- 1 Biomedical Research, The University of Texas Health Science Center, Tyler, TX, USA
- 2 Center for Tuberculosis Research, Johns Hopkins School of Medicine, Baltimore, MD, USA
Mycobacterium tuberculosis lysX mutant, defective for production of lysinylated phosphatidylglycerol, is sensitive to cationic antimicrobial peptides, is not proficient for proliferation in mice lungs, and exhibits altered membrane potential (Maloney et al., 2009). In the present study we show that a lysX complement strain expressing lysX from inducible tet promoter is proficient in restoring lysX phenotypes, confirming that the observed phenotypes are specific to lysX. To evaluate the correlation between changes in membrane potential and lysX activity, we visualized regions of cardiolipin (CL), one of the abundant phospholipids of mycobacteria, by staining with fluorescent dye 10-N-nonyl acridine orange and found that CL is localized as bright spots at septal regions and poles of actively dividing cells, but not in stationary phase cells. lysX mutants were elongated and showed more numerous and brighter CL staining at both mid cell and quarter cell septa, compared with wild type, indicating a defect in the cell division process. Evaluation of 14C-acetic acid incorporation into major phospholipids such as CL, phosphatidylethanolamine (PE), phosphatidylinositol (PI), and their degradation between lysX mutant and its parent revealed differences in the turnover of PE and PI. Our results favor a hypothesis that alterations in phospholipid metabolism could be contributing to changes in membrane potential, hence the observed phenotype of lysX mutant.
Introduction
Mycobacterium tuberculosis is a pathogenic bacterium that causes the infectious disease tuberculosis. Estimates indicate that approximately one-third of the world’s population is infected with M. tuberculosis and approximately 1.8 million deaths were attributable to tuberculosis in 2008 (WHO, 2010)1. Survival of M. tuberculosis following its uptake by alveolar macrophages and subsequent replication and multiplication in the hostile environment is a challenging task to the pathogen (Smith, 2003; Tischler and McKinney, 2010). Within the hostile macrophage environment M. tuberculosis is believed to face reactive oxygen and nitrogen species (ROS, RNS), acidic pH of phagolysosome compartments and possibly antimicrobial peptides. ROS and RNS damage DNA, lipids, and proteins and thereby promote pathogen killing. Mycobacterium tuberculosis multiplies in this hostile environment by efficiently operating multiple stress resistance pathways (reviewed in (Smith, 2003; Tischler and McKinney, 2010). One of these elegant strategies includes the operation of proteasome machinery for processing RNS damaged proteins (Darwin et al., 2003). This pathway includes the activities of proteasomal core subunit PrcBA, accessory factors Mpa and PafA that recognize specific proteins for targeted degradation by the activity of prokaryotic ubiquitin-like protein, Pup.
It is expected that the net charge or membrane potential of the bacterial cell wall is determined by its composition, including the ratio of acidic to basic phospholipids. Thus, another survival strategy involves the modulation of membrane surface charge so that the pathogen can weather the action of host cationic antimicrobial peptides (CAMPs; Peschel, 2002; Kraus and Peschel, 2006). Gram positive pathogens, such as Staphylococcus aureus, decrease net membrane charge by adding lysine groups on acidic phospholipid phosphatidylglycerol (PG) and thereby resist the action of CAMPs (Peschel et al., 2001). It is also known that other acidic phospholipids, such as cardiolipin (CL), are subject to such modification (Thedieck et al., 2006). We recently showed that a two-domain lysyltransferase and lysyl-tRNA-synthetase protein encoded by lysX gene of M. tuberculosis is necessary for PG lysinylation, optimal survival in lungs of mice and guinea pigs, resistance to the action of CAMPs and for maintaining optimal membrane potential (Maloney et al., 2009).
While the above study shows that lysX plays an important role in M. tuberculosis survival upon infection, several questions remain with respect to lysX in vivo phenotype. For example, these studies revealed that lysinylated phosphatidylglycerol (L-PG) is a constituent of M. tuberculosis, although it is unknown if other mycobacterial species also produce lysinylated phospholipids (L-PL). Also, the lysX complement strain, wherein lysX expression was achieved from M. smegmatis amidase promoter (Triccas et al., 1998), was partially proficient in restoring lysX defect (Maloney et al., 2009). While studies based on fluorescent probes allowed the visualization of acidic phospholipids such as PG and CL in Escherichia coli and other bacteria as defined domains (Kawai et al., 2004; Romantsov et al., 2007; Mileykovskaya and Dowhan, 2009), it is unknown whether the absence of L-PG production is associated with changes in membrane lipid domain organization and possibly their turnover thereby contributing to the observed changes in membrane potential (Maloney et al., 2009). The present study is undertaken to address some of these issues.
We found that the domain organization of the major acidic phospholipid CL, as revealed by 10-nonyl acridine orange (NAO) staining, is not significantly affected in lysX mutants. However, differences in the turnover of select membrane phospholipids were found between lysX and its parent. Furthermore, our results indicate that expression of lysX from a strong inducible tet promoter allowed full complementation in vivo and that radiolabeled lysine is incorporated in other mycobacterial phospholipids. Our studies favor a hypothesis that alterations in phospholipid turnover are in part responsible for the observed lysX phenotype.
Materials and Methods
Cloning
Unless otherwise noted all genes used in this study were generated by polymerase chain reaction (PCR) using genomic DNA as a template and Phusion DNA polymerase (New England Biolabs). As needed, PCR products were Taq polymerase treated, cloned into pGEM T-easy (Promega) prior to subcloning in other plasmids. Escherichia coli TOP10 cells were used to transform plasmids and the transformants were selected on LB agar containing 100 μg/mL ampicillin, 0.1 M IPTG, and 40 μg/mL X-gal.
Construction of lysX Complement Strains
We wished to express lysX from tetracycline (Ptet) and nitrile (PnitA) inducible promoters (Ehrt et al., 2005; Pandey et al., 2009) and created respective complement strains (see below). For placing lysX coding region under Ptet oligonucleotide primers bearing PacI and SwaI sites, 5′-AGAACCTTAATTAAAGCCCCACCAGGGAGGAAGCCGAACGATGGGACTCCACTTAA-CTG-3′ and 5′-ATCGGATTTAAATCTAATGCGGTTTTGCCAAC-3′, respectively, were used to amplify lysX gene, cloned into pLR56 (Dziedzic et al., 2010), which is a pMV306 based integration proficient vector bearing tetR- and tetO-containing mycobacterial promoter. Plasmids containing tetR- and tetO-containing mycobacterial promoter are a kind gift from Dr. Sabine Ehrt (Ehrt et al., 2005). Similarly, oligonucleotide primers 5′-CACCTCGTACTGTCCTGCCAA-3′ and 5′-CGCCGCGTTGAA-GTCGGC-3′ were used to clone lysX gene under PnitA. Both recombinant plasmids were used to transform the previously constructed lysX full gene Pami complement strain. In this strain, the chromosomal copy of lysX was replaced with a copy that has been disrupted by a gentamycin resistance gene; a second copy of lysX was integrated at the attB locus using a hygromycin resistant integrating vector. The Ptet construct or the PnitA construct bearing the lysX gene replaces the integrated hygromycin resistant vector at the attB site via swapping (Rajagopalan et al., 2005), thus creating Ptet and PnitA controlled complement strains designated as Ptet-lysX and PnitA-lysX, respectively. As previously described (Maloney et al., 2009), the lysX mutant strain was created by replacing the integrated copy of Pami-lysX with an empty vector at the attB site. These strains were confirmed by PCR.
Construction of M. smegmatis Overexpression Strains
For overexpression of M. tuberculosis lysX and Rv1639c genes in M. smegmatis, oligonucleotide primer combinations mvm530lysXF (5′-GGCGAATTCCATATGGGACTCCACTTAACTG-3′) and mvm531lysXR (5′-CCTACGGTCTAGACTAATGCGGTTTTGCAACGG-3′) for lysX and MR149F (5′-GGAATTCCATATGGCTCAGAATGAACTGTGAC-3′) and MR150R (5′-ATCGGTCTAGAATTTAAATCTATCTGCCGGTACCGGCGATCTG-3′) for Rv1639c, were used and the respective genes were cloned under Pami promoter. The resulting constructs were transformed into M. smegmatis MC2155 and the merodiploid strains were characterized further.
Growth of Bacterial Strains
Escherichia coli strains were grown in Luria–Bertani media, whereas M. smegmatis MC2155 and M. tuberculosis H37Rv strains were grown in Middlebrook 7H9 media (Becton–Dickenson) supplemented with albumin (5 g/L) and dextrose (2 g/L); in addition, M. tuberculosis cultures were supplemented with oleic acid (60 μL/L) and catalase (3 mg/L). Kanamycin (Sigma-Aldrich) at 25 μg/mL for M. smegmatis and 10 μg/mL for M. tuberculosis were used whereas hygromycin (Roche) at a final concentration of 50 μg/mL for both strains. In some experiments, Polymyxin B (PMNB; Sigma-Aldrich) was added to cultures at a final concentration of 500 units/mL for M. tuberculosis. For growth attenuation studies in M. smegmatis, PMNB was added to either 130 or 195 U/mL, vancomycin to either 5 or 10 μg/mL and lysozyme to 20 μg/mL; these concentrations were chosen using published MICs for M. smegmatis strains as a guideline. (Peteroy et al., 2000; Yasue, 2000; Raymond et al., 2005). For evaluation of lysX expression from the PnitA and Ptet promoters, tetracycline (25 ng/mL) or isovaleronitrile (10 nM) was added to growth media. Growth curves were generated by reading the optical density (600 nm).
Reverse Transcription and Real-Time PCR
Extraction of RNA using RNA Bee (Tel-Test, Inc.) and chloroform and quantitative real-time PCR were essentially as previously described (Chauhan et al., 2006a; Fol et al., 2006; Maloney et al., 2009). cDNA transcripts of interest were reverse transcribed at 42°C from total isolated RNA using the SuperScript II RT kit (Invitrogen); each reaction contained 0.25 mM each dNTP, 1× First Strand Buffer, 10 mM DTT, RNaseOUT (4 U/μL, Invitrogen), 0.6 μM gene specific RT primer, and 100 units SuperScript II reverse transcriptase. Gene specific RT primers are listed in Table 1. For each sample, identical reactions were performed without reverse transcriptase to confirm the absence of contaminating genomic DNA as described previously (Chauhan et al., 2006a; Fol et al., 2006; Maloney et al., 2009). The transcribed cDNA was used as template for real-time PCR reactions containing 0.5 μM each of the primer and probe set for the transcript of interest, 0.15 μM each of primers and probe for the control transcript 16S rRNA, 0.25 mM each dNTP, 1.5 mM magnesium chloride, 1× Taq buffer, and 0.2 units/15 μL Taq polymerase (New England Biolabs). Primers and Taqman probes are listed in Table 1.
Fluorescence Microscopy Methods
10-Nonyl acridine orange was added directly to media during growth of cultures for either 1 h (for M. smegmatis) or 24 h (for M. tuberculosis) prior to harvesting cells for microscopy. Mycobacterium tuberculosis pellets were suspended in 4% paraformaldehyde and stored at 4°C for 24 h before imaging. For NAO staining and visualization M. smegmatis and M. tuberculosis cells were resuspended in fresh media. Stained cells were examined by brightfield and fluorescence microscopy using a Nikon Eclipse E600 microscope with an attached CoolSnap ES CCD camera (Photometrics) and high-pressure mercury lamp (Nikon). NAO-stained cells were imaged using a custom NAO filter set (Ex450-490/Em645, Chroma Technology) for red fluorescence and the Nikon B2A filter set for green fluorescence. Images were analyzed using MetaMorph 6.2 software (Universal Imaging Corporation).
Aerosol Infection of Mice
All animal procedures were approved by the Johns Hopkins University Animal Care and Use Committee. Six-week-old female BALB/c mice (Charles River) were aerosol infected with M. tuberculosis lysX mutant, Ptet complement, and the wild-type strain using the Middlebrook inhalation exposure system (Glas-Col) with log-phase broth cultures. To induce the expression of Ptet complement and for fair comparison across groups, all mice were given medicated water containing 1 mg/mL of doxycycline and 5% sucrose in light protected bottle (Gandotra et al., 2007). Water was changed two times per week. At days 1, 7, 14, 28, 56, 84, and 112 post-infection, four mice from each group were sacrificed and the lungs and spleens were collected. The lung and spleen gross pathology, lung and spleen weights and body weights were recorded. From each mouse, one lung was homogenized and the supernatants were collected for cytokine profiling; the other lung was collected for colony-forming unit (CFU) enumeration with small portions collected and fixed with 10% formalin for histopathology study. The CFUs per lung or spleen were determined by plating diluted lung or spleen homogenates onto 7H11 selective agar.
14C-Acetic Incorporation and Phospholipid Analysis
Mycobacterium tuberculosis cultures (10 mL × 8 flasks) were grown to OD600 0.2 and 14C-acetate was added to a final concentration of 0.5 μCi/mL and cultures were grown for 24 h. At the end of incubation, cultures were harvested, washed with fresh prewarm media two times. One set of culture was removed for lipid extraction. Other culture pellets following washing in radioactive media were resuspended in fresh media containing 12 mM sodium acetate. After 12-, 24-, and 48-h incubation, cultures in duplicate were harvested, washed, and processed for lipid extraction. Mycobacterium tuberculosis cultures grown without radiolabeled acetic acid were mixed with radiolabeled cultures prior to chloroform–methanol extraction. The amount of 14C-acetate incorporated into lipid fractions was determined by scintillation counting. Approximately, equal counts of polar lipid fractions were loaded on to thin-layer chromatography (TLC) plates (Gel 60 F254, 20 cm × 20 cm, 250 μm, EMD #5715-7, 20 cm × 20 cm) resolved by TLC in a solvent system of chloroform:methanol:water (65:25:4 v/v/v) along with phosphatidylethanolamine (PE), PI, and CL of M. tuberculosis standards. Lipids were visualized followed by ninhydrin staining, bands corresponding to individual lipids were identified, scraped, extracted with chloroform:methanol, dried, and counted. Each spot in duplicate from two independent cultures were averaged and SD was estimated.
Statistical Analysis
Differences between data sets were analyzed using Student’s t-test. A P value of <0.01 was considered to be significant.
Results and Discussion
Evaluation of lysX Expression from the Tetracycline Inducible Promoter
Earlier experiments designed to evaluate lysX phenotype used a complement strain wherein lysX gene was expressed from a constitutively active M. smegmatis ami promoter and this strain was designated as ami-lysX (Maloney et al., 2009). It is known that M. smegmatis ami promoter is inducible in M. smegmatis, but is rather constitutively expressed in M. tuberculosis (Chauhan et al., 2006a,b; Fol et al., 2006). As reviewed, absence of lysX function led to membrane hyperpolarization, which in turn sensitized the pathogen to lipophilic antibiotics and CAMPs. The lysX mutant was also found to be defective for proliferation in macrophages and mice lungs (Maloney et al., 2009). While the ami-lysX complement strain was proficient in reversing lysX defect in most of the assays tested, albeit partially, it was defective in restoring lysX defect in vivo. Global gene expression profile studies by other investigators also indicated that lysX is one of the upregulated genes during acute and chronic infections (Talaat et al., 2004). Hence, we considered a possibility that lysX expression from heterologous amidase promoter is not optimal in vivo and the use of strong alternate promoters, such as tetracycline inducible promoters, could restore lysX phenotype. Another alternative is to express lysX from its native promoter to avoid issues associated with gene expression. The promoter elements controlling lysX gene have not been defined, and thus in the absence of this information, we chose to proceed with the best characterized Ptet promoter. Alternatively, disruption of lysX at its native locus could be affecting expression of downstream genes, thereby contributing to the observed phenotype. Both possibilities are not mutually exclusive.
To begin addressing these issues we first examined if lysX expression was elevated upon infection by quantitative real-time PCR. Analysis of lysX transcript levels relative to 16S rRNA from broth and intracellularly grown M. tuberculosis revealed that lysX expression was elevated about fourfold (4.1 ± 0.42) relative to broth during intracellular growth in monocyte derived macrophages or in THP-1 macrophages (not shown). Next, in an attempt to express lysX from strong inducible promoters, we constructed two plasmids wherein lysX expression was induced from tet and nitA promoters and created appropriate complement strains, Ptet-lysX and PnitA-lysX, respectively (see Materials and Methods) and evaluated lysX expression levels relative to control and that expressed from Pami (Figure 1). As can be seen, lysX expression in Ptet-lysX complement was about 10-fold (P = 0.003) more than that in the wild-type strain when tetracycline was added to the growth media and about fourfold (P = 0.002) higher when cells were grown without tetracycline. The latter result also indicated that the Ptet promoter under our experimental conditions is leaky. We also found that lysX expression in PnitA-lysX complement strain following the addition of isovaleronitrile (10 nM) was moderately less as compared to wild type indicating that the inducer isovaleronitrile was inhibiting lysX expression. However, there was no growth inhibition at the concentration of inducer used (data not shown). Expression of lysX in the Pami-lysX complement was similar to that of the wild-type strain (P = 0.211) both in the absence and presence of acetamide. It has been shown that select gene expression from the Pami in M. tuberculosis is constitutive and is not robust (Chauhan et al., 2006a,b; Fol et al., 2006). Hence, we conclude from these results that lysX expression from Ptet could lead to elevated or optimal levels of lysX and hopefully could restore lysX phenotype in vivo. Accordingly, the Ptet-lysX strain was further characterized in vitro and in vivo.
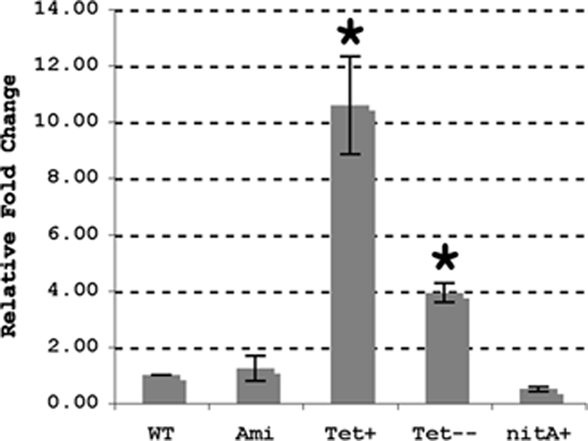
Figure 1. Expression of lysX in M. tuberculosis strains. Wild-type M. tuberculosis (WT) and the Pami (ami), Ptet (Tet), and PnitA (nitA) complement strains were used for RNA extraction. The Ptet complement was grown with (Tet+) and without (Tet−) tetracycline whereas the PnitA strain was grown with isovaleronitrile. Expression of lysX was evaluated by real-time PCR using RNA isolated from each strain; these results were normalized against expression of 16s rRNA. lysX expression in the wild-type strain is assigned a value of 1 and all other results are shown as fold change relative to this value. All values are the means ± SD from two independent experiments with triplicate samples; single star represents P < 0.01 versus WT (Student’s t-test). Note lysX expression from PnitA promoter was moderately less as compared to that in wild type (P = 0.068).
The Effect of Pmnb on Survival of Ptet-lysX Complement Strain
We have previously shown that the lysX mutant is sensitive to PMNB, a CAMP whereas Pami-lysX complement is as proficient for survival as the wild type at 100 units/mL of PMNB (Maloney et al., 2009). However, the Pami-lysX is only partially proficient for survival at higher doses of PMNB (data not shown). To test if Ptet-lysX complement reverses lysX sensitivity to PMNB at higher doses, we grew M. tuberculosis strains in the presence of 500 units/mL PMNB and recorded change in optical density as a measurement of growth (Figure 2). The Ptet-lysX complement grew in a manner similar to wild type whereas lysX mutant showed moderate growth defect in the absence of PMNB. However, in the presence of 500 units/mL PMNB Ptet-lysX complement grew better than that of the Pnit-lysX and Pami-lysX, indicating that the Ptet-lysX reverses growth defect significantly due to PMNB.
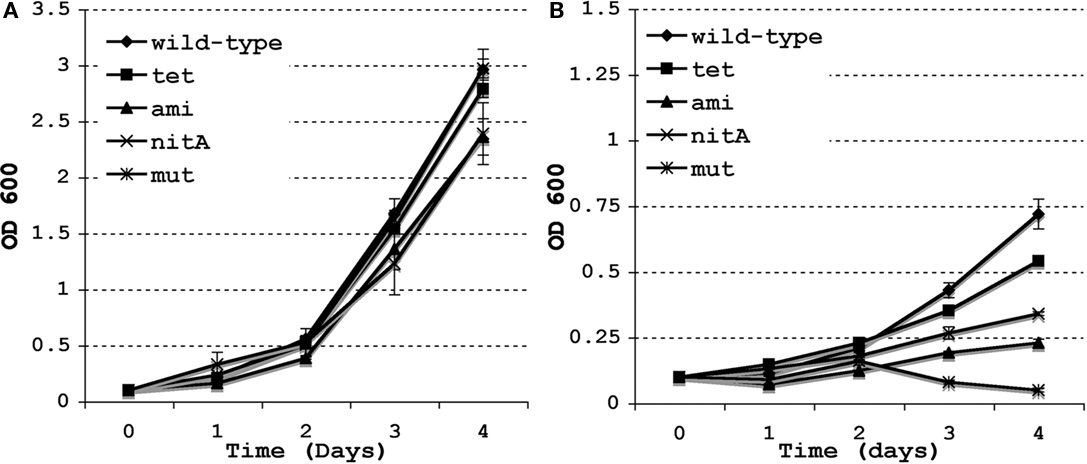
Figure 2. Growth of M. tuberculosis lysX strains and the effect of Polymyxin B. The M. tuberculosis wild type (WT), lysX mutant (mut), and the lysX Pami (ami), lysX Ptet (tet), and lysX PnitA (nitA) complement strains were grown in media alone (A) and in media supplemented with Polymyxin B 500 units/mL (B); optical density measured at 24 h intervals. All values are the means ± SD from three independent experiments.
Expression of Downstream Gene Rv1639c in lysX Mutant
The lysX mutant strain we created by homologous recombination lacks most of the lysX coding region. Genome sequence analysis revealed that the immediate downstream gene, Rv1639c, is separated from lysX by less than 60 nucleotides and the Rv1639c-lysX intergenic region lacks typical mycobacterial promoter features. Although not proven, lysX and Rv1639c genes could be polycistronic. Accordingly, we evaluated the expression levels of Rv1639c in lysX mutant relative to wild type and its complement derivatives Pami-lysX and Ptet-lysX strains. As can be seen, removal of lysX did not inhibit Rv1639c expression and in fact upregulated its expression approximately sevenfold (P = 0.0007; Figure 3). The Rv1639c expression levels restored to wild-type levels in Ptet-lysX complement strain (P = 0.128). Rv1639c expression was elevated two- to threefold in pami-lysX (P = 0.004; Figure 3). The function of Rv1639c gene product in M. tuberculosis metabolism is unknown, but Tuberculist gene annotation data indicate that it contains PS00904 protein phenyltransferase alpha subunit signature motif. It is tempting to speculate that the two-domain lysyltransferase-lysyl-tRNA synthetase protein negatively regulates Rv1639c expression. It remains to be evaluated how lysX activity is involved in regulating Rv1639c expression.
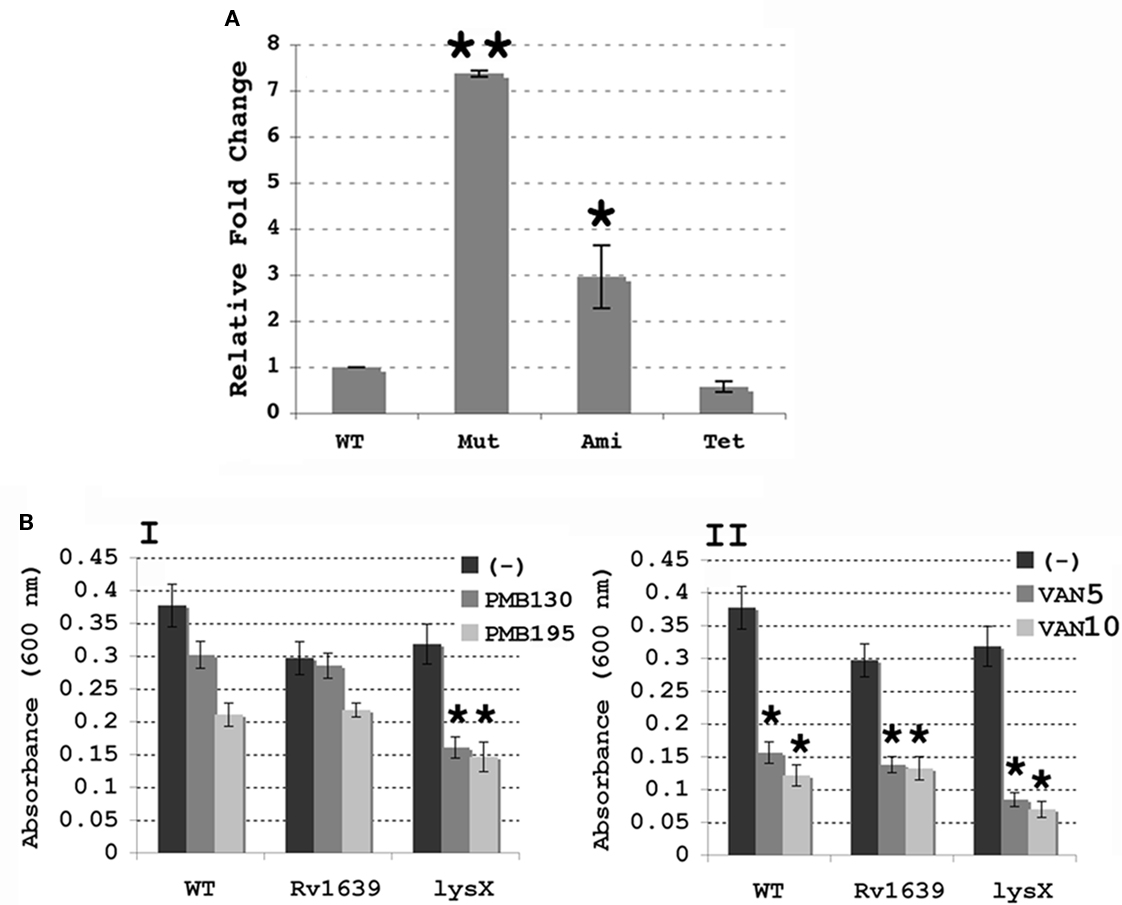
Figure 3. (A) Expression of Rv1639c in M. tuberculosis lysX strains. Expression of Rv1639c was evaluated by real-time PCR using RNA isolated from Wild-type M. tuberculosis (WT), the lysX mutant (Mut) and both the Pami (Ami) and Ptet (Tet) complement strains, and results were normalized against expression of 16s rRNA as described above. All values are the means ± SD from two independent experiments with triplicate samples; single star represents P < 0.01 and double stars indicate P < 0.001 versus WT (Student’s t-test). (B) Antibiotic sensitivity of M. smegmatis merodiploid strains overproducing Rv1639 and LysX proteins. Cultures were grown in the presence or absence of either (A) Polymyxin B 130 and 195 U/mL, or (B) vancomycin 5 μg/mL, 10 μg/mL (VAN5, VAN10). Bars represent absorbance values for each culture. The values represented are the means ± SD for four independent samples; stars represent P < 0.01 compared with the control culture (-) for each (Student’s t-test).
The above results, although unlikely, raise a question as to whether overexpression of Rv1639c is responsible for the observed lysX phenotype (Maloney et al., 2009). Bioinformatics analysis indicated that both lysX gene and the organization of its immediate flanking regions including Rv1639c are very conserved in M. smegmatis and other mycobacterial members. Furthermore, Rv1639c gene product and its M. smegmatis counterpart MSMEG3797 are 58% identical and 83% similar. Thus, to address the above issue we created M. smegmatis merodiploids overexpressing Rv1639c and lysX and evaluated their sensitivity toward CAMP relative to wild type (Figure 3B). If overexpression of Rv1639c has deleterious effect, then M. smegmatis overexpressing Rv1639c would be expected to be sensitive to CAMP. As can be seen, M. smegmatis merodiploid overexpressing Rv1639c is no more sensitive to PMNB and Van than the wild type (Figure 3B). Similar results were also noted with lysozyme (data not shown). We also found that M. smegmatis merodiploid overexpressing lysX is some what more sensitive to PMNB than that of the Rv1639c and wild type. We have not investigated the consequences of artificial overexpression of Rv1639c on M. tuberculosis survival in mice lungs. Nonetheless, our results suggest that overexpression of Rv1639c does not contribute to the observed lysX phenotype.
Survival of M. tuberculosis lysX (Rv1640c) and Ptet-lysX in Mouse Lung
In order to evaluate if Ptet-lysX is proficient in restoring lysX virulence phenotype, we infected Balb/c mice with an inoculum of similar size for wild type, lysX mutant and Ptet-lysX (10 mL of log-phased cultures containing 6.90-, 7.00-, and 6.91-log CFU/mL). After aerosol infection, CFUs were compared for bacteria isolated from mouse lung. Both wild-type and Ptet-lysX complement grew approximately 2 logs, while only minor growth was seen for the lysX deletion mutant (Figure 4A). At days 14, 28, and 112, CFU in the lungs of lysX deletion mutant was significantly lower than that of wild type or Ptet-lysX. The lysX mutant also showed reduced dissemination to the spleen, as shown by the lower CFU counts relative to wild type (Figure 4B). Partial restoration of the CFU counts in the spleen was seen for Ptet-lysX complement. Gross pathology showed slightly more apparent granulomas in the wild-type and Ptet-lysX complement lungs compared with the deletion mutant (Figure 5A). However, there was no significant difference in body weight, lung weight, and spleen weight (data not shown). Histopathology studies showed more functional alveolar sacs in the lysX mutant lungs than in the wild-type and Ptet-lysX complement lungs, with the wild-type lungs showing the most severe lesions as exemplified by consolidation, inflammatory cell infiltration, and nodule-like loci formation (Figure 5B).
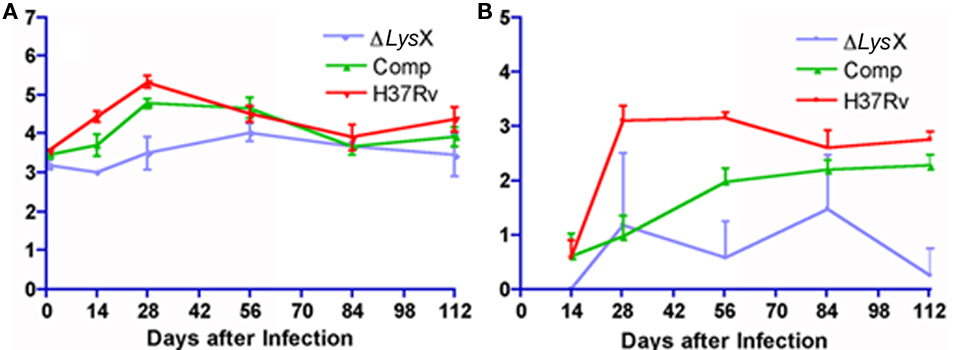
Figure 4. (A) Growth of Mycobacterium tuberculosis lysX deletion mutant (ΔlysX), lysX tet complement (Comp), and wild-type (H37Rv) in mouse lungs after low-dose aerosol infection. Groups of 28 mice were implanted at day 1 with 3.2- (ΔlysX), 3.5- (Comp), and 3.6-log (H37Rv) colony-forming units (CFU) in the lungs. (B) Growth of Mycobacterium tuberculosis in mouse spleens after low-dose implantation. Animals were infected via aerosol with M. tuberculosis lysX deletion mutant (ΔlysX), lysX tet complement (Comp), and wild-type (H37Rv) strains. Groups of 28 mice were implanted at day 1 with 3.2- (ΔlysX), 3.5- (Comp), and 3.6-log (H37Rv) CFU in the lungs.
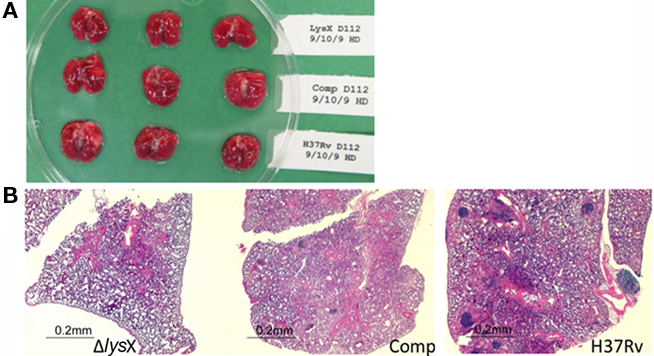
Figure 5. Mouse lung gross pathology and histopathology at day 112 after infection. (A) Gross pathology of the lungs infected with the M. tuberculosis lysX mutant (top row), lysX tet complement (middle), and wild-type (bottom) strains. (B) Lungs from mice were excised, stored in 10% formalin, embedded, and stained with hematoxylin and eosin for histopathological analysis. Hematoxylin-eosin staining confirmed that the lungs infected with wild-type and the lysX tet complement had extensive inflammation, but not those infected with the lysX mutant.
These results showing reduced lung pathology in lysX mutant relative to wild type suggest that lysX is required for full virulence in the mouse infection model. Introduction of a wild-type copy of this gene expressing from tet promoter restored most of the phenotype, unlike complementation with Pami-lysX (Maloney et al., 2009). We also found an overall lower than expected bacterial growth in the mouse lungs of all strains. One possibility is that the continuous propagation of M. tuberculosis H37Rv strain decreases overall ability to replicate in mice lungs and possibly its virulence potential. In all our experiments, doxycycline was added to all cultures throughout the infection period; doxycycline, has shown to be required for optimal gene expression from the Tet-ON promoter in vivo (Gandotra et al., 2007). Presumably, addition of doxycycline could have impacted the growth of all strains. It is not evident whether the concentration of doxycycline used to affect gene expression has any effect on immunity and this has not been addressed in this study. This limitation, however, did not impact our overall finding that lysX mutant showed lower CFU counts in mice lungs as compared to wild-type and Ptet-lysX complement strains. Together, these results showing the restoration of lysX defect by Ptet-lysX confirm that the observed growth defect phenotypes are specific to lysX.
LysX Mutant and Phospholipid Domains
Next, to evaluate the correlation between altered membrane potential and lysX phenotype, we characterized lysX strain with respect to organization of major membrane phospholipids and their turnover. Earlier studies showed that L-PG is a minor phospholipid constituent and that its absence is associated with an increase in membrane potential (Maloney et al., 2009). In mycobacteria PG is a minor phospholipid species and is the biosynthetic intermediate for diphosphatidylglycerol, CL, which is one of the most abundant phospholipid (Brennan and Nikaido, 1995). Recent studies on the interactions of bacterial membranes with fluorescent dyes indicate that membranes are organized into distinct domains containing lipids (Kawai et al., 2004; Romantsov et al., 2007; Mileykovskaya and Dowhan, 2009). Among these dyes, NAO, a hydrophobic fluorescent dye, has been generally used to mark polar acidic PL and specifically CL. NAO has been shown to bind CL in mitochondrial membranes with a ratio of 2:1, creating a stacked pair that is dimer-like. As the monomer form becomes less abundant and the number of stacked pairs increases, absorption shifts from 495 to 474 nm; this is accompanied by a shift in the emission from green (525 nm) to red (640 nm). Thus, binding of NAO to CL, but not to other acidic phospholipids, promotes a green to red shift in fluorescence (Petit et al., 1992; Gallet et al., 1995; Mileykovskaya et al., 2001; Mileykovskaya and Dowhan, 2009).
To visualize CL membrane lipid organization in lysX mutant we stained M. tuberculosis cells with NAO. It has been reported that in actively growing bacterial cells CL enriched domains are present at poles and near potential cell division sites and that changes in CL levels affect CL domain organization in membranes (Kawai et al., 2004; Mileykovskaya and Dowhan, 2009). Membrane lipid domains in mycobacteria have not been analyzed using NAO dye, hence, we first asked if NAO is a suitable marker stain for visualizing membrane lipid domains in M. tuberculosis. Since visualization of CL by NAO staining requires culturing cells in growth media containing dye, it was necessary to evaluate the effects of NAO on M. tuberculosis growth. We found concentrations up to 500 nM NAO did not interfere with growth (Figure 6A). We then examined NAO-stained cells of M. tuberculosis (Figure 6B) following paraformaldehyde fixation by fluorescence microscopy. Although staining was diffuse, distinct bright patterns of fluorescent spots are evident at poles and mid cell septa when visualized using both green and red filter sets (see Materials and Methods). Interestingly, red spots overlap with green indicating that these regions are where CL is enriched. The diffuse staining pattern could be a consequence of paraformaldehyde fixation, which is a required step to visualize M. tuberculosis in BSL3 setting. To visualize NAO-stained CL under non-fixation conditions, we examined NAO-stained cells of M. smegmatis, a non-pathogenic bacteria (Figure 6C). These cells showed rather sharp green and red spots primarily localized at mid cell septa and poles confirming that the localization patterns of CL in both mycobacterial species are similar. We also found that stationary phase grown cells of M. smegmatis were poorly stained (Figure 6D). Presumably, NAO readily stains newly synthesized CL. It is unlikely that the stationary phase grown cells lack CL, but is likely that CL turn over in actively growing cells is relatively high as compared to that in stationary phase cells.
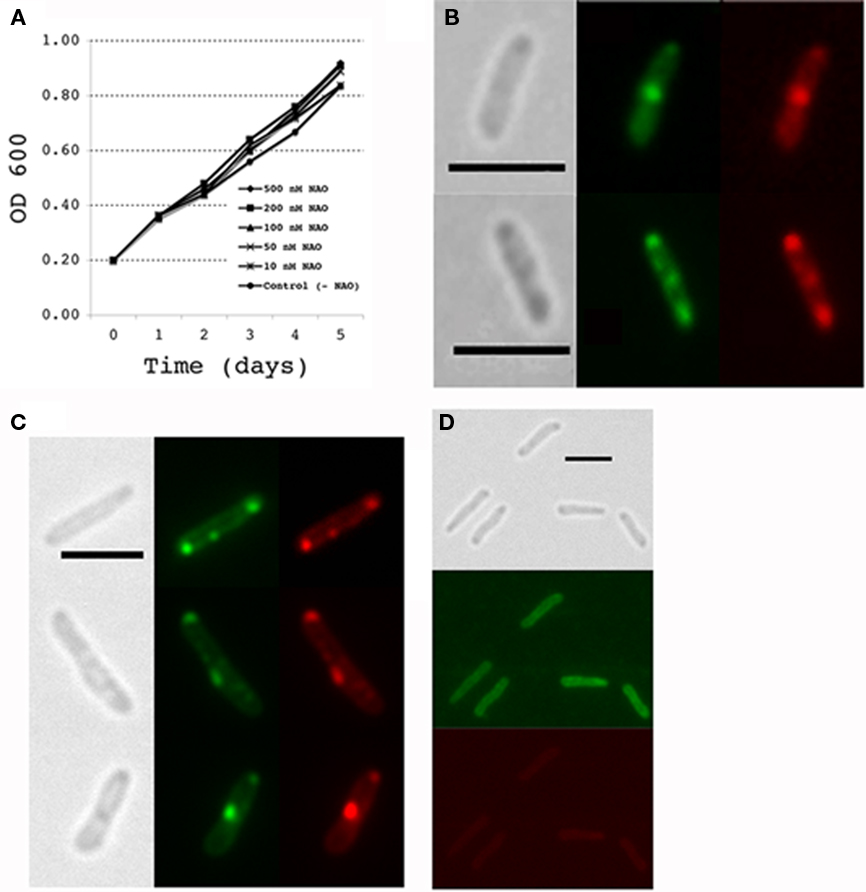
Figure 6. NAO staining in mycobacteria. (A) Effect of NAO addition to culture media during growth of M. tuberculosis. Mycobacterium tuberculosis cultures were grown in media with and without the NAO stain and optical density (OD 600 nM) measured at 24 h intervals. All values are the means ± SD. (B) M. tuberculosis cultures were stained with the lipid dye nonyl acridine orange (NAO) during active growth by adding the dye to the growth media (100 nM final concentration) for 24 h prior to harvest. Cells were then fixed in paraformaldehyde and observed by fluorescence microscopy. Grayscale images show brightfield illumination of cells; green fluorescence images show NAO staining of monoacidic phospholipids and red fluorescence images represent NAO staining in regions where cardiolipin is present. (C,D) NAO staining of M. smegmatis during active growth and stationary phase, respectively. Active cultures growing at an OD600 of 0.2 were supplemented with 100 nM NAO and incubated for 2 h at 37°C. Mycobacterium smegmatis cultures grown for overnight to an OD600 of 3.5 are referred to as stationary phase cells. NAO was added and incubated for 2 h at 37°C; live cells were removed from the cultures and directly observed by fluorescence microscopy. All images were taken at 100× magnification with scale bars representing 3 μm.
NAO Staining of lysX Mutant
Next we stained lysX mutant with NAO and visualized by microscopy (Figure 7). The following results are evident. First, visualization of cells by bright field microscopy revealed that lysX mutant cells were elongated compared to wild type. The average cell length determined from 200 cells of lysX mutant is 3.1 μM, whereas that for wild-type cells is 2.5 μM. Differences in cell length tend to suggest defects in cell division. Second, we found that more lysX cells were stained presumably due to increased permeability. Third, there appear be differences in the NAO staining pattern between the wild-type and lysX mutant. Some lysX mutant cells contained distinct spots not only at presumptive mid cell positions, but also at the three-quarter or daughter cell position. The number of NAO spots at poles in wild-type and lysX mutants were similar, however, those at mid cells and quarter position are more for lysX mutant relative to wild type (Figure 7). Also, NAO spots in lysX mutant are generally brighter as compared to wild type. Increased permeability of lysX mutant cannot explain the differences in the NAO staining patterns between the strains because a general increase in permeability would be expected to show a corresponding increase in the frequency of polar staining as well as that seen for mid and quarter cell.
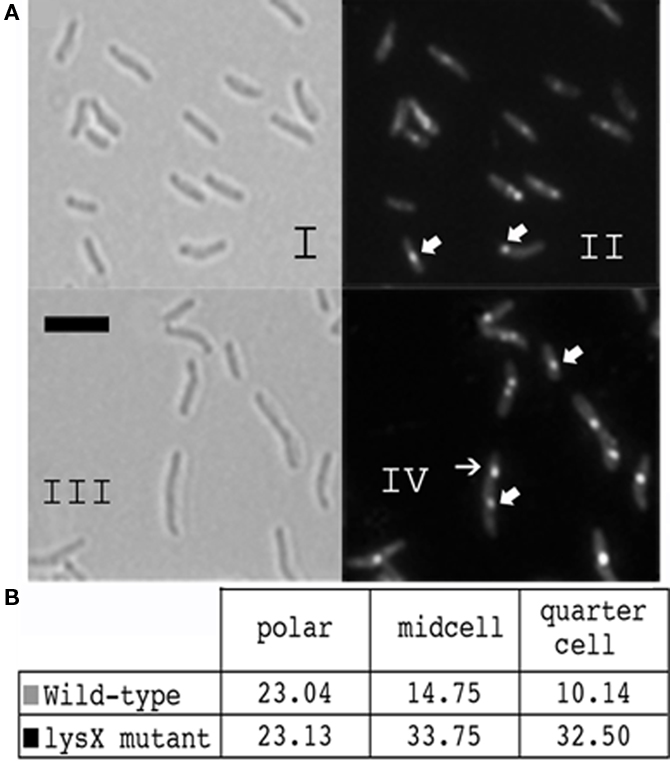
Figure 7. Cardiolipin staining of the M. tuberculosis lysX mutant. (A) Cultures were stained with NAO as described above, cells were fixed in paraformaldehyde and observed using fluorescence microscopy. Images I and III are brightfield 100× magnification, with scale bars representing 3 μm. Images II and IV show NAO bound to regions where cardiolipin is present. (B) Fluorescence images of NAO-stained cells were analyzed and type of staining was recorded for about 200 cells from each strain.
Major Phospholipid Turnover in lysX Mutant
Next we evaluated the degradation and turnover of major phospholipids in lysX mutant following incorporation of 14C-acetic acid into chloroform–methanol soluble lipids. Wild-type and lysX mutant cells were cultured with 14C-acetate, relative levels of CL, PE, and PI were determined following TLC analysis and autoradiography (Maloney et al., 2009). Significant reduction in the levels of PI between lysX (5.2%) and wild-type parent (7.2%) was noted (P value ≤0.02) whereas no significant differences in the levels of CL and PE were noted between the two (data not shown). We did however find differences in the turnover of PE and PI between the strains. While both lysX and wild type showed similar rates of turnover for CL, the turnover rates for PE and PI were slower in lysX mutant as compared to wild type (see Figure 8 and legend for P values). Notably, PE in lysX mutant did not exhibit appreciable turnover over the time period followed. Presumably, decreased turnover rates of PE and PI relative to wild type could be contributing to the changes in the membrane potential and possibly affecting the cell division process.
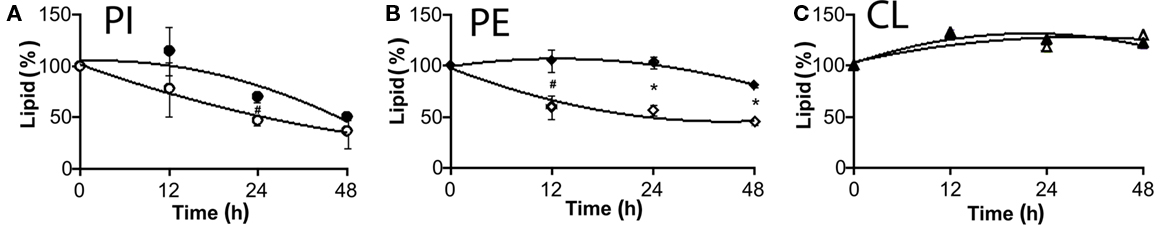
Figure 8. Turnover of M. tuberculosis phospholipids following 14C-acetic acid incorporation. Total lipids were extracted and analyzed by TLC. Radioactive spots corresponding to each phospholipid were identified, lipids were extracted and radioactivity present in each spot was measured by scintillation counting. Closed symbols stand for ΔlysX and open ones correspond to RvWT. (A) represents changes in the level of PI, (B) PE, and (C) CL. Data are presented as percentage of total lipids, relatively to time zero. Values were obtained from two independent experiments in duplicate. The star represents P < 0.02 and hash P < 0.05 (t-test method). The values represent averages and bars indicate the SD.
Lysinylated PL are also Present in Other Mycobacteria
LysX appears to be a conserved protein in all mycobacterial species that have been examined to date. LysX is a two-domain fusion protein containing N-terminal lysyltransferase (626 aa) and C-terminal lysyl-tRNA synthetase. Amino acid sequence analysis, including closely related family member Corynebacterium glutamicum, revealed that the LysX protein in mycobacteria shows significant sequence similarity, although differences in the N-terminal portion of these proteins (data not shown). Considering the similarities in the primary sequence, we asked if there are any differences in the distribution of L-PL in mycobacteria. To address this question, we cultured M. smegmatis, M. avium, and avirulent M. tuberculosis H37Ra in the presence of 14C-lysine, extracted total lipids and were analyzed as described (Maloney et al., 2009; Figure 9). As can be seen, 14C-lysine was incorporated in all mycobacterial species, however, the mobility of L-PL of M. tuberculosis strains is different from M. avium and M. smegmatis. Interestingly, one additional spot unique to M. avium was identified (Figure 9, see lane corresponding to M. avium). Presumably, differences in the N-terminal region of the LysX protein could contribute to these differences. It is likely the activities and possibly turnover of L-PG are different from virulent and avirulent mycobacterial species.
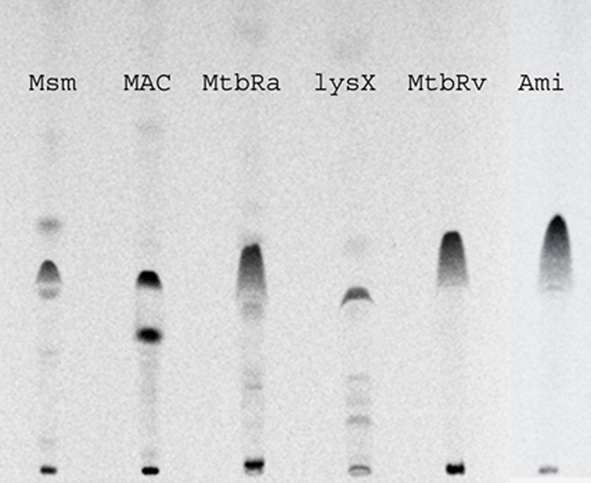
Figure 9. Polar lipid profiles of mycobacterial strains. Strains were grown in the presence and absence of 14C-lysine, total lipids were extracted in chloroform:methanol (2:1 v/v) and resolved by TLC in solvent system of chloroform:methanol:water (65:25:4 v/v/v).
In summary, our results confirm that lysX activity is required for resisting the action of CAMP and for expression of full virulence in mice. Since lysX transcript levels are elevated during intracellular growth, we reason that optimal survival of M. tuberculosis upon infection requires elevated expression levels of lysX and that this higher level expression could be attained in Ptet-lysX, but not in Pami-lysX. This could explain why Pami-lysX is not competent at restoring lysX phenotype optimally (Maloney et al., 2009).
There are three interesting and possibly related findings in this study. First, CL enriched regions are found to be present at mid cells and poles of actively dividing mycobacterial cells, but not in stationary phase cells. Second, the turnover rates of PE and PI, but not CL, in lysX mutant are found to be significantly different from that of wild type. Third, lysX mutant is elongated indicating a defect in cell division. It is pertinent to know that localization of CL near nucleoid-free poles and growing septa has been reported in other bacteria as well (Mileykovskaya and Dowhan, 2000, 2009; Romantsov et al., 2007). Although, we have not examined in this study, it is known that zwitterionic phospholipid PE, also localizes in the polar septa and septal membranes of other bacteria that have been examined (Nishibori et al., 2005). Mutations in genes affecting phospholipid metabolism have been shown to delay cell division and produce filamentous cells (Nishijima et al., 1988; DeChavigny et al., 1991). Thus, it is likely that septal localization of CL and possibly other phospholipids and cell division go together in mycobacteria. It remains to be explored how lysX activity affects phospholipid turnover rates and cell division in M. tuberculosis, and if both processes are connected. Presumably, optimal turnover of membrane phospholipids is necessary for regulated cell cycle progression and the maintenance of membrane potential necessary for M. tuberculosis multiplication upon infection and other stressful growth conditions. It is known that PG is accumulated as a consequence of CL catabolism (Bright-Gaertner and Proulx, 1972; Short and White, 1972). Presumably, optimal PG lysinylation mediated by LysX and its delysinylation step by hitherto unidentified enzymes could modulate membrane lipid turnover and this may in turn influence membrane potential and cell division. Finally, it is known that the complex lipid-rich M. tuberculosis cell envelope enables the pathogen to resist host defense mechanisms (Brennan and Nikaido, 1995). Mycobacterium tuberculosis phospholipids are known to function as immune modulators (Grant et al., 1999); often released during intracellular growth and are transported to lysosomes for possibly modulating their function (Fischer et al., 2001; Geisel et al., 2005; Rhoades et al., 2005). Accordingly, one can also envision that changes in phospholipid turnover affects phospholipid ability to function as effective immune modulators, which could in turn influence M. tuberculosis multiplication upon infection. Detailed studies on phospholipid metabolism in lysX mutant and its contribution in cell division could provide valuable clues as to how lysX activity promotes M. tuberculosis proliferation upon infection.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by NIH grants AI73966, AI084734 to Murty V. Madiraju, AI48417 to Malini Rajagoapalan, and AI30036, AI37856, and AI36973 to William R. Bishai. We thank Dr. Paul Converse for his interest and support at various stages of work, and Drs. Sabine Ehrt and Chris Sassetti for sharing Ptet and Pnit promoter plasmids, respectively.
Footnote
References
Brennan, P. J., and Nikaido, H. (1995). The envelope of mycobacteria. Annu. Rev. Biochem. 64, 29–63.
Bright-Gaertner, E., and Proulx, P. (1972). Metabolism of phosphoglycerides in Escherichia coli during growth at 37 degrees C and during a cold-induced lag phase. Biochim. Biophys. Acta 270, 40–49.
Chauhan, A., Lofton, H., Maloney, E., Moore, J., Fol, M., Madiraju, M. V., and Rajagopalan, M. (2006a). Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol. Microbiol. 62, 132–147.
Chauhan, A., Madiraju, M. V., Fol, M., Lofton, H., Maloney, E., Reynolds, R., and Rajagopalan, M. (2006b). Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J. Bacteriol. 188, 1856–1865.
Darwin, K. H., Ehrt, S., Gutierrez-Ramos, J. C., Weich, N., and Nathan, C. F. (2003). The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302, 1963–1966.
DeChavigny, A., Heacock, P. N., and Dowhan, W. (1991). Sequence and inactivation of the pss gene of Escherichia coli. Phosphatidylethanolamine may not be essential for cell viability. J. Biol. Chem. 266, 10710.
Dziedzic, R., Kiran, M., Plocinski, P., Ziolkiewicz, M., Brzostek, A., Moomey, M., Vadrevu, I. S., Dziadek, J., Madiraju, M., and Rajagopalan, M. (2010). Mycobacterium tuberculosis ClpX interacts with FtsZ and interferes with FtsZ assembly. PLoS ONE 5, e11058. doi: 10.1371/journal.pone.0011058
Ehrt, S., Guo, X. V., Hickey, C. M., Ryou, M., Monteleone, M., Riley, L. W., and Schnappinger, D. (2005). Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33, e21.
Fischer, K., Chatterjee, D., Torrelles, J., Brennan, P. J., Kaufmann, S. H., and Schaible, U. E. (2001). Mycobacterial lysocardiolipin is exported from phagosomes upon cleavage of cardiolipin by a macrophage-derived lysosomal phospholipase A2. J. Immunol. 167, 2187–2192.
Fol, M., Chauhan, A., Nair, N. K., Maloney, E., Moomey, M., Jagannath, C., Madiraju, M. V., and Rajagopalan, M. (2006). Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol. Microbiol. 60, 643–657.
Gallet, P. F., Maftah, A., Petit, J. M., Denis-Gay, M., and Julien, R. (1995). Direct cardiolipin assay in yeast using the red fluorescence emission of 10-N-nonyl acridine orange. Eur. J. Biochem. 228, 113–119.
Gandotra, S., Schnappinger, D., Monteleone, M., Hillen, W., and Ehrt, S. (2007). In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat. Med. 13, 1515–1520.
Geisel, R. E., Sakamoto, K., Russell, D. G., and Rhoades, E. R. (2005). In vivo activity of released cell wall lipids of Mycobacterium bovis bacillus Calmette–Guerin is due principally to trehalose mycolates. J. Immunol. 174, 5007–5015.
Grant, E. P., Degano, M., Rosat, J. P., Stenger, S., Modlin, R. L., Wilson, I. A., Porcelli, S. A., and Brenner, M. B. (1999). Molecular recognition of lipid antigens by T cell receptors. J. Exp. Med. 189, 195–205.
Kawai, F., Shoda, M., Harashima, R., Sadaie, Y., Hara, H., and Matsumoto, K. (2004). Cardiolipin domains in Bacillus subtilis marburg membranes. J. Bacteriol. 186, 1475–1483.
Kraus, D., and Peschel, A. (2006). Molecular mechanisms of bacterial resistance to antimicrobial peptides. Curr. Top. Microbiol. Immunol. 306, 231–250.
Maloney, E., Stankowska, D., Zhang, J., Fol, M., Cheng, Q. J., Lun, S., Bishai, W. R., Rajagopalan, M., Chatterjee, D., and Madiraju, M. V. (2009). The two-domain LysX protein of Mycobacterium tuberculosis is required for production of lysinylated phosphatidylglycerol and resistance to cationic antimicrobial peptides. PLoS Pathog. 5, e1000534. doi: 10.1371/journal.ppat.1000534
Mileykovskaya, E., and Dowhan, W. (2000). Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 182, 1172–1175.
Mileykovskaya, E., and Dowhan, W. (2009). Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim. Biophys. Acta 1788, 2084–2091.
Mileykovskaya, E., Dowhan, W., Birke, R. L., Zheng, D., Lutterodt, L., and Haines, T. H. (2001). Cardiolipin binds nonyl acridine orange by aggregating the dye at exposed hydrophobic domains on bilayer surfaces. FEBS Lett. 507, 187–190.
Nishibori, A., Kusaka, J., Hara, H., Umeda, M., and Matsumoto, K. (2005). Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes. J. Bacteriol. 187, 2163–2174.
Nishijima, S., Asami, Y., Uetake, N., Yamagoe, S., Ohta, A., and Shibuya, I. (1988). Disruption of the Escherichia coli cls gene responsible for cardiolipin synthesis. J. Bacteriol. 170, 775–780.
Pandey, A. K., Raman, S., Proff, R., Joshi, S., Kang, C. M., Rubin, E. J., Husson, R. N., and Sassetti, C. M. (2009). Nitrile-inducible gene expression in mycobacteria. Tuberculosis (Edinb.) 89, 12–16.
Peschel, A. (2002). How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10, 179–186.
Peschel, A., Jack, R. W., Otto, M., Collins, L. V., Staubitz, P., Nicholson, G., Kalbacher, H., Nieuwenhuizen, W. F., Jung, G., Tarkowski, A., van Kessel, K. P., and van Strijp, J. A. (2001). Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193, 1067–1076.
Peteroy, M., Severin, A., Zhao, F., Rosner, D., Lopatin, U., Scherman, H., Belanger, A., Harvey, B., Hatfull, G. F., Brennan, P. J., and Connell, N. D. (2000). Characterization of a Mycobacterium smegmatis mutant that is simultaneously resistant to D-cycloserine and vancomycin. Antimicrob. Agents Chemother. 44, 1701–1704.
Petit, J. M., Maftah, A., Ratinaud, M. H., and Julien, R. (1992). 10N-nonyl acridine orange interacts with cardiolipin and allows the quantification of this phospholipid in isolated mitochondria. Eur. J. Biochem. 209, 267–273.
Rajagopalan, M., Maloney, E., Dziadek, J., Poplawska, M., Lofton, H., Chauhan, A., and Madiraju, M. V. (2005). Genetic evidence that mycobacterial FtsZ and FtsW proteins interact, and colocalize to the division site in Mycobacterium smegmatis. FEMS Microbiol. Lett. 250, 9–17.
Raymond, J. B., Mahapatra, S., Crick, D. C., and Pavelka, M. S. Jr. (2005). Identification of the namH gene, encoding the hydroxylase responsible for the N-glycolylation of the mycobacterial peptidoglycan. J. Biol. Chem. 280, 326–333.
Rhoades, E. R., Geisel, R. E., Butcher, B. A., McDonough, S., and Russell, D. G. (2005). Cell wall lipids from Mycobacterium bovis BCG are inflammatory when inoculated within a gel matrix: characterization of a new model of the granulomatous response to mycobacterial components. Tuberculosis (Edinb.) 85, 159–176.
Romantsov, T., Helbig, S., Culham, D. E., Gill, C., Stalker, L., and Wood, J. M. (2007). Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol. Microbiol. 64, 1455–1465.
Short, S. A., and White, D. C. (1972). Biosynthesis of cardiolipin from phosphatidylglycerol in Staphylococcus aureus. J. Bacteriol. 109, 820–826.
Smith, I. (2003). Mycobacterium tuberculosis: pathogenesis and molecular determinants of virulence. Clin. Microbial. Rev. 16, 463–496.
Talaat, A. M., Lyons, R., Howard, S. T., and Johnston, S. A. (2004). The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. U.S.A. 101, 4602–4607.
Thedieck, K., Hain, T., Mohamed, W., Tindall, B. J., Nimtz, M., Chakraborty, T., Wehland, J., and Jansch, L. (2006). The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol. Microbiol. 62, 1325–1339.
Tischler, A. D., and McKinney, J. D. (2010). Contrasting persistence strategies in Salmonella and Mycobacterium. Curr. Opin. Microbiol. 13, 93–99.
Keywords: tuberculosis, mycobacteria, lysX, phospholipids, cardiolipin, cell division
Citation: Maloney E, Lun S, Stankowska D, Guo H, Rajagoapalan M, Bishai WR and Madiraju MV (2011) Alterations in phospholipid catabolism in Mycobacterium tuberculosis lysX mutant. Front. Microbio. 2:19. doi: 10.3389/fmicb.2011.00019
Received: 20 September 2010;
Accepted: 28 January 2011;
Published online: 11 February 2011.
Edited by:
Rey Carabeo, Imperial College London, UKReviewed by:
Tanya Parish, Queen Mary University of London, UKJose A. Bengoechea, Fundacion Caubet-CIMERA Illes Balears, Spain
Copyright: © 2011 Maloney, Lun, Stankowska, Guo, Rajagoapalan, Bishai and Madiraju. This is an open-access article subject to an exclusive license agreement between the authors and Frontiers Media SA, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Murty V. Madiraju, Biomedical Research, The University of Texas Health Science Center, 11937 US Highway 271, Tyler, TX 75708-3154, USA. e-mail: murty.madiraju@uthct.edu
 Erin Maloney1
Erin Maloney1