- 1Department of Biology, Faculty of Sciences, University of Porto, Porto, Portugal
- 2CIMAR/CIIMAR – Interdisciplinary Centre for Marine and Environmental Research, University of Porto, Porto, Portugal
Planctomycetes have been known since the description of Planctomyces bekefii by Gimesi at the beginning of the twentieth century (1924), although the first axenic cultures were only obtained in the 1970s. Since then, 11 genera with 14 species have been validly named and five candidatus genera belonging to the anaerobic ammonium oxidation, anammox bacteria have also been discovered. However, Planctomycetes diversity is much broader than these numbers indicate, as shown by environmental molecular studies. In recent years, the authors have attempted to isolate and cultivate additional strains of Planctomycetes. This paper provides a summary of the isolation work that was carried out to obtain in pure culture Planctomycetes from several environmental sources. The following strains of planctomycetes have been successfully isolated: two freshwater strains from the sediments of an aquarium, which were described as a new genus and species, Aquisphaera giovannonii; several Rhodopirellula strains from the sediments of a water treatment recycling tank of a marine fish farm; and more than 140 planctomycetes from the biofilm community of macroalgae. This collection comprises several novel taxa that are being characterized and described. Improvements in the isolation methodology were made in order to optimize and enlarge the number of Planctomycetes isolated from the macroalgae. The existence of an intimate and an important relationship between planctomycetes and macroalgae reported before by molecular studies is therefore supported by culture-dependent methods.
The Planctomycetes
Planctomycetes are a fascinating group of Bacteria, due to their unique and peculiar characteristics. Their distinct cell wall without the characteristic peptidoglycan, their nucleoid, their cell structure with compartmentalization, their reproduction, and their metabolism and ecological ubiquity make them an exciting group of organisms to study. The first planctomycete observed was morphologically similar to a planktonic fungus, Planctomyces bekefii (Gimesi, 1924), giving the name to this phylum. Curiously, it was never isolated in pure culture. The first report of the isolation of a planctomycete in axenic cultures is due to the work of James T. Staley (Staley, 1973). This organism was renamed taxonomically several times and was finally designated Pirellula staleyi (Schlesner and Hirsch, 1987). In 1986, a new order, Planctomycetales, and family, Planctomycetaceae, was proposed to accommodate the several members of this group that had been described, based on 16S rRNA gene sequence analysis and distinctive morphological characteristics (Schlesner and Stackebrandt, 1986). The phylum Planctomycetes was only proposed in 2001 (Garrity and Holt, 2001) and in 2006, a superphylum was designated to incorporate the phyla Planctomycetes, Verrucomicrobia, and Chlamydiae (superphylum PVC; Wagner and Horn, 2006). At present, the Planctomycetes, comprise 3 orders (Planctomycetales, Phycisphaerales, and “Candidatus Brocadiales”), 11 described genera and 14 species, and 5 candidatus genera with 14 candidatus species (Fukunaga et al., 2009; Jetten et al., 2010; Ward, 2010; Bondoso et al., 2011; Kulichevskaya et al., 2012). Although still small, these numbers are increasing. In the last 4 years, one new order, five new genera, and six species have been described. The cultured strains (see Schlesner, 1994; Wang et al., 2002; Pimentel-Elardo et al., 2003; Gade et al., 2004; Elshahed et al., 2007) are not at all representative of the great diversity and ubiquity that has been revealed by molecular microbial ecology techniques (Kirkpatrick et al., 2006; Penton et al., 2006; Schmid et al., 2007; Woebken et al., 2007; Lachnit et al., 2011; Pizzetti et al., 2011a; Pollet et al., 2011; Fuchsman et al., 2012; Ivanova and Dedysh, 2012). In fact, of the 11045 clone sequences belonging to Planctomycetes in Ribosomal Database Project (RDP; Cole et al., 2009) only 291 (2%) have been isolated in pure culture.
The remarkable characteristics of this group, together with the need to isolate new strains in pure cultures to extend our knowledge of their physiological role in microbial communities, prompted the authors to investigate Planctomycetes. During isolation experiments, methodological improvements were achieved and different environmental sources were attempted. In initial work, isolation of a single strain of Planctomycetes was achieved from the sediments of an ornamental freshwater aquarium. The bacteria obtained represent a novel genus within the Planctomycetales and was taxonomically described as Aquisphaera giovannonii (Bondoso et al., 2011). Subsequent work was focused on the marine environment and isolates were successfully obtained from the epiphytic community of macroalgae and from the sediment of a treatment water recycling tank of a marine fish farm. This work allowed the isolation of a collection of more than 150 Planctomycetes that represents new taxa that are being, or have already been, characterized.
Improvements in Isolation Methodologies
Knowledge of bacterial diversity and physiology was long based only on cultured organisms. With the golden era of the molecular revolution, the abundance, diversity, and ecology of microorganisms gained another dimension. This was especially relevant in marine aquatic environments, as was the case for the discovery of the oligotrophic ubiquitous bacterioplankton SAR11 cluster (Giovannoni et al., 1990). However, knowledge on certain aspects of the biology of microorganisms cannot be reached unless the organisms are available in culture. As an example, the cultivation of Pelagibacter ubique, a member of the SAR11 clade, by Rappé et al.(2002) enabled many unanswered questions to be addressed (Joint, 2008), such as the need of exogenous reduced sulfur compounds for a strain without the genes for assimilatory sulfate reduction.
For a longtime, only a few cultivable Planctomycetes were available and knowledge of this group was scarce. But several cultivation methods and media formulations for the isolation of planctomycetes were achieved in the last few decades (Schlesner, 1994; Zengler et al., 2002; Winkelmann and Harder, 2009). These advances were especially due to Dr. Heinz Schlesner’s work (Schlesner, 1994), leading to the isolation of a great number of isolates. More recently, several attempts have been made to bring “Candidatus Brocadiales” into culture. However, their isolation in pure culture has not succeeded. One main difficulty relies on their slow growth rate, with a doubling time of 10 days. After great technological accomplishments, they are, presently grown in membrane bioreactors with a purity of enrichment of 97.6% (van der Star et al., 2008; Kartal et al., 2010).
Planctomycetes are comparatively slow growing organisms with low demand for carbon and nitrogen sources. This makes them difficult to isolate in common media because they are easily outgrown by bacteria with faster growth rates. The utilization of media that have a relatively low content in yeast extract and peptone (usually less than 0.5%) and with the addition of glucose as a carbon source is useful for the isolation of these organisms. The addition of vitamin B12 to the isolation media, required by some members of the Planctomycetes, and micro- and macronutrients, also produced favorable results. The selective isolation of Planctomycetes is very much based on the capacity of these bacteria to grow in the presence of β-lactam antibiotics that affect peptidoglycan biosynthesis in dividing cells of the majority of bacteria (Schlesner, 1994). Some Planctomycetes are also resistant to the antibiotic streptomycin. The inhibition of bacterial growth by the action of these two antibiotics gives the relatively slow growing Planctomycetes the possibility to form colonies on the isolation plates. Another isolation strategy is to provide N-acetylglucosamine (NAG) as the only carbon and nitrogen source (Schlesner, 1994). Chitin composed of NAG monomers is the second most abundant organic compound in nature, and is present in fungi and several animals, namely copepods, that produce billions of tons of this compound annually (Yu et al., 1991). Ultimately, NAG ends up covering the ocean floor, where it is metabolized by bacteria. Planctomycetes are commonly present in marine sediments (Rusch et al., 2003; Musat et al., 2006; Chipman et al., 2010; Hu et al., 2010) where the availability of NAG favors the selection of the metabolic pathways for its degradation.
Besides the overgrowth of rapid growing bacteria, another problem, commonly faced when isolating bacteria from environmental samples, is the rapid and invasive growth of fungi. To inhibit fungal growth, cycloheximide or amphotericin B, are commonly added to the growth media (Schlesner, 1994; Wang et al., 2002; Winkelmann and Harder, 2009). However, these antifungal compounds have not always proven to be effective and fungicides like pevaryl (econazole nitrate; 1%) and benlate (benomyl, or methyl l-(butylcarbamoyl)-2-benzimidazolecarbamate; 4 mg∙mL–1) appear to be more adequate in inhibiting fungal growth (Lage and Bondoso, 2011). Another improvement for planctomycetes isolation from the surface of portions of macroalgae was to pre-wash them in a mixture of pevaryl and benlate before their introduction in the culture medium. With this, fungal overgrowth can be notably reduced. For the isolation of epiphytic planctomycetes from macroalgae diverse inocula, as the macerated macroalgae, the resuspended biofilm obtained from scrapping the surface of the macroalgae and the direct use of portions of the macroalgae on the isolation media were tested. This has enabled the isolation of a larger number of strains. These improvements, associated with the use of antibiotics (200 mg∙mL–1 ampicillin and 1000 mg∙mL–1 streptomycin), allowed the authors to obtain a large collection of culturable Planctomycetes, essentially from the surface of macroalgae (Lage and Bondoso, 2011).
Diversity of Rhodopirellula Isolated from Marine Environments
The majority of Planctomycetes isolates recovered belong to the genus Rhodopirellula. These organisms are widespread and have been described in many marine environments, including brackish and marine water from the Kiel Fjord on the Baltic Sea (Schlesner et al., 2004), meso-eutrophic lakes (Pizzetti et al., 2011b), other European seas (Winkelmann and Harder, 2009; Pizzetti et al., 2011a), marine snow (DeLong et al., 1993; Vergin et al., 1998), diatom blooms in Oregon coastal waters (Morris et al., 2006), tissues of the Mediterranean sponge Aplysina aerophoba (Pimentel-Elardo et al., 2003; Gade et al., 2004), and the giant tiger prawn, Penaeus monodon (Fuerst et al., 1991) as well as the biofilm of Laminaria hyperborea (Bengtsson and Ovreas, 2010). However, the only taxonomically described organism is Pirellula sp. strain 1 (presently Rhodopirellula baltica SH1T), isolated from the water column of the Kiel Fjord as a free-living bacterium (Schlesner, 1994; Schlesner et al., 2004). The great heterogeneity within the genus Pirellula led Schlesner et al. (2004) to emend this genus and create two new genera, Rhodopirellula and Blastopirellula. The 16S rRNA gene sequence analysis, DNA–DNA hybridization and MLSA analysis (Schlesner et al., 2004; Winkelmann et al., 2010) revealed that R. baltica is a cosmopolitan species with a great genetic diversity.
The isolation experiments, both from the biofilm community of macroalgae (Lage and Bondoso, 2011) and the marine fish farm environments (Lage et al., 2012), revealed that the mono-speciated Rhodopirellula genus was dominant among the isolates and was clearly diverse (Figure 1). The biofilm community of the macroalgae sampled provided a total of 138 Planctomycetes isolates. The genus Rhodopirellula represented 92% of the total isolates, of which 71% are strains (more than 99.5% 16S rRNA gene similarity) within the species R. baltica (group A). The remaining 29% are subdivided in two groups (groups B and C), with about 97–98% of similarity in the 16S rRNA gene to the type species. Rhodopirellula sp. was also present in the sediment of a water treatment recycling tank of a marine fish farm, from which eleven isolates were obtained representing two main phylogenetic clusters as defined by the 16S rRNA gene sequence similarity. Six of the isolates are intimately related to Rhodopirellula baltica (in group A) with more than a 99% 16S rRNA gene sequence similarity and the other five isolates, with a 97.7–97.9% 16S rRNA gene sequence similarity to this species, belong to group B. In total, 149 isolates within the genus Rhodopirellula from the two marine environments of the north Atlantic coast of Portugal were obtained in pure culture. These results confirmed the presence of this genus in the biofilm community of ten of the three major macroalgae groups (Gelidium pulchellum, Chondrus crispus, Corallina sp., Gracilaria bursa-pastoris, Grateloupia turuturu, Fucus spiralis, Mastocarpus stellatus, Porphyra dioica, Sargassum muticum, and Ulva sp.) and in marine fish farm environments. The Rhodopirellula sp. groups B and C are less numerous than R. baltica. Group B was isolated from the sediment of a treatment water recycling tank of a marine fish farm, from five macroalgae (G. bursa-pastoris, F. spiralis, Laminaria sp., M. stellatus, and Ulva sp.), from Sweden (from algae surface), the Baltic Sea and sediments from Italy and Mallorca (Winkelmann and Harder, 2009). Group C was mainly isolated from macroalgae (C. crispus, Corallina sp., G. bursa-pastoris, F. spiralis, M. stellatus, S. muticum, and Ulva sp.) sampled on the Carreço rocky beach in the north cost of Portugal and two more isolates from sediments in Brest, France (Winkelmann and Harder, 2009). So far, culture-independent methods did not detect the presence of this group in other habitats or sources.
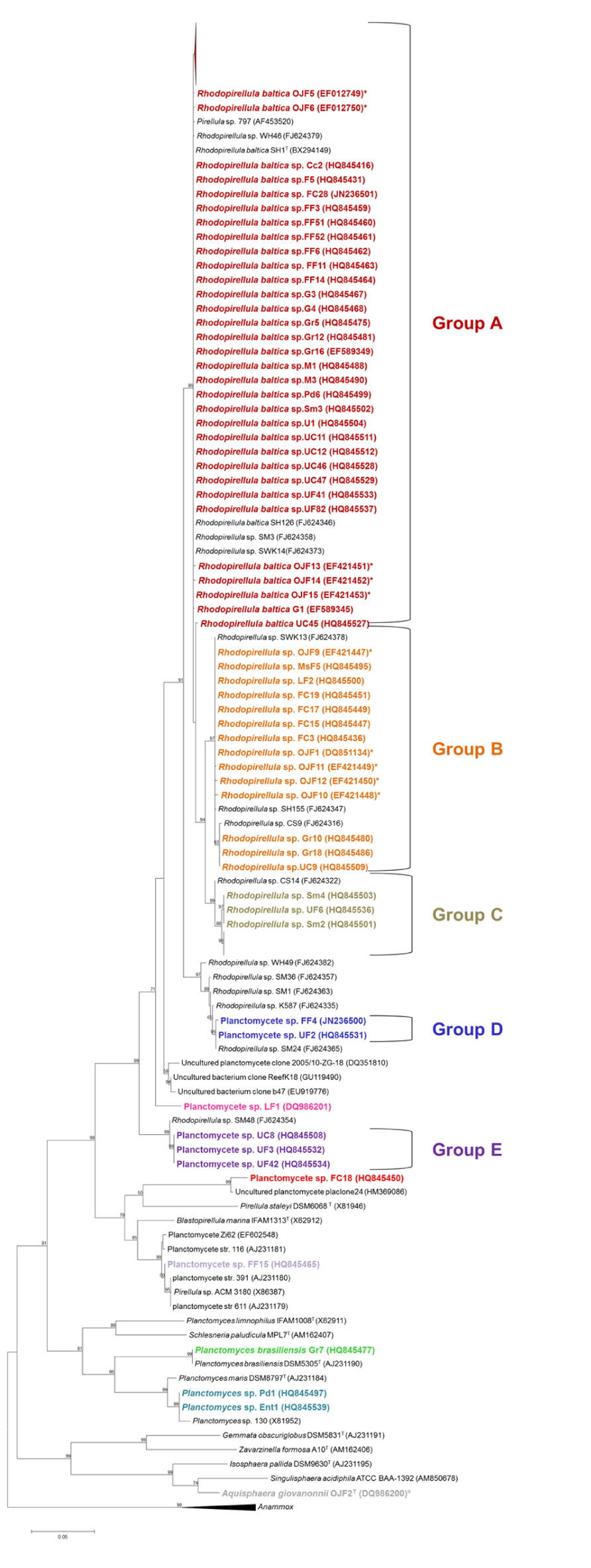
FIGURE 1. Phylogenetic 16S rRNA gene tree generated by a maximum-likelihood analysis based on the Jukes–Cantor model, indicating the relationship of the isolates reported in this article (shown in bold) to members of the Planctomycetes. GenBank accession numbers are shown in parentheses. The Anammox genera were used as an outgroup. The numbers shown beside nodes are the percentages for bootstrap analyses; only values above 50% are shown. *Strains isolated from the fish farm sediments; ¤Novel genera isolated from an ornamental freshwater aquarium. Scale bar = 0.05 substitutions per 100 nucleotides.
Presently, the RDP (Cole et al., 2009) accounts for 593 sequences classified in the genus Rhodopirellula, of which 416 are from uncultured organisms. Even though only one species was described in the genus Rhodopirellula, intrageneric diversity points to the existence of new species in this genus. An ERIC-PCR fingerprinting study performed with the strains isolated from the fish farm environment revealed a great intraspecies genetic variability. It was possible to differentiate 9 genotypes within the 11 isolates (Lage et al., 2012). This was also observed by Winkelmann et al. (2010) by multi-locus typing and Box-PCR and by Schlesner et al. (2004) by DNA-DNA hybridization.
Planctomycetes Diversity Hidden in Macroalgae Biofilms
Other taxa of Planctomycetes, in addition to Rhodopirellula, were isolated from the biofilm community of macroalgae (Figure 1). Three potential new genera (groups D, E and strain LF1) are phylogenetically more closely related to R. baltica (93–96% 16S rRNA gene sequence similarity), forming three clear separate clusters within the Planctomycetales. One isolate (strain FF15) is related to Blastopirellula marina (95% 16S rRNA gene sequence similarity) and isolate FC18 is phylogenetically more closely related to Pirellula staleyi (85.4% similarity in the 16S rRNA gene sequence) and shares an 87% 16S rRNA gene sequence similarity to B. marina and R. baltica. Both represent potential new genera isolated from Fucus spiralis. Strains Pd1 and UiF1 (isolated, respectively, from Porphyra dioica and Ulva intestinalis) sharea 98% 16S rRNA gene sequence similarity to P. maris. Curiously, one strain from G. bursa-pastoris, Gr7, which has 100% similarity to Planctomyces brasiliensis DSM5305T from Lagoa Vermelha, a salt pit environment from Brazil (Schlesner, 1989), was also isolated. Besides the three OTUs related to Rhodopirellula, the diversity of Planctomycetes associated with macroalgae represents more seven different OTUs based on a 98% cut-off (Stackebrandt and Ebers, 2006).
Macroalgae harbor a great diversity of Planctomycetes in their biofilm community. The relationship between Planctomycetes and macroalgae had been unveiled by Bengtsson and Ovreas (2010) and Bengtsson et al. (2010), who found that Planctomycetes account for 51–53% of the bacterial biofilm cells in July and September and 24% in February in the kelp Laminaria hyperborean from southwestern Norway. Clone libraries of the kelp revealed 23 OTUs at 98% sequence similarity, with the majority being species of Rhodopirellula and Blastopirellula. Lachnit et al. (2011) also found a temporal fluctuation of Planctomycetes in association with Fucus vesiculosus, Gracilaria vermiculophylla, and Ulva intestinalis. The denaturing gradient gel electrophoresis (DGGE) bands were related to Rhodopirellula, Planctomyces, and Blastopirellula. Planctomycetes were also found in Ulva australis and accounted for 3.4% of the total clones (Burke et al., 2011). A new order of Planctomycetes, Phycisphaerales, was proposed to accommodate a novel genus isolated from the surface of Porphyra sp., Phycisphaera (Fukunaga et al., 2009). These studies, together with our results (Lage and Bondoso, 2011), show that macroalgae provide a suitable eutrophic environment that supports the growth of heterotrophic Planctomycetes. Macroalgae produce organic sulfur compounds (Michel et al., 2006) and excrete photosynthetically derived dissolved exudates. Planctomycetes colonies recovered from the surface of small portions of macroalgae (Lage and Bondoso, 2011) proved that the growth of Planctomycetes can be supported by macroalgae compounds. Further support for the hypothesis of a nutritional role of macroalgae for Planctomycetes comes from growth experiments (Lage and Bondoso, 2011). Water-soluble extracts of Ulva sp. and F. spiralis enabled the growth of some Planctomycetes. However, in an enrichment study that used kelp constituent carbon sources to support the cultivation of bacterial populations associated with the kelp Laminaria hyperborean, Planctomycetes, and Verrucomicrobia, the two most frequently detected bacterial lineages, were not identified among the cultured bacteria (Bengtsson et al., 2011). Nevertheless, Planctomycetes are able to grow on several carbohydrates (mono and disaccharides and complex ones) as a carbon source and on the chitin monomer NAG (Schlesner, 1994; Rabus et al., 2002). R. baltica possesses several genes encoding for 110 sulfatases (Glöckner et al.,2003) and presumably could be involved in the recycling of carbon from complex sulfated heteropolysaccharides (Wecker et al., 2009). Macroalgae may benefit by hosting Planctomycetes biofilm communities through competitive exclusion of potentially undesirable microbes, the production of secondary metabolites and a role in avoiding desiccation, as Planctomycetes can possess extracellular matrices.
Conclusion
Planctomycetes are far from being a well-known group of Bacteria. Although their first observation goes back to the beginning of the last century, it is only since the 1970–1980s that more regular publications began to appear. In this mini review, culture aspects of this group have been highlighted, providing a summary of the work developed over recent years with the aim of isolating in pure culture Planctomycetes from several environmental sources. Methodological improvements through the use of a new combination of fungicides and antibiotics, the use of low organic media and portions of macroalgae that were essential for successful isolations have been pointed out. A large collection of Planctomycetes was, thus, obtained from the biofilm community of macroalgae, from the sediments of the water treatment tank of a fish farm, as well as from the sediments of a freshwater aquarium. As some of the isolates are novel taxa, their description is imperative. With these characterizations, knowledge on the morphology, metabolism, and ecology of this group will be enlarged. This phylogenetic diversity is the base for future ecological work namely on the comprehension of the interaction macroalgae–Planctomycetes and on the application of molecular approaches to the study of the biogeographic distribution of Planctomycetes in coastal environments. With this work the frontiers of diversity are being pushed forward, as indeed they should be.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Fundação para a Ciência e Tecnologia (FCT, PEst-C/MAR/LA0015/2011). Joana Bondoso was financed by FCT (PhD grant SFRH/BD/35933/2007). The authors are grateful to the Frontiers Foundation for supporting the publication of this paper and to Prof. Milton S. da Costa for the revision of the English.
References
Bengtsson, M. M., and Ovreas, L. (2010). Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol. 10:261. doi: 10.1186/1471-2180-10-261
Bengtsson, M. M., Sjøtun, K., and Øvreås, L. (2010). Seasonal dynamics of bacterial biofilms on the kelp Laminaria hyperborea. Aquat. Microb. Ecol. 60, 71–83.
Bengtsson, M. M., Sjøtun, K., Storesund, J. E., and øvreås, L. (2011). Utilization of kelp-derived carbon sources by kelp surface-associated bacteria. Aquat. Microb. Ecol. 62, 191–199.
Bondoso, J., Albuquerque, L., Nobre, M. F., Lobo-Da-Cunha, A., Da Costa, M. S., and Lage, O. M. (2011). Aquisphaera giovannonii gen. nov., sp. nov. A novel planctomycete isolated from a freshwater aquarium. Int. J. Syst. Evol. Microbiol. 61, 2844–2850.
Burke, C., Thomas, T., Lewis, M., Steinberg, P., and Kjelleberg, S. (2011). Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 5, 590–600.
Chipman, L., Podgorski, D., Green, S., Kostka, J., Cooper, W., and Huettel, M. (2010). Decomposition of plankton-derived dissolved organic matter in permeable coastal sediments. Limnol. Oceanogr. 55, 857–871.
Cole, J. R., Wang, Q., Cardenas, E., Fish, J., Chai, B., Farris, R. J., et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145.
DeLong, E. F., Franks, D. G., and Alldredge, L. (1993). Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38, 924–934.
Elshahed, M. S., Youssef, N. H., Luo, Q., Najar, F. Z., Roe, B. A., Sisk, T. M., et al. (2007). Phylogenetic and metabolic diversity of Planctomycetes from anaerobic, sulfide- and sulfur-rich Zodletone Spring, Oklahoma. Appl. Environ. Microbiol. 73, 4707–4716.
Fuchsman, C. A., Staley, J. T., Oakley, B. B., Kirkpatrick, J. B., and Murray, J. W. (2012). Free-living and aggregate-associated Planctomycetes in the Black Sea. FEMS Microbiol. Ecol. 80, 402–416.
Fuerst, J. A., Sambhi, S. K., Paynter, J. L., Hawkins, J. A., and Atherton, J. G. (1991). Isolation of a bacterium resembling Pirellula species from primary tissue culture of the giant tiger prawn (Penaeus monodon). Appl. Environ. Microbiol. 57, 3127–3134.
Fukunaga, Y., Kurahashi, M., Sakiyama, Y., Ohuchi, M., Yokota, A., and Harayama, S. (2009). Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J. Gen. Appl. Microbiol. 55, 267–275.
Gade, D., Schlesner, H., Glöckner, F. O., Amann, R., Pfeiffer, S., and Thomm, M. (2004). Identification of planctomycetes with order-, genus-, and strain-specific 16s rRNA-targeted probes. Microb. Ecol. 47, 243–251.
Garrity, G. M., and Holt, J. G. (2001). “The road map to the Manual,” in Bergey’s Manual of Systematic Bacteriology, 2nd Edn, eds D. R. Boone, R. W. Castenholz, and G. M. Garrity (New York: Springer), 119–166.
Gimesi, N. (1924). Hydrobiologiai Tanulmanyok (Hydrobiologische Studien). I: Planctomyces bekefii Gim. nov. gen. et sp. [in Hungarian, with German translation]. Budapest: Kiadja a Magyar Ciszterci Rend.
Giovannoni, S. J., Britschgi, T. B., Moyer, C. L., and Field, K. G. (1990). Genetic diversity in Sargasso Sea bacterioplankton. Nature 345, 60–63.
Glöckner, F. O., Kube, M., Bauer, M., Teeling, H., Lombardot, T., Ludwig, W., et al. (2003). Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. U.S.A. 100, 8298–8303.
Hu, Y. F., Fu, C. Z., Yin, Y. S., Cheng, G., Lei, F., Yang, X., et al. (2010). Construction and preliminary analysis of a deep-sea sediment metagenomic fosmid library from Qiongdongnan Basin, South China Sea. Mar. Biotechnol. 12, 719–727.
Ivanova, A. O., and Dedysh, S. N. (2012). Abundance, diversity, and depth distribution of planctomycetes in acidic northern wetlands. Front. Microbiol. 3:5. doi: 10.3389/fmicb.2012.00005
Jetten, M. S. M., Camp, H. J. M. O. D., Kuenen, J. G., and Strous, M. (2010). “Order II. “Candidatus Brocadiales” ord. nov.,” in The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes, 2nd Edn, eds N. R. Krieg, W. Ludwig, W. B. Whitman, B. P. Hedlund, B. J. Paster, J. T. Staley, N. Ward, and D. Brown (New York: Springer), 918–925.
Kartal, B., Kuenen, J. G., and Van Loosdrecht, M. C. M. (2010). Sewage treatment with anammox. Science 328, 702–703.
Kirkpatrick, J., Oakley, B., Fuchsman, C., Srinivasan, S., Staley, J. T., and Murray, J. W. (2006). Diversity and distribution of Planctomycetes and related bacteria in the suboxic zone of the Black Sea. Appl. Environ. Microbiol. 72, 3079–3083.
Kulichevskaya, I. S., Detkova, E. N., Bodelier, P. L., Rijpstra, W. I., Damste, J. S., and Dedysh, S. N. (2012). Singulisphaera rosea sp. nov., a planctomycete from acidic Sphagnum peat, and emended description of the genus Singulisphaera. Int. J. Syst. Evol. Microbiol. 62, 118–123.
Lachnit, T., Meske, D., Wahl, M., Harder, T., and Schmitz, R. (2011). Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13, 655–665.
Lage, O. M., and Bondoso, J. (2011). Planctomycetes diversity associated with macroalgae. FEMS Microbiol. Ecol. 78, 366–375.
Lage, O. M., Bondoso, J., and Viana, F. (2012). Isolation and characterisation of Planctomycetes from the sediments of a fish farm wastewater treatment tank. Arch. Microbiol. 194, 879–885.
Michel, G., Nyval-Collen, P., Barbeyron, T., Czjzek, M., and Helbert, W. (2006). Bioconversion of red seaweed galactans: a focus on bacterial agarases and carrageenases. Appl. Microbiol. Biotechnol. 71, 23–33.
Morris, R. M., Longnecker, K., and Giovannoni, S. J. (2006). Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ. Microbiol. 8, 1361–1370.
Musat, N., Werner, U., Knittel, K., Kolb, S., Dodenhof, T., Van Beusekom, J. E. E., et al. (2006). Microbial community structure of sandy intertidal sediments in the North Sea, Sylt-Rømø Basin, Wadden Sea. Syst. Appl. Microbiol. 29, 333–348.
Penton, C. R., Devol, A. H., and Tiedje, J. M. (2006). Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl. Environ. Microbiol. 72, 6829–6832.
Pimentel-Elardo, S., Wehrl, M., Friedrich, A. B., Jensen, P. R., and Hentschel, U. (2003). Isolation of planctomycetes from Aplysina sponges. Aquat. Microb. Ecol. 33, 239–245.
Pizzetti, I., Fuchs, B. M., Gerdts, G., Wichels, A., Wiltshire, K. H., and Amann, R. (2011a). Temporal variability of coastal Planctomycetes clades at Kabeltonne station, North Sea. Appl. Environ. Microbiol. 77, 5009–5017.
Pizzetti, I., Gobet, A., Fuchs, B. M., Amann, R., and Fazi, S. (2011b). Abundance and diversity of Planctomycetes in a Tyrrhenian coastal system of central Italy. Aquat. Microb. Ecol. 65, U129–U139.
Pollet, T., Tadonleke, R. D., and Humbert, J. F. (2011). Spatiotemporal changes in the structure and composition of a less-abundant bacterial phylum (Planctomycetes) in two perialpine lakes. Appl. Environ. Microbiol. 77, 4811–4821.
Rabus, R., Gade, D., Helbig, R., Bauer, M., Glöckner, F. O., Kube, M., et al. (2002). Analysis of N-acetylglucosamine metabolism in the marine bacterium Pirellula sp. strain 1 by a proteomic approach. Proteomics 2, 649–655.
Rappè, M. S., Connon, S. A., Vergin, K. L., and Giovannoni, S. J. (2002). Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418, 630–633.
Rusch, A., Huettel, M., Reimers, C. E., Taghon, G. L., and Fuller, C. M. (2003). Activity and distribution of bacterial populations in Middle Atlantic Bight shelf sands. FEMS Microbiol. Ecol. 44, 89–100.
Schlesner, H. (1989). Planctomyces brasiliensis sp. nov., a halotolerant bacterium from a salt pit. Syst. Appl. Microbiol. 12, 159–161.
Schlesner, H. (1994). The development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from various aquatic habitats using dilute media. Syst. Appl. Microbiol. 17, 135–145.
Schlesner, H., and Hirsch, P. (1987). Rejection of the genus name Pirella for pear-shaped budding bacteria and proposal to create the genus Pirellula gen. nov. Int. J. Syst. Evol. Microbiol. 37, 441.
Schlesner, H., Rensmann, C., Tindall, B. J., Gade, D., Rabus, R., Pfeiffer, S., et al. (2004). Taxonomic heterogeneity within the Planctomycetales as derived by DNA–DNA hybridization, description of Rhodopirellula baltica gen. nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula. Int. J. Syst. Evol. Microbiol. 54, 1567–1580.
Schlesner, H., and Stackebrandt, E. (1986). Assignment of the genera Planctomyces and Pirella to a new family Planctomycetaceae fam. nov. and description of the order Planctomycetales ord. nov. Syst. Appl. Microbiol. 8, 174–176.
Schmid, M. C., Risgaard-Petersen, N., Van De Vossenberg, J., Kuypers, M. M., Lavik, G., Petersen, J., et al. (2007). Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ. Microbiol. 9, 1476–1484.
Stackebrandt, E., and Ebers, J. (2006). Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33, 152–155.
Staley, J. T. (1973). Budding bacteria of the Pasteuria-Blastobacter group. Can. J. Microbiol. 19, 609–614.
van der Star, W. R. L., Miclea, A. I., Van Dongen, U. G. J. M., Muyzer, G., Picioreanu, C., and Van Loosdrecht, M. C. M. (2008). The membrane bioreactor: a novel tool to grow anammox bacteria as free cells. Biotechnol. Bioeng. 101, 286–294.
Vergin, K. L., Urbach, E., Stein, J. L., Delong, E. F., Lanoil, B. D., and Giovannoni, S. J. (1998). Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl. Environ. Microbiol. 64, 3075–3078.
Wagner, M., and Horn, M. (2006). The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 17, 241–249.
Wang, J., Jenkins, C., Webb, R. I., and Fuerst, J. A. (2002). Isolation of Gemmata-like and Isosphaera-like planctomycete bacteria from soil and freshwater. Appl. Environ. Microbiol. 68, 417–422.
Ward, N. L. (2010). “Family I. Planctomycetaceae Schlesner and Stackebrandt 1987, 179VP (Effective publication: Schlesner and Stackebrandt 1986, 175) emend. Ward (this volume),” in The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes, 2nd Edn, eds N. R. Krieg, W. Ludwig, W. B. Whitman, B. P. Hedlund, B. J. Paster, J. T. Staley, N. Ward, and D. Brown (New York: Springer), 879–925.
Wecker, P., Klockow, C., Ellrott, A., Quast, C., Langhammer, P., Harder, J., et al. (2009). Transcriptional response of the model planctomycete Rhodopirellula baltica SH1(T) to changing environmental conditions. BMC Genomics 10:410. doi: 10.1186/1471-2164-10-410
Winkelmann, N., and Harder, J. (2009). An improved isolation method for attached-living Planctomycetes of the genus Rhodopirellula. J. Microbiol. Methods 77, 276–284.
Winkelmann, N., Jaekel, U., Meyer, C., Serrano, W., Rachel, R., Rossello-Mora, R., et al. (2010). Determination of the diversity of Rhodopirellula isolates from European seas by multilocus sequence analysis. Appl. Environ. Microbiol. 76, 776–785.
Woebken, D., Teeling, H., Wecker, P., Dumitriu, A., Kostadinov, I., Delong, E. F., et al. (2007). Fosmids of novel marine Planctomycetes from the Namibian and Oregon coast upwelling systems and their cross-comparison with planctomycete genomes. ISME J. 1, 419–435.
Yu, C., Lee, A. M., Bassler, B. L., and Roseman, S. (1991). Chitin utilization by marine bacteria. A physiological function for bacterial adhesion to immobilized carbohydrates. J. Biol. Chem. 266, 24260–24267.
Keywords: Planctomycetes, isolation, Rhodopirellula, phylogeny, macroalgae biofilm, Aquisphaera giovannonii
Citation: Lage OM and Bondoso J (2012) Bringing Planctomycetes into pure culture. Front. Microbio. 3:405. doi:10.3389/fmicb.2012.00405
Received: 22 May 2012; Accepted: 11 November 2012;
Published online: 03 December 2012.
Edited by:
Naomi L. Ward, University of Wyoming, USAReviewed by:
Boran Kartal, Radboud University, NetherlandsCarl J. Yeoman, Montana State University, USA
Copyright: © 2012 Lage and Bondoso. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Olga M. Lage, Department of Biology, Faculty of Sciences, University of Porto, Rua do Campo Alegre, s/n, 4169-007 Porto, Portugal. e-mail: olga.lage@fc.up.pt
