- School of Chemistry and Molecular Biosciences, Centre for Metals in Biology, The University of Queensland, St. Lucia, QLD, Australia
Sulfite dehydrogenases (SDHs) are enzymes that catalyze the oxidation of the toxic and mutagenic compound sulfite to sulfate, thereby protecting cells from adverse effects associated with sulfite exposure. While some bacterial SDHs that have been characterized to date are able to use cytochrome c as an electron acceptor, the majority of these enzymes prefer ferricyanide as an electron acceptor and have therefore been termed “atypical” SDHs. Identifying the natural electron acceptor of these enzymes, however, is crucial for understanding how the “atypical” SDHs are integrated into cell metabolism. The SorT sulfite dehydrogenase from Sinorhizobium meliloti is a representative of this enzyme type and we have investigated the interactions of SorT with two small redox proteins, a cytochrome c and a Cu containing pseudoazurin, that are encoded in the same operon and are co-transcribed with the sorT gene. Both potential acceptor proteins have been purified and characterized in terms of their biochemical and electrochemical properties, and interactions and enzymatic studies with both the purified SorT sulfite dehydrogenase and components of the respiratory chain have been carried out. We were able to show for the first time that an “atypical” sulfite dehydrogenase can couple efficiently to a cytochrome c isolated from the same organism despite being unable to efficiently reduce horse heart cytochrome c, however, at present the role of the pseudoazurin in SorT electron transfer is unclear, but it is possible that it acts as an intermediate electron shuttle between. The SorT system appears to couple directly to the respiratory chain, most likely to a cytochrome oxidase.
Introduction
Sulfite dehydrogenases (SDHs) are enzymes that catalyze the oxidation of sulfite, which can cause damage to important cellular components such as proteins, DNA, and lipids, to the inert and non-toxic compound sulfate (Kappler and Dahl, 2001; Kappler, 2011). They belong to the sulfite oxidase family of molybdenum enzymes that comprises three distinct groups of enzymes, two of which are largely unstudied, while the third and best studied group contains the sulfite oxidases (SO) and SDHs that oxidize sulfite to sulfate (Kappler, 2008).
One of the distinctive features of molybdenum enzymes is that they can catalyze the transfer of an oxygen atom to a substrate using water as the oxygen donor (Hille, 1996). The reaction catalyzed by SOs and SDHs proceeds according to the general equation
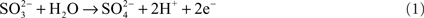
For SO [EC1.8.3.1], such as the enzyme from Arabidopsis thaliana (Eilers et al., 2001), oxygen serves as the electron acceptor, leading to the production of hydrogen peroxide, while for most SDHs [EC1.8.2.1], which include the vertebrate “SO,” two molecules of cytochrome c serve as the natural electron acceptor (Rajagopalan, 1980; Kappler, 2011).
Sulfite-oxidizing enzymes (SOEs) have been studied for over 40 years in vertebrates and humans, where SOEs are essential and their absence causes the lethal sulfite oxidase deficiency syndrome, however, significant progress in studying bacterial SOEs has only been made in the last 10 years (Kappler and Dahl, 2001; Kappler, 2011). SOEs have been identified in a wide variety of bacteria including sulfur-oxidizing chemolithotrophs, organosulfonate degraders, pathogenic, and extremophilic bacteria (Kappler and Dahl, 2001; Myers and Kelly, 2005; Di Salle et al., 2006; D’Errico et al., 2006; Denger et al., 2008; Wilson and Kappler, 2009; Kappler, 2011), suggesting an important role for these enzymes outside dissimilatory sulfur metabolism.
Vertebrate SOEs occur in the mitochondrial intermembrane space and appear to have a uniform overall structure. They are homodimers with a mobile heme b domain fused to the main body of the enzyme which consists of a molybdenum binding and a dimerization domain. As far as is known, this combination of domains is the common “core structure” of all SOEs, and, e.g., the plant SO are homodimers of this core structure (Schrader et al., 2003).
In contrast to the conserved architecture of the vertebrate and plant enzymes, bacterial SOEs are much more diverse in terms of their overall structure. Of the bacterial SOEs studied in sufficient detail to date one is a heterodimer of the core Moco/dimer subunit and a cytochrome c subunit (Kappler et al., 2000; Kappler and Bailey, 2005), while SOEs from other bacteria are either homodimers or even monomers of the core two domain structure (Denger et al., 2008; D’Errico et al., 2006; Di Salle et al., 2006; Wilson and Kappler, 2009; Kappler, 2011).
Intriguingly however, with the exception of the heterodimeric, cytochrome c containing enzyme isolated from the soil bacterium Starkeya novella (Kappler et al., 2000), almost all the bacterial SOEs isolated to date display higher activities when assayed with the artificial electron acceptor ferricyanide than when assayed with cytochrome c (Kappler, 2011), which raises the question how these enzymes are integrated into cell metabolism.
This is of particular interest as bacterial SOEs have been found in bacteria with widely differing lifestyles, including thermophilic bacteria, pathogens, and environmental bacteria, and thus might be fulfilling very different metabolic roles. The ferricyanide-linked bacterial SOEs have, in fact, been referred to as “atypical” SOEs due to their low activity with cytochrome c and were first described in organosulfonate degrading bacteria (Reichenbecher et al., 1999). In contrast, bacterial SOEs that prefer cytochrome c as the natural electron acceptor are directly linked to the respiratory chain and thus energy generation via the natural electron acceptor cytochromes (mitochondrial cytochrome c in vertebrates, a cytochrome c550 in Starkeya novella; Kappler et al., 2000) that are able to transfer electrons directly to the respective cytochrome c oxidases (Rajagopalan, 1980; Yamanaka et al., 1981).
As “atypical” SDHs, like any other redox enzyme, require an electron acceptor in order to retain their functionality, a variety of possibilities exist that could explain the high activity of these SDHs, with the artificial electron acceptor ferricyanide. The natural electron acceptor could be molecular oxygen, however, this would lead to the formation of hydrogen peroxide that would then have to be detoxified in additional enzymatic reactions. Alternatively, the natural acceptor could be an electron transfer protein occurring within the cells, but this protein might have redox properties that differ from those of horse heart cytochrome c (e.g., a different redox potential might be required), or might have different structural properties that enable efficient docking to these SDHs (Wilson and Kappler, 2009). Another option would be that the “atypical” SDHs can transfer electrons directly to, e.g., the cytochrome oxidase, thus by passing the need for a separate electron carrier.
We have recently purified a ferricyanide-linked SOE from the plant symbiontic soil bacterium Sinorhizobium meliloti grown on the organosulfonate taurine. The SorT SDH is a periplasmic, homodimeric SOEs with a clear preference for ferricyanide over horse heart cytochrome c in in vitro assays (Wilson and Kappler, 2009). The SorT SDH also displayed some interesting catalytic properties, including an almost pH invariant KM sulfite, which differs clearly from all the well studied cytochrome c linked SDHs, where KM sulfite increases almost exponentially above pH 8–8.5 (Wilson and Kappler, 2009). The sorT gene was found to be co-transcribed with two genes encoding for redox proteins, namely a cytochrome c (smc04048) and a pseudoazurin, azu2 (Wilson and Kappler, 2009). The smc04048 gene encodes a single heme cytochrome and, same as the pseudoazurin, did not appear to be part of the purified enzyme, but it was postulated that one or both of these proteins might be natural electron acceptors for SorT.
Using the SorT SDH from S. meliloti as the model system, we have investigated how atypical SDHs might be linked to cell metabolism (Figure 1). There are a variety of options, including the direct interaction of SorT with the respiratory chain, involvement of either or both of Smc04048 and Azu2. We have already shown previously that native SorT does not have sulfite oxidase activity (Wilson and Kappler, 2009), and thus investigations focused on the interactions of SorT with the two redox proteins that are co-transcribed with it the enzyme’s structural gene.
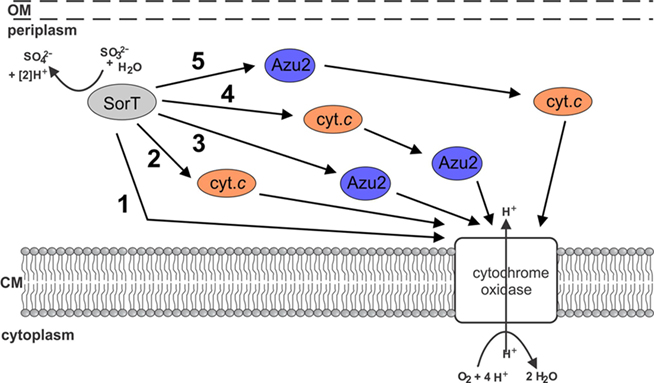
Figure 1. Schematic representation of potential electron transfer pathways between SorT and the respiratory chain. 1 direct transfer of electron transfer between SorT and the cytochrome oxidase 2 using the Smc04048 cytochrome c as an electron carrier, 3 using the pseudoazurin Azu2 as an electron carrier, 4 + 5 using both the Smc04048 cytochrome c and pseudoazurin Azu2 the as electron carriers.
Materials and Methods
Bacterial Strains and Growth Media Sinorhizobium meliloti
Rm 1021 was routinely grown on liquid or solidified DSMZ medium 69 supplemented with 20 mM taurine or TYS medium (Beringer, 1974) at 30°C. Liquid or solidified LB medium was used to cultivate Escherichia coli strains DH5α (Invitrogen), TP1000 (Palmer et al., 1996), and BL21(DE3; Novagen) at 37°C. Growth media were supplemented with antibiotics where appropriate: ampicillin 100 μg/mL, kanamycin 50 μg/mL, chloramphenicol 25 μg/mL, S. meliloti streptomycin 25 μg/mL.
Molecular Biological Methods
Standard methods were used throughout (Ausubel et al., 2005). Standard PCR reactions used GoTaqGreen Mastermix (Promega), restriction enzymes were from Invitrogen. Protein expression plasmids were constructed using pProex Htb (Invitrogen) for the sorT gene, and pET22b+ (Novagen) for the Smc04048, azu2, and Snovc550 (acc.no ADH88353) genes. The genes were amplified using the listed primers (SM_SDH4049f_bam AAAAGGATCCAAGGAGACGAAGCCTCTGCC; SM_SDH4049r_pst AAAACTGCAGTCAGGCAACGGTGAGTTTGACG; SM_04048F_nco AAAACCATGGAAGAAGACAAGCTTGCGCT; SM_4048r_tag AAAAGAATTCTTGGCTTTTCCGGCGAC; SM_azu2f-nco AAAACCATGGAGGAATATCGTGTCGAAATGCTG; SM_azu2r_tag AAAAGAATTCTCGCCGCCGCTCTCGATTTC; SNc550petF AAAACCATGGCACAGAGCCCGGCCGGCGATC; SNc550pETrevtag AAAAAAGCTTCTTCTTGGTGCCGTCGGCG) and Phusion HF polymerase (Finnzymes) before cloning into the appropriate expression vectors. Plasmids pProex-SorT, pET22b-Smc04048tag, pET22b-SNCtag, and pET22b-azu2tag were verified by DNA sequencing. The plasmid pProex-SorT contains only the coding region for the mature SorT enzyme but lacks the coding region for the signal peptide, leading to a cytoplasmic expression of recombinant SorT (rSorT). In contrast, the constructs pET-Smc04048tag, pET22b-SNCtag, and pEt22b-azu2tag contain the coding regions for the mature proteins (sequence parts encoding native signal peptides have been removed) fused to a sequence encoding the E. coli-specific PelB signal peptide, leading to protein expression in the periplasm. The choice of cytoplasmic and periplasmic expression was made depending on which cellular compartment the target protein matures in, i.e., folds and has its cofactor inserted. The location of production for the recombinant proteins should have no influence on the protein interaction studies, all of which used purified proteins.
Protein Expression Protocols
pProex-SorT was transformed into E. coli TP1000. Expression cultures (4 × 1 L in 2.5 L flasks) were grown at 37°C, 200 rpm in LB medium supplemented with ampicillin, kanamycin, and 1 mM sodium molybdate. At an OD600 of ∼0.8, protein expression was induced by the addition of 250 μM IPTG and cultures were grown overnight at 30°C before being harvested by centrifugation at 4°C and 5000 × g.
pET22b-Smc04048tag and pET22b-SNCtag were transformed into E. coli BL21(DE3) carrying plasmid pEC86 (Arslan et al., 1998). Expression cultures (4 × 1 L in 2 L flasks) were grown in autoinduction medium (Hill et al., 2007) in the presence of ampicillin and chloramphenicol at 20°C, 200 rpm for 48 h before harvesting by centrifugation. pET22b-azu2Tag was transformed into E. coli BL21(DE3) and expression cultures grown on LB medium supplemented with 1 mM copper(II) sulfate and ampicillin at 37°C, 200 rpm until an OD600 of 0.6–0.8 was reached. Protein expression was induced by addition of 50 μM IPTG and cultures incubated at 30°C for 3 h before harvesting by centrifugation and isolation of periplasmic proteins using the osmotic shock method (Ausubel et al., 2005).
Protein Purification Methods
Native SorT was purified from S. meliloti as in Wilson and Kappler (2009). For the purification of recombinant proteins, cell extracts from whole cells were prepared by passing the resuspended cells three times through a French Pressure Cell (Thermo) at 14000 psi. Cell debris was removed by centrifugation at 15000 × g, 4°C, 20 min resulting in a crude extract. rSorT was purified by preparing the crude extract in 20 mM potassium phosphate, 0.5 M NaCl pH 7.4, 4 mM Pefabloc SC (Roche) and, after adding imidazole (final conc. 20 mM), loading this extract onto a 5-mL HisTrap column (GE Healthcare Biosciences) equilibrated in the same buffer containing 20 mM imidazole. rSorT was eluted in a linear gradient from 20 to 500 mM imidazole. SorT containing fractions were pooled and desalted using a HiPrep 26/10 column (GE Healthcare Biosciences) equilibrated in 20 mM Tris–Cl pH 7.8, 5% (w/v) glycerol before concentrating the samples (Vivaspin 20 concentrators, MWCO 30 kDa, GE Healthcare Biosciences) and further purification on a Superdex75 16/60 column (GE Healthcare Biosciences) equilibrated in 20 mM Tris–Cl pH 7.8, 150 mM NaCl, 2.5% (w/v) glycerol.
Recombinant Smc04048 and SNCtag were purified from crude extracts using a 5-mL HisTrap column and the buffer system already described above. Cytochrome c containing fractions were pooled, 15% (w/v) ammonium sulfate was added and samples separated on a Phenyl Sepharose column (1.6 cm × 21 cm) equilibrated in 20 mM Tris–Cl, pH 7.8, 15% (w/v) ammonium sulfate using a linear, decreasing gradient of ammonium sulfate (100-0%, 5 CV). Fractions were pooled according to protein purity and desalted using dialysis before storing. If necessary, proteins were subjected to size exclusion chromatography on a Superdex 75 (16/60) column for further purification.
Following isolation of the periplasmic protein fraction recombinant Azu2 was purified using anion exchange chromatography (DEAE-Sepharose, buffer: 20 mM Tris–Cl pH 8.8) and a linear elution gradient (0–25% NaCl, 7 CV). Azu2 containing fractions were pooled and 0.5 mM CuSO4 added before concentrating of samples and separation on a Superdex 75 (16/60) in 20 mM Tris–Cl pH 7.8, 150 mM NaCl). If necessary, size exclusion chromatography was repeated to improve protein purity.
Biochemical Methods
SDS PAGE used the method of (Laemmli, 1970). Protein determinations were performed using either the BCA kit (Sigma-Aldrich) or 2D Quant kit (GE Healthcare Biosciences). Hemochrome spectra were carried out as described in (Berry and Trumpower, 1987), in gel heme stains used the method of (Thomas et al., 1976). UV–Vis electronic absorption spectra and kinetic assay data were recorded on a Cary50 spectrophotometer (Varian) at 25°C unless otherwise specified. Sulfite dehydrogenase activity assays were performed as in (Wilson and Kappler, 2009) and data fitted using SigmaPlot 9.0 (Sysstat). Temperature stability of SorT was tested by pre-incubating samples of purified SorT in the presence (1 mM) or absence of sulfite for 0, 5, 10, and 20 min at temperatures between 20 and 60°C before quick-chilling the sample on ice, followed by brief centrifugation and determination of SDH activity. pH stability of rSorT was determined by a similar method, where purified, concentrated rSorT was incubated in buffers between pH 6 and 11 and samples were taken after 5, 10, and 20 min and immediately assayed for SDH activity under standard conditions (pH 8.0, 25°C). Interaction assays used a combination of electronic absorption spectra and kinetic assays, in brief electronic absorption spectra were recorder between 700 and 250 nm before the start of the kinetic run (“oxidized”), immediately following the end of the kinetic run (“part reduced”) and after addition of dithionite (“fully reduced”). Experiments involving an oxygen electrode used a Hansatech Oxygraph, membrane fractions for use in these experiments were isolated from S. meliloti cells grown on medium 69 supplemented with taurine by subjecting a crude extract to ultracentrifugation (145,000 × g, 4°C, 90 min) and resuspension of the membrane pellet in 20 mM Tris–Cl pH 8.0. Mass fingerprints of tryptic peptides and whole proteins were determined as in Wilson and Kappler (2009).
Cyclic Voltammetry
All electrochemical experiments were performed with a BAS100B/W potentiostat coupled to a BAS C3 cell stand. The reference electrode was Ag/AgCl and the counter electrode was a Pt wire. The electrolyte solution comprised phosphate buffer (0.1 M) titrated to the desired pH with dilute NaOH or AcOH solution. The two proteins rAzu2 and SMC04048 were examined at different working electrodes as described below.
rAzu2: a Au disk working electrode was cleaned and polished as described (Tkac and Davis, 2008) and then chemically modified by immersion in a 1-mM ethanolic solution of 11-mercaptoundecanoic acid using standard procedures (Kalimuthu et al., 2010). To the inverted thiol-modified Au working electrode was added a 10-μL aliquot of rAzu2 (56 μM). The droplet was allowed to dry to a film. To prevent protein loss, the electrode surface was carefully covered with a perm-selective dialysis membrane (MW cutoff ca. 3.5 kDa), presoaked in water. The membrane was fastened with a rubber O-ring to prevent leakage. The pH-dependence of the rAzu2 redox potential was modeled with Eq. 1 which is applicable for a coupled single electron/single proton redox reaction where only the reduced (CuI) form exhibits a pKa.
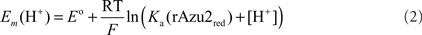
rSmc04048: A glassy carbon disk electrode was polished with alumina suspension (50 nm). Eastman AQ 29D polymer (28% w/v) was diluted 1:20 v/v and a 10-μL droplet of this solution was carefully deposited onto the inverted electrode surface and allowed to dry to a film at room temperature (ca. 1 h). A solution of rSmc04048 (40 μL, 250 μM) was diluted 1:10 v/v with 0.1 M phosphate buffer to give a volume of ca. 0.5 mL. The polymer-coated glassy carbon electrode was inserted into the protein solution and allowed to stand for approximately 3 h prior to measurement to allow the protein to accumulate within the polymer modified electrode. Voltammograms were recorded in the presence of dioxygen but at potentials above that at which dioxygen reduction is significant.
Phylogenetic analyses BLASTP was used to identify sequences related to either SorT, Azu2, or SMC04048. Phylogenetic relationships were analyzed using MEGA4.0 (Tamura et al., 2007) using neighbor-joining, maximum parsimony and minimum evolution algorithms. Bootstrap analysis was carried out with 500 replicates.
Results
Characterization of Recombinant SorT
The S. meliloti SDH, SorT, can only be purified with very low yields as several proteins co-purify with SorT (Wilson and Kappler, 2009), and this precludes detailed characterization of this protein both in terms of enzyme kinetics and structure as well as spectroscopic characterization of the protein. rSorT was produced in E. coli TP1000 as a cytoplasmic protein and purified to homogeneity using a combination of affinity chromatography and size exclusion chromatography. Molybdoenzymes are known to undergo folding and cofactor-insertion in the cytoplasm, which means that in the case of periplasmic enzymes such as SorT, the enzyme is fully functional before it is exported to the periplasmic space, allowing for expression of these enzymes in the E. coli cytoplasm. This type of expression strategy has been used successfully for other molybdenum enzymes located in extracytoplasmic cell compartments (Hilton et al., 1999; Temple et al., 2000).
The purified rSorT protein had a subunit mol. mass of 42669 ± 2 Da as determined by mass spectrometry. The small difference between the observed mass and the calculated molecular mass of rSorT (42791 Da) could indicate the loss of the N-terminal methionine residue. rSorT can appear as a double band when analyzed by gel electrophoresis during purification, and this is due to the loss of the cleavable His-Tag in some protein subunit during the early stages of purification as shown by mass fingerprinting (data not shown), use of protease inhibitor during purification minimizes this effect. rSorT contained 0.69 molecules of Mo per protein subunit, which is comparable to the value of 0.59 obtained for the native SorT by (Wilson and Kappler, 2009). Using size exclusion chromatography, the molecular mass of rSorT was estimated to be 71 kDa, indicating that just like the native SorT (est. mol mass 68 kDa by gel filtration, calculated subunit mol mass 39.42 kDa; Wilson and Kappler, 2009), the recombinant protein forms a homodimeric complex. Optical spectra of rSorT showed features typically associated with molybdenum proteins (Figure 2), with the “as prepared” protein being mostly in the oxidized state.
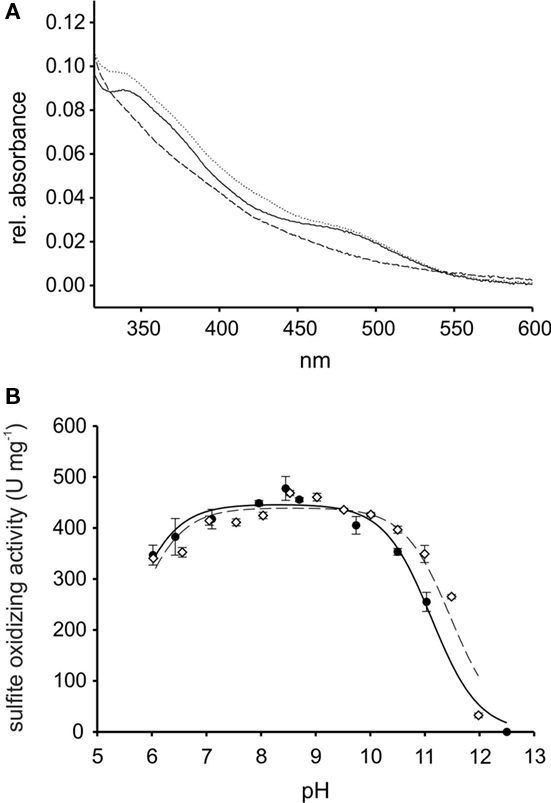
Figure 2. Properties of recombinant SorT (rSorT) (A) electronic absorption spectra of rSorT (solid line: as prepared, dotted line: oxidized and dashed line: sulfite reduced) (B) comparison of the pH dependency of SorT activity for the native and rSorT.
At pH 8.0, rSorT had a KM sulfite app of 13.4 ± 0.5 μM and a kcat of 338 ± 3/s, which is in very good agreement with the values obtained for the native SorT (KM sulfite app: 15.5 ± 1.1 μM; kcat: 343 ± 11/s). At pH 7 and pH 6, rSorT had KM sulfite app values of 23.4 ± 1.2 and 33.3 ± 6.5 μM, which is also very close to what was reported for the native enzyme (Wilson and Kappler, 2009). In addition, the rSorT activity profile between pH 6 and 12 also closely resembles the profile of the native SorT (Figure 2). To ascertain that changes in kinetic parameters and the enzyme’s catalytic performance were not caused by enzyme inactivation during the assay (duration: ∼2 min), the pH stability of rSorT was determined. rSorT was stable for at least 20 min between pH 6 and 10. At pH 11 the enzyme lost about 25% of activity within the first 5 min, and after 20 min an activity loss of ∼50% was apparent. Although typical SorT enzyme assays only take about 2 min to complete it therefore cannot be excluded that the significantly lower activities recorded at pH 11 (Figure 2) are at least in part due to enzyme inactivation.
SorT occurs in a mesophilic microorganism and standard assays are therefore carried out at 25°C, however, many of the Mo-containing SDHs are known to be resistant to thermal inactivation by temperature well above the growth temperature of the source organism (Kappler, 2011). To determine the temperature stability of rSorT, the enzyme was preincubated at temperatures between 20 and 60°C for up to 20 min followed by activity assays under standard conditions at 25°C and pH 8. In these experiments rSorT was stable for at least 20 min at temperatures of up to 40°C, while at 50 and 60°C the enzyme had a half-life of 2 min and under 1 min respectively (Figure 3). Interestingly, if rSorT was reduced by addition of sulfite before incubation, no significant loss of activity was observed at 50°C after 10 min of incubation, and at 60°C the half-life improved to an estimated 20–25 min (Figure 3). This effect is similar to what has been observed for both the bacterial SorAB SDH and for vertebrate SO (Southerland and Rajagopalan, 1978; Kappler et al., 2000). While for the latter the effect was explained by a tighter packing of the two mobile domains of the reduced vertebrate sulfite oxidase, it is unclear what is causing the increased temperature stability of the reduced bacterial enzymes neither of which has any mobile domains.
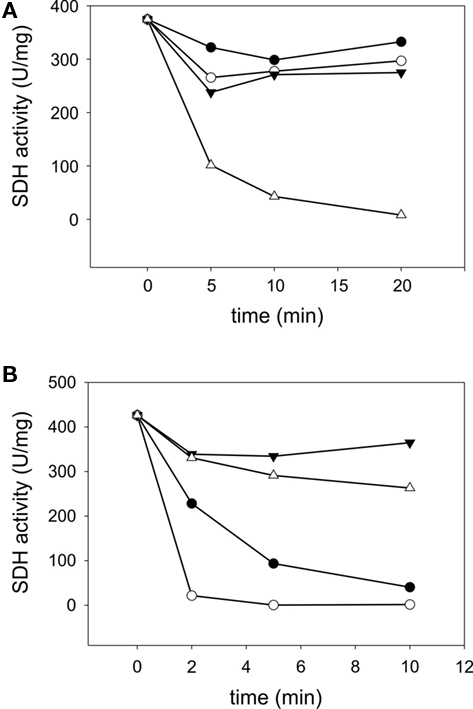
Figure 3. Temperature stability of rSorT (A) changes in rSorT activity following incubation at 20°C (● closed circles), 30°C (○ open circles), 40°C (▼ closed triangles), and 50°C (∆ open triangles) for 5, 10, and 20 min. (B) rSorT inactivation at 50 (● closed circles) and 60°C (○ open circles) and protective effect of sulfite reduction on rSorT temperature stability at these temperatures. 50°C + sulfite (▼ closed triangles) and 60°C + sulfite (∆ open triangles).
Properties of Purified Recombinant Cytochromes and Azu2
Both recombinant Smc04048 (rSmc04048) and recombinant Snc550 (rSnc550) were expressed in E. coli as a periplasmic protein using the helper plasmid pEC86 (Arslan et al., 1998) and purified to homogeneity from whole cell extracts. Purified rSmc04048 had a molecular mass of 12004 ± 2 Da, which corresponds to the calculated mass of the protein. rSmc04048 eluted from a calibrated gel filtration column in two peaks corresponding to 19.5 and 41 kDa, indicating that this cytochrome exists both as a monomer and as a dimer. rSmc04048 contained a full complement of the redox cofactor as determined by hemochrome spectra.
Redox potentiometry of rSmc04048 gave inconsistent results and protein precipitation during the experiment was a particular problem. Therefore, we turned to cyclic voltammetry as a means of determining the heme redox potential. A number of different working electrodes were tried including an Au electrode modified with several different long chain thiols but none gave a redox response. Pyrolytic graphite electrodes similarly were ineffective in facilitating a redox signal. Finally, we turned to using a polymer-coated glassy carbon electrode into which the protein had been soaked over a period of hours. This method has been successful with cyclic voltammetry of other cytochromes we have studied in the past (Santini et al., 2007; Creevey et al., 2008). A pair of well defined redox peaks were identified with an average peak potential of +205 mV vs NHE at pH 6.8 (Figure 4). This technique is limited to relatively slow sweep rates (<20 mV/s) to ensure that the protein is able to diffuse through the polymer matrix. At higher sweep rates the waves became distorted. In the range 2–20 mV/s the average peak potential remained the same (Figure 4).
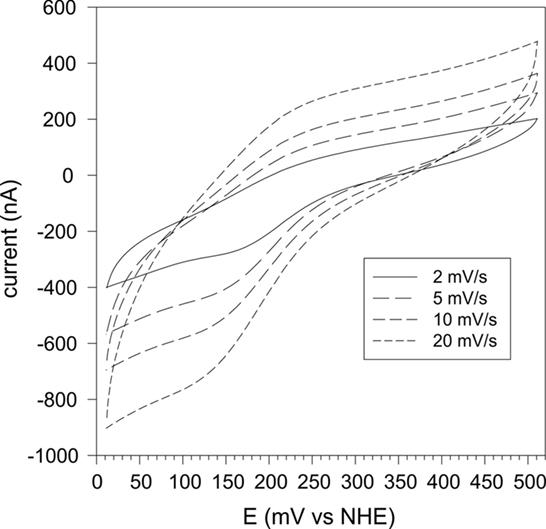
Figure 4. Cyclic voltammetry of rSmc04048 at sweep rates of 2, 5, 10, and 20 mV/s and at pH 6.9. The protein was soaked into a polymer film cast on a glassy carbon electrode (see Materials and Methods).
In contrast to the two cytochromes which could be purified with high yields, the pseudoazurin Azu2 was much more difficult to produce in soluble and cofactor-containing form. Most of the recombinant S. meliloti Azu2 was found in inclusion bodies, however, with optimized expression conditions some soluble protein could be obtained from periplasmic extracts (total yield of purified protein: ∼2 mg). Fractions containing purified rAzu2 had a blue color, and rAzu2 was found to be a monomeric protein with a molecular mass of 21 kDa as estimated by gel filtration and optical spectra reminiscent of other azurin proteins with peaks at 452, 591, and 758 nm. The relatively low ratio of the 591-nm absorbance peak to the protein peak at 280 nm (∼0.1) suggests however, that only some of the purified rAzu2 protein contained the copper cofactor. The calculated molecular mass of mature, Histidine-tagged rAzu2 is 15677.85 Da, mass spectrometry revealed a molecular mass of 15678.26 ± 2 Da for rAzu2.
Cyclic voltammetry of rAzu2 was straightforward. After a range of different thiol modifiers were trialed, the most successful one was the long chain acid 11-mercaptoundecanoic acid which forms a self-assembled monolayer on the Au surface which is highly biocompatible with terminal carboxylic acid/carboxylate groups interacting with the surface of the protein. At this electrode surface a pair of anodic and cathodic peaks were observed (Figure 5A) whose potentials were markedly pH-dependent (Figure 5B). Between pH 8.3 and 6.8 this potential was invariant at ca. +200 mV vs NHE, however, below pH 6.8 the redox potential became pH-dependent with a −59/pH variation in potential. This is indicative of a proton coupled redox reaction (single electron/single proton; Clark, 1960). The pH-dependence was fit to such a model with a pKa for the reduced (CuI) form of 6.3 (Figure 4B). It is also noteworthy that the redox response became weaker at the pH was raised above ca. 6.5. This is indicative of a weakening of the interaction between the terminal carboxylate groups at the electrode and the protein rAzu2 due to deprotonation. Presumably these key surface amino acid residues that undergo deprotonation (and are then repelled by the negatively charged electrode) are in proximity to the Cu center.
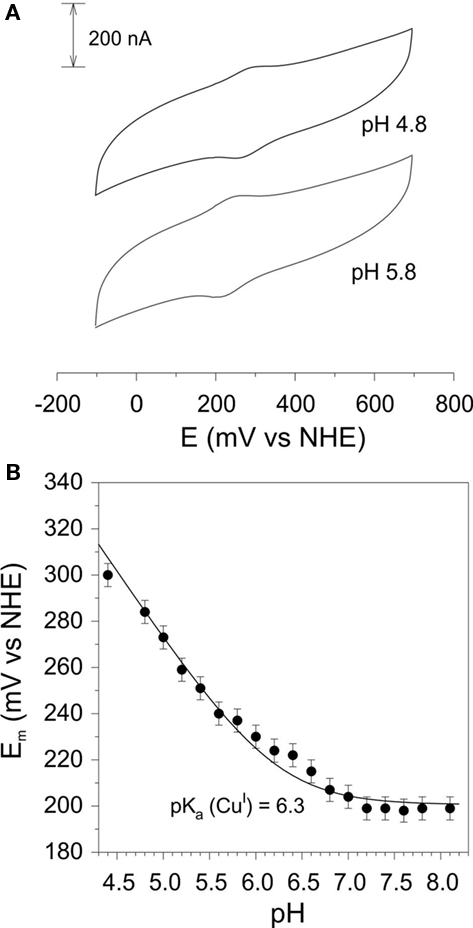
Figure 5. (A) Cyclic voltammograms of rAzu2 at a 11-mercaptoundecanoic acid-modified Au working electrode at pH 4.5 and 5.5 (sweep rate 50 mV/s) (B) pH-dependence of the redox potential. Curve is the best fit to the data using Eq. 2 which gave a pKa of 6.3 for the reduced (CuI) form of rAzu2. Experimental conditions: sweep rate 50 mV/s, 0.1 M phosphate titrated with NaOH or acetic acid to the desired pH.
Interactions between SorT and Potential Electron Acceptors
It had been previously reported that at pH 8.0 the native SorT only showed ∼13% of the activity observed with ferricyanide when horse heart cytochrome was used as the electron acceptor. The activity of rSorT was tested with three different cytochromes, horse heart cytochrome c, the S. meliloti cytochrome rSmc04048 and the recombinant Starkeya novella cytochrome c rSnc550, which is known to be the natural electron acceptor for the SorAB sulfite dehydrogenase (Kappler et al., 2000). Under standard assay conditions at pH 8.0, rSorT had activities of 7.4 ± 0.7, 33.4 ± 1.9, and 0.6 U/mg respectively with the three cytochromes. However, in all three cases only about 40% of the cytochrome added to the assay was reduced during the assay, which is in contrast to the reaction of the SorAB sulfite dehydrogenase which leads to a near complete reduction of the horse heart or S. novella cytochrome c present in the assay. For rSmc04048 the fraction of cytochrome reduced in the assay could be increased to >95% if the monomeric form of the protein was separated from the dimeric form by size exclusion chromatography, indicating that the rSmc04048 monomeric form interacts with rSorT. As the horse heart and S. novella cytochromes were in monomeric form, this excludes dimerization as a cause of their poor interaction with rSorT.
Further investigations of the interaction of rSmc04048 with rSorT showed that activity increased at lower pH with activities of 44.9 ± 3.7 and 57.7 ± 1.5 U/mg being observed at pH 7.0 and 6.0 respectively. This is in clear contrast to what is observed with ferricyanide as the electron acceptor, where activities are nearly pH invariant between pH 6.5 and 10, and then decrease at the extreme ends of the pH range tested.
No reduction of rAzu2 by rSorT was observed when rAzu2 was substituted for cytochrome c in assays conducted at pH 8.0, however, only low concentrations of rAzu2 could be used in these assays (standard: 8 μM, max: 56 μM) due to the limited amounts of protein available. Using a 56-μM protein solution, reduction of rAzu2 by sulfite (0.4 mM), and a combination of sulfite (0.4 mM) and rAzu2T was tested. Addition of sulfite produced no change in the rAzu2 absorption spectrum after incubation for 5 min, while following incubation with sulfite and rAzu2 for 5 min a 22% reduction of the absorption at 591 nm was observed, indicating that some reduction of rAzu2 had occurred. However, the absorbance change per minute for this reduction was only −0.002 ΔE595/min Reduction of rAzu2 with dithionite led to a marked change in the absorbance spectrum with all absorbance features being significantly reduced to near baseline levels (data not shown).
Does SorT Require More than One Redox Protein for Efficient Electron Transfer?
To investigate whether more than one redox carrier was required for efficient electron removal from rSorT the effect of inclusion of ferricyanide or purified rAzu2 in cytochrome c containing assay mixtures on enzyme activities was investigated. Inclusion of 50 μM ferricyanide in assays 40 μM containing any of the three cytochromes tested resulted in complete reduction of each cytochrome and reduction rates that increased ∼15–20 times over the rates detected without ferricyanide. A small (1–8 s) initial lag that was not seen in assays that did not contain ferricyanide was observed in these assays.
This indicated that that ferricyanide might be acting as the primary electron acceptor and in turn could be reducing the cytochrome c. Another explanation could be that, like Rhodobacter sphaeroides DMSO reductase expressed in E. coli (Hilton et al., 1999), rSorT needs to undergo “redox cycling” to become fully active. If ferricyanide treatment of rSorT were required for full activity, this would not be apparent in the ferricyanide containing activity assays routinely used for SorT enzymes as an excess of the activating agent would always be present.
To test whether rSorT required redox activation, an aliquot of the enzyme was incubated in the presence of 16 μM ferricyanide for 10 min, before assaying activity (ferricyanide concentration in the assay: 0.08 μM). For horse heart cytochrome c, the activity with redox cycled rSorT showed very limited increase from 7 to 10 U/mg, however, addition of 50 μM ferricyanide to such an assay mixture immediately increased the cytochrome reduction rate to values similar to those observed with untreated rSorT. rSmc04048 based assays using redox treated SorT did not show increased cytochrome reduction rates. This suggests that ferricyanide may have acted as a primary electron acceptor, and then subsequently transferred electrons to the different cytochromes c. The apparent “preference” of rSorT for ferricyanide over the cognate rSmc04048 cytochrome could be due to the difference in molecular mass of the two electron acceptors.
Inclusion of 8 μM Azu2 in the assays did not change the rSmc04048 reduction rates relative to the rates observed when only rSmc04048 was present at either pH 6, 7, or 8. Due to the strong absorbance of the heme group in rSmc04048 it was not possible to monitor whether absorbance changes occur in the rAzu2 spectrum, and it could therefore not be determined whether the cytochrome or rAzu2 is reduced first, or whether rAzu2 participates in the reaction at all (Figure 1).
Interactions of rSorT with Components of the Respiratory Chain
In addition to using a small redox protein as its natural electron acceptor, it is also possible that rSorT might either use molecular oxygen as an electron acceptor or interact directly with the cytochrome c oxidase or other respiratory complexes present in the cell membrane (Figure 1). We investigated these possibilities using an oxygen electrode and cell membranes isolated from S. meliloti grown on taurine-containing medium.
Addition of sulfite to the buffer in the oxygen electrode chamber led to a background oxygen consumption rate of −1.05 ± 0.07 nmol/mL/min. This rate was subtracted from all “enzymatic” oxygen consumption rates observed. With this correction, rSorT gave rise to a negligible oxygen consumption rate of −0.2 ± 0.21 nmol/mL/min, indicating that like the native enzyme, rSorT does not have significant sulfite oxidase activity.
When sulfite was added to membrane preparations a corrected respiratory rate of −1.81 ± 0.5 nmol/min/mL was observed while membrane preparations with sulfite and rSorT added resulted in a rate of −1.21 ± 0.07 nmol/mL/min. This suggests that in the presence of its substrate, sulfite, rSorT did not enhance respiratory activity, indicating that it was unlikely that rSorT interacted directly with any respiratory chain complexes.
When either rSmc04048 or rAzu2 were added to assays containing membrane preparations and sulfite at concentrations similar to those used in standard assays, oxygen consumption rates of −64 ± 6 nmol/min/mL and −62 nmol/min/mL were recorded. This clearly shows that both redox proteins were capable of interaction with the S. meliloti respiratory chain components, although we were unable to demonstrate an increase in the observed oxygen consumption rates following addition of purified rSorT to rSmc04048 containing assays. As the only substrate added to the assay was sulfite, and neither rSmc04048 nor rAzu2 are reduced by sulfite in standard photometric assays at a significant rate. These results suggest that some component of the membrane preparation must have been able to use sulfite as a substrate and then to reduce either the azurin or the cytochrome, which in turn were passing on electrons to a respiratory chain component, most likely a cytochrome oxidase. It is possible that the membrane preparations may have contained some SorT, as this protein is highly abundant in S. meliloti grown on taurine and some soluble proteins may have been present in the preparation. During the purification of SorT from S. melioti no SorT-independent SOE activity was noted (Wilson and Kappler, 2009).
Discussion
With the exception of the plant sulfite oxidase, which is a true sulfite oxidase, all sulfite-oxidizing molybdenum enzymes for which the natural electron acceptor is known couple sulfite oxidation to the reduction of a cytochrome c. Although the SorT sulfite dehydrogenase from S. meliloti is a representative of the “atypical” bacterial SDHs that show highest activities with the artificial electron acceptor ferricyanide, our data suggest that in vivo the reaction of SorT is coupled to a cognate cytochrome c that is found in the sorT operon.
In order to carry out this work it was first necessary to establish an efficient overexpression system for SorT, which increased the yields of purified protein from about 70 μg for the native enzyme to over 15 mg/L of bacterial culture for the rSorT. The low yields for the native enzyme are not due to a low abundance of SorT in S. meliloti, but to the fact that several proteins co-purify with SorT and cannot be removed from the majority of the enzyme present in cell extracts (Wilson and Kappler, 2009). Our data show that rSorT is fully active and its subunit structure and catalytic properties are very similar to those of the native enzyme, demonstrating that the cytoplasmic expression system used for the production of rSorT does not have an impact on enzyme performance or structure. In S. meliloti the mature SorT and the two acceptor proteins are located in the periplasmic space, i.e., in the same, extracellular compartment.
Like the native enzyme, rSorT had low activity (7 U/mg) with horse heart cytochrome c, however, rSorT showed good activities with the cognate rSmc04048 cytochrome (33.4 U/mg). In contrast, rSorT only showed negligible activity (0.6 U/mg) with the S. novella cytochrome c550, which is known to be the natural electron acceptor of the SorAB sulfite dehydrogenase.
A possible cause of this could be differences in the redox potentials of the three cytochromes. As SorT interacts very well with ferricyanide, which has a very high redox potential of +430 mV vs NHE, it could be possible that SorT requires an acceptor protein with a very high redox potential. The reported redox potentials for horse heart cytochrome c and the S. novella cytochrome c550 are relatively high with +263 and +302 mV vs NHE respectively (Kappler et al., 2000; Battistuzzi et al., 2002), however, the redox potential of rSmc04048 was only +205 mV (pH 6.8). It thus appears that rSorT interaction with the three cytochromes is not governed primarily by the redox potential of the acceptor protein.
Analysis of the three cytochromes on a molecular level uncovered further. Both the horse heart cytochrome and the S. novella cytochrome have basic isoelectric points (computed pI 9.59 and 8.86, respectively) and thus are positively charged at pH 7. In contrast, the Smc04048 cytochrome has an isoelectric point of 4.29 (pI 4.78 for His-tagged rSmc04048), and thus is negatively charged at physiological pH values. This difference in the charge state is likely to be a major factor in determining the interactions of rSorT with the cytochromes. However, in addition to the overall charge of the protein, structural determinants likely also play a role in shaping the interactions of rSorT with the cytochromes, as activity with horse heart cytochrome c which has the highest pI and is strongly positively charged (calculated total charge of +9 at pH 7) was 7 U/mg, while with the less strongly charged S. novella cytochrome (computed charge state +3.45 at pH 7) SDH activity was only 0.6 U/mg. The differences in the cytochrome fold and surface charge distribution should be investigated further in the future.
An interesting observation was that inclusion of ferricyanide in cytochrome c based assays led to a very strong increase of reduction rates for all three cytochromes tested, with the small lag observed initially being likely to indicate that ferricyanide was being reduced first, before cytochrome reduction started to take place. Ferricyanide is clearly not a natural electron acceptor for this enzyme, however, relative to any electron accepting proteins it is a very small compound, which leads to faster diffusion rates which in turn would enable a more efficient interaction with SorT relative to the cytochromes. The positive redox potential of ferricyanide (+430 mV vs NHE) could also contribute to a favorable interaction with SorT, however, it does not explain how subsequently the cytochromes, all of which have less positive redox potentials, are being reduced. We suggest that the interaction between the cytochromes and ferrocyanide might be charge-driven. In this case, interaction between positively charged cytochrome molecules and the strongly negatively charged ferricyanide would lead to the formation of a strong outer sphere complex that would favor the reduction of the cytochromes despite the unfavorable redox potentials. This has been experimentally observed for ferrocyanide and cobalt complexes (Bernhardt et al., 2002, 2005). However, studies of the electron transfer pathways in SorT and the respective sites of interaction with the acceptor cytochrome and ferri-/ferrocyanide will be required to fully understand this phenomenon.
The physiological role of the pseudoazurin, Azu2, which is also part of the sorT operon, remains, at this stage, elusive, which is partly due to the low protein yield obtained for rAzu2 that prevented more extensive investigations. rAzu2 did not appear to be reduced directly by rSorT at pH 8.0, and caused no significant changes in observed rSorT activity when it was added to assays in addition to rSmc04048. If anything, rSorT activities were slightly lower in assays containing rAzu2 as well as rSmc04048 although this difference was within the experimental error. If rAzu2 were participating in electron transfer within the SorT system, this might be indicative of rAzu2 accepting electrons from rSmc04048, which would lead to an apparent lowering of the cytochrome c reduction rate through reoxidation of reduced cytochrome molecules. However, insufficient rAzu2 was available to investigate this possibility in detail.
Based on our results we propose that SorT uses the Smc04048 cytochrome c as its in vivo electron acceptor, and that electrons get passed on to the cytochrome oxidase either directly or potentially via Azu2 (Figure 6).
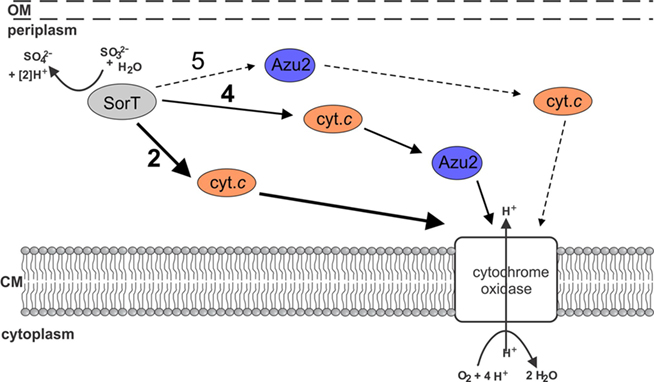
Figure 6. Current working model of electron transfer pathways between SorT and the respiratory chain. The main pathway appears to be no. 2 using the Smc04048 cytochrome c as an electron carrier. It is also possible that both Smc04048 and Azu2 participate in electron transfer (pathways 4 + 5), however, based on present data it seems unlikely that pathway 5 would play a significant role in electron transfer.
Do All Ferricyanide-Linked SDHs Use Smc04048-Like Cytochromes as Electron Acceptors?
SorT is a representative of the “atypical,” “ferricyanide-linked” SDHs which have been isolated from a variety of microorganisms including extremophiles such as Thermus thermophilus, Deinococcus radiodurans and organosulfonate degraders such as Cupriavidus necator, Delftia acidovorans (Kappler, 2011). Since SorT is the first SDH of its type for which a natural electron acceptor has been identified we investigated whether Smc04048 and Azu2-like protein were associated with any of the other ferricyanide-linked SOEs. With the exception of the enzyme from Deinococcus radiodurans, all genes encoding characterized SDHs are found upstream of genes encoding c-type cytochromes (Kappler, 2011), however, only the cytochrome c encoded downstream of the Delftia acidovorans SDH (Daci_0054) showed significant homology to Smc04048 (52% amino acid sequence similarity). For all other SDH associated cytochromes amino acid sequence similarities with Smc04048 were between 18 and 24%.
Protein sequences related to Smc04048 were identified using BLASTP searches (Figure 7). In some cases, Smc04048 related sequences were observed as domains of larger proteins (e.g., Ixodes scapularis, Conexibacter woesei), and such fusion proteins were excluded from further analysis. The only fusion proteins that were retained in later analyses were from the marine gamma Proteobacteria HTCC2080 and HTCC2148, where the Smc04048-like protein had been fused to a SorT-like protein. Smc04048 is distantly related to cytochrome c6 from Synechococcus sp. JA-2-3Ba(2-13) and to the SoxX cytochromes. Twenty-two of the Smc04048-like proteins were encoded by genes that were located adjacent to genes encoding SorT-like proteins (Figure 7). In some other cases, e.g., Marinobacter hydrocarbonoclasticus and Marinosulfomonas methylotropha only limited sequence data was available, and thus it could not be ascertained whether a similar gene arrangement exists in these organisms. For Verminephrobacter eiseniae a pseudogene (Veis_1600) encoding an Smc04048 related cytochrome was identified adjacent to the gene encoding the SorT-like SDH (Veis_1599) from this organism, however, this protein sequence was not included in the analysis due to its classification as a pseudogene.
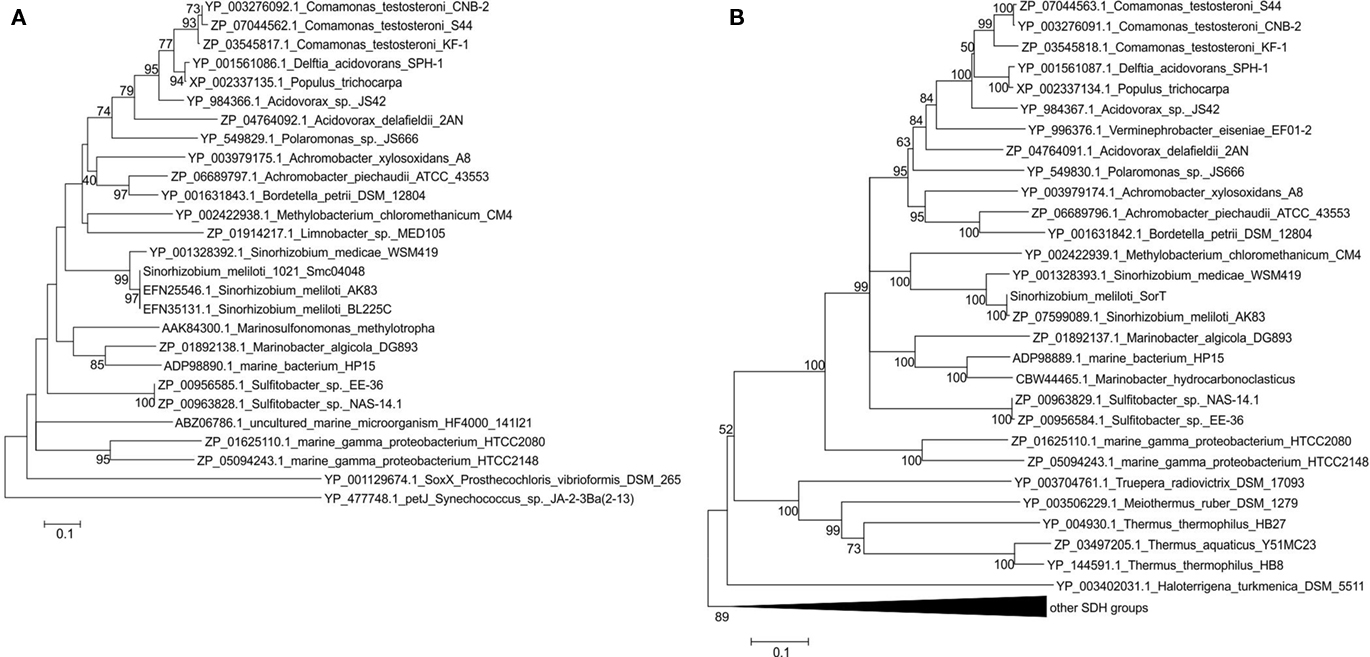
Figure 7. Phylogenetic analysis of Smc04048 and SorT related protein sequences available in the database. (A) Neighbor-joining tree or Smc04048 related protein sequences (B) neighbor-joining tree of SorT (Smc04049). Bootstrapping used 500 samples, for clarity bootstrap values below 40 are not shown. Minimum Evolution and Maximum Parsimony algorithms were also used and generated trees with the same topology and very similar bootstrap values for both sequences setc. Sequence accession numbers are shown in the figure. The sequences from the two marine gamma Proteobacteria are fusions between a smc04048 and a sorT genes, the cytochromes from Sulfitobacter species are diheme cytochromes. The pseudogene Veis_1600 that encodes an Smc04048 related protein from Verminephrobacter eiseniae was not included in the dataset.
Phylogenetic analysis of the SorT and Smc04048 related sequences clearly shows the similarity of the respective clades (Figure 7). In contrast, Azu2-like proteins only occurred in sorT operons of species of Sinorhizobium; although many other Azu2 related sequences were found in the database these genes did not occur in operons also containing SDH-encoding genes (data not shown).
This analysis confirms what had already been observed as part of a general analysis of the SO enzyme family (Kappler, 2008, 2011), namely that while the majority of the genes encoding the SOE Mo-binding subunits occur adjacent to genes encoding c-type cytochromes, these c-type cytochromes are specific to particular subtypes of SDHs and are not conserved outside their immediate phylogenetic grouping. Examples of this are the genes encoding the SorB subunit of the SorAB-type SDH from S. novella, the soxC and soxD subunits of the sulfur dehydrogenases (Kappler, 2008) and those encoding SorT and Smc04048 that were studied here. Our results also underline the catalytic and structural plasticity of the SDH proteins, as it should be extremely interesting to study the SDH/ cytochrome fusion proteins found in the two marine bacteria and to observed the structural and kinetic properties of these proteins.
Conclusion
We have for the first time identified a natural electron acceptor for one of the “atypical,” ferricyanide-linked SDHs, and found it to be a cytochrome c that is capable of transferring electrons to the respiratory chain and most likely directly to the cytochrome oxidase. Although this may seem unsurprising as other SDHs have been shown to interact with c-type cytochromes in this manner, the Smc04048 cytochrome was shown to have molecular properties that differed significantly from those of other characterized SDH electron acceptor cytochromes in terms of its charge state. The function of the Azu2 protein for the SDH reaction remains unclear at this stage and its role needs to be investigated further, especially as Azu2 proteins are only encoded in SDH operons of Sinorhizobium species. Future work will focus on the kinetics of the cytochrome linked SorT activity will also be studied in more detail, including the significance of the Smc04048 monomer/dimer interconversion. Furthermore the structures of these proteins will be determined to enable the identification of surface areas involved in the SorT/Smc04048 interactions, and to reveal any structural differences that may exist between Smc04048 and other cytochromes known to accept electrons from SOEs.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant (DP0878525) and fellowship from the Australian Research Council to UK and grant DP0880288 to Paul V. Bernhardt.
References
Arslan, E., Schulz, H., Zufferey, R., Kunzler, P., and Thony-Meyer, L. (1998). Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb(3) oxidase in Escherichia coli. Biochem. Biophys. Res. Comm. 251, 744–747.
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (2005). Current Protocols in Molecular Biology, ed. K Janssen. Hoboken, NJ: John Wiley & Sons Inc.
Battistuzzi, G., Borsari, M., Cowan, J. A., Ranieri, A., and Sola, M. (2002). Control of cytochrome c redox potential: axial ligation and protein environment effects. J. Am. Chem. Soc. 124, 5315–5324.
Beringer, J. E. (1974). R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84, 188–198.
Bernhardt, P. V., Bozoglian, F., Macpherson, B. P., and Martinez, M. (2005). Molecular mixed-valence cyanide bridged coiii-feii complexes. Coord. Chem. Rev. 249, 1902–1916.
Bernhardt, P. V., Macpherson, B. P., and Martinez, M. (2002). The influence of cis/trans isomerism on the physical properties of a cyano-bridged dinuclear mixed valence complex. J. Chem. Soc. Dalton Trans. 1435–1441.
Berry, E. A., and Trumpower, B. L. (1987). Simultaneous determination of hemes a, b and c from pyridine hemochrome spectra. Anal. Biochem. 161, 1–15.
Clark, W. M. (1960). The Oxidation-Reduction Potentials of Organic Systems. Baltimore: The Williams and Wilkins Company.
Creevey, N. L., McEwan, A. G., Hanson, G. R., and Bernhardt, P. V. (2008). Thermodynamic characterization of the redox centers within dimethylsulfide dehydrogenase. Biochemistry 47, 3770–3776.
D’Errico, G., Di Salle, A., La Cara, F., Rossi, M., and Cannio, R. (2006). Identification and characterization of a novel bacterial sulfite oxidase with no heme binding domain from Deinococcus radiodurans. J. Bacteriol. 188, 694–701.
Denger, K., Weinitschke, S., Smits, T. H. M., Schleheck, D., and Cook, A. M. (2008). Bacterial sulfite dehydrogenases in organotrophic metabolism: separation and identification in Cupriavidus necator h16 and in Delftia acidovorans sph-1. Microbiology 154, 256–263.
Di Salle, A., D’Errico, G., La Cara, F., Cannio, R., and Rossi, M. (2006). A novel thermostable sulfite oxidase from Thermus thermophilus: characterization of the enzyme, gene cloning and expression in Escherichia coli. Extremophiles 10, 587–598.
Eilers, T., Schwarz, G., Brinkmann, H., Witt, C., Richter, T., Nieder, J., Koch, B., Hille, R., Haensch, R., and Mendel, R. R. (2001). Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase – a new player in plant sulfur metabolism. J. Biol. Chem. 276, 46989–46994.
Hill, J. M., Kobe, B., Guss, M., and Huber, T. (2007). “Nmr screening for rapid protein characterisation in structural proteomics,” in Structural Proteomics: High-Throughput Methods, eds B. Kobe, M. Guss and T. Huber (Totowa, NJ: Humana Press).
Hilton, J. C., Temple, C. A., and Rajagopalan, K. V. (1999). Re-design of Rhodobacter sphaeroides dimethyl sulfoxide reductase. Enhancement of adenosine n1-oxide reductase activity. J. Biol. Chem. 274, 8428–8436.
Kalimuthu, P., Tkac, J., Kappler, U., Davis, J. J., and Bernhardt, P. V. (2010). Highly sensitive and stable electrochemical sulfite biosensor incorporating a bacterial sulfite dehydrogenase. Anal. Chem. 82, 7374–7379.
Kappler, U. (2008). “Bacterial sulfite dehydrogenases – enzymes for chemolithtrophs only?” in Microbial Sulfur Metabolism, eds C. Dahl and C. G. Friedrich (Berlin: Springer), 151–169.
Kappler, U., and Bailey, S. (2005). Molecular basis of intramolecular electron transfer in sulfite-oxidizing enzymes is revealed by high resolution structure of a heterodimeric complex of the catalytic molybdopterin subunit and a c-type cytochrome subunit. J. Biol. Chem. 280, 24999–25007.
Kappler, U., Bennett, B., Rethmeier, J., Schwarz, G., Deutzmann, R., McEwan, A. G., and Dahl, C. (2000). Sulfite: cytochrome c oxidoreductase from Thiobacillus novellus – purification, characterization and molecular biology of a heterodimeric member of the sulfite oxidase family. J. Biol. Chem. 275, 13202–13212.
Kappler, U., and Dahl, C. (2001). Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS Microbiol. Lett. 203, 1–9.
Laemmli, U. K. (1970). Cleavage of structural protein during the assembly of the head of bacteriophage t4. Nature 227, 680–685.
Myers, J. D., and Kelly, D. J. (2005). A sulphite respiration system in the chemoheterotrophic human pathogen Campylobacter jejuni. Microbiology 151, 233–242.
Palmer, T., Santini, C. L., Iobbi Nivol, C., Eaves, D. J., Boxer, D. H., and Giordando, G. (1996). Involvement of the narj and mob gene products in distinct steps in the biosynthesis of the molybdoenzyme nitrate reductase in Escherichia coli. Mol. Microbiol. 20, 875–884.
Rajagopalan, K. V. (1980). “Sulfite oxidase (sulfite: ferricytochrome c oxidoreductase),” in Molybdenum and Molybdenum-Containing Enzymes, ed. M. P. Coughlan (Oxford: Pergamon Press), 243–272.
Reichenbecher, W., Kelly, D. P., and Murrell, J. C. (1999). Desulfonation of propanesulfonic acid by Comamonas acidovorans strain p53: evidence for an alkanesulfonate sulfonatase and an atypical sulfite dehydrogenase. Arch. Microbiol. 172, 387–392.
Santini, J. M., Kappler, U., Ward, S. A., Honeychurch, M. J., vanden Hoven, R. N., and Bernhardt, P. V. (2007). The nt-26 cytochrome c552 and its role in arsenite oxidation. Biochim. Biophys. Acta 1767, 189–196.
Schrader, N., Fischer, K., Theis, K., Mendel, R. R., Schwarz, G., and Kisker, C. (2003). The crystal structure of plant sulfite oxidase provides insights into sulfite oxidation in plants and animals. Structure 11, 1251–1263.
Southerland, W. M., and Rajagopalan, K. V. (1978). Domain interactions in oxidized and reduced forms of rat liver sulfite oxidase. J. Biol. Chem. 253, 8753–8758.
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). Mega4: molecular evolutionary genetics analysis (mega) software version 4.0. Mol. Biol. Evol. 24, 1596–1599.
Temple, C. A., Graf, T. N., and Rajagopalan, K. V. (2000). Optimization of expression of human sulfite oxidase and its molybdenum domain. Arch. Biochem. Biophys. 383, 281–287.
Thomas, P. E., Ryan, D., and Levit, M. (1976). An improved staining procedure for the detection of peroxidase activity of cytochrome p-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75, 168–176.
Tkac, J., and Davis, J. J. (2008). An optimised electrode pre-treatment for sam formation on polycrystalline gold. J. Electroanal. Chem. 621, 117–120.
Keywords: sulfite dehydrogenase, sulfite oxidation, molybdoenzyme, cytochrome, redox protein, pseudoazurin
Citation: Low L, Kilmartin JR, Bernhardt PV and Kappler U (2011) How are “atypical” sulfite dehydrogenases linked to cell metabolism? Interactions between the SorT sulfite dehydrogenase and small redox proteins. Front. Microbio. 2:58. doi: 10.3389/fmicb.2011.00058
Received: 01 February 2011;
Paper pending published: 13 March 2011;
Accepted: 15 March 2011;
Published online: 25 March 2011.
Edited by:
Martin G. Klotz, University of Louisville, USAReviewed by:
Martin G. Klotz, University of Louisville, USAChristiane Dahl, Rheinische Friedrich-Wilhelms-Universität Bonn, Germany
Copyright: © 2011 Low, Kilmartin, Bernhardt and Kappler. This is an open-access article subject to an exclusive license agreement between the authors and Frontiers Media SA, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Ulrike Kappler, School of Chemistry and Molecular Bioscience, The University of Queensland, St. Lucia, QLD 4072, Australia. e-mail: u.kappler@uq.edu.au
 Louie Low
Louie Low