- Department of Viral Infections, Research Institute for Microbial Diseases, Osaka University, Suita, Osaka, Japan
Human immunodeficiency virus (HIV) has a very narrow host range. HIV type 1 (HIV-1) does not infect Old World monkeys, such as the rhesus monkey (Rh). Rh TRIM5α was identified as a factor that confers resistance, intrinsic immunity, to HIV-1 infection. Unfortunately, human TRIM5α is almost powerless to restrict HIV-1. However, human TRIM5α potently restricts N-tropic murine leukemia viruses (MLV) but not B-tropic MLV, indicating that human TRIM5α represents the restriction factor previously designated as Ref1. African green monkey TRIM5α represents another restriction factor previously designated as Lv1, which restricts both HIV-1 and simian immunodeficiency virus isolated from macaque (SIVmac) infection. TRIM5 is a member of the tripartite motif family containing RING, B-box2, and coiled-coil domains. The RING domain is frequently found in E3 ubiquitin ligase, and TRIM5α is thought to degrade viral core via ubiquitin–proteasome-dependent and -independent pathways. The alpha isoform of TRIM5 has an additional C-terminal PRYSPRY domain, which is a determinant of species-specific retrovirus restriction by TRIM5α. On the other hand, the target regions of viral capsid protein (CA) are scattered on the surface of core. A single amino acid difference in the surface-exposed loop between α-helices 6 and 7 (L6/7) of HIV type 2 (HIV-2) CA affects viral sensitivity to human TRIM5α and was also shown to be associated with viral load in West African HIV-2 patients, indicating that human TRIM5α is a critical modulator of HIV-2 replication in vivo. Interestingly, L6/7 of CA corresponds to the MLV determinant of sensitivity to mouse factor Fv1, which potently restricts N-tropic MLV. In addition, human genetic polymorphisms also affect antiviral activity of human TRIM5α. Recently, human TRIM5α was shown to activate signaling pathways that lead to activation of NF-κB and AP-1 by interacting with TAK1 complex. TRIM5α is thus involved in control of viral infection in multiple ways.
Introduction
The acquired immune response, both humoral and cellular immunity, requires lymphocyte differentiation and education for effective protection of the host from invasive infection. It requires priming and takes time. On the other hand, innate immunity provides antiviral defenses that can be deployed more rapidly. It does not require education, but most innate immune effectors generally require intracellular and intercellular signaling events, including receptor–ligand binding, adaptor protein phosphorylation, and interferon release from infected cells as well as the interferon signaling pathway to induce an antiviral state in bystander cells. Most toll-like receptors (TLRs), which play a critical role in pattern recognition of invaders, such as double-stranded RNA, lipopolysaccharide (LPS), and CpG DNA, are expressed on macrophages and dendritic cells.
Aside from these conventional immunological definitions, many pieces of evidence provide a new concept of potent protection from viral infection designated as intrinsic immunity. It is constitutively expressed and active in many cells, and does not require any virus-triggered signaling or intercellular communication. The molecules involved in intrinsic immunity are called restriction factors. Two major cellular defense mechanisms against retrovirus infection are Fv1 and TRIM5α that target incoming retroviral core and the Rfv3/APOBEC3 family that causes viral genome hypermutation. This review focuses on the roles of Fv1 and TRIM5α in intrinsic and innate immunity.
The Prototype Restriction Factor Fv1
Mammalian cells show differences in susceptibility to retrovirus infection. The idea that cellular genes could encode constitutive inhibitors of retroviral replication was first suggested in genetic studies of laboratory mice (Odaka and Yamamoto, 1965; Lilly, 1967). Susceptibility of mouse cells to murine leukemia virus (MLV) infection is determined by a restriction factor called Fv1 (Lilly, 1970; Pincus et al., 1971, 1975). The virus resistance induced by Fv1 is genetically dominant over susceptibility, and is evident in cells in vitro (Goff, 2004). Two major allelic variants of Fv1, called Fv1n and Fv1b, were shown to restrict infection by specific strains of MLV (Pincus et al., 1971). The Fv1b allele present in BALB/c mice blocks infection by so-called N-tropic MLV (N-MLV). The Fv1n allele present in NIH/Swiss mice blocks infection by B-tropic MLV (B-MLV). NB-tropic viruses are blocked by neither Fv1b nor Fv1n (Hartley et al., 1970). A less common third allele, Fv1nr, restricts B-MLVs and certain strains of N-MLV (Kozak, 1985). N-MLVs that are not restricted by Fv1nr are called NR-tropic MLVs (Jung and Kozak, 2000; Stevens et al., 2004). The inhibition of a particular virus infection could be abrogated by prior or simultaneous infection by other virus particles. Abrogating particles themselves do not need to be infectious, but they do need to be derived from a restrictive viral strain (Bassin et al., 1978; Boone et al., 1990). These data indicated that Fv1 encodes a unique inhibitor that targets the incoming viral capsid but could be saturated and overwhelmed by simultaneous challenge by multiple virion particles.
The Fv1 gene was successfully isolated by a positional cloning strategy (Best et al., 1996). The Fv1 gene product is a retroviral Gag-like protein, with sequence similarity to the HERV-L family of endogenous retroviral DNAs in the human genome, and to the MuERV-L family in the mouse (Benit et al., 1997). The B and N alleles differ in positions 358 and 399 and the C-terminal portion, all of which seem to contribute to the phenotype (Bock et al., 2000; Bishop et al., 2001). Fv1nr is identical to Fv1n, except for a single point mutation at position 352 (Stevens et al., 2004). A predicted coiled-coil region containing a dimerization domain is located in the N-terminus, and there is a second multimerization domain in the C-terminal half of the molecule (Yap and Stoye, 2003; Bishop et al., 2006). It is likely that multimerization is important for Fv1 function.
Infection of non-permissive cells by a restricted virus is blocked after reverse transcription. The virus enters the cell and synthesizes the viral cDNA by reverse transcription, but the DNA does not enter the nucleus and integrated proviral DNAs are not found (Pryciak and Varmus, 1992; Figure 1). Genetic studies have shown that the viral target of Fv1 is the MLV capsid protein (DesGroseillers and Jolicoeur, 1983) and subsequent work identified position 110 as the major determinant of susceptibility to Fv1 restriction (Kozak and Chakraborti, 1996). B-MLV has a glutamine (Q) at this position, and N-MLV has an arginine (R). More recently, many other residues in CA have been implicated in NB- and NR-tropism (Jung and Kozak, 2000; Stevens et al., 2004). Direct allele-specific binding between Fv1 and MLV CA has not been observed. Most recently, Hilditch et al. (2011) developed a method for the ordered assembly of MLV CA protein on the surface of lipid nanotubes and succeeded in showing specific binding between Fv1 and MLV CA protein. However, the mechanism of action remains unclear.
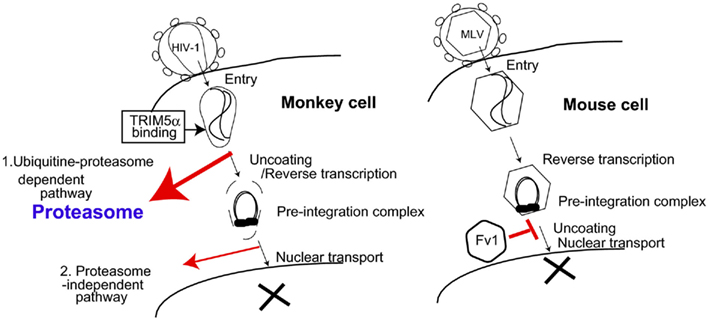
Figure 1. Proposed models of TRIM5α and Fv1 restriction pathways. (Left) Proteasome-dependent and -independent pathways of TRIM5α-mediated human immunodeficiency virus (HIV) restriction in monkey cells. 1. Ubiquitin–proteasome-dependent pathway. Oligomerized TRIM5α recognizes the incoming HIV-1 core. Subsequently, TRIM5α is polyubiquitinated, and ubiquitinated TRIM5α along with HIV-1 core complex are degraded in the proteasome (bold red arrow). 2. Proteasome-independent pathway. Direct binding of TRIM5α with HIV-1 core causes destruction of the viral core without any cellular factors (thin red arrow). (Right) Fv1 inhibits nuclear transport of pre-integration complex of murine leukemia virus (MLV). The precise mechanism of Fv1-mediated restriction is unclear.
Fv1 Like Restriction Factors
Cells from several mammalian species, including humans, acted as if they were homozygous for Fv1b in that they specifically resisted N-MLV infection (Towers et al., 2000). In humans, the postulated inhibitor was designated as Ref1 (for restriction factor 1) and the same capsid residue at the 110th position that controlled sensitivity to Fv1 also controlled sensitivity to Ref1 (Towers et al., 2000). The equine infectious anemia virus (EIAV) was also restricted in human cells, and this was abrogated by both EIAV itself and N-MLV particles (Towers et al., 2002). As analysis of the human genome revealed no intact Fv1 like endogenous retroviral Gag sequences that seemed likely to be responsible for Fv1 like activity (Best et al., 1996), Ref1 was thought to be independent from Fv1. Interest in these restriction systems increased markedly with the finding that several non-human primates restrict human immunodeficiency virus type 1 (HIV-1; Shibata et al., 1995; Himathongkham and Luciw, 1996) in a saturable manner (Hofmann et al., 1999; Towers et al., 2000). HIV-1 infects humans and chimpanzees but not Old World monkeys (OWMs), such as rhesus monkey (Rh) and cynomolgus monkey (CM). HIV-1 efficiently enters cells of OWMs but encounters a block before reverse transcription, and the resistance is dominant over sensitivity in human–monkey heterokaryons (Cowan et al., 2002; Munk et al., 2002). The gene responsible was named Lv1, for lentivirus restriction factor 1. Several primate species were shown to restrict a broader or different range of viruses than just HIV-1. African green monkey (AGM) cells, for example, restrict HIV-1, HIV-2, EIAV, and simian immunodeficiency virus isolated from macaque (SIVmac; Besnier et al., 2002; Hatziioannou et al., 2003).
In 2004, the screening of a Rh cDNA library identified TRIM5α as a cellular antiviral factor (Stremlau et al., 2004; Figure 1). Rh TRIM5α shows strong restriction of HIV-1, is less effective against SIVmac and N-MLV, and does not restrict B-MLV (Hatziioannou et al., 2004; Stremlau et al., 2004). CM TRIM5α restricts HIV-1 but not SIVmac (Nakayama et al., 2005). Human TRIM5α shows little restriction of HIV-1, has a slight effect against SIVmac, and is potently restrictive against N-MLV but shows no effect on B-MLV. It is now widely accepted that human TRIM5α represents the restriction factor Ref1 (Hatziioannou et al., 2004; Keckesova et al., 2004; Perron et al., 2004; Yap et al., 2004). On the other hand, AGM cells have been shown to possess Lv1, which restricts HIV-1, HIV-2, N-MLV, EIAV, and SIVmac infection, and our group and others identified the factor as AGM TRIM5α (Hatziioannou et al., 2004; Keckesova et al., 2004; Nakayama et al., 2005). AGM TRIM5α fails to restrict SIV isolated from AGM (SIVagm) and B-MLV (Song et al., 2005b; Figure 2). It is now known that type I interferon upregulates the transcription of TRIM5α in human (Asaoka et al., 2005) and monkey cells (Carthagena et al., 2008), and this in turn enhances restriction activity against N-MLV (Sakuma et al., 2007a; Carthagena et al., 2008).
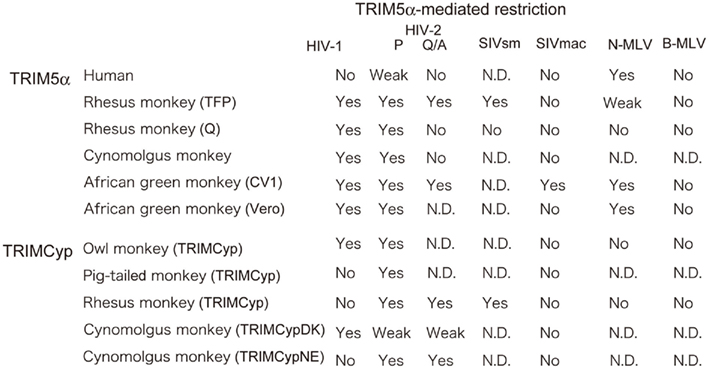
Figure 2. Species-specific restriction by TRIM5α. “Yes” denotes restriction. “Weak” denotes weak restriction. “No” denotes no restriction. “N. D.” denotes no result has yet been published. P and Q/A indicate human immunodeficiency virus type 2 (HIV-2) with proline and glutamine/alanine residues at position 120 of the capsid protein, respectively (Song et al., 2007). HIV-1: human immunodeficiency virus type 1 (Yap et al., 2004; Song et al., 2005a; Stremlau et al., 2005); SIVmac: simian immunodeficiency virus isolated from a macaque; SIVsm: simian immunodeficiency virus isolated from sooty mangabey (Kirmaier et al., 2010); N-MLV: N-tropic murine leukemia virus (Ohkura et al., 2006); B-MLV: B-tropic murine leukemia virus (Ohkura et al., 2006); AGM: CV1 (Nakayama et al., 2005); or Vero cells (Kim et al., 2011) from African green monkey. Rhesus monkey TFP and Q alleles (Stremlau et al., 2004; Ylinen et al., 2005; Ohkura et al., 2006; Kono et al., 2008; Wilson et al., 2008a; Kirmaier et al., 2010), cynomolgus monkey (Nakayama et al., 2005; Song et al., 2007), owl monkey TRIMCyp (Nisole et al., 2004; Sayah et al., 2004; Virgen et al., 2008), pig-tailed monkey TRIMCyp (Brennan et al., 2008; Virgen et al., 2008; Kuang et al., 2009), rhesus monkey TRIMCyp (Wilson et al., 2008b; Kirmaier et al., 2010), and the major and minor haplotypes of CM TRIMCyp (TRIMCypDK and TRIMCypNE, respectively; Ylinen et al., 2010; Dietrich et al., 2011; Saito et al., 2012) are also included.
TRIM5α
TRIM5α is a member of the tripartite motif family containing RING, B-box2, and coiled-coil domains (Figure 3). The RING domain is frequently found in E3 ubiquitin ligase and TRIM5α degrades incoming viral core via the ubiquitin–proteasome-dependent (Stremlau et al., 2006) and -independent pathways leading to potent suppression of HIV-1 reverse transcription (Anderson et al., 2006; Wu et al., 2006; Maegawa et al., 2010; Kim et al., 2011; Figure 1). The levels of HIV-1 late reverse transcription products recovered in the presence of the proteasome inhibitor MG132. However, the resultant HIV-1 cDNA still could not enter the nucleus, suggesting the presence of a proteasome-independent pathway of HIV-1 restriction. The distinct molecular mechanism of the proteasome-independent pathway has yet to be elucidated. TRIM5α has been shown to form a dimer via the coiled-coil region (Kar et al., 2008; Langelier et al., 2008), while the B-box2 domain mediates higher-order self-association of Rh TRIM5α oligomers (Li and Sodroski, 2008; Diaz-Griffero et al., 2009; Ganser-Pornillos et al., 2011). The α-isoform of TRIM5 has an additional C-terminal PRYSPRY (B30.2) domain. The sequence variations in variable regions of the PRYSPRY domain among different monkey species affect species-specific retrovirus infection, while differences in amino acid sequences in the viral capsid protein determine viral sensitivity to restriction (Nakayama and Shioda, 2010). TRIM5α recognizes the multimerized capsid (viral core) of an incoming virus by its PRYSPRY domain and is thus believed to control retroviral infection. Biochemical studies have shown that TRIM5α associates with CA in detergent-stripped N-MLV virions (Sebastian and Luban, 2005) or with an artificially constituted HIV-1 core structure composed of the capsid–nucleocapsid (CA–NC) fusion protein in a PRYSPRY domain-dependent manner (Stremlau et al., 2006). The PRYSPRY domain is thus thought to recognize viral cores.
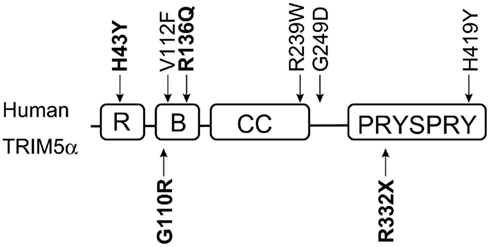
Figure 3. Domains of human TRIM5α and single nucleotide polymorphisms (SNPs). The RING (R), B-box2 (B), coiled-coil (CC), and PRYSPRY domains of human TRIM5α are indicated by squares. Polymorphisms are shown outside the squares. Downward and upward arrows show common and rare SNPs, respectively. SNPs discussed in this review are shown in bold.
Studies on human and Rh recombinant TRIM5αs have shown that the determinant of species-specific restriction against HIV-1 infection resides in variable region 1 (V1) of the PRYSPRY domain (Perez-Caballero et al., 2005; Sawyer et al., 2005). We found that 17 amino acid residues and the adjacent 20-amino acid duplication in the V1 of AGM TRIM5α determined species-specific restriction against SIVmac (Nakayama et al., 2005). Interestingly, a study comparing human and Rh TRIM5α showed that a single amino acid change from R to proline (P) at position 332 in the V1 of human TRIM5α (R332P) conferred potent restriction ability against not only HIV-1 but also SIVmac strain 239 (SIVmac239; Stremlau et al., 2005; Yap et al., 2005). In the case of human immunodeficiency virus type 2 (HIV-2) infection, we found that three amino acid residues of threonine, phenylalanine, and proline (TFP) at positions 339–341 of Rh TRIM5α V1 are important for restricting particular HIV-2 strains that are still resistant to CM TRIM5α (Kono et al., 2008).
Furthermore, a comparison of human and Rh TRIM5α restriction of N-MLV showed that the amino acid residues of human TRIM5α at positions 409 and 410 in variable region 3 (V3) of the PRYSPRY domain are important for restricting N-MLV (Perron et al., 2006).
TRIM5α on Viral Production
Sakuma et al. (2007b) reported that Rh but not human TRIM5α blocks HIV-1 production through rapid degradation of HIV-1 Gag polyproteins. They reported that the RING structure was essential for this activity. Subsequently, Zhang et al. (2008) at Aaron Diamond AIDS Research Center argued against this new pathway of TRIM5α-mediated restriction. Both groups found reduced HIV-1 Gag expression when they cotransfected high levels of Rh TRIM5α expression plasmid (1 μg) with HIV-1 proviral plasmids (0.1 μg) in 293T cells. However, Zhang et al. did not observe increased yield of virus production from TRIM5α knockdown Rh FRhK4 cells, even though they succeeded in almost complete knockdown of endogenous Rh TRIM5α by siRNA transfection as shown by Western blotting analysis. They transfected siRNA first and then transfected siRNA again together with plasmid expressing HIV-1 24 h later. In contrast, Sakuma et al. (2007b) showed increased levels of Gag precursor protein in cell lysates and 10-fold increased virus titers in the culture supernatant of siRNA-treated FRhK4 cells, in which TRIM5 mRNA was knocked down by siRNA. The results of these two studies were inconsistent, although both groups clearly showed TRIM5 knockdown in the same cell line. In the author reply to Zhang et al., Sakuma et al. (2008) suggested that the discrepancies in the results were due to differences in the method of siRNA transfection, in that they transfected HIV-1 plasmid first and siRNAs were transfected 6 h later. However, it is still unclear why the different transfection protocols led to different results in HIV-1 production even though both methods led to complete knockdown of TRIM5α expression. We feel that the importance of late-phase inhibition by Rh TRIM5α is limited, as it is widely accepted that Rh TRIM5α potently inhibits HIV-1 infection at the early phase before HIV-1 particle production (Stremlau et al., 2004, 2006). Consistent with this, Uchil et al. (2008) analyzed 55 TRIM family proteins along with Rh TRIM5α but failed to find an inhibitory effect of Rh TRIM5α on the late-phase of HIV-1 infection, while they did detect a potent inhibitory effect of Rh TRIM5α on the early phase of HIV-1 infection.
Sakuma et al. (2010) speculated that the species specificity for late-phase infection was determined by the coiled-coil region, as introduction of human TRIM5α-specific amino acid residues to Rh TRIM5α, M113T, and/or T146A, abrogated late-phase inhibition activity of Rh TRIM5α, while chimeric Rh TRIM5α containing PRYSPRY of human TRIM5α still inhibited HIV-1 production. On the other hand, the same group showed that the effects of CM and AGM TRIM5α on viral production were lower than that of Rh TRIM5α (Ohmine et al., 2011), consistent with the fact that AGM derived COS7 cells were widely used to recover HIV-1 stock by transfection with proviral plasmid. The experiment of chimeric TRIM5α showed that the C-terminal halves of CM and AGM TRIM5α are responsible for the weakened late-phase inhibition, in contrast to chimeric TRIM5α between Rh and human described above. Finally, Zhang et al. (2010) at Hokkaido University confirmed that human TRIM5α used in the first study had no effect on HIV-1 production and demonstrated that this human TRIM5α contained R437C substitution at the PRYSPRY domain. R437C substitution was not found in the NCBI single nucleotide polymorphism (SNP) database. In addition, they found that a human TRIM5α with authentic R at position 437 reduced HIV-1 production to the same extent as Rh TRIM5α in the high-dose cotransfection experiments (Zhang et al., 2010), consistent with the findings of Zhang et al. (2008). Furthermore, Zhang et al. (2008) found that high-level expression of Rh TRIM5α reduced production of virus with CA derived from SIVmac, while the first and third groups did not. The species specificity of the inhibition of viral production by TRIM5α is therefore controversial.
Viral Determinant of TRIM5α Sensitivity
To determine the CA region that interacts with TRIM5α, we focused on HIV-2, which highly resembles SIVmac (Hahn et al., 2000). Previous studies have shown that HIV-2 strains vary widely in their ability to grow in OWM cells such as baboon, Rh, and CM cells (Castro et al., 1990, 1991; Locher et al., 1998, 2003; Fujita et al., 2003), and HIV-2 isolates with various growth capabilities in OWM cells were evaluated for their sensitivity to CM TRIM5α (Song et al., 2007). We found that viral sensitivity to CM TRIM5α was inversely correlated with growth capability in OWM cells. Sequence analysis showed that the CM TRIM5α-sensitive viruses had proline (P) at position 119 or 120 of CA, while the CM TRIM5α-resistant viruses had either alanine (A) or glutamine (Q) at the same position (Figure 4). Replacing the P of a CM TRIM5α-sensitive HIV-2 molecular clone with A, Q, or glycine (G) changed the phenotype from sensitive to resistant and the mutant viruses replicated well in the presence of CM TRIM5α (Song et al., 2007; Miyamoto et al., 2011). Similar results, although to a lesser extent, were observed when human TRIM5α was used (Song et al., 2007). In the case of Rh TRIM5α, multiple regions of CA including the N-terminal region, L4/5, and amino acid 120 were shown to affect recognition by Rh TRIM5α (Ylinen et al., 2005; Lin and Emerman, 2008; Kono et al., 2010; Pacheco et al., 2010; Ohkura et al., 2011).
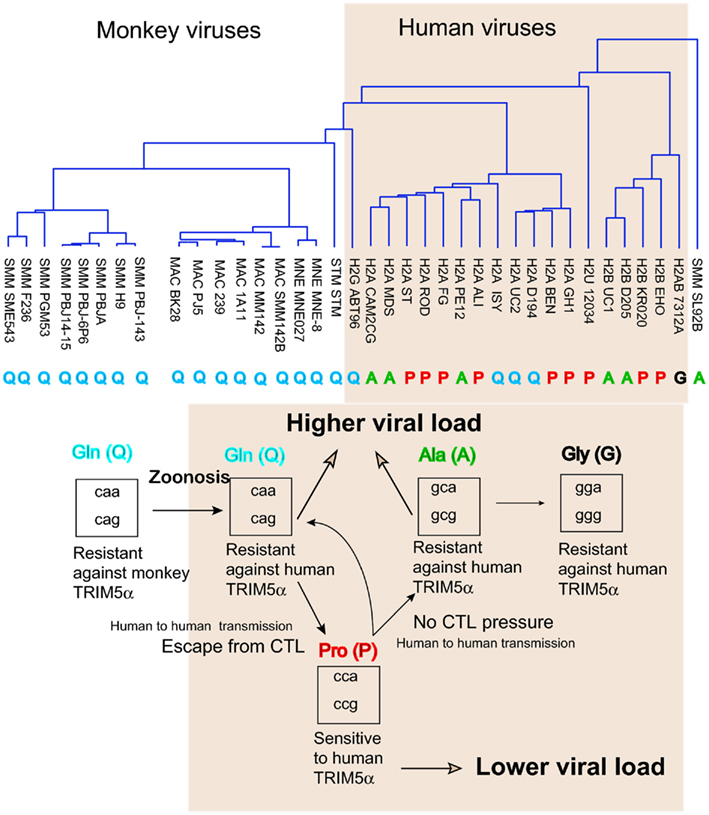
Figure 4. Amino acid variation in HIV-2/SIV capsid (CA). (Upper) A phylogenetic tree of amino acid sequences of capsid of the HIV-2 (shaded area) or SIV isolates obtained from the Los Alamos database. P, Q, A, and G indicate amino acid residue 120 of GH123 or the corresponding position of each virus. (Lower) Filled arrows indicate the possible evolution of amino acid residue 120 of SIV or HIV-2 capsid proteins in humans (shaded area). Open arrows indicate the effects on viral load. Boxes show the codons of glutamine (Gln, Q), proline (Pro, P), alanine (Ala, A), and glycine (Gly, G).
Positions 119 and 120 are located in the loop between α-helices 6 and 7 (L6/7; Figure 5). Previously, a single amino acid substitution at position 110 of MLV CA had been shown to determine viral susceptibility to Fv1 (Kozak and Chakraborti, 1996). The recently published 3-D structure of MLV CA (Mortuza et al., 2004, 2008) revealed that position 110 of N-MLV CA is located at a position in the surface-exposed loop analogous to position 119 or 120 of HIV-2 CA. HIV-2 is assumed to have originated from SIV isolated from sooty mangabey (SIVsm) as a result of zoonotic events involving monkeys and humans (Hahn et al., 2000). Almost all the SIV isolates in the Los Alamos database contain Q at the position corresponding to position 119 or 120 of HIV-2 CA (Figure 4). In contrast, HIV-2 strains possess a mixture of Q, A, and P at the corresponding position.
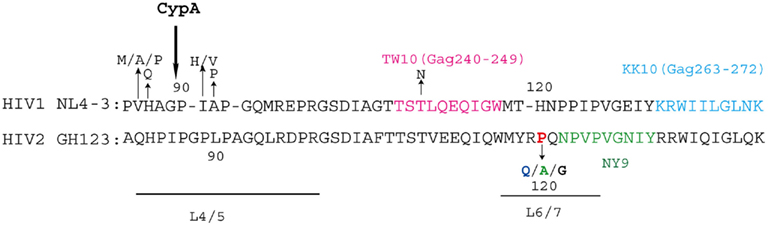
Figure 5. Human immunodeficiency virus (HIV)-1/-2 capsid sequence variations and epitopes of cytotoxic T lymphocytes (CTL). The amino acid sequences of the NL4-3 CA (amino acids 85–140) and GH123 CA (amino acids 82–140) are shown. The loop between α-helices 4 and 5 (L4/5) and the loop between α-helices 6 and 7 (L6/7) are underlined. The CTL epitopes are indicated in pink (TW10), sky blue (KK10), and green (NY9). Amino acid residues that are commonly mutated and were mentioned in this review are indicated. The amino acid residues are numbered both according to the amino acid residues in CA and the whole Gag precursor polyprotein (numbers in parentheses). Cyclophilin A (CypA) catalyzes the 90th proline residue of HIV-1 CA, and alanine residue at the 88th position is critical for CypA binding (Price et al., 2009).
Does amino acid residue at position 119 or 120 in HIV-2 CA affect HIV diseases in infected individuals? It is known that HIV-1 and HIV-2 have distinct natural histories, levels of viremia, transmission rates, and disease associations despite strong sequence homology between the two viruses (Rowland-Jones and Whittle, 2007). Although some HIV-2-infected patients progress to AIDS as rapidly as HIV-1-infected patients, virus replication is controlled in the majority of HIV-2 patients (Poulsen et al., 1989; Berry et al., 2001) and those with low viral load (VL) achieve much longer survival than those with high VL (Ariyoshi et al., 2000). Detailed sequence analysis of HIV-2 CA variations within a large community cohort in Guinea-Bissau comprised of both high- and low-VL patients indicated that CA from viruses in low-VL patients had P residues at position 119 or 120, but in patients with higher VL, position 119 or 120 was frequently occupied by non-P residues. Stratification of the subjects according to the presence or absence of P at position 119 or 120 showed a threefold difference in the median VL of the two groups. These results indicated that HIV-2 replication in infected individuals can be linked to CA variation and human TRIM5α sensitivity (Onyango et al., 2010).
CTL Escape, Drug Resistance, Compensatory Mutation, and TRIM5α Resistance
Recently, Leligdowicz et al. (2010) reported that HLA-B*3501 was associated with HIV-2 with P at position 119 or 120 in the community cohort in Guinea-Bissau. The cytotoxic T cell (CTL) NY9-epitope (NPVPVGNIY) was located two amino acids downstream of position 119 or 120. It is thus possible that viruses were forced to change Q to P at position 119/120 to escape from HLA-B*3501-specific immune response, even though the virus became more sensitive to human TRIM5α due to this substitution (Figure 5). After transmission to individuals lacking HLA-B*3501, viruses may have evolved from the P virus to become more resistant to human TRIM5α (Figure 4). Moreover, several patients with HIV-2 that had a high VL and developed AIDS rapidly have recently been identified in Japan. Sequence analysis of viruses isolated from these patients indicated that they carried G at position 119 or 120. The selection pressure for G substitution is not clear at present but it is worth noting that G was found only in clade A/B recombinants (Ibe et al., 2010).
In the case of HIV-1, Kootstra et al. proposed that a histidine (H)-to-Q substitution at position 87 (H87Q; H219Q in Gag) was a result of escape from human TRIM5α, as the H87Q mutation occurred in 7 of 30 HIV-1-infected individuals in the late-phase of the asymptomatic period and ultimately became the dominant virus population. They also showed that H87Q mutation was associated with resistance to human TRIM5α-mediated inhibition (Kootstra et al., 2007), although the restriction activity of human TRIM5α is much weaker than that of monkey TRIM5α. H87Q mutation was previously observed in HIV-1 variants isolated from HLA-B57-positive individuals. In these individuals, escape mutations in the HLA-B57-restricted CTL epitope TW10 (Figure 5) were observed and it was suggested that H87Q was a compensatory mutation to restore replicative capacity of the otherwise attenuated phenotype of the TW10 escape mutant (Leslie et al., 2004). Amino acid residue 87H is located in the L4/5 and H87Q mutation reduces incorporation of cyclophilin A (CypA) into HIV-1 virions (Gatanaga et al., 2006). H87Q was also observed in protease inhibitor-resistant viruses (Gatanaga et al., 2002) as well as non-nucleoside reverse transcriptase inhibitor-resistant viruses (Ibe et al., 2008). It remains to be elucidated whether mutations in CTL escape or drug-resistant viruses and compensatory mutations in revertant viruses affect viral sensitivity to human TRIM5α. From this point of view, Battivelli et al. (2011) recently reported that some Gag CTL escape mutations indeed increased sensitivity to human TRIM5α. In addition to the H87Q mutation, valine (V)-to-A or V-to-P at position 86, I-to-H or I-to-V at position 91, and A-to-P at position 92 were frequently found in the CypA binding site of HIV-1 in infected individuals, resulting in decreased binding affinity to CypA (Figure 5). Furthermore, Pacheco et al. (2010) adapted HIV-1 to cells expressing Rh TRIM5α and found that a mutant with V-to-M at position 86 showed reduced affinity for Rh TRIM5α but retained the ability to bind CypA efficiently. The relationship between CypA binding and TRIM5α sensitivity should also be evaluated.
Polymorphisms in the Human TRIM5 Gene
Human immunodeficiency virus-1 infection in humans is generally characterized by a long-term chronic disease course gradually progressing to AIDS. Polymorphisms in human CCR5 and other genes have been reported to affect susceptibility to HIV-1 transmission and/or the rate of disease progression to AIDS (O’Brien and Nelson, 2004; Shioda and Nakayama, 2006). Sawyer et al. (2006) reported a common H-to-tyrosine (Y) polymorphism at amino acid residue 43 (H43Y, rs3740996) of the human TRIM5 gene. This SNP is located in the RING domain (Figure 3) and was shown to greatly reduce the ability of TRIM5α to restrict N-MLV (Sawyer et al., 2006). Several studies have indicated that the anti-HIV-1 activity of TRIM5α with 43Y was lower than that with 43H in vitro (Javanbakht et al., 2006; Sawyer et al., 2006; Nakayama et al., 2007), although the difference in anti-HIV-1 activity was very small.
Associations of H43Y with the rate of progression to AIDS have been tested in several studies, but with inconsistent results (Javanbakht et al., 2006; Speelmon et al., 2006; Nakayama et al., 2007; van Manen et al., 2008). Despite the lower anti-N-MLV and anti-HIV-1 activities of TRIM5α with 43Y (Sawyer et al., 2006), Javanbakht et al. (2006) reported a paradoxical protective effect of TRIM5α with 43Y against HIV-1 transmission in African-Americans. Interestingly, we also found that the 43Y-allele was found less frequently in Japanese and Indian HIV-1-infected subjects than in ethnicity-matched controls (Nakajima et al., 2009). Furthermore, Liu et al. (2011) reported that the frequency of H43Y homozygotes was higher in seronegative intravenous drug users than in HIV-infected drug users. The reasons for the discrepancy between the epidemiological and functional effects of H43Y remain unclear, and further studies are required to clarify the impact of H43Y on susceptibility to HIV-1 transmission and/or rate of progression to AIDS. H43Y polymorphism was frequently found in humans but not in monkey species (Johnson and Sawyer, 2009).
In the B-box2 domain, we recently found a novel and rare G-to-R substitution at position 110 of TRIM5α (G110R, rs146215995) in Japan, and this 110R allele was observed more frequently in HIV-1-infected subjects than in controls. As observed epidemiologically, this substitution weakened the anti-HIV-1 and anti-HIV-2 activity in vitro (Nakajima et al., 2009). Price et al. (2010) sequenced exon 2 of the TRIM5 gene in 1032 women enrolled in a long-term monitored Pumwani sex worker cohort, and found that women with the R136Q polymorphism (rs10838525) were less likely to seroconvert despite heavy exposure to HIV-1 through active sex work. The B-box2 domain is important for higher-order multimerization, which is required to form the hexagonal structure to stabilize the interaction between TRIM5α and the capsid (Ganser-Pornillos et al., 2011). It is likely that R136Q substitution affects higher-order multimerization.
Position 332 in the V1 region of the PRYSPRY domain is critical for species-specific recognition of capsid by TRIM5α (Stremlau et al., 2005; Yap et al., 2005). There is no human SNP in this region except for a rare null allele 332X (Figure 3). Torimiro et al. reported that 332R changed to a stop codon in Baka pygmies at an allele frequency of 0.02. This rare allele encoded a truncated form of TRIM5α lacking part of the PRYSPRY domain and showed a dominant negative effect against authentic TRIM5α in vitro (Torimiro et al., 2009). These findings suggest that anti-HIV-1 activity of human TRIM5α may affect HIV-1 transmission although it can hardly protect humans from an HIV-1 pandemic.
Evolution of the TRIM5 Gene
TRIM5 homologs have been found in the genomes of primates, mice, rats, rabbits, dogs, cows, and pigs, but not in chickens (Sawyer et al., 2007; Schaller et al., 2007; Tareen et al., 2009). TRIM5 homolog genes are found in several copies in cows, rats, and mice, but the human genome contains only a single copy of the TRIM5 gene, and the canine homolog is inactivated by a transposon (Johnson and Sawyer, 2009). TRIM5 mRNA expressed in cat cells lacks the PRYSPRY domain (McEwan et al., 2009). No antiviral activity against eight retroviruses, i.e., HIV-1, SIVmac, EIAV, N-MLV, B-MLV, NB-MLV, feline immunodeficiency virus (FIV), and Mason-Pfizer monkey virus, has been reported for the mouse TRIM5 homologs (TRIM12 and TRIM30; Tareen et al., 2009) and mouse TRIM30 targets TAK1-binding protein (TAB) 2 for degradation (Shi et al., 2008).
The TRIM5 gene sequence varies considerably among primate species. The distribution of positively selected amino acid site is located in the PRYSPRY domain and coiled-coil domains (Sawyer et al., 2005; Song et al., 2005a; Newman et al., 2006). It is not surprising that the beginning of the PRYSPRY domain (V1) is highly variable because TRIM5α interacts with several different retroviral cores through this region, as discussed above. Interestingly, in Rh, there is a 339-threonine–phenylalanine–proline (TFP)-341-to-Q polymorphism in TRIM5α (Newman et al., 2006), which reduces the anti-HIV-2 (Kono et al., 2008) and anti-SIVsm (Kirmaier et al., 2010) activity. In the case of SIVsm challenge in vivo, Rh TRIM5αTFP/TFP homozygotes markedly diminished viral replication compared to Rh TRIM5αQ/Q homozygotes (Kirmaier et al., 2010; Reynolds et al., 2011; Yeh et al., 2011). Position 332 in human TRIM5α is arginine (R). Kaiser et al. (2007) showed that a 4-million-year-old endogenous retrovirus from the chimpanzee genome (ptERV1) was suppressed by chimpanzee and human TRIM5α bearing R at position 332 but not gorilla, gibbon, or orangutan TRIM5α bearing Q at the same position. Although Perez-Caballero et al. (2008) failed to reproduce the sensitivity of ptERV1 to human TRIM5α, the main positive selection pressure for TRIM5α is likely to be endogenous retroviruses.
Among New World monkeys, owl monkeys possess CypA as a fusion protein with TRIM5 (TRIMCyp) as a result of LINE-1-mediated retrotranspositional insertion in addition to the authentic CypA (Nisole et al., 2004; Sayah et al., 2004; Figure 6). CypA can bind to the CA of HIV-1, so that the TRIMCyp expressed in owl monkey cells recognizes the HIV-1 core and shows an anti-HIV-1 effect. Retrotransposition of CypA into the TRIM5 gene also occurred independently in OWM, an ancestor of Rh, CM and the pig-tailed monkey (PM; Brennan et al., 2007; Newman et al., 2008; Virgen et al., 2008; Wilson et al., 2008b; Kuang et al., 2009; Figure 6). Dietrich et al. and our group also found major and minor haplotypes of CM TRIMCyp with SNPs in the CypA domain. The major haplotype of CM TRIMCyp bears aspartic acid (D) and lysine (K) at positions 66 and 143 of the CypA domain, respectively. In contrast, the minor haplotype of CM TRIMCyp encodes asparagine (N) and glutamic acid (E) at positions 66 and 143, respectively (Dietrich et al., 2011; Saito et al., 2012). N66 and E143 were also found in PM and Rh TRIMCyps, and the CypA portion of the minor haplotype of CM TRIMCyp has the same amino acid sequence as that of Rh TRIMCyp. Rh, PM, and minor haplotype of TRIMCyp restrict infection of HIV-2, SIVsm, and FIV, but not HIV-1 or SIVmac, while the major haplotype of CM TRIMCyp restricts infection by HIV-1 but not HIV-2 or SIVmac (Brennan et al., 2007; Virgen et al., 2008; Wilson et al., 2008b; Kuang et al., 2009; Dietrich et al., 2011; Saito et al., 2012; Figure 2). As we reviewed recently, genotyping of the monkey TRIM5 gene is important to control animal experiments (Nakayama and Shioda, 2012).
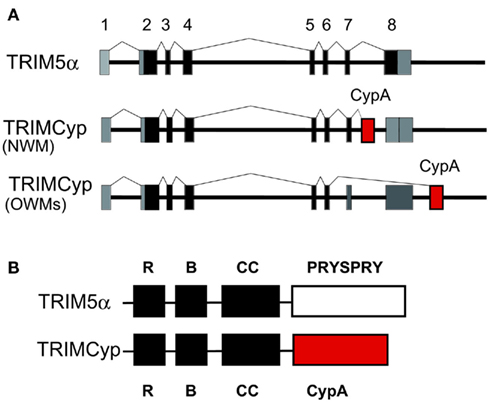
Figure 6. TRIM5α and TRIMCyp. (A) Diagram indicating splicing of TRIM5α or TRIMCyp in New World monkey (NWM) and Old World monkeys (OWMs). Non-coding and coding exons and cyclophilin A (CypA) sequences are shown in gray, black, and red, respectively. (B) The RING (R), B-box2 (B), coiled-coil (CC), PRYSPRY, and CypA domains of TRIM5α and TRIMCyp proteins are indicated by squares.
TRIM5α and TAK1 Complex
Ovyannikova et al. genotyped healthy children receiving rubella-containing vaccine for 14 candidate genes, including TLR3, TLR4, RIG-I, TRIM22, and TRIM5. They measured 6 interleukins, INF-γ, TNF-α, and GM-CSF secretion levels in peripheral blood mononuclear cell culture before and after rubella virus stimulation. An allelic dose-related decrease was observed between H43Y of TRIM5 and TNF-α secretion in response to stimulation, as the medians of 553 HH homozygotes, 131 HY heterozygotes, and 8 YY homozygotes were 34.7 pg/ml (IQR: −3.6 to 95.6), 16.2 pg/ml (IQR: −15.1 to 65.9), and −13.8 pg/ml (IQR: −37.5 to 61.5), respectively. They concluded that TRIM5 gene polymorphism could influence adaptive cytokine responses to rubella vaccination (Ovsyannikova et al., 2010).
How does TRIM5α affect immunological response against non-retroviruses? There have been several reports that TRIM5α has additional activities that are uncoupled from retroviral capsid recognition (Pertel et al., 2011; Tareen and Emerman, 2011). The observation that mouse TRIM30, one of the mouse TRIM5 homologs described above, inhibits NF-κB activation by targeting TLR4 signaling intermediates TAB2 and TAB3 for lysosome-mediated degradation (Shi et al., 2008) prompted Tareen and Emerman (2011) to evaluate the interaction between TRIM5α and TAB2. They showed that human TRIM5α was able to decrease the expression levels of human, mouse and Rh TAB2, while Rh TRIM5α was unable to affect the levels of either Rh or human TAB2 (Tareen and Emerman, 2011). Using an NF-κB-inducible luciferase reporter gene, they assessed the effects of overexpressing human or Rh TRIM5α in 293T cells; however, they found that the expression of human TRIM5α by itself resulted in activation of NF-κB-driven transcription, which was not the case with the mouse TRIM30. At a higher concentration (1.5 μg DNA of TRIM5α vs. 0.5 μg of TAB2), human but not Rh TRIM5α reached saturation and resulted in a drop in NF-κB activation, as human but not Rh TRIM5α degraded TAB2. Both abilities of TRIM5α to target TAB2 and to upregulate NF-κB were independent of the PRYSPRY domain, which is critical for capsid recognition. The RING domain of TRIM5α was necessary to activate NF-κB, while RING and B-box2 of human TRIM5α were sufficient to degrade TAB2 (Tareen and Emerman, 2011).
Subsequently, Pertel et al. (2011) reported similar upregulation of NF-κB and AP-1 activation in human TRIM5α-transfected HEK-293 cells and further confirmed interaction with TAB2, TAB3, and TAK1 complex by immunoprecipitation; however, they did not mention TAB2 degradation reported by Tareen and Emerman (2011). LPS recognized by TLR4 activates AP-1 and NF-κB-signaling, and this culminates in the expression of inflammatory genes. The knockdown of human TRIM5α in THP-1 cells attenuated the induction of AP-1 and NF-κB-dependent genes, indicating that TRIM5α makes a major contribution to LPS-signaling. Acting with the ubiquitin-conjugating enzyme UBC13–UEV1A, human TRIM5α catalyzed the synthesis of unattached K63-linked ubiquitin chains that activate the TAK1 complex. Anti-HIV-1 activity of LPS (Kornbluth et al., 1989) and Escherichia coli infection (Ahmed et al., 2010) were previously reported in macrophages. TRIM5, UBC13, or TAK1 knockdown in THP-1 macrophages rescued HIV-1, SIV, rhabdovirus vesicular stomatitis virus, and paramyxovirus Newcastle disease virus from LPS-induced antiviral state. Finally, they compared induced cytokine levels between stimulation with restricted (e.g., SIVmac) and unrestricted (e.g., HIV-1) virus by human TRIM5α in THP-1 and concluded that antiviral potency was correlated with TRIM5α avidity for the retrovirion capsid lattice, although it is not clear whether the induced cytokines are sufficient to protect macrophages themselves and bystander T cells from viral infection (Figure 7). Especially in HIV-1 infection, it has been speculated that LPS-signaling caused by microbial translocation stimulates cells non-specifically and chronically, resulting in exhaustion of immunity (Brenchley and Douek, 2008). As HIV-1 prefers stimulated T cells, it is reasonable that H43Y RING mutation of TRIM5α showed the paradoxical protective effect on HIV-1 transmission described above.
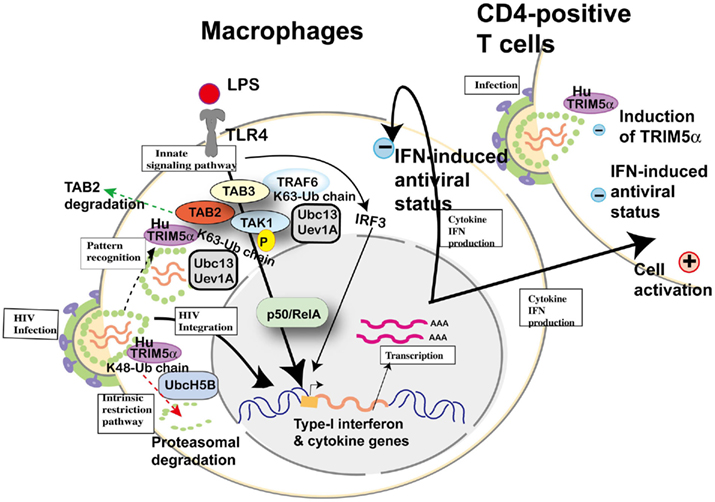
Figure 7. Cellular factors involved in toll-like receptor (TLR) 4-mediated innate signaling and possible involvement of human TRIM5α in HIV-1 infection. Upon lipopolysaccharide (LPS) stimulation, TLR4 recruits tumor necrosis factor receptor-associated factor 6 (TRAF6) to activate the TGF-β-activated kinase 1 (TAK1) complex (TAK1, TAK1-binding protein (TAB) 2 and TAB3) for NF-κB (p50/RelA heterodimer) activation. TRAF6 is polyubiquitinated by the ubiquitin-conjugating enzyme UBC13–UEV1A. TRIM5α is ubiquitinated by UbcH5B (Xu et al., 2003), but the recognition of HIV-1 core by human TRIM5α and proteasomal degradation (dotted arrow in red) cannot inhibit HIV-1 integration into the human genome. When human TRIM5α recognizes an invasive pathogen (dotted arrow in black), human TRIM5α catalyzes the synthesis of unattached K63-linked ubiquitin chains that activate the TAK1 complex (Pertel et al., 2011). On the other hand, TAB2 is degraded by human TRIM5α (dotted arrow in green; Tareen and Emerman, 2011). Activation of IRF3 and NF-κB-dependent gene expression causes both (+) positive status favorable for viral replication and (−) negative status suppressive for viral replication in macrophages and T cells.
Conclusion
The mechanism of antiviral intrinsic immunity via capsid recognition of monkey TRIM5α has been elucidated, although it is still unclear how the prototype antiviral factor Fv1 in mice suppresses nuclear import of MLV. Many TRIM family members, including TRIM21, TRIM23, TRIM27, and TRIM30α, were found to be involved in the TLR4 signaling pathway in mice (Kawai and Akira, 2011). Human TRIM5α has also recently been shown to be involved in this innate immunity (Pertel et al., 2011), and therefore the significance of human TRIM5α in vivo must be clarified in future studies. As the function of mouse TRIM30α is not identical to that of human TRIM5α, it would be interesting to perform human genetic association study with other infections, including bacterial infection.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology, and the Ministry of Health, Labour, and Welfare, Japan.
References
Ahmed, N., Hayashi, T., Hasegawa, A., Furukawa, H., Okamura, N., Chida, T., Masuda, T., and Kannagi, M. (2010). Suppression of human immunodeficiency virus type 1 replication in macrophages by commensal bacteria preferentially stimulating Toll-like receptor 4. J. Gen. Virol. 91, 2804–2813.
Anderson, J. L., Campbell, E. M., Wu, X., Vandegraaff, N., Engelman, A., and Hope, T. J. (2006). Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J. Virol. 80, 9754–9760.
Ariyoshi, K., Jaffar, S., Alabi, A. S., Berry, N., Schim Van Der Loeff, M., Sabally, S., N’Gom, P. T., Corrah, T., Tedder, R., and Whittle, H. (2000). Plasma RNA viral load predicts the rate of CD4 T cell decline and death in HIV-2-infected patients in West Africa. AIDS 14, 339–344.
Asaoka, K., Ikeda, K., Hishinuma, T., Horie-Inoue, K., Takeda, S., and Inoue, S. (2005). A retrovirus restriction factor TRIM5alpha is transcriptionally regulated by interferons. Biochem. Biophys. Res. Commun. 338, 1950–1956.
Bassin, R. H., Duran-Troise, G., Gerwin, B. I., and Rein, A. (1978). Abrogation of Fv-1b restriction with murine leukemia viruses inactivated by heat or by gamma irradiation. J. Virol. 26, 306–315.
Battivelli, E., Migraine, J., Lecossier, D., Yeni, P., Clavel, F., and Hance, A. J. (2011). Gag cytotoxic T lymphocyte escape mutations can increase sensitivity of HIV-1 to human TRIM5alpha, linking intrinsic and acquired immunity. J. Virol. 85, 11846–11854.
Benit, L., De Parseval, N., Casella, J. F., Callebaut, I., Cordonnier, A., and Heidmann, T. (1997). Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol. 71, 5652–5657.
Berry, N., Ariyoshi, K., Balfe, P., Tedder, R., and Whittle, H. (2001). Sequence specificity of the human immunodeficiency virus type 2 (HIV-2) long terminal repeat u3 region in vivo allows subtyping of the principal HIV-2 viral subtypes a and b. AIDS Res. Hum. Retroviruses 17, 263–267.
Besnier, C., Takeuchi, Y., and Towers, G. (2002). Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. U.S.A. 99, 11920–11925.
Best, S., Le Tissier, P., Towers, G., and Stoye, J. P. (1996). Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382, 826–829.
Bishop, K. N., Bock, M., Towers, G., and Stoye, J. P. (2001). Identification of the regions of Fv1 necessary for murine leukemia virus restriction. J. Virol. 75, 5182–5188.
Bishop, K. N., Mortuza, G. B., Howell, S., Yap, M. W., Stoye, J. P., and Taylor, I. A. (2006). Characterization of an amino-terminal dimerization domain from retroviral restriction factor Fv1. J. Virol. 80, 8225–8235.
Bock, M., Bishop, K. N., Towers, G., and Stoye, J. P. (2000). Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74, 7422–7430.
Boone, L. R., Innes, C. L., and Heitman, C. K. (1990). Abrogation of Fv-1 restriction by genome-deficient virions produced by a retrovirus packaging cell line. J. Virol. 64, 3376–3381.
Brenchley, J. M., and Douek, D. C. (2008). The mucosal barrier and immune activation in HIV pathogenesis. Curr Opin HIV AIDS 3, 356–361.
Brennan, G., Kozyrev, Y., and Hu, S. L. (2008). TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc. Natl. Acad. Sci. U.S.A. 105, 3569–3574.
Brennan, G., Kozyrev, Y., Kodama, T., and Hu, S. L. (2007). Novel TRIM5 isoforms expressed by Macaca nemestrina. J. Virol. 81, 12210–12217.
Carthagena, L., Parise, M. C., Ringeard, M., Chelbi-Alix, M. K., Hazan, U., and Nisole, S. (2008). Implication of TRIM alpha and TRIMCyp in interferon-induced anti-retroviral restriction activities. Retrovirology 5, 59.
Castro, B. A., Barnett, S. W., Evans, L. A., Moreau, J., Odehouri, K., and Levy, J. A. (1990). Biologic heterogeneity of human immunodeficiency virus type 2 (HIV-2) strains. Virology 178, 527–534.
Castro, B. A., Nepomuceno, M., Lerche, N. W., Eichberg, J. W., and Levy, J. A. (1991). Persistent infection of baboons and rhesus monkeys with different strains of HIV-2. Virology 184, 219–226.
Cowan, S., Hatziioannou, T., Cunningham, T., Muesing, M. A., Gottlinger, H. G., and Bieniasz, P. D. (2002). Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. U.S.A. 99, 11914–11919.
DesGroseillers, L., and Jolicoeur, P. (1983). Physical mapping of the Fv-1 tropism host range determinant of BALB/c murine leukemia viruses. J. Virol. 48, 685–696.
Diaz-Griffero, F., Qin, X. R., Hayashi, F., Kigawa, T., Finzi, A., Sarnak, Z., Lienlaf, M., Yokoyama, S., and Sodroski, J. (2009). A B-box 2 surface patch important for TRIM5alpha self-association, capsid-binding avidity and retrovirus restriction. J. Virol. 83, 10737–10751.
Dietrich, E. A., Brennan, G., Ferguson, B., Wiseman, R. W., O’Connor, D., and Hu, S. L. (2011). Variable prevalence and functional diversity of the antiretroviral restriction factor TRIMCyp in Macaca fascicularis. J. Virol. 85, 9956–9963.
Fujita, M., Yoshida, A., Sakurai, A., Tatsuki, J., Ueno, F., Akari, H., and Adachi, A. (2003). Susceptibility of HVS-immortalized lymphocytic HSC-F cells to various strains and mutants of HIV/SIV. Int. J. Mol. Med. 11, 641–644.
Ganser-Pornillos, B. K., Chandrasekaran, V., Pornillos, O., Sodroski, J. G., Sundquist, W. I., and Yeager, M. (2011). Hexagonal assembly of a restricting TRIM5alpha protein. Proc. Natl. Acad. Sci. U.S.A. 108, 534–539.
Gatanaga, H., Das, D., Suzuki, Y., Yeh, D. D., Hussain, K. A., Ghosh, A. K., and Mitsuya, H. (2006). Altered HIV-1 Gag protein interactions with cyclophilin A (CypA) on the acquisition of H219Q and H219P substitutions in the CypA binding loop. J. Biol. Chem. 281, 1241–1250.
Gatanaga, H., Suzuki, Y., Tsang, H., Yoshimura, K., Kavlick, M. F., Nagashima, K., Gorelick, R. J., Mardy, S., Tang, C., Summers, M. F., and Mitsuya, H. (2002). Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J. Biol. Chem. 277, 5952–5961.
Hahn, B. H., Shaw, G. M., De Cock, K. M., and Sharp, P. M. (2000). AIDS as a zoonosis: scientific and public health implications. Science 287, 607–614.
Hartley, J. W., Rowe, W. P., and Huebner, R. J. (1970). Host-range restrictions of murine leukemia viruses in mouse embryo cell cultures. J. Virol. 5, 221–225.
Hatziioannou, T., Cowan, S., Goff, S. P., Bieniasz, P. D., and Towers, G. J. (2003). Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22, 385–394.
Hatziioannou, T., Perez-Caballero, D., Yang, A., Cowan, S., and Bieniasz, P. D. (2004). Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. U.S.A. 101, 10774–10779.
Hilditch, L., Matadeen, R., Goldstone, D. C., Rosenthal, P. B., Taylor, I. A., and Stoye, J. P. (2011). Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc. Natl. Acad. Sci. U.S.A. 108, 5771–5776.
Himathongkham, S., and Luciw, P. A. (1996). Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology 219, 485–488.
Hofmann, W., Schubert, D., Labonte, J., Munson, L., Gibson, S., Scammell, J., Ferrigno, P., and Sodroski, J. (1999). Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73, 10020–10028.
Ibe, S., Shigemi, U., Sawaki, K., Fujisaki, S., Hattori, J., Yokomaku, Y., Mamiya, N., Hamaguchi, M., and Kaneda, T. (2008). Analysis of near full-length genomic sequences of drug-resistant HIV-1 spreading among therapy-naive individuals in Nagoya, Japan: amino acid mutations associated with viral replication activity. AIDS Res. Hum. Retroviruses 24, 1121–1125.
Ibe, S., Yokomaku, Y., Shiino, T., Tanaka, R., Hattori, J., Fujisaki, S., Iwatani, Y., Mamiya, N., Utsumi, M., Kato, S., Hamaguchi, M., and Sugiura, W. (2010). HIV-2 CRF01_AB: first circulating recombinant form of HIV-2. J. Acquir. Immune Defic. Syndr. 54, 241–247.
Javanbakht, H., An, P., Gold, B., Petersen, D. C., O’Huigin, C., Nelson, G. W., O’Brien, S. J., Kirk, G. D., Detels, R., Buchbinder, S., Donfield, S., Shulenin, S., Song, B., Perron, M. J., Stremlau, M., Sodroski, J., Dean, M., and Winkler, C. (2006). Effects of human TRIM5alpha polymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Virology 354, 15–27.
Johnson, W. E., and Sawyer, S. L. (2009). Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics 61, 163–176.
Jung, Y. T., and Kozak, C. A. (2000). A single amino acid change in the murine leukemia virus capsid gene responsible for the Fv1(nr) phenotype. J. Virol. 74, 5385–5387.
Kaiser, S. M., Malik, H. S., and Emerman, M. (2007). Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science 316, 1756–1758.
Kar, A. K., Diaz-Griffero, F., Li, Y., Li, X., and Sodroski, J. (2008). Biochemical and biophysical characterization of a chimeric TRIM21-TRIM5alpha protein. J. Virol. 82, 11669–11681.
Kawai, T., and Akira, S. (2011). Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol. Med. 3, 513–527.
Keckesova, Z., Ylinen, L. M., and Towers, G. J. (2004). The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. U.S.A. 101, 10780–10785.
Kim, J., Tipper, C., and Sodroski, J. (2011). Role of TRIM5alpha RING domain E3 ubiquitin ligase activity in capsid disassembly, reverse transcription blockade, and restriction of simian immunodeficiency virus. J. Virol. 85, 8116–8132.
Kirmaier, A., Wu, F., Newman, R. M., Hall, L. R., Morgan, J. S., O’Connor, S., Marx, P. A., Meythaler, M., Goldstein, S., Buckler-White, A., Kaur, A., Hirsch, V. M., and Johnson, W. E. (2010). TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 8, e1000462. doi:10.1371/journal.pbio.1000462
Kono, K., Song, H., Shingai, Y., Shioda, T., and Nakayama, E. E. (2008). Comparison of anti-viral activity of rhesus monkey and cynomolgus monkey TRIM5alphas against human immunodeficiency virus type 2 infection. Virology 373, 447–456.
Kono, K., Song, H., Yokoyama, M., Sato, H., Shioda, T., and Nakayama, E. E. (2010). Multiple sites in the N-terminal half of simian immunodeficiency virus capsid protein contribute to evasion from rhesus monkey TRIM5alpha-mediated restriction. Retrovirology 7, 72.
Kootstra, N. A., Navis, M., Beugeling, C., Van Dort, K. A., and Schuitemaker, H. (2007). The presence of the Trim5alpha escape mutation H87Q in the capsid of late stage HIV-1 variants is preceded by a prolonged asymptomatic infection phase. AIDS 21, 2015–2023.
Kornbluth, R. S., Oh, P. S., Munis, J. R., Cleveland, P. H., and Richman, D. D. (1989). Interferons and bacterial lipopolysaccharide protect macrophages from productive infection by human immunodeficiency virus in vitro. J. Exp. Med. 169, 1137–1151.
Kozak, C. A. (1985). Analysis of wild-derived mice for Fv-1 and Fv-2 murine leukemia virus restriction loci: a novel wild mouse Fv-1 allele responsible for lack of host range restriction. J. Virol. 55, 281–285.
Kozak, C. A., and Chakraborti, A. (1996). Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology 225, 300–305.
Kuang, Y. Q., Tang, X., Liu, F. L., Jiang, X. L., Zhang, Y. P., Gao, G., and Zheng, Y. T. (2009). Genotyping of TRIM5 locus in northern pig-tailed macaques (Macaca leonina), a primate species susceptible to human immunodeficiency virus type 1 infection. Retrovirology 6, 58.
Langelier, C. R., Sandrin, V., Eckert, D. M., Christensen, D. E., Chandrasekaran, V., Alam, S. L., Aiken, C., Olsen, J. C., Kar, A. K., Sodroski, J. G., and Sundquist, W. I. (2008). Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J. Virol. 82, 11682–11694.
Leligdowicz, A., Onyango, C., Yindom, L. M., Peng, Y., Cotten, M., Jaye, A., Mcmichael, A., Whittle, H., Dong, T., and Rowland-Jones, S. (2010). Highly avid, oligoclonal, early-differentiated antigen-specific CD8+ T cells in chronic HIV-2 infection. Eur. J. Immunol. 40, 1963–1972.
Leslie, A. J., Pfafferott, K. J., Chetty, P., Draenert, R., Addo, M. M., Feeney, M., Tang, Y., Holmes, E. C., Allen, T., Prado, J. G., Altfeld, M., Brander, C., Dixon, C., Ramduth, D., Jeena, P., Thomas, S. A., St John, A., Roach, T. A., Kupfer, B., Luzzi, G., Edwards, A., Taylor, G., Lyall, H., Tudor-Williams, G., Novelli, V., Martinez-Picado, J., Kiepiela, P., Walker, B. D., and Goulder, P. J. (2004). HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10, 282–289.
Li, X., and Sodroski, J. (2008). The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J. Virol. 82, 11495–11502.
Lilly, F. (1967). Susceptibility to two strains of Friend leukemia virus in mice. Science 155, 461–462.
Lilly, F. (1970). Fv-2: identification and location of a second gene governing the spleen focus response to Friend leukemia virus in mice. J. Natl. Cancer Inst. 45, 163–169.
Lin, T. Y., and Emerman, M. (2008). Determinants of cyclophilin A-dependent TRIM5 alpha restriction against HIV-1. Virology 379, 335–341.
Liu, F. L., Qiu, Y. Q., Li, H., Kuang, Y. Q., Tang, X., Cao, G., Tang, N. L., and Zheng, Y. T. (2011). An HIV-1 resistance polymorphism in TRIM5alpha gene among Chinese intravenous drug users. J. Acquir. Immune Defic. Syndr. 56, 306–311.
Locher, C. P., Blackbourn, D. J., Herndier, B. G., Reyes-Teran, G., Barnett, S. W., Murthy, K. K., and Levy, J. A. (1998). Transient virus infection and pathogenesis of a new HIV type 2 isolate, UC12, in baboons. AIDS Res. Hum. Retroviruses 14, 79–82.
Locher, C. P., Witt, S. A., Herndier, B. G., Abbey, N. W., Tenner-Racz, K., Racz, P., Kiviat, N. B., Murthy, K. K., Brasky, K., Leland, M., and Levy, J. A. (2003). Increased virus replication and virulence after serial passage of human immunodeficiency virus type 2 in baboons. J. Virol. 77, 77–83.
Maegawa, H., Miyamoto, T., Sakuragi, J., Shioda, T., and Nakayama, E. E. (2010). Contribution of RING domain to retrovirus restriction by TRIM5alpha depends on combination of host and virus. Virology 399, 212–220.
McEwan, W. A., Schaller, T., Ylinen, L. M., Hosie, M. J., Towers, G. J., and Willett, B. J. (2009). Truncation of TRIM5 in the Feliformia explains the absence of retroviral restriction in cells of the domestic cat. J. Virol. 83, 8270–8275.
Miyamoto, T., Yokoyama, M., Kono, K., Shioda, T., Sato, H., and Nakayama, E. E. (2011). A single amino acid of human immunodeficiency virus type 2 capsid protein affects conformation of two external loops and viral sensitivity to TRIM5alpha. PLoS ONE 6, e22779. doi:10.1371/journal.pone.0022779
Mortuza, G. B., Dodding, M. P., Goldstone, D. C., Haire, L. F., Stoye, J. P., and Taylor, I. A. (2008). Structure of B-MLV capsid amino-terminal domain reveals key features of viral tropism, gag assembly and core formation. J. Mol. Biol. 376, 1493–1508.
Mortuza, G. B., Haire, L. F., Stevens, A., Smerdon, S. J., Stoye, J. P., and Taylor, I. A. (2004). High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 431, 481–485.
Munk, C., Brandt, S. M., Lucero, G., and Landau, N. R. (2002). A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. U.S.A. 99, 13843–13848.
Nakajima, T., Nakayama, E. E., Kaur, G., Terunuma, H., Mimaya, J. I., Ohtani, H., Mehra, N., Shioda, T., and Kimura, A. (2009). Impact of novel TRIM5alpha variants, Gly110Arg and G176del, on the anti-HIV-1 activity and the susceptibility to HIV-1 infection. AIDS 23, 2091–2100.
Nakayama, E. E., Carpentier, W., Costagliola, D., Shioda, T., Iwamoto, A., Debre, P., Yoshimura, K., Autran, B., Matsushita, S., and Theodorou, I. (2007). Wild type and H43Y variant of human TRIM5alpha show similar anti-human immunodeficiency virus type 1 activity both in vivo and in vitro. Immunogenetics 59, 511–515.
Nakayama, E. E., Miyoshi, H., Nagai, Y., and Shioda, T. (2005). A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5alpha determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J. Virol. 79, 8870–8877.
Nakayama, E. E., and Shioda, T. (2010). Anti-retroviral activity of TRIM5 alpha. Rev. Med. Virol. 20, 77–92.
Nakayama, E. E., and Shioda, T. (2012). TRIM5α and species tropism of HIV/SIV. Front. Microbiol. 3:13. doi:10.3389/fmicb.2012.00013
Newman, R. M., Hall, L., Connole, M., Chen, G. L., Sato, S., Yuste, E., Diehl, W., Hunter, E., Kaur, A., Miller, G. M., and Johnson, W. E. (2006). Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc. Natl. Acad. Sci. U.S.A. 103, 19134–19139.
Newman, R. M., Hall, L., Kirmaier, A., Pozzi, L. A., Pery, E., Farzan, M., O’Neil, S. P., and Johnson, W. (2008). Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 4, e1000003. doi:10.1371/journal.ppat.1000003
Nisole, S., Lynch, C., Stoye, J. P., and Yap, M. W. (2004). A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. U.S.A. 101, 13324–13328.
Odaka, T., and Yamamoto, T. (1965). Inheritance of susceptibility to Friend mouse leukemia virus. 11. Spleen foci method applied to test the susceptibility of crossbred progeny between a sensitive and a resistant strain. Jpn. J. Exp. Med. 35, 311–314.
Ohkura, S., Goldstone, D. C., Yap, M. W., Holden-Dye, K., Taylor, I. A., and Stoye, J. P. (2011). Novel escape mutants suggest an extensive TRIM5alpha binding site spanning the entire outer surface of the murine leukemia virus capsid protein. PLoS Pathog. 7, e1002011. doi:10.1371/journal.ppat.1002011
Ohkura, S., Yap, M. W., Sheldon, T., and Stoye, J. P. (2006). All three variable regions of the TRIM5alpha B30.2 domain can contribute to the specificity of retrovirus restriction. J. Virol. 80, 8554–8565.
Ohmine, S., Sakuma, R., Sakuma, T., Thatava, T., Takeuchi, H., and Ikeda, Y. (2011). The antiviral spectra of TRIM5alpha orthologues and human TRIM family proteins against lentiviral production. PLoS ONE 6, e16121. doi:10.1371/journal.pone.0016121
Onyango, C. O., Leligdowicz, A., Yokoyama, M., Sato, H., Song, H., Nakayama, E. E., Shioda, T., De Silva, T., Townend, J., Jaye, A., Whittle, H., Rowland-Jones, S., and Cotten, M. (2010). HIV-2 capsids distinguish high and low virus load patients in a West African community cohort. Vaccine 28S2, B60–B67.
Ovsyannikova, I. G., Dhiman, N., Haralambieva, I. H., Vierkant, R. A., O’Byrne, M. M., Jacobson, R. M., and Poland, G. A. (2010). Rubella vaccine-induced cellular immunity: evidence of associations with polymorphisms in the Toll-like, vitamin A and D receptors, and innate immune response genes. Hum. Genet. 127, 207–221.
Pacheco, B., Finzi, A., Stremlau, M., and Sodroski, J. (2010). Adaptation of HIV-1 to cells expressing rhesus monkey TRIM5alpha. Virology 408, 204–212.
Perez-Caballero, D., Hatziioannou, T., Yang, A., Cowan, S., and Bieniasz, P. D. (2005). Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J. Virol. 79, 8969–8978.
Perez-Caballero, D., Soll, S. J., and Bieniasz, P. D. (2008). Evidence for restriction of ancient primate gammaretroviruses by APOBEC3 but not TRIM5alpha proteins. PLoS Pathog. 4, e1000181. doi:10.1371/journal.ppat.1000181
Perron, M. J., Stremlau, M., and Sodroski, J. (2006). Two surface-exposed elements of the B30.2/SPRY domain as potency determinants of N-tropic murine leukemia virus restriction by human TRIM5alpha. J. Virol. 80, 5631–5636.
Perron, M. J., Stremlau, M., Song, B., Ulm, W., Mulligan, R. C., and Sodroski, J. (2004). TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11827–11832.
Pertel, T., Hausmann, S., Morger, D., Zuger, S., Guerra, J., Lascano, J., Reinhard, C., Santoni, F. A., Uchil, P. D., Chatel, L., Bisiaux, A., Albert, M. L., Strambio-De-Castillia, C., Mothes, W., Pizzato, M., Grutter, M. G., and Luban, J. (2011). TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365.
Pincus, T., Hartley, J. W., and Rowe, W. P. (1975). A major genetic locus affecting resistance to infection with murine leukemia viruses. IV. Dose-response relationships in Fv-1-sensitive and resistant cell cultures. Virology 65, 333–342.
Pincus, T., Rowe, W. P., and Lilly, F. (1971). A major genetic locus affecting resistance to infection with murine leukemia viruses. II. Apparent identity to a major locus described for resistance to friend murine leukemia virus. J. Exp. Med. 133, 1234–1241.
Poulsen, A. G., Kvinesdal, B., Aaby, P., Molbak, K., Frederiksen, K., Dias, F., and Lauritzen, E. (1989). Prevalence of and mortality from human immunodeficiency virus type 2 in Bissau, West Africa. Lancet 1, 827–831.
Price, A. J., Marzetta, F., Lammers, M., Ylinen, L. M., Schaller, T., Wilson, S. J., Towers, G. J., and James, L. C. (2009). Active site remodeling switches HIV specificity of antiretroviral TRIMCyp. Nat. Struct. Mol. Biol. 16, 1036–1042.
Price, H., Lacap, P., Tuff, J., Wachihi, C., Kimani, J., Ball, T. B., Luo, M., and Plummer, F. A. (2010). A TRIM5alpha exon 2 polymorphism is associated with protection from HIV-1 infection in the Pumwani sex worker cohort. AIDS 24, 1813–1821.
Pryciak, P. M., and Varmus, H. E. (1992). Fv-1 restriction and its effects on murine leukemia virus integration in vivo and in vitro. J. Virol. 66, 5959–5966.
Reynolds, M. R., Sacha, J. B., Weiler, A. M., Borchardt, G. J., Glidden, C. E., Sheppard, N. C., Norante, F. A., Castrovinci, P. A., Harris, J. J., Robertson, H. T., Friedrich, T. C., Mcdermott, A. B., Wilson, N. A., Allison, D. B., Koff, W. C., Johnson, W. E., and Watkins, D. I. (2011). The TRIM5{alpha} genotype of rhesus macaques affects acquisition of simian immunodeficiency virus SIVsmE660 infection after repeated limiting-dose intrarectal challenge. J. Virol. 85, 9637–9640.
Rowland-Jones, S. L., and Whittle, H. C. (2007). Out of Africa: what can we learn from HIV-2 about protective immunity to HIV-1? Nat. Immunol. 8, 329–331.
Saito, A., Kono, K., Nomaguchi, M., Yasutomi, Y., Adachi, A., Shioda, T., Akari, H., and Nakayama, E. E. (2012). Geographic, genetic and functional diversity of antiretroviral host factor TRIMCyp in cynomolgus macaque (Macaca fascicularis). J. Gen. Virol. 93, 594–602.
Sakuma, R., Mael, A. A., and Ikeda, Y. (2007a). Alpha interferon enhances TRIM5alpha-mediated antiviral activities in human and rhesus monkey cells. J. Virol. 81, 10201–10206.
Sakuma, R., Noser, J. A., Ohmine, S., and Ikeda, Y. (2007b). Rhesus monkey TRIM5alpha restricts HIV-1 production through rapid degradation of viral Gag polyproteins. Nat. Med. 13, 631–635.
Sakuma, R., Ohmine, S., and Ikeda, Y. (2010). Determinants for the rhesus monkey TRIM5alpha-mediated block of the late phase of HIV-1 replication. J. Biol. Chem. 285, 3784–3793.
Sakuma, R., Ohmine, S., Mael, A. A., Noser, J. A., and Ikeda, Y. (2008). Reply to: no effect of endogenous TRIM5alpha on HIV-1 production. Nat. Med. 14, 236–238.
Sawyer, S. L., Emerman, M., and Malik, H. S. (2007). Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 3, e197. doi:10.1371/journal.ppat.0030197
Sawyer, S. L., Wu, L. I., Akey, J. M., Emerman, M., and Malik, H. S. (2006). High-frequency persistence of an impaired allele of the retroviral defense gene TRIM5alpha in humans. Curr. Biol. 16, 95–100.
Sawyer, S. L., Wu, L. I., Emerman, M., and Malik, H. S. (2005). Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U.S.A. 102, 2832–2837.
Sayah, D. M., Sokolskaja, E., Berthoux, L., and Luban, J. (2004). Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430, 569–573.
Schaller, T., Hue, S., and Towers, G. J. (2007). An active TRIM5 protein in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J. Virol. 81, 11713–11721.
Sebastian, S., and Luban, J. (2005). TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology 2, 40.
Shi, M., Deng, W., Bi, E., Mao, K., Ji, Y., Lin, G., Wu, X., Tao, Z., Li, Z., Cai, X., Sun, S., Xiang, C., and Sun, B. (2008). TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat. Immunol. 9, 369–377.
Shibata, R., Sakai, H., Kawamura, M., Tokunaga, K., and Adachi, A. (1995). Early replication block of human immunodeficiency virus type 1 in monkey cells. J. Gen. Virol. 76(Pt 11), 2723–2730.
Shioda, T., and Nakayama, E. E. (2006). Human genetic polymorphisms affecting HIV-1 diseases. Int. J. Hematol. 84, 12–17.
Song, B., Gold, B., O’Huigin, C., Javanbakht, H., Li, X., Stremlau, M., Winkler, C., Dean, M., and Sodroski, J. (2005a). The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J. Virol. 79, 6111–6121.
Song, B., Javanbakht, H., Perron, M., Park, D. H., Stremlau, M., and Sodroski, J. (2005b). Retrovirus restriction by TRIM5alpha variants from old world and new world primates. J. Virol. 79, 3930–3937.
Song, H., Nakayama, E. E., Yokoyama, M., Sato, H., Levy, J. A., and Shioda, T. (2007). A single amino acid of the human immunodeficiency virus type 2 capsid affects its replication in the presence of cynomolgus monkey and human TRIM5alphas. J. Virol. 81, 7280–7285.
Speelmon, E. C., Livingston-Rosanoff, D., Li, S. S., Vu, Q., Bui, J., Geraghty, D. E., Zhao, L. P., and Mcelrath, M. J. (2006). Genetic association of the antiviral restriction factor TRIM5alpha with human immunodeficiency virus type 1 infection. J. Virol. 80, 2463–2471.
Stevens, A., Bock, M., Ellis, S., Letissier, P., Bishop, K. N., Yap, M. W., Taylor, W., and Stoye, J. P. (2004). Retroviral capsid determinants of Fv1 NB and NR tropism. J. Virol. 78, 9592–9598.
Stremlau, M., Owens, C. M., Perron, M. J., Kiessling, M., Autissier, P., and Sodroski, J. (2004). The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853.
Stremlau, M., Perron, M., Lee, M., Li, Y., Song, B., Javanbakht, H., Diaz-Griffero, F., Anderson, D. J., Sundquist, W. I., and Sodroski, J. (2006). Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. U.S.A. 103, 5514–5519.
Stremlau, M., Perron, M., Welikala, S., and Sodroski, J. (2005). Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J. Virol. 79, 3139–3145.
Tareen, S. U., and Emerman, M. (2011). Human Trim5alpha has additional activities that are uncoupled from retroviral capsid recognition. Virology 409, 113–120.
Tareen, S. U., Sawyer, S. L., Malik, H. S., and Emerman, M. (2009). An expanded clade of rodent Trim5 genes. Virology 385, 473–483.
Torimiro, J. N., Javanbakht, H., Diaz-Griffero, F., Kim, J., Carr, J. K., Carrington, M., Sawitzke, J., Burke, D. S., Wolfe, N. D., Dean, M., and Sodroski, J. (2009). A rare null allele potentially encoding a dominant-negative TRIM5alpha protein in Baka pygmies. Virology 391, 140–147.
Towers, G., Bock, M., Martin, S., Takeuchi, Y., Stoye, J. P., and Danos, O. (2000). A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. U.S.A. 97, 12295–12299.
Towers, G., Collins, M., and Takeuchi, Y. (2002). Abrogation of Ref1 retrovirus restriction in human cells. J. Virol. 76, 2548–2550.
Uchil, P. D., Quinlan, B. D., Chan, W. T., Luna, J. M., and Mothes, W. (2008). TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 4, e16. doi:10.1371/journal.ppat.0040016
van Manen, D., Rits, M. A., Beugeling, C., Van Dort, K., Schuitemaker, H., and Kootstra, N. A. (2008). The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathog. 4, e18. doi:10.1371/journal.ppat.0040018
Virgen, C. A., Kratovac, Z., Bieniasz, P. D., and Hatziioannou, T. (2008). Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc. Natl. Acad. Sci. U.S.A. 105, 3563–3568.
Wilson, S. J., Webb, B. L., Maplanka, C., Newman, R. M., Verschoor, E. J., Heeney, J. L., and Towers, G. J. (2008a). Rhesus macaque TRIM5 alleles have divergent antiretroviral specificities. J. Virol. 82, 7243–7247.
Wilson, S. J., Webb, B. L., Ylinen, L. M., Verschoor, E., Heeney, J. L., and Towers, G. J. (2008b). Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc. Natl. Acad. Sci. U.S.A. 105, 3557–3562.
Wu, X., Anderson, J. L., Campbell, E. M., Joseph, A. M., and Hope, T. J. (2006). Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc. Natl. Acad. Sci. U.S.A. 103, 7465–7470.
Xu, L., Yang, L., Moitra, P. K., Hashimoto, K., Rallabhandi, P., Kaul, S., Meroni, G., Jensen, J. P., Weissman, A. M., and D’Arpa, P. (2003). BTBD1 and BTBD2 colocalize to cytoplasmic bodies with the RBCC/tripartite motif protein, TRIM5delta. Exp. Cell Res. 288, 84–93.
Yap, M. W., Nisole, S., Lynch, C., and Stoye, J. P. (2004). Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. U.S.A. 101, 10786–10791.
Yap, M. W., Nisole, S., and Stoye, J. P. (2005). A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 15, 73–78.
Yeh, W. W., Rao, S. S., Lim, S. Y., Zhang, J., Hraber, P. T., Brassard, L. M., Luedemann, C., Todd, J. P., Dodson, A., Shen, L., Buzby, A. P., Whitney, J. B., Korber, B. T., Nabel, G. J., Mascola, J. R., and Letvin, N. L. (2011). The TRIM5 gene modulates penile mucosal acquisition of simian immunodeficiency virus in rhesus monkeys. J. Virol. 85, 10389–10398.
Ylinen, L. M., Keckesova, Z., Wilson, S. J., Ranasinghe, S., and Towers, G. J. (2005). Differential restriction of human immunodeficiency virus type 2 and simian immunodeficiency virus SIVmac by TRIM5alpha alleles. J. Virol. 79, 11580–11587.
Ylinen, L. M., Price, A. J., Rasaiyaah, J., Hue, S., Rose, N. J., Marzetta, F., James, L. C., and Towers, G. J. (2010). Conformational adaptation of Asian macaque TRIMCyp directs lineage specific antiviral activity. PLoS Pathog. 6, e1001062. doi:10.1371/journal.ppat.1001062
Zhang, F., Perez-Caballero, D., Hatziioannou, T., and Bieniasz, P. D. (2008). No effect of endogenous TRIM5alpha on HIV-1 production. Nat. Med. 14, 235–236; author reply 236–238.
Keywords: Fv1, TRIM5α, TAB2, HIV-1, HIV-2, SIV, capsid, TRIMCyp
Citation: Nakayama EE and Shioda T (2012) Role of human TRIM5α in intrinsic immunity. Front. Microbio. 3:97. doi: 10.3389/fmicb.2012.00097
Received: 17 January 2012; Paper pending published: 17 February 2012;
Accepted: 28 February 2012; Published online: 15 March 2012.
Edited by:
Atsushi Koito, Kumamoto University, JapanReviewed by:
Elisa Vicenzi, San Raffaele Scientific Institute, ItalyAkatsuki Saito, Kyoto University, Japan
Copyright: © 2012 Nakayama and Shioda. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Tatsuo Shioda, Department of Viral Infections, Research Institute for Microbial Diseases, Osaka University, 3-1, Yamada-oka, Suita, Osaka 565-0871, Japan. e-mail: shioda@biken.osaka-u.ac.jp
