- 1 Marine Chemistry and Geochemistry Department, Woods Hole Oceanographic Institution, Woods Hole, MA, USA
- 2 Biology Department, Woods Hole Oceanographic Institution, Woods Hole, MA, USA
- 3 Monterey Bay Aquarium Research Institute, Moss Landing, CA, USA
Improvements in temporal and spatial sampling frequency have the potential to open new windows into the understanding of marine microbial dynamics. In recent years, efforts have been made to allow automated samplers to collect microbial biomass for DNA/RNA analyses from moored observatories and autonomous underwater vehicles. Measurements of microbial proteins are also of significant interest given their biogeochemical importance as enzymes that catalyze reactions and transporters that interface with the environment. We examined the influence of five preservatives solutions (SDS-extraction buffer, ethanol, trichloroacetic acid, B-PER, and RNAlater) on the proteome integrity of the marine cyanobacterium Synechococcus WH8102 after 4 weeks of storage at room temperature. Four approaches were used to assess degradation: total protein recovery, band integrity on an SDS detergent polyacrylamide electrophoresis (SDS-PAGE) gel, and number of protein identifications and relative abundances by 1-dimensional LC–MS/MS proteomic analyses. Total protein recoveries from the preserved samples were lower than the frozen control due to processing losses, which could be corrected for with internal standardization. The trichloroacetic acid preserved sample showed significant loss of protein band integrity on the SDS-PAGE gel. The RNAlater preserved sample showed the highest number of protein identifications (103% relative to the control; 520 ± 31 identifications in RNAlater versus 504 ± 4 in the control), equivalent to the frozen control. Relative abundances of individual proteins in the RNAlater treatment were quite similar to that of the frozen control (average ratio of 1.01 ± 0.27 for the 50 most abundant proteins), while the SDS-extraction buffer, ethanol, and B-PER all showed significant decreases in both number of identifications and relative abundances of individual proteins. Based on these findings, RNAlater was an effective proteome preservative, although further study is warranted on additional marine microbes.
Introduction
It is anticipated that higher spatial and temporal sampling of the oceans provided by deployment of a combination of in situ sensors and autonomous sample collectors will greatly improve our understanding of marine processes. This is likely to be particularly true for coupled microbiological and chemical processes that scale from genomic potential to global biogeochemical impacts on virtually all biologically utilized elements (Morel and Price, 2003; Falkowski et al., 2008; Saito et al., 2008). Multiple large scale programs are underway that aim to incorporate microbiological and/or biogeochemical observations in high temporal or spatial resolution, including the Ocean Observatories Initiative (OOI1) and the GEOTRACES trace element and isotope global survey section program2. The development of autonomous samplers and their deployment on moorings and underwater vehicles offers these increases in sampling resolution over the duration of deployment (Bell et al., 2002; Greenfield et al., 2006; Paul et al., 2007; Breier et al., 2009; Scholin et al., 2009). In addition, autonomous sample collection may be valuable during ocean section survey cruises, where the ship’s wire-time for sampling equipment is the limiting operational resource, and introduction of autonomous sampling systems could greatly increase biological and biochemical sample collection capabilities. Yet a major concern with automated sample collection is the potential for sample degradation during storage until instrument recovery and analysis. Standard laboratory and field sampling approaches for DNA, RNA, and protein storage involve filtration to concentrate biomass and immediate freezing in liquid nitrogen. While use of preservatives can be incorporated into current sampling platforms being developed, freezing in situ is likely beyond the power, space, and design criteria that are desirable for environmental microbial samplers. The ability of preservatives that maintain sample integrity without freezing over both the long durations associated with mooring or vehicle deployments, or the short-term station-time during survey cruises is an important design criterion. While preservation of RNA molecules has recently been successfully demonstrated on the Environmental Sample Processor (an autonomous sampler and analyzer) deployed in a coastal environment for laboratory-based metatranscriptomic analysis (Ottesen et al., 2011), marine protein preservation has been much less studied in this context.
Marine proteomics is a relatively new technique that has significant potential to contribute to the understanding of microbial biogeochemistry. Four potential applications include: (1) the direct quantitative measurement of enzymes responsible for the catalysis of biogeochemical reactions and their incorporation of this data within global ecosystem–circulation models (Saito et al., 2011), (2) the measurement of transporters and biomarkers for assessment of nutrient limitation status of key phytoplankton and bacterial communities, (3) the characterization of the community diversity and functional gene expression, and (4) the use of proteomic mass spectral data to assist in genome annotation, an application known as proteogenomics (Ansong et al., 2008). Mass spectrometry-based proteomics methods have recently been applied to important marine microbes such as the cyanobacteria Crocosphaera watsonii and Synechococcus, and the heterotrophic bacterium Pelagibacter ubique (Gonzales et al., 2005; Barrios-Llerena et al., 2006; Sowell et al., 2008a; Saito et al., 2011). In addition, community proteome analyses have begun to be applied to the natural environments such as acid-mine drainage microbial communities, and open-ocean, and coastal marine water columns environments (Ram et al., 2005; Sowell et al., 2008b; VerBerkmoes et al., 2009; Morris et al., 2010). One important emerging capability of proteomics is the ability for absolute quantification of target proteins (Wolf-Yadlin et al., 2007; Lange et al., 2008). Using isotopically labeled peptide standards, it is now possible to measure very low quantities of proteins on an absolute scale. This has significant potential for application to oceanographic biogeochemical studies where concentrations of key biogeochemical enzymes can be quantified and using their measured ranges of activities from laboratory studies, estimates of potential in situ biogeochemical reaction rates could then be calculated for environmental samples (Bertrand et al., 2011; Saito et al., 2011). Critical to the development of this capability is confidence in protein sample collection and preservation.
In this study we examined the influence of a suite of preservatives on protein integrity, as observed by total protein recovery, 1-dimensional (1-D) SDS polyacrylamide gel qualitative comparisons, and mass spectrometry-based protein identifications and relative abundances. A culture of marine cyanobacterium Synechococcus WH8102 was used for model proteinaceous material because of the ubiquity and abundance of this and other marine cyanobacteria in the oceans (Waterbury et al., 1986; Partensky et al., 1999; Saito et al., 2005).
Materials and Methods
Culturing
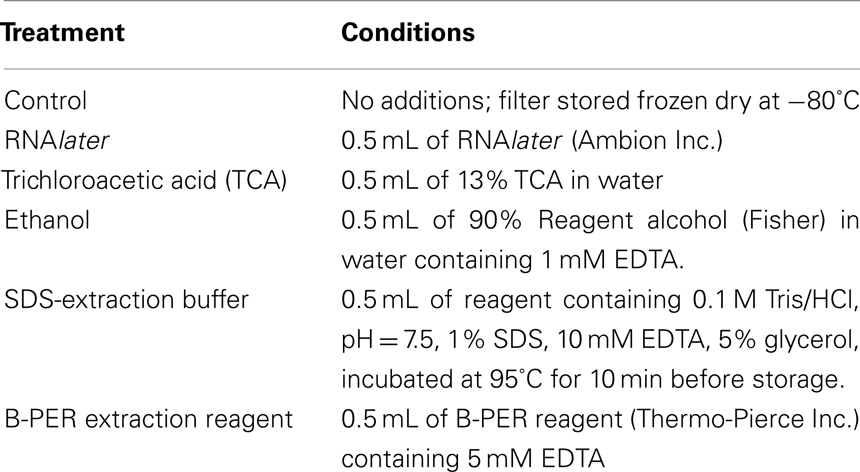
One liter of marine Synechococcus WH8102 culture was grown in SN media at 23°C to a final cell yield of 1.9 × 107 cells mL−1, as determined by epifluorescence microscopy. Six 150 mL aliquots were filtered onto 25 mm 0.22 μm Durapore membrane (hydrophilic polyvinylidene fluoride, Millipore) filters by gentle vacuum filtration. A control filter was frozen immediately without a preservative solution at −80°C, while five preservative solutions, RNAlater (Ambion Inc.), trichloroacetic acid (TCA), ethanol with 5 mM ethylenediamine tetraacetic acid (EDTA; preservative described as ethanol hereon), B-PER bacterial extraction reagent (Thermo-Pierce Inc.), and an SDS-based extraction buffer were applied to the remaining filters with specific recipes listed in Table 1. While the specific recipe of the commercial reagents is not published, RNAlater contains a metal chelator and high salt content, while B-PER contains a mild non-ionic detergent in 20 mM Tris HCl, pH 7.53. The filters were folded twice with the biomass facing inward, completely covered in each preservative solution in 1.5 mL microcentrifuge tubes, and incubated for 4 weeks in darkness at room temperature (∼20°C).
Extraction
Proteins were extracted from the sample filters with some tailoring of protocols to remove incompatible reagents such as solvents and salts found in the preservatives. The protocols share a detergent-based extraction and solubilization procedure with either an SDS-extraction buffer (0.1 M Tris/HCL pH 7.5, 5% glycerol, 10 mM EDTA, and 1% SDS detergent with heating at 95°C for 15 min) or the detergent already present within the preservative (B-PER), followed by centrifugation and protein precipitation with 50:50 acetone/methanol in the supernatant (with a 4:1 ratio of the acetone/methanol solution to the supernatant). Specific protocols are described by preservative type below through the acetone/methanol precipitation step, after which point all sample processing followed the same protocol.
The control sample was stored at −80°C for the 4 weeks, then resuspended in 0.5 mL of the SDS-extraction buffer, mixed well with a spatula on ice and left for 10 min on ice. The sample was then heated at 95°C for 15 min, incubated for 1 h at room temperature (RT hereon) with 350 rpm mixing using a benchtop Thermomixer (Eppendorf), vortexed, the supernatant removed from the filter and centrifuged at 14,000 × g for 20 min. The supernatant was passed through a 5 μm filter needle (Becton Dickinson Inc., filter needle hereon), and precipitated with cold 50:50 acetone/methanol solution at −20°C overnight.
In the ethanol–EDTA preserved sample, the ethanol was first evaporated by speed vacuum until dry. SDS-extraction buffer was added and allowed to incubate for 10 min prior to 15 min incubation at 95°C, and 1 h at RT with 350 rpm mixing. The sample was gently vortexed, the supernatant removed from the filter, centrifuged for 5 min at 14,000 × g, and precipitated with a 50:50 acetone/methanol solution overnight at −20°C.
The TCA preserved sample was gently mixed with a spatula, the filter pressed to the bottom of the vial and centrifuged for 30 min at 14,000 × g. The supernatant was removed and 1.4 mL of ice-cold 100% ethanol was added to the filter/pellet, mixed, and incubated for 3 h at −20°C, centrifuged for 30 min at 14,000 × g at 4°C, after which the supernatant was carefully removed. Another 1.4 mL of ethanol was added and incubated overnight at −20°C, followed by centrifugation for 30 min at 4°C, removal of 1 mL of ethanol, and concentration by speed vacuum on low heat setting until the remaining ethanol evaporated. 0.5 mL of SDS-extraction buffer was added to the filter/pellet and incubated with the sample for 10 min at RT, followed by 15 min at 95°C. After cooling, the sample was incubated for 1 h at room temperature. The sample was then separated from the filter by decanting with a filter needle syringe, and pH adjusted with 0.4 M Tris/HCl (pH 7.8) to a final pH of ∼7.5. The sample was then acetone/methanol precipitated overnight at −20°C.
The SDS-extraction buffer sample was incubated at 95°C for 10 min prior to the month-long incubation treatment, similar to an approach that might be used during in situ sample collection on the Environmental Sample Processor device, which is capable of in situ lysis (Greenfield et al., 2006). After storage, the sample was mixed, separated from the filter, and centrifuged for 20 min at 14,000 × g at RT. Supernatants were passed through a filter needle, split into two aliquots and precipitated with a 50:50 acetone/methanol solution, and incubated overnight at −20°C.
After the 4 weeks of storage, the B-PER preserved sample was mixed then centrifuged for 20 min at 14,000 × g at RT. Supernatants were collected with a filter needle syringe, split into two aliquots and precipitated with four volumes of a 50:50 acetone/methanol solution, and incubated overnight at −20°C.
The RNAlater sample was extracted by adding two volumes (1 mL) of the SDS-extraction buffer and mixing for 10 min at 350 rpm at RT. The sample was then heated to 95°C for 15 min, cooled, and mixed at 350 rpm for 1 h at room temperature. Three milliliters of Tris–EDTA buffer (100 mM Tris–HCl, 10 mM EDTA, pH 7.5, TE buffer hereon) were added for a total volume of 4.5 mL. The sample was mixed and passed through a filter needle. The syringe and sample filter were rinsed with an additional 0.5 mL of TE buffer to remove remaining sample from the filter (final volume ∼5 mL). A 5 kDa MWCO centrifuge concentrator (VivaSpin 6, Sartorius Stedim Inc.) was used to concentrate the sample at 6,000 × g for 2 h, to a final volume of 0.6 mL. The sample was acetone/methanol precipitated overnight at −20°C, resulting in a relatively large pellet due to salt carryover from the RNAlater. The sample was centrifuged at 14,000 × g at 4°C for 30 min, aspirated, and concentrated by speed vacuum for 10 min. To remove excess salt from the RNAlater preservative the following additional dilution and concentration steps were conducted: 1 mL of extraction buffer was added, and the sample was sonicated for 30 s using a microtip on constant setting (Branson Inc.). Four milliliters of extraction buffer were added and mixed by vortex, and the sample was concentrated by 5 kDa MWCO centrifuge concentrators at 6,000 × g at RT. Another 4 mL of extraction buffer was added and the sample concentrated again as described above to a volume of ∼0.6 mL, followed by acetone/methanol precipitation overnight at −20°C. The resulting precipitation was free of the excess salt observed earlier.
Using the samples extracts described for the control and each preservative described above, all acetone/methanol precipitated samples were centrifuged at 14,000 × g for 30 min at 4°C. Samples were aspirated and evaporated for 10 min by speed vacuum, and resuspended in SDS-extraction buffer. Samples were incubated with the buffer at room temperature for 30 min without mixing, followed by resuspension by pipetting.
Total protein was measured by DC-assay (BioRad) using bovine serum albumin for calibration as a protein standard (linear regression r2 value of 0.994) and diluting the sample 1:5 into SDS-extraction buffer, with Milli-Q water used as a blank solution. Each sample was analyzed in quadruplicate, and error bars represent the SD of the quadruplicate technical replicates. A 1-D SDS detergent polyacrylamide electrophoresis gel (SDS-PAGE) with a 4–20% gradient (BioRad) was loaded with 60 μg of protein loaded per well and a protein standard ladder (10–220kDa range Benchmark, Invitrogen Inc.).
Samples were suspended within a small tube gel for alkylation, reduction, trypsin digestion, and detergent removal. A total of 115 μg of the protein extracted prior was suspended within each tube gel prepared in microcentrifuge tube, as described by Lu et al. (Lu and Zhu, 2006) with some minor modifications. Briefly, samples were immobilized in 15% acrylamide in pH 7.5 Tris buffer, incubated twice with 10% acetic acid and 50% ethanol for 20 min and 1 h, then with 10% acetic acid and 50% methanol for 2 h at room temperature and mixing at 350 rpm, decanting between. Gel samples were cut into ∼1 mm3 pieces and incubated with 50% acetonitrile 50% 25 mM ammonium bicarbonate solution (pH 8.0) for 1 h and again overnight, shaking at 350 rpm at 16°C and decanting between. Proteins immobilized within the gel were reduced with 10 mM dithiothreitol (DTT) at 56°C for 1 h, decanted, and alkylated with 30 mM iodoacetamide for 1 h at room temperature in the dark, washed in 25 mM ammonium bicarbonate for 20 min, dehydrated twice with 100% acetonitrile for 10 min, and dried for 20 min by speed vacuum. Gel pieces were rehydrated and digested with trypsin in 25 mM ammonium bicarbonate for 16 h at 37°C (1:30 ratio trypsin to total protein, Promega Gold Mass Spectrometry Grade, Promega Inc., Madison, WI, USA). The peptides were extracted by three successive additions of 50% acetonitrile (Fisher Optima) with 5% formic acid (Michrom Biosciences, Ultra Pure). The extracted peptides were combined and concentrated by speed vacuum to less than 5 μL, diluted with 2% acetonitrile and 0.1% formic acid in water (Fisher Optima), sonicated for 10 min in a sonicator bath, and stored at −80°C until analysis.
Proteomic Analysis by LC–MS
Each sample was analyzed in technical duplicate using a Thermo LTQ mass spectrometer (Thermo Scientific Inc., San Jose, CA, USA), an advance electrospray source (Michrom Inc.), and a Michrom Paradigm MS4 HPLC (Michrom Inc.). The HPLC was outfitted with a C18 Cap Trap in-line for additional salt removal with a reversed phase Magic C18 AQ column (0.2 mm × 150 mm, 3 μm particle size, 200 Å pore size, Michrom Inc. Auburn, CA, USA). The chromatography consisted of a hyperbolic gradient from 5% buffer A to 95% buffer B for 300 min, where A was 0.1% formic acid (Michrom, Ultra Pure) in LC–MS grade water (Fisher Optima) and B was 0.1% formic acid in acetonitrile (Fisher Optima) at a flow rate of 2 μL min−1. Four micrograms of total protein was diluted into buffer A for each LC–MS/MS injection. The mass spectrometer was set to perform MS/MS on the top seven ions using data-dependent settings and a dynamic exclusion window of 30 s. Ions were monitored over the range of 400–2000 m/z.
Proteomic Data Analysis
Mass spectra were searched with SEQUEST in Bioworks 3.3 (Thermo Inc.) for protein identifications using a forward database and reverse database of the genome of Synechococcus WH8102 (Palenik et al., 2003). SEQUEST parameters were set at 30% ions required per peptide, ΔCN of 0.1, Xcorr versus CS 1.9, 2.4, 2.9, and 1e-3 protein probability. Database search results were further processed using the PeptideProphet statistical model (Keller et al., 2002) within Scaffold 3.0 (Proteome Software Inc., Portland OR) operating on a 64 bit Ubuntu Linux workstation, using a 99.9% protein probability, and 95% peptide probability, resulting in a peptide false discovery rate (FDR) of 0.6% (Peng et al., 2003). Five hundred twenty proteins were identified in this study accounting for 20% of the 2,528 protein coding genes within the genome (of 2,588 total genes4). A significantly higher numbers of protein identifications were acquired for this cyanobacterium using multi-dimensional chromatography and will be combined with a biological and biochemical interpretation elsewhere (Saito et al., in preparation). For the purposes of this degradation study, highly robust, and repeatable protein identification numbers and spectral counts (SpC) were found with 1-D chromatography (Saito et al., 2011), and hence this approach was more useful for our immediate the needs of this study than the deeper proteome depth allowed by multi-dimensional chromatography (strong cation exchange/reverse phase), which we have found can be more variable in both number of protein identifications and spectral counts for technical replicates. Relative protein abundances were calculated using spectral counting, where counts were normalized across samples in each experiment (including technical replicates), to allow comparison of relative protein abundance across all preservative treatments.
Results and Discussion
Five preservative solutions were applied to storage of replicate filters of marine Synechococcus strain WH8102 as described in Table 1. The survival of proteins after 1 month at room temperature in darkness was highly dependent on the type of preservative used. Four techniques were used to examine protein recovery after extraction: total protein concentration, a qualitative examination of band integrity on 1-D-SDS-PAGE gel, total number of proteins identified by LC–MS, and ratios of relative abundance of each individual protein between each preservative and the control as determined by spectral counting.
Total Protein Recoveries
Total protein recovery was quite variable between preservation approaches (Figure 1), with the highest recovery observed in the immediately frozen control, followed by the SDS-extraction sample, ethanol, RNAlater, TCA, B-PER. However, total protein recovery is likely not the most useful metric of protein preservation because losses were likely due to sample processing recovery rather than actual degradation during storage. In particular, each preservative had differing compatibility with downstream extraction protocols, and additional handling steps could result in additional losses of total protein. While these processing losses would be important when trying to measure absolute abundances of proteins, they likely can be easily accounted for by addition of internal standards such as isotopically labeled or exogenous proteins. In the future, extraction protocols could also be optimized to minimize losses and increase throughput.
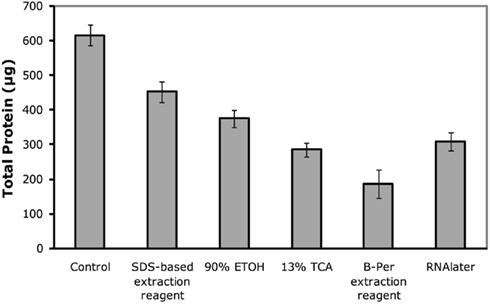
Figure 1. Total protein recovery as measured by the DC-assay on extracted filters. Control refers to the immediately frozen control and preservative details are listed in Table 1. Error bars correspond to quadruplicate technical replicates.
Protein Integrity by SDS-PAGE
The 1-D-SDS-PAGE gel provided qualitative information about sample integrity (Figure 2). Despite loading equivalent total protein in each gel lane, there was variability in the intensity of protein bands across the lanes. On a qualitative level, all samples with the exception of the TCA sample, had distinct protein bands suggesting good overall protein integrity by this assay. The TCA sample showed a broad smear across the entire lane, indicative of significant protein hydrolysis and degradation. With the total protein concentration measurements observed to be relatively precise (see error bars in Figure 1), the likely explanation for the difference in band intensity between lanes is protein degradation. This is evident in the B-PER lane, where this preservative was observed to have the smallest number of protein identifications (Figure 3, see Protein Identifications by LC–MS), and this preservative also showed a large enrichment in some of the most abundant proteins (phycoerythrin, swmA, and a porin; see Table 2 and see Relative Protein Abundances by LC–MS), both of which are consistent with the loss of the minor proteins relative to the major protein bands in the gel image.
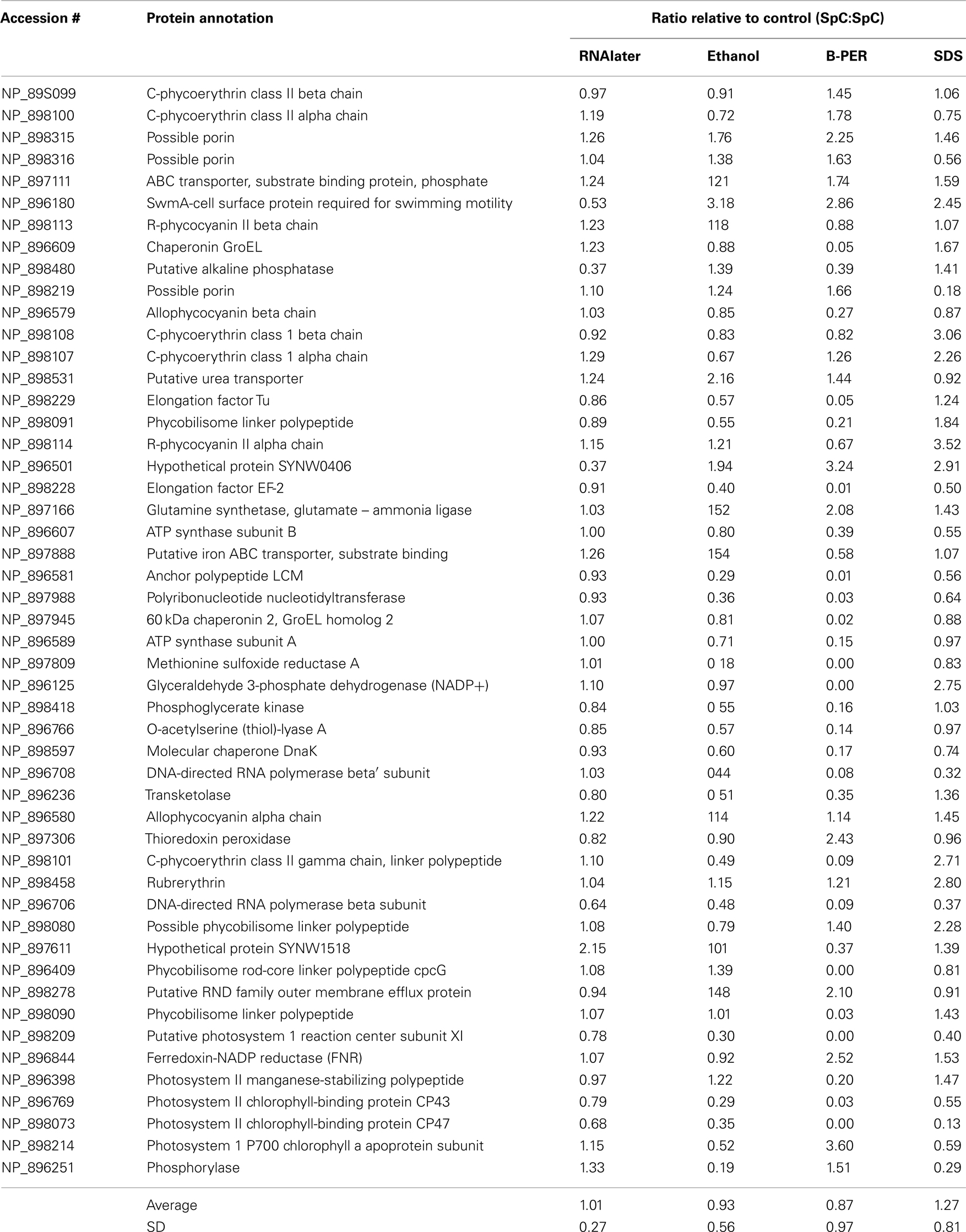
Table 2. Changes in relative abundance of 50 most abundant proteins during preservation (linear scale).
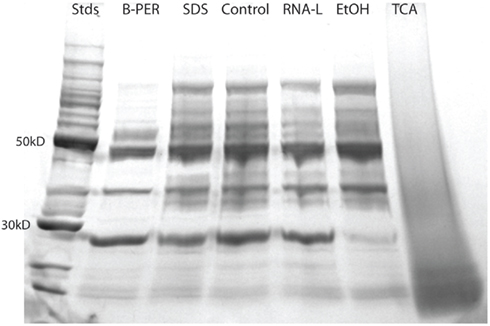
Figure 2. A 1-D SDS-PAGE gel of the control and preservation samples. The gel was a 4–20% gradient with 60 μg of protein loaded in each sample lane. Standards refers to the 10–220 kDa protein ladder, with intense 50 and 30 kDa bands.
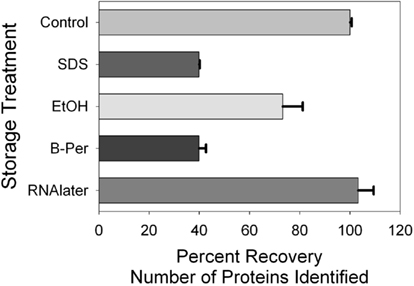
Figure 3. Comparison of number of protein identifications acquired from the control and four preservation techniques. Each sample was analyzed twice by LC–MS and the values reflect the average and SD, normalized to the control treatment (504 ± 4 protein identifications in each technical replicate of the control sample, the control = 100%).
Protein Identifications by LC–MS
The mass spectrometry-based proteomic analyses provided the most realistic assessment of sample preservation success (Figures 3–5), since they replicated the desired proteomic analyses from field samples. Technical replicate LC–MS injections were conducted for the control and each preservation sample, with the exception of the degraded TCA sample. Interestingly, the largest number of proteins identified was from the RNAlater preserved sample, yielding approximately 103% of the proteins relative to those identified in the control (Figure 3, frozen immediately without buffer/preservative; 520 ± 31 in RNAlater versus 504 ± 4 in the control). This surprising result implies that this preservative is as good as or perhaps more successful than freezing dry, despite storage at room temperature. This approach of using a salt/metal chelator preservative has been successfully applied to freezing of marine cyanobacterium for transcriptomic analysis (Zinser et al., 2009). Our findings suggest that freezing after preserving with RNAlater would be an optimal laboratory preservation technique in culture experiments of marine cyanobacterium as well, although the increased desalting effort required makes this preservative more labor intensive to work with. Other preservatives yielded fewer identifications: with ethanol yielding 369 protein identification (73 ± 8% of the control) and B-PER and SDS-Extraction buffer both yielding 201 unique protein identification (40 ± 3% and 40 ± 0.3%, respectively), suggesting that despite the clear bands seen in the 1-D gel (Figure 1), degradation is nonetheless likely occurring in those samples among the less abundant proteins.
Relative Protein Abundances by LC–MS
Comparison of the relative abundances of specific proteins allows an assessment of the extent of protein degradation on specific abundant and moderately abundant proteins. For this approach we employed spectral counting that involves a normalized count of all mass spectra associated with the peptides that correspond to each protein. For this study data was collected from the 1-D LC–MS/MS global proteome used for protein identifications described above. This 1-D approach has the advantage of minimizing variability and maximizing precision of replicate analyses to allow quantitative comparisons between preservation treatments. To demonstrate the precision of this approach, Figure 4A shows technical replicate injections for all five samples, with replicates on opposing axes. Coherence to the 1:1 line for each protein using the normalized spectral counting approach demonstrates reproducibility in this measurement. While spectral counts have been weighted within each injection and across all samples, if the overall number of peptide-associated spectra is varying due to degradation, some overall systematic shift in protein abundance may be observed. Also to demonstrate reproducibility of this approach, SDs of technical duplicate spectral counts are shown with the corresponding mean spectral counts for each of the 200 most abundant proteins (Figure 4B), where the relative SD is less than 26% for the top 100 proteins.
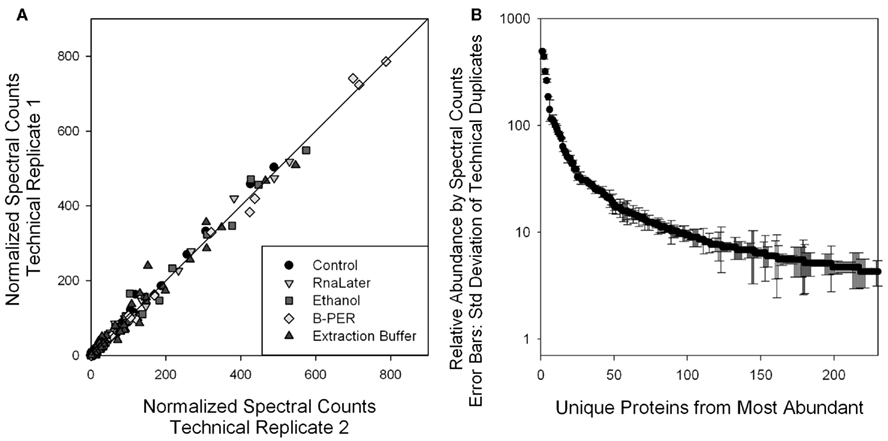
Figure 4. (A) Comparison of technical replicate injections of each of the four preservative samples and the control. Precision of the more abundant proteins using this 1-D LC/MS spectral counting relative quantitation approach was evident in the coherence with the 1:1 line. This reproducibility of protein spectral counts scores enabled the comparison of relative abundances of individual proteins with different preservation techniques used in Figure 5. (B) Average and SD of the technical duplicate normalized spectral counts from the control sample. Relative SD is 26% or less for the 100 highest abundance proteins, as determined by spectral counts. This error is smaller than the several fold variation observed in degradation studies shown in Figure 5.
We observed significant degradation of individual proteins, as measured by losses in normalized spectral counts, in the ethanol, B-PER, and the SDS-extraction buffer samples relative to the control. In contrast, the RNAlater preservative largely displayed little to no sign of degradation relative to the control. These results are visualized in Figure 5 as the Log2 ratios of relative abundance of proteins (where each point represents the average of the technical duplicates for a specific protein’s spectral counts) in each of four preservatives (RNAlater, ethanol, B-PER, and SDS-extraction buffer) normalized relative to the control treatment (also the average of technical duplicates). The proteins are presented in order of the highest number of normalized spectral counts in the control treatment (e.g., from the left side of x-axis), which should generally coincide with the most abundant proteins, with the caveat that spectral counting tends to compress the dynamic range of protein abundances. Log2 ratios close to zero (blank horizontal lines) reflect similar normalized spectral count scores for each protein between that preservative and the control, while deviation below the line indicates lower abundance in the preserved sample than the control presumably due to degradation. The RNAlater treatment (A) showed the least scatter around the zerofold change line indicating little degradation relative to the control. Most preservatives also showed a higher relative abundance of rarer proteins relative to the control (far right), although this interpretation is tentative since the relative error is larger at low spectral counts (Figure 4B). However, for the RNAlater sample this interpretation is consistent with the slightly higher number of protein identifications with RNAlater in Figure 3. The ethanol (B) and SDS-extraction buffer (D) preservatives showed good recovery of the most abundant proteins, but significant degradation of the remaining proteins, as well as large scatter around the zerofold change line. The B-PER treatment (C) showed degradation throughout including the most abundant proteins, but also had the largest scatter around the zerofold line of all the treatments.
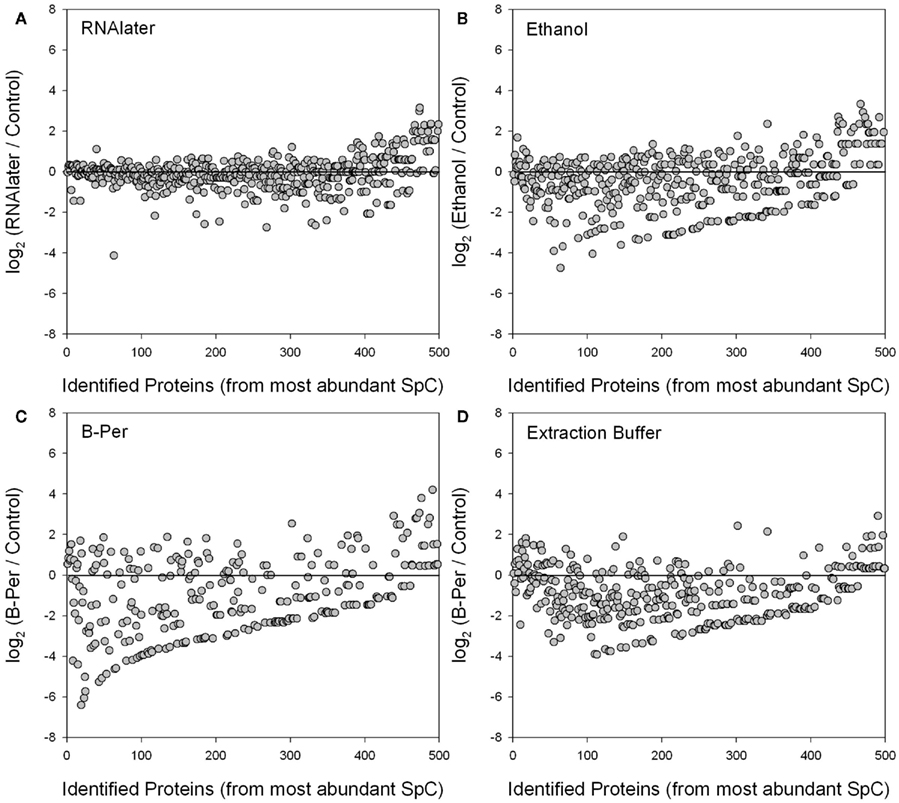
Figure 5. Log2 ratios of individual protein relative abundances (average of technical duplicates) identified in each of four preservatives (RNAlater, ethanol, B-PER, and SDS-extraction buffer) relative to the control treatment (average of technical duplicates). The proteins are ordered on the horizontal axis from those with the highest number of normalized spectral counts (SpC). Log2 ratios close to zero (horizontal lines) reflect similar normalized spectral count scores for each protein between that preservative and the control, while deviation below the line indicates lower abundance in the preserved sample than the control due to degradation. Normalization and weighting of spectral counts results involves normalizing to the total number of spectra within each sample, and weighting across samples (including replicates). The RNAlater treatment (A) showed the least degradation relative to the control, and perhaps even shows a higher relative abundance of rarer proteins in the RNAlater sample (far right). Ethanol (B) and SDS-extraction buffer (D) showed good recovery of the most abundant proteins, but degradation of many of the less abundant ones. The B-PER treatment (C) showed degradation throughout including the most abundant proteins. The upward-sloping line of data points in the bottom of (B–D) were caused by a combination of the abundance ordering on the horizontal axes and the low spectral counts of degraded proteins in the preserved treatment.
The influence of degradation on the 50 most abundant proteins (as determined by overall normalized spectral count scores) was compared quantitatively in Table 2. The average spectral count value for RNAlater preservative relative to the control was 1.01 + 0.27 (error reflects the SD), while the other three preservative treatments’ averages were farther from unity and had larger SD. The alkaline phosphatase enzyme, a protein of biogeochemical importance, was one of the few highly abundant proteins to show significant variability between the RNAlater and the control methods. This enzyme had low recovery in both RNAlater and B-PER relative to the control treatment (Table 2). Studies on E. coli alkaline phosphatase have found that this enzyme resides in the periplasm and can be released from unlysed cells by treatment with high concentrations of EDTA (Malamy and Horecker, 1961). It seems likely that a similar localization of alkaline phosphatase is occurring in Synechococcus WH8102. This result suggests that care should be taken in the choice of preservation methods used for cells intended for quantitative periplasm and membrane proteomic analyses.
Coloration of Extracted Material
The samples showed distinct coloration prior to acetone/methanol precipitation, indicative of differential extraction of chlorophyll and phycourobilin–phycoeryrthrin light harvesting molecules (Ong and Glazer, 1991; Blot et al., 2009). The TCA sample was slightly blue/green, ethanol was yellow/green, the B-PER sample was pink, the SDS-extraction buffer was yellow/green, RNAlater was yellow/brown, and the control was the most intense yellow/green. The pink color of the B-PER sample suggests that the phycourobilin–phycoerythrin complexes were particularly well extracted during the month-long incubation in its mixture of detergents. This is consistent with the high spectral count scores of some phycoerythrin proteins, in particular, the alpha and beta class II proteins are the two most abundant proteins detected in all samples and were most enriched in the B-PER treatment (Figure 4; Table 2).
Distinctions between Total Protein Recovery and Proteome Composition
Only one sample was used for each preservative, due to the expense and effort in conducting this preservative survey study with replication on six treatments. While we observed variability associated with the individual protein extractions that caused the total protein recovery results to diverge, we have little reason to think the protein extractions used here changed the proteome composition and hence the preservation results observed here. In other words, each extraction may have varying amounts of total protein recovery due to handling effects, but these influences should not alter the proteome composition that the mass spectrometry methodology is capable of characterizing. The main reason for this is that the protein extraction methods in this study all used a detergent-based extraction (SDS or B-PER) that solubilize proteins efficiently. For example, the ethanol, TCA, SDS, and RNAlater preserved samples all used an SDS-extraction protocol to solubilize and extract proteins, but the RNAlater treatment was far more effective in the number of protein identifications (Figure 3) and relative abundance of individual proteins compared to the immediately frozen control (Figure 5; Table 2). We have also previously observed that protein extractions produce consistent proteome composition with biological duplicates and during a diel cycle on a marine cyanobacterium (in relative and absolute abundance of individual proteins), despite the influence of extraction variability on the total protein yields (Saito et al., 2011). The data in the present study also emphasizes this distinction, where the variability in total protein recovery (Figure 1) showed little relationship to that of the global proteome composition (Figures 3 and 5). This difference reflects the fact that the handling processes that cause losses of total protein are likely distinct from any processes that might enrich or deplete individual proteins from the mixture. While future studies are needed to examine the potential fractionation of proteins in extraction protocols and to optimize their efficacy, these goals are distinct from our focus on preservative effectiveness here.
Implications and Degradation Mechanisms
Together, these results demonstrate that RNAlater is highly effective at preventing degradation of Synechococcus WH8102 culture biomass at ∼20°C over a duration and temperature range that are comparable to an actual field deployment. Other preservative approaches resulted in greater protein degradation. The potential use of RNAlater in simultaneously preserving DNA, RNA, and protein is particularly promising, allowing all three analytes to be potentially extracted from a single preserved sample (Ottesen et al., 2011). One limitation of this study is that only a single species was used to examine sample preservation, and it remains to be determined if other bacterial and eukaryotic algal species, including natural assemblages, preserve with comparable success. In addition, this study only investigated degradation over 1 month of storage; given the success of RNAlater in protein preservation, it seems quite likely that longer deployments would be successful and should be investigated. One factor that may be important is the abundance, localization, and types of proteases within each organism. RNAlater contains an organic metal chelating molecule that is known to deactivate metalloproteases by sequestering their zinc or cobalt (Huston et al., 2004), and the presence of this ingredient likely contributed to the preservation capability. The ethanol treatment also contained the organic metal chelator EDTA (1 mM, Table 1), yet this treatment did not show similar preservative properties, perhaps due to difficulties of metal chelation within this solvent. Marine Synechococcus has a significant complement of enzymes with proteolytic activity: the genome of WH8102 has 19 proteases and 26 peptidases (see text footnote 4), and extracellular proteolytic activity has been measured in marine Synechococcus strain WH7803 (Martinez and Azam, 1993). While these enzymes are likely involved in numerous cellular processes such as organic nitrogen acquisition and protein localization, they could contribute to protein degradation during storage and processing. The high salt content of the RNAlater likely also contributes to the protein preservation capabilities of the preservative. High salt solutions are commonly used to precipitate proteins and also dehydrates cells, likely also inhibiting proteolytic activity. Additional experimentation with protease inhibitors could also improve recoveries.
Conclusion
This study demonstrates successful proteome preservation of marine Synechococcus WH8102, a representative of one of the highly abundant marine cyanobacteria in the oceans. If this level of preservation can be achieved with other bacterial, archaeal, and eukaryotic microbes, autonomous remote sampling of the marine water column microbial community without freezing appears to be a tractable goal.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Erin Bertrand and Matthew McIlvin for assistance with mass spectrometry maintenance, data processing, and comments on the manuscript. We thank John Waterbury for cultures of Synechococcus WH8102 and helpful discussion. We are grateful to four anonymous reviewers for their comments on this manuscript. This work was funded by the National Science Foundation Chemical and Biological Oceanography, Center for Microbial Oceanography Research and Education (C-MORE), and the Gordon and Betty Moore Foundation.
Footnotes
References
Ansong, C., Purvine, S. O., Adkins, J. N., Lipton, M. S., and Smith, R. D. (2008). Proteogenomics: needs and roles to be filled by proteomics in genome annotation. Brief. Funct. Genomic. Proteomic. 7, 50–62.
Barrios-Llerena, M. E., Chong, P. K., Gan, C. S., Snijders, A. P. L., Reardon, K. F., and Wright, P. C. (2006). Shotgun proteomics of cyanobacteria – applications of experimental and data-mining techniques. Brief. Funct. Genomic. Proteomic. 5, 121–132.
Bell, J., Betts, J., and Boyle, E. (2002). MITESS: a moored in-situ trace element serial sampler for deep-sea moorings. Deep Sea Res. I 49, 2103–2118.
Bertrand, E. M., Saito, M. A., Jeon, Y. J., and Neilan, B. A. (2011). Vitamin B12 biosynthesis gene diversity in the Ross Sea: the identification of a new group of putative polar B12 biosynthesizers. Environ. Microbiol. 13, 1285–1298.
Blot, N., Wu, X.-J., Thomas, J.-C., Zhang, J., Garczarek, L., Bohm, S., Tu, J. M., Zhou, M., Plöscher, M., Eichacker, L., Partensky, F., Scheer, H., and Zhao, K. H. (2009). Phycourobilin in trichromatic phycocyanin from oceanic cyanobacteria is formed post-translationally by a phycoerythrobilin lyase-isomerase. J. Biol. Chem. 284, 9290–9298.
Breier, J. A., Rauch, C. G., McCartney, K., Toner, B. M., Fakra, S. C., White, S. N., and German, C. R. (2009). A suspended-particle rosette multi-sampler for discrete biogeochemical sampling in low-particle-density waters. Deep Sea Res. 56, 1579–1589.
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive earth’s biogeochemical cycles. Science 320, 1034–1039.
Gonzales, A. D., Light, Y. K., Zhang, Z., Iqbal, T., Lane, T. W., and Martino, A. (2005). Proteomic analysis of the CO2-concentrating mechanism in the open-ocean cyanobacterium Synechococcus WH8102. Can. J. Bot. 83, 735–745.
Greenfield, D. I. III. R. M., Jensen, S., Massion, E., Roman, B., Feldman, J., and Scholin, C. (2006). Application of the environmental sample processor (ESP) methodology for quantifying Pseudo-nitzschia australis using ribosomal RNA-targeted probes in sandwich and fluorescent in situ hybridization. Limnol. Oceanogr. Methods 4, 426–435.
Huston, A. L., Methe, B., and Deming, J. W. (2004). Purification, characterization, and sequencing of an extracellular cold-active aminopeptidase produced by marine psychrophile Colwellia psychrerythraea Strain 34H. Appl. Environ. Microbiol. 70, 3321–3328.
Keller, A., Nesvizhskii, A. I., Kolker, E., and Aebersold, R. (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392.
Lange, V., Malmstrom, J. A., Didion, J., King, N. L., Johansson, B. P., Schafer, J., Rameseder, J., Wong, C. H., Deutsch, E. W., Brusniak, M. Y., Bühlmann, P., Björck, L., Domon, B., and Aebersold, R. (2008). Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol. Cell Proteomics. 7, 1489–1500.
Malamy, M., and Horecker, B. L. (1961). The localization of alkaline phosphatase in, K12. Biochem. Biophys. Res. Commun. 5, 104–108.
Martinez, J., and Azam, F. (1993). Aminopeptidase activity in marine chroococcoid cyanobacteria. Appl. Environ. Microbiol. 59, 3701–3707.
Morel, F. M. M., and Price, N. M. (2003). The biogeochemical cycles of trace metals in the oceans. Science 300, 944–947.
Morris, R. M., Nunn, B. L., Frazar, C., Goodlett, D. R., Ting, Y. S., and Rocap, G. (2010). Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME J. 4, 673–685.
Ong, L. J., and Glazer, A. N. (1991). Phycoerythrins of marine unicellular cyanobacteria. I. Bilin types and locations and energy transfer pathways in Synechococcus spp. phycoerythrins. J. Biol. Chem. 266, 9515–9527.
Ottesen, E. A., Marin, R., Preston, C. M., Young, C. R., Ryan, J. P., Scholin, C. A., and DeLong, E. F. (2011). Metatranscriptomic analysis of autonomously collected and preserved marine bacterioplankton. ISME J. doi: 10.1038/ismej.2011.70. [Epub ahead of print].
Palenik, B., Brahamsha, B., Larimer, F. W., Land, M., Hauser, L., Chain, P., Lamerdin, J., Regala, W., Allen, E. E., McCarren, J., Paulsen, I., Dufresne, A., Partensky, F., Webb, E. A., and Waterbury, J. (2003). The genome of a motile marine Synechococcus. Nature 424, 1037–1042.
Partensky, F., Blanchot, J., and Vaulot, D. (1999). Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bull. Inst. Oceanogr. 19, 457–475.
Paul, J., Scholin, C. A., Engh, G. V. D., and Perry, M. J. (2007). In situ instrumentation. Oceanography 20, 70–78.
Peng, J., Elias, J. E., Thoreen, C. C., Licklider, L. J., and Gygi, S. P. (2003). Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50.
Ram, R. J., VerBerkmoes, N. C., Thelen, M. P., Tyson, G. W., Baker, B. J., Blake, R. C. II., Shah, M., Hettich, R. L., and Banfield, J. F. (2005). Community proteomics of a natural microbial biofilm. Science 308, 1915–1920.
Saito, M. A., Bertrand, E. M., Dutkiewicz, S., Bulygin, V. V., Moran, D. M., Monteiro, F. M., Follows, M. J., Valois, F. W., and Waterbury, J. B. (2011). Iron conservation by reduction of metalloenzyme inventories in the marine diazotroph Crocosphaera watsonii. Proc. Natl. Acad. Sci. U.S.A. 108, 2184–2189.
Saito, M. A., Goepfert, T. J., and Ritt, J. T. (2008). Some thoughts on the concept of colimitation: three definitions and the importance of bioavailability. Limnol. Oceanogr. 53, 276–290.
Saito, M. A., Rocap, G., and Moffett, J. W. (2005). Production of cobalt binding ligands in a Synechococcus feature at the Costa Rica upwelling dome. Limnol. Oceanogr. 50, 279–290.
Scholin, C., Doucette, G., Jensen, S., Roman, B., Pargett, D., Marin, R., Preston, C., Jones, W., Feldman, J., Everlove, C., Harris, A., Alvarado, N., Massion, E., Birch, J., Greenfield, D., Vrijenhoek, R., Mikulski, C., and Jones, K. (2009). Remote detection of marine microbes, small invertebrates, harmful algae, and biotoxins using the environmental sample processor (ESP). Oceanography 22, 158–167.
Sowell, S. M., Norbeck, A. D., Lipton, M. S., Nicora, C. D., Callister, S. J., Smith, R. D., Barofsky, D. F., and Giovannoni, S. J. (2008a). Proteomic analysis of stationary phase in the marine bacterium Candidatus Pelagibacter ubique. Appl. Environ. Microbiol. 74, 4091–4100.
Sowell, S. M., Wilhelm, L. J., Norbeck, A. D., Lipton, M. S., Nicora, C. D., Barofsky, D. F., Carlson, C. A., Smith, R. D., and Giovanonni, S. J. (2008b). Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 3, 93–105.
VerBerkmoes, N. C., Denef, V. J., Hettich, R. L., and Banfield, J. F. (2009). Systems Biology: functional analysis of natural microbial consortia using community proteomics. Nat. Rev. Microbiol. 7, 196–205.
Waterbury, J. B., Watson, S. W., Valois, F. W., and Franks, D. G. (1986). “Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus,” in Photosynthetic Picoplankton, ed. W. K. W. Li (Ottawa: Dept. of Fisheries and Oceans), 71–120.
Wolf-Yadlin, A., Hautaniemi, S., Lauffenburger, D. A., and White, F. M. (2007). Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A. 104, 5860–5865.
Zinser, E. R., Lindell, D., Johnson, Z. I., Futschik, M. E., Steglich, C., Coleman, M. L., Wright, M. A., Rector, T., Steen, R., McNulty, N., Thompson, L. R., and Chisholm, S. W. (2009). Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS ONE 4, e5135. doi: 10.1371/journal.pone.0005135
Keywords: proteome, preservation, autonomous sampling, cyanobacteria, alkaline phosphatase, proteomics, Synechococcus WH8102
Citation: Saito MA, Bulygin VV, Moran DM, Taylor C and Scholin C (2011) Examination of microbial proteome preservation techniques applicable to autonomous environmental sample collection. Front. Microbio. 2:215. doi: 10.3389/fmicb.2011.00215
Received: 13 July 2011;
Accepted: 10 October 2011;
Published online: 07 November 2011.
Edited by:
Julie LaRoche, Leibniz Institute of Marine Sciences (IFM-GEOMAR), GermanyReviewed by:
Claire Mahaffey, University of Liverpool, UKTom Bibby, University of Southampton, UK
Rachael Marie Morgan-Kiss, Miami University, USA
Heather Bouman, University of Oxford, UK
Copyright: © 2011 Saito, Bulygin, Moran, Taylor and Scholin. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Mak A. Saito, Marine Chemistry and Geochemistry Department, Woods Hole Oceanographic Institution, Woods Hole, MA 02543, USA. e-mail: msaito@whoi.edu