- 1 Department of Infection, Immunity and Inflammation, University of Leicester, Leicester, UK
- 2 The Roslin Institute and Royal (Dick) School of Veterinary Studies, University of Edinburgh, Easter Bush, UK
Burkholderia pseudomallei and Burkholderia mallei are closely related Gram-negative bacteria responsible for the infectious diseases melioidosis and glanders, respectively. Autotransporters (ATs) comprise a large and diverse family of secreted and outer membrane proteins that includes virulence-associated invasins, adhesins, proteases, and actin-nucleating factors. The B. pseudomallei K96243 genome contains 11 predicted ATs, eight of which share homologs in the B. mallei ATCC 23344 genome. This review distils key findings from in silico, in vitro, and in vivo studies on the ATs of B. pseudomallei and B. mallei. To date, the best characterized of the predicted ATs of B. pseudomallei and B. mallei is BimA, a predicted trimeric AT mediating actin-based motility which varies in sequence and mode of action between Burkholderia species. Of the remaining eight predicted B. pseudomallei trimeric autotransporters, five of which are also present in B. mallei, two (BoaA and BoaB), have been implicated in bacterial adhesion to epithelial cells. Several predicted Burkholderia ATs are recognized by human humoral and cell-mediated immunity, indicating that they are expressed during infection and may be useful for diagnosis and vaccine-mediated protection. Further studies on the mode of secretion and functions of Burkholderia ATs will facilitate the rational design of control strategies.
Burkholderia pseudomallei and Burkholderia mallei
Melioidosis, a febrile illness with disease states ranging from acute pneumonia or septicemia to chronic or localized abscess formation, was first documented by Whitmore and Krishnaswami (1912). The causative agent, Burkholderia pseudomallei, was subsequently identified as a motile, Gram-negative bacillus, which is principally a saprophyte found in soil and water in subtropical areas. The disease is acquired by inhalation, aspiration, sub-cutaneously, or occasionally via ingestion. Most cases occur in patients with immunosuppression, especially those with diabetes mellitus. Melioidosis has become an increasingly important disease in endemic areas such as South-East Asia and Northern Australia causing a significant number of deaths despite antibiotic treatment. Furthermore, B. pseudomallei is also recognized as a pathogen of numerous animal species (reviewed in Lazar Adler et al., 2009; Galyov et al., 2010).
Burkholderia mallei is closely related to B. pseudomallei and is the etiological agent of Glanders, an infectious equine disease that can be transmitted to humans. The disease may be acute or chronic, with the chronic form usually occurring in horses and the acute form occurring in mules and donkeys. Clinical signs include swollen lymphatics, a chronic nasal discharge, and ulcers. Contact with nasal discharge or infected ulcers is a major risk factor for infection. B. mallei is adapted to a narrower range of hosts than B. pseudomallei and is not considered able to persist for protracted periods in the environment. Horses are the primary carriers of disease and are largely responsible for transmission. Infections in humans frequently involve sepsis, pneumonia and abscess formation and have a high mortality rate. The incubation period ranges from days to weeks, and animals can die within a week of the onset of clinical symptoms (reviewed in Whitlock et al., 2007; Galyov et al., 2010).
The Autotransporter Family
Autotransporters (ATs) are a large and diverse family of bacterial secreted and outer membrane (OM) proteins that are translocated from the bacterial cytoplasm via the Type V pathway, one of seven recognized secretion pathways in Gram-negative bacteria. ATs play diverse roles, and many such proteins influence pathogenesis and immunity, for example by acting as invasins, adhesins, proteases, or actin-nucleating factors. ATs typically consist of a 20–400-kDa passenger domain that contains the effector functions and a 10–30-kDa β domain facilitating translocation across the OM (Dautin and Bernstein, 2007). ATs can be further divided into classical ATs and trimeric autotransported adhesins (TAAs), referred to as Type Va and Type Vc respectively. TAAs have a coiled-coil motif of unknown function, and are therefore referred to as oligomeric coiled-coil adhesins (Oca). Trimerization of TAAs is essential for their translocation and function as a single β barrel is formed from the three shorter 4-stranded β barrel domains which is superimposable with the classical ATs 12-stranded β barrel. While classical ATs have diverse effector functions, all characterized TAAs, with the notable exception of BimA from Burkholderia spp., are adhesins (Cotter et al., 2005; Dautin and Bernstein, 2007). The threefold symmetry provides the potential for a multivalent interaction to enhance avidity, which could overcome mechanical forces and provide stability against proteases and detergents within extracellular secretions (Cotter et al., 2005).
Despite diversity in sequence, length, and function of the passenger domains, most ATs are variations on a single structural theme: an elongated solenoid (a coil wound into a tightly packed helix) which may contribute to protein folding after transport and/or allow effector domains to traverse LPS and capsule layers. This conserved shape creates a scaffold for various loops and domains without disturbing structural integrity and allows multiple interfaces to mediate interactions (Wells et al., 2007; Nishimura et al., 2010). Classical ATs form a right-handed β helical structure while TAAs have a β roll (two β strands contributing to a single superhelical turn); these structural differences affect curvature and binding specificity (Dautin and Bernstein, 2007). However, the recent full structure for the Pseudomonas aeruginosa classical AT EstA demonstrated an exception to the solenoid structure, with a α-β-α globular fold (Van Den Berg, 2010), which may relate to its function as a lipase rather than an adhesin.
ATs must cross both plasma membranes of Gram-negative bacteria. Classical ATs are then predominantly cleaved for secretion into the extracellular milieu whereas TAAs remain integrated in the bacterial OM via their β barrel domain. Firstly, ATs are targeted to the IM by their N-terminal signal sequence, where translocation occurs via the general secretory (Sec) pathway. The N-terminal signal sequence ranges from 20 to 60 aa, with the most elongated signal peptides being associated with large (>1000 aa) passenger domains in TAAs (Dautin and Bernstein, 2007). Such extended signal peptides exhibit reduced interaction with the Sec complex promoting delayed post-translational translocation across the inner membrane which is proposed to allow partial folding for subsequent OM secretion (Peterson et al., 2006; Desvaux et al., 2007). Once in the periplasm, ATs can undergo modifications including glycosylation and lipidation and may also form disulfide bonds. As implied in their name, ATs were originally believed to mediate their own transport across the OM. However, recent research suggests that they are chaperoned by periplasmic proteins, such as SurA and DegP to the Bam (Omp85) complex located within the OM which may assist in translocation (Oomen et al., 2004; Dautin and Bernstein, 2007). Several ATs have been shown to interact directly with the Bam complex (Ieva and Bernstein, 2009; Sauri et al., 2009; Lehr et al., 2010), as well as periplasmic chaperones including SurA and DegP (Ieva and Bernstein, 2009; Ruiz-Perez et al., 2009). Furthermore, depletion of the essential BamA protein significantly affects AT secretion (Jain and Goldberg, 2007). BamA forms a 16-strand β barrel with a pore size of ~2.5 nm which is significantly larger than the AT β barrel (~1.2 nm) and would allow for translocation of partially folded ATs. Additionally, BamA oligomerizes in vitro and could possibly form oligomer pores capable of translocating folded ATs. These observations have led to two hypotheses: translocation of the unfolded passenger domain through the Bam-stabilized AT β barrel via a hairpin curvature or translocation of the entire AT, possibly partially folded, via the Bam complex (Sauri et al., 2009). Structural analysis indicates that sequential folding of the passenger domain may act as the driving force for translocation of AT (Van Den Berg, 2010), although evidence suggests that partially folded or misfolded ATs are still capable of translocation (Dautin and Bernstein, 2007). Therefore, the translocation of ATs is dependent on periplasmic chaperones and the Bam complex via a mechanism which remains to be elucidated but which appears to be mediated by interaction with the β domain of the ATs. This would account for the observation that the C-terminal domains of TAAs from diverse bacterial species are functionally interchangeable (Ackermann et al., 2008).
Autotransporters of B. pseudomallei and B. mallei
The B. pseudomallei K96243 genome contains 11 predicted ATs, annotated as such based on sequence similarity to known ATs (Table 1). Additionally, 10 of these ATs are conserved across the other three completed B. pseudomallei genomes, while BPSL1705 is missing from both the 1106a and 688 strains. The B. mallei ATCC 23344 genome has eight homologs of the B. pseudomallei ATs (Table 1). Six of these are conserved across the three additional B. mallei completed genomes, with BMA1027 being present only in B. mallei ATCC 23344, while the avirulent B. mallei SAVPI strain also lacks BMAA0649. Homologs for seven of the B. pseudomallei ATs are also found in avirulent B. thailandensis (Table 1). However, we caution that conservation of loci in pathogenic Burkholderia and B. thailandensis does not necessarily imply that the genes play no role in virulence or host cell-interactions, as demonstrated by our analysis of the bsa Type III secretion locus shared by such strains (Stevens et al., 2002, 2004). Only two ATs are predicted to be classical ATs: BPSS0962/BMAA1263 (a putative serine protease) and BPSL2237/BMA1647 (a putative lipase/esterase). The remainder are predicted TAAs (Tiyawisutsri et al., 2007). An in silico subtractive hybridization of pathogenic versus non-pathogenic Burkholderia species to generate a 650 gene virulome, identified six ATs (BPSS0962, BPSL2237, BPSL1631, BPSL2063, BPSS796, BPSS0908; Schell et al., 2008). Furthermore, a “gain of function” genomic library screen identified five ATs as having a possible role in invasion and survival within macrophages (BPSL1705, BPSL2237, BPSS0088, BPSS0796, BPSS1492; Dowling et al., 2010). Neither approach identified a common set of dominant ATs, which may indicate that the ATs have a cumulative importance. However, direct experimental evidence for a role of ATs in virulence or intramacrophage survival, as proposed by Schell et al. (2008) and Dowling et al. (2010), is lacking at the time of writing. Several predicted ATs are recognized by human convalescent sera, in some cases with a specificity adequate for serodiagnosis, and such data indicate that the proteins are expressed in vivo (Tiyawisutsri et al., 2007; Felgner et al., 2009; Suwannasaen et al., 2011). To date, the best characterized of the predicted ATs of B. pseudomallei and B. mallei is a factor influencing intracellular motility, BimA.
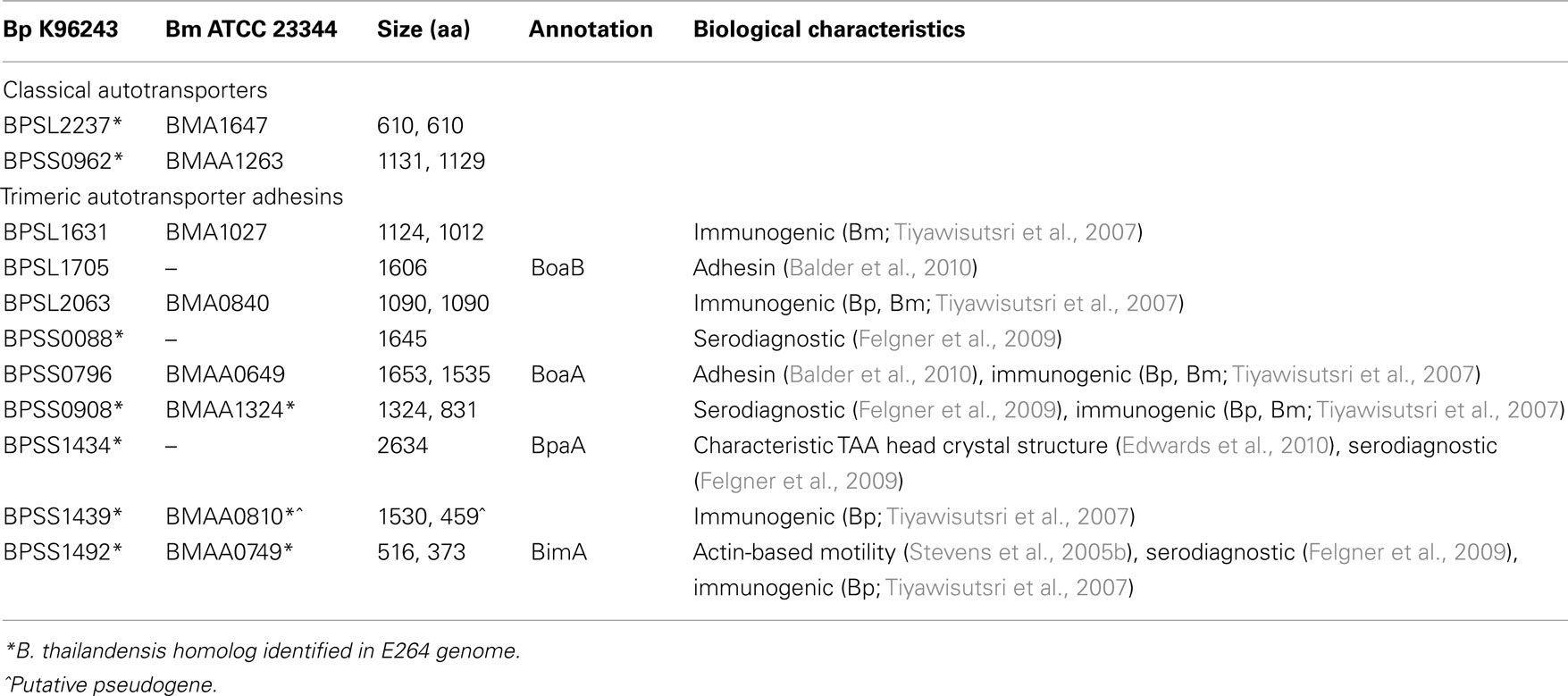
Table 1. Autotransporters of B. pseudomalle i K96243 and B. mallei ATCC 23344. The 11 B. pseudomalle i ATs and their eight B. malle i homologs are listed below alongside relevant functional information.
Burkholderia Intracellular Motility and the BimA Protein
In common with selected species of Listeria, Shigella, Rickettsia, and Mycobacterium marinum, B. pseudomallei shares the ability to induce polymerization of actin at one pole of the bacterial cell to promote its movement within and between host cells (Figure 1). The formation of actin-rich bacteria-containing membrane protrusions was first described in B. pseudomallei-infected HeLa and J774.1 cells (Kespichayawattana et al., 2000), but is also a feature of B. mallei and B. thailandensis infection (Stevens et al., 2005a). The bacterial factor(s) required for actin-based motility of B. pseudomallei have been sought by screening random mutants for a small plaque phenotype (Pilatz et al., 2006) and by targeted inactivation of candidate factors (Stevens et al., 2005b). One such candidate (BPSS1492, BimA) belongs to the Oca/type Vc family of TAAs with a β barrel sequence and linker region with 48% similarity to the Y. enterocolitica YadA C-terminal (pfam domain 03895). This was targeted on the basis that Shigella and Rickettsia use ATs for actin-based motility (IcsA and Sca2, respectively). The predicted BPSS1492 protein also contained domains associated with actin binding, including proline-rich motifs and Wiskott–Aldrich syndrome protein (WASP) homology-2 (WH2) domains that suggested it may be involved in actin assembly (Stevens et al., 2005b). Actin-based motility of B. pseudomallei in J774.2 cells was abolished by inactivation of bimA and could be restored by transient expression of the gene in trans (Stevens et al., 2005b). BimA is located at the pole of the bacterial cell at which actin polymerization occurs and polar localization is frequently seen in only one daughter cell at division (Figure 1), implying that polar targeting of BimA may rely on distinguishing the new and old poles of the cell (Stevens et al., 2005b).
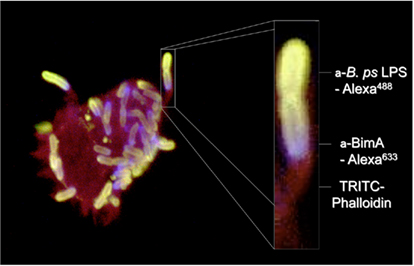
Figure 1. Actin-based motility of B. pseudomallei in a J774.2 murine macrophage-like cell 8 h post-infection. B. pseudomallei was detected with antibodies to its lipopolysaccharide (green) and BimA was detected with a panel of specific monoclonal antibodies (blue). Polymerized (F-) actin was stained red with fluorophore-labeled phalloidin.
Burkholderia pseudomallei BimA possesses an unusually long signal sequence typical of autotransporters and a passenger domain containing several putative functional domains. Within this effector domain, the two WH2 motifs have each been found to influence actin binding and polymerization in vitro (Sitthidet et al., 2011). In addition, B. pseudomallei BimA contains between 2 and 7 predicted target sites for host cell casein kinase II (PDASX), with variation across sequenced strains (Sitthidet et al., 2008). The PDASX repeats have since been established to act in an additive manner to enhance actin polymerization in vitro (Sitthidet et al., 2011). It remains unclear if such repeats are phosphorylated in vivo, or what the functional consequences of post-translational modification of BimA may be. Interestingly a 13 aa repeat region of BimA was found to be required for intercellular spread of B. pseudomallei but was dispensable for actin binding and polymerization (Sitthidet et al., 2011). Other bacterial factors involved in actin-based motility have been shown to promote intracellular survival through evasion of cytosolic killing by autophagy and further studies with defined mutants are required to determine if BimA or its domains confer such activity.
BimA orthologs exist in B. mallei ATCC23344 (BMAA0749) and B. thailandensis E264 (BTH_II0875) and can restore actin-based motility of a B. pseudomallei bimA mutant (Stevens et al., 2005a). The BimA passenger domain of the three species differ markedly in primary sequence resulting in differences in the number of WH2 domains, sequence of proline-rich motifs and the presence of a central and acidic (CA) domain, indicating that they may initiate actin assembly by distinct mechanisms (Stevens et al., 2005a). Many mechanisms of bacterial actin-based motility converge on activation of the cellular Arp (actin-related protein) 2/3 complex. Activation of the Arp2/3 complex requires cellular nucleation-promoting factors (NPFs) such as WASP-family members, and pathogens capable of actin-based motility often mimic the activity of NPFs or recruit and activate them at the bacterial pole (reviewed in Stevens et al., 2006). In recent years however, a number of virulence-associated factors have been identified in bacteria that initiate actin assembly in an Arp2/3-independent manner. For example, the AT Sca2 facilitates actin-based motility of certain Rickettsia species in an Arp2/3-independent manner akin to cellular formin proteins (Haglund et al., 2010; Kleba et al., 2010). It is notable that truncated recombinant B. pseudomallei BimA purified from E. coli polymerizes actin in vitro in the absence of any other cellular and bacterial co-factor in an Arp2/3-independent manner (Stevens et al., 2005b; Sitthidet et al., 2010); however, it remains unclear if such intrinsic activity would be adequate to propel the bacteria inside cells. In contrast, the ability of the B. thailandensis BimA ortholog to assemble actin in vitro is Arp2/3-dependent and requires the Arp2/3-recruiting CA domain (Sitthidet et al., 2010). Little is presently known about the mechanism of action of the B. mallei BimA ortholog; however, it is considered unlikely to be Arp2/3-dependent as it lacks an invariant tryptophan residue within an acidic stretch of residues required for Arp2/3 binding and activation (Stevens et al., 2005b). Intra-species conservation of BimA in natural populations of B. pseudomallei and B. thailandensis is high, with the exception of a geographically restricted subset of B. pseudomallei isolates harboring a B. mallei-like BimA variant (Sitthidet et al., 2008, 2010). The impact of sequence variation within BimA on pathogenesis and recognition by host immunity remains to be elucidated.
BimA has attracted much interest as a diagnostic tool. Recently, researchers have developed PCR-based methods to discriminate between Burkholderia species in a clinical setting (Ulrich et al., 2006a,b; Sitthidet et al., 2011), and protein microarray studies led to inclusion of BimA in a 10-protein multiplex classifier to improve diagnosis of human B. pseudomallei infection (Felgner et al., 2009; Suwannasaen et al., 2011). Curiously the B. mallei BimA protein was non-immunogenic in a test with serum from B. mallei-infected horses (Tiyawisutsri et al., 2007), yet B. pseudomallei BimA is one of the most immunogenic proteins recognized by sera from convalescent melioidosis patients (Felgner et al., 2009; Suwannasaen et al., 2011) and also serves as a T cell antigen (Felgner et al., 2009; Suwannasaen et al., 2011). The role of BimA in the pathogenesis and virulence of B. mallei and B. pseudomallei also appears to differ, with B. mallei BimA being dispensable for virulence in a Syrian hamster model of glanders (Schell et al., 2007) whilst B. pseudomallei BimA contributes to pathogenesis in murine models of melioidosis and confers protection against homologous challenge (Galyov et al., unpublished data). This may reflect differences in the animal species, dose and route of inoculation. BimA in B. mallei is regulated by the VirAG two-component system that is encoded nearby and which also controls expression of a virulence-associated Type VI secretion system (Schell et al., 2007). BimA appears to be poorly expressed in vitro and the stimuli for its expression are ill-defined.
Characterization of Other Candidate B. pseudomallei and B. mallei autotransporters
In addition to BimA, two predicted TAAs have been the subject of further characterization: BPSS0796/BMAA0649 and BPSL1705, re-annotated as BoaA and BoaB respectively, were expressed in a heterologous E. coli host where they were displayed on the bacterial surface and promoted attachment of the recombinant E. coli strains to epithelial cell lines (Balder et al., 2010). In common with the role of other TAA family members in adherence, B. pseudomallei and/or B. mallei mutants with disruptions in boaA or boaB exhibited reduced adherence to epithelial cells and a double mutant, but neither single mutant, displayed a growth defect in phagocytic cell lines (Balder et al., 2010). Though suggestive of a role in virulence, studies with defined mutant and repaired or complemented strains in animals are required to establish the role of BoaA and BoaB in pathogenesis.
Evidence of expression of selected TAAs has been obtained by screening of expression libraries with convalescent sera. Serum from B. mallei-infected horses identified multiple independent clones for four TAAs (BMA0840, BMA1027, BMA0649, and BMAA1324) suggestive of a strong antibody response (Tiyawisutsri et al., 2007). A similar screen with convalescent serum from human melioidosis patients identified five TAAs (BPSL2063, BPSS0908, BPSS0796, BPSS1439, and BPSS1492); however, these were found at a low frequency indicative of a relatively poor immune response (Tiyawisutsri et al., 2007). Further studies are required to determine if this reflects genuine differences in immunogenicity of the predicted proteins, or the distinct hosts, nature of exposure and/or sampling times. Additionally, a protein microarray probed with pooled melioidosis patients sera identified 4 ATs (BPSS0088, BPSS0796, BPSS1434/BpaA, BPSS1492/BimA) as having serodiagnostic potential due to their ability to interact with melioidosis-specific antibodies (Felgner et al., 2009). The ATs identified by these two studies demonstrate minimal overlap although this may reflect the variation in in vivo bacterial gene expression or the host immune response. Microarray studies under 82 in vitro different conditions demonstrated that the 11 ATs of B. pseudomallei are constitutively expressed (Tan, personal comm.), while qRT-PCR analysis of the transcription of two AT genes (boaA, boaB) demonstrated low expression (Balder et al., 2010).
Alignment of the C-terminal β barrel domain regions of the putative B. pseudomallei and B. mallei TAAs demonstrates considerable conservation indicative of amplification of these adhesins through gene duplication events (Figure 2A). This is potentially significant given the importance of C-terminal regions of TAAs in translocation and trimer stability, as evidenced by the ability of C-terminal domains of YadA family members from diverse bacteria to functionally replace the cognate domain in Yersinia (Ackermann et al., 2008). A phylogenetic tree of the β barrel domains is supportive of this idea as the TAAs form two clusters suggesting that these genes may have been amplified from two original ancestral genes (Figure 2B). Both clusters appear to be derived from a common ancestor to the prototypic Yersinia YadA TAA. Additionally, the two outliers in the alignment, BPSS0908/BMAA1324 and BPSS1434, display early separation in the phylogenetic tree. Interestingly, of the three unique B. pseudomallei TAAs, two (BPSS1434 and BPSS0088) are early branches in the phylogenetic tree suggesting that their absence from B. mallei is due to gene loss as later branching genes are present. Meanwhile BPSL1705, which is located within a genomic island (GI8), is very recently branched from BPSS0796/BMAA0649 and has been proposed to have originated as a duplication of BPSS0796/BMAA0649 (Tiyawisutsri et al., 2007). This duplication event may explain why BPSL1705 is the only AT not conserved across the four completed B. pseudomallei genomes. Furthermore, the β barrel domains of the B. pseudomallei and B. mallei TAA homologs show 100% identity although little similarity is seen between the passenger domains. However, the effector domains of TAAs are generally not conserved at a sequence level and the crystal structure analysis of a section of the passenger domain of BPSS1434 (re-annotated as BpaA, B. pseudomallei adhesion A) demonstrated quaternary structure similar to that of other TAAs (Edwards et al., 2010). This new annotation gives BPSS1434 the same name as the unrelated Type V two-partner secreted BpaA (Brown et al., 2004) for which no homolog exists in either B. pseudomallei K96243 or B. mallei ATCC 23344. The B. pseudomallei and B. mallei genomes contain homologs of BamA, BamB, and SurA which are proposed to assist in translocation of ATs across the OM (Ieva and Bernstein, 2009; Ruiz-Perez et al., 2009; Sauri et al., 2009; Lehr et al., 2010), and further studies are needed to evaluate the role of such factors in AT secretion and pathogenesis.
Figure 2. Alignment and phylogenetic analysis of the β barrel domains of the putative B. pseudomallei and B. mallei TAAs: (A) the nine B. pseudomallei C-terminal 70–72 aa β barrel domains (B. mallei homologs 100% identical at this region) were aligned; identical residues are highlighted in red, those with high homology (at least 66%) in blue while those with highly conserved substitutions in green. The consensus sequence is listed below. (B) The phylogenetic analysis of these domains demonstrates two distinct clusters both of which derived from a common ancestor to the prototypical Yersinia YadA TAA.
Concluding Remarks
On the basis of homology and the precedent of phenotypes for AT mutants in other bacteria, it is reasonable to infer that ATs predicted to be encoded in the genomes of pathogenic Burkholderia species may play a role in pathogenesis and immunity. In the short time since annotation of the genomes of B. pseudomallei and B. mallei, it has been established that several putative ATs mediate bacterial interactions with host cells. For example, BimA mediates intracellular actin-based motility, which in the case of B. pseudomallei is necessary for intercellular spread and full virulence, whereas BoaA and BoaB appear to function as adhesins. One should note that significant sequence divergence in effector domains can occur even among closely related species, and within species, as we have noted for BimA (Sitthidet et al., 2008). Such variation in BimA can markedly alter the mode of action (Sitthidet et al., 2011) and future studies will need to consider the extent to which the repertoire and sequence of ATs in prototype strains is typical of wider populations. Very few published studies have probed the role of ATs in pathogenesis or immunity during B. pseudomallei and B. mallei infection in model hosts, and further studies with defined mutants and complemented strains are needed. Attenuated mutants may be suitable as live-attenuated vaccines, and based on the ability of some ATs to act as B- and T-cell antigens (Tiyawisutsri et al., 2007), purified ATs may have merit as subunit vaccines or as diagnostic tools. Importantly, the mode of translocation of Burkholderia ATs is inferred only from homologous systems, and studies are needed to address outstanding questions regarding their biological role. Future experiments should evaluate the role of AT domains and specific residues in protein folding, oligomerization, interaction and translocation across the OM, with particular reference to the requirement for periplasmic chaperones and the Bam complex. Analysis of the structure of Burkholderia ATs may also provide clues to the mechanistic basis of their secretion and function.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ackermann, N., Tiller, M., Anding, G., Roggenkamp, A., and Heesemann, J. (2008). Contribution of trimeric autotransporter C-terminal domains of oligomeric coiled-coil adhesin (Oca) family members YadA, UspA1, EibA, and Hia to translocation of the YadA passenger domain and virulence of Yersinia enterocolitica. J. Bacteriol. 190, 5031–5043.
Balder, R., Lipski, S., Lazarus, J. J., Grose, W., Wooten, R. M., Hogan, R. J., Woods, D. E., and Lafontaine, E. R. (2010). Identification of Burkholderia mallei and Burkholderia pseudomallei adhesins for human respiratory epithelial cells. BMC Microbiol. 10, 250. doi:10.1186/1471-2180-10-250
Brown, N. F., Logue, C. A., Boddey, J. A., Scott, R., Hirst, R. G., and Beacham, I. R. (2004). Identification of a novel two-partner secretion system from Burkholderia pseudomallei. Mol. Genet. Genomics 272, 204–215.
Cotter, S. E., Surana, N. K., and St Geme, J. W. R. (2005). Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13, 199–205.
Dautin, N., and Bernstein, H. (2007). Protein secretion in Gram-negative bacteria via the autotransporter pathway. Annu. Rev. Microbiol. 61, 89–112.
Desvaux, M., Scott-Tucker, A., Turner, S. M., Cooper, L. M., Huber, D., Nataro, J. P., and Henderson, I. R. (2007). A conserved extended signal peptide region directs posttranslational protein translocation via a novel mechanism. Microbiology 153, 59–70.
Dowling, A. J., Wilkinson, P. A., Holden, M. T., Quail, M. A., Bentley, S. D., Reger, J., Waterfield, N. R., Titball, R. W., and Ffrench-Constant, R. H. (2010). Genome-wide analysis reveals loci encoding anti-macrophage factors in the human pathogen Burkholderia pseudomallei K96243. PLoS ONE 5, e15693. doi: 10.1371/journal.pone.0015693
Edwards, T. E., Phan, I., Abendroth, J., Dieterich, S. H., Masoudi, A., Guo, W., Hewitt, S. N., Kelley, A., Leibly, D., Brittnacher, M. J., Staker, B. L., Miller, S. I., Van Voorhis, W. C., Myler, P. J., and Stewart, L. J. (2010). Structure of a Burkholderia pseudomallei trimeric autotransporter adhesin head. PLoS ONE 5, e12803. doi:10.1371/journal.pone.0012803
Felgner, P. L., Kayala, M. A., Vigil, A., Burk, C., Nakajima-Sasaki, R., Pablo, J., Molina, D. M., Hirst, S., Chew, J. S., Wang, D., Tan, G., Duffield, M., Yang, R., Neel, J., Chantratita, N., Bancroft, G., Lertmemongkolchai, G., Davies, D. H., Baldi, P., Peacock, S., and Titball, R.W. (2009). A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc. Natl. Acad. Sci. U.S.A. 106, 13499–13504.
Galyov, E. E., Brett, P. J., and DeShazer, D. (2010). Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu. Rev. Microbiol. 64, 495–517.
Haglund, C. M., Choe, J. E., Skau, C. T., Kovar, D. R., and Welch, M. D. (2010). Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat. Cell Biol. 12, 1057–1063.
Ieva, R., and Bernstein, H. D. (2009). Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 19120–19125.
Jain, S., and Goldberg, M. B. (2007). Requirement for YaeT in the outer membrane assembly of autotransporter proteins. J. Bacteriol. 189, 5393–5398.
Kespichayawattana, W., Rattanachetkul, S., Wanun, T., Utaisincharoen, P., and Sirisinha, S. (2000). Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect. Immun. 68, 5377–5384.
Kleba, B., Clark, T. R., Lutter, E. I., Ellison, D. W., and Hackstadt, T. (2010). Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect. Immun. 78, 2240–2247.
Lazar Adler, N. R., Govan, B., Cullinane, M., Harper, M., Adler, B., and Boyce, J. D. (2009). The molecular and cellular basis of pathogenesis in melioidosis: how does Burkholderia pseudomallei cause disease? FEMS Microbiol. Rev. 33, 1079–1099.
Lehr, U., Schütz, M., Oberhettinger, P., Ruiz-Perez, F., Donald, J. W., Palmer, T., Linke, D., Henderson, I. R., and Autenrieth, I. B. (2010). C-terminal amino acid residues of the trimeric autotransporter adhesin YadA of Yersinia enterocolitica are decisive for its recognition and assembly by BamA. Mol. Microbiol. 78, 932–946.
Nishimura, K., Tajima, N., Yoon, Y. H., Park, S. Y., and Tame, J. R. (2010). Autotransporter passenger proteins: virulence factors with common structural themes. J. Mol. Med. 88, 451–458.
Oomen, C. J., Van Ulsen, P., Van Gelder, P., Feijen, M., Tommassen, J., and Gros, P. (2004). Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23, 1257–1266.
Peterson, J. H., Szabady, R. L., and Bernstein, H. D. (2006). An unusual signal peptide extension inhibits the binding of bacterial presecretory proteins to the signal recognition particle, trigger factor, and the SecYEG complex. J. Biol. Chem. 281, 9038–9048.
Pilatz, S., Breitbach, K., Hein, N., Fehlhaber, B., Schulze, J., Brenneke, B., Eberl, L., and Steinmetz, I. (2006). Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence. Infect. Immun. 74, 3576–3586.
Ruiz-Perez, F., Henderson, I. R., Leyton, D. L., Rossiter, A. E., Zhang, Y., and Nataro, J. P. (2009). Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J. Bacteriol. 191, 6571–6583.
Sauri, A., Soprova, Z., Wickström, D., De Gier, J. W., Van Der Schors, R. C., Smit, A. B., Jong, W. S., and Luirink, J. (2009). The Bam (Omp85) complex is involved in secretion of the autotransporter haemoglobin protease. Microbiology 155, 3982–3991.
Schell, M. A., Lipscomb, L., and DeShazer, D. (2008). Comparative genomics and an insect model rapidly identify novel virulence genes of Burkholderia mallei. J. Bacteriol. 190, 2306–2313.
Schell, M. A., Ulrich, R. L., Ribot, W. J., Brueggemann, E. E., Hines, H. B., Chen, D., Lipscomb, L., Kim, H. S., Mrázek, J., Nierman, W. C., and DeShazer, D. (2007). Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 64, 1466–1485.
Sitthidet, C., Korbsrisate, S., Layton, A. N., Field, T. R., Stevens, M. P., and Stevens, J. M. (2011). Identification of motifs of Burkholderia pseudomallei BimA required for intracellular motility, actin binding and actin polymerization. J. Bacteriol. 193, 1901–1910.
Sitthidet, C., Stevens, J. M., Chantratita, N., Currie, B. J., Peacock, S. J., Korbsrisate, S., and Stevens, M. P. (2008). Prevalence and sequence diversity of a factor required for actin-based motility in natural populations of Burkholderia species. J. Clin. Microbiol. 46, 2418–2422.
Sitthidet, C., Stevens, J. M., Field, T. R., Layton, A. N., Korbsrisate, S., and Stevens, M. P. (2010). Actin-based motility of Burkholderia thailandensis requires a central acidic domain of BimA that recruits and activates the cellular Arp2/3 complex. J. Bacteriol. 192, 5249–5252.
Stevens, J. M., Galyov, E. E., and Stevens, M. P. (2006). Actin-dependent movement of bacterial pathogens. Nat. Rev. Microbiol. 4, 91–101.
Stevens, J. M., Ulrich, R. L., Taylor, L. A., Wood, M. W., DeShazer, D., Stevens, M. P., and Galyov, E. E. (2005a). Actin-binding proteins from Burkholderia mallei and Burkholderia thailandensis can functionally compensate for the actin-based motility defect of a Burkholderia pseudomallei bimA mutant. J. Bacteriol. 187, 7857–7862.
Stevens, M. P., Stevens, J. M., Jeng, R. L., Taylor, L. A., Wood, M. W., Hawes, P., Monaghan, P., Welch, M. D., and Galyov, E. E. (2005b). Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol. Microbiol. 56, 40–53.
Stevens, M. P., Haque, A., Atkins, T., Hill, J., Wood, M. W., Easton, A., Nelson, M., Underwood-Fowler, C., Titball, R. W., Bancroft, G. J., and Galyov, E. E. (2004). Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis. Microbiology 150, 2669–2676.
Stevens, M. P., Wood, M. W., Taylor, L. A., Monaghan, P., Hawes, P., Jones, P. W., Wallis, T. S., and Galyov, E. E. (2002). An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46, 649–659.
Suwannasaen, D., Mahawantung, J., Chaowagul, W., Limmathurotsakul, D., Felgner, P. L., Davies, H., Bancroft, G. J., Titball, R. W., and Lertmemongkolchai, G. (2011). Human immune responses to Burkholderia pseudomallei characterized by protein microarray analysis. J. Infect. Dis. 203, 1001–1011.
Tiyawisutsri, R., Holden, M. T., Tumapa, S., Rengpipat, S., Clarke, S. R., Foster, S. J., Nierman, W. C., Day, N. P., and Peacock, S. J. (2007). Burkholderia Hep_Hag autotransporter (BuHA) proteins elicit a strong antibody response during experimental glanders but not human melioidosis. BMC Microbiol. 7, 19. doi:10.1186/1471-2180-7-19
Ulrich, M. P., Norwood, D. A., Christensen, D. R., and Ulrich, R. L. (2006a). Using real-time PCR to specifically detect Burkholderia mallei. J. Med. Microbiol. 55, 551–559.
Ulrich, R. L., Ulrich, M. P., Schell, M. A., Kim, H. S., and DeShazer, D. (2006b). Development of a polymerase chain reaction assay for the specific identification of Burkholderia mallei and differentiation from Burkholderia pseudomallei and other closely related Burkholderiaceae. Diagn. Microbiol. Infect. Dis. 55, 37–45.
Van Den Berg, B. (2010). Crystal structure of a full-length autotransporter. J. Mol. Biol. 396, 627–633.
Wells, T. J., Tree, J. J., Ulett, G. C., and Schembri, M. A. (2007). Autotransporter proteins: novel targets at the bacterial cell surface. FEMS Microbiol. Lett. 274, 163–172.
Keywords: Burkholderia pseudomallei, Burkholderia mallei, autotransporter, secreted, protein
Citation: Adler NR, Stevens JM, Stevens MP and Galyov EE (2011) Autotransporters and their role in the virulence of Burkholderia pseudomallei and Burkholderia mallei. Front. Microbio. 2:151. doi: 10.3389/fmicb.2011.00151
Received: 28 April 2011;
Paper pending published: 31 May 2011;
Accepted: 28 June 2011;
Published online: 15 July 2011.
Edited by:
Alfredo G. Torres, University of Texas Medical Branch, USAReviewed by:
Peggy Cotter, University of North Carolina, Chapel Hill, USAFernando Navarro-Garcia, Center for Research and Advanced Studies, Mexico
Copyright: © 2011 Adler, Stevens, Stevens and Galyov. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Edouard E. Galyov, Department of Infection, Immunity and Inflammation, University of Leicester, Medical Sciences Building, Leicester, LE1 9HN, UK. e-mail: eg98@leicester.ac.uk
 Natalie R. Lazar Adler1
Natalie R. Lazar Adler1