- 1 Federal Institute for Geosciences and Natural Resources, Hannover, Germany
- 2 Faculty of Natural Sciences, Leibniz Universität Hannover, Hannover, Germany
A cold methane seep was discovered in a forearc sediment basin off the island Sumatra, exhibiting a methane-seep adapted microbial community. A defined seep center of activity, like in mud volcanoes, was not discovered. The seep area was rather characterized by a patchy distribution of active spots. The relevance of anaerobic oxidation of methane (AOM) was reflected by 13C-depleted isotopic signatures of dissolved inorganic carbon. The anaerobic conversion of methane to CO2 was confirmed in a 13C-labeling experiment. Methane fueled a vital microbial community with cell numbers of up to 4 × 109 cells cm−3 sediment. The microbial community was analyzed by total cell counting, catalyzed reporter deposition–fluorescence in situ hybridization (CARD–FISH), quantitative real-time PCR (qPCR), and denaturing gradient gel electrophoresis (DGGE). CARD–FISH cell counts and qPCR measurements showed the presence of Bacteria and Archaea, but only small numbers of Eukarya. The archaeal community comprised largely members of ANME-1 and ANME-2. Furthermore, members of the Crenarchaeota were frequently detected in the DGGE analysis. Three major bacterial phylogenetic groups (δ- Proteobacteria, candidate division OP9, and Anaerolineaceae) were abundant across the study area. Several of these sequences were closely related to the genus Desulfococcus of the family Desulfobacteraceae, which is in good agreement with previously described AOM sites. In conclusion, the majority of the microbial community at the seep consisted of AOM-related microorganisms, while the relevance of higher hydrocarbons as microbial substrates was negligible.
Introduction
Anaerobic oxidation of methane (AOM) has been described for decades, but became accepted as a key process in anaerobic carbon cycling only during the last 15 years (Davis and Yarbrough, 1966; Barnes and Goldberg, 1976; Reeburgh, 1976, 2007; Knittel and Boetius, 2009). So far, it has been observed in many marine environments contributing significantly to carbon cycling in the sediments and the reduction of methane emissions. Continental margins and their forelands were examined in numerous biogeochemical studies (e.g., Bohrmann et al., 1998; Reed et al., 2002; Inagaki et al., 2006) and AOM was associated mainly to gas seeps and mud volcanoes (Aloisi et al., 2000; Joye et al., 2004; Valentine et al., 2005; Niemann et al., 2006b; Lösekann et al., 2007). Such cold seep ecosystems, especially in the Eel River Basin (Orphan et al., 2002), the Hydrate Ridge (Bohrmann et al., 1998), the Black Sea (Michaelis et al., 2002), and the Gulf of Mexico (Roberts and Aharon, 1994), have been intensively studied regarding the geochemistry and microbiology of AOM. Within these AOM ecosystems, ANaerobic MEthanotrophs (ANME) populations are often the prevailing constituents and drive the biogeochemical processes.
In sediment systems with diffusive methane fluxes, the distribution of ANME is restricted to the sulfate–methane transition zone (SMTZ), the only place where both methane and sulfate are available. The ANME populations and their sulfate-reducing partner bacteria are principally the same as those at cold seeps. However, cell numbers and activities of AOM-related populations are significantly lower (Knittel and Boetius, 2009).
Although the importance of AOM in a global context has been widely recognized, the process is on a mechanistical and physiological level still not very well understood (Thauer, 2011). In the initial AOM reports, methanogenesis and sulfate reduction were believed to be mutually exclusive processes (Martens and Berner, 1974). However, AOM coincides with methanogenesis (Krüger et al., 2005; Niemann et al., 2006b), and was therefore proposed to be reverse methanogenesis (Krüger et al., 2003; Scheller et al., 2010). So far, it was demonstrated that sulfate and nitrite reduction couple to AOM as a joint process of specialized methane oxidizers and sulfate- or nitrite-reducing microorganisms (Boetius et al., 2000; Raghoebarsing et al., 2006; Ettwig et al., 2010). Metal reduction coupled to AOM was also suggested (Beal et al., 2009). Phylogenetic analysis of AOM-sediments identified three novel groups of so-called ANME-Archaea, ANME-1, ANME-2, and ANME-3. These ANME are distantly related to cultivated methanogenic members from the orders Methanosarcinales and Methanomicrobiales (Orphan et al., 2002; Knittel et al., 2005; Niemann et al., 2006b). FISH techniques showed that ANME occur in aggregates (Boetius et al., 2000; Michaelis et al., 2002; Knittel et al., 2005) with bacteria related to Desulfosarcina–Desulfococcus or Desulfobulbus. These findings suggest that AOM coupled to sulfate reduction is a syntrophic process, in which ANME convert methane to a metabolite which is used as electron donor by the sulfate-reducing bacterial partner.
The Sumatra forearc is spatially remote from previous study sites. Therefore, our main objective on the R/V Sonne cruise SO189-2 into the Sumatra forearc basins was to detect and investigate methane seeps on the seafloor, and to describe the related geochemical and microbiological features in comparison to background sediments of nearby sites. After the successful first-time discovery of a methane-driven cold seep in this geographical region, we analyzed different biogeochemical proxies for AOM to determine related microbial activities. This was combined with molecular biological methods to study the involved microbial populations, to allow a comparison of the present results with those already published on other seep sites worldwide.
Materials and Methods
Site Description and Sampling
General features of the Simeulue and the Nias Basin in the Sumatra forearc are described in Sieh and Natawitjaja (2000) and Schippers et al. (2010). Focus of this study is an area without seismic reflections (“seismic blanking”) discovered SE of the Simeulue basin, indicating free gases in the sediment (Figures 1 and 2). Measurements of dissolved gas of samples taken from the water column indicated elevated methane concentrations in the water column in this area (Wiedicke-Hombach et al., 2006). Additionally, pictures and samples from the seafloor revealed a typical seep fauna comprising mainly bivalves and white crabs (Martin and Haney, 2005), colonizing outcropping carbonate precipitations, and the surrounding sediments.
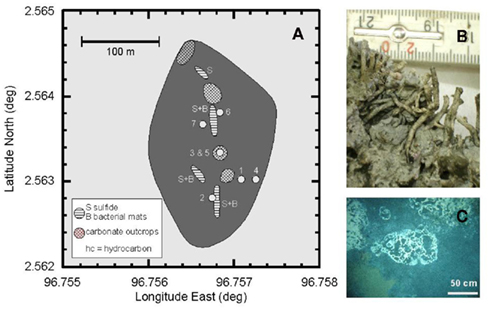
Figure 1. (A) Map of the methane seep in the Simeulue basin off Sumatra. The area without seismic reflections in the sediment is shaded in dark gray. Circles indicate sampling stations according to the ship’s position. Wire mesh areas depict carbonate outcrops and horizontal lines sulfide-rich surfaces “S” with microbial mats “B.” (B) Unclassified tubeworms found at the carbonate outcrops. (C) White bacterial mats near the sediment surface of the station 2.
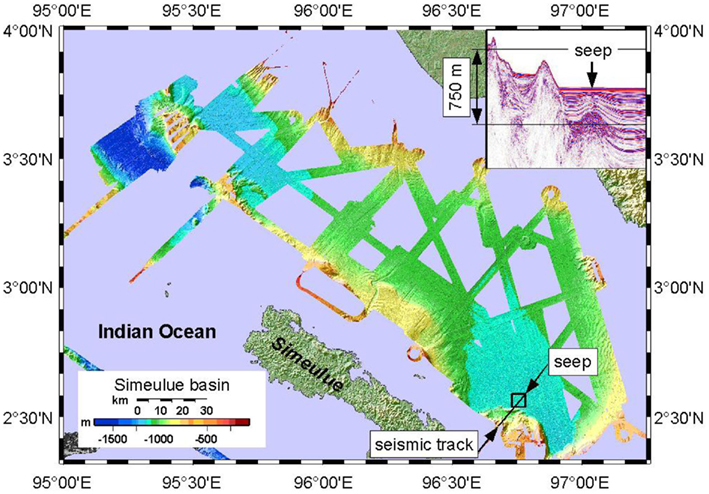
Figure 2. Bathymetric map of the Simeulue basin with the seep area in the SE. The insert (covering the island Sumatra) shows a seismic profile of the track drawn in the SE. 750 m is an approximate value.
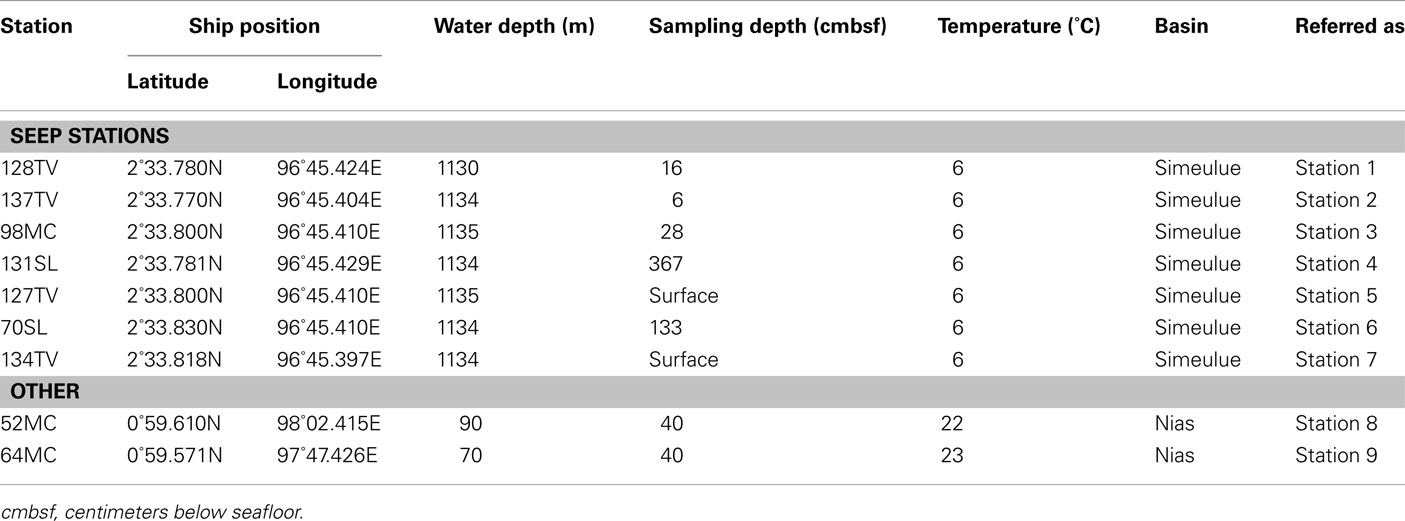
During the R/V Sonne cruise SO189-2 in 2006, nine sediment stations were sampled at the suspected seep area in the Simeulue basin, a reference site, and also in the Nias basin as summarized in Table 1. The sediment was sampled using a 6-m long gravity corer (SL), a 0.5 m long multicorer (MC), and a TV camera guided 0.9 m × 0.9 m grab (TV). The sediment in the TV-grab was further sampled using hand-held small pushcores.
Biological Samples
Immediately after porewater sampling (see Geochemical Analyses), approx. 300–400 ml sediment samples of multicorer or 20–100 ml sediment of gravity cores were collected in glass bottles. Bottles were sealed with butyl rubber stoppers and plastic screw caps. The headspace was flushed with nitrogen gas. These “live” samples for sediment microcosm were stored and transported at 4°C until further on-shore processing. For quantitative real-time PCR (qPCR) and denaturing gradient gel electrophoresis (DGGE) community analysis, parallel samples were frozen onboard at −20°C. For fluorescence microscopic methods, samples were treated as described below.
Incubation Experiments to Determine Microbial Activity in Sediment Microcosms
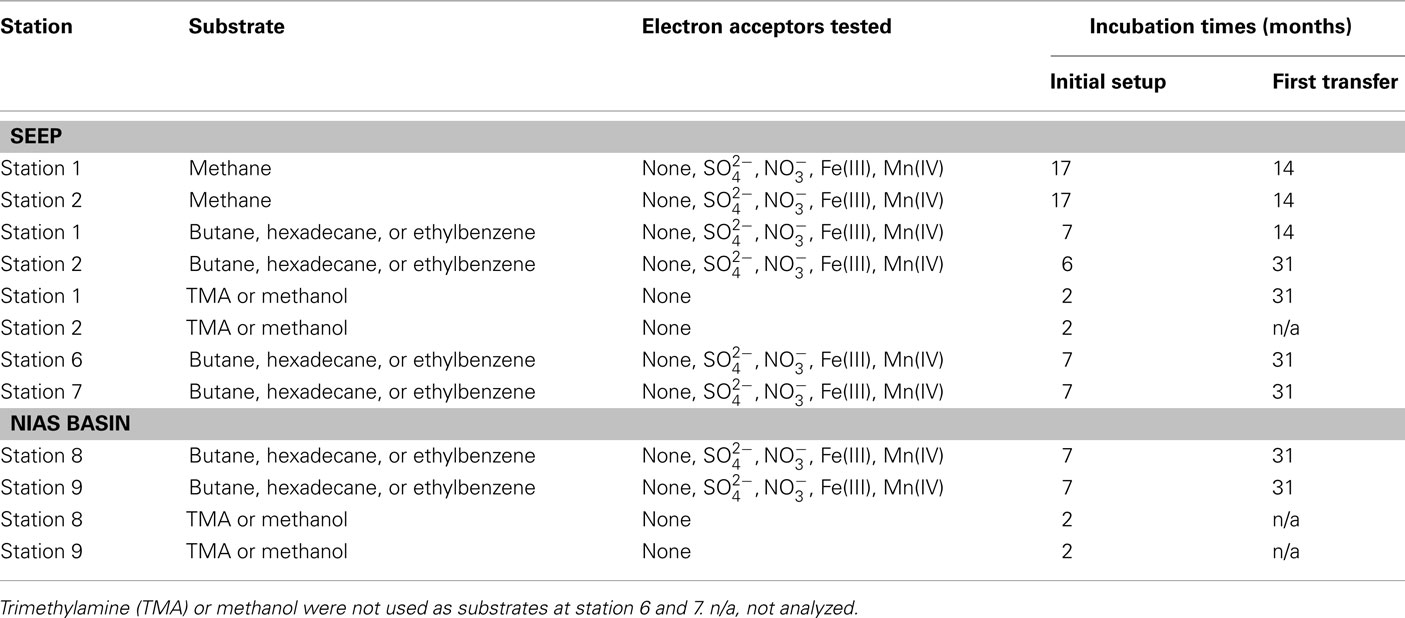
An overview over the experimental setup of the microcosms is displayed in Table 2.
General slurry preparation and microcosm inoculation
To prepare slurries for inoculation, sediment samples were mixed 1:1 with artificial seawater medium (Widdel and Bak, 1992). Subsequently, 10 ml of sediment slurry were added to 20 ml of medium, resulting in 5 ml sediment per bottle. Serum bottles of 60 ml volume were used. All manipulations were carried out under dinitrogen atmosphere in an anoxic glove box at room temperature. The headspace of the incubation tubes consisted of either methane (100%) or N2/CO2 (90%/10%v/v). These microcosms were incubated horizontally in the dark without shaking at in situ temperature which was either 6 or 23°C (Table 1).
Monitoring of AOM microcosms and determination of sulfate reduction rates
Anaerobic oxidation of methane was measured in vitro by methane-dependent sulfate reduction unless stated otherwise (Nauhaus et al., 2002; Krüger et al., 2005; Treude et al., 2005). Sulfide concentrations were determined spectrophotometrically by the formation of copper sulfide (Cord-Ruwisch, 1985). Sulfide concentrations of methane amended microcosms were subtracted from sulfide concentrations of controls without methane before rates were calculated. Rates are given in μmol cm−3 day−1 wet sediment and deviations are expressed as 95% confidence intervals unless stated otherwise.
AOM labeling experiment with 13C-methane
For further verification of AOM activity, 13C-labeled methane (17%v/v) was added to two first transfer microcosms (stations 1 and 2). After 14 months of incubation of the transfers, a 10-ml headspace sample was precipitated in 1 ml concentrated BaOH solution. The obtained BaCO3 suspension was transferred to a helium flushed vial and acidified with 100 μl concentrated HCl. To determine the δ13C value, a nearly methane-free headspace sample was injected into a MAT 252 GC-IRMS. Mean δ13C values were obtained from three individual batch cultures.
Determination of methanogenesis from TMA, methanol, and different hydrocarbons
Potential rates of methane production were determined in microcosms without substrates, with addition of typical marine substrates trimethylamine (TMA) and methanol, or selected hydrocarbons. Final concentrations in the microcosms for TMA and methanol were 1 and 2.4 mM. To demonstrate hydrocarbon-dependent methanogenesis, anaerobic microcosms with butane, hexadecane, and ethylbenzene were prepared. Butane was added by replacing the microcosm headspace completely by gaseous butane. Hexadecane or ethylbenzene were added directly to the medium in concentrations of 0.1%v/v. Methane was measured using a GC–FID equipped with a silica gel column (SRI 8610 C, SRI Instruments, USA). Rates are given in μmol cm−3 day−1. To simulate marine in situ conditions, all initial setups contained sulfate (28 mM) as electron acceptor. Alternatively applied electron acceptors to sulfate were nitrate (1 mM), ferrihydrite (2.5 mmol−1), or manganese dioxide (1.2 mmol−1). Stock solutions of the metal oxides were prepared as follows: ferrihydrite was precipitated by neutralization of a FeCl3 solution (Lovley and Phillips, 1986). Manganese dioxide was prepared by oxidation of a MnCl2 solution with KMnO4 (Lovley and Phillips, 1988). Hydrocarbon degradation was not measured directly. Methanogenesis served as proxy for anaerobic hydrocarbon degradation (Zengler et al., 1999; Siegert et al., 2011). Methane concentrations were measured after 2 months (TMA and methanol) or 7 months (hydrocarbons).
Determination of Cell Numbers by SYBR green® and CARD–FISH
For SYBR Green® total cell counts and catalyzed reporter deposition–fluorescence in situ hybridization (CARD–FISH), fixations were carried out immediately after sampling. Two fixatives, either formaldehyde (FA) or ethanol, were applied each. A volume of 1 cm3 wet sediment was treated for 10–15 h at 4°C with 1 ml fixative solution [4%w/v FA in phosphate buffered saline (PBS)], removed in two centrifugation steps by washing with 1 ml PBS and stored at −20°C in 50%v/v ethanol/PBS. Samples were transported to the home laboratory on dry ice. For total cell counts, in each sample 800–1000 cells were counted after SYBR Green® staining according to Weinbauer et al. (1998). CARD–FISH counts were conducted as described in Pernthaler et al. (2002) and Schippers et al. (2005).
Gene Quantification by qPCR
DNA extraction was carried out using a Fast DNA for Soil Kit (BIO 101, MP Biomedicals, Germany). To block sedimentary nucleic acid binding capacities, 10 μl of a 1% polyadenylic acid solution were added in the initial step (Webster et al., 2003). Directly before PCR, 125 μl of 0.3% bovine serum albumine (BSA) in ultra pure water were added as blocking agent to the Taqman master mix (Applied Biosystems, Germany) or the SYBR green® master mix (Eurogenetec, Germany). A real-time PCR instrument (ABI Prism 7000, Applied Biosystems) was employed to determine the 16S rRNA gene copy numbers of Archaea (Takai and Horikoshi, 2000) and Bacteria (Nadkarni et al., 2002). Eukaryotic 18S rRNA genes were quantified according to the manual’s instructions of the kit and cited literature therein (Applied Biosystems, 2002). The functional genes dsrA and mcrA were investigated according to Schippers and Nerretin (2006) and Nunoura et al. (2006) respectively. Specific functional mcrA genes from anaerobic methanotrophic Archaea ANME-1 and ANME-2 were quantified using assays of Nunoura et al. (2006). Values are expressed in copy numbers per cm3 wet sediment.
Denaturing Gradient Gel Electrophoresis
For DGGE, 16S rRNA genes of Bacteria were amplified using the primer set 533F (5′-TTACCGCGGCTGCTGGCAC-3′)/907R (5′-AATTCCTTTGAGTTT-3′) with a GC-clamp (5′-CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCC-3′) attached to the reverse primer 907R at the 5′-end (Weisburg et al., 1991; Muyzer et al., 1995). A thermocycler was programmed for a hot start PCR and 35 repetitions of each cycle: 94°C for 1 min, 63°C for 0.75 min, and 72°C for 1.5 min. Archaeal 16S rRNA genes were amplified with the primer set 340F (5′-CCCTACGGGGYGCASCAG-3′) and 915R (5′-GTGCTCCCCCGCCAATTCCT-3′; Øvreås et al., 1997; Coolen et al., 2002), a GC-clamped (as for 907R) 340F forward primer and a cycler program with 35 cycles each: 94°C for 1 min, 60°C for 1 min, 72°C for 1.5 min. A total of 2 μg of amplified and purified DNA (QIA quick PCR Purification Kit, Qiagen, Germany) was applied to each pocket of the gel and separated in a DCode System (BioRad, Germany) for 20 h at 60°C. Denaturing gradients of urea/formamide were 30–70%w/v (20–80%w/v for Archaea) and the acrylamide concentration was 6.5%w/v. Dominant bands were excised and stored at −20°C, vacuum-dried at 60°C for 3 h (Eppendorf Concentrator 5301, Eppendorf, Germany), extracted with 30 μl PCR grade water (4°C, over night), re-amplified without GC-clamps, and purified as described above. Sequencing reactions were conducted at Seqlab GmbH (Göttingen, Germany). Primers were 533F, 550F (5′-CGGAATTATTGGGCGTAAAG-3′), and 906R (5′-CCCCAATTCCTTTGAGTTTC-3′) for bacterial 16S rRNA genes and 340F and 915R for archaeal 16S rRNA genes. The obtained DNA sequences were aligned online by ARB-SILVA1 and assembled in an existing Parsimony tree of a recent reference database (SSURef 102, February 2010) according to (Ludwig et al., 2004; Pruesse et al., 2007). All sequence data were submitted to the GenBank database2 under accession numbers HQ231693 (see text footnote 2) to HQ231746 (see text footnote 2).
Bathymetry and Parasound
A Kongsberg SMRAD EM 120 multibeam echosounder was operated continuously 24 h day−1 during the cruise. The device enabled mapping to full ocean depth with a sonar frequency of 12 kHz, an angular coverage of 150° and 191 beams per ping. The parasound device vessel integrate (Krupp Atlas Electronics, Germany) was used as a low frequency sediment echosounder at 18.0 and 21.5 kHz to map seismic profiles of basin sediments. The transparent character of the seafloor (seismic blanking) was found to be indicative for defining the extent of sediments affected by methane seepage and authigenic carbonate precipitation (Figure 2). However, seismic blanking may as well be indicative for high density gas hydrates overlaying low density free gas, unveiled by a bottom simulating reflector (BSR; Hyndman and Spence, 1992; Berndt et al., 2004). Nevertheless, seismic blanking does not always indicate free gas in the sediment (Zühlsdorff et al., 1999).
Geochemical Analyses
Duplicate sediment core samples were taken for microbiological and geochemical analyses. TOC was measured with the instrument LECO CS 200 (LECO Corporation, USA). Porewater was squeezed from sediment slices of up to 125 cm3 using a porewater press at atmospheric pressure. Between 20 and 40 ml of porewater were obtained and filtered through a 0.45 μm cellulose nitrate filter (Sartorius, Germany) and directly collected in polypropylene-vials. Polypropylene-vials were acid prewashed and dried to remove possible traces of carbonate from the vial surface. To remove particles and microorganisms, the collected porewater was filtered again through 0.2 μm polyethersulfone filters (Sartorius, Germany). For metal ion and sulfate concentrations, 5 ml porewater were acidified with 50 μl HNO3 and measured using an ICP-MS instrument (Perkin Elmer Sciex Elan 5000, USA) as published by Dekov et al. (2006). After preservation of the sediment samples with ZnCl2, porewater sulfide was determined spectrophotometrically as described by Cline (1969).
To determine carbon stable isotopes of dissolved inorganic carbon (DIC), 2 ml of porewater, treated with 10 μl saturated HgCl2 solution on board, were analyzed in a Finnigan MAT 252 (Thermo Electron, USA) connected to a Finnigan Kiel III (Thermo Electron) carbonate preparation device (Wachter and Hayes, 1985). Free gases were measured using 5 ml fresh sediment from the center of the core which was immediately transferred into 10 ml 2 M NaOH in a 56 ml serum bottle. The bottle was sealed quickly with a butyl rubber stopper and the sediment was suspended by shaking the flask. Immediately, 5 ml headspace were removed and stored upside down in 20 ml injection vials over saturated NaCl solution for on-shore measurement. This procedure allowed a separation of free from adsorbed gases but some adsorbed gas may have evaporated as well (Faber and Stahl, 1983). δ13C values for free gases were obtained by direct injection into a Finnigan MAT 253 (Thermo Electron, USA) connected to a gas chromatograph (GC 6890, Agilent, USA).
To determine the carbon stable isotopic composition of the macro fauna, the soft tissues of bivalves and crabs were freeze dried, milled, weighed into tin capsules, and stored in a desiccator until measurement. Samples were taken from one individual bivalve and two individual crabs. One crab was sampled twice and the other one was sampled once. For TOC carbon isotope ratios, dried sediment was treated with 10% HCl in acid washed tin capsules and stored in a desiccator until measurement. Isotope ratios were determined by a coupled system of an elemental analyzer and a MAT 252 isotope ratio mass spectrometer (Thermo Electron, USA) via a Finnigan ConFlo III open split interface. Concentrations and isotopic data are given as the arithmetic mean of at least two measurements. Isotopic carbon values are expressed in ‰ relative to Vienna PeeDeeBelemnite (VPDB). Concentrations are relative to 1 cm3 of fresh (wet) sediment.
Results
Seafloor Observations
Areas covered with white-colored microbial mats were discovered and sampled in the Simeulue seep area using a TV-guided grab (Table 1; Figure 1). In the seep area, parts of the sediment were characterized by a black color and a strong sulfidic odor. The higher seafloor biota comprised tubeworms, bivalves, and crabs. The latter colonized carbonate outcrops and could be identified as members of the species Shinkaia crosnieri (Enrique Macpherson, personal communication). Their soft tissue stable carbon isotopic signature ranged from −31 to −45‰. Soft tissue of one mytilid bivalve individual had a δ13C of −31‰.
Free Gases and Their Isotopic Composition
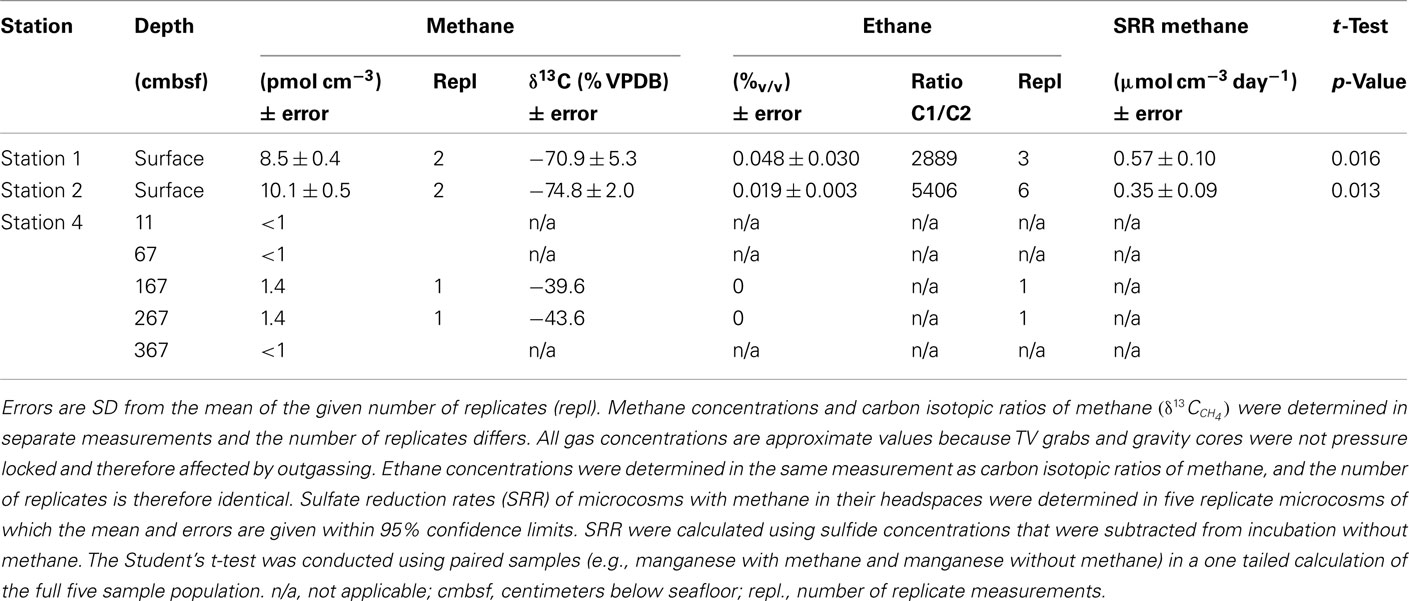
Free methane concentrations in the water column ranged from 10 to 200 nmol l−1 at the seep location as shown in Figure 1. In the southeast boundary of the seep, methane concentration never reached values above 5 nmol l−1. At the sediment surface of the station 1 and 2, methane accounted for a total of >99.9% of all hydrocarbon gases. Isotopic data of gases sampled at the methane seep are listed in Table 3.
Porewater Composition and Stable Carbon Isotopes
All geochemical porewater data of the Simeulue seep are summarized in Figure 3. At stations 1 and 2, δ13CDIC values were mainly below −40‰. At the station 3, δ13CDIC values were always above −8‰. At station 4, δ13CDIC values ranged from −2‰ at 6 cmbsf to −24‰ at 293 cmbsf. However, a clearly decreasing trend was observed with depth down to 392 cmbsf (Figure 3). Clear trends were missing in case of the solid phase δ13CTOC values. However, there was a slight decrease from −22.3‰ at 6 cmbsf to −27.6‰ at 153 cmbsf of the station 4. The values remained stable below −27.0‰ up to a depth of 293 cmbsf (Figure 3). At the stations 1 and 2, sulfate concentrations decreased with depth while sulfide concentrations increased. At the stations 3 and 4, neither such a decline of porewater sulfate concentrations was found, nor was sulfide detected. Elevated concentrations of reduced manganese were found at the station 2 as well as at the surface of the station 4 (Figure 3). Except for the station 1, no higher amounts of reduced iron in the porewater were observed (Figure 3).
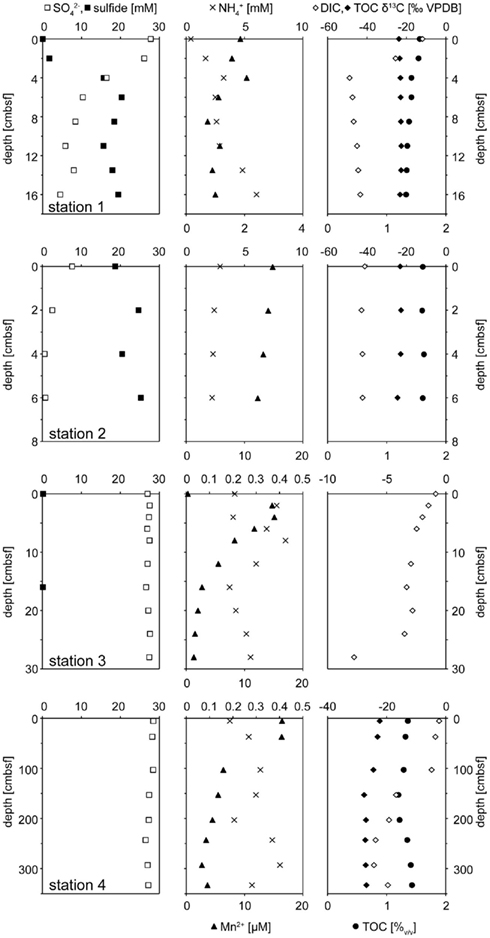
Figure 3. Geochemical profiles of stations 1 – 4. Stations 1 and 2 comprised ANME rich sediments actively performing AOM. The left panel shows sulfate (□) and sulfide (■) profiles. When sulfide concentrations were below the detection limit of 0.1 they are not sown. The central panel depicts dissolved manganese (▲) and ammonium (×). δ13C isotopic compositions of DIC (◊) and TOC (♦) are displayed on the right. The concentration of TOC (•) in the dry mass is given in (%).
Anaerobic Oxidation of Methane
Results obtained from microcosm experiments do not necessarily reflect the in situ situation but provide an estimate of microbial activity (Krüger et al., 2005; Treude et al., 2005). Sulfate reduction rates (SRR) in Simeulue seep sediment microcosms were higher with methane as electron donor than in controls without methane and the difference is shown in Table 3. The observed differences can be explained by AOM. However, sulfate reduction at station 1 and 2 without methane was 0.13 and 0.11 μmol cm−3 day−1, respectively. To confirm this, the conversion of methane into carbon dioxide was tested in 1:10 diluted subcultures of the initial microcosms incubated with 17%v/v 13C-methane. The label was recovered as 13C-enriched carbon dioxide. The detected 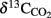 values were +503 ± 331‰ (station 1) and +319 ± 279‰ (station 2) for microcosms prepared from sulfidic sediments of the Simeulue methane seep. This provides evidence for the oxidation of 13C-methane to 13CO2 at both stations. In controls with unlabeled methane,
values were +503 ± 331‰ (station 1) and +319 ± 279‰ (station 2) for microcosms prepared from sulfidic sediments of the Simeulue methane seep. This provides evidence for the oxidation of 13C-methane to 13CO2 at both stations. In controls with unlabeled methane, 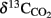 values were −15.7 and −17.4‰ respectively. The corresponding
values were −15.7 and −17.4‰ respectively. The corresponding 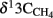 values of unlabeled methane were −36.6 and −34.9‰ respectively.
values of unlabeled methane were −36.6 and −34.9‰ respectively.
In 1:10 diluted subcultures of the initial microcosms, containing sulfate, nitrate, FeOOH, or MnO2, methanotrophy was observed after 14 months incubation as well. The methanotrophic rates, measured by headspace analysis of methane, were similar and ranged from 0.8 μmol cm−3 day−1 (with MnO2) at the station 1 to 1.8 μmol cm−3 day−1 (with sulfate) at station 2.
Methanogenesis from Trimethylamine (TMA), Methanol, Hydrocarbons
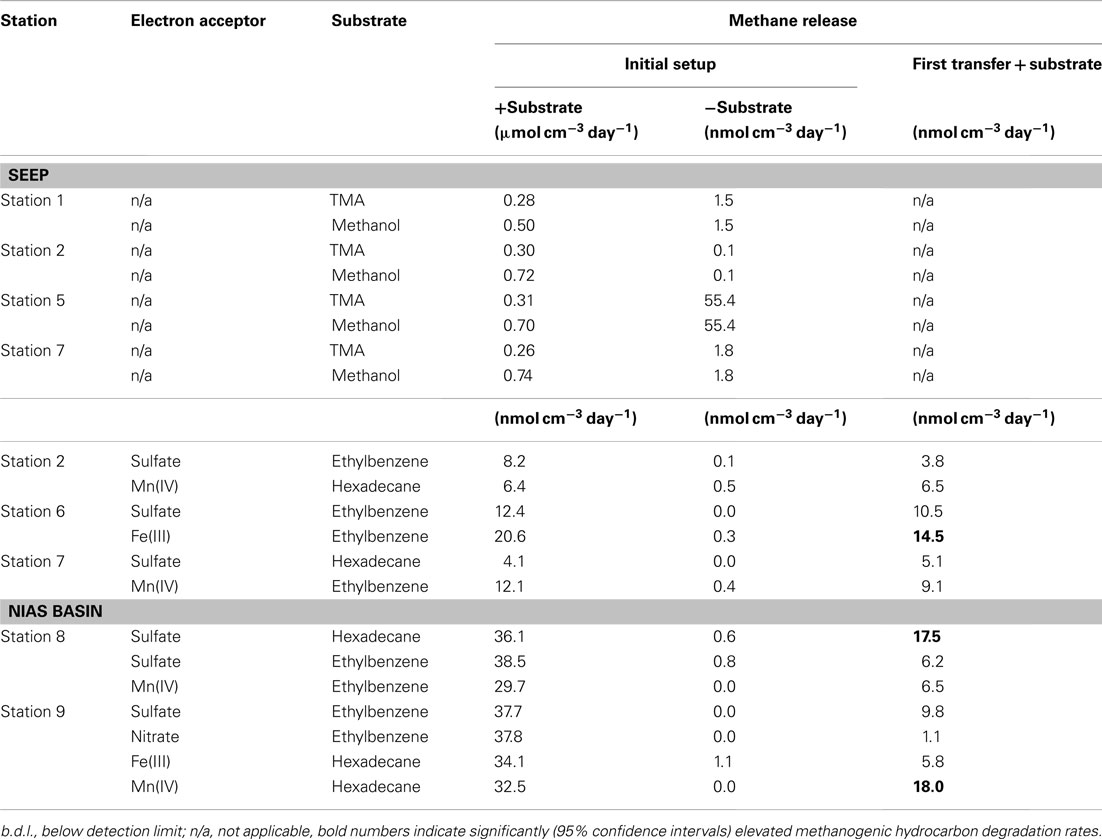
Except station 5, methanogenesis in microcosms without added methanogenic substrates (TMA, methanol, or hydrocarbons) was less than 1.8 nmol methane cm−3 day−1 (Table 4). However, when TMA or methanol were added to microcosms of the seep stations (1, 2, 5, and 7), methanogenesis increased by two orders of magnitude. The other seep stations (3, 4, and 6) were not tested for TMA or methanol methanogenesis. In microcosms prepared from sediments of the Nias basin, where no methane seepage was observed (stations 8 and 9), TMA or methanol did not stimulate methanogenesis. For the hydrocarbon substrates butane, hexadecane, and ethylbenzene, little methanogenesis was observed after 7 months incubation in 27 microcosms out of 280 initial microcosms (Table 4). To exclude false positive signals by means of stimulation of methanogenic TOC digesting microorganisms the hydrocarbon microcosms were compared with microcosms without hydrocarbons. For this, first 1:10 transfers of the methanogenic microcosms were prepared. In first transfers, the hydrocarbon-dependent methanogenesis rate at the seep station 2 was 6.5 nmol cm−3 day−1 (hexadecane and manganese dioxide) and 14.5 nmol cm−3 day−1 at station 6 (ethylbenzene and ferrihydrite). In the Nias basin, it was 17.5 nmol cm−3 day−1 at station 8 (hexadecane and sulfate) and 18.0 nmol cm−3 day−1 at station 9 (hexadecane and manganese dioxide).
Cell Numbers and Quantification of Functional Genes
Results of CARD–FISH and total cell counts for the stations 1, 3, and 4 are shown in Figure 4. At station 2, only the top and bottom layer were investigated and data for this station are therefore not depicted in Figure 4 but described in the following. At this station, total cell numbers of 1 × 109 cells cm−3 at the top (0 cmbsf) and 4 × 109 cells cm−3 at the bottom (8 cmbsf) were counted. CARD–FISH cell counts of active cells revealed cell numbers of 2 × 108 cells cm−3 for Bacteria and 4 × 107 cm−3 for Archaea at the station 2. At the stations 3 and 4, CARD–FISH indicated no living archaeal cells (Figure 4).
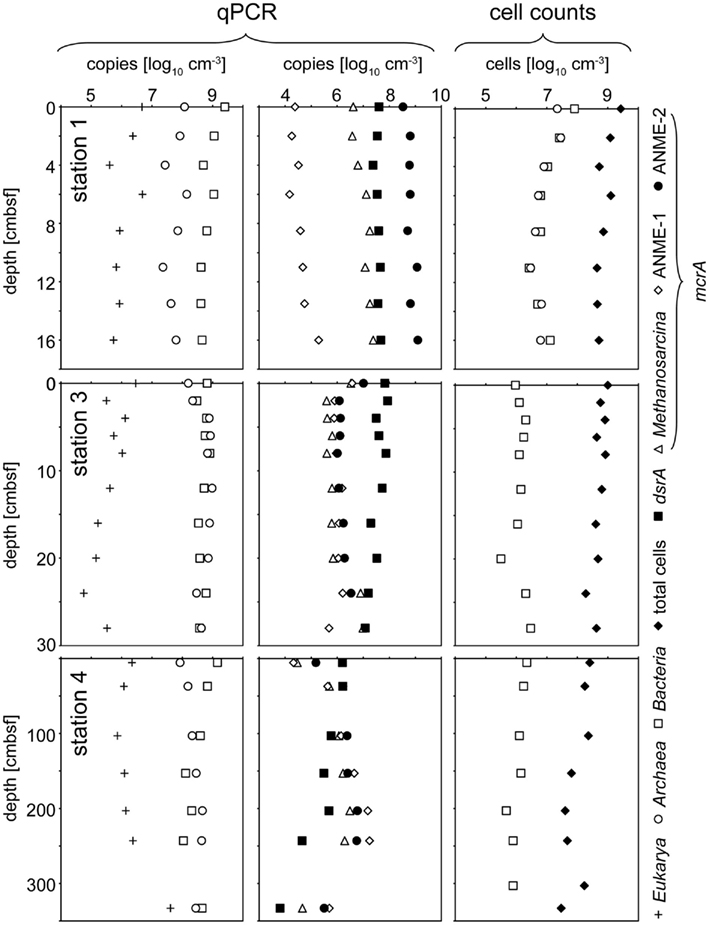
Figure 4. Depth trend of quantitative community composition of three different stations from the Simeulue seep. The left panel illustrates the composition of the three domains Bacteria, Archaea and Eukarya according to qPCR quantification of 16S or 18S rRNA genes, respectively. The middle panel gives an overview over the distribution of the functional gene numbers. In the right panel, total cell counts are compared to CARD–FISH cell counts for Bacteria and Archaea detecting active cells.
Domain specific 16S rRNA gene copies, obtained from qPCR measurements, and selected functional genes at three different stations are displayed in Figure 4. In general, a clear trend was observed only for Eukarya and only at the station 4 (Figure 4). The copy numbers increased with depth (Figure 4).
For functional genes, a trend with depth was observed only at the deeper sampled station 4. Its most prominent feature was the increase of all mcrA gene copy numbers, beginning at 103 cmbsf down to 267 cmbsf, and dropping again to the values at the surface further below. An opposite tendency was observed for dsrA gene copy numbers showing a clear decrease with depth. At station 2, ANME-2 mcrA gene copies increased from 6 × 106 copies cm−3 in the top layer to 2 × 108 copies cm−3 at the bottom (8 cmbsf). An opposite trend appeared for the ANME-1 mcrA gene with copy numbers of 2 × 108 copies cm−3 at the surface layer and 7 × 107 copies cm−3 at the bottom. No change with depth was observed for the Methanosarcina mcrA group where copy numbers of 5 × 108 copies cm−3 were detected. The dsrA gene measurement resulted in copy numbers of 3 × 106 copies cm−3 at the top and 6 × 107 copies cm−3 at the bottom of this core.
Microbial Diversity
Parsimony trees of 16S rRNA gene sequences obtained from DGGE separation of extracted DNA from Simeulue sediment stations are depicted in Figures 5 and 6. PCR products of the station 3 were run on a separate gel without reference to the stations 1 and 2. Therefore, band patterns were not comparable and a DGGE gel photograph is not shown. Stations 1, 2, and 4 are shown in Figure 5.
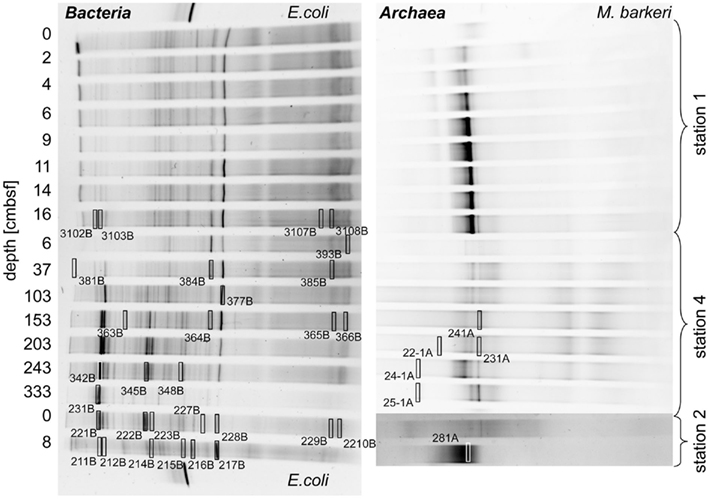
Figure 5. Denaturing gradient gel electrophoresis photographs showing the 16S rRNA gene sequence diversity of Bacteria (left) and Archaea (right) of the stations 1, 2, and 4 of the Simeulue seep. Escherichia coli and Methanosarcina barkeri served as positive controls. Excised bands used for 16S rRNA gene sequencing are highlighted by open rectangles. The numbers enumerate gels, lanes, and bands and refer to the phylogenetic trees in this figure and Figure 6. Multiple gels were prepared and bands excised from different gels could be relocated in the shown photographs. Bands resulting in unsuccessful PCR re-amplifications or sequencing reactions are not shown.
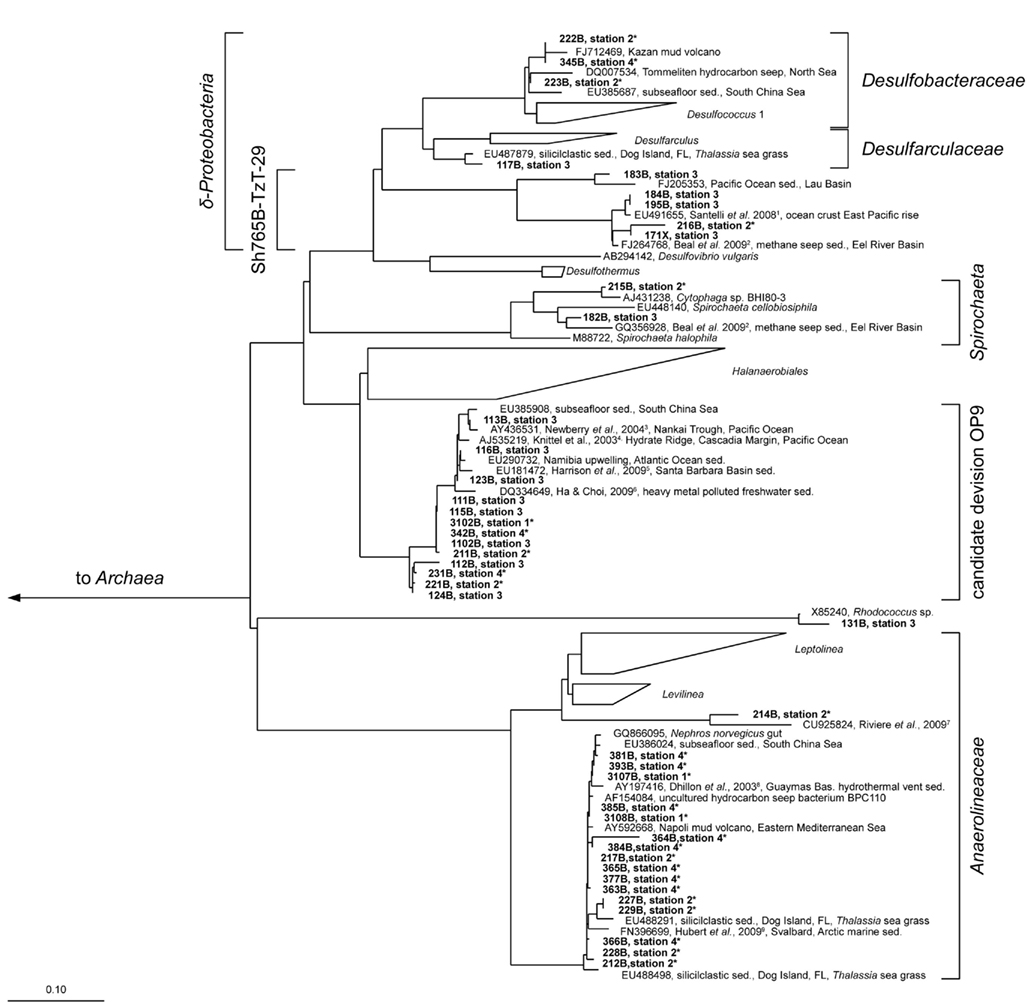
Figure 6. Parsimony tree of bacterial 16S rRNA gene sequences with all sampled stations. The tree is based on the SILVA tree version 102 (Pruesse et al., 2007). Sequences obtained from the Simeulue seep are in bold letters. Bold numbers describe bands excised and used for 16S rRNA gene sequencing from three different DGGE gels. On the right, the deepest branching affiliations are shown. References are indicated in upper case: 1 Santelli et al. (2008), 2 Beal et al. (2009), 3 Newberry et al. (2004), 4 Knittel et al. (2003), 5 Harrison et al. (2009), 6 Ha and Choi (2009), 7 Rivière et al. (2009), 8 Dhillon et al. (2003), 9 Hubert et al. (2009). Asterisks (*) indicate bands from DGGE gels in Figure 5. sed., Sediment.
Three major bacterial phylogenetic groups (δ-Proteobacteria, candidate division OP9, and Anaerolineaceae) were abundant across the study area. According to their band thickness in the DGGE gel, OP9 bacteria, and relatives of Desulfobacteraceae seemed to become increasingly important with depth at the station 4 (Figures 4 and 5). Nonetheless, since band thickness is an imprecise measure for abundance, this result indicates only a trend, despite the fact that when the total amount of amplified DNA products was equal for each layer. Besides δ- Proteobacteria, other Proteobacteria sequences were not recovered from the DGGE gels. Sequences affiliated to the clusters Desulfobacteraceae, Desulfarculaceae, and Sh765B-TzT-29 dominated the δ-Proteobacteria (Figure 5). One deeply branching sequence of the station 2 was more closely related to sequences obtained from municipal wastewater sludge (Rivière et al., 2009) than to the genus Leptolinea (Figure 6). Another deeply branching sequence of the station 3 was closely related to the genus Rhodococcus (Figure 6). Two bands found at stations 2, 3, and 4 were relatively close relatives of the genus Spirochaeta.
Simeulue seep sequences of all stations were related to three prominent archaeal groups (ANME-1, ANME-2, and Crenarchaeota; Figure 7). One sequence of the station 3 was assigned into the Deep Hydrothermal Vent Euryarchaeotal Group 6 (DHVEG6). The remaining sequences (stations 3 and 4) belonged to not further specified Crenarchaeota. Except for the Crenarchaeota, members of all other clusters were identified in stations 1–4. In summary, the bacterial diversity was greater than the archaeal one, with most sequences related to organisms typically found at methane and hydrocarbon seeps or mud volcanoes.
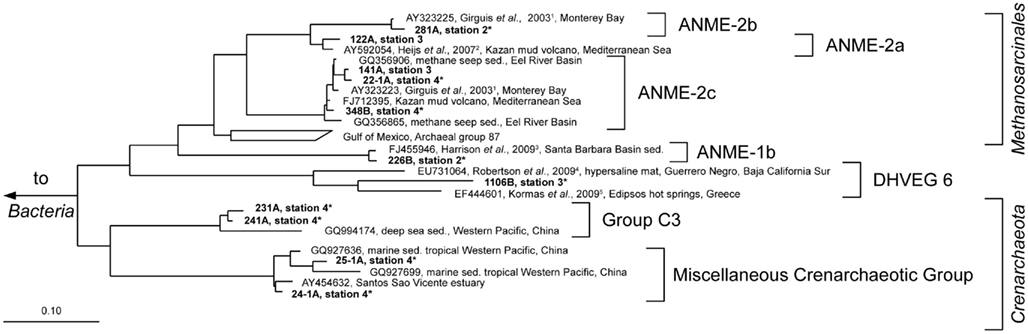
Figure 7. Parsimony tree of archaeal 16S rRNA gene sequences summarizing all sampled stations. Sequences obtained from the Simeulue seep are in bold letters. Numbers describe bands which were excised and used for 16S rRNA gene sequencing from three different DGGE gels with identical patterns. On the right, the deepest branching affiliations are shown. DHVEG6, Deep Hydrothermal Vent Euryarchaeotal Group 6; MHVG, Marine Hydrothermal Vent Group. References are indicated in upper case: 1 Girguis et al. (2003), 2 Heijs et al. (2007), 3 Harrison et al. (2009), 4 Robertson et al. (2009), 5 Kormas et al. (2009). Asterisks (*) indicate bands from DGGE gels in Figure 5. sed., Sediment.
Discussion
For the first time, an active methane seep was discovered in the Indian Ocean. The discovered seep comprised highly active (stations 1 and 2) and less active or inactive (stations 3 and 4) AOM-influenced areas (Table 1; Figure 4). The highly active stations 1 and 2 were characterized by black sulfidic surface sediments, depleted sulfate, and high sulfide concentrations and light δ13CDIC values of the porewater, the presence of ANME-1 and ANME-2 representatives, as well as high cell and high copy numbers of 16S rRNA and functional genes related to AOM, methanogenesis, and sulfate reduction. A defined seep center of activity, like in mud volcanoes, was not discovered, the seep area was rather characterized by a patchy distribution of active spots. Carbonate- or sulfide-rich spots were distributed randomly over the surface. A reason for the patchiness might be tectonic activity. While some gas conduits might have been shut, other could have opened over time. An apparent feature of the active parts at the seep was the strong depletion of 13C in DIC, which was also observed for TOC of the guts of the seep’s macro fauna. This confirms the importance of methane as carbon source for the benthos at this location. In addition, methanogenic activity was confirmed in sediment microcosms of the Simeulue seep area as well as in the Nias basin, where AOM activity was absent.
Methanotrophy and Sulfate Reduction Activities at the Seep Stations
Methane is an indirect electron source for dissimilatory microbial sulfate reducers in the syntrophic process of AOM (Knittel and Boetius, 2009). The terminal reaction products are carbonate and sulfide. The produced sulfide in turn may be oxidized at the oxic/anoxic interface near the sediment surface. White, sometimes filamentous sulfide-oxidizing bacteria are typical indicators for this interface (Niemann et al., 2006b). Areas covered with such white-colored microbial mats were discovered in the Simeulue basin and sampled in their proximity using a TV-guided grab (station 2, Figure 1). In contrast, microbial mats were absent near the station 1. Most likely, the white color of such mats is a result of intracellular sulfur inclusions as observed in Thioploca, Beggiatoa, or Thiomargarita aggregates, regularly found on the surface of sulfide-rich marine sediments (Gallardo, 1977; Jannasch et al., 1989; Schulz et al., 1999).
At the Simeulue seep, methane was probably a carbon source for higher biota, as indicated by their 13C-depleted carbon signatures. However, the δ13C (−31 to −45‰) values of the sampled crab guts indicate a mixed carbon source originating from AOM and water column carbon (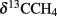 −70.9 to 74.8‰, δ13CDIC −0.8 to −48.8‰, δ13CTOC −22.3 to 27.6‰). Heterotrophic processes as well as symbiosis between methanotrophic microorganisms and macrofauna are well described for several hot and cold deep marine vents (Childress et al., 1986; Duperron et al., 2005; Petersen and Dubilier, 2009). AOM was clearly a carbon donating process. It has been previously demonstrated that more than 99% of the methane in AOM systems is used for energy metabolism (Wegener et al., 2008). The oxidized carbon is than excreted as carbonate and probably reassimilated into biomass. The same seems likely for the Simeulue invertebrate community as previously shown for symbiotic CO2-fixing microorganisms and the gutless worm Olavius (Blazejak et al., 2005). Symbiosis of higher benthos and methanotrophic microorganisms is also often associated with aerobic methanotrophy (Childress et al., 1986; Duperron et al., 2005; Niemann et al., 2006b; Petersen and Dubilier, 2009). However, the DGGE 16S rRNA gene analyses, focusing on dominant bands, did not reveal aerobic methane oxidizers in the seep sediments (Figures 5 and 6).
−70.9 to 74.8‰, δ13CDIC −0.8 to −48.8‰, δ13CTOC −22.3 to 27.6‰). Heterotrophic processes as well as symbiosis between methanotrophic microorganisms and macrofauna are well described for several hot and cold deep marine vents (Childress et al., 1986; Duperron et al., 2005; Petersen and Dubilier, 2009). AOM was clearly a carbon donating process. It has been previously demonstrated that more than 99% of the methane in AOM systems is used for energy metabolism (Wegener et al., 2008). The oxidized carbon is than excreted as carbonate and probably reassimilated into biomass. The same seems likely for the Simeulue invertebrate community as previously shown for symbiotic CO2-fixing microorganisms and the gutless worm Olavius (Blazejak et al., 2005). Symbiosis of higher benthos and methanotrophic microorganisms is also often associated with aerobic methanotrophy (Childress et al., 1986; Duperron et al., 2005; Niemann et al., 2006b; Petersen and Dubilier, 2009). However, the DGGE 16S rRNA gene analyses, focusing on dominant bands, did not reveal aerobic methane oxidizers in the seep sediments (Figures 5 and 6).
The anoxic nature of the sediment was confirmed by porewater data, showing in particular high sulfide concentrations, reduced iron, and manganese as well as ammonium to be present (Figure 3). While sulfide concentrations increased downward into the sediment, sulfate decreased to micromolar concentrations. However, a clear SMTZ was not observed.
Large amounts of dissolved gas with strong sulfidic odor evaporated from the sediment during sampling of the stations 1 and 2. Indicators for ongoing AOM at stations 1 and 2 were the low δ13CDIC values in the porewater apparently derived from 13C-depleted methane (Tables 2 and 3). These values were comparable to other methane influenced seeps, as in the Gulf of Mexico (Coffin et al., 2008) or at the Hydrate Ridge (Valentine et al., 2005). Since ocean water δ13CDIC values are usually between 0 and −10‰ (Deuser et al., 1968), DIC at the Simeulue seep was obviously derived from the anaerobic oxidation of upward migrating methane (Figure 3). 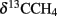 values were below −70‰ at the stations 1 and 2 (Table 3). It is commonly agreed that biogenic methane exhibits
values were below −70‰ at the stations 1 and 2 (Table 3). It is commonly agreed that biogenic methane exhibits 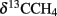 values below −70‰ (Whiticar et al., 1986). Hence, the observed methane at the Simeulue seep was likely biogenic methane as well.
values below −70‰ (Whiticar et al., 1986). Hence, the observed methane at the Simeulue seep was likely biogenic methane as well.
The AOM rates observed for the Simeulue seep area were slightly lower than maximum rates reported for other methane seeps, but higher than rates observed for mud volcanoes or sediments from various marine SMTZ (Knittel and Boetius, 2009).
That methane rather than TOC was the carbon source for microorganisms is supported by low δ13CTOC values at station 1 and 2. These values were in a narrow range between −22.7 and −24.5‰ which are typical for marine cellular carbon (Deuser et al., 1968; Rice, 1993). These values contrast δ13CDIC values between −11.8 and −48.8‰ at the stations 1 and 2, most of them below −40.0‰ (Figure 3). Since the carbon isotopic composition of methane at both sites was below −70.0‰ (Table 3), it is obvious that AOM contributed to the DIC budget at these active AOM sites. The calculated SRR in the equilibrium zone, i.e., the zone of sulfur input from sulfate does not exceed the SRR, and was between 0.43 μmol cm−1 day−1 at 4 cmbsf and 0.56 μmol cm−1 day−1 at 8 cmbsf. The fluid flux was assumed to be 10 cm year−1 (Girguis et al., 2003) and the equilibrium was reached when the sum of sulfide and sulfate concentrations did not exceed the bottom water concentration of sulfate any more (27.79 mM, Figure 3). Moreover, the seep sediments were highly methane laden, as indicated by intensive gas emission during sampling. Huge discrepancies between sulfate reduction and AOM rates are usually observed only when methane plays a minor role in the investigated system (Niemann et al., 2006a), which is not the case here.
However, one may expect that there would have been a greater contribution of AOM derived carbon to TOC as observed, as methane was apparently a carbon source for the AOM performing microorganisms. Wegener et al. (2008) reported the assimilation of methane derived carbon into AOM performing microbial consortia of various geographic origins via CO2 fixation. Furthermore, they could show that methane mostly serves microbial catabolism and to little extent microbial anabolism. This is also reflected in carbon stable isotopic signatures of DIC and TOC of the Simeulue seep, where stronger methane signals were detected in DIC and low δ13CTOC values do not support a significant impact of methane derived carbon to TOC (Table 3; Figure 3). On the other hand, ammonium concentrations decrease between 6 and 11 cmbsf at station 1. This could either be due to (i) heterotrophic TOC degradation for energy metabolism, (ii) hypothetical anaerobic ammonium oxidation with sulfate (Schrum et al., 2009), or (iii) ammonium uptake due to the increase of biomass upon AOM. TOC degradation would liberate ammonium and can be excluded (e.g., Wehrmann et al., 2011). Anaerobic ammonium oxidation with sulfate was suggested based on geochemical sediment profiles (Schrum et al., 2009; Wehrmann et al., 2011) but has not been convincingly demonstrated in vitro yet. However, it would have a stoichiometry of 1 mol sulfate reduced to 2.67 mol ammonium oxidized. From 4 to 6 cmbsf, the stoichiometry is 1–6.47 mol (Figure 3). That would indicate either an inefficient ammonium oxidation or a contribution of AOM to the ammonium decline. Given a one to one stoichiometry of sulfate driven AOM, the remaining 3.8 mol of sulfate could have been reduced by AOM. The measured AOM rate in the microcosm of station 1 was 0.57 μmol cm−3 day−1 (Table 3). During AOM, only 1% of the methane derived carbon is reassimilated into biomass (Wegener et al., 2008). That means 5.7 nmol carbon cm−3 day−1 were assimilated into biomass due to AOM. Given a relation of carbon to nitrogen in living organisms of 6 to 1, the ammonium uptake was 1.0 nmol nitrogen cm−3 day−1. Given the ammonium influx from the water column was constant, the ammonium uptake exceeded the input between 4 and 6 cmbsf (Figure 3). Assuming a fluid flux of about 10 cm year−1 (Girguis et al., 2003), the ammonium uptake was about 20 μmol cm−3 day−1. This exceeds the estimated ammonium uptake due to AOM by more than four orders of magnitude and therefore can not account for the decrease in this zone. As alternative explanation we propose ammonium oxidation with an unknown electron acceptor for this zone. Sulfate may be a candidate as suggested by Schrum et al. (2009).
Since at station 4 AOM was not reflected by the porewater chemistry we postulate a former, recently ceased AOM activity, which has left its imprint in the carbon isotope signatures and ANME-related 16S rRNA gene biomarkers. Consequently, the δ13CTOC values below −27‰ are suggestive for a greater contribution of methane to TOC compared to DIC than observed for the stations 1 and 2 (Figure 3). AOM driven microbial activity, as indicated by an increase of ANME members with sediment depth (Figure 4), could have caused a stronger methane signal in TOC when methane was the prevailing carbon and energy source. Moreover, the impact of TOC of the water column is presumably much lower at a depth of 150–300 cmbsf than closer to the sediment surface. Methane was relatively enriched in 13C at this station, which could also be well explained by AOM activity. Extremely low methane concentrations could have led to a less pronounced isotopic fractionation effect (Holler et al., 2009) – caused by rate limiting methane concentrations (Table 3).
A number of in vitro experiments could confirm AOM as an important process at the discovered methane seep. Anaerobic methanotrophy was observed in initial and in transferred microcosms, as indicated by labeling experiments as well as by methane-dependent SRR (Table 3). That sulfate reduction was mainly driven by methane was further supported by the observation of comparable TOC values in the sediments of both, stations 1 and 2 as well as station 4 (Figure 3). If TOC degraders significantly accounted for sulfate reduction, depleted sulfate and high sulfide concentration would have been measured for station 4 as well, and not only for the stations 1 and 2 for which sulfate reduction via active AOM could be demonstrated. The higher rates in the first transferred microcosms indicated an enrichment of methanotrophs in these assays after incubation for more than 1 year as indicated by labeling experiments using 13C-methane.
Moreover, relatively high concentrations of reduced manganese were detected at station 2 (Figure 3). The presence of reduced manganese may be an indication for sulfide dependent or direct microbial manganese reduction. Sulfide dependent or direct microbial manganese reduction was probably a result of AOM activity at the Simeulue seep as suggested for the Eel river basin by Beal et al. (2009). Additionally, one sequence of the station 2 and four sequences of the station 3 were assigned to the Sh765B-TzT-29 cluster, a cluster that was initially believed to belong to the family Geobacteraceae and was first described by Geißler (2003). This cluster contains heavy metal associated Bacteria which where originally found in uranium mill tailings at Shiprock (NM, USA). A recent study described metal associated AOM consortia (Beal et al., 2009). Members of the Sh765B-TzT-29 cluster may also be metal associated at the Simeulue seep. Ádditionally, our culturing experiments with oxidized metals showed oxidation of methane, possibly associated to the Sh765B-TzT-29 cluster. Moreover, 16S rRNA gene sequences reported by Beal et al. (2009) were found in a close phylogenetic neighborhood of operational taxonomic units (OTUs) identified in the Simeulue seep (Figure 6). In conjunction with the presence of reduced iron and manganese in the porewater (Figure 3), it seems possible that metal reduction played a role as electron acceptor, besides sulfate, for AOM.
Microbial Community Composition at the Seep Stations
Anaerobic oxidation of methane supported a vital microbial community as demonstrated by 13C-methane labeling experiments, rate measurements in vitro, rate estimations in situ, the presence of ANME-related mcrA genes, and an active archaeal and bacterial community at stations 1 and 2 (Figure 4). Many archaeal sequences obtained from the Simeulue seep stations 1 to 4, were distributed over the ANME-1 and -2 clusters (Figure 7). OTUs from the stations 1 and 2 were assigned to ANME-1b and ANME-2b clusters. Also at the station 3, ANME-2a/c members were identified. Furthermore, members of the Crenarchaeota were frequently detected in the DGGE analysis at both AOM-stations and the station 3. The occurrence of Crenarchaeota at a methane seep is not unusual and has been reported for other sites (Knittel et al., 2005; Knittel and Boetius, 2009).
Most bacterial groups belonged to the family Anaerolineaceae (phylum Chloroflexi), the candidate division OP9 (Webster et al., 2004) and the class δ-Proteobacteria. These groups have been described to be dominant in marine sediments (Teske, 2006; Blazejak and Schippers, 2010). Sequences affiliated to the family Anaerolineaceae were derived from stations 1 to 4. The nearest cultured genus Leptolinea (distance matrix: 84.5% identity with the nearest Leptolinea member) has been described as saccharolytic, including pectin and cellulose degrading species (Yamada et al., 2006; Ishii et al., 2008). In these studies, members of the genus Leptolinea were not able to reduce sulfate or other sulfur species (Yamada et al., 2006). Leptolinea and Levilinea species largely comprise taxa discovered in anaerobic waste water sludge (Rivière et al., 2009), indicating the possibility of active heterotrophic processes at the AOM seep. However, sequences from the Simeulue seep were closer related to sequences which could not be assigned to these genera (Figure 6). Three sequences of the stations 1 and 2 were closely related to the genus Desulfococcus of the family Desulfobacteraceae, which is in good agreement with previously described AOM sites (Knittel and Boetius, 2009).
At the stations 1 and 2 (only data for station 1 are shown in Figure 4), mcrA genes encoding for the enzyme methyl-CoM-reductase of the anaerobic methanotrophic ANME-2 group dominated over ANME-1. ANME-1 mcrA genes prevailed only at the surface of the station 2. The Methanosarcina (“methanogenic”) type of the mcrA gene was detected throughout the whole seep area, while it was completely absent in other sediments of the Sumatra forearc (Schippers et al., 2010). The simultaneous occurrence of both, methane production and oxidation in the seep area, underscores the important role of the methane cycle for this system. The dsrA gene, an indicator for the presence of sulfate reducers, was found in slightly higher gene copy numbers at the station 1–3 (Figure 4).
Catalyzed reporter deposition–fluorescence in situ hybridization cell counts, assessing the active community, indicated that living Bacteria were present with at least two orders of magnitude more cells than Archaea at the station 2. At the station 1, active bacterial and archaeal cells were distributed equally. In contrast, results obtained by qPCR, targeting also inactive microorganisms, indicate a dominance of Bacteria over Archaea by one order of magnitude.
The total cell numbers of 107 to 109 cells cm−3 at the Simeulue seep were quite similar to those at sites of the Sumatra forearc basins not influenced by methane seepage (Schippers et al., 2010). These are also comparable to cell numbers reported for the arctic, methane emitting, Haakon Mosby mud volcano, hosting mainly aerobic methylotrophs (Niemann et al., 2006b). Also the decrease of cell numbers with sediment depth was similar compared to this mud volcano. CARD–FISH cell counts, and qPCR measurements showed the presence of Bacteria and Archaea, but only small numbers of Eukarya. This observation is in agreement with previous marine sediment studies (Schippers and Nerretin, 2006; Schippers et al., 2010).
Methanogenesis as a Concomitant Process
Another goal of this research was to demonstrate methanogenic hydrocarbon degradation (Zengler et al., 1999; Head et al., 2003). From a geological point of view, the Sumatra forearc seems a promising location for hydrocarbon generation in the deep subsurface, a potential source for upward migrating of complex hydrocarbons. Hence, all stations were screened for such processes. Hydrocarbon-dependent methanogenesis was observed in microcosms of the Simeulue seep and the Nias basin. After a first transfer of sediment microcosms of five stations (Table 2), three stations showed sustained methanogenesis in the presence of higher hydrocarbons. Only one of these stations was located in the seep area (station 6), the other two in the Nias basin. The rates estimated in the initial setups as well as in the first transfers, were in the same range compared to a hydrocarbon adapted community of contaminated harbor mud in the North Sea (Siegert et al., 2011).
Methanogenesis was absent in microcosms without added substrates. Possibly, the presence of 28 mM sulfate and other electron acceptors inhibited methanogenesis from TOC, but not from higher hydrocarbons. In contrast, it seems likely that the addition of higher hydrocarbons stimulated activity of TOC and hydrocarbon utilizing microorganisms. Positive controls containing the substrates TMA or methanol with 28 mM sulfate confirmed that methanogenic activity in spite of present sulfate. Methanogenesis from these substrates evolved rapidly within the first weeks of incubation and was in the same order with the AOM rates.
Desulfobacteraceae species may be indicative for AOM consortia (Knittel and Boetius, 2009), but one member of this family is the hexadecane degrader Desulfococcus oleovorans strain Hxd3 (Aeckersberg et al., 1991; So et al., 2003). Hence, the occurrence of this family suggests the presence of consortia capable of anaerobic degradation of higher hydrocarbons. The finding of other closely related hydrocarbon seep associated sequences, e.g., from mud volcanoes or contaminated sites, confirmed this (Figure 6). However, hydrocarbons are abundant substances in nature and our culturing experiments show that the presence of hydrocarbon degraders does not necessarily depend on the presence of hydrocarbons in higher concentrations. In summary, hydrocarbon utilizing methanogenic communities were present in the Sumatra forearc sediments irrespective of methane seepage, and their presence does not infer that higher hydrocarbons played a significant role in the carbon cycle. Nonetheless, this is the first report of stable microcosms of hydrocarbon-dependent methanogenic microbial communities from the deep ocean.
Conclusion
The first-time discovery of an AOM-influenced methane seep in the Indian Ocean was confirmed by the presence of dissolved methane as well as methane-dependent pro- and eukaryotic communities. Methane δ13C signatures indicate a microbial origin of methane. The released methane was oxidized by an active microbial community, sharing features with other seep communities. Albeit negligible in situ, it was also observed that higher hydrocarbons were converted to methane in vitro.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank G. Mengel-Jung, D. Zoch, U. Günther, H.-E. Gäbler, T. M. Segl, C. Ostertag-Henning, S. Schlömer, and H. Probst for technical support and fruitful discussions. We thank the officers, crew and shipboard scientific party of R/V SONNE especially A. Lückge for excellent support during expedition SO189-2. This work was funded by the BMBF research grant 03G0189A and the DFG grants KR 3311/5-1 and 5-2.
Footnotes
References
Aeckersberg, F., Bak, F., and Widdel, F. (1991). Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch. Microbiol. 156, 5–14.
Aloisi, G., Pierre, C., Rouchy, J.-M., Foucher, J.-P., Woodside, J., and Party, M. S. (2000). Methane-related authigenic carbonates of eastern Mediterranean Sea mud volcanoes and their relation to gas hydrate destabilisation. Earth Planet. Sci. Lett. 184, 321–338.
Barnes, R. O., and Goldberg, E. D. (1976). Methane production and consumption in anoxic marine sediments. Geology 4, 297–300.
Beal, E. J., House, C. H., and Orphan, V. J. (2009). Manganese- and iron-dependent marine methane oxidation. Science 325, 184–187.
Berndt, C., Bünz, S., Clayton, T., Mienert, J., and Saunders, M. (2004). Seismic character of bottom simulating reflectors: examples from the mid-Norwegian margin. Mar. Petrol. Geol. 21, 723–733.
Blazejak, A., Erséus, C., Amann, R., and Dubilier, N. (2005). Coexistence of bacterial sulfide oxidizers, sulfate reducers, and spirochetes in a gutless worm (Oligochaeta) from the Peru margin. Appl. Environ. Microbiol. 71, 1553–1561.
Blazejak, A., and Schippers, A. (2010). High abundance of JS-1- and Chloroflexi-related bacteria in deeply buried marine sediments revealed by quantitative, real-time PCR. FEMS Microbiol. Ecol. 72, 198–207.
Boetius, A., Ravenschlag, K., Schubert, C. J., Rickert, D., Widdel, F., Gieseke, A., Amann, R., Jørgensen, B. B., Witte, U., and Pfannkuche, O. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 423–426.
Bohrmann, G., Greinert, J., Suess, E., and Torres, M. (1998). Authigenic carbonates from the Cascadia subduction zone and their relation to gas hydrate stability. Geology 26, 647–650.
Childress, J. J., Fisher, C. R., Brooks, J. M., Kennicutt, M. C., Bidigare, R., and Anderson, A. E. (1986). A methanotrophic marine molluscan (Bivalvia, Mytilidae) symbiosis: mussels fueled by gas. Science 233, 1306–1308.
Cline, J. D. (1969). Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14, 454–458.
Coffin, R., Hamdan, L., Plummer, R., Smith, J., Gardner, J., Hagen, R., and Wood, W. (2008). Analysis of methane and sulfate flux in methane-charged sediments from the Mississippi Canyon, Gulf of Mexico. Mar. Petrol. Geol. 25, 977–987.
Coolen, M. J. L., Cypionka, H., Sass, A. M., Sass, H., and Overmann, J. (2002). Ongoing modification of mediterranean pleistocene sapropels mediated by prokaryotes; online supplementary material. Science 296, 2407–2410.
Cord-Ruwisch, R. (1985). A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J. Microbiol. Methods 4, 33–36.
Davis, J. B., and Yarbrough, H. F. (1966). Anaerobic oxidation of hydrocarbons by Desulfovibrio desulfuricans. Chem. Geol. 1, 137–144.
Dekov, V. M., Kamenov, G. D., Savelli, C., Stummeyer, J., and Marchig, V. (2006). Origin of basal dolomitic claystone in the Marsili Basin, Tyrrhenian Sea. Mar. Geol. 236, 121–141.
Deuser, W. G., Degens, E. T., and Guillard, R. R. L. (1968). Carbon isotope relationships between plankton and sea water. Geochim. Cosmochim. Acta 32, 657–660.
Dhillon, A., Teske, A., Dillon, J., Stahl, D. A., and Sogin, M. L. (2003). Molecular characterization of sulfate-reducing bacteria in the Guaymas basin. Appl. Environ. Microbiol. 69, 2765–2772.
Duperron, S., Nadalig, T., Caprais, J.-C., Sibuet, M., Fiala-Médioni, A., Amann, R., and Dubilier, N. (2005). Dual symbiosis in a Bathymodiolus sp. mussel from a methane seep on the Gabon continental margin (Southeast Atlantic): 16S rRNA phylogeny and distribution of the symbionts in gills. Appl. Environ. Microbiol. 71, 1694–1700.
Ettwig, K. F., Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M. M., Schreiber, F., Dutilh, B. E., Zedelius, J., De Beer, D., Gloerich, J., Wessels, H. J. C. T., Van Alen, T., Luesken, F., Wu, M. L., Van De Pas-Schoonen, K. T., Op Den Camp, H. J. M., Janssen-Megens, E. M., Francoijs, K.-J., Stunnenberg, H., Weissenbach, J., Jetten, M. S. M., and Strous, M. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548.
Faber, E., and Stahl, W. (1983). Analytic procedure and results of an isotope geochemical surface survey in an Area of the British North Sea. Geol. Soc. Lond. Spec. Publ. 12, 51–63.
Gallardo, V. A. (1977). Large benthic microbial communities in sulphide biota under Peru-Chile subsurface countercurrent. Nature 268, 331–332.
Geißler, A. (2003). Molekulare Analyse der bakteriellen Diversität in Uranabraumhalden. Freiberg: Technische Universität Bergakademie Freiberg.
Girguis, P. R., Orphan, V. J., Hallam, S. J., and Delong, E. F. (2003). Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl. Environ. Microbiol. 69, 5472–5482.
Ha, M.-H., and Choi, J. (2009). Effects of environmental contaminants on hemoglobin gene expression in Daphnia magna: a potential biomarker for freshwater quality monitoring. Arch. Environ. Contam. Toxicol. 57, 330–337.
Harrison, B. K., Zhang, H., Berelson, W., and Orphan, V. J. (2009). Variations in archaeal and bacterial diversity associated with the sulfate-methane transition zone in continental margin sediments (Santa Barbara, California). Appl. Environ. Microbiol. 75, 1487–1499.
Head, I. M., Jones, D. M., and Larter, S. R. (2003). Biological activity in the deep subsurface and the origin of heavy oil. Nature 426, 344–352.
Heijs, S. K., Haese, R. R., Van Der Wielen, P. W. J. J., Forney, L. J., and Van Elsas, J. D. (2007). Use of 16S rRNA gene based clone libraries to assess microbial communities potentially involved in anaerobic methane oxidation in a mediterranean cold seep. Microb. Ecol. 53, 384–398.
Holler, T., Wegener, G., Knittel, K., Boetius, A., Brunner, B., Kuypers, M. M. M., and Widdel, F. (2009). Substantial 13C/12C and D/H fractionation during anaerobic oxidation of methane by marine consortia enriched in vitro. Environ. Microbiol. Rep. 1, 370–376.
Hubert, C., Loy, A., Nickel, M., Arnosti, C., Baranyi, C., Bruchert, V., Ferdelman, T., Finster, K., Christensen, F. M., Rosa De Rezende, J., Vandieken, V., and Jorgensen, B. B. (2009). A constant flux of diverse thermophilic bacteria into the cold arctic seabed. Science 325, 1541–1544.
Hyndman, R. D., and Spence, G. D. (1992). A seismic study of methane hydrate marine bottom simulating reflectors. J. Geophys. Res. 97, 6683–6698.
Inagaki, F., Nunoura, T., Nakagawa, S., Teske, A., Lever, M., Lauer, A., Suzuki, M., Takai, K., Delwiche, M., Colwell, F. S., Nealson, K. H., Horikoshi, K., D’Hondt, S., and Jørgensen, B. B. (2006). Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. U.S.A. 104, 2815–2820.
Ishii, S., Shimoyama, T., Hotta, Y., and Watanabe, K. (2008). Characterization of a filamentous biofilm community established in a cellulose-fed microbial fuel cell. BMC Microbiol. 8, 6. doi:10.1186/1471-2180-8-6
Jannasch, H. W., Nelson, D. C., and Wirsen, C. O. (1989). Massive natural occurrence of unusually large bacteria (Beggiatoa sp.) at a hydrothermal deep-sea vent site. Nature 342, 834–836.
Joye, S. B., Boetius, A., Orcutt, B. N., Montoya, J. P., Schulz, H. N., Erickson, M. J., and Lugo, S. K. (2004). The anaerobic oxidation of methane and sulfate reduction in sediments from Gulf of Mexico cold seeps. Chem. Geol. 205, 219–238.
Knittel, K., and Boetius, A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334.
Knittel, K., Boetius, A., Eilers, H., Lochte, K., Pfannkuche, O., Linke, P., and Amann, R. (2003). Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrate (Cascadia Margin, Oregon). Geomicrobiol. J. 20, 269–294.
Knittel, K., Lösekann, T., Boetius, A., Kort, R., and Amann, R. (2005). Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71, 467–479.
Kormas, K. A., Tamaki, H., Hanada, S., and Kamagata, Y. (2009). Apparent richness and community composition of bacteria and archaea in geothermal springs. Aquat. Microb. Ecol. 57, 113–122.
Krüger, M., Meyerdierks, A., Glöckner, F. O., Amann, R., Widdel, F., Kube, M., Reinhardt, R., Kahnt, J., Böcher, R., Thauer, R. K., and Shima, S. (2003). A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426, 878–818.
Krüger, M., Treude, T., Wolters, H., Nauhaus, K., and Boetius, A. (2005). Microbial methane turnover in different marine habitats. Palaeogeogr. Palaeoclimatol. Palaeoecol. 227, 6–17.
Lösekann, T., Knittel, K., Nadalig, T., Fuchs, B., Niemann, H., Boetius, A., and Amann, R. (2007). Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby Mud Volcano, Barents Sea. Appl. Environ. Microbiol. 73, 3348–3362.
Lovley, D. R., and Phillips, E. J. P. (1986). Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51, 683–689.
Lovley, D. R., and Phillips, E. J. P. (1988). Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54, 1472–1480.
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, Buchner, A., Lai, T., Steppi, S., Jobb, G., Förster, W., Brettske, I., Gerber, S., Ginhart, A. W., Gross, O., Grumann, S., Hermann, S., Jost, R., König, A., Liss, T., Lüßmann, R., May, M., Nonhoff, B., Reichel, B., Strehlow, R., Stamatakis, A., Stuckmann, N., Vilbig, A., Lenke, M., Ludwig, T., Bode, A., and Schleifer, K.-H. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371.
Martens, C. S., and Berner, R. A. (1974). Methane production in the interstitial waters of sulfate-depleted marine sediments. Science 185, 1167–1169.
Martin, J. W., and Haney, T. A. (2005). Decapod crustaceans from hydrothermal vents and cold seeps: a review through 2005. Zool. J. Linn. Soc. 145, 445–522.
Michaelis, W., Seifert, R., Nauhaus, K., Treude, T., Thiel, V., Blumenberg, M., Knittel, K., Gieseke, A., Peterknecht, K., Pape, T., Boetius, A., Amann, R., Jørgensen, B. B., Widdel, F., Peckmann, J., Pimenov, N. V., and Gulin, M. B. (2002). Microbial reefs in the black sea fueled by anaerobic oxidation of methane. Science 297, 1013–1015.
Muyzer, G., Teske, A., Wirsen, C. O., and Jannasch, H. W. (1995). Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164, 165–172.
Nadkarni, M. A., Martin, F. E., Jacques, N. A., and Hunter, N. (2002). Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148, 257–266.
Nauhaus, K., Boetius, A., Krüger, M., and Widdel, F. (2002). In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediments from a marine gas hydrate area. Environ. Microbiol. 4, 296–305.
Newberry, C. J., Webster, G., Cragg, B. A., Parkes, R. J., Weightman, A. J., and Fry, J. C. (2004). Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, ocean drilling program leg 190. Environ. Microbiol. 6, 274–287.
Niemann, H., Duarte, J., Hensen, C., Omoregie, E., Magalhães, V. H., Elvert, M., Pinheiro, L. M., Kopf, A., and Boetius, A. (2006a). Microbial methane turnover at mud volcanoes of the Gulf of Cadiz. Geochim. Cosmochim. Acta 70, 5336–5355.
Niemann, H., Lösekann, T., De Beer, D., Elvert, M., Nadalig, T., Knittel, K., Amann, R., Sauter, E. J., Schluter, M., Klages, M., Foucher, J. P., and Boetius, A. (2006b). Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443, 854–858.
Nunoura, T., Oida, H., Toki, T., Ashi, J., Takai, K., and Horikoshi, K. (2006). Quantification of mcrA by quantitative fluorescent PCR in sediments from methane seep of the Nankai Trough. FEMS Microbiol. Ecol. 57, 149–157.
Orphan, V. J., House, C. H., Hinrichs, K.-H., Mckeegan, K. D., and Delong, E. F. (2002). Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. U.S.A. 99, 7663–7668.
Øvreås, L., Forney, L., Daae, F. L., and Torsvik, V. (1997). Distribution of bacterioplankton in meromictic lake saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63, 3367–3373.
Pernthaler, A., Pernthaler, J., and Amann, R. (2002). Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68, 3094–3101.
Petersen, J. M., and Dubilier, N. (2009). Methanotrophic symbioses in marine invertebrates. Environ. Microbiol. Rep. 1, 319–335.
Pruesse, E., Quast, C., Knittel, K., Fuchs, B., Ludwig, W., Peplies, J., and Glöckner, F. O. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196.
Raghoebarsing, A. A., Arjan Pol, A., Van De Pas-Schoonen, K. T., Smolders, A. J. O., Ettwig, K. F., Rijpstra, W. I. C., Schouten, S., Damste, J. S. S., Op Den Camp, H. J. M., Jetten, M. S. M., and Strous, M. (2006). A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440, 918–921.
Reeburgh, W. S. (1976). Methane consumption in Cariaco Trench waters and sediments. Earth Planet. Sci. Lett. 28, 337–344.
Reed, D. W., Fujita, Y., Delwiche, M. E., Blackwelder, D. B., Sheridan, P. P., Uchida, T., and Colwell, F. S. (2002). Microbial communities from methane hydrate-bearing deep marine sediments in a forearc basin. Appl. Environ. Microbiol. 68, 3759–3770.
Rice, D. D. (1993). “Composition and origins of coalbed gas,” in AAPG Studies in Geology, Hydrocarbons from Coal, eds B. E. Law and D. D. Rice (Tulsa: American Association of Petroleum Geologists), 159–184.
Rivière, D., Desvignes, V., Pelletier, E., Chaussonnerie, S., Guermazi, S., Weissenbach, J., Li, T., Camacho, P., and Sghir, A. (2009). Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J 3, 700–714.
Roberts, H. H., and Aharon, P. (1994). Hydrocarbon-derived carbonate buildups of the northern Gulf of Mexico continental slope: a review of submersible investigations. Geol. Mar. Lett. 14, 135–148.
Robertson, C. E., Spear, J. R., Harris, J. K., and Pace, N. R. (2009). Diversity and stratification of archaea in a hypersaline microbial mat. Appl. Environ. Microbiol. 75, 1801–1810.
Santelli, C. M., Orcutt, B. N., Banning, E., Bach, W., Moyer, C. L., Sogin, M. L., Staudigel, H., and Edwards, K. J. (2008). Abundance and diversity of microbial life in ocean crust. Nature 253, 653–656.
Scheller, S., Goenrich, M., Boecher, R., Thauer, R. K., and Jaun, B. (2010). The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 465, 606–608.
Schippers, A., Köweker, G., Höft, C., and Teichert, B. M. A. (2010). Quantification of microbial communities in three forearc sediment basins off Sumatra. Geomicrobiol. J., 1–13.
Schippers, A., Neretin, L. N., Kallmeyer, J., Ferdelman, T. G., Cragg, B. A., Parkes, J. R., and Jørgensen, B. B. (2005). Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 433, 861–864.
Schippers, A., and Nerretin, L. N. (2006). Quantification of microbial communities in near-surface and deeply buried marine sediments on the Peru continental margin using real-time PCR. Environ. Microbiol. 8, 1251–1260.
Schrum, H. N., Spivack, A. J., Kastner, M., and D’Hondt, S. (2009). Sulfate-reducing ammonium oxidation: a thermodynamically feasible metabolic pathway in subseafloor sediment. Geology 37, 939–942.
Schulz, H. N., Brinkhoff, T., Ferdelman, T. G., Mariné, M. H., Teske, A., and Jørgensen, B. B. (1999). Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science 284, 493–495.
Siegert, M., Cichocka, D., Herrmann, S., Gründger, F., Feisthauer, S., Richnow, H.-H., Springael, D., and Krüger, M. (2011). Accelerated methanogenesis from aliphatic and aromatic hydrocarbons under iron and sulfate reducing conditions. FEMS Microbiol. Lett. 315, 6–16.
Sieh, K., and Natawitjaja, D. (2000). Neotectonics of the Sumatran fault, Indonesia. J. Geophys. Res. 105, 28295–28326.
So, C. M., Phelps, C. D., and Young, L. Y. (2003). Anaerobic transformation of alkanes to fatty acids by a sulfate-reducing bacterium, Hxd3. Appl. Environ. Microbiol. 69, 3892–3900.
Takai, K., and Horikoshi, K. (2000). Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 6, 5066–5072.
Teske, A. (2006). “Microbial community composition in deep marine subsurface sediments of ODP Leg 201: sequencing surveys and cultivations,” in Proc. ODP, Sci. Results 201, ed. B. B. Jørgensen College Station, Texas, 1–20.
Thauer, R. K. (2011). Anaerobic oxidation of methane with sulfate: on the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr. Opin. Microbiol. 14, 292–299.
Treude, T., Krüger, M., Boetius, A., and Jørgensen, B. B. (2005). Environmental control on anaerobic oxidation of methane in the gassy sediments of Eckernförde Bay (German Baltic). Limnol. Oceanogr. 50, 1771–1786.
Valentine, D. L., Solem, R. C., Kastner, M., Wardlaw, G. D., Boone, D. R., Kendall, M., Wang, X., Hill, T. M., Purdy, A., and Bartlett, D. H. (2005). Biogeochemical investigations of marine methane seeps, Hydrate Ridge, Oregon. J. Geophys. Res. 110, 1–17.
Wachter, E. A., and Hayes, J. M. (1985). Exchange of oxygen in carbon dioxide-phosphoric acid systems. Chem. Geol. 52, 365–374.
Webster, G., Newberry, C. J., Fry, J. C., and Weightman, A. J. (2003). Assessment of bacterial community structure in the deep sub-seafloor biosphere by 16S rDNA-based techniques: a cautionary tale. J. Microbiol. Methods 55, 155–164.
Webster, G., Parkes, R. J., Fry, J. C., and Weightman, A. J. (2004). Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Appl. Environ. Microbiol. 70, 5708–5713.
Wegener, G., Niemann, H., Elvert, M., Hinrichs, K.-U., and Boetius, A. (2008). Assimilation of methane and inorganic carbon by microbial communities mediating the anaerobic oxidation of methane. Environ. Microbiol. 10, 2287–2298.
Wehrmann, L. M., Risgaard-Petersen, N., Schrum, H. N., Walsh, E. A., Huh, Y., Ikehara, M., Pierre, C., D’Hondt, S., Ferdelman, T. G., Ravelo, A. C., Takahashi, K., and Zarikian, C. A. (2011). Coupled organic and inorganic carbon cycling in the deep subseafloor sediment of the northeastern bering sea slope (IODP Exp. 323). Chem. Geol. 284, 251–261.
Weinbauer, M. G., Beckmann, C., and Höfle, M. G. (1998). Utility of green fluorescent nucleic acid dyes and aluminum oxide membrane filters for rapid epifluorescence enumeration of soil and sediment bacteria. Appl. Environ. Microbiol. 64, 5000–5003.
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703.
Whiticar, M. J., Faber, E., and Schoell, M. (1986). Biogenic methane formation in marine and freshwater environments: CO2 reduction vs. acetate fermentation –isotope evidence. Geochim. Cosmochim. Acta 50, 693–709.
Widdel, F., and Bak, F. (1992). “Gram-negative mesophilic sulfate-reducing bacteria,” in The Prokaryotes, ed. M. Dworkin (New York: Springer), 3352–3372.
Wiedicke-Hombach, M., Ardhayastuti, S., Bruns, A., Delisle, G., Goergens, R., Hermawan, T., Kanamatsu, T., Lückge, A., Mohtadi, M., Mühr, P., Rahadyan, T., Riyadi, A., Rühlemann, C., Schippers, A., Schlömer, S., Taufik, M., Teichert, B., Vink, A., Weiss, W., Wijaya, H., Wöhrl, C., Zeibig, M., and Zoch, D. (2006). SUMATRA –The Hydrocarbon System of the Sumatra Forearc – SO189-2 Cruise Report. Hannover: Bundesanstalt für Geowissenschaften und Rohstoffe.
Yamada, T., Sekiguchi, Y., Hanada, S., Imachi, H., Ohashi, A., Harada, H., and Kamagata, Y. (2006). Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 56, 1331–1340.
Zengler, K., Richnow, H. H., Rossello-Mora, R., Michaelis, W., and Widdel, F. (1999). Methane formation from long-chain alkanes by anaerobic microorganisms. Nature 401, 266–269.
Keywords: DGGE, quantitative PCR, CARD–FISH, methane seep, stable isotopes, AOM, hydrocarbon-dependent methanogenesis
Citation: Siegert M, Krüger M, Teichert B, Wiedicke M and Schippers A (2011) Anaerobic oxidation of methane at a marine methane seep in a forearc sediment basin off Sumatra, Indian Ocean. Front. Microbio. 2:249. doi: 10.3389/fmicb.2011.00249
Received: 09 August 2011; Accepted: 23 November 2011;
Published online: 21 December 2011.
Edited by:
Andreas Teske, University of North Carolina at Chapel Hill, USAReviewed by:
Nils Risgaard-Petersen, Aarhus University, DenmarkHeide N. Schulz-Vogt, Max-Planck-Institute for Marine Microbiology, Germany
Copyright: © 2011 Siegert, Krüger, Teichert, Wiedicke and Schippers. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Martin Krüger, Federal Institute for Geosciences and Natural Resources, Stilleweg 2, 30655 Hannover, Germany. e-mail: martin.krueger@bgr.de
† Present address: Barbara Teichert, Institut für Geologie und Paläontologie, Universität Münster, 48149 Münster, Germany.