- 1 Faculté de Médecine Necker-Enfants Malades, Université Paris Descartes, Paris, France
- 2 INSERM, Unit of Pathogenesis of Systemic Infections, Paris, France
Francisella tularensis is a Gram-negative bacterium capable of causing the zoonotic disease tularaemia in a large number of mammalian species and in arthropods. F. tularensis is a facultative intracellular bacterium that infects and replicates in vivo mainly inside macrophages. During its systemic dissemination, F. tularensis must cope with very different life conditions (such as survival in different target organs or tissues and/or survival in the blood stream…) and may thus encounter a broad variety of carbon substrates, nitrogen, phosphor, and sulfur sources, as well as very low concentrations of essential ions. The development of recent genome-wide genetic screens have led to the identification of hundreds of genes participating to variable extents to Francisella virulence. Remarkably, an important proportion of the genes identified are related to metabolic and nutritional functions. However, the relationship between nutrition and the in vivo life cycle of F. tularensis is yet poorly understood. In this review, we will address the importance of metabolism and nutrition for F. tularensis pathogenesis, focusing specifically on amino acid and carbohydrate requirements.
Intracellular Pathogens and Metabolic Requirements
The in vivo metabolism of pathogenic bacteria constitutes an important, and yet insufficiently studied, aspect of host–pathogen interactions. Metabolic pathways comprise: (i) degradative pathways (catabolism) of organic molecules, processes generally accompanied by the production of energy; and (ii) biosynthetic pathways (anabolism) that uses energy to build-up molecules. Both metabolic pathways require the sequential action of dedicated enzymes whose expression and activity may be tightly regulated in response to environmental changes. In order to survive and efficiently replicate in host cells, intracellular pathogens must adapt their metabolism to the available nutrients and physical conditions (including pH, oxygen availability, osmotic pressure, etc.). Metabolism is tightly associated with nutritional capacities, which involve nutrient availability and dedicated nutrient uptake systems. Indeed, pathogenic bacteria use the host organism as a macronutrient system that comprises many different specialized microenvironments. In particular, bacteria capable of systemic dissemination like Francisella tularensis have to cope with very different life conditions (such as survival in different target organs or tissues and/or survival in the blood stream, etc.) and may thus encounter a broad variety of carbon substrates, nitrogen, phosphor, and sulfur sources, as well as very low concentrations of essential ions such as magnesium, manganese, and iron (Eisenreich et al., 2010).
Many intracellular bacteria reside in a vesicular compartment (e.g., Salmonella, Legionella, Brucella, Mycobacteria, etc.). These bacteria encounter stressful conditions in these membrane-bound vacuoles (low pH, free-radicals, nutrient deprivation, antimicrobial compounds, etc.) and have therefore developed efficient defense mechanisms. For example, S. enterica is able to survive and replicate for extended periods in Salmonella-containing vacuoles (SVCs) of infected cells (preferentially macrophages) and thus can cause chronic infections. Notably, S. enterica, seems to undergo only limited replication cycles in SVCs. Although the nutritional content of SVCs is still poorly defined, transcriptomic and proteomic analyses have suggested that ions such as magnesium, manganese, and iron, could be limited in the SCV. Furthermore, in vivo studies have indicated that sugars, fatty acids, and acetate, could be used as carbon sources (García-del Portillo et al., 2008). Other intracellular bacteria, like Francisella, Listeria, Shigella, and Rickettsia, have chosen the cytosol as a replication niche (Casadevall, 2008). The host cell cytosol is generally viewed as a more permissive milieu than the phagosomal compartment.
In this review, we shall try to understand the importance of metabolism and nutrition for F. tularensis pathogenesis, focusing specifically on amino acid and carbohydrate requirements.
F. Tularensis Nutritional Requirements
Francisella tularensis is a Gram-negative bacterium capable of causing the zoonotic disease tularaemia in a large number of mammalian species and in arthropods such as ticks, flies, and mosquitoes (Keim et al., 2007). It is a highly infectious bacterium that can be transmitted to humans in numerous ways, including contact with infected animals, inhalation, ingestion of contaminated water or food, or insect bites (Sjostedt, 2007). Four different subspecies (subsp.) of F. tularensis are generally recognized that differ in virulence and geographic distribution. These are four designated subsps. tularensis (type A), holarctica (type B), novicida, and mediasiatica, respectively. However, the classification of novicida as a subspecies is still a matter of debate (Huber et al., 2010; Johansson et al., 2010). F. tularensis subsp. tularensis is the most virulent subspecies causing a severe disease in humans, whereas F. tularensis subsp. holarctica causes a similar disease but of less severity (McLendon et al., 2006). Because of its high infectivity and lethality, F. tularensis is considered a potential bioterrorism agent (Oyston and Griffiths, 2009).
Francisella tularensis is a facultative intracellular bacterium that infects and replicates in vivo mainly inside macrophages, but which can also infect and survive in a variety of non-phagocytic mammalian cells such as hepatocytes, endothelial cells, epithelial cells, and fibroblasts (Santic et al., 2010). Remarkably, F. tularensis is also one of the rare bacteria that can survive within neutrophils (McCaffrey and Allen, 2006). F. tularensis subsp. holarctica live vaccine strain (LVS) has been shown to inhibit the respiratory burst by preventing NADPH oxidase assembly at the phagosomal membrane (McCaffrey and Allen, 2006). Attempts to identify LVS genes that affect neutrophil function have only led to the selection of uracil auxotrophs (carA, carB, and pyrB) whose intracellular growth defect are most likely pleiotropic.
Francisella tularensis strains, including highly virulent species, have also been reported to survive and multiply in amebae in the environment (Abd et al., 2003; El-Etr et al., 2009), suggesting a potential link between ameba–Francisella interactions and environmental persistence. The specific nutritional requirements of F. tularensis sensu lato in the ameba have not yet been studied.
The recent availability of complete genome sequences and the development of numerous genome-scale genetic methods have led to the identification of hundreds of genes participating to variable extents to Francisella virulence (Ahlund et al., 2010; Akimana et al., 2010; Asare and Abu Kwaik, 2010; Asare et al., 2010; Lai et al., 2010; Meibom and Charbit, 2010,and references therein;Moule et al., 2010; Peng and Monack, 2010). However, the specific contribution of only a limited number of these genes is currently understood at the molecular level. Although an important proportion of the genes identified are related to metabolic and nutritional functions, the relationship between nutrition and the in vivo life cycle of F. tularensis is yet poorly understood.
F. Tularensis Biosynthetic Pathways and Virulence
Let us first consider the general relationship between the presence or absence of a biosynthetic pathway and its associated nutritional requirement. By definition, in a synthetic (or minimal) medium, a prototrophic (heterotrophic) facultative intracellular bacterium is able to synthesize all its components from the carbohydrate source provided. When one or several genes of a biosynthetic pathway are missing, or have been inactivated, supplementation of the medium by the substrate is required for growth (the bacterium is auxotroph for this substrate). In infected cells, two outcomes exist when a biosynthetic pathway is impaired in a mutant strain: (i) if the mutant bacteria require supplementation of the medium by the cognate substrate for intracellular growth, it is deduced that the intracellular milieu is depleted (or limited) for that substrate; (ii) alternatively, if the mutant bacteria grow like the wild-type strain, this is deduced the intracellular milieu is replete in that substrate (and implies that the bacterium is able to take-up enough substrate for growth).
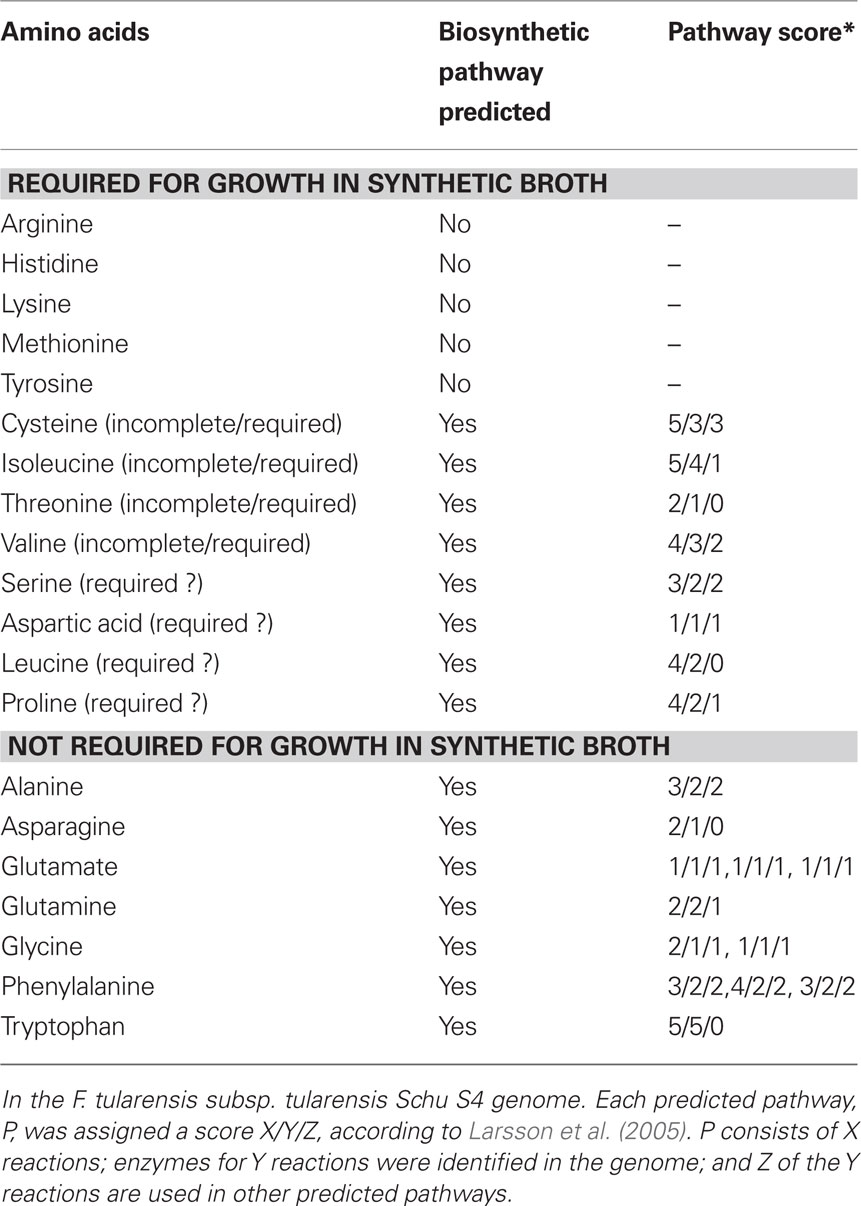
A chemically defined growth medium was developed to support the growth of F. tularensis, which includes 13 amino acids (Traub et al., 1955; Nagle et al., 1960). Later, Chamberlain optimized the concentrations of the different components (amino acids, vitamins, ions) of this medium that is still widely used (Chamberlain, 1965). F. tularensis strains Schu S4 (subsp. tularensis) and LVS (subsp. holarctica) both require cysteine for growth, most likely due to a non-functional pathway for sulfate assimilation (Larsson et al., 2005). The absolute requirement for growth of the other 12 amino acids contained in the medium has not been experimentally confirmed. Functional biosynthetic pathways have been identified in the Schu S4 genome for the seven non-essential amino acids (alanine, asparagine, glutamate, glycine, glutamine, phenylalanine, and tryptophan). Evidence was also found in the Schu S4 genome for biosynthetic pathways for 8 of the 13 amino acids that are supplied in the synthetic medium (see Table 1; Larsson et al., 2005). However, of these eight, the biosynthetic pathways for isoleucine, valine, and threonine are predicted to have missing steps and therefore to be non-functional (due to presence of pseudogenes encoding enzymes catalyzing the missing steps in the pathways). Altogether, the pathways for arginine, histidine, lysine, tyrosine, methionine, cysteine, threonine, valine, and isoleucine biosynthesis seem to be incomplete or absent. F. tularensis subsp. tularensis strain Schu S4 is hence auxotroph for these amino acids. It remains to be determined whether one or more of the other four amino acids for which pathways were predicted to be present, but supplied by the synthetic medium (i.e., serine, aspartate, leucine, and proline), are absolutely required for growth.
Aromatic amino acid biosynthetic pathways
The shikimate pathway (Figure 1) is the common pathway for the biosynthesis of chorismate, which is the precursor for the generation of aromatic amino acids, para-aminobenzoic acid (pABA, folate biosynthesis), 2,3-dihydroxybenzoic acid (DHB, biosynthesis of siderophores), ubiquinone, and menaquinone (Bentley, 1990; Figure 1). Notably, F. tularensis sensu lato has been shown to express a siderophore under iron limiting conditions. This siderophore, structurally similar to rhizoferrin, promotes the growth of both LVS and Schu S4 strains under iron limitation. The siderophore locus, designated fsl in F. tularensis subsp. tularensis (Schu S4) and F. tularensis subsp. holarctica (LVS), or fig in F. tularensis subsp. novicida, is involved in both synthesis and uptake of the siderophore. (Sullivan et al., 2006; Kiss et al., 2008; Ramakrishnan et al., 2008; Crosa et al., 2009).
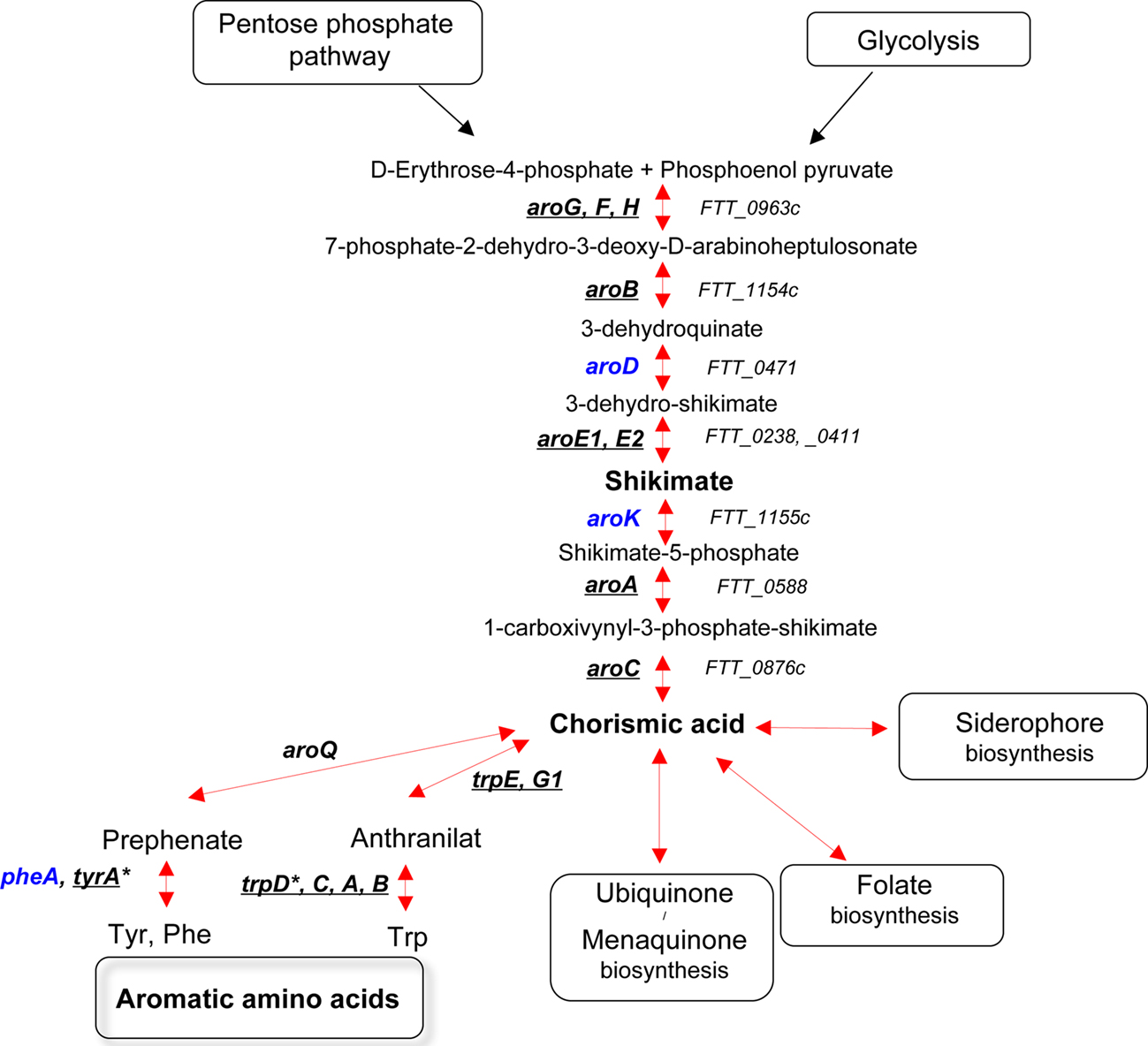
Figure 1. The shikimate pathway. Genes that have been identified in genetic screens (in vivo or in vitro) are underlined. Genes that have not been hit in any screen are in blue. TrpD*, the gene is absent in the Schu S4 and LVS strains but present in the subsp. novicida (FTA_2078). TyrA*, the gene is absent in the Schu S4 strain but present in both LVS (FTL_048) and subsp. novicida (FTN_0055).
Bruce Stocker’s pioneer work on the genetics of Salmonella enterica serovar Typhimurium (S. typhimurium) has demonstrated the crucial importance of the aromatic biosynthetic pathway for bacterial virulence. Mutations in different genes coding for the biosynthesis of aromatic amino acids have been used to reduce the virulence of Salmonella strains and for immunization of various animal species (Chatfield et al., 1994, 1995). The attenuation of aro mutants of S. typhimurium is thought to be due to the inability of the bacterium to generate pABA and DHB from chorismate (Hoiseth and Stocker, 1981). Remarkably, aroA and aroD mutants of S. enterica serovar Typhi were also successfully tested as a vaccine against human typhoid (Tacket and Levine, 2007).
All of the necessary genes encoding the chorismate pathway enzymes are present in the genome of F. tularensis Schu S4 (i.e., aroG, aroB, aroD, aroE, aroK, aroA, and aroC). The genes (pabA and pabB) encoding the two components of para-aminobenzoate synthase have also been identified in the F. tularensis Schu S4 genome as well as the gene encoding FolP, FolC, and FolA, involved in folate biosynthesis. In recent screens, we and others (Su et al., 2007; Alkhuder et al., 2009) have selected mutants in gene FTL_1240 (aroG), encoding DAHP synthase. This is the first enzyme of the aromatic amino acid biosynthetic pathway, which converts erythro-4-phosphate and phosphoenolpyruvate (PEP) to 3-deoxy-D-arabino-heptuosonate-7-phosphate. We have shown that an aroG mutant of LVS had only a slightly reduced intracellular growth capacity, in both J774 cells and in bone marrow-derived macrophages (BMM). Still, in the mouse model, the mutant strain was very severely attenuated. Of interest, wild-type Escherichia coli has been shown to produce three feedback inhibitor-sensitive DAHP synthase isoenzymes: a tyrosine-sensitive, a phenylalanine-sensitive, and a tryptophan-sensitive enzyme (encoded by genes genes aroF, aroG, and aroH, respectively). Hence, the functionality of a biosynthetic pathway does not only depend on the presence of intact genes but may also rely on the amount and activity of the enzymes.
This interplay between the availability of an amino acid and the activity of its cognate biosynthetic pathway might exist for other types of nutrients that the bacterium can either biosynthesize or acquire from its growth medium.
Additional genes of the F. tularensis aromatic amino acid biosynthetic pathway have been identified in genome-wide screens: aroA, aroB, aroC, aroE1, tyrA, trpA, trpB, trpC, and trpE. The gene aroA is responsible for the sixth step of the biosynthetic pathway (converting shikimate-3-phosphate and PEP to 5-enolpyruvyl-shikimate-3-phosphate). An aroA mutant of F. tularensis Schu S4 strain was selected in a screen of transposon insertion mutants performed in the human hepatic carcinoma cell line HepG2 (Qin and Mann, 2006). The same screen also led to the identification of purine and pyrimidine auxotrophs (see below).
An aroB mutant of F. tularensis subsp. novicida (FTN_1135), encoding a putative 3-dehydroquinate synthetase, has been very recently identified in a screen in human macrophages (Asare and Abu Kwaik, 2010). The mutant strain, which was also deficient for growth in Drosophila melanogaster-derived S2 cells, localized to the cytosol in macrophages, indicating a cytosolic growth defect.
The gene aroC encodes chorismate synthase, the seventh step of the aromatic biosynthetic pathway. It was identified in a screen of transposon insertion mutants performed in F. tularensis subsp. novicida, searching for genes required for pulmonary and systemic infection in mice (Kraemer et al., 2009). The gene tyrA (FTN_0055), which encodes the enzyme prephenate dehydrogenase converting chorismate to tyrosine, was also hit in this in vivo screen, as was aroC. Notably, the tyrA gene is absent in the Schu S4 strain but present in LVS (FTL_0048). aroC encodes the enzyme performing the last step of chorismic acid synthesis. The gene aroE1, which encodes shikimate-5-dehydrogenase, the fourth step of the biosynthetic pathway, was identified in two screens of transposon insertion mutants. One screen was performed in F. tularensis subsp. holarctica LVS searching for auxotrophic mutants unable to grow on chemically defined medium (Maier et al., 2006) and the other was a screen of F. tularensis subsp. novicida for mutants unable to replicate intracellularly in macrophages (Asare and Abu Kwaik, 2010). Finally, five genes (trpA, trpB, trpC, trpE, and trpG1) encoding enzymes involved in the conversion of chorismic acid to tryptophan were identified in three in vivo selections using F. tularensis subsp. novicida (Weiss et al., 2007; Kraemer et al., 2009; Peng and Monack, 2010).
Altogether, almost every gene in the aromatic amino acids biosynthetic pathway has been identified in mutant screens, highlighting the importance this pathway for F. tularensis virulence. Interestingly, several of the genes have also been found to be important for growth/virulence in a non-mammalian model, in vivo in D. melanogaster or in vitro in D. melanogaster-derived cells (Asare et al., 2010; Moule et al., 2010).
Purine and pyrimidine biosynthetic pathways
In silico analysis reveals that the F. tularensis genomes encode all the enzymes necessary for the de novo synthesis of purines and pyrimidines. The two pathways are virtually identical to those in E. coli.
Pur pathway. Several pur auxotrophic mutants of F. tularensis (Figure 2), selected from banks of mutants or genetically engineered, have already been obtained and tested for virulence (Gray et al., 2002; Qin and Mann, 2006; Tempel et al., 2006; Quarry et al., 2007; Titball et al., 2007; Weiss et al., 2007; Kadzhaev et al., 2009; Asare and Abu Kwaik, 2010; Asare et al., 2010; Peng and Monack, 2010). The pur mutants with single gene mutations showed variable degrees of attenuation in vivo and growth defects in vitro, suggesting that the step at which the pathway is inactivated may have a distinct impact on virulence. Remarkably, Pechous et al. showed that a triple mutant ΔpurMCD in both LVS (Pechous et al., 2006) and Schu S4 (Pechous et al., 2008) strains led to severe intracellular growth defects and strong attenuation in the mouse model. The fact that inability of these strains to synthesize purines de novo leads to a severe intracellular growth defect supports the notion that macrophages contain limiting concentrations of purines (Appelberg, 2006).
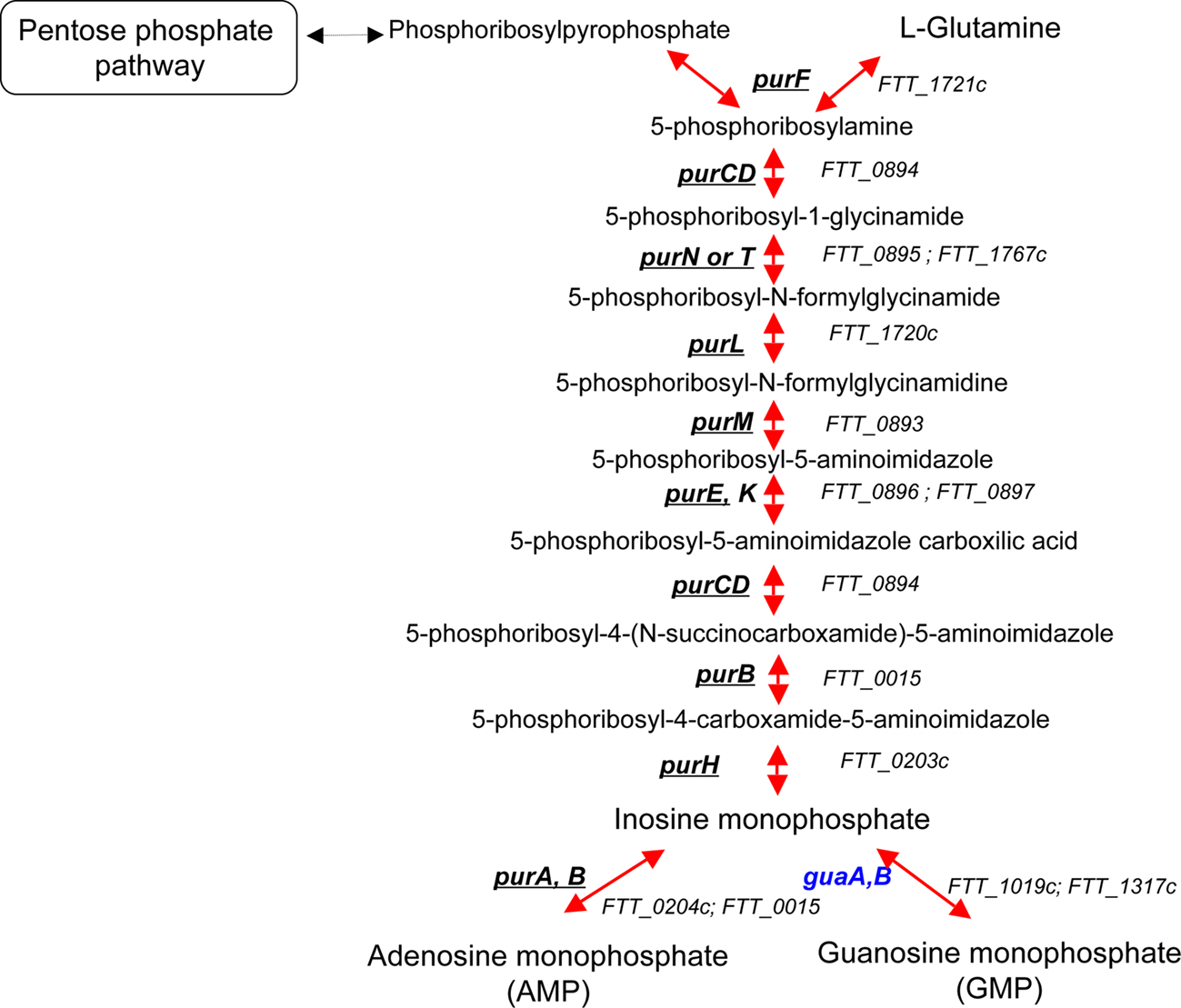
Figure 2. The purine biosynthetic pathway. Genes that have been identified in genetic screens (in vivo or in vitro) are underlined. Genes that have not been hit in any screen are in blue.
Pyr pathway. The pathway converting L-glutamine to uridine monophosphate (UMP) comprises six steps (Figure 3). Several mutants in this pathway have been identified in genetic screens (Qin and Mann, 2006; Weiss et al., 2007; Schulert et al., 2009; Asare and Abu Kwaik, 2010; Peng and Monack, 2010). Mutants unable to perform the first step, i.e., bacteria with mutations in genes carA or carB (encoding the two subunits of carbamoyl phosphate synthetase, converting L-glutamine to carbamoyl-P) were severely impaired in intramacrophage growth. However, conflicting results were reported regarding the phenotype of pyrB mutants. On one hand, a pyrB transposon insertion mutant of Schu S4 (Qin and Mann, 2006) showed reduced growth in HepG2 hepatocytes but normal growth in J774 cells, and was attenuated in mice and a similar mutant in subsp. novicida was shown to have a growth defect in human macrophages (Asare and Abu Kwaik, 2010). In contrast, another recent study (Kadzhaev et al., 2009) reports that a pyrB deletion mutant of Schu S4 is barely attenuated. In addition, mutants with transposon insertions in the carA, carB, and pyrB genes were selected upon screening of a bank of transposon mutants in LVS for mutants that failed to prevent the oxidative burst (Schulert et al., 2009). The three mutants appeared to grow normally in HepG2 and J774 cells but were killed by human monocytes and monocyte-derived macrophages. Transposon mutants of subsp. novicida with insertions in pyrF (encoding orotidine-5P decarboxylase, converting orotidine-5P to UMP) were identified in two in vivo screens (Weiss et al., 2007; Peng and Monack, 2010) and recently, a pyrF deletion mutant was generated in both F. tularensis subsp. holarctica (LVS) and tularensis (Schu S4). These F. tularensis ΔpyrF mutants were unable to replicate in primary human macrophages but retained full virulence in the mouse model (Horzempa et al., 2010).
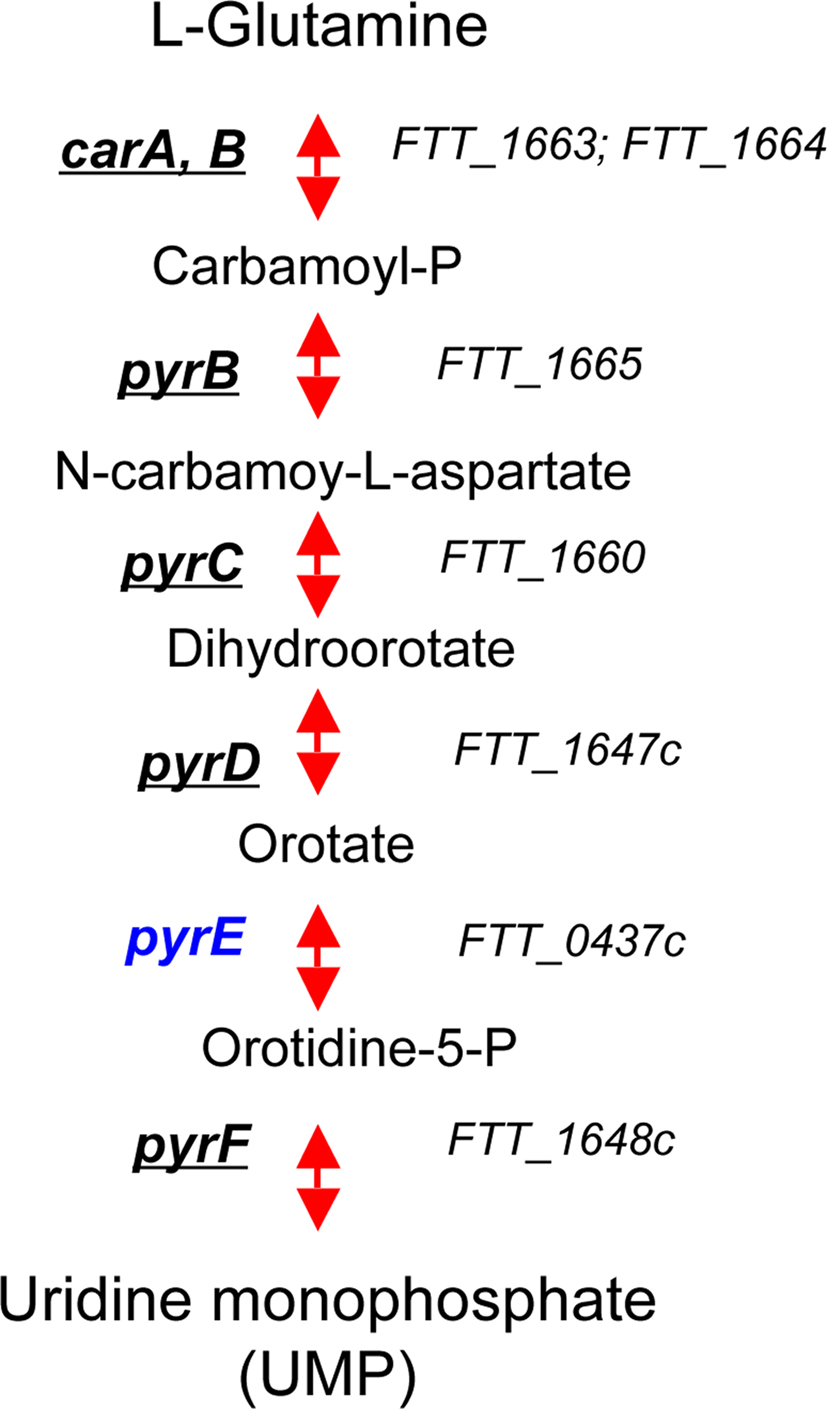
Figure 3. The pyrimydine biosynthetic pathway. Genes that have been identified in genetic screens (in vivo or in vitro) are underlined. Genes that have not been hit in any screen are in blue.
These data clearly indicate that defects in the pyrimidine biosynthetic pathway affect Francisella virulence in a cell type- and strain-specific manner and suggest that in vivo the pyr pathway is not as important as the pur pathway for F. tularensis virulence.
F. Tularensis Carbon Metabolism and Virulence
Each intracellular pathogen has adapted its intracellular metabolism to the nutrient supply of the host cell. Nevertheless, two bacterial species using the same host cell compartment (facing thus the same nutritional environment) may have quite different preferred carbon sources (and hence carbon metabolisms). Carbon catabolism provides the bacterial cell with energy and essential biosynthetic precursors such as glucose-6-phosphate (G6-P), fructose-6-phosphate (F6-P), 3-phosphoglycerate, PEP, and acetyl-CoA.
Glycolysis and gluconeogenesis pathways
Hexoses such as glucose are the preferred carbon and energy sources for many bacteria. The three best-characterized pathways of sugar catabolism in bacteria are glycolysis (Figure 4), the pentose phosphate pathway and the Entner–Doudoroff pathway. Each of these different pathways can be the preferred, or exclusive, carbon utilization pathway in a given pathogenic bacterial species. In the following we will discuss only genes encoding enzymes in glycolysis (and gluconeogenesis) and shown in Figure 4 and not other carbohydrate metabolism pathways.
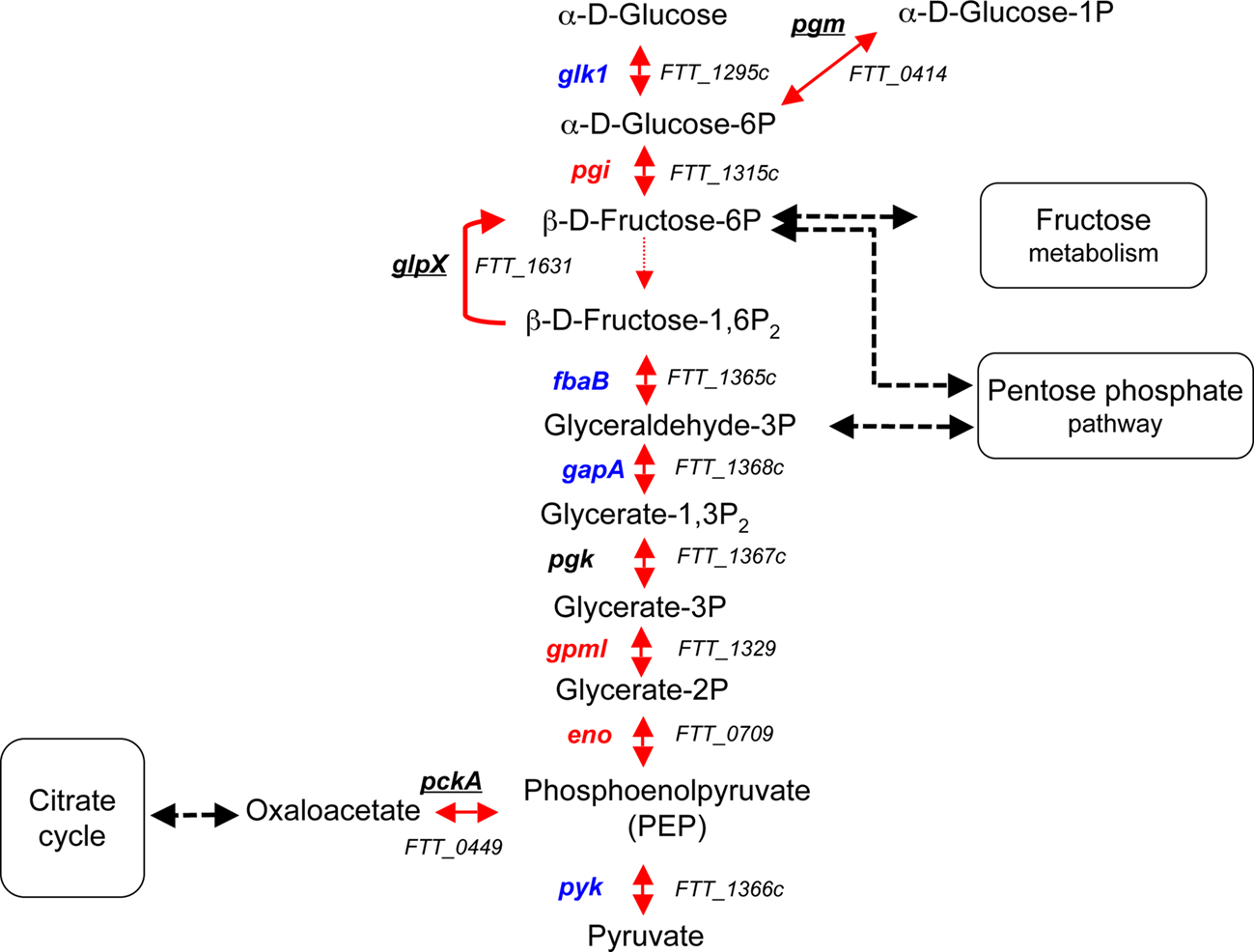
Figure 4. The glycolytic and gluconeogenic pathways. GlpX is the only enzyme not promoting a reversible reaction. Genes that have been identified in genetic screens (in vivo or in vitro) are underlined. Genes that have not been hit in any screen are in blue. Genes predicted to be essential (according to Gallagher et al., 2007) are in red. The black dotted arrows indicate the connection between the glycolytic/gluconeogenic pathways and the pentose phosphate and fructose pathways. In each pathway, the gene name is indicated to the left of each reaction and the corresponding FTT number, to the right.
pckA. The enzyme PEP carboxykinase, encoded by the gene pckA, catalyzes the conversion of oxaloacetate to PEP. In Mycobacterium bovis BCG, a pckA mutant is attenuated both in vitro and in vivo (Liu et al., 2003), while this gene is dispensable in Salmonella (Tchawa Yimga et al., 2006). In F. tularensis subsp. tularensis Schu S4, Kadzhaev et al. (2009) observed no attenuation of a ΔpckA mutant in mice. The biological activity of this enzyme has not been experimentally established.
glpX. The glycolytic and gluconeogenic pathways comprise essentially the same set of enzymes that catalyze reversible reactions, except between F6-P and fructose-1,6-bisphosphate (F1,6-P2). The glycolytic reaction leading to the production of F1,6-P2 is catalyzed by phosphofructokinase, while the gluconeogenic reaction yielding F6P is catalyzed by fructose-1,6-bisphosphatase (FBP). The F. tularensis subsp. tularensis Schu S4 genome encodes apparently only a FBP (encoded by glpX) and lacks a pfkA gene encoding phosphofructokinase (Raghunathan et al., 2010). This suggests that F. tularensis uses the Embden–Meyerhof–Parnas pathway for gluconeogenesis rather than for glycolysis. However, the exact biological activity of the FBP enzyme remains to be experimentally established in F. tularensis. Mutants in the glpX gene have been repeatedly obtained in genetic screens, in vivo as well as in vitro (Maier et al., 2007; Su et al., 2007; Weiss et al., 2007; Kraemer et al., 2009; Peng and Monack, 2010). Moreover, a glpX deletion mutant of F. tularensis subsp. tularensis Schu S4 has been shown recently to be almost avirulent in the mouse model (Kadzhaev et al., 2009). Altogether, these observations strongly suggest that gluconeogenesis is critical for the full virulence of F. tularensis. At this stage, it cannot be excluded that the virulence defect of the glpX mutant could be due to another function of this enzyme, possibly regulatory. Supporting this hypothesis, none of the other genes involved in the conversion of glucose to PEP has been identified in previous genetic screens, but it is possible that some of these (pgi, gpmI, eno) are essential genes and therefore will not be found in screens (Gallagher et al., 2007). It remains to be determined by a systematic mutagenesis approach whether other enzymes in this pathway may also be involved in virulence.
pgm. The pgm gene encodes a predicted phosphoglucomutase, a glyconeogenic enzyme involved in the reversible conversion of glucose-1-phosphate (G1-P) to G6-P. A mutant in the pgm gene has been identified after an in vivo genetic screen in the F. tularensis subsp. novicida (Weiss et al., 2007).
The intracellular transcriptome of F. tularensis reveals that a number of genes involved in carbohydrate metabolism were up-regulated in BMM (Wehrly et al., 2009). In particular, genes of the glycolytic/gluconeogenic pathway have been found (pgm, glk, fbaB, and pgk; Figure 4), suggesting that this pathway is used during intracellular growth.
Very recently, a systems biology approach was applied to identify F. tularensis metabolic networks (Raghunathan et al., 2010). Integration of in silico metabolic reconstitutions and experimental data (including metabolic profiling and transcriptomic analyses) suggested that significant changes in carbohydrate metabolism occur during the intracellular growth phase. Gene expression profiling further supported the prediction that F. tularensis preferentially utilizes specific amino acids for energy and fatty acids as gluconeogenic substrates rather than relying on carbohydrate sources like glucose and fructose during infection.
Nutrient Uptake Systems of F. Tularensis
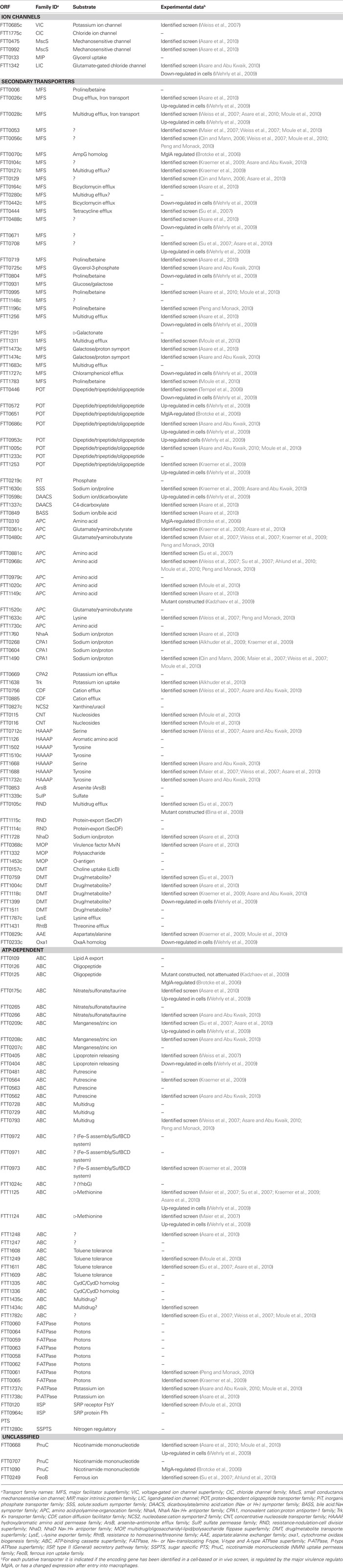
Intracellular bacteria must possess dedicated nutrient uptake systems to capture their necessary host-derived nutrients. These systems must be particularly efficient for substrates available in limiting concentrations. The majority of the predicted transport systems present in F. tularensis are secondary carriers (Table 2). Secondary transporters encompass several major families, including: (i) the major facilitator superfamily (MFS, 31 proteins), predicted to participate in various functions including drug efflux, amino acids and sugar uptake; (ii) the amino acid-polyamine-organocation transporters (APC, 11 proteins); (iii) the hydroxy/aromatic amino acid permeases (HAAAP, 7 proteins); and (iv) the proton-dependent oligopeptide transporters (POT, 8 proteins). Several mutants in secondary transporters were identified in various genetic screens. For example, the gene xasA, encoding a predicted glutamate/γ-aminobutyrate transporter of the APC family (FTT_0480c, Table 2), has been identified in four different screens (Maier et al., 2007; Weiss et al., 2007; Kraemer et al., 2009; Peng and Monack, 2010), supporting a functional role in F. tularensis virulence. The gene FTT_0708, encoding a transporter of the MFS family has also been identified in an in vivo screen (Su et al., 2007), as were other transporters of this family.
The F. tularensis subsp. tularensis Schu S4 genome also encodes 15 complete transport ABC-type carriers, consisting of a membrane-spanning permease and an ATP-binding subunit (Atkins et al., 2006). These ABC transporters are predicted to participate in diverse functions, ranging from amino acid/peptide and ion uptake to multidrug efflux. Remarkably, mutants in one ABC transporter (FTT_1125) have been identified in several in vitro (Maier et al., 2007) and in vivo (Su et al., 2007; Kraemer et al., 2009) screens for attenuated mutants, indicating a direct contribution to F. tularensis virulence.
Supporting a role of transport systems in intracellular survival, transcriptional profiling of the F. tularensis subsp. tularensis Schu S4 strain in BMMs (Wehrly et al., 2009) revealed that genes encoding various transporters showed significantly altered expression (either up- or down-regulated) after host cell entry. Notably, five of the eight POT family members were up-regulated intracellularly. The amino acid identity between the POT family members does not exceed 47% (ranging from 22 to 31%, in most cases), suggesting that they might have distinct transport properties. Also, four of the POT transporters have been found in mutant screens (Tempel et al., 2006; Kraemer et al., 2009; Asare and Abu Kwaik, 2010), further indicating that these oligopeptide transporters and therefore amino acid metabolism is important during infection.
In addition, the F. tularensis subsp. tularensis Schu S4 genome encodes six putative ion channels but is devoid of any PEP-dependent phosphotransferase (PTS) system. At present, no biochemical data are available on any of the transporters present in F. tularensis.
One example of amino acid supply provided by the host cytosol
The cytosol of eukaryotic cells contains a high concentration (10 mM) of the tripeptide γ-glu-gly-cys named glutathione (in its reduced form, GSH; Alkhuder et al., 2009). GSH plays a pleiotropic and major role in mammalian cell homeostasis and GSH-deficiency has been associated with various severe diseases (Griffith, 1999; Wu et al., 2004; Franco et al., 2007). Biosynthesis of GSH is dependent on the availability of the amino acid precursors glutamate, glycine, and cysteine. The intracellular pool of cysteine is relatively small (0.10–0.25 mM) and cysteine is generally the limiting amino acid for GSH synthesis. The other two precursors, glycine and glutamate, are found in considerable higher intracellular concentration. As mentioned earlier, F. tularensis subspecies requires cysteine for growth. We have recently demonstrated that gene FTL_0766 encodes a genuine γ-glutamyl transpeptidase (GGT) involved in the metabolism of γ-glutamyl-containing peptides. GGT allows the utilization of γ-glutamyl peptides as a source of cysteine during intracellular multiplication of the F. tularensis subsp. holarctica strain LVS, and is thus critical for its virulence (Alkhuder et al., 2009). This work represents the only direct experimental evidence of nutrient utilization by intracellular F. tularensis. The transporter of GSH and the molecular mechanism of crossing the bacterial envelope remain to be discovered.
Starving the Invading Bacteria as a Host Cell Defense Mechanism
The capacity of a macrophage to deprive intracellular pathogens of required nutrients (Appelberg, 2006) can be viewed as an intrinsic antimicrobial innate immune defense mechanism. Thus, microbial killing may not rely only on a toxic environment (such as low pH and oxidative stress in the phagosomal compartment) but also may result from the scarcity of nutrients in the cellular compartment it occupies (transiently or permanently). At any rate, one must keep in mind that the notion of a limiting concentration of nutrient may vary considerable from one intracellular pathogen to another.
Two distinct mechanisms of nutrient deprivation exist: (i) constitutive mechanisms, such as that mediated by the divalent cation transporter Nramp1, present in the membrane of endosomal compartment and participating to iron depletion (Cellier et al., 2007); and (ii) induced mechanisms, such as those triggered by the cytokine IFN-γ in activated macrophages, which may also affect iron availability (Mulero and Brock, 1999).
Notably, another pathway triggered by IFN-γ has been shown to play a role in the nutritional control of several intracellular pathogens (Taylor and Feng, 1991). Activation of the enzyme indoleamine 2,3-dioxygenase (ID) which degrades L-tryptophan by IFN-γ thus leads to tryptophan deprivation. Monack and co-workers very recently showed that tryptophan auxotrophs of F. tularensis subsp. novicida were severely affected in intracellular survival and multiplication and were attenuated in the mouse model (Peng and Monack, 2010). Interestingly, tryptophan metabolism appeared to be important only for bacterial colonization of the lungs, suggesting an organ specificity of this metabolic need. The authors found that this antimicrobial starvation mechanism mediated by the enzyme ID was effective against both auxotrophic and prototrophic microbes. These observations support the notion that, for bacteria, amino acid biosynthesis is more energetically costly than their capture from the environment. The necessity to biosynthesize amino acid in a depleted environment may thus reduce the capacity of the bacterium to replicate.
Concluding Remarks
Genetic screens have clearly established the critical importance of the aromatic amino acids biosynthetic pathway for F. tularensis virulence. The fact that inactivation of almost every gene in this pathway lead to reduced virulence suggests that the available pool of aromatic amino acids is limiting in the infected host. Similarly, the severe intracellular growth defect of mutants unable to synthesize purines de novo, also suggest that macrophages contain limiting concentrations of purines.
Altogether, experimental data and predictive models favor the notion that F. tularensis preferentially utilizes specific amino acids for energy and fatty acids as gluconeogenic substrates rather than carbohydrate sources during cytosolic multiplication. Intracellular transcriptomic studies revealed that significant changes occurred in the expression of genes encoding enzymes involved in carbohydrate metabolism as well as in genes encoding putative amino acid and carbohydrate transporters. Hence, the importance of nutrition in F. tularensis virulence can be seen as a fine balance between its ability to capture nutrients from the host (transport) and the regulation of its metabolism in response to the amounts of nutrients available (metabolism). Mutations affecting either one or the other (or both) of these two functions lead to an impaired fitness and are likely to cause a reduced virulence.
Intracellular pathogens often co-regulate their metabolic needs and the production of dedicated virulence factors by using pleiotropic (mainly transcriptional) regulators. The stringent response is a stress response that occurs in bacteria in reaction to amino acid or carbon starvation. The stringent response is signaled by the alarmone ppGpp. Notably, the expression of many virulence regulators is mediated by ppGpp, thereby coupling pathogenesis to metabolism (Dalebroux et al., 2010). F. tularensis also uses ppGpp to control the activity of its major regulator of virulence, MglA (Charity et al., 2009). In particular, ppGpp was shown recently to promote physical interactions between the MglA–SspA complex and the putative DNA binding factor PigR (also designated FevR) to control the PigR-dependent activation of the Francisella pathogenicity island (Charity et al., 2009). These data suggest a link between nutrient availability and virulence. They are in agreement with the transciptomic analysis of a mglA knock-out mutant of F. tularensis subsp. novicida (Brotcke et al., 2006). Indeed, among the 102 MglA-regulated genes identified, 20 were predicted to play a role in metabolism, particularly in amino acid metabolism.
A systematic mutational analysis of F. tularensis metabolic pathways, coupled to thorough biochemical and biophysical characterization of its metabolic capacities will be required to fully understand the complex interplays between metabolism and virulence.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
These studies were supported by the “Institut National de la Santé et de la Recherche Médicale” (INSERM), the “Centre National de la Recherche Scientifique” (CNRS), and the “Université René Descartes Paris V”.
References
Abd, H., Johansson, T., Golovliov, I., Sandstrom, G., and Forsman, M. (2003). Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69, 600–606.
Ahlund, M. K., Ryden, P., Sjostedt, A., and Stoven, S. (2010). Directed screen of Francisella novicida virulence determinants using Drosophila melanogaster. Infect. Immun. 78, 3118–3128.
Akimana, C., Al-Khodor, S., and Abu Kwaik, Y. (2010). Host factors required for modulation of phagosome biogenesis and proliferation of Francisella tularensis within the cytosol. PLoS ONE 5, e11025. doi: 10.1371/journal.pone.0011025
Alkhuder, K., Meibom, K. L., Dubail, I., Dupuis, M., and Charbit, A. (2009). Glutathione provides a source of cysteine essential for intracellular multiplication of Francisella tularensis. PLoS Pathog. 5, e1000284. doi: 10.1371/journal.ppat.1000284
Alkhuder, K., Meibom, K. L., Dubail, I., Dupuis, M., and Charbit, A. (2010). Identification of trkH, encoding a potassium uptake protein required for Francisella tularensis systemic dissemination in mice. PLoS One. 5(1): e8966. doi:10.1371/journal.pone.0008966
Appelberg, R. (2006). Macrophage nutriprive antimicrobial mechanisms. J. Leukoc. Biol. 79, 1117–1128.
Asare, R., and Abu Kwaik, Y. (2010). Molecular complexity orchestrates modulation of phagosome biogenesis and escape to the cytosol of macrophages by Francisella tularensis. Environ. Microbiol. 12, 2559–2586.
Asare, R., Akimana, C., Jones, S., and Abu Kwaik, Y. (2010). Molecular bases of proliferation of Francisella tularensis in arthropod vectors. Environ. Microbiol. 12, 2587–2612.
Atkins, H. S., Dassa, E., Walker, N. J., Griffin, K. F., Harland, D. N., Taylor, R. R., Duffield, M. L., and Titball, R. W. (2006). The identification and evaluation of ATP binding cassette systems in the intracellular bacterium Francisella tularensis. Res. Microbiol. 157, 593–604.
Bentley, R. (1990). The shikimate pathway – a metabolic tree with many branches. Crit. Rev. Biochem. Mol. Biol. 25, 307–384.
Bina, X. R., Lavine, C. L., Miller, M. A., and Bina, J. E. (2008). The AcrAB RND efflux system from the live vaccine strain of Francisella tularensis is a multiple drug efflux system that is required for virulence in mice. FEMS Microbiol. Lett. 279, 226–233.
Brotcke, A., Weiss, D. S., Kim, C. C., Chain, P. Malfatti, S., Garcia, E., and Monack, D. M. (2006). Identification of MglA regulated genes reveals novel virulence factors in Francisella tularensis. Infect. Immun. 74, 6642–6655.
Cellier, M. F., Courville, P., and Campion, C. (2007). Nramp1 phagocyte intracellular metal withdrawal defense. Microbes Infect. 9, 1662–1670.
Chamberlain, R. E. (1965). Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13, 232–235.
Charity, J. C., Blalock, L. T., Costante-Hamm, M. M., Kasper, D. L., and Dove, S. L. (2009). Small molecule control of virulence gene expression in Francisella tularensis. PLoS Pathog. 5, e1000641. doi: 10.1371/journal.ppat.1000641
Chatfield, S., Roberts, M., Li, J., Starns, A., and Dougan, G. (1994). The use of live attenuated Salmonella for oral vaccination. Dev. Biol. Stand. 82, 35–42.
Chatfield, S. N., Roberts, M., Dougan, G., Hormaeche, C., and Khan, C. M. (1995). The development of oral vaccines against parasitic diseases utilizing live attenuated Salmonella. Parasitology 110(Suppl.), S17–S24.
Crosa, L. M., Crosa, J. H., and Heffron, F. (2009). Iron transport in Francisella in the absence of a recognizable TonB protein still requires energy generated by the proton motive force. Biometals 22, 337–344.
Dalebroux, Z. D., Svensson, S. L., Gaynor, E. C., and Swanson, M. S. (2010). ppGpp conjures bacterial virulence. Microbiol. Mol. Biol. Rev. 74, 171–199.
Eisenreich, W., Dandekar, T., Heesemann, J., and Goebel, W. (2010). Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8, 401–412.
El-Etr, S. H., Margolis, J. J., Monack, D., Robison, R. A., Cohen, M., Moore, E., and Rasley, A. (2009). Francisella tularensis type A strains cause the rapid encystment of Acanthamoeba castellanii and survive in amoebal cysts for three weeks postinfection. Appl. Environ. Microbiol. 75, 7488–7500.
Franco, R., Schoneveld, O. J., Pappa, A., and Panayiotidis, M. I. (2007). The central role of glutathione in the pathophysiology of human diseases. Arch. Physiol. Biochem. 113, 234–258.
Gallagher, L. A., Ramage, E., Jacobs, M. A., Kaul, R., Brittnacher, M., and Manoil, C. (2007). A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc. Natl. Acad. Sci. U.S.A. 104, 1009–1014.
García-del Portillo, F., Núñez-Hernández, C., Eisman, B., and Ramos-Vivas, J. (2008). Growth control in the Salmonella-containing vacuole. Curr. Opin. Microbiol. 11, 46–52.
Gray, C. G., Cowley, S. C., Cheung, K. K., and Nano, F. E. (2002). The identification of five genetic loci of Francisella novicida associated with intracellular growth. FEMS Microbiol. Lett. 215, 53–56.
Griffith, O. W. (1999). Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic. Biol. Med. 27, 922–935.
Hoiseth, S. K., and Stocker, B. A. (1981). Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239.
Horzempa, J., Shanks, R. M., Brown, M. J., Russo, B. C., O’Dee, D. M., and Nau,G. J. (2010). Utilization of an unstable plasmid and the I-SceI endonuclease to generate routine markerless deletion mutants in Francisella tularensis. J. Microbiol. Methods 80, 106–108.
Huber, B., Escudero, R., Busse, H. J., Seibold, E., Scholz, H. C., Anda, P., Kampfer, P., and Splettstoesser,W. D. (2010). Description of Francisella hispaniensis sp. nov., isolated from human blood, reclassification of Francisella novicida (Larson et al. 1955). Olsufiev et al. 1959 as Francisella tularensis subsp. novicida comb. nov. and emended description of the genus Francisella. Int. J. Syst. Evol. Microbiol. 60, 1887–1896.
Johansson, A., Celli, J., Conlan, W., Elkins, K. L., Forsman, M., Keim, P. S., Larsson, P., Manoil, C., Nano, F. E., Petersen, J. M., and Sjostedt, A. (2010). Objections to the transfer of Francisella novicida to the subspecies rank of Francisella tularensis. Int. J. Syst. Evol. Microbiol. 60, 1717–1718; author reply 1718–1720.
Kadzhaev, K., Zingmark, C., Golovliov, I., Bolanowski, M., Shen, H., Conlan, W., and Sjostedt, A. (2009). Identification of genes contributing to the virulence of Francisella tularensis SCHU S4 in a mouse intradermal infection model. PLoS ONE 4, e5463. doi: 10.1371/journal.pone.0005463
Keim, P., Johansson, A., and Wagner, D. M. (2007). Molecular epidemiology, evolution, and ecology of Francisella. Ann. N. Y. Acad. Sci. 1105, 30–66.
Kiss, K., Liu, W., Huntley, J. F., Norgard, M. V., and Hansen, E. J. (2008). Characterization of fig operon mutants of Francisella novicida U112. FEMS Microbiol. Lett. 285, 270–277.
Kraemer, P. S., Mitchell, A., Pelletier, M. R., Gallagher, L. A., Wasnick, M., Rohmer, L., Brittnacher, M. J., Manoil, C., Skerett, S. J., and Salama, N. R. (2009). Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infect. Immun. 77, 232–244.
Lai, X. H., Shirley, R. L., Crosa, L., Kanistanon, D., Tempel, R., Ernst, R. K., Gallagher, L. A., Manoil, C., and Heffron, F. (2010). Mutations of Francisella novicida that alter the mechanism of its phagocytosis by murine macrophages. PLoS ONE 5, e11857. doi: 10.1371/journal.pone.0011857
Larsson, P., Oyston, P. C., Chain, P., Chu, M. C., Duffield, M., Fuxelius, H. H., Garcia, E., Halltorp, G., Johansson, D., Isherwood, K. E., Karp, P. D., Larsson, E., Liu, Y., Michell, S., Prior, J., Prior, R., Malfatti, S., Sjostedt, A., Svensson, K., Thompson, N., Vergez, L., Wagg, J. K., Wren, B. W., Lindler, L. E., Andersson, S. G., Forsman, M., and Titball, R. W. (2005). The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37, 153–159.
Liu, K., Yu, J., and Russell, D. G. (2003). pckA-deficient Mycobacterium bovis BCG shows attenuated virulence in mice and in macrophages. Microbiology 149, 1829–1835.
Maier, T. M., Casey, M. S., Becker, R. H., Dorsey, C. W., Glass, E. M., Maltsev, N., Zahrt, T. C., and Frank, D. W. (2007). Identification of Francisella tularensis Himar1-based transposon mutants defective for replication in macrophages. Infect. Immun. 75, 5376–5389.
Maier, T. M., Pechous, R., Casey, M., Zahrt, T. C., and Frank, D. W. (2006). In vivo Himar1-based transposon mutagenesis of Francisella tularensis. Appl. Environ. Microbiol. 72, 1878–1885.
McCaffrey, R. L., and Allen, L. A. (2006). Francisella tularensis LVS evades killing by human neutrophils via inhibition of the respiratory burst and phagosome escape. J. Leukoc. Biol. 80, 1224–1230.
McLendon, M. K., Apicella, M. A., and Allen, L. A. (2006). Francisella tularensis: taxonomy, genetics, and immunopathogenesis of a potential agent of biowarfare. Annu. Rev. Microbiol. 60, 167–185.
Meibom, K. L., and Charbit, A. (2010). The unraveling panoply of Francisella tularensis virulence attributes. Curr. Opin. Microbiol. 13, 11–17.
Moule, M. G., Monack, D. M., and Schneider, D. S. (2010). Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response. PLoS Pathog. 6, e1001065. doi: 10.1371/journal.ppat.1001065
Mulero, V., and Brock, J. H. (1999). Regulation of iron metabolism in murine J774 macrophages: role of nitric oxide-dependent and -independent pathways following activation with gamma interferon and lipopolysaccharide. Blood 94, 2383–2389.
Nagle, S. C. Jr., Anderson, R. E., and Gary, N. D. (1960). Chemically defined medium for the growth of Pasteurella tularensis. J. Bacteriol. 79, 566–571.
Oyston, P. C., and Griffiths, R. (2009). Francisella virulence: significant advances, ongoing challenges and unmet needs. Expert Rev. Vaccines 8, 1575–1585.
Pechous, R., Celli, J., Penoske, R., Hayes, S. F., Frank, D. W., and Zahrt, T. C. (2006). Construction and characterization of an attenuated purine auxotroph in a Francisella tularensis live vaccine strain. Infect. Immun. 74, 4452–4461.
Pechous, R. D., McCarthy, T. R., Mohapatra, N. P., Soni, S., Penoske, R. M., Salzman, N. H., Frank, D. W., Gunn, J. S., and Zahrt, T. C. (2008). A Francisella tularensis Schu S4 purine auxotroph is highly attenuated in mice but offers limited protection against homologous intranasal challenge. PLoS ONE 3, e2487. doi: 10.1371/journal.pone.0002487
Peng, K., and Monack, D. M. (2010). Indoleamine 2,3-dioxygenase 1 is a lung-specific innate immune defense mechanism that inhibits growth of Francisella tularensis tryptophan auxotrophs. Infect. Immun. 78, 2723–2733.
Qin, A., and Mann, B. J. (2006). Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 6, 69. doi: 10.1186/1471-2180-6-69
Quarry, J. E., Isherwood, K. E., Michell, S. L., Diaper, H., Titball, R. W., and Oyston, P. C. (2007). A Francisella tularensis subspecies novicida purF mutant, but not a purA mutant, induces protective immunity to tularemia in mice. Vaccine 25, 2011–2018.
Raghunathan, A., Shin, S., and Daefler, S. (2010). Systems approach to investigating host-pathogen interactions in infections with the biothreat agent Francisella. Constraints-based model of Francisella tularensis. BMC Syst. Biol. 4, 118. doi: 10.1186/1752-0509-4-118
Ramakrishnan, G., Meeker, A., and Dragulev, B. (2008). fslE is necessary for siderophore-mediated iron acquisition in Francisella tularensis Schu S4. J. Bacteriol. 190, 5353–5361.
Santic, M., Pavokovic, G., Jones, S., Asare, R., and Kwaik, Y. A. (2010). Regulation of apoptosis and anti-apoptosis signalling by Francisella tularensis. Microbes Infect. 12, 126–134.
Schulert, G. S., McCaffrey, R. L., Buchan, B. W., Lindemann, S. R., Hollenback, C., Jones, B. D., and Allen, L. A. (2009). Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infect. Immun. 77, 1324–1336.
Sjostedt, A. (2007). Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann. N. Y. Acad. Sci. 1105, 1–29.
Su, J., Yang, J., Zhao, D., Kawula, T. H., Banas, J. A., and Zhang, J. R. (2007). Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 75, 3089–3101.
Sullivan, J. T., Jeffery, E. F., Shannon, J. D., and Ramakrishnan, G. (2006). Characterization of the siderophore of Francisella tularensis and role of fslA in siderophore production. J. Bacteriol. 188, 3785–3795.
Tacket, C. O., and Levine, M. M. (2007). CVD 908, CVD 908-htrA, and CVD 909 live oral typhoid vaccines: a logical progression. Clin. Infect. Dis. 45(Suppl. 1), S20–S23.
Taylor, M. W., and Feng, G. S. (1991). Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 5, 2516–2522.
Tchawa Yimga, M., Leatham, M. P., Allen, J. H., Laux, D. C., Conway, T., and Cohen, P. S. (2006). Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect. Immun. 74, 1130–1140.
Tempel, R., Lai, X. H., Crosa, L., Kozlowicz, B., and Heffron, F. (2006). Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect. Immun. 74, 5095–5105.
Titball, R. W., Sjostedt, A., Pavelka, M. S. Jr., and Nano, F. E. (2007). Biosafety and selectable markers. Ann. N. Y. Acad. Sci. 1105, 405–417.
Traub, A., Mager, J., and Grossowicz, N. (1955). Studies on the nutrition of Pasteurella tularensis. J. Bacteriol. 70, 60–69.
Wehrly, T. D., Chong, A., Virtaneva, K., Sturdevant, D. E., Child, R., Edwards, J. A., Brouwer, D., Nair, V., Fischer, E. R., Wicke, L., Curda, A. J., Kupko, J. J. 3rd, Martens, C., Crane, D. D., Bosio, C. M., Porcella, S. F., and Celli, J. (2009). Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell. Microbiol. 11, 1128–1150.
Weiss, D. S., Brotcke, A., Henry, T., Margolis, J. J., Chan, K., and Monack, D. M. (2007). In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U.S.A. 104, 6037–6042.
Keywords: Francisella tularensis, pathogenesis, metabolism
Citation: Meibom KL and Charbit A (2010) Francisella tularensis metabolism and its relation to virulence. Front. Microbio. 1:140. doi: 10.3389/fmicb.2010.00140
Received: 26 October 2010;
Accepted: 13 December 2010;
Published online: 24 December 2010.
Edited by:
Anders Sjostedt, Umeå University, SwedenReviewed by:
Antje Flieger, Robert Koch Institute, GermanyJürgen Heesemann, Max von Pettenkofer-Institute, Germany
Copyright: © 2010 Meibom and Charbit. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Alain Charbit, Faculté de Médecine Necker, 156, rue de Vaugirard, 75730 Paris Cedex 15, France. e-mail: alain.charbit@inserm.fr