- School of Biotechnology and Biomolecular Sciences, University of New South Wales, Randwick, NSW, Australia
Novel cultivation strategies for bacteria are widespread and well described for recovering greater diversity from the “hitherto” unculturable majority. While similar approaches have not yet been demonstrated for fungi it has been suggested that of the 1.5 million estimated species less than 5% have been recovered into pure culture. Fungi are known to be involved in many degradative processes, including the breakdown of petroleum hydrocarbons, and it has been speculated that in Polar Regions they contribute significantly to bioremediation of contaminated soils. Given the biotechnological potential of fungi there is a need to increase efforts for greater species recovery, particularly from extreme environments such as sub-Antarctic Macquarie Island. In this study, like the yet-to-be cultured bacteria, high concentrations of nutrients selected for predominantly different fungal species to that recovered using a low nutrient media. By combining both media approaches to the cultivation of fungi from contaminated and non-contaminated soils, 91 fungal species were recovered, including 63 unidentified species. A preliminary biodegradation activity assay on a selection of isolates found that a high proportion of novel and described fungal species from a range of soil samples were capable of hydrocarbon degradation and should be characterized further.
Introduction
The isolation of fungi into pure culture has been severely limited. Of the estimated 1.5 million species that are thought to exist less than 5% have been successfully recovered into pure culture (Hawksworth, 2001; O’Brien et al., 2005). Given the biotechnological potential of fungal enzymes there is a need to increase efforts for greater species recovery. Limitations in cultivation methods are thought to be attributed to fast-growing fungal species outcompeting slower growing species, many fungi are not adapted to growth in high nutrient media, and a proportion of fungi will not sporulate in artificial culture media (Taylor et al., 2000; O’Brien et al., 2005). Additionally, traditional cultivation methods result in biases toward culturing predominantly from the Ascomycota phylum. Novel cultivation methods for the recovery of yet-to-be cultured species have been well described for bacteria and include the application of simulated natural environments, extended incubation times, and/or the use of limited nutrients (Janssen et al., 2002; Kaeberlein et al., 2002; Rappe et al., 2002; Ferrari et al., 2005). The development of a high-throughput dilution-to-extinction approach has been developed for fungi that increased species richness significantly (Collado et al., 2007). However, novel approaches to culturing fungi that utilize low levels of nutrients or simulated natural environments have not yet been adopted.
Macquarie Island is a sub-Antarctic Island located in the southern ocean, halfway between Antarctica and Australia. It is a world heritage listed area and is a breeding site for millions of migrating mammals and birds. The main features of Macquarie Islands climatic conditions include small variations in temperature ranging between an average of 3.25 in July to 7.05 in January, strong prevailing winds and a high number of days with precipitation. A permanent Australian Antarctic Research station has been present on the northern isthmus of the island since 1948. Consequently, the island has experienced a legacy of contamination events resulting from petroleum hydrocarbon fuel spills in the form of Special Antarctic Blend (SAB) diesel fuel (Rayner et al., 2007).
Fungi play a central role in biodegradation or decomposition and are producers of an array of extracellular enzymes. In particular, filamentous fungi have been implicated in the biodegradation of a wide range of aromatic hydrocarbons and thus they could contribute significantly to bioremediation efforts (Prenafeta-Boldu et al., 2006; Hughes et al., 2007; Hughes and Bridge, 2010). Petroleum contamination is a global problem and in Polar Regions these spills result in extensive damage to ecosystems as cold region ecosystem recovery is a much slower process than in warmer regions (Aislabie et al., 2001). In the Antarctic, over 1,000 accepted fungal species have been described, the majority of which are widely distributed or cosmopolitan species (Hughes and Bridge, 2010). Contamination of Antarctic soils with hydrocarbons has been reported to result in soils becoming dominated by high numbers of hydrocarbon degrading filamentous fungi such as Phialophora sp. and Hormoconis resinae (Kerry, 1990; Aislabie et al., 2001). It is expected that in sub-Antarctic Macquarie Island similar species would also appear following petroleum hydrocarbon contamination.
Little information exists on the fungal diversity of Macquarie Island with only leaf and litter-colonizing fungi such as Geomyces sp., Mortierella sp., Penicillium sp., Phoma sp., and Acremonium sp. previously isolated (Kerry, 1990). Climatic conditions on the island are well suited to fungal growth, which suggests they should dominate the soil ecosystem. Until now, the Islands soil fungal diversity, the effect of diesel fuel spills on natural fungal diversity, and the potential role of local fungal species in hydrocarbon degradation has not yet been investigated. In this study, a traditional high nutrient and a low nutrient approach were used to recover greater fungal species diversity from both pristine and SAB diesel fuel contaminated soil from Macquarie Island. The result was the recovery of 91 species including a high number of previously unidentified species from both types of nutrient media used. A dramatic shift in fungal diversity was observed with increasing concentrations of SAB diesel fuel and several presumptive hydrocarbon degrading species were isolated. Of the 63 potentially novel species recovered, those isolated from high concentrations of SAB and those which exhibit the ability to grow in the presence of petroleum hydrocarbons will be targeted for further characterization to determine their role in hydrocarbon degradation.
Materials and Methods
Field Collection
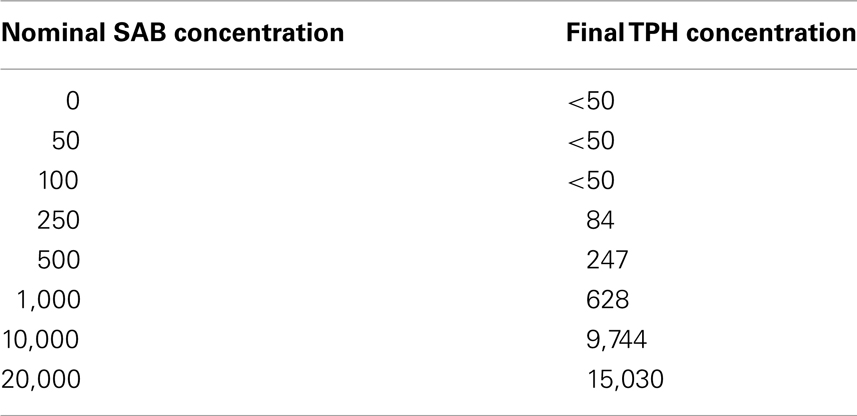
Soil samples were collected near the Australian Antarctic Research station at sub-Antarctic Macquarie Island (54°37′53″S, 158°52′15″) during February 2009. Approximately 1 kg of soil was excavated from the top 0–30 cm of 1 m × 1 m plot of similar terrain and 170 m away from the largest contaminated site without SAB contamination. The carbon and water content were determined to be 3.2 and 24% respectively (Rayner et al., 2007). The soil was divided into eight subsamples and spiked with SAB diesel fuel over the range of 0–20000 mg kg−1, homogenized and incubated at 10°C for 10 days. To determine final total petroleum hydrocarbon (TPH) concentrations a portion (10 g) from each spiked soil sample was extracted with hexane and analyzed by gas chromatography (Rayner et al., 2007). Table 1 provides the nominal verses final TPH concentrations obtained after SAB diesel fuel spiking.
Cultivation of Fungi from Soil
Fungi were isolated from soil following incubation of each soil subsample with SAB diesel fuel. In each case, 1 g of each soil sample were suspended into 10 ml of sterile distilled water in a 15-ml sterile plastic tube. The suspension was vortexed for 5 min and then large particles were allowed to settle for a further 5 min. Serial dilutions were then prepared up to a final concentration of 10−5. For each soil sample, 1 ml of the 10−3, 10−4 and 10−5 dilutions were spread onto large agar plates in triplicate.
Two forms of Cooke Rose Bengal agar (CRBA) were used for the selective isolation of fungi. Standard 1× CRBA contained 10 g glucose, 5 g Bacto soytone enzymatic digest of soybean meal, 1 g KH2PO4, 0.5 g MgSO4·7H2O, 35 mg Rose Bengal (Sigma-Aldrich) and 20 g agar (Oxoid, Sydney, Australia) per 1 L sterile distilled water (Atlas, 2010). The low nutrient media or 0.1× CRBA was prepared consisting of 1 g glucose, 0.5 g Bacto soytone enzymatic digest of soybean meal, 0.1 g KH2PO4, 0.05 g MgSO4·7H2O, 3.5 mg Rose Bengal and 20 g agar (Oxoid) per 1 L sterile distilled water. In order to inhibit bacteria growth, streptomycin sulfate (Sigma-Aldrich, Sydney, Australia) was also added to both types of media at a final concentration to 50 mg/L at 46°C following sterilization by autoclaving.
The agar plates were sealed and incubated at 15°C in the dark for 3 weeks as this temperature has been used previously for sub-Antarctic fungi (Kerry, 1990). Following incubation, each agar plate dilution contained diverse fungal colony morphotypes for both media. Subsequently, all three dilutions of soil from both the 1× and 0.1× CRBA agar plates were used for sub-cultivation onto potato dextrose agar (PDA). PDA consisted of 24 g potato dextrose broth (Sigma-Aldrich, Castle Hill, Australia) and 15 g agar per 1 L of sterile distilled water. Each fungal morphotype observed was sub-cultured by transferring hyphal tips, colonies, or spores to PDA plates for incubation for a further 3 weeks at 15°C. Sub-cultivation was repeated at least three times until pure cultures were obtained.
Isolate Preservation
A 15% glycerol solution was prepared for long-term preservation of fungal isolates at −80°C. For spore producing isolates or fungal mycelia growing on agar surface cultures, 2 ml of the sterile preserving fluid was placed on the surface of agar plates and fungi were gently scraped off with a pipette. Then a portion of the spore or mycelial suspension was placed into two 2 ml screw cap polypropylene vials. For non-sporulating isolates or fungal mycelia that grew deep within the agar cultures, several five agar plugs were cut from agar plates using a sterilized surgical blade and were placed into a screw cap polypropylene vial containing 0.5–1.0 ml of the 15% glycerol solution. The vials were frozen at −20°C overnight, then transferred to storage at −80°C.
DNA Extraction
Two DNA extraction methods were used for extraction of total genomic DNA from pure fungal isolates. For the first round, the prepGEM™ Tissue kit (ZyGEM, Hamilton, New Zealand) previously used for parasite DNA extractions was modified for fungi (Ferrari et al., 2007). For each isolate, a portion of a colony was scraped from the agar plates using an inoculating loop and transferred into a 1.5-ml microcentrifuge tube containing 89 μl ultra pure water, 10 μl of 10× buffer, and 1 μl prepGEM™ enzyme. Samples were then incubated at 75°C for 15 min followed by 95°C for 5 min using a MyCycler Thermocycler (Bio-rad, Gladesville, Australia). The DNA lysates were then transferred into fresh 1.5 ml tubes and centrifuged at 14 000 rpm for 1 min. Finally, DNA lysates were removed from the precipitated residue and placed into a 500-μl tube and stored at −20°C until required.
If DNA or PCR products were not detectable following prepGEM™ extractions a second round of DNA extracts were prepared using a modified bead beating method. For each isolate, five agar plugs containing a fungal colony were cut from agar plates using sterile surgical blades. Each piece of agar was then placed into a screw cap 2 ml microcentrifuge tube containing 0.5 g of 0.1 and 0.5 mm glass beads (Mo bio, Carlsbad, USA) and 1, 000 μl ultra pure water. Tubes were homogenized using the FastPrep 120 instrument (MP Biomedicals, Seven Hills, Australia) for 40 s at a speed setting of 6.0. Finally, DNA extracts were incubated at 95°C for 5 min and centrifuged at 14000 rpm for 1 min. The DNA lysate was removed and placed in a 500-μl tube for storage at −20°C until required.
ITS Sequence Amplification
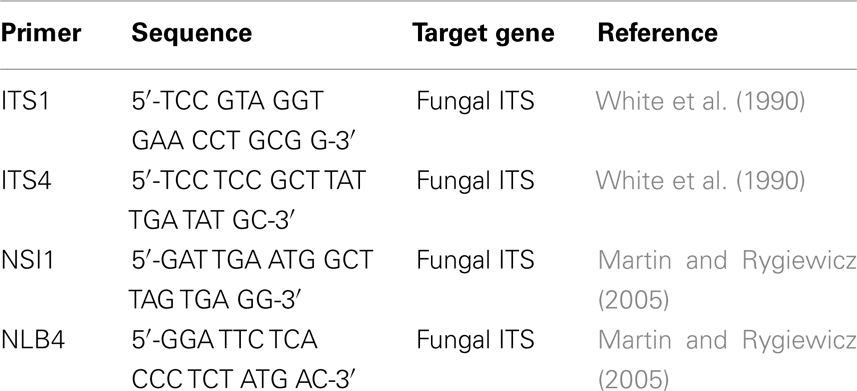
The fungal internal transcribed spacer (ITS) gene sequences were amplified using previously published ITS specific primers (Table 2). All PCR primer sets were obtained from Integrated DNA Technologies (McLeans Ridges, Australia) and PCR’s were carried out using the MyCycler Thermocycler.
For the majority of samples the fungal ITS sequence was amplified using the ITS1/ITS4 primer set (White et al., 1990). The PCR amplification was performed in 50 μl reaction mix containing 22 μl ultra pure water, 25 μl GoTaq hot start Green Master Mix (Promega, Doncaster, Australia), 0.5 μl of each primer, and 2 μl DNA template. The PCR program consisted of initial denaturation step at 94°C for 5 min, followed by 40 cycles of 94°C for 1 min, 55°C for 30 s, 72°C for 2 min followed by a final extension step at 72°C for 5 min. The PCR products were visualized by gel electrophoresis on a 2% agarose gel with the addition of SYBR safe DNA gel stain (Invitrogen, Sydney, Australia) at a concentration of 0.01% and exposure to UV light. The expected size of ITS region was 500–700 bp. PCR products were then purified using the QIAquick PCR Purification Kit (Qiagen, Castle Hill, Australia).
The NSI1/NLB4 primer set was also used which targets the ITS region and was used if the ITS1/ITS4 primer set did not produce a PCR amplicon (Table 2). The PCR reaction mix for the NSI1/NLB4 primer set was identical to the ITS1/ITS4 primer set. The PCR program consisted of an initial denaturation step at 95°C for 5 min, 35 cycles of 95°C for 30 s, 60°C for 40 s, 72°C for 40 s followed by a final extension at 72°C for 5 min (Martin and Rygiewicz, 2005).
Restriction Fragment Length Polymorphism Analysis and DNA Sequencing
Internal transcribed spacer PCR amplicons were digested with the restriction enzymes HaeIII and RsaI (Promega) as described by Imad et al. (2010) and Korabecna (2007) respectively (Korabecna, 2007; Imad et al., 2010). Briefly, 15 μl PCR product, 2.63 μl ultra pure water, 2 μl 10× buffer C, 0.2 μl 100× BSA (Promega) and 0.17 μl of each restriction enzyme were placed in 96 well PCR plates (Bio-rad), sealed with Microseal Foil (Bio-rad), and incubated overnight at 37°C. The digested products were visualized on a 2% agarose gel with SYBR safe DNA gel stain. Based on the DNA fingerprint for both restriction enzymes, each single ribotype was selected for DNA sequencing.
DNA sequencing was performed for each strand of all positive amplification products using the ITS1 and ITS4 primers using the Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Mulgrave, Australia) as described by the manufacturer. The sequencing analysis was carried out using the 3730 sequence scanner (Applied Biosystems) in the Ramaciotti Centre Sequencing Facility (School of Biotechnology and Biomolecular Sciences, UNSW, Sydney).
Identification and Phylogenetic Analysis of Isolates
The partial sequencing results were analyzed using appropriate programs within the NCBI database; nucleotide blast and ClustalX. For identification of isolates, morphologic information was combined with ITS sequence matches obtained within the GenBank database. Fungal ITS sequences for all 91 isolates were deposited into GenBank database under accession numbers JN104511–JN104578 (Table 3).
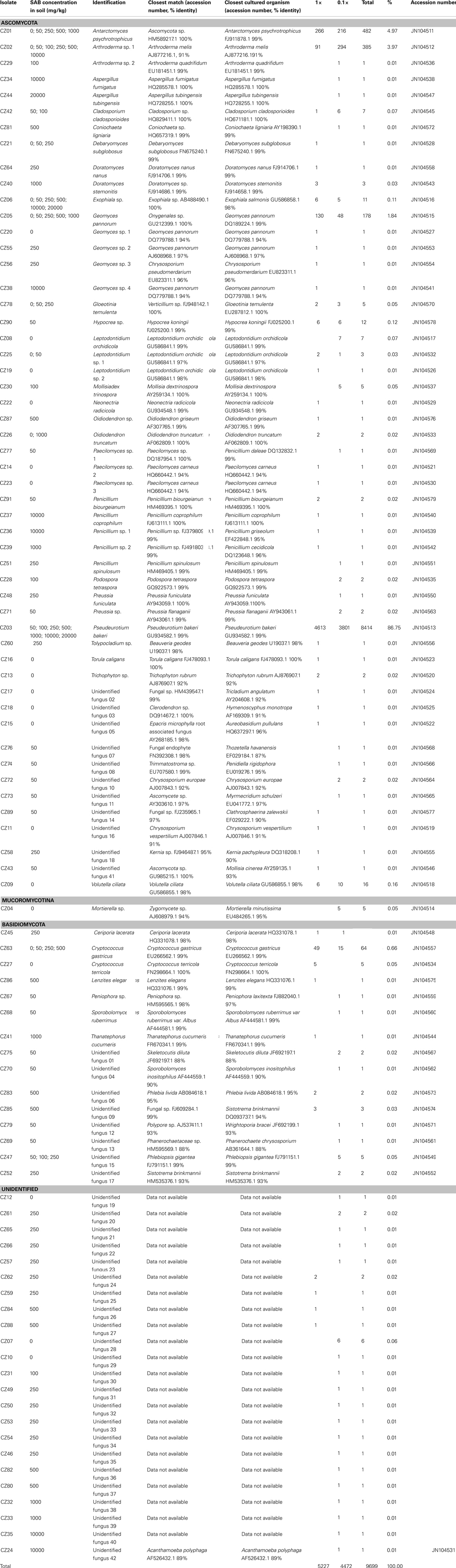
Table 3. Molecular and morphologic identification, ITS sequence matches, and isolation frequency for fungal isolates recovered from contaminated and non-contaminated Macquarie Island soil.
Petroleum Hydrocarbon Biodegradation Screening
Bushnell Haas broth was selected for petroleum hydrocarbon biodegradation screening of fungal isolates. Prior to screening, a selection of pure cultures was tested for the ability to grow in liquid medium. Twenty fungal isolates from both pristine and contaminated soils were incubated in 50 ml of special nutrient broth (SNB) in 250 ml flasks for 1 week at 15°C. The twelve isolates that were capable of growth in broth were next examined for the ability for growth and activity on petroleum hydrocarbons in the form of SAB diesel fuel.
Bushnell Haas broth was prepared by adding 0.2 g MgSO4·7H2O (Ajax Finechem), 0.02 g CaCl2 (Ajax Finechem), 1 g KH2PO4 (Ajax Finechem), 1 g K2HPO4 (Ajax Finechem), 0.05 g FeCl3 (Ajax Finechem), 1 g NH4NO3 (Ajax Finechem), and sterile distilled water to final volume of 1 L. After autoclaving for 30 min at 15 psi for 121°C, 1% v/v SAB diesel fuel (10 ml/L), 1 ml Tween 80 (Sigma-Aldrich) and 1 mg redox indicator 2,6-dichlorophenol indophenol (Sigma-Aldrich) were added into the Bushnell Haas broth for activity screening (Hanson et al., 1993; George-Okafor et al., 2009). For screening, 10 ml of Bushnell Haas (w/out SAB diesel fuel) medium was distributed into each well of a six well culture plate (Sarstedt). A portion (50 μl) of each pure fungal culture was inoculated into screening medium containing SAB (in triplicate) into the six well plate and into media without SAB which served as the negative control (in triplicate). Incubation was carried out at 15°C for 3 weeks. A color change in the media from deep–blue to colorless indicated redox reactions were occurring in the media and monitoring was performed daily.
Statistical Analysis
A two-way analysis of similarity test (ANOISM) test was carried out using Primer v6 software (Primer-E Ltd, Ivybridge, United Kingdom) to determine if the fungal diversity recovered between the low and high nutrient approach was significantly different. Primer v6 software is used for multivariate statistical analysis in ecological and environmental science, particularly for the analysis of species distribution and abundance. After raw data from each SAB diesel fuel concentration and culture medium was collected, the data were compiled into a data matrix within Primer v6 and multivariate data analysis was conducted using Plymouth routines in multivariate ecology research as described in the software manual. The Fungal assemblage data was square root transformed to balance the relative contribution of dominant species. After creating a resemblance matrix the similarity between samples was calculated on the basis of the Bray–Curtis coefficient (Clarke and Green, 1998). The similarity results representing the biological relationship were then examined using hierarchical clustering with group-average linkage and non-metric multi-dimensional scaling (NMDS) ordination plots. The distances between pairs of samples on the NMDS ordination plot reflect the relative similarity of species composition. Clusters (at various levels of similarity) superimposed on the NMDS ordination plot, ascertain the level of agreement between the clustering with group-average linkage and the NMDS methods. Global R values that result from these analyses provides an absolute measure of how separated the groups are, on a scale of 0 for groups that are indistinguishable, to a value of 1, where all similarities within groups is less than any similarity between groups. To determine the significance of the variables between samples, fuel concentration, and media type were tested against 999 permutations.
Results
Fungal Cultivation Using Both a High and a Low Nutrient Media
In total, 9,699 fungal isolates were recovered from the range of soil samples investigated. Based on the distinct macroscopic morphological characteristics of these isolates, 156 fungal isolates were selected for sub-cultivation on PDA until pure cultures were obtained for molecular identification. Following DNA extraction, 134 PCR amplification products were obtained yielding products between 500 and 700 bp in length for distinct isolates. Following RFLP analysis, 69 different ribotypes were selected for DNA sequencing. In an attempt to identify as many isolates as possible, DNA sequencing results were then combined with macroscopic and microscopic morphologic information. Finally, of the 69 different ribotypes sequenced; 28 isolates were identified to the species; and 21 to the genus level (Table 3). Due to the low match similarity or unique morphological characteristics, 20 isolates were not able to be identified to the genus level, using each available primer set and were termed unidentified fungus 1–18, 24, and 41–42. Of these, twenty were identified to phylum level based on microscopic morphological information only and one additional isolate (CZ24) could not be classified into any fungal phylum due to it’s unusual ITS gene sequence and morphological characteristics. The remaining 22 isolates did not yield PCR amplification products using primer sets ITS1/ITS4 and NSI1/NLB4 and were termed unidentified fungus 19–23 and 25–40.
Comparison of Species Diversity Recovered between the High and Low Nutrient Media Approach
Of the 91 fungal species recovered, the high nutrient culture approach recovered 52 species (57% of total species) from among 5,227 isolates and the low nutrient approach recovered 50 species (55% of total species) from among 4,472 isolates. Both culture media recovered a high proportion of singletons, which in total represented 67% of total species diversity. Additionally, a high number of unidentified species were recovered from both culture media used, 34 species (37% of total species) were isolated using the high nutrient media and 33 species (36% of total species) were isolated using the low nutrient media. Statistical results confirmed that the selection of media did significantly alter the recovered species distribution due to a global R value of 0.542 and P value of 0.001 (Figure 1). The SAB diesel fuel concentration within the spiked soil samples used for cultivation resulted in an even greater effect on downstream species distribution as revealed by a statistically significant global R value of 0.916 and P value of 0.001. Furthermore, a greater number of novel species were recovered from the low nutrient media, including two unidentified species from the 10,000-mg/kg SAB spiked soil samples (Tables 3 and 4).
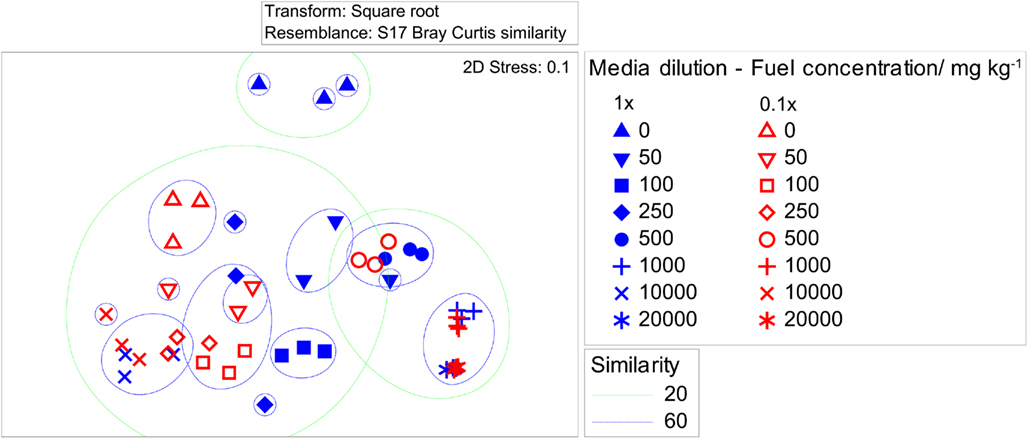
Figure 1. Non-metric MDS configuration plot comparing the fungal diversity recovered from low or high nutrient media. Non-metric multi-dimensional scaling (NMDS) plot of similarities calculated using the bray–curtis coefficient using square root transformed fungal diversity data. The NMDS plot includes superimposed clusters at similarity levels of 20% (blue line) and 60% (green line). Results confirmed that the selection of media did significantly alter the recovered species distribution due to a global R value of 0.542 and P value of 0.001. The SAB diesel fuel concentration within spiked soil samples resulted in an even greater effect on recovered species distribution as revealed by a global R value of 0.916 and P value of 0.001.
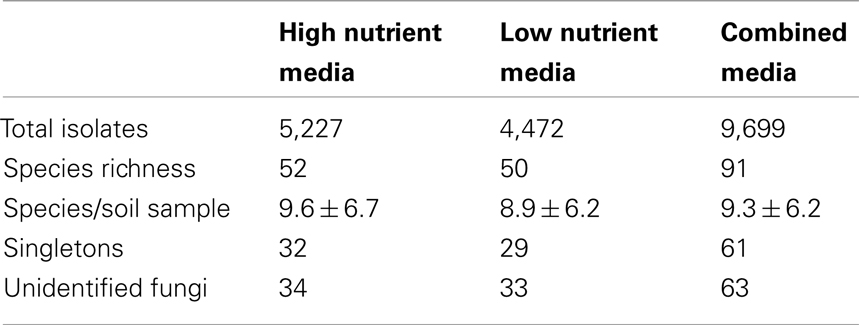
Table 4. Diversity and abundance of fungal species recovered from soil using both a high and a low nutrient media approach to cultivation.
The Distribution and Abundance of Recovered Fungal Diversity
The distribution and abundance data revealed that 52 species (57% of total species) belonged to phylum Ascomycota, 15 species (16% of total species) belonged to phylum Basidiomycota and one species (1% of total species) was identified within the Mucoromycotina, sub-phylum incertae sedis. Of these, only 11 species were recovered by both the high and low nutrient media approach (Tables 3 and 4). These species were the most dominant recovered from a range of both pristine and contaminated soil samples and represented both indigenous; Antarctomyces psychrotrophicus, Geomyces pannorum (4.97 and 1.8% of isolates respectively) and cosmopolitan species; Pseudeurotium bakeri, and Arthroderma sp. 1. (87 and 3.4% of isolates respectively; Table 3). Furthermore, 4 of the 11 dominant species; Arthroderma sp. 1 (CZ02), Exophiala sp. (CZ06), Leptodontidium sp. 1 (CZ25), and Hypocrea sp. (CZ90) represented potentially novel species based on low ITS sequence identity similarity combined with distinct morphological characteristics to their closest cultured organism. Arthroderma sp. 1 and Exophiala sp. were prevalent in pristine and highly contaminated soil samples. Exophiala sp. (CZ06) exhibited 98% similarity to Exophiala salmonis while Arthroderma sp. 1(CZ02) exhibited 91% similarity to Arthroderma melis.
Fungal abundance data showed that there were a couple of dominant species present in sub-Antarctic soil with a high proportion of singletons recovered (Tables 3 and 4). Pseudeurotium bakeri (CZ03) was isolated from all of the SAB diesel contaminated soil samples and was the most abundant fungal species present in the range of soil samples examined. This species increased in abundance by 100-fold as the SAB diesel concentration reached 1,000 and 20,000 mg/kg of soil compared with low levels of contamination (data not shown).
The Effect of Increasing Concentrations of SAB Diesel Fuel on Recovered Fungal Diversity
The 91 fungal isolates were classified to class and sub-phylum levels to investigate the species distribution over the range of SAB diesel fuel spiked soil samples. In summary, these fungi belonged to eight class, one sub-phylum incertae sedis and four unclassified groups (Table 2 and Figure 2). The effect of increasing concentration of SAB diesel fuel on diversity at the class-level showed that fungal diversity increased at lower concentrations and decreased at higher concentrations of SAB diesel fuel. Moreover, with increasing concentrations of SAB diesel fuel, the population shifted toward an increase in unidentified fungal species (Figure 2).
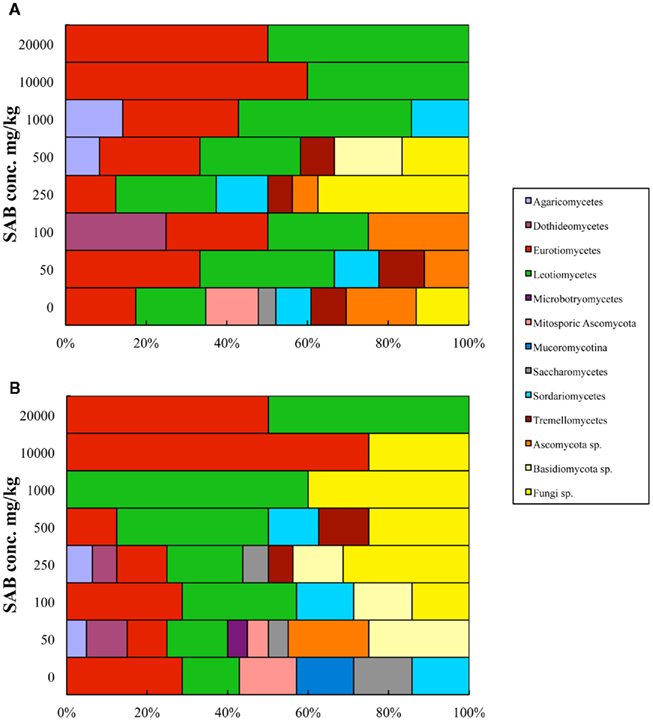
Figure 2. Proportional distribution of fungal ITS sequences at the Class-level isolated from a range of SAB diesel fuel spiked soil samples. (A) The distribution of OTUs recovered on high nutrient media. (B) The distribution of OTUs recovered on low nutrient media. The fungal diversity recovered changed significantly as the concentration of SAB diesel fuel increased. The greatest level of diversity was observed in the uncontaminated soils for both media used. At low concentrations of SAB diesel fuel, a shift in fungal diversity was observed including a small proportion of unidentified species. While in the highly contaminated soil samples the diversity decreased and was limited to the Eurotiomycetes and Leotiomycetes class only.
At the class-level the greatest diversity was recovered from within pristine soil using both the dilute and high nutrient media, with a high proportion of species from within Eurotiomycetes, Leotiomycetes, Saccharomycetes, and Sordariomycetes recovered. With increasing concentrations of SAB diesel fuel, the population shifted toward an overall increase in unidentified fungal species and an emergence of fungi from within the Dothideomycetes and Microbotryomycetes class. At 250–500 mg/kg SAB diesel fuel a second shift in diversity was observed with a higher proportion of unidentified fungi recovered as well as species belonging to the Agaricomycetes, Sordariomycetes, and Tremellomycetes class. At high concentrations of SAB diesel fuel exceeding 1,000 mg/kg SAB, the diversity became more limited to the Eurotiomycetes and Leotiomycetes class as well as unidentified species from the dilute media only (Figure 2). This reduction in diversity was combined with an overall increase in total colony forming units with one species Pseudeurotium bakeri (Leotiomycetes; CZ03) highly abundant in the 10,000 and 20,000-mg/kg samples (Table 3).
Isolation of Sub-Antarctic Fungi from Soil Spiked with SAB Diesel Fuel Biodegradation Screening
Medium to high levels of SAB diesel fuel contamination selected for a change in fungal diversity toward isolates similar to previously described hydrocarbon degrading species (Table 3, Figure 2). Most notable was Pseudeurotium bakeri (CZ03), which was found to dominate the soil samples as the concentration of SAB diesel fuel increased above 1,000-mg/kg soil (Table 2). A diverse range of fungi were isolated from soil spiked with high concentrations of SAB diesel fuel exceeding 10,000-mg/kg, including Arthroderma sp., Aspergillus sp., Geomyces sp., Exophiala sp., Penicillium sp., and two previously unidentified species (CZ40 and CZ42; Table 3).
A selection of 12 of an initial 20 fungal isolates recovered from pristine and contaminated soils was capable of growth in liquid media. These 12 isolates were examined for the ability to grow in the presence SAB diesel fuel as the only carbon source. The addition of a redox indicator was also used to screen isolates for hydrocarbon biodegradation activity (Table 5). A range of biodegradation activity was observed for the fungal isolates tested. Novel species from within the Exophiala (CZ06), Arthroderma (CZ02), and Penicillium (CZ36) genera did exhibit the ability to grow in the presence of SAB diesel fuel and require further characterization. Amongst those capable of the greatest biodegradation activity observed, Trichophyton sp. (CZ13) was isolated from non-contaminated soil. Despite Penicillium coprophilum (CZ37) and Neonectria radicicola (CZ22) both being isolated from soil spiked with high concentrations of SAB diesel fuel no biodegradation activity was observed for these isolates.
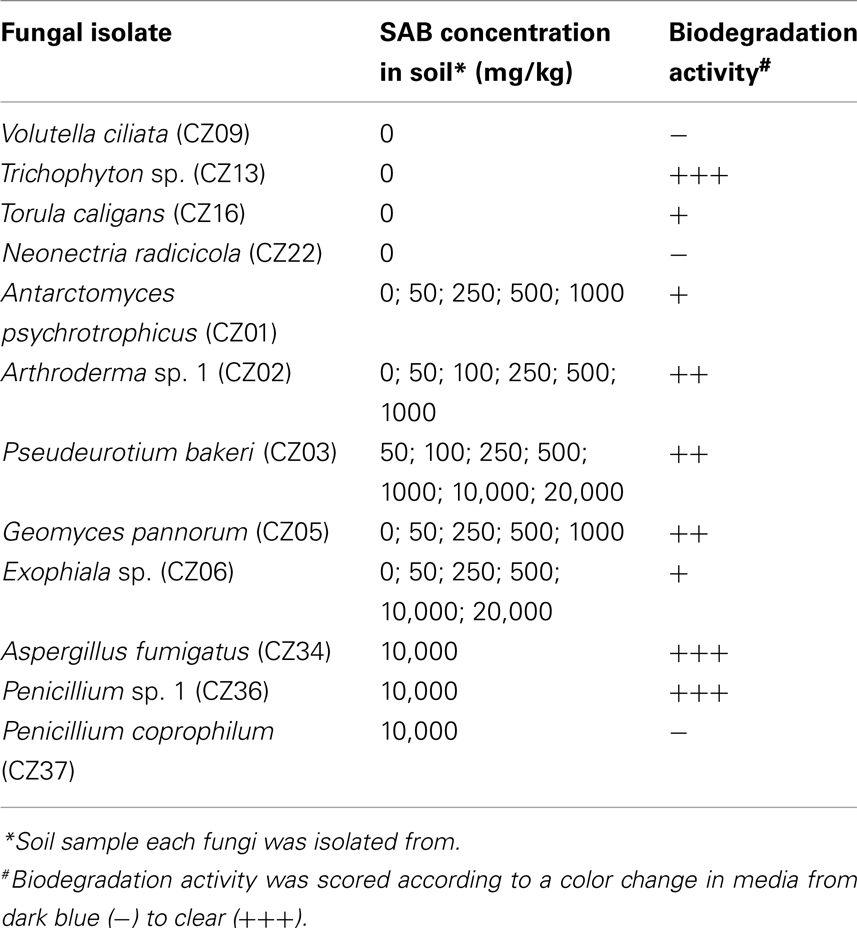
Table 5. Biodegradation activity of a selection of isolates with SAB diesel fuel as the sole carbon source.
Discussion
A re-emergence in the development of bacterial cultivation strategies over the last decade has resulted in the successful cultivation of predominantly slow-growing, previously undescribed species from a range of bacterial phylum (Janssen et al., 2002; Kaeberlein et al., 2002; Rappe et al., 2002; Ferrari et al., 2005). Here, the combination of a low nutrient and a traditional high nutrient approach to the cultivation of fungi resulted in 91 yeast and filamentous fungi being isolated from Macquarie Island soil. The application of the low nutrient approach to the cultivation of soil fungi resulted in a significantly different population being recovered from both non-contaminated and highly contaminated soils then that recovered using the high nutrient media approach only (Figure 1). This agrees with previous findings when a high-throughput dilution-to-extinction technique originally developed for bacteria was adapted for the cultivation of fungi from environmental samples (Collado et al., 2007). The diversity of fungal species recovered from Macquarie Island soil here highlights the fact that novel culturing approaches should be widely adopted for fungi. Their exists the potential to adapt the cultivation approach further to include a range of media and to investigate the effect of lowering the concentration of nutrients further (Watve et al., 2000; Ferrari and Gillings, 2009). Moreover, simulating natural environments using novel microcosm-based approaches may also increase cultivable diversity further and should also be developed for fungi.
Macquarie Island hosts a diverse group of yeast and filamentous fungi. Yet a high proportion of isolates (69%) could not been identified to the species level in this study using standard molecular techniques targeting the highly variable ITS region (Table 3). While the ITS region has been the most widely adopted molecular marker for fungal taxonomic and phylogenetic studies, of the 1.5 million extant fungal species, only 14,000 sequences have been deposited into publicly available sequence databases (Begerow et al., 2010). Like bacterial phylogenetics, this is not surprising as widely utilized fungal primer sets were designed using a limited diversity of sequences that were available over two decades ago (O’Brien et al., 2005; Begerow et al., 2010; Mohamed and Martiny, 2011). Molecular targets such as the 18S rDNA, 28S rDNA, and mitochondrial cytochrome C oxidase I gene (COI) genes offer potential alternatives for the identification of a range of isolates but there are even greater limitations on the diversity of sequence data available in public databases. Therefore, there is a requirement to design updated ITS primer sets that target a greater range of species belonging to the fungal domain (Guarro et al., 1999).
Fungal biodiversity obtained from sub-Antarctic Macquarie Island soil confirmed the presence of both indigenous and widely distributed cosmopolitan species that have been identified in Antarctic regions (Ruisi et al., 2007). Both yeasts and filamentous fungi were recovered from predominantly the Ascomycota phylum, as well as a small proportion from the Basidiomycota and the Mucoromycotina sub-phylum incertae sedis. The isolation of Geomyces pannorum, Antarctomyces psychrotrophicus, and Cryptococcus sp. supports the hypothesis that these species are endemic or indigenous to Antarctic and sub-Antarctic regions (Arenz et al., 2006, 2011; Hughes et al., 2007; Ruisi et al., 2007). Antarctomyces psychrotrophicus has been repeatedly reported as an Antarctic indigenous fungal species and this is the first report of the species being isolated from a sub-Antarctic region. While cosmopolitan species, those found widely distributed in a range of temperate environments, were also isolated including the filamentous fungus Cladosporium cladosporioides and the relatively common Basidiomycete yeast Sporobolomyces sp. (Blanchette et al., 2010; Hughes and Bridge, 2010).
The majority of fungi that have been reported to be involved in aromatic hydrocarbon breakdown have been found to belong to two major genera, Cladophialophora and Exophiala (Prenafeta-Boldu et al., 2001, 2006). While there have also been limited reports on the tolerance and potential for petroleum hydrocarbon biodegradation by the Aspergillus, Penicillium, Phialophora, Pseudeurotium, Trichophyton, and Geomyces genera following their isolation from contaminated soils (Leahy and Colwell, 1990; Prenafeta-Boldu et al., 2006; Hughes et al., 2007). A major shift in fungal diversity toward Geomyces pannorum, Pseudeurotium bakeri, Arthroderma sp., and Penicillium sp. in SAB diesel fuel contaminated soils here combined with an increase in total colony abundance is a phenomenon that has been observed previously in Antarctic soils (Aislabie et al., 2001). Most notable was the dominant species Pseudeurotium bakeri, which thrived in the presence of high concentrations of SAB diesel fuel with 100-fold more colonies detected compared to that found in soil containing lower levels of contamination.
At mid-high levels of SAB diesel fuel contamination (500–10,000-mg/kg soil), Arthroderma sp. 1 and 2 (CZ02 and CZ29), and Exophiala sp. (CZ06) were isolated in higher numbers than found at lower levels of contamination, and both Exophialia sp. and Arthroderma sp. 1 exhibited biodegradation activity. Isolates within the Arthoderma sp. and Exophiala sp. genera have previously been isolated from a range of environments polluted with both monoaromatic hydrocarbons and alkanes, yet they have also been shown to play a role in clinical infections (Cox et al., 1993; Prenafeta-Boldu et al., 2001, 2006; Ulfig et al., 2002; De Hoog et al., 2006). Two novel species; Penicillium sp. 1 (CZ36) and Geomyces sp. 4 (CZ38) and two unidentified fungi (CZ40 and CZ42) were recovered from highly contaminated soils and are considered here to be candidates for characterization for a role in hydrocarbon biodegradation. Interestingly, several novel isolates were observed to be inhibiting bacterial growth and will also be examined for their potential for the production of antibacterial agents (Zhang pers comm.).
Antarctic and sub-Antarctic regions are becoming increasingly important for bioprospecting for novel fungal species, as the potential exists for the isolation of cold-adapted enzymes for the biotechnological industry. Cold-active enzymes provide energy savings to processes at low temperature, as they are more productive than their mesophilic or thermophilic homologs (Cavicchioli et al., 2011). In this study, several novel and previously described hydrocarbon degrading fungal species were isolated from Macquarie Island. A selection of isolates that exhibited the ability to assimilate petroleum hydrocarbons in the form of SAB diesel fuel will now be characterized further to determine their role in hydrocarbon degradation. The potential now exists for the identification and future exploitation of cold-adapted enzymes from these isolates for applications into the biotechnology industry.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Professor Ian Snape and the Australian Antarctic Division (AAD) for their invaluable support, involving financial assistance, access to soil samples through expeditions to Macquarie Island, and the analysis of TPH concentrations in spiked soil samples. We would also like to thank UNSW for financial support from internal grant applications and Lily Zhang for her technical assistance.
References
Aislabie, J., Fraser, R., Duncan, S., and Farrell, R. E. (2001). Effects of oil spills on microbial heterotrophs in Antarctic soils. Polar Biol. 24, 308–313.
Arenz, B. E., Held, B. W., Jurgens, J. A., and Blanchette, R. A. (2011). Fungal colonization of exotic substrates in Antarctica. Fungal Divers. 49, 13–22.
Arenz, B. E., Held, B. W., Jurgens, J. A., Farrell, R. L., and Blanchette, R. A. (2006). Fungal diversity in soils and historic wood from the Ross Sea Region of Antarctica. Soil Biol. Biochem. 38, 3057–3064.
Begerow, D., Nilsson, H., Unterseher, M., and Maier, W. (2010). Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl. Microbiol. Biotechnol. 87, 99–108.
Blanchette, R. A., Held, B. W., Arenz, B. E., Jurgens, J. A., Baltes, N. J., Duncan, S. M., and Farrell, R. L. (2010). An Antarctic hot spot for fungi at Shackleton’s historic hut on Cape Royds. Microb. Ecol. 60, 29–38.
Cavicchioli, R., Charlton, T., Ertan, H., Mohd Omar, S., Siddiqui, K. S., and Williams, T. J. (2011). Biotechnological uses of enzymes from psychrophiles. Microb. Biotechnol. 4, 449–460.
Clarke, K. R., and Green, R. H. (1998). Statistical design and analysis for a “biological effect” study. Mar. Ecol. Prog. Ser. 46, 213–226.
Collado, J., Platas, G., Paulus, B., and Bills, G. F. (2007). High-throughput culturing of fungi from plant litter by a dilution-to-extinction technique. FEMS Microbiol. Ecol. 60, 521–533.
Cox, H. H. J., Houtman, J. H. M., Doddema, H. J., and Harder, W. (1993). Growth of the black yeast exophiala jeanselmei on styrene and styrene-related compounds. Appl. Microbiol. Biotechnol. 39, 372–376.
De Hoog, G. S., Zeng, J. S., Harrak, M. J., and Sutton, D. A. (2006). Exophiala xenobiotica sp. nov., an opportunistic black yeast inhabiting environments rich in hydrocarbons. Antonie van Leeuwenhoek 90, 257–268.
Ferrari, B., Binnerup, S. J., and Gillings, M. R. (2005). Microcolony cultivation on a soil substrate membrane system recovers previously unculturable bacteria. Appl. Environ. Microbiol. 71, 8174–8720.
Ferrari, B. C., and Gillings, M. (2009). Viability selection combined with the SSMS and micromanipulation results in the growth of fastidious bacteria from soil. Appl. Environ. Microbiol. 75, 3352–3354.
Ferrari, B. C., Power, M. L., and Bergquist, P. L. (2007). Closed-tube DNA extraction using a thermostable proteinase is highly sensitive, capable of single parasite detection. Biotechnol. Lett. 29, 1831–1837.
George-Okafor, U., Tasie, F., and Muotoe-Okafor, F. (2009). Hydrocarbon degradation potentials of indigenous fungal isolates from petroleum contaminated soils. J. Phys. Nat. Sci. 3, 1–6.
Guarro, J., Gene, J., and Stchigel, A. M. (1999). Developments in fungal taxonomy. Clin. Microbiol. Rev. 12, 454–500.
Hanson, K. G., Desai, J. D., and Desai, A. J. (1993). A rapid and simple screening technique for potential crude oil degrading microorganism. Biotechnol. Tech. 7, 745–748.
Hawksworth, D. L. (2001). The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol. Res. 105, 1422–1432.
Hughes, K. A., and Bridge, P. (2010). “Tolerance of Antarctic soil fungi to hydrocarbons and their potential role in soil bioremediation,” in Polar Microbiology: The Ecology, Biodiversity and Bioremediation Potential of Microorganisms in Extremely Cold Environments, eds A. K. Bej, J. Aislabie, and R. M. Atlas (Boca Raton, FL: Taylor and Francis), 277–300.
Hughes, K. A., Bridge, P., and Clark, M. S. (2007). Tolerance of Antarctic soil fungi to hydrocarbons. Sci. Total Environ. 372, 539–548.
Imad, Z., Mohammed, J., and Mohammed, I. E. A. (2010). Efficiency of IRAP and ITS-RFLP marker systems in accessing genetic variation of Pyrenophora graminea. Genet. Mol. Biol. 33, 328–332.
Janssen, P. H., Yates, P. S., Grinton, B. E., Taylor, P. M., and Sait, M. (2002). Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68, 2391–2396.
Kaeberlein, T., Lewis, K., and Epstein, S. (2002). Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296, 1127–1129.
Kerry, E. (1990). Effects of temperature on growth rates of fungi from subantarctic Macquarie Island and Casey, Antarctica. Polar Biol. 10, 293–299.
Korabecna, M. (2007). “The variability in the fungal ribosomal DNA (ITS1, ITS2, and 5.8S rRNA gene): its biological meaning and application in medical mycology,” in Communicating Current Research and Educational Topics and Trends in Applied Microbiology, ed. A. Méndez-Vilas (Badajoz: Formatex), 783–787.
Leahy, J. G., and Colwell, R. R. (1990). Microbial degradation of hydrocarbons in the environment. Microbiol. Rev. 54, 305–315.
Martin, K. J., and Rygiewicz, P. T. (2005). Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 5, 28. doi:10.1186/1471-2180-5-28
Mohamed, D. J., and Martiny, J. B. H. (2011). Patterns of fungal diversity and composition along a salinity gradient. ISME J. 5, 379–388.
O’Brien, H. E., Parrent, J. L., Jackson, J. A., Moncalvo, J., and Vilgalys, R. (2005). Fungal community analysis by large-scale sequencing of environmental samples. Appl. Environ. Microbiol. 71, 5544–5550.
Prenafeta-Boldu, F. X., Kuhn, A., Luykx, D. M. A. M., Anhe, H., Van Groenestijn, J. W., and De Bont, J. A. M. (2001). Isolation and characterisation of fungi growing on volatile aromatic hydrocarbons as their sole carbon and energy source. Mycol. Res. 105, 477–484.
Prenafeta-Boldu, F. X., Summerbell, R., and Sybren De Hoog, G. (2006). Fungi growing on aromatic hydrocarbons: biotechnology’s unexpected encounter with biohazard? FEMS Microbiol. Rev. 30, 109–130.
Rappe, M. S., Connon, S. A., Vergin, K. L., and Giovannoni, S. L. (2002). Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418, 630–633.
Rayner, J. L., Snape, I., Walworth, J. L., Harvey, P. M., and Ferguson, S. H. (2007). Petroleum-hydrocarbon contamination and remediation by micro-bioventing at sub-Antarctic Macquarie Island. Cold Reg. Sci. Technol. 48, 139–153.
Ruisi, S., Barreca, D., Selbmann, L., Zucconi, L., and Onofri, S. (2007). Fungi in Antarctica. Rev. Environ. Sci. Biotechnol. 6, 127–141.
Taylor, J. W., Jacobson, D. J., Kroken, S., Kasuga, T., Geiser, D. M., Hibbett, D. S., and Fischer, M. C. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31, 21–32.
Ulfig, K. G. P., Worsztynowicz, A., Manko, T., Tien, A. J., and Brigmon, R. L. (2002). Keratinolytic fungi as indicators of hydrocarbon contamination and bioremediation progess in a petroleum refinery. Pol. J. Environ. Stud. 12, 245–250.
Keywords: novel cultivation, fungi, sub-Antarctic, soil, diversity, hydrocarbon degradation
Citation: Ferrari BC, Zhang C and van Dorst J (2011) Recovering greater fungal diversity from pristine and diesel fuel contaminated sub-Antarctic soil through cultivation using both a high and a low nutrient media approach. Front. Microbio. 2:217. doi: 10.3389/fmicb.2011.00217
Received: 23 June 2011;
Accepted: 11 October 2011;
Published online: 15 November 2011.
Edited by:
Aurelio Briones, University of Idaho, USAReviewed by:
Liang Shi, Wright State University, USAZiyu Dai, Pacific Northwest National Laboratory, USA
Copyright: © 2011 Ferrari, Zhang and van Dorst. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Belinda C. Ferrari, School of Biotechnology and Biomolecular Sciences, University of New South Wales, Randwick, NSW 2052, Australia. e-mail: b.ferrari@unsw.edu.au