- Institute for Biological Sciences, National Research Council Canada, Ottawa, ON, Canada
Francisella tularensis is the causative agent of a spectrum of diseases collectively known as tularemia. The extreme virulence of the pathogen in humans, combined with the low infectious dose and the ease of dissemination by aerosol have led to concerns about its abuse as a bioweapon. Until recently, nothing was known about the virulence mechanisms and even now, there is still a relatively poor understanding of pathogen virulence. Completion of increasing numbers of Francisella genome sequences, combined with comparative genomics and proteomics studies, are contributing to the knowledge in this area. Tularemia may be treated with antibiotics, but there is currently no licensed vaccine. An attenuated strain, the Live Vaccine Strain (LVS) has been used to vaccinate military and at risk laboratory personnel, but safety concerns mean that it is unlikely to be licensed by the FDA for general use. Little is known about the protective immunity induced by vaccination with LVS, in humans or animal models. Immunoproteomics studies with sera from infected humans or vaccinated mouse strains, are being used in gel-based or proteome microarray approaches to give insight into the humoral immune response. In addition, these data have the potential to be exploited in the identification of new diagnostic or protective antigens, the design of next generation live vaccine strains, and the development of subunit vaccines. Herein, we briefly review the current knowledge from Francisella comparative proteomics studies and then focus upon the findings from immunoproteomics approaches.
Introduction
The intracellular pathogen, Francisella tularensis, is the etiological agent of tularemia in humans and animals. It is increasingly being isolated in the United States and several European countries (Eliasson et al., 2006; Vorou et al., 2007), although humans are an accidental host. The genus Francisella contains the species F. tularensis, F. novicida, and F. philomiragia. F. tularensis is further subdivided into three subspecies: F. tularensis subspecies tularensis (type A), F. tularensis subspecies holarctica (type B), and F. tularensis subspecies mediaasiatica (Sjostedt, 2001). Strains of subspecies tularensis and holarctica are primarily responsible for human disease, whilst F. philomiragia and F. novicida are avirulent in healthy humans. F. tularensis subsp. mediaasiatica is a restricted entity with unique biochemical characteristics that has only been isolated in Kazakhstan and Turkmenistan in Central Asia and exhibits virulence in rabbits similar to Type B strains. In recent years, F. tularensis has gained significant attention as one of six organisms designated as high priority agents that could be exploited as agents of bioterror (category A pathogens) by the US Center for Disease Control and Prevention. Combined, the low infectious dose and ease of dissemination of type A F. tularensis have made it a threat to both military personnel and civilians alike (Dennis et al., 2001).
There is currently no licensed vaccine available in North America, although an attenuated type B strain, known as the Live Vaccine Strain (LVS), has been used to vaccinate military personnel and laboratory workers. This strain was derived from a Soviet strain in the 1960s and was not rationally attenuated, leading to concerns regarding residual virulence. In addition, the lack of knowledge of the attenuating mutations and mechanisms of protection means that LVS is unlikely to be licensed for use in the general population in the near future. Despite these concerns, LVS remains the gold standard against which new tularemia vaccine candidates will be judged. LVS is also currently the only tularemia vaccine candidate to have been evaluated and shown to be effective in humans, and after the terrorist attacks of 2001, there has been renewed interest in improving the manufacturing and testing of LVS (Pasetti et al., 2008). In addition, LVS remains virulent in animals, making it an attractive surrogate for virulent strains of the pathogen, which require use of specialized containment facilities.
Despite the recent surge of interest in F. tularensis, there remain many unknowns, for example in the mechanisms of pathogen virulence or the host immune response. The prevailing dogma is that humoral immunity plays a critical role in defense against extracellular pathogens, whilst cell-mediated immunity is more important for clearance of intracellular pathogens, such as F. tularensis. It is unclear whether this is true for Francisella, and whether the roles of the immune system are different for type A and B strains. Recent studies have confirmed the role of cell-mediated immunity in protection against tularemia, and in addition the importance of humoral immunity is also now recognized (reviewed in Kirimanjeswara et al., 2008). Many laboratories have reported that a robust anti-Francisella antibody response is generated in humans within 2 weeks of LVS vaccination or actual infection (Koskela and Herva, 1982; Koskela and Salminen, 1985; Tarnvik, 1989; Waag et al., 1995; Janovska et al., 2007b), but the role of these antibodies in protective immunity remains unclear. A review of host immunity toward F. tularensis described the current knowledge in more depth and suggests that a synergy between humoral and cell-mediated immunity is required to induce effective protection (Kirimanjeswara et al., 2008). If either LVS is to be licensed, or next generation tularemia vaccines are to be successfully developed, there needs to be an understanding of the immune mechanisms in the host that need to be activated to induce protective immunity. This in turn requires a fundamental understanding of the mechanisms of virulence of F. tularensis.
For many years knowledge of Francisella virulence factors has been lacking, with no classical bacterial virulence factors having been identified. Despite a marked increase in the intensity of research surrounding Francisella, there is still not a complete explanation of how the pathogen can disseminate and proliferate so readily in the mammalian host. The completion of increasing numbers of Francisella genome sequences, commencing with that of strain SCHU S4 (Karlsson et al., 2000; Prior et al., 2001; Larsson et al., 2005), has propelled comparative Francisella genomics (Samrakandi et al., 2004; Rohmer et al., 2006, 2007; Champion et al., 2009) and proteomics studies (Hubalek et al., 2004; Twine et al., 2005a; Pavkova et al., 2006) toward identification of putative virulence genes and proteins. Genome comparisons (reviewed in Titball and Petrosino, 2007) have aided in the mapping of Francisella evolution and adaptation to different environmental niches (Karlsson et al., 2000; Prior et al., 2001; Larsson et al., 2005), whilst comparisons of the proteomes of in vitro grown strains of Francisella differing in virulence have revealed both similarities and differences in the profile of expressed proteins (Hubalek et al., 2004; Twine et al., 2005a; Pavkova et al., 2006). Such differences may shed light upon the molecular mechanisms of the marked virulence differences between the subspecies.
Proteomics studies have also extended to the characterization of the repertoire of proteins reactive with sera from convalescent or vaccinated subjects. The broad study of large sets of proteins involved in the host immune response has been termed “immunoproteomics,” and provides information regarding Francisella immunodominant antigens. This is increasing knowledge of Francisella proteins that stimulate the immune system, which can then be used in the development of subunit vaccines and diagnostics, in addition to determining potential correlates of protection. Given the relative rarity of human cases of tularemia, many of the studies of the host response to vaccination or infection with tularemia have been carried out in animal models of tularemia. The murine model of tularemia is widely used (animal models of tularemia were recently reviewed in Rick and Wu, 2007). Animal studies in the past and more recently have used rabbits (Larson, 1946; Bell et al., 1955; Skrodzki, 1961), rats (Downs and Coriell, 1947; Downs et al., 1949; Raymond and Conlan, 2009; Wu et al., 2009; Ray et al., 2010), guinea pigs (Bell et al., 1955; Eigelsbach and Downs, 1961; Eigelsbach et al., 1961), and non-human primates (Eigelsbach et al., 1962; Nelson et al., 2010).
In this article we briefly give an overview of comparative studies of Francisella proteomes, approaches used to study the immunoproteome of Francisella, and the characteristics of the reported immunoreactive proteins.
The Francisella Proteome
Comparative Proteomics of Francisella
Comparative genomics studies and molecular typing methods provide evidence that F. novicida, could be the common ancestor of F. tularensis subspecies holarctica, subspecies mediaasiatica and subspecies tularensis (Svensson et al., 2005). Type A strains have recently been further divided into type AI and AII, based largely on their geographical distribution (Svensson et al., 2005; Staples et al., 2006) and have been shown to differ in virulence in the mouse model of tularemia (Twine et al., 2006c; Molins et al., 2010). Genomic comparisons have allowed advances in explaining the differing virulence and infectivity of the four F. tularensis subspecies, but are insufficient to offer a complete understanding. Thus, proteomics studies have also been carried out, comparing the in vitro protein expression profiles of all four subspecies in hopes of addressing the differences in virulence that genomic studies alone, cannot (Hubalek et al., 2004; Pavkova et al., 2006).
The results of such studies have shown that very few differences exist at the proteome level between strains within the same subspecies, but a much greater number of differences were observed in proteome maps comparing the subspecies (Hubalek et al., 2004). The authors concluded that the proteomes of strains from subspecies tularensis and mediaasiatica showed greater similarity to one another than to the proteomes of strains from subspecies holarctica (Hubalek et al., 2004; Pavkova et al., 2006). The variations observed included differences in protein abundance, as well as the apparent presence and absence of specific proteins. Charge variants of certain proteins may account for some of the detected differences in protein abundance between subspecies, but it is also thought that the amount and method of gene expression and regulation may vary between subspecies of F. tularensis.
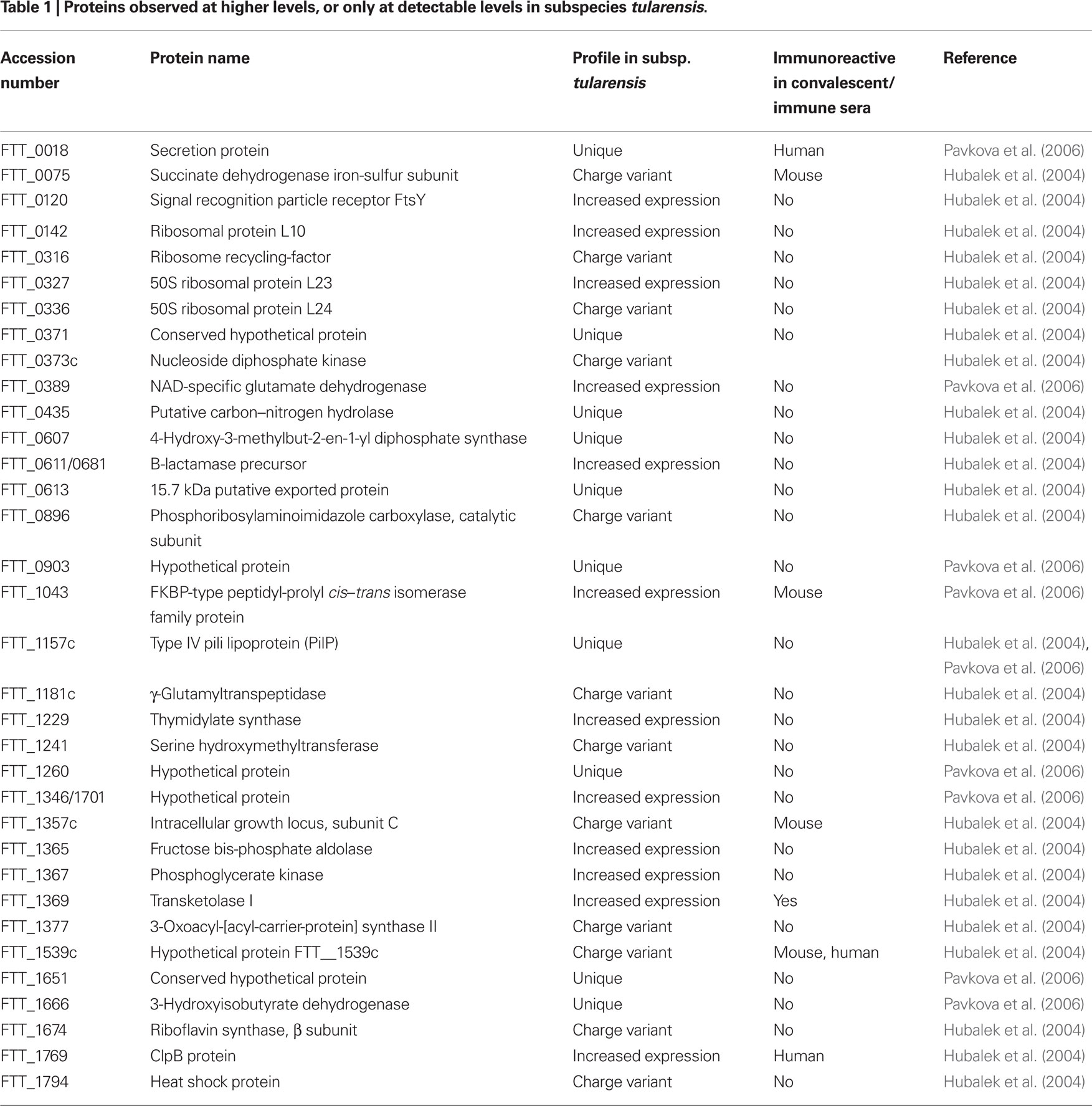
Species novicida and mediaasiatica are essentially avirulent in humans, therefore, of particular interest are proteins that are uniquely expressed, or up-regulated in both subspecies tularensis and holarctica, but not in novicida or mediaasiatica. Such differences in the proteomes of these subspecies may provide insight into the mechanisms used by virulent subspecies of F. tularensis. Using two-dimensional electrophoresis (2DE), Hubalek et al. (2004) compared the proteomes of representative strains of subspecies tularensis, holarctica, and mediaasiatica and identified 27 proteins that were either unique to, or produced in at least a twofold greater abundance in subspecies tularensis. These proteins are listed in Table 1, and the similarities and differences summarized in Figure 1. Seventeen of these proteins, included in Table 1, were found to have charge or mass variants present in the less virulent subspecies, possibly as a result of strain specific amino acid substitutions, or differences in the post-translational modification of the proteins in question (Hubalek et al., 2004). An additional nine proteins; signal recognition particle receptor FtsY (FTT_0120), ribosomal protein L10 (FTT_0142), 50S ribosomal protein L23 (FTT_0327), B-lactamase precursor (FTT_0611/0681), thymidylate synthase (FTT_1229), fructose bis-phosphate aldolase (FTT_1365), phosphoglycerate kinase (FTT_1367), transketolase I (FTT_1369), ClpB protein (FTT_1769), also shown in Table 1, were reported to be produced with at least twofold greater abundance in subspecies tularensis when compared to either mediaasiatica or holarctica. Heat shock protein ClpB has been shown to play a vital role in the multiplication of Francisella in target organs during infection, and thus the overall virulence and infectivity of the bacteria (Meibom et al., 2008). Finally, three proteins reported in this study were only detected in 2DE of subspecies tularensis; FTT_0607, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, FTT_0435, a carbon–nitrogen hydrolase and FTT_1157, a type IV pili lipoprotein. Type IV pilin proteins, such as PilA have been shown to play a significant role as virulence factors of Francisella (Forslund et al., 2006). The loss of the pilA gene also represents one of the major events that has led to the marked attenuation of the type B LVS strain (Salomonsson et al., 2009). While the pilA gene is present in F. novicida, and in pathogenic strains of subspecies holarctica, it does not appear that it functions as a component of a functional type IV pilin protein, but rather serves as a secretion system (Hager et al., 2006).
Figure 1. Using 2D-PAGE coupled with mass spectrometric methods for protein identification, Hubalek et al. (2004) identified proteins that were unique to specific subspecies, or differentially expressed between them.
A small number of proteins that were expressed at detectable levels only in subspecies tularensis, or appear to be expressed in greater amounts in this subspecies have also been found to be immunoreactive in immune or convalescent sera, as highlighted in Table 1. These include the putative virulence factor IglC, which is expressed as a unique charge variant in subspecies tularensis, as well as an FKBP-type peptidyl-prolyl cis–trans isomerase family protein (FTT_1043). Whilst it has not been conclusively shown to be true in Francisella, homologs of the latter protein in other intracellular pathogens such as Legionella, Neisseria, Chlamydia, and Trypanosomes play a key role in virulence and ensuring the uptake of these pathogens by host cells (Hacker et al., 1993; Ludwig et al., 1994; Moro et al., 1995; Leuzzi et al., 2005).
Also using gel-based comparative proteomics, the proteome of an attenuated, spontaneous mutant of SCHU S4, was compared with the parent strain, SCHU S4 (Twine et al., 2005a). The attenuated strain, denoted FSC043, was found to be avirulent in mice and outperformed LVS as a live vaccine strain (Twine et al., 2005a). The study determined that the strain had undergone a recombination event, resulting in the expression of a fusion protein, comprising the N-terminal half of the hypothetical protein FTT_0918 and the C-terminal half of hypothetical FTT_0919 as a single protein (Twine et al., 2005a). A similar defect, resulting in the expression of a homologous protein in LVS was also observed (Twine et al., 2005a), and has been shown to be a key cause of the attenuation of LVS in mice (Salomonsson et al., 2009). Early studies did not determine the role of the protein, FTT_0918, but show the utility of the corresponding deletion mutant as a rationally attenuated live vaccine strain in the mouse model of tularemia (Twine et al., 2005a). Since then, there have been several studies investigating the role of these proteins, which have no homology to any other proteins listed in the NCBInr. They have been proposed to be a novel family of potentially membrane associated proteins and involved in iron uptake and regulation (Lindgren et al., 2009). One study has denoted FTT_0918 as fupA, and the hybrid gene in LVS as fupA/B, with fupAB having a significant role in the siderophore-mediated iron uptake mechanism of LVS (Sen et al., 2010). The functionality of the fusion protein in the attenuated SCHU S4 strain has not been determined. However, genomics studies alone would have been unable to predict the expression of the fusion protein. This information has since been exploited in tularemia vaccine development for the construction of rationally attenuated live vaccine strains (Kadzhaev et al., 2009).
In order to work toward more complete proteome coverage, many complementary approaches need to be undertaken, including subfractionation of the proteome into, for example, the membrane subproteome or glycoproteome.
The Francisella Membrane Proteome
Membrane proteins, particularly those exposed to the extracellular environment, can play important roles in the initial infection stages and the overall virulence and survival of the bacteria within a host (Haake et al., 2000). Membrane proteins have also been viewed as having the potential to act as effective subunit vaccine candidates given the frequency with which they are able to promote an immune response in a host cell (Sjostedt et al., 1992a; Huntley et al., 2007). Thus, efforts have been placed on identifying and characterizing Francisella’s membrane proteins (Pavkova et al., 2005, 2006; Melillo et al., 2006; Huntley et al., 2007). Early studies aimed at characterizing outer membrane proteins (OMPs) from F. tularensis studies carried out with LVS used bulk membrane extraction techniques, including sonication of cells followed by ultracentrifugation and/or detergent extraction. These studies did identify OMPs, and assessed their potential as subunit vaccines (Sandstrom et al., 1987; Surcel et al., 1989; Ericsson et al., 1994a; Fulop et al., 1996), but also suffered low level contamination with periplasmic and/or cytoplasmic components (Sandstrom et al., 1987; Surcel et al., 1989; Ericsson et al., 1994a; Fulop et al., 1996). More recent studies have used sodium carbonate enrichment of membrane proteins, in which the addition of sodium carbonate preferentially causes membranes to form open membrane sheets, whilst cytoplasmic and peripheral membrane proteins are released in soluble form (Fujiki et al., 1982). Combined with gel-based proteomics approaches, two studies attempted to characterize the membrane enriched fraction of LVS (Pavkova et al., 2005; Twine et al., 2005b). Studies identified numerous candidate OMPS, although using the PSORT1b algorithm as a predictor of subcellular protein location, fewer than 10 proteins were predicted to be OMPs. One group then went further to identify immunogenic OMPs, using sera from LVS vaccinated mice. Of the 36 identified immunoreactive proteins, PSORTb analyses showed that the majority of the proteins identified in this study were more closely associated with the cytoplasm and non-membranous regions of the cell, thereby reflecting the relatively low specificity of the sodium carbonate enrichment for membrane proteins (Havlasova et al., 2005).
Using density gradient centrifugation, OMPs from LVS and SCHU S4 were purified, leading to the identification of 12 OMPs, which are listed in Table 2 (Huntley et al., 2007). Interestingly, some of the proteins in the enriched native OMP preparation were then shown to be immunogenic with F. tularensis infected C3H/HeN mice, with antibodies produced toward KatG, PilQ, GroEL, ATP synthase, OmpA, FopA, and Tul4-A (Huntley et al., 2007).
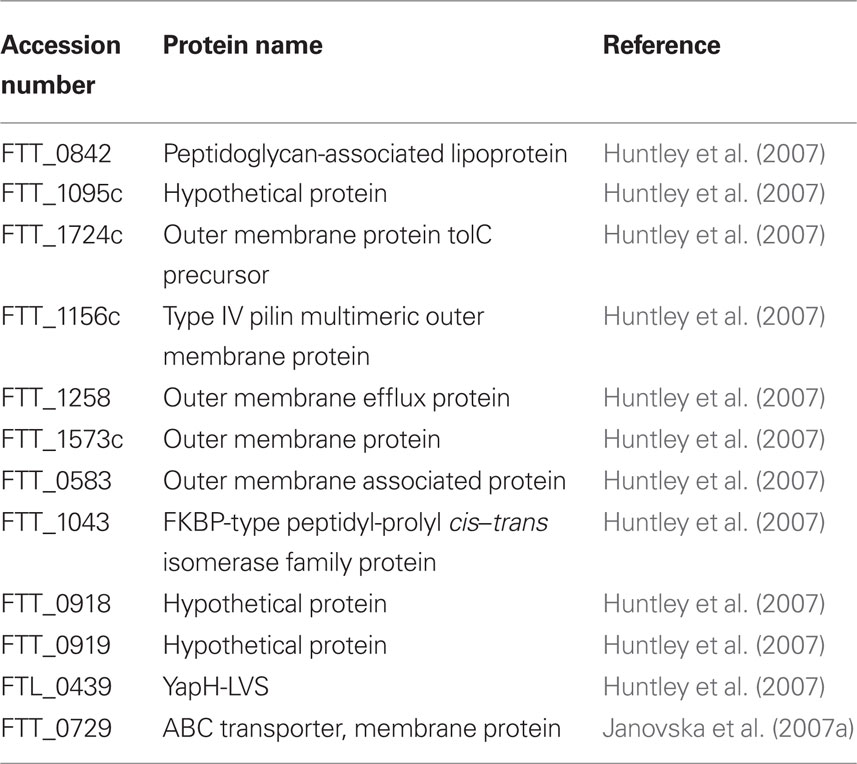
Table 2. Francisella tularensis LVS outer membrane proteins, isolated using density gradient centrifugation.
Putative outer membrane or cell surface exposed proteins have been identified in Francisella as possibly playing a role in the adherence of the bacteria to human lung cells. Using surface biotinylation and protein purification by the use of immobilized streptavidin beads, one study identified seven putative surface exposed proteins; Chaperone protein DnaK (FTT_1269), chaperone protein GroEL (FTT_1696), hypothetical membrane protein (FTT_0119), Outer membrane associated protein FopA (FTT_0583), intracellular growth locus, subunit A (FTT_1714), conserved hypothetical lipoprotein (FTT_1347), and hypothetical protein (FTT_1441), also identified as bacterioferritin in LVS (Melillo et al., 2006). Of particular interest to this study was hypothetical membrane protein FTT_0119, also annotated as FsaP in LVS. When expressed in a non-adhering Escherichia coli strain, this protein enabled E. coli to adhere to A549 human lung epithelial cells. Furthermore, it has been shown that FsaP is present on the cell surface of F. tularensis subsp. holarctica, which readily adheres to this same cell line. However, it is absent from the cell surface of F. novicida, which is unable to effectively adhere to A549 cells. Combined with the observed increased expression this protein in vivo (Twine et al., 2006a), and the production of antibodies in response to FsaP during tularemia infection, FTT_0119 could become a point of interest in exploring the pathogenesis of F. tularensis (Melillo et al., 2006).
The Francisella Glycoproteome
The long prevailing dogma that bacteria cannot modify proteins with carbohydrate moieties is being met with an increasing amount of evidence to the contrary. Discovery and characterization of bacterial glycoproteins presents many challenges, including the often unknown nature of the modifying carbohydrate moiety. Many studies focus upon characterization of a single purified protein whilst others are attempting to conduct more encompassing studies of the entire bacterial glycoproteome. Preliminary work strongly suggests that F. tularensis strains modify multiple proteins with unknown glycan moieties, although the covalent attachment of carbohydrate moieties to specific proteins has yet to be demonstrated.
Often the first indication that a protein is modified by glycan is aberrant migration on 1D- or 2D-PAGE. A type IV pilin protein, pilA (FTT_0890) was shown using 2DE to migrate to 18 kDa; a higher molecular weight than that predicted by the pilA gene sequence (14 kDa), suggesting post-translational modification (Forslund et al., 2006). Additionally, pilA from Francisella was cloned into a strain of P. aeruginosa that is known not to glycosylate type IV pilin proteins. Francisella PilA was expressed, but further analysis showed that the approximate molecular weight was 14 kDa; close to the mass predicted from the translated pilA gene sequence. Evidence of homologous pilin protein glycosylation in other intracellular, bacterial pathogens such as Neisseria and some strains of P. aeruginosa (Stimson et al., 1995; Banerjee and Ghosh, 2003) also supports the hypothesis that this protein may be glycosylated in Francisella.
Another study, exploited glycoproteomics approaches developed and optimized for the detection of eukaryotic glycans. Balonova et al. (2010) reported the identification of 31 putative Francisella glycoproteins. Detection and enrichment of putative glycoproteins was carried out using an in-gel glycostaining kit, and a lectin based glycan differentiation. A total of 11 proteins were observed to be reactive with the gel-based glycostain, which relies upon carbohydrate diol groups, which are oxidized to aldehydes, and subsequently form a stable hydrozone in reaction to a fluorescently labeled hydrazide. Lectin affinity resulted in the enrichment and identification of 20 putative glycoproteins, although none have to date been conclusively reported as glycoproteins and the role of protein glycosylation in Francisella pathogenesis is unknown.
The Francisella in vivo Proteome
The capacity of F. tularensis to cause disease appears to be a reflection of its ability to multiply intracellularly and damage various host organs rather than its ability to produce any specific toxins. This requires F. tularensis to subvert or otherwise avoid a variety of host defenses that possess the potential to kill it. In particular, F. tularensis multiplies extensively in macrophages in vitro and in vivo (Golovliov et al., 1997; Conlan et al., 2002). Studies have been carried out which attempt to mimic one or more of the stress responses to which F. tularensis may be exposed during proliferation within the host. Changes in the bacterial proteome in response to exposure to hydrogen peroxide, hoping to mimic the an oxidative stress response showed elevated levels of chaperonins such as GroEL, DnaK and stress response proteins such as ClpB, SodB (Ericsson et al., 1994b; Lenco et al., 2005; Twine et al., 2006a). Virulence factors, such as the pathogenicity island protein, IglC were also identified in some of these studies. Other studies have grown strains under conditions of iron restriction (Lenco et al., 2007), and manipulated growth temperatures (Lenco et al., 2009) hoping to mimic conditions to which the pathogen will be exposed within the host environment. Such in vitro studies offer a facile means of dissecting the response of the bacterial proteome to individual stress conditions, and some groups are working toward developing in vitro growth conditions that more closely mimic the host environment (Hazlett et al., 2008). Others have studied bacteria during intracellular growth in a macrophage model of tularemia and identified induced proteins (Golovliov et al., 1997; Kovarova et al., 2002). However, these studies still cannot necessarily accurately represent the myriad of stimuli to which the bacterium is exposed in the host environment. Attempting to study the proteome of bacteria growing within host tissues is extremely challenging, with limited reports of proteomes of pathogens isolated from host tissues (Becker et al., 2006; Twine et al., 2006a). An immunomagnetic separation approach was used to rapidly isolate bacteria from spleen tissues of mice infected with type A F. tularensis strain FSC033. In total, 78 proteins were shown to be differentially expressed when compared to the proteomes of the strain grown in liquid media. Of the proteins increased in expression some, such as IglC and chaperonins are also observed at higher levels in bacteria exposed to oxidative stress conditions. There were, however, a small number of proteins that were only observed at detectable levels in bacteria isolated from spleen tissues, including a cobalamin synthesis protein, universal stress protein and glycine cleavage system protein. These proteins are likely to be factors required for intracellular adaptation of the bacterium, rather than virulence factors. This study was the first attempt to determine the repertoire of proteins expressed by F. tularensis during proliferation in the hostile host environment, yet only provides a snapshot of the bacterial proteome, toward the latter stages of tularemia. Of greater interest, but also much more challenging, would be the isolation and proteome characterization of bacteria at the site of infection, during dissemination to host organs.
Francisella Virulence Factors
Also included in comparative proteomics studies, has been the analysis of the Francisella pathogenicity island (FPI); a highly conserved 16–19 gene cluster found in each of the four Francisella subspecies. In all subspecies the FPI is present in two identical copies and has been shown to code for several putative virulence factors. Within the FPI are 16 genes that have been found to be highly conserved across each of the four subspecies, leaving an additional 2–3 genes that display greater variability. These variable genes; pmcA and pdpD in F. novicida, may be absent from subspecies such as holarctica or present but interrupted or abbreviated, as in subspecies tularensis. However, the levels at which the FPI gene products are produced can vary between subspecies. PigA (FTT_1701/1346), for example, is present in a two- to sixfold greater abundance in SCHU S4, a subspecies tularensis strain, than in other subspecies (Pavkova et al., 2006). It has also been suggested however, that virulence factors coded for by the FPI can be regulated by genes not contained in the FPI itself. MglA, for example, has been found to be a major regulator of several virulence factors encoded both within and outside of the FPI (Lauriano et al., 2004).
The iglABCD operon is one of the most highly studied regions within the FPI with IglC being among the first of the FPI proteins to be elucidated. In a study by Golovliov et al. (1997), protein induction of F. tularensis during growth in macrophages was explored. Using protein radiolabeling coupled with 1D- and 2D-PAGE, a novel 23 kDa protein, later identified as IglC, was determined to be significantly induced during growth in macrophages and was sequenced using Edman degradation (Golovliov et al., 1997). Here, the gene products represent putative virulence factors, and their absence renders Francisella either avirulent or extremely attenuated (Golovliov et al., 2003b; Nix et al., 2006). Without IglA, F. tularensis has been shown to be entirely avirulent in a chicken embryo model, due to its inability to grow within macrophages (Nix et al., 2006). This growth has been found to be an intrinsic part of the pathogenicity of F. tularensis (Nix et al., 2006). Additional research has speculated that IglA associates with IglB in the cytoplasm, as they have been found to co-precipitate and insertions into the iglB gene results in the loss of detection of IglA as well (de Bruin et al., 2007). Such observations have led to the suggestion that the removal of either IglA or IglB may allow for the intracellular degradation of the other (de Bruin et al., 2007).
Modifications to IglC and its expression also result in a mutant that is severely hindered in its ability to grow in macrophages (Golovliov et al., 2003a). IglD too, is essential for intracellular replication in both mouse and monocyte derived macrophage studies (Santic et al., 2005).Where growth is possible, deletions of the iglC gene in LVS, for example, often resulted in a mutant unable to escape from phagosomes and it was therefore unable to effectively disseminate from the original site of infection (Telepnev et al., 2005). Regardless of the subspecies in which IglC was removed, mutants exhibited avirulence in the models they were tested in Golovliov et al. (2003a). It does not appear, however, that iglC plays a significant role in regulating protein expression during oxidative stress (Lenco et al., 2005). A ΔiglC LVS mutant showed very similar changes in protein expression to that of LVS when in the presence of hydrogen peroxide; namely the upregulation of approximately 10 proteins. The only significant difference between protein expression of LVS and ΔiglC in the presence of hydrogen peroxide was the increased expression of AhpC/TSA family protein in the ΔiglC mutant (Lenco et al., 2005).
The FPI itself contains several putative virulence factors, many of which may be common to all four Francisella subspecies, but it is not the only region of the genome that contains potential virulence factors.
The list of proteins with a potential role in pathogenesis is extensive. Despite the apparent diversity amongst these proteins though, there are several main classes into which these factors can be divided, including the FPI-encoded virulence factors that have already been discussed. Often, possible virulence factors are surface associated and may be part of a capsule, the lipopolysaccharide (LPS) or having to do with a type IV pili system. Proteins, such as the pilin subunit have been shown in Francisella strains, to have a role in host-cell recognition, virulence (Forslund et al., 2006, 2010), protein secretion (Hager et al., 2006), and adherence to host cells (Chakraborty et al., 2008). FTT_0918, a hypothetical protein present in both subsp. tularensis and in the LVS strain of holarctica, is thought to be membrane associated (Huntley et al., 2007). Transcriptional regulators such as MglA have also been shown to have an essential role in the virulence and pathogenesis of Francisella (Lauriano et al., 2004). Francisella also possesses large numbers of hypothetical proteins and lipoproteins, such as conserved hypothetical protein FTT_1103, that have been implicated as virulence factors (Qin et al., 2009). The virulence factors described here were elucidated through various proteomics studies and only begin to unravel Francisella’s mechanisms of virulence.
Approaches to Study Reactivity of Francisella Proteins with Antibodies
Scientists have been studying the antibody response to vaccination or infection with Francisella for many years. This has been accomplished by methods such as agglutination (Engelfried and Spear, 1966), ELISA (Carlsson et al., 1979), and 1D-Western blotting (Bevanger et al., 1988). Early work was often unable to definitively identify the immunoreactive proteins, but more recently 2D-Western blotting combined with protein identification by mass spectrometry has been exploited. The term “immunoproteomics” has been coined to describe such studies. A newer approach, proteome microarray technology, prints recombinant proteins on glass slides to allow high throughput screening of immune sera. Each approach is outlined briefly here, including a discussion of the strengths and weaknesses of each.
Western Blotting
The 2D-Western blotting approach, combines 2D separation of the antigen with Western blotting. The antigen used in these studies is usually a bacterial cell lysate, or subproteome fraction (e.g., membrane) of in vitro grown bacteria. 2D-PAGE resolves the majority of bacterial proteins to a single protein spot, and retains the native protein processing and post-translational modifications. Proteins are transferred to nitrocellulose or PVDF membrane, and probed with primary sera and conjugated secondary antibody, as per traditional Western blotting (protocol is outlined in Gallagher et al., 2008). Proteins may be stained after transfer to a membrane, and the captured image used to align regions of immunoreactivity with areas of protein staining. Excising the identified immunoreactive proteins from a second protein stained 2D-PAGE, and subsequent digestion with trypsin allows identification of proteins using mass spectrometry based techniques (e.g., MS/MS).
2D-Western blotting is one of the most accessible immunoproteomics approaches, that can be carried out in any laboratory equipped with gel-based electrophoresis and electroblotting equipment. It has, therefore, been adopted in many immunoproteomics studies. Despite its wide use, the approach has some disadvantages, which are largely related to limitations of gel-based 2D protein separations, including difficulties in resolving very large, small, hydrophobic or basic proteins. The analysis is also limited to those proteins expressed by bacteria under in vitro growth conditions, which may not be representative of the proteome expressed by the pathogen when proliferating in the host environment. 2D-Western blotting using bacteria isolated from host tissues has been carried out in tularemia proteomics work (Twine et al., 2006a), however the limited amount of antigen isolated makes this challenging to do with more than a few serum samples. In addition, the abundance of expressed proteins, whatever the growth conditions, can vary from a few copy numbers per cell to millions, therefore the observed intensity of immunoreactivity may not be representative of the immunogenicity of the protein, but its abundance. The relatively low throughput of 2D-Western blotting and mass spectrometry can be a bottleneck in immunoproteomics studies. Large format 2D gels offer superior proteome resolution, but the complete 2D-Western blotting experiment typically takes 4–5 days to perform. Experiments can be multiplexed, depending upon the resources of the lab, but this is still a relatively slow and laborious process. Despite the limitations of this approach, it remains by far the most accessible approach for most laboratories.
Immunoproteomics Using a Protein Microarray
The bacterial proteome microarray has been described in a number of reports, including two studies conducted with Francisella immune sera (Eyles et al., 2007; Sundaresh et al., 2007). The construction of the bacterial proteome microarray consists of three steps; a single-round PCR to amplify each open reading frame (ORF), followed by in vivo recombination cloning and in vitro protein expression and microarray printing (Eyles et al., 2007). Specifically, the PCR primers are designed with a gene specific portion, and an adaptor sequence. The adaptor sequences flank the amplified gene and are homologous to portions of the cloning vector (pXT7). The PCR products are cloned into the expression vector by in vivo homologous recombination using competent E. coli DH5α cells. The resulting clone also harbors an ATG start codon, polyhistidine and hemagglutinin tags. Proteins are expressed using commercially available in vitro transcription/translation kits and the resulting proteins printed directly onto nitrocellulose coated glass slides. This method allows the entire proteome of F. tularensis SCHU S4, comprising 1741 ORFs onto a single slide (Eyles et al., 2007). The chips are then treated in a manner analogous to traditional Western blotting; the chips are incubated with blocking buffer, then primary immune sera, washed, incubated with a conjugated secondary antibody, with detection of the fluorescent conjugate performed by a microarray scanner. It is reported that upwards of 800 sera could be process in a single experiment.
The proteome microarray has many advantages compared with 2D-Western blotting approach. Firstly, the antigens are presumably present at equal concentrations in contrast with gel-based approaches, where the concentration of each protein is dependent upon expression levels during in vitro bacterial growth. In addition, if desired the entire theoretical proteome of the organism can be interrogated. Proteome microarrays also offer multiplexing, high throughput, and reduced serum volume requirements (2 μl versus ∼50–100 μl for large 2D-Western blot). However, the proteins printed on the chip likely do not harbor native post-translational modifications, and may not be post-translationally processed in the same manner as native Francisella proteins, a drawback common to most recombinant protein expression systems. Other disadvantages include the cost and feasibility; many labs which are equipped to perform standard proteomic experiments are not able to fabricate proteome microarrays, and analysis, at present, is limited to specialist laboratories.
Immunoproteomics of Francisella Tularensis
In the following sections, we will review the immunoproteomics studies reported in the literature to date, including characteristics of the identified immunoreactive proteins and a brief comparison of the proteins identified using gel-based and proteome microarray studies.
Immunoproteomics in the Murine Model of Tularemia
The majority of our recent knowledge on the pathogenesis of F. tularensis infection has been derived from studies of mice infected with either the attenuated live vaccine strain (LVS) or virulent strains of F. tularensis by the intradermal (i.d.) or respiratory route (Golovliov et al., 1996; Conlan et al., 2003; Elkins et al., 2003; Wu et al., 2005) and more recently oral route (Kuolee et al., 2007). Studies have included mice of various genetic backgrounds, including immunodeficient mice (Chen et al., 2004), mouse strains deficient in Toll-like receptor 4 (TLR4) signaling (Macela et al., 1996). In the following sections, we review the current knowledge of Francisella immunoproteomics, carried out using the murine model of tularemia.
Successful versus unsuccessful vaccination of mice
TLR4, the signal transducing element of the LPS receptor complex, is thought to play an important role in innate immunity against Gram-negative bacteria (Underhill, 2004). TLR4-defective (TLR4d) mice (C3H/HeJ) are reported to be more susceptible to subcutaneous challenge with LVS than wild type (TLR4+/+) mice (C3H/HeN). The LD50 of the pathogen was 100-fold lower for C3H/HeJ mice compared with that for C3H/HeN mice (Macela et al., 1996). A follow-up study used gel-based immunoproteomics to compare the repertoire of immunoreactive proteins generated by C3H/HeN and C3H/HeJ mice in response to infection with LVS (Havlasova et al., 2005). The immunoreactive proteins identified in this study are summarized in Table 3. Of particular interest, this study monitored the immunoblotting patterns of sera drawn from mice over a 28-day period post-LVS infection, and reported that sera from infected C3H/HeJ had higher antibody titers, compared with C3H/HeN mice, up to 21 days post-infection. Despite this observation, the antibody patterns observed for each mouse strain were directed toward an almost identical subset of antigens (Havlasova et al., 2005). This study suggests, that in these mouse strains, protective immunity may not be dependent upon antibodies.
Table 3. Proteins which are recognized by sera from convalescent humans or from LVS vaccinated mice.
Another study exploited the differing susceptibility of four mouse strains to protective vaccination with an LVS strain derived from ATCC LVS, in order determine whether a subset of immunoreactive proteins would be predictive of protective vaccination. LVS inoculated intradermally elicits a similar sub-lethal infection in the skin, liver, and spleen of both BALB/c and C57BL/6 mice that persists for approximately 2 weeks (Chen et al., 2003). However, whereas this infection renders BALB/c and CH3/HeN mice immune to a subsequent systemic challenge with a virulent type A strain of F. tularensis, it fails to protect C57BL/6 mouse strains from a 100-fold smaller challenge (Chen et al., 2003). The repertoire of proteins reactive only with sera from mouse strains that were successfully vaccinated with LVS (BALB/c, CH3/HeN) were compared with immunoreactive proteins from strains that cannot be successfully vaccinated with LVS (C57BL/6, DBA). The difference in protective immunity of these mouse strains was hoped to reveal specific antigenic markers of protective immunity. The mouse strains successfully vaccinated with LVS, harbored antibodies toward a small set of proteins, that were not reactive with immune sera from vaccinated but unprotected mice, but no overall conclusion regarding patterns of protective immunoreactivity were drawn (Twine et al., 2006b; Table 3).
A more recent study used an almost identical approach of exploiting the varying ability of LVS vaccination to protect BALB/c and C57/BL6 mouse strains against type A challenge with LVS. This study differed in the LVS preparation, using a new lot of LVS, denoted lot 17. This LVS vaccine lot was produced in compliance with Current Good Manufacturing Practice (CGMP) guidelines and this new vaccine formulation was evaluated in a recent Phase 1 clinical study in humans (El Sahly et al., 2009). When compared to the previous work (Twine et al., 2006b), there are distinct differences in the observed immunoreactive proteins, which could potentially be attributed to the vaccinating strain of LVS. However, this study, identified nine proteins that were reactive with sera from lot 17 LVS vaccinated BALB/c mice, that were not observed to be reactive with sera from unsuccessfully vaccinated C57/BL6 mice (Twine et al., 2010). The proteins are listed in Table 3 and in addition, Figure 2 shows the similarities and differences in the profile of immunoreactive proteins identified in this study. The majority of the nine proteins have been observed to be reactive with sera from other murine or human tularemia studies and could have potential as markers of successful vaccination.
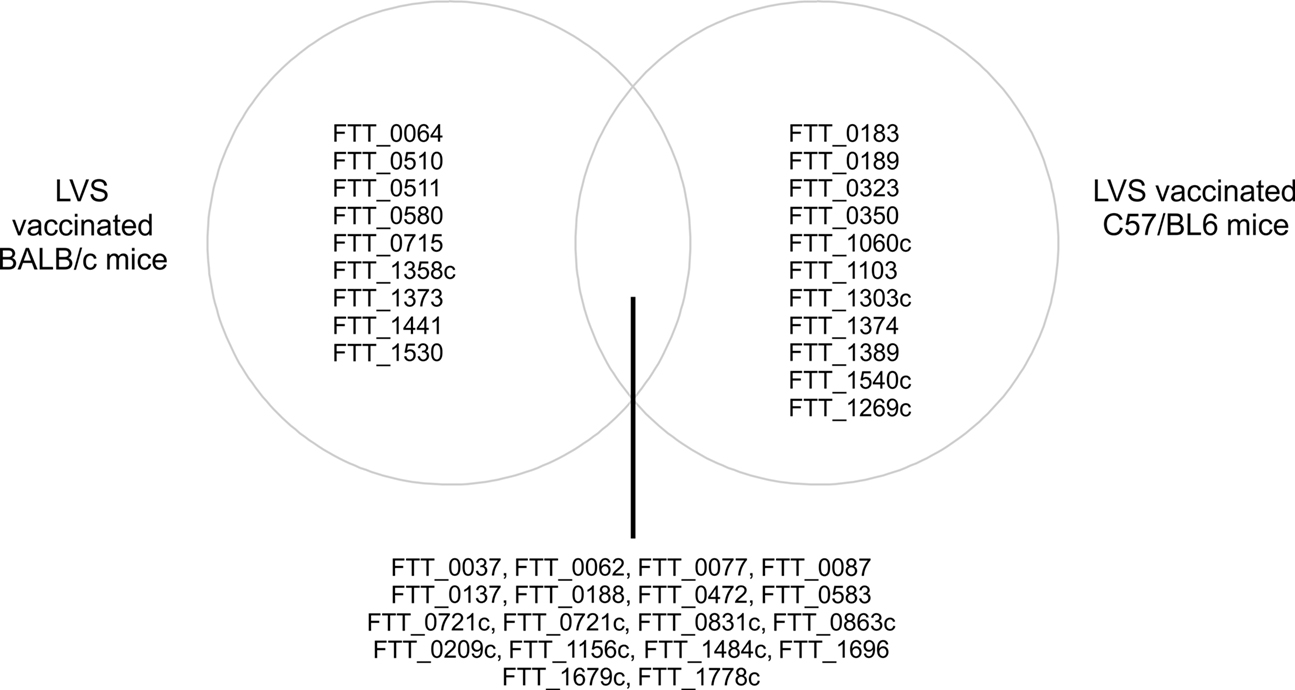
Figure 2. Venn diagram showing the intersection of immunoreactive proteins observed to be reactive with sera from LVS vaccinated and protected BALB/c mice and LVS vaccinated and unprotected C57/BL6 mice (Twine et al., 2010). Left hand side lists immunoreactive proteins only observed to be reactive with sera from LVS vaccinated BALB/c mice, right hand side shows proteins only reactive with sera from C57/BL6 mice.
Vaccination of mice with killed LVS
During the 1920s and 1930s there was a significant effort to develop killed F. tularensis vaccines, due to the reduced intrinsic safety and liability concerns, compared with live vaccines. Studies with heat or formalin killed Francisella strains showed that vaccines made using these methods are generally ineffective (Foshay et al., 1942; Kadull et al., 1950). However, recent work showed that LVS killed by irradiation, and administered to mice in combination with either alum, and immunostimulating complexes (ISCOMs) or CpG afforded some protection against challenge with virulent type B or A strains (Eyles et al., 2007). In addition, the authors used proteome microarray to determine the repertoire of antibodies generated in response to vaccination of mice with killed LVS and adjuvants. Overall, the murine antibody response was limited to a small subset of antigens within the total SCHU S4 proteome, 48 in total (Table 3). Of these, 11 of the top 12 immunoreactive proteins had previously been identified from studies using 2D-Western blotting approach. Interestingly, the study observed few differences in the profile of immunoreactive proteins generated in response to vaccination of mice with (1) viable LVS or (2) killed LVS combined with ISCOMs or (3) killed LVS combined with ISCOMs and CpG (Eyles et al., 2007; Table 3). This suggests that combining killed LVS with adjuvant may stimulate the humoral immune system similarly to live LVS. Killed LVS without any adjuvant produced a profile similar to killed LVS in alum or CpG, but the antibody titers were lower and reactive against just eight antigens. Overall, the data presented showed that killed LVS with a Th-1 promoting adjuvant (e.g., ISCOMS or CpG) confers protection against ID challenge with virulent type B strains and some protection against SCHU S4 challenge. Comparing the immunoreactive profiles of sera from mice vaccinated with the various preparations, it is interesting to note that the proteins FTT_1296, and FTT_0119 were reactive with sera from mice immunized with viable LVS, and killed LVS combined with ISCOMS or ISCOMS and CPG, perhaps suggesting that these antigens might be indicative of successful protection against challenge with virulent type B strains. In addition, some antigens, for example FTT_1696, and FTT_1484 were reactive in all live and killed LVS preparations and are likely not predictive of protective immunity in this case (Eyles et al., 2007).
Combined, the murine immunoproteomics studies have raised the possibility of using seroconversion to a particular subset of antigens of LVS as surrogates of or correlates of protection. The most challenging task in this regard is demonstrating that findings in mice are applicable to humans.
Immunoproteomics of Human Tularemia
The relative rarity of natural cases of type A tularemia have until recently, hampered the investigation of the humoral immune response in humans, including immunoproteomic studies. At the time of writing, only two studies describe the reactivity of human sera after type A infections, the first using a 2D-Western blotting approach (Janovska et al., 2007b) and the second, using a proteome microarray to screen a large number of sera (Sundaresh et al., 2007). The first study screened sera from a laboratory worker accidentally infected with type A strain SCHU S4 (Janovska et al., 2007b). The subject suffered oculoglandular tularemia, which recurred 17 months after the original infection. Sera collected 2, 5, and 16 years after infection, were used to screen a membrane enriched subproteome of LVS, and 10 immunoreactive proteins were identified (Table 3). Interestingly, the intensity of immunoreactivity toward these proteins showed no apparent decline over the three serum samples screened, with the exception of a loss of reactivity toward the hypothetical protein FTT_0918, which was only observed to be reactive with sera collected 2 years post-infection (Janovska et al., 2007b). This is in contrast with studies of patients who have recovered from type B infections. In such studies, a decline in overall antibody titers has been observed in the 25-year period post-infection (Ericsson et al., 1994a). Overall, only 3 of the 10 immunoreactive proteins observed in the study of the infected laboratory worker; intracellular growth locus, subunit B (FTT_1713c), pyruvate dehydrogenase, E1 component (FTT_1485c), and Succinyl-CoA synthetase subunit beta (FTT_0504c), have not thus far been observed to be reactive with sera from vaccinated mice, or convalescent sera from type B infected individuals.
The Francisella proteome microarray was used to screen sera from patients recovering from either type A or B Francisella infections contracted in North America (Sundaresh et al., 2007). Of the 46 sera from infected individuals, 10 were from confirmed cases of type A tularemia and 5 from confirmed type B tularemia. The immunodominant proteins were reported in this study (Table 3), and of the top 10 antigens, many have also been reported in immunoproteomics studies carried out with sera from LVS infected mice. In addition, a sufficient number of sera were screened, that a comparison of the repertoire of proteins reactive with sera from patients recovering from infection with different subtypes of Francisella and different routes of infection could be carried out. For example, authors noted that the protein FTT_1484 was more reactive with sera from patients suffering with ulceroglandular tularemia, compared with the sera from patients with the pneumonic form of the disease. Also, the protein FTT_0975 is more reactive with sera from type B infected individuals compared to type A infected individuals, although the authors note that a rigorous statistical assessment of these observations was outside the scope of their study.
Immunoproteomics was used to survey the repertoire of immunoreactive proteins with sera of human type B convalescents (Janovska et al., 2007a). This study described the reactivity of serum collected from nine tularemia patients with a membrane enriched fraction of LVS, and the proteome of LVS grown under oxidative stress conditions. Details of the type of tularemia, or the time after diagnosis that sera were drawn were not provided, presumably due to patient confidentiality. The immunoproteomics profiles of patient sera showed marked heterogeneity, with a limited number of commonly reactive proteins observed. This variability in profile of immunoreactive proteins has also been noted with human patient sera from other diseases, such as Helicobacter pylori (Kimmel et al., 2000; Krah et al., 2004). Of the 35 immunoreactive proteins identified (Table 3), a small proportion were reactive with the majority of sera screened, as illustrated in Figure 3; the antigens pyruvate dehydrogenase E2 component (FTT_1484), dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydrogenase component (FTT_077), chaperonin protein GroEL (FTT_1696), acetyl-CoA carboxylase (FTT_0472), hypothetical protein (FTT_1441) and 50S ribosomal protein L7/L12 (FTT_0143) were reactive with six or more of the sera screened. Of note, although the protein FTT_0077 was reactive with eight of the nine patient sera screened, no single protein was observed to be reactive with all of the patient sera in this study.
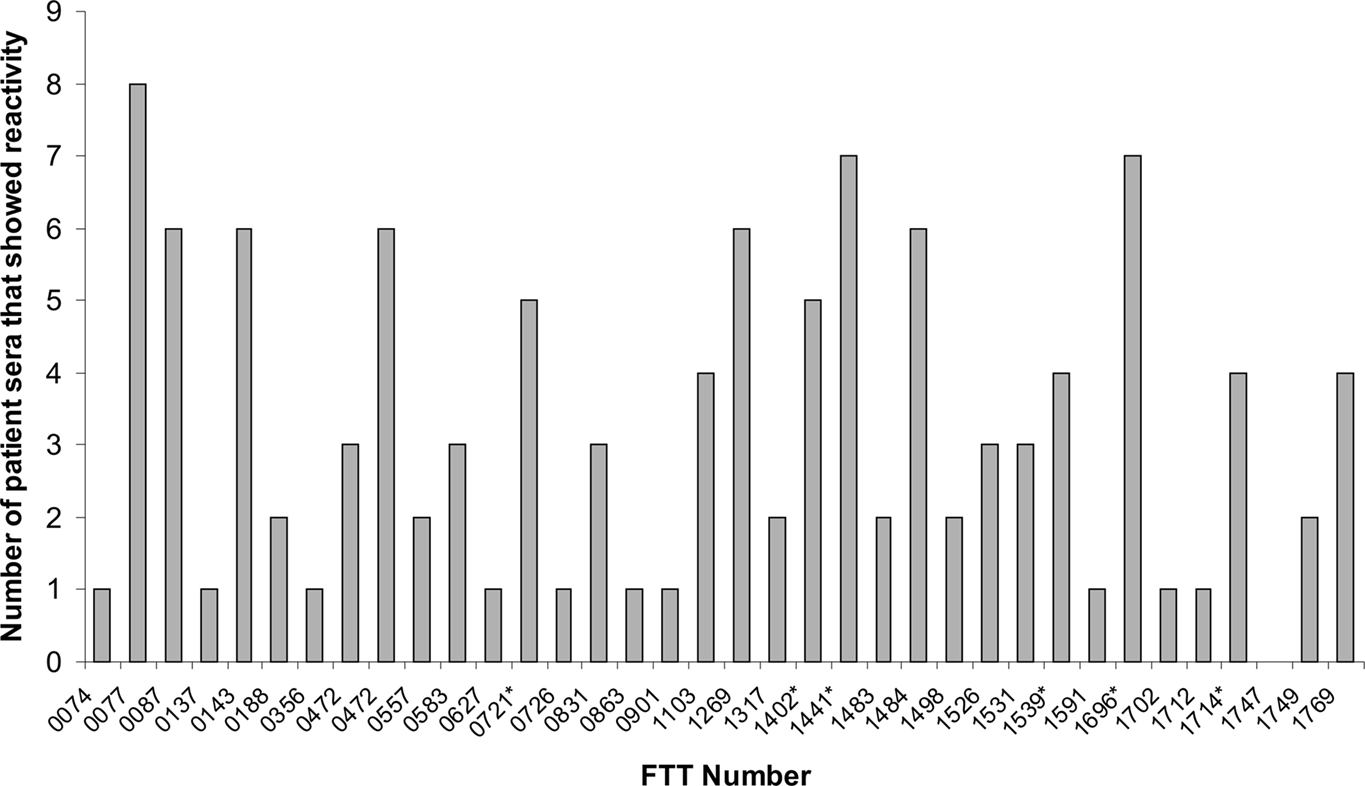
Figure 3. Frequency with which immunoreactive proteins from LVS proteome were observed to react with sera from patients recovering from type B Francisella tularensis infections. FTT_number refers to the locus tag of the ORF within the SCHU S4 genome sequence. *Indicates that sera reacted with isoforms of the same protein, shown is the highest observed frequency of reactivity. These data were originally reported by and tabulated in Havlasova et al. (2002).
Characteristics of the Total Reported Repertoire of Immunoreactive Proteins
Table 3 summarizes the antigenic proteins reported in recent immunoproteomics studies. Commonalities in the identified proteins across various studies can be noted and will be reviewed here, but comprehensive comparisons of the immunoproteomics data are hampered by distinct differences in the studies, including the immunoproteomics approach used, the host organism, the bacterial strain, source of strain, or treatment of the strain used for either infection or immunization, and the bacterial strain or subproteome used as the antigen. In total, 143 antigenic proteins were identified in 11 separate studies. Thirteen of these proteins were reported to be immunoreactive in half of the studies (FTT_1696, FTT_1269c, FTT_1696, FTT_0077, FTT_0137, FTT_0472, FTT_0583, FTT_0721c, FTT_0143, FTT_1103, FTT_0863, FTT_1484c). Of these proteins, FTT_2369c, FTT_1696, FTT_0583, FTT_0721c, and FTT_1103 were also noted to be components of LVS and SCHU S4 outer membrane preparations (Huntley et al., 2007).
Examining the immunoreactive proteins by Clusters of Orthologous groups, antigenic proteins are observed to belong to a diverse array of functional categories (Figure 4), but proteins involved in energy production, chaperonins and proteins of unknown function were highly represented. Using the PSORT1b algorithm as a predictor of protein localization within the bacterial cell (Nakai and Horton, 1999; Gardy et al., 2003; Rey et al., 2005), the vast majority of the identified immunoreactive proteins were predicted to be cytoplasmic (44%) or of unknown location (42%; Figure 4). This is in contrast with the results from proteome microarray studies alone, where half of the identified proteins were classified as membrane or surface associated (Eyles et al., 2007). This is likely a reflection of the number of immunoproteomics studies carried out using gel-based techniques, which are well documented to underrepresent very large or hydrophobic membrane proteins. The high proportion of proteins that have no predicted location reflects the high number of hypothetical proteins within the Francisella proteome, which have little or no homology to known proteins.
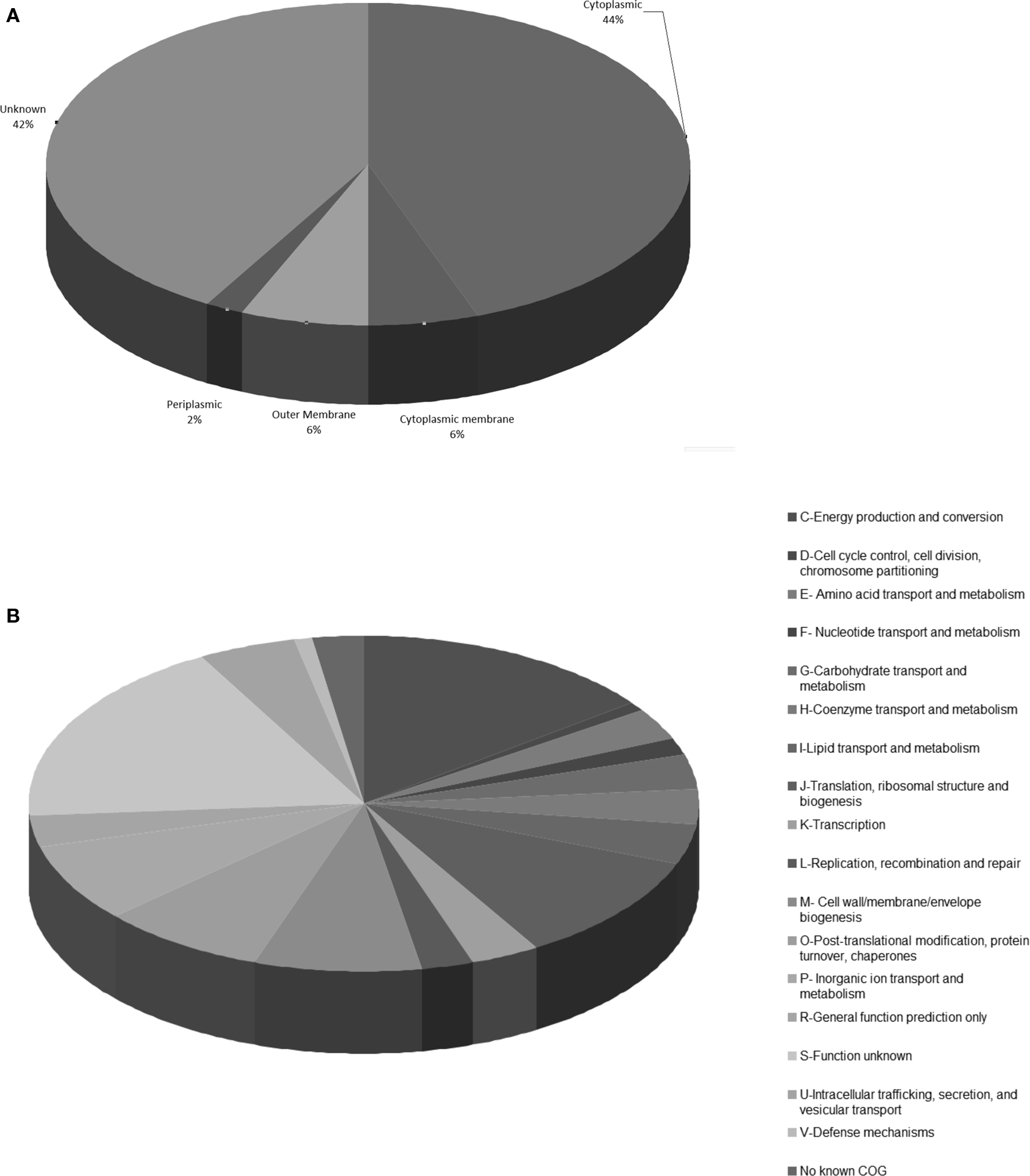
Figure 4. Basic bioinformatic characterization of the repertoire of reported Francisella immunoreactive proteins. Graphical representation of (A) clusters of orthologous groups (COGs) and (B) predicted subcellular location using PSORT algorithm, for total reported immunoreactive proteins from Francisella immunoproteomics studies. Taken from the reported immunoreactive proteins listed in Table 3.
Although underrepresented in the entire dataset, membrane proteins are of some interest in immunoproteomics studies. Exposed on the outer bacterial surface, OMPs are generally involved in pathogen evasion of the host, intracellular survival and immune evasion. Characterization of LVS outer membrane proteins, listed 12 OMPs (Huntley et al., 2008). Of these, 10 were observed to be immunoreactive with sera from either murine or human immunoproteomics studies. This LVS outer membrane preparation has been reported to show potential as an acellular vaccine; vaccination of mice with LVS native OMPs resulted in 50% survival rate over 20 days post-challenge with SCHU S4 (Huntley et al., 2008). Also observed was a three log reduction in bacterial burdens in the liver and spleen compared with sham vaccinated animals. This clearly suggests a link between OMPs and protective immunity.
Of the OMPs identified by a proteomics approach (Huntley et al., 2008), some have been identified previously, with demonstrated immunoreactivity. For example, the first presumed F. tularensis OMP was a 43-kDa protein identified by probing lithium chloride extracts of bacteria with antisera collected from individuals involved in an outbreak of tularemia in Norway (Ericsson et al., 1994a). This protein was named FopA, for Francisella outer membrane protein. Two separate vaccine trials demonstrated that FopA was not protective against type A F. tularensis or LVS challenge, despite its induction of antibodies (Fulop et al., 1995, 1996). FopA was observed to be immunoreactive in many of the immunoproteomics studies described herein.
The second reported presumed F. tularensis OMP was a 17-kDa T lymphocyte-reactive protein originally identified from an N-lauroylsarcosinate-insoluble protein preparation; this 17-kDa polypeptide was later named TUL4 (Sjostedt et al., 1990). TUL4 is conserved in F. tularensis and F. novicida strains, whereas an immunologically related protein is present in F. philomiragia. Additional studies confirmed the ability of TUL4 to stimulate lymphocyte proliferation, primarily CD4 T cells, and noted marked production of interleukin-2 (IL-2) and gamma interferon (IFN-γ) in response to TUL4 (Sjostedt et al., 1992a,b; Golovliov et al., 1995; Valentino et al., 2009). Initial [3H]palmitate radiolabeling and detergent extraction of F. tularensis suggested that TUL4 was an integral membrane lipoprotein (LP; Sjostedt et al., 1991). Bacterial lipoproteins have an inflammatory capacity and ability to stimulate cells of the innate immune system via TLR2 (Thakran et al., 2008). Although the lipoprotein TUL4 is not essential for virulence of F. tularensis (Forestal et al., 2008), amino acids 86–99 of the protein were identified as an immunodominant epitope of CD4T cells in B6 mice (Valentino et al., 2009). A recent study showed that the hypothetical lipoprotein, denoted FTT_1416c or FTL0645 of LVS, was enriched in a sarkosyl membrane extraction. Expression of the equivalent recombinant protein in E. coli produced a lipoprotein that was able to activate TLR2 and induce an immunogenic response in mice (Parra et al., 2009). The lipoprotein was also shown to be immunoreactive with immune sera from mice in other work.
Chaperonin proteins feature frequently in tularemia immunoproteomics studies. The chaperonin protein DnaK and GroEL (Cpn60) were reactive with the vast majority of sera screened. Chaperonins, also known as heat shock proteins are among the most highly conserved protein families in nature and are expressed in both prokaryotes and eukaryotes (Ranford and Henderson, 2002). These proteins facilitate the non-covalent assembly of proteins and when cells are exposed to stress conditions, such as temperature or pH, chaperones protect cellular proteins from aggregation and also promote refolding (Stewart et al., 2004). Chaperonins have also been observed to be activators of the innate immune system (Kol et al., 1999; Wallin et al., 2002), in addition to stimulating monocytes, macrophages and dendritic cells to produce nitric oxide and a variety of cytokines (Kol et al., 1999; Wallin et al., 2002). Although most heat shock proteins are widely accepted to be cellular proteins some, for example GroEL can also be released from cells, and stimulate a immune response (Henderson et al., 2006). Other studies have reported that GroEL and DnaK of Mycobacterium tuberculosis and F. tularensis are located either on the cell surface (Hickey et al., 2009) or partition with membrane protein subproteome (Huntley et al., 2007). GroEL of F. tularensis, has been observed at increased levels under conditions of stress (Ericsson et al., 1994b; Twine et al., 2006a) and has been detected in the cytosol of host cells infected with the bacterium (Lee et al., 2006). It has been suggested that the predominant source of Francisella chaperonins in the host are derived from bacteria that are either extracellular in the blood (Forestal et al., 2007) or lungs (Bosio et al., 2007) or from bacteria that fail to proliferate in the host environment. GroEL has also been reported to have an inflammatory role, stimulating macrophages via TLR4, but not human endothelial cells (Noah et al., 2010). In addition, GroEL and LPS derived from LVS acted together to elicit a synergistic response in macrophages (Noah et al., 2010). Similarly, another Francisella heat shock protein, DnaK, has been reported to mediate activation of dendritic cells via TLR4 (Ashtekar et al., 2008).
Heat shock proteins (Hsps) have been reported to be dominant antigens for the host immune response to various pathogens, and have been attempted to be used in subunit vaccines against some pathogens (Khan et al., 2009). The potential of Hsps to elicit both cell-mediated and humoral immune responses even in the absence of exogenous adjuvants (Lowrie et al., 1997; Harmala et al., 2002), their requirement in small quantities and ability to elicit memory T cell response, make them attractive vaccine candidates against infectious disease.
As noted in this article, the relative rarity of naturally occurring human tularemia and the ethical issues with human LVS vaccination and challenge studies, means that much of our recent knowledge stems from the murine model of tularemia. A significant challenge in this regard, is determining whether findings in mice are applicable to humans. Reviewing the complement of identified immunoreactive proteins listed in Table 3 and also in Figure 5, shows the overlap between antigens reactive with both murine and human sera. This shows that the 43 antigens have been observed to be reactive with LVS vaccinated murine and human convalescent sera.
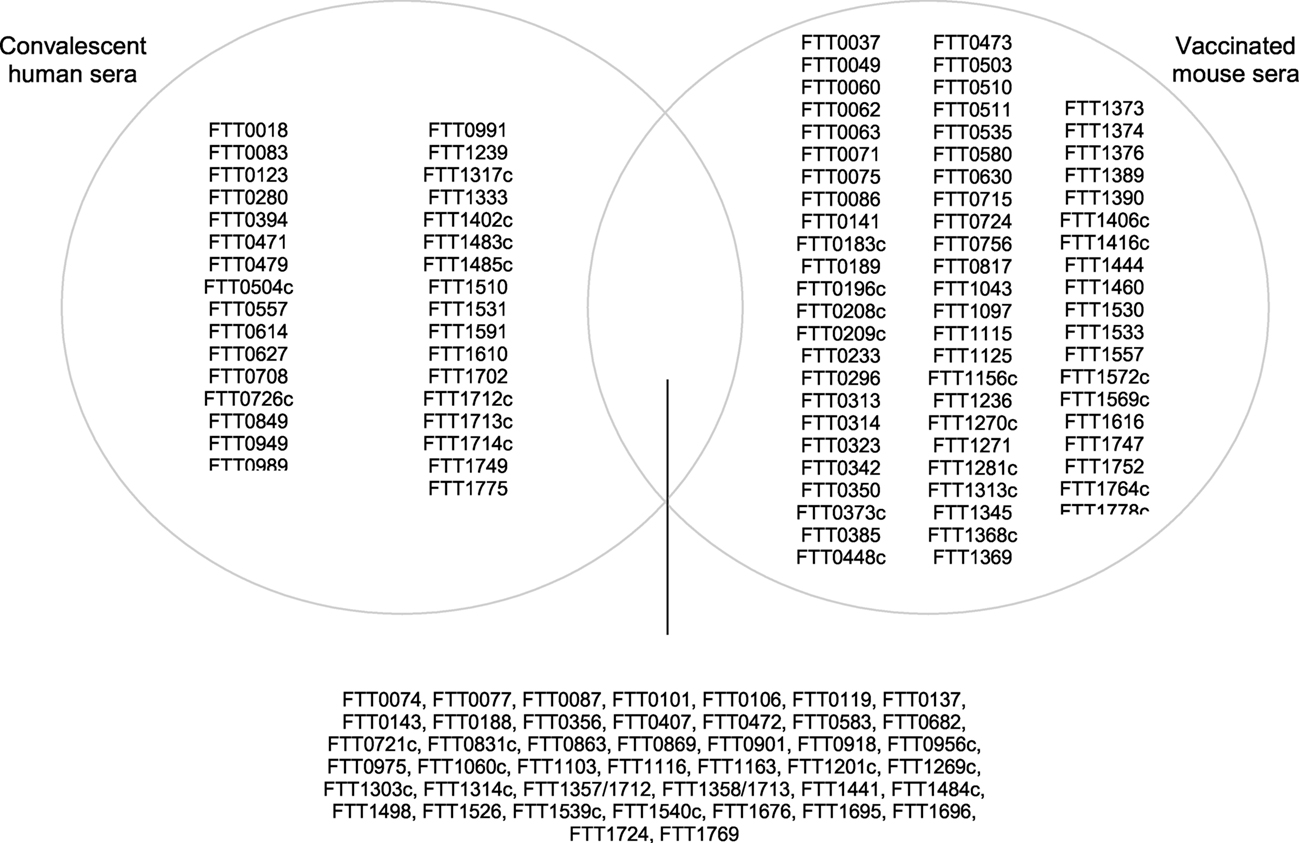
Figure 5. Venn diagram showing the intersection of immunoreactive proteins observed to be reactive with sera from LVS vaccinated mice and human convalescent. From Table 3.
Conclusion
There continues to be a significant focus upon the development of efficacious, safe and licensable tularemia vaccines. Acellular or subunit vaccines carry far less risk than attenuated live vaccines, but there are limited reports of success with subunit tularemia vaccine development. The lack of knowledge of both Francisella virulence factors, protective antigens and the complex nature of immunity to Francisella is inhibiting vaccine development. Further studies of the humoral immune response, especially those in other animal models of tularemia that will bridge the differences between mice and humans, will be required to determine those antigens that can be predicted to be protective in humans. This will aid greatly in the development of subunit vaccines and selection of antigens for incorporation into diagnostic tests.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
The authors thank Kelly Fulton, for critical reading of the manuscript.
References
Ashtekar, A. R., Zhang, P., Katz, J., Deivanayagam, C. C., Rallabhandi, P., Vogel, S. N., and Michalek, S. M. (2008). TLR4-mediated activation of dendritic cells by the heat shock protein DnaK from Francisella tularensis. J. Leukoc. Biol. 84, 1434–1446.
Balonova, L., Hernychova, L., Mann, B. F., Link, M., Bilkova, Z., Novotny, M. V., and Stulik, J. (2010). Multimethodological approach to identification of glycoproteins from the proteome of Francisella tularensis, an intracellular microorganism. J. Proteome Res. 9, 1995–2005.
Banerjee, A., and Ghosh, S. K. (2003). The role of pilin glycan in neisserial pathogenesis. Mol. Cell. Biochem. 253, 179–190.
Becker, D., Selbach, M., Rollenhagen, C., Ballmaier, M., Meyer, T. F., Mann, M., and Bumann, D. (2006). Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440, 303–307.
Bell, J. F., Owen, C. R., and Larson, C. L. (1955). Virulence of Bacterium tularense. I. A study of the virulence of Bacterium tularense in mice, guinea pigs, and rabbits. J. Infect. Dis. 97, 162–166.
Bevanger, L., Maeland, J. A., and Naess, A. I. (1988). Agglutinins and antibodies to Francisella tularensis outer membrane antigens in the early diagnosis of disease during an outbreak of tularemia. J. Clin. Microbiol. 26, 433–437.
Bosio, C. M., Bielefeldt-Ohmann, H., and Belisle, J. T. (2007). Active suppression of the pulmonary immune response by Francisella tularensis Schu4. J. Immunol. 178, 4538–4547.
Carlsson, H. E., Lindberg, A. A., Lindberg, G., Hederstedt, B., Karlsson, K. A., and Agell, B. O. (1979). Enzyme-linked immunosorbent assay for immunological diagnosis of human tularemia. J. Clin. Microbiol. 10, 615–621.
Chakraborty, S., Monfett, M., Maier, T. M., Benach, J. L., Frank, D. W., and Thanassi, D. G. (2008). Type IV pili in Francisella tularensis: roles of pilF and pilT in fiber assembly, host cell adherence, and virulence. Infect. Immun. 76, 2852–2861.
Champion, M. D., Zeng, Q., Nix, E. B., Nano, F. E., Keim, P., Kodira, C. D., Borowsky, M., Young, S., Koehrsen, M., Engels, R., Pearson, M., Howarth, C., Larson, L., White, J., Alvarado, L., Forsman, M., Bearden, S. W., Sjostedt, A., Titball, R., Michell, S. L., Birren, B., and Galagan, J. (2009). Comparative genomic characterization of Francisella tularensis strains belonging to low and high virulence subspecies. PLoS Pathog. 5:e1000459. doi: 10.1371/journal.ppat.1000459
Chen, W., Kuolee, R., Shen, H., and Conlan, J. W. (2004). Susceptibility of immunodeficient mice to aerosol and systemic infection with virulent strains of Francisella tularensis. Microb. Pathog. 36, 311–318.
Chen, W., Shen, H., Webb, A., Kuolee, R., and Conlan, J. W. (2003). Tularemia in BALB/c and C57BL/6 mice vaccinated with Francisella tularensis LVS and challenged intradermally, or by aerosol with virulent isolates of the pathogen: protection varies depending on pathogen virulence, route of exposure, and host genetic background. Vaccine 21, 3690–3700.
Conlan, J. W., Chen, W., Shen, H., Webb, A., and Kuolee, R. (2003). Experimental tularemia in mice challenged by aerosol or intradermally with virulent strains of Francisella tularensis: bacteriologic and histopathologic studies. Microb. Pathog. 34, 239–248.
Conlan, J. W., Kuolee, R., Shen, H., and Webb, A. (2002). Different host defences are required to protect mice from primary systemic vs pulmonary infection with the facultative intracellular bacterial pathogen, Francisella tularensis LVS. Microb. Pathog. 32, 127–134.
de Bruin, O. M., Ludu, J. S., and Nano, F. E. (2007). The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 7, 1. doi: 10.1186/1471-2180-7-1
Dennis, D. T., Inglesby, T. V., Henderson, D. A., Bartlett, J. G., Ascher, M. S., Eitzen, E., Fine, A. D., Friedlander, A. M., Hauer, J., Layton, M., Lillibridge, S. R., McDade, J. E., Osterholm, M. T., O’Toole, T., Parker, G., Perl, T. M., Russell, P. K., and Tonat, K. (2001). Tularemia as a biological weapon: medical and public health management. JAMA 285, 2763–2773.
Downs, C. M., Buchele, L., and Edgar, E. P. (1949). Studies on pathogenesis and immunity in tularemia; the pathogenesis of tularemia in the white rat. J. Immunol. 63, 117–133.
Downs, C. M., and Coriell, L. L. (1947). Studies on tularemia; immunization of white rats. J. Immunol. 56, 229–243.
Eigelsbach, H. T., and Downs, C. M. (1961). Prophylactic effectiveness of live and killed tularemia vaccines. I. Production of vaccine and evaluation in the white mouse and guinea pig. J. Immunol. 87, 415–425.
Eigelsbach, H. T., Tulis, J. J., McGavran, M. H., and White, J. D. (1962). LIVE TULAREMIA VACCINE I. : host–parasite relationship in monkeys vaccinated intracutaneously or aerogenically. J. Bacteriol. 84, 1020–1027.
Eigelsbach, H. T., Tulis, J. J., Overholt, E. L., and Griffith, W. R. (1961). Aerogenic immunization of the monkey and guinea pig with live tularemia vaccine. Proc. Soc. Exp. Biol. Med. 108, 732–734.
El Sahly, H. M., Atmar, R. L., Patel, S. M., Wells, J. M., Cate, T., Ho, M., Guo, K., Pasetti, M. F., Lewis, D. E., Sztein, M. B., and Keitel, W. A. (2009). Safety, reactogenicity and immunogenicity of Francisella tularensis live vaccine strain in humans. Vaccine 27, 905–911.
Eliasson, H., Broman, T., Forsman, M., and Back, E. (2006). Tularemia: current epidemiology and disease management. Infect. Dis. Clin. North Am. 20, 289–311, ix.
Elkins, K. L., Cowley, S. C., and Bosio, C. M. (2003). Innate and adaptive immune responses to an intracellular bacterium, Francisella tularensis live vaccine strain. Microbes Infect. 5, 135–142.
Engelfried, J. J., and Spear, F. (1966). Modified agglutination test for Pasteurella tularensis. Appl. Microbiol. 14, 267–270.
Ericsson, M., Sandstrom, G., Sjostedt, A., and Tarnvik, A. (1994a). Persistence of cell-mediated immunity and decline of humoral immunity to the intracellular bacterium Francisella tularensis 25 years after natural infection. J. Infect. Dis. 170, 110–114.
Ericsson, M., Tarnvik, A., Kuoppa, K., Sandstrom, G., and Sjostedt, A. (1994b). Increased synthesis of DnaK, GroEL, and GroES homologs by Francisella tularensis LVS in response to heat and hydrogen peroxide. Infect. Immun. 62, 178–183.
Eyles, J. E., Unal, B., Hartley, M. G., Newstead, S. L., Flick-Smith, H., Prior, J. L., Oyston, P. C., Randall, A., Mu, Y., Hirst, S., Molina, D. M., Davies, D. H., Milne, T., Griffin, K. F., Baldi, P., Titball, R. W., and Felgner, P. L. (2007). Immunodominant Francisella tularensis antigens identified using proteome microarray. Proteomics 7, 2172–2183.
Forestal, C. A., Gil, H., Monfett, M., Noah, C. E., Platz, G. J., Thanassi, D. G., Benach, J. L., and Furie, M. B. (2008). A conserved and immunodominant lipoprotein of Francisella tularensis is proinflammatory but not essential for virulence. Microb. Pathog. 44, 512–523.
Forestal, C. A., Malik, M., Catlett, S. V., Savitt, A. G., Benach, J. L., Sellati, T. J., and Furie, M. B. (2007). Francisella tularensis has a significant extracellular phase in infected mice. J. Infect. Dis. 196, 134–137.
Forslund, A. L., Kuoppa, K., Svensson, K., Salomonsson, E., Johansson, A., Bystrom, M., Oyston, P. C., Michell, S. L., Titball, R. W., Noppa, L., Frithz-Lindsten, E., Forsman, M., and Forsberg, A. (2006). Direct repeat-mediated deletion of a type IV pilin gene results in major virulence attenuation of Francisella tularensis. Mol. Microbiol. 59, 1818–1830.
Forslund, A. L., Salomonsson, E. N., Golovliov, I., Kuoppa, K., Michell, S., Titball, R., Oyston, P., Noppa, L., Sjostedt, A., and Forsberg, A. (2010). The type IV pilin, PilA, is required for full virulence of Francisella tularensis subspecies tularensis. BMC Microbiol. 10, 227. doi: 10.1186/1471-2180-10-227
Foshay, L., Hesselbrock, W. H., Wittenberg, H. J., and Rodenberg, A. H. (1942). Vaccine prophylaxis against tularemia in man. Am. J. Public Health Nations Health 32, 1131–1145.
Fujiki, Y., Hubbard, A. L., Fowler, S., and Lazarow, P. B. (1982). Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93, 97–102.
Fulop, M., Manchee, R., and Titball, R. (1995). Role of lipopolysaccharide and a major outer membrane protein from Francisella tularensis in the induction of immunity against tularemia. Vaccine 13, 1220–1225.
Fulop, M., Manchee, R., and Titball, R. (1996). Role of two outer membrane antigens in the induction of protective immunity against Francisella tularensis strains of different virulence. FEMS Immunol. Med. Microbiol. 13, 245–247.
Gallagher, S., Winston, S. E., Fuller, S. A., and Hurrell, J. G. (2008). Immunoblotting and immunodetection. Curr. Protoc. Immunol. Chapter 8:Unit 8.10.
Gardy, J. L., Spencer, C., Wang, K., Ester, M., Tusnady, G. E., Simon, I., Hua, S., deFays, K., Lambert, C., Nakai, K., and Brinkman, F. S. (2003). PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 31, 3613–3617.
Golovliov, I., Baranov, V., Krocova, Z., Kovarova, H., and Sjostedt, A. (2003a). An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 71, 5940–5950.
Golovliov, I., Sjostedt, A., Mokrievich, A., and Pavlov, V. (2003b). A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 222, 273–280.
Golovliov, I., Ericsson, M., Akerblom, L., Sandstrom, G., Tarnvik, A., and Sjostedt, A. (1995). Adjuvanticity of ISCOMs incorporating a T cell-reactive lipoprotein of the facultative intracellular pathogen Francisella tularensis. Vaccine 13, 261–267.
Golovliov, I., Ericsson, M., Sandstrom, G., Tarnvik, A., and Sjostedt, A. (1997). Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect. Immun. 65, 2183–2189.
Golovliov, I., Kuoppa, K., Sjostedt, A., Tarnvik, A., and Sandstrom, G. (1996). Cytokine expression in the liver of mice infected with a highly virulent strain of Francisella tularensis. FEMS Immunol. Med. Microbiol. 13, 239–244.
Haake, D. A., Chao, G., Zuerner, R. L., Barnett, J. K., Barnett, D., Mazel, M., Matsunaga, J., Levett, P. N., and Bolin, C. A. (2000). The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 68, 2276–2285.
Hacker, J., Kestler, H., Hoschutzky, H., Jann, K., Lottspeich, F., and Korhonen, T. K. (1993). Cloning and characterization of the S fimbrial adhesin II complex of an Escherichia coli O18:K1 meningitis isolate. Infect. Immun. 61, 544–550.
Hager, A. J., Bolton, D. L., Pelletier, M. R., Brittnacher, M. J., Gallagher, L. A., Kaul, R., Skerrett, S. J., Miller, S. I., and Guina, T. (2006). Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 62, 227–237.
Harmala, L. A., Ingulli, E. G., Curtsinger, J. M., Lucido, M. M., Schmidt, C. S., Weigel, B. J., Blazar, B. R., Mescher, M. F., and Pennell, C. A. (2002). The adjuvant effects of Mycobacterium tuberculosis heat shock protein 70 result from the rapid and prolonged activation of antigen-specific CD8+ T cells in vivo. J. Immunol. 169, 5622–5629.
Havlasova, J., Hernychova, L., Brychta, M., Hubalek, M., Lenco, J., Larsson, P., Lundqvist, M., Forsman, M., Krocova, Z., Stulik, J., and Macela, A. (2005). Proteomic analysis of anti-Francisella tularensis LVS antibody response in murine model of tularemia. Proteomics 5, 2090–2103.
Havlasova, J., Hernychova, L., Halada, P., Pellantova, V., Krejsek, J., Stulik, J., Macela, A., Jungblut, P. R., Larsson, P., and Forsman, M. (2002). Mapping of immunoreactive antigens of Francisella tularensis live vaccine strain. Proteomics 2, 857–867.
Hazlett, K. R., Caldon, S. D., McArthur, D. G., Cirillo, K. A., Kirimanjeswara, G. S., Magguilli, M. L., Malik, M., Shah, A., Broderick, S., Golovliov, I., Metzger, D. W., Rajan, K., Sellati, T. J., and Loegering, D. J. (2008). Adaptation of Francisella tularensis to the mammalian environment is governed by cues which can be mimicked in vitro. Infect. Immun. 76, 4479–4488.
Henderson, B., Allan, E., and Coates, A. R. (2006). Stress wars: the direct role of host and bacterial molecular chaperones in bacterial infection. Infect. Immun. 74, 3693–3706.
Hickey, T. B., Thorson, L. M., Speert, D. P., Daffe, M., and Stokes, R. W. (2009). Mycobacterium tuberculosis Cpn60.2 and DnaK are located on the bacterial surface, where Cpn60.2 facilitates efficient bacterial association with macrophages. Infect. Immun. 77, 3389–3401.
Hubalek, M., Hernychova, L., Brychta, M., Lenco, J., Zechovska, J., and Stulik, J. (2004). Comparative proteome analysis of cellular proteins extracted from highly virulent Francisella tularensis ssp. tularensis and less virulent F. tularensis ssp. holarctica and F. tularensis ssp. mediaasiatica. Proteomics 4, 3048–3060.
Huntley, J. F., Conley, P. G., Hagman, K. E., and Norgard, M. V. (2007). Characterization of Francisella tularensis outer membrane proteins. J. Bacteriol. 189, 561–574.
Huntley, J. F., Conley, P. G., Rasko, D. A., Hagman, K. E., Apicella, M. A., and Norgard, M. V. (2008). Native outer membrane proteins protect mice against pulmonary challenge with virulent type A Francisella tularensis. Infect. Immun. 76, 3664–3671.
Janovska, S., Pavkova, I., Hubalek, M., Lenco, J., Macela, A., and Stulik, J. (2007a). Identification of immunoreactive antigens in membrane proteins enriched fraction from Francisella tularensis LVS. Immunol. Lett. 108, 151–159.
Janovska, S., Pavkova, I., Reichelova, M., Hubaleka, M., Stulik, J., and Macela, A. (2007b). Proteomic analysis of antibody response in a case of laboratory-acquired infection with Francisella tularensis subsp. tularensis. Folia Microbiol. (Praha) 52, 194–198.
Kadull, P. J., Reames, H. R., Coriell, L. L., and Foshay, L. (1950). Studies on tularemia. V. Immunization of man. J. Immunol. 65, 425–435.
Kadzhaev, K., Zingmark, C., Golovliov, I., Bolanowski, M., Shen, H., Conlan, W., and Sjostedt, A. (2009). Identification of genes contributing to the virulence of Francisella tularensis SCHU S4 in a mouse intradermal infection model. PLoS ONE 4, e5463. doi: 10.1371/journal.pone.0005463
Karlsson, J., Prior, R. G., Williams, K., Lindler, L., Brown, K. A., Chatwell, N., Hjalmarsson, K., Loman, N., Mack, K. A., Pallen, M., Popek, M., Sandstrom, G., Sjostedt, A., Svensson, T., Tamas, I., Andersson, S. G., Wren, B. W., Oyston, P. C., and Titball, R. W. (2000). Sequencing of the Francisella tularensis strain Schu 4 genome reveals the shikimate and purine metabolic pathways, targets for the construction of a rationally attenuated auxotrophic vaccine. Microb. Comp. Genomics 5, 25–39.
Khan, M. N., Shukla, D., Bansal, A., Mustoori, S., and Ilavazhagan, G. (2009). Immunogenicity and protective efficacy of GroEL (hsp60) of Streptococcus pneumoniae against lethal infection in mice. FEMS Immunol. Med. Microbiol. 56, 56–62.
Kimmel, B., Bosserhoff, A., Frank, R., Gross, R., Goebel, W., and Beier, D. (2000). Identification of immunodominant antigens from Helicobacter pylori and evaluation of their reactivities with sera from patients with different gastroduodenal pathologies. Infect. Immun. 68, 915–920.
Kirimanjeswara, G. S., Olmos, S., Bakshi, C. S., and Metzger, D. W. (2008). Humoral and cell-mediated immunity to the intracellular pathogen Francisella tularensis. Immunol. Rev. 225, 244–255.
Kol, A., Bourcier, T., Lichtman, A. H., and Libby, P. (1999). Chlamydial and human heat shock protein 60s activate human vascular endothelium, smooth muscle cells, and macrophages. J. Clin. Invest. 103, 571–577.
Koskela, P., and Herva, E. (1982). Cell-mediated and humoral immunity induced by a live Francisella tularensis vaccine. Infect. Immun. 36, 983–989.
Koskela, P., and Salminen, A. (1985). Humoral immunity against Francisella tularensis after natural infection. J. Clin. Microbiol. 22, 973–979.
Kovarova, H., Halada, P., Man, P., Golovliov, I., Krocova, Z., Spacek, J., Porkertova, S., and Necasova, R. (2002). Proteome study of Francisella tularensis live vaccine strain-containing phagosome in Bcg/Nramp1 congenic macrophages: resistant allele contributes to permissive environment and susceptibility to infection. Proteomics 2, 85–93.
Krah, A., Miehlke, S., Pleissner, K. P., Zimny-Arndt, U., Kirsch, C., Lehn, N., Meyer, T. F., Jungblut, P. R., and Aebischer, T. (2004). Identification of candidate antigens for serologic detection of Helicobacter pylori-infected patients with gastric carcinoma. Int. J. Cancer 108, 456–463.
Kuolee, R., Harris, G., Conlan, J. W., and Chen, W. (2007). Oral immunization of mice with the live vaccine strain (LVS) of Francisella tularensis protects mice against respiratory challenge with virulent type A F. tularensis. Vaccine 25, 3781–3791.
Larson, C. L. (1946). A skin reaction in rabbits produced by intradermal inoculation of suspensions of killed Pasteurella tularensis. Public Health Rep. 61, 1797–1806.
Larsson, P., Oyston, P. C., Chain, P., Chu, M. C., Duffield, M., Fuxelius, H. H., Garcia, E., Halltorp, G., Johansson, D., Isherwood, K. E., Karp, P. D., Larsson, E., Liu, Y., Michell, S., Prior, J., Prior, R., Malfatti, S., Sjostedt, A., Svensson, K., Thompson, N., Vergez, L., Wagg, J. K., Wren, B. W., Lindler, L. E., Andersson, S. G., Forsman, M., and Titball, R. W. (2005). The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37, 153–159.
Lauriano, C. M., Barker, J. R., Yoon, S. S., Nano, F. E., Arulanandam, B. P., Hassett, D. J., and Klose, K. E. (2004). MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. U.S.A. 101, 4246–4249.
Lee, B. Y., Horwitz, M. A., and Clemens, D. L. (2006). Identification, recombinant expression, immunolocalization in macrophages, and T-cell responsiveness of the major extracellular proteins of Francisella tularensis. Infect. Immun. 74, 4002–4013.
Lenco, J., Hubalek, M., Larsson, P., Fucikova, A., Brychta, M., Macela, A., and Stulik, J. (2007). Proteomics analysis of the Francisella tularensis LVS response to iron restriction: induction of the F. tularensis pathogenicity island proteins IglABC. FEMS Microbiol. Lett. 269, 11–21.
Lenco, J., Link, M., Tambor, V., Zakova, J., Cerveny, L., and Stulik, A. J. (2009). iTRAQ quantitative analysis of Francisella tularensis ssp. holarctica live vaccine strain and Francisella tularensis ssp. tularensis SCHU S4 response to different temperatures and stationary phases of growth. Proteomics 9, 2875–2882.
Lenco, J., Pavkova, I., Hubalek, M., and Stulik, J. (2005). Insights into the oxidative stress response in Francisella tularensis LVS and its mutant DeltaiglC1 + 2 by proteomics analysis. FEMS Microbiol. Lett. 246, 47–54.
Leuzzi, R., Serino, L., Scarselli, M., Savino, S., Fontana, M. R., Monaci, E., Taddei, A., Fischer, G., Rappuoli, R., and Pizza, M. (2005). Ng-MIP, a surface-exposed lipoprotein of Neisseria gonorrhoeae, has a peptidyl-prolyl cis/trans isomerase (PPIase) activity and is involved in persistence in macrophages. Mol. Microbiol. 58, 669–681.
Lindgren, H., Honn, M., Golovlev, I., Kadzhaev, K., Conlan, W., and Sjostedt, A. (2009). The 58-kilodalton major virulence factor of Francisella tularensis is required for efficient utilization of iron. Infect. Immun. 77, 4429–4436.
Lowrie, D. B., Silva, C. L., Colston, M. J., Ragno, S., and Tascon, R. E. (1997). Protection against tuberculosis by a plasmid DNA vaccine. Vaccine 15, 834–838.
Ludwig, B., Rahfeld, J., Schmidt, B., Mann, K., Wintermeyer, E., Fischer, G., and Hacker, J. (1994). Characterization of Mip proteins of Legionella pneumophila. FEMS Microbiol. Lett. 118, 23–30.
Macela, A., Stulik, J., Hernychova, L., Kroca, M., Krocova, Z., and Kovarova, H. (1996). The immune response against Francisella tularensis live vaccine strain in Lps(n) and Lps(d) mice. FEMS Immunol. Med. Microbiol. 13, 235–238.
Meibom, K. L., Dubail, I., Dupuis, M., Barel, M., Lenco, J., Stulik, J., Golovliov, I., Sjostedt, A., and Charbit, A. (2008). The heat-shock protein ClpB of Francisella tularensis is involved in stress tolerance and is required for multiplication in target organs of infected mice. Mol. Microbiol. 67, 1384–1401.
Melillo, A., Sledjeski, D. D., Lipski, S., Wooten, R. M., Basrur, V., and Lafontaine, E. R. (2006). Identification of a Francisella tularensis LVS outer membrane protein that confers adherence to A549 human lung cells. FEMS Microbiol. Lett. 263, 102–108.
Molins, C. R., Delorey, M. J., Yockey, B. M., Young, J. W., Sheldon, S. W., Reese, S. M., Schriefer, M. E., and Petersen, J. M. (2010). Virulence differences among Francisella tularensis subsp. tularensis clades in mice. PLoS ONE 5, e10205. doi: 10.1371/journal.pone.0010205
Moro, A., Ruiz-Cabello, F., Fernandez-Cano, A., Stock, R. P., and Gonzalez, A. (1995). Secretion by Trypanosoma cruzi of a peptidyl-prolyl cis–trans isomerase involved in cell infection. EMBO J. 14, 2483–2490.
Nakai, K., and Horton, P. (1999). PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–36.
Nelson, M., Lever, M. S., Dean, R. E., Savage, V. L., Salguero, F. J., Pearce, P. C., Stevens, D. J., and Simpson, A. J. (2010). Characterization of lethal inhalational infection with Francisella tularensis in the common marmoset (Callithrix jacchus). J. Med. Microbiol. 59, 1107–1113.
Nix, E. B., Cheung, K. K., Wang, D., Zhang, N., Burke, R. D., and Nano, F. E. (2006). Virulence of Francisella spp. in chicken embryos. Infect. Immun. 74, 4809–4816.
Noah, C. E., Malik, M., Bublitz, D. C., Camenares, D., Sellati, T. J., Benach, J. L., and Furie, M. B. (2010). GroEL and lipopolysaccharide from Francisella tularensis live vaccine strain synergistically activate human macrophages. Infect. Immun. 78, 1797–1806.
Parra, M. C., Shaffer, S. A., Hajjar, A. M., Gallis, B. M., Hager, A., Goodlett, D. R., Guina, T., Miller, S., and Collins, C. M. (2009). Identification, cloning, expression, and purification of Francisella lpp3: an immunogenic lipoprotein. Microbiol. Res. 165, 531–545.
Pasetti, M. F., Cuberos, L., Horn, T. L., Shearer, J. D., Matthews, S. J., House, R. V., and Sztein, M. B. (2008). An improved Francisella tularensis live vaccine strain (LVS) is well tolerated and highly immunogenic when administered to rabbits in escalating doses using various immunization routes. Vaccine 26, 1773–1785.
Pavkova, I., Hubalek, M., Zechovska, J., Lenco, J., and Stulik, J. (2005). Francisella tularensis live vaccine strain: proteomic analysis of membrane proteins enriched fraction. Proteomics 5, 2460–2467.
Pavkova, I., Reichelova, M., Larsson, P., Hubalek, M., Vackova, J., Forsberg, A., and Stulik, J. (2006). Comparative proteome analysis of fractions enriched for membrane-associated proteins from Francisella tularensis subsp. tularensis and F. tularensis subsp. holarctica strains. J. Proteome Res. 5, 3125–3134.
Prior, R. G., Klasson, L., Larsson, P., Williams, K., Lindler, L., Sjostedt, A., Svensson, T., Tamas, I., Wren, B. W., Oyston, P. C., Andersson, S. G., and Titball, R. W. (2001). Preliminary analysis and annotation of the partial genome sequence of Francisella tularensis strain Schu 4. J. Appl. Microbiol. 91, 614–620.
Qin, A., Scott, D. W., Thompson, J. A., and Mann, B. J. (2009). Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect. Immun. 77, 152–161.
Ranford, J. C., and Henderson, B. (2002). Chaperonins in disease: mechanisms, models, and treatments. Mol. Pathol. 55, 209–213.
Ray, H. J., Chu, P., Wu, T. H., Lyons, C. R., Murthy, A. K., Guentzel, M. N., Klose, K. E., and Arulanandam, B. P. (2010). The Fischer 344 rat reflects human susceptibility to Francisella pulmonary challenge and provides a new platform for virulence and protection studies. PLoS ONE 5, e9952. doi: 10.1371/journal.pone.0009952
Raymond, C. R., and Conlan, J. W. (2009). Differential susceptibility of Sprague-Dawley and Fischer 344 rats to infection by Francisella tularensis. Microb. Pathog. 46, 231–234.
Rey, S., Acab, M., Gardy, J. L., Laird, M. R., deFays, K., Lambert, C., and Brinkman, F. S. (2005). PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res. 33, D164–D168.
Rick, L. C., and Wu, T. H. (2007). Animal models of Francisella tularensis infection. Ann. N. Y. Acad. Sci. 1105, 238–265.
Rohmer, L., Brittnacher, M., Svensson, K., Buckley, D., Haugen, E., Zhou, Y., Chang, J., Levy, R., Hayden, H., Forsman, M., Olson, M., Johansson, A., Kaul, R., and Miller, S. I. (2006). Potential source of Francisella tularensis live vaccine strain attenuation determined by genome comparison. Infect. Immun. 74, 6895–6906.
Rohmer, L., Fong, C., Abmayr, S., Wasnick, M., Larson Freeman, T. J., Radey, M., Guina, T., Svensson, K., Hayden, H. S., Jacobs, M., Gallagher, L. A., Manoil, C., Ernst, R. K., Drees, B., Buckley, D., Haugen, E., Bovee, D., Zhou, Y., Chang, J., Levy, R., Lim, R., Gillett, W., Guenthener, D., Kang, A., Shaffer, S. A., Taylor, G., Chen, J., Gallis, B., D’Argenio, D. A., Forsman, M., Olson, M. V., Goodlett, D. R., Kaul, R., Miller, S. I., and Brittnacher, M. J. (2007). Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 8, R102. doi: 10.1186/gb-2007-8-6-r102
Salomonsson, E., Kuoppa, K., Forslund, A. L., Zingmark, C., Golovliov, I., Sjostedt, A., Noppa, L., and Forsberg, A. (2009). Reintroduction of two deleted virulence loci restores full virulence to the live vaccine strain of Francisella tularensis. Infect. Immun. 77, 3424–3431.
Samrakandi, M. M., Zhang, C., Zhang, M., Nietfeldt, J., Kim, J., Iwen, P. C., Olson, M. E., Fey, P. D., Duhamel, G. E., Hinrichs, S. H., Cirillo, J. D., and Benson, A. K. (2004). Genome diversity among regional populations of Francisella tularensis subspecies tularensis and Francisella tularensis subspecies holarctica isolated from the US. FEMS Microbiol. Lett. 237, 9–17.
Sandstrom, G., Tarnvik, A., and Wolf-Watz, H. (1987). Immunospecific T-lymphocyte stimulation by membrane proteins from Francisella tularensis. J. Clin. Microbiol. 25, 641–644.
Santic, M., Molmeret, M., Klose, K. E., Jones, S., and Kwaik, Y. A. (2005). The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7, 969–979.
Sen, B., Meeker, A., and Ramakrishnan, G. (2010). The fslE homolog, FTL_0439 (fupA/B), mediates siderophore-dependent iron uptake in Francisella tularensis LVS. Infect. Immun. 78, 4276–4285.
Sjostedt, A. (2001). “Family XVII. FRANCISELLACEAE Genus I. Francisella,” in Bergey’s Manual of Systemic Bacteriology, Vol. 1, ed. D. J. Brenner (New York: Springer), 200–210.
Sjostedt, A., Eriksson, M., Sandstrom, G., and Tarnvik, A. (1992a). Various membrane proteins of Francisella tularensis induce interferon-gamma production in both CD4+ and CD8+ T cells of primed humans. Immunology 76, 584–592.
Sjostedt, A., Sandstrom, G., and Tarnvik, A. (1992b). Humoral and cell-mediated immunity in mice to a 17-kilodalton lipoprotein of Francisella tularensis expressed by Salmonella typhimurium. Infect. Immun. 60, 2855–2862.
Sjostedt, A., Sandstrom, G., Tarnvik, A., and Jaurin, B. (1990). Nucleotide sequence and T cell epitopes of a membrane protein of Francisella tularensis. J. Immunol. 145, 311–317.
Sjostedt, A., Tarnvik, A., and Sandstrom, G. (1991). The T-cell-stimulating 17-kilodalton protein of Francisella tularensis LVS is a lipoprotein. Infect. Immun. 59, 3163–3168.
Skrodzki, E. (1961). Studies of the pathogenesis of tularemia. III. Survival of B. tularense in normal and immunized rabbits. Arch. Immunol. Ther. Exp. (Warsz.) 9, 509–517.
Staples, J. E., Kubota, K. A., Chalcraft, L. G., Mead, P. S., and Petersen, J. M. (2006). Epidemiologic and molecular analysis of human tularemia, United States, 1964–2004. Emerging Infect. Dis. 12, 1113–1118.
Stewart, G. R., Robertson, B. D., and Young, D. B. (2004). Analysis of the function of mycobacterial DnaJ proteins by overexpression and microarray profiling. Tuberculosis (Edinb) 84, 180–187.
Stimson, E., Virji, M., Makepeace, K., Dell, A., Morris, H. R., Payne, G., Saunders, J. R., Jennings, M. P., Barker, S., and Panico, M. (1995). Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol. Microbiol. 17, 1201–1214.
Sundaresh, S., Randall, A., Unal, B., Petersen, J. M., Belisle, J. T., Hartley, M. G., Duffield, M., Titball, R. W., Davies, D. H., Felgner, P. L., and Baldi, P. (2007). From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics 23, i508–i518.
Surcel, H. M., Sarvas, M., Helander, I. M., and Herva, E. (1989). Membrane proteins of Francisella tularensis LVS differ in ability to induce proliferation of lymphocytes from tularemia-vaccinated individuals. Microb. Pathog. 7, 411–419.
Svensson, K., Larsson, P., Johansson, D., Bystrom, M., Forsman, M., and Johansson, A. (2005). Evolution of subspecies of Francisella tularensis. J. Bacteriol. 187, 3903–3908.
Tarnvik, A. (1989). Nature of protective immunity to Francisella tularensis. Rev. Infect. Dis. 11, 440–451.
Telepnev, M., Golovliov, I., and Sjostedt, A. (2005). Francisella tularensis LVS initially activates but subsequently down-regulates intracellular signaling and cytokine secretion in mouse monocytic and human peripheral blood mononuclear cells. Microb. Pathog. 38, 239–247.
Thakran, S., Li, H., Lavine, C. L., Miller, M. A., Bina, J. E., Bina, X. R., and Re, F. (2008). Identification of Francisella tularensis lipoproteins that stimulate the toll-like receptor (TLR) 2/TLR1 heterodimer. J. Biol. Chem. 283, 3751–3760.
Titball, R. W., and Petrosino, J. F. (2007). Francisella tularensis genomics and proteomics. Ann. N. Y. Acad. Sci. 1105, 98–121.
Twine, S., Bystrom, M., Chen, W., Forsman, M., Golovliov, I., Johansson, A., Kelly, J., Lindgren, H., Svensson, K., Zingmark, C., Conlan, W., and Sjostedt, A. (2005a). A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58-kilodalton protein is attenuated for virulence and is an effective live vaccine. Infect. Immun. 73, 8345–8352.
Twine, S. M., Mykytczuk, N. C., Petit, M., Tremblay, T. L., Conlan, J. W., and Kelly, J. F. (2005b). Francisella tularensis proteome: low levels of ASB-14 facilitate the visualization of membrane proteins in total protein extracts. J. Proteome Res. 4, 1848–1854.
Twine, S. M., Mykytczuk, N. C., Petit, M., Shen, H., Sjostedt, A., Conlan, J. W., and Kelly, J. F. (2006a). In vivo proteomic analysis of the intracellular bacterial pathogen, Francisella tularensis, isolated from mouse spleen. Biochem. Biophys. Res. Commun. 345, 1621–1633.
Twine, S. M., Petit, M. D., Shen, H., Mykytczuk, N. C., Kelly, J. F., and Conlan, J. W. (2006b). Immunoproteomic analysis of the murine antibody response to successful and failed immunization with live anti-Francisella vaccines. Biochem. Biophys. Res. Commun. 346, 999–1008.
Twine, S. M., Shen, H., Kelly, J. F., Chen, W., Sjostedt, A., and Conlan, J. W. (2006c). Virulence comparison in mice of distinct isolates of type A Francisella tularensis. Microb. Pathog. 40, 133–138.
Twine, S. M., Petit, M. D., Fulton, K. M., House, R. V., and Conlan, J. W. (2010). Immunoproteomics analysis of the murine antibody response to vaccination with an improved Francisella tularensis live vaccine strain (LVS). PLoS ONE 5, e10000. doi: 10.1371/journal.pone.0010000
Underhill, D. M. (2004). Toll-like receptors and microbes take aim at each other. Curr. Opin. Immunol. 16, 483–487.
Valentino, M. D., Hensley, L. L., Skrombolas, D., McPherson, P. L., Woolard, M. D., Kawula, T. H., Frelinger, J. A., and Frelinger, J. G. (2009). Identification of a dominant CD4 T cell epitope in the membrane lipoprotein Tul4 from Francisella tularensis LVS. Mol. Immunol. 46, 1830–1838.
Vorou, R. M., Papavassiliou, V. G., and Tsiodras, S. (2007). Emerging zoonoses and vector-borne infections affecting humans in Europe. Epidemiol. Infect. 135, 1231–1247.
Waag, D. M., McKee, K. T. Jr., Sandstrom, G., Pratt, L. L., Bolt, C. R., England, M. J., Nelson, G. O., and Williams, J. C. (1995). Cell-mediated and humoral immune responses after vaccination of human volunteers with the live vaccine strain of Francisella tularensis. Clin. Diagn. Lab. Immunol. 2, 143–148.
Wallin, R. P., Lundqvist, A., More, S. H., von, B. A., Kiessling, R., and Ljunggren, H. G. (2002). Heat-shock proteins as activators of the innate immune system. Trends Immunol. 23, 130–135.
Wu, T. H., Hutt, J. A., Garrison, K. A., Berliba, L. S., Zhou, Y., and Lyons, C. R. (2005). Intranasal vaccination induces protective immunity against intranasal infection with virulent Francisella tularensis biovar A. Infect. Immun. 73, 2644–2654.
Keywords: Francisella tularensis, LVS, immunoproteomics, proteome microarray, 2D-PAGE, antibody, vaccine, mouse vaccination
Citation: Kilmury SLN and Twine SM (2011) The Francisella tularensis proteome and its recognition by antibodies. Front. Microbio. 1:143. doi: 10.3389/fmicb.2010.00143
Received: 29 October 2010;
Accepted: 18 December 2010;
Published online: 07 January 2011.
Edited by:
Anders Sjostedt, Umeå University, SwedenReviewed by:
Barbara Mann, University of Virginia, USAJiri Stulik, University of Defence, Czech Republic
Copyright: © 2011 Kilmury and Twine. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Susan M. Twine, Institute for Biological Sciences, National Research Council Canada, 100 Sussex Drive, Ottawa, ON, Canada K1A 0R6. e-mail: susan.twine@nrc-cnrc.gc.ca
 Sara L. N. Kilmury
Sara L. N. Kilmury