorus was analyzed colorimetrically with the molybdate ascorbic acid method using a 10 cm path length cell.
Phylogenetic Analysis
PhnD sequences obtained in this (GenBank accession numbers GU724612–GU724682) and previous (EU362636–EU362729, FJ172179–FJ172204; Ilikchyan et al., 2009) studies were aligned together with PhnD sequences from available genomes of Synechococcus and Prochlorococcus spp. using ClustalW2 at http://www.ebi.ac.uk/Tools/clustalw2/ (Larkin et al., 2007). The amino acid sequences that were further included in phylogenetic analysis differed among each other by at least 2%. Neighbor-joining phylogenetic analysis was done in ARB (Ludwig et al., 2004).
Results
Picocyanobacterial phnD Expression in the Sargasso Sea, May and October 2008
The presence and expression of Synechococcus spp. and Prochlorococcus spp. phnD was assessed in samples collected during Sargasso Sea cruises in May and October 2008. Samples were analyzed from three depths (surface, 40, and 100 m) in May and from four depths (surface, 40 m; deep chlorophyll maximum, DCM, and 200 m) in October. Additionally, an on-deck nutrient amendment incubation experiment was conducted on board during the October cruise.
DNA extracted from samples taken at three different depths in the Sargasso Sea in May 2008 all yielded PCR amplicons for both Synechococcus spp. phnD and Prochlorococcus spp. phnD (Figure 2A). The mixed layer depth was 80 m. Expression of Prochlorococcus spp. phnD was observed in almost all 24 samples (Figure 2B). The samples yielding no Prochlorococcus RT-PCR phnD amplicon included the Bermuda Atlantic Time Series station (BATS, also designated as Station 2) at 40 m, Station 3 at 100 m, and Station 4 at 40 m. These samples also did not yield a picocyanobacterial rnpB RT-PCR amplicon (Figure 2B). Low RNA yield in samples from BATS at 40 m, Station 3 at 100 m, and Station 4 at 40 m due to errors in RNA extraction might be a reason for the failure to detect a Prochlorococcus spp. RT-PCR phnD amplicon. By contrast, no samples from any station and depth from the Sargasso Sea May 2008 cruise revealed expression of Synechococcus spp. phnD (Figure 2B), despite the fact that all corresponding DNA samples yielded a PCR amplicon with Synechococcus spp. specific phnD primers (Figure 2A).
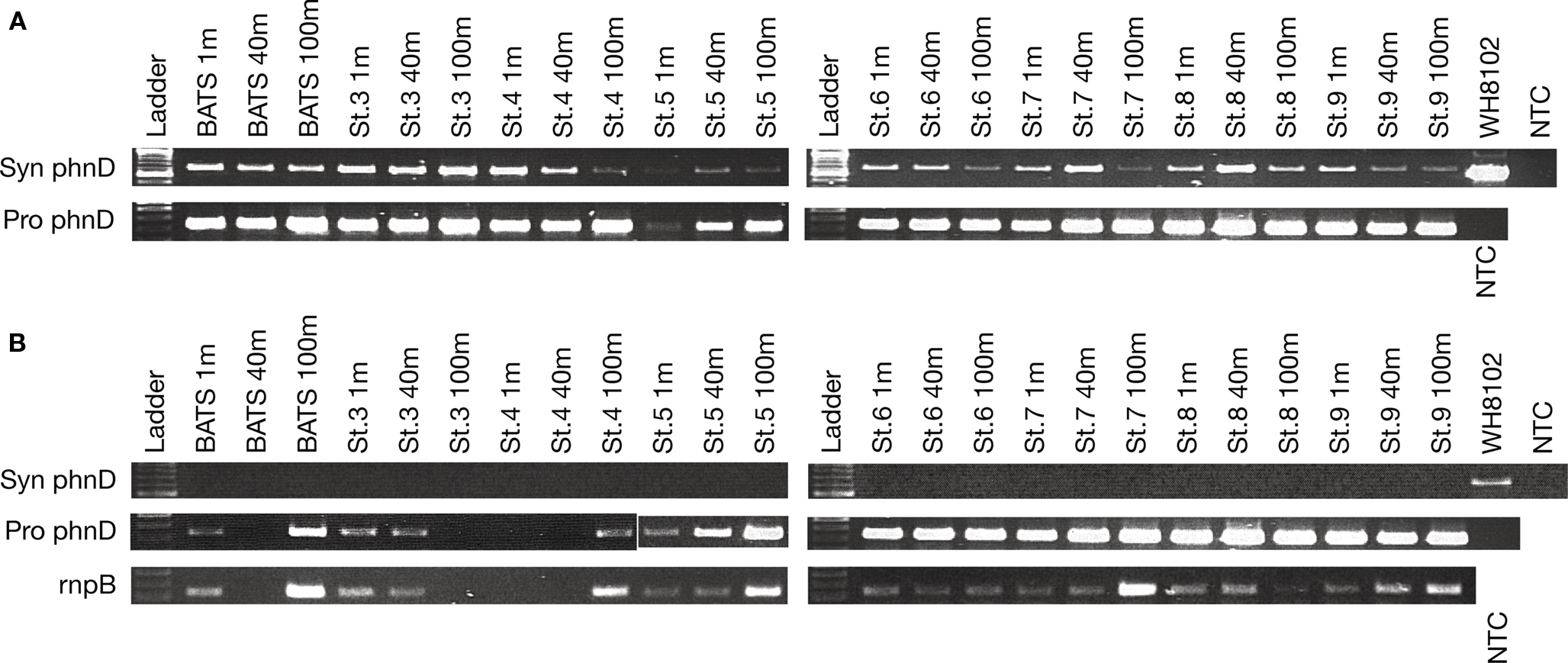
Figure 2. (A) PCR of Synechococcus and Prochlorococcus spp. phnD from the Sargasso Sea samples taken in at several stations during May 2008. The mixed layer was 80 m, and the DCM was at 100 m. (B), top rows: RT-PCR of environmental RNA extracted from the same stations during the May cruise. (B), bottom: RT-PCR of picocyanobacterial rnpB transcripts as a positive control for picocyanobacteria. NTC, no template control.
Samples from two stations, Station 1 and BATS (Station 2), were processed from the October 2008 cruise. All DNA samples yielded Prochlorococcus and Synechococcus phnD amplicons, although only faint bands were observed in the BATS sample at 200 m (Figure 3A). Flow cytometry confirmed low abundance of both taxa at this depth (<100 cells mL−1). Synechococcus spp. phnD expression was detected in the samples taken at the surface and 40 m depth, but not at the DCM (100 m), or 200 m, both below the mixed layer depth of 55 m (Figure 3B). Prochlorococcus spp. phnD was expressed in all samples except BATS 200 m, a sample found to be negative for picocyanobacterial RNA (Figure 3B).
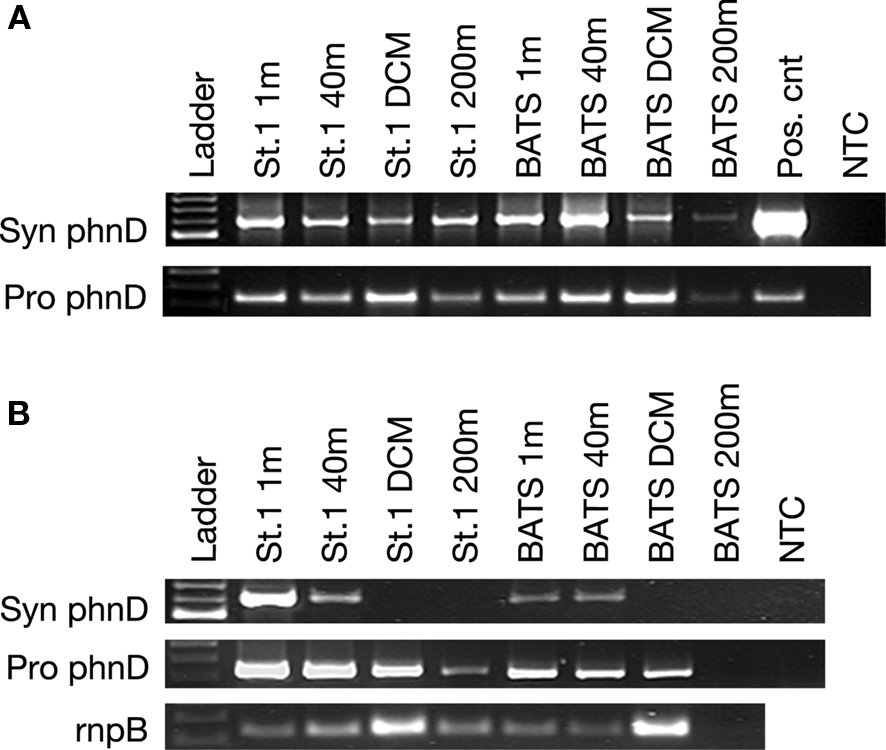
Figure 3. (A) PCR of Synechococcus and Prochlorococcus spp. phnD from the Sargasso Sea samples taken in at Station 1 and BATS (Station 2) during October 2008. The mixed layer was 50 m, and the DCM was at 100 m. (B) Top rows: RT-PCR of environmental RNA extracted from the same stations. (B) Bottom: RT-PCR of picocyanobacterial rnpB transcripts as a positive control for picocyanobacterial RNA. NTC, no template control.
Despite the fact that PCR as performed here is not a quantitative assay, the intensity of bands corresponding to phnD and rnpB reflects the abundance of picocyanobacteria at depth with highest number of Prochlorococcus spp. cells at the DCM and Synechococcus spp. cells at the surface (DuRand et al., 2001).
phnD Expression: DIP and 2-AEP
The patterns of Synechococcus spp. phnD expression were examined in light of parallel determination of inorganic phosphorus concentration during May and October 2008. In both seasons, a depth-dependent gradient of DIP was observed, with depleted P at the surface, increasing at depth yielding maximum values below the mixed layer. Near the surface, spring P was elevated compared to October measurements, with May samples yielded average DIP of 2.62 nmol L−1 in the mixed layer samples assayed by RT-PCR (n = 16 samples), ranging from 0.5 (detection limit) to 17.64 nmol L−1. By contrast, all October mixed layer values remained at or below the detection limit of 0.5 nmol L−1. Since phnD expression was only observed in the October samples, yet spring DIP was at or below detection in 9 of 16 mixed layer samples, expression patterns cannot be ascribed to DIP concentration alone. This raises the possibility that DOP (phosphonates and organic phosphates) may directly influence the accumulation of phn transcripts.
A nutrient amendment incubation experiment was performed with water collected from BATS at the surface in October 2008. The RT-PCR assay revealed that an addition of 500 nmol L−1 phosphate quenched expression of the Synechococcus spp. phnD in 48 h while not affecting expression of the Prochlorococcus spp. phnD (Figure 4). The amendments with 0.5 μM 2-AEP yielded both Synechococcus spp. and Prochlorococcus spp. phnD amplicons. The control sample failed to amplify Synechococcus spp. phnD and yielded weak bands for Prochlorococcus spp. phnD and picocyanobacterial rnpB (Figure 4). This suggests overall low picocyanobacterial cell abundance in the control at 48 h. Analysis of POP in the control, as well as in the phosphate and 2-AEP-amended bottles revealed a significant increase in both the amended samples after 48 h. (Table 2). Bottles amended with phosphate or with 2-AEP accumulated nearly threefold more POP than did unamended control bottles (one-way ANOVA; p < 0.005). There was no treatment effect discernible between the phosphate or phosphonate treatments (Tukey’s honest significance test). Addition of nutrients stimulated the phytoplankton community, as chlorophyll increased from 0.51 to 0.69 μg L−1 in phosphate amended and 0.74 μg L−1 in the 2-AEP treatments. Overall, these data suggest that phnD expression in Synechococcus spp. was regulated by phosphorus availability, and that 2-AEP was assimilated by the microbial community. Additionally, the experiment raises the possibility that the presence of 2-AEP stimulates phnD expression among the endemic Synechococcus spp. By contrast, Prochlorococcus spp. phnD expression was not influenced by either 2-AEP or phosphate availability. The activity of alkaline phosphatase was measured before and 48 h after amendments with activities ranging from 0.7 to 1.31 pmol h−1 ml−1, with no significant difference between the samples. Since eubacteria may contribute the largest component of total APA in environmental samples (Hoppe, 2003), this assay was likely not an accurate indication of picocyanobacterial P status.
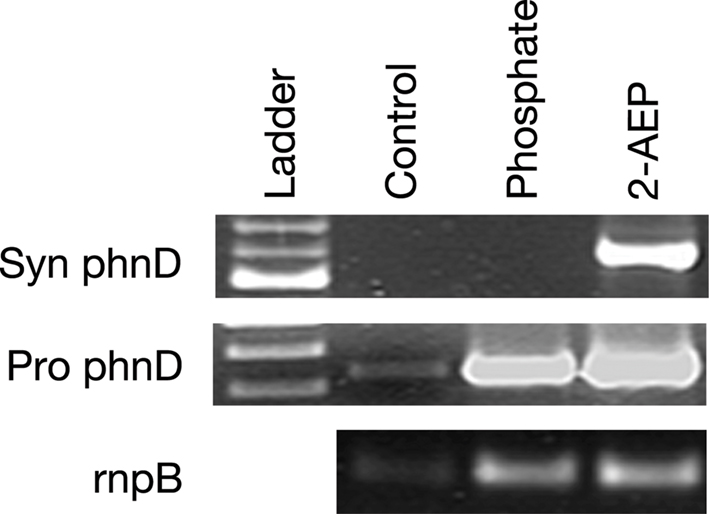
Figure 4. RT-PCR from the growout experiment RNA extracted in 48 h after the addition of nutrients. Control – no amendments, Phosphate – addition of 0.5 μM of phosphate, and 2-AEP – addition of 500 nM of 2-aminoethylphosphonate. Top row, Synechococcus phnD expression; middle row, Prochlorococcus phnD expression; bottom row, rnpB expression.

Table 2. Particulate organic phosphorus content of BATS surface water samples amended with 2-AEP and inorganic phosphate.
Phylogenetic Analysis of Picocyanobacterial PhnD from Sargasso Sea Samples
The majority of Synechococcus PhnD sequences from May and October 2008 clustered within clade III similar to our previous study (Figure 5; Ilikchyan et al., 2009). Synechococcus Clade III is relatively abundant throughout the open oceans between 40N and 40S latitudes (Zwirglmaier et al., 2008). Ahlgren and Rocap (2006) showed that Synechococcus clades II, IV, and a novel clade XV (closely related to III) dominated in the western Sargasso Sea in March. However, a comprehensive study on diversity and temporal variation (Tai and Palenik, 2009) of Synechococcus groups has not yet been performed for the Sargasso Sea. Synechococcus sequences from the Sargasso Sea May and October cruises formed an additional cluster (sequence ADE58334) closely related to the cluster III (Figure 5A). Synechococcus cluster IV also includes sequences from Monterey Bay obtained from the previous study (Ilikchyan et al., 2009).
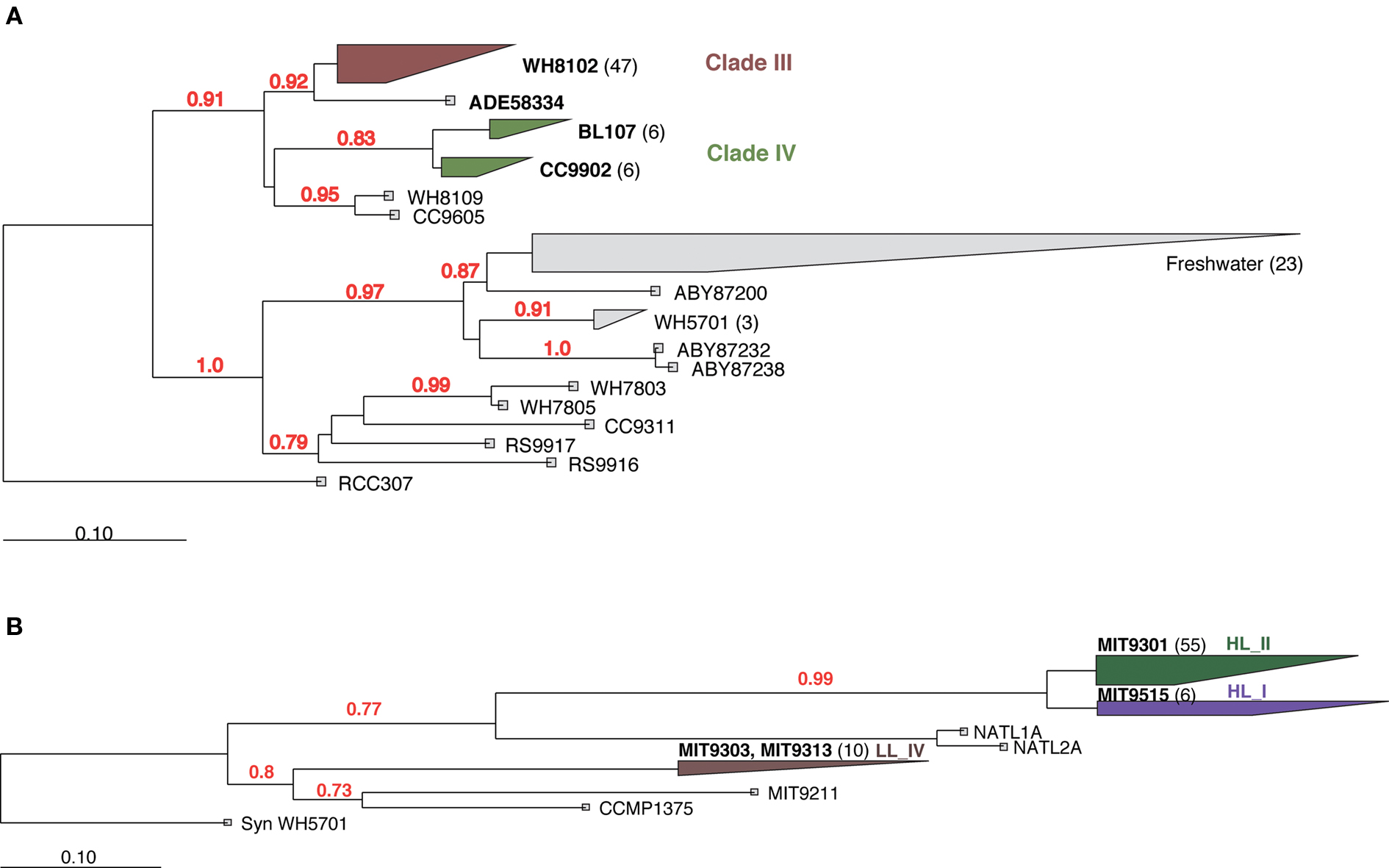
Figure 5. Neighbor joining phylogenetic analysis of Synechococcus (A) and Prochlorococcus (B) PhnD protein regions. A region of 187 aa for Synechococcus and 64 aa for Prochlorococcus was analyzed. Bootstrap values >70% are shown. Number of unique (less than 98% aa similarity) sequences obtained in this and previous studies are shown in parentheses.
Most Prochlorococcus PhnD sequences were clustered within the High Light II clade similar to what has been observed with primers for 16S rRNA and 16S–23S ITS region (Moore et al., 1998; Ahlgren et al., 2006; Zinser et al., 2006). A DNA sample taken at depth (200 m) at BATS in October 2008 yielded Prochlorococcus PhnD sequences closely related to Low Light clade IV (Figure 5B). Considering the observed diversity among PhnD sequences obtained in this and previous studies, we are confident that the specificity of the phnD primers is broad enough to capture representative members of endemic picocyanobacterial community worldwide. This will enable more detailed studies of phosphonate utilization genes in diverse oceanic regions.
Discussion
P limitation in the surface waters of the Sargasso Sea surface waters is well documented (Cotner et al., 1997; Karl, 2000; Wu et al., 2000), with a significant depletion in P occurring during late summer and early fall when the Sargasso Sea is stably stratified (DuRand et al., 2001; Ahlgren et al., 2006). In this study, we have assessed spatiotemporal expression of phnD in the photic zone of the Sargasso Sea. Prochlorococcus spp. phnD expression was observed in all samples from the Sargasso Sea drawn in May and October 2008, except BATS at 40 m, St. 3 at 100 m, and St. 4 at 40 m in May and BATS 200 m in October. These negative samples likely contained prohibitively low total picocyanobacterial RNA. The constitutive presence of the Prochlorococcus spp. phnD RNA is consistent with the data obtained earlier for the Sargasso Sea samples collected in October 2007 (Ilikchyan et al., 2009). In contrast, whereas a PCR amplicon was detected for Synechococcus spp. phnD in all samples at all depths, expression of this gene was not observed in May when the concentration of phosphate was generally higher throughout the water column due to recent winter mixing (DuRand et al., 2001). In October 2007 and 2008, Synechococcus spp. expressed phnD in the mixed water layer but not at the DCM (ca. 100 m) or at 200 m. This is consistent with lower DIP levels typically observed in the mixed layer, which spans only 50 m in depth in the fall. Overall, the lack of springtime phosphonate gene expression can be partially explained by increased inorganic phosphate availability below the mixed layer depth, but the absence of phnD expression in samples lacking detectable DIP suggests that organic phosphate or phosphonate availability may also be important in regulating phn encoded functions. Furthermore, higher competition for DIP and organic phosphates in the fall may lead to phnD expression, so both P flux and total P concentration need to be considered.
In a nutrient amendment experiment, addition of phosphate yielded no Synechococcus spp. phnD expression at 48 h, whereas the biomass increased as suggested by chlorophyll a measurements and RT-PCR with the rnpB primers. An addition of 2-AEP resulted in increased picocyanobacterial biomass as well, although Synechococcus spp. phnD expression was evident in this sample. The control showed no detectable expression for Synechococcus spp. phnD in 48 h; Prochlorococcus spp. phnD and picocyanobacterial rnpB expression was barely detectable as well suggesting an overall decrease in picocyanobacterial biomass. Synechococcus spp. expressed phnD at time zero because the water for the experiment was taken from the surface at the BATS station where in an identical sample, expression of the Synechococcus spp. phnD was detected in the mixed layer (Figure 4). Additionally, amendment with DIP repressed phnD expression. The results confirmed that expression of phnD is influenced by P availability in Synechococccus, but not in Prochlorococcus spp. The experiment also raises the possibility that 2-AEP can induce phnD within the endemic Synechococcus spp. similar to what is observed in some heterotrophic bacteria (Quinn, 2002). However, experiments with cultured Synechococcus sp. WH8102 suggest otherwise, because added 2-AEP does not yield enhanced phnD transcription. Additionally, BLAST searches (http://blast.ncbi.nlm.nih.gov) reveal that the gene for LysR type transcriptional regulator, which is adjacent to the genes for phosphonate utilization in these heterotrophic bacteria (Kulakova et al., 2001, 2003; Quinn, 2002), is lacking in picocyanobacterial genomes (data not shown). The fact that DIP repressed the level of phnD transcripts likely differs from what has been observed for the N-fixing cyanobacterium Trichodesmium IMS101, as DIP addition did not inhibit phosphonate utilization in culture. This strain is capable of utilizing multiple P sources simultaneously (Beversdorf et al., 2010).
Complicating our understanding of expression of phosphonate utilization genes is a recent study describing phn gene expression in thermophilic Synechococcus spp. (Gomez-Garcia et al., 2010). The authors compare two related strains, one of which (OS-B′) harbors both the phnCDE transporter and C–P lyase genes. The OS-A strain lacks genes for the C–P lyase, but instead harbors phosphonatase genes that are expressed in the presence of phosphonates (Gomez-Garcia et al., 2010). These data suggest that multiple strategies for phosphonate utilization exist in picocyanobacteria and that P deficiency, DIP/DOP ratios and the presence or absence of phosphonates may influence phn transcription in different picocyanobacterial ecotypes. This could explain the pattern of Synechococcus spp. phnD expression observed in this paper, in which the DIP concentration alone does not predict the community-wide phnD transcription detected in our RT-PCR assay.
Prochlorococcus spp. detected via this assay express phnD independently from P limitation. This corresponds to a reduced genome size of Prochlorococcus HL clade species targeted primarily by our primer set (Ilikchyan et al., 2009). The HL clade genomes indicate the loss of many functional and regulatory genes in comparison to the LL clade and to Synechococcus spp. (Moore et al., 2002, 2005; Scanlan and West, 2002; Dufresne et al., 2003; Palenik et al., 2003; Rocap et al., 2003). Nonetheless, the number and distribution of P assimilation genes in picocyanobacteria varies depending on the marine environment (Moore et al., 2005; Martiny et al., 2006; Scanlan et al., 2009).
Conclusion
The fact that picocyanobacteria are utilizing phosphonates in marine environments underscores the significance of phosphonates to the bioavailable DOP pool. Synechococcus spp. expressed phnD under seasonal (autumn) P-limited conditions in samples from the Sargasso Sea, and Prochlorococcus spp. phnD expression was constitutive independent of season. Constitutive expression may be a consequence of genomic reduction in Prochlorococcus spp. as some regulatory functions may have been lost through evolution (e.g., Rocap et al., 2003; Moore et al., 2005). In both taxa, the same patterns of phnD expression were observed previously during October 2007 (Ilikchyan et al., 2009). Development of clade specific molecular probes for phosphonate utilization genes in picocyanobacteria will bring insight into differences in regulation of phosphonate assimilation among the clades and improve our overall understanding of P physiology in picocyanobacteria. Overall, the pattern of Synechococcus spp. phnD expression observed in natural water samples herein and earlier (Ilikchyan et al., 2009) together with the results of a nutrient amendment experiment conducted in the field suggest applicability of the developed RT-PCR method in assessing the P status of Synechococcus spp. populations.
It is clear that much remains to be known about how cyanobacteria and bacterioplankton recruit phosphonates as a P source. Multiple pathways for phosphonate degradation and phn gene activation have been documented in diverse taxa, and many more mechanisms may yet to be described. Future work should focus in the distribution of phosphonate degradation pathways and their regulatory networks in all taxa capable of recruiting phosphonates from the DOP pool, while simultaneously monitoring DIP and the speciation of the DOP.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the captain and crew of the R/V Bank of Bermuda Atlantic Explorer for assistance in sample collection, Mark Rozmarynowycz for sample preparation during the May 2008 cruise, and Nigel D’Souza for conducting the POP assays. Special thanks are due Dr. Michael Lomas (BIOS) for offering space on the May and October 2008 cruises. This material is based upon work supported by the National Science Foundation Grant No. OCE-0727644 and Ohio Sea Grant R/ER-080 to Robert Michael L. McKay and George S. Bullerjahn.
References
Ahlgren, N. A., and Rocap, G. (2006). Culture isolation and culture-independent clone libraries reveal new marine Synechococcus ecotypes with distinctive light and N physiologies. Appl. Environ. Microbiol. 72, 7193–7204.
Ahlgren, N. A., Rocap, G., and Chisholm, S. W. (2006). Measurement of Prochlorococcus ecotypes using real-time polymerase chain reaction reveals different abundances of genotypes with similar light physiologies. Environ. Microbiol. 8, 441–454.
Benitez-Nelson, C. (2004). Phosphonates and particulate organic phosphorus cycling in an anoxic marine basin. Limnology 49, 1593–1604.
Beversdorf, L. J., White, A. E., Bjorkman, K. M., Letelier, R. M., and Karl, D. M. (2010). Phosphonate metabolism of Trichodesmium IMS101 and the production of greenhouse gases. Limnol. Oceanogr. 55, 1755–1767.
Casey, J. R., Lomas, M. W., Mandecki, J., and Walker, D. E. (2007). Prochlorococcus contributes to new production in the Sargasso Sea deep chlorophyll maximum. Geophys. Res. Lett. 34, L10604.
Clark, L. L., Ingall, E. D., and Benner, R. (1998). Marine phosphorus is selectively remineralized. Nature 393, 426.
Cotner, J. B., Ammerman, J. W., Peele, E. R., and Bentzen, E. (1997). Phosphorus-limited bacterioplankton growth in the Sargasso Sea. Aquat. Microb. Ecol. 13, 141–149.
Dufresne, A., Salanoubat, M., Partensky, F., Artiguenave, F., Axmann, I. M., Barbe, B., Duprat, S., Galperin, M. Y., Koonin, E. V., Le Gall, F., Makarova, K. S., Ostrowski, M., Oztas, S., Robert, S., Rogozin, I. B., Scanlan, D. J., Tandeau de Marsac, N., Weissenbach, J., Wincker, P., Wolf, Y. I., and Hess, W. R. (2003). Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. U.S.A. 100, 10020–10025.
DuRand, M. D., Olson, R. J., and Chisholm, S. W. (2001). Phytoplankton population dynamics at the Bermuda Atlantic time-series station in the Sargasso Sea. Deep Sea Res. Part II 48, 1983–2003.
Dyhrman, S. T., Benitez-Nelson, C. R., Orchard, E. D., Haley, S. T., and Pellechia, P. J. (2009). A microbial source of phosphonates in oligotrophic marine systems. Nat. Geosci. 2, 696–699.
Dyhrman, S. T., Chappell, P. D., Haley, S. T., Moffett, J. W., Orchard, E. D., Waterbury, J. B., and Webb, E. A. (2006). Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature 439, 68–71.
Gilbert, J. A., Thomas, S., Cooley, N. A., Kulakova, A. N., Field, D., Booth, T., McGrath, J. W., Quinn, J. P., and Joint, I. (2009). Potential for phosphonoacetate utilization by marine bacteria in temperate coastal waters. Environ. Microbiol. 11, 111–125.
Gomez-Garcia, M. R., Davison, M., Blain-Hartnung, M., Grossman, A. R., and Bhaya, D. (2010). Alternative pathways for phosphonate metabolism in thermophilic cyanobacteria from microbial mats. ISME J. doi: 10.1038/ismej.2010.96 [Epub ahead of print].
Hood, J. M., Brovold, S., Sterner, R. W., Villar-Argaiz, M., and Zimmer, K. D. (2006). Near-infrared spectrometry (NIRS) for the analysis of seston carbon, nitrogen, and phosphorus from diverse sources. Limnol. Oceanogr. Methods 4, 96–104.
Hoppe, H.-G. (1993). “Use of fluorogenic model substrates for extracellular enzyme activity (EEA) measurement of bacteria,” in Handbook of Methods in Aquatic Microbial Ecology, eds P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (Boca Raton, FL: Lewis Publishers), 423–431.
Ilikchyan, I. N., McKay, R. M., Zehr, J. P., Dyhrman, S. T., and Bullerjahn, G. S. (2009). Detection and expression of the phosphonate transporter gene phnD in marine and freshwater picocyanobacteria. Environ. Microbiol. 11, 1314–1324.
Karl, D. M., Beversdorf, L., Björkman, K. M., Church, M. J., Martinez, A., and DeLong, E. F. (2008). Aerobic production of methane in the sea. Nat. Geosci. 1, 473–478.
Kolowith, L. C., Ingall, E. D., and Benner, R. (2001). Composition and cycling of marine organic phosphorus. Limnol. Oceanogr. 46, 309–320.
Kulakova, A. N., Kulakov, L., Villarreal-Chiu, J., Gilbert, J. A., McGrath, J. W., and Quinn, J. P. (2009). Expression of the phosphonoalanine degradative gene cluster from Variovorax sp. Pal2 is induced by growth on phosphonoalanine and phosphonopyruvate. FEMS Microbiol. Lett. 292, 100–106.
Kulakova, A. N., Kulakov, L. A., Akulenko, N. V., Ksenzenko, V. N., Hamilton, J. T. G., and Quinn, J. P. (2001). Structural and functional analysis of the phosphonoacetate hydrolase (phnA) gene region in Pseudomonas fluorescens 23F. J. Bacteriol. 183, 3268–3275.
Kulakova, A. N., Wisdom, G. B., Kulakov, L. A., and Quinn, J. P. (2003). The purification characterization of phosphonopyruvate hydrolase, a novel carbon–phosphorus bond cleavage enzyme from Variovorax sp Pal2. J. Biol. Chem. 278, 23426–23431.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2007). ClustalW and ClustalX version 2. Bioinformatics 23, 2947–2948.
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, Buchner, A., Lai, T., Steppi, S., Jobb, G., Förster, W., Brettske, I., Gerber, S., Ginhart, A.W., Gross, O., Grumann, S., Hermann, S., Jost, R., König, A., Liss, T., Lüßmann, R., May, M., Nonhoff, B., Reichel, B., Strehlow, R., Stamatakis, A., Stuckmann, N., Vilbig, A., Lenke, M., Ludwig, T., Bode, A., and Schleifer, K.-H. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371.
Martinez, A., Tyson, G. W., and DeLong, E. F. (2010). Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ. Microbiol. 12, 222–238.
Martiny, A. C., Coleman, M. L., and Chisholm, S. W. (2006). Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc. Natl. Acad. Sci. U.S.A. 103, 12552–12557.
Menzel, D. W., and Ryther, J. H. (1960). The annual cycle of primary production in the Sargasso Sea off Bermuda. Deep Sea Res. I 6, 351–367.
Menzel, D. W., and Ryther, J. H. (1961). Annual variations in primary production of the Sargasso Sea off Bermuda. Deep Sea Res. I 7, 282–288.
Moore, L. R., Ostrowski, M., Scanlan, D. J., Feren, K., and Sweetsir, T. (2005). Ecotypic variations in phosphorus-acquisition mechanisms within marine picocyanobacteria. Aquat. Microb. Ecol. 39, 257–269.
Moore, L. R., Post, A. F., Rocap, G., and Chisholm, S. W. (2002). Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol. Oceanogr. 47, 989–996.
Moore, L. R., Rocap, G., and Chisholm, S. W. (1998). Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393, 464–467.
Palenik, B., Brahamsha, B., Larimer, F. W., Land, M., Hauser, L., Chain, P., Lamerdin, J., Regala, W., Allen, E. E., McCarren, J., Paulsen, I., Dufresne, A., Partensky, F., Webb, E. A., and Waterbury, J. (2003). The genome of a motile marine Synechococcus. Nature 424, 1037–1042.
Quinn, J. P. (2002). A Genetic Analysis of Bacterial Enzymes Involved in the Degradation of Organophosphonates. Ph.D. Thesis, Queen’s University, Belfast, UK.
Quinn, J. P., Kulakova, A. N., Cooley, N. A., and McGrath, J. W. (2007). New ways to break an old bond: the bacterial carbon–phosphorus hydrolases and their role in biogeochemical phosphorus cycling. Environ. Microbiol. 9, 2392–2400.
Rocap, G., Larimer, F. W., Lamerdin, J., Malfatti, S., Chain, P., Ahlgren, N. A., Arellano, A., Coleman, M., Hauser, L., Hess, W. R., Johnson, Z. I., Land, M., Lindell, D., Post, A. F., Regala, W., Shah, M., Shaw, S. L., Steglich, C., Sullivan, M. B., Ting, C. S., Tolonen, A., Webb, E. A., Zinser, E. R., and Chisholm, S. W. (2003). Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424, 1042–1047.
Scanlan, D. J., Ostrowski, M., Mazard, S., Dufresne, A., Garczarek, L., Hess, W. R., Post, A. F., Hagemann, M., Paulsen, I., and Partensky, F. (2009). Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73, 249–299.
Scanlan, D. J., and West, N. J. (2002). Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol. Ecol. 40, 1–12.
Schroeder, E., and Stommel, H. (1969). How representative is the series of Palinuris stations of monthly mean conditions on Bermuda? Prog. Oceanogr. 5, 31–40.
Sterner, R. W., Smutka, T. M., McKay, R. M. L., Qin, X., Brown, E. T., and Sherrell, R. M. (2004). Phosphorus and trace metal limitation of algae and bacteria in Lake Superior. Limnology 49, 495–507.
Tai V., and Palenik, B. (2009). Temporal variation of Synechococcus clades at a coastal Pacific Ocean monitoring site. ISME J. 3, 903–915.
Thomas, S., Burdett, H., Temperton, B., Wick, R., Snelling, D., McGrath, J. W., Quinn, J. P., Munn, C., and Gilbert, J. A. (2010). Evidence for phosphonate usage in the coral holobiont. ISME J. 4, 459–461.
Welschmeyer, N. A. (1994). Fluorometric analysis of chlorophyll-a in the presence of chlorophyll-b and pheopigments. Limnol. Oceanogr. 39, 1985–1992.
Wu, J., Sunda, W., Boyle, E. A., and Karl, D. M. (2000). Phosphate depletion in the western North Atlantic Ocean. Science 289, 759–762.
Zinser, E. R., Coe, A., Johnson, Z. I., Martiny, A. C., Fuller, N. J., Scanlan, D. J., and Chisholm, S. W. (2006). Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl. Environ. Microbiol. 72, 723–732.
Zwirglmaier, K., Jardillier, L., Ostrowski, M., Mazard, S., Garczarek, L., Vaulot, D., Not, F., Massana, R., Ulloa, O., and Scanlan, D. J. (2008). Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10, 147–161.
Keywords: picocyanobacteria, phosphonates, Synechococcus, Prochlorococcus, Sargasso Sea, phosphorus utilization
Citation: Ilikchyan IN, McKay RML, Kutovaya OA, Condon R and Bullerjahn GS (2010) Seasonal expression of the picocyanobacterial phosphonate transporter gene phnD in the Sargasso Sea. Front. Microbio. 1:135. doi: 10.3389/fmicb.2010.00135
Received: 24 September 2010;
Accepted: 02 December 2010;
Published online: 14 December 2010.
Edited by:
Julie Laroche, Leibniz Institute of Marine Sciences, GermanyReviewed by:
Eric A. Webb, University of Southern California, USAJack A. Gilbert, Plymouth Marine Laboratory, UK
Copyright: © 2010 Ilikchyan, McKay, Kutovaya, Condon and Bullerjahn. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: George S. Bullerjahn, Department of Biological Sciences, Bowling Green State University, Life Sciences 516, Bowling Green, OH 43403, USA. e-mail: bullerj@bgsu.edu
 Irina N. Ilikchyan1
Irina N. Ilikchyan1