- 1Department of Earth Sciences, University of Southern California, Los Angeles, CA, USA
- 2Department of Biological Sciences, University of Southern California, Los Angeles, CA, USA
- 3Department of Molecular Ecology, Max Planck Institute for Marine Microbiology, Bremen, Germany
- 4Geomicrobiology, Center for Applied Geosciences, University of Tübingen, Tübingen, Germany
- 5Department of Microbiology and Environmental Toxicology, University of California, Santa Cruz, Santa Cruz, CA, USA
- 6Department of Geochemistry and Hydrogeology, University of Bremen, Bremen, Germany
Phase separation is a ubiquitous process in seafloor hydrothermal vents, creating a large range of salinities. Toxic elements (e.g., arsenic) partition into the vapor phase, and thus can be enriched in both high and low salinity fluids. However, investigations of microbial diversity at sites associated with phase separation are rare. We evaluated prokaryotic diversity in arsenic-rich shallow-sea vents off Milos Island (Greece) by comparative analysis of 16S rRNA clone sequences from two vent sites with similar pH and temperature but marked differences in salinity. Clone sequences were also obtained for aioA-like functional genes (AFGs). Bacteria in the surface sediments (0–1.5 cm) at the high salinity site consisted of mainly Epsilonproteobacteria (Arcobacter sp.), which transitioned to almost exclusively Firmicutes (Bacillus sp.) at ~10 cm depth. However, the low salinity site consisted of Bacteroidetes (Flavobacteria) in the surface and Epsilonproteobacteria (Arcobacter sp.) at ~10 cm depth. Archaea in the high salinity surface sediments were dominated by the orders Archaeoglobales and Thermococcales, transitioning to Thermoproteales and Desulfurococcales (Staphylothermus sp.) in the deeper sediments. In contrast, the low salinity site was dominated by Thermoplasmatales in the surface and Thermoproteales at depth. Similarities in gas and redox chemistry suggest that salinity and/or arsenic concentrations may select for microbial communities that can tolerate these parameters. Many of the archaeal 16S rRNA sequences contained inserts, possibly introns, including members of the Euryarchaeota. Clones containing AFGs affiliated with either Alpha- or Betaproteobacteria, although most were only distantly related to published representatives. Most clones (89%) originated from the deeper layer of the low salinity, highest arsenic site. This is the only sample with overlap in 16S rRNA data, suggesting arsenotrophy as an important metabolism in similar environments.
Introduction
Geochemical gradients often dictate microbial community structures, and metabolic processes directly influence these gradients. In hydrothermal systems, steep gradients generate redox disequilibria that can provide the necessary energy for a wide diversity of Archaea and Bacteria. Since their discovery in the 1970s, deep-sea hydrothermal environments have garnered much attention in this regard, with a number of studies linking geochemical composition, reaction energetics, and metabolic diversity (e.g., Flores et al., 2011; Meyer-Dombard et al., 2011). Their shallow-sea counterparts provide highly complementary research opportunities that are often overlooked. These systems are ubiquitous, readily accessible, and geochemically diverse; many exhibit geological, chemical, and biological characteristics similar to those found at deep-sea vents.
Shallow-sea and deep-sea hydrothermal systems also differ in several key ways (Dando et al., 1999; Pichler, 2005; Tarasov et al., 2005; Price et al., 2012). For example, the evolved discharging hydrothermal fluids in shallow-sea systems may have originally been seawater, meteoric water, or a mixture of the two. They often occur within the photic zone and thus provide the opportunity for both chemosynthetic and photosynthetic microbial metabolisms. They are most often associated with arc volcanism, which provides the heat source that drives hydrothermal circulation. Well known examples of shallow-sea hydrothermal systems are found near the Tabar-Feni (Pichler et al., 1999, 2006), the Aeolian (Italiano and Nuccio, 1991; Amend et al., 2003), the South Aegean (or Hellenic; Varnavas and Cronan, 1988; Dando et al., 1995), the Caribbean (McCarthy et al., 2005), and the Kurile-Kamchatka island arcs (Tarasov et al., 1990). In addition, some shallow-sea vent systems also occur in other areas related to faulting (e.g., Bahía Concepción; Forrest et al., 2005) or serpentinization reactions (e.g., New Caledonia; Cox et al., 1982). Many of these shallow-sea systems are sediment covered, allowing steep vertical and horizontal geochemical gradients to evolve as reduced, low pH, high temperature fluids mix with overlying seawater (e.g., Price et al., 2007). Across the geochemical gradients, “niches” of potential energy develop that constrain the resident microbial communities. While most shallow-sea hydrothermal fluids seem to be sulfur- and/or iron-rich, they can also be elevated in potentially toxic elements such as As, Sb, Cr, Pb, Cd, Cu, Zn, and Hg (Varnavas and Cronan, 1988; Koski et al., 2001; Pichler et al., 2006; Price et al., 2012).
Many seafloor hydrothermal systems are modified by phase separation (German and Von Damm, 2003; Von Damm et al., 2003), resulting in vent fluids that can vary drastically in salinity, from <6 to ~200% of seawater values (German and Von Damm, 2003). The resultant high and low salinity fluids differ markedly in the concentrations of many solutes, but the relation to microbial community structures has not been adequately considered. Previous investigations indicate that the vapor phase fluids may be enriched in important electron donors, such as H2, relative to the high salinity brine phase, and may contribute to stratification of microbial communities (Nakagawa et al., 2005; Nunoura and Takai, 2009). Arsenic is known to partition into the vapor phase (Pokrovski et al., 2002; Price et al., 2012). If the hydrothermal reservoir is enriched in arsenic and undergoing phase separation, this element can therefore be elevated in both high and low salinity discharging hydrothermal fluids. The highest As levels in a marine hydrothermal fluid were reported for the shallow-sea hydrothermal vent system in Palaeochori Bay, Milos Island, Greece (Price et al., 2012). There, high arsenic concentrations were reported for high and low salinity fluids; typically in the range of 30 μ M, but as high as 78 μ M in the low salinity fluids, suggesting As partitions into the vapor phase in this system.
The purpose of this investigation was to identify the dominant bacterial and archaeal lineages in Palaeochori Bay in an attempt to understand the link between geochemical gradients and microbial groups, including arsenic oxidizers. These relationships were investigated at two sites with similar pH and temperature, but different salinities.
Site Characteristics
Gas discharge defines an ~35 km2 area of hydrothermal venting around Milos, making it one of the largest shallow-sea hydrothermal systems described to date (Figures 1A,B; Dando et al., 1995). The most intense venting occurs in Palaeochori Bay, where CO2-rich gases and hydrothermal fluids discharge through sand (Figures 1A–C, 2). Nearby—but little explored—Spathi Bay, to the east, also features abundant hydrothermal venting, although free gas emissions are less abundant. The high salinity hydrothermal fluids in Palaeochori Bay were slightly acidic (pH ~5), sometimes highly sulfidic (up to 3 mM), and warm (ambient to ~110°C; Stüben and Glasby, 1999; Valsami-Jones et al., 2005; Price et al., 2012).
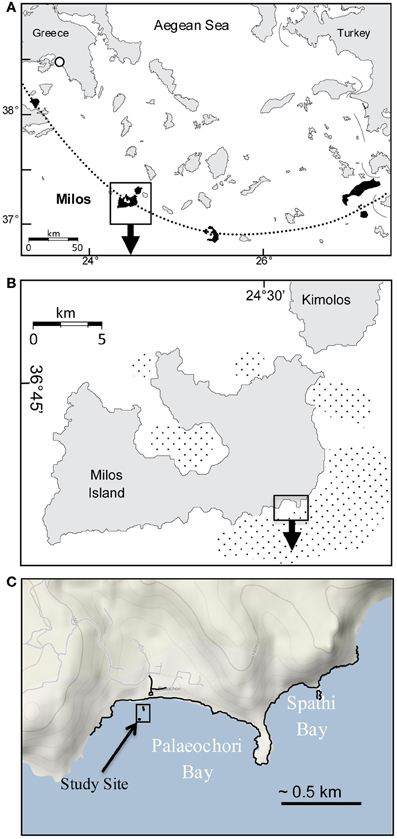
Figure 1. (A) Location of Milos Island (box) and other calc-alkaline volcanoes (shaded) along the Aegean island arc (dotted line). (B) Milos Island and the location of Palaeochori and Spathi Bays (box). Stippled offshore areas around the island are mapped gas emissions by echo sounding (Dando et al., 1995). (C) Location of our study site (box) in Palaeochori Bay.
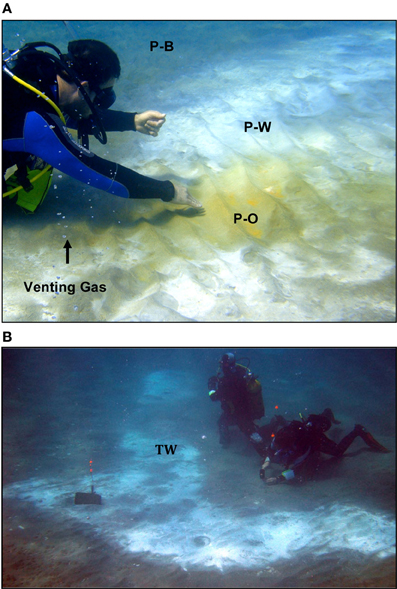
Figure 2. (A) Photograph of the RP site investigated in this study. P-O, Palaeochori orange; P-W, Palaeochori white; and P-B, Palaeochori background. Geochemical profiles and sediment cores were sampled at ~P-W. (B) Photograph of the TW site. Geochemical and microbiological samples for this study are indicated at the location of TW.
The occurrence of orange, white, or brown microbial mats and hydrothermal precipitates on the seafloor sediments are a major feature of the hydrothermal system. The different colors correlate to different sediment temperatures, where the hottest areas (>90°C) are typically covered by bright yellow native sulfur or orange-colored arsenic sulfide deposits (Figure 2A). White mats make up the largest areas of hydrothermal venting throughout the bay, and display mid-range temperatures (e.g., ~45–85°C). In the lower temperature areas (~30–35°C), brown colored manganese and/or iron oxide deposits are present (Figure 2A).
Several microbiological investigations were carried out for the high salinity sites in Palaeochori Bay, although none to date targeted low salinity environments. They included culture-dependent and molecular-based microbiological studies, as well as the isolation of several new bacterial and archaeal species (Jochimsen et al., 1997; Dando et al., 1998; Brinkhoff et al., 1999; Sievert et al., 1999, 2000a,b; Arab et al., 2000; Sievert and Kuever, 2000; Schlesner et al., 2001; Bayraktarov et al., 2013). One study investigated the microbial ecology and arsenic functional genes in lower temperature 0–6 cm pooled sediments at the same high salinity site investigated here (Nitzsche, 2010). Results indicated a high diversity in the bacterial community sequences of Gamma-, Delta-, and Epsilonproteobacteria, Bacteroidetes, and Cyanobacteria. Closest cultivated relatives were associated with photo- and chemotrophic lifestyle and seemed mostly to be involved in sulfur cycling. Archaeal sequences predominantly affiliated with Thermoplasmatales or Thermococcales. Screening for arsenic functional genes indicated affiliation with either Alpha- or Betaproteobacteria, although affiliation with Gammaproteobacteria was noted. These studies indicated that metabolisms based on sulfur, either oxidation of H2S or reduction of SO2−4, may be dominant at the high salinity sites, although dissimilatory iron reducing (DIR) Bacteria are abundant in some FeIII-rich sediments. They also suggest that both heterotrophic and chemotrophic metabolisms are possible, with a range from mesophilic to hyperthermophilic functioning, and that arsenite oxidase genes are present at the high-salinity, lower temperature sites. This study expands on these investigations by studying the microbial ecology at greater sediment depth than the previous investigations and for the first time compares community structure between a high salinity and low salinity site.
Materials and Methods
Site Description
Two sites in Palaeochori Bay [Rocky Point (RP) and Twinkie (TW)] were investigated in this study; both are ~100 m offshore in ~4.5 m deep water (Figures 1C, 2A,B). Preliminary data indicate nearly identical pH and temperature profiles, although the RP site was demonstrably more saline than the TW site. The RP site features concentric rings of colored sediments, with orange in the center, transitioning to white and then brown (Figure 2A). The TW site is located ~8 m due east of the RP site and consisted primarily of white mats with some gas bubbles at the north side of the patch. Samples for this study were collected from the white areas from both sites (“P-W” in Figure 2A and “TW” in Figure 2B).
Field sampling protocols
Pore fluids and sediments for geochemical and microbiological analyses were collected by SCUBA. Sediments were cored with polycarbonate tubes (Arthur Krüger GmbH) and capped underwater with rubber end caps. Pore fluids were either collected in situ into a BD™ (Becton, Dickinson and Company) 60 mL syringe from a tube inserted into the sediments, or extracted through holes in the core tube once onshore using 10 mL syringes attached to rhizons (long filters which can be inserted into the core; Rhizosphere) following Seeberg-Elverfeldt et al. (2005). Sediment cores were sliced for subsequent microbiological analyses. Each core slice was aseptically placed into sterile bags and frozen on dry ice. Samples were then shipped on dry ice back to the laboratory and kept frozen at −80°C.
Samples for free gas geochemistry were obtained by inverting a serum bottle under water and holding it over the streaming gas bubbles. Once the gas had completely filled the bottle, it was capped and crimped underwater with little to no seawater inside the bottle. Pore fluids for dissolved gases were sampled at 10 cm depth in the sediments by inserting a tube into the sediments, and filling a syringe. Subsequently, these pore fluids were put into N2-filled 60 mL serum bottles until a reverse meniscus formed, then capped and crimped.
Geochemistry
Temperatures were measured in situ using a temperature probe in a custom-built underwater housing (constructed at the Max Planck Institute for Marine Microbiology, Bremen, Germany). The pH was measured on shore using a WTW pH meter 3210 with Mic-D electrode with temperature compensation. Analytical uncertainties were approximately ±0.1°C for temperature and ±0.1 for pH. Pore fluid concentrations of Fe2+, NO−3, NO−2, and NH3 were analyzed in the field using a HACH spectrophotometer. Samples for free (CO2, O2, N2, He, H2, CO, and CH4) and dissolved (He, H2, O2, N2, CO, CH4, and CO2) gases were shipped on ice to the Istituto Nazionale di Geofisica e Vulcanologia, Palermo (Italy), for analysis. Pore fluid samples for anion analysis (Br, Cl, and SO2−4) were filtered in the field (0.2 μm), placed on dry ice, and kept frozen until measurement in the laboratory. Samples for analysis of major seawater cations (Na, Ca, K, B, Sr), minor elements (Si, Ba, Mn, Fe), and trace elements (Li, Rb, Cs) were preserved in the field by filtering (0.2 μm) and acidification with 0.1% ultrapure HNO3.
Laboratory Protocols
Geochemistry
Major anions were analyzed using a Dionex ion chromatography system, whereas major cations were measured by inductively coupled plasma-optical emission spectrometry (ICP-OES; Perkin-Elmer Optima 3300). Lithium, rubidium, and cesium were measured by inductively coupled plasma-mass spectrometry (ICP-MS; High-resolution double-focusing ICP-MS Thermo Finnigan Element 2). Free (CO2, O2, H2, CO, and CH4) and dissolved (H2, O2, CO, CH4, and CO2) gases were measured on a Perkin Elmer 8500 gas chromatograph equipped with a double-detector (TCD-FID) using argon as carrier gas (Italiano et al., 2009).
Microbiology
DNA extraction, PCR, and Cloning. Bulk environmental (genomic) DNA was extracted from ~0.5 g of sediment according to the protocol outlined in the MPBio FastDNA® spin kit, and stored at −20°C. Amplification of 16S rRNA genes for both Archaea and Bacteria was performed by polymerase chain reaction (PCR) using primer sets 27F/1492R and 21F/1391R, respectively (Lane, 1991), on a Hybaid PCR express thermocycler. A total reaction volume of 20 or 50 μ L was prepared, containing 1× PCR buffer, 0.25 mM each dNTPs, 0.5 μ M each forward and reverse primer, 0.5–2 μ L DNA template depending on sample concentration, and 5 U Taq DNA polymerase (5 PRIME). DMSO was added to some samples to increase reaction yield. The final volume was adjusted with autoclaved milli-Q water. The amplification was initiated with denaturation at 95°C for 5 min, followed by denaturation (at 95°C for 1 min), annealing (at 55°C for Archaea, 52°C for Bacteria, for 1 min), and elongation (at 72°C for 1 min) for 30–35 cycles. A final extension step was carried out at 72°C for 10 min. Agarose gel electrophoresis was performed to verify amplification.
Degenerate primer sets used for arsenite oxidase functional gene (aioA-like) amplification were AOX-F-A2 (5′-TGC ATCGTCGGCT GYGGNTAY-3′) and AOX-R-E2 (5′-TTCGGAGTTATAG GCCGGNCKRTTRTG-3′) (Zargar et al., 2012) on a Hybaid PCR express thermocycler. These primers target the aoxB gene sequence of the arsenite oxidase operon and typically result in ~670 bp product. A total reaction volume of 50 μL was prepared, containing 1× PCR buffer, 0.25 mM each dNTPs, 0.5 μ M each forward and reverse primer, 0.5–2 μ L DNA template depending on sample concentration, and 5 U Taq DNA polymerase (5 PRIME). DMSO was added for some samples to increase reaction yield, and the final volume was adjusted with autoclaved milli-Q water. The amplification was initiated with denaturation at 95°C for 1 min, followed by denaturation (at 95°C for 0.5 min), annealing (at 57°C for 0.5 min), and elongation (at 72°C for 1 min) for 35 cycles. A final extension step was carried out at 72°C for 5 min. Agarose gel electrophoresis was performed to verify amplification.
PCR products were cloned separately using the pCR4-TOPO® Plasmid Vector Kit with One Shot TOP10 chemically competent Escherichia coli cells. Volumes of 25, 50, and 100 μ L of each transformation reaction were spread on selective LB agar plates containing ampicillin and incubated overnight at 37°C. White colonies were transferred with sterile toothpicks into 2.5 mL, 96 well microtiter plates containing a final volume of 200 μ L of LB medium. After an overnight incubation at 37°C, 100 μ L aliquots of liquid cultures were pipetted into a 96 well standard microplate with 20 % glycerol and shipped frozen to Beckman Coulter for sanger sequencing.
16S rRNA gene clone library analysis. Forward and reverse sequence fragments were trimmed and assembled using Geneious version 5.6.3, created by Biomatters (http://www.geneious.com/). Contigs were checked for chimeras using Mothur (Schloss et al., 2009) and Bellerophon (Huber et al., 2004). Sequences were classified using Mothur's Bayesian classifier and the RDP training set version 9 (Wang et al., 2007). Sequences were also aligned, compared, and clustered into operational taxonomic units (OTUs) using the default settings in Mothur. Representative OTUs at the 97 % sequence identity were selected and used to search for similar sequences in NCBI's nr/nt database using nucleotide BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Organisms with complete genomes, closest cultured representatives, or sequences isolated from relevant hydrothermal or arsenic-rich environments were added as references to the 16S rRNA and aioA-like phylogenetic trees. The 97% OTU representatives and the database reference sequences where realigned in Mothur, and imported into Geneious. The PHYML plugin (Guindon and Gascuel, 2003) was used to construct a maximum likelihood tree for bacterial and archaeal sequences.
Several richness and evenness calculations were performed using their respective commands in Mothur. These include calculations for rarefaction curve data, Sobs (the observed richness reported, OTU), chao (the Chao1 estimator for richness), and the simpsoneven and shannoneven (both index-based measures of evenness), all at 97% cutoff.
For aioA-like gene sequences, assembly and vector trimming was also done in Geneious. Trimmed sequences were searched against Ref_Seq using BLASTx. Sequences that hit arsenite oxidase subjects with an e-value lower than e-40 were used to build the tree. Selected bacterial aioA-like genes where taken from Genbank and also added to the tree for reference. Sequences were aligned and compared in Geneious using both nucleotide and amino acid translation. The aioA tree was built using PHYML.
Submission of Nucleotide Sequence and Accession Numbers
Sequence data for 16S rRNA bacterial and archaeal clone libraries were submitted to GenBank under accession numbers KF278514-KF278557 and KF278485-KF278513, respectively. Partial nucleotide sequences for the arsenite oxidase genes were submitted under accession numbers KF303544-KF303564.
Results
Geochemistry
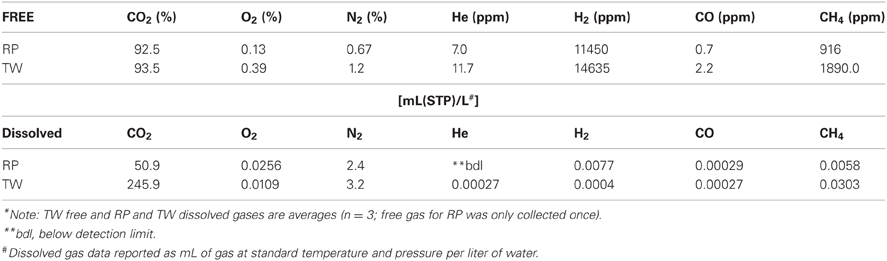
Table 1 presents the geochemical data, including temperature, pH, Fe2+, NO−3, NO−2, NH3, and major and minor elements, collected from RP and TW sites. For comparison, seawater values, as well as a depth profile from a control site, were included. Temperature and pH were very similar between sites, but Mg2+ and SO2−4 concentrations were depleted in both RP and TW samples. Furthermore, major cations, such as Na, Ca, K, B, and Sr, were enriched in the RP samples, but depleted in the TW samples, relative to seawater. Major anions, such as Cl and Br, are also enriched in RP samples but depleted in TW samples, relative to seawater. Most minor elements, including Si, Li, Rb, Fe, and Ba, were enriched in both fluids, although only slightly so in the TW fluids. Mn was only enriched in the RP samples. Arsenic concentrations followed a different pattern compared to all other elements. Both sites were highly enriched in this element, but in this case, the TW site was much more so, with a maximum concentration of 31.7 μ M compared to 9.4 μ M at RP. Depth profiles of temperature, pH, and concentrations of Cl and As are also shown in Figure 3. Note that the values of temperature and pH for RP and TW were similar with depth, but values for Cl and As differed markedly between sites (Figure 3). Table 2 presents the dissolved and free gas data collected for this study. Dissolved gases were sampled at 10 cm depths at equivalent temperature and pH. Both RP and TW free gas was mostly CO2, followed by N2, O2, H2, CH4, He, and CO, although H2S concentrations in the free gas phase were not analyzed. Dissolved gases followed a similar trend.
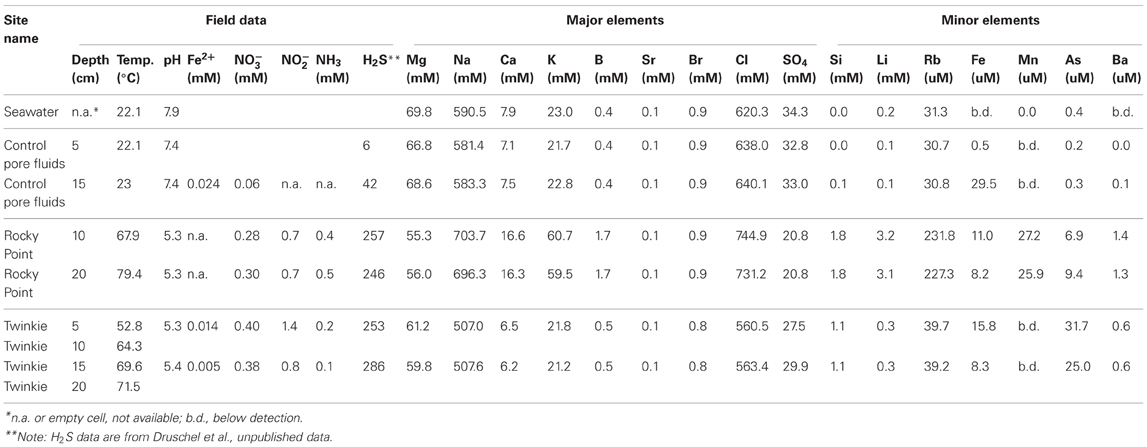
Table 1. Major physicochemical parameters, major and minor elements in hydrothermal fluids collected from Paleochori Bay hydrothermal system.
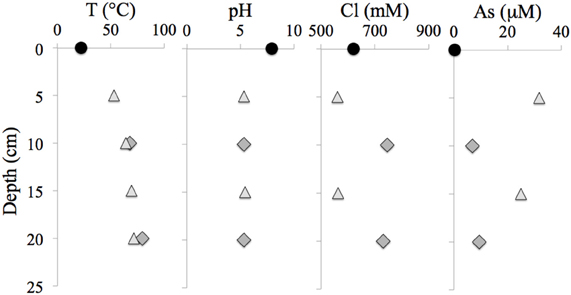
Figure 3. Depth profiles of temperature, pH, chloride and arsenic concentrations at the RP and TW sites. Note (1) similarities between the RP and TW site with respect to T and pH, and (2) differences between these sites with respect to Cl and As concentration. Seawater values are indicated with a filled circle, RP data are indicated with a dark gray filled diamond, and TW data are indicated with a light gray filled triangle.
Microbiology
Richness and evenness
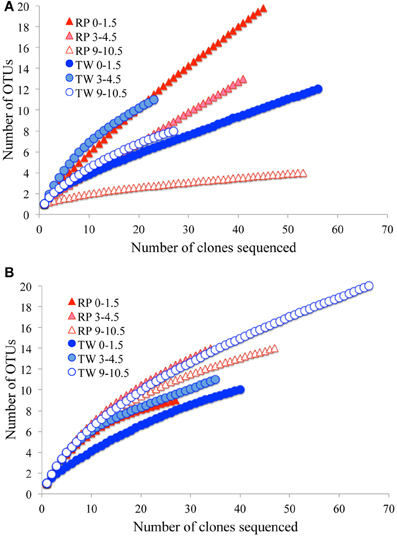
The total number of clones generated for Bacteria per site is as follows: RP 0–1.5 (51), RP 3–4.5 (41), RP 9–10.5 (53), TW 0–1.5 (56), TW 3–4.5 (23), and TW 9–10.5 (27). The total number of clones generated for Archaea per site was: RP 0–1.5 (27), RP 3–4.5 (34), RP 9–10.5 (47), TW 0–1.5 (40), TW 3–4.5 (35), and TW 9–10.5 (66). Rarefaction analyses of the 16S rRNA gene sequences for Bacteria (Figure A1A) and Archaea (Figure A1B) indicated that the RP 0–1.5 sample displayed the highest diversity, with 22 OTUs out of 51 sequences. The steep slope for the RP 0–1.5 sample indicates that a large fraction of the species diversity remains to be discovered. The same is true for the RP 3–4.5 layer, which had 13 OTUs out of 41 sequences. The RP 9–10.5 sample had the lowest bacterial diversity for all samples analyzed. However, the TW 0–1.5 sample had the lowest bacterial diversity of all TW samples, with 12 OTUs out of 56 sequences. The TW 9–10.5 layer diversity was similar to TW 0–1.5, only slightly more diverse, and a slight development of an asymptotic curve. The TW 3–4.5 layer had the highest bacterial diversity of all TW samples, with a slope similar to the RP 0–1.5 sample. The RP site showed a clear decrease in bacterial diversity vs. depth, although the TW site did not follow this pattern. Hydrothermal environments, compared with many other marine environments, are often dominated by relatively few taxa (Polz and Cavanaugh, 1995; Schrenk et al., 2004). Therefore, in theory, fewer clones may be necessary for adequate sampling and evaluation of diversity. However, it is important to note that these curves may be most useful for assessing relative differences in the diversity of major groups between sites and depths.
For the archaeal OTUs, both sites showed increasing diversity vs. depth (Figure A1B). Archaeal OTU diversity for the TW 0–1.5 layer was the lowest, sequences from TW 3–4.5 slightly more so, with the TW 9–10.5 layer having the highest diversity. The RP 0–1.5, RP 3–4.5, and RP 9–10.5 samples followed a similar trend, with increasing archaeal diversity vs. depth. Each of the archaeal rarefaction curves level off to the right of the plot, suggesting the archaeal diversity may have been sampled adequately.
Richness indices for Bacteria and Archaea for each site vs. depth are presented in Table 3. The Chao1 index indicated that generally the bacterial communities were more diverse in the surface layers of the RP site relative to any other sample, and both RP and TW bacterial richness decreased with depth. Both the Shannon index and Simpson index indicated that the most diverse samples were RP 0–1.5 and TW 3–4.5, and that the RP site bacterial richness decreases vs. depth, whereas the TW site has the highest bacterial richness in the 3–4.5 depth sample. Each of these three indices generally indicated that archaeal richness was highest in the deepest samples with overall richness increasing vs. depth.
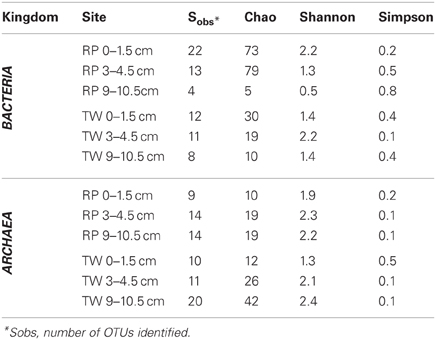
Table 3. Microbial diversity richness indices for bacterial and archaeal depth profiles from RP and TW sites in Palaeochori Bay.
Bacterial affiliations
The diversity and distribution of bacterial 16S rRNA clones at the phylum (Figure 4A) and class (Figure 4B) levels were investigated for three depths at RP and TW. The RP 0–1.5 cm sediments were dominated by the phylum Proteobacteria (40 out of 51 clones), followed by representatives from the Firmicutes (6), Bacteroidetes (2), Actinobacteria (1), Aquificae (1), and Deferribacteres (1) (Figure 4A). Within the Proteobacteria, the Epsilon class dominated (Figure 4B), with nearly all clones identified as Arcobacter spp. Minor classes include Deltaproteobacteria (7 clones), Bacilli (6), and 1–2 clones each affiliated with Actinobacteria, Aquificae, Flavobacteria, and Deferribacteres (Figure 4B). In the 3–4.5 cm layer, the Firmicutes phylum dominated (33/41), followed by Proteobacteria (6), and one clone each of Bacteroidetes and Actinobacteria (Figure 4A). The Firmicutes consisted almost entirely of Bacillus spp. (100% confidence); the Proteobacteria were composed of the Epsilon (4) and Gamma classes (2) (Figure 4B). The deepest layer investigated at RP (9–10.5 cm) was also dominated by the Firmicutes phylum (51/53; Figure 4A), with 47 clones identified as Bacilli (Figure 4B), predominantly within the genus Bacillus.
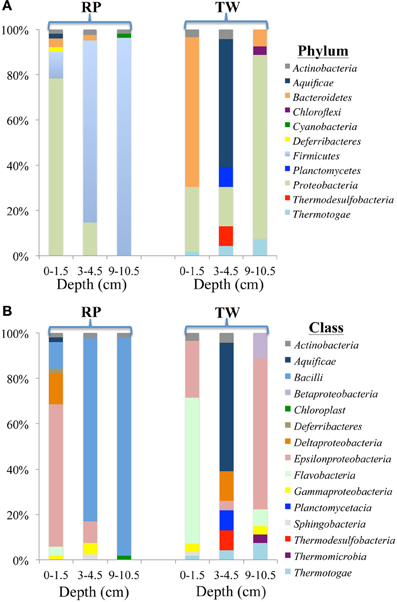
Figure 4. Diversity and distribution of clones within bacterial phylum (A) and class (B) clone libraries for three depths of RP and TW sediment layers investigated in this study. Each color represents the percentages of each taxon in the total respective clone library, as determined by screening of full length 16S rRNA gene sequences. Depths are 0–1.5, 3–4.5, and 9–10.5 cm, and indicated at the bottom of site designation.
At TW in the 0–1.5 cm layer, Bacteroidetes dominated (37/56; Figure 4A), with 36 of those affiliating with the Flavobacteria (Figure 4B). The other phyla include Proteobacteria (16), Actinobacteria (2), and Thermotogae (1) (Figure 4A). Within the Proteobacteria, 14 affiliated with the Epsilon class (genus Arcobacter), and 2 with the Gamma class (Figure 4B). The TW 3–4.5 cm layer was dominated by the Aquifiacae (13 of 23 clones; Figures 4A,B), especially Thermosulfidibacter spp. (often with 100% confidence). The other phyla in this sample included Actinobacteria, Planctomycetes, Proteobacteria, Thermodesulfobacteria, and Thermotogae (Figure 4A). The deepest (9–10.5 cm) sediment sample at TW was dominated by the Proteobacteria, followed by Bacteroidetes, Thermotogae, and Chloroflexi (Figure 4A). Within the Proteobacteria, the Epsilon class (Arcobacter spp.; 18 of 27) dominated, followed by the Beta (3) and Gamma classes (2) (Figure 4B).
The bacterial 16S rRNA phylogenetic tree (Figure 5) includes the OTUs from our 16S rRNA clone libraries and some of their closest relatives, both isolates and other uncultured clones. Three OTUs represented >5 clones each, which are noted in the Figure 5 with a green star (173 total clones), and 50 OTUs represented ≤5 clones (78 total clones). OTU 23 represents 80 clones from the RP site (4 in the top layer, 29 in the middle, and 47 in the deepest layer). This OTU closely affiliates with the Firmicutes class (Bacillus spp.; Figure 5). OTU 30 (54 clones) affiliates with the Epsilonproteobacteria and was identified in all samples, except RP 9–10.5 cm. OTU 39 (39 clones) affiliated with Bacteroidetes and was identified almost exclusively in the TW 0–1.5 cm layer.
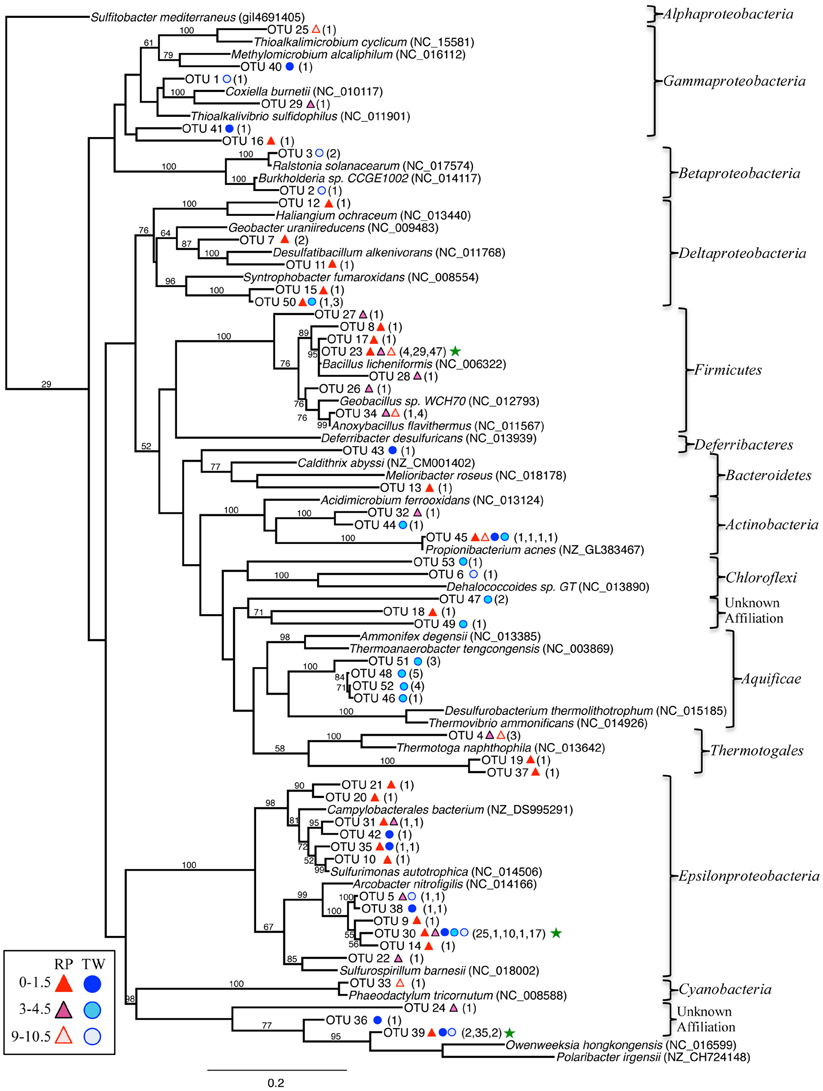
Figure 5. Maximum-likelihood phylogenetic tree of closest relatives found for representatives from the 16S rRNA bacterial clone libraries. OTUs are defined by a 97% similarity cutoff. Symbols next to the OTUs from this study represent the samples from which they were recovered, as indicated by the sample key at the bottom left. The number of clones belonging to each OTU per site is in parentheses. Bootstrap values of >50% are shown at nodes. Sulfitobacter mediterraneus was used as an outgroup. Green stars indicate the OTUs with the highest number of clones.
Archaeal affiliations
The diversity and distribution of archaeal clones at the three depths of RP and TW sediments is shown in Figures 6A (phylum) and B (class). At both sites, Euryarchaeota dominated in the surface layers (Figure 6A). At RP, most of the clones affiliated with the Archaeoglobi (16/27; specifically the genus Ferroglobus) and Thermococci (8/27; specifically the genera Palaeococcus and Thermococcus) (Figure 6B). At TW, clones affiliating with Thermoplasmata (Euryarchaeota) dominated (31 of 40 clones; Figure 6B; specifically the genus Thermogymnomonas). Two clones affiliated with Thermoprotei (Crenarchaota). At both sites, with increasing depth, the phyla changed from predominantly Euryarcharota to more and mostly Crenarchaeota. The middle layer at RP (3–4.5 cm) was dominated by Archaeoglobi (Euryarchaeota), followed by Thermoprotei (Crenarchaeota; Figure 6B). The Archaeoglobi were again affiliated with genus Ferroglobus, while the Thermoprotei genera included Staphylothermus, Pyrodictium, and Thermofilum. The deepest RP site (9–10.5 cm) was dominated by Thermoprotei (Crenarchaeota; Figure 6B), with 42 of 47 clones affiliating with 5 genera including Thermofilum (25) and Staphylothermus (10). In the middle TW layer (3–4.5 cm), Euryarchaeota affiliated predominantly with Archaeoglobi (15; Ferroglobus sp.), and Thermoplasmata (9; Thermogymnomonas sp.), while the Crenarchaeota were entirely composed of Thermoprotei (5) (Figure 6B; specifically the genera Hyperthermus and Thermofilum). The TW 9–10.5 sequences affiliated with Thermoprotei (Crenarchaeota; 30 clones; genus Thermofilum), Archaeoglobi (Euryarchaeota; 11 clones; genus Ferroglobus) and Thermoplasmata (Euryarchaeota; 22 clones; genus Thermogymnomonas) (Figure 6B). Note, only one clone in all the libraries (RP 9–10.5) affiliated with the Korarchaeota.
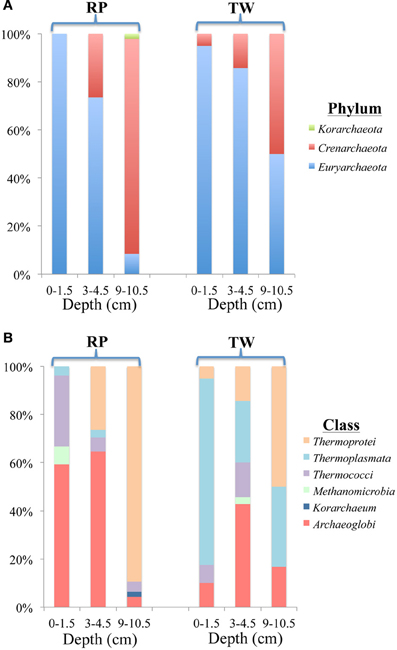
Figure 6. Diversity and distribution of clones within archaeal phylum (A) and class (B) clone libraries for three depths of RP and TW sediment layers investigated in this study. Each color represents the percentages of each taxon in the total respective clone library, as determined by screening of full length 16S rRNA gene sequences. Depths are 0–1.5, 3–4.5, and 9–10.5 cm, and indicated at the bottom of site designation.
The 16S rRNA archaeal phylogenetic tree (Figure 7) includes the OTUs from our 16S rRNA clone libraries and some of their closest relatives, other clones, and related isolates. Twelve OTUs represented >5 clones each, and are indicated by a green star in Figure 7 (212 total clones), and 20 OTUs represented ≤5 clones (37 total clones). Primary OTUs affiliated with Thermoprotei (Crenarchaeota), and Thermococci (Euryarchaeota; Figure 7). Many sequences were <90% similar to other archaeal representatives found in public databases and therefore do not cluster with any major group. A considerable fraction of our archaeal sequences (~20%) contained large ~26–80 bp “inserts,” possibly introns (Burggraf et al., 1993). The OTU sequences containing these inserts (OTUs 8, 1, 30, 5, 3, 27, 2, 33, and 4) passed comprehensive quality and chimera checks, and are therefore included in the phylogenetic tree, and indicated with an asterisk, “*” (Figure 7). BLAST hits indicate they are related to the Euryarchaeota (Archaeoglobi or Thermoplasmata), and Crenarchaeota (Thermoprotaeles or Desulfurococcales). Five of the top six OTUs with the highest number of clones contained these inserts (8, 1, 30, 3, and 27). OTU 27 (39 clones) was found in all samples except RP 0–1.5 (Figure 7), and is identifiable to Thermoprotei according to the RDP classification. Three OTUs (8, 30, and 1; 38, 38, and 13 clones, respectively) were distantly related to Archaeoglobi or Thermoplasmata. OTU 12 (25 clones) poorly affiliated with Archaeoglobi (38% ID), which results in it being separate from the other groups in the phylogenetic tree (Figure 7, top left).
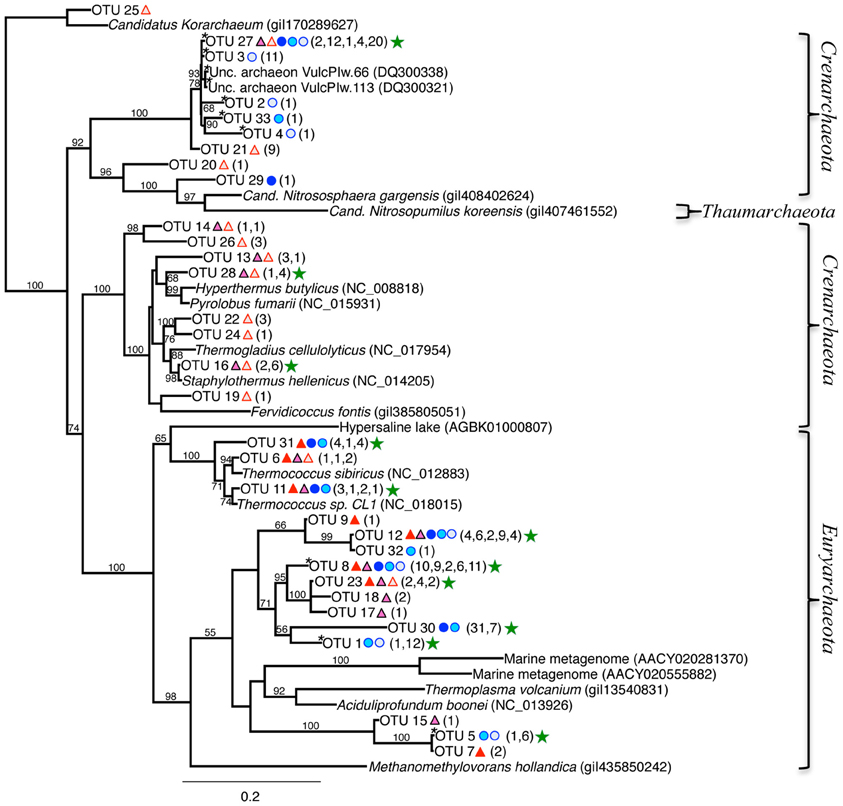
Figure 7. Maximum-likelihood phylogenetic tree of closest relatives found for representatives from the 16S rRNA archaeal clone libraries. OTUs are defined by a 97% similarity cutoff. Symbols next to the OTUs from this study represent the samples from which they were recovered, as indicated by the sample key in Figure 5. The number of clones belonging to each OTU per site is in parentheses. Bootstrap values of >50% are shown at nodes. Methanomethylovorans hollandica was used as an outgroup. Sequences containing inserts are noted with an “*.” Green stars indicate the OTUs with the highest number of clones.
Arsenic functional genes
A phylogenetic tree of the aioA-like functional genes (AFGs) is shown in Figure 8, and includes sequences from our clone library, related isolates and other uncultured clones. Sequences affiliated with those detected in Beta (Ralstonia and Burkholderia genera), and Alpha classes of the Proteobacteria (Polymorphum, Roseovarius and Bradyrhizobium genera) cluster into two distinct lineages (Figure 8). Most of the AFGs associated with Beta class had no closely related cultured representative. A total of 105 clones across 22 taxa contained the AFG. Of these, 93 were from the TW site, with 57, 24, and 12 clones from the surface, middle, and deepest layers, respectively. At the RP site, the AFG was found only in the surface (5 clones) and middle (7 clones) layers.
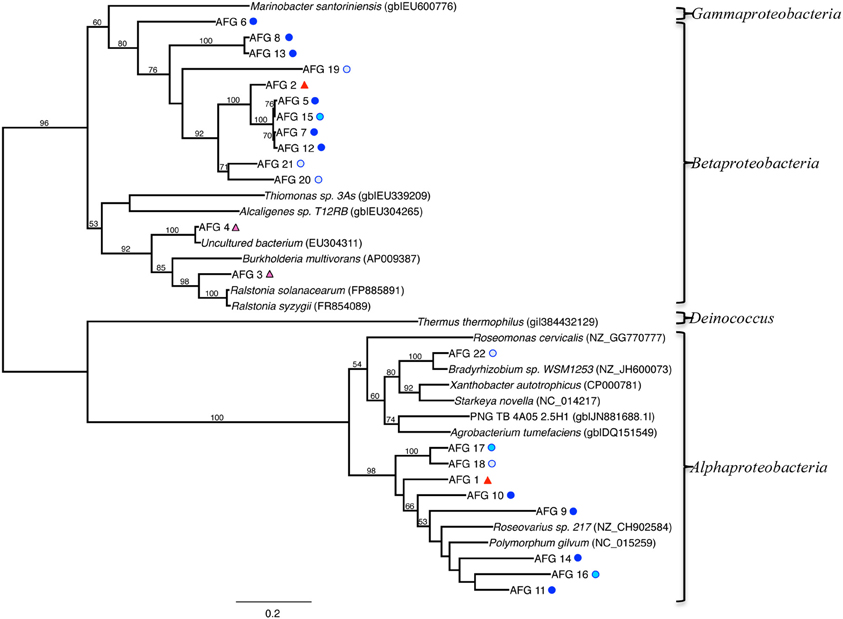
Figure 8. Maximum-likelihood phylogenetic tree of closest relatives found for representatives from the aioA-like functional gene (AFG) clone libraries. AFGs are defined by a 97% similarity cutoff. Symbols next to the OTUs from this study represent the samples from which they were recovered, as indicated by the sample key in Figure 5. Bootstrap values of >50% are shown at nodes. Marinobacter santoriniensis was used as an outgroup.
Discussion
Microbial Diversity
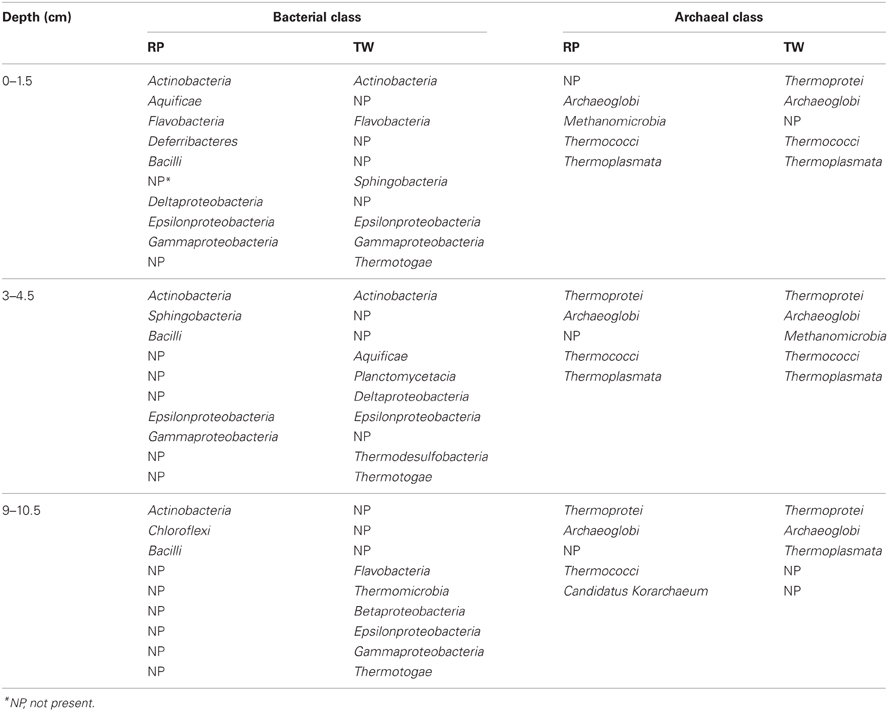
Our 16S rRNA gene sequence data indicate that bacterial and archaeal communities in the hydrothermally influenced sediments of Palaeochori Bay differ significantly when comparing the same depths of the RP high salinity and TW low salinity sites, although they have approximately the same temperature and pH (Figures 4–8, Table 1). Archaeal and bacterial class distribution between each site and vs. depth is shown in Table 4, and indicates that several classes were exclusive to either the RP or TW site. For example, comparing the 0–1.5 cm sediment depth, Aquificae, Deferribacteres, Bacilli, and Deltaproteobacteria were only present in the RP site, whereas Sphingobacteria and Thermotogae were only present in the TW sample (Table 4; Figures 4A,B). The same is true for the other sediment horizons, with distinct bacterial affiliations only occurring at one or the other site. Of the major bacterial groups, the Firmicutes were only present in the RP samples, while none were present in any of the TW sample depths. The 9–10.5 layer has zero overlapping bacterial classes between RP and TW sites (Table 4). Betaproteobacteria were identified in the TW sample from this depth, the only time this group was seen in any of the samples.
Differences in the archaeal communities in surface samples are also quite distinct; e.g., Archaeoglobi is dominant at the RP site while Thermoplasmata was dominant at the TW site (Figures 6A,B). As was the case for the Bacteria, there exist some groups at RP, which were not found at the TW site, and vice versa (Table 4). For example, the RP surface sample did not contain Thermoprotei, a dominant archaeal group for the TW site. In the deepest layer, the RP site contained Thermococci, but no Thermoplasmata (Table 4). These results suggest that the major archaeal and bacterial groups in Palaeochori Bay are distinctly different in the high vs. low salinity sites.
The Role of Phase Separation
Subcritical phase separation is a ubiquitous process in hydrothermal systems, but its effect on microbial communities is rarely evaluated. Investigations of the Iheya North Field back-arc hydrothermal system south of Japan, suggested that subseafloor phase-separation and segregation may influence the supply of energy and carbon to vent-associated chemolithoautotrophs and related microbial communities (Nakagawa et al., 2005). When comparing microbial communities of gas-depleted “normal” fluids (i.e., those related to the remaining fluid phase after phase separation has taken place) to gas-enriched fluids (i.e., those which are related to the vapor phase; German and Von Damm, 2003), chemolithotrophs associated with gas-dependent energy metabolism, such as hydrogenotrophic methanogenesis, were more abundant. Another study investigated differences in microbial communities in phase separated hydrothermal fluids from the Yonaguni Knoll IV hydrothermal field in the southern Okinawa Trough (Nunoura and Takai, 2009). While fluids showed variability in the physical and chemical compositions (e.g., gas content), the microbial communities were relatively similar. There, chemolithotrophic hydrogen oxidation was the primary metabolism, and it was suggested that similarities in microbial communities could have been due to elevated concentrations of hydrogen in both types of fluids. Thus, microbial diversity may be influenced by a change in the concentration of potential electron donors as a result of phase separation (i.e., dissolved gases such as H2), not necessarily from salinity differences alone, although halotolerance cannot be ruled out. As we discuss below, metabolisms based on dissolved gases such as H2 are possible at each site investigated here, but do not seem to play a dominant role in influencing major groups.
Our geochemical results indicate that salinity and arsenic concentrations are among the most variable parameters when comparing RP with TW pore fluid compositions (Tables 1, 2; Figure 3). Free gas data indicate that O2, N2, He, H2, CO, and CH4 were only slightly higher for TW compared to RP (Table 2). Dissolved gas data suggest that no significant differences between sites exists for O2, CO, and H2 concentrations; the dissolved CH4 is slightly higher in the TW pore fluids (Table 2). For the redox sensitive species analyzed in this study, dissolved Fe2+, NO−3, and NO−2 were slightly higher, while NH3 was slightly lower, in fluids from the TW site (Table 1). Hydrogen sulfide concentrations can be very elevated (up to 3 mM) in high salinity fluids, but were essentially equal (~0.25 mM) in both high- and low-salinity hydrothermal fluids analyzed for this study (Druschel et al., unpublished data; Table 1). These geochemical parameters suggest that the phase separation process at Milos may create differences in concentrations of dissolved gases, as well as some other important redox couples, which could ultimately be contributing to the variability in microbial communities when comparing high- vs. low-salinity sites. However, we suggest that these differences are not significant, and therefore differences in microbial diversity may be influenced more by salinity and/or arsenic concentrations.
Dominant Bacterial Metabolisms
The genus Arcobacter, which dominates the RP 0–1.5 cm sample, can grow at moderately thermophilic and halophilic conditions, utilizing nitrogen or sulfur, including sulfide oxidation (Wirsen, 2004; Donachie et al., 2005; Pati et al., 2010). For example, A. nitrofigilis, isolated from the roots of a salt marsh plant, can fix nitrogen (Pati et al., 2010). “Candidatus Arcobacter sulfidicus” has been shown to produce large white sulfur mats in hydrothermal environments (Wirsen, 2004), and related organisms may be responsible for the large white mats observed in Palaeochori Bay (Sievert et al., 1999). The white sulfur mats contain elemental sulfur, which may be the end product of sulfide oxidation. However, a comparison of our representative 16S rRNA gene sequence (OTU 30; Figure 5) against this and other Arcobacter species indicated only a 91–93% sequence similarity (BLAST comparison Max ID).
In the deeper RP sediments, Firmicutes dominate, in particular, Bacillus sp. This group is phenotypically highly diverse, and includes obligate aerobes, facultative anaerobes, chemotrophs, and heterotrophs, halotolerant members, and organisms that tolerate a wide range of temperatures (Graumann, 2012). Our representative sequence (OTU 23) is most closely related to B. licheniformis DSM 13 (Figure 5) which, along with many other members of the Bacilli, can reduce nitrate to N2 (Nakano and Zuber, 1998). Other investigations suggest that Mn(II) oxidation (Dick et al., 2006) and arsenate reduction may also be mediated by Bacillus spp. (Switzer et al., 1998). Four new strains of Bacillus were isolated from shallow-sea hydrothermal vents—two from Panarea and Vulcano Islands (Italy) (Maugeri et al., 2002; Spano et al., 2013), and two from the sediments in Palaeochori Bay (Milos) (Sievert et al., 2000a; Schlesner et al., 2001). The Milos isolates are Halothiobacillus kellyi, a mesophilic, obligately autotrophic, strictly aerobic, chemolithoautotrophic, sulfur-oxidizing bacterium (Sievert et al., 2000a), and Filobacillus milensis, a halophilic, faculatively anaerobic, endospore-forming heterotroph (Schlesner et al., 2001). In addition, 39 of 80 aerobic mesophiles isolated from Milos by Dando et al. (1998) are phenotypically similar to Bacillus sp.
The most abundant bacterial group in the surface sample at Twinkie (TW 0–1.5) was Bacteroidetes. However, the RDP classification confidence was quite low even at the class level (average 74%), suggesting many possibilities for representatives from this phylum. The representative OTU for these sequences is OTU 39, which had 35 out of 39 clones in the TW 0–1.5 cm sample. Due to the low confidence identification of this group, it is not affiliated with any other main group in the bacterial phylogentic tree (Figure 5, bottom left). BLAST results most closely identified with uncultured clones, making identification of primary metabolisms difficult. The phylum Bacteroidetes contains among the most abundant microbes in coastal marine waters (Alonso et al., 2007). Cultured representatives from this phylum are typically heterotrophs, non-thermophilic, and use complex organic substrates, either by aerobic respiration or fermentation (Kirchman, 2002). They can also be found in hydrothermal environments. For example, Kormas et al. (2006) detected Bacteroidetes spp. on a white smoker chimney at 9° N, East Pacific Rise. Recently, Sylvan et al. (2012) suggested that Bacteroidetes replaced “normal” microbial communities once smoker chimneys become inactive. Sievert et al. (2000b) and Nitzsche (2010) both identified Bacteroidetes as dominant members in the white areas from Milos sediments, but temperatures were in both cases quite lower than for our study (~20°C less). This suggests the possibility that Bacteroidetes activity is limited to lower temperature environments. Temperatures were comparable between the RP and TW sites (i.e., < ~40°C), but salinity was quite different, suggesting the lack of Arcobacter in place of Bacteroidetes at the TW site may also be influenced by halotolerance. However, as discussed, dissolved gases and other redox couple concentrations may play a role, as they are slightly elevated in the TW pore fluids.
The TW 3–4.5 site was dominated by Thermosulfidibacter sp., whose closest cultured representative is Thermosulfidibacter takaii, a thermophilic, sulfur-reducing chemolithoautotroph from a deep-sea hydrothermal vent at the Yonaguni Knoll IV, Southern Okinawa Trouph (Nunoura et al., 2008). Growth was observed within the temperature range of 55–78°C, and 0.5–4.5% NaCl. Temperatures at this sediment depth should be near 50°C, and salinity should be slightly lower than seawater (Table 1; Figure 3).
The TW 9–10.5 was dominated by Arcobacter spp. (Epsilonproteobacteria; 18 out of 27 clones). As discussed, the group could be conducting nitrate reduction or sulfide oxidation. Comparing the OTU representative sequence (OTU 5) from TW 9–10.5 to the 16S rRNA gene sequence from A. nitrofigilus and “Ca. A. sulfidicus” indicates BLAST max ID of 93% and 93%, respectively. Indeed, the representative OTUs from RP 0–1.5 (OTU 14) and TW 9–10.5 (OTU 5) were only 96% max ID when compared to each other. This suggests that the Arcobacter species in both the RP 0–15 and TW 9–10.5 samples are only distantly related, and it is probable that they are different species or genera with different environmental tolerances.
These data indicate that bacterial communities are stratified by depth, and that sulfur- and nitrogen-based metabolisms may dominate at the RP site, whereas phototrophy, sulfur, and or nitrogen-based metabolisms may be dominant at the TW site. Temperature may be the dominant parameter which influences microbial community structure as a function of depth at both RP and TW sites, as vertical gradients are more pronounced for this parameter compared to any other (Table 1; Figure 4). Sunlight may also play a role, particularly for the surface TW site where Bacteroidetes is present.
Dominant Archaeal Metabolisms
The dominant archaeal genera for RP 0–1.5 were Ferroglobus (Archaeoglobi) and Palaeococcus/Thermococcus (Thermococci). These groups become replaced mostly by the Thermoprotei (Staphylothermus and Thermophilum) at depth. However, poor RDP classification confidence makes identification beyond the phylum level difficult. Further complicating matters, the OTU chosen to represent the sequences associated with the Archaeoglobi (OTU 8) contained a 44 bp insert. BLAST analysis of OTU 8 returned no cultured representatives, and identified most closely with uncultured clones from the Logatchev hydrothermal field (LHF) and terrestrial hydrothermal vents on Ambitle Island, PNG. Removing the insert from OTU 8 and conducting a BLAST analysis for the remaining sequence indicates Aciduliprofundum spp. as the closest cultured match (100% query coverage; 86% max ID). This organism belongs to the “Deep-sea Hydrothermal Vent Euryarchaeota 2” (DHVE2) lineage (Takai and Horikoshi, 1999), which are widespread at deep-sea hydrothermal vents (Reysenbach et al., 2006; Flores et al., 2012). Unfortunately, relatively little is known about their distribution and phylogenetic diversity (Flores et al., 2012). The only described species from the Aciduliprofundum genus is A. boonei T469T, which is an obligate thermoacidophilic sulfur- or iron-reducing heterotroph capable of growing from pH 3.3–5.8 and between 55 and 75°C (Reysenbach et al., 2006). This organism is included in the phylogenetic tree, but does not affiliate closely with our sequences (Figure 7).
The second most abundant archaeal group in the RP 0–1.5 sample was Thermococci, consisting of both Thermococcus spp. and Palaeococcus spp., with ~100% RDP confidence. Our OTUs for this group (31, 6, and 11) did not contain insertions, and identified most closely with Thermococcus spp. (Figure 7). There are numerous species related to this genus; most are considered extreme thermophiles, typically identified as organotrophic anaerobes which grow above 70°C, sometimes requiring elemental S, producing H2S and CO2, and have the ability to use H+ (Arab et al., 2000).
The RP 9–10.5 sample was dominated by the Crenarchaeota–Thermoprotei, was composed of several genera, which were dominated by Thermofilum spp., although mostly with very poor confidence, followed by Staphylothermus spp., often with 100% confidence, and several others. Our OTUs chosen to represent the Thermofilum spp. identified by the RDP classification (21, 26, and 27) most closely affiliated with Thermoprotei (Figure 7). However, OTU 26 is within a slightly different cluster, and had much higher RDP % confidence than the other two with Thermofilum spp. BLAST analysis of this OTU most closely matched with Thermofilum pendens, a thermophilic, anaerobic sulfur-respirer. OTU 27 contained an insert, and BLAST analysis indicated the closest representative was the uncultured clone VulcPlw.113, from the shallow-sea hydrothermal vents off Vulcano, Italy. The sequence from this clone, as noted, also contains an insert. Removing the insert from OTU 27 and conducting a BLAST analysis indicates it does not affiliate with any cultured representatives, and is most closely related to various uncultured Archaea from the Mariana Trough, East Pacific Rise, and the shallow-sea vents off Ambitle Island, Papua New Guinea (Meyer-Dombard et al., 2011).
The TW 0–1.5 was by far dominated by the Thermoplasmata class, Thermogymnomonas genus. This genus has one heterotrophic species, T. acidicola, and can grow at temperatures in the range 38–68°C (optimally at 60°C), at pH 1.8–4.0 (optimally at around pH 3.0), and is obligately aerobic and heterotrophic, requiring yeast extract for growth (Itoh et al., 2007). These clones are represented by OTU 30, which according to RDP, poorly affiliated with Thermoprotei. This OTU contained an insert, and BLAST analysis indicated affiliation to uncultured clones VulcPlw.76, and those from terrestrial vents from Papua New Guinea. The closest cultured representative was Aciduliprofundum sp. MAR08-237A. BLAST analysis of this sequence with the insert removed indicates similar results.
In the deeper 9–10.5 sample for TW, dominant members transition to Thermoprotei (mostly Thermofilum spp.; OTUs 2 and 3), although abundant Thermoplasmata (Thermogymnomonas spp.; OTUs 1 and 5) were also present. Metabolisms for these organisms have been discussed previously. Each of these OTUs contained inserts. OTUs 2 and 3 identified most closely with VulcPLw.76 uncultured clone and previously obtained clone sequences from Milos, with no affiliation to a cultured representative. OTUs 1 and 5 identified with the isolate A. boonei. OTU 4 in this sample identified with Stetteria sp., from the Thermoprotei–Desulfurococcaceae family, by the RDP classification. BLAST analysis of this sequenced indicates affiliation with the previously described uncultured clones from the shallow-sea vents off Vulcano and Milos.
These results indicate that archaeal metabolisms in both surface and deeper sediments are dominated by sulfur oxidizers, can be heterotrophic, and may also have the ability to conduct Fe oxidation. These poorly classified groups, particularly those which contain an insert, could represent new genera or new higher order taxa.
Although we cannot say for sure these extra nucleotide sequence insertions are introns, we believe the extra nucleotides in our sequences are real for three main reasons: (1) The quality scores for most of these sequences were very high. (2) Multiple chimera checks suggested they are not chimeras (see Materials and Methods section), (3) They are found not only in our dataset, but also in several other datasets from Milos [Brehmer, 2000 (unpublished data, NCBI database); Nitzsche, 2010], and the near-shore hydrothermal vent system of Vulcano (Italy) (Rogers and Amend, 2005). The other datasets were obtained from completely different laboratories using different methods, making it highly unlikely that there is an error. All of the other introns found in this study, when blasted independently after exons were removed, consistently returned affiliations with clone sequences from these previously reported groups as the closest representatives. An intron is an additional piece of nucleotide sequence found in the genes of most organisms and many viruses, and can be located in a wide range of genes, including those that generate proteins, rRNA, and tRNA (Itoh et al., 2003). Introns in the 16S rRNA gene of Archaea are rare, and primarily restricted to the Crenarchaeota, within the families Thermoproteaceae and Desulfurococcales (Morinaga et al., 2002; Itoh et al., 2003). Although outside the current scope of this paper, it is important to note that if the extra nucleotide sequence contained within our archaeal sequences for OTU 8, as well as OTUs 1, 30, and 5, are in fact introns, it is a significant finding as introns contained within the 16S rRNA gene sequences of Euryarchaeota have not, to our knowledge, been reported.
Arsenic Functional Gene Survey
Elevated arsenic concentrations may clearly influence microbial community structure and function, since the concentrations encountered can be toxic and may also provide additional redox couples for microbial metabolism (e.g., arsenotrophy). For example, in reducing hydrothermal fluids, arsenic exists most commonly as arsenite, with a +3 oxidation state (AsIII, H2AsO−3). However, in surface seawater and other oxidizing environments, arsenic is dominantly in the +5 oxidation state (arsenate, AsV, HAsO2−4). Thus, microorganisms can play an important role in the arsenic cycle as they catalyze the transformation of different arsenic redox species (Handley et al., 2009; Akerman et al., 2011; Meyer-Dombard et al., 2011, 2013). The three main types of arsenotrophs thus far known to occur are heterotrophic arsenite oxidizers (HAOs), chemotrophic arsenite oxidizers (CAOs), and dissimilatory arsenate-reducing prokaryotes (DARPs; Oremland and Stolz, 2003). Genes involved in these arsenic transformations include detoxification via the ArsC system, and energy gain via the arsenate respiratory reductase (two subunits, ArrA and ArrB) or the arsenite oxidase (two subunits, AioA and AioB, formerly referred to as AroA and/or AoxB; Oremland and Stolz, 2003; Zargar et al., 2010). Recently, an alternative AsIII-oxidation gene was discovered (arxA-type; Zargar et al., 2010). To date, most studies of As microbial cycling have focused on various naturally or anthropogenically impacted environments, primarily located in terrestrial (non-marine) settings. Next to nothing is known about As cycling in coastal marine environments.
At the two sites investigated here, analysis of the AFGs revealed affiliation with only two classes within the Proteobacteria phylum, although we must keep in mind that lateral gene transfer may have occurred (Figure 8). The Alphaproteobacteria containing the arsenite oxidase gene were predominantly affiliated with the genera Polymorphum, Bradyrhizobium and Roseovarious, while the Betaproteobacteria were affiliated with the Burkholderia or Ralstonia genera. The primers used for screening of aioA-like genes were based on a range of groups, and therefore should be broadly applicable (i.e., should not be specific to one or two groups). For example, primer design relied on the sequence alignment of 16 arsenite oxidase amino acid sequences consisting of 5 Alpha-, 4 Beta-, 3 Gammaproteobacteria, 2 Deinococci, and one Chloroflexi (Zargar et al., 2012). Thus, there could be a bias toward Alpha- and Betaproteobacteria, but many published arsenite oxidizers are in these groups (e.g., the most updated phylogenetic tree for arsenotrophs has many of the aioA-like gene containing microbes as Alpha- and Betaproteobacteria). There are some newer isolates being reported within the Gammaproteobacteria, and various genome sequencing projects are also starting to show the presence of aioA-like genes (Handley et al., 2009; Cavalca et al., 2013). The primers used here were the best available at the time of their design.
AFG distribution was primarily associated with the TW samples, which is somewhat expected given the higher concentrations of arsenic at this site (Figure 8). Phylogenetic analysis revealed that both the Beta- and Alphaproteobacteria relationships can be further subdivided into two distinct lineages (Figure 8). While some of the Betaproteobacteria were most closely related to Ralstonia and Burkholderia genera (AFGs 3 and 4), nearly all of the other AFGs associated with Betaproteobacteria had no closely related cultured representative (Figure 8). Four main clusters exist for these Betaproteobacteria. They consist of (1) AFG 6, (2) AFGs 2, 5, 15, 7, 12, 20, and 21, (3) AFG 19, and (4) AFGs 8 and 13. BLAST analysis of these AFGs indicate they are associated with either Beta- or Alphaproteobacteria, mostly Burkholderiales, but cannot be constrained beyond the Proteobacteria phylum. The Alphaproteobacteria were most closely related either to Polymorphum, Roseovarius, or Bradyrhizobium spp. AFG 11 was closest to Polymorphum gilvum in the phylogenetic analysis (Figure 8). Only Burkholderiales occurred in the low salinity 9–10.5 cm depth in the 16S rRNA gene clone libraries. BLAST analysis of these AFG sequences indicates similarity to aioA-like genes from various terrestrial arsenic-rich environments.
These species are only distantly related, and therefore discussion of their arsenic oxidation strategy is limited. For example, Polymorphum gilvum was isolated from a crude oil-polluted saline soil in Shengli Oilfield, China, and was able to use the crude oil as the sole carbon source, and is not known to conduct arsenic metabolism, although its genome contains genes for both arsenite oxidation and arsenate reduction (detoxification) (Li et al., 2011). All of the related groups in the AFG analysis can be confirmed to contain an arsenite oxidase, but none of them have been shown to oxidize or reduce arsenic for detoxification or for their metabolism.
Overlap in AFG and 16S rRNA gene sequence data occurred only in the low salinity TW 9–10.5 sediments. For example, two OTUs (2 and 3) from the bacterial 16S rRNA data set were identified most closely with the Betaproteobacteria, specifically Ralstonia sp. and Burkholderia sp. This suggests arsenotrophy may be an important metabolism in deep, low-salinity, As-rich environments similar to those found in the TW 9–10.5 layer.
Other molecular investigations of the genes responsible for As redox transformations have revealed intriguing evidence for As microbial transformations in marine hydrothermal environments. For example, Nitzsche (2010) used arsenic primers for aioA-like gene amplification of 0–6 cm pooled sediments from Milos, and obtained similar results to our data (i.e., Alpha- and Betaproteobacteria). At a marine shallow-water hydrothermal vent off Ambitle Island, Papua New Guinea, microorganisms may be key in the oxidation of As(III) in areas immediately surrounding the vent source (Akerman, 2009; Meyer-Dombard et al., 2011, 2013). Using PCR primers designed to amplify arsenic functional genes, biofilm communities at the site were shown to possess the genetic capacity for the oxidation of As(III). The aroA-like (now known as aioA-like) clone sequences analyzed belonged to relatives of Alpha- and Betaproteobacteria, Thermus, and Pyrobaculum, but were only 71–80% similar to these sequences available in GenBank, suggesting that many of the Ambitle Island As-functional genes are unique. Thus, in marine environments, arsenite oxidation may be limited to the Alpha- and Betaproteobacteria, but much more research is necessary, particularly cultivation and molecular-based (next generation metagenomic) approaches focusing on arsenotrophy.
Conclusions and Outlook
Our results indicate that differences in archaeal and bacterial community structure exist at the site as a function of not only depth, but also when comparing high vs. low salinity environments with similar redox potential. A wide range of metabolisms seem possible, with affiliated members likely involved in the nitrogen or sulfur cycle. However, it should be noted that rare members of the community can also be very active and function as “keystone species,” yet not be detected in clone libraries. Differences in bacterial and archaeal communities between the RP high-salinity and TW low-salinity sites suggest geochemical signatures resulting from phase separation may be a controlling factor in hydrothermal vent microbial ecology (e.g., salinity, arsenic). Our screening of AFGs responsible for arsenite oxidation indicate that both the Alpha- and Betaproteobacteria may be responsible for arsenite oxidation in sediments from both sites, although the most abundant clusters are from the higher-arsenic TW site. These results are similar to previously reported AFG screening in marine environments, suggesting that Alpha- and Betaproteobacteria are the key players in marine hydrothermal arsenic metabolism. Since these groups are only distantly related to known arsenite oxidizers, novel genera may be present at the site; much more research is necessary including targeted cultivation of arsenotrophs. These results also provide incentive for metagenomic investigation of these sites to identify potential novel shallow vent species, and confirm arsenite metabolism is present primarily in the Alpha- and Betaproteobacteria. Affiliations from the arsenic gene screening were often not reflected in the 16S rRNA results, suggesting that arsenite may only be an important electron donor in the deeper, low salinity, high arsenic sediments. However, this could also be the result of a lack of sufficient database representation, relatively shallow sequencing depth or potential bias in the AFG primers. Finally, many of the archaeal 16S rRNA sequences contained putative introns, including members of Euryarchaeota, which has not been reported previously.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Funding for this project was provided by a University of Bremen MARUM Post-doc Fellowship (to Roy E. Price), MARUM Unforeseen Incentive Funding (to Roy E. Price and Solveig Buehring), NSF-EAR Grant 1222533 and NSFOCE Grant 1061476 (to David Fike, Gregory Druschel, and Jan P. Amend), and NSF-EAR Grant 0951947 (to Chad Saltikov). We thank Paul Dando for his unselfish discussions about the Milos system, Wolfgang Bach and Heike Anders for the ICP-MS analyses. We thank David Fike, William Gilhooly, and Gregory Druschel for help in the field, and Gregory Druschel for providing H2S data for this manuscript. Thanks to Katrina Edwards for providing laboratory space. Special thanks to Athanasios Godelitsas, Antonios Vichos and the Artemis Bungalows, and the Sirocco Restaurant for logistical support in Athens and Milos.
References
Akerman, N. (2009). Microbial diversity and geochemical energy sources of Tutum Bay, Ambitle Island, Papua New Guinea, an arsenic-rich shallow-sea hydrothermal system. Ph.D. dissertation, Washington University in St. Louis.
Akerman, N., Price, R., Pichler, T., and Amend, J. P. (2011). Energy sources for chemlithotrophs in an arsenic- and iron-rich shallow-sea hydrothermal system. Geobiology 9, 436–445.
Alonso, C., Warnecke, F., Amann, R., and Pernthaler, J. (2007). High local and global diversity of Flavobacteria in marine plankton. Environ. Microbiol. 9, 1253–1266. doi: 10.1111/j.1462-2920.2007.01244.x
Amend, J. P., Rogers, K. L., Shock, E. L., Gurrieri, S., and Inguaggiato, S. (2003). Energetics of chemolithoautotrophy in the hydrothermal system of Vulcano Island, southern Italy. Geobiology 1, 37–58. doi: 10.1046/j.1472-4669.2003.00006.x
Arab, H., Voelker, H., and Thomm, M. (2000). Thermococcus aegaeicus sp. nov. and Staphylothermus hellenicus sp. nov., two novel hyperthermophilic archaea isolated from geothermally heated vents off Palaeochori Bay, Milos, Greece. Int. J. Syst. Evol. Microbiol. 50, 2101–2108. doi: 10.1099/00207713-50-6-2101
Bayraktarov, E., Price, R. E., Ferdelman, T. G., and Finster, K. (2013). The pH and pCO2 dependence of sulfate reduction in shallow-sea hydrothermal CO2-venting sediments (Milos Island, Greece). Front. Microbiol. 4, 1–10. doi: 10.3389/fmicb.2013.00111
Brinkhoff, T., Sievert, S., Kuever, J., and Muyzer, G. (1999). Distribution and diversity of sulfur-oxidizing Thiomicrospira spp. at a shallow-water hydrothermal vent in the Aegean Sea (Milos, Greece). Appl. Environ. Microbiol. 65, 3843–3849.
Burggraf, S., Larsen, N., Woese, C. R., and Stetter, K. (1993). An intron within the 16S ribosomal RNA gene of the archaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. U.S.A. 90, 2547–2550. doi: 10.1073/pnas.90.6.2547
Cavalca, L., Corsini, A., Zaccheo, P., Andreoni, V., and Muyzer, G. (2013). Microbial transformations of arsenic: perspectives for biological removal of arsenic from water. Future Microbiol. 8, 753–768. doi: 10.2217/fmb.13.38
Cox, M. E., Launay, J., and Paris, J. P. (1982). “Geochemistry of low temperature geothermal systems in New Caledonia,” in Pacific Geothermal Conference, (Auckland: University of Auckland), 453–459.
Dando, P. R., Thomm, M., Arab, H., Brehmer, M., Hooper, L. E., Jochimsen, B., et al. (1998). Microbiology of shallow hydrothermal sites off Palaeochori Bay, Milos (Hellenic Volcanic Arc). Cah. Biol. Mar. 39, 369–372.
Dando, P. R., Hughes, J. A., Leahy, Y., Niven, S. J., Taylor, L. J., and Smith, C. (1995). Gas venting rates from the submarine hydrothermal areas around the island of Milos, Hellenic volcanic arc. Cont. Shelf Res. 15, 913–929. doi: 10.1016/0278-4343(95)80002-U
Dando, P. R., Stüben, D., and Varnavas, S. P. (1999). Hydrothermalism in the Mediterranean Sea. Prog. Oceanogr. 44, 333–367. doi: 10.1016/S0079-6611(99)00032-4
Dick, G. J., Lee, Y. E., and Tebo, B. M. (2006). Manganese(II)-oxidizing Bacillus spores in Guaymas Basin hydrothermal sediments and plumes. Appl. Environ. Microbiol. 72, 3184–3190. doi: 10.1128/AEM.72.5.3184-3190.2006
Donachie, S. P., Bowman, J. P., On, S. L. W., and Alam, M. (2005). Arcobacter halophilus sp. nov., the first obligate halophile in the genus Arcobacter. Int. J. Syst. Evol. Microbiol. 55, 1271–1277. doi: 10.1099/ijs.0.63581-0
Flores, G. E., Campbell, J. H., Kirshtein, J. D., Meneghin, J., Podar, M., Steinberg, J. I., et al. (2011). Microbial community structure of hydrothermal deposits from geochemically different vent fields along the Mid-Atlantic Ridge. Environ. Microbiol. 13, 2158–2171. doi: 10.1111/j.1462-2920.2011.02463.x
Flores, G. E., Wagner, I. D., Liu, Y., and Reysenbach, A. L. (2012). Distribution, abundance, and diversity patterns of the thermoacidophilic “deep-sea hydrothermal vent euryarchaeota 2”. Front. Microbiol. 3:47. doi: 10.3389/fmicb.2012.00047
Forrest, M. J., Ledesma-Vazquez, J., Ussler, I, W., Kulongoski, J. T., Hilton, D. R., and Greene, H. G. (2005). Gas geochemistry of a shallow submarine hydrothermal vent associated with the El Requeson fault zone, Bahia Concepcion, Baja California Sur, Mexico. Chem. Geol. 224, 82–95. doi: 10.1016/j.chemgeo.2005.07.015
German, C. R., and Von Damm, K. L. (2003). “Hydrothermal processes,” in Treatise on Geochemistry, eds H. D. Holland and K. K. Turekian (Amsterdam: Elsevier), 145–180.
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Handley, K. M., Hery, M., and Lloyd, J. R. (2009). Redox cycling of arsenic by the hydrothermal marine bacterium Marinobacter santoriniensis. Environ. Microbiol. 11, 1601–1611. doi: 10.1111/j.1462-2920.2009.01890.x
Huber, T., Faulkner, G., and Hugenholtz, P. (2004). Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20, 2317–2319. doi: 10.1093/bioinformatics/bth226
Italiano, F., Bonfanti, P., Ditta, M., Petrini, R., and Slejko, F. (2009). Helium and carbon isotopes in the dissolved gases of Friuli Region (NE Italy): geochemical evidence of CO2 production and degassing over a seismically active area. Chem. Geol. 266, 76–85. doi: 10.1016/j.chemgeo.2009.05.022
Italiano, F., and Nuccio, F. (1991). Geochemical investigations of submarine volcanic exhalations to the east of Panarea, Aeolian Islands, Italy. J. Volcanol. Geother. Res. 46, 125–141. doi: 10.1016/0377-0273(91)90079-F
Itoh, T., Nomura, N., and Sako, Y. (2003). Distribution of 16S rRNA introns among the family Thermoproteaceae and their evolutionary implications. Extremophiles 7, 229–233.
Itoh, T., Yoshikawa, N., and Takashina, T. (2007). Thermogymnomonas acidicola gen. nov., sp. nov., a novel thermoacidophilic, cell wall-less archaeon in the order Thermoplasmatales, isolated from a solfataric soil in Hakone, Japan. Int. J. Syst. Evol. Microbiol. 57, 2557–2561. doi: 10.1099/ijs.0.65203-0
Jochimsen, B., Peinemann-Simon, S., Volker, H., Stuben, D., Botz, R., Stoffers, P., et al. (1997). Stetteria hydrogenophila gen. nov. and sp. nov., a novel mixotrophic sulfur-dependent crenarchaeote isolated from Milos, Greece. Extremophiles 1, 67–73. doi: 10.1007/s007920050016
Kirchman, D. L. (2002). The ecology of Cytophata-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39, 91–100. doi: 10.1016/S0168-6496(01)00206-9
Kormas, K. A., Tivey, M. K., Von Damm, K. L., and Teske, A. (2006). Bacterial and archaeal phylotypes associated with distinct mineralogical layers of a white smoker spire from a deep-sea hydrothermal vent site (9°N, East Pacific Rise). Environ. Microbiol. 8, 909–920. doi: 10.1111/j.1462-2920.2005.00978.x
Koski, R. A., Pichler, T., and Foster, A. L. (2001). “The fate of arsenic in submarine hydrothermal environments: a summary of recent data,” in USGS Workshop on Arsenic in the Environment (Denver, CO).
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (Chichester: John Wiley and Sons), 115–175.
Li, S.-G., Tang, Y.-Q., Nie, Y., Cai, M., and Wu, X.-L. (2011). Complete genome sequence of Polymorphum gilvum SL003B-26A1T, a crude oil-degrading bacterium from oil polluted saline soil. J. Bacteriol. 193, 2894–2895. doi: 10.1128/JB.00333-11
Maugeri, T. L., Gugliandolo, C., Caccamo, D., Panico, A., Lama, L., Gambacorta, A., et al. (2002). A halophilic thermotolerant Bacillus isolated from a marine hot spring able to produce a new exopolysaccharide. Biotechnol. Lett. 24, 515–519. doi: 10.1023/A:1014891431233
McCarthy, K. T., Pichler, T., and Price, R. E. (2005). Geochemistry of Champagne Hot Springs shallow hydrothermal vent field and associated sediments, Dominica, Lesser Antilles. Chem. Geol. 224, 55–68. doi: 10.1016/j.chemgeo.2005.07.014
Meyer-Dombard, D., Amend, J. P., and Osburn, M. R. (2013). Microbial diversity and potential for arsenic and iron biogeochemical cycling at an arsenic-rich, shallow-sea hydrothermal vent (Tutum Bay, Papua New Guinea). Chem. Geol. 348, 37–47. doi: 10.1016/j.chemgeo.2012.02.024
Meyer-Dombard, D. R., Price, R. E., Pichler, T., and Amend, J. P. (2011). Prokaryotic populations in arsenic-rich shallow-sea hydrothermal sediments of Ambitle Island, Papua New Guinea. Geomicrobiol. J. 29, 1–17.
Morinaga, Y., Nomura, N., and Sako, Y. (2002). Population dynamics of archaeal mobile introns in natural environments: a shrewd invasion strategy of the latent parasitic DNA. Microbes Environ. 17, 153–163. doi: 10.1264/jsme2.17.153
Nakagawa, S., Takai, K., Inagaki, F., Chiba, H., Ishibashi, J.-I., Kataoka, S., et al. (2005). Variability in microbial community and venting chemistry in a sediment-hosted backarc hydrothermal system: impacts of subseafloor phase-separation. FEMS Microbiol. Ecol. 54, 141–155. doi: 10.1016/j.femsec.2005.03.007
Nakano, M. M., and Zuber, P. (1998). Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52, 165–190. doi: 10.1146/annurev.micro.52.1.165
Nitzsche, K. (2010). Microbial Diversity of Hydrothermally Influenced Arsenic-Rich Sediments off the Coast of Milos Island, Greece. Diploma thesis, TU Bergakademie, Freiberg, Germany.
Nunoura, T., Oida, H., Miyazaki, M., and Suzuki, Y. (2008). Thermosulfidibacter takaii gen. nov., sp. nov., a thermophilic, hydrogen-oxidizing, sulfur-reducing chemolithoautotroph isolated from a deep-sea hydrothermal field in the Southern Okinawa Trough. Int. J. Syst. Evol. Microbiol. 58, 659–665. doi: 10.1099/ijs.0.64615-0
Nunoura, T., and Takai, K. (2009). Comparison of microbial communities associated with phase-separation-induced hydrothermal fluids at the Yonaguni Knoll IV hydrothermal field, the Southern Okinawa Trough. FEMS Microbiol. Ecol. 67, 351–370. doi: 10.1111/j.1574-6941.2008.00636.x
Oremland, R. S., and Stolz, J. F. (2003). The ecology of arsenic. Science 300, 939–944. doi: 10.1126/science.1081903
Pati, A., Gronow, S., Lapidus, A., Copeland, A., Glavina Del Rio, T., Nolan, M., et al. (2010). Complete genome sequence of Arcobacter nitrofigilis type strain (CI). Stand. Genomic. Sci. 2, 300–308. doi: 10.4056/sigs.912121
Pichler, T. (2005). Stable and radiogenic isotopes as tracers for the origin, mixing and subsurface history of fluids in submarine shallow-water hydrothermal systems. J. Volcanol. Geother. Res. 139, 211–226. doi: 10.1016/j.jvolgeores.2004.08.007
Pichler, T., Amend, J. P., Garey, J., Hallock, P., Hsia, N. P., Karlen, D. J., et al. (2006). A natural laboratory to study arsenic geobiocomplexity. EOS 87, 221–225. doi: 10.1029/2006EO230002
Pichler, T., Giggenbach, W. F., McInnes, B. I. A., and Duck, B. (1999). Fe-sulfide formation due to seawater-gas-sediment interaction in a shallow-water hydrothermal system, Lihir Island, Papua New Guinea. Econ. Geol. 94, 281–288. doi: 10.2113/gsecongeo.94.2.281
Pokrovski, G. S., Zakirov, I. V., Roux, J., Testemale, D., Hazemann, J. L., Bychkov, A., et al. (2002). Experimental study of arsenic speciation in vapor phase to 500°C: implications for As transport and fractionation in low-density crustal fluids and volcanic gases. Geochim. Cosmochim. Acta 66, 3453–3480. doi: 10.1016/S0016-7037(02)00946-8
Polz, M. F., and Cavanaugh, C. M. (1995). Dominance of one bacterial phylotype at a Mid-Atlantic ridge hydrothermal vent site. Proc. Natl. Acad. Sci. U.S.A. 92, 7232–7236. doi: 10.1073/pnas.92.16.7232
Price, R. E., Amend, J. P., and Pichler, T. (2007). Enhanced geochemical gradients in a marine shallow-water hydrothermal system: unusual arsenic speciation in horizontal and vertical pore water profiles. Appl. Geochem. 22, 2595–2605. doi: 10.1016/j.apgeochem.2007.06.010
Price, R. E., Savov, I., Planer-Friedrich, B., Bühring, S., Amend, J. P., and Pichler, T. (2012). Processes influencing extreme As enrichment in shallow-sea hydrothermal fluids of Milos Island, Greece. Chem. Geol. 348, 15–26. doi: 10.1016/j.chemgeo.2012.06.007
Reysenbach, A. L., Liu, Y., Banta, A. B., Beveridge, T. J., Kirshtein, J. D., Schouten, S., et al. (2006). A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nat. Lett. 442, 444–447. doi: 10.1038/nature04921
Rogers, K. L., and Amend, J. P. (2005). Archaeal diversity and geochemical energy yields in a geothermal well on Vulcano Island, Italy. Geobiology 3, 319–332. doi: 10.1111/j.1472-4669.2006.00064.x
Schlesner, H., Lawson, P. A., Collins, M. D., Weiss, N., Wehmeyer, U., Voelker, H., et al. (2001). Filobacillus milensis gen. nov., sp. nov., a new halophilic spore-forming bacterium with Orn-D-Glu-type peptidoglycan. Int. J. Syst. Evol. Microbiol. 51, 425–431.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schrenk, M. O., Kelley, D. S., Bolton, S. A., and Baross, J. A. (2004). Low archaeal diversity linked to subseafloor geochemical processes at the Lost City hydrothermal field, Mid-Atlantic Ridge. Environ. Microbiol. 6, 1086–1095. doi: 10.1111/j.1462-2920.2004.00650.x
Seeberg-Elverfeldt, J., Schlüter, M., Feseker, T., and Kölling, M. (2005). Rhizon sampling of pore waters near the sediment/water interface of aquatic systems. Limnol. Oceanogr. Methods 3, 361–371. doi: 10.4319/lom.2005.3.361
Sievert, S. M., Brinkhoff, T., Muyzer, G., Ziebis, W., and Kuever, J. (1999). Spatial heterogeneity of bacterial population along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island, Greece. Appl. Environ. Microbiol. 65, 3834–3842.
Sievert, S. M., Heidorn, T., and Kuever, J. (2000a). Halothiobacillus kellyi sp. nov., a mesophilic obligately chemolithoautotrophic sulfur-oxidizing bacterium isolated from a shallow-water hydrothermal vent in the Aegean Sea and emended description of the genus Halothiobacillus. Int. J. Syst. Evol. Microbiol. 50, 1229–1237. doi: 10.1099/00207713-50-3-1229
Sievert, S. M., Kuever, J., and Muyzer, G. (2000b). Identification of 16S ribosomal DNA-defined bacterial populations at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 66, 3102–3109. doi: 10.1128/AEM.66.7.3102-3109.2000
Sievert, S. M., and Kuever, J. (2000). Desulfacinum hydrothermale sp. nov., a thermophilic sulfate-reducing bacterium from geothermally heated sediments near Milos island, Greece. Int. J. Syst. Evol. Microbiol. 50, 1239–1246. doi: 10.1099/00207713-50-3-1239
Spano, A., Gugliandolo, C., Lentini, V., Maugeri, T. L., Anzelmo, G., Annarita, P., et al. (2013). A novel EPS-producing strain of Bacillus licheniformis isolated from a shallow vent off Panarea Island (Italy). Curr. Microbiol. 67, 21–29. doi: 10.1007/s00284-013-0327-4
Stüben, D., and Glasby, G. P. (1999). Geochemistry of shallow submarine hydrothermal fluids of Paleochori Bay, Milos, Aegean Sea. Explor. Mining Geol. 8, 273–287.
Switzer, B. J., Burns, B. A., Buzzelli, J., Stolz, J. F., and Oremland, R. S. (1998). Bacillus arsenicoselenatis sp. nov., and Bacillus selentireducens sp. nov.: two haloalkaliphilies from Mono Lake, California that respire oxyanions of selenium and arsenic. Arch. Microbiol. 171, 19–30. doi: 10.1007/s002030050673
Sylvan, J. B., Toner, B. M., and Edwards, K. J. (2012). Life and death of deep-sea vents: bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. mBio 3, 1–10. doi: 10.1128/mBio.00279-11
Takai, K., and Horikoshi, K. (1999). Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152, 1285–1297.
Tarasov, V. G., Gebruk, A. V., Mironov, A. N., and Moskalev, L. I. (2005). Deep-sea and shallow-water hydrothermal vent communities: two different phenomena? Chem. Geol. 224, 5–39. doi: 10.1016/j.chemgeo.2005.07.021
Tarasov, V. G., Propp, M. V., Propp, L. N., Zhirmunsky, A. V., Namsaraev, B. B., Gorlenko, V. M., et al. (1990). Shallow-water gasohydrothermal vents of Ushishir Volcano and the ecosystem of Kraternaya Bight (The Kurile Islands). Mar. Ecol. 11, 1–23. doi: 10.1111/j.1439-0485.1990.tb00225.x
Valsami-Jones, E., Baltatzis, E., Bailey, E. H., Boyce, A. J., Alexander, J. L., Magganas, A., et al. (2005). The geochemistry of fluids from an active shallow submarine hydrothermal system: Milos island, Hellenic Volcanic Arc. J. Volcanol. Geothermal Res. 148, 130–151. doi: 10.1016/j.jvolgeores.2005.03.018
Varnavas, S. P., and Cronan, D. S. (1988). Arsenic, antimony and bismuth in sediments and waters from the Santorini hydrothermal field, Greece. Chem. Geol. 67, 295–305. doi: 10.1016/0009-2541(88)90135-0
Von Damm, K. L., Lilley, M. D., Shanks, W. C., Brockington, M., Bray, A. M., O'Grady, K. M., et al. (2003). Extraordinary phase separation and segregation in vent fluids from the southern East Pacific Rise. Earth Planet. Sci. Lett. 206, 365–378. doi: 10.1016/S0012-821X(02)01081-6
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wirsen, C. (2004). Is life thriving deep beneath the seafloor? Oceanus 42, 1–6. Available online at: http://oceanusmag.whoi.edu/v42n2/wirsen.html
Zargar, K., Conrad, A., Bernick, D. L., Lowe, T. M., Stolc, V., Hoeft, S., et al. (2012). ArxA, a new clade of arsenite oxidase within the DMSO reductase family of molybdenum oxidoreductases. Environ. Microbiol. 14, 1635–1645. doi: 10.1111/j.1462-2920.2012.02722.x
Zargar, K., Hoeft, S. E., Oremland, R. S., and Saltikov, C. (2010). Identification of a novel arsenite oxidase gene, arxA, in the haloalkaliphilic, arsenite-oxidizing bacterium Alkalilimnicola ehrlichii strain MLHE-1. J. Bacteriol. 192, 3755–3762. doi: 10.1128/JB.00244-10
Appendix
Keywords: microbial diversity, arsenic, hydrothermal, gradients, milos, phase separation
Citation: Price RE, Lesniewski R, Nitzsche KS, Meyerdierks A, Saltikov C, Pichler T and Amend JP (2013) Archaeal and bacterial diversity in an arsenic-rich shallow-sea hydrothermal system undergoing phase separation. Front. Microbiol. 4:158. doi: 10.3389/fmicb.2013.00158
Received: 15 March 2013; Accepted: 30 May 2013;
Published online: 09 July 2013.
Edited by:
Anna-Louise Reysenbach, Portland State University, USACopyright © 2013 Price, Lesniewski, Nitzsche, Meyerdierks, Saltikov, Pichler and Amend. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Roy E. Price, Department of Earth Sciences, University of Southern California, 3651 Trousdale Pkwy, Los Angeles, 90089 CA, USA e-mail: royprice@usc.edu