- Department of Civil and Environmental Engineering, The University of Michigan, Ann Arbor, MI, USA
Microbially mediated bioremediation of polluted sites has been a subject of much research over the past 30 years, with many different compounds shown to be degraded under both aerobic and anaerobic conditions. Aerobic-mediated bioremediation commonly examines the use of methanotrophs, microorganisms that consume methane as their sole source of carbon and energy. Given the diverse environments in which methanotrophs have been found, the range of substrates they can degrade and the fact that they can be easily stimulated with the provision of methane and oxygen, these microorganisms in particular have been examined for aerobic degradation of chlorinated hydrocarbons. The physiological and phylogenetic diversity of methanotrophy, however, has increased substantially in just the past 5 years. Here in this review, the current state of knowledge of methanotrophy, particularly as it applies to pollutant degradation is summarized, and suggestions for future research provided.
Introduction
Methanotrophs are distinguished from other microorganisms by their ability to utilize methane as their sole carbon and energy source, yet are physiologically and phylogenetically diverse, maintaining affiliations with the bacterial phyla Γ-Proteobacteria, α-Proteobacteria Verrucomicrobia, and NC10, as well within the Euryarchaeota phylum of the Archaea. Initially, only aerobic methanotrophs were found, but it is now known that methane can also be oxidized by different microorganisms anaerobically by being coupled to sulfate, nitrite, iron, or manganese reduction (Boetius et al., 2000; Michaelis et al., 2002; Knittel et al., 2005; Beal et al., 2009; Knittel and Boetius, 2009; Ettwig et al., 2010; Semrau et al., 2010).
As a result, methanotrophs are ubiquitous and play a major role in the global cycling of carbon and nitrogen and, for the case of aerobic methanotrophs, have also been found to be useful for the biodegradation of hazardous organic materials (Anthony, 1982; DiSpirito et al., 1992; Hanson and Hanson, 1996; Lontoh and Semrau, 1998; Lontoh et al., 2000). All aerobic methanotrophs employ the enzyme methane monooxygenase (MMO) to convert methane to methanol in the first step of methane oxidation to CO2 (Semrau et al., 2010). It is intriguing that methane oxidation coupled to nitrite oxidation also uses the methane monooxygenase (Ettwig et al., 2010). Here, “Candidatus Methylomirabilis oxyfera” reduces nitrite to nitric oxide, which is then converted to dinitrogen and dioxygen using nitric oxide dismutase, i.e., M. oxyfera generates its own oxygen for methane oxidation.
Other types of anaerobic methane oxidation do not employ the MMO, and the biochemical pathway that allows for such anaerobic methane oxidation is as yet unknown. In the case of methane oxidation coupled to sulfate reduction, three different groups of archaea are commonly, yet not always found in association with sulfate-reducing bacteria (Beal et al., 2009; Knittel and Boetius, 2009; Knittel et al., 2005) and reverse methanogenesis is the only hypothesis for such anaerobic methane oxidation that explains all available data (Knittel et al., 2005). Anaerobic methane oxidation coupled to iron or manganese reduction is also proposed to be due to concerted activity of archaea and bacteria, although the possibility that bacteria may be solely responsible for anaerobic oxidation of methane coupled to manganese reduction cannot be ruled out (Beal et al., 2009). It is also unclear how methane oxidation coupled to metal reduction proceeds, although reverse methanogenesis is a possibility.
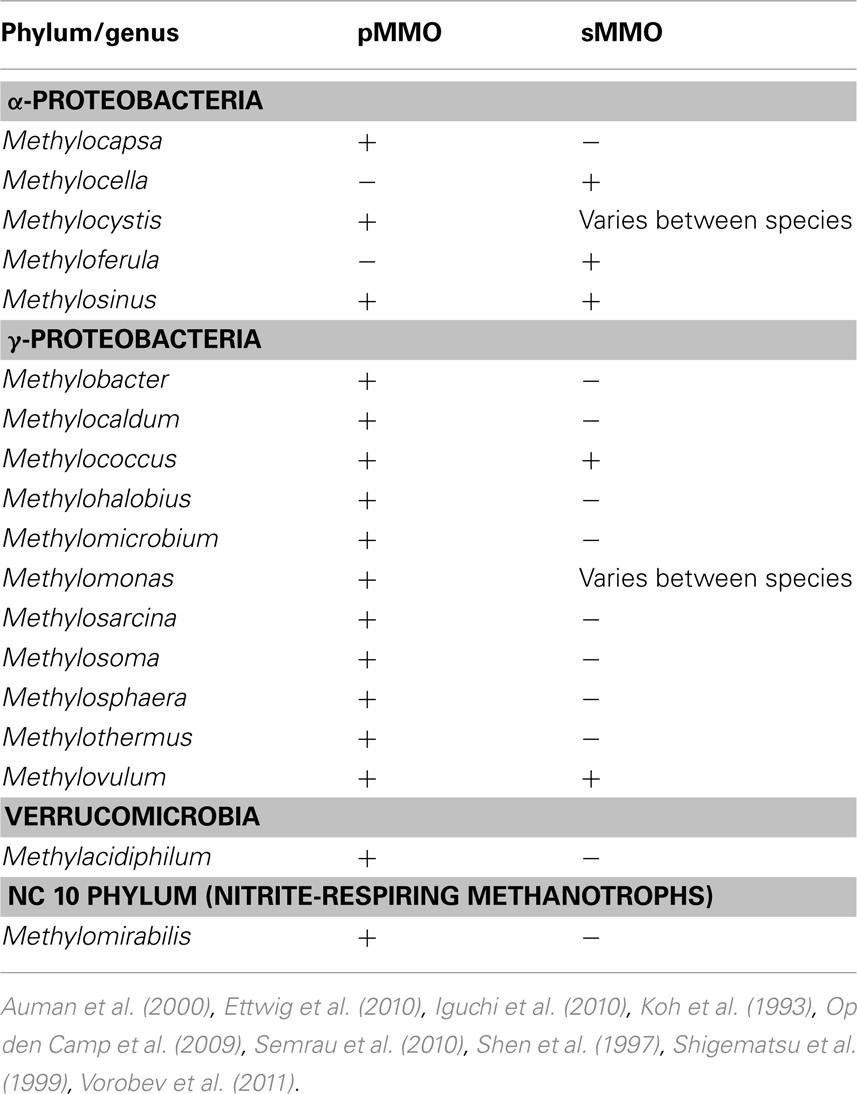
As mentioned earlier, for the aerobic methanotrophs, methane is initially transformed into methanol by the MMO. Methanol is then further oxidized to formaldehyde by the methanol dehydrogenase. Formaldehyde can then either be assimilated into biomass via the serine cycle or the ribulose monophosphate pathway (RuMP), or further oxidized to formate and carbon dioxide to generate reducing equivalents needed in the initial oxidation of methane (Trotsenko and Murrell, 2008; Semrau et al., 2010). MMO is known to exist in at least two forms. One form, the particulate methane monooxygenase (pMMO) is found in most known aerobic methanotrophs as well as M. oxyfera and is located in the cytoplasmic membrane (Op den Camp et al., 2009; Ettwig et al., 2010; Semrau et al., 2010). Another form, the soluble methane monooxygenase (sMMO) is found in some aerobic methanotrophs (so far only in a handful of Γ- and α-Proteobacteria) and is located in the cytoplasm (Semrau et al., 2010). A summary of the current genera of methanotrophs known to synthesize MMO and the forms expressed is shown in Table 1.
Both forms of MMO have been shown to oxidize a range of pollutants, particularly halogenated hydrocarbons (Semrau et al., 2010). Given the ubiquity of aerobic methanotrophs (found wherever air:methane interfaces develop), these microorganisms have been extensively used for pollutant degradation. The utility of methanotrophy for pollutant degradation, however, is complicated by three general issues as noted earlier (Semrau et al., 2010). First, overall pollutant degradation by methanotrophs can be limited by the toxicity of both the pollutant itself and oxidative product(s) formed by both forms of MMO. Second, pollutant degradation by either form of MMO requires a source of reducing equivalents for the reduction of dioxygen. Third, pollutant degradation by methanotrophs is typically co-metabolic, i.e., it can only occur over an extended period in the presence of the growth substrate, methane, which acts as an inhibitor of pollutant degradation by both forms of the MMO. As such, overall pollutant degradation by methanotrophs can be limited by the need to ensure adequate methane levels necessary for methanotrophic survival.
Here, the use of methanotrophs that express the methane monooxygenase for pollutant degradation is reviewed, with a particular discussion on how methanotrophic-mediated bioremediation can be improved.
History of Pollutant Degradation via Methanotrophs
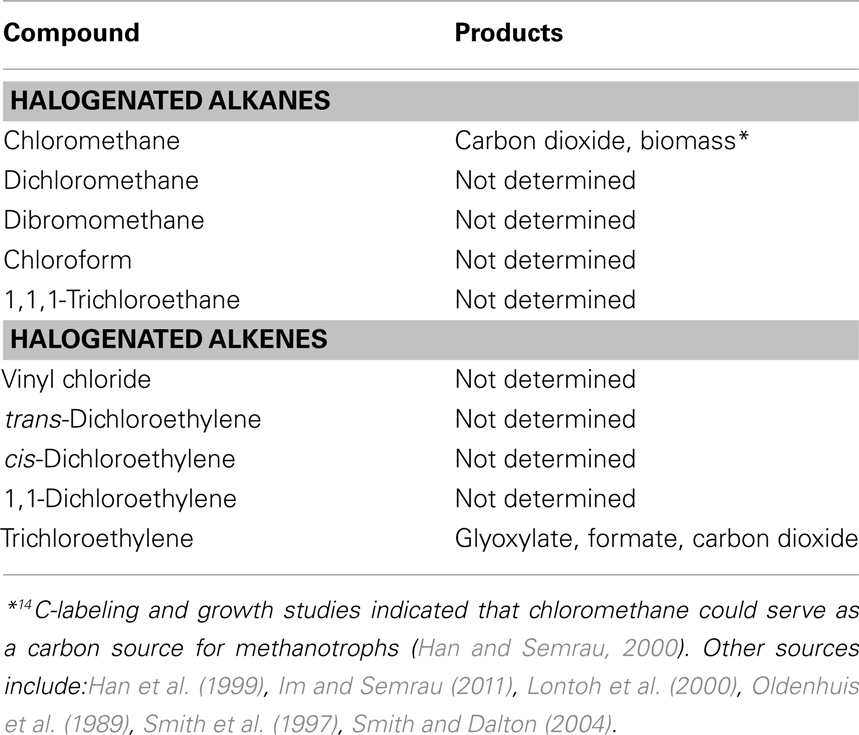
Many compounds have been shown to be degraded to varying degrees by aerobic methanotrophs, including halogenated alkanes, alkenes, and aromatic compounds. In methanotrophs, such degradation is due to MMO activity, and the rate and range of pollutants degraded is dependent on the form of MMO expressed. sMMO-expressing cells typically degrade more compounds than pMMO-expressing cells, and degrade such compounds at faster initial rates (Colby et al., 1977; Hou et al., 1979; Burrows et al., 1984). A partial list of halogenated hydrocarbons shown to be degraded by pMMO or sMMO is provided in Tables 2 and 3. It should be noted that the data presented in Tables 2 and 3 is based on studies of methanotrophic genera within the Γ-Proteobacteria and α-Proteobacteria phyla, e.g., Methylobacter, Methylococcus, Methylocystis, Methylomicrobium, and Methylosinus (Colby et al., 1977; Oldenhuis et al., 1989; Fox et al., 1990; Sullivan and Chase, 1996; van Hylckama Vleig et al., 1996; Smith et al., 1997; Bosma and Janssen, 1998; Grosse et al., 1999; Han et al., 1999; Han and Semrau, 2000; Lindner et al., 2000; Lontoh et al., 2000; Jechorek et al., 2003; Smith and Dalton, 2004; Im and Semrau, 2011). Further investigation will likely extend not only the list of such compounds for methanotrophs expressing either form of MMO, but also the range of phyla shown to degrade halogenated hydrocarbons.
Soluble methane monooxygenase and pMMO-expressing cells degrade the compounds shown in Tables 2 and 3 via oxidation, typically to the corresponding alcohol (for alkanes) and epoxide (for alkenes) and, in the case of sMMO-expressing cells, through the hydroxylation of halogenated aromatic compounds. In many cases, these initial products can undergo further abiotic and biotic transformations, with the stable products shown in Tables 2 and 3. It should be noted that for pMMO-expressing cells oxidizing chloromethane and trichloroethylene, mineralization of these compounds (i.e., conversion to carbon dioxide), was observed, suggesting that these compounds may provide some, albeit small, benefit to methanotrophs (Han and Semrau, 2000; Lontoh et al., 2000). Further, it was found that carbon in 14C-labeled chloromethane was assimilated into biomass, indicating that chloromethane also served as a carbon source (Han and Semrau, 2000). Here it was postulated that chloromethane was first transformed to chloromethanol, which then abiotically broke down to formaldehyde, and thus was able to be used for generation of both biomass and reducing equivalents by methanotrophs.
The finding that aerobic methanotrophs can degrade priority pollutants such as halogenated hydrocarbons coupled with the fact that aerobic methanotrophs are ubiquitous generated a great deal of interest in using methanotrophy to remediate polluted soils and aquifers. In particular, field demonstrations at Moffett Naval Air Station and the Savannah River Site of the Department of Energy clearly showed that aerobic methanotrophs could be stimulated through the provision of methane and air with subsequent pollutant degradation (Semprini et al., 1990; Pfiffner et al., 1997).
Use of sMMO vs. pMMO-Expressing Cells for Pollutant Degradation
Given the finding that sMMO-expressing aerobic methanotrophs degraded more compounds more quickly than pMMO-expressing aerobic methanotrophs, it is not surprising that much of the early literature focused on the utility of sMMO-expressing aerobic methanotrophs for pollutant degradation. It should be noted, however, that these studies typically focused on simple systems where one methanotrophic culture (either pure or mixed) was exposed to one pollutant. More recent work suggests that pMMO-expressing aerobic methanotrophs may be preferable as these cells are able to survive more easily in the presence of relatively complex mixtures of chlorinated ethenes that are more representative of what could be found in situ (Lee et al., 2006). Here it was found that under certain conditions (i.e., at relatively high initial concentrations of chlorinated ethenes), the faster degradation of chlorinated ethenes by sMMO-expressing methanotrophs led to a rapid buildup of toxic products that reduced overall methanotrophic growth, and ultimately less overall pollutant degradation than that observed for pMMO-expressing microorganisms.
From these observations, a simple model, the “Δ Model” was developed based on Michaelis–Menten kinetics to predict when either sMMO or pMMO-expressing systems would be preferred. Δ is calculated as the difference between the rate of growth substrate turnover (vG in Eq. 1below) and the sum of the rate(s) of competing pollutant degradation (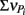 in Eq. 1below):
in Eq. 1below):
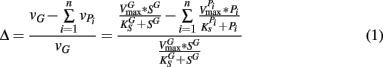
It should be noted that the values of vG and 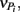 and thus Δ depends on the concentrations of growth substrate and pollutant(s) and can vary from 1 [no pollutant(s) present] to less than zero (the total rate of pollutant turnover is greater than that of growth substrate turnover). From previous studies (Lee et al., 2006; Yoon and Semrau, 2008), it was found that when Δ values were less than zero, little if any methanotrophic growth was observed with limited concomitant pollutant degradation. Further, it was found that the Δ value of sMMO-expressing methanotrophs, due to the relatively broad substrate range and fast pollutant turnover kinetics of these microorganisms, decreased much more quickly as pollutant concentrations increased than that calculated for pMMO-expressing methanotrophs. As such, pMMO-expressing methanotrophs may be preferred over sMMO-expressing methanotrophs over a broad range of pollutant concentrations as those microorganisms are better able to bind and turnover methane in the presence of competing compounds, and thus increase cell numbers and generate reducing equivalents, both of which enhance pollutant degradation.
and thus Δ depends on the concentrations of growth substrate and pollutant(s) and can vary from 1 [no pollutant(s) present] to less than zero (the total rate of pollutant turnover is greater than that of growth substrate turnover). From previous studies (Lee et al., 2006; Yoon and Semrau, 2008), it was found that when Δ values were less than zero, little if any methanotrophic growth was observed with limited concomitant pollutant degradation. Further, it was found that the Δ value of sMMO-expressing methanotrophs, due to the relatively broad substrate range and fast pollutant turnover kinetics of these microorganisms, decreased much more quickly as pollutant concentrations increased than that calculated for pMMO-expressing methanotrophs. As such, pMMO-expressing methanotrophs may be preferred over sMMO-expressing methanotrophs over a broad range of pollutant concentrations as those microorganisms are better able to bind and turnover methane in the presence of competing compounds, and thus increase cell numbers and generate reducing equivalents, both of which enhance pollutant degradation.
Pollutant Degradation by Facultative Methanotrophs
Although the use of the Δ model can help determine which MMO should be utilized for pollutant degradation under any specific set of circumstances, the fact that pollutant(s) compete with the growth substrate, methane, for binding complicates the use of methanotrophs for bioremediation. This issue, when considered in conjunction with the fact that methane is sparingly soluble in water, can make methanotrophic-mediated bioremediation challenging despite the advantages that methanotrophs provide, i.e., these microorganisms are ubiquitous, can degrade a wide range of substrates, and can be stimulated with the provision of a readily available and non-toxic substrate (methane).
Recently, however, facultative methanotrophs have been characterized that can utilize a variety of multi-carbon substrates such as ethanol and acetate for growth (Dedysh et al., 2005; Dunfield et al., 2010; Belova et al., 2011; Im et al., 2011; Semrau et al., 2011). Facultative methanotrophy is intriguing as it has been found that some, but not all facultative methanotrophs constitutively express the MMO regardless of the growth substrate (e.g., methane vs. acetate or ethanol). As a result, it may be possible to utilize methanotrophs for pollutant degradation without the need for the provision of methane, and thus minimizing competition for binding to the MMO. At least one facultative methanotroph, Methylocystis strain SB2, that groups with the α-Proteobacteria and constitutively expresses pMMO, has been shown to degrade a variety of chlorinated hydrocarbons when grown on acetate or ethanol (Im and Semrau, 2011; Yoon et al., 2011).
One interesting aspect of pollutant degradation by facultative methanotrophs is the possibility that these microorganisms may actually generate some benefit from oxidizing these compounds. Such a phenomenon is not unknown, for as described earlier, chloromethane has been found to stimulate methanotrophic growth when methanol was provided as the growth substrate (Han and Semrau, 2000). It is still unclear how facultative methanotrophs assimilate carbon from multi-carbon compounds, but all facultative methanotrophs to date group with the α-Proteobacteria, and use the serine cycle for carbon assimilation from formaldehyde. Many known pathways for acetate assimilation (e.g., the glyoxylate shunt of the tricarboxylic acid cycle, the ethylmalonyl-CoA pathway, the citramalate cycle, and the methylaspartate cycle) make malate and glyoxylate, which are intermediates of the serine cycle (Semrau et al., 2011). Of interest here is that glyoxylate has been shown to be a primary product of trichloroethylene oxidation by pMMO-expressing methanotrophs as well as by purified sMMO (Fox et al., 1990; Lontoh et al., 2000), suggesting that for facultative methanotrophs, and for methanotrophs that utilize the serine cycle for carbon assimilation, compounds such as trichloroethylene may provide some, albeit likely limited, benefit.
Given this, it is possible that facultative methanotrophs could be grown on a multi-carbon substrate such as acetate, with MMO activity focused on pollutant degradation that could have some, as yet unknown benefit. In this scenario, competition for binding to MMO is avoided, and the transformation of select pollutants into intermediates of the serine cycle and/or pathways for multi-carbon assimilation consumption may provide carbon compounds that can be used to support growth. Here, toxicity of the pollutant and its products is still possible, and this should be kept in mind when attempting to utilize facultative methanotrophy for pollutant degradation.
Is Methanotrophic-Mediated Pollutant Degradation Possible Under Anaerobic Conditions?
To date, only aerobic methanotrophs have been extensively studied for pollutant degradation, although anaerobic methane oxidizing microbial systems could also be utilized, particularly nitrite-driven methane oxidation by M. oxyfera. This very intriguing microorganism surprisingly uses pMMO for methane oxidation, which requires oxygen. As noted above, M. oxyfera generates its own oxygen from nitrite by first reducing it to nitric oxide, and then via the activity of a nitric oxide dismutase, converts nitric oxide to dinitrogen and dioxygen (Ettwig et al., 2010). Given that M. oxyfera utilizes pMMO for methane oxidation, it is very possible that this microorganism could also oxidize a variety of halogenated hydrocarbons in anaerobic conditions. Such activity, however, will not only be affected by competition with methane for binding to pMMO as well as possible toxicity of the pollutant and its oxidative products, but also could be limited by the necessity of M. oxyfera to generate oxygen for pMMO activity. It is not clear if such activity would be purposely stimulated in situ as nitrite would have to be added, but nonetheless, it is possible that these microorganisms could have some use for pollutant degradation in anaerobic zones.
Can Acidophilic or Thermoacidophilic Methanotrophs be Used for Pollutant Degradation?
Most known aerobic methanotrophs are mesophilic, growing best at near-neutral pH and 30°C. Some moderate aerobic acidophilic methanotrophs that grow best at pH between ∼5 and 6 have been characterized that group with the α-Proteobacteria (Dedysh et al., 1998a,b, 2000, 2002, 2004, 2005; Dunfield et al., 2010). Given their ability to survive in acidic conditions, it is possible that these strains could be used to degrade various pollutants under a wider range of environmental conditions currently considered feasible. It should be noted, however, that many of these strains, particularly species within the Methylocella genera are only capable of expressing sMMO, and none have been examined for their ability to degrade pollutants. Further, it is interesting that many, but not all of these strains are also facultative. Of the facultative acidophilic strains, it is known that the Methylocella silvestris does not express sMMO in the presence of acetate (Theisen et al., 2005), while another facultative acidophilic methanotroph, Methylocapsa aureus, only expresses pMMO, but it is not known if pMMO is constitutively expressed (Dunfield et al., 2010). Clearly, more work is needed in this area if these microorganisms are to be utilized for pollutant degradation.
Although the finding of acidophilic methanotrophy provides interesting possibilities for pollutant degradation, the finding of thermoacidophilic methanotrophs within the Verrucomicrobia phylum dramatically expands the potential use of methanotrophs for pollutant degradation. These microorganisms, simultaneously described by three independent groups are capable of growth in environments with pH values of approximately 1 and temperatures greater than 50°C (Dunfield et al., 2007; Pol et al., 2007; Islam et al., 2008). These microorganisms utilize pMMO for methane oxidation (Hou et al., 2008; Semrau et al., 2008, 2010; Op den Camp et al., 2009), and have been recently shown to be able to utilize carbon dioxide as the sole carbon source via the Calvin–Benson–Bassham cycle (Khadem et al., 2011). As with the moderate acidophilic methanotrophs, there have been no reported data in the open literature that indicates that these thermoacidophilic methanotrophs can degrade various pollutants via pMMO activity, although such a possibility appears to be likely. Further, the finding that carbon dioxide is fixed by these microorganisms suggests that, like facultative methanotrophs, the use of carbon sources other than methane may enhance the utility of these microorganisms for pollutant degradation, although it should be noted that methane is used as the energy source by these microorganisms (Khadem et al., 2011).
Suggestions for Future Research
Recent findings have extended the possible utility of methanotrophic-mediated pollutant degradation to include thermally active and acidic environments as well in anaerobic regimes, particularly where nitrite levels may be high. Much more can and should be done, however, to fully maximize the use of methanotrophic activity for pollutant degradation, including:
• How can complex aerobic methanotrophic communities exposed to multiple pollutants be effectively used for in situ bioremediation? As described above, the form of MMO expressed has a significant effect on overall pollutant degradation. More effort should be put forward to characterize how methanotrophic communities respond to the presence of pollutants, both in terms of community structure (i.e., do specific groups of methanotrophs predominate?) and activity (i.e., which form of MMO is primarily expressed?).
• What is the extent and range of facultative methanotrophy? That is, how widespread are these microorganisms and can multi-carbon substrates such as acetate or ethanol be effectively added in situ to stimulate their activity? Can facultative methanotrophs effectively compete with heterotrophs for any added acetate or ethanol? Further, given that these microorganisms likely have multiple pathways for carbon assimilation, can any specific pollutant serve to help support growth through the production of intermediates such as glyoxylate?
• How can novel methanotrophic physiologies such as acidophilic, thermoacidophilic, and nitrite-utilizing methanotrophs be best used for pollutant degradation? How widespread are these types of methanotrophs? Can they be easily stimulated in situ? Much information is lacking here as to the range of compounds that these strains can bind via the sMMO or pMMO, as well as the rates and products of such degradation.
Answers to these issues will not only facilitate the use of methanotrophs for pollutant degradation, but also help address other interesting issues in methanotrophy, e.g., more complete knowledge on the diversity of methanotrophs and how these microorganisms help regulate elemental cycling in various environments.
Finally, although not explicitly considered in this review, it is possible that anaerobic methane oxidation coupled to sulfate or metal reduction could also be utilized for pollutant degradation, although it is likely that in these conditions, reductive dehalogenation would be the primary mechanism for pollutant removal. Such activity has been predominantly found in marine systems, but has also been shown to exist in terrestrial systems, specifically landfill leachate (Grossman et al., 2002). In such environments, pollutants such as chlorinated solvents can also be expected, and the possibility of anaerobic methane oxidation helping to remove various pollutants is intriguing. It should be stressed, however, that it is unknown what, if any, pollutant degradation ability such microorganisms have.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the Office of Science (BER), US Department of Energy.
References
Auman, A. J., Stolyar, S., Costello, A. M., and Lidstrom, M. E. (2000). Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl. Environ. Microbiol. 66, 5259–5266.
Beal, E. J., House, C. H., and Orphan, V. J. (2009). Manganese- and iron-dependent marine methane oxidation. Science 325, 184–187.
Belova, S. E., Baani, M., Suzina, N. E., Bodelier, P. L. E., Liesack, W., and Dedysh, S. N. (2011). Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ. Microbiol. Rep. 3, 36–46.
Boetius, A., Ravenschlag, K., Schubert, C., Rickert, D., Widdel, F., Gieseke, A., Amann, R., Jørgensen, B. B., Witte, U., and Pfannkuche, O. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 6236–6626.
Bosma, T., and Janssen, D. B. (1998). Conversion of chlorinated propanes by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl. Microbiol. Biotechnol. 50, 105–112.
Burrows, K. J., Cornish, A., Scott, D., and Higgins, I. J. (1984). Substrate specificities of the soluble and particulate methane mono-oxygenases of Methylosinus trichosporium OB3b. J. Gen. Microbiol. 130, 3327–3333.
Colby, J., Stirling, D. I., and Dalton, H. (1977). The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Biochem. J. 165, 395–402.
Dedysh, S. N., Berestovskaya, Y. Y., Vasylieva, L. V., Belova, S. E., Khmelenina, V. N., Suzina, N. E., Trotsenko, Y. A., Liesack, W., and Zavarzin, G. A. (2004). Methylocella tundrae sp. nov., a novel methanotrophic bacterium from acidic tundra peatlands. Int. J. Syst. Evol. Microbiol. 54, 151–156.
Dedysh, S. N., Khmelenina, V. N., Suzina, N. E., Trotsenko, Y. A., Semrau, J. D., Liesack, W., and Tiedje, J. M. (2002). Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 52, 251–261.
Dedysh, S. N., Knief, C., and Dunfield, P. F. (2005). Methylocella species are facultatively methanotrophic. J. Bacteriol. 187, 4665–4670.
Dedysh, S. N., Liesack, W., Khmelenina, V. N., Suzina, N. E., Trotsenko, Y. A., Semrau, J. D., Bares, A. M., Panikov, N. S., and Tiedje, J. M. (2000). Methylocella palustris gen nov., a new methane-oxidizing acidophilic bacterium from pat bogs, representing a novel subtype of serine pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50, 955–969.
Dedysh, S. N., Panikov, N. S., and Tiedje, J. M. (1998a). Acidophilic methanotrophic communities from Sphagnum peat bogs. Appl. Environ. Microbiol. 64, 922–929.
Dedysh, S. N., Panikov, N. S., Liesack, W., Großkopf, R., Zhou, J., and Tiedje, J. M. (1998b). Isolation of acidophilic methane-oxidizing bacteria from northern peat wetlands. Science 282, 281–284.
DiSpirito, A. A., Gulledge, J., Shiemke, A. K., Murrell, J. C., Lidstrom, M. E., and Krema, C. L. (1992). Trichloroethylene oxidation by the membrane-associated methane monooxygenase in Type I, type II, and type X methanotrophs. Biodegradation 2, 151–164.
Dunfield, P. F., Belova, S. E., Vorob’ev, A. V.Cornish, S. L., and Dedysh, S. N. (2010). Methylocapsa aurea sp. nov., a facultative methanotroph possessing a particulate methane monooxygenase and emended description of the genus Methylocapsa. Int. J. Syst. Evol. Microbiol. 60, 2659–2664.
Dunfield, P. F., Yuryev, A., Senin, P., Smirnova, A. V., Stott, M. B., Hou, S., Ly, B., Saw, J. H., Zhou, A., Ren, Y., Wang, J., Mountain, B. W., Crowe, M. A., Weatherby, T. M., Bodelier, P. L. E., Liesack, W., Feng, L., and Alam, M. (2007). Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450, 879–883.
Ettwig, K. F., Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M. M., Schreiber, F., Dutilh, B. E., Zedelius, J., de Beer, D., Gloerich, J., Wessels, H. J. C. T., van Alen, T., Luesken, F., Wu, M. L., can de Pas-Schoonen, K. T., Op den Camp, H. J. M., Janssen-Megens, E. M., Francoijs, K.-J., Stunnenberg, H., Weissenbach, J., Jetten, M. S. M., and Strous, M. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548.
Fox, B. G., Borneman, J. G., Wackett, L. P., and Lipscomb, J. D. (1990). Haloalkene oxidation by the soluble methane monooxygenase from Methylosinus trichosporium OB3b: mechanistic and environmental implications. Biochemistry 29, 6419–6427.
Grosse, S., Laramee, L., Wendlandt, K.-D., McDonald, I. R., Miguez, C. B., and Kleber, H.-P. (1999). Purification and characterization of the soluble methane monooxygenase of the type II methanotrophic bacterium Methylocystis sp. Strain WI 14. Appl. Environ. Microbiol. 65, 3929–3935.
Grossman, E. L., Cifuentes, L. A., and Cozzarelli, I. M. (2002). Anaerobic methane oxidation in a landfill-leachate plume. Environ. Sci. Technol. 36, 2436–2442.
Han, J.-I., Lontoh, S., and Semrau, J. D. (1999). Degradation of chlorinated and brominated hydrocarbons by Methylomicrobium album BG8. Arch. Microbiol. 172, 393–400.
Han, J.-I., and Semrau, J. D. (2000). Chloromethane stimulates growth of Methylomicrobium album BG8 on methanol. FEMS Microbiol. Lett. 187, 77–81.
Hou, C. T., Patel, R., Laskin, A. I., and Barnabe, N. (1979). Microbial oxidation of gaseous hydrocarbons: epoxidation of C2 to C4 n-alkenes by methylotrophic bacteria. Appl. Environ. Microbiol. 38, 127–134.
Hou, S., Makarova, K. S., Saw, J. H. W., Senin, P., Ly, B. V., Zhou, Z., Ren, Y., Wang, J., Galperin, M. Y., Omelchenko, M. V., Wolf, Y. I., Yutin, N., Koonin, E. V., Stott, M. B., Mountain, B. W., Crowe, M. A., Smirnova, A. V., Dunfield, P. F., Feng, L., Wang, L., and Alam, W. (2008). Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol. Direct 3, 26.
Iguchi, H., Yurimoto, H., and Sakai, Y. (2010). Soluble and particulate methane monooxygenase gene clusters of the type I methanotroph Methylovulum miyakonense HT12. FEMS Microbiol. Lett. 312, 71–76.
Im, J., Lee, S.-W., Yoon, S., DiSpirito, A. A., and Semrau, J. D. (2011). Characterization of a novel facultative Methylocystis species capable of growth on methane, ethanol, and acetate. Environ. Microbiol. Rep. 3, 174–181.
Im, J., and Semrau, J. D. (2011). Pollutant degradation by Methylocystis strain SB2 grown on ethanol: bioremediation via facultative methanotrophy. FEMS Microbiol. Lett. 318, 137–142.
Islam, T., Jensen, S., Reigstad, L. J., Larsen, Ø., and Birkeland, N.-K. (2008). Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. U.S.A. 105, 300–304.
Jechorek, M., Wendlandt, K.-D., and Beck, M. (2003). Cometabolic degradation of chlorinated aromatic compounds. J. Biotechnol. 102, 93–98.
Khadem, A. F., Pol, A., Wieczorek, A., Mohammadi, S. S., Francoijs, K.-J., Stunnenberg, H. G., Jetten, M. S. M., and Op den Camp, H. J. M. (2011). Autotrophic methanotrophy in Verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J. Bacteriol. 193, 4438–4446.
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334.
Knittel, K., Lösekann, T., Boetius, A., Kort, R., and Amann, R. (2005). Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71, 467–479.
Koh, S.-C., Bowman, J. P., and Sayler, G. S. (1993). Soluble methane monooxygenase production and trichloroethylene degradation by a type I methanotroph, Methylomonas methanica 68-1. Appl. Environ. Microbiol. 59, 960–967.
Lee, S.-W., Keeney, D. R., Lim, D.-H., DiSpirito, A. A., and Semrau, J. D. (2006). Mixed pollutant degradation by Methylosinus trichosporium OB3b expressing either soluble or particulate methane monooxygenase: can the tortoise beat the hare? Appl. Environ. Microbiol. 72, 7503–7509.
Lindner, A. S., Adriaens, P., and Semrau, J. D. (2000). Transformation of ortho-subtituted biphenyls by Methylosinus trichosporium OB3b: substituent effects on oxidation kinetics and product formation. Arch. Microbiol. 174, 35–41.
Lontoh, S., and Semrau, J. D. (1998) Methane and trichloroethylene oxidation by the particulate methane monooxygenase of methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 64, 1106–1114.
Lontoh, S., Zahn, J. A., DiSpirito, A. A., and Semrau, J. D. (2000). Identification of intermediates of in vivo trichloroethylene oxidation by the membrane-associated methane monooxygenase. FEMS Microbiol. Lett. 186, 109–113.
Michaelis, W., Seifert, R., Nauhaus, K., Treude, T., Thiel, V., Blumenberg, M., Knittel, K., Gieseke, A., Peterknecht, K., Pape, T., Boetius, A., Amann, R., Jørgensen, B. B., Widdel, F., Peckmann, J., Pimenov, N. V., and Gulin, M. B. (2002). Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297, 1013–1015.
Oldenhuis, R., Vink, R. L. J. M., Janssen, D. B., and Witholt, B. (1989). Degradation of chlorinated aliphatic hydrocarbons by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl. Environ. Microbiol. 55, 2819–2826.
Op den Camp, H. J. M., Islam, T., Stott, M. B., Harhangi, H. R., Hynes, A., Schouten, S., Jetten, M. S. M., Birkeland, N.-K., Pol, A., and Dunfield, P. F. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 5, 293–306.
Pfiffner, S. M., Palumbo, A. V., Phelps, T. J., and Hazen, T. C. (1997). Effects of nutrient dosing on subsurface methanotrophic populations and trichloroethylene degradation. J. Ind. Microbiol. Biotechnol. 18, 204–212.
Pol, A., Heijmans, K., Harhangi, H. R., Tedesco, D., Jetten, M. S. M., and Op den Camp, H. J. M. (2007). Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450, 874–878.
Semprini, L., Roberts, P. V., Hopkins, G. D., and McCarty, P. L. (1990). A field evaluation of in-situ biodegradation of chlorinated ethenes: part 2, results of biostimulation and biotransformation experiments. Ground Water 28, 715–727.
Semrau, J. D., DiSpirito, A. A., and Murrell, J. C. (2008). Life in the extreme: thermoacidophilic methanotrophy. Trends Microbiol. 16, 190–193.
Semrau, J. D., DiSpirito, A. A., and Vuilleumier, S. (2011). Facultative methanotrophy: false leads, true results, and suggestions for future research. FEMS Microbiol. Lett. 323, 1–12.
Semrau, J. D., DiSpirito, A. A., and Yoon, S. (2010). Methanotrophs and copper. FEMS Microbiol. Rev. 34, 496–531.
Shen, R.-N., Yu, C.-L., Ma, Q.-Q., and Li, S.-B. (1997). Direct evidence for a soluble methane monooxygenase from a type I methanotrophic bacteria: purification and properties of a soluble methane monooxygenase from Methylomonas sp. GYJ3. Arch. Biochem. Biophys. 345, 223–229.
Shigematsu, T., Hanada, S., Eguchi, M., Kamagata, Y., Kanagawa, T., and Kurane, R. (1999). Soluble methane monooxygenase gene clusters from trichloroethylene-degrading Methylomonas sp. strains and detection of methanotrophs during in situ bioremediation. Appl. Environ. Microbiol. 65, 5198–5206.
Smith, K. S., Costello, A. M., and Lidstrom, M. E. (1997). Methane and trichloroethylene oxidation by an estuarine methanotroph, Methylobacter sp. strain BB5.1. Appl. Environ. Microbiol. 63, 4617–4620.
Smith, T. J., and Dalton, H. (2004). Biocatalysis by methane monooxygenase and its implications for the petroleum industry. Stud. Surf. Sci. Catal. 151, 177–192.
Sullivan, J. P., and Chase, H. A. (1996). 1,2,3-Trichlorobenzene transformation by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl. Microbiol. Biotechnol. 45, 427–433.
Theisen, A. R., Ali, M. H., Radajewski, S., Dumont, M. G., Dunfield, P. F., McDonald, I. R., Dedysh, S. N., Miguez, C. B., and Murrell, J. C. (2005). Regulation of methane oxidation in the facultative methanotroph Methylocella silvestris BL2. Mol. Microbiol. 58, 682–692.
Trotsenko, Y. A., and Murrell, J. C. (2008). Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 63, 183–229.
van Hylckama Vleig, J. E. T., de Koning, W., and Janssen, D. B. (1996). Transformation kinetics of chlorinated ethenes by Methylosinus trichosporium OB3b and detection of unstable epoxides by on-line gas chromatography. Appl. Environ. Microbiol. 62, 3304–3312.
Vorobev, A. V., Baani, M., Doronina, N. V., Brady, A. L., Liesack, W., Dunfield, P. F., and Dedysh, S. N. (2011). Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium possessing only a soluble methane monooxygenase. Int. J. Syst. Evol. Microbiol. doi: 10.1099/ijs.0.028118-0
Yoon, S., Im, J., Bandow, N., DiSpirito, A. A., and Semrau, J. D. (2011). Constitutive expression of pMMO by Methylocystis strain SB2 when grown on multi-carbon substrates: implications for biodegradation of chlorinated ethenes. Environ. Microbiol. Rep. 3, 182–188.
Keywords: methanotrophy, bioremediation, facultative methanotrophy
Citation: Semrau JD (2011) Bioremediation via methanotrophy: overview of recent findings and suggestions for future research. Front. Microbio. 2:209. doi: 10.3389/fmicb.2011.00209
Received: 11 August 2011;
Accepted: 24 September 2011;
Published online: 12 October 2011.
Edited by:
Qiaoyun Huang, Huazhong Agricultural University, ChinaReviewed by:
Wensheng Lan, Shenzhen Entry-Exit Inspection and Quarantine Bureau, ChinaXu Han, Tianjin Polytechnic University, China
Copyright: © 2011 Semrau. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Jeremy D. Semrau, Department of Civil and Environmental Engineering, The University of Michigan, 1351 Beal Avenue, Ann Arbor, MI 48109-2125, USA. e-mail: jsemrau@umich.edu