- 1Paul G. Allen School for Global Animal Health, Washington State University, Pullman, WA, USA
- 2Department of Veterinary Microbiology and Pathology, Washington State University, Pullman, WA, USA
- 3Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, Scotland, UK
- 4Department of Veterinary Integrative Biosciences, Texas A&M University, College Station, TX, USA
When we consider factors that contribute to the emergence, amplification, and persistence of antibiotic resistant bacteria, the conventional assumption is that antibiotic use is the primary driver in these processes and that selection occurs primarily in the patient or animal. Evidence suggests that this may not always be the case. Experimental trials show that parenteral administration of a third-generation cephalosporin (ceftiofur) in cattle has limited or short-term effects on the prevalence of ceftiofur-resistant bacteria in the gastrointestinal tract. While this response may be sufficient to explain a pattern of widespread resistance to cephalosporins, approximately two-thirds of ceftiofur metabolites are excreted in the urine raising the possibility that environmental selection plays an important additive role in the amplification and maintenance of antibiotic resistant E. coli on farms. Consequently, we present a rationale for an environmental selection hypothesis whereby excreted antibiotic residues such as ceftiofur are a significant contributor to the proliferation of antibiotic resistant bacteria in food animal systems. We also present a mathematical model of our hypothesized system as a guide for designing experiments to test this hypothesis. If supported for antibiotics such as ceftiofur, then there may be new approaches to combat the proliferation of antibiotic resistance beyond the prudent use mantra.
Introduction
Since first being introduced in the 1940's, antibiotics have improved or saved the lives of countless millions of people either directly through disease prevention and treatment, or indirectly by enabling greater food production capacity. Unfortunately, antibiotic use of any kind invariably selects for the emergence, amplification, and persistence of resistant bacteria. The evolutionary dynamics of antibiotic resistance differ depending on how different resistance traits function. For example, when antibiotic resistance involves active efflux or enzymatic degradation pathways, we might expect longer periods to elapse between the introduction of a new antibiotic and adaptation of resistance mechanisms. Of course, naturally produced antibiotics have been selecting for emergence of resistance mechanisms as long as antibiotics have existed in nature (Allen et al., 2010). Consequently, if resistance mechanisms exist, these toolboxes will eventually be co-opted by pathogens. In contrast, antibiotics such as rifamycins, quinolones, and fluoroquinolones bind specific proteins where simple chromosomal mutations are sufficient to alter the binding sites and produce clinically relevant resistance. Acquisition of these mutations can occur during antibiotic exposure, and resistant organisms can subsequently sweep through populations that experience drug selection pressure (Humphrey et al., 2005). Despite the relative ease with which these chromosomal mutations can arise for quinolones, and by extension fluoroquinolones, more complex resistance mechanisms of resistance have already been documented in the field (Xia et al., 2010).
There are several strategies to address the challenge of antibiotic resistance with introduction of new antibiotics being an important avenue in the past 50 years. Aside from the multitude of challenges that now limit development of new antibiotic products (Spellberg et al., 2008), there are fewer completely novel antibiotics being developed and thus we can expect less time to pass between introduction of new products and emergence of widespread antibiotic resistance. For example, a recent review identified 20 new antibiotics that are currently working through product development pipelines (Butler and Cooper, 2011). Of these, 9 are synthetic compounds and 8 of these are quinolones for which we can predict relatively rapid emergence of resistance. Consequently, it is unlikely that we will stay ahead of this problem in the future through new drug development alone.
“Prudent use” is an important policy-based strategy to combat the growing antibiotic resistance challenge by ensuring that antibiotics are only used when they are actually needed, that the most appropriate antibiotic is used for a given disease agent, and that exacting dosage guidelines are followed (Gyssens, 2011). While emergence of antibiotic resistance is inevitable, prudent use practices are likely to limit the overall level of drug selection pressure and consequently limit the equilibrium prevalence of resistant pathogens (Austin et al., 1999). Greater attention to biosecurity and infection prevention measures, use of rapid diagnostics, and use of effective vaccines and probiotics should also reduce disease incidence and thus reduce demand for antibiotics.
Attention to prudent use invariably brings scrutiny to antibiotic use practices in food animal production. From a “mass-action” perspective, more antibiotics are used in food animal production than in human medicine and consequently this sector may contribute disproportionately to development of antibiotic resistance (Sarmah et al., 2006). In the U.S. an estimated 3.3 million kg of antibiotics were sold for human use in 2010 (FDA, 2012b) while 12.2 million kg (FDA, 2011) were sold for use in food animal applications. There are a number of reasons why we should be cautious about directly comparing these numbers (FDA, 2012a), but it is important to note that monensins account for 28.9% of the total sales for use in food animals. These ionophores are entirely unique from antibiotics that are used in people and they function by transporting ions across bacterial cell membranes thereby disrupting ion gradients and killing susceptible bacteria (Callaway et al., 2003). Resistance to monensins has been suggested, but it is not clear if this is a function of intrinsically resistant populations dominating in a community exposed to monensins, or due to emergence of novel resistance mechanisms. To date, no genetically-encoded, horizontally transmissible resistance traits have been described for monensins (Callaway et al., 2003) and there is no recognized means by which use of monensins contribute to selection for other antibiotic resistance traits in pathogenic or commensal bacteria.
The largest component of antibiotic sales for food animals includes oxytetracycline and chlortetracycline (42.2%) (FDA, 2011) and these are mostly used as in-feed additives to promote animal health and growth. It could be argued that because tetracyclines make-up a relatively small percentage of demand in human medicine (3.9%) (FDA, 2012a), resistance to these drugs has a limited potential to impact human health and thus these compounds are a “good” choice relative to other options that might be employed. An important caveat to this conclusion is that tetracycline resistance is commonly associated with multidrug resistant bacteria (FDA, 2012c) and thus selection that favors tetracycline resistance will also co-select for other genetically-linked antibiotic resistance traits. Co-selection of this nature has been described for other antibiotics and toxins such as heavy metals (Stepanauskas et al., 2006; Tremblay et al., 2012).
The therapeutic use of veterinary antibiotics, while representing a lower total mass of antibiotics, is also heavily scrutinized as a contributor to the antibiotic resistance crisis. In the U.S. this is probably best exemplified by fluoroquinolone use in poultry production. Flock-wide treatment (metaphylactic) with an antibiotic such as enrofloxacin (veterinary antibiotic) exerts selective pressure for resistance to the antibiotic ciprofloxacin (human antibiotic) in Campylobacter jejuni, which is a non-target bacterium in this application. Because simple chromosomal mutations quickly lead to ciprofloxacin resistance and because poultry is a major reservoir for transmission of C. jejuni to people, fluoroquinolones are no longer approved for use as a metaphylactic treatment via poultry water (FDA, 2005). Unfortunately, despite this withdrawal in 2005, the prevalence of ciprofloxacin resistant C. jejuni has remained >20% by 2010 for poultry and human clinical isolates alike in the U.S. (FDA, 2012c). It is unclear if this persistence is due to alternative exposure routes or due to resistant strains disseminating via food and travel. Some mutations that convey resistance to ciprofloxacin might also be fitness neutral or they might convey a fitness advantage to the bacteria. If so, the prevalence of ciprofloxacin resistant C. jejuni is unlikely to decrease significantly even after cessation of enrofloxacin use in poultry medicine (Luo et al., 2005).
Another important therapeutic antibiotic in production medicine is ceftiofur. This third-generation cephalosporin is widely used as an injectable antibiotic to treat respiratory infections, metritis, and pododermatitis in cattle, but it has also been used in swine, small ruminants, and poultry. Ceftiofur gained rapid acceptance since the 1990's, particularly in the dairy sector (Zwald et al., 2004; Sawant et al., 2005; Sarmah et al., 2006; Saini et al., 2012) because it is a very effective antibiotic with no withholding time for milk production. Nevertheless, increasing resistance to third-generation cephalosporins in Salmonella recently led the US Food and Drug Administration to adopt new rules banning non-therapeutic and extra-label uses of this important veterinary antibiotic (FDA, 2012d). There is debate as to whether or not these regulatory changes will have a significant impact on use practices (Wittum, 2012), and as we outline below these changes are likely to have little impact on the amplification of resistant enteric bacteria if amplification of resistant populations is primarily driven by excreted ceftiofur metabolites in the environment.
Environmental Fate of Antibiotics
Ceftiofur poses an interesting conundrum because while resistance is prevalent in E. coli and Salmonella in the U.S. (Winokur et al., 2000; Donaldson et al., 2006; Daniels et al., 2007, 2009; Sawant et al., 2007; Heider et al., 2009; Lindsey et al., 2009), empirical studies show either no treatment effect (Singer et al., 2008; Daniels et al., 2009; Mann et al., 2011) or transient and relatively short-term amplification of resistant populations (Jiang et al., 2006; Lowrance et al., 2007) after administration of ceftiofur. Lowrance et al. (2007) provided the most convincing evidence that selection occurs in the gastrointestinal tract where there was a proportional increase in the number of ceftiofur-resistant E. coli following 1, 2, or 3 days administration of a ceftiofur crystalline-free acid product. This study included 61 feedlot steers and resistant E. coli were found between 1 and 2 log higher densities compared with control animals, and the effects lasted ~2 weeks. Two-weeks might be considered a relatively short-term impact because it is also consistent with a failure of survey studies to find a relationship between the percentage of cows with ceftiofur-resistant E. coli and the percentage of cows treated at the herd level (Tragesser et al., 2006; Daniels et al., 2009; Heider et al., 2009). Jiang et al. (2006) reported an ~0.5 log increase in the number of ceftiofur-resistant E. coli immediately following treatment of three calves with a ceftiofur hydrochloride product. In contrast, Mann et al. (2011) reported no significant effects on the proportion of ceftiofur-resistant E. coli after administration of a ceftiofur hydrochloride product (n = 42 animals). Singer et al. (2008) also reported no change in the number of ceftiofur resistant E. coli (n = 10 animals). These latter studies did report a significant decrease in total E. coli following ceftiofur administration. Thus, if ceftiofur administration impacts the intestinal flora, amplification of ceftiofur-resistant E. coli populations is variable with outcomes ranging from no detectable effect to 1–2 log increases probably depending, in part, on the dose (e.g., 2.2 mg/kg vs. 6.6 mg/kg) and number of sequential treatments.
Given the inconsistent and relatively short-term effects of ceftiofur treatment on E. coli populations in the gut, this raises the question of whether selection in vivo represents the entire story. For example, resistant strains may be originating from other sources and are being transported to cattle via feed or water supplies. Until recently (FDA, 2012d), ceftiofur could be used as an intramammary prophylactic treatment and this practice has been associated with an increased prevalence of ceftiofur-resistant fecal coliform bacteria (Mollenkopf et al., 2010). The authors of this latter study noted that we normally would not expect local administration of an antibiotic in cow udder to affect enteric flora, but the authors proposed that amplification of resistant bacteria and subsequent shedding into the environment and contact transmission could explain this result. In a similar manner, resistant populations of bacteria such as E. coli could be exposed to antibiotic residues in urine and feces resulting in selective amplification of the resistant populations in the environment with a subsequent increased risk of contact-dependent transmission and colonization of livestock with resistant E. coli.
For an environmental selection process to be important a sufficient concentration of biologically active compound must be present for a sufficient duration to impact the bacterial community. Furthermore, animals exposed to higher densities of resistant bacteria must be at a greater risk of contact transmission and, ultimately, the resistant bacteria must colonize new hosts. Environmental surveillance studies have typically reported that the concentration of antibiotic residues in soil and water varies between parts per trillion (ppt) and parts per billion (ppb) (Winokur et al., 2000; Tolls, 2001; Koplin et al., 2002; Thiele-Bruhn, 2003; Sawant et al., 2007; Ji et al., 2012); values typically well below the concentration needed (ppm) to demonstrate effects from antibiotics in vitro. There is speculation that bacteria could be impacted by additive or synergistic effects or that non-target organisms could be impacted at very low concentrations, but with the exception of possible effects on algae there is little evidence that ppt and ppb concentrations of antibiotics have any impact in the environment (Ji et al., 2012).
Another way to address this question is to mix combinations of antibiotics at low doses and determine if there is an additive or synergistic effect. Using a simple experiment we show here that there is no evidence for inhibition of bacterial growth in vitro until the antibiotics being used reach a concentration where we should expect inhibition of a sensitive strain (i.e., ppm) (Figure 1). There is also reason to expect that any subtle additive or synergistic effects on fitness will be even less prominent in the environment where bacteria typically reside within biofilms that are recalcitrant to antibiotics (Lewis, 2007) and where antibiotic bioavailability may be compromised (Subbiah et al., 2011).
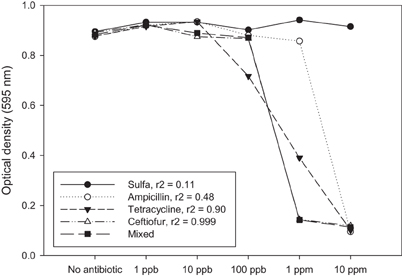
Figure 1. Antibiotic residues only affect bacteria from a fitness standpoint when the concentration of residue approaches a minimum inhibitory concentration (MIC). MIC values are typically > 1 ppm, whereas most environmental residue reported in the literature are found at ppb or ppt. These might be significant if there is an additive or synergistic effect attributed to exposure to multiple low-dose antibiotic residues. In this example, single antibiotics and a mixture of four antibiotics at the same concentration were tested against a sensitive strain of E. coli (K-12). The response to the mixture is most closely associated with ceftiofur (r2 = 0.999). If there was an additive or synergistic effect on fitness we would expect to see the mixed antibiotic (solid square) as having the lowest optical density at 100 ppb, which is not the case in this experiment. Each data point represents the optical density after 24 h growth in LB as measured using a Bioscreen plate reader. Average for three independent replicates is shown.
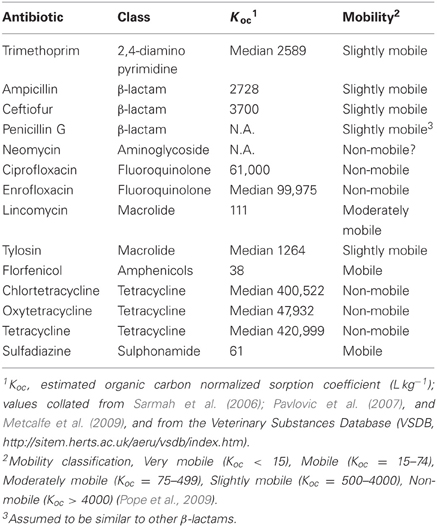
Studies of environmental residues typically measure antibiotics using analytic tools such as ELISA, HPLC, and mass spectrometry (Koplin et al., 2002; Sczesny et al., 2003; Thiele-Bruhn, 2003; Kumar et al., 2004; Aga et al., 2005; Berendsen et al., 2012); methods that can be exquisitely well-suited for detecting minimal concentrations of antibiotics, but these methods do not tell us if the antibiotic residues being detected are biologically available. To be biologically available, antibiotics need to be physically available to interact with target cells. Antibiotics such as tetracyclines, however, are known to rapidly adsorb to clay surfaces (Tolls, 2001) rendering them unavailable to exert biological effects in soils. Subbiah et al. (2011) explored this process further by mixing high concentrations of antibiotics (200 ppm) with soil slurries that differed in clay content, pH and other properties. After mixing, supernatant was recovered, filter sterilized, and added to a broth culture of an antibiotic sensitive E. coli strain. If the strain grew, this indicated that there was an insufficient concentration of antibiotic in the liquid phase to have any impact on the test strain because the antibiotic had adsorbed to the soil materials. Using this assay, tetracycline, neomycin, and ciprofloxacin had no effect on bacterial growth after mixing with sand-loam and silt-loam soils; tetracycline, ciprofloxacin, and two sulfonamide antibiotics retained at least partial activity in sand. Even when antibiotic sensitive E. coli were added to a viscous soil slurry with adsorbed tetracycline, there was no evidence of an impact on fitness (Subbiah et al., 2011). Florfenicol and β-lactams, however, were mostly available in the liquid phase and these antibiotics still affected the test bacteria. These results are mostly consistent with what could be predicted from the organic normalized dissociation constants (Koc) for these compounds (Table 1). Koc provides an imperfect prediction of how tightly antibiotics adsorb to soil (Tolls, 2001), but at the extremes tetracycline adsorbs very tightly (Koc > 420,999) while florfenicol and sulfadiazine adsorb very weakly (Koc < 38 and 61, respectively). Ceftiofur is considered slightly mobile by this measure (Table 1), but ceftiofur may exist as an anion in soils and this would increase mobility thus biological availability.
In a subsequent study Subbiah et al. (2012) focused on the fate of ceftiofur metabolites in soils. When injected into cattle, ceftiofur is rapidly metabolized with most of the metabolite being biologically active desfuroylceftiofur (Hornish and Kotarski, 2002). Jaglan et al. (1989) estimated that 70 and 30% of the metabolites are then excreted through urine and feces, respectively. Ceftiofur metabolites are mostly excreted within 24 h and the concentration varies between ~ 7 and 160 ppm in the urine (El-Gendy et al., 2007; Subbiah et al., 2012); values well within the range needed to affect sensitive bacteria in vitro. When urine containing ceftiofur metabolites was added to a soil:feces microcosm (25:1), inhibition from the antibiotic was evident for 3 days at room temperature. Biological degradation appeared to explain the loss of the residues in part because at 4°C the residues were biologically available up to 3 weeks from the start of the experiment. Importantly, exposure to urine containing ceftiofur metabolites was sufficient to produce a log greater increase in growth for a resistant strain of E. coli compared to the same strain that was exposed to urine without ceftiofur metabolites. Exposure to ceftiofur metabolites in urine also resulted in > 2 month longer retention of the resistant E. coli strain compared to the same strain in control microcosms.
In the case of ceftiofur, biologically active metabolites are excreted at a sufficient concentration that remains available to affect sensitive bacteria in soil. Clearly, the distribution of excreted residues will be very heterogeneous in a cattle herd depending on where treated animals urinate and how materials are disturbed and moved. Furthermore, for this environmental selection scenario to be feasible the amplified populations of resistant E. coli on the soil surface must present a greater risk of transmission back to naïve animals compared with conditions where resistant populations undergo no selective amplification. Subbiah et al. (2012) used a controlled experiment to show that bedding contaminated with a ceftiofur resistant strain of E. coli is sufficient to colonize naïve calves by contact transmission alone, although a wider range of concentrations and environmental conditions need to be explored to validate this component of an environmental selection scenario.
Lowrance et al. (2007) demonstrated a clear in vivo selection effect from ceftiofur that was dependent on the number injections of a ceftiofur product (a high dose per injection, 6.6 mg/kg compared to most studies, 2.2 mg/kg). Importantly, the untreated control animals in this study mingled freely with the treated animals and yet did not show a strong response to the presence of the treated animals. The control animals were clearly colonized and apparently they shed a higher concentration at the study outset (app. 3.5 Log10/g feces) compared to 2 weeks later (app. 3.0 Log10/g feces), but cause and effect for this response is confounded, in part, by prior colonization and there was no isolated control group for comparison. We further surmise that a number of factors enhance or diminish an environmental effect such as the season, floor substrate, dose delivered, number of injections, density of livestock, waste management practices, animal age, and animal behavior. As a simple example, Lowrance et al. (2007) studied beef cattle (steers) that probably bed down in contaminated substrates less frequently than young calves thereby limiting an important transmission pathway. If environmental selection is an important factor in the amplification and persistence of antibiotic resistant bacteria, and if this process only makes a significant contribution under certain environmental conditions, then it is critical to determine which factors can be modified to limit selection and transmission as much as possible.
Environmental Selection Hypothesis
Volkova et al. (2012) provided a comprehensive mathematical model of the dynamics of ceftiofur-sensitive and resistant commensal E. coli in the cow large intestine. This model considered population size and growth rate in vivo, fitness cost of plasmid carriage by resistant bacteria, and the effect of ceftiofur metabolites in the intestine (entering via bile salts). Volkova and colleagues focused on the in vivo selection compartment, but their model included an “in flow” component from the environment through which resistant and sensitive bacteria enter the cow. They concluded, in part, that the rate of replacement E. coli acquired through ingestion is an important factor when these strains are ceftiofur resistant. Volkova et al. (2013) explicitly addressed approaches for controlling plasmid-mediated resistance in enteric commensal bacteria and their model also included multiple pathways by which resistant bacteria can be ingested. Our proposed model focuses on selection in the environment based, in part, on the fact that approximately two-thirds of biologically active metabolite after ceftiofur administration is excreted in the urine and thus may be a significant factor leading to amplification of resistant bacteria that subsequently enter the Volkova model via the “in flow” parameter. Notably, neither Volkova et al. (2012) nor Volkova et al. (2013) considered the effects of excreted antibiotic in their scenarios. From a “mass action” perspective, selection in the environment may play a more significant role in this process. Nonetheless, it is possible that a more accurate model will be a hybrid of both the in vivo and ex vivo selection compartments.
We hypothesize that excreted ceftiofur contributes to amplification and persistence of resistant enteric bacteria in soils, and consequently use of these antibiotics increases the risk that resistant bacteria will be transmitted back to cattle (Figure 2). While our focus is centered on ceftiofur in these discussions, if the environmental selection hypothesis is valid then it likely applies to other excreted antibiotics that remain biologically available after contact with the ground substrate (Subbiah et al., 2011). A mathematical representation of this hypothesis can be formulated as follows:
where SH and RH represent susceptible and resistant strains in the host, and SE and RE represent susceptible and resistant strains in the urine contaminated environment, respectively. We are initially assuming that there is no selection by ceftiofur in the host, that the host population ingests bacteria from the environment at a rate α and excretes bacteria into the whole environment at rate λ, and that a proportion f of bacteria is excreted into areas contaminated with urine.
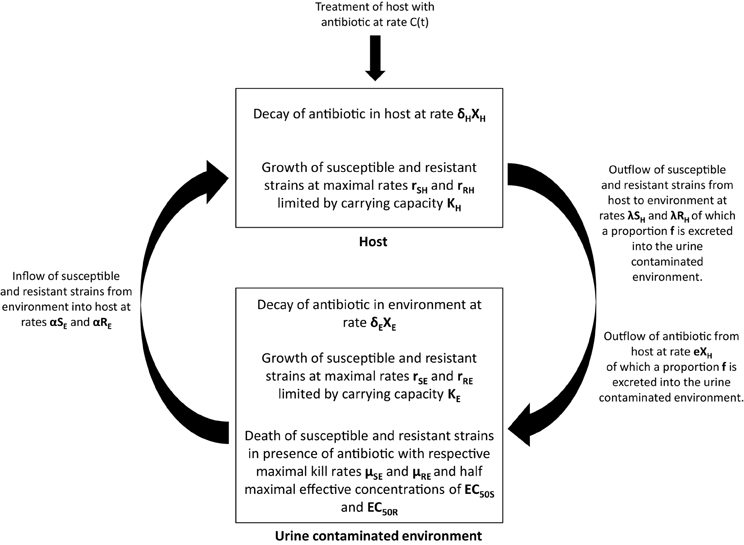
Figure 2. Graphical depiction of the environmental selection hypothesis by which excreted antibiotic metabolites selectively amplify resistant bacteria in the environment. Amplification of resistant bacteria subsequently increases the probability of transmission back to the host. See text for details about model parameters.
Within the host, the net replication rate of the strains is given by rSH and rRH, and the total size of the resistant and susceptible population is limited by a carrying capacity KH. Within the urine contaminated environment, the dynamics of the susceptible and resistant populations are similarly dependent on the ingestion and excretion of strains by the host and by growth that is limited by a carrying capacity KE. If in vivo selection proves to be a more significant selection factor than we presume based on the literature, then an in vivo selection parameter can be added to the model.
In addition, the dynamics in the urine contaminated environment also include terms that capture antibiotic induced death of the resistant and susceptible strains, where μSE and μRE are the maximal kill rates, XE, the concentration of antibiotic in the environment, γ the Hill coefficient, and EC50S and EC50R the half maximal effective concentration for the susceptible and resistant strains, respectively (Vinks, 2002). The concentration of antibiotic in the host is determined by a treatment rate, C(t), a decay rate in the host, δH, and an excretion rate into the environment, e.
To test the environmental selection hypothesis for ceftiofur we propose to compare the rates of acquisition of resistance among calves in groups including both ceftiofur treated and untreated animals. The calves would initially be presumptively free of ceftiofur-resistant E. coli and they would be introduced into an environment where ceftiofur resistant E. coli are present. We propose to then compare acquisition rates in groups that differ in the proportions of treated animals. Our hypothesis predicts a more rapid acquisition of resistant strains in groups with a higher proportion of treated animals because greater excretion of ceftiofur will allow greater environmental selection, but no difference between the treated and untreated animals within a group. Specifically, by monitoring animals for colonization, we can obtain estimates for αRE, the rate of acquisition of resistant strains, and compare these estimates between groups and between treated and untreated animals. Additionally, comparing environmental concentrations of resistant strains between groups with high and low proportions of treated animals, and relating these concentrations to acquisition rates, would enable us to estimate the uptake parameter α.
Volkova et al. (2012) describes the mechanism by which resistance may persist within the host via plasmid mediated transfer, whilst the model of Volkova et al. (2013) additionally captures plasmid-mediated transfer in the environment. Our goal, however, is to specifically assess whether environmental selection of strains in the presence of ceftiofur is a more important source of resistance acquisition than within host selection.
Conclusion
Antibiotic resistance is an increasing challenge to public health worldwide. Currently, our only tools to combat this challenge include developing new antibiotics and preserving the utility of existing antibiotics as long as possible through prudent use principles. Our work suggests that blocking environmental selection could be another important avenue to combat resistance to some antibiotics that are used in food animal medicine. If environmental selection proves to be a significant contributor to maintenance of antibiotic resistance for drugs like ceftiofur (e.g., florfenicol), then it is likely that management and engineered solutions can be devised to limit this component of the selection problem. Importantly, by finding solutions to address unappreciated components of the selection process such as the one described here, it should be possible to extend the utility of important drugs like ceftiofur for use in food animal production. Doing so benefits animal welfare and public health while helping to maintain lower costs for producing food for the burgeoning human population.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Technical assistance was provided by Lisa Orfe. This work was supported in part by the Paul G. Allen School for Global Animal Health, Washington State University, Pullman, WA, and by the Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, Scotland.
References
Aga, D. S., O'Connor, S., Ensley, S., Payero, J. O., Snow, D., and Tarkalson, D. (2005). Determination of the persistence of tetracycline antibiotics and their degradates in manure-amended soil using enzyme-linked immunosorbent assay and liquid chromatography-mass spectrometry. J. Agric. Food Chem. 53, 7165–7171. doi: 10.1021/jf050415
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
Austin, D. J., Kristinsson, K. G., and Anderson, R. M. (1999). The relationship between the volume of antimicrobial consumption in human communities and the frequency of resistance. Proc. Natl. Acad. Sci. U.S.A. 96, 1152–1156. doi: 10.1073/pnas.96.3.1152
Berendsen, B. J., Stolker, L. A., and Nielen, M. W. (2012). Assessment of liquid chromatography-tandem mass spectrometry approaches for the analysis of ceftiofur metabolites in poultry muscle. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 29, 197–207. doi: 10.1080/19440049.2011.635348
Butler, M. S., and Cooper, M. A. (2011). Antibiotics in the clinical pipeline in (2011). J. Antibiot. (Tokyo) 64, 413–425. doi: 10.1038/ja.2011.44
Callaway, T. R., Edrington, T. S., Rychlik, J. L., Genovese, K. J., Poole, T. L., Jung, Y. S., et al. (2003). Ionophores: their use as ruminant growth promotants and impact on food safety. Curr. Issues Intest. Microbiol. 4, 43–51.
Daniels, J. B., Call, D. R., and Besser, T. E. (2007). Molecular epidemiology of blaCMY − 2 plasmids carried by Salmonella enterica and Escherichia coli isolates from cattle in the Pacific Northwest. Appl. Environ. Microbiol. 73, 8005–8011. doi: 10.1128/AEM.01325-07
Daniels, J. B., Call, D. R., Hancock, D., Sischo, W. M., Baker, K., and Besser, T. E. (2009). Role of ceftiofur in selection and dissemination of blaCMY − 2-mediated cephalosporin resistance in Salmonella enterica and commensal Escherichia coli isolates from cattle. Appl. Environ. Microbiol. 75, 3648–3655. doi: 10.1128/AEM.02435-08
Donaldson, S. C., Straley, B. A., Hegde, N. V., Sawant, A. A., DebRoy, C., and Jayarao, B. M. (2006). Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves. Appl. Environ. Microbiol. 72, 3940–3948. doi: 10.1128/AEM.02770-05
El-Gendy, A. A. M., Tohamy, M. A., and Ismail, M. (2007). Comparative pharmacokinetic and renal clearance study of ceftiofur in cross breed Friesian and buffalo calves. BS Vet. Med. J. 17, 69–77.
FDA. (2005). Enrofloxacin for poultry; final decision on withdrawal of new animal drug application following formal evidentiary public hearing. Fed. Regist. 70, 44105.
FDA. (2011). 2010 Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals. Available online at: http://www.fda.gov/downloads/ForIndustry/UserFees/AnimalDrugUserFeeActADUFA/ucm277657.pdf: Department of Health and Human Services, Food and Drug Administration, Center for Veterinary Medicine
FDA. (2012a). Drug Use Review. Available online at: http://www.fda.gov/downloads/Drugs/DrugSafety/InformationbyDrugClass/UCM319435.pdf: Department of Health and Human Services, Public Health Service, Food and Drug Administration, Center for Drug Evaluation and Research, Office of Surveillance and Epidemiology
FDA. (2012b). Estimates of Antibacterial Drug Sales in Human Medicine. http://www.fda.gov/Drugs/DrugSafety/InformationbyDrugClass/ucm261160.htm: Department of Health and Human Services, Food and Drug Administration
FDA. (2012c). National Antimicrobial Resistance Monitoring System—Enteric Bacteria (NARMS): 2010 Executive Report. Rockville, MD: Department of Health and Human Services, Food and Drug Administration. Available online at: http://www.fda.gov/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/ucm312356.htm
FDA. (2012d). New animal drugs; cephalosporin drugs; extralabel animal drug use; order of prohibition. Fed. Regist. 77, 735–745.
Gyssens, I. C. (2011). Antibiotic policy. Int. J. Antimicrob. Agents 38(Suppl.), 11–20. doi: 10.1016/j.ijantimicag.2011.09.002
Heider, L. C., Funk, J. A., Hoet, A. E., Meiring, R. W., Gebreyes, W. A., and Wittum, T. E. (2009). Identification of Escherichia coli and Salmonella enterica organisms with reduced susceptibility to ceftriaxone from fecal samples of cows in dairy herds. Am. J. Vet. Res. 70, 389–393. doi: 10.2460/ajvr.70.3.389
Hornish, R. E., and Kotarski, S. F. (2002). Cephalosporins in veterinary medicine—ceftiofur use in food animals. Curr. Top. Med. Chem. 2, 717–731. doi: 10.2174/1568026023393679
Humphrey, T. J., Jorgensen, F., Frost, J. A., Wadda, H., Domingue, G., Elviss, N. C., et al. (2005). Prevalence and subtypes of ciprofloxacin-resistant Campylobacter spp. in commercial poultry flocks before, during, and after treatment with fluoroquinolones. Antimicrob. Agents Chemother. 49, 690–698. doi: 10.1128/AAC.49.2.690-698.2005
Jaglan, P. S., Kubicek, M. F., Arnold, T. S., Cox, B. L., Robins, R. H., Johnson, D. B., et al. (1989). Nature of urinary and plasma metabolites in rates and cattle. J. Agric. Food Chem. 37, 1112–1118. doi: 10.1021/jf00088a066
Ji, K., Kim, S., Han, S., Seo, J., Lee, S., Park, Y., et al. (2012). Risk assessment of chlortetracycline, oxytetracycline, sulfamethazine, sulfathiazole, and erythromycin in aquatic environment: are the current environmental concentrations safe? Ecotoxicology 21, 2031–2050. doi: 10.1007/s10646-012-0956-6
Jiang, X., Yang, H., Dettman, B., and Doyle, M. P. (2006). Analysis of fecal microbial flora for antibiotic resistance in ceftiofur-treated calves. Foodborne Pathog. Dis. 3, 355–365. doi: 10.1089/fpd.2006.3.355
Koplin, D. W., Furlong, E. T., Meyer, M. T., Thurman, E. M., Zaugg, S. D., Barber, L. B., et al. (2002). Pharmaceuiticals, hormones, and other organic wastewater contaminants in streams, U.S., 1999–2000, a national reconnaissance. Environ. Sci. Technol. 36, 1202–1211. doi: 10.1021/es011055j
Kumar, K., Thompson, A., Singh, A. K., Chander, Y., and Gupta, S. C. (2004). Enzyme-linked immunosorbent assay for ultratrace determination of antibiotics in aqueous samples. J. Environ. Qual. 33, 250–256. doi: 10.2134/jeq2004.0250
Lewis, K. (2007). Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5, 48–56. doi: 10.1038/nrmicro1557
Lindsey, R. L., Fedorka-Cray, P. J., Frye, J. G., and Meinersmann, R. J. (2009). Inc A/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl. Environ. Microbiol. 75, 1908–1915. doi: 10.1128/AEM.02228-08
Lowrance, T. C., Loneragan, G. H., Kunze, D. J., Platt, T. M., Ives, S. E., Scott, H. M., et al. (2007). Changes in antimicrobial susceptibility in a population of Escherichia coli isolated from feedlot cattle administered ceftiofur crystalline-free acid. Am. J. Vet. Res. 68, 501–507. doi: 10.2460/ajvr.68.5.501
Luo, N., Pereira, S., Sahin, O., Lin, J., Huang, S., Michel, L., et al. (2005). Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. U.S.A. 102, 541–546. doi: 10.1073/pnas.0408966102
Mann, S., Siler, J. D., Jordan, D., and Warnick, L. D. (2011). Antimicrobial susceptibility of fecal Escherichia coli isolates in dairy cows following systemic treatment with ceftiofur or penicillin. Foodborne Pathog. Dis. 8, 861–867. doi: 10.1089/fpd.2010.0751
Metcalfe, C., Boxall, A. B., Fenner, K., Kolpin, D., Servos, M., Silberhorn, E., et al. (2009). “Exposure assessment of veterinary medicines in aquatic systems,” in Veterinary Medicines in the Environment, eds M. Crane, A. B. Boxall, and K. Barrett (New York, NY: CRC Press), 57–96.
Mollenkopf, D. F., Glendening, C., Wittum, T. E., Funk, J. A., Tragesser, L. A., and Morley, P. S. (2010). Association of dry cow therapy with the antimicrobial susceptibility of fecal coliform bacteria in dairy cows. Prev. Vet. Med. 96, 30–35. doi: 10.1016/j.prevetmed.2010.05.014
Pavlovic, D. M., Babic, S., Horvat, A. J. M., and Kastelan-Macan, M. (2007). Sample preparation in analysis of pharmaceuticals. Trends Anal. Chem. 26, 1062–1075. doi: 10.1016/j.trac.2007.09.010
Pope, L., Boxall, A., Corsing, C., Halling-Sørensen, B., Tait, A., and Topp, E. (2009). “Exposure assessment of veterinary medicines in terrestrial systems,” in Veterinary Medicines in the Environment, eds M. Crane, A. B. Boxall, and K. Barrett (New York, NY: CRC Press), 129–153.
Saini, V., McClure, J. T., Leger, D., Dufour, S., Sheldon, A. G., Scholl, D. T., et al. (2012). Antimicrobial use on Canadian dairy farms. J. Dairy Sci. 95, 1209–1221. doi: 10.3168/jds.2011-4527
Sarmah, A. K., Meyer, M. T., and Boxall, A. B. (2006). A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65, 725–759. doi: 10.1016/j.chemosphere.2006.03.026
Sawant, A. A., Hegde, N. V., Straley, B. A., Donaldson, S. C., Love, B. C., Knabel, S. J., et al. (2007). Antimicrobial-resistant enteric bacteria from dairy cattle. Appl. Environ. Microbiol. 73, 156–163. doi: 10.1128/AEM.01551-06
Sawant, A. A., Sordillo, L. M., and Jayarao, B. M. (2005). A survey on antibiotic usage in dairy herds in Pennsylvania. J. Dairy Sci. 88, 2991–2999. doi: 10.3168/jds.S0022-030272979-9
Sczesny, S., Nau, H., and Hamscher, G. (2003). Residue analysis of tetracyclines and their metabolites in eggs and in the environment by HPLC coupled with a microbiological assay and tandem mass spectrometry. J. Agric. Food Chem. 51, 697–703. doi: 10.1021/jf0258407
Singer, R. S., Patterson, S. K., and Wallace, R. L. (2008). Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Appl. Environ. Microbiol. 74, 6956–6962. doi: 10.1128/AEM.01241-08
Spellberg, B., Guidos, R., Gilbert, D., Bradley, J., Boucher, H. W., Scheld, W. M., et al. (2008). The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 46, 155–164. doi: 10.1086/524891
Stepanauskas, R., Glenn, T. C., Jagoe, C. H., Tuckfield, R. C., Lindell, A. H., King, C. J., et al. (2006). Coselection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ. Microbiol. 8, 1510–1514. doi: 10.1111/j.1462-2920.2006.01091.x
Subbiah, M., Mitchell, S. M., Ullman, J. L., and Call, D. R. (2011). Beta-lactams and florfenicol antibiotics remain bioactive in soils while ciprofloxacin, neomycin, and tetracycline are neutralized. Appl. Environ. Microbiol. 77, 7255–7260. doi: 10.1128/AEM.05352-11
Subbiah, M., Shah, D. H., Besser, T. E., Ullman, J. L., and Call, D. R. (2012). Urine from treated cattle drives selection for cephalosporin resistant Escherichia coli in soil. PLoS ONE 7:e48919. doi: 10.1371/journal.pone.0048919
Thiele-Bruhn, S. (2003). Pharmaceutical antibiotic compounds in soils – a reivew. J. Plant Nutr. Soil Sci. 166, 145–167. doi: 10.1002/jpln.200390023
Tolls, J. (2001). Sorption of veterinary pharmaceuticals in soils: a review. Environ. Sci. Technol. 35, 3397–3406. doi: 10.1021/es0003021
Tragesser, L. A., Wittum, T. E., Funk, J. A., Winokur, P. L., and Rajala-Schultz, P. J. (2006). Association between ceftiofur use and isolation of Escherichia coli with reduced susceptibility to ceftriaxone from fecal samples of dairy cows. Am. J. Vet. Res. 67, 1696–1700. doi: 10.2460/ajvr.67.10.1696
Tremblay, C. L., Letellier, A., Quessy, S., Daignault, D., and Archambault, M. (2012). Antibiotic-resistant Enterococcus faecalis in abattoir pigs and plasmid colocalization and cotransfer of tet(M) and erm(B) genes. J. Food Prot. 75, 1595–1602. doi: 10.4315/0362-028X.JFP-12-047
Vinks, A. A. (2002). The application of population pharmacokinetic modeling to individualized antibiotic therapy. Int. J. Antimicrob. Agents 19, 313–322. doi: 10.1016/S0924-857900023-7
Volkova, V. V., Lanzas, C., Lu, Z., and Grohn, Y. T. (2012). Mathematical model of plasmid-mediated resistance to ceftiofur in commensal enteric Escherichia coli of cattle. PLoS ONE 7:e36738. doi: 10.1371/journal.pone.0036738
Volkova, V. V., Lu, Z., Lanzas, C., and Grohn, Y. T. (2013). Evaluating targets for control of plasmid-mediated antimicrobial resistance in enteric commensals of beef cattle: a modelling approach. Epidemiol. Infect. 1–19. doi: 10.1017/S0950268812002993. [Epub ahead of print].
Winokur, P. L., Brueggemann, A., DeSalvo, D. L., Hoffmann, L., Apley, M. D., Uhlenhopp, E. K., et al. (2000). Animal and human multidrug-resistant, cephalosporin-resistant salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob. Agents Chemother. 44, 2777–2783. doi: 10.1128/AAC.44.10.2777-2783.2000
Wittum, T. E. (2012). The challenge of regulating agricultural ceftiofur use to slow the emergence of resistance to extended-spectrum cephalosporins. Appl. Environ. Microbiol. 78, 7819–7821. doi: 10.1128/AEM.01967-12
Xia, L. N., Li, L., Wu, C. M., Liu, Y. Q., Tao, X. Q., Dai, L., et al. (2010). A survey of plasmid-mediated fluoroquinolone resistance genes from Escherichia coli isolates and their dissemination in Shandong, China. Foodborne Pathog. Dis. 7, 207–215. doi: 10.1089/fpd.2009.0378
Keywords: ceftiofur, cephalosporin, antibiotic resistance, environmental selection
Citation: Call DR, Matthews L, Subbiah M and Liu J (2013) Do antibiotic residues in soils play a role in amplification and transmission of antibiotic resistant bacteria in cattle populations? Front. Microbiol. 4:193. doi: 10.3389/fmicb.2013.00193
Received: 17 April 2013; Accepted: 23 June 2013;
Published online: 11 July 2013.
Edited by:
Axel Cloeckaert, Institut National de la Recherche Agronomique, FranceReviewed by:
Harvey M. Scott, Kansas State University, USARandall Singer, University of Minnesota, USA
Copyright © 2013 Call, Matthews, Subbiah and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Douglas R. Call, Paul G. Allen School for Global Animal Health, Washington State University, PO Box 647090, 240 SE Ott Road, Pullman, WA 99164-7090, USA e-mail: drcall@wsu.edu