- 1 Department of Marine Sciences, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
- 2 Center for Geomicrobiology, Aarhus University, Aarhus, Denmark
- 3 Bigelow Laboratory for Ocean Sciences, East Boothbay, ME, USA
- 4 Rosenstiel School of Marine and Atmospheric Science, University of Miami, Miami, FL, USA
The microbiology of subsurface, hydrothermally influenced basaltic crust flanking mid-ocean ridges has remained understudied, due to the difficulty in accessing the subsurface environment. The instrumented boreholes resulting from scientific ocean drilling offer access to samples of the formation fluids circulating through oceanic crust. We analyzed the phylogenetic diversity of bacterial communities of fluid and microbial mat samples collected in situ from the observatory at Ocean Drilling Program Hole 896A, drilled into ~6.5 million-year-old basaltic crust on the flank of the Costa Rica Rift in the equatorial Pacific Ocean. Bacterial 16S rRNA gene sequences recovered from borehole fluid and from a microbial mat coating the outer surface of the fluid port revealed both unique and shared phylotypes. The dominant bacterial clones from both samples were related to the autotrophic, sulfur-oxidizing genus Thiomicrospira. Both samples yielded diverse gamma- and alphaproteobacterial phylotypes, as well as members of the Bacteroidetes, Planctomycetes, and Verrucomicrobia. Analysis of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) genes (cbbL and cbbM) from the sampling port mat and from the borehole fluid demonstrated autotrophic carbon assimilation potential for in situ microbial communities; most cbbL genes were related to those of the sulfur-oxidizing genera Thioalkalivibrio and Thiomicrospira, and cbbM genes were affiliated with uncultured phylotypes from hydrothermal vent plumes and marine sediments. Several 16S rRNA gene phylotypes from the 896A observatory grouped with phylotypes recovered from seawater-exposed basalts and sulfide deposits at inactive hydrothermal vents, but there is little overlap with hydrothermally influenced basaltic boreholes 1026B and U1301A on the Juan de Fuca Ridge flank, suggesting that site-specific characteristics of Hole 896A (i.e., seawater mixing into borehole fluids) affect the microbial community composition.
Introduction
The deep-sea subsurface is characterized by relatively low organic carbon input, elevated pressures, geographically variable temperatures, and sparse nutrient availability. Together, these conditions create unique and challenging habitats for microorganisms. Sub-seafloor sediments are estimated to constitute a large proportion of Earth’s biomass (Whitman et al., 1998; Parkes et al., 2000), although local cell densities and microbial activities are low compared to surficial sediments (D’Hondt et al., 2004, 2009; Jørgensen, 2007). Less is known, however, about a biosphere potentially hosted in subsurface oceanic basalt crust, largely due to difficulties is accessing this environment. Chemical and microscopic evidence coupled with theoretical models of potential metabolic reactions (Bach and Edwards, 2003) suggest that microbial activity in oceanic crust occurs and may have a significant impact on global biogeochemical cycles. Specifically, gradual microbial oxidation of reduced metals and sulfur species within the basalt crusts results in increased oxidation state and biocatalyzed weathering of ocean crust, as it ages and moves from the center of a mid-ocean ridge to the outer flanks (Bach and Edwards, 2003). Microaerophilic, autotrophic iron-oxidizing, or nitrate-reducing bacteria from ridge flank surfaces have been isolated, and include alpha- and gammaproteobacteria (Edwards et al., 2003); seafloor basalts from the 9°N East Pacific Rise harbor mostly gamma- and alphaproteobacteria, and Planctomycetes (Santelli et al., 2008, 2009). The question is whether similar bacteria are active in the deep basaltic subsurface.
Scientific ocean drilling provides one potential mechanism for the collection of deep subsurface samples for microbiological analysis. Such drilling is often conducted within the framework of the Integrated Ocean Drilling Program (IODP) and its precursor, the Ocean Drilling Program (ODP). Subsurface drilling can work well in sedimentary layers, since contamination monitoring protocols allow for stringent control of seawater and drilling fluid contamination (Lever et al., 2006). By contrast, collection of pristine hard rock crustal samples through drilling is often problematic (Santelli et al., 2010), and rock sample surfaces are generally prone to seawater and drilling fluid contamination (Lever et al., 2006). Installation of long-term observatories within the resulting borehole offers an alternative for exploring subsurface crustal environments. Such borehole observatories, known as Circulation Obviation Retrofit Kits or “CORKs” (Davis et al., 1992; Becker and Davis, 2005), have been used successfully in the past to study hydrogeologic properties of crustal aquifers (Fisher et al., 2008) and to examine the biogeochemical composition of fluids circulating within oceanic crust (Wheat et al., 2010).
More recently, the composition of microbial communities inhabiting deep basaltic crust has been investigated through the collection of observatory fluids (Cowen et al., 2003), mineral crusts formed on the surface on observatories (Nakagawa et al., 2006), and mineral chip colonization experiments conducted within the observatories (Orcutt et al., 2011). Those experiments have focused on a series of CORK observatories, mostly at ODP Hole 1026B and IODP Hole U1301A (Fisher et al., 2005), installed on the eastern flank of the Juan de Fuca Ridge in the northeastern Pacific Ocean. Here, reduced, anoxic, sulfate-rich hydrothermal (~64°C) fluids flow within 3.5 million-year-old basaltic crust. Analysis of borehole fluids and mineral crusts from these sites revealed diverse microbial communities containing novel Firmicutes bacteria, some of which are distantly related to Ammonifex degensii, a chemolithoautotrophic, thermophilic bacterium that oxidizes hydrogen, formate or pyruvate with nitrate, sulfate or elemental sulfur (Cowen et al., 2003; Nakagawa et al., 2006; Orcutt et al., 2011). Furthermore, enrichment experiments conducted on mineral crusts from the Hole 1026B observatory yielded anaerobic thermophiles, including methanogens, fermenters, and sulfate reducers (Nakagawa et al., 2006; Steinsbu et al., 2010). Archaeal communities in these observatories appear to be less diverse and align more closely with cultivated members of thermophilic, hydrogenotrophic methanogens of the genus Methanothermococcus (Nakagawa et al., 2006). Similar microbial communities of deep basalt origin might permeate adjacent layers of the overlying sediment column, as indicated by elevated counts of bacteria and archaea in sediments just above the basalt/sediment interface at Juan de Fuca (Engelen et al., 2008). The microbial community composition of the Juan de Fuca Ridge flank crustal subsurface differs significantly from the composition of communities harbored in seafloor-exposed basalts (Santelli et al., 2008, 2009), near-surface hydrothermal fluids circulating within basaltic outcrops (Huber et al., 2006), and hydrothermal fluids collected from sulfide- and/or iron-rich hydrothermal vents (Suzuki et al., 2004; Perner et al., 2007; Takai et al., 2008; Rassa et al., 2009). This disparity suggests that, although the crustal subsurface shares similar mineralogical and geochemical characteristics with the other habitats, it nonetheless harbors a unique subsurface crustal biosphere (Orcutt et al., 2011). Investigations of other subsurface crustal habitats are necessary to evaluate this hypothesis.
Here, we report the results of culture-independent 16S rRNA and RuBisCO gene molecular surveys conducted on opportunistic borehole fluid and mineral crust samples collected from an observatory in ODP Hole 896A. Hole 896A is located on the southern flank of the Costa Rica Rift in the equatorial Pacific Ocean at a water depth of 3,440 m (Shipboard Scientific Party, 1993). The borehole was drilled in 1993 to a maximum depth of 469 meters below seafloor (mbsf), with the lowermost 290 m penetrating roughly 6.8 million-year-old altered basaltic basement. Many features of the Hole 896A crustal aquifer are similar to those observed at the Juan de Fuca Ridge flank sites mentioned earlier. For instance, both sites are characterized by pillow and sheet-flow plagioclase-olivine phyric basalts; and vein filling with saponite and celadonite alteration minerals indicates complex hydrothermal evolutions of the systems (Alt et al., 1996; Hunter et al., 1999). Furthermore, formation fluids at Hole 896A are roughly 58°C (Becker et al., 2004), which is similar to those observed in the Juan de Fuca boreholes. Chemically, the basement fluids at both sites are characterized by reduced, high sulfate, low alkalinity fluids with relatively high calcium and low magnesium contents, indicating hydrothermal fluid-rock interactions (Table 1). Finally, both sites are located on buried basement highs (Alt et al., 1996; Fisher et al., 2005) with inferred fluid flow on the order of hundreds of liters per hour (Becker et al., 2004). As at the Juan de Fuca Ridge flank sites, microbial alteration textures, elemental analyses, cell counts, extractable DNA, and carbon and oxygen isotope data from basalts of Hole 896A indicate microbial activity (Giovannoni et al., 1996; Fisk et al., 1998; Torsvik et al., 1998; Furnes et al., 2001).
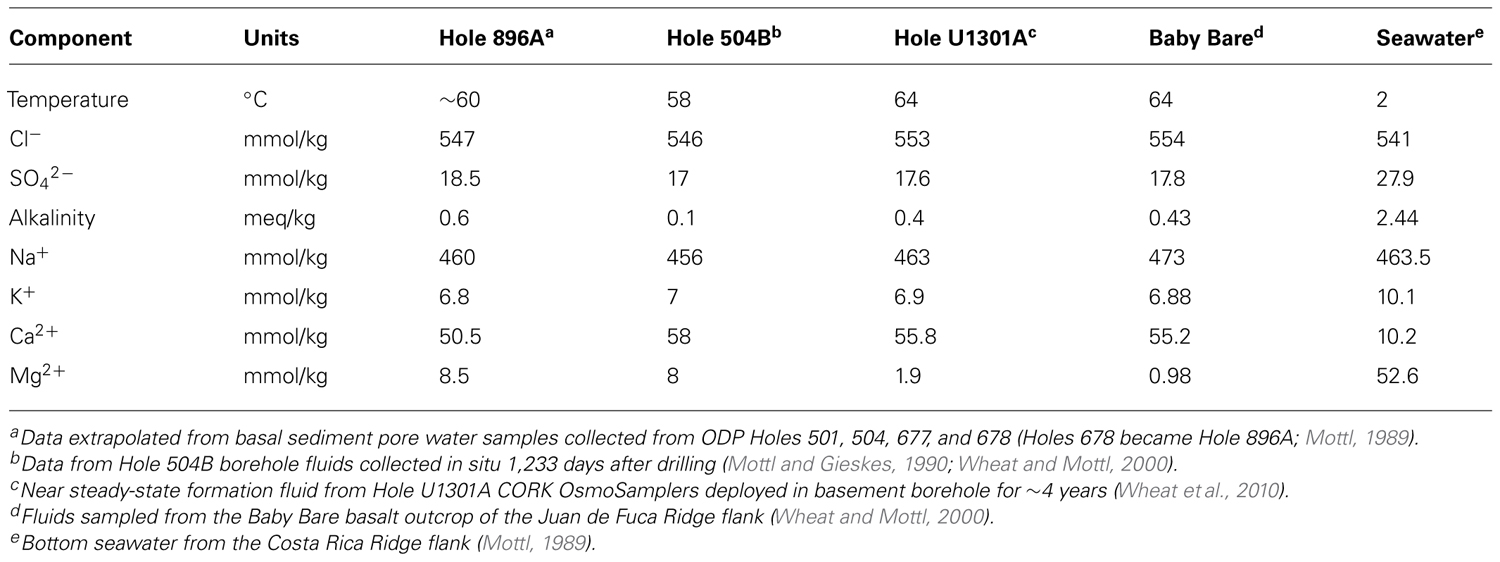
TABLE 1. Comparative chemistry of fluids from Costa Rica Ridge flank Holes 896A and 504B with Juan de Fuca Ridge flank fluids, with seawater for comparison.
In this study, samples were collected from an observatory placed at Hole 896A in 2001 and revisited in 2002 (Becker et al., 2004) to conduct comparative phylogenetic analysis for evaluating the in situ microbial communities.
Materials and Methods
Study Site
ODP Hole 896A on the Costa Rica Rift (1°13′ N, 83°43′ W; Figure 1) is 3,463 m below sea level and drilled to a depth of 469 mbsf (Figure 1). The lower 290 m of the hole consists of altered basaltic oceanic crust. During drilling in 1993, the upper 196 m section of Hole 896A was cased, thus sealing out sediment pore water and allowing the influx and accumulation of hydrothermal subsurface fluids from the basaltic crust. In 2001, roughly 8 years after the borehole was originally drilled, a wireline packer seal apparatus similar to a CORK observatory was deployed with the intent to plug the hole, record the pressure and temperature in the sealed zones, sample borehole formation fluids, and monitor the return to in situ hydrogeological conditions (Becker et al., 2004). The wireline CORK apparatus was constructed primarily from mild steel, and it included one packer in the cased section and a second packer intended to be set about 50 m into the open-hole section, as well as steel tubing umbilicals to bring formation fluids to sampling ports at the wellhead. However, on deployment, the lower packer became stuck in the hole about 20 m above the intended setting depth. In the attempts to deal with this, it is likely that the tubing umbilicals were damaged and the packers could not be inflated to seal the hole.
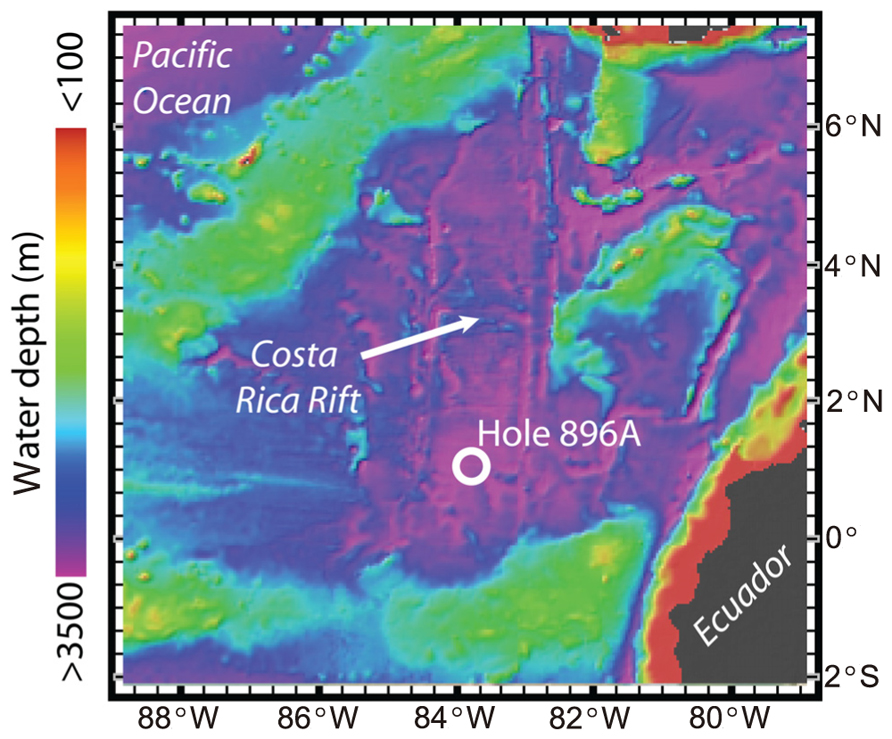
FIGURE 1. Map of the Ocean Drilling Program Hole 896A study site in the Pacific Ocean in relation to the Costa Rica Rift. Color gradient scale provided for water depth in meters. Maps were created using the GeoMapApp software (www.geomapapp.org; Ryan et al., 2009).
Immediately prior to the attempt to deploy the wireline packer, a video and temperature log had been conducted to verify that the hole was open down to the intended setting depth of the lower packer about 50 m into open hole. The temperature log indicated that upper basement formation fluids were being produced up the hole, with an average temperature of 57.8°C and total fluid flow rate through the casing of 12 m/h, equivalent to a volume flux of 800 l/h (Becker et al., 2004). In situ video monitoring revealed the presence of thick whitish crusts resembling sulfur-oxidizing bacterial mats on both the cased and open-hole sections of Hole 896A, and flocculent material seems to be entering the borehole around 220 mbsf (Becker et al., 2004).
Sample Collection
Roughly 15 months after the installation of the observatory, fluid and microbial mat samples were collected on November 18, 2002 during submersible operations with DSV Alvin (Dive 3840; Woods Hole Oceanographic Institution). Because the wireline CORK packers had not been inflated, it is likely that the uphole flow of upper basement fluids revealed by the 2001 temperature log was continuing up around the wellhead. The microbial mat sample, referred to as mat, consisted of a flocculent grey crust growing on the seawater exposed exterior of Port L that was swabbed with a fresh green plastic sponge, which was exposed to seawater and therefore not sterile (Figure 2A). The sponge was stored on the submersible in a closed plastic biobox filled with site bottom water before return to the ship. The fluid sample, referred to as bore, consisted of fluids collected at a wellhead sampling port about 10 min after the valve (Port L; Figure 2A) was opened, into an ethanol-sterilized titanium bottle using a flexible sampling hose (Figure 2B). The sampling port was connected to the tubing leading from just below the lower packer. However, because of the possibility of damage to the umbilical and lack of packer seal, it is likely that a mix of bottom water and true formation fluids was being sampled. No warm water venting and efflux of borehole water was observed at Port L. After return to the ship, approximately 0.75 l of borehole fluid was filtered, using a handpump, through a 90-mm diameter 0.22 μm mesh nylon filter to collect particulate matter and microorganisms. Both samples were returned to the ship (RV Atlantis, Woods Hole Oceanographic Institution) within a few hours and immediately frozen at -80°C for preservation of DNA. Samples were then transported frozen to a shore-based laboratory.
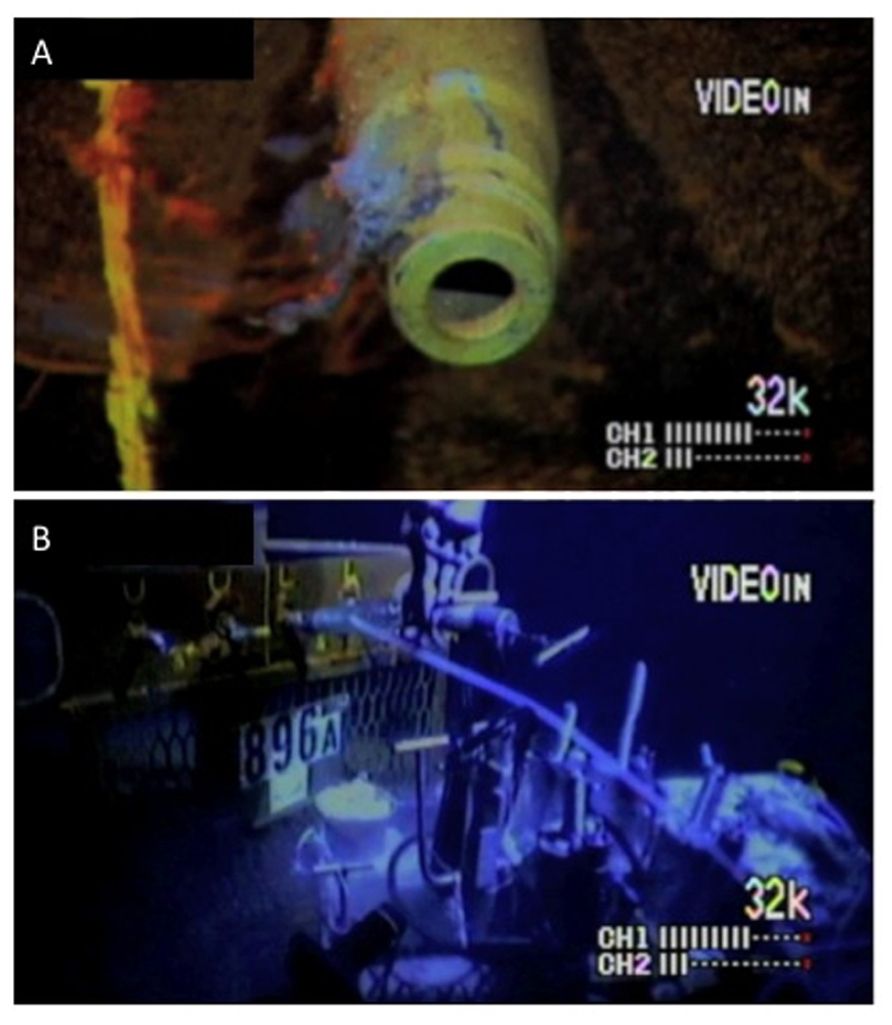
FIGURE 2. Microbial mat and fluid sampling at ODP Hole 896A made from still images captured during Alvin operations. (A) The L port with attached biofilm, removed with the first scrubber and sequenced as “mat” sample; (B) water sampling (the material used here) by tube coupling to the L port.
DNA Extraction, Amplification, and Sequencing
Environmental DNA was extracted from samples following a protocol modified from two published procedures for extracting DNA from deep subsurface sediments (Juniper et al., 2001; Kormas et al., 2003). Using sterilized tools, the filter and sponge were cut into pieces and then transferred into replicate vials of 6 ml DNA extraction buffer (200 mM NaCl, 200 mM Tris, 2 mM sodium citrate, 10 mM CaCl2, 50 mM EDTA, pH 8). The filter pieces were vortexed, and subjected to two freeze-thaw cycles (alternating -80°C and +65°C). Fifty microliters of Proteinase K (20 mg/ml) and 100 μl of 20% [w/v] sodium dodecyl sulfate (SDS) were added, and the samples were incubated for 30 min at 37°C. About 1.5 ml of 20% SDS was added and the samples were incubated at 55°C for 2 h in a shaking chamber. Then, the samples were centrifuged at 6,000×g for 10 min, the supernatant was harvested, an equal amount of 24:25:1 phenol/chloroform/isoamyl alcohol was added, and they were centrifuged again at 6,000×g for 10 min. The upper phase was harvested and 2.5× volume of ice-cold ethanol and 0.1× volume of ice-cold 5 M NaCl were added. The samples were placed in the -80°C freezer overnight to allow for DNA precipitation. The DNA was harvested by centrifuging at 10,000×g for 30 min, removing the supernatant, washing with 500 μl of 70% EtOH, and centrifuging again at 10,000×g for an additional 30 min. The remaining pellet was allowed to air dry before re-dissolving in 50 μl sterile water. To obtain more sequences, additional filter and mat samples were extracted with the Powersoil DNA Isolation Kit (Mobio) according to the manufacturer’s instructions and final volume of 50 μl elution solution.
Near full-length 16S rRNA genes were amplified by polymerase chain reaction (PCR) using bacterial primers BAC-8F (5′-AGRGTTTGATCCTGGCTCAG-3′) and BAC-1492R (5′-CGGCTACCTTGTTACGACTT-3′; Lane, 1991). The PCR mixture for the phenol/chloroform/isoamyl alcohol extracted samples consisted of 1 μl of the environmental DNA, 2 μl of each primer (0.5 mM), 0.5 μl of enzyme included in the FailsafeTM PCR System kit (EpiCentre Biotechnologies), 25 μl of FailsafeTM Premix B and a balance of water for a total reaction volume of 50 μl. The PCR cycle conditions involved an initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 3 min. These 30 cycles were followed by a final extension at 72°C for 10 min. Samples extracted with the Mobio Powersoil DNA Isolation Kit were amplified with Speedstar HS DNA Polymerase (Takara) using 2 μl of DNA sample and the manufacturer’s recommendations concentrations of buffer, dNTPs, and polymerase in a final volume of 25 μl. Thermal cycling was performed as previously described above with the exception that cycling times were 95°C for 10 s, 52°C for 15 s, and 72°C for 20 s over 28 cycles.
To examine the potential for autotrophy using the Calvin cycle, ccbL and ccbM genes involved in the formation of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) from the mat sample environmental DNA were PCR amplified as described previously (Elsaied and Nagamuna, 2001). An initial round of PCR, cloning and sequencing yielded only one cbbL clone and three cbbM clones (mat clones A06, E03, H09, and F03). Additional DNA was amplified from the Powersoil Isolation Kit extractions with the GoTaq Flexi DNA Polymerase (Promega) using a final concentration of 3 mM MgCl2, 1 μM of each primer, and the manufacturer’s recommendation of dNTP, buffer, and polymerase concentration. Primers for the cbbL gene included cbbL1AF172 (5′-ACNTGGACNACNGTNTGG-3′) and cbbL1AR1382 (5′-TCRAAYTTGATYTCBTTCCA-3′). Primers for the cbbM gene included cbbM337F (5′-AACCARGYATGGGYGAY-3′) and cbbM1126R (5′-TCATRCCVCCVGADAT-3′). In the bore sample, non-specific priming was extensive in one cbbL PCR sample and an additional primer, cbbL1AR1142 (5′-GGCATRTGCCANACRTGRAT-3′), was used with primer cbbLAF172 in a nested PCR using 0.2 μl of the bore PCR sample and the same mixture and cycling conditions described above, with the exception that the number of cycles was reduced to 20. We note that the cbbL primers did not fully target the total diversity of the “red-like” IC form of RuBisCO (Badger and Bek, 2008) due to the need to limit the number of degeneracies; some diversity may have been missed.
The PCR products were subjected to 1.5% agarose gel electrophoresis, stained with 0.5 μg/ml of ethidium bromide or 1× GelRed (Biotium), and visualized by UV excitation for bands indicating successful DNA amplification. PCR products were either excised from the agarose gel or directly purified using the Wizard® PCR preparation kit (Promega) or the Qiagen Minelute Gel Extraction kit, and then cloned using TOPO XL PCR cloning kit (Invitrogen). From each clone library, clones were selected randomly for sequencing at the Josephine Bay Paul Center of the Marine Biological Laboratory in Woods Hole, MA, USA or Genewiz (Research Triangle Park, NC, USA).
Phylogenetic Analysis
Raw 16S rRNA gene and RuBisCO sequences were edited using Chromas or Sequencher (Genecodes) software to remove cloning vector sequences, and potential chimeric sequences were evaluated using Pintail1. Closely related sequences were identified using the Basic Local Alignment Search Tool (BLAST) program searches in GenBank2 (Altschul et al., 1997). 16S rRNA gene sequences were aligned against a curated sequence database (using the NAST alignment tool available at greengenes.org; DeSantis et al., 2006a) and manually checked for alignment accuracy against reference sequences from the Greengenes database (DeSantis et al., 2006b) using the ARB software package (Ludwig et al., 2004). A phylogenetic tree of the nearly full-length 16S rRNA gene sequences of this study, plus key reference sequences, was calculated in ARB based on Jukes-Cantor distances via neighbor joining. The cbbL and cbbM gene sequences were translated to amino-acid sequences and aligned with Clustal W in MEGA 4.0 (Kumar et al., 2008) using a Gonnet protein weight matrix. The RuBisCO phylogeny was constructed in MEGA using a neighbor-joining method based on Poisson-corrected distances. Statistical support of tree topology was estimated by bootstrapping with 1,000 replicates in MEGA. The 16S rRNA gene sequences reported here are available in GenBank under the accession numbers GQ903340–GQ903342, GQ903344, GQ903346–GQ903350, and GQ903352–GQ903376. The RuBisCO genes have GenBank accession numbers HQ856238–HQ856241 and JQ795724–JQ795729.
The similarity of the bore and mat 16S rRNA gene clone libraries to previously published clone libraries was evaluated by comparing sequence similarity distance matrices with the programs DOTUR and SONS (Schloss and Handelsman, 2005, 2006) using methods described previously (Santelli et al., 2008). In particular, the nearly full-length Hole 896A 16S rRNA gene sequences were compared to sequence datasets generated from seafloor-exposed basalts from the East Pacific Rise and the Loihi Seamount (Santelli et al., 2008); from subsurface (3 m) hydrothermal basalt formation fluids from the Baby Bare outcrop on the Juan de Fuca Ridge flank (Huber et al., 2006); from fluids freely venting from the ODP Hole 1026B borehole (Huber et al., 2006); from bottom water samples collected above the Juan de Fuca Ridge flank (Huber et al., 2006); and from seafloor-exposed inactive massive sulfides from the East Pacific Rise (Sylvan et al., 2012). Sequence datasets were compared to identify the number and type of shared “species” between samples, with “species” operationally defined as operational taxonomic units (OTUs) with 97% or greater sequence similarity, as defined previously (Santelli et al., 2008). Sequence distance matrices were generated in ARB using the neighbor-joining method, the Jukes–Cantor correction, and application of an in-program filter for bacteria, of E. coli base pair positions 228–1420. Diversity estimators were calculated in DOTUR under standard settings at the 97% or greater sequence similarity definition level, and shared richness estimates were generated using standard settings in the SONS program.
Results
16S rRNA Gene Phylogeny
A total of 66 and 60 nearly full-length 16S rRNA gene clones were successfully sequenced from the bore and mat sample clone libraries, respectively. Phylogenetic analysis of the clone libraries revealed 40 phylotypes from several bacterial phylum-level groups including Bacteroidetes, aerobic and anaerobic heterotrophs widespread in soil and water (Kirchman, 2002); Cyanobacteria; Actinobacteria (OM1 group; Rappé et al., 1997), oxygenic marine phototrophs; the phylum-level lineages Verrucomicrobia and Planctomycetes, often detected in oxygen-depleted marine habitats (Kirkpatrick et al., 2006) and with relatively few cultured chemoorganotrophic isolates (Wagner and Horn, 2006); and diverse Alpha-, Gamma-, and Epsilonproteobacteria (Figures 3 and 4). The seven most abundant phylotypes (highlighted in purple in Figures 3 and 4) were found in both the borehole fluids and the microbial mat.
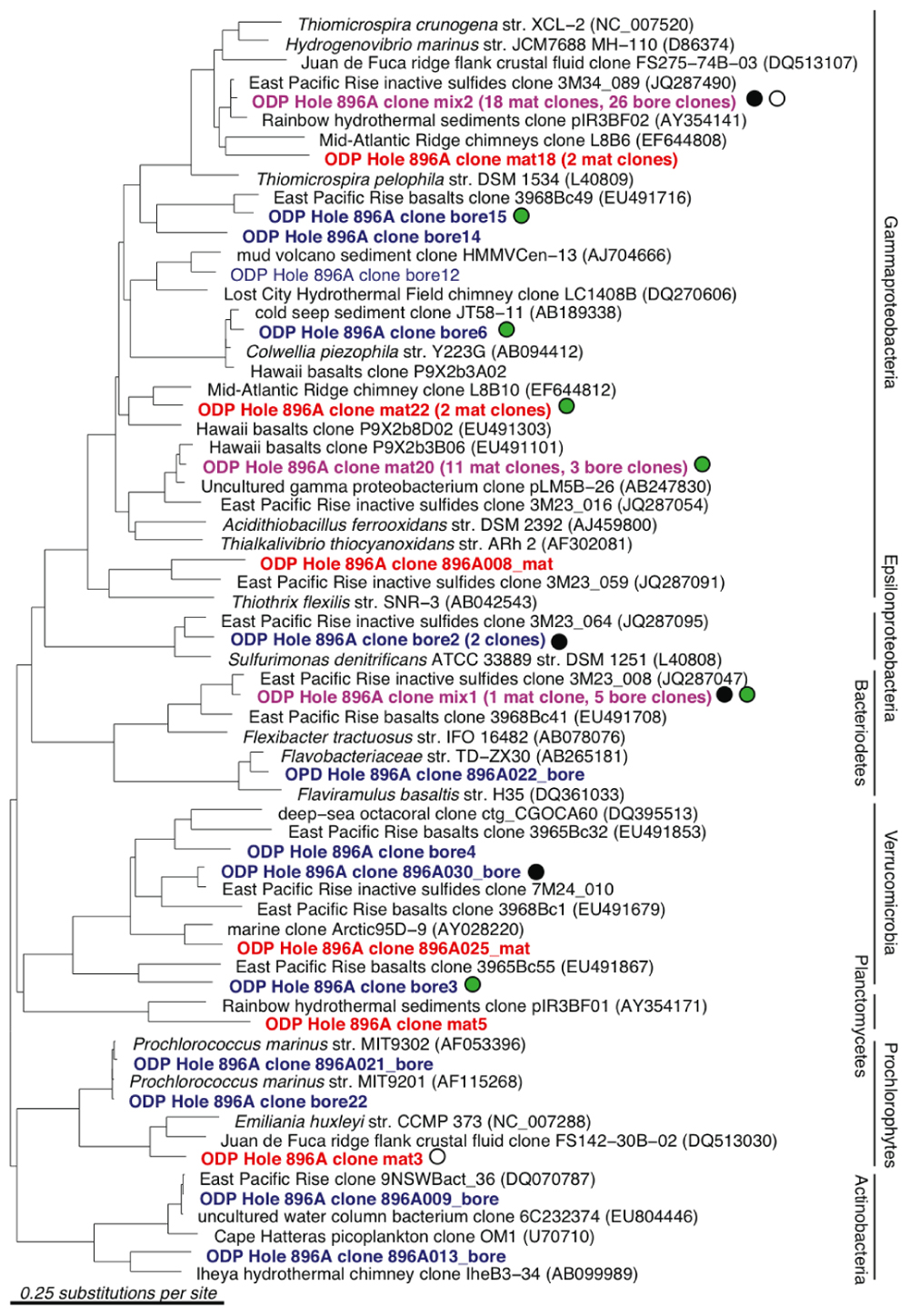
FIGURE 3. 16S rRNA gene neighbor-joining phylogenetic tree for Hole 896A observatory bore and mat sample sequences in comparison to close relatives from the GenBank database, excluding the Alphaproteo- bacteria phylum. Clones from borehole fluids collected at the observatory spigot are labeled as bore in blue, and clones from the microbial mat on the exterior of the borehole are labeled as mat in red. Identical clones from both samples are labeled in purple. Short sequences are indicated with asterisks. Sequences that are shared with other clone libraries are indicated with colored dots: East Pacific Rise inactive massive sulfides (black); East Pacific Rise and Loihi Seamount basalts (green); Juan de Fuca Ridge flank crustal fluids (white); Juan de Fuca CORK Hole 1301A (yellow).
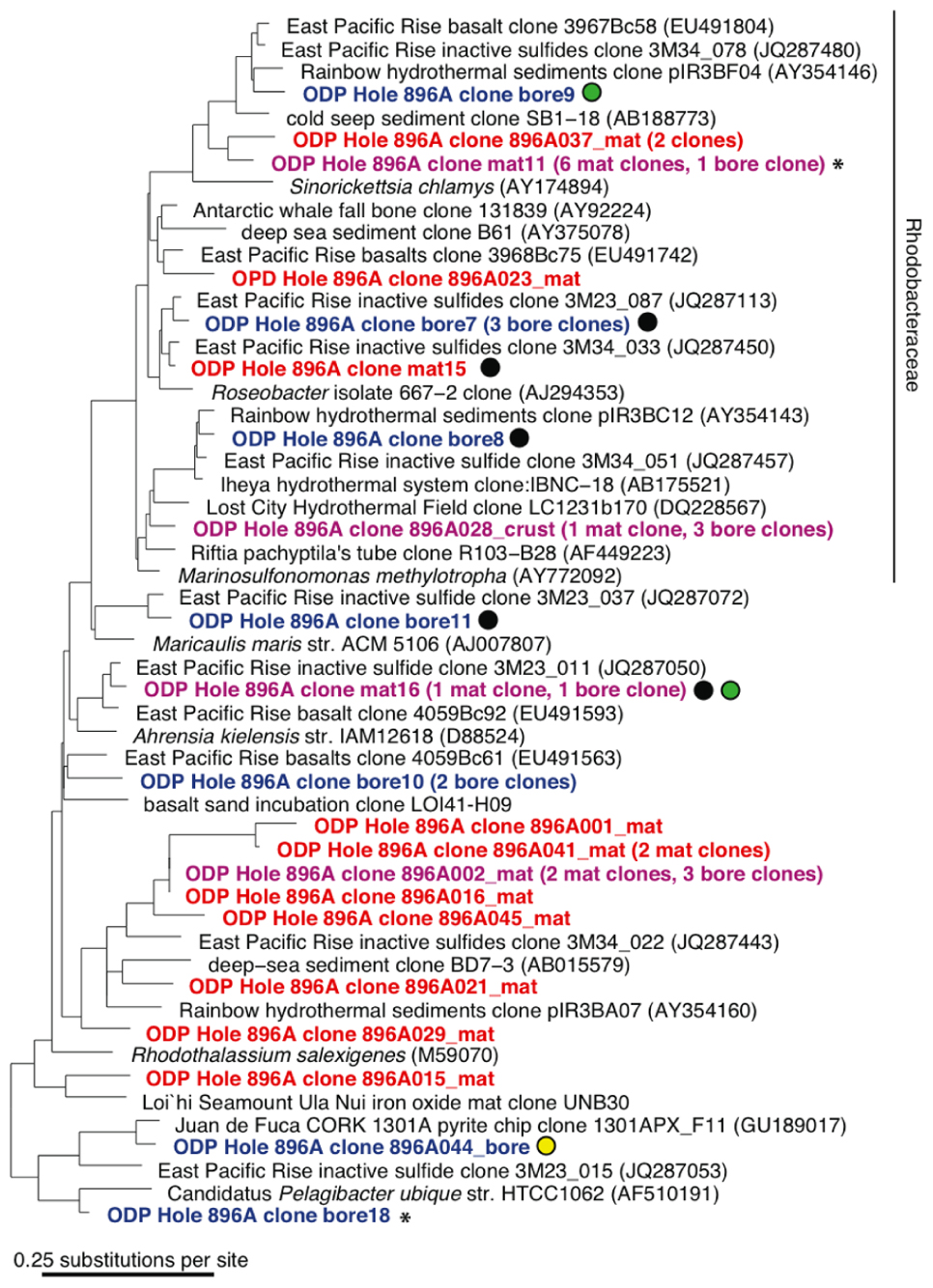
FIGURE 4. 16S rRNA gene neighbor-joining phylogenetic tree for Hole 896A observatory bore and mat sample Alphaproteobacteria sequences in comparison to close relatives from the GenBank database. Same details as for Figure 3.
Despite using bacteria-specific 16S rRNA gene primers, one archaeal clone was also recovered from the mat sample. This clone, mat1, was related to Thermocladium modestius, an extremely thermophilic crenarchaeote isolated from acidic hot spring areas in Japan (Itoh et al., 1998). The presence of an archaeal clone in a bacterial 16S rRNA gene clone library was unexpected, given that the bacterial and archaeal versions of the forward primers had five nucleotide mismatches and thus strong PCR bias against archaeal gene amplification.
A number of sequences grouped near known sulfur cycling microorganisms. Phylotype mix2 was related to the free-living, autotrophic, oxygen- and nitrate-respiring, sulfur-oxidizing genus Thiomicrospira of the Gammaproteobacteria; this phylotype comprised nearly half of the sequences of both the bore and mat clone libraries (Figure 3). This phylotype has also been observed in clone libraries from seafloor-exposed massive sulfides (Sylvan et al., 2012) and in fluids sampled from the ODP Hole 1026B observatory on the Juan de Fuca Ridge flank (Huber et al., 2006). One bore phylotype affiliated with the sulfur-oxidizing chemolithoautotrophic Epsilonproteobacteria Sulfurimonas denitrificans and S. autotrophica. Generally, hydrogen- and sulfur-oxidizing, chemolithoautotrophic Epsilonproteobacteria have a wide environmental distribution at hydrothermal vents and in marine surficial sediments (Campbell et al., 2006; Nakagawa and Takai, 2008) and employ an alternative pathway of autotrophic CO2 fixation, the reverse tricarboxylic acid (TCA) cycle (Hügler et al., 2005). The whitish mats deposited on the walls of the observatory and recorded in video logs of the Hole 896A borehole (Becker et al., 2004) bear a conspicuous resemblance to the sulfur precipitates and flocs produced by sulfur-oxidizing bacteria (Kuenen and Veldkamp, 1972; Taylor and Wirsen, 1997; Wirsen et al., 2002).
Several other bore and mat sequences also grouped most closely with uncultivated environmental sequences recovered from seawater-exposed basalts and inactive sulfides from the East Pacific Rise and Hawaii (Santelli et al., 2009; Sylvan et al., 2012), from hydrothermal fluids from a basaltic outcrop (Huber et al., 2006) and from cold seep and hydrothermally influenced deep marine sediments. These phylotypes are highlighted with colored circles in Figures 3 and 4. Nine of the forty observed bacterial phylotypes, marked with black circles, grouped closely with phylotypes from inactive massive sulfide samples collected from the East Pacific Rise (Sylvan et al., 2012), whereas eight phylotypes, marked with green circles, were shared with clone libraries from seafloor-exposed basalts (Santelli et al., 2009). Three phylotypes, marked with white and yellow circles, were similar to clones from samples from other deep basalt observatories. Of these, one alphaproteobacterial sequence (Figure 4) grouped closely with a sequence from a biofilm formed on pyrite incubated in the subsurface for 4 years (Orcutt et al., 2011); the Thiomicrospira-related clone mix 2 and Prochlorophyte clone mat 3 are close to clones from venting basalts at Baby Bare on the Juan de Fuca ridge (Huber et al., 2006). No 16S rRNA clone library phylotypes were shared between the Hole 896A samples and background bottom water collected above basaltic lavas the Pacific Ocean (Santelli et al., 2008). While the identification of shared phylotypes is robust, shared diversity estimates between the sample sets were tested, but they were skewed due to the difference in clone library sizes between the Hole 896A samples and the comparison studies, and the possibility of seawater entrainment in the Hole 896A samples.
Several sequences grouped most closely to environmental sequences commonly found in seawater (Figures 3 and 4). For example, the bore18 phylotype grouped near Pelagibacter ubique within the SAR 11 cluster, a cosmopolitan clade of marine oligotrophic bacteria (Rappé et al., 2002). Several alphaproteobacterial phylotypes from each sample grouped within the Rhodobacteraceae or marine Roseobacter group, a cosmopolitan group of marine bacteria that often metabolize and oxidize organosulfur compounds (Buchan et al., 2005; Brinkhoff et al., 2008). Two phylotypes were most closely related (99% identity) to sequences from the marine cyanobacterial genus Prochlorococcus, which is found at varying depths in the water column in oceans worldwide (West et al., 2001). The presence of these phylotypes is unexpected in samples of oceanic basement formation fluids, and indicates seawater entrainment and contamination during sampling of the bore and mat samples at the observatory platform, or seawater entrainment into the mixed borehole fluids itself. Moreover, the occasional close phylogenetic association of some phylotypes with known aerobic chemoorganotrophic isolates, such as Flaviramulus basaltis and Marivirga tractuosus (Figure 3), suggests chronic oxic seawater mingling with basement fluids at the Hole 896A observatory.
RuBisCO Phylogeny
Amplification of the cbbM and cbbL genes of RuBisCO yielded sequences of sufficient length and quality for phylogenetic analysis from the mat sample and from the borehole fluid sample. BLAST search based on amino-acid sequence, and subsequent phylogenetic analysis (Figure 5) revealed that most cbbL sequences (mix clone ODP3_4_1) recovered from the mat (45 clones) as well as the borehole sample (16 clones) were most closely related to the obligately chemolithoautotrophic, sulfur-oxidizing gammaproteobacterial genera Thioalkalivibrio, isolated from Siberian and East African soda lakes (Sorokin et al., 2001; Thioalkalivibrio thiocyanoxidans, GenBank accession ZP_08930733, 93% identity), to sequences from hypersaline soda lake sediment in Kulunda Steppe (Russia; Kovaleva et al., 2011; GenBank ADN96557, 97% identity), and to the facultatively phototrophic sulfur oxidizer Thiocapsa (Guyoneaud et al., 1998), capable of chemolithoautotrophic growth with reduced sulfur compounds under microoxic conditions (Caumette, 1986). Other clones (mix clone ODP 6_1_9; five bore and one mat clone) were most closely related to Thiomicrospira (Thiomicrospira crunogena, GenBank YP_391108, 94% identity) isolated from marine sediments and hydrothermal vents (Jannasch et al., 1985; Scott et al., 2006) and the obligately autotrophic hydrogen oxidizer Hydrogenovibrio marinus, phylogenetically a lineage within the genus Thiomicrospira (GenBank BAD15312, 93% identity; Nishihara et al., 1998). Two other borehole sequences were most closely related to those from a pogonophoran bacterial endosymbiont from a cold methane seep in the Japan Trench (Nagamuna et al., 2007).
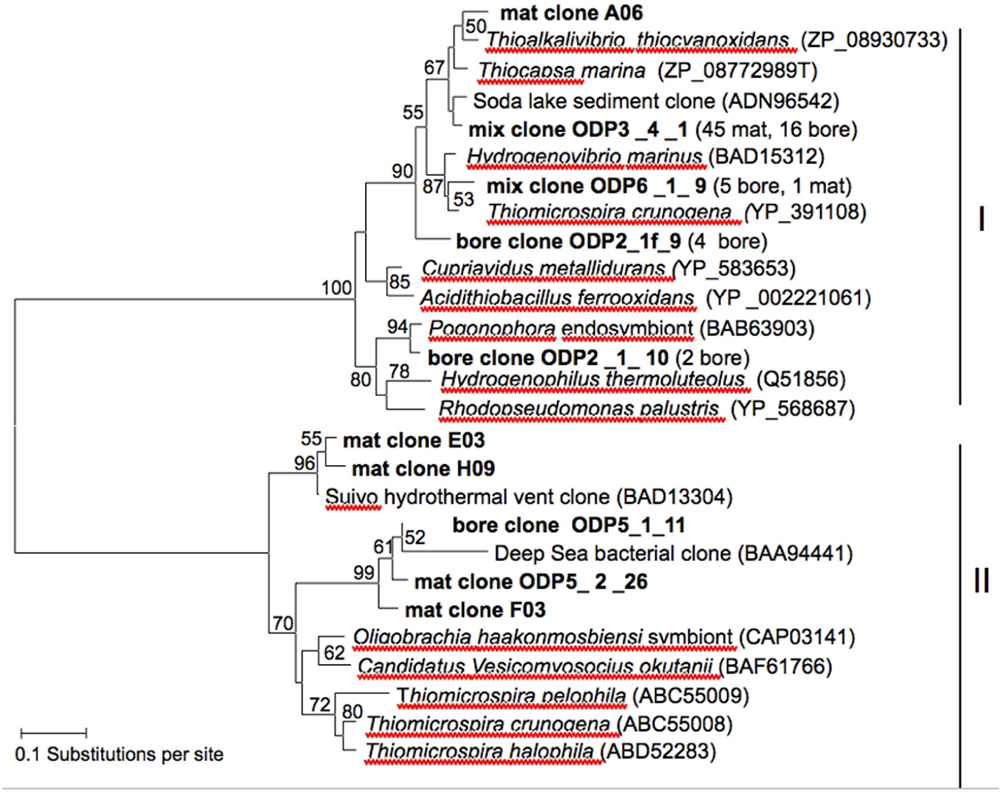
FIGURE 5. Neighbor-joining tree based on amino-acid sequences translated from RuBisCO cbbL and cbbM large subunit gene sequences. Bootstrap values (1,000 replicates) above 50% are displayed beside the nodes. Clone sequences are indicated in bold.
Only five cbbM sequences from the mat and bore sample were obtained in total despite attempts to amplify the gene with published (Elsaied and Nagamuna, 2001) and newly designed primers (this study). The cbbM sequences recovered from the Hole 896A samples were related to cbbM sequences of uncultivated bacteria from the hydrothermally active Suiyo Seamount, a submarine black smoker volcano in the Izu-Bonin trench off Japan (Clone Suiyo II-5, nucleotide accession number AB174751, protein ID BAD13304; Elsaied et al., 2007) and to cbbM sequences obtained from reducing sediments near the deepest known chemosynthetic microbial community, in deep-sea sediments of the Japan Trench at 7,434 m depth (Clone JT-Sed(II)-5, nucleotide accession number AB040517, protein ID BAD94441; Elsaied and Nagamuna, 2001; Figure 5).
Discussion
The phylogenetic data presented here from Hole 896A on the Costa Rica Rift flank represents the second dataset from a basaltic crust borehole observatory, providing the first comparison to the available data from the Holes 1026B and U1301A CORK observatories on the Juan de Fuca Ridge flank (Cowen et al., 2003; Nakagawa et al., 2006; Orcutt et al., 2011). Such comparisons face important limitations; the two samples from the Hole 896A observatory were not pristine, and the sample type (formation fluids from the damaged observatory, and a scraping of a mat on the observatory exterior – both with likely seawater admixture) differs from previous samples, for example the enrichments on mineral surfaces in Hole U1301A (Orcutt et al., 2011), the CORK mineral crusts at Hole U1301A (Nakagawa et al., 2006), the vent fluids at Hole 1026B (Huber et al., 2006), or the biocolumn enrichments at 1026B (Cowen et al., 2003). However, we believe that the samples still allow broad comparisons toward a more complete picture of the biosphere hosted in subsurface oceanic crust. Our analysis also highlights operational issues that need to be addressed for future efforts to investigate the subsurface deep biosphere.
Comparison of Hole 896A Microbial Communities to Those from Other Habitats
Based on 16S rRNA gene clone libraries, the microbial communities observed in the Hole 896A samples (Figures 3 and 4) bore little resemblance to the communities described in formation fluids and mineral crusts from Holes 1026B and U1301A, despite the previously reported similarities in mineralogy, basement fluid chemistry, and temperature (Table 1). Holes 1026B and U1301A on the Juan de Fuca ridge flank reveal subsurface microbial communities characterized by an abundance of Firmicutes bacteria (Cowen et al., 2003; Nakagawa et al., 2006; Orcutt et al., 2011). The predominant bacterial phylotypes in the Hole 1026B clone libraries were distantly related to thermophilic, nitrate-, and sulfate-reducing bacteria, such as the hydrogen-oxidizing nitrate-reducing ammonia producer Ammonifex degensii, and the gram-positive, spore-forming sulfate reducing genus Desulfotomaculum. In contrast, the Hole 896A observatory samples were dominated by sequences grouping with Gammaproteobacteria related to the chemolithoautotrophic, sulfur-oxidizing genus Thiomicrospira that is predominantly isolated from sulfidic marine sediments and hydrothermal vents, and hydrothermal plumes; these bacteria were not detected in other borehole surveys (Cowen et al., 2003; Nakagawa et al., 2006; Orcutt et al., 2011).
One explanation for these differences in the dominant members of the microbial communities, with suspected metabolic differences as well, may be different redox regimes within the boreholes, despite the similarity in major ion concentrations of the formation fluids (Table 1). Namely, the extensive accumulation of white flocculent crusts in Hole 896A (Becker et al., 2004) suggests microoxic or nitrate-reducing conditions, while an anaerobic environment is known to prevail in Holes 1026B and U1301A (Wheat et al., 2010). The formation of extensive white flocs is also known from bioreactor experiments, where microaerobic sulfur-oxidizing bacteria produce extracellular sulfur in large amounts (Taylor and Wirsen, 1997); sulfur precipitation is also a characteristic by-product of aerobic, sulfur-oxidizing Thiomicrospira spp. growing in laboratory culture (for an instructive example, see Figure 2 in Kuenen and Veldkamp, 1972). The in situ observation of the flocculent mat-like material within the Hole 896A borehole (Becker et al., 2004) indicates in situ production of biomass and flocculent mats by sulfur-oxidizing bacteria within the borehole. Seawater influence at the Hole 896A CORK observatory is consistent with this interpretation. If seawater were entrained or mixed with the borehole fluids, which are presumably rich in reduced substrates such as sulfur and iron, this might create an ideal niche for the enrichment for the sulfide-oxidizing and biofilm-forming phylotypes observed. The sulfur-oxidizing bacteria that dominate the borehole could ultimately be derived from bottom water mixed with highly dilute hydrothermal plumes and microbial populations (Huber et al., 2006, 2007). The observation of seawater-related phylotypes such as SAR11, Roseobacter, Prochlorophytes, and OM1 Actinobacteria in the bore hole sample clone libraries also supports the argument for seawater entrainment. In consequence, the in situ enrichment of bacteria growing under likely conditions of seawater entrainment in the borehole and within the CORK distorts the assessment of potential indigenous microbial diversity in basaltic basement fluids (Cowen, 2004).
Autotrophic Potential
Based on theoretical models of the energy available from sulfur and iron oxidation in basaltic crust, significant levels of primary production should occur in the subsurface (Bach and Edwards, 2003), a prediction testable by RuBisCO genes analysis. RuBisCO catalyzes the assimilation of carbon dioxide to organic carbon via the Calvin–Benson–Bassham cycle. Of its currently four known forms, form I is oxygen tolerant and found predominantly in cyanobacteria, chloroplasts, and aerobic chemolithoautotrophic bacteria, while form II is adapted to high CO_2 conditions and found predominantly in microaerobic or anaerobic bacteria (Delwiche and Palmer, 1996; Badger and Bek, 2008). We observed both forms of RuBisCO in the Hole 896A mat and borehole samples (Figure 5). RuBisCO sequences were phylogenetically related to Thiomicrospira, the closest cultured relative of the most frequently recovered 16S rRNA sequences at the 896A CORK. The form I RuBisCO sequences obtained from the bore hole and from the mat sample were most closely related to the sulfur-oxidizing chemolithoautotrophic genera Thioalkalivibrio, Thiocapsa, and Thiomicrospira, within the form IA “green-type” clade that is associated with proteobacteria and cyanobacteria (Badger and Bek, 2008). This pattern is consistent with the 16S rRNA sequencing results, and with the interpretation that the borehole fluid and the mat sample contain autotrophic, sulfur-oxidizing bacteria related to these gammaproteobacterial genera. The presence of additional autotrophic bacteria (or of bacteria that contain form II in addition to one of the form I sequences found here) is indicated by the form II sequences in the mat; these sequences were related to RuBisCO of uncultured marine bacteria, not from the open water column but from methane seep sediments and hydrothermal plumes (Figure 5).
As a note of caution, RuBisCO has a high rate of horizontal gene transfer events (Delwiche and Palmer, 1996). The gammaproteobacterial form I types are also found in cyanobacteria, for example, the common marine cyanobacterium Prochlorococcus which most likely acquired its RuBisCO genes by horizontal gene transfer (Hess et al., 2001). Thus, the RuBisCO sequences do not rule out seawater contamination, as indicated by the two Prochlorococcus 16S rRNA gene sequences found in the borehole fluid (Figure 3). A certain degree of seawater contamination is obvious, but it does not invalidate the abundance of phylotypes most closely related to sulfur-oxidizing bacteria (Thiomicrospira, Sulfurimonas) and to basalt-associated bacterial phylotypes. With this caveat, the 16S rRNA gene and RuBisCO sequence data are consistent with autotrophic, most likely sulfur-oxidizing bacterial populations growing within the borehole and on the sampling ports of the CORK; this interpretation is fully consistent with in situ borehole observations of microbial mat growth within the borehole (Becker et al., 2004).
Recommendations for Future Observatory Studies
The Hole 896A observatory was installed for primarily geophysical experiments (Becker et al., 2004) and was not designed for high-quality sampling for microbiological analysis of the subsurface crustal biosphere. Nevertheless, samples were collected opportunistically and analyzed despite potential contamination pitfalls, as they represented a unique chance to evaluate the crustal biosphere. Our analysis indicates that fluids from the crustal subsurface mixed with seawater can support microbial communities that appear to form biofilms, and that some of these biofilm-forming species are related to known sulfide oxidizing microbial groups like Thiomicrospira. Given the stark differences in the microbial communities observed between subsurface observatories placed in similar crustal settings (i.e., between the Hole 896A observatory and the Juan de Fuca Ridge flank Holes 1026B and U1301A), enhanced sample characterization may shed light on the underlying environmental conditions that could explain these differences. For example, analysis of borehole fluid oxygen, nitrate and redox conditions may help resolve whether the presence of these electron acceptors may have influenced the Hole 896A community. A time series analysis of the Hole 896A observatory would provide background information on whether the observed community was representative of “steady state” formation fluids or if it instead represented a transitory evolution of the community post-observatory installation, as has been observed elsewhere for microbial communities in other boreholes (Orcutt et al., 2011). Most importantly, improvements in the Hole 896A observatory infrastructure to allow cleaner microbiological sampling would reduce the confounding influence of seawater contamination.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Steve D’Hondt and Eddie Roggenstein for obtaining samples from the ODP Hole 896A observatory during DSV Alvin dives in November 2002, and C. Geoff Wheat for helpful discussion regarding borehole fluid chemistry. Installation of the wireline CORK and the 2002 Alvin dives were supported by the National Science Foundation under grant OCE-9819316 to Keir Becker. Funding for this project was provided by the American Society for Microbiology Undergraduate Research Fellowship to Kate Harris, and by the NASA Astrobiology Institutes “Subsurface Biospheres” at the University of Rhode Island, and “Environmental Genomes” at the Marine Biological Laboratory. Andreas Teske was further supported through the Center for Dark Energy Biosphere Investigations. We thank the reviewers for their constructive comments that improved this manuscript significantly.
Footnotes
References
Alt, J. C., Teagle, D. A. H., Laverne, C., Vanko, D., Bach, W., Honnorez, J., Becker, K., Ayadi, M., and Pezard, P. A. (1996). “Ridge flank alteration of upper ocean crust in the eastern Pacific: a synthesis of results for volcanic rocks of holes 504B and 896A,” in Proceedings of the ODP, Scientific Results, Vol. 148, eds J. C. Alt, H. Kinoshita, L. B. Stokking, and P. J. Michael (College Station, TX: Ocean Drilling Program), 434–452.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997). Gapped BLAST and PSI-blast: a new generation of protein data-base search programs. Nucleic Acids Res. 25, 3389–3402.
Bach, W., and Edwards, K. J. (2003). Iron and sulfide oxidation within the basaltic ocean crust: implications for chemolithoautotrophic microbial mass production. Geochim. Cosmochim. Acta 67, 3871–3887.
Badger, M. R., and Bek, E. J. (2008). Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 59, 1525–1541.
Becker, K., and Davis, E. E. (2005). “A review of CORK designs and operations during the ocean drilling program,” in Proceedings of the IODP 301, eds A. T. Fisher, T. Urabe, A. Klaus, and the Expedition 301 Scientists (College Station, TX: Integrated Ocean Drilling Program Management International, Inc.), 1–28.
Becker, K., Davis, E. E., Spiess, F. N., and de Moustier, C. (2004). Temperature and video logs from the upper oceanic crust, Holes 504B and 896A, Costa Rica Rift flank: implications for the permeability of the upper oceanic crust. Earth Planet. Sci. Lett. 222, 881–896.
Brinkhoff, T., Giebel, H.-A., and Simon, M. (2008). Diversity, ecology and genomics of the Roseobacter clade: a short overview. Arch. Microbiol. 189, 531–539.
Buchan, A., González, J. M., and Moran, A. M. (2005). Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71, 5665–5677.
Campbell, B. J., Engel, A. S., Porter, M. L., and Takai, K. (2006). The versatile ε-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4, 458–468.
Caumette, P. (1986). Phototrophic sulphur bacteria and sulphate-reducing bacteria causing red waters in a shallow brackish coastal lagoon (Prévost Lagoon, France). FEMS Microbiol. Ecol. 38, 113–124.
Cowen, J. P. (2004). The microbial biosphere of sediment-buried oceanic basement. Res. Microbiol. 155, 497–506.
Cowen, J. P., Giovannoni, S. J., Kenig, F., Johnson, H. P., Butterfield, D., Rappe, M. S., Hutnak, M., and Lam, P. (2003). Fluids from aging ocean crust that support microbial life. Science 299, 120–123.
Davis, E. E., Becker, K., Pettigrew, T., Carson, B., and MacDonald, R. (1992). CORK: a hydrologic seal and downhole observatory for deep-ocean boreholes. Proc. Ocean Drill. Prog. Init. Rep. 139, 43–53.
Delwiche, C., and Palmer, J. (1996). Rampant horizontal gene transfer and duplication of RuBisCO genes in eubacteria and plastids. Mol. Biol. Evol. 13, 873–882.
DeSantis, T. Z., Hugenholtz, P., Keller, K., Brodie, E. L., Larsen, N., Piceno, Y. M., Phan, R., and Andersen, G. L. (2006a). NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34, W394–W399.
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., Huber, T., Dalevi, D., Hu, P., and Andersen, G. L. (2006b). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 75, 5069–5072.
D’Hondt, S., Jørgensen, B. B., Miller, D. J., Batzka, A., Blake, R., Cragg, B. A., Cypionka, H., Dickens, G. R., Ferdelman, T., Hinrichs, K.-U., Holm, N. G., Mitterer, R., Spivack, A., Wang, G., Bekins, B., Engelen, B., Ford, K., Gettemy, G., Rutherford, S., Sass, H., Skilbeck, C. G., Aiello, I. W., Guèrin, G., House, C. H., Inagaki, F., Meister, P., Naher, T., Niitsuma, S., Parkes, R. J., Schippers, A., Smith, D. C., Teske, A., Wiegel, J., Padilla, C. N., and Acosta, J. L. S. (2004). Distributions of microbial activities in deep subseafloor sediments. Science 306, 2216–2221.
D’Hondt, S., Spivack, A. J., Pockalny, R., Ferdelman, T. G., Fischer, J. P., Kallmeyer, J., Abrams, L. J., Smith, D. C., Graham, D., Hasiuk, F., Schrum, H., and Stancin, A. M. (2009). Subseafloor sedimentary life in the South Pacific Gyre. Proc. Nat. Acad. Sci. U.S.A. 106, 11651–11656.
Edwards, K. J., Rogers, D. R., and Wirsen, C. O. (2003). Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing chemolithoautotrophic alpha- and gamma-proteobacteria from the deep-sea. Appl. Environ. Microbiol. 69, 2906–2913.
Elsaied, H., and Nagamuna, T. (2001). Phylogenetic diversity of ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes from deep-sea microorganisms. Appl. Environ. Microbiol. 67, 1751–1765.
Elsaied, H. E., Kimura, H., and Nagamuna, T. (2007). Composition of archaeal, bacterial, and eukaryal RuBisCO genes in three Western Pacific arc hydrothermal vent systems. Extremophiles 11, 191–202.
Engelen, B., Ziegelmüller, K., Wolf, L., Köpke, B., Gittel, A., Cypionka, H., Treude, T., Nakagawa, S., Inagaki, F., Lever, M. A., and Steinsbu, B. O. (2008). Fluids from the ocean crust support microbial activities within deep biosphere. Geomicrobiol. J. 25, 56–66.
Fisher, A. T., Davis, E. E., and Becker, K. (2008). Borehole-to-borehole hydrologic response across 2.4 km in the upper oceanic crust: implications for crustal-scale properties. J. Geophys. Res. 113, B07106.
Fisher, A. T., Wheat, C. G., Becker, K., Davis, E. E., Jannasch, H., Schroeder, D., Dixon, R., Pettigrew, T. L., Meldrum, R., McDonald, R., Nielsen, R., Fisk, M., Cowen, J., Bach, W., and Edwards, K. (2005). “Scientific and technical design and deployment of long-term, subseafloor observatories for hydrogeologic and related experiments, IODP Expedition 301, eastern flank of Juan de Fuca Ridge,” in Proceedings of the IODP 301, eds A. T. Fisher, T. Urabe, A. Klaus, and the Expedition 301 Scientists (College Station, TX: Integrated Ocean Drilling Program Management International, Inc.), 1–39.
Fisk, M. R., Giovannoni, S. J., and Thorseth, I. H. (1998). Alteration of oceanic volcanic class: textural evidence of microbial activity. Science 281, 978–980.
Furnes, H., Muehlenbachs, K., Torsvik, T., Thorseth, I. H., and Tumyr, O. (2001). Microbial fractionation of carbon isotopes in altered basaltic glass from the Atlantic Ocean, Lau Basin, and Costa Rica Rift. Chem. Geol. 173, 313–330.
Giovannoni, S. J., Fisk, M. R., Mullins, T. D., and Furnes, H. (1996). “Microbial activity in the alteration of glass from pillow lavas from ODP Hole 896A,” in Proceedings of the ODP Science Results, Vol. 148, eds J. C. Alt, H. Kinoshita, L. B. Stokking, and P. J. Michael (College Station, TX: Ocean Drilling Program), 191–206.
Guyoneaud, R., Süling, J., Petri, R., Matheron, R., Caumette, P., Pfennig, N., and Imhoff, J. F. (1998). Taxonomic rearrangements of the genera Thiocapsa and Amoebobacter on the basis of 16S rDNA sequence analyses, and description of Thiolamprovum gen. nov. Int. J. Syst. Bacteriol. 48, 957–964.
Hess, W. R., Rocap, G., Ting, C. S., Larimer, F., Stilwagen, S., Lamerdin, J., and Chrisholm, W. (2001). The photosynthetic apparatus of Prochlorococcus: insights through comparative genomics. Photosynth. Res. 70, 53–71.
Huber, J. A., Johnson, H. P., Butterfield, D. A., and Baross, J. A. (2006). Microbial life in ridge flank crustal fluids. Environ. Microbiol. 8, 88–99.
Huber, J. A., Welch, D. B. M., Morrison, H. G., Huse, S. M., Neal, P. R., Butterfield, D. A., and Sogin, M. L. (2007). Microbial population structure in the deep marine biosphere. Science 318, 97–100.
Hügler, M., Wirsen, C. O., Fuchs, G., Taylor, C. D., and Sievert, S. M. (2005). Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the epsilon subdivision of proteobacteria. J. Bacteriol. 187, 3020–3027.
Hunter, A. G., Kempton, P. D., and Greenwood, P. (1999). Low-temperature fluid-rock interaction – an isotopic and mineralogical perspective of upper crustal evolution, eastern flank of the Juan de Fuca Ridge (JdFR), ODP Leg 168. Chem. Geol. 155, 3–28.
Itoh, T., Suzuki, K., and Nakase, T. (1998). Thermocladium modestius gen. nov., sp. nov., a new genus of rod-shaped, extremely thermophilic crenarchaeote. Int. J. Syst. Evol. Microbiol. 48, 879–887.
Jannasch, H. W., Wirsen, C. O., Nelson, D. C., and Robertson, L. A. (1985). Thiomicrospira crunogena, sp. nov., a colorless, sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Bacteriol. 35, 422–424.
Jørgensen, B. B., and Boetius, A. (2007). Feast and famine – microbial life in the deep-sea bed. Nat. Rev. Microbiol. 5, 770–781.
Juniper, S. K., Cambon, M.-A, Lesongeur, F., and Barbier, G. (2001). Extraction and purification of DNA from organic rich subsurface sediments (ODP Leg 169S). Mar. Geol. 174, 241–247.
Kirchman, D. L. (2002). The ecology of Cytophaga–Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39, 91–100.
Kirkpatrick, J., Oakley, B., Fuchsman, C., Srinivasan, S., Staley, J. T., and Murray, J. W. (2006). Diversity and distribution of Planctomycetes and related bacteria in the suboxic zone of the Black Sea. Appl. Environ. Microbiol. 72, 3079–3083.
Kormas, K., Smith, D. C., Edgcomb, V., and Teske, A. (2003). Molecular analysis of deep subsurface microbial communities in Nankai Trough sediments (ODP Leg 190, Site 1176). FEMS Microbiol. Ecol. 45, 115–125.
Kovaleva, O. L., Tourova, T. P., Muyzer, G., Kolganova, T. V., and Sorokin, D. Y. (2011). Diversity of RuBisCO and ATP citrate lyase genes in soda lake sediments. FEMS Microbiol. Ecol. 75, 37–47.
Kuenen, J. G., and Veldkamp, H. (1972). Thiomicrospira pelophila, gen. nov., sp. nov., a new obligately chemolithotrophic colourless sulfur bacterium. Antonie van Leeuwenhoek 38, 241–256.
Kumar, S., Dudley, J., Nei, M., and Tamura, K. (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9, 299–306.
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (New York, NY: John Wiley & Sons), 115–175.
Lever, M. A., Alperin, M., Inagaki, F., Nakagawa, S., Steinsbu, B. O., Teske, A., and IODP Expedition 301 Scientists. (2006). Trends in basalt and sediment core contamination during IODP Expedition 301. Geomicrobiol. J. 23, 517–530.
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, Buchner, A., Lai, T., Steppi, S., Jobb, G., Förster, W., Brettske, I., Gerber, S., Ginhart, A., Gross, O., Grumann, S., Hermann, S., Jost, R., König, A., Liss, T., Lüssmann, R., May, M., Nonhoff, B., Reichel, B., Strehlow, R., Stamatakis, A., Stuckmann, N., Vilbig, A., Lenke, M., Ludwig, T., Bode, A., and Schleifer, K.-H. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371.
Mottl, M. J. (1989). Hydrothermal convection, reaction, and diffusion in sediments on the Costa Rica Rift flank: pore-water evidence from ODP Sites 677 and 678. Proc. ODP. Sci. Results 111, 195–213.
Mottl, M. J., and Gieskes, J. M. (1990). Chemistry of waters sampled from oceanic basement boreholes, 1979–1988. J. Geophys. Res. 95, 9227–9242.
Nagamuna, T., Elsaied H. E., Hoshii, G., and Kimura, H. (2007). Bacterial endosymbioses of gutless tube-dwelling worms in nonhydrothermal vent habitats. Mar. Biotechnol. 6, 416–428.
Nakagawa, S., Inagaki, F., Suzuki, Y., Steinsbu, B. O., Lever, M. A., Takai, K., Engelen, B, Sako, Y., Wheat, C. G., Horikoshi, K., and Integrated Ocean Drilling Program Expedition 301 Scientists. (2006). Microbial community in black rust exposed to hot ridge flank crustal fluids. Appl. Environ. Microbiol. 72, 6789–6799.
Nakagawa, S., and Takai, K. (2008). Deep-sea vent chemoautotrophs: diversity, biochemistry and ecological significance. FEMS Microbiol. Ecol. 65, 1–14.
Nishihara, H., Yaguchi, T., Chung, S. Y., Suzuki, K.-I., Yanagi, M., Yamasato, K., Kodama, T., and Igarashi, Y. (1998). Phylogenetic position of an obligately chemoautotrophic, marine hydrogen-oxidizing bacterium, Hydrogenovibrio marinus, on the basis of 16S rRNA gene sequences and two form I RubisCO gene sequences. Arch. Microbiol. 169, 364–368.
Orcutt, B., Bach, W., Becker, K., Fisher, A. T., Hentscher, M., Toner, B. M., Wheat, C. G., and Edwards, K. J. (2011). Colonization of subsurface microbial observatories deployed in young ocean crust. ISME J. 5, 692–703.
Parkes, R. J., Cragg, B. A., and Wellsbury, P. (2000). Recent studies on bacterial populations and processes in subseafloor sediments: a review. Hydrogeol. J. 8, 160.
Perner, M., Kuever, J., Seifert, R., Pape, T., Koschinsky, A., Schmidt, K., Strauss, H., and Imhoff, J. F. (2007). The influence of ultramafic rocks on microbial communities at the Logatchev hydrothermal field, located 15°N on the Mid-Atlantic Ridge. FEMS Microbiol. Ecol. 61, 97–109.
Rappé, M. S., Connon, S. A., Vergin, K. L., and Giovannoni, S. J. (2002). Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418, 630–633.
Rappé, M. S., Kemp, P. F., and Giovannoni, S. J. (1997). Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol. Oceanogr. 45, 811–826.
Rassa, A. C., McAllister, S. M., Safran, S. A., and Moyer, C. L. (2009). Zeta-proteobacteria dominate the colonization and formation of microbial mats in low-temperature hydrothermal vents at Loihi Seamount, Hawaii. Geomicrobiol. J. 26, 623–638.
Ryan, W. B. F., Carbotte, S. M., Coplan, J. O., O’Hara, S., Melkonian, A., Arko, R., Weissel, R. A., Ferrini, V., Goodwillie, A., Nitsche, F., Bonczkowski, J., and Zemsky, R. (2009). Global multi-resolution topography synthesis, Geochem. Geophys. Geosyst. 10, Q03014.
Santelli, C. M., Banarjee, N., Bach, W., and Edwards, K. J. (2010). Tapping into the subsurface ocean crust biosphere: low biomass and drilling-related contamination calls for improved quality controls. Geomicrobiol. J. 27, 158–169.
Santelli, C. M., Edgcomb, V. P., Bach, W., and Edwards, K. J. (2009). The diversity and abundance of bacteria in habiting seafloor laves positively correlate with rock alteration. Environ. Microbiol. 11, 86–98.
Santelli, C. M., Orcutt, B. N., Banning, E., Bach, W., Moyer, C. L., Sogin, M. L., Staudigel, H., and Edwards, K. J. (2008). Abundance and diversity of microbial life in ocean crust. Nature 453, 653–656.
Schloss, P. D., and Handelsman, J. (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71, 1501–1506.
Schloss, P. D., and Handelsman, J. (2006). Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 72, 6773–6779.
Scott, K. M., Sievert S. M., Abril, F. N., Ball, L. A., Barrett, C. J., Blake, R. A., Boller, A. J., Chain, P. S., Clark, J. A., Davis, C. R., Detter, C., Do, K. F., Dobrinski, K. P., Faza, B. I., Fitzpatrick, K. A., Freyermuth, S. K., Harmer, T. L., Hauser, L. J., Hügler, M., Kerfeld, C. A., Klotz, M. G., Kong, W. W., Land, M., Lapidus, A., Larimer, F. W., Longo, D. L., Lucas, S., Malfatti, S. A., Massey, S. E., Martin, D. D., McCuddin, Z., Meyer, F., Moore, J. L., Ocampo, L. H. Jr., Paul, J. H., Paulsen, I. T., Reep, D. K., Ren, Q., Ross, R. L., Sato, P. Y., Thomas, P., Tinkham, L. E., and Zeruth, G. T. (2006). The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena XCL-2. PLoS Biol. 4, 2196–2212. doi: 10.1371/journal.pbio.0040383
Shipboard Scientific Party. (1993). “Site 896,” in Proceedings of the ODP, Scientific Results, eds J. C. Alt, H. Kinoshita, L. B. Stokking, and P. J. Michael (College Station, TX: Ocean Drilling Program), 123–192.
Sorokin, D. Y., Lysenko, A. M., Mityushina, L. L., Tourova, T. P., Jones, E., Rainey, F. A., Robertson, L. A., and Kuenen, J. G. (2001). Thioalkalimicrobium aerophilum gen. nov., sp. nov., and Thioalkalimicrobium sibericum sp. nov., and Thioalkalivibrio versutus gen. nov., sp. nov. Thioalkalivibrio nitratis sp. nov., and Thioalkalivibrio denitrificans sp. nov., novel obligately alkaliphilic and obligately chemolithoautotrophic sulfur-oxidizing bacteria from soda lakes. Int. J. Syst. Evol. Microbiol. 51, 565–580.
Steinsbu, B. O., Thorseth, I. H., Nakagawa, S., Inagaki, F., Lever, M. A., Engelen, B., Øvreas, L., and Pedersen, R. B. (2010). Archaeoglobus sulfaticallidus sp. nov., a novel thermophilic and facultative lithoautotrophic sulfate-reducer isolated from black rust exposed to hot ridge flank crustal fluids. Int. J. Syst. Evol. Microbiol. 60, 2745–2752.
Suzuki, Y., Inagaki, F., Takai, K., Nealson, K. H., and Horikoshi, K. (2004). Microbial diversity in inactive chimney structures from deep-sea hydrothermal systems. Microb. Ecol. 47, 186–196.
Sylvan, J. B., Toner, B. M., and Edwards, K. J. (2012). Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. MBio 3, e00279-11. doi: 10.1128/mBio.00279-11
Takai, K., Nunoura, T., Ishibashi, J., Lupton, J., Suzuki, R., Hamasaki, H., Ueno, Y., Kawagucci, S., Gamo, T., Suzuki, Y., Hirayama, H., and Horikoshi, K. (2008). Variability in the microbial communities and hydrothermal fluid chemistry at the newly discovered Mariner hydrothermal field, southern Lau Basin. J. Geophys. Res. 113, G02031.
Taylor, C. D., and Wirsen, C. O. (1997). Microbiology and ecology of filamentous sulfur formation. Science 277, 1483–1485.
Torsvik, T., Furnes, H., Muehlenbachs, K., Thorseth, T. H., and Tumyr, O. (1998). Evidence for microbial activity at the glass-alteration interface in oceanic basalts. Earth Planet. Sci. Lett. 162, 165–176.
Wagner, M., and Horn, M. (2006). The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 17, 241–249.
West, N. J., Schonhuber, W. A., Fuller, N. J., Amann, R. I., Rippka, R., Post, A. F., and Scanlan, D. J. (2001). Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology 147, 1731–1744.
Wheat, C. G., Jannasch, H. W., Fisher, A. T., Becker, K., Sharkey, J., and Hulme, S. (2010). Subseafloor seawater-basalt-microbe reactions: continuous sampling of borehole fluids in a ridge flank environment. Geochem. Geophys. Geosyst. 11, Q07011.
Wheat, C. G., and Mottl, M. J. (2000). Composition of pore and spring waters from Baby Bare: global implications of geochemical fluxes from a ridge flank hydrothermal system. Geochim. Cosmochim. Acta 64, 629–642.
Whitman, W. B., Coleman, D. C., and Wiebe, W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583.
Keywords: basalt, chemolithoautotrophic bacteria, CORKs, Costa Rica rift, formation fluids, ocean drilling program, subsurface, thiomicrospira
Citation: Nigro LM, Harris K, Orcutt BN, Hyde A, Clayton-Luce S, Becker K and Teske A (2012) Microbial communities at the borehole observatory on the Costa Rica Rift flank (Ocean Drilling Program Hole 896A). Front. Microbio. 3:232. doi: 10.3389/fmicb.2012.00232
Received: 22 March 2011; Accepted: 07 June 2012;
Published online: 29 June 2012.
Edited by:
Axel Schippers, Federal Institute for Geosciences and Natural Resources, GermanyReviewed by:
Julie A. Huber, Marine Biological Laboratory, USAJames Cowen, University of Hawaii, USA
Copyright: © 2012 Nigro, Harris, Orcutt, Hyde, Clayton-Luce, Becker and Teske. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Andreas Teske, Department of Marine Sciences, University of North Carolina at Chapel Hill, 340 Chapman Hall, CB 3300, Chapel Hill, NC 27599, USA. e-mail: teske@email.unc.edu
 Lisa M. Nigro1
Lisa M. Nigro1