- 1Aquatic Microbial Biogeochemistry Laboratory, Department of Marine Sciences, University of Georgia, Athens, GA, USA
- 2Department of Marine Sciences, The University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
We measured potential nitrate removal and denitrification rates in hydrothermally altered sediments inhabited by Beggiatoa mats and adjacent brown oil stained sediments from the Guaymas Basin, Gulf of California. Sediments with Beggiatoa maintained slightly higher rates of potential denitrification than did brown sediments at 31.2 ± 12.1 versus 21.9 ± 1.4 µM N day−1, respectively. In contrast, the nitrate removal rates in brown sediments were higher than those observed in mat-hosting sediments at 418 ± 145 versus 174 ± 74 µM N day−1, respectively. Additional experiments were conducted to assess the responses of denitrifying communities to environmental factors [i.e., nitrate, sulfide, and dissolved organic carbon (DOC) concentration)]. The denitrifying community had a high affinity for nitrate (Km = 137 ± 91 µM ), in comparison to other environmental communities of denitrifiers, and was capable of high maximum rates of denitrification (Vmax = 1164 ± 153 µM N day−1). The presence of sulfide resulted in significantly lower denitrification rates. Microorganisms with the potential to perform denitrification were assessed in these sediments using the bacterial 16S rRNA gene and nitrous oxide reductase (nosZ) functional gene libraries. The bacterial 16S rRNA gene clone library was dominated by Epsilonproteobacteria (38%), some of which (e.g., Sulfurimonas sp.) have a potential for sulfide-dependent denitrification. The nosZ clone library did not contain clones similar to pure culture denitrifiers; these clones were most closely associated with environmental clones.
Introduction
In anoxic environments, nitrate serves as an energy-rich electron acceptor for microbial terminal metabolism and its complete reduction generates either dinitrogen or ammonium. In the hydrothermally altered surficial sediments of Guaymas Basin, nitrate is abundant (highest 40 µM; McHatton et al., 1996 and references therein) and nitrate-concentrating Beggiatoa mats cover vast areas of sediment surface. Mat-hosting sediments are sites of rigorous coupled nitrogen–carbon–sulfur cycling (Teske and Nelson, 2006 and references therein). Dissimilatory processes, such as sulfate reduction and anaerobic methane oxidation, have been studied frequently in Guaymas Basin sediments, including mat-hosting sites (Elsgard et al., 1994; Weber and Jø, 2002; Kallmeyer and Boetius, 2004; Holler et al., 2011; Biddle et al., 2012). However, nitrate dynamics and dissimilatory pathways of nitrate reduction, such as denitrification, have not been explored.
Environmental conditions are postulated to control the dominant dissimilatory nitrate reduction pathways expressed in sediments and therefore which product(s) accumulate (Brunet and Garcia-Gil, 1996; Burgin and Hamilton, 2007; Porubsky et al., 2009). In Guaymas Basin, abundant stocks of thermally-derived organic carbon and reduced substrates, including sulfide, can influence nitrate reduction pathways. Complex organic carbon can support heterotrophic dissimilatory denitrification (DNF) and fermentative dissimilatory nitrate reduction to ammonium (DNRA; Burgin and Hamilton, 2007; Widdel and Rabus, 2001). Sulfide can have a stimulatory or inhibitory effect on nitrate reduction – depending on the pathway employed – and hence may affect the products of nitrate reduction (Joye, 2002). Sulfide can also fuel the activity of some autotrophic denitrifiers (e.g., Sulfurimonas denitrificans) and some nitrate reducers capable of DNRA use sulfide as an electron donor. Sulfide inhibition can reduce denitrification rates and stimulate nitrous oxide production because nitrous oxide reductase, the enzyme catalyzing the reduction of nitrous oxide to dinitrogen, is sensitive to sulfide (Brunet and Garcia-Gil, 1996; Joye, 2002; Porubsky et al., 2009). The absence of the nitrous oxide reductase gene could also result in N2O production in the environment. The abundance of reduced carbon substrates and sulfide are not considered conducive for anaerobic ammonium oxidation (ANAMMOX; Burgin and Hamilton, 2007), so this pathway was not examined in this study.
Microorganisms capable of complete or incomplete DNF exist across the microbial tree of life. Previous studies of the molecular ecology of Guaymas Basin sediments focused on general prokaryotic, sulfate reducing, or methane cycling populations (Teske et al., 2002; Dhillon et al., 2003; Dhillon et al., 2005; Biddle et al., 2012). In Guaymas bacterial clone libraries, potential nitrogen cycling members of Gammaproteobacteria related to bacterial mat species (e.g., Beggiatoa) and of Epsilonproteobacteria (Teske et al., 2002; Dhillon et al., 2003) were observed. These groups of nitrate-utilizing bacteria can use sulfide as an electron donor, and are thought to produce dinitrogen as the end product of their metabolism (Sievert et al., 2003; Sweerts et al., 1990). However, some members of Gammaproteobacteria reduce nitrate to ammonium (Høgslund et al., 2009; Otte et al., 1999). The functional gene responsible for the final step of DNF (nosZ) reflects the dinitrogen producers present.
Working under the assumption that DNF and nitrate removal were active in Guaymas Basin sediments, we tested the impact of varying nitrate, sulfide, and dissolved organic carbon concentrations on cycling processes, and describe the diversity of the nosZ functional genes in Guaymas sediments. First, we describe and compare potential nitrate removal and DNF rates in Beggiatoa mat hosting sediments versus adjacent brown sediments lacking Beggiatoa mat biomass. Then, from similar Beggiatoa mat hosting sediments, we report how denitrifying bacteria responded to changes in nitrate, sulfide, and dissolved organic carbon (DOC) concentration. Finally, we describe the microbial communities performing nitrous oxide reduction using specific functional genes for this step (nosZ), complemented by bacterial 16S rRNA gene surveys.
Materials and Methods
Sample Collection and Treatment
Sediments were collected from Guaymas Basin, Gulf of California, by the manned submersible DSV Alvin during research cruises in 2008 and 2009. From the 2008 expedition, cores were collected from a dense Beggiatoa mat and surrounding sediments, within the area named “Megamat” (DSV Alvin-Dives 4490 and 4492; 27°0.459N, 111°24.526W; December 14 and 16, 2008). During the 2009 expedition, several sediment cores within a dense Beggiatoa mat were collected (DSV Alvin- Dive 4572; December 3, 2009). All sediments were collected at approximately 2000 meters below sea level with upper sediment horizons near 4°C, but typically temperatures increased to >80°C by 20 cm below the sediment water interface. Sediment cores were stored at 4°C until use in laboratory experiments (within 2 weeks).
For sediments sampled in 2008, experiments were conducted on cores either hosting Beggiatoa mat (hereafter, mat) at the surface or brown sediment lacking visible mat (hereafter, brown). The brown sediment was sulfidic within millimeters of the sediment surface and was oil-stained over its entire depth. Rates of potential DNF and nitrate removal were measured as described by Bowles and Joye (2011). Briefly, sediment was collected from the 0–6 cm horizon and mixed with an Ar purged artificial porewater mixture [APW (mM): NaCl, 491, MgCl2*6H2O, 24, CaCl2*2H2O, 1.6, KH2PO4, 0.03, KCl, 11, NaHCO3, 5] in a 2:1 ratio. The APW and sediment mixture was then centrifuged. The supernatant APW was poured off, fresh APW was added to reconstitute the initial volume, then the sample was gently mixed. This process was repeated three times to obtain a known amount of dissolved constituents in the porewater prior to starting the rate experiments. Before dispensing the slurry into individual culture tubes, the slurry was bubbled with 0.09% Ar and a balance of He for 1 h. Individual culture tubes containing 15 mL of slurry were sealed with butyl rubber stoppers without a headspace (Orcutt et al., 2005). All samples were next injected with 100 µL of concentrated DOC with equimolar carbon from lactate and acetate to achieve a final concentration of 2 mM DOC. Then samples were injected with enriched nitrate (K15NO3, 99% Cambridge Isotope Laboratories®) to a final concentration of 100 µM. Samples were constantly shaken (75 rpm) and incubated at 40°C, a temperature representing the approximate average for the upper 6 cm. Prior to injection with 15 and DOC, samples were pre-incubated at 40°C for 48 h to reacclimatize microorganisms to quasi in situ temperatures. Incubations were terminated (for triplicate sub-samples) at each time point (0, 5, 13, and 26 h) by centrifugation at 500 rpm for 15 minutes, after which porewater was sampled immediately for dissolved gases (i.e., dinitrogen as 29N2 and 30N2). Briefly, dinitrogen was sampled on a membrane inlet mass spectrometer (MIMS) with minimum detection limit <5 nM and precision of standard measurement of <0.5% (Kana et al., 1998). After gas sampling, the remaining porewater was filtered (0.2 µm), and samples for determination of dissolved components nitrate (), and nitrite (), were collected and subsequently measured. The preservation and analytical methods used for dissolved constituents were described previously (Bowles and Joye, 2011). Briefly, samples for dissolved constituents were refrigerated (4°C) until measurement on an Antek 7050 with chemiluminescence detection (NOx) and on a spectrophotometer (; Joye et al., 2004). The detection limit for NOx was approximately 150 nM and precision was <5%, while the detection limit was 50 nM and the precision was 2%.
Nitrate, Sulfide, and Doc Experiments
To investigate the response of nitrogen cycle dynamics to environmental factors, we used cores collected from a site occupied by dense Beggiatoa mat. Prior to these experiments, the mat was gently removed from the sediment surface to reduce background nitrate levels because Guaymas Beggiatoa vacuoles often contain 50–100 mM (McKay et al., 2012). For this sampling the upper 3 cm of the sediment from three sediment cores was collected and a slurry was generated, as described above. We used three separate treatments to analyze the physiological response of the mixed community of nitrate reducers: (1) nitrate, (2) sulfide, and (3) DOC treatments at concentrations of 0, 100, 500, 1000, 5000 µM. The sediment slurry was mixed with APW, purged, and rinsed as described previously for potential DNF rate assays. All treatments were run in triplicate. For the individual treatments, 100 µL of the concentrated respective component was added to achieve the target concentration. In the nitrate treatment, samples were injected with 100 µL of a DOC solution (equimolar C from lactate, acetate, and glucose) to yield 1 mM DOC. After purging the nitrate (K15NO3, 99% Cambridge Isotope Laboratories®) with 0.09% Ar and a balance of He, an aliquot was injected into samples so that the target concentration was reached. The pH of the sulfide treatment was adjusted by addition of an equimolar amount of He purged hydrochloric acid applied directly to sulfide solution, and subsequently the sample was tested to ensure that the desired pH was maintained (Teske and Nelson, 2006). For the DOC treatment, carbon was added as equimolar C from lactate, acetate, and glucose. In sulfide and DOC treatments, 100 µL of nitrate (15N) was injected to equal the final concentration of 1 mM. Immediately following the injection, samples were placed on a shaker table at 75 rpm at 4°C and allowed to incubate for 5 h. A lower temperature was used on these experiments because the temperature of the surficial sediments used was comparable to the ambient bottom seawater.
After the incubation, tubes were centrifuged and sampled as described above. Immediately after the incubation subsamples for dissolved gases were collected (30N2, 29N2, and N2O). Measurement of N2O was conducted for the nitrate and DOC treatments; the N2O samples from the sulfide treatment were unfortunately contaminated. The concentration of N2O was measured with a gas chromatograph equipped with an electron capture detector with a detection limit of around 20 ppb in the headspace analysis, which gave a detection limit of ~9 nM at 5% precision (Porubsky et al., 2009). Briefly, 1 mL of media sample was collected after centrifugation and injected into a He purged headspace vial closed with a butyl rubber stopper and containing a pellet of NaOH. After gas sampling, the artificial porewater was filtered (0.2 µm) and a sample was collected for NOx and concentration determination. Methods used to preserve and analyze the dissolved constituents were described previously.
Molecular Analysis
DNA extraction, primer design, and polymerase chain reaction
Approximately 0.5 g of wet sediment was used for DNA extraction with the MOBIO® (Carlsbad, CA) Ultrapure Soil DNA extraction kit following the manufacturer’s protocol. The PCR master mix consisted of (25 µL): 2µL of template DNA, 0.25 µL of forward, and reverse primers (100 µmol L−1), 0.75 µL of bovine serum albumin (10 mg/mL), 0.12 µL of GoTaq Taq polymerase (PromegaTM), and the balance of the volume as sterile H2O. The PCR cycle for 16S rRNA bacterial gene region began with an initial denaturation of 10 min at 94°C followed by 30 cycles of 1.5 min of denaturation at 94°C, 30 s of annealing at 55°C, 30 s of elongation at 72°C, ending with 7 min of elongation at 72°C. The 16S rRNA gene region of bacteria was amplified using the primer B27f (AGAGTTTGATCCTGGCTCAG) and UNI1392r (ACGGGCGGTGTGTRCA; Orphan et al., 2001a,b).
The gene catalyzing the reduction of N2O to N2, nitrous oxide reductase, nosZ is highly diverse (Scala and Kerkhof, 1999). Primers nosZ661f (CGGCTGGGGGCTGACCAA) and nosZ1773r (ATRTCGATCARCTGBTCGTT) were used to amplify the region 661–1773, with positions relative to Pseudomonas stutzeri (Scala and Kerkhof, 1999). For PCR amplification of the ~1,100 base pair nosZ gene, a denaturation of 5 min at 94°C was used and followed by 35 cycles of 30 s of denaturation at 94°C, 1.5 min of annealing at 56°C, 2 min of elongation at 72°C, ending with 7 min of elongation at 72°C (Scala and Kerkhof, 1999).
Cloning
All PCR products were verified to contain the gene of interest based on the size of the amplicon, and subsequently purified using a Qiagen Gel Extraction Kit, as specified by the manufacturer. The PCR products were then ligated into a pCR4 vector (Invitrogen) and transformed into Escherichia coli according to manufacturer’s specifications. All colonies were screened for ampicillin resistance and lacZ expression. Sequencing was commenced at the M13F primer within the pCR4 vector.
Phylogenetic analysis
The 16S rRNA gene sequences were screened first using blastn (NCBI), and next were aligned using the Silva Incremental Aligner (SINA; Pruesse et al., 2007). For nosZ gene analysis, sequences were converted to amino acids and verified to be on the correct reading frame using the open reading frame (ORF) finder (NCBI). The nosZ functional gene was aligned using Clustal W and the alignment (196 amino acids) was then manually edited (Larkin et al., 2007). After alignment, all 16S rRNA gene clones (600 nucleotides) were tested for chimeras using Bellerophon (Huber et al., 2004). Following the chimera check, sequences were imported to ARB and the quality of the alignment was verified and manually adjusted in ARB_EDIT (Ludwig et al., 2004). Mega5 was used to create 16S rRNA gene and nosZ phylogenetic trees (Tamura et al., 2011). Phylogenetic trees for 16S rRNA were made by neighbor joining, with a Jukes–Cantor model for distance correction, and the tree was verified by bootstrap analysis (n = 1000). The nosZ tree was created using amino acid translations (197 amino acids) using minimum evolution with complete deletion and verification by bootstrap analysis (n = 1000). The cutoff for operational taxonomic units (OTUs) for bacterial 16S rRNA genes was 97%, and since divergence and hence OTUs are not established for nosZ, sequences are shown with identical sequences removed. All bacterial 16S rRNA gene OTUs were established using mothur (Schloss et al., 2009).
Results
Potential Denitrification Rates in mat and Brown Sediments
Over the course of a 26-h incubation, mat and brown sediments consumed all added 15 (110 ± 33 µM at time zero and <1 µM at 26 h); nitrite was not detected at any time point (Figure 1A). In brown sediments without mat, nitrate concentrations at the start of the incubation were 108 (±29) µM and the amended nitrate was exhausted (<1 µM) after only 13 h (Figure 1B). Potential denitrification, tracked as accumulation of 30N2 and 29N2, was observed in as little as 5 h in both types of sediment. The majority of 15 was converted to 30N2, with only a minor fraction (<2 µM) ending up as 29N2. In mat sediments, 16.2 (±6.4) µM 30N2 accumulated by 13 h. Relatively more 30N2, 22.4 (±1.4) µM, accumulated in brown sediments after 26 h.
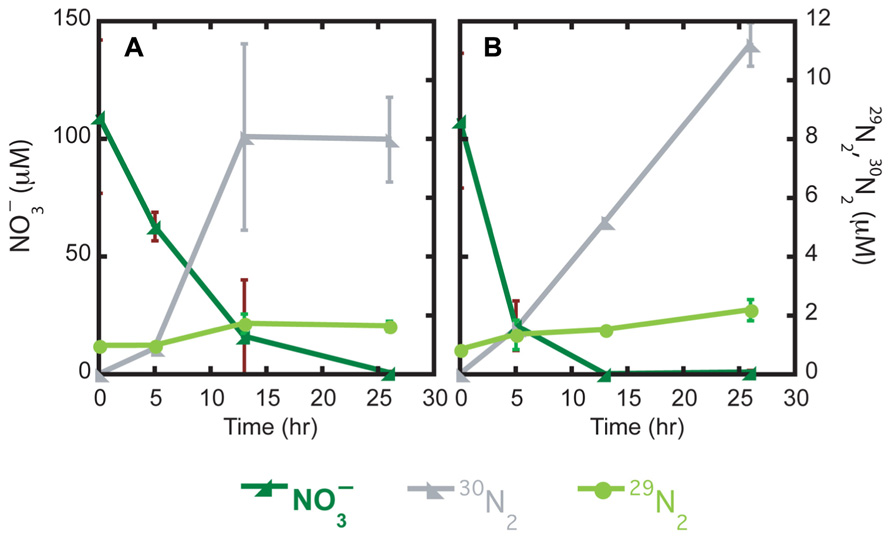
FIGURE 1. Time series incubations of mat (A) and brown (B) sediments with the substrate (, µM) and products (29N2 and 30N2, µM) plotted against time (h).
Potential denitrification is defined as the sum of 30N2 and 29N2 formation rates. Linear portions of nitrate consumption and 30N2 and 29N2 formation curves were used to estimate nitrate removal rates and potential denitrification rates, respectively. In mat sediments, over the first 13 h of the incubation, the rate of nitrate removal was 174 (±74) µM N day−1. In brown sediments, the first 5 h were used to estimate a nitrate removal rate of 418 (±145) µM N day−1. Integrated areal rates of nitrate removal were 435 and 1045 µmol m−2 h−1 in mat and brown sediments, respectively. Potential denitrification accounted for only a small fraction of nitrate removal in both mat and brown sediments. In mat sediments, potential denitrification rates were estimated over the first 13 h to be 31.2 (±12.1) µM N day−1. Potential denitrification rates were slightly lower in brown sediments at 21.9 (±1.4) µM N day−1, but this difference was not statistically significant. Integrated areal rates of potential denitrification were 78 and 54 µmol m−2 h−1 in mat and brown sediments, respectively.
Denitrification Kinetics and Influences of Sulfide and Doc
Nitrate, sulfide, and DOC amendment generated different responses of the nitrate utilizing populations in Guaymas Basin sediments. The consumption levels were the highest in the nitrate treatment of 5000 µM at 675 µM consumed (∆; Figure 2A). In the nitrate treatment at 1000 µM and at all DOC concentration treatments, the nitrate removal levels were roughly ~300 µM (Figures 2A,C). With respect to increases in concentrations of nitrate, sulfide, and DOC, nitrate consumption levels increased, decreased, or did not change, respectively (Figure 2).
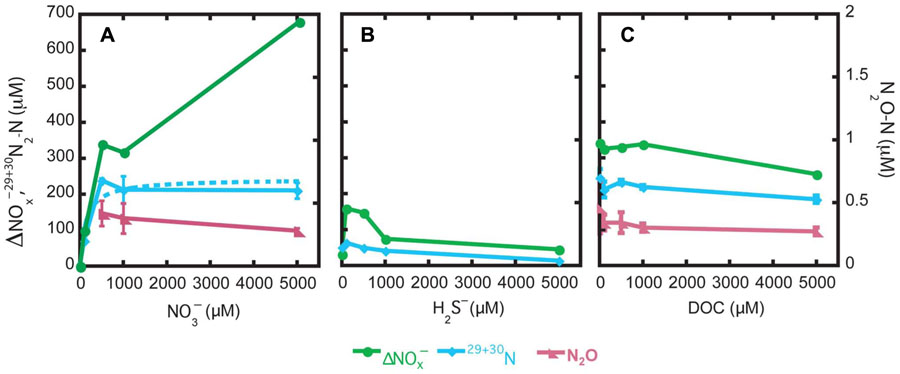
FIGURE 2. Environmental influences of nitrate (A), sulfide (B), and DOC (C) on denitrification and nitrogen species end products.
The proportion of 15 converted to 30N2 and 29N2 was summed to generate the µM N as N2 and is presented as the potential denitrification rate (Figure 2). In the nitrate treatment, maximum denitrification was observed at 500 µM with 243 (±6.1) µM N converted to dinitrogen, corresponding to a DNF rate of 1166 µM N d−1. A Michaelis–Menten equation was fit to the data (r2 = 0.91) from the nitrate treatments resulting in a maximum denitrification rate (Vmax) of 1164 (±153) µM N day−1 and half-saturation constant (Km) of 137 (±91) µM for the endogenous denitrifying community (Figure 2A). In the sulfide amended samples, much less N2 formation was observed relative to the nitrate or DOC treatments. The most N2 formed in the sulfide treatment was at the concentration of 100 µM, at 61 (±1) µM N as N2. Denitrification slowed as sulfide concentration rose above 100 µM H2S. In the DOC treatment, rates of denitrification to N2 varied little, from 178 (±10) to 238 (±24) µM N day−1, with no observable trends in response versus DOC concentration.
In the nitrate and DOC treatments, N2O was measured to determine if there was N2O accumulation in response to the specific treatments. In the nitrate and DOC treatments, N2O accumulation was high relative to typical environmental levels (range of 0.2–0.4 µM N as N2O; Figures 2A,C). The exception was the nitrate treatment with no nitrate added and with 100 µM nitrate wherein N2O was not detected.
In order to assess the pathways or fates of nitrate in Guaymas Basin sediments, percentages of total nitrate consumption converted to N2 or N2O relative to nitrate removed (e.g., assimilation, storage, or other processes DNRA and anaerobic ammonium oxidation, ANAMMOX) were determined. The fraction of nitrate converted to N2 represents the sum of 30N2 and 29N2 and the proportion classified as nitrate removed is the balance of nitrate consumption not occurring as N2 or N2O (where N2O data was available). In general, as nitrate concentrations increased, the fraction of nitrate converted to N2 decreased, from ~75 to 32% (Table 1). Systematic changes in the fraction of N2 formation observed were not influenced by sulfide or DOC treatments; note that gaseous N formation in the sulfide treatment does not include N2O. In the DOC treatment the fraction of nitrate converted to N2O appeared to decrease slightly with respect to DOC concentration from 0.14 to 0.11% (Table 1).
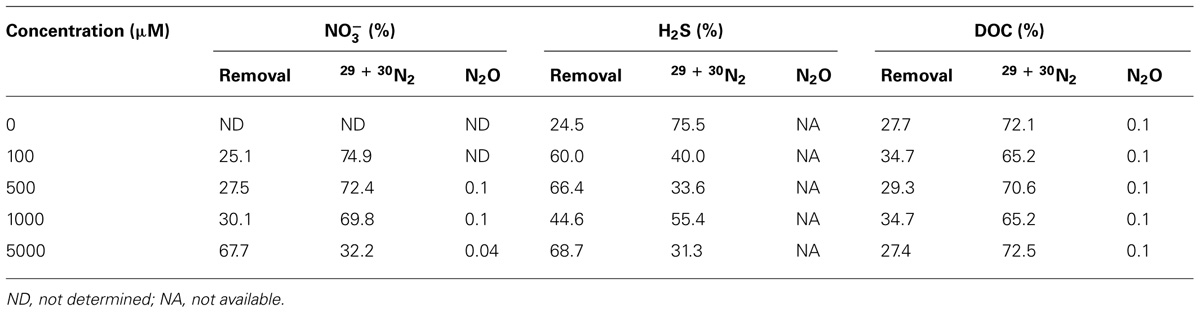
TABLE 1. Percent of end products observed in samples amended with nitrate, sulfide, and DOC for all concentration levels.
Molecular Census of Bacteria and Possible Denitrifiers
Bacterial 16S rRNA gene and nosZ functional genes
Cloning and sequencing of PCR-amplified 16S rRNA and nosZ genes from sediments used in the nitrate, sulfide, and DOC experiments suggested that the sediments maintained a microbiological capacity for denitrification. The 16S rRNA bacterial gene clone library (n = 77 clones total) was dominated by Epsilonproteobacteria (38%), Bacteroidetes (21%), Deltaproteobacteria (8%), and Gammaproteobacteria (7%). Within the Epsilonproteobacteria most clones were affiliated with the genus Sulfurovum (27%) or Sulfurimonas (6%; percentages are relative to the entire 16S library). Many of the clones were highly similar to Epsilonproteobacteria clones previously detected in Guaymas Basin sediments (Figure 3). In addition to the occurrence of 16S rRNA gene clones potentially relevant to denitrification, we also characterized the nitrous oxide reductase functional gene from these sediments. A total of 20 nosZ functional genes were retrieved. Of these, two sequences reflected multiple identical clones, leaving 10 unique sequences (Figure 4). The Guaymas Basin sequences were dissimilar from all pure culture nosZ, and similar to nosZ functional genes extracted from continental margin sediments (Scala and Kerkhof, 1999).
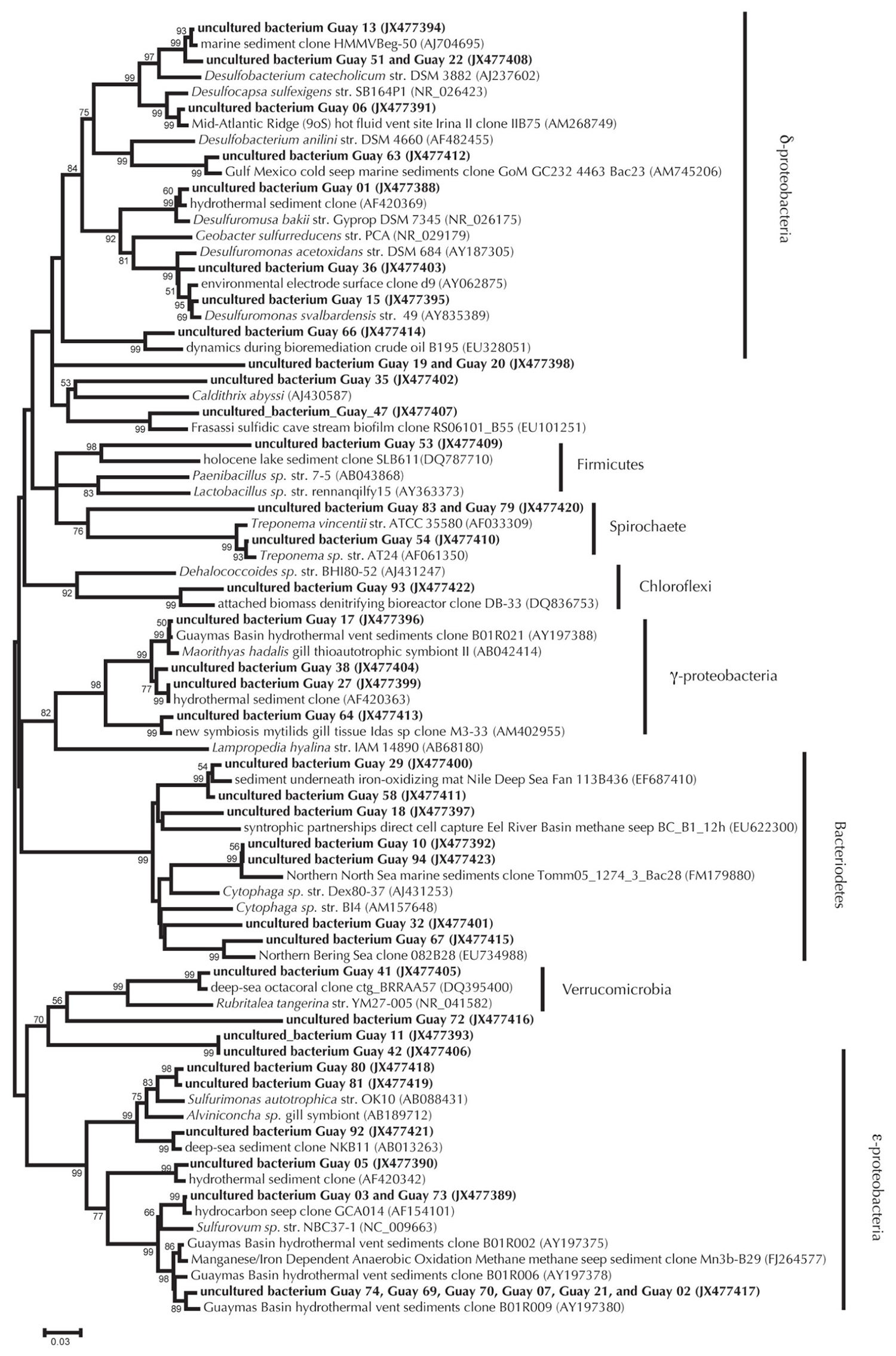
FIGURE 3. 16S rRNA bacterial gene phylogeny of Guaymas Basin environmental clones relative to pure culture and other environmental clones. Environmental clones from this work appear as bold text, pure cultures are italicized, and environmental clones from other environments appear as normal text. Neighbor joining method was used to generate a tree with a Jukes–Cantor correction of evolutionary distance. Bootstrap values for branches occurring for >50% of 1000 iterations are reported. Scaling of the phylogenetic tree is based on an evolutionary distance of 0.03.
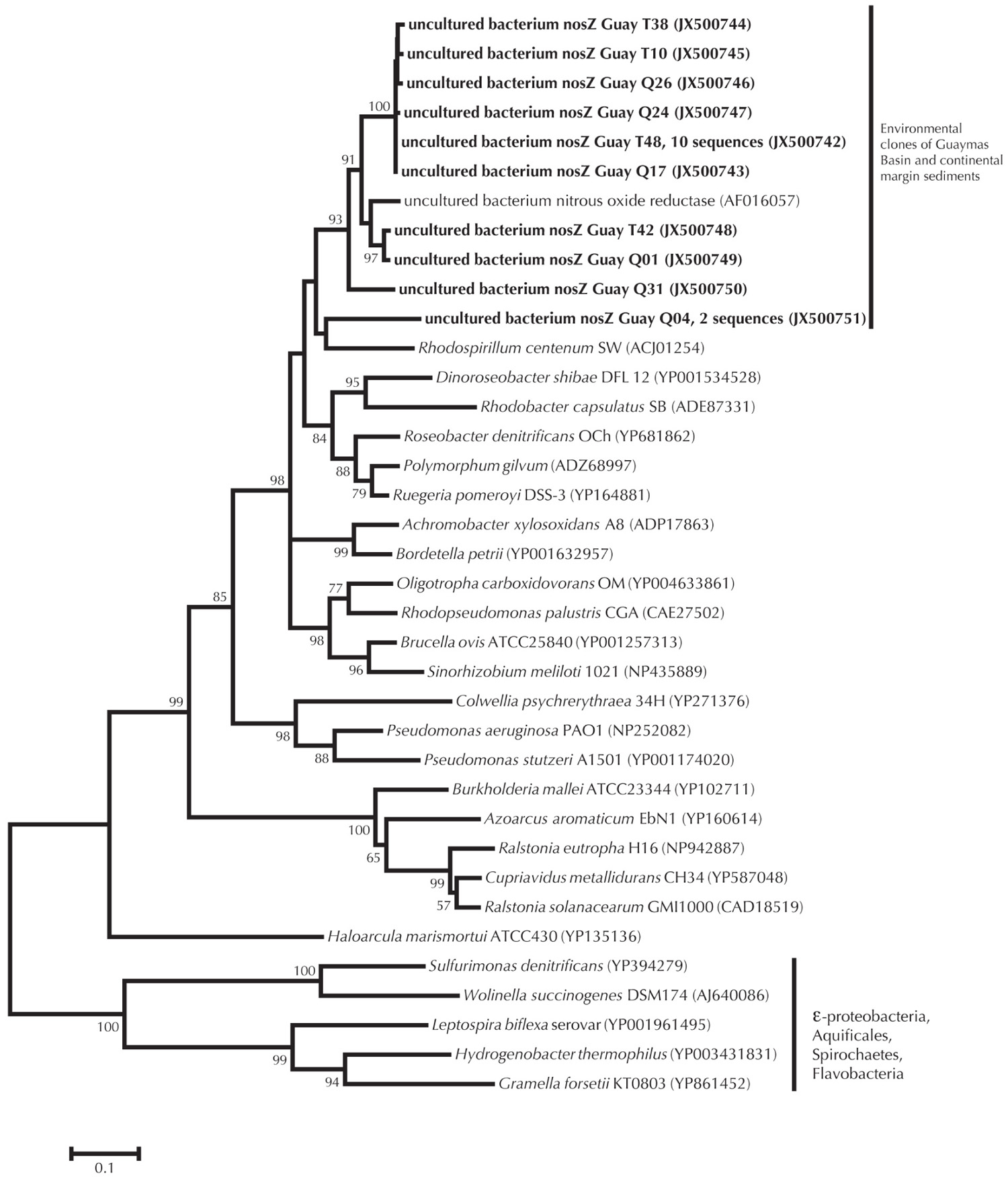
FIGURE 4. nosZ phylogeny of Guaymas Basin environmental clones relative to environmental clones from continental margin sediments and pure culture data. Minimum evolution method was used to generate the phylogeny, with a Poisson correction of evolutionary distance. Bootstrap values for branches occurring for >50% of 1000 iterations are reported. Scaling of the phylogenetic tree is based on an evolutionary distance of 0.10.
Discussion
Bacterial Mats as Nitrogen Cycle Hot Spots
Bacterial mats are considered areas of intensified nitrogen cycling (Teske and Nelson, 2006; Bourbonnais et al., 2012). Significant accumulations of thick (~3 cm) bacterial mats (Beggiatoa sp.) thrive at the surface of Guaymas Basin hydrothermal sediments and these mats concentrate up to mM levels of nitrate inside their vacuoles (Jannasch et al., 1989; McHatton et al., 1996; McKay et al., 2012). Given the high concentration of nitrate in mat-forming vacuolate sulfur bacteria and their abundance along surficial sediments in the Guaymas Basin, we postulated that nitrate cycling processes like nitrate reduction would be stimulated in mat-hosting sediments. Surprisingly, denitrification and nitrate removal rates were high in both Guaymas Basin mat-hosting sediments and brown sediments that lacked mats. Rates of potential denitrification were somewhat higher in sediments that hosted bacterial mats, versus the brown sediments adjacent to mats. However, nitrate removal rates were faster in brown sediments relative to mat sediments. Collectively these data suggest that while denitrifying communities might be more prominent in mat sediments, other nitrate removal processes are predominant in brown sediments. Other nitrate removal processes in brown sediments could include DNRA, ANAMMOX, and assimilation by endogenous prokaryotes. The lack of any nitrite or 29N2 accumulation, which would be the reactant and product of ANAMMOX, respectively, suggests that its occurrence is limited in these sediments, assuming of course that there was no stored nitrate or nitrite present in Beggiatoa during experiments. Furthermore, available evidence suggests that in sulfidic sediments, DNRA rather than ANAMMOX is the favored process (Burgin and Hamilton, 2007). Nitrogen assimilation can be performed by numerous heterotrophic and autotrophic organisms and might also contribute to nitrate removal (Allen et al., 2001). Therefore we postulate that DNRA and possibly assimilation are most likely the leading nitrate removal processes in brown sediments.
Few measurements of rates of nitrogen related processes exist in deep-sea sediments (Zopfi et al., 2001; Preisler et al., 2007; Høgslund et al., 2009; Bowles and Joye, 2011). In mat-hosting Gulf of Mexico cold seep sediments, potential denitrification rates were 32 µM N day−1; virtually the same rate as observed in Guaymas Basin mat sediments. Deeper (>6 cm) cold seep sediments not directly associated with mat material had a lower rate that was similar to brown sediments from Guaymas Basin. However, nitrate removal from Guaymas Basin sediments were an order magnitude faster than those measured in Gulf of Mexico cold seep sediments (Bowles and Joye, 2011). Integrated areal rates of potential denitrification (78 and 54 µmol m−2 h−1) in Guaymas Basin sediments were similar to rates measured in coastal and eutrophic environments (range: 3.5–1067 µmol m−2 h−1; Seitzinger, 1988). The areal integrated rate of nitrate removal in Guaymas Basin of 1045 µmol m−2 h−1 was very high and similar to the highest reported denitrification rate from sediments in the Tejo estuary (1067 µmol m−2 h−1; Seitzinger, 1988). In highly eutrophic Eckernförde Bay on the German Baltic coast, Preisler et al. (2007) performed a stable isotopic tracer rate (15) analyses and found Beggiatoa-associated nitrate removal was only about 6.5 µmol m−2 h−1, which amounts to a minor fraction of the rates observed in sediments of Guaymas Basin. Collectively these data suggest that in addition to the activity of Beggiatoa, other sediment microorganisms rigorously metabolize nitrate in organic carbon rich sediments.
Environmental Factors: Nitrate, Sulfide, and Doc
Nitrate, sulfide, and DOC concentrations influenced the rates and end products of nitrate reduction (Figure 2; Table 1). Nitrate utilizing communities in Guaymas Basin sediments reduced nitrate rapidly (high Vmax) to N2, with a high affinity for nitrate (relatively low Km), in comparison to other endogenous communities in other environments (Vmax = 422 µM N2-N cm−3 day−1, Km = 344 µM; Oren and Blackburn, 1979). Sulfide clearly influenced the production of dinitrogen; N2 production rates plummeted and may have resulted in the production of substantial amounts of N2O (Figure 2B). But, we were unfortunately unable to measure N2O in the sulfide treatments. Finally, microbial communities displayed a limited response to the availability of labile DOC, as observed in consistent potential DNF rates and N2O accumulation over a range of DOC concentrations.
Denitrification Kinetics and Nitrate Concentration Effects
Data from kinetic experiments on mixed environmental sediment populations performing denitrification are not common (e.g., Oren and Blackburn, 1979; Garcia-Ruiz et al., 1998). In the most relevant dataset from Kysing Fjord (Denmark) sediments, the community Km was 344 µM nitrate and Vmax was 422 µM N day−1 (Oren and Blackburn, 1979). The microorganisms within Guaymas Basin sediments maintained a higher affinity for nitrate and were able to perform denitrification at a higher maximum rate. This feature may be related to the presence of autotrophic, or sulfide-dependent denitrifiers and heterotrophic populations that utilize endogenous particulate organic carbon. For example, low Km values (3–161 µM) for nitrate in sulfidic bioreactors have been observed (Zeng and Zhang, 2005). In enrichments for sulfide oxidizing communities of denitrifiers, Shao et al. (2011) observed Vmax values up to 700 µM h−1. Molecular results also support our findings, as Guaymas sediments contained functional genes associated with heterotrophic denitrification (nosZ) as well as some Epsilonproteobacteria typically associated with sulfide dependent denitrification within 16S clone libraries.
Another environmental ramification of enhanced nitrate availability is the potential for a higher proportion of N2O production relative to N2 during denitrification (Tiedje, 1988). Increasing amounts of nitrous oxide production as a function of nitrate concentrations have been observed in environmental samples and in pure cultures (Sacks and Barker, 1952; Blackmer and Bremner, 1978; Joye, 2002). However Shao et al. (2011) recently observed no relationship between extremely high levels of nitrate (80 mM) and N2O production. We did not observe substantial increases in the proportion of N2O formed in Guaymas Basin sediments up to 5000 µM .
Sulfide and Denitrification
Nitrogen related processes are often subject to substantial influences from sulfides; nitrogen fixation (e.g., Marino et al., 2003), denitrification (e.g., Joye, 2002), and nitrification (Joye and Hollibaugh, 1995) are all influenced by sulfide concentration. Denitrification to N2 often slows substantially or ceases at relatively low sulfide concentrations, and sulfide might be a contributing factor to the observed accumulation of N2O in anoxic marine sediments (Sørensen et al., 1978; Joye, 2002). In Guaymas Basin sediments we observed low rates of potential denitrification in the presence of sulfide. The production of dinitrogen is limited by the sulfide-induced inhibition of nitrous oxide reductase (nosZ; Sørensen et al., 1980) or by limitation of denitrifiers by sulfide in general (Joye, 2002). Some pure culture data (Aeromonas sp., Vibrio sp., and Pseudomonas fluorescens) and environmental assays suggest that nitrous oxide reductase is already significantly inhibited at low sulfide concentrations of 100–300 µM (Sørensen et al., 1980; Senga et al., 2006). In contrast to relatively low inhibitory concentrations of sulfide observed in pure cultures, some environmental studies have reported evidence of nitrous oxide reductase inhibition (nitrous oxide accumulation) at much higher sulfide concentrations of 1–2.5 mM (Brunet and Garcia-Gil, 1996; Senga et al., 2006).
A paradoxical feature of denitrification in sulfidic sediments is that some microorganisms are able to use sulfide as an electron donor for denitrification. Perhaps it is the rigorous activity of sulfide utilizing denitrifying microorganisms that supports denitrification at high sulfide concentrations (2.5 mM) in some environments, such as Lake Shinji (Japan; Senga et al., 2006). The Guaymas sediments investigated here and previously investigated sediments from the same site hosted Epsilonproteobacteria (here ~38%; Teske et al., 2002; Dhillon et al., 2003) and cultured representatives of this group can likely use sulfide as an electron donor (Takai et al., 2003). Considering the high environmental concentrations of sulfide and nitrate, it is surprising that the endogenous denitrifier community in Guaymas Basin sediments was not more tolerant of sulfide.
Dissolved Organic Carbon Addition
The addition of labile DOC is thought to support high denitrification rates and might also support production of N2O (Tiedje, 1988). Though functional gene analysis of nosZ suggests the activity of heterotrophic denitrifying communities, we did not observe any stimulation of denitrification rates in response to increases in labile carbon concentration (DOC). These data suggest that heterotrophic metabolism to N2 was important, though other denitrification pathways (autotrophic) must also contribute substantially to the total observed rate. Another distinct possibility is that these communities are adapted to high DOC levels in the natural environment and the addition here was insufficient to stimulate potential denitrification rates. In soils, additions of glucose decreased N2O production (Weier et al., 1993). In Guaymas sediments, despite large additions of DOC, N2O accumulation did not systematically change.
Complex Nitrogen Cycling Communities
Molecular microbiological data are indicative of autotrophic and heterotrophic denitrification (Figures 3 and 4). Though not quantitative, the domination of clone libraries by Epsilonproteobacteria related to Sulfurovum and Sulfurimonas implies the presence and activity of an autotrophic denitrifying population in Guaymas sediments (Shao et al., 2010). The Epsilonproteobacteria clones outnumbered Deltaproteobacteria, which are largely sulfate reducers. Though elevated rates of sulfate reduction are often measured in this environment, the quantitative and qualitative data presented here (e.g., measured rates of denitrification and nitrate removal and molecular data) underscore the importance of nitrogen related processes (Elsgard et al., 1994; Figure 3) in this habitat as well.
The amino acid composition of nosZ within sulfide utilizing microorganisms is dissimilar to that of other organisms, corresponding to the outgroup in Figure 4. Traditional primers used for nosZ do not capture the Epsilonproteobacteria nosZ functional genes (see Sievert et al., 2003 and discussion therein). However utilizing the traditional nosZ primers (Scala and Kerkhof, 1999), we observed an outgroup of environmental samples similar to nosZ isolated from continental margin sediments (Scala and Kerkhof, 1999; Figure 4). The relationship of Guaymas nosZ clones with continental margins could be because the primers developed by Scala and Kerkhof (1999) target specific types of denitrifiers or because the recovered sequences reflect microorganisms that are general heterotrophic denitrifiers.
Nitrate Removal
An observation similar between Guaymas and cold seep sediments (Bowles and Joye, 2011) is that a great fraction of added is not recovered or accounted for. Though termed nitrate removal we speculate that this component is largely composed of DNRA and assimilation. Detecting both processes can be difficult and made more complex by the fact that DNRA and assimilation can be related. Ammonium generated by DNRA can be directly assimilated into biomass representing an interconnected series of pathways, from nitrate to biomass. Measuring DNRA in Guaymas Basin sediments was complicated by high background levels of ammonium (100s µM to >10 mM; Simoneit et al., 1992). Though sediments were rinsed prior to the experiments, ammonium levels during these incubations were still 100s of µM owing to high initial concentrations and potentially ammonium absorption to sediment particles (data not shown). In high activity sediments such as Guaymas Basin sediments we speculate that assimilation alone could also play a large role in nitrate removal. Carbon assimilation has been directly measured in Guaymas Basin sediments and these rates are substantial at about 4000 µM C day−1 (Joye and Samarkin, unpublished results). If we assume stoichiometric uptake of nitrogen by prokaryotes (C:N ~1:0.24; Whitman et al., 1998), then the demand for nitrogen comes to approximately 960 µM N day−1. This estimate from previous measurements of C assimilation shows that nitrate assimilation is the correct order of magnitude to account for all of the nitrate that was not recovered. In addition, this estimate would only include nitrogen incorporation from autotrophic microorganisms, and has to be regarded as conservative since it does not include heterotrophic nitrogen assimilation processes in carbon rich environments (Allen et al., 2001).
Conclusion
Denitrification is carried out by the endogenous prokaryotic communities in Guaymas Basin sediments. Denitrification rates are high in comparison to those observed in other environments and rates and endpoints are influenced by environmental factors, most notably sulfide. In molecular surveys of denitrifying bacteria from Guaymas Basin the potential for both heterotrophic and autotrophic, sulfide based, denitrification was observed. Future studies should further document the presence of nitrate in these environments, contained within Beggiatoa and free nitrate, in order to understand the importance of denitrification in this type of environment, relative to other processes. Additionally, quantification of heterotrophic and autotrophic gene transcripts should be performed to better constrain carbon and sulfur interrelations in these complex settings.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential con-flict of interest.
Acknowledgments
We thank the R/V Atlantis and HOV Alvin crews for exemplary work and unflagging support during our dives in Guaymas Basin, Jennifer Biddle, Vladimir Samarkin, and all the participants of the Guaymas Basin cruise science parties for collecting and providing samples for these analyses. We also thank the two reviewers whose suggestions greatly improved this manuscript. This research was supported by grants from the U.S. National Science Foundation (OCE 0959337 to Samantha B. Joye and OCE 0647633 to Andreas P. Teske).
References
Allen, A. E., Booth, M. G., Frischer, M. E., Verity, P. G., Zehr, J. P., and Zani, S. (2001). Diversity and detection of nitrate assimilation genes in marine Bacteria. Appl. Environ. Microbiol. 67, 5343–5348.
Biddle, J. F., Cardman, Z., Mendlovitz, H., Albert, D., Lloyd, K. G., Boetius, A., et al. (2012). Anaerobic oxidation of methane at different temperature regimes in Guaymas Basin hydrothermal sediments. ISME J. 6, 1018–1031.
Blackmer, M., and Bremner, J. M. (1978). Inhibitory effect of nitrate on reduction of N2O to N2 by soil microorganisms. Soil Biol. Biochem. 10, 187–191.
Bourbonnais, A., Lehmann, D. A., and Juniper, S. K. (2012). Subseafloor nitrogen transformations in diffuse hydrothermal vent fluids of the Juan de Fuca Ridge evidenced by the isotopic composition of nitrate and ammonium. Geochem. Geophys. Geosysts. 13, Q02T01. doi: 10.1029/2011GC003863 [Epub ahead of print].
Bowles, M. W., and Joye, S. B. (2011). High rates of denitrification and nitrate removal in cold seep sediments. ISME J. 5, 565–567.
Brunet, R. C., and Garcia-Gil, L. J. (1996). Sulfide-induced dissimilatory nitrate reduction to ammonia in anaerobic freshwater sediments. FEMS Microbiol. Ecol. 21, 131–138.
Burgin, A. J., and Hamilton, S. K. (2007). Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front. Ecol. Environ. 5, 89–96.
Dhillon, A., Lever, M., Lloyd, K. G., Albert, D. B., Sogin, M. L., and Teske, A. (2005). Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A (mcrA) genes in hydrothermal sediments of the Guaymas Basin. Appl. Environ. Microbiol. 71, 4592–4601.
Dhillon, A., Teske, A., Dillon, J., Stahl, D. A., and Sogin, M. L. (2003). Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 69, 2765–2772.
Elsgard, L., Isaksen, M. I., Jørgensen, B. B., Alayse, A.-M., and Jannasch, H. W. (1994). Microbial sulfate reduction in deep-sea sediments at the Guaymas basin hydrothermal vent area: influence of temperature and substrates. Geochim. Cosmochim. Acta 58, 3335–3343.
Garcia-Ruiz, R., Pattinson, S. N., and Whitton, B. A. (1998). Kinetic parameters of denitrification in a River Continuum. Appl. Environ. Microbiol. 64, 2533–2538.
Høgslund, S., Revsbech, N. P., Kuenen, G., Jørgensen, B. B., Gallardo, V. A., Vossenberg, J. V. D., et al. (2009). Physiology and behaviour of marine Thioploca. ISME J. 3, 647–657.
Holler, T., Widdel, F., Knittel, K., Amann, R., Kellermann, M. Y., Hinrichs, K.-U., et al. (2011). Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J. 5,1946–1956.
Huber, T., Faulkner, G., and Hugenholtz, P. (2004). Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20, 2317–2319.
Jannasch, H. W., Nelson, D. C., and Wirsen, C. O. (1989). Massive natural occurrence of unusually large bacteria (Beggiatoa sp.) at a hydrothermal deep-sea vent site. Nature 342, 834–836.
Joye, S. B. (2002). “Denitrification in the marine environment,” in Encyclopedia of Environmental Microbiology, ed. G. Collins (New York: John Wiley & Sons, Inc.), 1010–1019.
Joye, S. B., Boetius, A., Orcutt, B. N., Montoya, J. P., Schulz, H. N., Erickson, M. J., et al. (2004). The anaerobic oxidation of methane and sulfate reduction in sediments from Gulf of Mexico cold seeps. Chem. Geol. 205, 219–238.
Joye, S. B., and Hollibaugh, J. T. (1995). Influence of sulfide inhibition of nitrification on nitrogen regeneration in sediments. Science 270, 623–625.
Kallmeyer, J., and Boetius, A. (2004). Effects of temperature and pressure on sulfate reduction and anaerobic oxidation of methane in hydrothermal sediments of Guaymas Basin. Appl. Environ. Microbiol. 70, 1231–1233.
Kana, T. M., Sullivan, M. B., Cornwell, J. C., and Groszkowski, K. M. (1998). Denitrification in estuarine sediments determined by membrane inlet mass spectrometry. Limnol. Oceanogr. 43, 334–339.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948.
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, A., et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371.
Marino, R., Howarth, R. W., Chan, F., Cole, J. J., and Likens, G. E. (2003). Sulfide inhibition of molybdenum-dependent nitrogen fixation by planktonic cyanobacteria under seawater conditions: a non-reversible effect. Aquat. Biodivers. 171, 277–293.
McHatton, S. C., Barry, J. P., Jannasch, H. W., and Nelson, D. C. (1996). High nitrate concentrations in vacuolate autotrophic marine Beggiatoa spp. Appl. Environ. Microbiol. 62, 954–958.
McKay, L. J., MacGregor, B. J., Biddle, J. F., Mendlovitz, H. P., Hoer, D., Lipp, J. S., et al. (2012). Spatial heterogeneity and underlying geochemistry of phylogenetically diverse orange and white Beggiatoa mats in Guaymas Basin hydrothermal sediments. Deep Sea Res. 67, 21–31.
Orcutt, B., Boetius, A., Elvert, M., Samarkin, V., and Joye, S. B. (2005). Molecular biogeochemistry of sulfate reduction, methanogenesis and the anaerobic oxidation of methane at Gulf of Mexico cold seeps. Geochim. Cosmochim. Acta 69, 4267–4281.
Oren, A., and Blackburn, T. H. (1979). Estimation of sediment denitrification rates at in situ nitrate concentration. Appl. Environ. Microbiol. 37, 174–176.
Orphan, V. J., Hinrichs, K. U., Ussler, W., Paull, C. K., Taylor, L. T., Sylva, S. P., et al. (2001a). Comparative analysis of methane-oxidizing Archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67, 1922–1934.
Orphan, V. J., House C. H., Hinrichs K.-U., McKeegan K. D., and DeLong E. F. (2001b). Methane-consuming Archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293, 484–487.
Otte, S., Kuenen, J. G., Nielsen, L. P., Paerl, H. P., Zopfi, J., Schulz, H. N., et al. (1999). Nitrogen, carbon, and sulfur metabolism in natural Thioploca samples. Appl. Environ. Microbiol. 65, 3148–3157.
Porubsky, W. P, Weston, N. B., and Joye, S. B. (2009). Interactions between benthic primary production, denitrification and dissimilatory nitrate reduction to ammonium in intertidal sediments. Estuar. Coast. Shelf Sci. 83, 392–402.
Preisler, A., de Beer, D., Lichschlag, A., Lavik, G., Boetius, A., and Jørgensen, B. B. (2007). Biological and chemical sulfide oxidation in a Beggiatoa inhabited marine sediment. ISME J. 1, 341–353.
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196.
Sacks, L. E., and Barker, H. A. (1952). Substrate oxidation and nitrous oxide utilization in denitrification. J. Bacteriol. 64, 247–252.
Scala, D. J., and Kerkhof, L. J. (1999). Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl. Environ. Microbiol. 65, 1681–1687.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541.
Seitzinger, S. P. (1988). Denitrification in freshwater and coastal marine ecosystems: ecological and geochemical significance. Limnol. Oceanogr. 33, 702–724.
Senga, Y., Mochida, K., Fukumori, R., Okamoto, N., and Seiki, Y. (2006). N2O accumulation in estuarine and coastal sediments: the influence of H2S on dissimilatory nitrate reduction. Estuar. Coast. Shelf Sci. 67, 231–238.
Shao, M.-F., Zhang, T., and Fang, H. (2010). Sulfur-driven autotrophic denitrification: diversity, biochemistry, and engineering applications. Appl. Microbiol. Biotechnol. 88, 1027–1042.
Shao, M.-F., Zhang, T., Fang, H., and Li, X. (2011). The effect of nitrate concentration on sulfide-driven autotrophic denitrification in marine sediment. Chemosphere 83, 1–6.
Sievert, S. M., Scott, K. M., Klotz, M. G., Chain, P. S. G., Hauser, L. J., Hemp, J., et al. (2003). Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl. Environ. Microbiol. 74, 1145–1156.
Simoneit, B. R. T., Leif, R. N., Sturz, A. A., Sturdivant, A. E., and Gieskes, J. M. (1992). Geochemistry of shallow sediments in Guaymas Basin, Gulf of California: hydrothermal gas and oil migration and effects of mineralogy. Org. Geochem. 18, 765–784.
Sørensen, J. (1978). Occurence of nitric and nitrous oxides in a coastal marine sediment. Appl. Environ. Microbiol. 36, 809–813.
Sørensen, J., Tiedje, J. M., and Firestone, R. B. (1980). Inhibition by sulfide of nitric and nitrous oxide reduction by denitrifying Pseudomonas fluorescens. Appl. Environ. Microbiol. 39, 105–108.
Sweerts, J. P. R. A., De Beer, D., Nielsen, L. P., Verdouw, H., Van den Heuvel, J. C., Cohen, Y., et al. (1990). Denitrification by sulfur oxidizing Beggiatoa spp. mats on freshwater sediments. Nature 344, 762–763.
Takai, K., Inagaki, F., Nakagawa, S., Hirayama, H., Nunoura, T., Sako, Y., et al. (2003). Isolation and phyloge-netic diversity of members of previously uncultivated ε-Proteobacteria in deep-sea hydrothermal vents. FEMS Microbiol. Lett. 218, 167–174.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Teske, A., Hinrichs, K.-U., Edgcomb, V., Gomez, A. V., Kysela, D., Sylva, S. P., et al. (2002). Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68, 1994–2007.
Teske, A., and Nelson, D. C. (2006). The Genera Beggiatoa and Thioploca Prokaryotes, Vol. 3. New York: Springer, 784–810.
Tiedje, J. M. (1988). “Ecology of denitrification and dissimilatory nitrate reduction to ammonium,” in Biology of Anaerobic Microorganisms, ed. A. J. B. Zehnder (New York: John Wiley), 197–244.
Weber, A., and Jørgensen, B. B. (2002). Bacterial sulfate reduction in hydrothermal sediments of the Guaymas Basin, Gulf of California, Mexico. Deep Sea Res. 49, 827–841.
Weier, K. L., Macrae, I. C., and Myers, R. J. K. (1993). Denitrification in a clay soil under pasture and annual crop: estimation of potential losses using intact soil cores. Soil Biol. Biochem. 25, 991–997.
Whitman, W. B., Coleman, D. C., and Wiebe, W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583.
Widdel, F., and Rabus, R. (2001). Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12, 259–276.
Zeng, H., and Zhang, C. (2005). Evaluation of kinetic parameters of a sulfur-limestone autotrophic denitrification biofilm process. Water Res. 39, 4941–4952.
Keywords: denitrification, nitrogen cycle, Beggiatoa
Citation: Bowles MW, Nigro LM, Teske AP and Joye SB (2012) Denitrification and environmental factors influencing nitrate removal in Guaymas Basin hydrothermally altered sediments. Front. Microbio. 3:377. doi: 10.3389/fmicb.2012.00377
Received: 08 July 2012; Paper pending published: 03 August 2012;
Accepted: 03 October 2012; Published online: 25 October 2012.
Edited by:
Bess B. Ward, Princeton University, USAReviewed by:
Xiao-Hua Zhang, Ocean University of China, ChinaAlyson E Santoro, University of Maryland Center for Environmental Science, USA
Copyright: © 2012 Bowles, Nigro, Teske and Joye. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Samantha B. Joye, Aquatic Microbial Biogeochemistry Laboratory, Department of Marine Sciences, University of Georgia, Room 220, Marine Sciences Building, Athens, GA 30602-3636, USA. e-mail: mjoye@uga.edu
†Present address: Marshall W. Bowles, MARUM Center for Marine Environmental Sciences, University of Bremen, Bremen, Germany.
