- Winogradsky Institute of Microbiology, Russian Academy of Sciences, Moscow, Russia
Members of the bacterial phylum Planctomycetes inhabit various aquatic and terrestrial environments. In this study, fluorescence in situ hybridization (FISH) was applied to assess the abundance and depth distribution of these bacteria in nine different acidic wetlands of Northern Russia. Planctomycetes were most abundant in the oxic part of the wetland profiles. The respective cell numbers were in the range 1.1–6.7 × 107 cells g−1 of wet peat, comprising 2–14% of total bacterial cells, and displaying linear correlation to the peat water pH. Most peatland sites showed a sharp decline of planctomycete abundance with depth, while in two particular sites this decline was followed by a second population maximum in an anoxic part of the bog profile. Oxic peat layers were dominated by representatives of the Isosphaera–Singulisphaera group, while anoxic peat was inhabited mostly by Zavarzinella- and Pirellula-like planctomycetes. Phylogenetically related bacteria of the candidate division OP3 were detected in both oxic and anoxic peat layers with cell densities of 0.6–4.6 × 106 cells g−1 of wet peat.
Introduction
The phylum Planctomycetes is a remarkable group of budding bacteria that possess highly distinctive cell morphology, peptidoglycan-less cell-walls, and a unique cell organization (Schlesner and Stackebrandt, 1986; Fuerst, 1995; Ward et al., 2006; Fuerst and Sagulenko, 2011). Members of this phylum are ubiquitous in a wide range of aquatic and terrestrial environments with diverse conditions. Recently, it was found that planctomycetes are highly abundant in acidic Sphagnum-dominated wetlands (Dedysh et al., 2006; Kulichevskaya et al., 2006). Several moderately acidophilic, peat-inhabiting planctomycetes were isolated in pure cultures and described as representatives of the novel genera Schlesneria, Singulisphaera, and Zavarzinella (Kulichevskaya et al., 2007b, 2008, 2009, 2012). Interestingly, fluorescence in situ hybridization (FISH) with the 16S rRNA-targeted, Planctomycete-specific oligonucleotide probes revealed two distinct population maxima of these bacteria within the profile of the peat bog Bakchar, West Siberia (Dedysh et al., 2006; Ivanova and Dedysh, 2006). The first population maximum was detected in the uppermost, oxic layer of the bog profile, while the second maximum was located at a depth of 30 cm below the water table level. Since all currently characterized peat-inhabiting planctomycetes are chemoorganotrophs capable of growth in aerobic or microaerobic but not in anoxic conditions (Kulichevskaya et al., 2007b, 2008, 2009, 2012), the present study was initiated in order to examine depth distribution of these bacteria in different types of northern wetlands and to compare planctomycete diversity in oxic and anoxic peat layers.
Materials and Methods
Sampling Sites
The peat samples were collected from the depths 0–10, 10–20, 20–30, 30–40, and 40–50 cm over the profiles of nine different acidic wetlands of West Siberia and European North Russia (Table 1). The trophic status of these wetlands varied from oligotrophic to eutrophic and the pH varied from 3.7 to 6.0. Six oligotrophic wetlands (Bakchar, Obukhovskoye, Sekirnoye, Torfjanoye, Dubrovskoye, and Valdayskoye) are dominated by Sphagnum mosses. Aapa-type mesotrophic fen Muksalma is covered by Calluna vulgaris and Carex spp., while two eutrophic fens, Obskoe and Blizhnee, are dominated by Carex spp. Three subsamples collected from each depth were combined to form one composite sample per site. The samples were transported to the laboratory in boxes containing icepacks, homogenized by cutting the peat material into small fragments (about 0.5 cm) with sterile scissors, and fixed for FISH or frozen at −20°C for DNA extraction within 1 day after sampling.
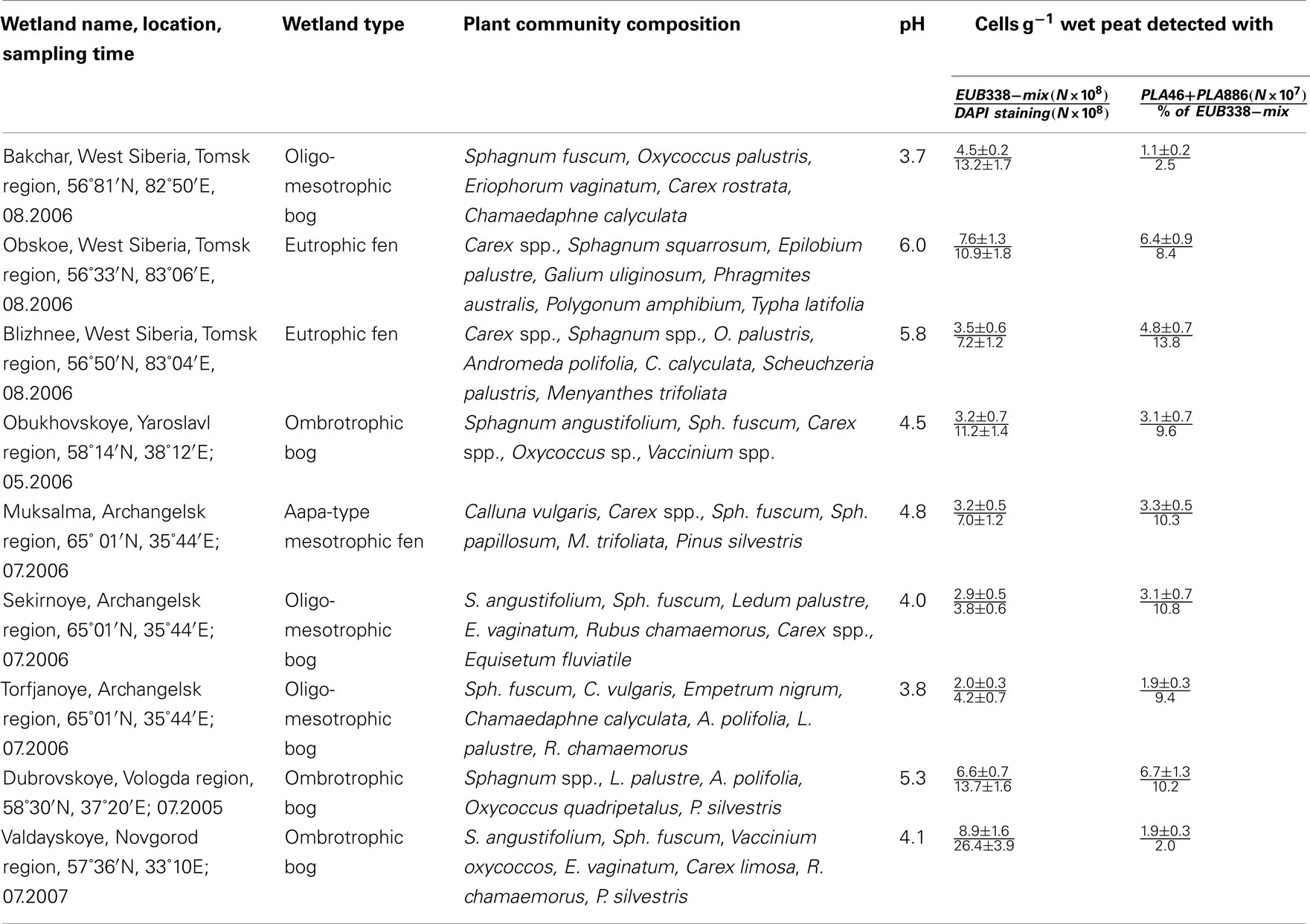
Table 1. The number of cells detected by FISH with the Planctomycetes-specific probes in acidic northern wetlands of different geographic location (data are presented for the uppermost (0–10 cm) peat bog layer corresponding to the maximal number of planctomycete cells detected over the bog profile).
Fluorescence in situ Hybridization
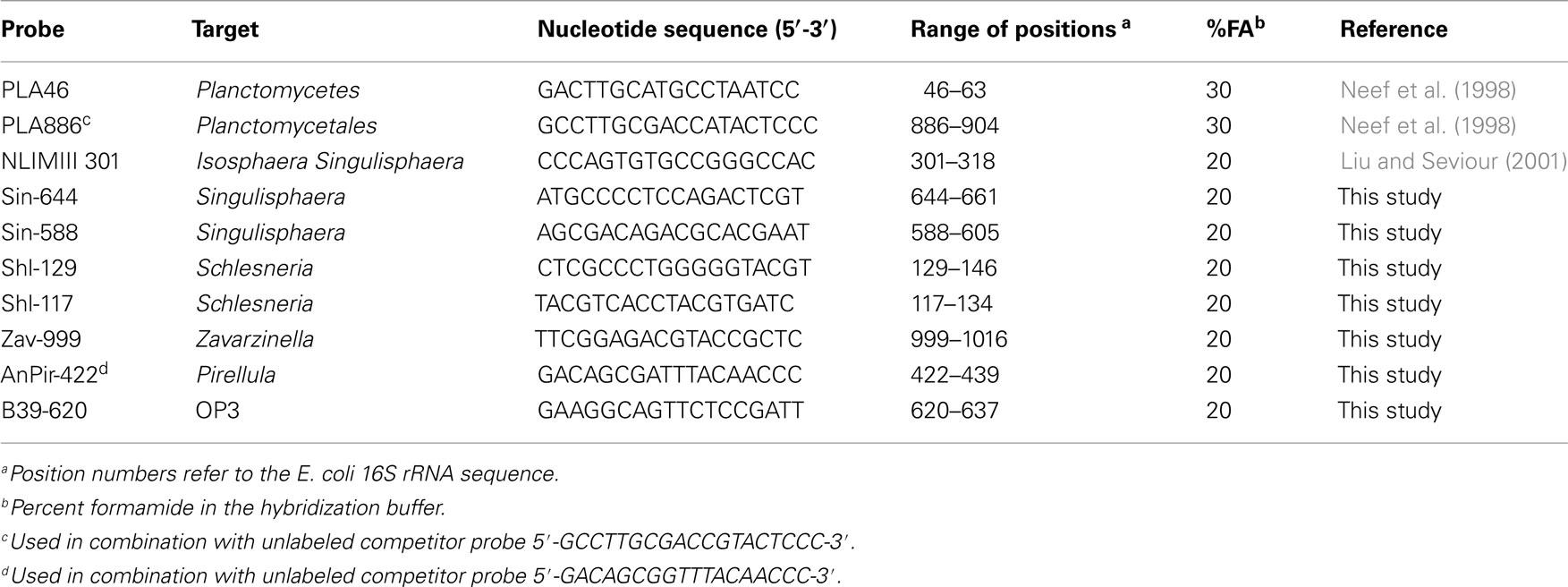
The fixation procedure was carried out according to Dedysh et al. (2001) and included (i) the separation of the peat water enriched with microbial cells and micro-particles of non-decomposed organic material from the rough (≥2–3 mm) Sphagnum debris by repeated stomacher treatments, (ii) recovery of the microbial cells from the peat water, and (iii) cell fixation with 4% (w/v) freshly prepared paraformaldehyde solution. Total bacterial cell numbers were determined by means of hybridization with Cy3-labeled oligonucleotide probes EUB338-mix (Daims et al., 1999). A combination of two Cy3-labeled oligonucleotide probes PLA46 and PLA886 (Neef et al., 1998) was applied for specific detection of planctomycetes. A set of eight oligonucleotide probes was used for differential detection and enumeration of specific sub-groups within the Planctomycetes (Table 2). This set included the probe NLMIII 301 designed by Liu and Seviour (2001) for members of the Isosphaera-group and seven newly developed probes for specific detection of peat-inhabiting planctomycetes. All oligonucleotide probes applied in this study were purchased from Syntol (Moscow, Russia). Hybridization was done on gelatin-coated (0.1%, w/v) and dried Teflon-laminated slides (MAGV, Germany) with eight wells for independent positioning of the samples. The fixed samples were applied to these wells, hybridized to the corresponding fluorescent probes, and stained with the universal DNA stain 4′,6-diamidino-2-phenylindole (DAPI, 1 μM) as described earlier (Dedysh et al., 2001). The cell counts were carried out with a Zeiss Axioplan 2 microscope (Zeiss, Jena, Germany) equipped with the Zeiss Filters No 20 and 02 for Cy3-labeled probes and DAPI staining, respectively. Cell counting was performed on 100 randomly chosen fields of view (FOV) for each test sample. The number of target cells per gram of wet peat was determined from the area of the sample spot, the FOV area, the volume of the fixed sample used for hybridization, and the volume of the peat water extracted from the sample.
DNA Extraction
Extracts of total DNA were obtained from two peat samples collected from the uppermost, oxic (0–10 cm depth) peat layers of Obukhovskoye and Bakchar peat bogs, and one peat sample collected from the anoxic (30–40 cm depth) layer of the peat bog Bakchar. Two subsamples, each of 0.5 g wet weight, were taken from each peat sample and processed separately. The extraction was performed using a FastDNA SPIN kit for Soil (Bio101, Carlsbad, USA) according to manufacturer’s instruction.
PCR Amplification, Cloning, and Sequencing
PCR-mediated amplification of the 16S rRNA gene fragments (~1350 bp) of peat-inhabiting planctomycetes was performed using the combination of the Planctomycete-specific forward primer Pla46 (5′-GGA TTA GGC ATG CAA GTC-3′) targeting positions 46–63 of Escherichia coli 16S rRNA gene (Neef et al., 1998) and the universal bacterial reverse primer Univ1390R (5′-GAC GGG CGG TGT GTA CAA-3′; Zheng et al., 1996). PCR mixtures (100 μl) contained 1 μl of template DNA, 50 μl of 2× MasterMix (Promega), and 0.3 μM of each primer (Syntol). PCR amplifications were performed in a DNA thermal cycler (model 9700; PE Applied Biosystems) under the following conditions: initial denaturation (4 min at 94°C), 33 cycles consisting of denaturation (1 min at 94°C), primer annealing (1 min at 59°C), and elongation (1.5 min at 72°C), with a final elongation step for 7 min at 72°C. Aliquots (5 μl) of the 16S rRNA gene amplicons were checked on a 1.2% (w/v) agarose gel and visualized after being stained with ethidium bromide. In order to reduce the potential bias of separate PCR reactions, the amplicons of two independent PCRs were combined before cloning. The mixed PCR product was cloned using a pGEM-T Easy Vector System II (Promega) according to the manufacturer’s instructions. Clones were screened for the correct insert with T7 and SP6 primers. Plasmid DNA was purified using Wizard Plus Minipreps DNA Purification System (Promega) and the cloned inserts were sequenced using an ABI 377A sequencer (Perkin–Elmer Applied Biosystems, USA).
The 16S rRNA gene sequences were checked for chimeras using Bellerophon 3.0 with the Huber–Hugenholtz correction and a 300 bp window size (Huber et al., 2004). Sequence alignments and phylogenetic analyses were carried out using the ARB software package (Ludwig et al., 2004). Trees were constructed using the neighbor-joining method. The significance levels of interior branch points obtained in neighbor-joining analysis were determined by bootstrap analyses (1000 data resamplings) using the PHYLIP program package (Felsenstein, 1989). The 16S rRNA gene sequences retrieved from Sphagnum peat in this study have been deposited in the GenBank, EMBL, and DDBJ nucleotide sequence databases under accession numbers JN867640 to JN867719.
Results
Abundance of Planctomycetes in Northern Wetlands
We applied a combination of two planctomycete-specific oligonucleotide probes PLA46 and PLA886 to determine the in situ abundance of these bacteria in peat sampled from nine acidic wetlands of different geographic locations within European North Russia and West Siberia (Table 1). The probes hybridized to numerous spherical or ellipsoid-shaped cells that were arranged in chains or in shapeless cell aggregates and were mostly attached to the particles of non-decomposed organic material (Figure 1). Highest abundances of cells targeted with the probes PLA46 and PLA886 were observed in the uppermost, oxic layers of the wetland profiles (Table 1; Figure 2). The cell numbers of planctomycetes in these layers were in the range 1.1–6.7 × 107 cells g−1 of wet peat, comprising 2–14% of cells detected in peat with the Bacteria-specific probe Eub338-mix. The abundance of planctomycetes in wetlands was strongly and positively correlated with pH value of peat water (r = 0.897; p value < 0.002; Figure 3). Highest cell numbers of these bacteria were detected in mildly acidic (pH 5.3–6.0) fens, whereas low planctomycete abundance was more typical for more acidic (pH 3.7–4.5) ombrotrophic bogs. By contrast, we did not observe any correspondence between the pH value of peat water and the cell numbers determined by hybridization with the Bacteria-specific probe Eub338-mix (Table 1).
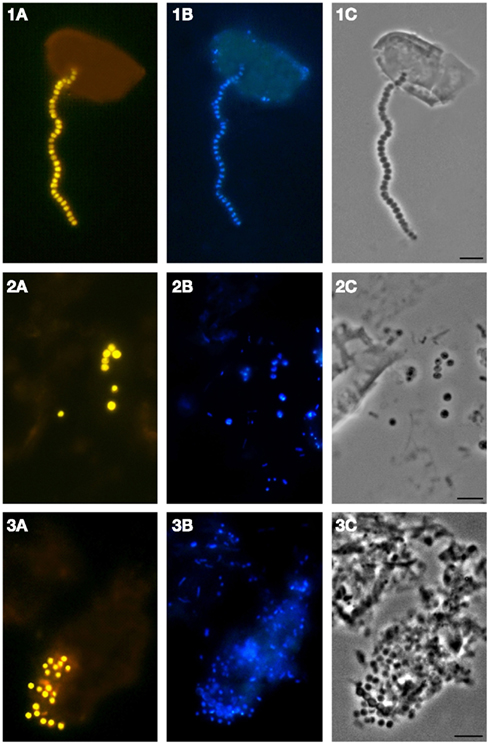
Figure 1. Specific detection of planctomycete cells in Sphagnum peat by FISH. Epifluorescent micrographs of in situ hybridizations with Cy3-labeled probes PLA46 and PLA886 (A), DAPI staining (B), and the phase-contrast images (C) are shown. Bar, 10 μm.
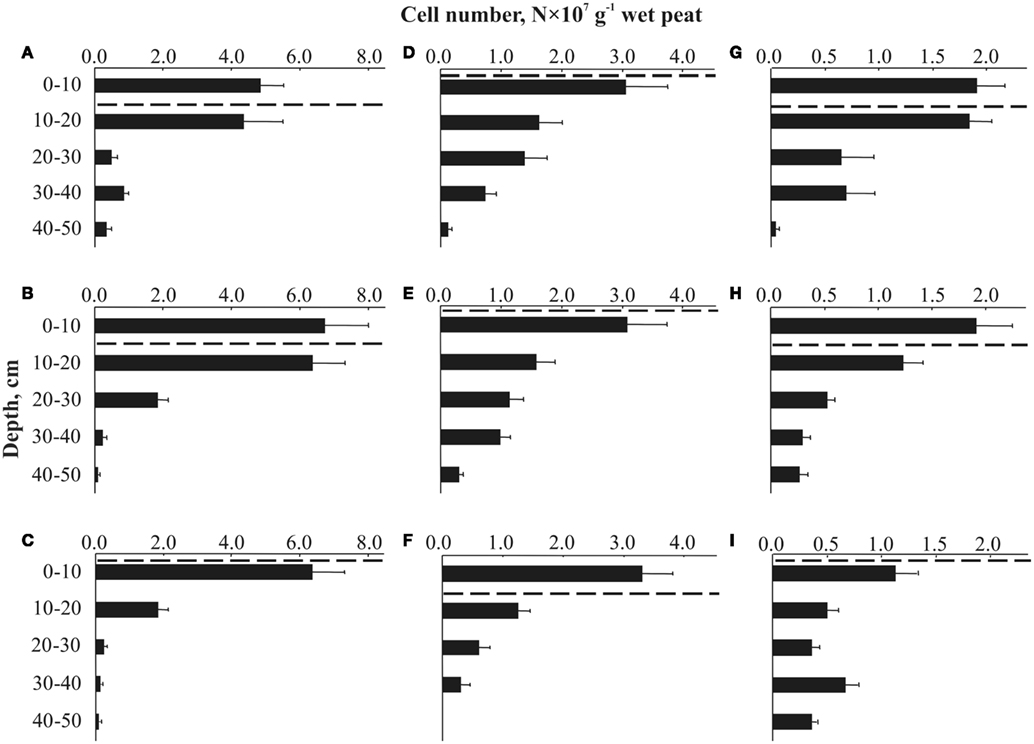
Figure 2. Depth distribution of cells targeted by FISH with the probes PLA46 and PLA886 in Sphagnum peat bogs located in different regions of Russia: (A) Blizhnee (West Siberia, Tomsk region); (B) Dubrovskoye (Vologda region); (C) Obskoe (West Siberia, Tomsk region); (D) Obukhovskoye (Yaroslavl region); (E) Sekirnoye (Archangelsk region); (F) Muksalma (Archangelsk region); (G) Torfjanoye (Archangelsk region); (H) Valdayskoye (Novgorod region); (I) Bakchar (West Siberia, Tomsk region). The dashed line indicates the water table level during the sampling. Values are means from 100 randomly chosen counting fields of view; error bars indicate SE.
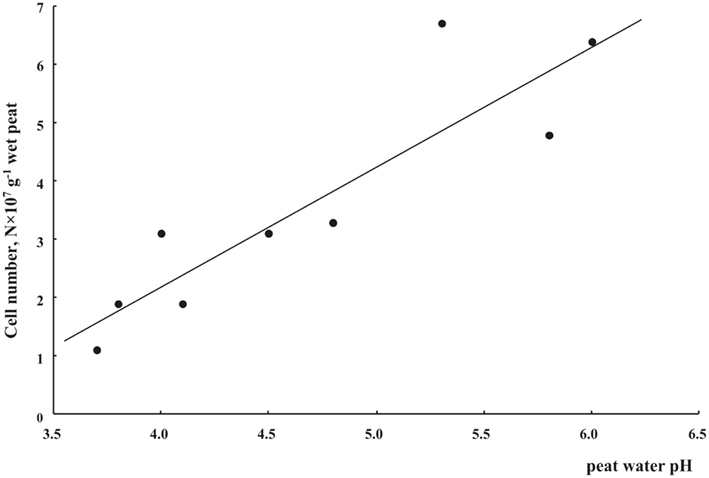
Figure 3. Cell numbers of planctomycetes in oxic (0–10 cm depth) Sphagnum peat as related to peat water pH.
Most peatland sites showed a sharp decline of planctomycete abundance with depth, while in two particular sites (wetlands Bakchar and Blizhnee) this decline was followed by a second population maximum in an anoxic part of the wetland profile (Tukey’s HSD test, p < 0.05; Figures 2A,I).
Culture-Independent Identification of Peat-Inhabiting Planctomycetes
A total of eighty 16S rRNA gene clones were randomly selected for sequencing and analyzed phylogenetically. The sequences retrieved from the oxic peat layer of the peat bog Obukhovskoye were abbreviated O-Ox. The sequences obtained from the oxic and anoxic peat from the peat bog Bakchar were abbreviated B-Ox and B-An (Figure 4). Comparative sequence analysis showed that 74 of eighty 16S rRNA gene sequences affiliate with the order Planctomycetales of the phylum Planctomycetes. None of the sequences retrieved in this study belonged to Anammox-planctomycetes (Jetten et al., 2005, 2010). Finally, six clones displayed high sequence similarity (97.3–99.3%) to the environmental clone sequence B39 (AM162486) retrieved from the West Siberian peat bog Bakchar in our previous study (Dedysh et al., 2006) and to two other environmental clone sequences, Molly56 (AY775515, 95–96% similarity) and Molly91 (AY775535, 93–94% similarity) retrieved from a Sphagnum peat bog in New England, USA (Morales et al., 2006). Taken together, these clones formed a separate phylogenetic cluster and belonged to the sub-lineage V within the candidate division OP3 (Glöckner et al., 2010).
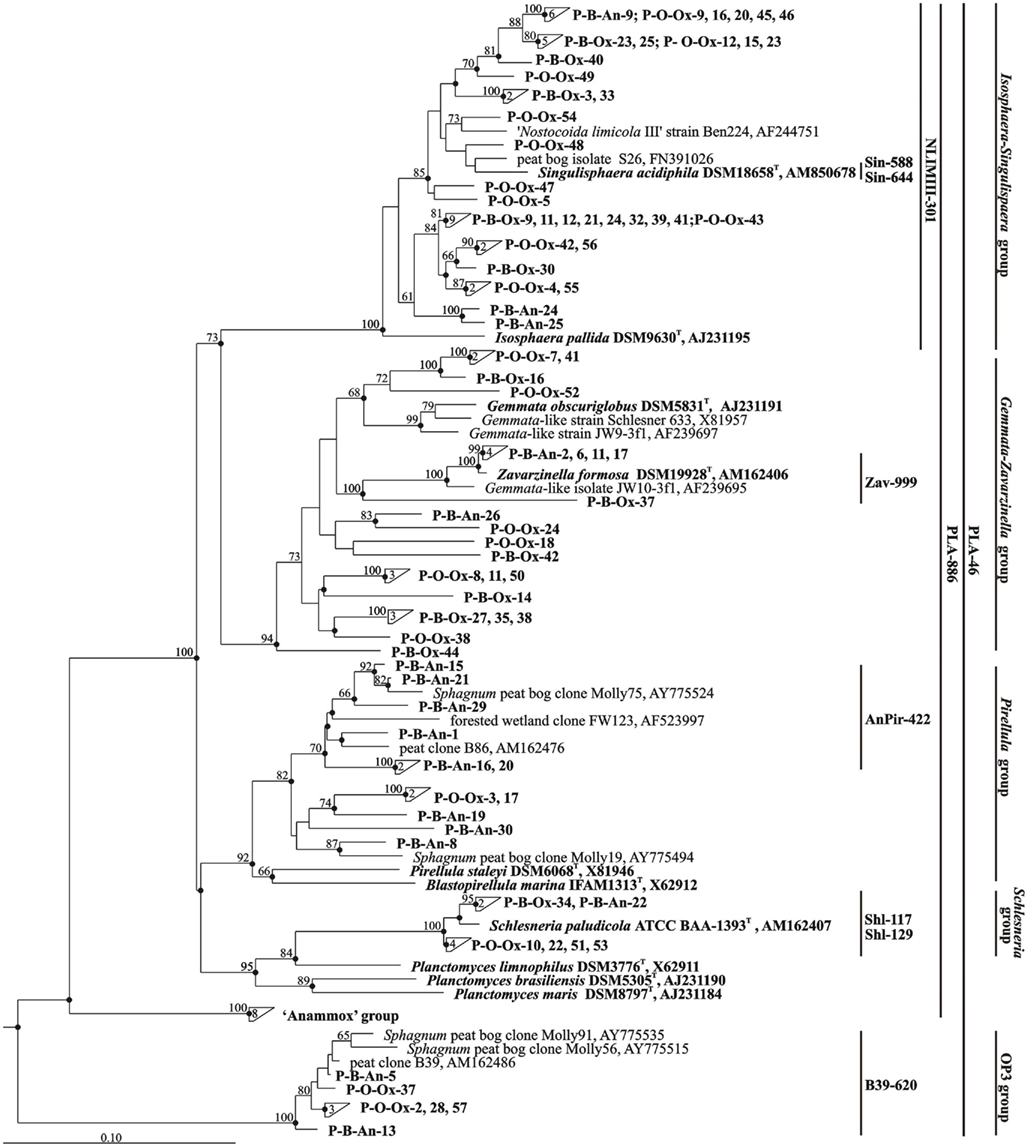
Figure 4. Neighbor-joining phylogenetic tree showing the relative positions of 80 peat-derived clones and 16S rRNA gene sequences of taxonomically described planctomycetes. Environmental sequences that were retrieved from oxic and anoxic peat layers of the peat bog Bakchar are labeled as B-Ox and B-An, respectively. The clones obtained from oxic peat layer of the peat bog Obukhovskoye are indicated as O-Ox. The numbers inside triangles indicate the number of sequences in these clusters. The brackets indicate the scope of each of the oligonucleotide probes applied in this study. The root was composed of ten 16S rRNA gene sequences from members of the Acidobacteria (accession numbers AM887757, AM887756, AM887758, AM887759, DQ660892, DQ528760, DQ528761, D26171, AM162405, and AY234728). Bootstrap values (1000 data resamplings) >50% are shown. Bar, 0.10 substitutions per nucleotide position.
The clone libraries constructed for two oxic peat samples from wetlands in West Siberia (peat bog Bakchar) and European North Russia (peat bog Obukhovskoye) had highly similar composition. More than half of all clones (53–61%) in both clone libraries affiliated with the phylogenetic lineage defined by the genera Isosphaera and Singulisphaera. None of the peat-derived sequences, however, displayed high similarity (≥97%) to those in taxonomically described members of this lineage. A group of 14 cloned 16S rRNA gene sequences showed only a distant relationship (88.8–92.8% sequence similarity) to 16S rRNA gene sequence from Isosphaera pallida, a neutrophilic filamentous planctomycete from a hot spring (Giovannoni et al., 1987; Figure 4). Another group of 18 clones displayed moderate similarity (93.8–95.5%) to 16S rRNA gene sequences of acidophilic peat-inhabiting planctomycete Singulisphaera acidiphila (Kulichevskaya et al., 2008) and of taxonomically uncharacterized planctomycete from activated sludge, “Nostocoida limicola III” (Liu et al., 2001). The second abundant group of sequences (25–35% of all clones) obtained from oxic peat belonged to the phylogenetic lineage defined by the genera Gemmata and Zavarzinella. Four clones belonged to the Planctomyces-like group and displayed high sequence similarity (98.0–98.8%) to the 16S rRNA gene sequence of another characterized peat-inhabiting planctomycete Schlesneria paludicola (Kulichevskaya et al., 2007b). Finally, only two clones affiliated with Pirellula-like planctomycetes.
By contrast, most 16S rRNA gene sequences (45% of all clones) retrieved from the anoxic peat sample clustered within the group defined by the genus Pirellula. These sequences displayed only a distant relationship (85.7–89.7% sequence similarity) to those from taxonomically described representatives of this planctomycete group, but were highly similar (95.2–98.7%) to the environmental clone sequences Molly75 (AY775524), Molly19 (AY775494), and B86 (AM162476) obtained from Sphagnum-dominated wetlands in the USA and Russia (Dedysh et al., 2006; Morales et al., 2006). One-fourth of all sequences retrieved from anoxic peat belonged to the Gemmata–Zavarzinella-related group of planctomycetes. Four of these sequences displayed high similarity (99.5–99.7%) to 16S rRNA gene sequence of Zavarzinella formosa (Kulichevskaya et al., 2009). This planctomycete was described as an aerobic organism, which is not capable of growth in micro-oxic or anoxic conditions. Minor groups of clones in a clone library made of anoxic peat were the sequences from Isosphaera–Singulisphaera-like (15% of all clones) and Schlesneria-like (5%) planctomycetes.
Differential Detection of Specific Sub-Groups of Planctomycetes Over the Bog Profile
Since the clone library composition may not adequately reflect the quantitative proportion of different bacterial groups within microbial communities, we attempted to assess the population structure of peat-inhabiting planctomycetes by means of FISH with the probes designed for differential detection of specific sub-groups within the Planctomycetes (Table 2). Six of these probes were developed in this study based on 16S rRNA gene sequences of the recently described acidophilic planctomycetes, S. paludicola, S. acidiphila, and Z. formosa, and related environmental clone sequences retrieved from acidic peat. One additional probe, B39–620, was designed to specifically target the OP3-related group of bacteria from Sphagnum-dominated wetlands. The target specificity of all probes applied in this study is indicated by brackets in Figure 4.
The set of probes listed in Table 2 was applied to assess the depth distribution of specific planctomycete groups in the peat bog Bakchar, West Siberia (Figure 5). The largest detectable population of indigenous planctomycetes hybridized with a probe targeting members of the Isosphaera–Singulisphaera group (Figure 5A). The population size of these planctomycetes declined from 3.7 × 107 cells g−1 wet peat in the uppermost, oxic peat layer to 0.1 × 107 cells g−1 wet peat at a depth of 40–50 cm. Representatives of the genus Schlesneria were significantly less abundant but showed the same distribution trend, with the population maximum in oxic peat (5.0 × 106 cells g−1 wet peat) versus population minimum (0.2 × 106 cells g−1 wet peat) in anoxic part of the bog profile (Figure 5B). The number of cells targeted with Pirellula-specific probe AnPir-422 was in the range of 1.0–2.1 × 106 cells g−1 wet peat and did not show significant variations within the bog profile. The most unexpected distribution pattern was observed for Zavarzinella-like planctomycetes, which peaked up to 4.4 × 106 cells g−1 wet peat at a depth of 30–40 cm. Finally, a minor but clearly detectable population of spherical cells hybridized with a probe targeting OP3-related organisms. These bacteria were enumerated at 0.6–4.6 × 106 cells g−1 wet peat, which accounted for 9–20% of all cells targeted with the probes PLA46 + PLA886 (shown by white in Figure 5A). Given that OP3-related bacteria do not belong to the Planctomycetes (discussed below), we have corrected our estimations of the planctomycete abundance in peat as follows: Nplanctomycetes = NPLA46 + PLA886 − NB39–620. The resulting range of planctomycete abundance within the profile of peat bog Bakchar was 0.6–4.6 × 107 cells g−1 wet peat. The estimated overall coverage of the group-specific probes used in our study was 92–98%.
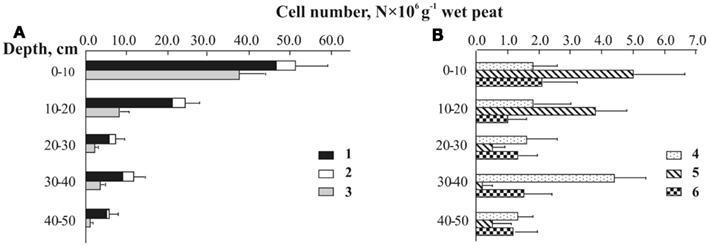
Figure 5. Depth distribution of planctomycete cells detected by means of in situ hybridization with group-specific probes in Sphagnum peat bog Bakchar: (A) 1-total cell number of planctomycetes, 2 – OP3-related group, 3 – Isosphaera–Singulisphaera group, (B) 4 – Zavarzinella-like planctomycetes, 5 – Schlesneria spp., 6 – Pirellula-like planctomycetes.
Discussion
As evidenced by a number of recent studies, planctomycetes densely colonize diverse acidic ecosystems, such as acidic northern wetlands (Dedysh et al., 2006; Kulichevskaya et al., 2006; this study), tank-forming bromeliads in tropical rainforests (Goffredi et al., 2011), and acidic hot springs (Bohorquez et al., 2012). Various water-saturated habitats with primarily decay-resistant organic matter seem to be one of the preferred econiches of these bacteria. The population size of planctomycetes in peatlands – 107 cells g−1 of wet peat – is especially high in comparison to many other ecosystems. For example, the peak of planctomycete abundance, which is usually observed in marine habitats during a diatom bloom, corresponds to 104 cells ml−1 (Morris et al., 2006; Pizzetti et al., 2011).
The highest population sizes of planctomycetes were revealed in mildly acidic peat sampled from two eutrophic fens, Obskoe (pH 6.0) and Blizhnee (pH 5.8), as well as in peat sampled from ombrotrophic bog Dubrovskoye (pH 5.3). Vice versa, the lowest numbers of cells targeted with the probes PLA46 + PLA886 were detected in more acidic (pH 3.7–4.1) peat bogs Bakchar, Torfjanoye, and Valdayskoye. These data suggest that most peat-inhabiting planctomycetes are moderately acidophilic bacteria with pH growth optima similar to those in Schlesneria, Singulisphaera, and Zavarzinella (Kulichevskaya et al., 2007b, 2008, 2009, 2012).
The culture-independent analysis of 16S rRNA gene sequences retrieved from acidic peat using Planctomycetes-specific primer pair revealed the presence of a highly diverse community, which consisted mostly of as-yet-uncultivated and uncharacterized planctomycetes. This highlights the need for further cultivation efforts in uncovering the Planctomycetes diversity in acidic northern wetlands. Notably, none of the sequences retrieved in our study belonged to Anammox-planctomycetes. This is not surprising since only one anoxic peat sample was examined in our study, and this particular sample was taken from an oligotrophic Sphagnum peat bog where available forms of nitrogen are mostly at very low or undetectable levels.
Based on the results of differential cell counts, we were able to compare the planctomycete community structure in oxic (0–10 cm depth) and anoxic (30–40 cm depth) peat layers (Figure 6). Oxic peat was dominated by members of the Isosphaera–Singulisphaera group, which accounted for up to 80% of all planctomycete cells detected by FISH. The second numerically significant planctomycete group in oxic peat was represented by members of the genus Schlesneria. By contrast, the most abundant planctomycete population in anoxic peat was Zavarzinella-like bacteria (45% of all planctomycete cells), while Isosphaera–Singulisphaera and Pirellula-like planctomycetes accounted for 30 and 20% of all detectable planctomycetes, respectively. High abundance of Zavarzinella-like planctomycetes in anoxic peat is difficult to explain based on the currently available information. Z. formosa was originally described as an obligately aerobic chemoheterotroph, which is not capable of growth in microaerobic or anoxic conditions (Kulichevskaya et al., 2009). Cultivation-independent retrieval of Zavarzinella-like 16S rRNA gene sequences from anoxic peat prompted us to re-analyze the ability of these planctomycetes to grow in anoxic conditions using the only currently available strain of these bacteria, Z. formosa A10T, but the result was negative. Possibly, other members of this genus may differ from the single taxonomically characterized representative by a number of traits. The occurrence of Pirellula-like planctomycetes in anoxic environments was well-documented before (Derakshani et al., 2001; Chouari et al., 2003; Kirkpatrick et al., 2006; Elshahed et al., 2007).
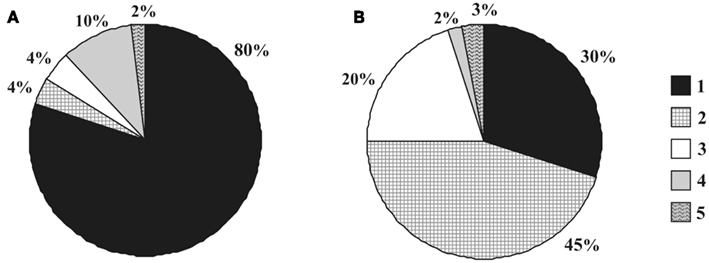
Figure 6. Community structure of planctomycetes in oxic (A) versus anoxic (B) peat: 1 – Isosphaera–Singulisphaera group, 2 – Zavarzinella-like planctomycetes, 3 – Pirellula-like planctomycetes, 4 – Schlesneria spp., 5 – others.
Based on our current knowledge, planctomycetes are slow-acting decomposers of plant-derived organic matter. They were identified as a numerically abundant component of a bacterial community that develops in the course of Sphagnum moss decomposition (Kulichevskaya et al., 2007a). Notably, the cell numbers of planctomycetes increased at the final stage of decomposition process. Given also that all currently characterized peat-inhabiting planctomycetes are capable of degrading heteropolysaccharides but not cellulose or chitin (Kulichevskaya et al., 2007b, 2008, 2009, 2012), the role of primary degraders is unlikely to be attributed to this group of bacteria. Yet, the biogeochemical role of planctomycetes in peatlands remains to be clarified since characterized representatives make only a minor part of the whole planctomycete diversity in acidic wetlands.
Our study provided the first quantitative estimation of the OP3-related bacteria in acidic wetlands. Candidate division OP3 (Obsidian Pool 3) was originally described based on a single 16S rRNA gene sequence retrieved from Obsidian Pool sediment (Hugenholtz et al., 1998). Later, OP3-related 16S rRNA gene sequences were obtained from a flooded paddy soil using a forward PCR primer PLA-40F that was designed for specific detection of the phyla Planctomycetes and Verrucomicrobia (Derakshani et al., 2001). Recent phylogenetic analysis of all currently available OP3-related 16S rRNA gene sequences showed that they represent at least five different subdivision level lineages, and the sequences retrieved from Sphagnum wetlands belong to the sub-lineage V (Glöckner et al., 2010). So far, OP3 bacteria were detected mostly in anoxic environments. The metagenomic analysis revealed that they share a high proportion of orthologs with members of the Deltaproteobacteria and appear to be capable of anaerobic respiration (Glöckner et al., 2010). In our study, we detected these bacteria in both oxic and anoxic parts of the bog profile, which suggests that at least members of the sub-lineage V may possess a facultatively anaerobic lifestyle.
In summary, we revealed the presence of a highly abundant and diverse community of Planctomycetes in acidic northern wetlands. These bacteria inhabit both oxic and anoxic peat layers and belong mostly to as-yet-undescribed taxa. Further work is needed, therefore, to characterize the unknown planctomycetes found in this habitat and to explore their physiology and functional role in wetlands.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the Program “Molecular and Cell Biology” of Russian Academy of Sciences and the Russian Fund of Basic Research (project No 06-04-49148). The authors thank Marc Dumont for editing this manuscript.
References
Bohorquez, L. C., Delgado-Serrano, L., López, G., Osorio-Forero, C., Klepac-Ceraj, V., Kolter, R., Junca, H., Baena, S., and Zambrano, M. M. (2012). In-depth characterization via complementing culture-independent approaches of the microbial community in an acidic hot spring of the Colombian Andes. Microb. Ecol. 63, 103–115.
Chouari, R., Le Paslier, D., Daegelen, P., Ginestet, P., Weissenbach, J., and Sghir, A. (2003). Molecular evidence for novel planctomycete diversity in a municipal wastewater treatment plant. Appl. Environ. Microbiol. 69, 7354–7363.
Daims, H., Brühl, A., Amann, R., Schleifer, K.-H., and Wagner, M. (1999). The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22, 434–444.
Dedysh, S. N., Derakshani, M., and Liesack, W. (2001). Detection and enumeration of methanotrophs in acidic Sphagnum peat by 16S rRNA fluorescence in situ hybridisation, including the use of newly developed oligonucleotide probes for Methylocella palustris. Appl. Environ. Microbiol. 67, 4850–4857.
Dedysh, S. N., Pankratov, T. A., Belova, S. E., Kulichevskaya, I. S., and Liesack, W. (2006). Phylogenetic analysis and in situ identification of bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 72, 2110–2117.
Derakshani, M., Lukow, T., and Liesack, W. (2001). Novel bacterial lineages at the (sub) division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms. Appl. Environ. Microbiol. 67, 623–631.
Elshahed, M. S., Youssef, N. H., Luo, Q., Najar, F. Z., Roe, B. A., Sisk, T. M., Bühring, S. I., Hinrichs, K.-U., and Krumholz, L. R. (2007). Phylogenetic and metabolic diversity of Planctomycetes from anaerobic, sulfide- and sulfur-rich Zodletone Spring, Oklahoma. Appl. Environ. Microbiol. 73, 4707–4716.
Fuerst, J. A. (1995). The Planctomycetes: emerging models for microbial ecology, evolution and cell biology. Microbiology 141, 1493–1506.
Fuerst, J. A., and Sagulenko, E. (2011). Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 9, 403–413.
Giovannoni, S. J., Schabtach, E., and Castenholz, R. W. (1987). Isosphaera pallida, gen. nov., and comb. nov., a gliding, budding eubacterium from hot springs. Arch. Microbiol. 147, 276–284.
Glöckner, J., Kube, M., Shrestha, P. M., Weber, M., Glöckner, F. O., Reinhardt, R., and Liesack, W. (2010). Phylogenetic diversity and metagenomics of candidate division OP3. Environ. Microbiol. 12, 1218–1229.
Goffredi, S. K., Kantor, A. H., and Woodside, W. T. (2011). Aquatic microbial habitats within a neotropical rainforest: bromeliads and pH-associated trends in bacterial diversity and composition. Microb. Ecol. 61, 529–542.
Huber, T., Faulkner, G., and Hugenholtz, P. (2004). Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20, 2317–2319.
Hugenholtz, P., Pitulle, C., Hershberger, K. L., and Pace, N. R. (1998). Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180, 366–376.
Ivanova, A. O., and Dedysh, S. N. (2006). High abundance of Planctomycetes in anoxic layers of a Sphagnum peat bog. Microbiology 75, 716–719.
Jetten, M. S. M., Cirpus, I., Kartal, B., van Niftrik, L., van de Pas-Schoonen, K. T., Sliekers, O., Haaijer, S., van der Star, W., Schmid, M., van de Vossenberg, J., Schmidt, I., Harhangi, H., van Loosdrecht, M., Kuenen, G. J., Op den Camp, H., and Strous, M. (2005). 1994–2004: 10 years of research on the anaerobic oxidation of ammonium. Biochem. Soc. Trans. 33, 119–123.
Jetten, M. S. M., Op den Camp, H., Kuenen, G. J., and Strous, M. (2010). “Order II. ‘Candidatus Brocadiales’ ord. nov.,” in Bergey’s Manual of Systematic Bacteriology, Vol. 4, eds N. R. Krieg, J. T. Staley, D. R. Brown, B. P. Hedlung, B. J. Paster, N. L. Ward, W. Ludwig, and W. B. Whitman (New York, NY: Springer), 918–925.
Kirkpatrick, J., Oakley, B., Fuchsman, C., Srinivasan, S., Staley, J. T., and Murray, J. W. (2006). Diversity and distribution of Planctomycetes and related bacteria in the suboxic zone of the Black Sea. Appl. Environ. Microbiol. 72, 3079–3083.
Kulichevskaya, I. S., Baulina, O. I., Bodelier, P. L. E., Rijpstra, W. I. C., Sinninghe Damsté, J. S., and Dedysh, S. N. (2009). Zavarzinella formosa gen. nov., sp. nov., a novel stalked, Gemmata-like planctomycete from a Siberian peat bog. Int. J. Syst. Evol. Microbiol. 59, 357–364.
Kulichevskaya, I. S., Belova, S. N., Kevbrin, V. V., Dedysh, S. N., and Zavarzin, G. A. (2007a). Analysis of the bacterial community developing in the course of Sphagnum moss decomposition. Microbiology 76, 621–629.
Kulichevskaya, I. S., Ivanova, A. O., Belova, S. E., Baulina, O. I., Bodelier, P. L. E., Rijpstra, W. I. C., Sinninghe Damsté, J. S., Zavarzin, G. A., and Dedysh, S. N. (2007b). Schlesneria paludicola gen. nov., sp. nov., the first acidophilic member of the order Planctomycetales from Sphagnum-dominated boreal wetlands. Int. J. Syst. Evol. Microbiol. 57, 2680–2687.
Kulichevskaya, I. S., Detkova, E. N., Bodelier, P. L. E., Rijpstra, W. I. C., Sinninghe Damsté, G. S., and Dedysh, S. N. (2012). Singulisphaera rosea sp. nov., a novel planctomycete from acidic Sphagnum peat. Int. J. Syst. Evol. Microbiol. 62, 118–123.
Kulichevskaya, I. S., Ivanova, A. O., Baulina, O. I., Bodelier, P. L. E., Sinninghe Damsté, J. S., and Dedysh, S. N. (2008). Singulisphaera acidiphila gen. nov., sp. nov., a non-filamentous, Isosphaera-like planctomycete from acidic northern wetlands. Int. J. Syst. Evol. Microbiol. 58, 1186–1193.
Kulichevskaya, I. S., Pankratov, T. A., and Dedysh, S. N. (2006). Detection of representatives of the Planctomycetes in Sphagnum peat bogs by molecular and cultivation approaches. Microbiology 75, 329–335.
Liu, J.-R., McKenzie, C. A., Seviour, E. M., Webb, R. I., Blackall, L. L., Saint, C. P., and Seviour, R. J. (2001). Phylogeny of the filamentous bacterium ‘Nostocoida limicola’ III from activated sludge. Int. J. Syst. Evol. Microbiol. 51, 195–202.
Liu, J.-R., and Seviour, R. J. (2001). Design and application of oligonucliotide probes for fluorescent in situ identification of the filamentous bacterial morphotype Nostocoida limicola in activated sludge. Environ. Microbiol. 3, 551–560.
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Kumar, Y., Buchner, A., Lai, T., Steppi, S., Jobb, G., Förster, W., Brettske, I., Gerber, S., Ginhart, A. W., Gross, O., Grumann, S., Hermann, S., Jost, R., König, A., Liss, T., Lüssmann, R., May, M., Nonhoff, B., Reichel, B., Strehlow, R., Stamatakis, A., Stuckmann, N., Vilbig, A., Lenke, M., Ludwig, T., Bode, A., and Schleifer, K. H. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371.
Morales, S. E., Mouser, P. J., Ward, N., Hudman, S. P., Gotelli, N. J., Ross, D. S., and Lewis, T. A. (2006). Comparison of bacterial communities in New England Sphagnum bogs using terminal restriction fragment length polymorphism. Microb. Ecol. 52, 34–44.
Morris, R. M., Longnecker, K., and Giovannoni, S. J. (2006). Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ. Microbiol. 8, 1361–1370.
Neef, A., Amann, R., Schlesner, H., and Schleifer, K.-H. (1998). Monitoring a widespread bacterial group: in situ detection of Planctomycetes with 16S rRNA-targeted probes. Microbiology 144, 3257–3266.
Pizzetti, I., Fuchs, B. M., Gerdts, G., Wichels, A., Wiltshire, K. H., and Amann, R. (2011). Temporal variability of coastal Planctomycetes clades at Kabeltonne station, North sea. Appl. Environ. Microbiol. 77, 5009–5017.
Schlesner, H., and Stackebrandt, E. (1986). Assignment of the genera Planctomyces and Pirella to a new family Planctomycetaceae fam. nov. and description of the order Planctomycetales ord. nov. Syst. Appl. Microbiol. 8, 174–176.
Ward, N., Staley, J. T., Fuerst, J. A., Giovannoni, S., Schlesner, H., and Stackebrandt, E. (2006). “The order Planctomycetales, including the genera Planctomyces, Pirellula, Gemmata and Isosphaera and the Candidatus genera Brocadia, Kuenenia and Scalindua,” in The Prokaryotes: A Handbook on the Biology of Bacteria, 3rd Edn, Vol. 7, eds M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (New York: Springer), 757–793.
Keywords: Planctomycetes, candidate division OP3, FISH, depth distribution, acidic northern wetlands
Citation: Ivanova AO and Dedysh SN (2012) Abundance, diversity, and depth distribution of Planctomycetes in acidic northern wetlands. Front. Microbio. 3:5. doi: 10.3389/fmicb.2012.00005
Received: 10 November 2011;
Paper pending published: 29 November 2011;
Accepted: 04 January 2012;
Published online: 18 January 2012.
Edited by:
Paul Bodelier, Netherlands Institute of Ecology, NetherlandsCopyright: © 2012 Ivanova and Dedysh. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Svetlana N. Dedysh, Winogradsky Institute of Microbiology, Russian Academy of Sciences, Prospect 60-Letya Octyabrya 7/2, Moscow 117312, Russia. e-mail: dedysh@mail.ru
 Anastasia O. Ivanova
Anastasia O. Ivanova