- 1 Agrivirology Laboratory, Institute of Plant Science and Bioresources, Okayama University, Kurashiki, Okayama, Japan
- 2 State Key Laboratory Breeding Base for Zhejiang Sustainable Pest and Disease Control, Ministry of Agriculture Key Laboratory of Biotechnology in Plant Protection, Institute of Virology and Biotechnology, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021, P. R. China
The family Reoviridae is one of the largest virus families with genomes composed of 9–12 double-stranded RNA segments. It includes members infecting organisms from protists to humans. It is well known that reovirus genomes are prone to various types of genome alterations including intragenic rearrangement and reassortment under laboratory and natural conditions. Recently distinct genetic alterations were reported for members of the genus Mycoreovirus, Mycoreovirus 1 (MyRV1), and MyRV3 with 11 (S1–S11) and 12 genome segments (S1–S12), respectively. While MyRV3 S8 is lost during subculturing of infected host fungal strains, MyRV1 rearrangements undergo alterations spontaneously and inducibly. The inducible MyRV1 rearrangements are different from any other previous examples of reovirus rearrangements in their dependence on an unrelated virus factor, a multifunctional protein, p29, encoded by a distinct virus Cryphonectria parasitica hypovirus 1 (CHV1). A total of 5 MyRV1 variants with genome rearranged segments (S1–S3, S6 and S10) are generated in the background of a single viral strain in the presence of CHV1 p29 supplied either transgenically or by coinfection. MyRV1 S4 and S10 are rearranged, albeit very infrequently, in a CHV1 p29 independent fashion. A variant of MyRV1 with substantial deletions in both S4 and S10, generated through a combined reassortment and rearrangement approach, shows comparable replication levels to the wild-type MyRV1. In vivo and in vitro interactions of CHV1 p29 and MyRV1 VP9 are implicated in the induction of MyRV1 rearrangements. However, the mechanism underlying p29-mediated rearrangements remains largely unknown. MyRV1 S4 rearrangements spontaneously occurred independently of CHV1 p29. In the absence of reverse genetics systems for mycoreoviruses, molecular and biological characterization of these MyRV1 and MyRV3 variants contribute to functional analyses of the protein products encoded by those rearranged segments.
Introduction
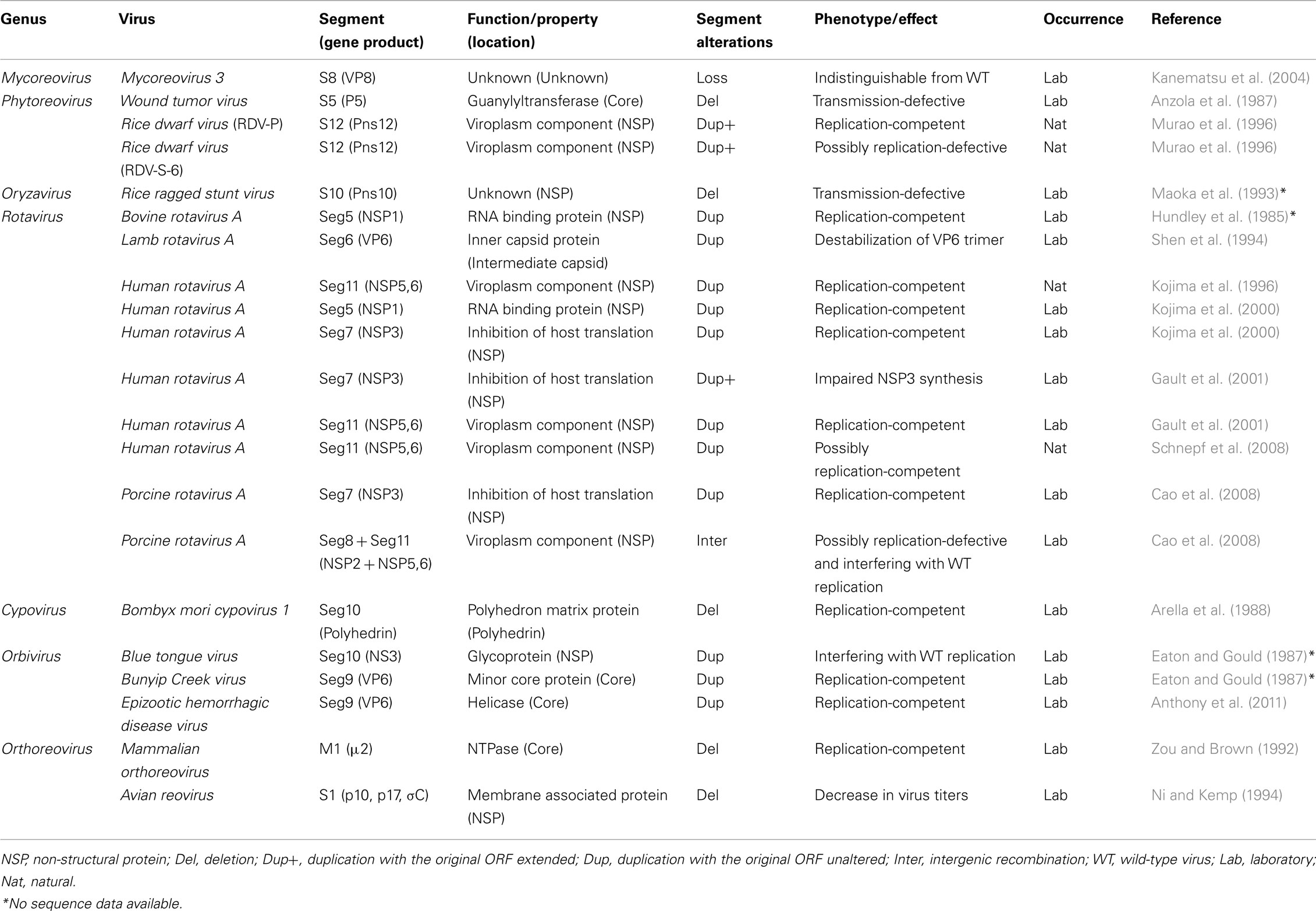
The family Reoviridae accommodates a wide range of members that infect protists, fungi, plants, invertebrates, and vertebrates, and are characterized by 9–12 segmented double-stranded RNA (dsRNA) genomes, multi-layered virion structures, and particle-associated enzymes for RNA synthesis. The family now consists of 15 genera divided into two subfamilies Spinareovirinae (“turreted” or spiked reoviruses) and Sedoreovirinae (non-spiked reoviruses) (Attoui et al., 2012). Among the 15 genera is a relatively recently established genus Mycoreovirus containing three members (Mycoreovirus 1–3, MyRV1-3) that were isolated in two phytopathogenic fungi, specifically, the chestnut blight fungus (MyRV1 and MyRV2) and the white root rot fungus (MyRV3) by the groups of Drs. Bradley I. Hillman, William MacDonald, and Naoyuki Matsumoto (Enebak et al., 1994; Hillman and Suzuki, 2004; Hillman et al., 2004; Suzuki et al., 2004; Wei et al., 2004). MyRV1 and MyRV2 have 11-segmented genomes (Figure 1; S1–S11, termed with an increasing order of mobility in SDS-polyacrylamide gel electrophoresis), while MyRV3 has a 12-segmented genome (S1–S12). The Mycoreovirus belongs to the Spinareovirinae subfamily and its members retain conserved NTP binding motif and di-histidine motif specifically conserved in the subfamily (Suzuki et al., 2004; Supyani et al., 2007; Spear et al., 2012).
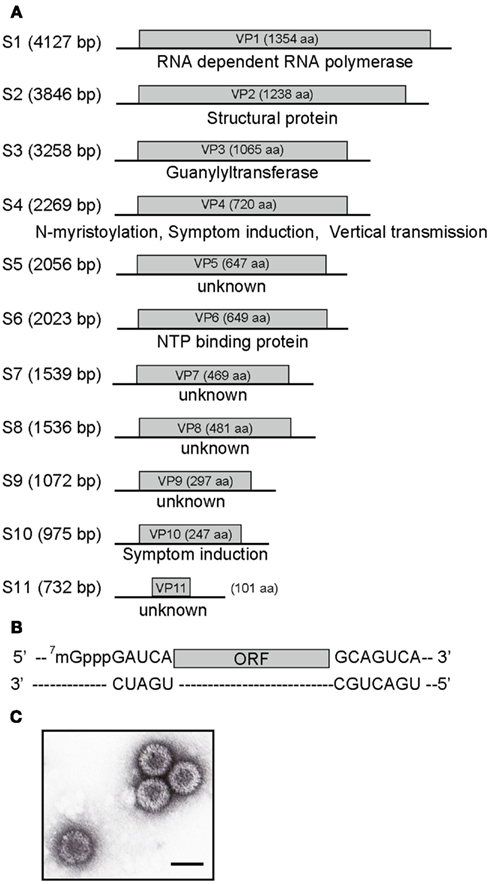
Figure 1. MyRV1 genome organization and virion morphology. (A) Diagram illustrating the organization of the MyRV1 genome segments S1–S11. Each segment has a large single ORF corresponding between 42 and 98% of its entire size. Light gray boxes refer to ORFs and the sizes of their protein products are shown. The locations and assigned functions of encoded proteins are denoted below. RdRp, guanylyltransferase, and NTP binding protein are assigned to S1, S3, and S6. VP4 and VP10 are dispensable for replication, while they are required for normal symptom induction. (B) Basic structure of the MyRV1 genome segment. The plus-sense strand of each dsRNA segment possesses the strictly conserved 5’ pentanucleotide (5’-GAUCA—), single ORF and 3’ heptanucleotide (—GCAGUCA-3’). (C) Electron microscopy of MyRV1 virions. MyRV1 has a double-shelled particle structure of approximately 80 nm in diameter. Bar represents 80 nm.
Like point mutations, rearrangements and reassortments are major driving forces for reovirus genome molecular diversity and evolution. Both could involve large extents of genome segment alterations, but they are different in the generation mechanism. Reassortment events, involving exchange of genome segments between two viruses, occur at varying rates during genome packaging in coinfected cells. Genome rearrangements, defined as “alterations of considerable tracts of sequence within single genome segments often in the form of deletions and extensions” (Taniguchi and Urasawa, 1995), are a common phenomenon in all major genera of the family Reoviridae (Table 1). They occur under natural conditions (Murao et al., 1996; Schnepf et al., 2008) and laboratory conditions, for example, by serial passage of rotaviruses at high multiplicities of infection (MOIs; Hundley et al., 1985; Kojima et al., 2000) and exclusive maintenance in one of their two hosts (plant, vector insect) in the case of plant reoviruses (Nuss, 1984). Rearrangements can be regarded as non-homologous, intramolecular RNA recombination and exclude intermolecular RNA recombinations that are documented frequently in plus-sense (+), single-stranded RNA (ssRNA) viruses but are infrequently reported for reoviruses (Taniguchi and Urasawa, 1995; Desselberger, 1996). Rearrangements are hypothesized to happen by copy choice (template switch) mechanism during RNA synthesis mediated by the RNA-dependent RNA polymerase (RdRp) complex, as is RNA recombination in ssRNA viruses.
In this review article we overview genome alterations, with a focus on unusual genome rearrangements observed in mycoreoviruses and discuss differences between other reovirus rearrangements in type of genome segment alterations and possible mechanisms underlying their occurrence. Readers are referred to excellent review articles on RNA recombination (Lai, 1992; Nagy and Simon, 1997; Simon-Loriere and Holmes, 2011; Sztuba-Solinska et al., 2011).
Three Types of Unusual Genome Segment Alterations in the Genus Mycoreovirus
Three types of very unusual genome alterations were reported to occur in mycoreoviruses that differ from those reported for other reoviruses. The first example is complete loss of one of the segment S8 reported for MyRV3, a very rare event for a reovirus. Reovirus genome segment sorting and assembly is tightly regulated during virus replication. An entire set of genome segments are believed to be incorporated and packaged into single core particles during replication (plus-sense strand synthesis). Therefore, all reovirus strains with rearranged segments still contain a set of all genome segments whether intact or altered. To our knowledge, the MyRV3 strain reported by Kanematsu et al. (2004) is the only example of reovirus that lacks a genome segment and is still viable. These observations allowed the authors to conclude that MyRV3 S8 is dispensable for maintenance under laboratory conditions.
The second type of mutations found in MyRV1 is unique among reovirus examples in their dependence on a distinct virus or a viral protein. Specifically, MyRV1 rearrangements are observed at extremely high rates in the fungal host either coinfected with the prototype hypovirus Cryphonectria hypovirus 1/EP713 (CHV1-EP713) or expressing a multifunctional protein p29 encoded by CHV1-EP713 (Sun and Suzuki, 2008). After repeated subculturing of those fungal strains for one to two months, several dozen percent of resulting MyRV1 isolates carry genome rearrangements. From doubly infected fungal mycelia, a MyRV1 variant, MyRV1/S10ss is isolated from 22.4% of subcultures, which harbors an altered segment S10ss, a deleted form of S10, in place of an intact S10. S10ss lacks approximately 75% of the internal ORF region while retaining the 5’ and 3’ terminal regions (Sun and Suzuki, 2008). In transformant fungal strains with the CHV1 p29 coding domain, a variety of rearrangements are generated in addition to S10ss that include S1L, S2L, S3L, and S6L (Figure 2). In contrast to S10ss, these rearranged segments involve ORF extensions, but are distinct from many previously reported animal reovirus examples. Extensions have been reported mostly from rotaviruses and those entail all head-to-tail tandem partial duplication occurring downstream of the termination codon of the authentic ORF. Thus, unaltered protein products are being synthesized from their cognate transcripts in infected cells. However, altered MyRV1 genome segments, i.e., S1L, S2L, S3L, and S6L are all with duplicated ORFs in-frame with their preceding ORFs, resulting in extension of ORFs by 1.4–1.9 fold. Importantly, the products of the expected sizes are detectable in mycelia infected with MyRV1 variants although S1L-encoded product remains to be detected (Sun and Suzuki, 2008; Tanaka et al., 2011). These extended segments S1L, S2L, S3L, and S6L are frequently concomitant with S10ss although its significance is unclear. S1, S3, and S6-encoded proteins carry sequence motifs characteristic of RdRp (Hillman et al., 2004), guanylyltransferase (Supyani et al., 2007), and NTP binding proteins (Suzuki et al., 2004). It should be noted that these sequence motifs are duplicated in the expected protein products. The capacity of core particles to encapsidate reovirus genomic segments is limited. The maximum size expansion of an orthoreovirus was predicted to be 10% of the entire size (approximately 2.0 kb) (Roner and Steele, 2007). Congruent with this notion, MyRV1/S1L + S10ss variant, with the largest extension per segment (approximately 2.3 kb), has a genome-based extension of 1.8 kb after deduction of the deleted S10 sequence in S10ss. MyRV1/S6L extends its entire genome-size by approximately 1.9 kb. These extensions correspond to approximately an 8% increase on a genome-size (23,433 bp) basis.
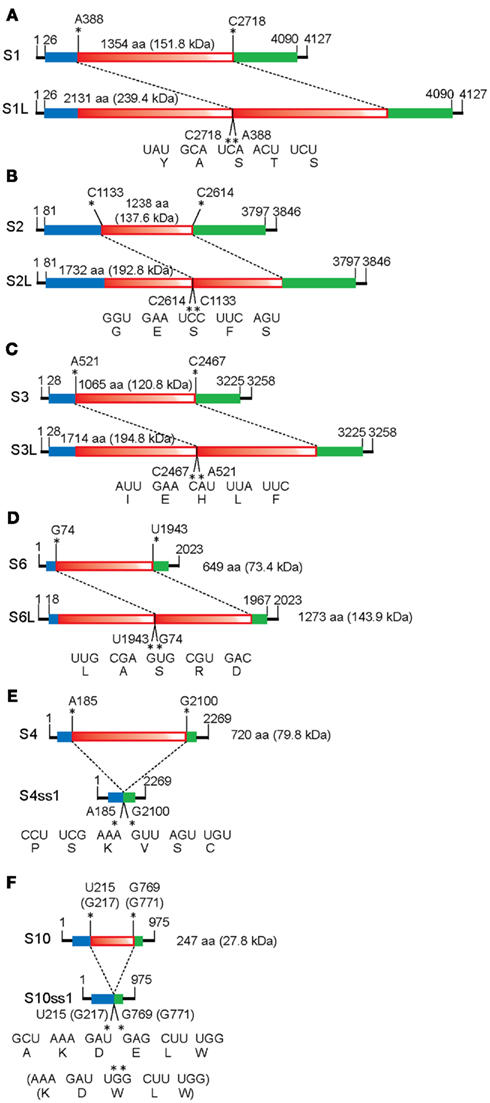
Figure 2. Schematic representation of the organization of MyRV1 rearranged segments. There are two types of MyRV1 rearrangements reported: extensions (S1L, S2L, S3L, and S6L) and deletions (S4ss and S10ss) that could happen by single events. S1L, S2L, and S3L result from in-frame ORF fusion but S4ss and S10ss do not always involve in-frame ORF deletions. Genetic organizations of the normal (top) and rearranged forms (bottom) of genome segments is shown. For extended segments S1L (A), S2L (B), S3L (C), and S6L (D), three fragments colored differently (blue, red, and green) are separated by break points. The red portions refer to duplicated sequence while red color gradation is to indicate orientation (head-to-tail for all rearrangements). In internal deletion mutations S4ss and S10ss (E,F), only 11–22% (for S4ss) and 10–43% (for S10ss) of the intact ORFs are retained. S4ss was generated spontaneously in a CHV1 p29 independent manner. Note that S4ss and S10ss each have at least 4 (S4ss1–S4ss4) and 2 variants (S10ss1 and 2) and one each of them (S4ss1 and S10ss1) are shown in (E) and (F), respectively. Solid lines denote the 5’ and 3’ UTRs, while blue and green boxes refer to the N and C terminal portions of ORF retained after each rearrangement event. Nucleotide positions for the start and stop codons, and rearrangement breakpoints are shown above the diagrams of each normal segments. Nucleotide and aa sequences adjacent to the altered sites on normal and rearranged segments are also shown below the diagrams of each altered segment. Neither sequence heterogeneity nor mismatch was detected at the junction sites for S1L, S2L, and S3L, while mismatches are found at or close to breakpoints of S4ss and S10ss (Sun and Suzuki, 2008; Eusebio-Cope et al., 2010).
The third example is obtained through a combined reassortment and rearrangement approach. MyRV1 S4 (Eusebio-Cope et al., 2010) and S10 (Suzuki, unpublished results), albeit very infrequently, are rearranged in a CHV1 p29 independent fashion. MyRV1/S4ss variants carry deletion mutations on S4 (S4ss1, S4ss2, S4ss3, and S4ss4) lacking approximately 80% of the S4 ORF but retaining its ability to be transcribed and replicated. The genome segments including S4ss, S4ss1–S4ss4 are different from each other in deletion endpoint. A reassortant variant of MyRV1 that contains substantial deletions in both S4 and S10 (MyRV1/S4ss1 + S10ss2) was generated in fungal colonies coinfected with MyRV1/S4ss1 and MyRV1/S10ss2. Surprisingly, MyRV1/S4ss + S10ss shows comparable replication levels to the wild-type MyRV1. This is the first example of a reovirus that is replication-competent without two viral proteins.
Implications in the Functional Roles of MyRV1 Genome Segments Undergoing Rearrangements
Unlike members of the genera Orthoreovirus (Kobayashi et al., 2007), Orbivirus (Boyce et al., 2008; Matsuo et al., 2010), and Rotavirus (Komoto et al., 2006), no reverse genetics system has been developed for mycoreoviruses. However, the MyRV1 rearrangements (shown in Figure 2) provide insights into the functional roles of the genome segments and their protein products. Common to all these altered rearrangements, they are competent to be packaged and transcribed given the comparable accumulation levels of their transcripts in infected cells. These results show that signals for packaging, transcription, and replication reside within the remaining genome sequence. The viability of MyRV1/S4ss + S10ss indicates that neither MyRV1 non-structural protein encoded by S4 (VP4) nor S10 (VP10) is required for virus replication. Furthermore, Eusebio-Cope et al. (2010) reported that VP4 contributes to mycelial growth and efficient vertical transmission through conidia. VP10 contributes inversely to the reduction of aerial mycelial growth induced by the virus (Sun and Suzuki, 2008). Recombination events altering protein sequences are often deleterious to its function and purified from the population (Simon-Loriere and Holmes, 2011; Sztuba-Solinska et al., 2011). In fact, most extensions reported previously for reoviruses occur downstream of the termination codon and thus result in no change of protein sequences (Kojima et al., 1996). In this sense, the fact that several MyRV1 variants with extension rearrangements are viable is surprising. Interpretation of functional roles for extended segments (S1L, S2L, S3L, and S6L) in virus replication is complex due to coexistence of the extended and normal unaltered genome segments in fungal colonies infected by MyRV1 variants. Therefore, heterocomplementation of rearranged segments by the normal segments cannot be ruled out. In this regard, an interesting observation was made by Tanaka et al. (2011), that fungal colonies infected with MyRV1/S1L + S10ss2, MyRV1/S2L + S10ss2, and MyRV1/S3L + S10ss2 are similarly restored in aerial mycelial growth to some extent (Figure 3). The restoration levels of aerial mycelial growth increase as the ratio of transcript levels of rearranged S1L to normal segment S1 rises (Tanaka et al., 2011).
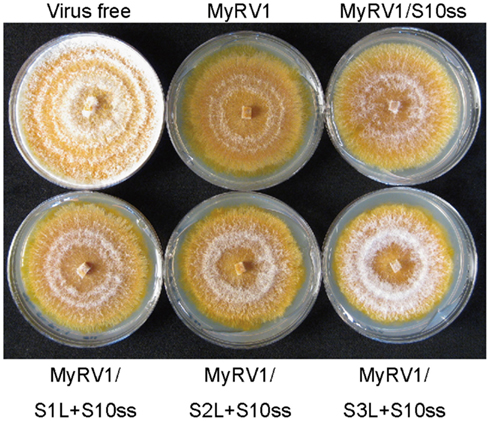
Figure 3. Effects of rearrangements on virus symptom induction. Fungal colonies were grown on PDA for 8 days under the bench top conditions approximately 24°C. C. parasitica strain EP155 was infected with wild-type MyRV1 (MyRV1), MyRV1/S10ss2 (MyRV1/S10ss), MyRV1/S1L + S10ss2 (MyRV1/S1L + S10ss), MyRV1/S2L + S10ss2 (MyRV1/S2L + S10ss), or MyRV1/S3L + S10ss2 (MyRV1/S3L + S10ss). Virus-free EP155 was cultured in parallel.
As in other reoviruses, rearrangements are considered to contribute to evolution and molecular diversity of mycoreoviruses, while mycoreoviruses with such rearrangements including the MyRV3 S8 loss are found under laboratory conditions, but not in the natural environment. MyRV1 variants with large deletions may have selective advantages in replication or packaging over wild-type MyRV1. MyRV1 variants with large extensions require CHV1 p29 for their maintenance, suggesting no selective advantage of these variants in the absence of p29. Maintenance of animal viruses in cultured host cells frequently induces mutations that confer selective advantages under laboratory conditions and lead to virulence attenuation in host animals. In this regard, MyRV1 with genome rearrangements are different because they retain the ability to induce symptoms, while distinct from those induced by wild-type MyRV1 (Figure 3), including the reduction of host virulence to plants. Rather, MyRV1 is reminiscent of plant reoviruses which undergo various mutations when they are maintained exclusively in one of their hosts, plant and insect vectors in laboratory conditions. Examples include those of would tumor virus (WTV) and rice dwarf virus (RDV) that are vectored by different species of leafhoppers (Table 1). In fact, transmission defective isolates of WTV emerge after being maintained for a long period of time in cultured insect cells that carry internal deletions on segments encoding structural and non-structural proteins (Nuss, 1984; Anzola et al., 1987). In the case of RDV maintained only in plants, point mutations rather than internal deletions accumulate resulting in little expression of protein products encoded by the mutated segments. Consequently, such isolates dominate the virus population. However, one single passage in cultured insect vector cell lines resulted in inverse population structures (Pu et al., 2011).
Mechanisms of General RNA Recombination and Reovirus Rearrangements
Rearrangements are a type of RNA recombination. Two major mechanisms are proposed for it, specifically, replication independent breakage-rejoining recombination, and replication dependent RdRp-mediated recombination (template switch or copy choice). The non-replicative, breaking-joining mechanism entails dsRNA or ssRNA fragmentation and re-ligation for which several variations are proposed such as self-recombination requiring no proteins, and RNase-catalyzed cleavage and ligation (Chetverin, 1999). The second model, replicase-mediated mechanism is more widely accepted (Lai, 1992; Nagy and Simon, 1997; Sztuba-Solinska et al., 2011). In this model, the nascent RNA and RdRp complex jumps after proceeding to a certain point from one template to another (intergenic or intermolecular recombination) or to another point of the same template (intragenic or intramolecular recombination). Template switches could occur in homologous and non-homologous (illegitimate) manners such that their crossover regions are of similar and unrelated sequences respectively. Replicative non-homologous recombination is sometimes considered to be facilitated by sequences features of template and acceptor strands, such as direct, inverted repeats and/or secondary structures. A number of viral RNA and protein elements as well as host factors are involved in RNA recombinations in different single-stranded, plus-sense RNA plant virus/host systems (Serviene et al., 2005; Sztuba-Solinska et al., 2011). Recently, RNA silencing was shown to contribute to the generation of defective-interfering (DI) RNAs of the prototype hypovirus (CHV1; Zhang and Nuss, 2008), associated with the picornavirus superfamily (Koonin et al., 1991), that infects the chestnut blight fungus and confers hypovirulence.
Rearrangements reported for members of the family Reoviridae could be accounted for by an intramolecular template switch, in which the replicase complex jumps backward (for extension) or forward (for deletions) on the same template molecule. For example, intragenic rearrangements of human rotavirus segment S11 are hypothesized to be generated in the process of plus-sense strand synthesis (transcription) mediated by short direct repeats of 4–6 nucleotides (Kojima et al., 1996; Schnepf et al., 2008; Table 1). Gault et al. (2001) inferred that duplications of human rotavirus genes 7 and 11 were presumed to occur by template switch mediated by an inverted repeat-involving secondary structure during the replication step (Gault et al., 2001; Table 1). A rare example is intergenic recombination with a crossover of two rotavirus segments encoding NSP2 and NSP5,6 (Cao et al., 2008; Table 1). Recent structural studies led to 3D modeling of RdRps from two reoviruses, human orthoreovirus and rotavirus. They are commonly characterized by 4-window structures which allow entry of substrates (rear window) and templates (left window), and exit of template RNA (front window) and nascent RNA (bottom window; Tao et al., 2002; Lu et al., 2008; Guglielmi et al., 2010). Based on this RdRp structural prediction, a loop model was proposed to explain how reovirus extension rearrangements occur (Matthijnssens et al., 2006). According to this model, extension rearrangements occur during the transcription step via intramolecular template switch from the transcribed region of the minus-strand to upstream regions of the same molecule re-entering from the left channel. It remains unknown how the mechanism of intergenic recombination or large deletion rearrangements fits into the RdRp 3D model.
Implication in the Mechanism of CHV1 P29-Mediated Generation of MyRV1 Rearrangements
Addressing how p29 is involved in the enhanced emergence of MyRV1 rearrangements is an interesting challenge. Before discussing this issue, replication of hypoviruses and reoviruses should be briefly explained. Hypovirus RNA are replicated in Golgi-derived vesicles of ∼80 nm whose production is enhanced in the cytoplasm upon viral infection (Fahima et al., 1994; Hillman and Suzuki, 2004; Jacob-Wilk et al., 2006). Reovirus RNAs are synthesized in viroplasms or virus factories formed in the cytoplasm of infected cells by viral and host components (Patton et al., 2006; Cheung et al., 2010). CHV1 p29 is a multifunctional protein acting as a symptom determinant (Choi and Nuss, 1992; Craven et al., 1993; Suzuki et al., 1999, 2003), protease (Choi et al., 1991a,b), an RNA silencing suppressor (Segers et al., 2006), and an enhancer of heterologous and homologous viruses (Suzuki et al., 2003; Sun et al., 2006). Some functional domains of p29 are mapped such as the N terminal aa 1–24 essential for virus viability, adjacent 25–74 for the symptom determinant and the C terminal half for the proteolytic catalytic domain (Suzuki and Nuss, 2002; Hillman et al., 2004).
Two possibilities are proposed to account for the p29-mediated generation of MyRV1 rearrangements. Firstly, p29 may interact the RNA synthesis machinery to perturb transcription (plus-sense RNA synthesis) and/or replication (minus-sense RNA synthesis), leading to enhanced rate of template switch of RdRp-mediated via RNA sequence features such as inverted or direct repeats. Another possibility is that p29 alters the physiological state of infected cells, which enhances selection of pre-existing mutant viruses with rearranged segments by unknown means. These two possibilities are not mutually exclusive. Regarding the first possibility, it remains unknown whether CHV1 p29 reside in MyRV1 core particles viewed as a nano-scale RNA synthesis factory (Guglielmi et al., 2010) to perturb MyRV1 RNA synthesis. Sun and Suzuki (2008) showed that CHV1 p29 interacts with MyRV1 VP9, considered to be a non-structural protein, in vitro and in vivo, providing a clue to the mechanism. As in cells infected with other reoviruses, viroplasm is produced in MyRV1-infected cells (Sun and Suzuki, unpublished data). One or two viral non-structural proteins are required as matrix proteins for production of reoviral viroplasms which are believed to be the site of virus replication where core particle assembly and RNA synthesis occur. Consistent with this notion, knocking-down of the genes responsible for functional viroplasm formation is detrimental for virus replication (Campagna et al., 2007; Shimizu et al., 2009, 2011). As discussed above, a MyRV1 variant shows comparable replication competency without VP4 and VP10. Therefore, VP4 and VP10 are unlikely to be matrix proteins of viroplasms given the essentiality of viroplasm. One of key points to be addressed is what the possible role of VP9 in MyRV1 replication is. It is of interest to infer that VP9 is a viroplasm matrix protein that is remodeled upon interaction with p29. Moreover, to explore whether CHV1 p29 is incorporated into MyRV1 core particles is a prerequisite.
The other possibility may be associated with the reason why almost a half of the MyRV1 segments, whether encoding structural or non-structural proteins, are generated at high rates. CHV1 p29, as a symptom determinant, slightly enhances mycelial growth and suppresses asexual sporulation and orange pigmentation, which involve complex regulatory pathways. Additionally, p29 is an RNA silencing suppressor which may potentially be able to perturb the cellular physiological state. The p29 protein might be involved in selection and maintenance of MyRV1 variants with rearrangements. Apparent “reversion” of S1L, S2L, S3L, and S6L to respective normal segments (Sun and Suzuki, 2008; Tanaka et al., 2011) in the absence of p29 suggests a role for the protein in the maintenance of the extended segments.
Conclusion
Three unusual types of genome alterations are found in the genus Mycoreovirus; a S8-deficient MyRV3 mutant, a MyRV1 variant with two rearranged segments (S4ss and S10ss) each lacking three fourths of the ORF, and MyRV1 variants with genome rearrangements (S1L, S2L, S3L, S6L, and S10ss) induced by a multifunctional protein p29 of an unrelated virus, CHV1. While S8-lacking MyRV3 strains have properties indistinguishable from wild-type MyRV3, most MyRV1 variants with rearranged segments induced different symptoms in the chestnut blight fungus than the wild-type virus. Therefore, these genome segment alterations are useful for gaining insights into functional roles of genome segments of mycoreoviruses for which a reverse genetics system is unavailable like many other Reoviridae members. The CHV1 p29-dependent rearrangements of MyRV1 are a novel type of intriguing virus/virus interactions, and accordingly there are a few important unanswered questions. What is the biological significance of the interactions between the two viruses? Do either of the viruses benefit from the interactions? What function of CHV1 p29 is related to rearrangements? Does p29 contribute to generate de novo genome rearrangements or to select pre-existing rearrangements by altering the cellular state, while it is poorly defined at present? Does CHV1 p29 induce genome rearrangements on other reoviruses such as MyRV2 and MyRV3? Experiments to address these issues are underway.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Yomogi Inc. and the Program for Promotion of Basic and Applied Researches for Innovations in Bio-Oriented Industry for financial support during this study. The authors wish to thank Drs. Bradley I. Hillman and William Macdonald for their generous gift of fungal strain Cryphonectria parasitica 9B21.
References
Anthony, S. J., Darpel, K. E., Belaganahalli, M. N., Maan, N., Nomikou, K., Sutton, G., Attoui, H., Maan, S., and Mertens, P. P. (2011). RNA segment 9 exists as a duplex concatemer in an Australian strain of epizootic haemorrhagic disease virus (EHDV): genetic analysis and evidence for the presence of concatemers as a normal feature of orbivirus replication. Virology 420, 164–171.
Anzola, J. V., Xu, Z. K., Asamizu, T., and Nuss, D. L. (1987). Segment-specific inverted repeats found adjacent to conserved terminal sequences in wound tumor virus genome and defective interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 84, 8301–8305.
Arella, M., Lavallee, C., Belloncik, S., and Furuichi, Y. (1988). Molecular cloning and characterization of cytoplasmic polyhedrosis virus polyhedrin and a viable deletion mutant gene. J. Virol. 62, 211–217.
Attoui, H., Mertens, P. P. C., Becnel, J., Belaganahalli, S., Bergoin, M., Brussaard, C. P., Chappell, J. D., Ciarlet, M., del Vas, M., Dermody, T. S., Dormitzer, P. R., Duncan, R., Fcang, Q., Graham, R., Guglielmi, K. M., Harding, R. M., Hillman, B., Makkay, A., Marzachì, C., Matthijnssens, J., Milne, R. G., Mohd Jaafar, F., Mori, H., Noordeloos, A. A., Omura, T., Patton, J. T., Rao, S., Maan, M., Stoltz, D., Suzuki, N., Upadhyaya, N. M., Wei, C., and Zhou, H. (2012). “Family Reoviridae,” in Virus Taxonomy: Ninth Report of the International Committee for the Taxonomy of Viruses, eds A. M. Q. King, M. J. Adams, E. B. Carstens, and E. J. Lefkowits (New York: Elsevier, Academic Press), 541–637.
Boyce, M., Celma, C. C., and Roy, P. (2008). Development of reverse genetics systems for bluetongue virus: recovery of infectious virus from synthetic RNA transcripts. J. Virol. 82, 8339–8348.
Campagna, M., Budini, M., Arnoldi, F., Desselberger, U., Allende, J. E., and Burrone, O. R. (2007). Impaired hyperphosphorylation of rotavirus NSP5 in cells depleted of casein kinase 1alpha is associated with the formation of viroplasms with altered morphology and a moderate decrease in virus replication. J. Gen. Virol. 88, 2800–2810.
Cao, D., Barro, M., and Hoshino, Y. (2008). Porcine rotavirus bearing an aberrant gene stemming from an intergenic recombination of the NSP2 and NSP5 genes is defective and interfering. J. Virol. 82, 6073–6077.
Cheung, W., Gill, M., Esposito, A., Kaminski, C. F., Courousse, N., Chwetzoff, S., Trugnan, G., Keshavan, N., Lever, A., and Desselberger, U. (2010). Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J. Virol. 84, 6782–6798.
Choi, G. H., and Nuss, D. L. (1992). A viral gene confers hypovirulence-associated traits to the chestnut blight fungus. EMBO J. 11, 473–477.
Choi, G. H., Pawlyk, D. M., and Nuss, D. L. (1991a). The autocatalytic protease p29 encoded by a hypovirulence-associated virus of the chestnut blight fungus resembles the potyvirus-encoded protease HC-Pro. Virology 183, 747–752.
Choi, G. H., Shapira, R., and Nuss, D. L. (1991b). Cotranslational autoproteolysis involved in gene expression from a double-stranded RNA genetic element associated with hypovirulence of the chestnut blight fungus. Proc. Natl. Acad. Sci. U.S.A. 88, 1167–1171.
Craven, M. G., Pawlyk, D. M., Choi, G. H., and Nuss, D. L. (1993). Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 67, 6513–6521.
Eaton, B. T., and Gould, A. R. (1987). Isolation and characterization of orbivirus genotypic variants. Virus Res. 6, 363–382.
Enebak, S. A., Hillman, B. I., and Macdonald, W. L. (1994). A hypovirulent isolate of Cryphonectria parasitica with multiple, genetically unique dsRNA segments. Mol. Plant Microbe Interact. 7, 590–595.
Eusebio-Cope, A., Sun, L., Hillman, B. I., and Suzuki, N. (2010). Mycoreovirus 1 S4-coded protein is dispensable for viral replication but necessary for efficient vertical transmission and normal symptom induction. Virology 397, 399–408.
Fahima, T., Wu, Y., Zhang, L., and Van Alfen, N. K. (1994). Identification of the putative RNA polymerase of cryphonectria hypovirus in a solubilized replication complex. J. Virol. 68, 6116–6119.
Gault, E., Schnepf, N., Poncet, D., Servant, A., Teran, S., and Garbarg-Chenon, A. (2001). A human rotavirus with rearranged genes 7 and 11 encodes a modified NSP3 protein and suggests an additional mechanism for gene rearrangement. J. Virol. 75, 7305–7314.
Guglielmi, K. M., McDonald, S. M., and Patton, J. T. (2010). Mechanism of intraparticle synthesis of the rotavirus double-stranded RNA genome. J. Biol. Chem. 285, 18123–18128.
Hillman, B. I., Supyani, S., Kondo, H., and Suzuki, N. (2004). A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the coltivirus genus of animal pathogens. J. Virol. 78, 892–898.
Hillman, B. I., and Suzuki, N. (2004). Viruses of the chestnut blight fungus, Cryphonectria parasitica. Adv. Virus Res. 63, 423–472.
Hundley, F., Biryahwaho, B., Gow, M., and Desselberger, U. (1985). Genome rearrangements of bovine rotavirus after serial passage at high multiplicity of infection. Virology 143, 88–103.
Jacob-Wilk, D., Turina, M., and Van Alfen, N. K. (2006). Mycovirus cryphonectria hypovirus 1 elements cofractionate with trans-Golgi network membranes of the fungal host Cryphonectria parasitica. J. Virol. 80, 6588–6596.
Kanematsu, S., Arakawa, M., Oikawa, Y., Onoue, M., Osaki, H., Nakamura, H., Ikeda, K., Kuga-Uetake, Y., Nitta, H., Sasaki, A., Suzaki, K., Yoshida, K., and Matsumoto, N. (2004). A reovirus causes hypovirulence of Rosellinia necatrix. Phytopathology 94, 561–568.
Kobayashi, T., Antar, A. A., Boehme, K. W., Danthi, P., Eby, E. A., Guglielmi, K. M., Holm, G. H., Johnson, E. M., Maginnis, M. S., Naik, S., Skelton, W. B., Wetzel, J. D., Wilson, G. J., Chappell, J. D., and Dermody, T. S. (2007). A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1, 147–157.
Kojima, K., Taniguchi, K., Kawagishi-Kobayashi, M., Matsuno, S., and Urasawa, S. (2000). Rearrangement generated in double genes, NSP1 and NSP3, of viable progenies from a human rotavirus strain. Virus Res. 67, 163–171.
Kojima, K., Taniguchi, K., Urasawa, T., and Urasawa, S. (1996). Sequence analysis of normal and rearranged NSP5 genes from human rotavirus strains isolated in nature: implications for the occurrence of the rearrangement at the step of plus strand synthesis. Virology 224, 446–452.
Komoto, S., Sasaki, J., and Taniguchi, K. (2006). Reverse genetics system for introduction of site-specific mutations into the double-stranded RNA genome of infectious rotavirus. Proc. Natl. Acad. Sci. U.S.A. 103, 4646–4651.
Koonin, E. V., Choi, G. H., Nuss, D. L., Shapira, R., and Carrington, J. C. (1991). Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. U.S.A. 88, 10647–10651.
Lu, X., McDonald, S. M., Tortorici, M. A., Tao, Y. J., Vasquez-Del Carpio, R., Nibert, M. L., Patton, J. T., and Harrison, S. C. (2008). Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure 16, 1678–1688.
Maoka, T., Omura, T., Harjosudarmo, J., Usugi, T., Hibino, H., and Tsuchizaki, T. (1993). Loss of vector-transmissibility by maintaining rice ragged stunt virus in rice plants without vector transmission. Nippon Shokubutsu Byori Gakkaiho 59, 185–187.
Matsuo, E., Celma, C. C., and Roy, P. (2010). A reverse genetics system of African horse sickness virus reveals existence of primary replication. FEBS Lett. 584, 3386–3391.
Matthijnssens, J., Rahman, M., and Van Ranst, M. (2006). Loop model: mechanism to explain partial gene duplications in segmented dsRNA viruses. Biochem. Biophys. Res. Commun. 340, 140–144.
Murao, K., Uyeda, I., Ando, Y., Kimura, I., Cabauatan, P. Q., and Koganezawa, H. (1996). Genomic rearrangement in genome segment 12 of rice dwarf phytoreovirus. Virology 216, 238–240.
Nagy, P. D., and Simon, A. E. (1997). New insights into the mechanisms of RNA recombination. Virology 235, 1–9.
Ni, Y., and Kemp, M. C. (1994). Subgenomic S1 segments are packaged by avian reovirus defective interfering particles having an S1 segment deletion. Virus Res. 32, 329–342.
Patton, J. T., Silvestri, L. S., Tortorici, M. A., Vasquez-Del Carpio, R., and Taraporewala, Z. F. (2006). Rotavirus genome replication and morphogenesis: role of the viroplasm. Curr. Top. Microbiol. Immunol. 309, 169–187.
Pu, Y., Kikuchi, A., Moriyasu, Y., Tomaru, M., Jin, Y., Suga, H., Hagiwara, K., Akita, F., Shimizu, T., Netsu, O., Suzuki, N., Uehara-Ichiki, T., Sasaya, T., Wei, T., Li, Y., and Omura, T. (2011). Rice dwarf viruses with dysfunctional genomes generated in plants are filtered out in vector insects-implications for the virus origin. J. Virol. 85, 2975–2979.
Roner, M. R., and Steele, B. G. (2007). Features of the mammalian orthoreovirus 3 Dearing l1 single-stranded RNA that direct packaging and serotype restriction. J. Gen. Virol. 88, 3401–3412.
Schnepf, N., Deback, C., Dehee, A., Gault, E., Parez, N., and Garbarg-Chenon, A. (2008). Rearrangements of rotavirus genomic segment 11 are generated during acute infection of immunocompetent children and do not occur at random. J. Virol. 82, 3689–3696.
Segers, G. C., van Wezel, R., Zhang, X., Hong, Y., and Nuss, D. L. (2006). Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryotic Cell 5, 896–904.
Serviene, E., Shapka, N., Cheng, C. P., Panavas, T., Phuangrat, B., Baker, J., and Nagy, P. D. (2005). Genome-wide screen identifies host genes affecting viral RNA recombination. Proc. Natl. Acad. Sci. U.S.A. 102, 10545–10550.
Shen, S., Burke, B., and Desselberger, U. (1994). Rearrangement of the VP6 gene of a group A rotavirus in combination with a point mutation affecting trimer stability. J. Virol. 68, 1682–1688.
Shimizu, T., Nakazono-Nagaoka, E., Akita, F., Uehara-Ichiki, T., Omura, T., and Sasaya, T. (2011). Immunity to rice black streaked dwarf virus, a plant reovirus, can be achieved in rice plants by RNA silencing against the gene for the viroplasm component protein. Virus Res. 160, 400–403.
Shimizu, T., Yoshii, M., Wei, T., Hirochika, H., and Omura, T. (2009). Silencing by RNAi of the gene for Pns12, a viroplasm matrix protein of rice dwarf virus, results in strong resistance of transgenic rice plants to the virus. Plant Biotechnol. J. 7, 24–32.
Simon-Loriere, E., and Holmes, E. C. (2011). Why do RNA viruses recombine? Nat. Rev. Microbiol. 9, 617–626.
Spear, A., Sisterson, M. S., and Stenger, D. C. (2012). Reovirus genomes from plant-feeding insects represent a newly discovered lineage within the family Reoviridae. Virus Res. 163, 503–511.
Sun, L., Nuss, D. L., and Suzuki, N. (2006). Synergism between a mycoreovirus and a hypovirus mediated by the papain-like protease p29 of the prototypic hypovirus CHV1-EP713. J. Gen. Virol. 87, 3703–3714.
Sun, L., and Suzuki, N. (2008). Intragenic rearrangements of a mycoreovirus induced by the multifunctional protein p29 encoded by the prototypic hypovirus CHV1-EP713. RNA 14, 2557–2571.
Supyani, S., Hillman, B. I., and Suzuki, N. (2007). Baculovirus expression of the 11 mycoreovirus-1 genome segments and identification of the guanylyl transferase-encoding segment. J. Gen. Virol. 88, 342–350.
Suzuki, N., Chen, B., and Nuss, D. L. (1999). Mapping of a hypovirus p29 protease symptom determinant domain with sequence similarity to potyvirus HC-Pro protease. J. Virol. 73, 9478–9484.
Suzuki, N., Maruyama, K., Moriyama, M., and Nuss, D. L. (2003). Hypovirus papain-like protease p29 functions in trans to enhance viral double-stranded RNA accumulation and vertical transmission. J. Virol. 77, 11697–11707.
Suzuki, N., and Nuss, D. L. (2002). Contribution of protein p40 to hypovirus-mediated modulation of fungal host phenotype and viral RNA accumulation. J. Virol. 76, 7747–7759.
Suzuki, N., Supyani, S., Maruyama, K., and Hillman, B. I. (2004). Complete genome sequence of Mycoreovirus-1/Cp9B21, a member of a novel genus within the family Reoviridae, isolated from the chestnut blight fungus Cryphonectria parasitica. J. Gen. Virol. 85, 3437–3448.
Sztuba-Solinska, J., Urbanowicz, A., Figlerowicz, M., and Bujarski, J. J. (2011). RNA-RNA recombination in plant virus replication and evolution. Annu. Rev. Phytopathol. 49, 415–443.
Tanaka, T., Sun, L., Tsutani, K., and Suzuki, N. (2011). Rearrangements of mycoreovirus 1 S1, S2 and S3 induced by the multifunctional protein p29 encoded by the prototypic hypovirus cryphonectria hypovirus 1 strain EP713. J. Gen. Virol. 92, 1949–1959.
Tao, Y., Farsetta, D. L., Nibert, M. L., and Harrison, S. C. (2002). RNA synthesis in a cage – structural studies of reovirus polymerase lambda3. Cell 111, 733–745.
Wei, C. Z., Osaki, H., Iwanami, T., Matsumoto, N., and Ohtsu, Y. (2004). Complete nucleotide sequences of genome segments 1 and 3 of Rosellinia anti-rot virus in the family Reoviridae. Arch. Virol. 149, 773–777.
Zhang, X., and Nuss, D. L. (2008). A host dicer is required for defective viral RNA production and recombinant virus vector RNA instability for a positive sense RNA virus. Proc. Natl. Acad. Sci. U.S.A. 105, 16749–16754.
Keywords: reovirus, rearrangement, mycoreovirus, Cryphonectria parasitica, chestnut blight, hypovirus, papain-like protease p29, dsRNA
Citation: Tanaka T, Eusebio-Cope A, Sun L and Suzuki N (2012) Mycoreovirus genome alterations: similarities to and differences from rearrangements reported for other reoviruses. Front. Microbio. 3:186. doi: 10.3389/fmicb.2012.00186
Received: 14 March 2012; Accepted: 04 May 2012;
Published online: 01 June 2012.
Edited by:
Joseph K. Li, Utah State University, USAReviewed by:
Dale L. Barnard, Utah State University, USAKwok-Yung Yuen, The University of Hong Kong, Hong Kong
Copyright: © 2012 Tanaka, Eusebio-Cope, Sun and Suzuki. This is an openaccess article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Nobuhiro Suzuki, Agrivirology Laboratory, Institute of Plant Science and Bioresources, Okayama University, Kurashiki, Okayama 710-0046, Japan. e-mail: nsuzuki@rib.okayama-u.ac.jp
 Toru Tanaka1
Toru Tanaka1