- Institut für Mikrobiologie und Biotechnologie, Rheinische Friedrich-Wilhelms-Universität Bonn, Bonn, Germany
In the purple sulfur bacterium Allochromatium vinosum, thiosulfate oxidation is strictly dependent on the presence of three periplasmic Sox proteins encoded by the soxBXAK and soxYZ genes. It is also well documented that proteins encoded in the dissimilatory sulfite reductase (dsr) operon, dsrABEFHCMKLJOPNRS, are essential for the oxidation of sulfur that is stored intracellularly as an obligatory intermediate during the oxidation of thiosulfate and sulfide. Until recently, detailed knowledge about the regulation of the sox genes was not available. We started to fill this gap and show that these genes are expressed on a low constitutive level in A. vinosum in the absence of reduced sulfur compounds. Thiosulfate and possibly sulfide lead to an induction of sox gene transcription. Additional translational regulation was not apparent. Regulation of soxXAK is probably performed by a two-component system consisting of a multi-sensor histidine kinase and a regulator with proposed di-guanylate cyclase activity. Previous work already provided some information about regulation of the dsr genes encoding the second important sulfur-oxidizing enzyme system in the purple sulfur bacterium. The expression of most dsr genes was found to be at a low basal level in the absence of reduced sulfur compounds and enhanced in the presence of sulfide. In the present work, we focused on the role of DsrS, a protein encoded by the last gene of the dsr locus in A. vinosum. Transcriptional and translational gene fusion experiments suggest a participation of DsrS in the post-transcriptional control of the dsr operon. Characterization of an A. vinosum ΔdsrS mutant showed that the monomeric cytoplasmic 41.1-kDa protein DsrS is important though not essential for the oxidation of sulfur stored in the intracellular sulfur globules.
Introduction
The ability to utilize reduced sulfur compounds as electron donors for anaerobic phototrophic and aerobic or anaerobic chemotrophic growth is phylogenetically wide-spread (Dahl et al., 2008a). It occurs in organisms residing in environments abundant with sulfide like organic nutrient-rich anoxic sediments or hydrothermal vents. Many environmentally important photo- and chemotrophic sulfur-oxidizing bacteria accumulate globules of polymeric, water-insoluble sulfur as an intermediary product during the oxidation of reduced sulfur compounds such as thiosulfate or sulfide. These sulfur globules are deposited intracellularly, inside the periplasm in many chemotrophic sulfur oxidizers (e.g., Beggiatoa species or the bacterial endosymbionts of marine invertebrates like Riftia pachyptila or Calyptogena okutanii) and also in phototrophic purple sulfur bacteria of the family Chromatiaceae. Allochromatium vinosum, a representative of the latter, has been especially well characterized on a molecular genetic level (Dahl, 2008; Frigaard and Dahl, 2009).
It is established that thiosulfate oxidation in this organism is strictly dependent on the presence of three periplasmic Sox proteins encoded by the soxB, soxXAK, and soxYZ genes (Hensen et al., 2006; Welte et al., 2009). These genes are organized in three different transcriptional units (Hensen et al., 2006) within the genome of A. vinosum (Acc. No. NC_013851). It is also well documented that proteins encoded by the A. vinosum dissimilatory sulfite reductase (dsr) genes, dsrABEFHCMKLJOPNRS, are essential for the oxidation of sulfur stored intracellularly as an obligatory intermediate during the oxidation of sulfide and thiosulfate (Pott and Dahl, 1998; Dahl et al., 2005; Sander et al., 2006).
Until recently, detailed knowledge about the regulation of these two important enzyme systems was not available. We started to fill this gap and showed that the dsr genes are expressed in a reduced sulfur compound-dependent manner (Grimm et al., 2010b). In A. vinosum, the expression of most of the 15 dsr genes is at a low basal level in the absence of reduced sulfur compounds and greatly enhanced in the presence of sulfide. Real-time-PCR experiments suggested that the genes dsrC and dsrS are not only expressed from the main dsr promoter but also from secondary internal promoters, pointing at a special function of the encoded proteins. DsrC was identified as a potential DNA-binding protein (Grimm et al., 2010b). An A. vinosum ΔdsrR deletion strain showed a significantly reduced sulfur oxidation rate that was fully restored upon complementation with dsrR in trans. Immunoblot analyses revealed a reduced level of DsrE and DsrL in the ΔdsrR strain. These proteins are absolutely essential for sulfur oxidation. Transcriptional and translational gene fusion experiments suggested a participation of DsrR in the post-transcriptional control of the dsr operon (Grimm et al., 2010a).
Much less is currently known about regulation of thiosulfate oxidation. The most detailed information regarding sox gene regulation is available for the chemotrophic sulfur oxidizer Paracoccus pantotrophus. It should be noted that in this organism thiosulfate oxidation does not proceed along exactly the same pathway as in A. vinosum. P. pantotrophus contains an additional Sox protein, the sulfane dehydrogenase SoxCD (Zander et al., 2010), and oxidizes thiosulfate to sulfate without the formation of sulfur globules as an intermediate. In P. pantotrophus, two genes, soxR and soxS, are divergently oriented to the other sox genes, soxVWXYZA–H. SoxR, a transcriptional regulator of the ArsR family, has been shown to act as a repressor of sox gene expression in the absence of thiosulfate. SoxS appears to be a periplasmic thioredoxin and is essential for full sox gene expression (Rother et al., 2005). In A. vinosum neither soxR nor soxS homologs are present. Information on sox gene regulation in this model organism has so far been restricted to the finding by Hensen et al. (2006) that SoxA is produced in higher amounts during photolithoautotrophic as compared to photoorganoheterotrophic growth.
In the present work, we therefore set out to provide more detailed information on sox gene regulation in a purple sulfur bacterium at the molecular biological level via RT-PCR and translational gene fusions. Furthermore, we analyzed dsr gene regulation in A. vinosum in more detail and assessed the role of the dsrS encoded protein in this context.
Materials and Methods
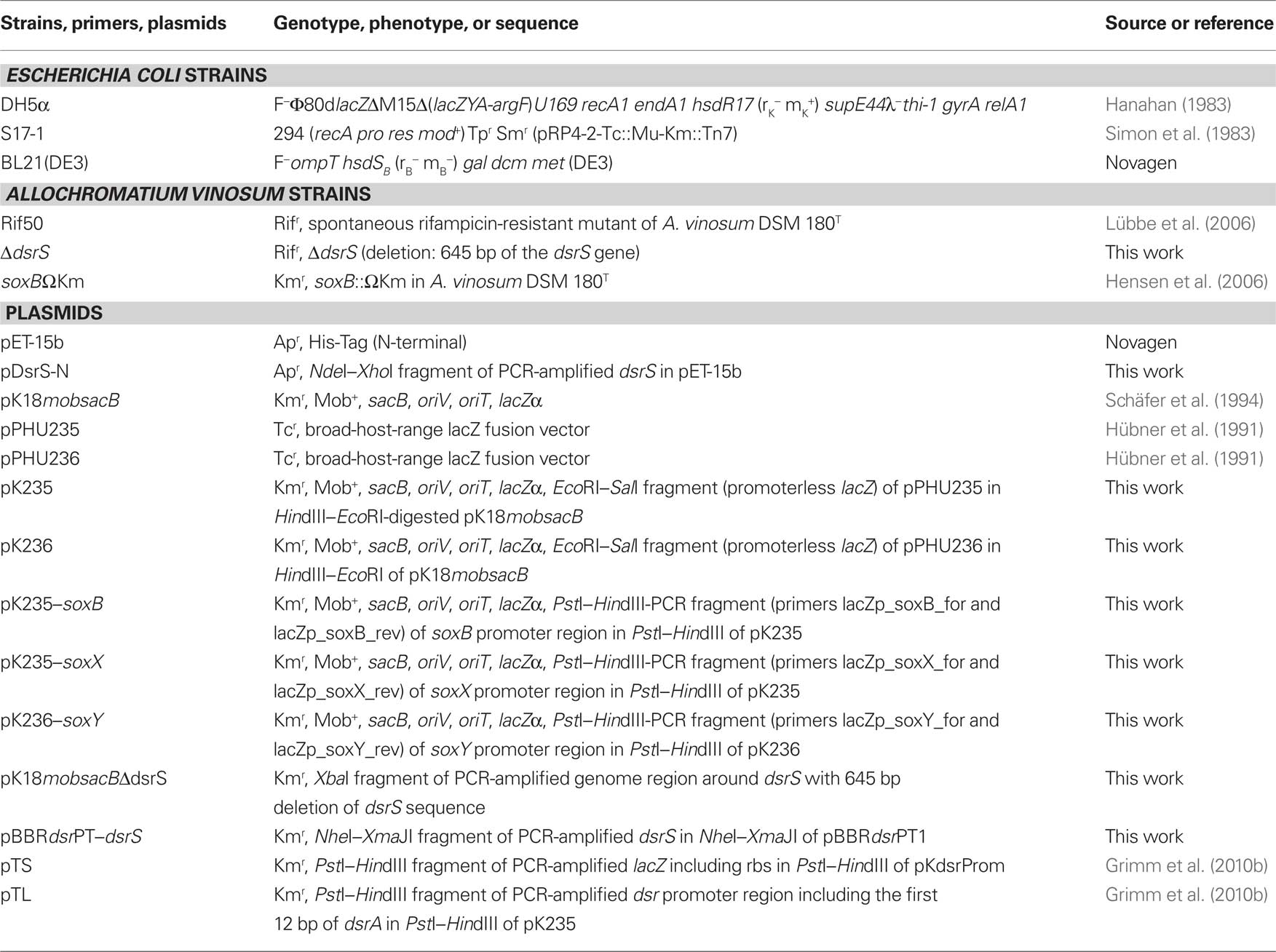
Bacterial Strains, Plasmids, Media, and Growth Conditions
The strains and plasmids used in this study are listed in Table 1. A. vinosum was grown and harvested as described (Dahl et al., 2008b). Antibiotics were used at the following concentrations (in μg mL−1): for Escherichia coli, kanamycin, 50; ampicillin, 100; for A. vinosum, kanamycin, 10; rifampicin, 50.
Overproduction and Purification of Recombinant DsrS
DsrS was overproduced with an amino-terminal His-tag in E. coli BL21(DE3) cells containing pDsrS-N (Table 1). The cells were cultured in 500 mL LB medium containing 100 μg ampicillin mL−1 at 25°C and 180 r.p.m. At an OD600 nm of 0.5, 2 μM IPTG was added and the cells were harvested after 3 h. The pellet was resuspended in 50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 7.5, including Complete protease inhibitor cocktail, EDTA-free (Roche), and 1 mg lysozyme mL−1. The cells were disrupted by sonication (2 min mL−1, Cell Disruptor B15, Branson) and centrifuged at 10000g for 30 min at 4°C. The N-terminally His-tagged DsrS was mainly found in the pellet, though solubility was improved by a low growth temperature (25°C). The supernatant containing soluble DsrS was purified using a nickel agarose column (Qiagen) followed by dialysis against 50 mM Tris–HCl (pH 7.5), 300 mM NaCl. The protein was concentrated to a final volume of no more than 2 mL via Centriprep-10 (Amicon). The state of oligomerization of the protein was investigated by gel filtration chromatography on a Superdex-200 column (GE Healthcare) equilibrated with 50 mM Tris–HCl (pH 7.5), 300 mM NaCl.
Construction, Characterization, and Complementation of an A. vinosum ΔdsrS in Frame Deletion Strain
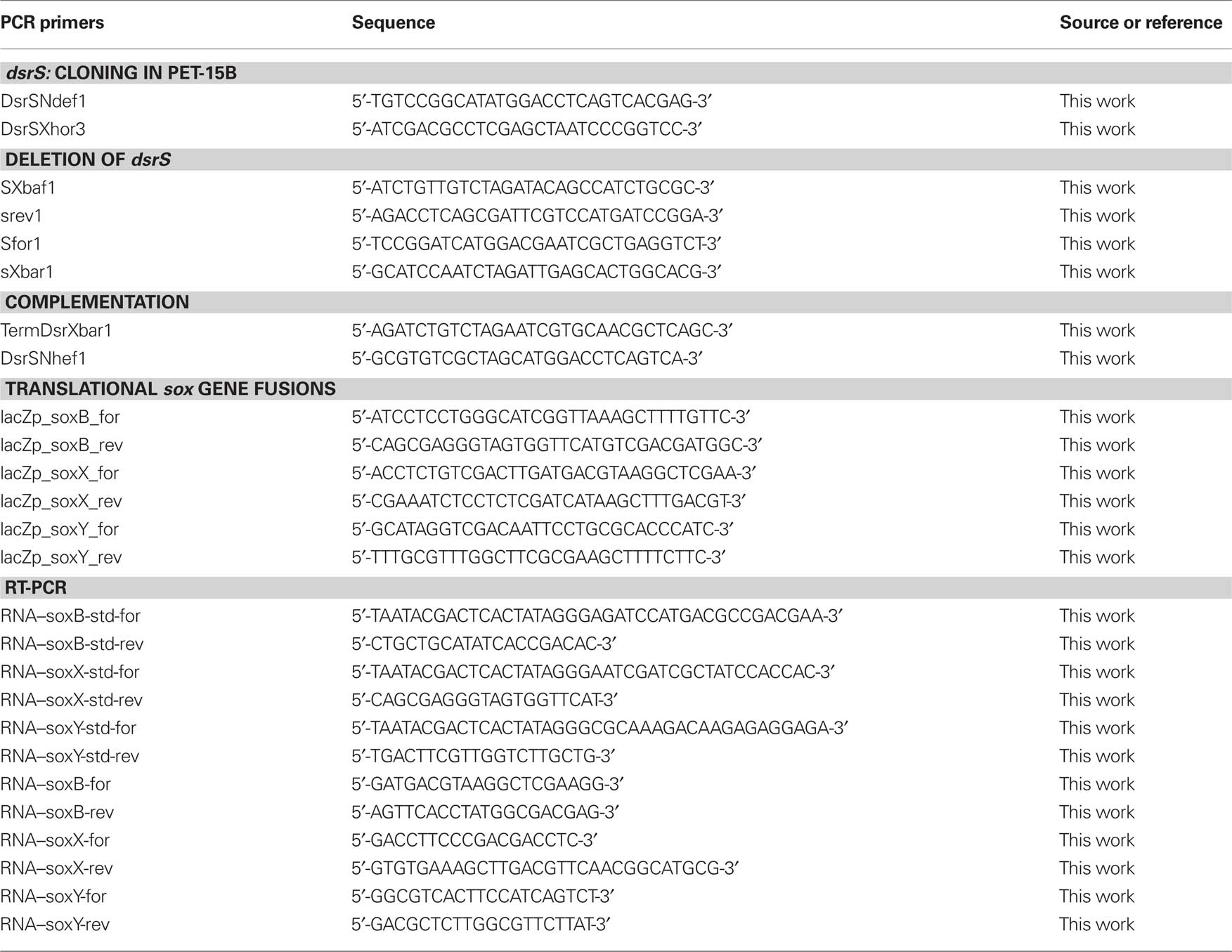
All general molecular genetic techniques, as well as the method for achieving and complementing in frame deletions in A. vinosum, were described earlier (Dahl et al., 2008b). The primers SXbaf1, srev1, Sfor1, and sXbar1 were utilized for the construction of the dsrS deletion and the primer pair DsrSNhef1 and TermDsrXbar1 was used to amplify the dsrS gene for the complementation of the deletion strain (Table 2). Photolithoautotrophic growth of A. vinosum strains was examined in batch culture under continuous illumination essentially as described by Prange et al. (2004) in a medium containing sulfide as the sole sulfur compound. 250 mL of a photoheterotrophically grown stationary-phase culture were harvested (5900g, 10 min) and the cell material was used to inoculate 1 L of modified Pfennig’s medium (Dahl et al., 2008b) in a thermostatted fermenter. Sulfur compounds were determined as described in (Dahl et al., 2008b). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Immunoblot (Western) analyses were performed as described in Dahl et al. (2005).
Expression Studies by RT-PCR
Cells were harvested in the stationary growth phase and used to inoculate the modified “Pfennig’s” medium described in Dahl et al. (2008b) supplemented with either 2 mM malate, 2 mM sulfide, 2 mM thiosulfate, or 5 mM sulfite. In case of the experiments concerning dsr genes, cells were harvested for RNA isolation 3 h after inoculation. At that time the maximum content of intracellular sulfur is achieved and its oxidation commences (Grimm et al., 2010b). For experiments concerning sox genes, cells were harvested 30 min, 1 h, and 3 h after inoculation into medium containing sulfide, thiosulfate, and sulfite, respectively because cells exhibited high and constant substrate oxidation rates around these time points. Total RNA of A. vinosum Rif50, A. vinosum ΔdsrS, and A. vinosum soxB::Ω-Km was isolated and the concentration was determined as described in Prange et al. (2004). 250 ng of total RNA were used as template in RT-PCR analysis via the QuantiTect SYBR Green RT-PCR kit (Qiagen) and the iCycler iQ real-time detection system (Bio-Rad) according to the manufacturers’ instructions. “No RT” control reactions were performed for each RNA sample. In case of DNA contamination, the RNA samples were digested with RNase-free DNase (Qiagen) and purified using RNeasy Mini Kit (Qiagen). Fragments of approximately 200 bp were amplified in all cases (dsrA: 215, dsrE: 217, dsrC: 196, dsrL: 186, dsrR: 181, dsrS: 172, soxB: 167, soxX: 163, soxY: 180 bp) following established protocols (Grimm et al., 2010b) and using primers listed by Grimm et al. (2010b) and in Table 2. RNA standards were generated as described in Fey et al. (2004). The samples were automatically quantified by the iCycler iQ software (Bio-Rad) based on the RNA standards. The absence of non-specific PCR products and primer–dimers that would otherwise contribute to the fluorescence signal was confirmed by melting curve analysis as described in Grimm et al. (2010b). The PCR products were furthermore analyzed by agarose gel electrophoresis. To guarantee comparability, the levels of dsr and sox gene expression were measured by absolute quantitative RT-PCR using gene-specific RNA standards in every run. It was assumed that the PCR efficiency did not vary in a single run between the samples and the in vitro transcribed RNA fragments, containing the target sequence that served as external standards. Variations in PCR efficiencies between different runs or different target genes were taken into account by quantifying the samples using the run-specific standard curve that allows for variations of reagents, primers, and sequence etc. The gene-specific RNA standards yielded calibration curves of high linearity in all cases (correlation coefficient >0.990).
Construction of Transcriptional and Translational Reporter Gene Fusions
DNA fragments of 1074, 1046, and 733 bp encompassing the probable soxB, soxX, and soxY promoter regions, respectively, including the first 12 or 15 bp of the respective gene were amplified using primers that introduced PstI and HindIII restriction sites. A. vinosum Rif50 chromosomal DNA served as a template. The fragments were introduced into plasmids pK235 or pK236 yielding plasmids pK235–soxB, pK235–soxX, and pK236–soxY (Table 1). Plasmids pK235 and pK236 were constructed by excising the promoterless lacZ gene with SalI and EcoRI from the translational fusion vectors pPHU235 or pPHU236 (Hübner et al., 1991), respectively. The SalI sites were filled in with the Klenow fragment of DNA polymerase. The fragments were then inserted into the 5670-bp EcoRI/HindIII fragment of pK18mobsacB.
Plasmids pK235–soxB, pK235–soxX, and pK236–soxY as well as the transcriptional gene fusion plasmid pTS (Table 1; Grimm et al., 2010b), containing a fusion of the lacZ gene to the dsr promoter dsrAP region without the Shine–Dalgarno sequence of dsrA, and the translational gene fusion plasmid pTL (Table 1; Grimm et al., 2010b), containing the dsrAP region including the first 12 bp of dsrA fused to the lacZ gene, were transferred into A. vinosum Rif50 by conjugation as described in Pattaragulwanit and Dahl (1995) and integrated into the genome via single-crossover. The plasmid carrying strains were grown on 12 mL modified Pfennig’s medium containing 2 mM thiosulfate, 2 mM sulfide, 2 mM malate, 5 mM sulfite, or combinations thereof for 24 h under continuous illumination before β-galactosidase activity was tested as described in Grimm et al. (2010b). A. vinosum wild type did not exhibit any β-galactosidase activity.
Sequence Analysis
Promoter prediction for prokaryotic sequence was achieved with Neural Network Promoter Prediction1 and BPROM2. The online version of the program REPuter3 was used for detection of inverted or direct repeats in the nucleotide sequence upstream of dsrA. All amino acid sequences were obtained from GenBank. PSI-BLAST was used with default parameters to generate the protein sequence family (Altschul et al., 1997). Putative helix-turn-helix motives were identified using GYM2.0 (Gao et al., 1999; Narasimhan et al., 2002) and Helix-turn-Helix Motif Prediction (Combet et al., 2000).
Results
Regulation of sox Genes in A. vinosum
In A. vinosum, the soxXAKL genes (Hensen et al., 2006; Welte et al., 2009) form a transcriptional unit that may also include gene Alvin_2172 (formerly ORFb) located immediately downstream of soxL (Figure 1). Alvin_2172 encodes a conserved hypothetical protein predicted to be soluble and to reside in the cytoplasm. A typical Cys-X2-Cys thioredoxin motif and a typical heme c-binding Cys-X2-Cys-His binding motif are present. An inverted repeat with a potential for formation of a hairpin loop structure in the corresponding mRNA was found within the nucleotide sequence of the downstream gene Alvin_2173 (formerly ORFc) by Hensen et al. (2006) and proposed to function as a site for transcription termination. A second predicted transcriptional unit comprises the gene soxB and probably also includes Alvin_2166 (formerly ORFa). These genes are located upstream of soxX and are divergently transcribed. The product of Alvin_2166 is a putative multi-sensor histidine kinase. The corresponding putative response regulator is encoded by gene Alvin_2165. The genes soxYZ (Alvin_2111 and 2112) are not found in the vicinity of the other sox genes and are located in a third independent transcriptional unit (Hensen et al., 2006; Figure 1).
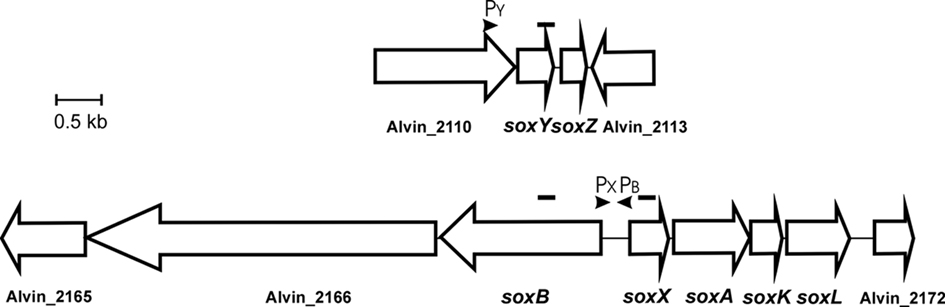
Figure 1. Schematic overview of the two sox gene loci in A. vinosum. The location of potential promoters for soxB, soxX, and soxY is indicated. Amplicons generated for RT-PCR are indicated by black bars above the genes.
Transcriptional regulation of sox genes
When reconstituted in vitro, the P. pantotrophus Sox proteins do not only oxidize thiosulfate but also accept hydrogen sulfide, sulfur (or polysulfide), and also sulfite as substrates. In addition, it has been shown for P. pantotrophus and for the phototrophic alphaproteobacterial Rhodovulum sulfidophilum that the Sox system is not only essential for thiosulfate oxidation but is also strictly required for the oxidation of sulfide in vivo (Chandra and Friedrich, 1986; Wodara et al., 1994; Appia-Ayme et al., 2001). This prompted us to investigate expression of the A. vinosum genes soxB, soxX, and soxY during photoorganoheterotrophic growth with malate and photolithoautotrophic growth with thiosulfate, sulfide, or sulfite using quantitative RT-PCR with absolute standards. Table 3 shows a constitutive expression for all three genes during growth with malate in the absence of reduced sulfur compounds. When A. vinosum was grown in the presence of 2 mM thiosulfate as electron donor, the expression levels increased six and fourfold for soxB and soxX, respectively, and 23-fold for soxY. The presence of sulfide also induced the expression of the sox genes, albeit to a lesser extent. It can currently not be excluded that this is a secondary effect caused by the formation of thiosulfate from sulfide. It has been repeatedly reported that A. vinosum produces thiosulfate during growth with sulfide as electron donor (Steudel et al., 1990; Franz et al., 2009). Expression levels for all three studied sox genes were not increased by the presence of 5 mM sulfite as compared to growth on malate (Table 3).

Table 3. Expression levels of three sox genes under photoorganoheterotrophic (malate) and photolithoautotrophic (thiosulfate, sulfide, or sulfite) conditions determined by RT-PCR.
Translational gene fusions
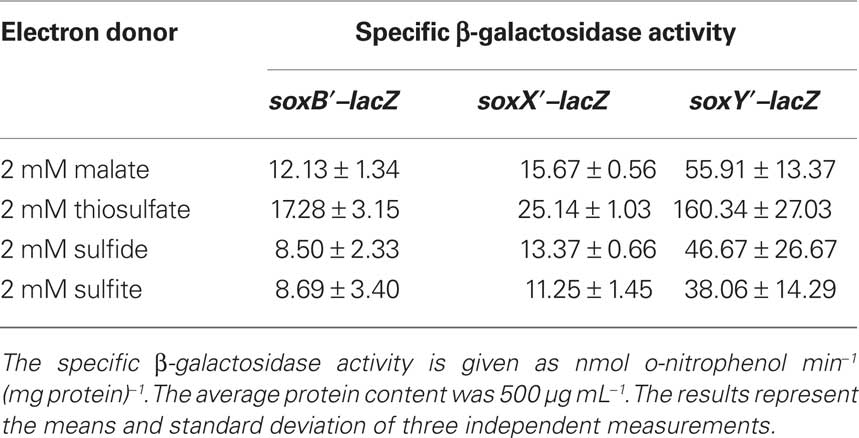
As the next step, we investigated expression of the three sox-genes also on the translational level by determining the specific β-galactosidase activity of Sox–LacZ fusion proteins (Table 4). All three Sox–LacZ fusion proteins showed a basal level activity during photoorganoheterotrophic growth with malate. When A. vinosum grew with 2 mM thiosulfate as an electron donor, specific β-galactosidase activity clearly increased and increasing thiosulfate concentrations led to even higher activities (Table 5). In contrast to the situation on the transcriptional level, sulfide did not induce significantly higher specific β-galactosidase activity of any of the Sox–LacZ fusion proteins. This effect might be caused by the different sensitivities of the assays. A formation of up to 0.15 mM thiosulfate has been documented when A. vinosum grows with 2 mM sulfide (Franz et al., 2009). While this rather low thiosulfate concentration may lead to an increase of transcription rates detectable by RT-PCR, the effect on the translational level may not cause increases of specific β-galactosidase activities significantly above the experimental error range. In coincidence with the RT-PCR results, the presence of 5 mM sulfite led to specific β-galactosidase activities comparable to those obtained during photoorganoheterotrophic growth, i.e., in contrast to thiosulfate sulfite does not induce sox gene expression in A. vinosum.
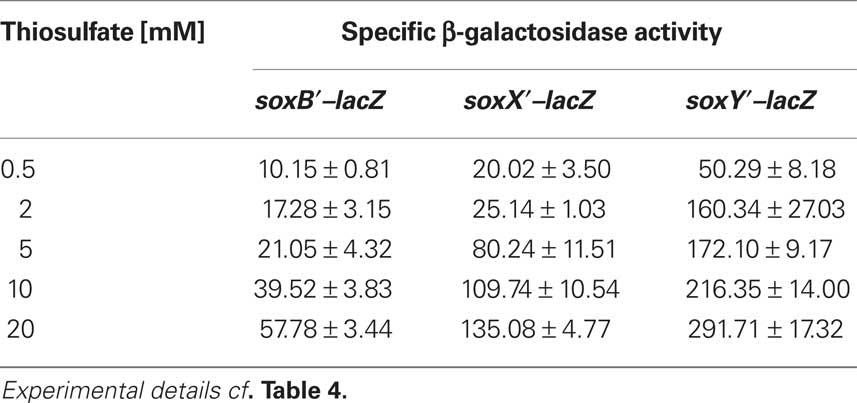
Table 5. Dependence on thiosulfate concentration of specific β-galactosidase activity of Sox–LacZ fusion proteins in A. vinosum.
Proteins encoded by Alvin_2166 and Alvin_2165 as potential regulators of sox gene expression
The protein encoded by A. vinosum gene Alvin_2166 is homologous to multi-sensor histidine kinases from various proteobacteria. The protein is predicted to reside in the cytoplasm and shows a complex organization with several predicted conserved domains specific for multi-sensor hybrid histidine kinases. At the N-terminus two PAS domains (cd00130) are located followed by a HisKA domain (cd00082), a HATPase_c domain (cd00075), and two carboxy-terminal REC domains (cd00156). Both PAS domains contain a Cys-X2-Cys motif indicating a redox signal as a stimulus for the sensing domain of the putative histidine kinase. The protein encoded by Alvin_2165 shows homology to response regulators of two-component systems from various proteobacteria. It is also predicted to be a cytoplasmic protein and contains a conserved REC domain at the N-terminus and a GGDEF domain typical for di-guanylate cyclases at the C-terminus (cd01949; Chan et al., 2004). A helix-turn-helix motif is not predicted for Alvin_2165 indicating a role of cyclic di-GMP as a second messenger (Römling and Amikam, 2006) in the regulation of the sox genes in A. vinosum. Homologs of Alvin_2166 and Alvin_2165 are also present in two other sulfur oxidizers, namely Halorhodospira halophila SL1, a purple sulfur bacterium of the family Ectothiorhodospiraceae, and Magnetococcus sp. MC-1. In both organisms, however, the corresponding genes are not located in immediate vicinity of sox genes.
In order to find some experimental evidence for a role of the proteins encoded by Alvin_2166 and Alvin_2165 in sox gene regulation, transcription of soxX and soxY was investigated in the A. vinosum mutant strain soxB::Ω-Km (Hensen et al., 2006). This strain carries an insertion of a polar Ω kanamycin resistance cassette (Frey and Krisch, 1985; Fellay et al., 1987) in soxB preventing the transcription of soxB and genes located downstream in the same transcriptional unit. Table 3 shows the transcription rates of soxX and soxY in mutant soxB::Ω-Km as compared to those in the wild type. The expression levels of both genes were found to be significantly higher (about 10-fold increase in each case) in mutant soxB::Ω-Km than in the wild type during growth with malate in the absence of thiosulfate. The presence of 2 mM thiosulfate did not lead to increased expression levels of soxY and soxX in the mutant strain, i.e., the induction by thiosulfate observed for the wild type was lost in the mutant very probably due to the deleterious effect of the interposon on the formation of the Alvin_2166-encoded regulatory protein. This interpretation gains further support in case of soxX gene regulation by our finding that the basal level of expression was unaffected as compared to the wild type and induction of transcription by thiosulfate still observable in strain A. vinosum ΔsoxY carrying an in frame deletion of the soxY gene (data not shown).
Further Insights into Regulation of Purple Bacterial dsr Genes: Role of DsrS
The only known gene region responsible for the oxidation of stored zero-valent sulfur is the dsr gene cluster (Dahl et al., 2008b; Grimm et al., 2008; Frigaard and Dahl, 2009; Sander and Dahl, 2009). In A vinosum, the first sulfur-oxidizing bacterium for which the dsr genes were described, this cluster encompasses 15 genes (dsrABEFHCMKJLOPNRS; Pott and Dahl, 1998; Dahl et al., 2005). The first two genes, dsrAB, encode a key enzyme of this pathway, the reverse-acting dsr. Comparison with the dsr sequences of other sulfur-oxidizing bacteria showed the genes dsrABEFHCMKLJOPN to be the core unit of the operon, present in all sulfur-oxidizing bacteria that form sulfur globules as an intermediate (Sander et al., 2006). While the proteins encoded by the core dsr genes have been the subject of a number of recent studies (Pott and Dahl, 1998; Dahl et al., 2005; Lübbe et al., 2006; Sander et al., 2006; Cort et al., 2008), hardly anything is known about dsrS, the last gene of the A. vinosum dsr operon.
Properties of DsrS and occurrence of the gene
The dsrS gene is predicted to encode a soluble cytoplasmic protein with a molecular mass of 41.1 kDa (Dahl et al., 2005). Neither conserved domains nor motifs are present in the sequence and significant similarities to proteins of known function are not apparent. Recombinant DsrS was purified from E. coli and its oligomerization state was investigated by gel filtration chromatography. DsrS eluted as a monomer and the sequence deduced mass of 43.3 kDa for the recombinant protein was confirmed (data not shown). UV–Vis spectra did not indicate the presence of any cofactors.
In fact, A. vinosum is so far the only organism in which dsrS is part of the dsr gene cluster. In other sulfur-oxidizing bacteria the gene is either absent [green sulfur bacetria (e.g. Chlorobium tepidum TLS, NC_002932, complete genome; Chlorobium limicola DSM 245, NC_010803, complete genome; Chlorobaculum parvum NCIB 8327, NC_011027, complete genome), Halorhodospira halophila SL1 (NC_008789, complete genome)] or located elsewhere in the genome [Thiobacillus denitrificans (NC_007404, complete genome), Beggiatoa sp. PS (NZ_ABBZ00000000, whole genome draft sequence), Thioalkalivibrio sp. HL-EbGR7 (NC_011901, complete genome), and endosymbionts Candidatus Vesicomyosocius okutanii HA (NC_009465, complete genome) and Candidatus Ruthia magnifica str. Cm (NC_008610, complete genome)]. Thus, the question arose whether the protein encoded by dsrS is at all involved in sulfur oxidation. We constructed and characterized an A. vinosum ΔdsrS in frame deletion mutant to answer this question and studied the effect of the gene deletion on transcription and translation of the dsr genes.
Construction, phenotypic characterization, and complementation of a ΔdsrS A. vinosum in frame deletion strain
To assess the importance of DsrS for sulfur oxidation we constructed an A. vinosum strain with in frame deletion of dsrS. In order to examine the phenotype of A. vinosum ΔdsrS, we cultivated the strain photolithoautotrophically in batch culture with 2 mM sulfide as electron source. As expected for a classical purple sulfur bacterium like A. vinosum (Brune, 1995), sulfide was immediately oxidized to zero-valent sulfur that was stored in periplasmic sulfur globules. During the oxidation of sulfide to sulfur of oxidation state zero, two different polysulfides are formed as intermediates by A. vinosum wild type (Prange et al., 2004). The exact chain length of the polysulfides formed is not known (Prange et al., 2004; Franz et al., 2009). The formation of both polysulfides was not affected in the ΔdsrS mutant (not shown). Neither the sulfide oxidation rate, the rate of sulfur globule formation nor the growth yield were affected by the deletion of dsrS (Table 6). When sulfide is depleted, A. vinosum further metabolizes stored sulfur to sulfate that is excreted into the medium. The ΔdsrS mutant was clearly still able to completely perform this pathway and formed sulfate as the end product, however, compared to the wild type it exhibited a specific sulfur oxidation rate that was significantly reduced by ∼30% (Table 6). Complementation of the A. vinosum ΔdsrS strain with dsrS in trans under the control of the main dsr promoter dsrAP did not restore the wild type oxidation rate, but further reduced the sulfur oxidation rate to 45% of the wild type rate (Table 6).
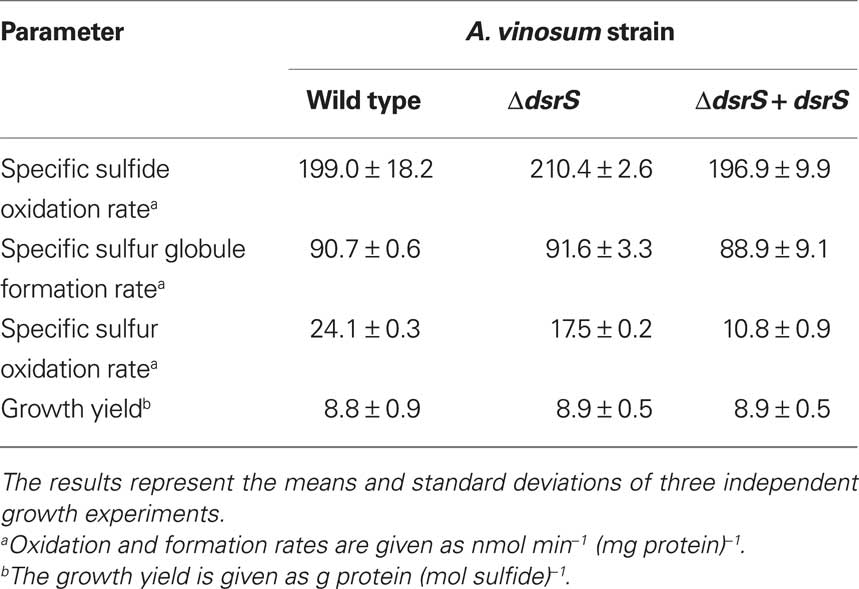
Table 6. Characteristics of the A. vinosum ΔdsrS deletion mutant compared to the wild type and the complementation mutant.
In order to find an explanation for the observed phenotypes we performed comparative immunoblot analysis of A. vinosum wild type and ΔdsrS soluble cell fractions (Figure 1). These did not reveal any apparent influence of the lack of DsrS on the formation of the proteins DsrE and DsrL. These proteins are known to be essential for sulfur oxidation (Lübbe et al., 2006; Dahl et al., 2008b). Interestingly, the formation of DsrE and DsrL appeared to be disturbed in the ΔdsrS + dsrS complementation strain. Both of these proteins were hardly detectable in cells harvested while they were still oxidizing internal sulfur globules (Figure 2). In a later phase of sulfur oxidation, however, when sulfur globules had essentially vanished from the cells, both proteins were apparently no longer adversely affected. At that point, the same or even higher amounts of both proteins were detected in the ΔdsrS + dsrS strain as compared to the wild type or the ΔdsrS mutant (Figure 2). One likely explanation for these observations is that formation of DsrE and DsrL and possibly also other Dsr proteins is delayed in the complementation strain.
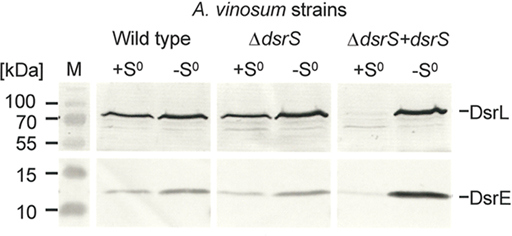
Figure 2. Immunological detection of DsrE (14.6 kDa) and DsrL (71.4 kDa) in dsrS deletion (ΔdsrS) and complementation (ΔdsrS + dsrS) strains in comparison to A. vinosum wild type. Cells were grown in batch culture on 2 mM sulfide and harvested either at the maximum content of intracellularly stored sulfur (+S0) or after the sulfur had been completely metabolized (−S0). Sixty-eight microgram protein of the soluble fraction were used per lane. Both antisera were raised against oligopeptides comprising a highly immunogenic epitope deduced from the nucleotide sequence and their specific reaction with DsrE and DsrL overproduced in E. coli has been proven earlier (Dahl et al., 2005).
Transcription of A. vinosum dsr genes is not affected by deletion of dsrS
We took a closer look at the transcript levels of several dsr genes via RT-PCR (Figure 3). In the wild type, the gene dsrS exhibited an enhanced expression under sulfur-oxidizing conditions, thus confirming the involvement of DsrS in sulfur oxidation. Interestingly, dsrS is transcribed at rates similar to those observed for the constitutively expressed gene dsrC (Figure 3). Furthermore, it has been previously observed that dsrC as well as dsrS are still transcribed in the A. vinosum interposon mutant 21D that carries a transcriptional and translational block in dsrB thus abolishing the expression of downstream genes transcribed from the dsrA promoter (Pott and Dahl, 1998; Grimm et al., 2010b). The high transcription levels of dsrS and the similarity to the transcription pattern of dsrC, for which a secondary promoter has already been postulated (Pott and Dahl, 1998), suggest a possible secondary internal promoter also for dsrS. Indeed, in silico analyses of the sequence upstream of dsrS using the online tools Neural Network Promoter Prediction and BPROM revealed potential promoter sequences in the region of dsrNR (Figure 3).
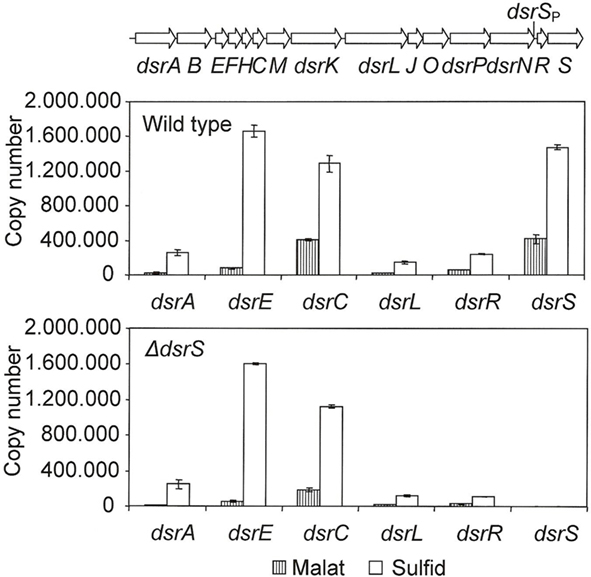
Figure 3. Expression levels of six dsr genes under photoorganoheterotrophic (malate) and photolithoautotropic (sulfide) conditions determined by RT-PCR. The RNAs were isolated from A. vinosum wild type and the ΔdsrS mutant. Quantified external RNA fragments containing the target sequence served as standards. The location of a potential secondary promoter for dsrS as determined by BPROM and Neural Network Promoter Prediction is indicated.
Effect of dsrS deletion on the transcriptional and post-transcriptional level
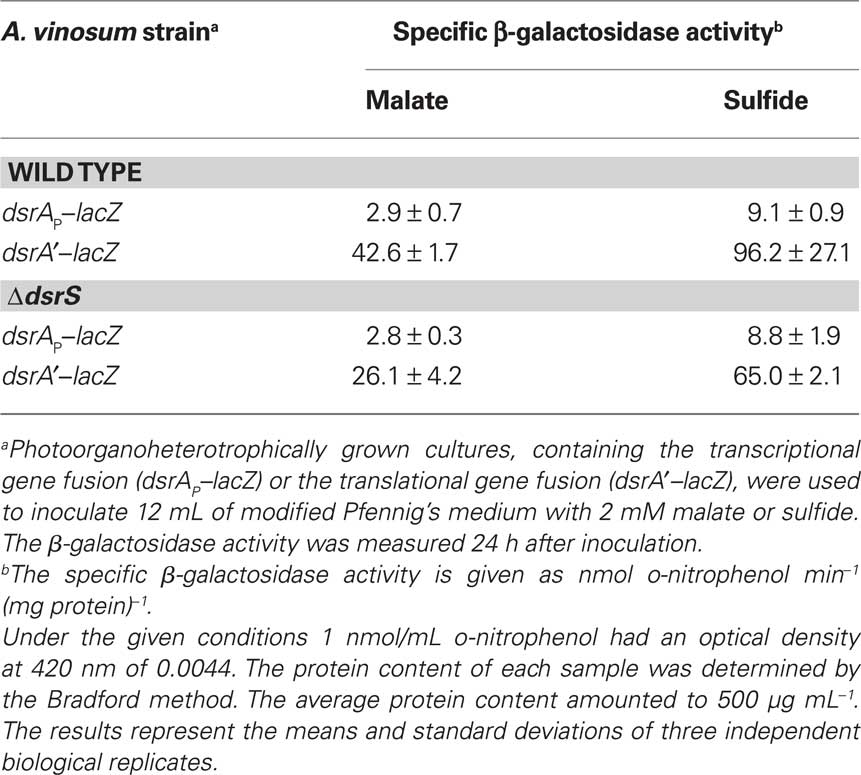
In an attempt to find an explanation for the reduced sulfur oxidation rate of the A. vinosum ΔdsrS strain we introduced lacZ reporter gene fusions into the ΔdsrS mutant strain and the wild type and examined the expression under photoorganoheterotrophic (malate) and photolithoautotrophic (CO2 and sulfide) growth conditions. The transcription rate of the dsrA gene was measured by determining the specific β-galactosidase activities of the lacZ fusion to the main dsr promoter dsrAP. A dsrA′–lacZ gene fusion, where both transcription and translation of the lacZ gene were dependent on dsrA gene expression, was used to determine the translation of the gene. The specific β-galactosidase activities were at a low basal level in malate-grown cells and increased approximately threefold under sulfur-oxidizing conditions (Table 7). In agreement with the RT-PCR results, the ΔdsrS mutant carrying the transcriptional fusion exhibited the same level of activities as the wild type. We deduce that DsrS has no effect on the transcription of dsrA. On the other hand, the ΔdsrS mutant carrying the translational fusion showed a ∼35% reduction of β-galactosidase activities as compared to the wild type. The effect was independent of the growth conditions.
Discussion
Here, we show that the sox genes which encode proteins essential for thiosulfate oxidation are constitutively expressed even in the absence of reduced sulfur compounds in the purple bacterium A. vinosum. A low basal expression has also been found for the dsr genes encoding the enzymes required for the oxidation of stored sulfur in the same organism under photoorganoheterotrophic growth conditions (Grimm et al., 2010b). Both, Dsr and Sox proteins are obviously constitutively formed to ensure a basal level, so that reduced sulfur compounds can be used as soon as they become available. This observations fits well with other lines of evidence that suggest a preferential utilization of reduced sulfur compounds over organic compounds as electron donors in A. vinosum, e.g., the reported repressive effect of thiosulfate on pyruvate utilization and the concomitant use of thiosulfate and pyruvate or acetate (Hurlbert and Lascelles, 1963; Hurlbert, 1968; Grimm et al., 2010b). Thiosulfate and possibly sulfide lead to an induction of sox gene transcription. Additional translational regulation was not apparent. Experiments with varying thiosulfate concentrations showed sox gene expression levels to be dependent on the strength of the inducing signal. Apparently, the expression can be modified according to demand.
Regulation of soxXAKL is probably performed by a two-component system encoded by Alvin_2166 and Alvin_2165 and consisting of a multi-sensor histidine kinase and a regulator with proposed di-guanylatecyclase activity. So far our experiments indicate a derepression of soxXAKL expression when the protein encoded by Alvin_2166 is not present in the cells. We base this conclusion on our finding that in the absence of thiosulfate transcript levels for soxX are lower in the wild type than in A. vinosum soxB::ΩKm carrying an interposon with an adverse effect also on Alvin_2166. The inductory effect of thiosulfate was lost in the mutant strain, i.e., the high transcript levels found in this strain were not further increased by the presence of the reduced sulfur compound. Sequence analyses indicate that Alvin_2166 has the potential to act as a redox sensor and the NADH/NAD+ ratio which is directly coupled to the oxidation of thiosulfate via the photosynthetic electron transport chain might be a possible sensing signal. However, this hypothesis has to be substantiated in the future.
Apparently, regulation of sox genes in A. vinosum follows pathways fundamentally different from those described for the very few other organisms for which sox gene regulation has been studied. In P. pantotrophus the transcriptional ArsR family regulator SoxR has been shown to bind via a helix-turn-helix motif at two positions within the sox genes (Rother et al., 2005). Homology modeling of SigE and ORF1 in Starkeya novella provided evidence that SigE functions as a repressor binding via a helix-turn-helix motif at the promoter region while presence of reduced sulfur compounds led to binding of ORF1 to SigE and finally detachment of SigE from the DNA (Kappler et al., 2001; Bagchi and Ghosh, 2006). In A. vinosum neither genes encoding homologs of SoxR nor genes encoding proteins resembling SigE or ORF1 appear to be present. When we take into account that homologs of Alvin_2165 and Alvin_2166 were found only in H. halophila SL1 and Magnetococcus MC-1 but closely related genes were not detected in the genomes of other sulfur-oxidizing prokaryotes harboring sox genes, sox gene regulation in A. vinosum appears to follow a quite unique mechanism.
In contrast to soxXAKL, expression of soxYZ is probably not regulated by Alvin_2166 and Alvin_2165. The copy numbers of soxY in A. vinosum soxB::Ω-Km indeed differ from the wild type ones in our RT-PCR experiments, but soxY shows the same expression pattern in a soxX::Ω-Km strain (data not shown). Therefore the changes in expression of soxY in A. vinosum soxB::ΩKm are unlikely due to the missing multi-sensor histidine kinase. In accordance with the fact that the three essential sox genes are located in three different transcription units (Figure 1) regulation of their expression seems to be quite complex and will require more attention in the future.
In the second part of this work we show that the dsrS encoded protein is relevant though not essential for the oxidation of sulfur stored in intracellular sulfur globules in the purple sulfur bacterium A. vinosum. We confirmed earlier sequence analyses by recombinant expression: the gene dsrS encodes a monomeric protein of the deduced 41.1-kDa molecular mass and does not contain cofactors. The gene is transcribed at a high level under photoorganoheterotrophic conditions in the absence of reduced sulfur compounds and the mRNA level further increases under sulfur-oxidizing conditions, indicating direct or indirect involvement of the encoded protein in the sulfur-oxidizing process. The transcript levels are similar to those observed for the constitutively and highly expressed gene dsrC for which a secondary promoter has been postulated. They are significantly higher than those for dsrA under photoorganoheterotrophic conditions in the absence of reduced sulfur compounds even though dsrA encodes a subunit of the key enzyme for intracellular sulfur oxidation. The presence of secondary promoters for dsrC and dsrS is in agreement with previous comparative analyses of dsr gene gene transcription in A. vinosum wild type and mutant 21D (Grimm et al., 2010b). Both, dsrC and dsrS were still expressed at a high level in the mutant strain. The mutant carries an insertion in dsrB of a kanamycin Ω interposon which abolishes transcription of any downstream genes in the same transcriptional unit unless secondary promoters are present. In addition, in silico analyses revealed potential promoter sequences in the regions upstream of dsrC and dsrS (Grimm et al., 2010b).
Characterization of a ΔdsrS mutant showed that DsrS is important though not essential for the oxidation of intracellular stored sulfur. Complementation in trans of the ΔdsrS strain with dsrS under control of the main dsr promoter dsrAP did not restore the sulfur oxidation rate to wild type levels even though comparable plasmids carrying a single dsr gene cloned immediately downstream of dsrAP have already been successfully used for complementation of A. vinosum mutants carrying deletions of the respective dsr gene. In all cases described so far, wild type oxidation rates were restored (Dahl et al., 2008b). The ΔdsrS + dsrS strain clearly behaved differently, indicating that the dsrA promoter may not be able to provide the cell with the necessary level of DsrS and pointing at the presence of a special secondary promoter for dsrS. When we consider that dsrS is not part of the dsr operon in other sulfur-oxidizing bacteria, it is not too surprising that dsrS is additionally regulated by a separate promoter in A. vinosum. This would explain why complementation of the A. vinosum ΔdsrS mutant with dsrS under control of the dsrA promoter did not cause the expected phenotype. The main dsr promoter dsrAP may not be the major factor in the expression of dsrS. In order to operate correctly, dsrS probably has to be under control of the correct regulating element(s). When we compared the transcription patterns of several dsr genes in the wild type with those in the ΔdsrS deletion mutant, major differences were not apparent, though the transcription of dsrC under organotrophic conditions appeared to be diminished in the ΔdsrS strain (Figure 3). In summary, the deletion of dsrS does not appear to have a major effect on the transcription of the dsr genes. This observation fits well with the results of the immunoblot analysis, as a perceptible reduction in the formation of DsrE and DsrL could not be observed.
Translational gene fusion experiments suggest a participation of DsrS in the post-transcriptional control of the dsr operon. The in frame deletion of dsrS lead to a reduced formation of DsrA′–LacZ and concomitantly to a reduced sulfur oxidation rate. In fact, a reduced production of DsrA protein is a straight forward explanation for the observed reduced sulfur oxidation rate of the ΔdsrS mutant. Apparently, less dsrAB-encoded sulfite reductase is formed when DsrS is lacking, than when it is not. DsrS could act either indirectly as part of a signal transducing reporter chain cascade or directly by stabilizing the ribosome–mRNA interaction and thus enhancing translation. Another possibility is that DsrS is involved in translational attenuation, i.e., induces a conformational change in the mRNA thereby permitting translational initiation. Interestingly, the entire Shine–Dalgarno sequence of dsrA is part of a possible stem-loop preventing ribosomal access.
Although we do not have enough direct evidence to clarify these possibilities at present, we showed that the region encompassing the dsrA ribosome binding site is required for the down-regulation of the accumulation of DsrA protein in the absence of DsrS. Additional studies are, however, necessary to elucidate the exact mechanism of the post-transcriptional-regulation of DsrA by DsrS and to explain the delayed formation of DsrE and DsrL in the complementation mutant.
Conclusion
Here we show, that the sox genes in A. vinosum are expressed on a low constitutive level even in the absence of reduced sulfur compounds. The extent of induction of sox gene transcription is dependent on the thiosulfate concentration. Additional translational regulation of the A. vinosum sox genes was not apparent. A two-component system consisting of a multi-sensor histidine kinase and a regulator with proposed di-guanylate cyclase activity probably exerts a regulatory effect on the soxXAK genes. In A. vinosum the expression of most dsr genes was also found to be at a low basal level in the absence of reduced sulfur compounds and to be enhanced in the presence of sulfide (Grimm et al., 2010b). In the present work, we demonstrate a participation of DsrS in the post-transcriptional control of the dsr operon. Previous work had suggested a participation in post-transcriptional control of the same operon also for DsrR, a protein encoded immediately upstream of the dsrS gene (Grimm et al., 2010a). Apparently the regulation of the dsr operon and the function of DsrS are more intricate than previously expected. Further studies are clearly necessary to obtain a complete picture of the regulation of genes involved in sulfur oxidation in purple sulfur bacteria.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants Da 351/3-4, Da 351/3-5,and Da 351/4-3 from the Deutsche Forschungsgemeinschaft (DFG). We thank Bernd Masepohl for his helpful hints concerning gene fusions.
Footnotes
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Appia-Ayme, C., Little, P. J., Matsumoto, Y., Leech, A. P., and Berks, B. C. (2001). Cytochrome complex essential for photosynthetic oxidation of both thiosulfate and sulfide in Rhodovulum sulfidophilum. J. Bacteriol. 183, 6107–6118.
Bagchi, A., and Ghosh, T. C. (2006). Structural study of two proteins SigE and ORF1 to predict their roles in the biochemical oxidation of sulfur anions via the global sulfur oxidation operon (sox). Comput. Biol. Chem. 30, 227–232.
Brune, D. C. (1995). “Sulfur compounds as photosynthetic electron donors,” in Anoxygenic Photosynthetic Bacteria, eds R. E. Blankenship, M. T. Madigan, and C. E. Bauer (Dordrecht: Kluwer Academic Publishers), 847–870.
Chan, C., Paul, R., Samoray, D., Amiot, N. C., Giese, B., Jenal, U., and Schirmer, T. (2004). Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. U.S.A. 101, 17084–17089.
Chandra, T. S., and Friedrich, C. G. (1986). Tn5-induced mutations affecting the sulfur-oxidizing ability (Sox) of Thiosphaera pantotropha. J. Bacteriol. 166, 446–452.
Combet, C., Blanchet, C., Geourjon, C., and Deleage, G. (2000). NPS@: network protein sequence analysis. Trends Biochem. Sci. 25, 147–150.
Cort, J. R., Selan, U. M., Schulte, A., Grimm, F., Kennedy, M. A., and Dahl, C. (2008). Allochromatium vinosum DsrC: solution-state NMR structure, redox properties and interaction with DsrEFH, a protein essential for purple sulfur bacterial sulfur oxidation. J. Mol. Biol. 382, 692–707.
Dahl, C. (2008). “Inorganic sulfur compounds as electron donors in purple sulfur bacteria,” in Sulfur in Phototrophic Organisms, eds R. Hell, C. Dahl, D. B. Knaff, and T. Leustek (Dordrecht: Springer), 289–317.
Dahl, C., Engels, S., Pott-Sperling, A. S., Schulte, A., Sander, J., Lübbe, Y., Deuster, O., and Brune, D. C. (2005). Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J. Bacteriol. 187, 1392–1404.
Dahl, C., Friedrich, C. G., and Kletzin, A. (2008a). “Sulfur oxidation in prokaryotes,” in Encyclopedia of Life Sciences (ELS) (Chichester: John Wiley & Sons, Ltd.), doi: 10.1002/9780470015902.a0021155. Available at: http://www.els.net/
Dahl, C., Schulte, A., Stockdreher, Y., Hong, C., Grimm, F., Sander, J., Kim, R., Kim, S.-H., and Shin, D. H. (2008b). Structural and molecular genetic insight into a wide-spread bacterial sulfur oxidation pathway. J. Mol. Biol. 384, 1287–1300.
Fellay, R., Frey, J., and Krisch, H. M. (1987). Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vivo insertional mutagenesis of Gram-negative bacteria. Gene 52, 147–154.
Fey, A., Eichler, S., Flavier, S., Christen, R., Höfle, M. G., and Guzmán, C. A. (2004). Establishment of a real-time PCR-based approach for accurate quantification of bacterial RNA targets in water, using Salmonella as a model organism. Appl. Environ. Microbiol. 70, 3618–3623.
Franz, B., Gehrke, T., Lichtenberg, H., Hormes, J., Dahl, C., and Prange, A. (2009). Unexpected extracellular and intracellular sulfur species during growth of Allochromatium vinosum with reduced sulfur compounds. Microbiology 155, 2766–2774.
Frey, J., and Krisch, H. M. (1985). Omega mutagenesis in Gram-negative bacteria: a selectable interposon which is strongly polar in a wide range of bacterial species. Gene 36, 143–150.
Frigaard, N.-U., and Dahl, C. (2009). Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 54, 103–200.
Gao, Y., Mathee, K., Narasimhan, G., and Wang, X. (1999). “Motif detection in protein sequences,” in Proceedings of 6th International Symposium on String Processing and Information Retrieval, Cancun, 63–72.
Grimm, F., Cort, J. R., and Dahl, C. (2010a). DsrR, a novel IscA-like protein lacking iron- and FeS-binding function involved in the regulation of sulfur oxidation in Allochromatium vinosum. J. Bacteriol. 192, 1652–1661.
Grimm, F., Dobler, N., and Dahl, C. (2010b). Regulation of dsr genes encoding proteins responsible for the oxidation of stored sulfur in Allochromatium vinosum. Microbiology 156, 764–773.
Grimm, F., Franz, B., and Dahl, C. (2008). “Thiosulfate and sulfur oxidation in purple sulfur bacteria,” in Microbial Sulfur Metabolism, eds C. Dahl and C. G. Friedrich (Berlin: Springer), 101–116.
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580.
Hensen, D., Sperling, D., Trüper, H. G., Brune, D. C., and Dahl, C. (2006). Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol. Microbiol. 62, 794–810.
Hübner, P., Willison, J. C., Vignais, P., and Bickle, T. A. (1991). Expression of regulatory nif genes in Rhodobacter capsulatus. J. Bacteriol. 173, 2993–2999.
Hurlbert, R. E. (1968). Effect of thiol-binding reagents on the metabolism of Chromatium D. J. Bacteriol. 95, 1706–1712.
Hurlbert, R. E., and Lascelles, J. (1963). Ribulose diphosphate carboxylase in Thiorhodaceae. J. Gen. Microbiol. 33, 445–458.
Kappler, U., Friedrich, C. G., Trüper, H. G., and Dahl, C. (2001). Evidence for two pathways of thiosulfate oxidation in Starkeya novella (formerly Thiobacillus novellus). Arch. Microbiol. 175, 102–111.
Lübbe, Y. J., Youn, H.-S., Timkovich, R., and Dahl, C. (2006). Siro(haem)amide in Allochromatium vinosum and relevance of DsrL and DsrN, a homolog of cobyrinic acid a,c diamide synthase for sulphur oxidation. FEMS Microbiol. Lett. 261, 194–202.
Narasimhan, G., Bu, C., Gao, Y., Wang, X., Xu, N., and Mathee, K. (2002). Mining for motifs in protein sequences. J. Comput. Biol. 9, 707–720.
Pattaragulwanit, K., and Dahl, C. (1995). Development of a genetic system for a purple sulfur bacterium: conjugative plasmid transfer in Chromatium vinosum. Arch. Microbiol. 164, 217–222.
Pott, A. S., and Dahl, C. (1998). Sirohaem-sulfite reductase and other proteins encoded in the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 144, 1881–1894.
Prange, A., Engelhardt, H., Trüper, H. G., and Dahl, C. (2004). The role of the sulfur globule proteins of Allochromatium vinosum: mutagenesis of the sulfur globule protein genes and expression studies by real-time RT PCR. Arch. Microbiol. 182, 165–174.
Römling, U., and Amikam, D. (2006). Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9, 218–228.
Rother, D., Orawski, G., Bardischewsky, F., and Friedrich, C. G. (2005). SoxRS-mediated regulation of chemotrophic sulfur oxidation in Paracoccus pantotrophus. Microbiology 151, 1707–1716.
Sander, J., and Dahl, C. (2009). “Metabolism of inorganic sulfur compounds in purple bacteria,” in Purple Bacteria, eds C. N. Hunter, F. Daldal, M. C. Thurnauer, and J. T. Beatty (Dordrecht: Springer), 595–622.
Sander, J., Engels-Schwarzlose, S., and Dahl, C. (2006). Importance of the DsrMKJOP complex for sulfur oxidation in Allochromatium vinosum and phylogenetic analysis of related complexes in other prokaryotes. Arch. Microbiol. 186, 357–366.
Schäfer, A., Tauch, A., Jäger, W., Kalinowski, J., Thierbach, G., and Pühler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73.
Simon, R., Priefer, U., and Pühler, A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1, 784–791.
Steudel, R., Holdt, G., Visscher, P. T., and van Gemerden, H. (1990). Search for polythionates in cultures of Chromatium vinosum after sulfide incubation. Arch. Microbiol. 155, 432–437.
Welte, C., Hafner, S., Krätzer, C., Quentmeier, A. T., Friedrich, C. G., and Dahl, C. (2009). Interaction between Sox proteins of two physiologically distinct bacteria and a new protein involved in thiosulfate oxidation. FEBS Lett. 583, 1281–1286.
Wodara, C., Kostka, S., Egert, M., Kelly, D. P., and Friedrich, C. G. (1994). Identification and sequence analysis of the soxB gene essential for sulfur oxidation of Paracoccus denitrificans GB17. J. Bacteriol. 176, 6188–6191.
Zander, U., Faust, A., Klink, B. U., de Sanctis, D., Panjikar, S., Quentmeier, A., Bardischewsky, F., Friedrich, C. G., and Scheidig, A. J. (2010). Structural basis for the oxidation of protein-bound sulfur by the sulfur cycle molybdohemo-enzyme sulfane dehydrogenase SoxCD. J. Biol. Chem. 286, 8349–8360.
Keywords: Allochromatium vinosum, sox genes, thiosulfate oxidation, anoxygenic phototrophic sulfur bacteria, dsr genes, regulation, dissimilatory sulfite reductase, sulfur globules
Citation: Grimm F, Franz B and Dahl C (2011) Regulation of dissimilatory sulfur oxidation in the purple sulfur bacterium Allochromatium vinosum. Front. Microbio. 2:51. doi: 10.3389/fmicb.2011.00051
Received: 31 January 2011;
Accepted: 08 March 2011;
Published online: 22 March 2011.
Edited by:
Martin G. Klotz, University of Louisville, USAReviewed by:
Niels-Ulrik Frigaard, University of Copenhagen, DenmarkJan Kuever, Bremen Institute for Materials Testing, Germany
Copyright: © 2011 Grimm, Franz and Dahl. This is an open-access article subject to an exclusive license agreement between the authors and Frontiers Media SA, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Christiane Dahl, Institut für Mikrobiologie und Biotechnologie, Rheinische Friedrich-Wilhelms-Universität Bonn, Meckenheimer Allee 168, D-53115 Bonn, Germany.e-mail: chdahl@uni-bonn.de
 Frauke Grimm
Frauke Grimm