- Division of Molecular Virology, Institute of Immunological Science, Hokkaido University, Sapporo, Japan
Retroviruses have evolved mechanisms for transporting their intron-containing RNAs (including genomic and messenger RNAs, which encode virion components) from the nucleus to the cytoplasm of the infected cell. Human retroviruses, such as human immunodeficiency virus (HIV) and human T cell leukemia virus type 1 (HTLV-1), encode the regulatory proteins Rev and Rex, which form a bridge between the viral RNA and the export receptor CRM1. Recent studies show that these transport systems are not only involved in RNA export, but also in the encapsidation of genomic RNA; furthermore, they influence subsequent events in the cytoplasm, including the translation of the cognate mRNA, transport of Gag proteins to the plasma membrane, and the formation of virus particles. Moreover, the mode of interaction between the viral and cellular RNA transport machinery underlies the species-specific propagation of HIV-1 and HTLV-1, forming the basis for constructing animal models of infection. This review article discusses recent progress regarding these issues.
Almost all eukaryote cellular messenger RNAs (mRNAs) undergo splicing to remove introns, and only fully spliced mRNAs are able to reach the cytoplasm (Legrain and Rosbash, 1989; Fischer et al., 1994). The nuclear retention of unspliced, or incompletely spliced, mRNAs is strictly regulated because incompletely spliced mRNAs yield proteins that are non-functional, or even deleterious to the cell. However, the expression of retroviral genes is a notable exception. Retroviruses are subdivided into two categories, simple and complex, according to their genome structure (Goff, 2001). Simple retroviruses encode mainly structural genes, whereas complex retroviruses harbor several accessory genes that play important roles in their replication and pathogenesis. All retroviruses share a common genome structure comprising long terminal repeat (LTR) sequences at either end and centrally located gag, pol, and env genes, which encode the component proteins of the virions (Figure 1). All except foamy viruses (Yu et al., 1996) utilize the 5′-LTR as the sole promoter, resulting in the synthesis of a single full-length viral RNA as the primary transcript. Since translation preferentially starts at the first AUG codon (methionine) in eukaryotes (Kozak, 1992), viruses must position the initiation codons for all their genes (which are located within the primary transcript) close enough to the 5′-cap to ensure appropriate and efficient translation. Retroviruses utilize alternative splicing of the full-length transcript to produce the appropriate mRNAs. This means that the introns of the full-length primary transcript encode the gag gene, for instance. The mRNA must then be exported and translated in the cytoplasm. In addition, viral genomic RNAs (gRNAs), which are apparently indistinguishable to gag mRNA, must be exported to the cytoplasm for viral replication. Therefore, retroviruses have evolved the means to circumvent cellular restrictions on the export of their intron-containing RNAs. Studies regarding this issue have not only shed light on the mechanisms underlying viral replication, but have also led to the discovery of the nucleocytoplasmic transport systems in higher eukaryotes. Moreover, recent findings indicate that the correct mode of nucleocytoplasmic transport of viral RNA and proteins forms the basis of subsequent cytoplasmic events involved in efficient viral replication and, in some cases, forms the basis for species-specific propagation.
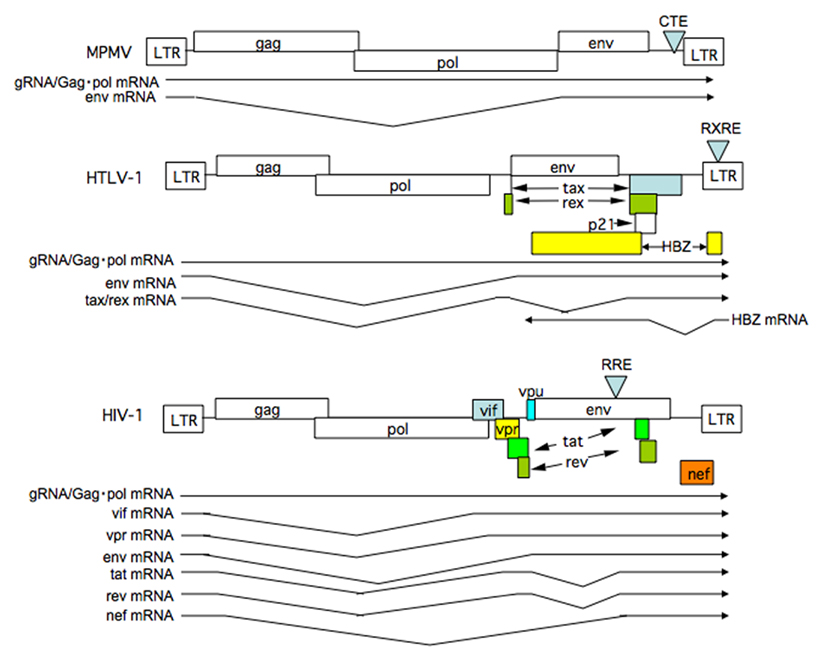
Figure 1. Structure of retrovirus genomes and RNAs synthesized. The genome structure of simple retrovirus exemplified by MPMV and complex retroviruses including HTLV-1 and HIV-1 are illustrated. Splicing patterns of viral mRNAs are also shown. Note complex retroviruses, but not a simple retrovirus, synthesize doubly spliced mRNAs.
Transport of HTLV-1 RNA
Human T cell leukemia virus type 1 (HTLV-1) is the causative agent of human adult T cell leukemia (ATL), a chronic progressive neurological disorder termed HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP; Hinuma et al., 1981; Kalyanaraman et al., 1982; Osame et al., 1986; Jacobson et al., 1988), and other diseases. HTLV-1 encodes an unspliced, intron-containing 9 kb RNA, which encodes Gag and Gag–Pol, a singly spliced 4 kb mRNA encoding Env, and multiply spliced, intron-free 2 kb mRNAs encoding Tax and Rex (Green and Chen, 2001). Recently, the HTLV-1 leucine zipper factor (HBZ) gene was identified, which is encoded by a complimentary strand of the HTLV-1 genome, driven by a promoter residing in the 3′ LTR (Figure 1). Tax and HBZ are implicated in oncogenesis (Satou et al., 2006; Matsuoka and Jeang, 2011). Historically, Rex was the first protein to be identified as a factor that induces the cytoplasmic expression of intron-containing 9 and 4 kb RNAs (Inoue et al., 1986; Hidaka et al., 1988). Rex binds directly to viral RNAs containing Rex response element (RXRE) located in the 3′ LTR. RXRE, which consists of a highly ordered stem-and-loop structure, functions as a Rex binding site (Ballaun et al., 1991; Bogerd et al., 1991). Since all HTLV-1 RNAs contain RXRE, Rex also enhances transport of fully spliced 2 kb mRNA, although this RNA can be transported via the normal route for cellular mRNAs that do not require Rex (Bai et al., 2012). The sequence elements that retain HTLV-1 unspliced RNAs in the nucleus have been identified in the pol–env and LTR regions of the HTLV-1 genome (Saiga et al., 1997; King et al., 1998).
An important property of Rex, which is essential for its role, is its ability to dynamically shuttle between the cytoplasm and nucleus, although Rex is predominantly localized in the nucleus and nucleolus (Bogerd et al., 1996; Kim et al., 1996; Siomi et al., 1988). Rex is synthesized in the cytoplasm and then imported into the nucleus. In the nucleus, Rex directly binds to RXRE, after which it escorts the viral RNAs into the cytoplasm (Bogerd et al., 1991). Finally, Rex is released from its cognate RNAs for recycling. Extensive mutational analyses show that Rex contains functional domains that are essential for its function (Figure 2). The basic domain is rich in arginine residues and maps between amino acids (aa) 1–19 in the 189 aa Rex protein. This domain serves as a nuclear/nucleolar localization signal (NLS), and foreign proteins containing these residues are localized to the nucleus/nucleolus (Siomi et al., 1988). Importantly, the same region mediates binding to RXRE, thereby determining its specific target RNA (Bogerd et al., 1991). The second essential domain, which maps between aa 84 and 94 of Rex, is a leucine-rich activation domain that functions as a nuclear export signal (NES; Bogerd et al., 1996). The third important property of Rex is multimerization. The domain responsible for this was first ascribed to a 60–70 aa region of Rex (Weichselbraun et al., 1992); however, subsequent in vivo two-hybrid assays revealed that a 100–120 aa region was also involved in multimerization (Heger et al., 1998). Multimerization of Rex on its cognate RNA is generally thought to be critical for its ability to export viral RNAs, although it is not required for export of the Rex protein itself (Heger et al., 1998). Although shuttling proteins have now been identified in a wide range of other viral and cellular proteins, multimerization may distinguish Rex from other shuttling proteins that are not involved in RNA export since their multimerization is not necessarily required for their functioning (Weichselbraun et al., 1992; Bogerd and Greene, 1993; Heger et al., 1998).
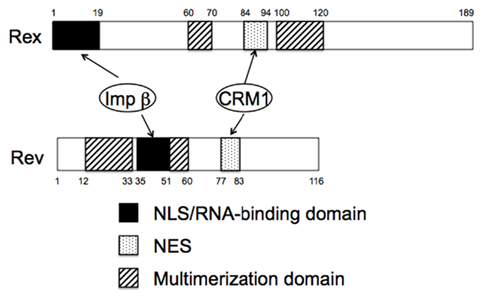
Figure 2. Domain structures of HTLV-1 Rex and HIV-1 Rev proteins. Note both proteins functionally similar domains and use the same cellular cofactors.
A detailed understanding of Rex shuttling was obtained from studies that identified the cellular factors binding to the functional domains, and the cellular machinery involved in protein transport into and out of the nucleus (Figure 3). Transport of all macromolecules occurs via nuclear pore complexes (NPCs), which comprise at least 50 different proteins, termed nucleoporins (Doye and Hurt, 1997). The import of proteins containing an NLS is mediated by the importin family of import receptors, which have affinity for nucleoporins, and the GTP-bound status of Ran (RanGTP; Mattaj and Englmeier, 1998). Rex directly binds to importin β through its NLS (Palmeri and Malim, 1999). Translocation through the pore is thought to be facilitated by sequential direct interactions between importin β and various nucleoporins. Ran is a small GTPase that can exist in either the GTP or GDP-bound state. RanGTP reflects the nuclear localization of chromatin binding protein, RCC1, which specifically catalyzes the exchange of guanine nucleotides on Ran, and RanGDP, the predominant form of Ran in the cytoplasm, reflects the cytoplasmic location of a GTPase-activating protein, RanGAP. One role of RanGTP is to promote cargo release in the nucleus by dissociating the imported receptor–cargo complexes; RanGDP has no affinity for importin β (Mattaj and Englmeier, 1998). Since the NLS of Rex overlaps with its RNA binding domains, binding to importin β in the cytoplasm may facilitate the release of viral RNAs from Rex (Bogerd et al., 1991; Siomi et al., 1988; Palmeri and Malim, 1999).
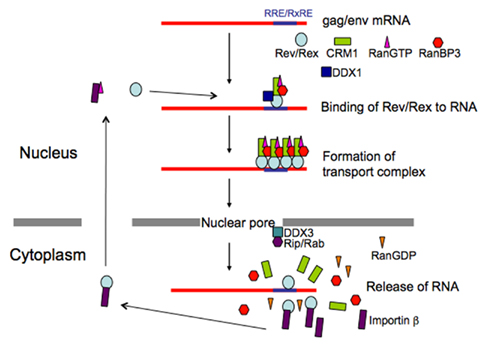
Figure 3. Schematic presentation of transport of HTLV-1 and HIV-1 RNAs. Action mechanism of viral transporter Rex/Rev and involvement of their cellular cofactors are illustrated.
The cellular cofactor that interacts with the NES of Rex is human (h)CRM1. CRM1 was originally identified as binding the NES of the human immunodeficiency virus (HIV)-1 Rev protein. Sequence similarity to importin β suggested that hCRM1 was an export receptor (Fornerod et al., 1997a; Fukuda et al., 1997). Subsequently, hCRM1 was shown to form a specific complex with NES; however, unlike the Rex-importin β complex, this only occurred in the presence of RanGTP (Fornerod et al., 1997a). The association between Rex and hCRM1 was confirmed in vivo using a two-hybrid assay (Hakata et al., 1998). Moreover, CRM1 directly binds to Ran-binding protein 3 (RanBP3), which binds to RCC1 in a Ran-dependent manner and increases the nucleotide exchange activity of RCC1, resulting in high local concentrations of RanGTP. Consequently, RanBP3 acts as a scaffold protein through which the components of the export complex are concentrated around the RCC1 site, thereby promoting complex assembly. RanBP3 continues to interact with hCRM1 to stabilize the hCRM1–substrate–RanGTP complex, thereby forming a quaternary complex (Englmeier et al., 2001; Nemergut et al., 2002). Since CRM1 associates with various nucleoporins (Fornerod et al., 1997b), the Rex–hCRM1–RanGTP–RanBP3 complex is translocated to the cytoplasm across the NPC; this occurs via possible hydrophobic interactions between hCRM1 and nucleoporins. Following this, the RanGTP within the complex is converted to RanGDP by RanGAP and RanBP1 (and probably RanBP2), leading to the dissociation of the complex and release of the substrate into the cytoplasm (Askjaer et al., 1999; Kehlenbach et al., 1999).
An important question is whether cellular cofactors, such as hCRM1, are involved in the multimerization of Rex proteins (Hakata et al., 1998). Therefore, we examined the multimerization of Rex proteins using a two-hybrid assay. First, we found that a Rex mutant, RexM90 (which harbors a mutation in the NES), lost the ability to bind to hCRM1 and to form multimers. Second, we found that the RNA transport activity of RexM64, which harbors a mutation in the multimerization domain, was partially restored by the overexpression of hCRM1, and this coincided with the partial restoration of multimerization. Third, a dominant-negative (DN) mutant of Rex, termed TAgRexM64, which sequesters Rex cofactors (Katahira et al., 1995), abrogated the multimerization and activity of the wild-type Rex protein. Both functions were simultaneously restored by the overexpression of hCRM1. Taken together, these results suggest that the multimerization of Rex in vivo requires both the intrinsic ability of Rex to form multimers, and the hCRM1 protein. Moreover, these results also suggest that the multimerization of Rex is not a prerequisite for interaction with hCRM1 but, rather, that Rex initially interacts with hCRM1, leading to the multimerization of Rex proteins in a step that is favored by its intrinsic capacity to form oligomers on RXRE. This, in turn, would favor a structure that facilitates interactions with other hCRM1 molecules on the RNA (Hakata et al., 1998). Comparison of human and rat CRM1 (rCRM1) mapped the CRM1 domain that induces Rex multimerization to the central region, particularly a region containing two residues (aa 411 and 414; Hakata et al., 2003); this domain is different from the region spanning aa 566–720 that is involved in binding to the NES (Ossareh-Nazari and Dargemont, 1999). These data suggest that CRM1 not only functions as an export receptor but also participates in the formation of the RNA export complex through a high-order interaction with Rex. Curiously, the CRM1 region responsible for the interaction with RanBP3, which comprises four residues (aa 411, 414, 478, and 484), and the region responsible for Rex multimerization form an overlapping domain (Hakata et al., 2003).
RNA Export Step as a Major Species-Specific Barrier for HTLV-1
Appropriate animal models of HTLV-1 infection would allow us to analyze the pathogenesis and oncogenesis of HTLV-1-associated diseases, which could lead to the development of therapeutic and preventative measures. HTLV-1 is able to infect experimental animals such as monkeys, rabbits, and rats (Akagi et al., 1985; Nakamura et al., 1987; Oka et al., 1992). The utility of monkey and rabbit models is limited, however, because of the difficulties in breeding a large number of these animals and the lack of inbred strains. Consequently, it would be more convenient to use small animal models. In particular, rat models have been extensively developed to study HTLV-1-associated diseases (Ishiguro et al., 1992; Ohashi et al., 2000). However, although the current rat models have certainly allowed us to understand HTLV-1-associated diseases better, they are still incomplete because of the poor replication of HTLV-1 in rats.
Human T cell leukemia virus type 1 can infect a number of rat cell types, which indicates that rat cells possess the receptors for viral attachment and penetration (Li et al., 1996; Sutton and Littman, 1996). Thus, the blockade of viral propagation within the rat cells must occur during subsequent steps. Identification of the blocking step and the responsible host factor(s) could lead to the construction of transgenic rats that express the critical human factor(s), which are highly susceptible to HTLV-1 infection.
The first hint of a blocking step came in a report showing that the viral mRNAs encoding Gag and Env proteins are produced only at low levels in rat cells despite the fact that the mRNA for Tax/Rex protein is abundant (Koya et al., 1999). This observation led us to hypothesize that Rex may function poorly in rat cells. Indeed, we found that the activity of Rex in rat cells is quite low compared with that in human cells. As the functions of Rex depend largely on the CRM1 protein, we examined whether rCRM1 can act as a cofactor for Rex activity as it does in human cells, although CRM1 is highly conserved from yeast to mammalians, and hCRM1 can function in yeast cells as an export receptor. Actually, only 24 out of 1027 amino acids are different between rat and human CRM1 (Hakata et al., 2003). Also, we found that both rCRM1 and hCRM1 could bind to and export the Rex protein to the cytoplasm with similar efficiency. However, unlike hCRM1, rCRM1 could not efficiently support Rex function because of its poor ability to induce the Rex–Rex interaction required for RNA export into the cytoplasm (Figure 4). These results underline the absolute requirement of hCRM1 for Rex multimerization. It was also concluded that the poor ability of rCRM1 to act as a cofactor for Rex is responsible for the poor replication of HTLV-1 in rats (Hakata et al., 2001). This notion was further supported by a study showing that a HTLV-1-transformed rat CD4+ T cell line, which was a poor producer of HTLV-1, efficiently synthesized functional Env and Gag proteins, which were then fully processed, and produced infectious HTLV-1 progeny viruses at the level comparable to HTLV-1-transformed human T cells when it was transduced with a retroviral vector expressing hCRM1 at physiological levels (Zhang et al., 2006).
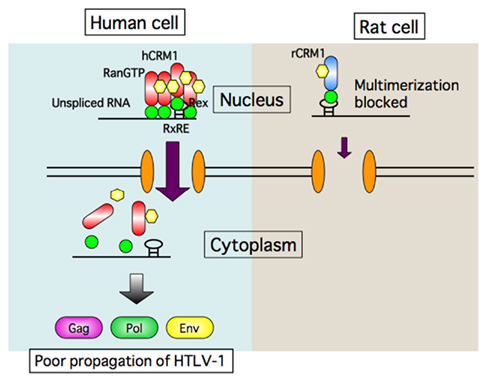
Figure 4. Schematic presentation of transport defect of HTLV-1 RNA caused by rat CRM1. Rex proteins multimerize with help of human CRM1 on HTLV-1 RNA to form transport complex in human cells, and then export it to the cytoplasm. In contrast Rex proteins can bind to rat CRM1, but not multimerize. Consequently Rex is not able to export HTLV-1 RNA in rat cells.
Transgenic (Tg) rats were constructed using an artificial bacterial chromosome clone containing the entire regulatory and coding regions of the CRM1 gene (Figure 5), since the expression of the CRM1 gene is elaborately regulated by a protein kinase C pathway during lymphocyte activation, initially in a post-transcriptional and subsequently in a transcriptional manner. Consequently, Tg rats expressed hCRM1 protein in a manner similar to that of intrinsic rCRM1 in various organs. HTLV-1-infected T cell lines derived from these Tg rats produced 100- to 10,000-fold more HTLV-1 than T cells from wild-type rats, and the absolute levels of HTLV-1 were similar to those produced by human T cells. We also observed increased dissemination of HTLV-1 into the thymuses of Tg rats after intraperitoneal inoculation, although the proviral loads were low in both wild-type and Tg rats (Takayanagi et al., 2007). Furthermore, rat regulatory T (Treg) cells were preferentially transformed by HTLV-1 (Takayanagi et al., 2009), as is the case for human Treg cells (Chen et al., 2006). Therefore, the Tg rat model forms the basis for an improved animal model for HTLV-1 infection.
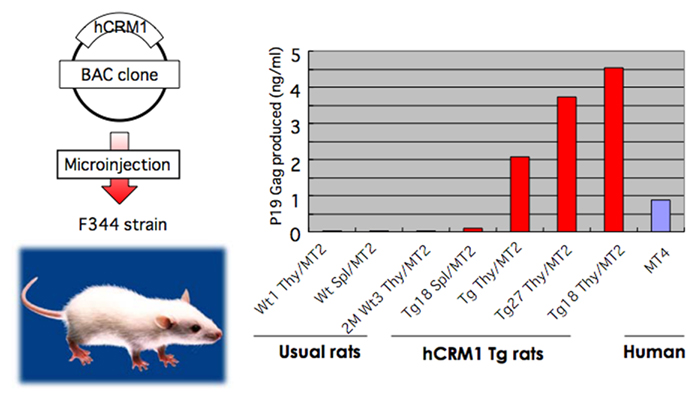
Figure 5. Restoration of HTLV-1 production in T cells prepared from human CRM1 transgenic rats. The BAC clone harboring entire human CRM1 genome was microinjected to eggs of rat F344 strain to construct transgenic rats. T cells prepared from normal and hCRM1 transgenic rats were transformed by mixed culture with mitomycin-treated HTLV-1+ human T cell MT2. Then the amounts of p19 Gag protein in the culture medium were quantified by ELISA.
RNA Transport of HIV
Human immunodeficiency virus-1 belongs to the subfamily of complex retroviruses and, like HTLV-1, also encodes accessory genes besides gag and env; however, HIV-1 shows more complex patterns of mRNA expression. HIV-1 produces one unspliced 9 kb RNA encoding Gag and Gag–Pol, five singly spliced 4 kb mRNAs encoding Vif, Vpr, Vpu, and Env, and 16 multiply spliced 2 kb mRNAs encoding Tat, Rev, and Nef (Figure 1; Freed and Martin, 2001). As is the case for HTLV-1 Rex, the 9- and 4-kb viral RNAs of HIV-1 require the viral protein Rev for export (Cullen, 1991). However, expression of 2 kb RNAs is not augmented by Rev, since the Rev response element (RRE), an approximately 300 nucleotide region with a highly ordered stem-and-loop structure that functions as a Rev binding site, is located in the env gene rather than the 3′ LTR (Malim et al., 1990; Mann et al., 1994). Consequently, during the early phase of infection when Rev expression is low, proteins encoded by the 2-kb class of mRNAs are expressed. During the late phase, when Rev is expressed in sufficient quantities, the remaining viral proteins are produced (Kim et al., 1989), including the virion constituents and infectivity enhancing and virion-releasing factors, all of which work toward the efficient production and dissemination of progeny virus.
Rev is a functional homolog of Rex, although Rev and Rex do not show any homology at the amino acid level. However, they do have similar domain structures and function in a similar manner (Figures 2 and 3), as exemplified by the fact that Rex can replace Rev (Ahmed et al., 1990). The basic domain rich in arginine residues, which maps between aa 35 and 50 in the 116 aa Rev protein, serves as an NLS, which recruits importin β and binds to the RRE (Daly et al., 1989; Malim et al., 1991; Tiley et al., 1992; Madore et al., 1994). Replacing the amino-terminal domain of the Rex protein with the RNA binding domain of the Rev protein yields a chimeric protein that can specifically interact with the RRE (Kubota et al., 1991). A leucine-rich activation domain, which maps between aa 73 and 83, acts as an NES that associates with CRM1 in the presence of RanGTP (Fisher et al., 1995; Wen et al., 1995). In vivo randomization selection experiments identified the consensus sequence, which contains relatively evenly spaced bulky hydrophobic residues (often leucine) that are crucial for NES function (Bogerd et al., 1996; Kim et al., 1996). In vitro studies of the multimerization of recombinant Rev proteins bound to the RRE-containing RNA defined two essential domains: one between aa 12 and 33 and one between aa 46 and 60 (Malim et al., 1991). The C-terminal region of Rev is highly conserved between HIV-1 strains and supports efficient multimerization and subsequent Rev function, probably by stabilizing its conformation. In contrast to Rex, Rev is able to dimerize without CRM1 molecules in vitro, although in vivo two-hybrid assays suggest that CRM1 improves dimerization (Chaitanya and Belasco, 2001; Hakata et al., 2002; Matthew et al., 2008). Early studies suggested that Rev binds initially as a monomer to a single high-affinity Rev binding site within the RRE (Battiste et al., 1996), but recent studies on the tertiary structure of Rev indicate that the dimerization of Rev induces conformational changes in the position of the arginine-rich domain to facilitate RNA binding (Daugherty et al., 2010). RNA bound to the RRE augments further Rev multimerization, recruiting up to eight Rev molecules on the viral RNA, which subsequently recruit several hCRM1 molecules (Mann et al., 1994; Chaitanya and Belasco, 2001). Thus, multimerization, RNA binding to Rev, and Rev–CRM1 interactions are highly cooperative processes. The process is further accelerated by the DEAD (D–E–A–D = Asp–Glu–Ala–Asp) box helicase, DDX1, which is an RNA-dependent ATPase that binds directly to Rev. Knockdown of DDX1 greatly reduces Rev function, supporting the important role of DDX1 (Fang et al., 2004; Robertson-Anderson et al., 2011; Edgcomb et al., 2012). After the transport complex is formed, which comprises HIV-1 RNA, Rev, CRM1, and other cellular components, it moves to the nuclear pores, possibly via the actin filament network (Kimura et al., 2000). Multiple CRM1 molecules, which have affinity for nucleoporins, may be required to pass through the nuclear pore because the transport complex could be longer than the depth of the nuclear pore; thus, the rear portion of the complex is still within the nucleus when the front portion reaches the cytoplasm. CRM1 molecules associated with Rev within the front portion of the ribonucleoprotein complex (in the cytoplasm) will dissociate due to the lack of RanGTP (Fornerod et al., 1997a). Thus, additional CRM1 molecules may be required to associate with the rear part of the RNA complex to enable passage of the whole RNA complex out of the nucleus. Another DEAD box protein, DDX3, which is located outside of the nuclear pore, associates with CRM1 directly and facilitates the passage of the complex through the nuclear pore and the release of RNA into the cytoplasm by inducing conformational changes in the ribonucleoprotein complex (Yedavalli et al., 2004). Additionally, Rip (also named Rab), which indirectly binds to Rev and has affinity for nuclear pores, is thought to function in releasing the viral RNA within the cytoplasm (Sanchez-Velar et al., 2004; Yu et al., 2005).
Suboptimal viral RNA splicing is also important for the function of Rev. Both inefficient branch point region and polypyrimidine tract are thought to be responsible for the low splicing efficiency of the HIV-1 tat/rev intron, which comprises most of the HIV-1 env-coding sequences (Staffa and Cochrane, 1994). Two additional elements, an exon-splicing enhancer (ESE) and an exon-splicing silencer (ESS), both of which modulate the overall efficiency of the 3′ tat–rev splice site, were identified within the 3′ terminal exon (Staffa and Cochrane, 1995). It has been proposed that inefficient splicing causes the retention of unspliced, or partially spliced, mRNAs since incomplete spliceosome formation at the splice sites inhibits export (Chang and Sharp, 1989).
Secondary sequence elements, which retain unspliced HIV-1 RNAs within the nucleus, are termed cis-acting repressive sequences (CRSs) or instability sequences (INSs; Cochrane et al., 1991; Maldarelli et al., 1991; Schneider et al., 1997). When appended in cis to heterologous reporter genes, these elements inhibit their expression. In situ hybridization studies demonstrated discrete localization of unspliced gag and env RNAs. In contrast to fully spliced mRNA, which is dispersed throughout both the nucleus and cytoplasm, the location of unspliced gag and env RNAs does not coincide with splicing factor SC35-containing nuclear speckles. Deletion of the intron sequence containing the CRS resulted in a shift from discrete regional localization within the nucleus to a diffuse localization throughout the cell. On the other hand, mutations that affect splicing efficiency do not alter the sequestration of unspliced RNAs (Seguin et al., 1998). Based on these results, one may hypothesize that CRSs (in conjunction with other elements that cause suboptimal splicing) cause significant amounts of full-length primary RNA, or partially spliced RNA, to accumulate. This RNA is inaccessible to subsequent splicing due to its localization in discrete regions within the nucleus. This sequestration of intron-containing RNAs may be a prerequisite for subsequent export by Rev. Indeed, addition of the RRE to β-globin mRNA, which contains efficient splice sites, does not make it responsive to Rev unless the splice sites are replaced by more inefficient ones, or a CRS is introduced into the mRNA. The finding that hnRNPA1 and a 50-kDa protein associate with the CRS suggests a connection between the CRS and RNA splicing, since hnRNPA1 controls the selection of splice sites (Najera et al., 1999).
RNA Export as a Species-Specific Barrier for HIV-1
Current animal models of HIV-1 disease use non-human primates (Veazey et al., 2005; Koff et al., 2006) or severe combined immunodeficiency (SCID) mice transplanted with human hemopoietic stem cells and fetal tissues (Shultz et al., 2007; Watanabe et al., 2007). These models have made significant contributions to our understanding of lentiviral pathogenesis and have assisted in the development of several therapeutic strategies. However, they also have significant shortcomings, such as limited availability and high cost (for non-human primates), they are permissive only for related retroviruses, and they often show aberrant immune responses. Therefore, new animal models are needed. Rodent models susceptible to infection by human viruses would be ideal, since the inbred strains are well characterized and can be genetically manipulated.
Studies on rodent cell-specific defects in the HIV-1 life cycle have facilitated the identification and characterization of host cell gene products that are essential for viral replication, and these may provide a molecular basis for generating fully permissive small animal models. The major block to HIV-1 replication is at the level of cell entry. It is thought that this hurdle may be overcome by introducing human CD4 and CCR5 into the animal cells, which would serve as receptors for HIV-1. However, Tg mice and rats expressing human CD4 and CCR5 do not appear to support HIV-1 replication (Browning et al., 1997; Keppler et al., 2002; Goffinet et al., 2007), suggesting the presence of additional blocks post-entry.
Studies of the viral transcription activator, Tat, indicate the existence of a profound block at the transcription stage in murine cells (Bieniasz et al., 1998; Garber et al., 1998) and in rat T cells but, curiously, not in rat macrophages or fibroblasts (Okada et al., 2009). Expression of the human CyclinT1 (hCycT1), a cellular cofactor for Tat, in mouse and rat T cells restored viral transcriptional activity (Okada et al., 2009). A single amino acid difference between human and mouse CyclinT1 (mCycT1), which has a tyrosine at residue 261 in place of the cysteine in hCycT1, caused almost complete loss of Tat cofactor activity (Bieniasz et al., 1998; Garber et al., 1998).
In contrast to the definitive results obtained for Rex in rat cells, the existence of a profound block in Rev function in rodent cells remains controversial, although a reduced level of the HIV-1 9 kb transcript has been reported (Bieniasz and Cullen, 2000; Keppler et al., 2001). Some studies have reported impaired Rev activity (Trono and Baltimore, 1990; Marques et al., 2003), whereas others have ascribed the reduced transcript levels to over splicing or the reduced stability of unspliced transcripts in murine cells compared with human cells (Malim and Cullen, 1991); a problem corrected by the expression of the human p32 protein (Zheng et al., 2003). However, Malim et al found that the Gag protein is still efficiently synthesized in murine cells, indicating that Rev is functional, although it is not transported to the plasma membrane, resulting in defective virion formation. When Gag mRNA is transported via a different route using the cellular transporter, Tap, instead of the Rev–CRM1 system, Gag protein was synthesized and transported to the surface membrane, leading to efficient virus production (Swanson et al., 2004). This indicates that the RNA export step is a one of the species-specific barriers to HIV-1 propagation, and that the mode of mRNA nucleocytoplasmic transport affects subsequent processes occurring in the cytoplasm.
The situation in rat cells is somewhat different to that in murine cells. Even when HIV-1 genes are expressed from transfected plasmids (which circumvents the block at the entry step), virus production in rat cells is still poor. However, it increases markedly in rat macrophages and fibroblasts expressing hCRM1 (but only marginally in rat T cells), which is in sharp contrast to cells transfected with hCycT1 (Okada et al., 2009). Further analyses revealed that hCRM1 enhanced Gag mRNA export a couple of times at most in rat fibroblasts. The amount of Gag protein within the cells increased two to three times, which paralleled the increase in Gag mRNA in the cytoplasm, although the trafficking of Gag to the plasma membrane did not increase. However, the number of viral particles shed into the culture medium increased 20- to 50-fold, accompanied by the increased processing of the p55Gag precursor to fully matured Gag proteins. Even a protease-deficient viral construct that caused accumulation of the p55 Gag precursor in the cytoplasm facilitated the release of viral-like particles in the presence of hCRM1, suggesting increased budding at the plasma membrane. The N-terminal half of hCRM1 was found to be responsible for this as the rCRM1–Rev interaction was much less efficient. These facts suggest that incomplete formation of transport complexes in the nucleus may affect the mechanisms underlying particle formation (Figure 6; Nagai-Fukataki et al., 2011). Indeed, the interaction between Rev and the RRE is essential for proper encapsidation of gRNA into virus particles (Blissenbach et al., 2010; Cockrell et al., 2011). Moreover, HIV-1 Gag proteins gain access to the nucleus because they have NLSs (Bukrinsky et al., 1993) and NESs that associate with CRM1 (Dupont et al., 1999). Thus, it would be interesting to examine the role of intra-nuclear Gag proteins in virus morphogenesis. More studies are required to conclude since contradictory results that NLS defective mutant Gag proteins still support viral-like particle formation has been also reported (Grewe et al., 2012).
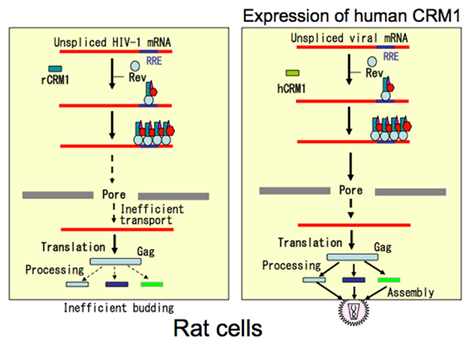
Figure 6. Schematic presentation of RNA transport and particle formation defect of HIV-1 caused by rat CRM1. In rat cells HIV-1 genomic RNA and unspliced mRNA are inefficiently transported to the cytoplasm. Additionally budding process is also affected. Expression of human CRM1 corrects both defects, suggesting linking of RNA nucleocytoplasmic transport to the subsequent cytoplasmic event.
RNA Export of Simple Retroviruses
Although simple retroviruses, exemplified by the Mason-Pfizer monkey virus (MPMV), encode only the gag, pol, and env genes located centrally between the two LTRs (Figure 1; Goff, 2001), they also require the means to circumvent cellular restrictions, since the gag coding region is located within the intron of the pre-mRNA encoding the envelope proteins. To express the intron-containing gag mRNA and gRNA, inefficient splicing is necessary (as in the case of HIV-1), which is demonstrated by the finding that the modification of their splice sites increases their efficiency, resulting in the over splicing of retroviral RNAs, which renders the virus replication incompetent (Katz and Skalka, 1990). However, inefficient splicing is not sufficient; a sequence known as the constitutive transport element (CTE; first identified in the 3′-untranslation region of MPMV by Bray et al., 1994) is also required for efficient export. Since none of the simple retroviruses encode accessory proteins such as Rex and Rev, CTE was thought to recruit cellular proteins. Subsequently, a cellular protein called transporter associated with antigen processing (TAP) was identified, which binds CTE directly, and TAP sequence similarity to yeast Mex67 (which is involved in mRNA transport) indicates that CTE accesses the cellular mRNA export pathway (Grüter et al., 1998; Katahira et al., 1999). The fact that the RNA export pathway involving CTE is independent from those involving RRE/Rev or RXRE/Rex was illustrated by its sensitivity to inhibition by a DN mutant, TAgRexM64, which inhibits Rev/Rex function by sequestering the cellular cofactor, CRM1 (Katahira et al., 1995; Kiyokawa et al., 1997). The ribonucleoprotein complex containing simple retroviral RNA must also undergo a conformational change during export, which is mediated by RNA helicase A (Tang et al., 1997). Release of viral RNA into the cytoplasm is mediated by another RNA helicase, Dbp5, which localizes outside the nuclear pores (LeBlanc et al., 2007).
An intriguing question is: are gRNA and unspliced gag mRNA exported via different pathways that determine whether they are encapsidated or translated, respectively? Rous sarcoma virus (RSV), a simple avian retrovirus, contains direct repeat (DR) elements (Ogert et al., 1996) that mediate the cytoplasmic accumulation of unspliced viral RNA in a manner similar to that of MPMV CTE, since DN TAP and Dbp5 mutants abrogate DR-dependent RNA expression (LeBlanc et al., 2007). On the other hand, RSV Gag is reported to gain access to the nucleus via its NLS, which interacts with importin α/β, transportin SR, and importin 11 (Butterfield-Gerson et al., 2006; Gudleski et al., 2010), and is exported via its NES, which is associated with CRM1 (Scheifele et al., 2002, 2005). Gag binds to gRNA at the packaging element, ψ, and another unidentified site (D’Souza and Summers, 2005), and then forms multimers on gRNA to assemble viral core particles (Scheifele et al., 2007). Thus, it is conceivable that Gag proteins drive the transport of viral particles formed in the nucleus. This notion is supported by the following observations: First, transient nuclear trafficking of Gag is required for efficient gRNA encapsidation (Garbitt-Hirst et al., 2009), suggesting that Gag may initially bind to gRNA in the nucleus. Second, formation of Gag:gRNA prevents the association between Gag and importins, while the binding of Gag to gRNA instead enhances its association with CRM1, consequently promoting the export of the Gag–gRNA complex to the cytoplasm (Gudleski et al., 2010; Parent, 2011). These facts suggest the intriguing possibility that gag mRNA is transported through the TAP pathway to be translated, whereas gRNA (which appears to be indistinguishable from gag mRNA), is exported by the Gag–CRM1 pathway for encapsidation.
Detailed analyses on mouse mammary tumor virus (MMTV) and human endogenous retrovirus K (HERV-K) that had been initially classified as simple retroviruses revealed doubly spliced mRNAs as the case of complex retroviruses. Moreover, such mRNAs have been found to encode Rev like proteins such as Rem in MMTV (Indik et al., 2005; Mertz et al., 2005) and K-Rev in HERV-K (Yang et al., 1999) that export their cognate viral RNAs through CRM1 pathway. The results suggest that these viruses belong to complex retrovirus family. The presence of K-Rev in HERV-K that integrated in primate genome ≥30 million years ago may evoke the possibility of evolutional relation in the acquisition of Rev by exogenous lentiviruses (Yang et al., 1999).
Conclusion
Studies on RNA transport of retroviruses originally stemmed from molecular biological interest on unusual function of Rex and Rev that break the cellular blockage against transport of intron-containing RNA. A great deal of investigation not only elucidated their own mechanisms but also led discovery of the cellular nucleocytoplasmic transport system. Moreover, recent studies showed that the mode of RNA export influences subsequent events in the cytoplasm, consequently affecting whole process of viral production including the translation of the cognate mRNA, transport of Gag proteins to the plasma membrane, and the formation of virus particles. Lastly, the discovery on the interaction between the viral and cellular RNA transport machinery as a species-specific barrier of HIV-1 and HTLV-1 forms the basis to construct convenient animal models of infection, which should facilitate development of more potent therapeutic and preventing means.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahmed, Y. F., Hanly, S. M., Malim, M. H., Cullen, B. R., and Greene, W. C. (1990). Structure-function analyses of the HTLV-I Rex and HIV-1 Rev RNA response elements: insights into the mechanism of Rex and Rev action. Genes Dev. 4, 1014–1022.
Akagi, T., Takeda, I., Oka, T., Ohtsuki, Y., Yano, S., and Miyoshi, I. (1985). Experimental infection of rabbits with human T-cell leukemia virus type I. Jpn. J. Cancer Res. 76, 86–94.
Askjaer, P., Bachi, A., Wilm, M., Bischoff, F. R., Weeks, D. L., Ogniewski, V., Ohno, M., Niehrs, C., Kjems, J., Mattaj, I. W., and Fornerod, M. (1999). RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol. 19, 6276–6285.
Bai, X. T., Sinha-Datta, U., Ko, N. L., Bellon, M., and Nicot, C. (2012). Nuclear export and expression of HTLV-I tax/rex mRNA is RxRE/Rex-dependent. J. Virol. 86, 4559–4565.
Ballaun, C., Farrington, G. K., Dobrovnik, M., Rusche, J., Hauber, J., and Bohnlein, E. (1991). Functional analysis of human T-cell leukemia virus type I rex-response element: direct RNA binding of Rex protein correlates with in vivo activity. J. Virol. 65, 4408–4413.
Battiste, J. L., Mao, H., Rao, N. S., Tan, R., Muhandiram, D. R., Kay, L. E., Frankel, A. D., and Williamson, J. R. (1996). Alpha helix-RNA major groove recognition in an HIV-1 Rev peptide-RRE RNA complex. Science 273, 1547–1551.
Bieniasz, P. D., and Cullen, B. R. (2000). Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol. 74, 9868–9877.
Bieniasz, P. D., Grdina, T. H., Bogerd, H. P., and Cullen, B. R. (1998). Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 17, 7056–7065.
Blissenbach, M., Grewe, B., Hoffmann, B., Brandt, S., and Uberla, K. (2010). Nuclear RNA export and packaging functions of HIV-1 Rev revisited. J. Virol. 84, 6598–6604.
Bogerd, H., and Greene, W. C. (1993). Dominant negative mutants of human T-cell leukemia virus type I Rex and human immunodeficiency virus type 1 Rev fail to multimerize in vivo. J. Virol. 67, 2496–2502.
Bogerd, H. P., Fridell, R. A., Benson, R. E., Hua, J., and Cullen, B. R. (1996). Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16, 4207–4214.
Bogerd, H. P., Huckaby, G. L., Ahmed, Y. F., Hanly, S. M., and Greene, W. C. (1991). The type I human T-cell leukemia virus (HTLV-I) Rex trans-activator binds directly to the HTLV-I Rex and the type 1 human immunodeficiency virus Rev RNA response elements. Proc. Natl. Acad. Sci. U.S.A. 88, 5704–5708.
Bray, M., Prasad, S., Dubay, J. W., Hunter, E., Jeang, K. T., Rekosh, D., and Hammarskjöld, M. L. (1994). A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. U.S.A. 91, 1256–1260.
Browning, J., Horner, J. W., Pettoello-Mantovani, M., Raker, C., Yurasov, S., DePinho, R. A., and Goldstein, H. (1997). Mice transgenic for human CD4 and CCR5 are susceptible to HIV infection. Proc. Natl. Acad. Sci. U.S.A. 94, 14637–14641.
Bukrinsky, M. I., Haggerty, S., Dempsey, M. P., Sharova, N., Adzhubel, A., Spitz, L., Lewis, P., Goldfarb, D., Emerman, M., and Stevenson, M. (1993). A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365, 666–669.
Butterfield-Gerson, K. L., Scheifele, L. Z., Ryan, E. P., Hopper, A. K., and Parent, L. J. (2006). Importin-beta family members mediate alpharetrovirus gag nuclear entry via interactions with matrix and nucleocapsid. J. Virol. 80, 1798–1806.
Chaitanya, J., and Belasco, J. G. (2001). Structural model for the cooperative assembly of HIV-1 Rev multimers on the RRE as deduced from analysis of assembly-defective mutants. Mol. Cell 7, 603–614.
Chang, D. D., and Sharp, P. A. (1989). Regulation by HIV Rev depends upon recognition of splice sites. Cell 59, 789–795.
Chen, S., Ishii, N., Ine, S., Ikeda, S., Fujimura, T., Ndhlovu, L. C., Soroosh, P., Tada, K., Harigae, H., Kameoka, J., Kasai, N., Sasaki, T., and Sugamura, K. (2006). Regulatory T cell-like activity of Foxp3+ adult T cell leukemia cells. Int. Immunol. 18, 269–277.
Cochrane, A. W., Jones, K. S., Beidas, S., Dillon, P. J., Skalka, A. M., and Rosen, C. A. (1991). Identification and characterization of intragenic sequences which repress human immunodeficiency virus structural gene expression. J. Virol. 65, 5305–5313.
Cockrell, A. S., van Praag, H., Santistevan, N., Ma, H., and Kafri, T. (2011). The HIV-1 Rev/RRE system is required for HIV-1 5’ UTR cis elements to augment encapsidation of heterologous RNA into HIV-1 viral particles. Retrovirology 8, 51.
Cullen, B. R. (1991). Regulation of human immunodeficiency virus replication. Annu. Rev. Microbiol. 45, 219–250.
Daly, T. J., Cook, K. S., Gray, G. S., Maione, T. E., and Rusche, J. R. (1989). Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature 342, 816–819.
Daugherty, M. D., Liu, B., and Frankel, A. D. (2010). Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 17, 1337–1342.
Doye, V., and Hurt, E. (1997). From nucleoporins to nuclear pore complexes. Curr. Opin. Cell Biol. 9, 401–411.
D’Souza, V., and Summers, M. F. (2005). How retroviruses select their genomes. Nat. Rev. Microbiol. 3, 643–655.
Dupont, S., Sharova, N., DéHoratius, C., Virbasius, C. M., Zhu, X., Bukrinskaya, A. G., Stevenson, M., and Green, M. R. (1999). A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature 402, 681–685.
Edgcomb, S. P., Carmel, A. B., Naji, S., Ambrus-Aikelin, G., Reyes, J. R., Saphire, A. C., Gerace, L., and Williamson, J. R. (2012). DDX1 is an RNA-dependent ATPase involved in HIV-1 Rev function and virus replication. J. Mol. Biol. 415, 61–74.
Englmeier, L., Fornerod, M., Bischoff, F. R., Petosa, C., Mattaj, I. W., and Kutay, U. (2001). RanBP3 influences interactions between CRM1 and its nuclear protein export substrates. EMBO Rep. 2, 926–932.
Fang, J., Kubota, S., Yang, B., Zhou, N., Zhang, H., Godbout, R., and Pomerantz, R. J. (2004). A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology 330, 471–480.
Fischer, U., Meyer, S., Teufel, M., Heckel, C., Luhrmann, R., and Rautmann, G. (1994). Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 13, 4105–4112.
Fisher, U., Huber, J., Boelens, W. C., Mattaj, I. W., and Luhemann, R. (1995). The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82, 475–483.
Fornerod, M., Ohno, M., Yoshida, M., and Mattaj, I. W. (1997a). CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051–1060.
Fornerod, M., van Deursen, J., van Baal, S., Reynolds, A., Davis, D., Murti, K. G., Fransen, J., and Grosveld, G. (1997b). The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 16, 807–816.
Freed, E. O., and Martin, M. A. (2001). “HIVs and their replication,” in Fields Virology, eds B. N. Fields, D. M. Knipe, and P. M. Howley (Philadelphia: Lippincott Williams & Wilkins), 1941–1970.
Fukuda, M., Asano, S., Nakamura, T., Adachi, M., Yoshida, M., Yanagida, M., and Nishida, E. (1997). CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390, 308–311.
Garber, M. E., Wei, P., KewalRamani, V. N., Mayall, T. P., Herrmann, C. H., Rice, A. P., Littman, D. R., and Jones, K. A. (1998). The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 15, 3512–3527.
Garbitt-Hirst, R., Kenney, S. P., and Parent, L. J. (2009). Genetic evidence for a connection between Rous sarcoma virus gag nuclear trafficking and genomic RNA packaging. J. Virol. 83, 6790–6797.
Goff, S. P. (2001). “Retroviridae: the retroviruses and their replication,” in Fields Virology, eds B. N. Fields, D. M. Knipe, and P. M. Howley (Philadelphia: Lippincott Williams & Wilkins), 1871–1940.
Goffinet, C., Michel, N., Allespach, I., Tervo, H. M., Hermann, V., Krausslich, H. G., Greene, W. C., and Keppler, O. T. (2007). Primary T-cells from human CD4/CCR5-transgenic rats support all early steps of HIV-1 replication including integration, but display impaired viral gene expression. Retrovirology 4, 53.
Green, P. L., and Chen, I. S. Y. (2001). “Human T-cell leukemia virus type-1 infection,” in Fields Virology, eds B. N. Fields, D. M. Knipe, and P. M. Howley (Philadelphia: Lippincott Williams & Wilkins), 1941–1970.
Grewe, B., Hoffmann, B., Ohs, I., Blissenbach, M., Brandt, S., Tippler, B., Grunwald, T., and Uberla, K. (2012). Cytoplasmic utilization of human immunodeficiency virus type 1 genomic RNA is not dependent on a nuclear interaction with gag. J. Virol. 86, 2990–3002.
Grüter, P., Tabernero, C., von Kobbe, C., Schmitt, C., Saavedra, C., Bachi, A., Wilm, M., Felber, B. K., and Izaurralde, E. (1998). TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1, 649–659.
Gudleski, N., Flanagan, J. M., Ryan, E. P., Bewley, M. C., and Parent, L. J. (2010). Directionality of nucleocytoplasmic transport of the retroviral gag protein depends on sequential binding of karyopherins and viral RNA. Proc. Natl. Acad. Sci. U.S.A. 107, 9358–9363.
Hakata, Y., Umemoto, T., Matsushita, S., and Shida, H. (1998). Involvement of human CRM1 (exportin 1) in the export and multimerization of the Rex protein of human T-cell leukemia virus type 1. J. Virol. 72, 6602–6607.
Hakata, Y., Yamada, M., Mabuchi, N., and Shida, H. (2002). The carboxy-terminal region of the human immunodeficiency virus type 1 protein Rev has multiple roles in mediating CRM1-related Rev functions. J. Virol. 76, 8079–8089.
Hakata, Y., Yamada, M., and Shida, H. (2001). Rat CRM1 is Responsible for the Poor Activity of Human T-Cell Leukemia Virus Type-1 Rex Protein in Rat Cells. J. Virol. 75, 11515–11525.
Hakata, Y., Yamada, M., and Shida, H. (2003). A multi-functional domain in human CRM1 (exportin 1) mediates RanBP3 binding and multimerization of human T-cell leukemia virus type 1 Rex protein. Mol. Cell. Biol. 23, 8751–8761.
Heger, P., Rosorius, O., Koch, C., Casari, G., Grassmann, R., and Hauber, J. (1998). Multimer formation is not essential for nuclear export of human T-cell leukemia virus type 1 Rex trans-activator protein. J. Virol. 72, 8659–8668.
Hidaka, M., Inoue, J., Yoshida, M., and Seiki, M. (1988). Post-transcriptional regulator (rex) of HTLV-1 initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J. 7, 519–523.
Hinuma, Y., Nagata, K., Hanaoka, M., Nakai, M., Matsumoto, T., Kinoshita, K. I., Shirakawa, S., and Miyoshi, I. (1981). Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc. Natl. Acad. Sci. U.S.A. 78, 6476–8640.
Indik, S., Günzburg, W. H., Salmons, B., and Rouault, F. (2005). A novel, mouse mammary tumor virus encoded protein with Rev-like properties. Virology 337, 1–6.
Inoue, J., Seiki, M., and Yoshida, M. (1986). The second pX product p27 chi-III of HTLV-1 is required for gag gene expression. FEBS Lett. 209, 187–190.
Ishiguro, N., Abe, M., Seto, K., Sakurai, H., Ikeda, H., Wakisaka, A., Togashi, T., Tateno, M., and Yoshiki, T. (1992). A rat model of human T lymphocyte virus type I (HTLV-I) infection. 1. Humoral antibody response, provirus integration, and HTLV-I-associated myelopathy/tropical spastic paraparesis-like myelopathy in seronegative HTLV-I carrier rats. J. Exp. Med. 176, 981–989.
Jacobson, S., Raine, C. S., Mingioli, E. S., and McFarlin, D. E. (1988). Isolation of an HTLV-1-like retrovirus from patients with tropical spastic paraparesis. Nature 331, 540–543.
Kalyanaraman, V. S., Sarngadharan, M. G., Nakao, Y., Ito, Y., Aoki, T., and Gallo, R. C. (1982). Natural antibodies to the structural core protein (p24) of the human T-cell leukemia (lymphoma) retrovirus found in sera of leukemia patients in Japan. Proc. Natl. Acad. Sci. U.S.A. 79, 1653–1657.
Katahira, J., Ishizaki, T., Sakai, H., Adachi, A., Yamamoto, K., and Shida, H. (1995). Effects of translation initiation factor eIF-5A on the functioning of human T-cell leukemia virus type I Rex and human immunodeficiency virus Rev inhibited trans dominantly by a Rex mutant deficient in RNA binding. J. Virol. 69, 3125–3133.
Katahira, J., Strässer, K., Podtelejnikov, A., Mann, M., Jung, J. U., and Hurt, E. (1999). The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18, 2593–2609.
Katz, R. A., and Skalka, A. M. (1990). Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol. Cell. Biol. 10, 696–704.
Kehlenbach, R. H., Dickmanns, A., Kehlenbach, A., Guan, T., and Gerace, L. (1999). A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 145, 645–657.
Keppler, O. T., Welte, F. J., Ngo, T. A., Chin, P. S., Patton, K. S., Tsou, C. L., Abbey, N. W., Sharkey, M. E., Grant, R. M., You, Y., Scarborough, J. D., Ellmeier, W., Littman, D. R., Stevenson, M., Charo, I. F., Herndier, B. G., Speck, R. F., and Goldsmith, M. A. (2002). Progress toward a human CD4/CCR5 transgenic rat model for de novo infection by human immunodeficiency virus type 1. J. Exp. Med. 195, 719–736.
Keppler, O. T., Yonemoto, W., Welte, F. J., Patton, K. S., Iacovides, D., Atchison, R. E., Ngo, T., Hirschberg, D. L., Speck, R. F., and Goldsmith, M. A. (2001). Susceptibility of rat-derived cells to replication by human immunodeficiency virus type 1. J. Virol. 5, 8063–8073.
Kim, F. J., Beeche, A. A., Hunter, J. J., Chin, D. J., and Hope, T. J. (1996). Characterization of the nuclear export signal of human T-cell lymphotropic virus type 1 Rex reveals that nuclear export is mediated by position-variable hydrophobic interactions. Mol. Cell. Biol. 16, 5147–5155.
Kim, S. Y., Byrn, R., Groopman, J., and Baltimore, D. (1989). Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63, 3708–3713.
Kimura, T., Hashimoto, I., Yamamoto, A., Nishikawa, M., and Fujisawa, J. I. (2000). Rev-dependent association of the intron-containing HIV-1 gag mRNA with the nuclear actin bundles and the inhibition of its nucleocytoplasmic transport by latrunculin-B. Genes Cells 5, 289–307.
King, J. A., Bridger, J. M., Lochelt, M., Lichter, P., Schulz, T. F., Schirrmacher, V., and Khazaie, K. (1998). Nucleocytoplasmic transport of HTLV-1 RNA is regulated by two independent LTR encoded nuclear retention elements. Oncogene 25, 3309–3316.
Kiyokawa, T., Umemoto, T., Watanabe, Y., Matsushita, S., and Shida, H. (1997). Two distinct pathways for intronless mRNA expression: one related, the other unrelated to human immunodeficiency virus Rev and human T cell leukemia virus type I Rex functions. Biol. Signals 6, 134–142.
Koff, W. C., Johnson, P. R., Watkins, D. I., Burton, D. R., Lifson, J. D., Hasenkrug, K. J., McDermott, A. B., Schultz, A., Zamb, T. J., Boyle, R., and Desrosiers, R. C. (2006). HIV vaccine design: insights from live attenuated SIV vaccines. Nat. Immunol. 7, 19–23.
Koya, Y., Ohashi, T., Kato, H., Hanabuchi, S., Tsukahara, T., Takemura, F., Etoh, K., Matsuoka, M., Fujii, M., and Kannagi, M. (1999). Establishment of a seronegative human T-cell leukemia virus type 1 (HTLV-1) carrier state in rats inoculated with a syngeneic HTLV-1-immortalized T-cell line preferentially expressing Tax. J. Virol. 73, 6436–6443.
Kozak, M. (1992). Regulation of translation in eukaryotic systems. Annu. Rev. Cell Biol. 8, 197–225.
Kubota, S., Nosaka, T., Cullen, B. R., Maki, M., and Hatanaka, M. (1991). Effect of chimeric mutants of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex on nucleolar targeting signals. J. Virol. 65, 2452–2456.
LeBlanc, J. J., Uddowla, S., Abraham, B., Clatterbuck, S., and Beemon, K. L. (2007). Tap and Dbp5, but not Gag, are involved in DR-mediated nuclear export of unspliced Rous sarcoma virus RNA. Virology 363, 376–386.
Legrain, P., and Rosbash, M. (1989). Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57, 573–583.
Li, Q. X., Camerini, D., Xie, Y., Greenwald, M., Kuritzkes, D. R., and Chen, I. S. (1996). Syncytium formation by recombinant HTLV-II envelope glycoprotein. Virology 218, 279–284.
Madore, S. J., Tiley, L. S., Malim, M. H., and Cullen, B. R. (1994). Sequence requirements for Rev multimerization in vivo. Virology 202, 186–194.
Maldarelli, F., Martin, M. A., and Strebel, K. (1991). Identification of posttranscriptionally active inhibitory sequences in human immunodeficiency virus type 1 RNA: novel level of gene regulation. J. Virol. 65, 5732–5743.
Malim, M. H., and Cullen, B. R. (1991). HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell 65, 241–248.
Malim, M. H., McCarn, D. F., Tiley, L. S., and Cullen, B. R. (1991). Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J. Virol. 65, 4248–4254.
Malim, M. H., Tiley, L. S., McCarn, D. F., Rusche, J. R., Hauber, J., and Cullen, B. R. (1990). HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell 60, 675–683.
Mann, D. A., Mikaélian, I., Zemmel, R. W., Green, S. M., Lowe, A. D., Kimura, T., Singh, M., Butler, P. J., Gait, M. J., and Karn, J. (1994). Molecular rheostat: Co-operative rev binding to stem I of the Rev-response element modulates human immunodeficiency virus type-1 late gene expression. J. Mol. Biol. 241, 193–207.
Marques, S. M., Veyrune, J. L., Shukla, R. R., and Kumar, A. (2003). Restriction of human immunodeficiency virus type 1 Rev function in murine A9 cells involves the Rev C-terminal domain. J. Virol. 77, 3084–3090.
Matsuoka, M., and Jeang, K. T. (2011). Human T-cell leukemia virus type 1 (HTLV-1) and leukemic transformation: viral infectivity, Tax, HBZ and therapy. Oncogene 30, 1379–1389.
Mattaj, I. W., and Englmeier, L. (1998). Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67, 265–306.
Matthew, D. D., D’Orso, I., and Frankel, A. D. (2008). A solution to limited genomic capacity: using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol. Cell 31, 824–834.
Mertz, J. A., Simper, M. S., Lozano, M. M., Payne, S. M., and Dudley, J. P. (2005). Mouse mammary tumor virus encodes a self-regulatory RNA export protein and is a complex retrovirus. J. Virol. 79, 14737–14747.
Nagai-Fukataki, M., Ohashi, T., Hashimoto, I., Kimura, T., Hakata, Y., and Shida, H. (2011). Nuclear and cytoplasmic effects of human CRM1 on HIV-1 production in rat cells. Genes Cells 16, 203–216.
Najera, I., Krieg, M., and Karn, J. (1999). Synergistic stimulation of HIV-1 rev-dependent export of unspliced mRNA to the cytoplasm by hnRNP A1. J. Mol. Biol. 285, 1951–1964.
Nakamura, H., Hayami, M., Ohta, Y., Ishikawa, K., Tsujimoto, H., Kiyokawa, T., Yoshida, M., Sasagawa, A., and Honjo, S. (1987). Protection of cynomolgus monkeys against infection by human T-cell leukemia virus type-I by immunization with viral env gene products produced in Escherichia coli. Int. J. Cancer 40, 403–407.
Nemergut, M. E., Lindsay, M. E., Brownawell, A. M., and Macara, I. G. (2002). Ran-binding protein 3 links Crm1 to the Ran guanine nucleotide exchange factor. J. Biol. Chem. 277, 17385–17388.
Ogert, R. A., Lee, L. H., and Beemon, K. L. (1996). Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J. Virol. 70, 3834–3843.
Ohashi, T., Hanabuchi, S., Kato, H., Tateno, H., Takemura, F., Tsukahara, T., Koya, Y., Hasegawa, A., Masuda, T., and Kannagi, M. (2000). Prevention of adult T-cell leukemia-like lymphoproliferative disease in rats by adoptively transferred T cells from a donor immunized with human T-cell leukemia virus type 1 Tax-coding DNA vaccine. J. Virol. 74, 9610–9616.
Oka, T., Sonobe, H., Iwata, J., Kubonishi, I., Satoh, H., Takata, M., Tanaka, Y., Tateno, M., Tozawa, H., Mori, S., Yoshiki, T., and Ohtsuki, Y. (1992). Phenotypic progression of a rat lymphoid cell line immortalized by human T-lymphotropic virus type I to induce lymphoma/leukemia-like disease in rats. J. Virol. 66, 6686–6694.
Okada, H., Zhang, X., Fofana, I. B., Nagai, M., Suzuki, H., Ohashi, T., and Shida, H. (2009). Synergistic effect of human CycT1 and CRM1 on HIV-1 propagation in rat T cells and macrophages. Retrovirology 6, 43.
Osame, M., Usuku, K., Izumo, S., Ijichi, N., Amitani, H., Igata, A., Matsumoto, M., and Tara, M. (1986). HTLV-I associated myelopathy, a new clinical entity. Lancet 1, 1031–1032.
Ossareh-Nazari, B., and Dargemont, C. (1999). Domains of Crm1 involved in the formation of the Crm1, RanGTP, and leucine-rich nuclear export sequences trimeric complex. Exp. Cell Res. 252, 236–241.
Palmeri, D., and Malim, M. H. (1999). Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol. Cell. Biol. 19, 1218–1225.
Parent, L. J. (2011). New insights into the nuclear localization of retroviral Gag proteins. Nucleus 2, 92–97.
Robertson-Anderson, R. M., Wang, J., Edgcomb, S. P., Carmel, A. B., Williamson, J. R., and Millar, D. P. (2011). Single-molecule studies reveal that DEAD box protein DDX1 promotes oligomerization of HIV-1 Rev on the Rev response element. J. Mol. Biol. 410, 959–971.
Saiga, A., Orita, S., Minoura-Tada, N., Maeda, M., Aono, Y., Asakawa, M., Nakahara, K., Kubota, R., Osame, M., and Igarashi, H. (1997). cis-Acting inhibitory elements within the pol-env region of human T-cell leukemia virus type 1 possibly involved in viral persistence. J. Virol. 71, 4485–4494.
Sanchez-Velar, N., Udofia, E. B., Yu, Z., and Zapp, M. L. (2004). hRIP, a cellular cofactor for Rev function, promotes release of HIV RNAs from the perinuclear region. Genes Dev. 18, 23–34.
Satou, Y., Yasunaga, J., Yoshida, M., and Matsuoka, M. (2006). HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Natl. Acad. Sci. U.S.A. 103, 720–725.
Scheifele, L. Z., Garbitt, R. A., Rhoads, J. D., and Parent, L. J. (2002). Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. U.S.A. 99, 3944–3949.
Scheifele, L. Z., Kenney, S. P., Cairns, T. M., Craven, R. C., and Parent, L. J. (2007). Overlapping roles of the Rous sarcoma virus Gag p10 domain in nuclear export and virion core morphology. J. Virol. 81, 10718–10728.
Scheifele, L. Z., Ryan, E. P., and Parent, L. J. (2005). Detailed mapping of the nuclear export signal in the Rous sarcoma virus Gag protein. J. Virol. 79, 8732–8741.
Schneider, R., Campbell, M., Nasioulas, G., Felber, B. K., and Pavlakis, G. N. (1997). Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 71, 4892–4903.
Seguin, B., Staffa, A., and Cochrane, A. (1998). Control of human immunodeficiency virus type 1 RNA metabolism: role of splice sites and intron sequences in unspliced viral RNA subcellular distribution. J. Virol. 72, 9503–9513.
Shultz, L. D., Ishikawa, F., and Greiner, D. L. (2007). Humanized mice in translational biomedical research. Nat. Rev. Immunol. 7, 118–130.
Siomi, H., Shida, H., Nam, S.-H., Nosaka, T., Maki, M., and Hatanaka, M. (1988). Sequence requirements for nucleolar localization of human T-cell leukemia virus type-I pX protein that regulate viral RNA processing. Cell 55, 197–209.
Staffa, A., and Cochrane, A. (1994). The tat/rev intron of human immunodeficiency virus type 1 is inefficiently spliced because of suboptimal signals in the 3’ splice site. J. Virol. 68, 3071–3079.
Staffa, A., and Cochrane, A. (1995). Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 15, 4597–4605.
Sutton, R. E., and Littman, D. R. (1996). Broad host range of human T-cell leukemia virus type 1 demonstrated with an improved pseudotyping system. J. Virol. 70, 7322–7326.
Swanson, C. M., Puffer, B. A., Ahmad, K. M., Doms, R. W., and Malim, M. H. (2004). Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 23, 2632–2640.
Takayanagi, R., Ohashi, M., Yamashita, M., Kurosaki, Y., Tanaka, K., Hakata, Y., Komoda, Y., Ikeda, S., Tsunetsugu-Yokota, Y., Tanaka, Y., and Shida, H. (2007). Enhanced replication of human T-cell leukemia virus type 1 in T cells from transgenic rats expressing human CRM1 that is regulated in a natural manner. J. Virol. 81, 5908–5918.
Takayanagi, R., Ohashi, T., and Shida, H. (2009). Functional analysis of Foxp3 and CTLA-4 expressing HTLV-1-infected cells in a rat model. Microbes Infect. 12, 964–972.
Tang, H., Gaietta, G. M., Fischer, W. H., Ellisman, M. H., and Wong-Staal, F. (1997). A cellular cofactor for the constitutive transport element of type D retrovirus. Science 276, 1412–1415.
Tiley, L. S., Malim, M. H., Tewary, H. K., Stockley, P. G., and Cullen, B. R. (1992). Identification of a high-affinity RNA-binding site for the human immunodeficiency virus type 1 Rev protein. Proc. Natl. Acad. Sci. U.S.A. 89, 758–762.
Trono, D., and Baltimore, D. (1990). A human cell factor is essential for HIV-1 Rev action. EMBO J. 9, 4155–4160.
Veazey, R. S., Klasse, P. J., Schader, S. M., Hu, Q., Ketas, T. J., Lu, M., Marx, P. A., Dufour, J., Colonno, R. J., Shattock, R. J., Springer, M. S., and Moore, J. P. (2005). Protection of macaques from vaginal SHIV challenge by vaginally delivered inhibitors of virus-cell fusion. Nature 438, 99–102.
Watanabe, S., Terashima, K., Ohta, S., Horibata, S., Yajima, M., Shiozawa, Y., Dewan, M. Z., Yu, Z., Ito, M., Morio, T., Shimizu, N., Honda, M., and Yamamoto, N. (2007). Hematopoietic stem cell-engrafted NOD/SCID/IL2Rgamma null mice develop human lymphoid systems and induce long-lasting HIV-1 infection with specific humoral immune responses. Blood 109, 212–218.
Weichselbraun, I., Berger, J., Dobrovnik, M., Bogerd, H., Grassmann, R., Greene, W. C., Hauber, J., and Bohnlein, E. (1992). Dominant-negative mutants are clustered in a domain of the human T-cell leukemia virus type I Rex protein: implications for trans dominance. J. Virol. 66, 4540–4545.
Wen, W., Meinkoth, J. L., Tsien, R. Y., and Taylor, S. S. (1995). Identification of a signal for rapid export of proteins from the nucleus. Cell 82, 463–473.
Yang, J., Bogerd, H. P., Peng, S., Wiegand, H., Truant, R., and Cullen, B. R. (1999). An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc. Natl. Acad. Sci. U.S.A. 96, 13404–13408.
Yedavalli, V. S., Neuveut, C., Chi, Y. H., Kleiman, L., and Jeang, K. T. (2004). Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119, 381–392.
Yu, S. F., Baldwin, D. N., Gwynn, S. R., Yendapalli, S., and Linial, M. L. (1996). Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271, 1579–1582.
Yu, Z., Sanchez-Velar, N., Catrina, I. E., Kittler, E. L., Udofia, E. B., and Zapp, M. L. (2005). The cellular HIV-1 Rev cofactor hRIP is required for viral replication. Proc. Natl. Acad. Sci. U.S.A. 102, 4027–4032.
Zhang, X., Hakata, Y., Tanaka, Y., and Shida, H. (2006). CRM1, an RNA transporter, is a major species-specific restriction factor of human T cell leukemia virus type 1 (HTLV-1) in rat cells. Microbes Infect. 8, 851–859.
Keywords: RNA export, HTLV-1, HIV-1, infection rat models for, CRM1, simple retrovirus, species-specific barrier, Rex/Rev
Citation: Shida H (2012) Role of nucleocytoplasmic RNA transport during the life cycle of retroviruses. Front. Microbio. 3:179. doi: 10.3389/fmicb.2012.00179
Received: 26 March 2012; Paper pending published: 16 April 2012;
Accepted: 26 April 2012; Published online: 18 May 2012.
Edited by:
Atsushi Koito, Kumamoto University, JapanReviewed by:
Yasuo Ariumi, Kumamoto University, JapanTominori Kimura, Ritsumeikan University, Japan
Copyright: © 2012 Shida. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Hisatoshi Shida, Division of Molecular Virology, Institute of Immunological Science, Hokkaido University, Sapporo 060-0815, Japan. e-mail: hshida@igm.hokudai.ac.jp
