- Department of Pathology and Laboratory Medicine, The Institute for Heart and Lung Health, St. Paul’s Hospital, University of British Columbia, Vancouver, BC, Canada
Many virus infections and stresses can induce endoplasmic reticulum (ER) stress response, a host self-defense mechanism against viral invasion and stress. During this event, viral and cellular gene expression is actively regulated and often encounters a switching of the translation initiation from cap-dependent to internal ribosome-entry sites (IRES)-dependent. This switching is largely dependent on the mRNA structure of the 5′ untranslated region (5′ UTR) and on the particular stress stimuli. Picornaviruses and some other viruses contain IRESs within their 5′ UTR of viral genome and employ an IRES-driven mechanism for translation initiation. Recently, a growing number of cellular genes involved in growth control, cell cycle progression and apoptosis were also found to contain one or more IRES within their long highly structured 5′ UTRs. These genes initiate translation usually by a cap-dependent mechanism under normal physiological conditions; however, in certain environments, such as infection, starvation, and heat shock they shift translation initiation to an IRES-dependent modality. Although the molecular mechanism is not entirely understood, a number of studies have revealed that several cellular biochemical processes are responsible for the switching of translation initiation to IRES-dependent. These include the cleavage of translation initiation factors by viral and/or host proteases, phosphorylation (inactivation) of host factors for translation initiation, overproduction of homologous proteins of cap-binding protein eukaryotic initiation factors (eIF)4E, suppression of cap-binding protein eIF4E expression by specific microRNA, activation of enzymes for mRNA decapping, as well as others. Here, we summarize the recent advances in our understanding of the molecular mechanisms for the switching of translation initiation, particularly for the proteins involved in cell survival and apoptosis in the ER stress pathways during viral infections.
Introduction
Translation initiation is a rate-limiting step of protein synthesis. It is highly regulated by different mechanisms, depending on the structural distinction of mRNAs. Most cellular mRNAs are translated by a cap-dependent mechanism that requires the binding of the trimeric complex eukaryotic initiation factors (eIF)4F, comprised of eIF4G, eIF4E, and eIF4A, to the 7-methyl GpppN cap structure at the 5′ end of the mRNA. Some viral and cellular mRNAs have evolved a cap-independent mechanism of translation initiation that uses the internal ribosome-entry site (IRES) sequence that is located in the 5′ untranslated region (5′ UTR) of mRNA (Holcik and Sonenberg, 2005). The IRES was discovered first in poliovirus (a typical member of picornaviruses) and later in other viruses such as hepatitis C virus (HCV), HIV, Herpesviruses, etc., and also in many cellular mRNAs (Jang et al., 1988; Pelletier and Sonenberg, 1988; Labadie et al., 2004; Locker et al., 2011). The cellular physiological conditions dictate when a given mRNA uses cap-dependent or IRES-dependent translation initiation. Under normal conditions, cellular mRNAs translation is initiated by a cap-dependent manner; however, under stress conditions, such as starvation, irradiation, heat shock, hypoxia, toxin, and viral infection, the translation initiation is switched from cap-dependent to an IRES-driven mechanism (Komar and Hatzoglou, 2005; Spriggs et al., 2005).
Many viral infections trigger endoplasmic reticulum (ER) stress responses in a variety of ways inside the host cell. One of the most significant effects is the shutting off cap-dependant translation, which results in activation of IRES-dependent translational mechanisms. This is quite apparent in picornaviruses because their viral mRNA does not contain a cap structure at the 5′ end and its IRES located in the 5′ UTR recruits ribosomes and other factors, which then scan to reach the initiation codon without the requirement of the eIF4E (Jang, 2006; Jang et al., 2009). These viruses are able to benefit from the ER stress response, enhancing protein synthesis, and thus enhancing their self-defense capability. The mechanisms by which the virus infections and other stress signals achieve inhibition of cap-dependent translation of cellular mRNAs include: (i) specific cleavage of cellular translational initiation factors, such as the eukaryotic translation initiation factor 4GI (eIF4GI) by picornaviral and HIV proteases (Etchison et al., 1982; Lamphear et al., 1993; Ohlmann et al., 2002; Chau et al., 2007; Castello et al., 2009) or by cellular caspases (Marissen and Lloyd, 1998). (ii) Phosphorylation of eIF2α and other co-factors of translation. The cleavage or modification of the translation factors does not affect IRES-driven translation, instead promotes IRES-containing mRNA to utilize specific IRES transacting factors (ITAF) for their translation (Morley et al., 2005; Raught and Gringas, 2007). (iii) Overproduction of homologous proteins of cap-binding protein eIF4E (e.g., 4E-BP), which competes with eIF4G for binding (Marcotrigiano et al., 1999) to eIF4E (iv) suppression of eIF4E expression by certain microRNAs (miRNA; Mathonnet et al., 2007; Ho et al., 2011).
The rapid inhibition of cellular cap-dependent protein synthesis has been demonstrated as a critical precursor to cell fate, in this context, it is noteworthy that the IRES-containing cellular mRNAs are found to be preferentially involved in the control of cell fate by functioning to promote cell growth and survival or apoptosis (Spriggs et al., 2005; Sonenberg and Hinnebusch, 2009; Jackson et al., 2010). These genes include; Beta-cell lymphoma 2 (Bcl-2) family proteins, apoptotic protease activating factor 1 (Apaf-1), checkpoint homolog kinase 1(Chk-1), eIF4GII, p53, p58IPK, and 78 kDa Glucose-regulated protein 78 or Binding immunoglobulin protein (GRP78/BiP), etc. (Komar and Hatzoglou, 2005; Spriggs et al., 2005). Cellular genes containing IRESs in their mRNA are continually being newly discovered. However, to date, all of these genes are involved in apoptosis/proliferation, stress response, and/or cell cycle regulation. It was therefore suggested that IRES-mediated translation plays critical roles in regulation of cell fate (Spriggs et al., 2005). Previous studies have indicated that the cell fate decision is made based on the severity and duration of the stress signal. Under a transient stress or infection, the IRES will mediate translation initiation of genes promoting cell survival/growth, which enhance cellular capability to combat viral infection. However, under a severe or prolonged stress such as persistent infection of picornaviruses and others, translation initiation will selectively express the genes responsible for inducing cell apoptosis (Henis-Korenblit et al., 2002; Lewis et al., 2008), effectively destroying the host cells, and limiting viral infection of surrounding cells. In any circumstance, the host cell will employ an alternate way to defend itself. In this review we will discuss the recent advances in the understanding of IRES-mediated translational control of genes under stress conditions, particularly focus on ER stress caused by picornaviral and other viral infections.
Viral Induced ER Stress Response Pathways
Endoplasmic reticulum stress response is a major component of disease (Tabas and Ron, 2011). Many viral infections induce ER stress and have adapted mechanisms to modulate the stress response and its effectors. On the cellular level, ER stress may be triggered by many factors, including serum starvation, hypoxia, changes in calcium homeostasis, and viral infections as well as other perturbations (Chakrabarti et al., 2011). In general, ER stress is triggered by the accumulation of misfolded or unfolded proteins in the ER lumen. In response to this stress, a coordinated adaptive program termed the unfolded protein response (UPR) is activated and serves to minimize the accumulation and aggregation of misfolded proteins (Chakrabarti et al., 2011). The molecules and signaling pathways of the UPR may vary slightly dependant upon cell type. The stress response or UPR is regulated by master regulatory protein, BiP or GRP78. The initial phase of the ER stress response acts to increase the removal and folding of misfolded or unfolded proteins. In its non-stressed state, BiP is bound to the ER-luminal domain of the transmembrane proteins including PKR-like ER kinase (PERK), inositol requiring enzyme 1 (IRE1), and activating transcription factor 6 (ATF6; Chakrabarti et al., 2011). These are the three major arms of the UPR. Viral infection causes the rapid accumulation of viral and other cellular proteins trafficked to the ER. When excess proteins accumulate in the ER lumen, BiP dissociates from its three transmembrane sensors, which results in the initiation of the functional activation of the three major arms of the UPR. PERK and IRE1 are activated and undergo homodimerization and auto-phosphorylation (Liu et al., 2000; Bollo et al., 2010; Oikawa and Kimata, 2011), triggering their downstream genes. The activation of the IRE1 pathway leads to the splicing of X box binding protein 1 (XBP1; Lee et al., 2002). This spliced form of XBP1 mRNA encodes an active transcription factor that binds to the promoter of unfold protein response element (UPRE) to induce expression of a subset of genes encoding protein degradation enzymes, resulting in ER-associated misfolded protein degradation (Lee et al., 2003). The activation of PERK results in the phosphorylation of eIF2 on its α subunit (Raven and Koromilas, 2008). This effectively shuts down global, cap-dependant protein synthesis, and causes a shift in translation to that of cellular mRNA containing IRESs, reducing the burden of accumulating proteins in the ER (Harding et al., 2002). This constitutes a translational switch to IRES-mediated translation initiation. UPR activation also involves ATF6 activation, resulting in its migration to the Golgi apparatus, where it is cleaved by S1P and S2P proteases, releasing a soluble fragment that enters the nucleus and bind to promoters containing the ER stress response elements (ERSE) and ATF/cAMP response elements (CREs) to activate ER chaperone genes, such as BiP, GRP94, and calreticulin (Yoshida et al., 2001). These newly synthesized chaperones refold misfolded proteins in the ER in an effort to relieve ER stress. ATF6 also promotes XBP1 splicing (Lee et al., 2002), which indicates the interconnectedness of the three branches of the UPR. The shift from cap-dependant to cap-independent translation mediated by ER stress is critical to both cell fate and viral infection productivity. Many viruses, particularly RNA viruses, such as members of the Picornaviridae family, have evolved to replicate through cap-independent mechanisms, thus the shut-off of global protein synthesis induced by ER stress is of major strategic importance.
Endoplasmic reticulum stress when chronic or prolonged may lead to the induction of ER mediated apoptosis (Tabas and Ron, 2011). As is the case in viral infection, viral proteases also inhibit select cellular translational components, which may be initiated by ER stress. Our group has demonstrated that coxsackievirus B3 (CVB3) protease 2A and 3C can cleave eIFGI and induce cell apoptosis (Chau et al., 2007). Viral proteins, such as picornaviral protein 2B, have been shown to contribute to the depletion of calcium stores within the ER (Wang et al., 2011a), furthering the viral life cycle by contributing to viral release. Prolonged and sustained severe ER stress eventually drives the cell to apoptosis (Mekahli et al., 2011). Although significant progress in our understanding of apoptosis initiated by ER stress has been made in recent years, the molecular mechanisms of ER induced apoptosis are yet to be fully elucidated. The functions of the three branches of the UPR (IRE1, ATF6, PERK) act in concert during prolonged/severe ER stress to induce apoptosis. During these conditions, the endonuclease activity of IRE1 becomes less specific. As a result IRE1 contributes to the degradation of membrane associated mRNA, termed regulated IRE1 dependant degradation (RIDD). RIDD activation and XBP1 splicing indicate two distinct functions for IRE1 during ER stress, the former being pro-apoptotic and the latter generally regarded as protective (Hollien et al., 2009). Previous studies indicate a correlation between enhanced ER stress-induced apoptosis and the induction of RIDD activity. RIDD activity requires the nuclease domain of IRE1 to be activated whereas IRE1 induced XBP1 splicing is modulated by IRE1 kinase domain activation (Hollien et al., 2009). IRE1 has also been shown to bind Bcl-2 homologous antagonist/killer (Bak) and Bcl-2 associated x protein (Bax; Hetz et al., 2006), two pro-apoptotic proteins from the Bcl-2 family previously described in mitochondria derived apoptosis. Recently, however, it was shown that Bax translocates not only to the mitochondria, but also to the ER membrane during prolonged ER stress (McCullough et al., 2001; Gotoh et al., 2004; Hetz et al., 2006; Wang et al., 2011b). Once on the ER membrane, Bax permeabilizes the membrane, and causes the translocation of ER-luminal proteins to the cytosol (Wang et al., 2011b). Normally anti-apoptotic in function, BiP, once in the cytoplasm translocates to the plasma membrane where it becomes an apoptotic inducing receptor for prostate apoptosis response-4 (Par-4; Wang et al., 2011a). Par-4 has been shown to co-localize with BiP in the ER. The binding of Par-4 to membrane bound BiP activates the extrinsic apoptotic cascade through FADD, caspase-8, and caspase-3 (Burikhanov et al., 2009). Interestingly, the secretion of Par-4 is activated by TRAIL (Hart and El-Deiry, 2009). Several viruses including avian H5N1 and HIV have been shown promote cell death through TRAIL activated apoptosis in macrophages by enhancing TRAIL induced caspase-10 activation (Ekchariyawat et al., 2011; Zhu et al., 2011).
In addition, during prolonged and severe ER stress, PERK also enhances the translation of specific downstream genes, including activating transcription factor 4 (ATF4; Fels and Koumenis, 2006). ATF4 is able to activate pro-apoptotic C/EBP homologous protein (CHOP) in conditions of prolonged, severe ER stress (Ma et al., 2002). CHOP acts to induce apoptosis by promoting constitutively expressed Bax translocation to the mitochondria through inhibition of anti-apoptotic Bcl-2 transcription, as Bcl-2 functions to inhibit Bax in pro-survival conditions (McCullough et al., 2001; Gotoh et al., 2004). Here we see a connection between apoptosis mediated by IRE1 (by binding to Bax/Bak) and by PERK-mediated CHOP activation through ATF4, stressing the importance of cross talk between the three arms of the UPR. Interestingly, CHOP acts as a negative regulator of eIF2α phosphorylation as well (Novoa et al., 2001). The importance of these pathways in both global translation attenuation and apoptosis has made them the target of manipulation of many viruses. For example, hepatitis E virus (HEV) open reading frame 2 protein (ORF-2) is able to modulate ER stress-induced apoptosis by increasing eIF2α phosphorylation and activation of CHOP, simultaneously (John et al., 2011). Our lab also obtained a similar result in studying CVB3-induced apoptosis through phosphorylation of eIF2α and activation of CHOP; however, this activation is not through ATF4 but through ATF6 (Zhang et al., 2011b). For HEV, during infection, CHOP, which normally induces apoptosis and translocation of Bax to the mitochondria, is unable to perform this pro-apoptotic function. This is due to the simultaneous activation and interaction of heat shock proteins Hsp70B, Hsp72, and Hsp40 by HEV protein ORF-2 (John et al., 2011). Several members of the heat shock protein family, including Hsp70, have been demonstrated to contain an IRES element in its long 5′ UTR region of mRNA (Ahmed and Duncan, 2004; Hernandez et al., 2004). This strategic modulation of pro-apoptosis and pro-survival proteins occurs presumably to delay apoptosis, while allowing the viral replication cycle to continue to completion. This demonstrates the careful strategic interplay between the virus and host translational factors as well as host cell components of the UPR. In doing so, the virus is able to modulate the delicate balance between apoptosis and survival.
Structures of IRES
Classification of Viral IRESs
Internal ribosome-entry sites-dependant translation initiation was first described in 1988 in the 5′ UTR of the RNA genome of poliovirus (PV; Pelletier and Sonenberg, 1988). Since this original discovery, IRES elements have been identified in the long, highly structured 5′ UTR of almost all picornaviruses, including encephalomyocarditis virus (EMCV; Lindeberg and Ebendal, 1999), Foot-and-mouth disease virus (FMDV; Ohlmann and Jackson, 1999), CVB3 (Yang et al., 1997) human rhinoviruses (HRV; Rojas-Eisenring et al., 1995), and other viruses, such as, Hepatitis A (Ali et al., 2001), HIV (Weill et al., 2010), and DNA viruses such as Kaposi’s sarcoma herpesvirus (KSHV; Bieleski et al., 2004). Inherit to viral strategy viruses must utilize cellular translational machinery in order for translation and replication occur. Translation initiation is the rate-limiting step of translation, which is the reason that it has evolved as a key strategic process, vital to viral strategy. Picornaviral mRNA, like many RNA viruses, is uncapped or lacks the 5′ terminal m7GpppN cap structure found in cellular mRNAs (Belsham, 2009). Instead, picornaviruses and other IRES translating viruses contain a small, virus-encoded peptide or VPg (Jang et al., 1990). The discovery of IRES elements across a variety of viruses also identified distinct structural and functional differences amongst them, leading to the implementation of an IRES classification scheme. Viral IRESs are subdivided into four categories based on their structure, function, and mechanism of initiation of translation. All four IRES types commonly share the necessity of (on some level) involving non-canonical translational factors that interact with IRES and replace the function of some canonical translation initiation factors. The factors involved vary, dependent upon the IRES, the structure, degree of interaction, and factors involved form the basis for IRES designation and classification.
Type I IRESs (Figure 1) comprise enteroviruses and rhinoviruses. These IRESs contain a tetra-loop, which is cloverleaf structure in stem-loop position I that resembles the four-way junction of tRNA. This structure interacts with host cellular protein poly(rC)-binding protein 2 (PCBP2) and viral protein 3CD to form a bridge between the 5′ and 3′ ends to facilitate multiple rounds of viral replication (Fernandez-Miragall et al., 2009). Downstream of the cloverleaf stem-loop at position I are three distinctive C-rich motifs that precede the stem-loop at position II. Two more C-rich regions are present in domain IV. There is also a pyrimidine tract motif located downstream of domain V, with a silent AUG region found 10–15 bases further downstream. The functional AUG initiation codon is traditionally further downstream from the silent AUG in type I IRESs, so the ribosome must scan downstream to the next AUG to begin translation initiation. Unlike cap-dependant translation initiation, there are binding sites for the eIF4G region absent the N-terminal, eIF4E-binding domain. It is this feature that allows the ribosome to be recruited independent of the cap structure, which is the hallmark of IRES-dependent translation. N-terminal deficient eIF4G is the integral translation initiation factor in the recruitment of the 43S ribosomal subunit, a process that is further enhanced by eIF4A. In fact, mutations made to the eIF4G-binding domain of the poliovirus IRES are the basis for the mutation of the PV strain given as the vaccine, further stressing the importance of translation initiation as a rate-limiting step (Malnou et al., 2004). All together, type I IRESs contain six stem loops termed stem loops I–VI. The authentic IRES structure is located in the stem loop II–VI region, which facilitates initiation and translation of the viral genome (Pelletier and Sonenberg, 1988). Many of the canonical translation initiation factors with the exception of eIF4E and the N-terminal region of eIF4G are necessary for type I and II IRES translation. For this reason, viral modulation of these cap-dependant translation initiation factors has been identified as a vital component to viral strategy. Type I and II IRESs also utilize non-canonical translation initiation factors, termed IRES transacting factors (ITAFs). Examples of ITAFs include La autoantigen, pyrimidine tract binding (PTB) protein, and upstream of N-Ras (UNR; Costa-Mattioli et al., 2004; Cornelis et al., 2005; Verma et al., 2010). ITAFs allow for the bypass of canonical translation initiation factors that are likely the target of viral strategy, either through direct proteolytic cleavage or modulation of pathways (such as UPR modulation) that negate their function.
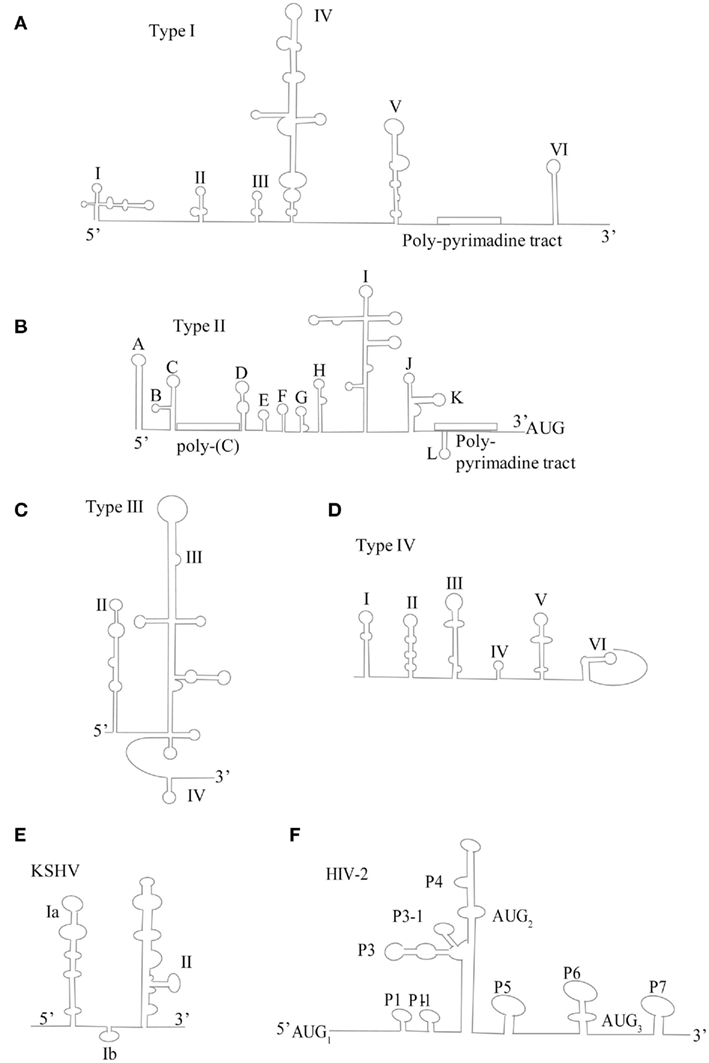
Figure 1. Schematic of proposed secondary structure of viral IRESs. (A) Type I IRES represented by PV-1 (adapted from Jang, 2006) (B) Type II IRES represented by EMCV (adapted from Jang, 2006) (C) Type III IRES represented by HCV (adapted from Beales et al., 2003(D) Type IV IRES represented by Plautia stali intestine virus (PSIV; adapted from Kanamori and Nakashima, 2001) (E) DNA virus IRES represented by Kaposi’s sarcoma herpesvirus (KSHV; adapted from Beales et al., 2003) (F) HIV IRES, represent by HIV-2 (adapted from Locker et al., 2011).
Type II IRESs (Figure 1) comprise the cardio- and aphthoviruses of the Picornaviridae family. There are several distinctive features of the IRES structure which differentiates the type II from that of the type I IRES. The 5′ UTR are significantly longer than their type I counterparts. In place of the cloverleaf structure at stem-loop position I, there is a hairpin or S structure. Just downstream of the S structure is an ∼200 bp C-tract that separates the S structure from the coding region. In between the C-rich tract and the coding region there are three structural distinct regions. The first are two to four pseudoknots, next is the cis-acting replication element (cre) and lastly the IRES element, which spans stem loops II–V, also termed H–L. Just downstream are two AUG triplets that actively initiate protein synthesis. Interestingly, each produces a unique version of the leader protein. Type II IRESs require many of the canonical translation initiation factors. eIF4G, eIF4A, and eIF4B have been demonstrated to interact with the SL J/K/L regions of the type II IRESs, with mutations to theses domains causing reductions in IRES activity (Jang, 2006). As mentioned above, IRES often utilizes ITAFs, which further enhance translation in the absence of the canonical translation factors. The variability of ITAFs and canonical translation factors seen amongst the four types of IRESs is indicative of differences amongst IRES structural components, which are able mimic the function of both.
Type III and IV IRES (Figure 1) structures demonstrate a new level of IRES-mediated translation initiation in which they are able to induce conformational changes directly to the ribosome that influence its entry, position, and stability (Hellen, 2009). Flaviviruses, such as HCV, IRES are prototypical representatives of type III IRESs. The HCV IRES contains three distinctive domains; II, III, and IV. Domain II is an irregular shaped, long stem-loop structure. Domain III is a pseudoknot that also contains several hairpin-structured sub-domains, IIIa–IIIf, whereas domain IV is a short hairpin structure containing the initiation codon. The HCV IRES, like all other type III IRESs, is able to directly and independently bind the 40S subunit, thereby bypassing the need for canonical eIFs 4A, 4B, 4F, 1, and 1A. HCV has been shown to require eIF3 and the 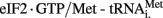 ternary complex to bind sequentially for translation initiation. However, some type III IRESs, such as the simian picornavirus type 9 (SPV9) IRES, have been shown to promote Met-tRNAMeti recruitment to the ribosome independent of eIF2 (de Breyne et al., 2008). Therefore negating the need for eIF2, which is quite often phosphorylated (i.e., translationally inactivated) during viral infection due to interferon activation of PKR or PERK, which induce subsequent phosphorylation of the eIF2α subunit. Type III IRES-containing viral mRNA has been demonstrated to be more resistant to translation inhibition caused by eIF2α phosphorylation than that of the cap-dependent cellular mRNAs (Pestova et al., 2008).
ternary complex to bind sequentially for translation initiation. However, some type III IRESs, such as the simian picornavirus type 9 (SPV9) IRES, have been shown to promote Met-tRNAMeti recruitment to the ribosome independent of eIF2 (de Breyne et al., 2008). Therefore negating the need for eIF2, which is quite often phosphorylated (i.e., translationally inactivated) during viral infection due to interferon activation of PKR or PERK, which induce subsequent phosphorylation of the eIF2α subunit. Type III IRES-containing viral mRNA has been demonstrated to be more resistant to translation inhibition caused by eIF2α phosphorylation than that of the cap-dependent cellular mRNAs (Pestova et al., 2008).
Type IV IRESs (Figure 1) initiate translation on the intergenic region (IGR) by direct binding of the 40S subunit or to the 80S ribosome. They are represented by the dicistroviruses, particularly the cricket paralysis virus (CrPV), which contain the smallest regions for internal ribosomal entry. Structurally, its IRES consist of three distinct domains. Each domain contains a pseudoknot and may or may not contain a hairpin like structure in stem-loop 3. Type IV IRESs translation initiation occurs without the involving any canonical initiation factors, initiator tRNA or a proper AUG start codon. In contrast to conventional AUG codon for IRES translation initiation, the start codon of type IV IRESs may be GCU, GCA, GCC, or CAA. In fact, studies have shown that translation initiation of CrPV IRES is impaired by the promotion of the 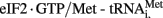 ternary complex to the 40S subunit. This may be an evolutionary advancement of conditions where the eIF2α is phosphorylated, such as during ER stress and viral infection (Hellen, 2009).
ternary complex to the 40S subunit. This may be an evolutionary advancement of conditions where the eIF2α is phosphorylated, such as during ER stress and viral infection (Hellen, 2009).
IRES of Lentiviruses
The HIV IRESs (Figure 1) represent yet another new class of IRES, not previously characterized. On one hand, it displays type III IRES properties possessing the ability to directly and indirectly bind to 40S and eIF3 (Locker et al., 2011). On the other hand, it requires all eIF’s except for eIF4E and eIF1, a property of class I and II IRESs (Locker et al., 2011). The structure of the HIV IRES is highly complex. It contains a long 5′ UTR harboring a Tar stem-loop, Poly-(A), PBS, DIS, SD, and Psi regions (Vallejos et al., 2011). Interestingly, in contrast to its type I, II, and III IRES counterparts, the HIV IRES appears to be resistant to structural mutations which to date have been unable to alter its function (Vallejos et al., 2011). Also unique is its ability to recruit three initiation complexes to a single RNA molecule (Locker et al., 2011). The translational requirements of HIV IRESs lend themselves to the notion that, while able to be translated cap-dependently, HIV RNA possesses and indeed utilizes IRESs as part of a tightly regulated and conserved method of cap-independent translation. The redundant of ability of HIV to translate through a variety of mechanisms highlights the importance of translation being a key, highly regulated process of the viral lifecycle. The utilization of the HIV IRESs takes place relatively late in the viral life cycle, in fact, it is regulated by the G2/M phase of the cell cycle and also activated by osmotic stress (Vallejos et al., 2011). This is particularly interesting given that cap-dependent translation is shut-off during the cell cycle, leading to the notion of a new level of evolutionary complexity exemplified by the ability of HIV to modulate translation between cap-dependant and independent translation based on cell physiology. The HIV IRES also utilizes a subset of ITAFs that are exclusively available during the G2/M phase (Vallejos et al., 2011). The utilization of its IRES is thought to regulate the transition between translation and encapsidation. The HIV-2 virus is only able to be encapsidated once the cognate form of it is translated, versus HIV-1 that can be either translated or propagated as a genome and encapsidated into virons (Locker et al., 2011). This is suggestive of a possible role of generation of structural/functional proteins in correlation with its IRES. In fact, the gag polyprotein encoded by the Gag IRES associates with 5′ UTR of HIV mRNA, forming a gRNA–Gag complex that inhibits ribosomal scanning, decreases translation, and increases encapsidation (Chamond et al., 2010). The ability to switch from cap-dependent to IRES-dependant translation by HIV is most closely related to that of cellular IRES-containing mRNA, which will be addressed in the next section.
IRES of Cellular mRNA
While many of the viral IRES-containing mRNAs have been studied quite extensively, much less is known about cellular IRES-containing mRNA. It is estimated that ∼10–15% of cellular mRNA possesses the ability to translate via cap-independent mechanisms (Johannes et al., 1999; Qin and Sarnow, 2004; Spriggs et al., 2008; Graber et al., 2010). The cellular genes that contain IRESs in their mRNAs usually code for proteins that are involved in growth, proliferation, apoptosis, stress response, differentiation, and cell cycle regulation (Komar and Hatzoglou, 2011). Cellular IRESs often are found in mRNA containing long 5′ UTRs that are rich in GC and have complex secondary structures (Holcik and Sonenberg, 2005). Often, in the mRNA structure there are also multiple short modules whose combined effects are IRES activation, as well as pseudoknots, that are believed to be inhibitory in function (Stoneley and Willis, 2004). However, to date there is no consensus structural or conformational motifs that are conserved among cellular IRES that would make them easily identifiable. Unlike their structurally stable viral counterparts, cellular IRESs identified to date follow a pattern of less structure corresponding to enhanced IRES activation (Filbin and Kieft, 2009). Like their viral counterparts, cellular IRESs are able to initiate translation without many of the canonical translational factors, particularly cap-binding factors such as eIF4E (Hellen and Sarnow, 2001). Cellular IRESs also utilize ITAFs to replace canonical translational factors rendered unavailable. Many of the ITAFs utilized by the cell are also utilized by viruses, including PTB, UNR, poly-(rC)-binding protein 1 (PCBP1), La autoantigen, and hnRNPC1/C2, many of which shuttle between the nucleus and cytoplasm (Stoneley and Willis, 2004). Dicistronic cellular mRNA containing IRESs were inactive when introduced directly into the cytoplasm, suggesting the possibility of prerequisite nuclear ITAF–IRES complex formation for IRES activation, at least for apoptotic genes (Spriggs et al., 2005). Interestingly, much like the highly evolved HIV IRES, the G2/M phase of the cell cycle (where cap-dependent protein synthesis is inhibited) is important for cell cycle regulatory gene’s IRES activation as well, including p58PITSLRE (Stoneley and Willis, 2004).
DNA Virus IRES
Much less studied are the DNA viruses, which transcribe mRNA containing an IRES that translates certain proteins independent of the cap structure, much like there cellular IRES counterparts. To date, there are six known DNA viruses known to contain IRESs, four of which belong to the Herpesviridae family (http://iresite.org/), particularly the latent gammaherpesviruses (Coleman et al., 2003). The most well documented DNA viral IRES is that of the Kaposi’s sarcoma herpes virus (KSHV; Figure 1; Bieleski and Talbot, 2001) while others include Herpes simplex virus (Griffiths and Coen, 2005) and Marek’s disease virus (Tahiri-Alaoui et al., 2009) to name a few. The KSHV IRES is representative of most IRESs in the Herpesviridae family in that it is similar in structure to that of HCV, containing two major stem loops (Beales et al., 2003). Although most IRESs identified are located in the 5′ UTR, the KSHV IRES is found in the coding sequence of the upstream cistron, vCyclin (Bieleski and Talbot, 2001). Interestingly, the KSHV IRES is translational active during viral latency and codes for a viral Fas-associated death domain (FADD)-like interleukin-1 beta-converting enzyme (FLICE)-inhibitory protein, vFLIP (Flice inhibitory protein homolog), which inhibits caspase activation and also promotes proliferation (Bieleski and Talbot, 2001). Again, the trend for IRES involved in cell growth/proliferation is consistent in DNA viruses as well. While there remains quite a bit yet to be discovered in our understanding of the structure and function of IRES elements in translation initiation, clearly, the stress-induced shift from cap-dependent to IRES-dependant translation is a vital strategy for the cell and virus to survive unfavorable conditions (for a comprehensive review of current known IRESs, the reader may refer to http://iresite.org/).
Mechanisms of Survival: Switching Translation Initiation from Cap-Dependent to IRES-Dependent
As discussed above, both the cells and viruses use the strategy for survival through switching translation initiation from cap-dependent to IRES-dependent. During this process, both the canonical translation factors and ITAFs utilized by a given virus are dependent upon IRES structure, as it is highly indicative of function. For example, structural components found in the mRNA of HCV IRES are able to mimic the function of certain canonical translational factors (Sonenberg and Hinnebusch, 2009). HCV also utilizes litagin and the oncogenes MCT-1/DENR as ITAFs, supplementing the function canonical factors of eIF1, eIF1A, eIF3, and eIF3 (Skabkin et al., 2010). Picornaviruses and others have demonstrated the capability of influencing the cell and manipulating its translational components, favoring its own translation and replication. Viral translation includes modulating not only canonical eukaryotic initiation factors, but also their binding proteins as well. The eukaryotic translation initiation components modulated during infection are specific to a given virus and can vary quite substantially. On the other hand, host cells utilize highly conserved mechanisms of defense to a variety of stimuli, including viral infection, osmotic shock, toxin, heat shock, etc. Here, we summarize some of the recent advances in our knowledge of the mechanisms utilized by viruses and cells to promote IRES-dependent translation allowing survival during unfavorable conditions.
Cleavage of Translation Initiation Factors by Viral Proteases
In order to influence cellular translation, viral proteases often target the cellular canonical translation initiation factors for cleavage. The early identified such factor is eIF4G (later called eIF4GI). Along with eIF4E, the scaffolding protein eIF4GI is a critical targeted translational factor during several viral infections. This is evident by the highly specific cleavage of eIF4GI during picornaviral infection, which generates a truncated C-terminal form that is unable to bind eIF4E (Svitkin et al., 2005). Another translation initiation factor eIF4GII as well as the Poly-(A) binding protein (PABP), a protein facilitating the formation of a closed translation initiation loop by interaction of the 5′ and 3′ ends of the mRNA, has been reported to be cleaved by picornaviral 2A (Gradi et al., 1998; Joachims et al., 1999). All these cleavages often correspond with a translational shift to IRES-dependent translation (Redondo et al., 2011; Welnowska et al., 2011). Another group also showed that the shift in translation seen during the later phase of poliovirus infection is not entirely due to phosphorylation (inactivation) of eIF2α (see Discussion in later session), but may also depend upon protease 3C activation and cleavage of another translation initiation factor, eIF5B, to a C-terminal truncated version thought to replace eIF2 during translation (White et al., 2011). In all these cleavage events, viral protein synthesis was increased during periods of global protein suppression caused by eIF2α phosphorylation, however the mechanism may likely be a combination of both 2A and 3C proteolytic activity. The apparent shift in translation occurs at times during infection when viral proteases are highly expressed. These observations are representative of viral evolution in correspondence to cellular anti-viral mechanisms. Other factors such as FMDV protease 3C mediated specific cleavage of eIF4AI but not eIF4AII highlight the target specificity that has quite often evolved to be viral specific (Li et al., 2001).
Cleavage of Translation Initiation Factors by Caspases
Like their viral counterparts, the cell utilizes a subset of proteases, the caspases, to cleave some translation initiation factors. The activation of the caspases often corresponds to the induction of apoptosis (Cohen, 1997). It has been demonstrated in cells committed to apoptosis that caspases cleave eIF4E-BP1, which enhances its capability to bind and inhibit eIF4E, thereby inhibiting cap-dependant translation (Tee and Proud, 2002). eIF2 is cleaved at its α subunit by caspase-3, further implicating its critical role in translational control (Satoh et al., 1999). Caspase-3 was also shown to cleave scaffolding protein eIF4GI, inhibiting its eIF4E-binding capabilities, as well as cleaving its homolog death associated protein 5 (DAP5, also called NAT1/p97), both during conditions of apoptosis (Marissen and Lloyd, 1998; Henis-Korenblit et al., 2000). Perhaps not surprisingly, viral strategy targets many of the same canonical translation initiation factors (including all of those mentioned here) and is reflective of a similar strategy used by the cell defense system, marking a translational switch to cap-independent translation during stress.
Phosphorylation of Eukaryotic Initiation Factors and Co-Factors
The cell has multiple signaling mechanisms that it utilizes to influence translation. Phosphorylation is perhaps the one of most common forms and conserved method utilized by the cell. Protein kinases involved in cellular stress response regulation such as PKR, PERK, GCN2, and HRT (heme-regulated kinases) all conservatively deactivate eIF2 on its α subunit in response to their respective stress stimulus, influencing the shift to cap-independent translation (Sonenberg and Hinnebusch, 2009). This multi-faceted capability of the cell to redundantly suppress cap-dependant translation initiation through phosphorylation of eIF2α is quite intriguing and spans multiple disease and stress conditions. This highlights the critical importance of translation initiation in cell fate and physiology. eIF4E also is a highly targeted translation factor during viral infection as well as during other conditions of stress, such as heat shock, ER stress, oxidative stress, etc. In fact, eIF4E and its regulatory protein eIF4E-BP have been utilized as predictive biomarkers in breast cancer (Coleman et al., 2009). This is because it functions as the cap-binding translation initiation factor thought to be the rate-limiting step of translation and therefore is a key component to cap-dependent translation (Gingras et al., 1999). The availability of eIF4E (which is highly cytoplasmic) to participate in cap-dependent translation is regulated by several factors, the most apparent being 4E-BP, which binds eIF4E and is involved in its localization to the nucleus and in stress granules, rendering it inactive (Sukarieh et al., 2009). 4E-BP is regulated by phosphorylation by the highly conserved serine/threonine kinase [mammalian target of rapamycin (mTOR), which decreases its affinity to eIF4E (Kimball and Jefferson, 2004], thus resulting in increased levels of protein translated cap-dependently due to increased availability of cap-binding protein eIF4E. However, hypophosphorylated 4E-BPs binds strongly to eIF4E and thus attenuates cap-dependent translation. Similarly, eIF4G has been shown to be phosphorylated by protein kinase C (PKCα) through the Ras–ERK pathway, resulting in increase affinity for eIF4E binding and enhanced eIF4E-mnk1 modulating capabilities (Dobrikov et al., 2011). Therefore, phosphorylation modulated by stress stimulus (i.e., heat shock, osmotic stress, ER stress, viral infection) results in stress pathway activation (ERK, MAPK, PKR, etc.) and subsequent phosphorylation of a translation initiation component (i.e., eIF4E, eIF4G, eIF2, 4E-BP) which repress or enhances its function and contributes to the translational switch between IRES and cap-dependant modes.
EIF4E-Binding Proteins and Other Associated Proteins Compete with EIF4E to Inhibit Cap-Dependent Translation
Another similar mechanism for controlling the shift of translation initiation is the up-regulation of 4E-BP production, which affects the mRNA 5′-cap recognition process of eIF4F. In cap-dependent translation, eIF4E forms the eIF4F complex along with translation initiation factors eIF4A, eIF4B, and eIF4G (Merrick, 1992). The interaction between eIF4G and eIF4E in the eIF4F complex is inhibited by 4E-BPs (also called eIF4E homolog). Recently, it was reported that Argonaut (Ago) protein, a core component of RNA induced silencing complex (RISC), binds directly to the cap structure and that this binding competes with eIF4E and results in inhibition of cap-dependent translation initiation (Kiriakidou et al., 2007). The central domain of Ago exhibits limited sequence homology to the eIF4E and contains two aromatic residues that could function in a similar manner to those in eIF4E in interaction with the cap structure. However, this conclusion has been questioned by another study (Eulalio et al., 2008). Another factor eIF6 has been reported to associate with Ago protein and the large ribosomal subunits (Chendrimada et al., 2007). By binding to the large ribosomal subunit, eIF6 prevents this subunit from prematurely joining with the small ribosomal subunit. Thus, if Ago2 recruits eIF6, then the large and small ribosomal subunits might not be able to associate, causing translation to be repressed (Chendrimada et al., 2007). Drosophila Cup also suppresses cap-dependent translation by binding eIF4E at the same conserved sequence utilized by 4E-BPs (Nakamura et al., 2004).
The Role of microRNAs in Translational Control
Many viruses also indirectly influence the availability of cellular translational components. miRNAs are small (∼20–24 nts) non-coding RNAs that bind partially complimentary mRNA sequences (mostly in the 3′ UTR and less so in the 5′ UTR and coding regions) resulting in translational repression and mRNA degradation or (in instances of cellular quiescence) translational activation (Sonenberg and Hinnebusch, 2009; Fabian et al., 2011). They are loaded onto target mRNA sequences by an RISC, whose major component proteins are the Ago protein family (Sonenberg and Hinnebusch, 2009). It was recently shown that Ago proteins are required for miR-122 activated translation during HCV infection (Roberts et al., 2011). In addition, as mention earlier, Ago binds competitively to the cap structure of mRNA to inhibit cap-dependent initiation of translation. It is not surprising that miRNA-mediated repression has been shown to be specific to a given mRNA containing both a cap structure and poly-(A) tail, in fact mRNA without a cap structure or poly-(A) tail were resistant to miRNA-mediated repression (Humphreys et al., 2005). miRNA modulated repression takes place in processing (P)-bodies that contain decapping enzymes (see Discussion in a later section), further supporting the role of miRNA in suppressing cap-dependent translation initiation (Sonenberg and Hinnebusch, 2009). Viruses have been shown to influence the expression of select miRNAs (Humphreys et al., 2005; Lei et al., 2010; Ho et al., 2011), which are often involved in the inhibition of cap-dependent translation (Humphreys et al., 2005; Walters et al., 2009) lending to a viral influenced a shift to IRES-mediated translation. In the early study of the mechanism of translation suppression using an artificial miRNA targeting CXCR4, the cap/4E-BP and the poly-(A) tail of mRNA were all found to play an important role because they are each necessary but not sufficient for full miRNA-mediated repression of translation. Replacing the cap with an IRES of virus impairs miRNA-mediated suppression. These results suggest that miRNAs interfere with the initiation step of translation and implicate 4E-BP as a molecular target (Humphreys et al., 2005). This finding was further solidified by a recent study, which demonstrated that enterovirus 71 (EV71) infection upregulated miR-141 expression and resulted in a shift from cap-dependent to cap-independent translation initiation through targeting 4E-BP. As EV71 RNA contains an IRES, this targeting enhanced EV71 replication (Ho et al., 2011). Another miRNA, miR-2, has also been reported to utilize a similar mechanism to target the cap structure (Zdanowicz et al., 2009). This study screened a library of chemical m7GpppN cap structures and identified defined modifications of the triphosphate backbone that augment miRNA-mediated inhibition of translation but are “neutral” toward to general cap-dependent translation. Interestingly, these caps also augment inhibition by 4E-BP, suggesting that miR-2’s cap targeting is through a mechanism related to the 4E-BP class of translation regulators (Zdanowicz et al., 2009).
The above studies clearly support the notion of a virally influenced translational shift favoring cap-independent translation. This is achieved through several mechanisms including indirectly, such as; up-regulating the expression of certain miRNAs that repress cap-binding canonical translation initiation factors in the eI4F complex (Mathonnet et al., 2007). Here, it is worth mentioning that viruses with a nuclear DNA phase, including HIV and Herpesviruses, during the infection cycle may generate virally derived miRNAs (Griffiths-Jones et al., 2008; Pilakka-Kanthikeel et al., 2011), however, miRNA derived from HIV is still contentious as other labs have not been able to verify them experimentally (Pfeffer et al., 2005; Lin and Cullen, 2007). Intriguingly, the cytoplasmic RNA tick-borne encephalitis virus (TBEV), a member of the Flaviviridae family, has been shown to encode its own viral miRNA when a heterlogous miRNA-precursor stem-loop was artificially introduced into the RNA viral genome (Rouha et al., 2010). This opens up the possibility of other cytoplasmic RNA viruses to have similar capabilities. Clearly miRNAs represent an exciting and newly emerging dimension to our study and understanding of viruses and their ability to manipulate cellular translation during infection and other conditions of stress.
Activation of Decapping Enzymes
Decapping of mRNA by decapping enzymes represents another modality by which cap-dependent translation is suppressed by the cell. To date, two decapping enzymes have been identified: Dcp2 which cleaves mRNA at the cap site and the scavenger decapping enzyme (DcpS) that hydrolyzes the cap structure, both function to facilitate the subsequent degradation of target cap-dependant mRNA (Li and Kiledjian, 2010). Enzymatic decapping of select mRNAs is influenced by miRNA. miRNA-mediated repression occurs in P bodies where Ago proteins have been shown to co-immunoprecipitate with decapping enzymes, suggesting their close association (Parker and Sheth, 2007). P bodies also contain other proteins including, GW182, the CAF1–CCR4–NOT deadenylase complex, the decapping activators (e.g., DCP1, EDC3, Ge-1), and the RNA helicase RCK/p54, all of which have been implicated in miRNA function (Eulalio et al., 2007; Parker and Sheth, 2007). Decapping enzymes functions may also be modulated by cell signaling pathways and are also found in stress granules. Indeed, the phosphorylation of the decapping enzyme DCP2 has been shown to influence stress granule formation and its availability in P bodies (Yoon et al., 2010). HCV has been shown to selectively disrupt P-body components during infection leaving the decapping enzyme DCP2, active and high jacking other translational machinery for the enhancement of its own translation (Ariumi et al., 2011). Therefore, not surprisingly, viruses modulate decapping enzyme activity to favor their translation.
Cellular IRES-Containing Genes and Their Roles in ER Stress Response
The cell too utilizes a subset of genes that have available in their mRNA an IRES, allowing translation to shift to cap-independent initiation when under stress. IRES-containing cellular genes typically code for proteins involved in growth, proliferation, responses to stress (i.e., virus infection, ER stress, heat shock, osmotic stress, etc.) cell cycle, and apoptosis. In fact, there is a subset of pro-survival genes that are activated during transient stress [e.g., Bcl-2, vascular endothelia growth factor (VEGF), BiP, p58IPK, etc.] that differs from the pro-apoptotic genes expressed during prolonged stress (e.g., Apaf-1, DAP5, p53, CHOP, etc.). The following will discuss several selected representatives from each subset.
Beta-Cell Lymphoma 2
The Bcl-2 family represents key regulators of cell proliferation and death, having both anti-apoptotic (Mcl-1, Bcl-xL, Bcl-2, etc.) and pro-apoptotic (Bax, Bak, Bad, Bcl-xs, BIM, etc.) protein family members. Its expression is tightly regulated at the transcriptional, post transcriptional, and post-translational levels by transcription factors (e.g., ATF-2), miRNAs, and other factors (i.e., kinases), all dependant upon the physiological conditions of the cell (Ma et al., 2007; Willimott and Wagner, 2010). miR-181a has been shown to repress pro and anti-apoptotic Bcl-2 family members (also shown to be repressed by miR-15a and miR-16-1), Mcl-1 (myeloid leukemia cell differentiation protein 1), and Bcl-2-11/Bim in ischemic astrocytes where decreasing its levels resulted in a reduction of apoptosis, oxidative stress, and preservation of mitochondrial integrity (Willimott and Wagner, 2010; Ouyang et al., 2012). Like many of the cellular genes that regulate growth, differentiation, proliferation and apoptosis, Bcl-2, and Bcl-xL also contains IRESs in their mRNA and is therefore able to switch modes of translation during conditions where cap-binding elements are inhibited. The IRES of Bcl-2 has been shown to be stimulated during mitosis by another IRES-containing cellular translation factor, DAP5 (Marash et al., 2008).
Death Associated Protein 5
Death associated protein 5, also termed p97 and NAT1, is an IRES-containing translation initiation factor (Henis-Korenblit et al., 2000). It shares homology with the central and C-terminal region of translation initiation factors of the eIFG family, specifically the region that binds eIF4A and eIF3 and the Mnk1 binding site and also contains an eIF2β bind site (Lee and McCormick, 2006; Lewis et al., 2008). Perhaps DAP5 can be thought of as performing the function of eIF4GI/eIF4GII without eIF4E-binding capabilities and is therefore specific to translation of IRES-containing mRNA. DAP5 is a key cellular regulator involved in apoptosis, growth, differentiation as well as involvement in ER stress response (Marash and Kimchi, 2005; Lewis et al., 2008; Marash et al., 2008). In its proform (p97), it has been shown to have a protective role in the cell from apoptosis during the cell cycle. This is accomplished by stimulating not only its own IRES, but also acting as a translation factor (ITAF) to stimulate protective Bcl-2 and Chk1 (Marash et al., 2008). DAP5 has been shown to undergo post-translational modification by proteolytic cleavage (cell and virus-mediated; Henis-Korenblit et al., 2000; Lee and McCormick, 2006; Chau et al., 2007). During caspase-mediated apoptosis, it is cleaved at the C-terminus into an 86-kDa form by caspase-3, which has been shown to stimulate its affinity for activating a different subset of apoptosis related genes (Henis-Korenblit et al., 2002). Studies showing the dual nature of pro and anti-apoptotic translational targets of DAP5 and the multiple mechanisms for its post-translational regulation are indicative of its tight regulation in response to changing physiological conditions caused by a variety of stimuli. Reports have shown the p86 version of DAP5 is capable of activating IRESs quite distinct from that of the protective p97 form, including anti-apoptotic human inhibitor of apoptosis protein 2 (HIAP2), x-linked inhibitor of apoptosis protein (XIAP), and the pro-apoptotic Apaf-1 (Henis-Korenblit et al., 2002; Warnakulasuriyarachchi et al., 2004). p86 is also able to generate a positive feedback loop by stimulating its own IRES and thereby enhancing the apoptotic cascade, lending to the notion that the 86-kDa form of DAP5 is the more potent isoform (Henis-Korenblit et al., 2000). Conditions of ER stress increase the translation of DAP5 and therefore its activity (Lewis et al., 2008). The target genes whose expression is regulated by DAP5 have not been fully elucidated, thus, newly identified translational effectors will provide novel insight into its function in regulation of translation.
P53
p53 is a tumor suppressor and a transcription factor, its up-regulation can trigger the altered expression of a number of responsive genes through its binding on the promoter of these genes (Das et al., 2008). These genes include two major classes, one that controls cell cycle and the other controls apoptosis. p53 is also a IRES-containing gene (Ray et al., 2006). Like other IRES-containing genes, its expression is also regulated by many stimuli such as viral infection. For example, during HIV infection, HIV protein vpu functions to stabilized p53, which is upregulated late in HIV infection, leading to p53-induced apoptosis (Verma et al., 2011). p53 is known to be upregulated during conditions of oxidative stress, which is induced by HIV infection (Kottilil et al., 2001; Puzio-Kuter, 2011). p53 is expressed in a time dependent manner during late HIV infection, likely just prior to G2/M arrest and subsequent apoptosis (Verma et al., 2011). Conversely, p53 expression is down-regulated during picornaviral (CVB3) infection through virus-induced down regulation of p53 transcriptional activator activating transcriptional factor 3 (ATF-3; Hwang et al., 2007). DNA viruses too, have been shown to regulate the expression of p53. MDV protein meq has been demonstrated to directly interact with p53, inhibiting its transcriptional and apoptotic inducing capabilities (Deng et al., 2010). Additionally, KSHV protein LANA binds to the DNA binding domain of p53, contributing to its ubiquitylation and proteasome degradation, subsequently maintaining viral latency by avoiding p53-induced apoptosis (Suzuki et al., 2010). p53 has also been shown to play an oncogenic role in some cancers, including human melanoma where it directly up regulates miR-191, resulting in increased expression of Mcl-1 (see Beta-Cell Lymphoma 2 above), thus contributing to apoptotic resistance (Jin et al., 2011). p53 also binds hypophosphorylated Bcl-2, rendering it incapable of performing its anti-apoptotic function and decreasing cell survival (Willimott and Wagner, 2010). Therefore, the function of p53 being pro- or anti-apoptotic is dependant upon cellular physiology, type of stress, and likely cell type. Understandably, much research has been and is currently being devoted to the study of p53 during various stress conditions (i.e., cancer, viral infection; Hwang et al., 2007).
p58IPK
p58IPK, an ER-luminal co-chaperone associated with BiP, is an another IRES-containing cellular gene actively involved during ER stress and selectively modulated as part of the strategy of several viruses during infections, both positively and negatively. During conditions of ER stress, it functions to negatively regulate the phosphorylation of eIF2α induced by both PKR and PERK, achieved through direct binding of their kinase domains (Yan et al., 2002). This acts to restore homeostasis and cellular global translation when a given ER stress is transient and easily corrected by cellular mechanisms initiated by the UPR (Yan et al., 2002). Knockdown of p58IPK during ER stress results in increased BiP and CHOP expression, as well as increased eIF2α phosphorylation (Chakrabarti et al., 2011). p58IPK has been shown to be regulated both positively and negatively during such viral infections as influenza and CVB3. Due to its ability to negatively regulate PKR, which elicits the dsRNA induced interferon response, p58IPK’s functions are indicative of a critical role in viral ability to escape immune evasion early in the viral lifecycle. The influenza virus has been shown to have a reduced translational capability in p58IPK −/− cells. This translational regulation was shown to be dependant on its negative regulatory role on PKR and showed little effect on PERK regulation (Yan et al., 2002). Interestingly, p58IPK has been shown to be down-regulated at both the protein and mRNA level in a temporal manner during CVB3 infection (Zhang et al., 2011b). This suggests that the need to regulate global protein synthesis shut-off and switch to IRES-dependant translation during CVB3 infection may be accomplished in part through inhibition of p58IPK (Zhang et al., 2011b). The inhibition of p58IPK during the later stages of infection would in theory inhibit cellular return to homeostasis and global protein synthesis, which may trigger a possible shift to apoptosis. The regulation (repression by CVB3 and activation by influenza) of p58IPK during viral infection is indicative of viral modulation of downstream components activated by the UPR, such as mediating the cross talk between ATF6, IRE1 through selective regulation of PERK (Jin et al., 2011; Merquiol et al., 2011; Zhang et al., 2011b). In the case of CVB3, down regulation of p58IPK has been demonstrated to activate pro-apoptotic genes CHOP, SREBP1, and ER mediated caspase-12, which may explain why it is time dependently suppressed during the latter stages of CVB3 infection (Zhang et al., 2011b). The time course of an increase in CHOP, SREBP1 and caspase-12 during CVB3 infection all correspond to increases in protease (both viral and cellular) expression promoting conditions of apoptosis (Chau et al., 2007). Clearly the modulation of the IRES containing, ER stress related p58IPK provides the critical ability to control other stress responsive pathways, as is evident here where it is utilized for viral evasion of host immune response.
Heat Shock Proteins
The heat shock family of proteins are a family of chaperone proteins that are translated from IRES-containing mRNAs such as BiP, heat shock protein 70 (Hsp70), Hsp22 and Hsp27, and non-IRES-containing Hsp90, which individually are highly functional and perform separate, integral roles during conditions, such as cancer (due to their pro-survival nature) and stress (i.e., ER stress, heat shock; Ahmed and Duncan, 2004; Hernandez et al., 2004; Behnsawy et al., 2011; Mymrikov et al., 2011). Heat shock proteins such as Hsp90 have also been implemented in the regulation of cap-dependent translation (Ahmed and Duncan, 2004; Mymrikov et al., 2011). Although their name might imply that they are expressed only as a heat shock responsive subset of proteins, many of them are in fact ubiquitously expressed (Mymrikov et al., 2011). Like many of the heat shock proteins, Hsp27 and Hsp22 function to assist in protein folding and preventing the accumulation of protein aggregates, respectively. Functional mutations to these proteins is believed to be involved in the development of neurodegenerative disorders (Mymrikov et al., 2011). BiP (the major chaperone protein of the ER) has been shown to play a critical role in SV-40 and human cytomegalovirus (HCMV) infections (Geiger et al., 2011; Zhang et al., 2011a) with its translation selectively enhanced directly through its IRES HCMV infection (Buchkovich et al., 2010). miR-181 has also been shown to decrease levels of BiP, further implicating its role in cellular stress regulation (Ouyang et al., 2012). Hsp70 has been identified as an anti-apoptotic chaperone protein, whose activities include recruitment of refolding of proteins in an ATP regulated manner (Johnson, 2011). Hsp70 has been shown to protect the cell against a wide of stresses. siRNA knockdown of Hsp70 resulted in activation of caspase-9 and caspase-3 (Behnsawy et al., 2011). Hsp70 has been shown during infection to modulate viral entry, transcription, translation, nuclear entry, and viron release (Halder et al., 2011; Sugiyama et al., 2011). A very recent study questioned the previous suggestion and indicated that Hsp70 may not have an IRES within its 5′ UTR (Sun et al., 2011). This is because these authors found that the 5′ UTR of Hsp70 mRNA has little effects in driving translation when placed in a bicistronic expression construct. However, other studies found that the 5′ UTR of Drosophila Hsp70 mRNA allows efficient translation at high temperature when other non-heat shock mRNAs are poorly translated (Klemenz et al., 1985; McGarry and Lindquist, 1985). This raises the question of how the 5′ UTR of Hsp70 mRNA drives the cap-independent translation without acting as an IRES. It is possible that the presence of a 5′ proximal mRNA structure prevents the direct recruitment of the ribosome by Hsp70 5′ UTR (Sun et al., 2011). Another possibility is that the expression of several decapping enzymes was enhanced during heat stress (Neef and Thiele, 2009) and this phenomenon could lead to the selective translation of Hsp70 mRNA due to the unique features of the Hsp70 5′ UTR in mediating cap-independent translation (Sun et al., 2011). Hsp90 has been also shown not to harbor an IRES in its extensive 5′ UTR secondary structure and is sensitive to inhibition of the cap-binding complex, eIF4F (Ahmed and Duncan, 2004). Hsp90 has been shown to enhance binding of eIF4E to eIF4G and inhibition of Hsp90 has also been shown to decrease the formation of P bodies (Suzuki et al., 2009). Therefore, Hsp90 is a critical chaperone protein that appears necessary for eIF4E dependant translation and localization. Clearly the heat shock family of proteins represents a subset of critical stress regulatory proteins. While some of these proteins have been studied quite extensively (i.e., BiP) it is apparent that novel functions for the heat shock protein family are continually to be discovered.
Vascular Endothelia Growth Factor
As mentioned previously, during some conditions of growth and differentiation, cellular translation initiation shifts from cap-dependent to an IRES-dependent mechanism. The VEGF proteins represent another family of cellular proteins translated through an IRES-driven mechanism. These proteins are involved in vasculogenesis and angiogenesis (Bornes et al., 2007). The mRNA of VEGF-A harbors two IRESs (A and B), where A translates through a traditional AUG start codon and B translates through a CUG start codon for a higher molecular weight isoform (L-VEGF; Bornes et al., 2004). The translation of VEGF through the IRES-dependant mechanism has been shown to be selectively activated during conditions of oxygen deprivation in ischemic muscle and are therefore activated by stress (Bornes et al., 2007). IRES B of VEGF has also been shown to be selectively repressed by miR-16 targeting, whereas IRES-A remained active, lending to the notion that VEGF isoforms can be selectively expressed from the same mRNA by miR specific targeting (Karaa et al., 2009). VEGF expression is upregulated in HIV-1 or HRV infection, both of which utilize IRES translation (Psarras et al., 2006; Korgaonkar et al., 2008) suggesting the possibility of related translational mechanisms of their respective IRESs.
Conclusion and Perspectives
It is clear that more and more newly discovered cellular mRNAs contain IRESs and can shift translation to IRES-driven initiation during ER stress. One of the most common causes of ER stress is viral infection, which can globally shut down cap-dependent translation initiation by different mechanisms. To adapt in unfavorable stress conditions, both cell and virus (e.g., HIV) need to adjust their mode translation initiation by switching from the cap-dependent to cap-independent mechanism. As picornaviruses do not have a cap structure, its RNA translation will not be inhibited; instead it will be enhanced because more translational machinery is available due to the shut-off of global cap-dependent translation are by a number of mechanisms. During transient ER stress, the IRES-containing cellular mRNAs that responsible for cell survival/growth, such as BiP, Bcl-2, VEGF, etc., will be selectively translated by the IRES-dependent mechanism using ITAFs. This mechanism allows cells to respond rapidly to the transient changes in growth conditions and to delay apoptosis. Once the stress condition is removed, the cells will resume the normal growth. However, during prolonged or severe stress, such as in persistent infection of picornaviruses, the pro-death genes, such as Apaf-1, DAP5, CHOP, p53, etc., are also selectively translated by the same IRES-driven mechanism, allowing the cells to fine-tune their responses to cellular stress and, if conditions for cell survival are not restored, to proceed with final execution of apoptosis (Figure 2).
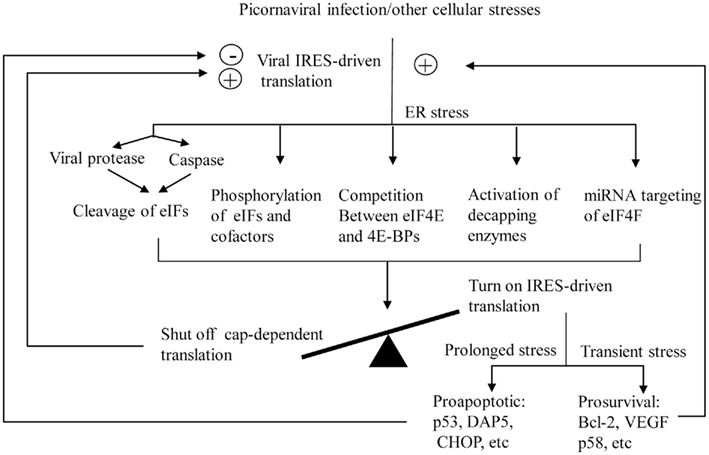
Figure 2. The proposed model for the switch of translation initiation from cap-dependent to IRES-dependent during picornaviral infection or other cellular stresses. Positive and negative feedback loops are indicated by plus and minus signs, respectively.
At the present, although some mechanisms on the switch of the translation initiation and subsequent selective translation have been described, many questions are still unanswered: for example, what are the regulators for selecting the pro-survival or pro-apoptotic genes? In other words, do these genes contain different binding sequences for their specific regulators? Previous studies using a polysome system predicted that ∼10–15% of the cellular mRNAs contain IRESs (Carter et al., 2000; Qin and Sarnow, 2004; Graber et al., 2010); thus more IRES-containing cellular mRNAs will need to be discovered to fully understand the underlying mechanisms of IRES-dependent translational control. In the shut-off of global cap-dependent translation, cleavages of cellular proteins are known to play an important role. In this regard, besides the viral proteases and the activated cellular caspases, other cellular proteases responsible for the cleavage need to be identified. In addition, efforts to discover other cellular target proteins that are specifically cleaved during cellular stress are another future direction. Identification of these target proteins may uncover the linkage between translational control and pathogenesis. Recently, miRNAs, as a group of new regulators of gene expression, were found to be involved in regulation of the shift of translation initiation. However, the research in this direction is just emerging. More studies on the interactions between miRNAs and their target mRNAs encoding translation initiation factors need to be carried out. Indeed, the biological implications of the selective translation of specific genes are clearly important. Since the IRES-mediated translation initiation links with many pathophysiological conditions, such as hypoxia, heat shock, toxin, metabolic disorder, viral infection, etc., the failure of maintaining the balance between the cap-dependent and cap-independent translation initiation may cause human diseases, such as heart disease, stroke, diabetes, and viral diseases. Similarly, dysregulated apoptosis has been associated with many human disorders, ranging from autoimmune diseases, neurodegeneration to cancers. Therefore, better understanding how the translational control determines the cellular response to stresses will provide novel insights into the molecular pathogenesis of human disorders and might eventually lead to the development of effective therapeutics.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research and the Heart and Stroke Foundation of BC and Yukon. Dr. Maged Gomaa Hemida is a recipient of the CIHR-IMPACT postdoctoral training fellowship and the Heart and Stroke foundation of Canada postdoctoral training fellowship. Xin Ye is supported by a UGF Award from the University of British Columbia.
References
Ahmed, R., and Duncan, R. F. (2004). Translational regulation of Hsp90 mRNA. AUG-proximal 5′-untranslated region elements essential for preferential heat shock translation. J. Biol. Chem. 279, 49919–49930.
Ali, I. K., McKendrick, L., Morley, S. J., and Jackson, R. J. (2001). Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 75, 7854–7863.
Ariumi, Y., Kuroki, M., Kushima, Y., Osugi, K., Hijikata, M., Maki, M., Ikeda, M., and Kato, N. (2011). Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J. Virol. 85, 6882–6892.
Beales, L. P., Holzenburg, A., and Rowlands, D. J. (2003). Viral internal ribosome entry site structures segregate into two distinct morphologies. J. Virol. 77, 6574–6579.
Behnsawy, H. M., Miyake, H., Kusuda, Y., and Fujisawa, M. (2011). Small interfering RNA targeting heat shock protein 70 enhances chemosensitivity in human bladder cancer cells. Urol. Oncol. doi: 10.1016/j.urolonc.2011.07.007
Bieleski, L., Hindley, C., and Talbot, S. J. (2004). A polypyrimidine tract facilitates the expression of Kaposi’s sarcoma-associated herpesvirus vFLIP through an internal ribosome entry site. J. Gen. Virol. 85, 615–620.
Bieleski, L., and Talbot, S. J. (2001). Kaposi’s sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J. Virol. 75, 1864–1869.
Bollo, M., Paredes, R. M., Holstein, D., Zheleznova, N., Camacho, P., and Lechleiter, J. D. (2010). Calcineurin interacts with PERK and dephosphorylates calnexin to relieve ER stress in mammals and frogs. PLoS ONE 5, e11925. doi:10.1371/journal.pone.0011925
Bornes, S., Boulard, M., Hieblot, C., Zanibellato, C., Iacovoni, J. S., Prats, H., and Touriol, C. (2004). Control of the vascular endothelial growth factor internal ribosome entry site (IRES) activity and translation initiation by alternatively spliced coding sequences. J. Biol. Chem. 279, 18717–18726.
Bornes, S., Prado-Lourenco, L., Bastide, A., Zanibellato, C., Iacovoni, J. S., Lacazette, E., Prats, A. C., Touriol, C., and Prats, H. (2007). Translational induction of VEGF internal ribosome entry site elements during the early response to ischemic stress. Circ. Res. 100, 305–308.
Buchkovich, N. J., Yu, Y., Pierciey, F. J. Jr., and Alwine, J. C. (2010). Human cytomegalovirus induces the endoplasmic reticulum chaperone BiP through increased transcription and activation of translation by using the BiP internal ribosome entry site. J. Virol. 84, 11479–11486.
Burikhanov, R., Zhao, Y., Goswami, A., Qiu, S., Schwarze, S. R., and Rangnekar, V. M. (2009). The tumor suppressor Par-4 activates an extrinsic pathway for apoptosis. Cell 138, 377–388.
Carter, M., Kuhn, K., and Sarnow, P. (2000). Translational Control of Gene Expression. New York: Cold Spring Harbor Laboratory Press.
Castello, A., Franco, D., Moral-Lopez, P., Berlanga, J. J., Alvarez, E., Wimmer, E., and Carrasco, L. (2009). HIV- 1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS ONE 4, e7997. doi:10.1371/journal.pone.0007997
Chakrabarti, A., Chen, A. W., and Varner, J. D. (2011). A review of the mammalian unfolded protein response. Biotechnol. Bioeng. 108, 2777–2793.
Chamond, N., Locker, N., and Sargueil, B. (2010). The different pathways of HIV genomic translation. Biochem. Soc. Trans. 38, 1548–1552.
Chau, D. H., Yuan, J., Zhang, H., Cheung, P., Lim, T., Liu, Z., Sall, A., and Yang, D. (2007). Coxsackievirus B3 proteases 2A and 3C induce apoptotic cell death through mitochondrial injury and cleavage of eIF4GI but not DAP5/p97/NAT1. Apoptosis 12, 513–524.
Chendrimada, T. P., Finn, K. J., Ji, X., Baillat, D., Gregory, R. I., Liebhaber, S. A., Pasquinelli, A. E., and Shiekhattar, R. (2007). MicroRNA silencing through RISC recruitment of eIF6. Nature 447, 823–828.
Coleman, H. M., Brierley, I., and Stevenson, P. G. (2003). An internal ribosome entry site directs translation of the murine gammaherpesvirus 68 MK3 open reading frame. J. Virol. 77, 13093–13105.
Coleman, L. J., Peter, M. B., Teall, T. J., Brannan, R. A., Hanby, A. M., Honarpisheh, H., Shaaban, A. M., Smith, L., Speirs, V., Verghese, E. T., McElwaine, J. N., and Hughes, T. A. (2009). Combined analysis of eIF4E and 4E-binding protein expression predicts breast cancer survival and estimates eIF4E activity. Br. J. Cancer 100, 1393–1399.
Cornelis, S., Tinton, S. A., Schepens, B., Bruynooghe, Y., and Beyaert, R. (2005). UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein. Nucleic Acids Res. 33, 3095–3108.
Costa-Mattioli, M., Svitkin, Y., and Sonenberg, N. (2004). La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 24, 6861–6870.
Das, S., Boswell, S. A., Aaronson, S. A., and Lee, S. W. (2008). P53 promoter selection: choosing between life and death. Cell Cycle 7, 154–157.
de Breyne, S., Yu, Y., Pestova, T. V., and Hellen, C. U. (2008). Factor requirements for translation initiation on the simian picornavirus internal ribosomal entry site. RNA 14, 367–380.
Deng, X., Li, X., Shen, Y., Qiu, Y., Shi, Z., Shao, D., Jin, Y., Chen, H., Ding, C., Li, L., Chen, P., and Ma, Z. (2010). The Meq oncoprotein of Marek’s disease virus interacts with p53 and inhibits its transcriptional and apoptotic activities. Virol. J. 7, 348.
Dobrikov, M., Dobrikova, E., Shveygert, M., and Gromeier, M. (2011). Phosphorylation of eukaryotic translation initiation factor 4G1 (eIF4G1) by protein kinase C{alpha} regulates eIF4G1 binding to Mnk1. Mol. Cell. Biol. 31, 2947–2959.
Ekchariyawat, P., Thitithanyanont, A., Sirisinha, S., and Utaisincharoen, P. (2011). Apoptosis induced by avian H5N1 virus in human monocyte-derived macrophages involves TRAIL-inducing caspase-10 activation. Innate Immun. doi: 10.1177/1753425911410998. [Epub ahead of print].
Etchison, D., Milburn, S. C., Edery, I., Sonenberg, N., and Hershey, J. W. (1982). Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257, 14806–14810.
Eulalio, A., Behm-Ansmant, I., and Izaurralde, E. (2007). P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8, 9–22.
Eulalio, A., Huntzinger, E., and Izaurralde, E. (2008). GW182 interaction with argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 15, 346–353.
Fabian, M. R., Sonenberg, N., and Filipowicz, W. (2011). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379.
Fels, D. R., and Koumenis, C. (2006). The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol. Ther. 5, 723–728.
Fernandez-Miragall, O., Lopez de Quinto, S., and Martinez-Salas, E. (2009). Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res. 139, 172–182.
Filbin, M. E., and Kieft, J. S. (2009). Toward a structural understanding of IRES RNA function. Curr. Opin. Struct. Biol. 19, 267–276.
Geiger, R., Andritschke, D., Friebe, S., Herzog, F., Luisoni, S., Heger, T., and Helenius, A. (2011). BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat. Cell Biol. 13, 1305–1314.
Gingras, A. C., Raught, B., and Sonenberg, N. (1999). eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913–963.
Gotoh, T., Terada, K., Oyadomari, S., and Mori, M. (2004). Hsp70-DnaJ chaperone pair prevents nitric oxide- and CHOP-induced apoptosis by inhibiting translocation of Bax to mitochondria. Cell Death Differ. 11, 390–402.
Graber, T. E., Baird, S. D., Kao, P. N., Mathews, M. B., and Holcik, M. (2010). NF45 functions as an IRES trans-acting factor that is required for translation of cIAP1 during the unfolded protein response. Cell Death Differ. 17, 719–729.
Gradi, A., Svitkin, Y. V., Imataka, H., and Sonenberg, N. (1998). Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. U.S.A. 95, 11089–11094.
Griffiths, A., and Coen, D. M. (2005). An unusual internal ribosome entry site in the herpes simplex virus thymidine kinase gene. Proc. Natl. Acad. Sci. U.S.A. 102, 9667–9672.
Griffiths-Jones, S., Saini, H. K., van Dongen, S., and Enright, A. J. (2008). miRBase: tools for microRNA genomics. Nucleic Acids Res. 36, D154–D158.
Halder, U. C., Bagchi, P., Chattopadhyay, S., Dutta, D., and Chawla-Sarkar, M. (2011). Cell death regulation during influenza A virus infection by matrix (M1) protein: a model of viral control over the cellular survival pathway. Cell Death Dis. 2, e197.
Harding, H. P., Calfon, M., Urano, F., Novoa, I., and Ron, D. (2002). Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18, 575–599.
Hellen, C. U. (2009). IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim. Biophys. Acta 1789, 558–570.
Hellen, C. U., and Sarnow, P. (2001). Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15, 1593–1612.
Henis-Korenblit, S., Shani, G., Sines, T., Marash, L., Shohat, G., and Kimchi, A. (2002). The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc. Natl. Acad. Sci. U.S.A. 99, 5400–5405.
Henis-Korenblit, S., Strumpf, N. L., Goldstaub, D., and Kimchi, A. (2000). A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol. Cell. Biol. 20, 496–506.
Hernandez, G., Vazquez-Pianzola, P., Sierra, J. M., and Rivera-Pomar, R. (2004). Internal ribosome entry site drives cap-independent translation of reaper and heat shock protein 70 mRNAs in Drosophila embryos. RNA 10, 1783–1797.
Hetz, C., Bernasconi, P., Fisher, J., Lee, A. H., Bassik, M. C., Antonsson, B., Brandt, G. S., Iwakoshi, N. N., Schinzel, A., Glimcher, L. H., and Korsmeyer, S. J. (2006). Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science 312, 572–576.
Ho, B. C., Yu, S. L., Chen, J. J., Chang, S. Y., Yan, B. S., Hong, Q. S., Singh, S., Kao, C. L., Chen, H. Y., Su, K. Y., Li, K. C., Cheng, C. L., Cheng, H. W., Lee, J. Y., Lee, C. N., and Yang, P. C. (2011). Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 9, 58–69.
Holcik, M., and Sonenberg, N. (2005). Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318–327.
Hollien, J., Lin, J. H., Li, H., Stevens, N., Walter, P., and Weissman, J. S. (2009). Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 186, 323–331.
Humphreys, D. T., Westman, B. J., Martin, D. I., and Preiss, T. (2005). MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. U.S.A. 102, 16961–16966.
Hwang, H. Y., Kim, J. Y., Lim, J. Y., Chung, S. K., Nam, J. H., and Park, S. I. (2007). Coxsackievirus B3 modulates cell death by downregulating activating transcription factor 3 in HeLa cells. Virus Res. 130, 10–17.
Jackson, R. J., Hellen, C. U., and Pestova, T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127.
Jang, C. J., Lo, M. C., and Jan, E. (2009). Conserved element of the dicistrovirus IGR IRES that mimics an E-site tRNA/ribosome interaction mediates multiple functions. J. Mol. Biol. 387, 42–58.
Jang, S. K. (2006). Internal initiation: IRES elements of picornaviruses and hepatitis c virus. Virus Res. 119, 2–15.
Jang, S. K., Krausslich, H. G., Nicklin, M. J., Duke, G. M., Palmenberg, A. C., and Wimmer, E. (1988). A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62, 2636–2643.
Jang, S. K., Pestova, T. V., Hellen, C. U., Witherell, G. W., and Wimmer, E. (1990). Cap-independent translation of picornavirus RNAs: structure and function of the internal ribosomal entry site. Enzyme 44, 292–309.
Jin, L., Hu, W. L., Jiang, C. C., Wang, J. X., Han, C. C., Chu, P., Zhang, L. J., Thorne, R. F., Wilmott, J., Scolyer, R. A., Hersey, P., Zhang, X. D., and Wu, M. (2011). MicroRNA-149*, a p53-responsive microRNA, functions as an oncogenic regulator in human melanoma. Proc. Natl. Acad. Sci. U.S.A. 108, 15840–15845.
Joachims, M., Van Breugel, P. C., and Lloyd, R. E. (1999). Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73, 718–727.
Johannes, G., Carter, M. S., Eisen, M. B., Brown, P. O., and Sarnow, P. (1999). Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. U.S.A. 96, 13118–13123.
John, L., Thomas, S., Herchenroder, O., Putzer, B. M., and Schaefer, S. (2011). Hepatitis E virus ORF2 protein activates the pro-apoptotic gene CHOP and anti-apoptotic heat shock proteins. PLoS ONE 6, e25378. doi:10.1371/journal.pone.0025378
Johnson, J. L. (2011). Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim. Biophys. Acta 1823, 607–613.
Kanamori, Y., and Nakashima, N. (2001). A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7, 266–274.
Karaa, Z. S., Iacovoni, J. S., Bastide, A., Lacazette, E., Touriol, C., and Prats, H. (2009). The VEGF IRESes are differentially susceptible to translation inhibition by miR-16. RNA 15, 249–254.
Kimball, S. R., and Jefferson, L. S. (2004). Regulation of global and specific mRNA translation by oral administration of branched-chain amino acids. Biochem. Biophys. Res. Commun. 313, 423–427.
Kiriakidou, M., Tan, G. S., Lamprinaki, S., De Planell-Saguer, M., Nelson, P. T., and Mourelatos, Z. (2007). An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell 129, 1141–1151.
Klemenz, R., Hultmark, D., and Gehring, W. J. (1985). Selective translation of heat shock mRNA in Drosophila melanogaster depends on sequence information in the leader. EMBO J. 4, 2053–2060.
Komar, A. A., and Hatzoglou, M. (2005). Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 280, 23425–23428.
Komar, A. A., and Hatzoglou, M. (2011). Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10, 229–240.
Korgaonkar, S. N., Feng, X., Ross, M. D., Lu, T. C., D’Agati, V., Iyengar, R., Klotman, P. E., and He, J. C. (2008). HIV-1 upregulates VEGF in podocytes. J. Am. Soc. Nephrol. 19, 877–883.
Kottilil, S., Bowmer, M. I., Trahey, J., Howley, C., Gamberg, J., and Grant, M. D. (2001). Fas/FasL-independent activation-induced cell death of T lymphocytes from HIV-infected individuals occurs without DNA fragmentation. Cell. Immunol. 214, 1–11.
Labadie, K., Pelletier, I., Saulnier, A., Martin, J., and Colbere-Garapin, F. (2004). Poliovirus mutants excreted by a chronically infected hypogammaglobulinemic patient establish persistent infections in human intestinal cells. Virology 318, 66–78.
Lamphear, B. J., Yan, R., Yang, F., Waters, D., Liebig, H. D., Klump, H., Kuechler, E., Skern, T., and Rhoads, R. E. (1993). Mapping the cleavage site in protein synthesis initiation factor eIF-4 gamma of the 2A proteases from human Coxsackievirus and rhinovirus. J. Biol. Chem. 268, 19200–19203.
Lee, A. H., Iwakoshi, N. N., and Glimcher, L. H. (2003). XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23, 7448–7459.
Lee, K., Tirasophon, W., Shen, X., Michalak, M., Prywes, R., Okada, T., Yoshida, H., Mori, K., and Kaufman, R. J. (2002). IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16, 452–466.
Lee, S. H., and McCormick, F. (2006). p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. EMBO J. 25, 4008–4019.
Lei, X., Bai, Z., Ye, F., Huang, Y., and Gao, S. J. (2010). Regulation of herpesvirus lifecycle by viral microRNAs. Virulence 1, 433–435.
Lewis, S. M., Cerquozzi, S., Graber, T. E., Ungureanu, N. H., Andrews, M., and Holcik, M. (2008). The eIF4G homolog DAP5/p97 supports the translation of select mRNAs during endoplasmic reticulum stress. Nucleic Acids Res. 36, 168–178.
Li, W., Ross-Smith, N., Proud, C. G., and Belsham, G. J. (2001). Cleavage of translation initiation factor 4AI (eIF4AI) but not eIF4AII by foot-and-mouth disease virus 3C protease: identification of the eIF4AI cleavage site. FEBS Lett. 507, 1–5.
Li, Y., and Kiledjian, M. (2010). Regulation of mRNA decapping. Wiley Interdiscip. Rev. RNA 1, 253–265.
Lin, J., and Cullen, B. R. (2007). Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J. Virol. 81, 12218–12226.
Lindeberg, J., and Ebendal, T. (1999). Use of an internal ribosome entry site for bicistronic expression of Cre recombinase or rtTA transactivator. Nucleic Acids Res. 27, 1552–1554.
Liu, C. Y., Schroder, M., and Kaufman, R. J. (2000). Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 275, 24881–24885.
Locker, N., Chamond, N., and Sargueil, B. (2011). A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res. 39, 2367–2377.
Ma, Q., Li, X., Vale-Cruz, D., Brown, M. L., Beier, F., and LuValle, P. (2007). Activating transcription factor 2 controls Bcl-2 promoter activity in growth plate chondrocytes. J. Cell. Biochem. 101, 477–487.
Ma, Y., Brewer, J. W., Diehl, J. A., and Hendershot, L. M. (2002). Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J. Mol. Biol. 318, 1351–1365.
Malnou, C. E., Werner, A., Borman, A. M., Westhof, E., and Kean, K. M. (2004). Effects of vaccine strain mutations in domain V of the internal ribosome entry segment compared in the wild type poliovirus type 1 context. J. Biol. Chem. 279, 10261–10269.
Marash, L., and Kimchi, A. (2005). DAP5 and IRES-mediated translation during programmed cell death. Cell Death Differ. 12, 554–562.
Marash, L., Liberman, N., Henis-Korenblit, S., Sivan, G., Reem, E., Elroy-Stein, O., and Kimchi, A. (2008). DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol. Cell 30, 447–459.
Marcotrigiano, J., Gingras, A. C., Sonenberg, N., and Burley, S. K. (1999). Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3, 707–716.
Marissen, W. E., and Lloyd, R. E. (1998). Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 18, 7565–7574.
Mathonnet, G., Fabian, M. R., Svitkin, Y. V., Parsyan, A., Huck, L., Murata, T., Biffo, S., Merrick, W. C., Darzynkiewicz, E., Pillai, R. S., Filipowicz, W., Duchaine, T. F., and Sonenberg, N. (2007). MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 317, 1764–1767.
McCullough, K. D., Martindale, J. L., Klotz, L. O., Aw, T. Y., and Holbrook, N. J. (2001). Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol. Cell. Biol. 21, 1249–1259.
McGarry, T. J., and Lindquist, S. (1985). The preferential translation of Drosophila hsp70 mRNA requires sequences in the untranslated leader. Cell 42, 903–911.
Mekahli, D., Bultynck, G., Parys, J. B., De Smedt, H., and Missiaen, L. (2011). Endoplasmic-reticulum calcium depletion and disease. Cold Spring Harb. Perspect. Biol. 3, doi: 10.1101/cshperspect.a004317
Merquiol, E., Uzi, D., Mueller, T., Goldenberg, D., Nahmias, Y., Xavier, R. J., Tirosh, B., and Shibolet, O. (2011). HCV causes chronic endoplasmic reticulum stress leading to adaptation and interference with the unfolded protein response. PLoS ONE 6, e24660. doi:10.1371/journal.pone.0024660
Merrick, W. C. (1992). Mechanism and regulation of eukaryotic protein synthesis. Microbiol. Rev. 56, 291–315.
Morley, S. J., Coldwell, M. J., and Clemens, M. J. (2005). Initiation factor modifications in the preapoptotic phase. Cell Death Differ. 12, 571–584.
Mymrikov, E. V., Seit-Nebi, A. S., and Gusev, N. B. (2011). Large potentials of small heat shock proteins. Physiol. Rev. 91, 1123–1159.
Nakamura, A., Sato, K., and Hanyu-Nakamura, K. (2004). Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev. Cell 6, 69–78.
Neef, D. W., and Thiele, D. J. (2009). Enhancer of decapping proteins 1 and 2 are important for translation during heat stress in Saccharomyces cerevisiae. Mol. Microbiol. 73, 1032–1042.
Novoa, I., Zeng, H., Harding, H. P., and Ron, D. (2001). Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 153, 1011–1022.
Ohlmann, T., and Jackson, R. J. (1999). The properties of chimeric picornavirus IRESes show that discrimination between internal translation initiation sites is influenced by the identity of the IRES and not just the context of the AUG codon. RNA 5, 764–778.
Ohlmann, T., Prevot, D., Decimo, D., Roux, F., Garin, J., Morley, S. J., and Darlix, J. L. (2002). In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. J. Mol. Biol. 318, 9–20.
Oikawa, D., and Kimata, Y. (2011). Experimental approaches for elucidation of stress-sensing mechanisms of the IRE1 family proteins. Meth. Enzymol. 490, 195–216.
Ouyang, Y. B., Lu, Y., Yue, S., Xu, L. J., Xiong, X. X., White, R. E., Sun, X., and Giffard, R. G. (2012). miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol. Dis. 45, 555–563.
Parker, R., and Sheth, U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646.
Pelletier, J., and Sonenberg, N. (1988). Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325.
Pestova, T. V., de Breyne, S., Pisarev, A. V., Abaeva, I. S., and Hellen, C. U. (2008). eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 27, 1060–1072.
Pfeffer, S., Sewer, A., Lagos-Quentana, M., Sheridan, R., Sander, C., Grasser, F. A., van Dyk, L. F., Ho, C. K., Shuman, S., Chien, M., Russo, J. J., Jy, J., Randall, G., Lindenbach, B. D., Rice, C. M., Simon, V., Ho, D. D., Zavolan, M., and Tuschi, T. (2005). Identification of microRNAs of the herpesvirus family. Nat. Methods 2, 269–276.
Pilakka-Kanthikeel, S., Saiyed, Z. M., Napuri, J., and Nair, M. P. (2011). MicroRNA: implications in HIV, a brief overview. J. Neurovirol. 17, 416–423.
Psarras, S., Volonaki, E., Skevaki, C. L., Xatzipsalti, M., Bossios, A., Pratsinis, H., Tsigkos, S., Gourgiotis, D., Constantopoulos, A. G., Papapetropoulos, A., Saxoni-Papageorgiou, P., and Papadopoulos, N. G. (2006). Vascular endothelial growth factor-mediated induction of angiogenesis by human rhinoviruses. J. Allergy Clin. Immunol. 117, 291–297.
Qin, X., and Sarnow, P. (2004). Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem. 279, 13721–13728.
Raught, B., and Gringas, A. C. (2007). Translational Control in Biology and Medicine. New York: Cold Springs Harbor Laboratory Press.
Raven, J. F., and Koromilas, A. E. (2008). PERK and PKR: old kinases learn new tricks. Cell Cycle 7, 1146–1150.
Ray, P. S., Grover, R., and Das, S. (2006). Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 7, 404–410.
Redondo, N., Sanz, M. A., Welnowska, E., and Carrasco, L. (2011). Translation without eIF2 promoted by poliovirus 2A protease. PLoS ONE 6, e25699. doi:10.1371/journal.pone.0025699
Roberts, A. P., Lewis, A. P., and Jopling, C. L. (2011). miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 39, 7716–7729.
Rojas-Eisenring, I. A., Cajero-Juarez, M., and del Angel, R. M. (1995). Cell proteins bind to a linear polypyrimidine-rich sequence within the 5′-untranslated region of rhinovirus 14 RNA. J. Virol. 69, 6819–6824.
Rouha, H., Thurner, C., and Mandl, C. W. (2010). Functional microRNA generated from a cytoplasmic RNA virus. Nucleic Acids Res. 38, 8328–8337.
Satoh, S., Hijikata, M., Handa, H., and Shimotohno, K. (1999). Caspase-mediated cleavage of eukaryotic translation initiation factor subunit 2alpha. Biochem. J. 342(Pt 1), 65–70.
Skabkin, M. A., Skabkina, O. V., Dhote, V., Komar, A. A., Hellen, C. U., and Pestova, T. V. (2010). Activities of ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 24, 1787–1801.
Sonenberg, N., and Hinnebusch, A. G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745.
Spriggs, K. A., Bushell, M., Mitchell, S. A., and Willis, A. E. (2005). Internal ribosome entry segment-mediated translation during Apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 12, 585–591.
Spriggs, K. A., Stoneley, M., Bushell, M., and Willis, A. E. (2008). Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell 100, 27–38.
Stoneley, M., and Willis, A. E. (2004). Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene 23, 3200–3207.
Sugiyama, R., Naganuma, H., Nishitsuji, H., and Takaku, H. (2011). Human immunodeficiency virus-1 Nef suppresses Hsp70-mediated Tat activation. FEBS Lett. 585, 3367–3371.
Sukarieh, R., Sonenberg, N., and Pelletier, J. (2009). The eIF4E-binding proteins are modifiers of cytoplasmic eIF4E relocalization during the heat shock response. Am. J. Physiol. Cell Physiol. 296, C1207–C1217.
Sun, J., Conn, C. S., Han, Y., Yeung, V., and Qian, S. B. (2011). PI3K-mTORC1 attenuates stress response by inhibiting cap-independent Hsp70 translation. J. Biol. Chem. 286, 6791–6800.
Suzuki, T., Isobe, T., Kitagawa, M., and Ueda, K. (2010). Kaposi’s sarcoma-associated herpesvirus-encoded LANA positively affects on ubiquitylation of p53. Biochem. Biophys. Res. Commun. 403, 194–197.
Suzuki, Y., Minami, M., Suzuki, M., Abe, K., Zenno, S., Tsujimoto, M., Matsumoto, K., and Minami, Y. (2009). The Hsp90 inhibitor geldanamycin abrogates colocalization of eIF4E and eIF4E-transporter into stress granules and association of eIF4E with eIF4G. J. Biol. Chem. 284, 35597–35604.
Svitkin, Y. V., Herdy, B., Costa-Mattioli, M., Gingras, A. C., Raught, B., and Sonenberg, N. (2005). Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell. Biol. 25, 10556–10565.
Tabas, I., and Ron, D. (2011). Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 13, 184–190.
Tahiri-Alaoui, A., Smith, L. P., Baigent, S., Kgosana, L., Petherbridge, L. J., Lambeth, L. S., James, W., and Nair, V. (2009). Identification of an intercistronic internal ribosome entry site in a Marek’s disease virus immediate-early gene. J. Virol. 83, 5846–5853.
Tee, A. R., and Proud, C. G. (2002). Caspase cleavage of initiation factor 4E-binding protein 1 yields a dominant inhibitor of cap-dependent translation and reveals a novel regulatory motif. Mol. Cell. Biol. 22, 1674–1683.
Vallejos, M., Deforges, J., Plank, T. D., Letelier, A., Ramdohr, P., Abraham, C. G., Valiente-Echeverria, F., Kieft, J. S., Sargueil, B., and Lopez-Lastra, M. (2011). Activity of the human immunodeficiency virus type 1 cell cycle-dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acids Res. 39, 6186–6200.
Verma, B., Bhattacharyya, S., and Das, S. (2010). Polypyrimidine tract-binding protein interacts with coxsackievirus B3 RNA and influences its translation. J. Gen. Virol. 91, 1245–1255.
Verma, S., Ali, A., Arora, S., and Banerjea, A. C. (2011). Inhibition of beta-TrcP-dependent ubiquitination of p53 by HIV-1 Vpu promotes p53-mediated apoptosis in human T cells. Blood 117, 6600–6607.
Walters, R. W., Bradrick, S. S., and Gromeier, M. (2009). Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA 16, 239–250.
Wang, K., Xie, S., and Sun, B. (2011a). Viral proteins function as ion channels. Biochim. Biophys. Acta 1808, 510–515.
Wang, X., Olberding, K. E., White, C., and Li, C. (2011b). Bcl-2 proteins regulate ER membrane permeability to luminal proteins during ER stress-induced apoptosis. Cell Death Differ. 18, 38–47.
Warnakulasuriyarachchi, D., Cerquozzi, S., Cheung, H. H., and Holcik, M. (2004). Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J. Biol. Chem. 279, 17148–17157.
Weill, L., James, L., Ulryck, N., Chamond, N., Herbreteau, C. H., Ohlmann, T., and Sargueil, B. (2010). A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res. 38, 1367–1381.
Welnowska, E., Sanz, M. A., Redondo, N., and Carrasco, L. (2011). Translation of viral mRNA without active eIF2: the case of picornaviruses. PLoS ONE 6, e22230. doi:10.1371/journal.pone.0022230
White, J. P., Reineke, L. C., and Lloyd, R. E. (2011). Poliovirus switches to an eIF2-independent mode of translation during infection. J. Virol. 85, 8884–8893.
Willimott, S., and Wagner, S. D. (2010). Post-transcriptional and post-translational regulation of Bcl2. Biochem. Soc. Trans. 38, 1571–1575.
Yan, W., Frank, C. L., Korth, M. J., Sopher, B. L., Novoa, I., Ron, D., and Katze, M. G. (2002). Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc. Natl. Acad. Sci. U.S.A. 99, 15920–15925.
Yang, D., Wilson, J. E., Anderson, D. R., Bohunek, L., Cordeiro, C., Kandolf, R., and McManus, B. M. (1997). In vitro mutational and inhibitory analysis of the cis-acting translational elements within the 5′ untranslated region of coxsackievirus B3: potential targets for antiviral action of antisense oligomers. Virology 228, 63–73.
Yoon, J. H., Choi, E. J., and Parker, R. (2010). Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J. Cell Biol. 189, 813–827.
Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., Negishi, M., and Mori, K. (2001). Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6alpha and 6beta that activates the mammalian unfolded protein response. Mol. Cell. Biol. 21, 1239–1248.
Zdanowicz, A., Thermann, R., Kowalska, J., Jemielity, J., Duncan, K., Preiss, T., Darzynkiewicz, E., and Hentze, M. W. (2009). Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol. Cell 35, 881–888.
Zhang, A., Williamson, C. D., Wong, D. S., Bullough, M. D., Brown, K. J., Hathout, Y., and Colberg-Poley, A. M. (2011a). Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection. Mol. Cell Proteomics 10, M111.009936.
Zhang, H. M., Ye, X., Su, Y., Yuan, J., Liu, Z., Stein, D. A., and Yang, D. (2011b). Coxsackievirus B3 infection activates the unfolded protein response and induces apoptosis through downregulation of p58IPK and activation of CHOP and SREBP1. J. Virol. 84, 8446–8459.
Keywords: internal ribosome-entry site, endoplasmic reticulum stress, microRNA, apoptosis, picornavirus, translation control, coxsackievirus
Citation: Hanson PJ, Zhang HM, Hemida MG, Ye X, Qiu Y and Yang D (2012) IRES-dependent translational control during virus-induced endoplasmic reticulum stress and apoptosis. Front. Microbio. 3:92. doi: 10.3389/fmicb.2012.00092
Received: 08 December 2011; Accepted: 23 February 2012;
Published online: 19 March 2012.
Edited by:
Akio Adachi, The University of Tokushima Graduate School, JapanReviewed by:
Chris Sullivan, University of Texas at Austin, USADahlene N. Fusco, Massachusetts General Hospital, USA
Copyright: © 2012 Hanson, Zhang, Hemida, Ye, Qiu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Decheng Yang, The James Hogg Research Center, St. Paul’s Hospital, University of British Columbia, 1081 Burrard Street, Vancouver, BC, Canada V6Z 1Y6. e-mail: decheng.yang@hli.ubc.ca
 Paul J. Hanson
Paul J. Hanson