- 1 Department of Chemistry, University of Warwick, Coventry, UK
- 2 School of Life Sciences, University of Warwick, Coventry, UK
- 3 Department of Biochemistry, Henry Wellcome Building, Leicester, UK
Zinc is a recognized essential element for the majority of organisms, and is indispensable for the correct function of hundreds of enzymes and thousands of regulatory proteins. In aquatic photoautotrophs including cyanobacteria, zinc is thought to be required for carbonic anhydrase and alkaline phosphatase, although there is evidence that at least some carbonic anhydrases can be cambialistic, i.e., are able to acquire in vivo and function with different metal cofactors such as Co2+ and Cd2+. Given the global importance of marine phytoplankton, zinc availability in the oceans is likely to have an impact on both carbon and phosphorus cycles. Zinc concentrations in seawater vary over several orders of magnitude, and in the open oceans adopt a nutrient-like profile. Most studies on zinc handling by cyanobacteria have focused on freshwater strains and zinc toxicity; much less information is available on marine strains and zinc limitation. Several systems for zinc homeostasis have been characterized in the freshwater species Synechococcus sp. PCC 7942 and Synechocystis sp. PCC 6803, but little is known about zinc requirements or zinc handling by marine species. Comparative metallo-genomics has begun to explore not only the putative zinc proteome, but also specific protein families predicted to have an involvement in zinc homeostasis, including sensors for excess and limitation (SmtB and its homologs as well as Zur), uptake systems (ZnuABC), putative intracellular zinc chaperones (COG0523) and metallothioneins (BmtA), and efflux pumps (ZiaA and its homologs).
Introduction
Zinc might be considered as one of the most inconspicuous trace elements. To some extent, this is due to its “boring” (Levi, 1984) chemistry – in its only biologically relevant oxidation state, Zn(II), it is colorless and does not display any redox chemistry of its own. Because Zn2+ is not redox-active, many authors tend to consider it as less important than iron or copper in terms of both essentiality and toxicity, even though between 5 and 9% of the predicted proteomes of most organisms correspond to zinc-requiring proteins – in most cases more than either the predicted iron or copper sub-proteomes (Andreini et al., 2009).
This is even true for prokaryotes which once were thought to “avoid the hidden cost of zinc homeostasis” (Luisi, 1992). Several recent bioinformatic approaches aimed at predicting metalloproteomes (Andreini et al., 2006, 2008; Dupont et al., 2010) found that although overall zinc utilization in bacteria is undoubtedly lower than in eukaryotes, Zn-binding domains are yet highly abundant in predicted bacterial proteomes. For the case of currently existing prokaryotes, there is clear evidence for widespread zinc utilization, in particular in hydrolytic enzymes (Decaria et al., 2010). A major reason for lower zinc utilization by bacteria is likely the much lower abundance of zinc finger domains in their proteomes. Furthermore, at least in heterotrophs, the cellular quotas for zinc and iron tend to be similar (Outten and O’Halloran, 2001), although it should be emphasized that metal quotas do not necessarily bear a direct relationship to metal requirements.
Virtually all organisms have elaborate mechanisms to control zinc levels and distribution (Hantke, 2005; Eide, 2006; Fukada and Kambe, 2011). Total cellular concentrations typically are in the high micromolar range; however, various lines of evidence have indicated that “free” cytosolic zinc concentrations are extremely low. Values given in the literature vary between nanomolar and femtomolar – with the true regulated value probably somewhere in the picomolar range (Krezel and Maret, 2006). The apparent need for the narrow range of tolerable zinc concentrations had initially puzzled some researchers, as there seems to be a widespread belief that zinc is not particularly toxic to cells. It could be argued that this is only true for cells that have efficient mechanisms to deal with zinc; otherwise free zinc concentrations as low as nanomolar can be toxic (Bozym et al., 2010). Deleterious effects of Zn2+ may, at least to some extent, be due to its high position in the Irving–Williams series (Irving and Williams, 1953), meaning that it outcompetes less competitive metal ions such as Fe2+ and Mn2+ for their protein binding sites, as demonstrated recently for the Mn-binding protein MncA (Tottey et al., 2008). This latter study illustrated why it is important to limit the amount of exchangeable zinc in the cytosol of cells, by the demonstration that the major periplasmic Mn-binding protein MncA of Synechocystis sp. PCC 6803 (a freshwater cyanobacterium) can only incorporate the essential cofactor Mn2+ to a significant extent if this is present in 100000-fold molar excess over Zn2+. Since MncA folds and is loaded with Mn2+ in the cytosol, this finding implies the need for a very low free cytosolic zinc concentration in Synechocystis sp. PCC 6803. Little information is available on zinc toxicity to marine cyanobacteria. A study on Synechococcus strains in the strait of Gibraltar noted that even micromolar concentrations of zinc had only a moderate effect on growth (Debelius et al., 2011), but zinc sensitivity may differ considerably depending on the natural habitat of a given cyanobacterium.
There are indications for an impact of zinc on major global biogeochemical cycles. A “zinc hypothesis” was put forward in 1994 in a study that demonstrated zinc and carbon co-limitation in marine phytoplankton (Morel et al., 1994). A link between zinc and carbon fixation is also reflected in the arctic ice-core record (Hong et al., 1996): during periods of glaciation, zinc levels were at least 10 times higher, whilst CO2 levels were significantly lower than in the intervening periods. Changes in zinc levels due to increased deposition of dust into the oceans are thought to have had an effect on marine microbial community structure, and the decrease in CO2 levels could be attributed to increased CaCO3 production by coccolithophores and a resulting decrease in atmospheric CO2 (Schulz et al., 2004). The amount of data on geochemical zinc fluxes is limited, and it is not clear whether dust deposition today does (Thuroczy et al., 2010) or does not (Bruland et al., 1994) significantly contribute. In terms of a direct biochemical link between zinc and organic CO2 fixation, there are of course the carbonic anhydrases, which operate in all marine phytoplankton, including cyanobacteria (Cannon et al., 2010), although substitution with either Co or Cd has been demonstrated for eukaryotic phytoplankton (Xu et al., 2008). There are also strong indications for links between Zn and phosphorus cycles (Jakuba et al., 2008), thought to be due to the requirement of Zn for alkaline phosphatase.
An absolute requirement for zinc has been clearly demonstrated for marine eukaryotic phytoplankton (Sunda and Huntsman, 1995, 2005), but the situation is less clear for marine cyanobacteria, as discussed below. With this review, we aim to make a case for intensifying studies into the relevance of zinc for marine cyanobacteria.
Marine cyanobacteria
Cyanobacteria are a group of phototrophic prokaryotes that all have the ability to perform oxygenic photosynthesis. In the marine environment a large diversity of both unicellular (e.g., Synechococcus, Prochlorococcus, Cyanobium, Acaryochloris, and Crocosphaera) and filamentous (e.g., Trichodesmium, Lyngbya, Oscillatoria, Nodularia, and Microcoleus) genera exist, occupying habitats ranging from intertidal microbial mats through to oligotrophic open-ocean waters (see Whitton and Potts, 2000).
The numerically dominant cyanobacteria in open-ocean waters are the unicellular genera Synechococcus and Prochlorococcus which contribute significantly to marine CO2 fixation (Li, 1994; Jardillier et al., 2010). Synechococcus are the more widely distributed, being found in waters covering a broad temperature range, from ca. 2–3°C to >30°C (Shapiro and Haugen, 1988; Waterbury et al., 1996; Fuller et al., 2006; Zwirglmaier et al., 2008), and including open-ocean, coastal and estuarine environments (Partensky et al., 1999; Scanlan, 2003). Prochlorococcus appears to be more constrained in its distribution occupying waters roughly between 45°N and 40°S but within these latitudes it is extremely abundant, routinely reaching concentrations of 105 cells per ml or higher (Partensky et al., 1999; Partensky and Garczarek, 2010). Prochlorococcus can be distinguished from Synechococcus by its lack of a phycobilisome light-harvesting antenna complex. Instead, it possesses thylakoid membrane proteins binding unique divinyl derivatives of chlorophyll a and b (Goericke and Repeta, 1992; Partensky and Garczarek, 2003). In stratified tropical and subtropical waters Prochlorococcus cells undergo vertical partitioning between distinct high light- and low light-adapted ecotypes (Moore et al., 1998; West and Scanlan, 1999). Both Synechococcus and Prochlorococcus have relatively small genomes ranging in size between 1.64 and 2.7 Mb in Prochlorococcus and from 2.2 to 2.86 Mb in Synechococcus (Kettler et al., 2007; Dufresne et al., 2008; Scanlan et al., 2009). In the case of Prochlorococcus, significant genome reduction has occurred during evolution of the genus, likely an adaptation to the oligotrophic gyre systems they inhabit, providing significant economies in energy and nutrients (Dufresne et al., 2005).
As well as contributing to marine carbon cycling, some cyanobacteria are also capable of nitrogen fixation (Zehr, 2011). Trichodesmium, a filamentous non-heterocystous genus, is ubiquitous in tropical and subtropical environments (Capone et al., 1997) and until recently was thought to be the dominant marine nitrogen-fixer. However, it is now clear that the unicellular UCYN-A and UCYN-B lineages, the latter encompassing the genera Crocosphaera and Cyanothece, also contribute significantly to this process (Zehr, 2011). Surprisingly, despite its global distribution in the Atlantic and Pacific Oceans Crocosphaera appears to have limited genetic diversity with high identity and synteny of the cultured genome sequence to environmental metagenomic datasets for this genus (Zehr et al., 2007). Interestingly, metabolic insights from the genome of the UCYN-A lineage reveals a cyanobacterium lacking photosystem II, RuBisCO, and a tricarboxylic acid cycle (Tripp et al., 2010) suggesting it requires a symbiotic partner.
The high diversity of marine cyanobacteria is epitomized by Acaryochloris marina, a cyanobacterium that uniquely utilizes chlorophyll d as its main photosynthetic pigment (Kuhl et al., 2005) trapping the far-red light that penetrates beneath the didemnid ascidians (sea squirts) upon which these organisms are found (Ohkubo et al., 2006). Curiously, the A. marina genome is considerably larger than other sequenced unicellular strains (Swingley et al., 2008) comprising a circular chromosome of 6.5 Mb and nine distinct plasmids giving a total DNA content of 8.3 Mb. Over 10% of the protein families contain duplicated copies in A. marina and this information, together with its utilization of far-red light that is not absorbed by other aerobic photoautotrophs, suggests that Acaryochloris species fill a non-competitive niche where they are apparently free to specialize their metabolic library, and potentially explains their expansive genome size (Swingley et al., 2008).
Biological and chemical co-evolution, and cyanobacterial metal requirements
It has been hypothesized that metal ion bioavailability presented an evolutionary selection pressure on the “choice” of metals within metalloenzymes (Williams and Da Silva, 2003). Conversely, biological evolution and the emergence of life has changed the chemical composition, or more precisely, the speciation of the atmosphere, the lithosphere, and of course the hydrosphere. Arguably, cyanobacteria might be deemed responsible for the greatest change of all by inventing oxygenic photosynthesis (Raymond and Blankenship, 2004). Consequently, they were amongst the first organisms that encountered, and had to cope with, the changes in the chemical composition of their environment that oxygenation brought about (Cavet et al., 2003; Saito et al., 2003). For metal ion speciation, both fundamental considerations (Williams and Da Silva, 2001) as well as detailed studies (Saito et al., 2003) agree that the presence of oxygen meant a drastic reduction in iron, cobalt, nickel, and manganese availability, and a significant increase in the concentrations of zinc, copper, and cadmium (Williams and Da Silva, 2001; Williams, 2011). It is reasonable to accept that these changes in chemistry and bioavailability directed biological evolution, including that of metal-binding biomolecules (Williams and Da Silva, 2000). Indeed, even though it has been suggested that microbial metalloproteomes are still largely uncharacterized (Cvetkovic et al., 2010), bioinformatic genome analyzes of known metal-binding protein domains (Dupont et al., 2010) as well as elemental analysis experiments on marine phytoplankton (Bertilsson et al., 2003; Heldal et al., 2003; Ho et al., 2003; Quigg et al., 2003, 2011; Morel, 2008) give a picture that is consistent with this idea. Thus, the “co-evolution of biology and chemistry” is imprinted on both the metallome and the metalloproteome. The interested reader is directed to a recent debate regarding the evolution of zinc-binding domains (Mulkidjanian and Galperin, 2009; Dupont and Caetano-Anolles, 2010).
In the case of cyanobacteria, metal ion requirements and sensitivities, as far as they have been experimentally determined, agree with the notion that they evolved in an environment with metal ion concentrations typical of a sulfidic or a ferrous ocean (Saito et al., 2003): both marine Synechococcus (Sunda and Huntsman, 1995) and Prochlorococcus (Saito et al., 2002) strains have been shown to be cobalt-limited, whereas the requirements for zinc are so low (Saito et al., 2003) that only mild reductions in growth rates were observed at the lowest possible free zinc concentrations (Saito et al., 2002).
Our previous genome-mining approaches have identified strong candidate genes for potentially zinc-requiring carboxysomal carbonic anhydrases, ABC-type zinc uptake systems, as well as for proteins involved in the intracellular handling of zinc (Blindauer, 2008b). Several other enzymes in cyanobacteria are also predicted to require zinc for function, including for example DNA ligase and alkaline phosphatase (Palenik et al., 2003), the latter leading to the suggestion that cyanobacteria may be Zn–P co-limited. Indeed, in certain cyanobacterial strains, alkaline phosphatase activity is elicited by phosphorus limitation (Moore et al., 2005), and direct crosstalk between P and Zn, mediated by the regulatory protein PtrA, has been found in Synechococcus sp. WH8102 (Ostrowski et al., 2010). PtrA responds to phosphorus depletion and its expression up-regulates not only the expression of phosphatases, but also that of proteins predicted to be involved in zinc acquisition and distribution – including ZnuABC and a member of the COG0523 family (see below). However, it has to be noted that the true metal requirements of each of these proteins has yet to be experimentally verified, and there is reason to be cautious, with some evidence for the in vivo replacement of zinc with cobalt (Sunda and Huntsman, 1995) and cadmium (Lee and Morel, 1995) in marine phytoplankton. Very recently though, utilization of an alternative calcium-requiring phosphatase (PhoX) has been shown for uncultured Prochlorococcus (Kathuria and Martiny, 2011), suggesting a further mechanism for reducing zinc requirements.
In conclusion, although cyanobacteria are at the root of what life and marine trace metal chemistry are like today, their metal requirements require further study; none of the predicted major destinations for zinc are experimentally confirmed, and information about if, and how, zinc requirements can be alleviated by either Co or Cd substitution, is limited.
Zinc speciation in sea water
In order to understand how cyanobacteria might acquire zinc from the marine environment, we must first understand its chemical nature in seawater. The concentration of zinc in oceanic waters follows typical nutrient-like depth profiles (Bruland, 1980; Butler, 1998), with the lowest concentrations found in the euphotic zone with dissolved zinc rapidly removed to lower depths as a constituent of colloidal particles (Bruland, 1989; Wells et al., 1998). Total dissolved zinc concentrations in surface waters of the North Atlantic Ocean range from just 0.1 to 0.3 nM (Ellwood and van den Berg, 2000), similar to values obtained from measurements taken in the North Pacific Ocean (Bruland, 1980). The vast majority of this zinc (∼98%) is found complexed to uncharacterized organic ligands (Bruland, 1989; Donat and Bruland, 1990; Ellwood and van den Berg, 2000) with conditional stability constants, log K′ZnL, of between 10.0 and 10.5 (Wells et al., 1998; Ellwood and van den Berg, 2000). This results in a concentration of free Zn2+ of 1–20 pM. The distribution of these metal complexing ligands suggests a surface source (Bruland, 1980) that may include phytoplankton including cyanobacteria. This contribution could be through the direct secretion of zinc-binding ligands into the ocean. There is already some evidence that cyanobacteria actively secrete ligands that complex other biologically important trace metals including copper, iron, and cobalt. The marine Synechococcus sp. strains WH8101 and WH7805 have both been found to produce siderophores (Wilhelm and Trick, 1994) for scavenging iron from nutrient-depleted environments, whilst Cu-complexing ligands are produced to mitigate the toxic effects of copper (Wiramanaden et al., 2008), with cyanobacteria particularly sensitive to this metal (Mann et al., 2002). Significant quantities of strong cobalt-binding ligands were produced by a Synechococcus-dominated microbial community in the Costa Rica upwelling dome (Saito et al., 2005).
Alternatively the Zn complexing ligands could be released from cyanobacterial cells indirectly, perhaps as a consequence of cell lysis by marine phages (Wells et al., 1998). Despite the fact that the vast majority of zinc in ocean waters is present in the form of organic metal complexes, there is evidence that free Zn2+ is the major form of this nutrient taken up by phytoplankton (Anderson et al., 1978; Sunda and Huntsman, 1992; Sunda et al., 2005). In the Pacific Ocean the concentration of free Zn2+ in surface waters ranges from only 1 to 14 pM (Bruland, 1980; Donat and Bruland, 1990), and in the Atlantic Ocean ranges from 6.8 to 20 pM (Brand et al., 1983). The concentration of free Zn2+ in surface waters is thought to be sufficiently low to limit the growth of some marine phytoplankton (Brand et al., 1983; Sunda and Huntsman, 1992) although one study found that phytoplankton growth was not limited by low Zn2+ concentrations even after iron limitation was alleviated (Coale et al., 1996). Conversely, productivity in the subtropical Atlantic was further boosted by addition of Zn or Co to water that had also been enriched with Fe and P (Dixon, 2008), providing evidence for Zn/Fe/P co-limitation. Similarly, the iron-depleted waters of the Southern Ocean and the sub-arctic Pacific are also extremely zinc-depleted (Sunda and Huntsman, 2000) which appears to induce high levels of Cd uptake and high Cd:P ratios in phytoplankton. It is thus clear that interactions between the cycles of different metal ions exist, and that co-limitation needs to be studied (Saito et al., 2008). How important such crosstalk is in cyanobacteria is not yet well understood, and further work is required to determine if mechanisms exist for the active uptake of zinc, and if so, in what form zinc is acquired by cyanobacteria, and to determine the impact that cyanobacteria have on trace metal speciation in ocean waters (Leao et al., 2007).
Systems for Zinc Homeostasis
Zinc homeostasis in bacteria is largely achieved through a balance of active uptake and efflux by specific membrane transporters (Hantke, 2005), plus proteins mediating intracellular zinc handling (Figure 1). In the freshwater cyanobacterium Synechocystis sp. PCC 6803, a zinc-specific high affinity ABC transporter termed ZnuABC has been identified for the active uptake of zinc from the periplasm (Cavet et al., 2003). Putative ZnuABC systems have also been identified in most strains of marine cyanobacteria (Blindauer, 2008b; Scanlan et al., 2009). The putative znuABC gene cluster of many of the marine strains also contains a putative zur gene (Blindauer, 2008b). Zur proteins (for zinc uptake regulator) are low-zinc sensors; their zinc-loaded forms repress the expression of znuABC under zinc-replete conditions (Patzer and Hantke, 2000).
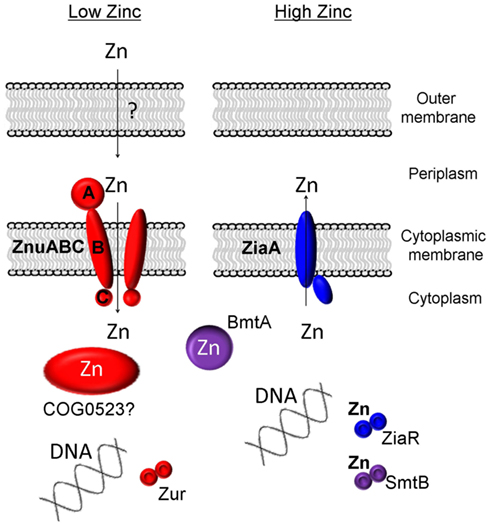
Figure 1. Known and proposed elements of zinc homeostasis in cyanobacteria. A requirement for zinc is sensed by the transcriptional regulator Zur, leading to the upregulation of the components of the ZnuABC uptake system. Some members of the putative metallochaperone family COG0523 are also regulated by Zur, and have, in other bacteria, been shown to be expressed in response to zinc deficiency. In most freshwater cyanobacteria, excessive levels of zinc are sensed by SmtB and its homologs (ZiaR, AztR, BxmR), and these sensors regulate either the expression of efflux pumps (ZiaA) or metallothioneins (SmtA and homologs).
To date no specific mechanisms for the active uptake of zinc across the outer cell membrane of cyanobacteria has been identified. It is generally considered that metal ions are small enough to diffuse freely through porins in the outer-membrane of Gram-negative bacteria; however, as described above, the concentration of free Zn2+ in surface layers of the world’s oceans is extremely low, with the vast majority of zinc complexed to as yet uncharacterized ligands of unknown structure and origin (Bruland, 1989). Whether at least some of these ligands are actively secreted to aid in zinc acquisition remains an open question; there is also the possibility that at least in coastal environments, ligands are synthesized in response to zinc excess and hence to avoid toxicity (Lohan et al., 2005; Leao et al., 2007).
In order to deal with excess zinc, Synechocystis sp. PCC 6803 has a zinc-specific efflux pump, ZiaA (Thelwell et al., 1998), similar to other P1-type ATPase metal ion transporters, that transports Zn2+ from the cytoplasm to the periplasmic space (Figure 1). Expression of this efflux system is induced by zinc and is regulated by ZiaR (Thelwell et al., 1998), a zinc-specific repressor protein. With the exception of Lyngbya sp. and Nodularia sp., most marine strains of cyanobacteria appear to lack zinc-specific efflux pumps (Blindauer, 2008b; Scanlan et al., 2009) reflecting the nutrient poor environments they occupy, with free Zn2+ concentrations being in the picomolar range in ocean waters (Hunter and Boyd, 1999). Instead, at least some marine cyanobacteria seem to rely on a mechanism of zinc sequestration by bacterial metallothioneins (BmtAs; Blindauer, 2008b) to deal with any eventual excess. Metallothioneins are small cytosolic proteins rich in cysteine residues that bind and sequester metal ions and thereby prevent any deleterious interactions (Blindauer and Leszczyszyn, 2010). The metallothionein SmtA of the freshwater cyanobacterium Synechococcus sp. PCC 7942 is induced by several metal ions but most prominently by zinc (Huckle et al., 1993) and its expression is controlled by the zinc sensor SmtB (Osman and Cavet, 2010) that is highly similar to ZiaR. Several marine strains of cyanobacteria appear to lack an SmtB/ZiaR type regulator (Blindauer, 2008b) despite the presence of one or more genes for a BmtA (Table 1). Furthermore, many also appear to lack any established mechanism for dealing with zinc excess, with all Prochlorococcus and some Synechococcus strains apparently lacking both a ZiaA efflux pump and a metallothionein encoding gene (Blindauer, 2008b). Presumably these bacteria never encounter toxic levels of zinc in the environment, although it also remains possible that they may employ novel mechanisms for zinc homeostasis that have yet to be discovered.
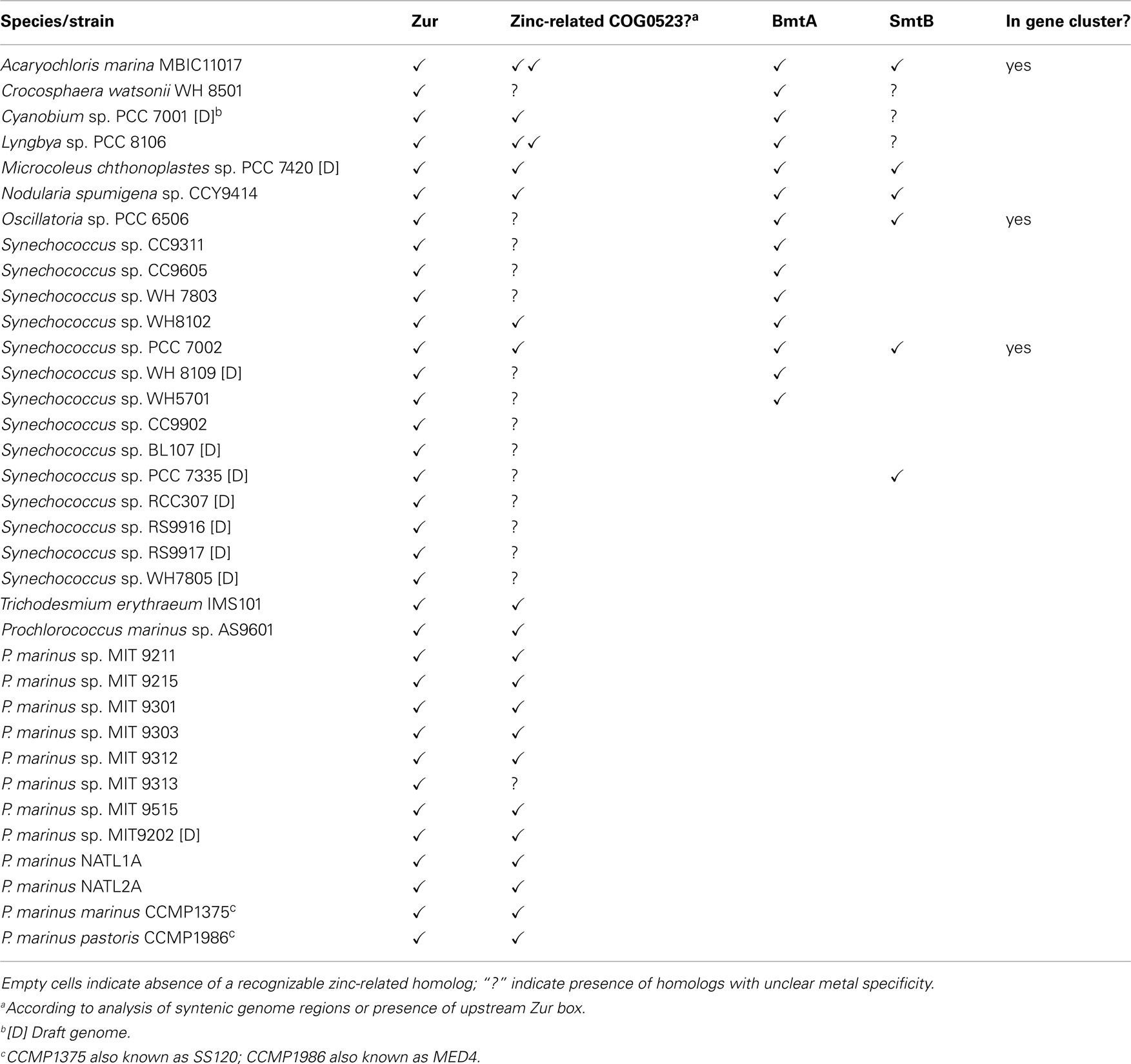
Table 1. Presence of selected genes predicted to be involved in zinc homeostasis in marine cyanobacterial genomes.
Sensing a requirement for zinc: zur transcription factors
In bacteria, the expression of proteins that deal with metal ion homeostasis is predominantly regulated at the transcriptional level (Finney and O’Halloran, 2003; Giedroc and Arunkumar, 2007; Waldron et al., 2009), and is mediated by sensor proteins for zinc excess (e.g., SmtB and its relatives; Giedroc and Arunkumar, 2007; Osman and Cavet, 2010) and zinc depletion (Zur and others). Seven major groups of bacterial metalloregulatory proteins have so far been defined: the Fur-family (for “ferric uptake regulator”) is one of them (COG0735; Bagg and Neilands, 1987), and also comprises paralogous sensors for zinc (Zur), nickel (Nur), manganese (Mur; Lee and Helmann, 2007), and hydrogen peroxide (PerR; Jacquamet et al., 2009).
Metal sensing in bacteria occurs overwhelmingly in the cytosol (Waldron and Robinson, 2009). Ultimately, the sensor proteins are the proteins that need to be the “most specific” – they should ideally either not bind any other metal ion, or if that is not possible, they should not respond to other metal ions in the same way as to their cognate metal. The key concepts of access and allostery, as well as the importance of relative affinities of different metalloproteins for different metal ions, have been highlighted by Waldron et al., 2009.
The dissociation constants of Zn2+–Zur complexes are in the femtomolar range; similar data are also available for zinc excess sensors (Outten and O’Halloran, 2001; Giedroc and Arunkumar, 2007) – these data also support the idea that the free Zn2+ concentration in the cytosol is extremely low. Several crystal structures of representatives of the COG0735 family have been determined (Pohl et al., 2003; Lucarelli et al., 2007; Jacquamet et al., 2009; Sheikh and Taylor, 2009; Shin et al., 2011). All members studied adopt a “winged-helix” fold (Figures 2A,C) and all assemblies are homo-dimeric, as are many other proteins that specifically recognize DNA sequences.
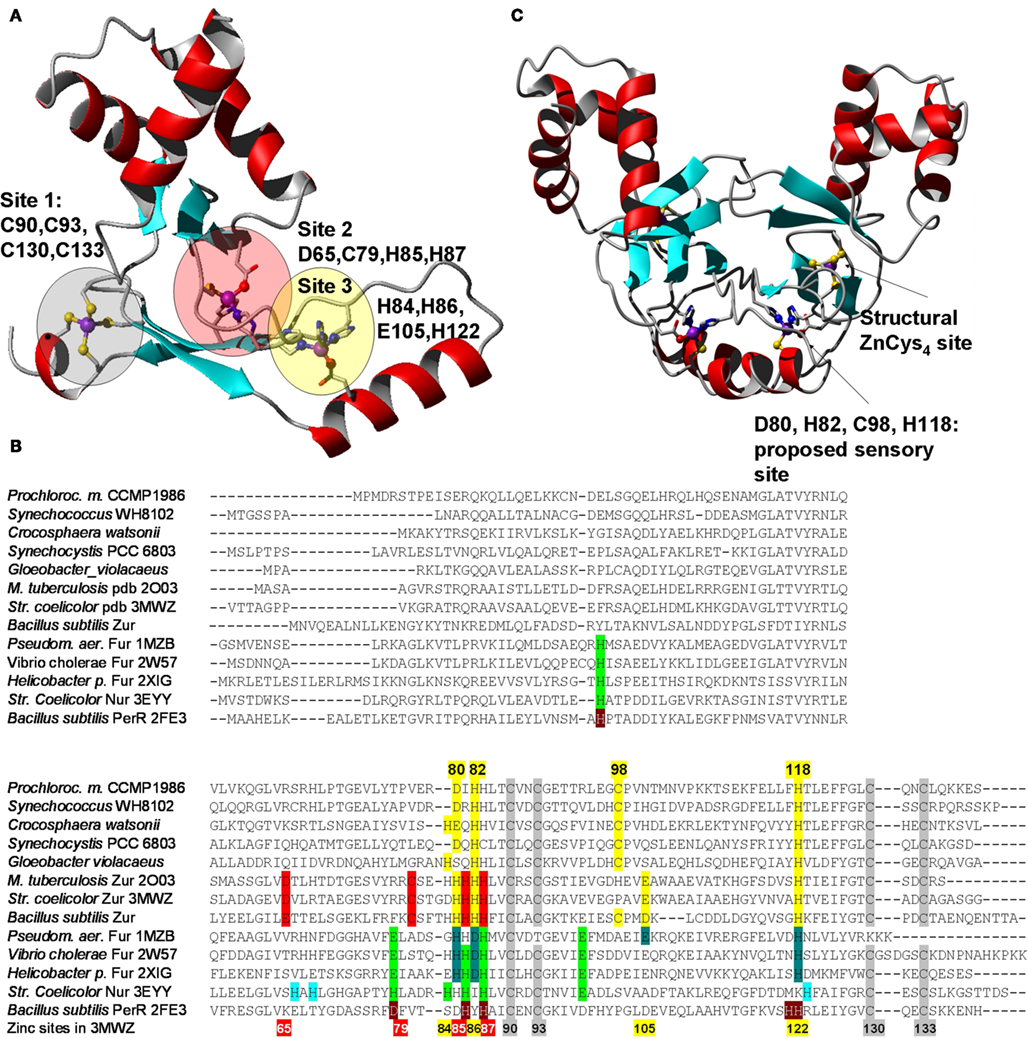
Figure 2. Structural features of Zur proteins. (A) Crystal structure of Zur from Streptomyces coelicolor (Shin et al., 2011; pdb 3MWM). Only one monomer is shown. The three zinc-binding sites are highlighted in red (site 2 – major sensory site), yellow (site 3), and gray (site 1 – structural site). (B) Sequential alignment of structurally characterized Fur-family proteins from various bacteria together with selected sequences from cyanobacteria. The color-coding for the Zur proteins from S. coelicolor and M. tuberculosis corresponds to that shown in (A). The consensus sensory site 2 is clearly not present in cyanobacterial sequences, but a variation of site 3, highlighted in yellow, can be discerned. Corresponding sites in Fur/Nur/PerR proteins are highlighted in dark and light green, and gray for the structural zinc site. (C) Homology model for Pro1502, a predicted Zur protein from Prochlorococcus marinus sp. CCPM1375. Inspection of initial metal-free models and conservation of potential ligands [see (B)] suggested that cyanobacterial Zurs contain only one sensory binding site that differs significantly from the sites in other Fur-family proteins including the two Zur proteins from S. coelicolor and M. tuberculosis, but the combination of donor atoms is the same as for site 2 (= N2OS). Further variations within the cyanobacterial Zur proteins are possible, as indicated for the sequences from Gloeobacter violaceus (a genus forming the earliest branch of the cyanobacterial phylogenetic tree) and Crocosphaera watsonii, which could form N3S sites.
All “urs” are thought to bind their cognate DNA in the presence of the entity to be sensed. Although no structures in the presence of DNA are available, it is thought that DNA-binding is mediated by the first ca. 80 residues, whilst dimerization, also a prerequisite of DNA-binding, is mediated by the C-terminal half, in particular by the formation of a six-stranded β-sheet formed by both monomers. The overall shape of the dimeric assembly can be described as an “arch” (Figure 2C), and the two DNA-binding domains are thought to “grip” the DNA using their DNA-recognition helices. It is likely that the interaction between protein and DNA requires a particular conformation that is stabilized by the presence of the sensed metal. The sensing appears to be mediated by two inter-domain hinges that are likely to be stabilized by metal-binding. In contrast to metal sensors of the SmtB family in which complete metal sites form between monomers, each metal site in Fur-family proteins is formed from residues from one monomer only. If the respective metal is absent, a different conformation may become more favorable, and DNA-binding no longer occurs, leading to the de-repression of gene transcription. Despite this general mechanistic idea, there is considerable ambiguity about the molecular detail of metal-binding, and how the binding of the “correct” metal mediates DNA-recognition. Despite the availability of X-ray structure for no less than seven Fur-family proteins, the identity of the residues involved in binding the metal ion to be sensed is unclear, in particular in those “urs” that contain three metal sites per monomer.
A structural zinc site formed by four conserved Cys residues is present in the majority of Fur-family members, independent of the sensed metal. One or two further sites participate in sensing. An inspection of various X-ray structures of Fur-family proteins suggested that sample preparation for such studies seems to be quite challenging; in particular, appropriate population with the correct complement of metal ions appears to be less than straightforward, and in several cases, workers appear to have resorted to populating all sites with Zn2+. Whilst this is certainly appropriate for Zurs, in other cases this may lead to ambiguous conclusions, as the coordination preferences of Zn2+ are rather different to those of Fe2+ or Ni2+. It has been demonstrated experimentally for SmtB/ArsR sensors that coordination geometry (Cavet et al., 2002) is an important discriminator in metal sensor proteins (via allostery). For Pseudomonas aeruginosa Fur, it has been shown in vitro that Zn2+-loaded Fur interacted with a Fur-binding DNA sequence in a different manner to that observed with Fe2+ (Ochsner et al., 1995). It should also be noted that despite full conservation of the respective residues, the crystallographically observed metal-binding sites in the Fur proteins from Pseudomonas aeruginosa on the one hand and Vibrio cholerae and Helicobacter pylori on the other (all structures contain only Zn2+ ions) are not identical (see Figure 2B), and that the domain orientations in the dimeric assemblies also differ significantly – likely as a consequence of the different coordination modes. It is conceivable that the binding mode for Fe2+ differs from both experimentally observed sites, with likely consequences for the structure that is competent to bind to Fur boxes.
Even in the case of the Zur sensors, metal population seems to be problematic, and as a consequence, there is some controversy over stoichiometry as well as the role of the various sites. For the Zurs from Streptomyces coelicolor (Shin et al., 2011) and Bacillus subtilis (Ma et al., 2011), there is agreement that site 2 (Figure 2A) is the major sensory site. Variations of site 2 are present in Fur, Nur, and PerR (Figure 2B), and in each case, this site has been identified as the sensory site. Site 3 was only partially occupied in the M. tuberculosis structure (Lucarelli et al., 2007), and the dimer displayed an open conformation, probably with reduced DNA-binding ability. In contrast, site 3 (site D) was fully occupied in the structure of S. coelicolor Zur, and the dimer showed a closed conformation, thought to be DNA-binding competent. This led to suggestions that site 3 fine-tunes the response to zinc (Shin et al., 2011). This assessment has been contested based on studies of mutants of B. subtilis Zur (Ma et al., 2011), which suggested that site 3 is not populated under physiological conditions at all, but that the two site 2s in the dimer have different affinities and show negative cooperativity. In either case, it was suggested that different metal affinities of the various sites allow the broadening of the operating range of the Zur proteins.
We believe that the preceding discussion demonstrates the challenges encountered in the study of metal-binding and -sensing proteins, but also highlights how important continued studies of metalloproteins are. Unfortunately, so far, no structure for any “ur” from a cyanobacterium has been elucidated, but in the following, we will explore what can be achieved using theoretical approaches.
BLAST searches in the genomes of marine cyanobacteria retrieved members of the COG0735 family as summarized in Table 1; Figure 3 and Figures S1 and S2 in Supplementary Material. They cluster into four distinct groups. Although it should be recognized that sequence similarity and metal specificity of sensors (or indeed other metalloproteins) need not be congruent (Campbell et al., 2007), comparisons with other “urs” of known specificity, first and foremost Synechocystis Zur, suggest that the branch highlighted in light purple corresponds to Zurs. Furthermore, an analysis of the genome environments of the Zurs from the majority of Prochlorococcus strains supports this idea (Figure 4).
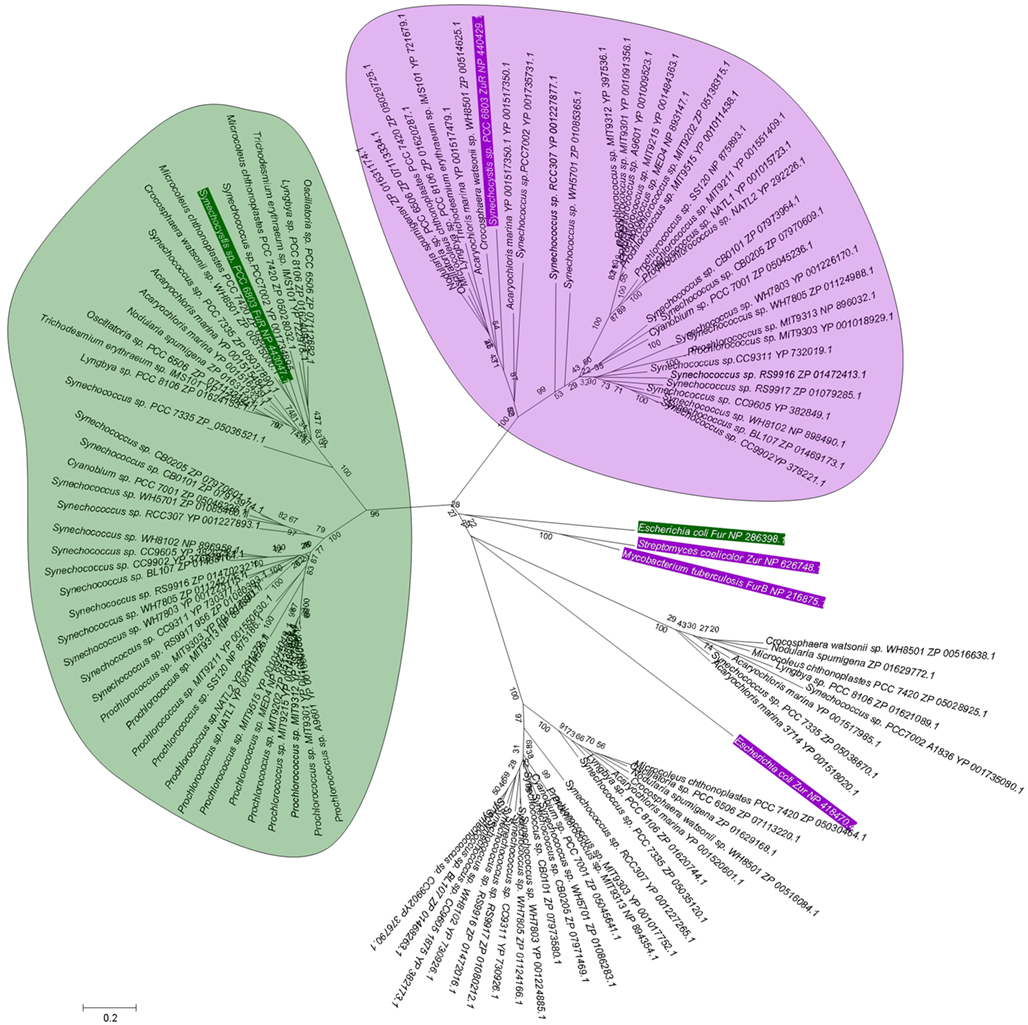
Figure 3. Phylogenetic relationship among Fur-like proteins from marine cyanobacteria. Sequences are labeled with species and protein accession number; a rectangular tree including bootstrap statistics can be found in Figure S1 in Supplementary Material, and the actual sequences are documented in Figure S2 in Supplementary Material. Additional proteins for the experimentally verified Fur from Escherichia coli and Synechocystis sp. PCC 6803 are also included (green boxes), along with the Zur proteins from Streptomyces coelicolor, Mycobacterium tuberculosis, and Synechocystis sp. PCC 6803 (purple boxes). Amino acid sequences (see Figure S2 in Supplementary Material) were aligned using CLUSTALW (Larkin et al., 2007), and manually edited prior to import into MEGA5 (Tamura et al., 2011). Phylogeny was inferred using the minimal evolution method. Bootstrap values are the result of 1000 replications. Evolutionary distance was estimated using the JTT model of substitution. There are four clear branches. Inclusion of Synechocystis Zur and Fur allows the suggestion that the branches containing these sequences correspond to zinc-responsive (light purple bubble) and iron-responsive (light green bubble) regulators. The Zur and Fur sequences from other bacteria do not cluster with any of the four branches, indicating that similarity between cyanobacterial sequences and those from other bacteria do not allow to infer metal specificity.
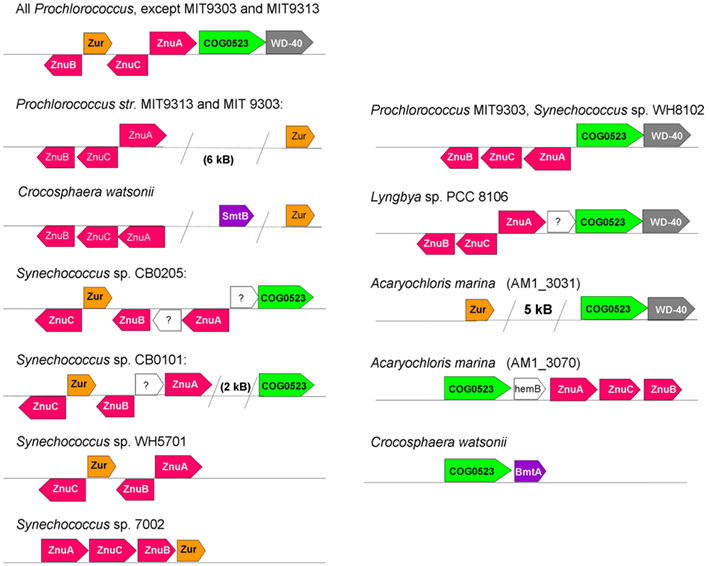
Figure 4. Selected variations in genome neighborhoods of elements of zinc homeostasis in marine cyanobacteria, with a focus on putative Zur (left) and COG0523 (right) proteins. ZnuA is the periplasmic binding protein, ZnuB the permease, and ZnuC the ATPase component of the ZnuABC uptake transporter (also see Figure 1). In other strains and species (e.g., other Synechococcus strains, Lyngbya sp. PCC 8106, Microcoleus chthonoplastes), the gene for Zur is not co-localized with recognizable elements of zinc homeostasis. Note that there appear to be two ZnuABC-type systems in Prochlorococcus marinus sp. MIT 9303. The Acaryochloris marina genome and plasmids harbor at least nine COG0523 family members; AM1_3070 (corresponding to UniProt entry B0CDJ7_ACAM1) and AM1_3031 (corresponding to UniProt entry B0CCJ8_ACAM1) are directly or indirectly associated with zinc homeostasis.
Intriguingly, sequence comparisons (Figure 2B) show that an equivalent of site 2 cannot be identified in the cyanobacterial Zur homologs, and neither are all residues of site 3 the same as in structurally characterized Zur proteins. We have constructed a homology model of the representative from Prochlorococcus sp. SS120 (CCMP1375) to test whether likely alternative binding sites might be predicted. The model obtained (Figure 2C) suggests that the cyanobacterial Zurs may contain a variation of site 3, comprising D80, H82, C98, and H118. Thus, the predicted zinc-sensing site is in a location corresponding to site 3, but has a ligand set that is similar to that of site 2, which may suggest that the zinc-binding affinity of this site is also closer to that of site 2. The four residues identified are almost fully conserved in putative Zur proteins from all cyanobacteria (Figure S2 in Supplementary Material). A notable exception is the homolog from Trichodesmium erythraeum, in which H82 is replaced by a Tyr residue.
From this analysis, we would predict that the molecular mechanism for sensing in cyanobacterial Zurs differs to some extent from that of other bacterial Zurs with three metal sites, but the general idea of zinc stabilizing domain orientation still holds. Notably, the recognition motifs (Zur boxes) for cyanobacterial Zurs differ somewhat from those of other bacteria (Haas et al., 2009), which may require an adapted mode of operation of the protein.
Using the Zur box motif developed by Haas et al. (2009) we have interrogated the genomes of Synechococcus sp. CC9311, Prochlorococcus sp. CCMP1375, and Prochlorococcus sp. CCMP1986. We have also examined the entries for cyanobacterial Zur regulons in the RegPrecise database (Novichkov et al., 2010). The motifs developed by Haas and by RegPrecise differ (Figure 5), but common features can still be discerned, and it is likely that the consensus Zur box in cyanobacteria corresponds to an (imperfect) inverted repeat, analogous to Zur and Fur boxes from other bacteria (Gabriel et al., 2008).
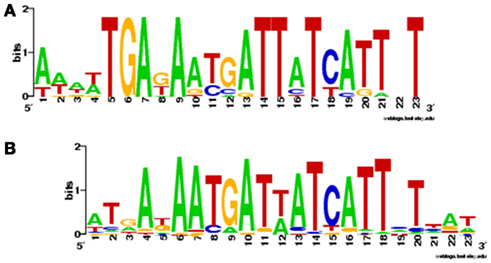
Figure 5. Sequence logos for Zur boxes in cyanobacteria. (A) Consensus sequence developed by Haas et al. (2009), using 14 sequences found upstream of COG0523 genes in Prochlorococcus marinus sp. CCMP1986, P. marinus sp. CCMP1375, Nostoc sp. PCC 7120, Cyanothece sp. PCC 7424, Cyanothece sp. PCC 8802, Cyanothece sp. PCC 8801, Cyanothece sp. ATCC 51142, and Cyanothece sp. PCC 7425. (B) Consensus sequence deposited in the RegPrecise database (Novichkov et al., 2010) using 75 sequences from the genomes of Synechocystis PCC 6803, Synechococcus sp. PCC 7002, Synechococcus sp. PCC 7942, Synechococcus JA-3.3Ab, Thermosynechococcus elongatus BP-1, Synechococcus sp. WH8102, Prochlorococcus sp. MIT 9313, Trichodesmium erythraeum IMS101, Cyanothece sp. PCC 7425, Cyanothece sp. PCC 8801, Cyanothece sp. ATCC 51142, Microcystis aeruginosa NIES-483, Nostoc sp. PCC 7120, and Gloeobacter violaceus PCC 7421. There are clear similarities between the two logos as indicated by the suggested alignment of parts A and B.
In the Prochlorococcus strains examined, a putative Zur box was found in the intergenic region between a putative znuA and znuC. All three components of the ABC transporter are arranged into one gene cluster that also comprises the respective putative zur gene (Figure 4). All other strains inspected (Trichodesmium erythraeum, Synechococcus sp. WH8102, Synechococcus sp. PCC 7002) also contain one or more Zur boxes in their znuABC gene cluster.
In the genome of Prochlorococcus sp. CCMP1986, Zur boxes were also identified upstream of several genes encoding ribosomal proteins (S7, S12, and S14p/S29e). This is of potential interest, because in other bacteria several ribosomal proteins, e.g., S14, L31, L33, and L36, occur in two versions, one requiring a zinc ion to stabilize a zinc-ribbon fold, and one version not requiring zinc (Makarova et al., 2001). The latter versions have been shown to be regulated by Zur in several bacterial species (Panina et al., 2003; Owen et al., 2007; Gabriel and Helmann, 2009). It has been suggested that the ribosome is, under zinc-replete conditions, a substantial store for cellular zinc, and that the “alternative” versions are expressed in response to zinc deprivation (Owen et al., 2007), operating as a backup for growth in Zn-poor environments, thus helping to reduce the overall requirement for zinc and the cellular zinc quota. The transcriptional response of E. coli to extreme zinc limitation (Graham et al., 2009) highlights that zinc limitation not only affects the transcription of genes encoding proteins involved in zinc homeostasis, and that are zinc-requiring, but also, importantly, zinc-independent proteins. Similarly, in other bacterial species it has been demonstrated that a variety of zinc-independent proteins including an alternative version of the global transcription factor DksA in Pseudomonas aeruginosa (Blaby-Haas et al., 2011), are under the control of Zur, and genome analyses of Zur regulons suggest that this is a widespread phenomenon (Haas et al., 2009). However, neither of the two versions of S14 present in the genome of Prochlorococcus sp. MED4 displays any salient signatures for zinc-binding, and neither S7 nor S12 are known to bind zinc or occur in duplicate, so the significance of their vicinity to Zur boxes is in need of further investigation.
Another set of potential Zur boxes were found in a cluster that comprises FutC, ferritin, and a Rieske iron–sulfur protein. According to the manually curated database of bacterial regulons RegPrecise, the cognate sequences of Furs and Zurs differ significantly (Novichkov et al., 2010). Hence, the detection of potential Zur boxes within a cluster related to iron homeostasis suggests that there is some crosstalk between the homeostasis of these two metal ions; this has been observed in other bacteria, e.g., in S. coelicolor, where the gene cluster responsible for the production of the siderophore coelibactin is regulated by Zur (Kallifidas et al., 2010).
The only Zur box that we were able to identify in the genome of Synechococcus sp. CC9311 was upstream of a predicted ZIP transporter protein (Sync_2443). ZIP (for “zinc–iron permeases”) proteins are involved in zinc uptake in a variety of organisms including plants, animals, fungi, and bacteria. However, ZIP proteins have as yet not been reported for cyanobacteria, and no recognizable homologs of Sync_2443 were found in any other cyanobacterium; the closest match in a BLAST search was a zinc transporter from the γ-proteobacterium Francisella novicida. If Sync_2443 really is a zinc transporter, this coastal strain has an even more remarkable repertoire for dealing with fluctuations in zinc concentrations than previously thought (Palenik et al., 2006). Two further notable RegPrecise entries were found for the genomes of Trichodesmium erythraeum, indicating Zur boxes upstream of a putative metallochaperone of the COG0523 family (Tery_4617), and Synechococcus sp. WH8102, which contains two Zur boxes upstream of a gene encoding a potential bacterial metallothionein (SYNW0359) – both groups of proteins are discussed below.
Very recently, Napolitano et al. (2012) studied the response of Anabaena sp. PCC7120 to zinc starvation. Aided by gel shift assays and a deletion mutant, the product of the all2473 gene, previously designated FurB and thought to be involved in the response to oxidative stress (López-Gomollón et al., 2009), was identified as a true Zur. Several gene clusters that contain putative Zur boxes were shown to be regulated by changes in zinc levels in this organism. The expression of four categories of proteins was regulated by Zur: (i) zinc-free paralogs of zinc proteins, (ii) putative metallochaperones of the COG0523 family, (iii) ABC transporters including a predicted ZnuABC system, and (iv) outer-membrane proteins, particularly TonB-dependent receptors. A 7-1-7 palindromic DNA sequence to which Zur bound with high specificity was also determined, and agrees well with the Zur box consensus motifs shown in Figure 5. The protein sequence of the all2473 product clusters with those predicted by us to be Zurs (Figure 3), and the four residues predicted to be involved in zinc-sensing are also conserved in the all2473 protein.
Members of the COG0523 family and zinc
In our earlier work (Blindauer, 2008b), we noted that the genome neighborhood of putative zur and znuABC genes in numerous genomes from cyanobacteria harbored genes that were annotated as “Putative GTPase, G3E family”, “Cobalamin synthesis protein/P47K”, or “CobW”. The latter protein is a cobalt chaperone that is part of the machinery for Vitamin B12 synthesis, and at the time, this raised the question whether our putative zinc-related genes might actually be involved in the regulation and transport of cobalt. In the meantime however, Haas et al. (2009) have conducted a thorough analysis of the COG0523 family (Leipe et al., 2002) to which CobW belongs. All members of COG0523 have a P-loop GTPase domain, and are thus also related to nickel-chaperones of the G3E family with the same domain that are involved in the maturation and assembly of urease (UreG), and hydrogenase (HypB), as well as to the iron-chaperone Nha3 required for maturation of nitrile hydratase. G3E family proteins, and by inference, COG0523 proteins, function as either insertases – proteins that perform energy-requiring metal insertion into target proteins – or as cytosolic storage and transport devices for metals (=metallochaperones), or both.
The COG0523 members are composed of two domains, a well-conserved N-terminal GTPase domain, and a more variable C-terminal domain. They have been categorized as segmentally variable genes (Haas et al., 2009), and this points toward a role in adaptation to environmental stresses and/or variability, and indicates development of binding specificity for other proteins or small molecules. Haas et al. note that “the COG0523 family is a striking example of systematic homology-based mis-annotation” – a specific function (in this case a role in cobalamin biosynthesis) being assigned even though the level of sequence similarity does not support this conclusion. We would add that such mis-annotation is certainly rife in the case of gene annotation for metal-binding proteins. The fact that many Zur proteins are annotated as Fur is another example of this problem, and issues surrounding the annotation of ABC-type transporters and metal-transporting ATPases have been discussed elsewhere (Blindauer, 2008b).
Importantly, the theoretical and experimental studies of Haas et al. as well as the work of other groups (Gabriel et al., 2008) strongly suggested that a subset of COG0523 proteins is involved in zinc homeostasis. The most prominent member of zinc-related COG0523 is probably Bacillus subtilis YciC (Gabriel et al., 2008); it has been established that its expression is under the control of Zur, leading to upregulation when zinc is scarce. It is suggested that the expression of zinc-related COG0523 proteins may provide advantages under conditions of poor zinc nutrition – obviously, these are conditions always present for open-ocean cyanobacteria. Unfortunately, no suitable studies at the protein level seem to be available for any COG0523 member, so although a link to zinc deprivation is solidly established at the transcriptional level, it is not known whether and which metal ion these proteins bind either in vitro or in vivo. Whilst the requirement of metallochaperones for copper, nickel, and cobalt can be easily understood, as the target proteins for whose assembly they are required are few and well identified, a similar scenario for zinc has been deemed unlikely, as it is thought that even in bacteria, there are too many destinations for zinc, so the tenet that there is one chaperone per metalloprotein is inconceivable. Hence, two hypotheses have been put forward regarding the role of the zinc-related COG0523 members (Haas et al., 2009): (i) they are up-regulated to function in the recruitment and supply of a metal ion that is not zinc to metalloproteins – the latter may be either normally zinc-requiring or a different paralog – (ii) these COG0523 proteins may be involved in the (re-)allocation of zinc when this becomes necessary.
Sequence analysis of cyanobacterial COG0523 proteins (Figure 7; Figures S3 and S4 in Supplementary Material) illustrates characteristic features, for example a well-conserved GCxCC motif which has been suggested as potential metal-binding site previously (Haas et al., 2009); however, its location in a β strand makes this less likely (see below and Figure 8). Another intriguing feature is the insertion of stretches with repetitive HHX, and HXH motifs of up to 70 amino acid residues in total. These His-rich stretches have been suggested to be hallmarks for metallochaperone activity of G3E GTPases. In the COG0523 subset we have analyzed 115 out of 150 proteins (77%) display such stretches. The phylogenetic tree for cyanobacterial COG0523 proteins (Figure 6) is roughly split into three major branches, each of which is divided into two sub-branches. Phylogenetic analysis, together with analyses of genome contexts (Figure 4), suggests that phylogeny and metal specificity are not congruent – in agreement with the findings of Haas et al. who defined 15 phylogenetic sub-groups for COG0523 members, with links to zinc homeostasis found in several sub-groups, and various sub-groups containing representatives with links to several different metal ions. A strong link to znuABC- and zur-containing gene clusters is present in one branch (1a) of the phylogenetic tree (Figure 6) for sequences from Prochlorococcus and Synechococcus sp. WH8102. Analysis of the genome environment for these sequences reveals co-localization of COG0523 with a WD-40 repeat gene (Figure 4). This association is also strongly conserved for other members in this branch, although the functional significance of the WD-40 repeat protein is unclear. Interestingly, COG0523 sequences of branch 1a contain long inserts (ca. 150 aa), distinct from those described above, as they are not particularly rich in His residues (Figure 7). Secondary structure prediction for these inserts suggests the presence of four to six β-strands and one or two short α-helices. It is conceivable that this insert forms an additional domain in its own right, though no similarity to any known domains was found in an InterPro-scan search.
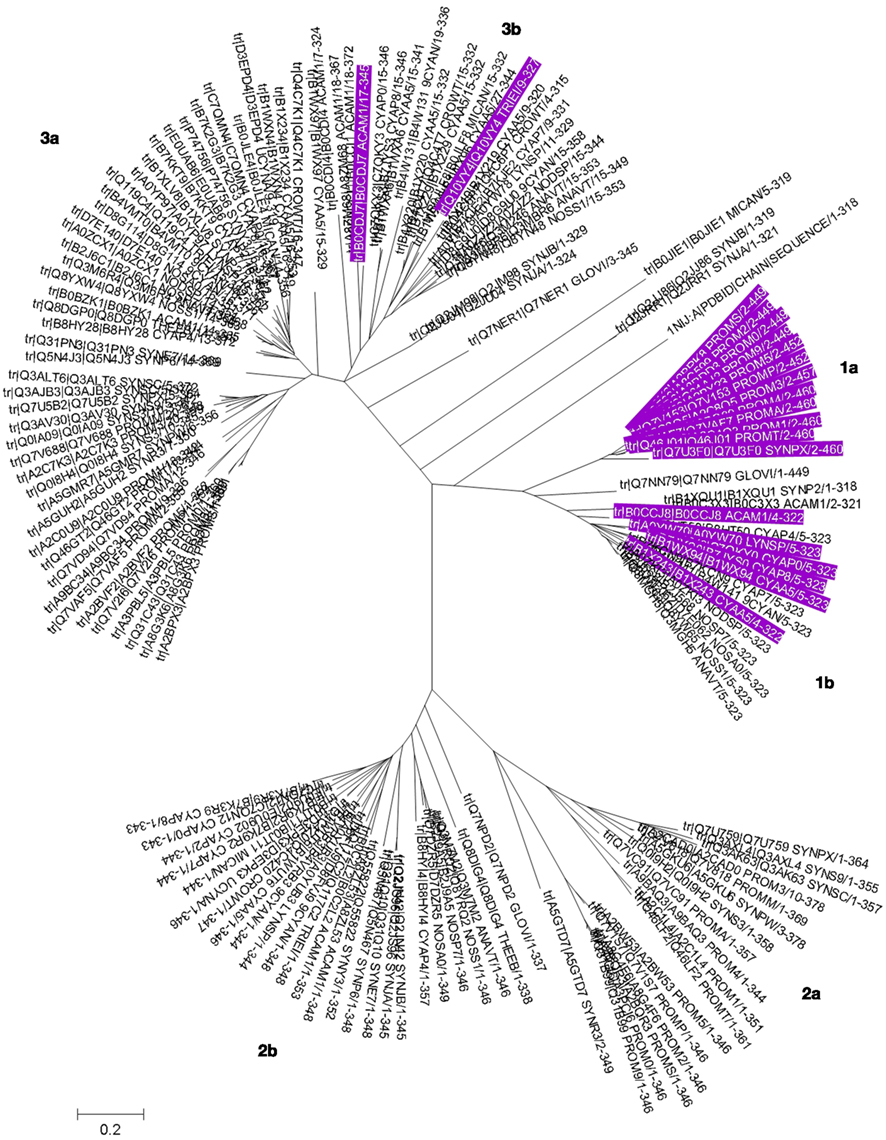
Figure 6. Phylogenetic tree for COG0523 family members. The phylogenetic tree for cyanobacterial COG0523 family members was generated in MEGA5 using the maximum likelihood method with the JJT substitution model. Bootstrap values are the result of 1000 replications. Sequences are labeled with UniProt accession numbers; the actual sequences are documented Figure S3 in Supplementary Material, and a rectangular tree including bootstrap statistics can be found Figure S4 in Supplementary Material. The sequence of E. coli YjiA, the only COG0523 member for which an experimentally determined structure is available, was also included in the tree. Entries with a genomic context linked to zinc homeostasis are highlighted in purple (also see Table 1). Sub-branches are labeled and discussed in the main text.

Figure 7. Examples of the six sub-groups of COG0523 family members. All four homologs from Prochlorococcus marinus CCMP1375 are shown, as well as two sequences from Acaryochloris marina, structural models of which are shown in Figure 8. A further A. marina representative from branch 2b and the potentially Zur-regulated representative from Trichodesmium erythraeum (locus tag Tery_4617, see text) have also been added. The sequences are labeled with their location in the phylogenetic tree (see Figure 6). The first three rows correspond to the GTPase domain. This is followed by a highly variable section. Group 1a is characterized by ca. 150 aa long inserts with no His residues. Inserts of medium length are found for some representatives of group 3. Very short linkers between the N-terminal GTPase domain and the C-terminal domain (which is relatively well-conserved again) are found in all three major groups.
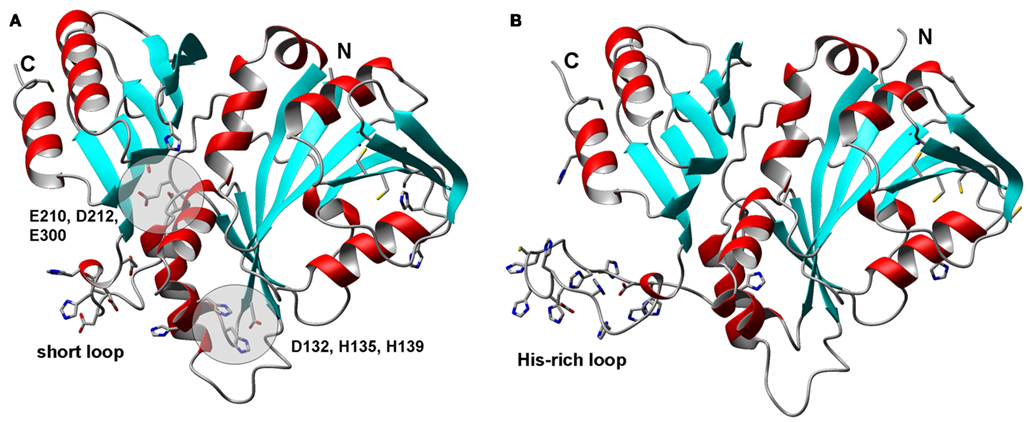
Figure 8. Structural models for zinc-related COG0523 proteins from Acaryochloris marina. (A) B0CCJ8_ACAM1 (AM1_3031). (B) B0CDJ7_ACAM1 (AM1_3070). The N-terminal GTPase domains are shown on the right hand side, the C-terminal extension that distinguishes COG0523 members from other metal-related G3E GTPases is shown on the left hand side. The cysteines forming CCXC motifs in the GTPase domain are also shown; their location in a β-strand makes their involvement in metal-binding unlikely in these structures, as their sulfurs are too far apart. C69 and H34 in B0CCJ8_ACAM1 are within >4 Å of each other, but no other potential metal-binding residues are nearby.
There is currently only one X-ray structure available for a COG0523 protein – the YjiA protein from E. coli (Khil et al., 2004). According to the analysis by Haas et al. (2009) YjiA belongs to their subgroup 9, for which no clear association with any particular metal ion was reported. The crystal structure (pdb 1NJI) also does not contain any metal ions, but considering that the structure is a product of a structural genomics effort, this may not be surprising. Analysis of potential metal sites in the structure using the CHED server (Levy et al., 2009) revealed the presence of two surface-exposed sites with potential for metal-binding; these are both in the GTPase domain and are composed of H23, E27, and H29, and D52, D79, and D82.
We were interested to see whether it would be possible to pinpoint distinguishing elements that would indicate metal-binding capacity, with the help of homology models for COG0523 proteins. We therefore chose B0CCJ8_ACAM1 from Acaryochloris marina that is present in branch 1 as a target for comparative modeling. The respective AM1_3031 gene is next to a WD-40 repeat gene and also in the neighborhood of a zur gene (Figure 4). In addition, we modeled B0CDJ7_ACAM1, since the gene for this protein (AM1_3070) is co-localized with a putative znuABC system (Figure 4). The sequence for this protein is located in branch 3b of the phylogenetic tree and is highlighted in Figure 6. Several other closely related sequences from the same organism (including some plasmid-encoded sequences) are present in all three branches, but none of these occurs in a zinc-related genomic context. The two models are shown in Figure 8. Beside a His-rich loop between the two domains that also contains two Asp and a Cys residue, there are no further recognizable metal sites in B0CDJ7_ACAM1. In contrast, B0CCJ8_ACAM1 has a much shorter loop comprising only two His and several Asp and Glu residues that might bind a metal. Other branch 1b members also contain this short loop, but so do some representatives from the other sub-branches. The model also displays two further potential metal sites composed of D132, H135, and H139, and E210, D212, and E300, as identified using the CHED server, but only D132 and H135 are conserved within the branch, questioning the significance of these sites. The quest for the true metal-binding sites and the mode of action of the zinc-related COG0523 proteins remains open.
Intracellular handling: Metallothioneins (BmtAs)
Metallothioneins, small proteins with a high content of cysteine with the capability to bind multiple metal ions in metal–sulfur clusters, were initially reported as cadmium-binding proteins in the livers and kidneys of mammals (Kägi, 1991). In the five decades since their discovery (Margoshes and Vallee, 1957), genes encoding metallothioneins have been identified in virtually all phyla, and numerous studies regarding their biophysical properties (Blindauer and Leszczyszyn, 2010) and biological functions (Davis and Cousins, 2000; Cobbett and Goldsbrough, 2002; Klaassen et al., 2009) have been carried out. It has become clear that their main function is not restricted to cadmium detoxification nor to responses to other chemical and physical stresses. At least for vertebrates, it is now accepted that they also play a more general and essential role in zinc homeostasis (Maret, 2009; Colvin et al., 2010), and that they constitute an important link between cellular redox state and zinc signaling networks (Maret, 2011).
The presence of metallothionein-like proteins in bacteria (for recent reviews see (Blindauer, 2009, 2011) was first indicated in 1979, namely in the marine cyanobacterium Synechococcus RRIMP N1 (Olafson et al., 1979). So-called “pseudo-thioneins” were also discovered in cadmium-adapted Pseudomonas putida (Higham et al., 1984). The first gene for a bacterial metallothionein, smtA, was isolated from Synechococcus sp. PCC 7942 (Robinson et al., 1990), and was shown to be regulated by the zinc-sensing transcriptional repressor SmtB. Phenotypically, smtA knock-out mutants are hypersensitive to Zn2+, and to a lesser extent to Cd2+, with no effect on tolerance to other metal ions (Turner et al., 1993), even though smtA transcription is stimulated not only by Zn2+ (which is by far the strongest inducer) and Cd2+, but also Hg2+, Cu2+, Co2+, Cr3+, and Ni2+ (Huckle et al., 1993). Moreover, SmtA expressed in E. coli was shown to bind not only zinc and cadmium, but also copper and mercury (Shi et al., 1992). Considering that SmtB is clearly a Zn2+-responsive metal sensor (Turner et al., 1996), the documented responses to other metal ions may be mediated indirectly, by displacement of Zn from proteins by these metal ions.
As soon as the protein sequence of what was later to be called SmtA was available, it was clear that apart from the high cysteine content, there was very little sequence similarity between previously characterized metallothioneins and their bacterial counterparts (Olafson et al., 1988). However, it should be made clear that this statement is essentially true for all MTs from different phyla (Blindauer and Leszczyszyn, 2010). For example, in the animal kingdom, the sequences from MTs from nematodes, snails, earthworms, and vertebrates are so divergent that it is not possible to demonstrate a clear evolutionary relationship. To some extent, this is due to their small size, their low level of complexity, and the absence of a defined protein fold. The latter feature is also reflected in the fact that the folding of MTs is dominated by the formation of the metal–sulfur clusters, and unless the “correct” complement of metal ions is bound, MTs do not adopt well-defined conformations.
In that sense, bacterial metallothioneins of the BmtA type are an exception – they contain a clearly identifiable zinc finger fold (Blindauer et al., 2001, 2002; Blindauer and Sadler, 2005; Figure 9A), and it has been demonstrated experimentally that the constituents of this fold (residues 7–38) form an ordered, folded structure, even if only one Zn2+ ion is bound to SmtA (Leszczyszyn et al., 2007a). Another “special feature” that is becoming less exceptional as more and more MTs from other phyla are being studied, is the presence of aromatic residues, including histidines, the latter often with a direct involvement in metal-binding (Blindauer et al., 2007; Leszczyszyn et al., 2007b; Blindauer, 2008a; Peroza et al., 2009; Zeitoun-Ghandour et al., 2010).
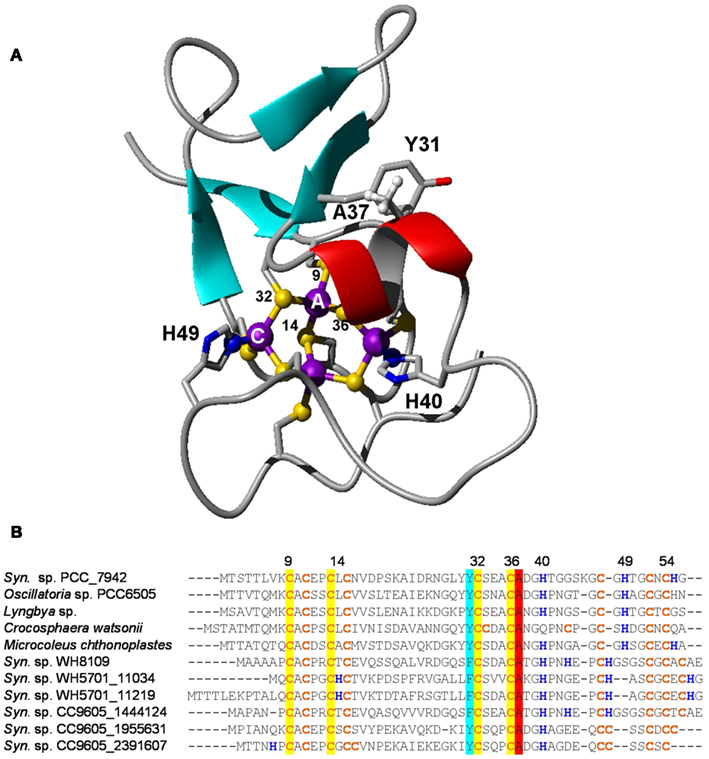
Figure 9. Structural features of bacterial metallothioneins. (A) NMR solution structure of Zn4SmtA. Residues important for the zinc finger fold and metal ligands are highlighted. (B) Selected primary sequences of BmtAs from marine cyanobacteria. The level of conservation is different for different metal sites. Site A, the zinc finger site, is fully conserved, as it is required for structural stability. Conservation is also high for residues Y31 and A37; these mediate a CH-π interaction between β hairpin and α-helix. The least conserved site is site C, which is also the “business end” of BmtAs, i.e., the site that (in SmtA) releases zinc most readily (Leszczyszyn et al., 2007a). It is envisaged that the high variability of this site may allow to fine-tune the metal-binding and -release properties of different SmtAs.
The high abundance of cysteine residues are the cause of the high thermodynamic stability of MT complexes with soft1 metal ions such as Cu+ and Cd2+, and these tend to bind more strongly to MTs than Zn2+, which is classified as a borderline metal ion. It should however be noted that thermodynamic stability is not necessarily a criterion to determine which metal ions are handled by a particular MT (or indeed a particular protein) in vivo. If the metallated protein has not been obtained in its natively metallated form from the natural source, it is important to take into account information on which metal ion(s) induce(s) MT gene transcription most strongly, whether the protein confers tolerance against a particular metal ion, and also on how well-folded the protein is in the presence of different metal ions. Bofill et al. (2009) have compiled large amounts of biophysical data on recombinantly expressed MTs from a variety of species, and have suggested that there are clear Cu-MTs and Zn/Cd-MTs, as well as MTs between these two extremes with less well-defined metal preferences.
SmtA is thought to be a prototypical Zn-MT. To some extent, this is indeed also reflected in the in vitro properties of the protein: the zinc finger site and fold require a four-coordinate metal ion – which excludes Cu+ as it prefers trigonal or linear coordination modes with thiolate ligands. In addition, the two His-containing metal sites augment the relative affinity for Zn2+ compared to Cd2+, although this is certainly not their sole purpose (Blindauer et al., 2007). Nevertheless, SmtA folds equally well in the presence of four Zn (Blindauer et al., 2001) or four Cd (Blindauer et al., 2008) ions, or any mixture thereof, a feature that greatly facilitated the determination of its 3D structure. Although no in-depth study of SmtA (or any other BmtA) loaded with copper ions has so far been carried out, recombinant expression of SmtA in the presence of added copper yielded samples with only ca. 1.5 copper ions bound, whereas contents of Cd2+, Hg2+, and Zn2+ varied between ca. 4 and 6 when the respective metal ion was added to the culture medium (Shi et al., 1992).
An analysis of the genomes of all marine cyanobacteria reveals that 14 out of 33 sequenced genomes contain one or more genes for a BmtA (Table 1; Figures 9B). The BmtA sequences were retrieved using a protein vs. DNA TBLASTN search with the Synechococcus sp. PCC 7942 sequence as query and the six-frame translations of the genomic nucleotide databases. This is necessary to ensure that all BmtAs are captured, as due to their small size, BmtA ORFs are easy to overlook. This is illustrated by the fact that bmtA genes are not annotated in the genomes of Microcoleus, Cyanobium, and Crocosphaera watsonii, all of which are in their “Draft” stage, but they have also been overlooked in the finished genome and plasmid sequences of Acaryochloris marina.
In a select few genomes (Synechococcus sp. PCC 7002, Oscillatoria sp. PCC 6506, and the genome as well as pREB2 and pREB6 plasmids of Acaryochloris marina) BmtAs occur in a cluster together with SmtB. On the latter two plasmids, a COG0523 homolog is also nearby. In the Synechococcus sp. WH5701 genome, both BmtAs are annotated, and WH5701_11219 (the latter number refers to the first base of the start codon) is located near a CsbD-like protein, thought to be involved in the general stress response, a Cd/Co/HG/Pg/Zn-transporting ATPase, and the permease and ATPase components of an ABC transporter, but since no periplasmic component is present in the vicinity, the specificity for this transporter cannot be predicted.
Several of the benthic strains also have SmtB homologs, for example Microcoleus chthonoplastes and Lyngbya (in a cluster with ZiaA), and Nodularia spumigena (intriguingly in a gene cluster with an ABC transporter), but even though several open-ocean and coastal marine Synechococcus (WH8102, WH8109, WH7803, CC9311, CC9605) have genes for BmtA, there are no identifiable SmtB or other known zinc excess sensors present. Therefore, it remains unclear how the expression of these metallothionein genes is regulated, although the discovery of two Zur boxes upstream of BmtA in Synechococcus sp. WH8102 gives rise to the suggestion that this strain might use its metallothionein in response to zinc limitation rather than excess.
We note that although the prototype smtA clearly responds most strongly to zinc, and the SmtA protein can be considered a “zinc-metallothionein,” this may not necessarily hold true for all homologs from marine strains. Since the concentrations of Cd2+ can be significant in various natural seawaters, the marine BmtAs may also help with dealing with this metal ion – potentially both for preventing toxicity as well as in preparation for utilization as a cofactor. Cd also has a nutrient-like profile in stratified marine waters, and there are several precedents where marine phytoplankton have been found to utilize Cd instead of Zn (Lane et al., 2005). If this was also common in marine cyanobacteria, BmtAs may be part of this utilization network. Furthermore, it can also be envisaged that some BmtAs may play a role in defense against oxidative stress, as shown for metallothioneins from other species (Zeitoun-Ghandour et al., 2011).
Finally, it should be highlighted that cyanobacteria are not the only marine microorganisms with metallothioneins. Several γ-proteobacteria of the genus Nitrosococcus also harbor bmtA genes in their genomes, and additional bmtA sequences can also be retrieved from marine metagenomes (Blindauer, 2008b), suggesting that many more marine bacterial species utilize these proteins.
Conclusion
At the time of writing this report (December 2011), there were 64 cyanobacterial genomes available, 35 of them from marine species. Genome annotation must, ultimately, be based on experimental evidence, and we have seen that there is a persistent shortfall of reliable information regarding function of predicted proteins. This is a general problem, witnessed by the large number of hypothetical proteins even in finished genomes, but even in the case of well-defined protein families, annotations regarding metal specificity have to be approached with caution. Our discussions emphasize that sequence similarity alone does not hold the key to determine metal specificity, but we believe that, amongst other approaches, the generation of more sound data at the protein level is central to refine our ideas about how metal homeostasis works.
With this caveat in mind, we are highlighting in the following some salient points from our analyses. The ubiquitous presence of bona-fide Zur and ZnuABC systems – both thought to be involved in responding to a lack of cellular zinc – in all strains we have studied suggests that these cyanobacteria are at the very least capable of utilizing zinc. The identification of putative Zur boxes in several genomes we and others have inspected also suggests that zinc levels are an integral part of the metabolic network of marine cyanobacteria. Nevertheless, an absolute requirement for zinc has not been demonstrated experimentally for any cyanobacterium – in the few instances where this has been studied, only small reductions in growth rates were observed. This may indicate that marine cyanobacteria have either very low zinc requirements, or extremely efficient uptake mechanisms, or both, enabling them to thrive at extremely low free zinc concentrations. It would hence be interesting to study whether cyanobacteria that are growing at such extremely low zinc concentrations actually contain any zinc, and if so, what the ratio of bio-accumulation is.
Genes for zinc excess sensors appear to be absent from the genomes of open-ocean strains (Blindauer, 2008b), but other strains including Microcoleus chthonoplastes, Synechococcus sp. PCC 7002, and Oscillatoria sp. PCC 6506 have clear SmtB homologs. Although the coastal strain Synechococcus sp. CC9311 does not appear to have SmtB, it not only has four BmtA homologs, but also a putative CzrA efflux pump that is thought to be able to transport zinc (and cobalt). Together with at least one ZnuABC system and a putative ZIP transporter for uptake, as well as at least one zinc-related COG0523 protein, this strain is expected to cope well with both zinc excess and scarcity.
The suggestion that metallothioneins in marine Synechococcus (e.g., WH8102) may be under the control of Zur is exciting. Whilst this finding may at first seem counterintuitive, it suggests that, also in (some) bacteria, metallothioneins are not just devices to combat metal toxicity, but may play a more central role in essential zinc homeostasis. This hypothesis has been raised before (Robinson et al., 2001; Blindauer, 2009), and this newly identified association between Zur and BmtA provides support.
Methods
Sequences of Fur-family proteins were retrieved from cyanobacterial genomes using BLAST (Altschul et al., 1997). Briefly, GenBank files were retrieved for each genome and all protein sequences were extracted using BioPerl (Stajich et al., 2002) and a custom BLAST database was created for each genome. Fur-like proteins were identified using the three Fur-like proteins previously identified in Synechococcus sp. CC9311 as queries (accessions YP_730377.1, YP_732019.1, and YP_730926.1) with an e value cutoff <10−5. Any duplications were removed and amino acid sequences were aligned using CLUSTALW (Larkin et al., 2007). The Fur amino acid sequence from Escherichia coli and Synechocystis sp. PCC 6803, along with Zur from Streptomyces coelicolor, Mycobacterium tuberculosis, and Synechocystis sp. PCC 6803 were extracted from the Protein Data Bank2. Phylogenetic analysis was carried out in MEGA version 5 (Tamura et al., 2011).
Sequences for COG0523 family proteins were collected by searching the fully sequenced and annotated cyanobacterial proteomes plus all sequences from Oscillatoria sp. PCC 6506, Lyngbya sp. PCC 8106, Microcoleus chthonoplastes sp. PCC 7420, Nodularia spumigena sp. CCY9414, Crocosphaera watsonii WH 8501, and Trichodesmium erythraeum IMS101 from UniProt with the Pfam PF07683 Hidden Markov Model using HMMER4 with standard cutoffs. The collated sequences were filtered for fragments and aligned in JALVIEW and MUSCLE v.3.8 (Edgar, 2004), and were manually adjusted where necessary.
Gene Ortholog Neighborhood analyses were performed using the respective resource on the Integrated Microbial Genome Server (Markowitz et al., 2010).
Comparative modeling was carried out using Modeller version 9.7 (Eswar et al., 2008). A dimeric assembly of Streptomyces coelicolor Zur was generated from the two B chains in the original pdb file (3MWM; Shin et al., 2011) using PISA3 (Krissinel and Henrick, 2007). This dimeric structure was used as a template for modeling the putative Zur from Prochlorococcus marinus CCMP1375 (locus tag Pro1502). Template and model sequences were aligned manually. Sidechains and improper dihedrals were optimized with SCWRL 3.0 (Wang et al., 2008). Inspection of initial models and sequence alignments suggested that Cys98 may be a zinc ligand; this residue is highly conserved in all putative Zur sequences from cyanobacteria (See Figure S2 in Supplementary Material). Hydrogens, Zn ions, and metal–ligand bonds were added in MOE v. 2004.03. Energy minimization of the final model was performed in MOE using a customized Amber94 force-field. Final structures were validated using the WHATIF server4. Structural images (Figures 2, 8, and 9) were generated with MOLMOL v.2K.2 (Koradi et al., 1996). A similar strategy was applied for modeling two putatively zinc-binding representatives of the COG0523 family based on the E. coli protein as a template (pdb-entry 1NIJ). The target template sequence alignment made use of the HHPRED (Soding et al., 2005) fold recognition server.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank the Leverhulme Trust (F/00 215/AY) and the Natural Environment Research Council (grants NE/F004249/1 and NE/G017948/1) for their support of this work, and the Al-Baath University (Homs, Syria) for a scholarship to Amira Z. Ksibe.
Supplementary Material
The Supplementary Material for this article can be found online at fmicb.2012.00142/abstract
Figure S1. Phylogenetic tree for Fur/Zur-like proteins extracted from genomes of marine cyanobacteria.
Figure S2. Sequence alignment of Fur/Zur-like proteins.
Figure S3. Phylogenetic tree for COG0523 family members.
Figure S4. Sequence alignment of members of the COG0523 protein family from the genomes of cyanobacteria.
Footnotes
- ^The terms “soft” and “borderline” refer to Pearson’s HSAB principle: Pearson (1990).
- ^http://www.rcsb.org/
- ^http://www.ebi.ac.uk/msd-srv/prot_int/cgi-bin/piserver
- ^http://swift.cmbi.ru.nl/servers/html/index.html
References
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Anderson, M. A., Morel, F. M. M., and Guillard, R. R. L. (1978). Growth limitation of a coastal diatom by low zinc ion activity. Nature 276, 70–71.
Andreini, C., Banci, L., Bertini, I., and Rosato, A. (2006). Zinc through the three domains of life. J. Proteome Res. 5, 3173–3178.
Andreini, C., Bertini, I., Cavallaro, G., Holliday, G. L., and Thornton, J. M. (2008). Metal ions in biological catalysis: from enzyme databases to general principles. J. Biol. Inorg. Chem. 13, 1205–1218.
Andreini, C., Bertini, I., and Rosato, A. (2009). Metalloproteomes: a bioinformatic approach. Acc. Chem. Res. 42, 1471–1479.
Bagg, A., and Neilands, J. B. (1987). Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26, 5471–5477.
Bertilsson, S., Berglund, O., Karl, D. M., and Chisholm, S. W. (2003). Elemental composition of marine Prochlorococcus and Synechococcus: implications for the ecological stoichiometry of the sea. Limnol. Oceanogr. 48, 1721–1731.
Blaby-Haas, C. E., Furman, R., Rodionov, D. A., Artsimovitch, I., and De Crecy-Lagard, V. (2011). Role of a Zn-independent DksA in Zn homeostasis and stringent response. Mol. Microbiol. 79, 700–715.
Blindauer, C. A. (2008a). Metallothioneins with unusual residues: histidines as modulators of zinc affinity and reactivity. J. Inorg. Biochem. 102, 507–521.
Blindauer, C. A. (2008b). Zinc handling in cyanobacteria – an update. Chem. Biodivers. 5, 1990–2013.
Blindauer, C. A. (2009). “Bacterial metallothioneins,” in Metallothioneins and Related Chelators, eds A. Sigel, H. Sigel, and R. Sigel (Cambridge: The Royal Society of Chemistry), 51–81.
Blindauer, C. A. (2011). Bacterial metallothioneins: past, present, and questions for the future. J. Biol. Inorg. Chem. 16, 1011–1024.
Blindauer, C. A., Harrison, M. D., Parkinson, J. A., Robinson, A. K., Cavet, J. S., Robinson, N. J., and Sadler, P. J. (2001). A metallothionein containing a zinc finger within a four-metal cluster protects a bacterium from zinc toxicity. Proc. Natl. Acad. Sci. U.S.A. 98, 9593–9598.
Blindauer, C. A., Harrison, M. D., Parkinson, J. A., Robinson, N. J., and Sadler, P. J. (2008). Isostructural replacement of zinc by cadmium in bacterial metallothionein. Met. Ions Biol. Med. 10, 167–173.
Blindauer, C. A., Harrison, M. D., Robinson, A. K., Parkinson, J. A., Bowness, P. W., Sadler, P. J., and Robinson, N. J. (2002). Multiple bacteria encode metallothioneins and SmtA-like zinc fingers. Mol. Microbiol. 45, 1421–1432.
Blindauer, C. A., and Leszczyszyn, O. I. (2010). Metallothioneins: unparalleled diversity in structures and functions for metal ion homeostasis and more. Nat. Prod. Rep. 27, 720–741.
Blindauer, C. A., Razi, M. T., Campopiano, D. J., and Sadler, P. J. (2007). Histidine ligands in bacterial metallothionein enhance cluster stability. J. Biol. Inorg. Chem. 12, 393–405.
Blindauer, C. A., and Sadler, P. J. (2005). How to hide zinc in a small protein. Acc. Chem. Res. 38, 62–69.
Bofill, R., Capdevila, M., and Atrian, S. (2009). Independent metal-binding features of recombinant metallothioneins convergently draw a step gradation between Zn- and Cu-thioneins. Metallomics 1, 229–234.
Bozym, R. A., Chimienti, F., Giblin, L. J., Gross, G. W., Korichneva, I., Li, Y. A., Libert, S., Maret, W., Parviz, M., Frederickson, C. J., and Thompson, R. B. (2010). Free zinc ions outside a narrow concentration range are toxic to a variety of cells in vitro. Exp. Biol. Med. 235, 741–750.
Brand, L. E., Sunda, W. G., and Guillard, R. R. L. (1983). Limitation of marine phytoplankton reproductive rates by zinc, manganese, and iron. Limnol. Oceanogr. 28, 1182–1198.
Bruland, K. W. (1980). Oceanographic distribution of cadmium, zinc, nickel, and copper in the North Pacific. Earth Planet. Sci. Lett. 47, 176–198.
Bruland, K. W. (1989). Complexation of zinc by natural organic ligands in the central North Pacific. Limnol. Oceanogr. 34, 269–285.
Bruland, K. W., Orians, K. J., and Cowen, J. P. (1994). Reactive trace metals in the stratified central North Pacific. Geochim. Cosmochim. Acta 58, 3171–3182.
Butler, A. (1998). Acquisition and utilization of transition metal ions by marine organisms. Science 281, 207–210.
Campbell, D. R., Chapman, K. E., Waldron, K. J., Tottey, S., Kendall, S., Cavallaro, G., Andreini, C., Hinds, J., Stoker, N. G., Robinson, N. J., and Cavet, J. S. (2007). Mycobacterial cells have dual nickel-cobalt sensors – sequence relationships and metal sites of metal-responsive repressors are not congruent. J. Biol. Chem. 282, 32298–32310.
Cannon, G. C., Heinhorst, S., and Kerfeld, C. A. (2010). Carboxysomal carbonic anhydrases: structure and role in microbial CO2 fixation. Biochim. Biophys. Acta 1804, 382–392.
Capone, D. G., Zehr, J. P., Paerl, H. W., Bergman, B., and Carpenter, E. J. (1997). Trichodesmium, a globally significant marine cyanobacterium. Science 276, 1221–1229.
Cavet, J. S., Borrelly, G. P. M., and Robinson, N. J. (2003). Zn, Cu and Co in cyanobacteria: selective control of metal availability. FEMS Microbiol. Rev. 27, 165–181.
Cavet, J. S., Meng, W. M., Pennella, M. A., Applehoff, R. J., Giedroc, D. P., and Robinson, N. J. (2002). A nickel-cobalt-sensing ArsR-SmtB family repressor – contributions of cytosol and effector binding sites to metal selectivity. J. Biol. Chem. 277, 38441–38448.
Coale, K. H., Johnson, K. S., Fitzwater, S. E., Gordon, R. M., Tanner, S., Chavez, F. P., Ferioli, L., Sakamoto, C., Rogers, P., Millero, F., Steinberg, P., Nightingale, P., Cooper, D., Cochlan, W. P., Landry, M. R., Constantinou, J., Rollwagen, G., Trasvina, A., and Kudela, R. (1996). A massive phytoplankton bloom induced by an ecosystem-scale iron fertilization experiment in the equatorial Pacific Ocean. Nature 383, 495–501.
Cobbett, C., and Goldsbrough, P. (2002). Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 53, 159–182.
Colvin, R. A., Holmes, W. R., Fontaine, C. P., and Maret, W. (2010). Cytosolic zinc buffering and muffling: their role in intracellular zinc homeostasis. Metallomics 2, 306–317.
Cvetkovic, A., Menon, A. L., Thorgersen, M. P., Scott, J. W., Poole, F. L., Jenney, F. E., Lancaster, W. A., Praissman, J. L., Shanmukh, S., Vaccaro, B. J., Trauger, S. A., Kalisiak, E., Apon, J. V., Siuzdak, G., Yannone, S. M., Tainer, J. A., and Adams, M. W. W. (2010). Microbial metalloproteomes are largely uncharacterized. Nature 466, 779–782.
Davis, S. R., and Cousins, R. J. (2000). Metallothionein expression in animals: a physiological perspective on function. J. Nutr. 130, 1085–1088.
Debelius, B., Forja, J. M., and Lubian, L. M. (2011). Toxicity of copper, nickel and zinc to synechococcus populations from the strait of gibraltar. J. Mar. Syst. 88, 113–119.
Decaria, L., Bertini, I., and Williams, R. J. P. (2010). Zinc proteomes, phylogenetics and evolution. Metallomics 2, 706–709.
Dixon, J. L. (2008). Macro and micro nutrient limitation of microbial productivity in oligotrophic subtropical Atlantic waters. Environ. Chem. 5, 135–142.
Donat, J. R., and Bruland, K. W. (1990). A comparison of 2 voltammetric techniques for determining zinc speciation in northeast Pacific-Ocean waters. Mar. Chem. 28, 301–323.
Dufresne, A., Garczarek, L., and Partensky, F. (2005). Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 6, R14.
Dufresne, A., Ostrowski, M., Scanlan, D. J., Garczarek, L., Mazard, S., Palenik, B. P., Paulsen, I. T., De Marsac, N. T., Wincker, P., Dossat, C., Ferriera, S., Johnson, J., Post, A. F., Hess, W. R., and Partensky, F. (2008). Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9, 16.
Dupont, C. L., Butcher, A., Valas, R. E., Bourne, P. E., and Caetano-Anolles, G. (2010). History of biological metal utilization inferred through phylogenomic analysis of protein structures. Proc. Natl. Acad. Sci. U.S.A. 107, 10567–10572.
Dupont, C. L., and Caetano-Anolles, G. (2010). Mulkidjanian and Galperin: Zn may have constrained evolution during the Proterozoic but not the Archean Reply. Proc. Natl. Acad. Sci. U.S.A. 107, E138.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Eide, D. J. (2006). Zinc transporters and the cellular trafficking of zinc. Biochim. Biophys. Acta 1763, 711–722.
Ellwood, M. J., and van den Berg, C. M. G. (2000). Zinc speciation in the Northeastern Atlantic Ocean. Mar. Chem. 68, 295–306.
Eswar, N., Eramian, D., Webb, B., Shen, M.-Y., and Sali, A. (2008). “Protein structure modeling with MODELLER,” in Methods in Molecular Biology, eds B. Kobe, M. Guss, and T. Huber (Totowa, NJ: Humana Press Inc), 145–159.
Finney, L. A., and O’Halloran, T. V. (2003). Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300, 931–936.
Fukada, T., and Kambe, T. (2011). Molecular and genetic features of zinc transporters in physiology and pathogenesis. Metallomics 3, 662–674.
Fuller, N. J., Tarran, G. A., Yallop, M., Orcutt, K. M., and Scanlan, D. J. (2006). Molecular analysis of picocyanobacterial community structure along an Arabian Sea transect reveals distinct spatial separation of lineages. Limnol. Oceanogr. 51, 2515–2526.
Gabriel, S. E., and Helmann, J. D. (2009). Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J. Bacteriol. 191, 6116–6122.
Gabriel, S. E., Miyagi, F., Gaballa, A., and Helmann, J. D. (2008). Regulation of the Bacillus subtilis yciC gene and insights into the DNA-binding specificity of the zinc-sensing metalloregulator zur. J. Bacteriol. 190, 3482–3488.
Giedroc, D. P., and Arunkumar, A. I. (2007). Metal sensor proteins: nature’s metalloregulated allosteric switches. Dalton Trans. 3107–3120.
Goericke, R., and Repeta, D. J. (1992). The pigments of Prochlorococcus marinus – the presence of divinyl chlorophyll-a and chlorophyll-b in a marine prokaryote. Limnol. Oceanogr. 37, 425–433.
Graham, A. I., Hunt, S., Stokes, S. L., Bramall, N., Bunch, J., Cox, A. G., Mcleod, C. W., and Poole, R. K. (2009). Severe zinc depletion of Escherichia coli roles for high affinity zinc binding by ZinT, zinc transport and zinc-independent proteins. J. Biol. Chem. 284, 18377–18389.
Haas, C. E., Rodionov, D. A., Kropat, J., Malasarn, D., Merchant, S. S., and De Crecy-Lagard, V. (2009). A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10, 470.
Heldal, M., Scanlan, D. J., Norland, S., Thingstad, F., and Mann, N. H. (2003). Elemental composition of single cells of various strains of marine Prochlorococcus and Synechococcus using X-ray microanalysis. Limnol. Oceanogr. 48, 1732–1743.
Higham, D. P., Sadler, P. J., and Scawen, M. D. (1984). Cadmium-resistant Pseudomonas putida synthesizes novel cadmium proteins. Science 225, 1043–1046.
Ho, T. Y., Quigg, A., Finkel, Z. V., Milligan, A. J., Wyman, K., Falkowski, P. G., and Morel, F. M. M. (2003). The elemental composition of some marine phytoplankton. J. Phycol. 39, 1145–1159.
Hong, S. M., Candelone, J. P., Turetta, C., and Boutron, C. F. (1996). Changes in natural lead, copper, zinc and cadmium concentrations in central Greenland ice from 8250 to 149,100 years ago: their association with climatic changes and resultant variations of dominant source contributions. Earth Planet. Sci. Lett. 143, 233–244.
Huckle, J. W., Morby, A. P., Turner, J. S., and Robinson, N. J. (1993). Isolation of a prokaryotic metallothionein locus and analysis of transcriptional control by trace metal ions. Mol. Microbiol. 7, 177–187.
Hunter, K. A., and Boyd, P. (1999). Biogeochemistry of trace metals in the ocean. Mar. Freshw. Res. 50, 739–753.
Irving, H., and Williams, R. J. P. (1953). The stability of transition-metal complexes. J. Chem. Soc. 3192–3210.
Jacquamet, L., Traore, D. A. K., Ferrer, J. L., Proux, O., Testemale, D., Hazemann, J. L., Nazarenko, E., El Ghazouani, A., Caux-Thang, C., Duarte, V., and Latour, J. M. (2009). Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol. Microbiol. 73, 20–31.
Jakuba, R. W., Moffett, J. W., and Dyhrman, S. T. (2008). Evidence for the linked biogeochemical cycling of zinc, cobalt, and phosphorus in the western North Atlantic Ocean. Global Biogeochem. Cycle 22, 13.
Jardillier, L., Zubkov, M. V., Pearman, J., and Scanlan, D. J. (2010). Significant CO2 fixation by small prymnesiophytes in the subtropical and tropical northeast Atlantic Ocean. ISME J. 4, 1180–1192.
Kallifidas, D., Pascoe, B., Owen, G. A., Strain-Damerell, C. M., Hong, H. J., and Paget, M. S. B. (2010). The Zinc-responsive regulator Zur controls expression of the coelibactin gene cluster in Streptomyces coelicolor. J. Bacteriol. 192, 608–611.
Kathuria, S., and Martiny, A. C. (2011). Prevalence of a calcium-based alkaline phosphatase associated with the marine cyanobacterium Prochlorococcus and other ocean bacteria. Environ. Microbiol. 13, 74–83.
Kettler, G. C., Martiny, A. C., Huang, K., Zucker, J., Coleman, M. L., Rodrigue, S., Chen, F., Lapidus, A., Ferriera, S., Johnson, J., Steglich, C., Church, G. M., Richardson, P., and Chisholm, S. W. (2007). Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 3, e231.
Khil, P. P., Obmolova, G., Teplyakov, A., Howard, A. J., Gilliland, G. L., and Camerini-Otero, R. D. (2004). Crystal structure of the Escherichia coli YjiA protein suggests a GTP-dependent regulatory function. Proteins 54, 371–374.
Klaassen, C. D., Liu, J., and Diwan, B. A. (2009). Metallothionein protection of cadmium toxicity. Toxicol. Appl. Pharmacol. 238, 215–220.
Koradi, R., Billeter, M., and Wuthrich, K. (1996). MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55.
Krezel, A., and Maret, W. (2006). Zinc-buffering capacity of a eukaryotic cell at physiological pZn. J. Biol. Inorg. Chem. 11, 1049–1062.
Krissinel, E., and Henrick, K. (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797.
Kuhl, M., Chen, M., Ralph, P. J., Schreiber, U., and Larkum, A. W. D. (2005). A niche for cyanobacteria containing chlorophyll d. Nature 433, 820.
Lane, T. W., Saito, M. A., George, G. N., Pickering, I. J., Prince, R. C., and Morel, F. M. M. (2005). A cadmium enzyme from a marine diatom. Nature 435, 42.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948.
Leao, P. N., Vasconcelos, M., and Vasconcelos, V. M. (2007). Role of marine cyanobacteria in trace metal bioavailability in seawater. Microb. Ecol. 53, 104–109.
Lee, J. G., and Morel, F. M. M. (1995). Replacement of zinc by cadmium in marine phytoplankton. Mar. Ecol. Prog. Ser. 127, 305–309.
Lee, J. W., and Helmann, J. D. (2007). Functional specialization within the Fur family of metalloregulators. Biometals 20, 485–499.
Leipe, D. D., Wolf, Y. I., Koonin, E. V., and Aravind, L. (2002). Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72.
Leszczyszyn, O. I., Evans, C. D., Keiper, S. E., Warren, G. Z. L., and Blindauer, C. A. (2007a). Differential reactivity of individual zinc ions in clusters from bacterial metallothioneins. Inorganica Chim. Acta 360, 3–13.
Leszczyszyn, O. I., Schmid, R., and Blindauer, C. A. (2007b). Toward a property/function relationship for metallothioneins: histidine coordination and unusual cluster composition in a zinc-metal lothionein from plants. Proteins 68, 922–935.
Levy, R., Edelman, M., and Sobolev, V. (2009). Prediction of 3D metal binding sites from translated gene sequences based on remote-homology templates. Proteins 76, 365–374.
Li, W. K. W. (1994). Primary production of prochlorophytes, cyanobacteria, and eukaryotic ultraphytoplankton – measurements from flow cytometric sorting. Limnol. Oceanogr. 39, 169–175.
Lohan, M. C., Crawford, D. W., Purdie, D. A., and Statham, P. J. (2005). Iron and zinc enrichments in the northeastern subarctic Pacific: ligand production and zinc availability in response to phytoplankton growth. Limnol. Oceanogr. 50, 1427–1437.
López-Gomollón, S., Sevilla, E., Bes, M. T., Peleato, M. L., and Fillat, M. F. (2009). New insights into the role of Fur proteins: FurB (All2473) from Anabaena protects DNA and increases cell survival under oxidative stress. Biochem. J. 418, 201–207
Lucarelli, D., Russo, S., Garman, E., Milano, A., Meyer-Klaucke, W., and Pohl, E. (2007). Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J. Biol. Chem. 282, 9914–9922.
Ma, Z., Gabriel, S. E., and Helmann, J. D. (2011). Sequential binding and sensing of Zn(II) by Bacillus subtilis Zur. Nucleic Acids Res. 39, 9130–9138.
Makarova, K. S., Ponomarev, V. A., and Koonin, E. V. (2001). Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2, R33.
Mann, E. L., Ahlgren, N., Moffett, J. W., and Chisholm, S. W. (2002). Copper toxicity and cyanobacteria ecology in the Sargasso Sea. Limnol. Oceanogr. 47, 976–988.
Maret, W. (2009). Molecular aspects of human cellular zinc homeostasis: redox control of zinc potentials and zinc signals. Biometals 22, 149–157.
Maret, W. (2011). Redox biochemistry of mammalian metallothioneins. J. Biol. Inorg. Chem. 16, 1079–1086.
Margoshes, M., and Vallee, B. L. (1957). A cadmium protein from equine kidney cortex. J. Am. Chem. Soc. 79, 4813–4814.
Markowitz, V. M., Chen, I. M. A., Palaniappan, K., Chu, K., Szeto, E., Grechkin, Y., Ratner, A., Anderson, I., Lykidis, A., Mavromatis, K., Ivanova, N. N., and Kyrpides, N. C. (2010). The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 38, D382–D390.
Moore, L. R., Ostrowski, M., Scanlan, D. J., Feren, K., and Sweetsir, T. (2005). Ecotypic variation in phosphorus acquisition mechanisms within marine picocyanobacteria. Aquat. Microb. Ecol. 39, 257–269.
Moore, L. R., Rocap, G., and Chisholm, S. W. (1998). Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393, 464–467.
Morel, F. M. M. (2008). The co-evolution of phytoplankton and trace element cycles in the oceans. Geobiology 6, 318–324.
Morel, F. M. M., Reinfelder, J. R., Roberts, S. B., Chamberlain, C. P., Lee, J. G., and Yee, D. (1994). Zinc and carbon co-limitation of marine phytoplankton. Nature 369, 740–742.
Mulkidjanian, A. Y., and Galperin, M. Y. (2009). On the origin of life in the zinc world. 2. Validation of the hypothesis on the photosynthesizing zinc sulfide edifices as cradles of life on Earth. Biol. Direct 4, 37.
Napolitano, M., Rubio, M. A., Santamaría-Gómez, J., Olmedo-Verd, E., Robinson, N. J., and Luque, I. (2012). Characterization of the response to zinc-deficiency in the cyanobacterium Anabaena sp. PCC 7120. J. Bacteriol.
Novichkov, P. S., Laikova, O. N., Novichkova, E. S., Gelfand, M. S., Arkin, A. P., Dubchak, I., and Rodionov, D. A. (2010). RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38, D111–D118.
Ochsner, U. A., Vasil, A. I., and Vasil, M. L. (1995). Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J. Bacteriol. 177, 7194–7201.
Ohkubo, S., Miyashita, H., Murakami, A., Takeyama, H., Tsuchiya, T., and Mimuro, M. (2006). Molecular detection of epiphytic Acaryochloris spp. on marine macroalgae. Appl. Environ. Microbiol. 72, 7912–7915.
Olafson, R. W., Abel, K., and Sim, R. G. (1979). Prokaryotic metallothionein – preliminary characterization of a blue green alga heavy metal-binding protein. Biochem. Biophys. Res. Commun. 89, 36–43.
Olafson, R. W., Mccubbin, W. D., and Kay, C. M. (1988). Primary- and secondary-structural analysis of a unique prokaryotic metallothionein from a Synechococcus sp. cyanobacterium. Biochem. J. 251, 691–699.
Osman, D., and Cavet, J. S. (2010). Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat. Prod. Rep. 27, 668–680.
Ostrowski, M., Mazard, S., Tetu, S. G., Phillippy, K., Johnson, A., Palenik, B., Paulsen, I. T., and Scanlan, D. J. (2010). PtrA is required for coordinate regulation of gene expression during phosphate stress in a marine Synechococcus. ISME J. 4, 908–921.
Outten, C. E., and O’Halloran, T. V. (2001). Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492.
Owen, G. A., Pascoe, B., Kallifidas, D., and Paget, M. S. B. (2007). Zinc-responsive regulation of alternative ribosomal protein genes in Streptomyces coelicolor involves Zur and σR. J. Bacteriol. 189, 4078–4086.
Palenik, B., Brahamsha, B., Larimer, F. W., Land, M., Hauser, L., Chain, P., Lamerdin, J., Regala, W., Allen, E. E., Mccarren, J., Paulsen, I., Dufresne, A., Partensky, F., Webb, E. A., and Waterbury, J. (2003). The genome of a motile marine Synechococcus. Nature 424, 1037–1042.
Palenik, B., Ren, Q. H., Dupont, C. L., Myers, G. S., Heidelberg, J. F., Badger, J. H., Madupu, R., Nelson, W. C., Brinkac, L. M., Dodson, R. J., Durkin, A. S., Daugherty, S. C., Sullivan, S. A., Khouri, H., Mohamoud, Y., Halpin, R., and Paulsen, I. T. (2006). Genome sequence of Synechococcus CC9311: insights into adaptation to a coastal environment. Proc. Natl. Acad. Sci. U.S.A. 103, 13555–13559.
Panina, E. M., Mironov, A. A., and Gelfand, M. S. (2003). Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. U.S.A. 100, 9912–9917.
Partensky, C., and Garczarek, L. (2003). “The photosynthetic apparatus of chlorophyll b- and d- containing oxychlorobacteria,” in Photosynthesis in Algae, Vol. 14, eds A. W. D. Larkum, S. E. Douglas, and J. A. Raven (Dordrecht: Kluwer Academic Publishers), 29–62.
Partensky, F., and Garczarek, L. (2010). Prochlorococcus: advantages and limits of minimalism. Ann. Rev. Mar. Sci. 2, 305–331.
Partensky, F., Hess, W. R., and Vaulot, D. (1999). Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63, 106–127.
Patzer, S. I., and Hantke, K. (2000). The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275, 24321–24332.
Pearson, R. G. (1990). Hard and soft acids and bases – the evolution of a chemical concept. Coord. Chem. Rev. 100, 403–425.
Peroza, E. A., Schmucki, R., Guntert, P., Freisinger, E., and Zerbe, O. (2009). The βE-domain of wheat E-c-1 metallothionein: a metal-binding domain with a distinctive structure. J. Mol. Biol. 387, 207–218.
Pohl, E., Haller, J. C., Mijovilovich, A., Meyer-Klaucke, W., Garman, E., and Vasil, M. L. (2003). Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47, 903–915.
Quigg, A., Finkel, Z. V., Irwin, A. J., Rosenthal, Y., Ho, T. Y., Reinfelder, J. R., Schofield, O., Morel, F. M. M., and Falkowski, P. G. (2003). The evolutionary inheritance of elemental stoichiometry in marine phytoplankton. Nature 425, 291–294.
Quigg, A., Irwin, A. J., and Finkel, Z. V. (2011). Evolutionary inheritance of elemental stoichiometry in phytoplankton. Proc. R. Soc. Lond. B Biol. Sci. 278, 526–534.
Raymond, J., and Blankenship, R. E. (2004). Biosynthetic pathways, gene replacement and the antiquity of life. Geobiology 2, 199–203.
Robinson, N. J., Gupta, A., Fordham-Skelton, A. P., Croy, R. R. D., Whitton, B. A., and Huckle, J. W. (1990). Prokaryotic metallothionein gene characterization and expression: chromosome crawling by ligation-mediated PCR. Proc. R. Soc. Lond. B Biol. Sci. 242, 241–247.
Robinson, N. J., Whitehall, S. K., and Cavet, J. S. (2001). Microbial metallothioneins. Adv. Microb. Physiol. 44, 183–213.
Saito, M. A., Goepfert, T. J., and Ritt, J. T. (2008). Some thoughts on the concept of colimitation: three definitions and the importance of bioavailability. Limnol. Oceanogr. 53, 276–290.
Saito, M. A., Moffett, J. W., Chisholm, S. W., and Waterbury, J. B. (2002). Cobalt limitation and uptake in Prochlorococcus. Limnol. Oceanogr. 47, 1629–1636.
Saito, M. A., Rocap, G., and Moffett, J. W. (2005). Production of cobalt binding ligands in a Synechococcus feature at the Costa Rica upwelling dome. Limnol. Oceanogr. 50, 279–290.
Saito, M. A., Sigman, D. M., and Morel, F. M. M. (2003). The bioinorganic chemistry of the ancient ocean: the co-evolution of cyanobacterial metal requirements and biogeochemical cycles at the Archean-Proterozoic boundary? Inorganica Chim. Acta 356, 308–318.
Scanlan, D. J. (2003). Physiological diversity and niche adaptation in marine Synechococcus. Adv. Microb. Physiol. 47, 1–64.
Scanlan, D. J., Ostrowski, M., Mazard, S., Dufresne, A., Garczarek, L., Hess, W. R., Post, A. F., Hagemann, M., Paulsen, I., and Partensky, F. (2009). Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73, 249–299.
Schulz, K. G., Zondervan, I., Gerringa, L. J. A., Timmermans, K. R., Veldhuis, M. J. W., and Riebesell, U. (2004). Effect of trace metal availability on coccolithophorid calcification. Nature 430, 673–676.
Shapiro, L. P., and Haugen, E. M. (1988). Seasonal distribution and temperature tolerance of Synechococcus in Boothbay Harbor, Maine. Estuar. Coast. Shelf Sci. 26, 517–525.
Sheikh, M. A., and Taylor, G. L. (2009). Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol. Microbiol. 72, 1208–1220.
Shi, J., Lindsay, W. P., Huckle, J. W., Morby, A. P., and Robinson, N. J. (1992). Cyanobacterial metallothionein gene expressed in Escherichia coli. Metal-binding properties of the expressed protein. FEBS Lett. 303, 159–163.
Shin, J. H., Jung, H. J., An, Y. J., Cho, Y. B., Cha, S. S., and Roe, J. H. (2011). Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc. Natl. Acad. Sci. U.S.A. 108, 5045–5050.
Soding, J., Biegert, A., and Lupas, A. N. (2005). The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248.
Stajich, J. E., Block, D., Boulez, K., Brenner, S. E., Chervitz, S. A., Dagdigian, C., Fuellen, G., Gilbert, J. G., Korf, I., Lapp, H., Lehvaslaiho, H., Matsalla, C., Mungall, C. J., Osborne, B. I., Pocock, M. R., Schattner, P., Senger, M., Stein, L. D., Stupka, E., Wilkinson, M. D., and Birney, E. (2002). The Bioperl toolkit: perl modules for the life sciences. Genome Res. 12, 1611–1618.
Sunda, W. G., and Huntsman, S. A. (1992). Feedback Interactions between zinc and phytoplankton in seawater. Limnol. Oceanogr. 37, 25–40.
Sunda, W. G., and Huntsman, S. A. (1995). Cobalt and zinc interreplacement in marine phytoplankton: biological and geochemical implications. Limnol. Oceanogr. 40, 1404–1417.
Sunda, W. G., and Huntsman, S. A. (2000). Effect of Zn, Mn, and Fe on Cd accumulation in phytoplankton: implications for oceanic Cd cycling. Limnol. Oceanogr. 45, 1501–1516.
Sunda, W. G., and Huntsman, S. A. (2005). Effect of CO2 supply and demand on zinc uptake and growth limitation in a coastal diatom. Limnol. Oceanogr. 50, 1181–1192.
Sunda, W. G., Price, N. M., and Morel, F. M. M. (2005). “Trace metal ion buffers and their use in culture studies,” in Algal Culturing Techniques, ed. R. A. Andersen (Burlington, MA: Academic Press), 35–63.
Swingley, W. D., Chen, M., Cheung, P. C., Conrad, A. L., Dejesa, L. C., Hao, J., Honchak, B. M., Karbach, L. E., Kurdoglu, A., Lahiri, S., Mastrian, S. D., Miyashita, H., Page, L., Ramakrishna, P., Satoh, S., Sattley, W. M., Shimada, Y., Taylor, H. L., Tomo, T., Tsuchiya, T., Wang, Z. T., Raymond, J., Mimuro, M., Blankenship, R. E., and Touchman, J. W. (2008). Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acarylochloris marina. Proc. Natl. Acad. Sci. U.S.A. 105, 2005–2010.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Thelwell, C., Robinson, N. J., and Turner-Cavet, J. S. (1998). An SmtB-like repressor from Synechocystis PCC 6803 regulates a zinc exporter. Proc. Natl. Acad. Sci. U.S.A. 95, 10728–10733.
Thuroczy, C. E., Boye, M., and Losno, R. (2010). Dissolution of cobalt and zinc from natural and anthropogenic dusts in seawater. Biogeosciences 7, 1927–1936.
Tottey, S., Waldron, K. J., Firbank, S. J., Reale, B., Bessant, C., Sato, K., Cheek, T. R., Gray, J., Banfield, M. J., Dennison, C., and Robinson, N. J. (2008). Protein-folding location can regulate manganese binding versus copper- or zinc-binding. Nature 455, 1138–1142.
Tripp, H. J., Bench, S. R., Turk, K. A., Foster, R. A., Desany, B. A., Niazi, F., Affourtit, J. P., and Zehr, J. P. (2010). Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature 464, 90–94.
Turner, J. S., Glands, P. D., Samson, A. C. R., and Robinson, N. J. (1996). Zn2+-sensing by the cyanobacterial metallothionein repressor SmtB: different motifs mediate metal-induced protein-DNA dissociation. Nucleic Acids Res. 24, 3714–3721.
Turner, J. S., Morby, A. P., Whitton, B. A., Gupta, A., and Robinson, N. J. (1993). Construction of Zn2+/Cd2+ hypersensitive cyanobacterial mutants lacking a functional metallothionein locus. J. Biol. Chem. 268, 4494–4498.
Waldron, K. J., and Robinson, N. J. (2009). How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 7, 25–35.
Waldron, K. J., Rutherford, J. C., Ford, D., and Robinson, N. J. (2009). Metalloproteins and metal sensing. Nature 460, 823–830.
Wang, Q., Canutescu, A. A., and Dunbrack, R. L. (2008). SCWRL and MolIDE: computer programs for side-chain conformation prediction and homology modeling. Nat. Protoc. 3, 1832–1847.
Waterbury, R. D., Byrne, R. H., Kelly, J., Leader, B., Mcelligott, S., and Russell, R. (1996). Development of an Underwater In-Situ Spectrophotometric Sensor for Seawater pH. Bellingham: Spie – Int Soc Optical Engineering.
Wells, M. L., Kozelka, P. B., and Bruland, K. W. (1998). The complexation of ‘dissolved’ Cu, Zn, Cd and Pb by soluble and colloidal organic matter in Narragansett Bay, RI. Mar. Chem. 62, 203–217.
West, N. J., and Scanlan, D. J. (1999). Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl. Environ. Microbiol. 65, 2585–2591.
Whitton, B. A., and Potts, M. (2000). The Ecology of Cyanobacteria: Their Diversity in Time and Space. Dordrecht: Kluwer Academic Publishers.
Wilhelm, S. W., and Trick, C. G. (1994). Iron-limited growth of cyanobacteria – multiple siderophore production is a common response. Limnol. Oceanogr. 39, 1979–1984.
Williams, R. J. P. (2011). Chemical advances in evolution by and changes in use of space during time. J. Theor. Biol. 268, 146–159.
Williams, R. J. P., and Da Silva, J. J. R. F. (2000). The distribution of elements in cells. Coord. Chem. Rev. 200, 247–348.
Williams, R. J. P., and Da Silva, J. J. R. F. (2001). The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. Oxford: Oxford University Press.
Williams, R. J. P., and Da Silva, J. J. R. F. (2003). Evolution was chemically constrained. J. Theor. Biol. 220, 323–343.
Wiramanaden, C. I. E., Cullen, J. T., Ross, A. R. S., and Orians, K. J. (2008). Cyanobacterial copper-binding ligands isolated from artificial seawater cultures. Mar. Chem. 110, 28–41.
Xu, Y., Feng, L., Jeffrey, P. D., Shi, Y. G., and Morel, F. M. M. (2008). Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms. Nature 452, U56–U53.
Zehr, J. P., Bench, S. R., Mondragon, E. A., McCarren, J., and Delong, E. F. (2007). Low genomic diversity in tropical oceanic N2–fixing cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 104, 17807–17812.
Zeitoun-Ghandour, S., Charnock, J., Hodson, M. E., Leszczyszyn, O. I., Blindauer, C. A., and Stürzenbaum, S. R. (2010). The two Caenorhabditis elegans metallothioneins (CeMT-1 and CeMT-2) discriminate between essential zinc and toxic cadmium. FEBS J. 277, 2531–2542.
Zeitoun-Ghandour, S., Leszczyszyn, O. I., Blindauer, C. A., Geier, F. M., Bundy, J. G., and Stürzenbaum, S. R. (2011). C. elegans metallothioneins: response to and defence against ROS toxicity. Mol. Biosyst. 7, 2397–2406.
Zwirglmaier, K., Jardillier, L., Ostrowski, M., Mazard, S., Garczarek, L., Vaulot, D., Not, F., Massana, R., Ulloa, O., and Scanlan, D. J. (2008). Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10, 147–161.
Keywords: zinc limitation, Zur, COG0523, SmtA, BmtA
Citation: Barnett JP, Millard A Ksibe AZ, Scanlan DJ, Schmid R and Blindauer CA (2012) Mining genomes of marine cyanobacteria for elements of zinc homeostasis. Front. Microbio. 3:142. doi: 10.3389/fmicb.2012.00142
Received: 21 December 2011; Accepted: 25 March 2012;
Published online: 11 April 2012.
Edited by:
Martha Gledhill, University of Southampton, UKReviewed by:
Mak Saito, Woods Hole Oceanographic Institution, USANigel John Robinson, Durham University, UK
Jennifer Cavet, University of Manchester, UK
Copyright: © 2012 Barnett, Millard, Ksibe, Scanlan, Schmid and Blindauer. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Claudia Andrea Blindauer, Department of Chemistry, University of Warwick, Coventry CV4 7AL, UK. e-mail: c.blindauer@warwick.ac.uk
