- O.N. Allen Laboratory for Soil Microbiology, Department of Soil Science, University of Wisconsin, Madison, WI, USA
Bacteria are key in the biodegradation of polycyclic aromatic hydrocarbons (PAH), which are widespread environmental pollutants. At least six genotypes of PAH degraders are distinguishable via phylogenies of the ring-hydroxylating dioxygenase (RHD) that initiates bacterial PAH metabolism. A given RHD genotype can be possessed by a variety of bacterial genera, suggesting horizontal gene transfer (HGT) is an important process for dissemination of PAH-degrading genes. But, mechanisms of HGT for most RHD genotypes are unknown. Here, we report in silico and functional analyses of the phenanthrene-degrading bacterium Delftia sp. Cs1-4, a representative of the phnAFK2 RHD group. The phnAFK2 genotype predominates PAH degrader communities in some soils and sediments, but, until now, their genomic biology has not been explored. In the present study, genes for the entire phenanthrene catabolic pathway were discovered on a novel ca. 232 kb genomic island (GEI), now termed the phn island. This GEI had characteristics of an integrative and conjugative element with a mobilization/stabilization system similar to that of SXT/R391-type GEI. But, it could not be grouped with any known GEI, and was the first member of a new GEI class. The island also carried genes predicted to encode: synthesis of quorum sensing signal molecules, fatty acid/polyhydroxyalkanoate biosynthesis, a type IV secretory system, a PRTRC system, DNA mobilization functions and >50 hypothetical proteins. The 50% G + C content of the phn gene cluster differed significantly from the 66.7% G + C level of the island as a whole and the strain Cs1-4 chromosome, indicating a divergent phylogenetic origin for the phn genes. Collectively, these studies added new insights into the genetic elements affecting the PAH biodegradation capacity of microbial communities specifically, and the potential vehicles of HGT in general.
Introduction
Bacteria are key agents affecting the fate and behavior of polycyclic aromatic hydrocarbons (PAH), which are widespread environmental pollutants. At least six genotypes of PAH degraders have been identified from phylogenies of the ring-hydroxylating dioxygenase (RHD) that initiates PAH metabolism (Moser and Stahl, 2001). The RHD genotypes also show conservation in the organization of other PAH degradation enzymes that are associated with the RHD (Moser and Stahl, 2001; Stolz, 2009). A given RHD genotype can be possessed by a variety of bacterial genera, suggesting horizontal gene transfer (HGT) is an important process for dissemination of PAH-degrading genes. Moreover, the RHD genotypes have characteristic patterns of distribution, and typically occur within genera of a single bacterial family (Moser and Stahl, 2001), possibly reflecting different pathways of gene flow that are operative within differing phylogenetic groups. Information on such pathways is limited, but their elucidation is essential to understand processes shaping the function of microbial communities.
Information on mechanisms of HGT in proteobacterial PAH degraders is mostly derived from analyses of sphingomonads (Romine et al., 1999; Demaneche et al., 2004; Stolz, 2009), and species of Pseudomonas (Dennis and Zylstra, 2004; Li et al., 2004; Basu and Phale, 2008; Heinaru et al., 2009). For both, studies have focused on plasmids. In Pseudomonas, PAH-degrading genes are characteristically of the nah phylotype, which are invariably located on IncP-9 plasmids. Complete sequencing of one such element, pDTG1, revealed all nah genes were contained within a Tn3-like transposon (Dennis and Zylstra, 2004), potentially providing a means of intracellular gene movement as well. But, although IncP-9 plasmids have a host range that spans the alpha-, beta-, and gamma-proteobacteria (Suzuki et al., 2010), these elements have not yet been identified to carry PAH-degrading genes beyond the genus Pseudomonas. Plasmids of the sphingomonads show a similar pattern, and are limited to that group. Thus, mechanisms of HGT for other RHD genotypes are largely unknown.
Many vehicles of HGT exist as elements integrated into bacterial chromosomes rather than autonomously replicating plasmids, and are termed integrating conjugative elements (ICE) or genomic islands (GEI). High throughput sequencing has revealed that GEI are wide spread in bacterial genomes, and can encode a variety of adaptive traits (van der Meer and Sentchilo, 2003; Dobrindt et al., 2004; Mario et al., 2009). However, while there has been extensive study of GEI associated with pathogenicity-related functions, comparatively little is known about GEI that confer other types of phenotypes, including biodegradation activities. To date, the clc element is the only biodegradation-related GEI examined in some detail, and it is a pKLC102/PAGI-type GEI (Toleman and Walsh, 2011) that encodes degradation of chlorocatechols/aminophenols (Gaillard et al., 2006). Involvement of GEI in PAH biodegradation has not been examined, although these are now being revealed by genome sequencing projects. For example, in Alteromonas sp. SN2 (Jin et al., 2011), genes for an nah-type RHD are carried on 100 kb Tn3-like transposon, a structure similar to that of the nah genes on pDTG1. Also, in Polaromonas naphthalenivorans CJ2, nag-type RHD genes are associated with an unclassified GEI (Yagi et al., 2009).
Recently, genomes were determined for two bacteria of the phnAFK2 genotype: Delftia sp. Cs1-4 (closed genome sequence) and Burkholderia sp. Ch1-1 (shotgun genome sequence). The phnAFK2 genotype is interesting as it has appeared as a predominant class of PAH degraders in surveys of some soils and marine sediments (Lozada et al., 2008; Ding et al., 2010), but relatively little is known about this group. To date, the most extensive genetic information about the phnAFK2 genotype has been limited to a ca. 24 kb GenBank entry (AB024945) from Alcaligenes faecalis AFK2 that included genes for the RHD and nine downstream enzymes. This GenBank entry, however, was not associated with any empirical evidence for the predicted functionality, which as yet remains un-established. Furthermore, while HGT of the phnAFK2 genotype is indicated by its structural conservation across multiple genera, the vehicle(s) mediating transfer are unknown.
Here, we present the first genomic and functional analyses of the phnAFK2 genotype. The studies focused on the closed sequence of Delftia sp. Cs1-4 and, where relevant, additional analyses were done with Burkholderia sp. Ch1-1. Our findings included: elucidation of a new ca. 232 kb GEI that encoded the entire pathway for PAH (phenanthrene) catabolism, discovery of four new genes associated with PAH degradation and elucidation of a novel operon structure for a key pathway enzyme (o-phthalate dioxygenase). Collectively, these studies provided new insights into the evolutionary and metabolic processes associated with PAH degradation and HGT in microbial communities.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
Bacterial strains and plasmids used in this study are listed in Table S1 in Supplementary Material. Burkholderia sp. strain Ch1-1 and Delftia sp. strain Cs1-4 were isolated from PAH-contaminated soil collected in Chippewa Falls, WI, USA, based on their abilities to grow on phenanthrene as sole carbon source (Vacca et al., 2005). Cells of strain Ch1-1 and Cs1-4 were routinely grown on mineral salt medium (MSM; Hickey and Focht, 1990) supplemented with phenanthrene as a sole carbon source (1 g/l) and incubated at 25°C with shaking at 150 rpm. When pyruvate was used as a substrate, it was added to MSM from a filter-sterilized stock to a final concentration of 50 mM. E. coli strains were grown at 28 or 37°C on Luria–Bertani (LB) medium supplemented as appropriate with antibiotics. The PAH were obtained from Aldrich Chemicals (Milwaukee, WI, USA), Packard (La Grange, IL, USA), ACROS Organics, and PFALTZ and Bauer (Waterbury, CT, USA) at the highest purity available.
Proteomics
Phenanthrene-grown cells (200 ml, OD600 = 0.6) were filtered through glass wool and then recovered by centrifugation at 3,800 × g for 5 min. The cells were washed twice with phosphate buffer and resuspended in phosphate buffered saline. Pyruvate-grown cells were prepared similarly, except for omission of the filtration step. “In Liquid” digestion and mass spectrometric analysis was done at the University of Wisconsin–Madison, Biotechnology Center, Mass Spectrometry Facility. Cells were incubated on ice (15 min) in 20% (w/v) trichloroacetic acid/acetone (10-fold excess) and precipitated proteins were collected by centrifugation (10 min, 16,000 × g). The protein pellets were washed sequentially with ice-cold acetone (2×), ice-cold methanol (1×), and then solubilized in 4 μl of 8 M urea in 100 mM NH4HCO3 (ABC). After a 10-min incubation, this denatured protein solution was diluted to 25 μl by addition of 1.25 μl 25 mM dithiothreitol, 2 μl acetonitrile (ACN), 15.12 μl 25 mM ABC, 0.12 μl 1 M Tris–HCl (pH 7.5), and 2.5 μl trypsin [100 ng/μl Trypsin Gold (Promega Corp.) in 25 mM ABC]. Following a 12-h of trypsin digestion (37°C) another 2 μl of trypsin was added and a second digestion done for 2 h at 42°C. Digestion was terminated by addition of trifluoroacetic acid to a final concentration of 0.3% (w/v).
Peptides were analyzed by nanoLC–MS/MS using an Agilent 1100 nanoflow system (Agilent Technologies, Palo Alto, CA, USA) connected to a hybrid linear ion trap-orbitrap mass spectrometer (LTQ-Orbitrap XL, Thermo Fisher Scientific, San Jose, CA, USA) equipped with a nanoelectrospray ion source. Capillary HPLC was done using an in-house fabricated column with integrated electrospray emitter essentially as described by (Martin et al., 2000) except that 360 μm × 75 μm fused silica tubing was used. The column was packed with Jupiter 4 μm C12 particles (Phenomenex Inc., Torrance, CA, USA) to ca. 12 cm. Sample loading (8 μl) and desalting were achieved using a trapping column in line with the autosampler (Zorbax 300SB-C18, 5 μm, 5 mm × 0.3 mm, Agilent Technologies). During sample loading and desalting, an isocratic mobile phase of 1% (v/v) ACN, 0.1% formic acid (w/v) was used, and run at 10 μl/min. Peptides were then eluted by a gradient (200 nl/min) of 0.1% (w/v) formic acid in water (Buffer A) and 95% (v/v) ACN, 0.1% (w/v) formic acid in water (Buffer B) with increasing Buffer A from 0 to 40% (75 min), 40 to 60% (20 min), and 60 to 100% (5 min). The LTQ-Orbitrap acquired MS/MS spectra in data-dependent mode with MS survey scans (m/z 300–2,000) collected in centroid mode at a resolving power of 100,000. Spectra were collected on the five most-abundant signals in each survey scan. Dynamic exclusion was used to increase dynamic range and maximize peptide identifications, and excluded precursors between 0.55 m/z below and 1.05 m/z above previously selected precursors; precursors remained on the exclusion list for 15 s. Singly charged ions and ions for which the charge state could not be assigned were rejected from consideration for MS/MS. Raw MS/MS data was searched against the non-redundant Delftia sp. Cs1-4 amino acid sequence database (5,867 protein entries; GenBank entry NC_015563) using an in-house Mascot search engine allowing variable modifications (methionine oxidation; glutamine, asparagine deamidation) a peptide mass tolerance of 15 ppm and 0.6 Da fragment mass.
Protein annotation and significance of protein identifications were done with Scaffold (version 3.00.07, Proteome Software Inc., Portland, OR, USA). Peptide identifications were accepted if identities of at least two unique, matching peptides were established at ≥90.0% probability via the Peptide Prophet algorithm (Keller et al., 2002). Protein identification probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
DNA Manipulations, Cloning, and Sequence Analysis
Standard procedures were used for DNA manipulations (Sambrook et al., 1989). Restriction enzymes were obtained from Promega (Madison, WI, USA) or New England Biolabs (Beverly, MA, USA). Genomic DNA was extracted using Wizard Genomic DNA Purification kit (Promega, Madison, WI, USA). Primers were synthesized by the UW–Madison Biotechnology Center. Ex Taq DNA polymerase (Takara, Madison, WI, USA) was used in PCR amplification reactions. DNA Fragments were purified from agarose gels with QIAquick spin columns (QIAGEN, Valencia, CA, USA) and sequenced at the Univ. Wisconsin Biotechnology Center. GenBank files of all sequences used in this study were imported into Geneious (v. 5.4.5; Biomatters Ltd., Auckland 1010, New Zealand) for manual curation, and alignment by Clustal, MUSCLE, or MAUVE, as appropriate. Whole genome sequences were uploaded to Islandviewer1 for identification of GEIs. Alignment figures were generated with Geneious v. 5.4.5 (http://www.geneious.com).
Generation of ΔphnAcAd Mutants and Mutant Complementation
Genes putatively encoding the phenanthrene dioxygenase large and small subunits (phnAcAd), upstream (Cs1-4 genome positions 1959578–1960237) and downstream (strain Cs1-4 genome positions 1962038–1962817) fragments were amplified with primers phnAcbglII/phnACKpnI and phnACsacII/phnAcsacI, respectively (Table S2 in Supplementary Material). The amplicons were gel purified and cloned into pGEM-T Easy (pSCH448 and pSCH449). These fragments were sequentially assembled on the same sites on pJK100 (pSCH453). Next, pSCH453 was introduced into E. coli BW19851 (λ pir) then conjugated into strain Cs1-4 and Tcs and Kmr transconjugants were selected (strain SCH455, ΔphnAcAd:Km). Replacement of phnAcAd with the kanamycin resistance gene was confirmed by PCR using primers phndectF and phndetcR.
For complementation, a 2,064-bp fragment containing phnAcAd (strain Cs1-4 genome positions 1960120–1962184) was amplified with primers PhnAcNdeF1 and PhnAcSacIIR1 (Table S2 in Supplementary Material) using strain Cs1-4 genomic DNA as a template. The amplicon was then cloned into pGEM-T easy (pSCH434). The insertion was released by SacII and cloned into the site on pSCH442 and under the control of a strong promoter (PnpdA) identified in strain Cs1-4 (Chen and Hickey, 2011). A construct with the correct insertion (pSCH462, PnpdA + phenAcAd) was identified by sequencing multiple random colonies, which was then introduced into the ΔphenAcAd:Km mutant, yielding strain SCH471. The phenAcAd expression vector pSCH453 was used in a similar manner to test for complementation of a Burkholderia sp. Ch1-1 ΔphnAcAd:Km mutant.
Heterologous Expression
The pET5a expression system (Promega, Madison, WI, USA) was used for gene expression in E. coli. Primers, phnF and phnR (Table S2 in Supplementary Material) were used for cloning of the 5.3-kb phn RHD (phnAb–Ad) cluster from genomic DNA of Burkholderia sp. Ch1-1. The purified fragment was cloned into pGEM-T Easy, yielding pTPHE2 and propagated in E. coli DH5α. The Fragment inserted into pTPHE2 was released by digestion with NdeI, and then purified and ligated into NdeI-digested pET5a to give pPHE2. The orientation and sequence of phnAb–Ad in pPHE2 was confirmed by sequencing. Plasmids were introduced by electroporation into E. coli strain BL21 AI strain for heterologous expression. Single colonies of E. coli BL21 AI (pPHE2) and the negative control E. coli BL21 AI (pET5a) were inoculated into separate tubes of LB (3 ml) and incubated overnight at 37°C. Aliquots of each culture (2 ml) were then inoculated into fresh LB (100 ml), incubated at 37°C. Upon reaching OD600 = 0.6, arabinose was added to a final concentration of 0.2% (w/v) for induction. Following overnight incubation at 25°C, cells were collected by centrifugation (3,800 × g, 5 min) then washed with, and resuspended in, M9 medium (43). For substrate specificity tests, PAH were added from hexane stocks to a final concentration of 100 μM. The cells were incubated at 25°C, and the reaction was stopped by addition of an equal volume of ethyl acetate. Analysis of phenanthrene and phenanthrene metabolites was done in a manner similar to that described by Kang et al. (2003).
Results
Structure of the phn Island
The Delftia sp. Cs1-4 genome consisted of a single, 6.8 Mb chromosome. MAUVE Alignment of the strain Cs1-4 chromosome with that of its closest relative with a complete genome sequence, Delftia acidovorans SPH1, indicated that genes encoding the entire pathway for phenanthrene catabolism were located within a 232,325-bp region absent in D. acidovorans SPH1 (Figure 1A). Analysis by GEI detection algorithms also identified this 232 kb region as a GEI (Figure 1B), now termed the phn island. The 3′ terminus of the island was located in a non-coding region, upstream of a gene putatively encoding an S fimbrial protein. This terminus was delimited by a 57-bp sequence, which also occurred nearby in the chromosomal region as an inverted repeat (Figures 2A,B).
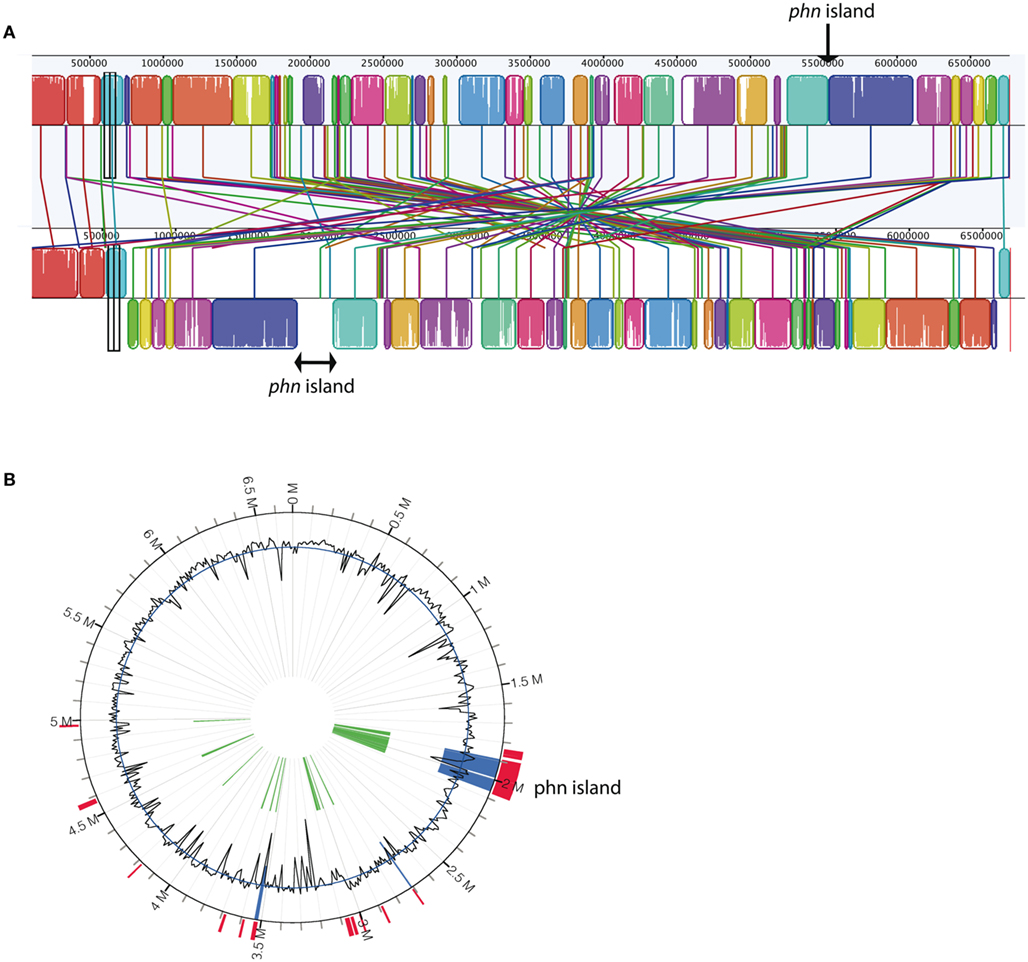
Figure 1. (A) MAUVE alignment of chromosomes from Delftia acidovorans SPH1 (top, NC_010002) and Delftia sp. Cs1-4 (bottom, NC_015563). The location of the phn island in strain Cs1-4 is indicated by the brackets and the insertion site in strain SPH1 indicated by the arrow. In strain Cs1-4, loci bordering the island are (5′–3′) DelCs14_1618 and DelCs14_1848; corresponding loci in strain SPH1 are Daci_4996 and Daci_4997, respectively. (B) IslandViewer analyses of the Delftia sp. Cs1-4 chromosome, with colored regions indicating genomic islands.
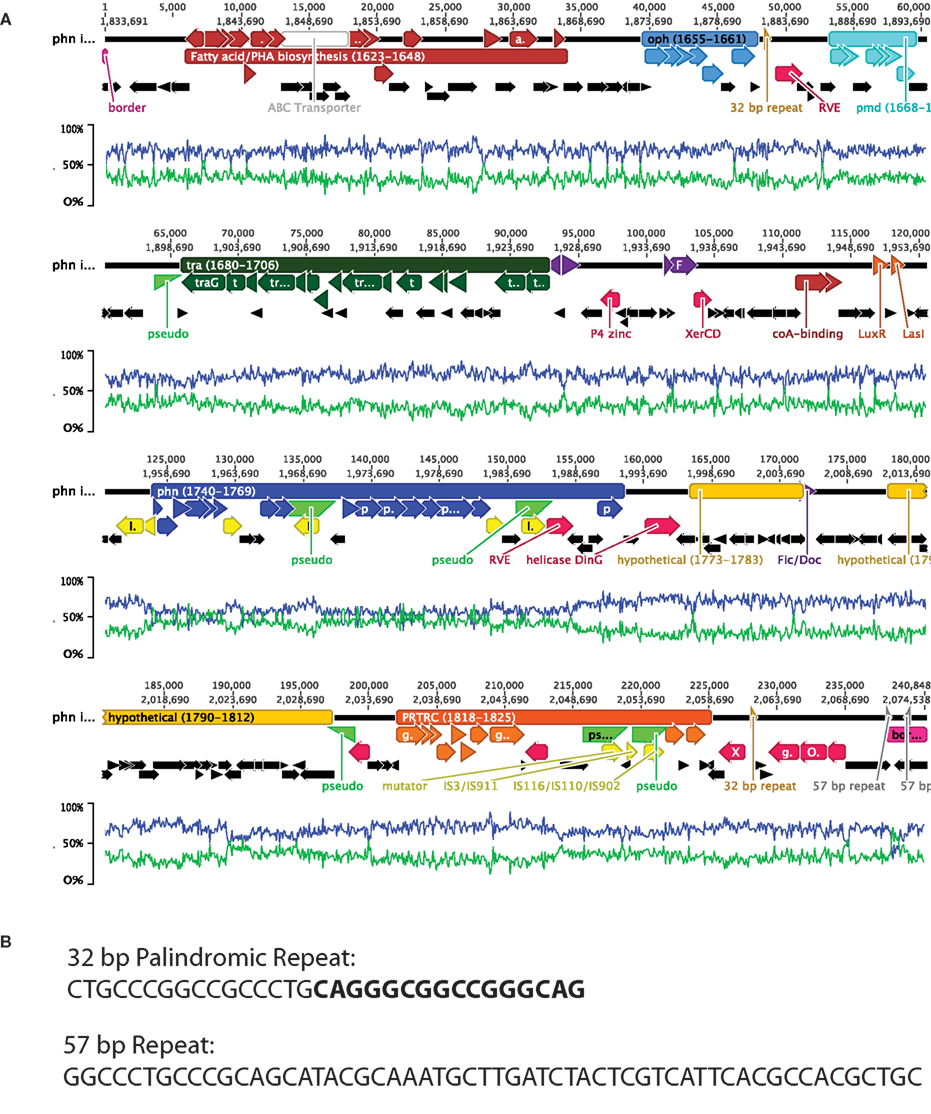
Figure 2. (A) Summary of phn island features. Major regions discussed in the text and the associated genes are indicated by colored bars and arrows, respectively. The values within the bars are the ranges of Delftia sp. Cs1-4 loci that are encompassed by each region. Highlighted regions are the: phn cluster (blue), oph cluster (light blue), pmd cluster (turquoise), tra region (dark green), PRTRC cluster (orange), fatty acid/polyhydroxyalkanoate (PHA) biosynthesis (brown). Other colored genes are: phage/plasmid integration/excision (red), phage/plasmid regulation/stabilization (purple), transposons (yellow), pseudogenes (bright green half arrows), and other genes including hypothetical (black). Additional genes and features discussed in the text are colored and labeled. Lines beneath the gene diagram indicate the average%G + C (blue) and average%A + T (green) of the overlaying region. (B) Sequences of the 32-bp palindromic and 57-bp repeats indicated in (A) and discussed in the text. The two halves of the palindrome are distinguished by normal vs. boldface type.
There were 10 clusters of genes related by encoding: (1) phenanthrene catabolism to o-phthalate, (2) o-phthalate degradation to protocatechuate, (3) protocatechuate catabolism to central intermediates, (4) fatty acid/polyhydroxyalkanoate biosynthesis, (5) a type IV secretory system, (6) a PRTRC system, (7) element integration, and (8–10) hypothetical proteins. The island also contained genes encoding quorum sensing signal molecules (Figure 2A). Genes encoding phage-like mobilization functions and prophage stabilization were distributed across the element, and a 32-bp palindromic sequence was present at two locations (Figures 2A,B). While the G + C content of the phn island as a whole was similar to that of the strain Cs1-4 chromosome (66.7%), that of the region encoding the upper pathway for phenanthrene degradation (phn cluster; Figure 2A) showed a marked divergence at ca. 50% G + C.
The cluster encoding phenanthrene catabolism to o-phthalate encompassed 17 phn genes (phnAa to phnI; Figures 3 and 4) interspersed with genes for glutathione S-transferase, regulatory elements, transposons/integrases and hypothetical proteins. Alignment of the phn clusters of strain Cs1-4 with those of A. faecalis AFK2 and Burkholderia sp. Ch1-1 revealed general conservation of phnAb–phnD, the exception being strain Ch1-1 in which a transposon was inserted between phnAb–Aa (Figure 4). Further downstream, there were two regulatory genes: tetR was present in strains Cs1-4 and Ch1-1, but fragmented in A. faecalis AFK2, while a marR regulator was conserved between the clusters. The gst gene was also conserved, as were phnH–G. But, the clusters diverged at phnI, which was interrupted by a transposase in strain Cs1-4. In strain Cs1-4, phnI was the last gene in the cluster (Figure 4).
Figure 3. Pathway of phenanthrene degradation through o-phthalate and meta-cleavage of protocatechuate. Compounds are: phenanthrene (I), phenanthrene-cis-3,4-dihydrodiol (II), 3,4-dihydroxyphenanthrene (III), 2-hydroxy-2H-benzo[h]chromene-2-carboxylate (IV), trans-4-(1′-hydroxynaphth-2′-yl)-2-oxobut-3-enoate (V), 1-hydroxy-2-naphthaldehyde (VI), 1-hydroxy-2-naphthoate (VII), trans-2′-carboxy-benzalpyruvate (VIII), 2-carboxy-benzaldehyde (IX), o-phthalate (X), phthalate-4,5-cis-dihydrodiol (XI), 4,5-dihydroxyphthalate (XII), protocatechuate (XIII), 4-carboxy-2-hydroxymuconate semialdehyde (XIV), 2-pyrone-4,6-dicarboxylate (XV), 4-carboxy-2-hydroxy-6-methoxy-6-oxohexa-2,4-dienoate (XVI), 4-oxalomesaconate (XVII), 4-oxalocitramalate (XVIII), pyruvate (XIX), oxaloacetate (XX). Numbers above arrows correspond to enzymes and cognate genes mediating each step and are: phnAb, ferredoxin reductase (1); phnAa, ferredoxin (1); phnAc, phenanthrene dioxygenase large subunit (1); phnAd, phenanthrene dioxygenase small subunit (1); phnB, phenanthrene dihydrodiol dehydrogenase (2); phnC, 3,4-dihydroxyphenanthrene dioxygenase (3); phnD, 2-hydroxychromene-2-carboxylate isomerase (4); phnE, trans-o-hydroxybenzylidenepyruvate hydratase-aldolase (5); phnF, 1-hydroxy-2-naphtoaldehyde dehydrogenase (6); phnG, 1-hydroxy-2-naphthoate dioxygenase (7); phnH, trans-2′-carboxy-benzalpyruvate hydratase-aldolase (8); phnI, 2-carboxybenzaldehyde dehydrogenase (9); ophA1, phthalate 4,5-dioxygenase reductase (10); ophA2, phthalate 4,5-dioxygenase oxygenase (10); ophB, cis-phthalate dihydrodiol dehydrogenase (11); ophC, 4,5-dihydroxyphthalate decarboxylase (12); pmdA, protocatechuate meta-dioxygenase large subunit (13); pmdB, protocatechuate meta-dioxygenase small subunit (13); pmdC, 2-hydroxy-4-carboxymuconate semialdehyde dehydrogenase (14); pmdD, 2-pyrone-4,6-dicarboxylic acid hydrolase (15); pmdE, 4-oxalomesaconate hydratase (16); pmdF, 4-oxalocitramalate aldolase (17).
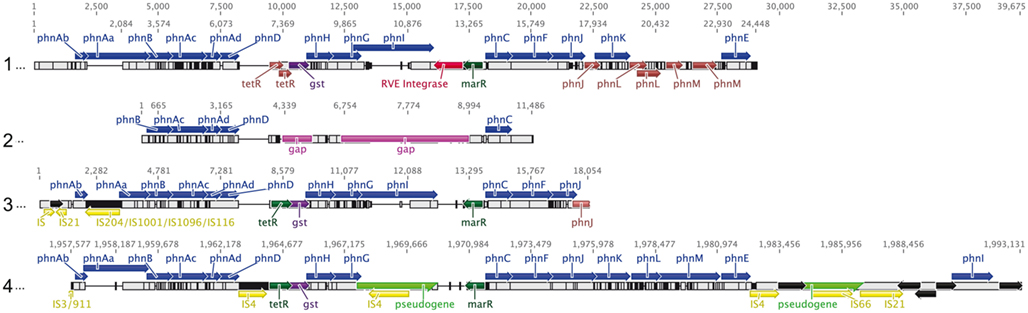
Figure 4. Alignment of all currently available phn clusters. Sequences are from: (1) A. facaelis AFK2 (AB024945), (2) Acidovorax sp. AN3 (EU910094), (3) Burkholderia sp. Ch1-1 (NZ_ADNR01000074), and (4) Delftia sp. Cs1-4 (NC_015563). Values across the top of the diagram indicate position (bp) within the alignment, while those above each sequence indicate position in the original source of that sequence. Colors indicate phn genes (blue), transposons (yellow), regulatory elements (dark green), integrases (red), fragmented genes (brown), pseudogenes (bright green half arrows), and regions of sequence gaps (purple). Reactions catalyzed for all gene products except phnJ, phnK, phnL, and phnM (function unknown) are summarized in Figure 3. For A. facaelis AFK2, features not annotated in the original GenBank entry included: phnJ, phnK, marR, the RVE-type integrase and all fragmented genes.
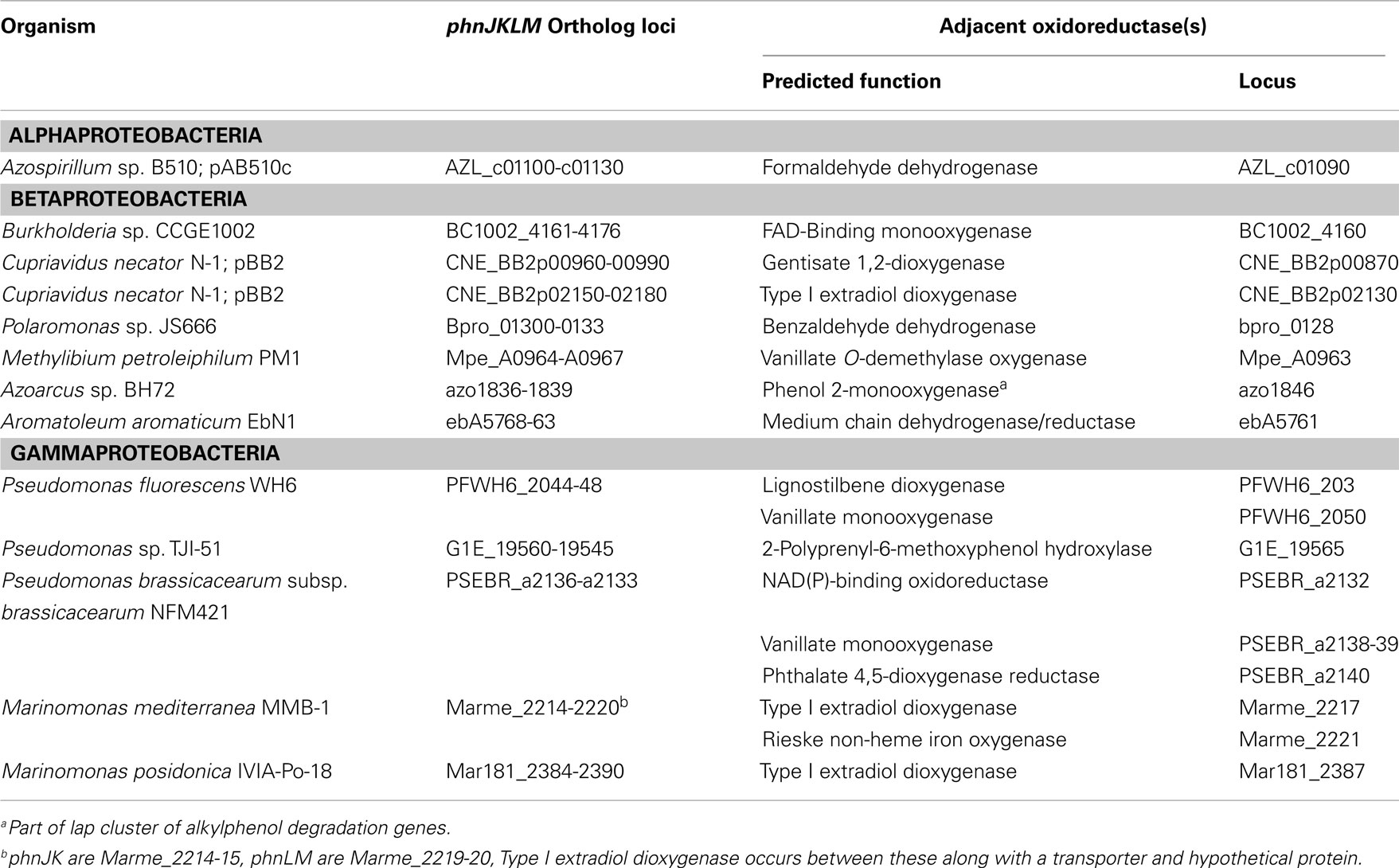
Four new genes (termed phnJKLM) were identified (Figure 4), which proteomics indicated were linked to phenanthrene degradation (see below). The first of these, phnJ, encoded a protein containing a signal peptide, and a conserved domain of unknown function (DUF) 1302. The product of the following gene, phnK, contained a signal peptide and DUF 1329. The phnL product was a Ycf48-like protein, which in phototrophs, functions in the assembly of Photosystem II (Komenda et al., 2008; Rengstl et al., 2011). Lastly, phnM, encoded an RND-type hydrophobe/amphiphile efflux protein. In the other phnAFK2 phylotypes, phnJ was either truncated (A. faecalis AFK2) or fragmented (Burkholderia sp. Ch1-1). Further comparison of the phnJKLM cluster could be done only with strain AFK2, which differed from strain Cs1-4 in that phnLM were fragmented and only phnK was intact. GenBank searches revealed the phnJKLM cluster was widely distributed across diverse alpha-, beta-, and gamma-proteobacteria, the latter with representatives from terrestrial and marine environments (Table 1). A common feature linking these diverse organisms was the location of phnJKLM adjacent to genes for oxidoreductases (Table 1).
In strain Cs1-4, the lower pathway of phenanthrene catabolism entails transformation of o-phthalate to protocatechuate (PCA), and PCA degradation to oxaloacetate and pyruvate via the meta pathway (Figure 3). The genes for o-phthalate and PCA degradation (oph and pmd, respectively) occurred in separate clusters (Figures 2A, 5, and 6). The organization of the oph cluster (Figure 5) was generally similar to that of Comamonas testosteroni KF-1. However, a significant difference between the oph clusters of strain Cs1-4 and that of C. testosteroni KF-1 was the presence in the strain Cs1-4 cluster of two non-identical copies of ophA2 (Figure 5), which encodes the oxygenase component of phthalate-4,5-dioxygenase (Batie et al., 1987).
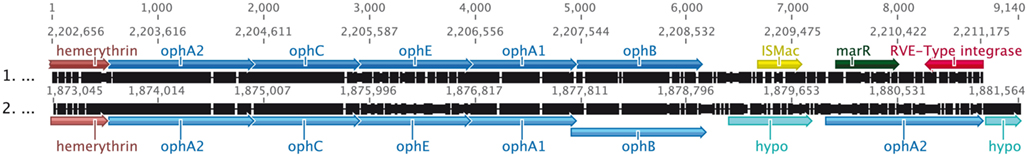
Figure 5. Alignment of oph clusters from Comamonas testosteroni KF-1 (top, NZ_AAUJ02000001) and Delftia sp. Cs1-4 (bottom, NC_015563). Values across the top of the diagram indicate position (bp) within the alignment, while those above each sequence indicate position in the original source of that sequence. Colors indicate oph genes (light blue), transposons (yellow), integrases (red), regulatory elements (dark green), hemerythrin (brown), and genes for hypothetical proteins (turquoise). Reactions catalyzed by oph gene products are summarized in Figure 3.
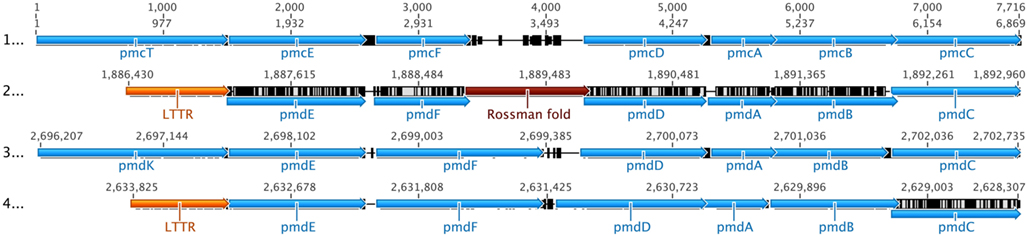
Figure 6. Alignment of pmd/pmc clusters. Sequences are from: (1) Comamonas sp. DJ-12 (AY866410), (2) Delftia sp. Cs1-4 Cluster 1 (NC_015563), (3) Delftia sp. Cs1-4 Cluster 2 (NC_015563), (4) Ramlibacter tataouinensis TTB310 (CP000245). Values across the top of the diagram indicate position (bp) within the alignment, while those above each sequence indicate position in the original source of that sequence. Colors indicate: pmd/pmc genes (light blue), genes for LTTR-Domain proteins (orange), genes for dehydrogenase-Rossmann fold-NAD(H) binding proteins (brown). Other genes including those encoding hypothetical proteins are black. Reactions catalyzed by pmd gene products are summarized in Figure 3.
The pmd cluster occurred in two loci, one located on the phn island (Cluster 1; Figures 2A and 6) and the other outside of it (Cluster 2; Figure 6). There was also a third locus in strain Cs1-4 where additional copies of pmdAB [protocatechuate 4,5 (meta)-dioxygenase] were located, but were not associated with any other PCA degradation genes (DelCs14_3008, 3009). The structure of pmd Cluster 1 was similar to that of Ramlibacter tataouinensis TTB310 while Cluster 2 organization was like that of other Comamonadaceae as exemplified by Comamonas sp. DJ-12 (Figure 6). The structures of the pmd clusters diverged in two ways (Figure 6). First, Cluster 1 lacked pmdK/pmcT (putatively encoding an aromatic acid transporter) and instead had a gene for a predicted LTTR-domain protein located in that position. Secondly, Cluster 1 encompassed a gene predicted to encode an NAD-binding dehydrogenase, the function of which in the meta pathway is unknown. The latter gene was conserved in the pmd clusters of strains Cs1-4 and TTB310, but the NAD-binding dehydrogenase was present only in strain Cs1-4 (Figure 6).
A cluster of genes near the 5′ end of the island-encoded enzymes were predicted to link biosynthesis of fatty acids with that of the carbon storage polymers, polyhydroxyalkanoates (PHA; Figure 2A). Key intermediates of fatty acid synthesis used in production of medium chain length PHA are enoyl-CoA, 3-ketoacyl-CoA, (S)-3-hydroxyacyl-CoA and 3-hydroxyacyl-acyl-carrier-protein (Witholt and Kessler, 1999). Enzymes linking these to PHA synthesis are enoyl-CoA hydratase (Fiedler et al., 2002; Tsuge et al., 2003), 3-ketoacyl-(acyl-carrier-protein) reductase (Ren et al., 2000), and epimerase (Madison and Huisman, 1999; Witholt and Kessler, 1999). Within the putative PHA cluster, three genes (DelCs14_1623, 1625, 1627) were predicted to encode MaoC-like proteins, which have been demonstrated as important in PHA biosynthesis (Park and Lee, 2003). These MaoC-like proteins were orthologs of the (R)-specific enoyl-CoA hydratase, PhaJ1 (Park and Lee, 2003). The product of DelCs14_1636 was a FabG ortholog, which is a 3-ketoacyl-(acyl-carrier-protein) reductase. Epimerase activity could be provided by two genes predicted to encode MmgE/PrpD family proteins (DelCs14_1624, DelCs14_1635) for which this function has recently been established (Lohkamp et al., 2006). Other genes with potential links to fatty acid biosynthesis were (locus): enoyl-CoA reductase (a.k.a. butyrl-CoA dehydrogenase; DelCs14_1626), acetyl-CoA synthetase (DelCs14_1645), crotonase (DelCs14_1643), and NAD(P)(H)-dependent oxidoreductases (DelCs14_1637) for the inter-conversion of aldehydes/ketones and alcohols.
Another function of the PHA cluster may be to synthesize pantothenate. This compound is essential for coenzyme-A (CoA), which in turn is required for fatty acid metabolism, and thus PHA biosynthesis as well. β-Alanine is required for pantothenate production, and is produced primarily by aspartate decarboxylation (Williamson and Brown, 1979); the enzyme catalyzing this reaction, L-aspartate decarboxylase, was the predicted product of DelCs14_1648. Aspartate formation in turn proceeds from oxaloacetic acid (OAA), a product of the phenanthrene lower pathway. Levels of OAA (and acetyl-CoA) could also be modulated by formyl-CoA transferase (DelCs14_1628), which catalyzes the reversible transfer of CoA between acetyl-CoA and OAA, and citryl-CoA lyase (DelCs14_1639, _1629), which transforms citryl-CoA into acetyl-CoA and OAA. The PHA cluster contained five proteins of a complete ABC transport system with similarity to that for branched chain amino acids. The function of the transporter is unknown, but the presence of a periplasmic binding protein (DelCs14_1630) suggested substrate import.
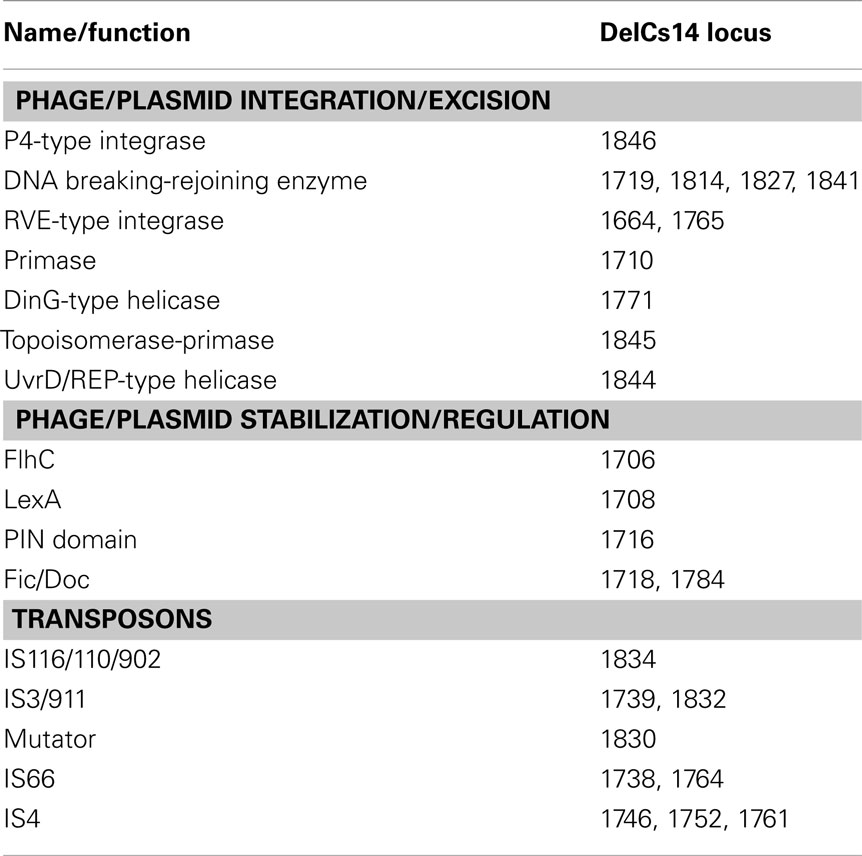
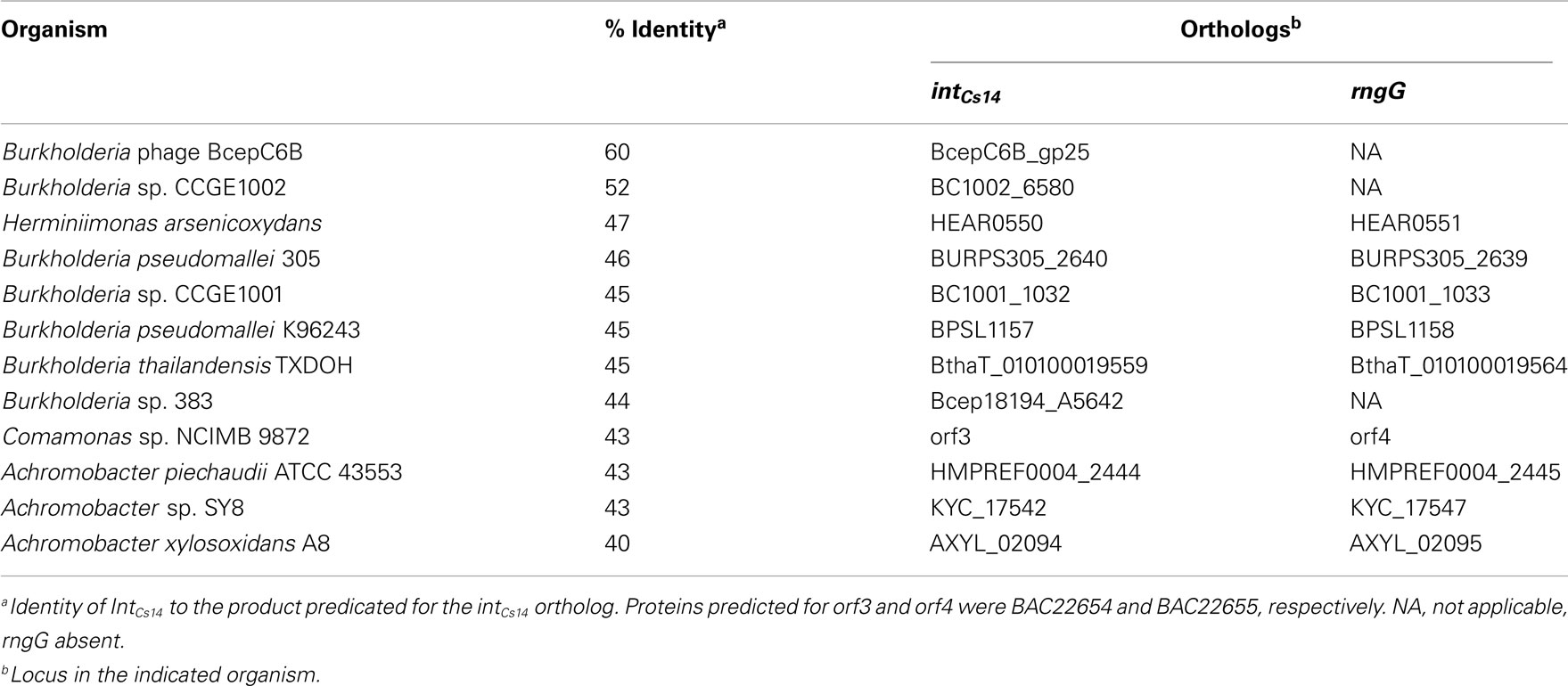
The phn island contained 25 genes predicted to be involved in some aspect of DNA mobilization (Table 2). Like other GEI, it possessed a bacteriophage P4-like integrase located near the site of insertion, which had the greatest similarity to an integrase from Burkholderia phage BcepC6B (Table 3). While additional close orthologs (≥60% homology; Bi et al., 2012) of IntCs14 were not identified, a number of low similarity hits were found in betaproteobacterial genera of the Burkholderiales, including the pathogens Burkholderia pseudomallei and Burkholderia thailandensis (Table 3). A feature common to most of these was integrase localization adjacent to genes predicted to encode RngG/CafA-orthologs (RNaseG; Table 3).
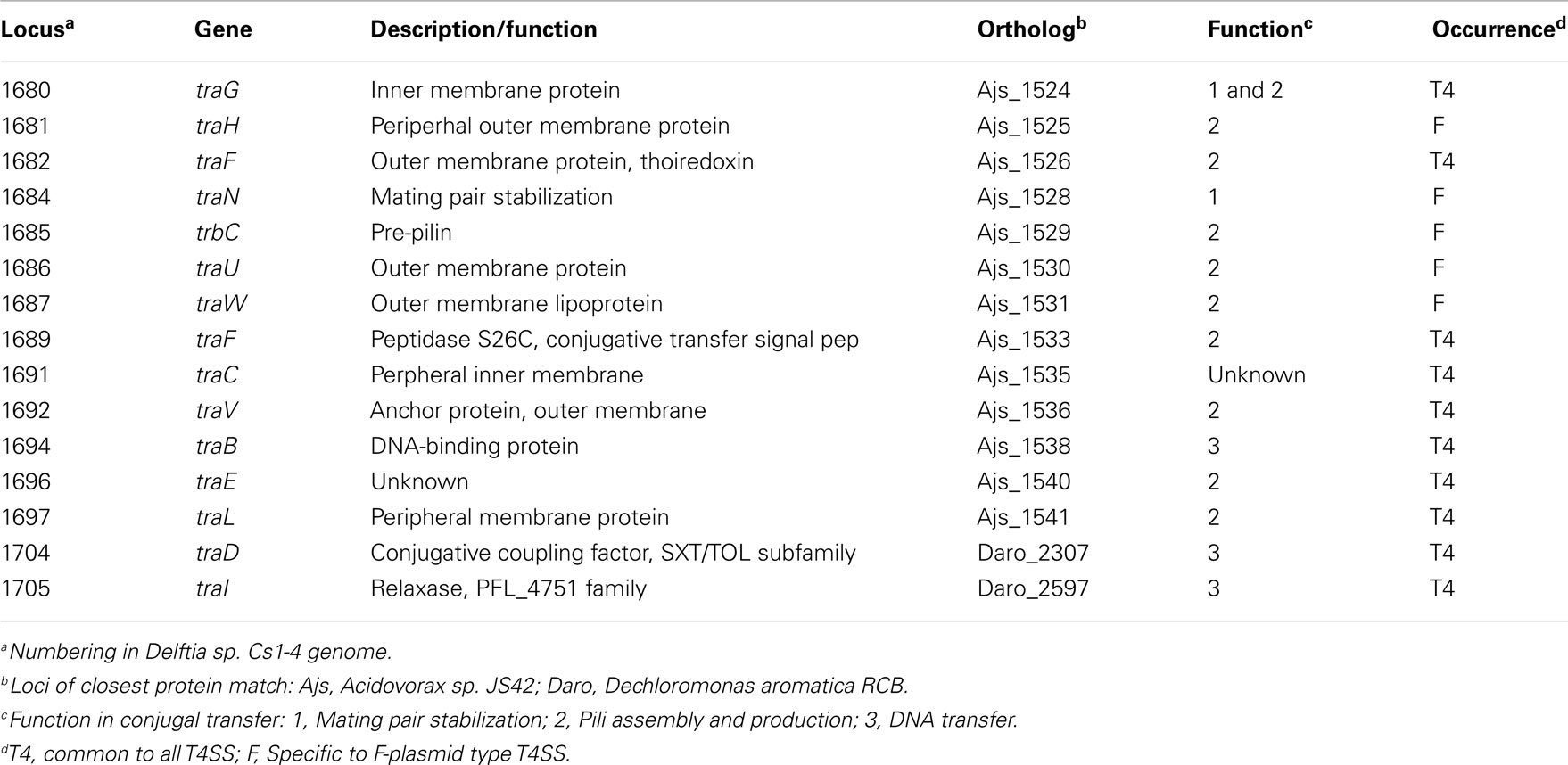
The type IV secretion system (T4SS) of the phn island was similar to that of the F-plasmid in overall arrangement of the tra functions, and possession of five tra genes specific to F-plasmid-like T4SS (Lawley et al., 2003; Table 4). But, the phn island T4SS lacked a number of the F-plasmid T4SS genes, and contained seven hypothetical genes the F-plasmid lacks. One gene notably absent from the phn island was traJ that, on the F-plasmid, controls T4SS gene transcription (Frost et al., 1994). The phn island TraI protein belonged to the PFL_4751 family of relaxases, which are required for transfer of SXT/R391 elements, as well as ICE of Pseudomonas fluorescens Pf-5 (Flannery et al., 2011).
Genes encoding other types of DNA integration/excision functions were dispersed throughout the island (Figure 2A; Table 2). There were 10 transposases, and those of the IS4- and IS66-type were closely associated with the phn cluster. Seven genes had predicted functions that could have roles in GEI stabilization or regulation of GEI excision. Potential regulators were a LexA-like protein and FlhC ortholog (Flagellar transcriptional activator). The LexA family includes bacterial prophage repressor proteins involved in the SOS response (Beaber et al., 2004) while FlhC (flagellar transcriptional activator) orthologs can function in global regulation of cellular processes. Both LexA and FlhC are part of a regulatory loop that is activated as part of the SOS response, which controls excision and transfer of SXT-type GEI (Beaber et al., 2002; Burrus and Waldor, 2003). Proteins that encoded potential toxin/antitoxin systems that could function in GEI stabilization included (Table 2): a protein with a PIN (PilT N terminus) domain, two Fic/Doc (Filamentation induced by cAMP/Death on curing)-like proteins and the OLD protein/UvrD pair (Overcome lysogeny defect/UV repair). The latter combination is commonly associated with putative GEI, and has been hypothesized to be either a toxin–antitoxin system (RNase toxin linked with an antitoxin), or a recombinase possibly involved in GEI integration (Khan et al, 2010). The phn island also contained all seven constituents of a “PRTRC” system, which is cataloged in TFAM as “a genetic system associated with mobile elements”2
Lastly, the lasI/luxR pair (Figure 2A) could direct the formation of N-acylhomoserine lactone (AHL) cell signaling molecules. The role of AHL as global regulators of many cellular functions is well established, and lasI/luxR orthologs have been identified on GEI in Burkholderia cepacia (Baldwin et al., 2004). However, in strain Cs1-4, deletion of lasI had no detectable effect on its growth or degradation of phenantherene (Chen and Hickey, 2011); thus, the function of a LasI-encoded AHL are unknown.
Comparative Proteomic Analysis
More than 600 proteins were identified in each of the proteomes of pyruvate- and phenanthrene-grown Delftia sp. Cs1-4, including 12 proteins of the phenantherene degradation upper pathway and 13 proteins of the lower pathway (Table 5). The relative abundance of PhnF was the greatest of all phenanthrene degradation pathway proteins detected, and ranked third overall in relative abundance in the phenanthrene proteome (Table S3 in Supplementary Material). In the pyruvate proteome, the same array of upper pathway proteins were identified, but at lower abundances. In contrast, no proteins of the lower pathway were detected in the pyruvate-grown cells. Based on comparative abundance of peptide scans in pyruvate vs. phenanthrene-grown cells, all phenanthrene degradation proteins detected exhibited induction during growth on phenanthrene, except perhaps PhnG (Table 5). Peptides were detected for all four of the new phenanthrene degradation genes described above (phnJKLM), and of these PhnJ ranked just behind PhnF in terms of overall abundance (Table S3 in Supplementary Material). Furthermore, all four proteins showed apparent induction by growth on phenanthrene. In the lower pathway, peptides matching both copies of OphA2 were identified, with those from locus DelCs14_1655 more abundant. Likewise, peptides matching gene products from both pmd clusters were detected, including both copies of pmdB,C,J,K. However, peptides matching the third copy of pmdAB (DelCs14_3008-09) were not detected.
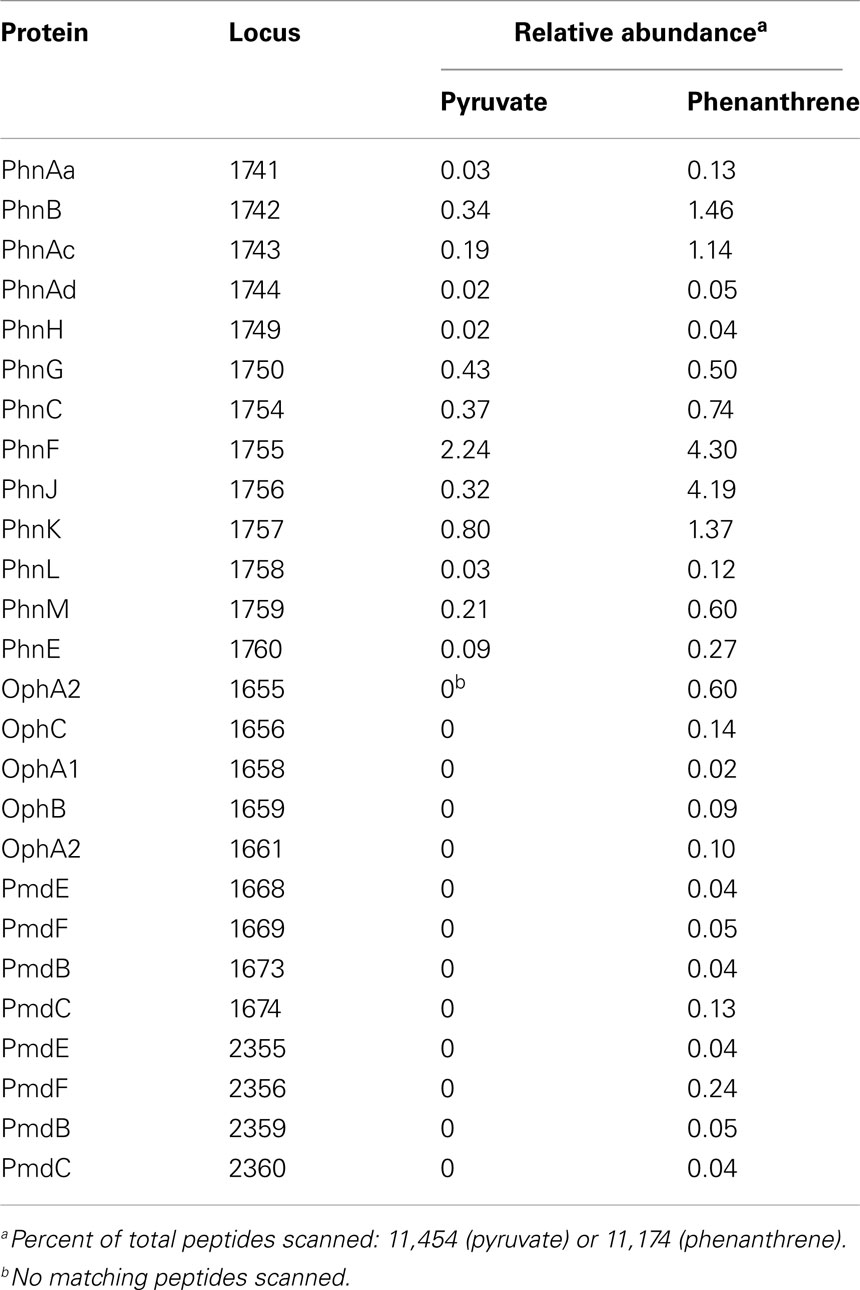
Table 5. Comparative proteomic analysis of pyruvate- and phenanthrene-grown cells of Delftia sp. Cs1-4.
Genetic Analyses of phnAa–d
The genes encoding the RHD large and small subunits, phnAcAd, were deleted by allelic exchange in strain Cs1-4, and the resultant ΔphnAcAd mutant was unable to grow on phenanthrene (Figure 7A). Complementation in trans with phnAcAd restored growth of the ΔphnAcAd mutant to levels similar to that of the wild type (Figure 7A). The growth impairment imposed upon the ΔphnAcAd mutant was specific to use of phenanthrene as a sole carbon source, as the mutant’s growth on pyruvate was indistinguishable from that of the wild type or complemented mutant (Figure 7B). The phnAa–d cluster was cloned from Burkholderia sp. Ch1-1, and expressed in E. coli; phnAa–d from Ch1-1 to determine if interruption by a transposon (see above) affected the ability of the cluster to produce a functional enzyme. Heterologous expression was successful, and E. coli harboring phnAa–d transformed all PAH tested (Figure 7C). No transformation was detected by the control E. coli BL21 (AI)(pET5a). Naphthalene and fluorene were most rapidly transformed, and the parent compounds were not detectable by HPLC after 1 h of incubation. For the remaining PAH tested, the apparent preference of the RHD decreased in the order of phenanthrene > anthracene > pyrene (Figure 7C). Notably, the RHD transformed ca. 50% of pyrene to pyrene dihydrodiols.
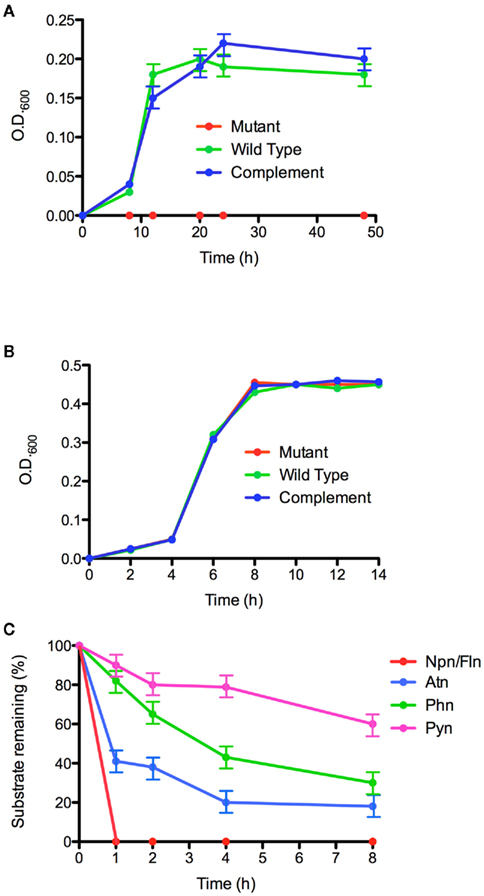
Figure 7. Functional analyses of the phn RHD genes. Growth on phenanthrene (A) and pyruvate (B) by Delftia sp. Cs1-4 wild type (wild type), ΔphnAcAd:Km mutant (mutant), and mutant complemented with phnAcAd (complement). (C) Heterologous expression in E. coli of phnAaAbAcAd and transformation of: naphthalene (Npn), fluorene (Fln), phenanthrene (Phn), anthracene (Atn), and pyrene (Pyn). Data points are the means of three biological replicates; error bars are the SD.
Discussion
The phn island represented a type of GEI that, to the best of our knowledge, has not been previously reported. Classes of GEI are defined by having an integrase homology of ≥60%, significant structural synteny and a common site of chromosomal integration (Bi et al., 2012). For the phn island, the only protein reaching the 60% homology threshold with IntCs14 was the integrase of the podophage BcepC68, an element with no additional similarities to the phn island. Thus, other GEI of the phn island class have not yet been revealed by genome sequencing projects.
The preferred site(s) of integration for phn island-like elements cannot be conclusively identified without orthologs of IntCs14 for comparison. But, one of the most common targets for GEI, tRNA genes, can probably be ruled out as the nearest such gene was ca. 60 kb downstream of the phn island. Integration of GEI at sites besides tRNA loci is most extensively studied with the SXT/R391-type elements, which target prfC (Peptide Chain Release Factor 3; Burrus and Waldor, 2003). Because tRNA genes and prfC encode essential functions, they are widely conserved and sequences embedded in these can serve as effective targets (attB sites) for homologous regions (attP site) in an ICE (Toleman and Walsh, 2011). It is unclear whether the fimbrial protein gene present in the chromosome at the 3′ end of the phn island had the broad conservation typical of ICE integration loci. However, the 57-bp sequence located in the 3′ terminus of the phn island and repeated in the adjacent chromosomal region strongly resembled an attP–attB pair, but was far larger than any such sequence recognition site yet reported. Further study is needed to elucidate the potential function of that 57-bp motif.
Processes governing GEI mobilization are key components of their biology and, while these are currently undetermined for the phn island, that element possessed genes known to mediate mobilization of SXT/R391-type GEI. Conjugal transfer of the SXT element occurs via a T4SS, and TraD (T4SS conjugal coupling factor) of the phn island was a protein of the SXT/TOL subfamily. Also, both the phn island and the SXT element possessed orthologs of FlhC and the lambda CI repressor (SetC and SetR, respectively in the SXT element), which are key (along with FlhD) in regulating SXT element’s excision and circularization (Burrus and Waldor, 2003). The phn island and SXT element differed in that the latter has an flhD ortholog (setD) adjoining setC, whereas the phn island did not. But, there was a second locus in strain Cs1-4 (DelCs14_4452, _4453) that could supply FlhD. With the SXT element, these regulatory proteins are part of a system that triggers its excision in response to DNA damage, which can result from exposure to UV light or reactive oxygen species (ROS). A similar system could operate with the phn island and ROS exposure might be particularly important, as ROS would be generated via activity of oxygenases the island encodes for phenanthrene catabolism, and/or catecholic metabolites that are produced during that process (Elstner, 1982; Dalton et al., 1999; Schweigert et al., 2001).
Genomic islands are emerging as key elements shaping the biodegradative capacity of bacterial communities. Other GEI that carry biodegradation functions are the (substrates degraded): 100 kb clc element (chlorocatechols, aminophenols; Gaillard et al., 2006), 90 kb bph–sal element (biphenyl; Nishi et al., 2000), 55 kb biphenyl catabolic transposon Tn4371 (biphenyl; Toussaint et al., 2003), 100 kb Tn3-like Alteromonas sp. SN2 transposon (naphthalene; Jin et al., 2011) and an unclassified GEI in P. naphthalenivorans CJ2 (naphthalene; Yagi et al., 2009). In terms of catabolic activity, all of these GEI (except for the clc element) were similar to the phn island in encoding both the “upper” and “lower” pathways for catabolism of a polyaromatic compound. However, the ca. 235 kb phn island was distinct from all of the above in encoding additional functions such as a potential link of catabolism to PHA/fatty acid biosynthesis, production of AHL signaling molecules and >50 hypothetical proteins. Thus, phn island-encoded functions likely extended to cellular processes beyond biodegradation. The phn island was also distinguished from other biodegradation GEI as it was the only element of this group with apparent similarity in its mobilization biology to that of SXT/R391-type GEI.
The present study provided the first insight into a potential mechanism of HGT for the phnAFK2 genotype. “Curing” experiments with A. faecalis AFK2 indicated possible association of the phenanthrene degradation phenotype with a ca. 43 kb plasmid (Kiyohara et al., 1990). But, which, if any, of the phn, oph, or pmd genes were carried by that plasmid was not reported. The divergence in G + C content of the phn cluster (ca. 50%) indicated a phylogenetic origin distinct from any host of the phn genes that is currently known. As noted above, the G + C content for Delftia sp. Cs1-4 was 66.7%, while that of A. faecalis AFK2 was 68% (Kiyohara et al., 1982) and 62.5% for Burkholderia sp. Ch1-13. The G + C content for Acidovorax sp. NA3 has not been reported, but in other Acidovorax genomes, it ranged from 64.9 to 66.8% (see text footnote 3). Thus, the phn cluster constituted a module mobilized independently of the phn island. Mechanisms for HGT of the phn cluster are unknown, but the bracketing by IS4- and IS66-type transposons may have implicated participation in HGT by those elements. Finally, although the phylogenetic origin of the phn cluster cannot be unequivocally determined, a betaproteobacterial source would be consistent with that of other phn island components (e.g., T4SS cluster, PRTRC cluster, etc.). Also, while the 50% G + C content of the phn cluster was divergent from the genomic G + C content of its known hosts, which were all of the order Burkholderiales, it would be in line with the median genomic G + C content for the betaproteobacteria as a whole (Lightfield et al., 2011).
Six other GEI were identified by BLAST searches that had a T4SS cluster with high homology to that of the phn island; three of these also contained a PRTRC region like that of the phn island. All of these GEI were located in betaproteobacteria of the Comamonadaceae (Acidovorax sp. JS42) or Rhodocyclaceae (Alicycliphilus denitrificans K601, Dechloromonas aromatica RCB), although none could be classed with the phn island because of low integrase homology with IntCs14 (≤23%). But, the GEI from strains JS42 and K601 were of the same class based on high integrase homology (96%), a common site of insertion (tRNAAla), and overall synteny.
Two significant findings regarding the phn cluster in strain Cs1-4 were the relocation of phnI, and the identification of four new genes, phnJKLM. The latter were not annotated in the GenBank record for strain AFK2, but were readily identified by ORF analysis of that sequence, and it is alignment with the phn region of strain Cs1-4. The degeneration of phnJLM in strain AFK2 suggested these genes were part of an ancestral phn module common to strains AFK2 and Cs1-4, but not utilized in the former. In contrast, while specific functions of phnJKLM in strain Cs1-4 were unknown, all exhibited apparent induction by growth on phenanthrene, suggesting some role in the utilization of that compound. Furthermore, since phnJKLM orthologs were associated with oxidoreductases for metabolism of a variety of hydrocarbons, their functions may be broadly applicable in hydrocarbon degradation, rather than specific to degradation of phenanthrene, or even PAH in general.
Segregation of phnI from the main body of the phn cluster could potentially have significant regulatory and metabolic implications. Singleton et al. (2009) demonstrated that in Acidovorax sp. AN3 phnAc, phnB, and phnC were co-transcribed and, since phnI was upstream of phnC, it too would be included in that transcript. But, in strain Cs1-4, such co-transcription of phnI is uncertain as ca. 8 kb and five genes separate it from the body of the phn cluster. In this case, phnI expression may be under separate transcriptional control, which could potentially affect the catabolic pathway. PhnI catalyzes the last step of the upper pathway (2-carboxybenzaldehyde conversion to o-phthalate), and a notable difference between strains Cs1-4 and Ch1-1 (in which phnI was included within the phn cluster) was that the former accumulated o-phthalate during growth on phenanthrene while the latter did not (Shetty, 2011). In strain Cs1-4, o-phthalate accumulation could reflect dis-synchronization of PhnI activity with that of the upstream enzymes in the phn catabolic pathway.
Growth on phenanthrene is known to induce phn expression (Singleton et al., 2009), but regulatory mechanisms controlling transcription of the phn genes are unknown. Conservation of the marR-like element within the phn cluster may have indicated it served a regulatory function in phenanthrene degradation that is common to all of the phn genotype bacteria. The marR elements are generally responsive to aromatic compounds in general (Alekshun and Levy, 1999), and in P. naphthalenivorans CJ2, regulates expression of naphthalene degradation genes (Jeon et al., 2006). Also, marR genes can be up-regulated by exposure to naphthenic acids (Zhang et al., 2011), chemicals that have some resemblance to metabolites produced during phenanthrene metabolism. Thus, a possible role for marR is regulation of the phn genes via interaction with one or more phenanthrene metabolites.
The oph and pmd clusters each presented interesting types of apparent redundancy. The multiple pmd clusters were similar to that in other bacteria, such as the multiple phenol degradation clusters (mhp genes) in Dechloromonas aromatica (Salinero et al., 2009). The physiological significance of such redundancy is as yet unknown. But, for Delftia sp. Cs1-4, proteomics data presented here proved both pmd paralogs were expressed, and apparently induced by growth on phenanthrene. A search of all currently available genomes possessing genes annotated as “phthalate 4,5-dioxygenase” yielded no other examples of an oph operon with two copies of ophA2. Two nearly adjacent copies of phthalate 4,5-dioxygenase oxygenase were located in the actinobacterium Pseudonocardia dioxanivorans CB1190 (Psed_3921, Psed_3923; NC_015312.1), but these were not associated with any other oph genes. Thus, the structure of the oph cluster in strain Cs1-4 is likely the fist example of its kind. Phthalate dioxygenase is composed of a reductase, OphA1, and a single alpha subunit, OphA2 (Batie et al., 1987; Kweon et al., 2008), and thus the roles of two non-identical copies of OphA2 is unknown. Proteomics data established expression of both copies, but the within-cluster copy (DelCs14_1655) appeared more abundant from spectral scans. We speculate that the second copy of OphA2 may be optimized for a substrate other than o-phthalate.
Heterologous expression and targeted mutagenesis established phnAa–d as encoding the RHD responsible for initiating metabolism of phenanthrene. As yet, evidence for this activity has been indirect, such as monitoring transcript production (Singleton et al., 2009). Heterologous expression demonstrated that, like other PAH RHD, the phnAa–d enzyme transformed a number of PAH including a four-ring compound, pyrene. However, a notable difference between the phn RHD and the nah-like RHD was that organisms possessing the latter typically grow on many of the PAH that are substrates for the nah RHD. But, phenanthrene is the only PAH that supports growth of all currently cultured bacteria with the phnAFK2 genotype (Acidovorax sp. NA3, Delftia sp. Cs1-4, and Burkholderia sp. Ch1-1).
In conclusion, the present analyses of the phn island have added new dimensions to our knowledge of PAH biodegradation, mechanisms of HGT that shape microbial communities and the nature of GEI in general. This study has provided starting points for investigations into new biodegradative functions, such as the roles of PhnJKLM or the two copies of OphA2, as well as identification of molecular mechanisms mediating phn island mobilization. The acquisition of complete genome sequences for additional bacteria of the phnAFK2 genotype and/or possessing close orthologs of IntCs14 would greatly facilitate future studies on the structure and function of phn island-type GEI.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
These studies were supported by funding (to William J. Hickey) from the Univ. of Wisconsin–Madison, College of Agricultural and Life Sciences (Hatch–McIntire-Stennis), National Science Foundation (MCB0920664), and the O.N. Allen Professorship in Soil Microbiology. Sequencing, assembly, and computational annotation of the Delftia sp. Cs1-4 and Burkholderia sp. Ch1-1 genomes was done by the U.S. Department of Energy, Joint Genome Institute, through the Community Sequencing Project (CSP795673 to William J. Hickey). The work conducted by the U.S. Department of Energy, Joint Genome Institute was supported by the U.S. Department of Energy, Office of Science under contract No. DE-AC02-05CH11231.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Microbiotechnology,_Ecotoxicology_and_Bioremediation/10.3389/fmicb.2012.00125/abstract
Table S1. Bacterial strains and plasmids used in this study.
Table S2. PCR Primers used in this study.
Table S3. Proteomes of Delftia sp. Cs1-4 grown on the indicated carbon source.
Footnotes
Abbreviations
AHL, N-acylhomoserine lactone; CoA, coenzyme-A; GEI, genomic island; HGT, horizontal gene transfer; ICE, integrating conjugative elements; OAA, oxaloacetic acid; oph, ortho-phthalate degradation genes; PAH, polycyclic aromatic hydrocarbon; PCA, protocatechuate; PHA, polyhydroxyalkanoate; phn, phenanthrene degradation genes; pmd, protocatechuate meta-dioxygenase and other protocatechuate degradation genes; RHD, ring-hydroxylating dioxygenase; ROS, reactive oxygen species; T4SS, type IV secretion system.
References
Alekshun, M. N., and Levy, S. B. (1999). The Mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7, 410–413.
Baldwin, A., Sokol, P. A., Parkhill, J., and Mahenthiralingam, E. (2004). The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 80, 1537–1547.
Basu, A., and Phale, P. S. (2008). Conjugative transfer of preferential utilization of aromatic compounds from Pseudomonas putida CSV86. Biodegradation 19, 83–92.
Batie, C. J., Lahaie, E., and Ballou, D. P. (1987). Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J. Biol. Chem. 262, 1510–1518.
Beaber, J. W., Hochhut, B., and Waldor, M. K. (2002). Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184, 4259–4269.
Beaber, J. W., Hochhut, B., and Waldor, M. K. (2004). SOS Response promotes horizontal dissemination of antibiotic resistance genes. Nature 427, 72–74.
Bi, D. X., Xu, Z., Harrison, E. M., Tai, C., Wei, Y. Q., He, X. Y., Jia, S. R., Deng, Z. X., Rajakumar, K., and Ou, H. Y. (2012). ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res. 40, D621–D626.
Burrus, V., and Waldor, M. K. (2003). Control of SXT integration and excision. J. Bacteriol. 185, 5045–5054.
Chen, S., and Hickey, W. J. (2011). Development of tools for genetic analysis of phenanthrene degradation and nanopod production by Delftia sp. Cs1-4. Front. Microbiol. 2:187. doi:10.3389/fmicb.2011.00187
Dalton, T. P., Shertzer, H. G., and Puga, A. (1999). Regulation of gene expression by reactive oxygen. Annu. Rev. Pharmacol. Toxicol. 39, 67–101.
Demaneche, S., Meyer, C., Micoud, J., Louwagie, M., Willison, J. C., and Jouanneau, Y. (2004). Identification and functional analysis of two aromatic-ring-hydroxylating dioxygenases from a Sphingomonas strain that degrades various polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70, 6714–6725.
Dennis, J. J., and Zylstra, G. J. (2004). Complete sequence and genetic organization of pDTG1, the 83 kilobase naphthalene degradation plasmid from Pseudomonas putida strain NCIB 9816-4. J. Mol. Biol. 341, 753–768.
Ding, G. C., Heuer, H., Zuhlke, S., Spiteller, M., Pronk, G. J., Heister, K., Kogel-Knabner, I., and Smalla, K. (2010). Soil type-dependent responses to phenanthrene as revealed by determining the diversity and abundance of polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenase genes by using a novel PCR detection system. Appl. Environ. Microbiol. 76, 4765–4771.
Dobrindt, U., Hochhut, B., Hentschel, U., and Hacker, J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424.
Fiedler, S., Steinbüchel, A., and Rehm, B. H. A. (2002). The role of the fatty acid beta-oxidation multienzyme complex from Pseudomonas oleovorans in polyhydroxyalkanoate biosynthesis: molecular characterization of the fadBA operon from P. oleovorans and of the enoyl-CoA hydratase genes phaJ from P. oleovorans and Pseudomonas putida. Arch. Microbiol. 178, 149–160.
Flannery, E. L., Antczak, S. M., and Mobley, H. L. T. (2011). Self-transmissibility of the integrative and conjugative element ICEPm1 between clinical isolates requires a functional integrase, relaxase, and type IV secretion system. J. Bacteriol. 194, 4104–4112.
Frost, L. S., Ippen-Ihler, K., and Skurray, R. A. (1994). Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58, 162–210.
Gaillard, M., Vallaeys, T., Vorholter, F. J., Minoia, M., Werlen, C., Sentchilo, V., Puhler, A., and Van Der Meer, J. R. (2006). The clc element of Pseudomonas sp strain B13, a genomic island with various catabolic properties. J. Bacteriol. 188, 1999–2013.
Heinaru, E., Vedler, E., Jutkina, J., Aava, M., and Heinaru, A. (2009). Conjugal transfer and mobilization capacity of the completely sequenced naphthalene plasmid pNAH20 from multiplasmid strain Pseudomonas fluorescens PC20. FEMS Microbiol. Ecol. 70, 563–574.
Hickey, W. J., and Focht, D. D. (1990). Degradation of mono-, di-, and tri-halogenated benzoates by Pseudomonas aeruginosa strain JB2. Appl. Environ. Microbiol. 56, 3842–3850.
Jeon, C. O., Park, M., Ro, H.-S., Park, W., and Madsen, E. L. (2006). The naphthalene catabolic (nag) genes of Polaromonas naphthalenivorans CJ2: evolutionary implications for two gene clusters and novel regulatory control. Appl. Environ. Microbiol. 72, 1086–1095.
Jin, H. M., Jeong, H., Moon, E. J., Math, R. K., Lee, K., Kim, H. J., Jeon, C. O., Oh, T. K., and Kim, J. F. (2011). Complete genome sequence of the polycyclic aromatic hydrocarbon-degrading bacterium Alteromonas sp. strain SN2. J. Bacteriol. 193, 4292–4293.
Kang, H., Hwang, S. Y., Kim, Y. M., Kim, E., Kim, Y. S., Kim, S. K., Kim, S. W., Cerniglia, C. E., Shuttleworth, K. L., and Zylstra, G. J. (2003). Degradation of phenanthrene and naphthalene by a Burkholderia species strain. Can. J. Microbiol. 49, 139–144.
Keller, A., Nesvizhskii, A. I., Kolker, E., and Aebersold, R. (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392.
Khan, F., Furuta, Y., Kawai, M., Kaminska, K. H., Ishikawa, K., Bujnicki, J. M., and Kobayashi, I. (2010). A putative mobile genetic element carrying a novel type IIF restriction-modification system (PluTI). Nucleic Acids Res. 38, 3019–3030.
Kiyohara, H., Nagao, K., Kouno, K., and Yano, K. (1982). Phenanthrene-degrading phenotype of Alcaligenes faecalis AFK2. Appl. Environ. Microbiol. 43, 458–461.
Kiyohara, H., Takizawa, N., Date, H., Torigoe, S., and Yano, K. (1990). Characterization of a phenanthrene degradation plasmid from Alcaligenes faecalis AFK2. J. Ferment. Bioeng. 69, 54–56.
Komenda, J., Nickelsen, J., Tichy, M., Prasil, O., Eichacker, L. A., and Nixon, P. J. (2008). The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp PCC 6803. J. Biol. Chem. 283, 22390–22399.
Kweon, O., Kim, S.-J., Baek, S., Chae, J.-C., Adjei, M. D., Baek, D. H., Kim, Y. C., and Cerniglia, C. E. (2008). A new classification system for bacterial Rieske non-heme iron aromatic ring-hydroxylating oxygenases. BMC Biochem. 9, 11. doi:10.1186/1471-2091-9-11
Lawley, T. D., Klimke, W. A., Gubbins, M. J., and Frost, L. S. (2003). F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224, 1–15.
Li, W., Shi, J. D., Wang, X. G., Han, Y. N., Tong, W., Ma, L., Liu, B., and Cai, B. L. (2004). Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp strain ND6. Gene 336, 231–240.
Lightfield, J., Fram, N. R., and Ely, B. (2011). Across bacterial phyla, distantly-related genomes with similar genomic GC content have similar patterns of amino acid usage. PLoS ONE 6, e17677. doi:10.1371/journal.pone.0017677
Lohkamp, B., Bäuerle, B., Rieger, P.-G., and Schneider, G. (2006). Three-dimensional structure of iminodisuccinate epimerase defines the fold of the MmgE/PrpD Protein. J. Mol. Biol. 362, 555–566.
Lozada, M., Mercadal, J. P. R., Guerrero, L. D., Di Marzio, W. D., Ferrero, M. A., and Dionisi, H. M. (2008). Novel aromatic ring-hydroxylating dioxygenase genes from coastal marine sediments of Patagonia. BMC Microbiol. 8, 50. doi:10.1186/1471-2180-8-50
Madison, L. L., and Huisman, G. W. (1999). Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63, 21–53.
Mario, J., Van Der Meer, J. R., Gaillard, M., Harding, R. M., Hood, D. W., and Crook, D. W. (2009). Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33, 376–393.
Martin, S., Shabanowitz, J., Hunt, D., and Marto, J. (2000). Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 72, 4266–4274.
Moser, R., and Stahl, U. (2001). Insights into the genetic diversity of initial dioxygenases from PAH-degrading bacteria. Appl. Microbiol. Biotechnol. 55, 609–618.
Nesvizhskii, A. I., Keller, A., Kolker, E., and Aebersold, R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658.
Nishi, A., Tominaga, K., and Furukawa, K. (2000). A 90-kilobase conjugative chromosomal element coding for biphenyl and salicylate catabolism in Pseudomonas putida KF715. J. Bacteriol. 182, 1949–1955.
Park, S. J., and Lee, S. Y. (2003). Identification and characterization of a new enoyl coenzyme a hydratase involved in biosynthesis of medium-chain-length polyhydroxyalkanoates in recombinant Escherichia coli. J. Bacteriol. 185, 5391–5397.
Ren, Q., Sierro, N., Witholt, B., and Kessler, B. (2000). FabG, an NADPH-dependent 3-ketoacyl reductase of Pseudomonas aeruginosa, provides precursors for medium-chain-length poly-3-hydroxyalkanoate biosynthesis in Escherichia coli. J. Bacteriol. 182, 2978–2981.
Rengstl, B., Oster, U., Stengel, A., and Nickelsen, J. (2011). An intermediate membrane subfraction in cyanobacteria is involved in an assembly network for photosystem II biogenesis. J. Biol. Chem. 286, 21944–21951.
Romine, M. F., Stillwell, L. C., Wong, K. K., Thurston, S. J., Sisk, E. C., Sensen, C., Gaasterland, T., Fredrickson, J. K., and Saffer, J. D. (1999). Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181, 1585–1602.
Salinero, K. K., Keller, K., Feil, W. S., Feil, H., Trong, S., Di Bartolo, G., and Lapidus, A. (2009). Metabolic analysis of the soil microbe Dechloromonas aromatica str. RCB: indications of a surprisingly complex life-style and cryptic anaerobic pathways for aromatic degradation. BMC Genomics 10, 351. doi:10.1186/1471-2164-10-351
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Schweigert, N., Zehnder, A. J. B., and Eggen, R. I. L. (2001). Chemical properties of catechols and their molecular modes of toxic action in cells, from microorganisms to mammals. Environ. Microbiol. 3, 81–91.
Shetty, A. R. (2011). Characterization of Nanopods Produced by Delftia sp. Cs1-4. Madison: University of Wisconsin.
Singleton, D. R., Ramirez, L. G., and Aitken, M. D. (2009). Characterization of a polycyclic aromatic hydrocarbon degradation gene cluster in a phenanthrene-degrading Acidovorax strain. Appl. Environ. Microbiol. 75, 2613–2620.
Stolz, A. (2009). Molecular characteristics of xenobiotic-degrading sphingomonads. Appl. Microbiol. Biotechnol. 81, 793–811.
Suzuki, H., Yano, H., Brown, C. J., and Top, E. M. (2010). Predicting plasmid promiscuity based on genomic signature. J. Bacteriol. 192, 6045–6055.
Toleman, M. A., and Walsh, T. R. (2011). Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol. Rev. 35, 912–935.
Toussaint, A., Merlin, C., Monchy, S., Benotmane, M. A., Leplae, R., Mergeay, M., and Springael, D. (2003). The biphenyl- and 4-chlorobiphenyl-catabolic transposon Tn4371, a member of a new family of genomic islands related to incP and Ti plasmids. Appl. Environ. Microbiol. 69, 4837–4845.
Tsuge, T., Taguchi, K., Taguchi, S., and Doi, Y. (2003). Molecular characterization and properties of (R)-specific enoyl-CoA hydratases from Pseudomonas aeruginosa: metabolic tools for synthesis of polyhydroxyalkanoates via fatty acid β-oxidation. Int. J. Biol. Macromol. 31, 195–205.
Vacca, D. J., Bleam, W. F., and Hickey, W. J. (2005). Isolation of soil bacteria adapted to degrade humic acid-sorbed phenanthrene. Appl. Environ. Microbiol. 71, 3797–3805.
van der Meer, J. R., and Sentchilo, V. (2003). Genomic islands and the evolution of catabolic pathways in bacteria. Curr. Opin. Biotechnol. 14, 248–254.
Williamson, J. M., and Brown, G. M. (1979). Purification and properties of L-Aspartate-alpha-decarboxylase, an enzyme that catalyzes the formation of beta-alanine in Escherichia coli. J. Biol. Chem. 254, 8074–8082.
Witholt, B., and Kessler, B. (1999). Perspectives of medium chain length poly(hydroxyalkanoates), a versatile set of bacterial bioplastics. Curr. Opin. Biotechnol. 10, 279–285.
Yagi, J. M., Sims, D., Brettin, T., Bruce, D., and Madsen, E. L. (2009). The genome of Polaromonas naphthalenivorans strain CJ2, isolated from coal tar-contaminated sediment, reveals physiological and metabolic versatility and evolution through extensive horizontal gene transfer. Environ. Microbiol. 11, 2253–2270.
Keywords: polynuclear aromatic hydrocarbons, phenanthrene, biodegradation, horizontal gene transfer, genomic island, Delftia, bioremediation, genomics
Citation: Hickey WJ, Chen S and Zhao J (2012) The phn island: a new genomic island encoding catabolism of polynuclear aromatic hydrocarbons. Front. Microbio. 3:125. doi: 10.3389/fmicb.2012.00125
Received: 08 February 2012; Accepted: 16 March 2012;
Published online: 04 April 2012.
Edited by:
Jeremy Semrau, The University of Michigan, USAReviewed by:
Carl James Yeoman, University of Illinois at Champaign–Urbana, USAChristopher L. Hemme, University of Oklahoma, USA
Copyright: © 2012 Hickey, Chen and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: William J. Hickey, Department of Soil Science, O.N. Allen Laboratory for Soil Microbiology, University of Wisconsin, 1525 Observatory Drive, Madison, WI 53706, USA. e-mail: wjhickey@wisc.edu