- Department of Veterinary Pharmacology and Toxicology, National Reference Laboratory of Veterinary Drugs Residues, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
Lincomycin is commonly used on swine farms for growth promotion as well as disease treatment and control. Consequently, lincomycin may accumulate in the environment adjacent to the swine farms in many ways, thereby influencing antibiotic resistance in the environment. Levels of lincomycin-resistance genes and lincomycin residues in water and soil samples collected from multiple sites near wastewater discharge areas were investigated in this study. Sixteen lincomycin-resistance and 16S rRNA genes were detected using real-time PCR. Three genes, lnu(F), erm(A), and erm(B), were detected in all water and soil samples except control samples. Lincomycin residues were determined by rapid resolution liquid chromatography-tandem mass spectrometry, with concentrations detected as high as 9.29 ng/mL in water and 0.97 ng/g in soil. A gradual reduction in the levels of lincomycin-resistance genes and lincomycin residues in the waters and soils were detected from multiple sites along the path of wastewater discharging to the surrounding environment from the swine farms. Significant correlations were found between levels of lincomycin-resistance genes in paired water and soil samples (r = 0.885, p = 0.019), and between lincomycin-resistance genes and lincomycin residues (r = 0.975, p < 0.01). This study emphasized the potential risk of dissemination of lincomycin-resistance genes such as lnu(F), erm(A), and erm(B), associated with lincomycin residues in surrounding environments adjacent to swine farms.
Introduction
Antibiotics are commonly used in large confined animal feeding operations (CAFOs) worldwide to promote animal growth and treat animal diseases. Many antibiotics are poorly absorbed in the gut of treated animals and therefore, up to 75% of them can be excreted in an unmetabolized form via feces and urine, allowing antibiotics to persist and accumulate in water and soil (Kumar et al., 2005). It was previously reported that a high concentration (7820 ng/mL) of lincomycin could be detected in liquid manure of swine following administration in feed. In liquid manure, ~84% of the lincomycin was in the dissolved phase, and 16% was associated with the solid components of the manure (Kuchta and Cessna, 2009b). Additionally, lincomycin could be detected in lagoon manure over a period of 5 months when applied as an amendment to agricultural land. When livestock manure from CAFOs is used as liquid fertilizer, antibiotics may transport to surface and ground water, as well as soil, and act as a reservoir (Hornish et al., 1987; Kuchta and Cessna, 2009b; Kwon, 2011). Therefore, lincomycin is one of the antibiotics that could easily accumulate in the environment adjacent to CAFOs (Hu et al., 2010).
A previous study reported that application of animal manure could lead to the potential spread of antibiotic resistance to environmental bacteria by lateral gene transfer (Ghosh and Lapara, 2007). Moreover, antibiotic resistance genes (ARGs) may transfer between pathogens and non-pathogens under selection pressure in the environment (Kruse and Sorum, 1994). Tetracycline-resistance genes, plasmid-mediated quinolone resistance genes, and chloramphenicol resistance genes were reportedly found in wastewater and soil adjacent to swine farms in China (Wu et al., 2010; Li et al., 2012, 2013). There are three known mechanisms responsible for resistance to lincomycin (Schmitz et al., 2000; Lozano et al., 2012): the 23S rRNA methylases [encoded by erm(A), erm(B), erm(C), erm(TR)]; O-nucleotidyltransferases [encoded by lnu(A), lnu(B), lnu(C), lnu(D)] and lincomycin export mediated by efflux [vga(A), vga(B), vga(C), vga(D), vga(E), lsa(A), lsa(B), lsa(C)]. vga(A), erm(B), and erm(A) were detected in swine farm manure and waste treatment systems and vga(A)-positive pathogens were recovered from swine and swine farmers (Chen et al., 2010; Mendes et al., 2011; Zhu et al., 2013). However, few investigations have searched for lincomycin-resistance genes (especially for erm, lnu, and vga) in the environment of swine farms.
China is the largest producer and consumer of antibiotics in the world, and almost half of them are used in livestock industries (Hvistendahl, 2012). Also, swine manure water has been used as fertilizers of fish ponds, which when connected with surrounding waterways will promote the growth of photosynthetic organisms in China. Once swine manure water was discharged to the environment lincomycin-resistance genes and lincomycin residues most likely occur in waters and soils, and subsequently could form a selection pressure to environmental ecology.
The aim of this study was to quantify lincomycin-resistance genes in relation to lincomycin residues in the surrounding environment (soils and waters), especially agricultural fields adjacent to swine farms. In addition, the association between the lincomycin residues and the development of lincomycin resistance was also determined. Water and soil samples were collected from multiple sites along the path of wastewater discharged to the environment from the swine farm (Figure 1), and a culture-independent method was used to investigate the levels of lincomycin-resistance genes.
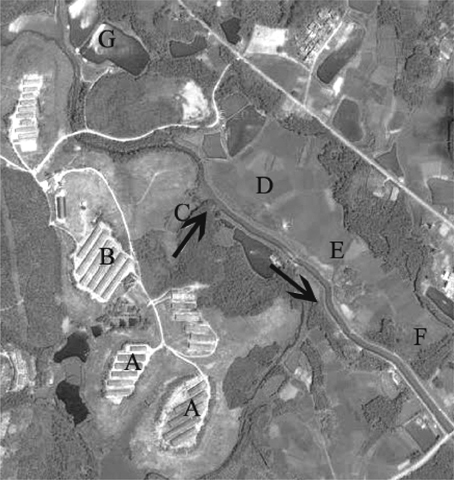
Figure 1. Geographical map of the swine farm and its surrounding environment. The black arrow indicates the direction of effluent of swine manure water. Water and soil samples were collected from sites A to G. A, farrowing pen; B, nursery house; C, fish pond; D–F, agriculture fields; G, reservoir.
Materials and Methods
Sampling
A total of 14 water and soil samples were collected in October 2012 from a swine farm located in Guangdong, China, with an animal density of 10,000 market hogs or more each year. This swine farm is representative of farms that are disposing swine manure to the environment. Water from swine manure was collected in trenches, and then discharged into a fish pond that connects to surrounding waterways through the ditches. Meanwhile, the water in waterways was subsequently applied to surrounding agriculture fields through irrigation (Figure 1). Methods of sample collection and preparation were described in detail by Li et al. (2012). Specifically, water samples (about 1 L for each site) were collected from six sites along the path of wastewater discharged to the environment from the swine farm (Figure 1): the farrowing pen (site A), the nursery house (site B), the fish pond (site C), and surrounding agriculture fields (sites D–F) (designated as A-w, B-w, C-w, D-w, E-w, and F-w for water samples). Soil samples (about 200 g for each site) were collected from the bottom of the trench which was used to discharge of wastewater adjacent to the water collection sites (site A and B), water-sediments of the fish pond (site C), and surrounding agriculture fields (sites D–F) (designated as A-s, B-s, C-s, D-s, E-s, and F-s for soil samples). In addition, control specimens were collected from a reservoir (site G) water (G-w) and surrounding agriculture fields soils (G-s), which received no animal wastes (thus presumably no antibiotics) upstream from the swine farm. For each site, four replicates taken from four different locations were pooled to form one composite sample. The samples were immediately kept in a cooler box during transportation and stored at –80°C before DNA extraction and quantitative analysis of lincomycin residues.
DNA Extraction
DNA extraction was carried out by a culture-independent method described previously (Li et al., 2012). Total DNA from water (about 200 mL) and soil samples (about 0.25 g) were extracted with the Power Water DNA Kit (MO BIO Laboratories Inc., Carlsbad, CA, USA) and Power Soil DNA Kit (MO BIO) according to the manufacturer's instructions, respectively.
PCR and qPCR Assays
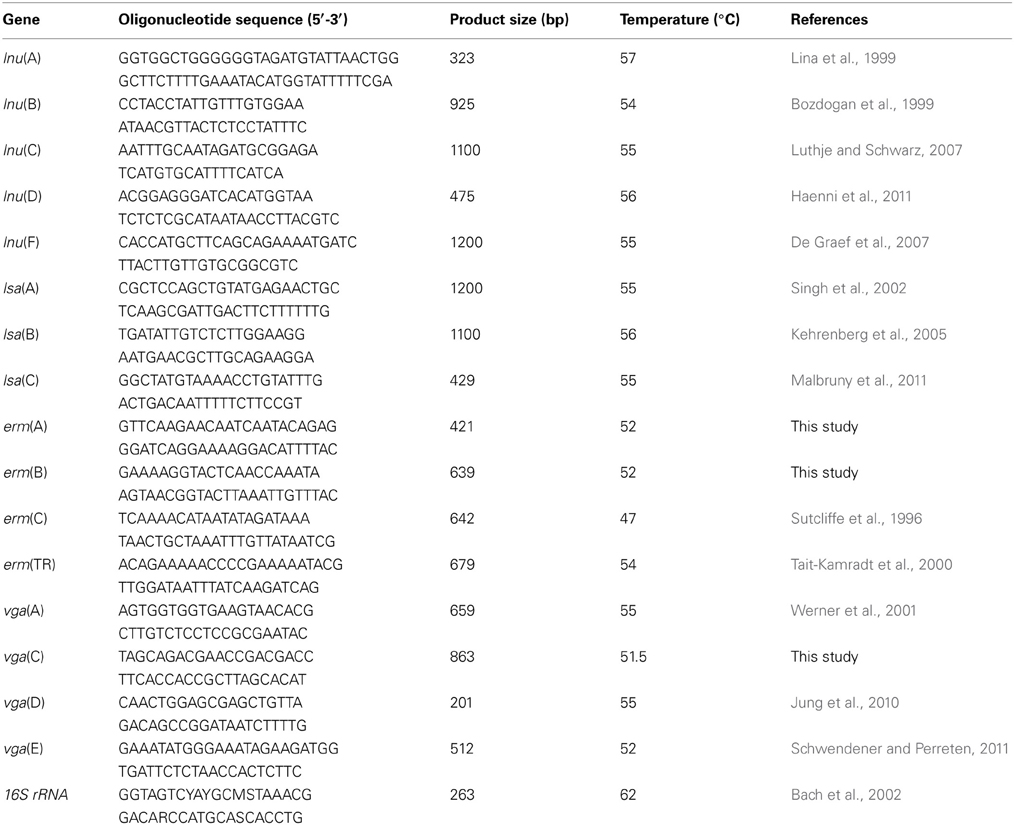
PCR assays were used to detect the presence of 16 lincomycin resistance genes [i.e., erm(A), erm(B), erm(C), erm(TR), lnu(A), lnu(B), lnu(C), lnu(D), lnu(F), lsa(A), lsa(B), lsa(C), vga(A),vga(C), vga(D), and vga(E)] in all of the environmental and control samples. Primers for these genes were either reported or newly designed (Table A1 of the Appendix). To ensure reproducibility, PCR for lincomycin-resistance genes and 16SrRNA genes was performed in triplicate using a thermal cycler (iQ5; Bio-Rad, Hercules, CA) and SYBR® Premix Ex Taq™ in parallel with a negative control in every run. All PCR products were directly sequenced, and the results were compared against those in the GenBank nucleotide database (http://www.ncbi.nlm.nih.gov/blast/). Primers used for real-time PCR for detecting lincomycin-resistance genes were the same as those used in the qualitative PCR, while primers for the 16S rRNA genes were reported previously (Li et al., 2012). Each reaction was performed in a 25 μL volume containing 12.5 μL of SYBR Premix Ex Taq, 0.5 μL of each primer, 9.5 μ L of ddH2O and 2 μ L of template, with the following conditions: denaturing at 94°C for 5 min, followed by 40 cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 5 min. The melt curve was read every 1°C, from 60 to 95°C toward the end of PCR reactions. All agents were supplied by TaKaRa (TaKaRa Bio, Dalian, China). The PCR efficiencies were examined to test inhibition. The R2-values were more than 0.9 for all calibration curves. In order to minimize variance caused by differential extraction and analytical efficiencies, and differences to the background bacterial abundance, the level of each lincomycin resistance gene was normalized with the 16S rRNA copy number using the method recommended previously (Livak and Schmittgen, 2001).
Quantification of Lincomycin
Extraction and quantitative analysis of lincomycin in water, soil, and control samples were performed according to methods described previously (Peru et al., 2006; Hu et al., 2010). Lincomycin residues were detected by rapid resolution liquid chromatography-tandem mass spectrometry (RRLC-MS/MS), comprising an Agilent liquid chromatography 1200 series RRLC system (Agilent Technologies, Palo Alto, CA, USA) coupled to an API 4000 triple quadrupole mass spectrometer (Applied Biosystems, Foster, CA, USA) with the software of Analyst 1.5 The analytical column was a 2.1 mm ID × 150 mm, 1.8 μm Zorbax SB-Aq (Agilent Technologies, Atlanta, GA, USA). The recoveries for lincomycin based on matrix-matched calibration were 90.1% in water samples and 78.4% in soil samples. The quantification limits were 10 pg/mL for water and 0.1 ng/g for soil, respectively.
Statistics
Statistical evaluation of the data was conducted by SPSS version 17.0. The homogeneity of values was assessed via a One-Way analysis of variance (ANOVA) test. A two-tailed Pearson's bivariate correlation analysis was used to compare levels of lincomycin resistance genes in paired water and soil samples and to correlate levels of lincomycin-resistance genes with concentrations of lincomycin residues. The relative quantification of lincomycin resistance genes lnu(F), erm(A), and erm(B), presented in all of the water and soil samples, were conducted by correlation analysis.
Results
Concentration of Lincomycin in Waters and Soils
Lincomycin residues were commonly detected in all water samples (ranged from 0.018 to 9.29 ng/mL) and soil samples (ranging from 0.024 to 0.97 ng/g) from sites A to F (Figure 2). In contrast, concentrations of lincomycin were below the detection limit in all control samples. Among both water and soil samples, the highest concentrations of lincomycin were observed in the A-w sample (9.29 ng/mL) and the A-s sample (0.97 ng/g), respectively, and then declined substantially downstream of the site A. Lincomycin residues detected in water samples were higher than those in soil samples.
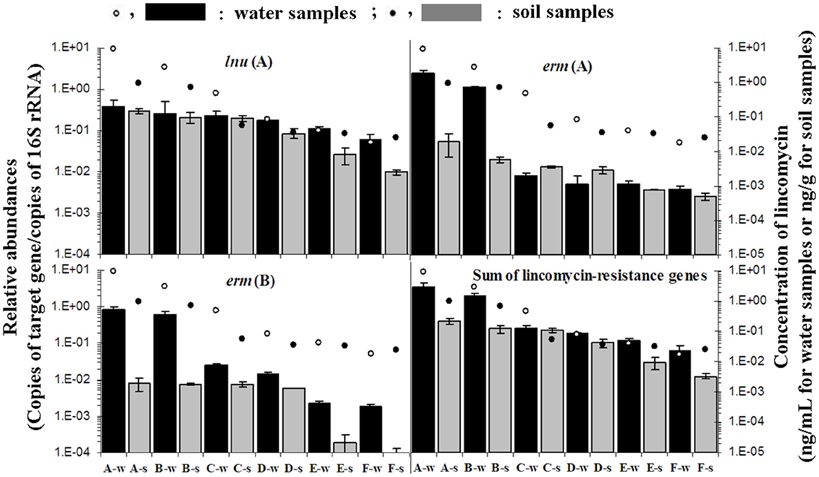
Figure 2. Relative abundance of lincomycin-resistance genes: lnu(F), erm(A), erm(B), and the sum of the nine lincomycin resistance genes [lnu(F), erm(A), erm(B), lnu(A), lnu(D), vga(C), vga(E), vga(A), and vga(D)] and concentration of lincomycin. Black bars and blank symbols indicate a relative abundance of lincomycin resistance genes and concentration of lincomycin in water samples, respectively; Gray bars and symbols indicate relative abundance of lincomycin resistance genes and concentration of lincomycin in soil samples, respectively. Error bars represent the standard deviation.
Occurrence and Levels of Lincomycin Resistance Genes in Waters and Soils
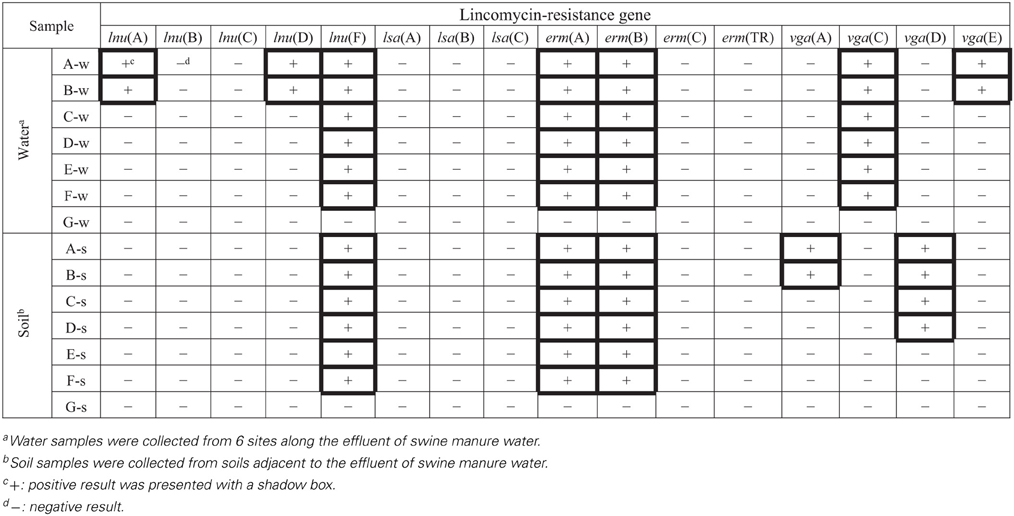
Prevalence of lincomycin resistance genes in each sample tested are presented in Table A2. Of the 16 lincomycin resistance genes investigated, lnu(F), erm(A), and erm(B) were detected in all water and soil samples except the control samples. The vga(C) was found in all water samples; while vga(E) was found only in two water samples (A-w and B-w). vga(A) and vga(D) were detected in two water (A-s and B-s) and four (A-s, B-s, C-s, and D-s) soil samples, respectively. No novel sequences were observed.
Relative quantification of nine lincomycin resistance genes is shown in Figure 2 and Table A3. The level of individual lincomycin resistance genes varied in samples from site to site when present. A gradual reduction in relative quantification of lincomycin resistance genes in water and soil samples was detected from site A to F. Moreover, levels of lincomycin resistance genes in water were higher than those in soil samples from each site.
Correlation Analysis
Significant positive correlation was observed for the concentration of lincomycin residues between paired water and soil samples (r = 0.925, p = 0.008). Additionally, quantification of lincomycin resistance genes lnu(F) (r = 0.981, p = 0.001), erm(A) (r = 0.958, p = 0.003), and total lincomycin-resistance genes [lnu(F), erm(A), erm(B), lnu(A), lnu(D), vga(C), vga(E), vga(A), and vga(D)] (r = 0.885, p = 0.019) in water samples was significantly correlated with soil samples, except erm(B) (r = 0.626, p = 0.184). Significant correlations were exhibited between the level of erm(A) (r = 0.982, p < 0.01), erm(B) (r = 0.919, p < 0.01), the sum of the nine lincomycin resistance genes (r = 0.975, p < 0.01) and lincomycin residues. The level of vga(C) was significantly correlated with lincomycin residues in water samples (r = 0.999, p < 0.01). However, moderately significant correlations between lincomycin residue and relative quantification of lnu(F) (r = 0.705, p = 0.01) (Table 1) were noted.
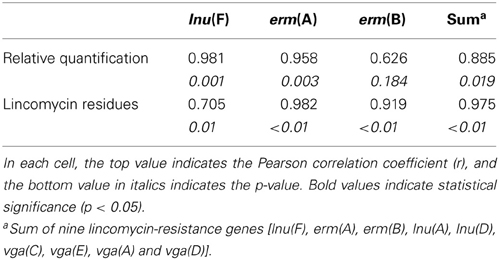
Table 1. Correlation analysis of lincomycin-resistance genes in paired water and soil samples as well as correlation between lincomycin-resistance genes and lincomycin residues.
Discussion
Since swine manure water was discharged from the site A, it was not surprising that the highest concentrations were observed in the A-w samples and the A-s samples. Concentrations of lincomycin in the A-w sample and B-w, C-w, D-w, E-w samples detected in this study were similar to previous studies of liquid swine manure and ground water from manure-amended cropland, respectively (Kuchta and Cessna, 2009a; Kuchta et al., 2009). However, lincomycin residues in soil samples were higher than that in manure-amended soil reported previously (Kuchta et al., 2009). Lincomycin residues declined substantially downstream of site A probably as a result of dilution. The accumulation of lincomycin in the environmental water and soil samples is likely attributed to the use of lincomycin as feed additives in the facility. Our findings indicated that the swine manure water was a lincomycin reservoir. Lincomycin residues in D-s, E-s, and F-s samples maintained at a relatively constant level which suggests that they are close to the detection limit of the residue detection method used.
A total of nine lincomycin resistance genes were detected in this study. The genes erm(A), erm(B), and lnu(F) were widespread. They were found in almost all of the water and soil samples. This is similar to a previous report where a high level of the erm gene was present in typical swine manures (Chen et al., 2010). Although water and soil bacterial communities are different, erm(A) and erm(B) could spread widely due to the discharge of swine manure water containing ARGs in the environment. In this study, it was noteworthy that levels of vga(A), and lnu(A) in swine manure water were 10-fold higher than previous reports in soil and water samples, respectively (Zhu et al., 2013). lnu(D), vga(C), vga(D), and vga(E) were sporadically detected in environmental samples from sites close to site A. Contrary to previous studies (Zhu et al., 2013), vga(B) was not detected in any sample in our study. To our knowledge, this is the first report of the presence of lnu(F), lnu(D), vga(C), vga(D), and vga(E) genes in surrounding environments adjacent to swine farms in China. Lincosamide O-nucleotidyltransferases encoded by lnu(A), lnu(D), lnu(F), and ATP-binding cassette transporters encoded by vga(A), vga(C), vga(D), and vga(E) could also be transferred by plasmids and transposons; thus it is posing a potential dissemination risk (Petinaki et al., 2008; Kadlec et al., 2010; Schwendener and Perreten, 2011).
Similar to the levels of plasmid-mediated quinolone resistance genes in environmental samples, we found that the levels of lincomycin resistance genes in water were higher than those in soil samples at each site (Li et al., 2012). Significant correlation between paired water and soil samples with regard to the relative levels of lincomycin resistance genes and lincomycin residues was found. Lincomycin resistance genes and lincomycin residues were not detected in control samples (site G). This indicates that the swine manure water is the reservoir of these contaminants. Similar reservoirs of ARGs and antibiotics are probably common in China and other countries (Zhu et al., 2013). Previous studies suggested that there was the exchange of ARGs between environmental bacterial and clinical pathogens, which might pose health risks to nearby residents exposed to contaminated field soil, fish pond water, and waterway water during farming (Forsberg et al., 2012; Li et al., 2012).
Considering that high levels of vga(C) and vga(D) were only detected in soil samples and the level of vga(C) was significantly correlated with lincomycin residues, it may be concluded that environmental quantification of lincomycin resistance genes was not only influenced by a dilution effect of contamination but also by a selective effect from lincomycin residues in the environment. Previous studies indicated that antibiotic residues in the environment (soil and water) could affect the selection and dissemination of resistance genes, and promote or inhibit ecological functions (Ghosh and Lapara, 2007; Naslund et al., 2008; Schauss et al., 2009; Ding and He, 2010). Therefore, long-term use of lincomycin, coupled with its slow degradation in soil and water (Kuchta et al., 2009; Williams and McLain, 2012) could potentially lead to the selection of resistant bacteria species and the transfer of ARGs located in transferable elements (Looft et al., 2012). This hypothesis will be investigated in future work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Science Fund for Distinguished Young Scholars (Grant No. 31125026), the Special Fund for Agro-scientific Research in the Public Interest (Grant No. 201203040), the National Natural Science Foundation of China (Grant No. U0631006), and Natural Science Foundation team projects of Guangdong Province (Grant No. S2012030006590).
References
Chen, J., Michel, F. C. Jr., Sreevatsan, S., Morrison, M., and Yu, Z. (2010). Occurrence and persistence of erythromycin resistance genes (erm) and tetracycline resistance genes (tet) in waste treatment systems on swine farms. Microb. Ecol. 60, 479–486. doi: 10.1007/s00248-010-9634-5
Ding, C., and He, J. (2010). Effect of antibiotics in the environment on microbial populations. Appl. Microbiol. Biotechnol. 87, 925–941. doi: 10.1007/s00253-010-2649-5
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O. A., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111. doi: 10.1126/science.1220761
Ghosh, S., and Lapara, T. M. (2007). The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 1, 191–203. doi: 10.1038/ismej.2007.31
Hornish, R. E., Gosline, R. E., and Nappier, J. M. (1987). Comparative metabolism of lincomycin in the swine, chicken, and rat. Drug Metab. Rev. 18, 177–214. doi: 10.3109/03602538708998305
Hu, X., Zhou, Q., and Luo, Y. (2010). Occurrence and source analysis of typical veterinary antibiotics in manure, soil, vegetables and groundwater from organic vegetable bases, northern China. Environ. Pollut. 158, 2992–2998. doi: 10.1016/j.envpol.2010.05.023
Hvistendahl, M. (2012). Public health. China takes aim at rampant antibiotic resistance. Science 336, 795. doi: 10.1126/science.336.6083.795
Kadlec, K., Pomba, C. F., Couto, N., and Schwarz, S. (2010). Small plasmids carrying vga(A) or vga(C) genes mediate resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-resistant Staphylococcus aureus ST398 from swine. J. Antimicrob. Chemother. 65, 2692–2693. doi: 10.1093/jac/dkq365
Kruse, H., and Sorum, H. (1994). Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl. Environ. Microbiol. 60, 4015–4021.
Kuchta, S. L., and Cessna, A. J. (2009a). Fate of lincomycin in snowmelt runoff from manure-amended pasture. Chemosphere 76, 439–446. doi: 10.1016/j.chemosphere.2009.03.069
Kuchta, S. L., and Cessna, A. J. (2009b). Lincomycin and spectinomycin concentrations in liquid swine manure and their persistence during simulated manure storage. Arch. Environ. Contam. Toxicol. 57, 1–10. doi: 10.1007/s00244-008-9229-z
Kuchta, S. L., Cessna, A. J., Elliott, J. A., Peru, K. M., and Headley, J. V. (2009). Transport of lincomycin to surface and ground water from manure-amended cropland. J. Environ. Qual. 38, 1719–1727. doi: 10.2134/jeq2008.0365
Kumar, K., Gupta, S. C., Chander, Y., and Singh, A. K. (2005). Antibiotic use in agriculture and its impact on the terrestrial environment. Adv. Agron. 87, 1–54. doi: 10.1016/S0065-2113(05)87001-4
Kwon, J. W. (2011). Mobility of veterinary drugs in soil with application of manure compost. Bull. Environ. Contam. Toxicol. 87, 40–44. doi: 10.1007/s00128-011-0297-9
Li, J., Shao, B., Shen, J., Wang, S., and Wu, Y. (2013). Occurrence of chloramphenicol-resistance genes as environmental pollutants from swine feedlots. Environ. Sci. Technol. 47, 2892–2897. doi: 10.1021/es304616c
Li, J., Wang, T., Shao, B., Shen, J., Wang, S., and Wu, Y. (2012). Plasmid-mediated quinolone resistance genes and antibiotic residues in wastewater and soil adjacent to swine feedlots: potential transfer to agricultural lands. Environ. Health Perspect. 120, 1144–1149. doi: 10.1289/ehp.1104776
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Looft, T., Johnson, T. A., Allen, H. K., Bayles, D. O., Alt, D. P., Stedtfeld, R. D., et al. (2012). In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. U.S.A. 109, 1691–1696. doi: 10.1073/pnas.1120238109
Lozano, C., Aspiroz, C., Rezusta, A., Gomez-Sanz, E., Simon, C., Gomez, P., et al. (2012). Identification of novel vga(A)-carrying plasmids and a Tn5406-like transposon in meticillin-resistant Staphylococcus aureus and Staphylococcus epidermidis of human and animal origin. Int. J. Antimicrob. Agents 40, 306–312. doi: 10.1016/j.ijantimicag.2012.06.009
Mendes, R. E., Smith, T. C., Deshpande, L., Diekema, D. J., Sader, H. S., and Jones, R. N. (2011). Plasmid-borne vga(A)-encoding gene in methicillin-resistant Staphylococcus aureus ST398 recovered from swine and a swine farmer in the United States. Diagn. Microbiol. Infect. Dis. 71, 177–180. doi: 10.1016/j.diagmicrobio.2011.06.009
Naslund, J., Hedman, J. E., and Agestrand, C. (2008). Effects of the antibiotic ciprofloxacin on the bacterial community structure and degradation of pyrene in marine sediment. Aquat. Toxicol. 90, 223–227. doi: 10.1016/j.aquatox.2008.09.002
Peru, K. M., Kuchta, S. L., Headley, J. V., and Cessna, A. J. (2006). Development of a hydrophilic interaction chromatography-mass spectrometry assay for spectinomycin and lincomycin in liquid hog manure supernatant and run-off from cropland. J. Chromatogr. A 1107, 152–158. doi: 10.1016/j.chroma.2005.12.057
Petinaki, E., Guerin-Faublee, V., Pichereau, V., Villers, C., Achard, A., Malbruny, B., et al. (2008). Lincomycin resistance gene lnu(D) in Streptococcus uberis. Antimicrob. Agents Chemother. 52, 626–630. doi: 10.1128/AAC.01126-07
Schauss, K., Focks, A., Leininger, S., Kotzerke, A., Heuer, H., Thiele-Bruhn, S., et al. (2009). Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ. Microbiol. 11, 446–456. doi: 10.1111/j.1462-2920.2008.01783.x
Schmitz, F. J., Sadurski, R., Kray, A., Boos, M., Geisel, R., Kohrer, K., et al. (2000). Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J. Antimicrob. Chemother. 45, 891–894. doi: 10.1093/jac/45.6.891
Schwendener, S., and Perreten, V. (2011). New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrob. Agents Chemother. 55, 4900–4904. doi: 10.1128/AAC.00528-11
Williams, C. F., and McLain, J. E. T. (2012). Soil persistence and fate of carbamazepine, lincomycin, caffeine, and ibuprofen from wastewater reuse. J. Environ. Qual. 41, 1473–1480. doi: 10.2134/jeq2011.0353
Wu, N., Qiao, M., Zhang, B., Cheng, W. D., and Zhu, Y. G. (2010). Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China. Environ. Sci. Technol. 44, 6933–6939. doi: 10.1021/es1007802
Zhu, Y. G., Johnson, T. A., Su, J. Q., Qiao, M., Guo, G. X., Stedtfeld, R. D., et al. (2013). Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. U.S.A. 110, 3435–3440. doi: 10.1073/pnas.1222743110
Appendix
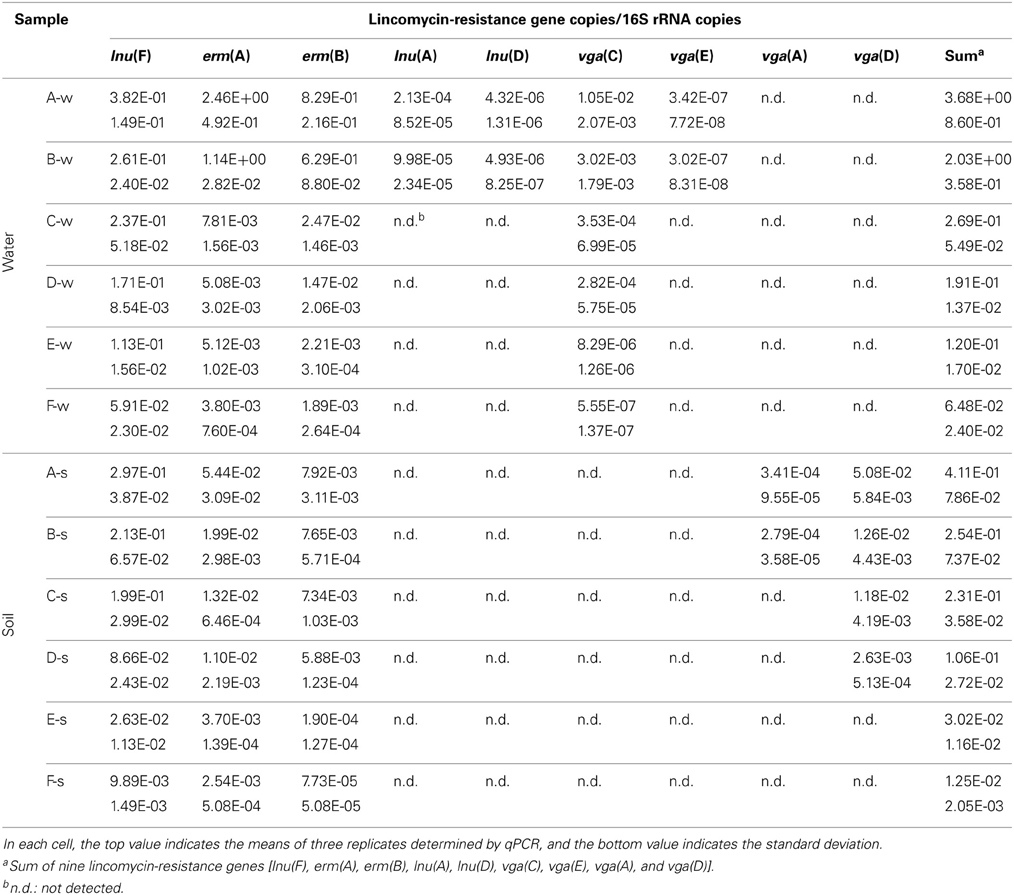
Table A3. Relative quantification of lincomycin-resistance genes in water and soil samples, normalized to corresponding 16S rRNA copies.
References
Bach, H. J., Tomanova, J., Schloter, M., and Munch, J. C. (2002). Enumeration of total bacteria and bacteria with genes for proteolytic activity in pure cultures and in environmental samples by quantitative PCR mediated amplification. J. Microbiol. Methods 49, 235–245. doi: 10.1016/S0167-7012(01)00370-0
Bozdogan, B., Berrezouga, L., Kuo, M. S., Yurek, D. A., Farley, K. A., Stockman, B. J., et al. (1999). A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob. Agents Chemother. 43, 925–929.
De Graef, E. M., Decostere, A., De Leener, E., Goossens, H., Baele, M., and Haesebrouck, F. (2007). Prevalence and mechanism of resistance against macrolides, lincosamides, and streptogramins among Enterococcus faecium isolates from food-producing animals and hospital patients in Belgium. Microb. Drug Resist. 13, 135–141. doi: 10.1089/mdr.2007.718
Haenni, M., Saras, E., Chaussiere, S., Treilles, M., and Madec, J. Y. (2011). ermB-mediated erythromycin resistance in Streptococcus uberis from bovine mastitis. Vet. J. 189, 356–358. doi: 10.1016/j.tvjl.2010.06.021
Jung, Y. H., Shin, E. S., Kim, O., Yoo, J. S., Lee, K. M., Il Yoo, J., et al. (2010). Characterization of two newly identified genes, vgaD and vatG, conferring resistance to streptogramin A in Enterococcus faecium. Antimicrob. Agents Chemother. 54, 4744–4749. doi: 10.1128/AAC.00798-09
Kehrenberg, C., Schwarz, S., Jacobsen, L., Hansen, L. H., and Vester, B. (2005). A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57, 1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x
Lina, G., Quaglia, A., Reverdy, M. E., Leclercq, R., Vandenesch, F., and Etienne, J. (1999). Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 43, 1062–1066.
Luthje, P., and Schwarz, S. (2007). Molecular basis of resistance to macrolides and lincosamides among staphylococci and streptococci from various animal sources collected in the resistance monitoring program BfT-GermVet. Int. J. Antimicrob. Agents 29, 528–535. doi: 10.1016/j.ijantimicag.2006.12.016
Malbruny, B., Werno, A. M., Murdoch, D. R., Leclercq, R., and Cattoir, V. (2011). Cross-resistance to lincosamides, streptogramins A, and pleuromutilins due to the lsa(C) gene in Streptococcus agalactiae UCN70. Antimicrob. Agents Chemother. 55, 1470–1474. doi: 10.1128/AAC.01068-10
Singh, K. V., Weinstock, G. M., and Murray, B. E. (2002). An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 46, 1845–1850. doi: 10.1128/AAC.46.6.1845-1850.2002
Sutcliffe, J., Grebe, T., Tait-Kamradt, A., and Wondrack, L. (1996). Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40, 2562–2566.
Tait-Kamradt, A., Davies, T., Cronan, M., Jacobs, M. R., Appelbaum, P. C., and Sutcliffe, J. (2000). Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44, 2118–2125. doi: 10.1128/AAC.44.8.2118-2125.2000
Keywords: culture-independent method, agriculture field, swine farm, lnu(F), erm(A), erm(B)
Citation: Li L, Sun J, Liu B, Zhao D, Ma J, Deng H, Li X, Hu F, Liao X and Liu Y (2013) Quantification of lincomycin resistance genes associated with lincomycin residues in waters and soils adjacent to representative swine farms in China. Front. Microbiol. 4:364. doi: 10.3389/fmicb.2013.00364
Received: 09 August 2013; Accepted: 16 November 2013;
Published online: 03 December 2013.
Edited by:
Mark Montforts, National Institute for Public Health and the Environment, NetherlandsReviewed by:
Patrick R. Butaye, Ghent University, BelgiumJulie Zilles, University of Illinois Urbana Champaign, USA
Copyright © 2013 Li, Sun, Liu, Zhao, Ma, Deng, Li, Hu, Liao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yahong Liu, Department of Veterinary Pharmacology and Toxicology, National Reference Laboratory of Veterinary Drug Residues, College of Veterinary Medicine, South China Agricultural University, Wushan Road 483, Guangzhou 510642, China e-mail: gale@scau.edu.cn
†These authors have contributed equally to this work.
 Liang Li
Liang Li Jian Sun
Jian Sun Baotao Liu
Baotao Liu