- The ithree Institute, University of Technology, Sydney, NSW, Australia
Spatial regulation of cell division in bacteria has been a focus of research for decades. It has been well studied in two model rod-shaped organisms, Escherichia coli and Bacillus subtilis, with the general belief that division site positioning occurs as a result of the combination of two negative regulatory systems, Min and nucleoid occlusion. These systems influence division by preventing the cytokinetic Z ring from forming anywhere other than midcell. However, evidence is accumulating for the existence of additional mechanisms that are involved in controlling Z ring positioning both in these organisms and in several other bacteria. In some cases the decision of where to divide is solved by variations on a common evolutionary theme, and in others completely different proteins and mechanisms are involved. Here we review the different ways bacteria solve the problem of finding the right place to divide. It appears that a one-size-fits-all model does not apply, and that individual species have adapted a division-site positioning mechanism that best suits their lifestyle, environmental niche and mode of growth to ensure equal partitioning of DNA for survival of the next generation.
Introduction
The location of proteins inside bacterial cells is tightly regulated, with different proteins having specific cellular addresses that can change dynamically with time (Shapiro et al., 2009; Rudner and Losick, 2010; Lenz and Sogaard-Andersen, 2011; Nevo-Dinur et al., 2012). This exquisite spatial organization has provided unique mechanistic insights into fundamental biological processes. Cell division, or cytokinesis, is one such process that is under strict spatial control and this ensures equal partitioning of DNA into newborn cells (Thanbichler, 2009; Lutkenhaus et al., 2012; Monahan and Harry, 2013). Just how bacterial cells identify the site of division is an important question that has not been fully resolved.
The first protein to localize to the division site in bacteria is the highly conserved tubulin-like protein, FtsZ. FtsZ polymerizes at this site to form a ring, called the Z ring, which then recruits all other known division proteins, about 20 in all, to form a protein complex called the divisome (Adams and Errington, 2009; De Boer, 2010). The divisome subsequently contracts, pulling in the cell envelope to facilitate cytokinesis. Importantly, the Z ring marks the site of cell division. Not surprisingly therefore, proteins that are known to influence division site placement within bacterial cells, actually do so via their influence on the position of the Z ring.
Nucleoid Occlusion and the Min System
In the model rod-shaped bacteria, Escherichia coli and Bacillus subtilis, Z ring formation occurs precisely at the cell midpoint (standard deviation of 2.6 and 2.2% off center, respectively; Yu and Margolin, 1999; Migocki et al., 2002), generating daughter cells of equal size. Research into the control of Z ring positioning in these organisms has centered mainly on two regulatory systems, nucleoid occlusion (NO) and the Min system, which inhibit Z rings forming anywhere in the cell other than midcell.
The Min system blocks Z ring assembly at the cell poles by inhibiting the polymerization of FtsZ (Barak and Wilkinson, 2007; Lutkenhaus, 2007; Bramkamp and van Baarle, 2009). In both E. coli and B. subtilis, the MinC protein serves as the primary inhibitor of Z ring formation by direct interaction and destablization of FtsZ polymers (Hu et al., 1999; Dajkovic et al., 2008). MinC is localized to the cytoplasmic membrane via association with the membrane-bound ATPase MinD, and is directed to the poles by fundamentally different mechanisms in E. coli and B. subtilis. In B. subtilis DivIVA pilots MinCD to the poles via the bridging protein MinJ (Bramkamp et al., 2008; Patrick and Kearns, 2008), while in E. coli the topological determinant MinE undergoes a dynamic pole-to-pole oscillation that actively displaces the MinCD complex from the membrane. The net result in both organisms is that the MinCD concentration is highest in the polar regions of the cell, blocking polar Z ring formation (Figure 1A).
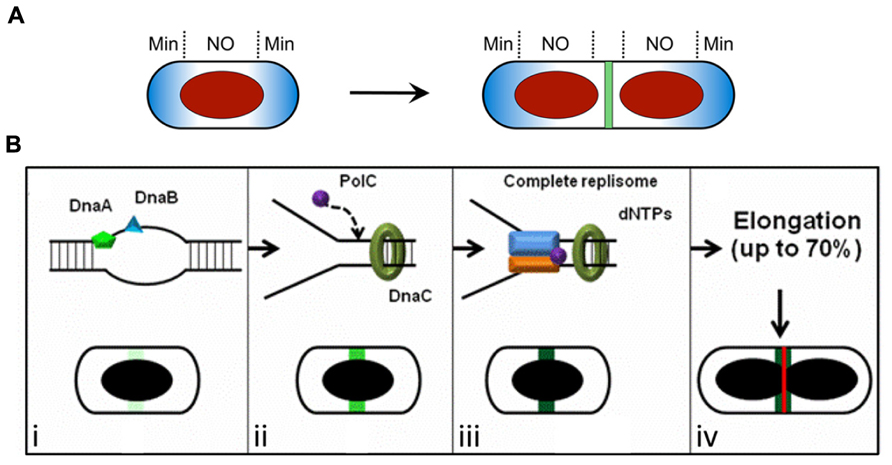
FIGURE 1. Spatial regulation of Z ring assembly in E. coli and B. subtilis. (A) The Min system and nucleoid occlusion (NO) inhibit Z ring formation at inappropriate sites. The Min proteins (blue) are concentrated to the cell poles, while the nucleoid (red) occupies the central region of the cell. The later stages of chromosome segregation result in a relief of nucleoid occlusion due to the removal of Noc/SlmA away from midcell, allowing Z ring formation at this site. (B) The “Ready-Set-Go” model proposes that identification of the division site is linked to the progress of DNA replication in B. subtilis, independently of both the Min system and nucleoid occlusion (Moriya et al., 2010). (i–iv) Progression of the initiation phase of DNA replication in B. subtilis, resulting in replisome assembly at oriC, promotes the maturation of a midcell site. This may occur due to the accumulation of a factor at midcell that activates FtsZ polymerization into a ring at this site (increasing darkness of green shading). (i) The binding of the early initiation protein DnaA to unwind the DNA at oriC starts midcell “potentiation.” Next, other early DNA replication initiation proteins, such as DnaB, bind this chromosomal region and increase midcell potentiation (light green area at midcell). (ii) DnaC helicase is then loaded, followed by PolC (the α-subunit of DNA polymerase III) and other replisome components, creating the replication fork at oriC. This further potentiates midcell (green area at midcell). (iii) Assembly of the remaining replisome components to complete initiation, ready for DNA synthesis, allows 100% potentiation of midcell (dark green area at midcell) for Z ring formation (red line). (iv) Midcell Z ring formation does not occur straight away since this requires ~70% of the chromosome to be replicated (Wu et al., 1995) to clear the bulk of the replicating DNA from midcell. (A) is reproduced from Monahan and Harry (2013) and (B) is adapted from Moriya et al. (2010), both with permission from Wiley Interscience.
Nucleoid occlusion prevents Z ring assembly over the nucleoid or chromosome (Wu and Errington, 2012). This effect is mediated at least in part by the non-homologous proteins Noc in B. subtilis (Wu and Errington, 2004; Wu et al., 2009) and SlmA in E. coli (Bernhardt and de Boer, 2005; Cho et al., 2011; Tonthat et al., 2011; Tonthat et al., 2013), which localize over nucleoids by binding to specific DNA sequences. SlmA affects FtsZ polymerization directly (Cho et al., 2011; Tonthat et al., 2011), while it is unclear how Noc influences Z ring formation (Wu and Errington, 2012). Interestingly, binding sites for both Noc and SlmA are sparse or absent near the terminus region of the chromosome (Wu et al., 2009; Cho et al., 2011; Tonthat et al., 2011), which occupies a midcell location during the late stages of chromosome replication and segregation. Thus chromosome segregation generates a relief of NO at midcell, allowing the Z ring to form there (Figure 1A).
A Link between DNA Replication and Z Ring Positioning
There is no doubt that in E. coli and B. subtilis Min and NO play an important role in influencing Z ring placement (Figure 1A; Rothfield et al., 2005; Harry et al., 2006; Barak and Wilkinson, 2007; Wu and Errington, 2012). However, there are now several lines of evidence that strongly implicate additional Z ring positioning mechanisms in these rod-shaped bacteria. First, cells are still viable in the absence of both the Min system and NO proteins, Noc/SlmA, although viability is conditional and division is much less efficient (Wu and Errington, 2004; Bernhardt and de Boer, 2005; Rodrigues and Harry, 2012). Second, overproduction of FtsZ in min noc B. subtilis and min slmA E. coli double mutants results in partial restoration of division, with Z rings assembling at the correct site between segregated chromosomes (Wu and Errington, 2004; Bernhardt and de Boer, 2005).
More recently, it was shown using outgrown spores of B. subtilis that Z rings can form precisely at midcell in this organism in the complete absence of both the Min system and Noc (Rodrigues and Harry, 2012). In these min noc outgrown cells, Z rings were positioned at midcell with wild-type precision, although their assembly was delayed and less efficient, with Z rings forming also at future division sites (1/4 and 3/4 positions) and the cell poles (Rodrigues and Harry, 2012). Overproduction of FtsZ in these cells significantly reduced the delay, and increased the proportion of midcell Z rings (Rodrigues and Harry, 2012). To test whether any NO is required for precise positioning of the Z ring at midcell, chromosomes in min noc outgrown cells were allowed to replicate and separate to the extent that a significant gap of DNA (no NO) occurred in the central region of the cell. FtsZ production was then switched on (Rodrigues and Harry, 2012). Remarkably, Z rings formed precisely at midcell under these conditions, and there was a high preference (87%) for Z ring assembly at midcell as opposed to any other DNA-free regions, including the cell poles (Rodrigues and Harry, 2012).
The above data argue for the identification of a specific site at midcell for Z ring assembly in B. subtilis that does not require Min or any NO, and have led to a model in which NO and Min do not identify the correct division site at midcell but rather ensure that the Z ring forms there and only there, at the right time in the cell cycle (Rodrigues and Harry, 2012). In other words, in B. subtilis at least, Min and NO do not appear to be the division “signpost,” but enable this signpost at midcell to be efficiently utilized.
So what does identify the division site? In B. subtilis it has been proposed that a positive signal links progress of the initiation phase of DNA replication with identification of the division site at midcell (Figures 1A,B; Moriya et al., 2010; Rodrigues and Harry, 2012). It has been known for some time that the early stages of DNA replication influence Z ring positioning in this organism (Harry et al., 1999; Regamey et al., 2000). More recently it has been shown that even in the absence of Noc the frequency of midcell Z rings increases with progression of the initiation phase of replication (Moriya et al., 2010). This has led to a model for Z ring positioning in B. subtilis, called the “Ready-Set-Go” model, in which midcell becomes increasingly “potentiated” for Z ring formation as initiation of DNA replication is progressively completed, much like a runner in a race changing position in readiness to race from the start line when the gun fires (Figure 1B; Moriya et al., 2010). The ordered, stable association of replication initiation proteins with oriC in B. subtilis (as opposed to all initiation proteins associating with oriC simultaneously) is consistent with a progressive step-wise potentiation of midcell (Smits et al., 2010). Importantly the observation that Z rings can form precisely at midcell even when there is no DNA synthesis (elongation) establishes that the midcell site is determined in B. subtilis very early in the cell cycle, much earlier than when Z ring formation actually occurs. However, its utilization is blocked [either by a NO protein other than Noc (Bernard et al., 2010) or some other mechanism] until the chromosome has been replicated beyond 70% completion (Figure 1B; Wu et al., 1995).
In E. coli, SlmA-, MinC-, and SOS-independent inhibition of midcell Z rings can occur over a partially replicated, unsegregated nucleoid indicating that additional Z-ring positioning mechanisms also exist in this organism (Bernard et al., 2010; Cambridge et al., 2013). Interestingly, the localization of the replisome protein, DnaX, to midcell prior to oriC and the Z ring in E. coli raises the possibility that DNA replication has a positive role in division site positioning in organisms other than B. subtilis (Bates and Kleckner, 2005).
Control of Z Ring Placement in Other Organisms: A Diversity of Mechanisms
The importance of Z ring positioning mechanisms other than Min and NO is further exemplified by the fact that many bacteria lack Noc/SlmA and/or Min protein homologues (Margolin, 2005; Harry et al., 2006). Recent studies on division site selection in several such bacteria have uncovered a number of novel Z-ring positioning mechanisms. These include both negative regulators (FtsZ inhibitors) as well as positive signals that actively promote Z ring formation at the correct location. Interestingly, many of the proteins involved in these systems are not highly conserved, suggesting that Z ring placement is controlled differently between different bacterial species. Cell shape is also emerging as an important factor in division site selection, particularly in spherical cells that must select the correct division plane from a theoretically infinite number of possible midcell planes (Margolin, 2000). Different coccal species select different midcell planes for division (Staphylococci divide in three alternating planes and Neisseria in two for example), further highlighting the species-specific nature of bacterial division site placement. Below we describe the different positioning mechanisms at play in a range of recently studied organisms.
Caulobacter crescentus: MipZ
In Caulobacter crescentus, which lacks both Min and NO proteins, Z ring positioning is governed by a bipolar gradient of the FtsZ inhibitor MipZ (Figure 2A; Thanbichler and Shapiro, 2006). Interestingly, MipZ belongs to the same family of ATPases as MinD, but unlike MinD it acts on FtsZ directly to block Z ring assembly (Thanbichler and Shapiro, 2006). MipZ is conserved across all α-proteobacteria that lack MinCD orthologs.
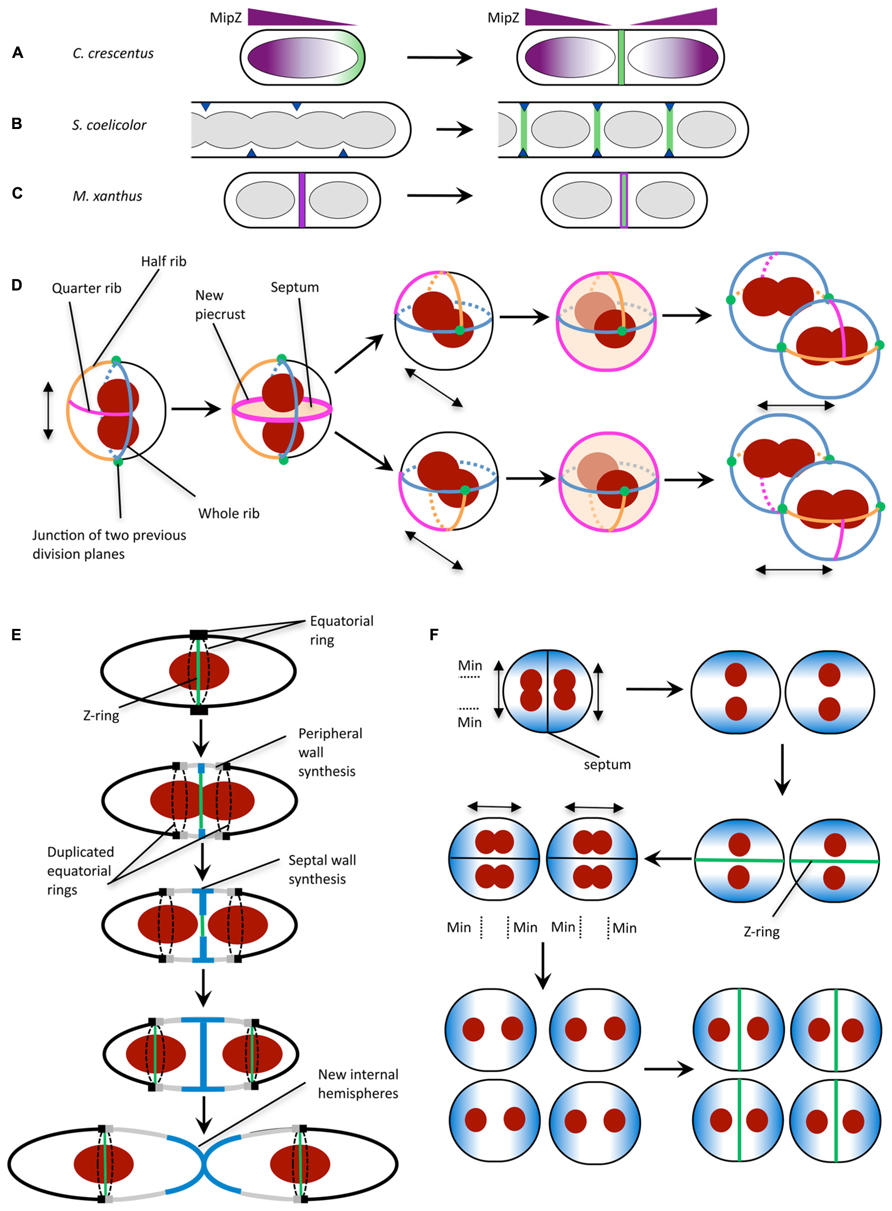
FIGURE 2. Mechanisms for division site selection in different bacterial species. (A) In C. crescentus, Z ring positioning is controlled by the FtsZ inhibitor MipZ (purple). MipZ associates with the chromosome, and forms a gradient of decreasing concentration with distance from the cell pole (see text). In newborn cells with one chromosome, the MipZ gradient is established from a single pole, confining FtsZ (green) to the opposite pole. Chromosome replication and segregation establishes a bipolar MipZ gradient that dislodges FtsZ from the pole and restricts Z ring assembly to midcell. (B) In sporulating hyphae of S. coelicolor, Z ring assembly is positively regulated by the SsgB protein (dark blue), which localizes to division sites (via SsgA) then recruits FtsZ (green). SsgB remains associated with the Z ring. (C) In M. xanthus, PomZ (magenta) localizes to the cell center following chromosome segregation, then recruits FtsZ to form the Z ring (green). (D) S. aureus has been proposed to utilize specific peptidoglycan features in conjunction with nucleoid occlusion for division in three perpendicular planes. Double-headed arrow indicates the axis of chromosome segregation. The plane for septum formation is marked by the presence of the quarter rib. The axis of chromosome segregation is determined by the movement of the nucleoids to junctions of two previous division planes. To determine the next division plane, Staphylococcal cells may recognize the quarter rib feature via a direct receptor-ligand type interaction (Turner et al., 2010). The plane containing the quarter rib also has the longest circumference which could be used for recognition by the cell (Turner et al., 2010). Division plane selection might also be aided by establishing the axis of chromosome segregation toward the junctions between division planes (green circles). (E) In S. pneumoniae, division plane selection has been suggested to rely on the presence of equatorial rings for cell division in consecutive parallel planes (see text). Equatorial rings are present at the cell equator and mark the site for septal cell wall synthesis through recruitment of divisome components such as FtsZ. The equatorial rings are then duplicated and move apart, via the synthesis of new peripheral peptidoglycan, until both rings are located at the equators of the new daughter cells. New peripheral and septal peptidoglycan synthesis are highlighted in gray and blue, respectively. (F) N. gonorrhoeae cell division in alternating perpendicular planes. At the onset of cell division, two temporarily asymmetric daughter cells are generated which have a short and long axis (Pinho et al., 2013). This leads to the oscillation of the Min protein complex as well as chromosome segregation along the long axis, which is parallel to the septal plane (Pinho et al., 2013). Note that N. gonorrhoeae does not contain a Noc/SlmA homolog, but the presence of the replisome machinery around the DNA may negatively regulate divisome assembly around the DNA, analogous to the action of Noc (Ramirez-Arcos et al., 2002). Double-headed arrow indicates axis of chromosome segregation. (A) is reproduced from Monahan and Harry (2013) and (B,C) are adapted from Monahan and Harry (2013) with permission from Wiley Interscience.
The bipolar MipZ gradient relies on interaction with the chromosome partitioning protein ParB, which itself localizes to cell poles via association with the chromosomal origin region. Interaction with ParB triggers the formation of MipZ dimers, which diffuse away and bind non-specifically to the chromosome (Kiekebusch et al., 2012). This establishes a gradient of decreasing MipZ concentration with increasing distance from the cell pole (the location of ParB). The intrinsic ATPase activity of MipZ releases MipZ monomers from the chromosome, which are then re-captured by ParB to continue the cycle (Kiekebusch et al., 2012).
Importantly, MipZ provides both spatial and temporal cues for Z ring formation. In newborn C. crescentus cells, which contain only one chromosomal origin, the MipZ gradient is established from a single pole, restricting FtsZ to the opposite pole (Figure 2A). Replication and segregation of the chromosomal origins then sets up a bipolar gradient in pre-divisional cells, which dislodges FtsZ from the pole and restricts Z ring formation to midcell (Figure 2A).
Streptomyces coelicolor: SsgAB
In sporulating cells of Streptomyces coelicolor, which also lacks the Min and NO systems, FtsZ is recruited and tethered to the division site directly via interaction with the membrane-associated protein SsgB (Figure 2B; Willemse et al., 2011). SsgB promotes FtsZ polymerization in vitro, and presumably stimulates Z ring assembly in S. coelicolor cells. The localization of SsgB is mediated by the orthologous protein SsgA (Traag and van Wezel, 2008; Willemse et al., 2011). Significantly, the SsgAB system was the first positive control mechanism to be reported for Z ring positioning (Willemse et al., 2011). However, both SsgA and SsgB are only present in Actinomycetes.
Myxococcus xanthus: PomZ
Another positive regulatory mechanism for Z ring placement has recently been reported in Myxococcus xanthus, a rod-shaped δ-proteobacterium that lacks all of the known FtsZ positioning proteins. In this organism, a novel protein called PomZ was shown to be required for efficient Z ring formation and for midcell Z ring placement (Treuner-Lange et al., 2013). Importantly, PomZ localizes to the division site, and does so both prior to and independently of FtsZ (Treuner-Lange et al., 2013). On this basis, it has been suggested that PomZ may not only identify the division site, but recruit FtsZ to this site and stabilize the Z ring (Figure 2C; Monahan and Harry, 2013; Treuner-Lange et al., 2013). Indeed, PomZ was shown to pull down FtsZ from whole-cell M. xanthus extracts (Treuner-Lange et al., 2013). However, it is not yet clear whether the two proteins bind directly or via an interaction partner.
Staphylococcus aureus
Staphylococcus aureus divides sequentially in orthogonal planes in three dimensional space (Tzagoloff and Novick, 1977), raising the question of how staphylococcal cells can re-orientate the division machinery and how these daughter cells then “remember” previous division events for subsequent divisions. Interestingly, S. aureus lacks a visible Min homolog but does contain a homolog of the nucleiod occlusion protein, Noc (Veiga et al., 2011). Noc co-localizes with the nucleoid and reduces the frequency of Z-ring formation over DNA (Veiga et al., 2011), consistent with its role in B. subtilis (Wu and Errington, 2004). The nucleoid in S. aureus fills a large region of the cytoplasmic space (Yu et al., 2010), which suggests that nucleiod occlusion plays a critical role in division plane selection (Pinho et al., 2013).
In order to divide in more than one plane with fidelity however, S. aureus needs to carry information from previous planes of division. This data is spatially affected by each round of division and so is unlikely to be encoded by DNA. It is possible that non-DNA cell components, such as the cell wall (Turner et al., 2010) spatially regulate division site selection by acting as a marker for previous and potential division planes. Early studies of S. aureus cell wall architecture revealed a heterogeneity in cell wall thickness (Gilbo et al., 1967; Touhami et al., 2004) which have been more recently defined as equatorial rings of thicker bands of peptidoglycan (“piecrusts”), as well as orthogonal bands of peptidoglycan which presumably represent remnants of previous piecrust features (Figure 2D; Turner et al., 2010). These differences in cell wall architecture may encode epigenetic information in S. aureus (that could be recognized by currently unidentified protein components; Veiga et al., 2011) to maintain planar division site selection with fidelity over many generations due to the presentation of specific cell wall architectural features denoting the division plane from two previous rounds of division (Turner et al., 2010).
Negative regulation of midcell selection by cell wall components may also play a role; a cryo-electron microscopy study revealed that wall teichoic acids (WTAs) may not be uniformly localized throughout the cell wall, at least during septum formation (Matias and Beveridge, 2007). WTA localization has been shown previously to affect the midcell localization of Atl and PBP4 in S. aureus (Atilano et al., 2010; Schlag et al., 2010) indicating that WTA distribution could regulate midcell localization. However, the role of cell wall architecture and composition in division site selection, and the mechanisms by which bacteria translate this architectural information to ensure correct division site selection still remains elusive.
Streptococcus pneumoniae
Ovococci, similar to rod-shaped organisms, divide in a single plane along the short axis of the cells (Margolin, 2000). However, the genomes of Streptococci and Enterococci show an absence of Noc and Min homologs (Pinho et al., 2013). The lack of NO is further illustrated by the observation that Z-rings in S. pneumoniae frequently form over nucleoids and that cell constriction occurs concurrently with the separation of the DNA (Land et al., 2013). Cell division in Streptococci begins with divisome assembly and initial in-growth of the septum at the cell equator which is marked by the equatorial ring (Higgins and Shockman, 1970; Wheeler et al., 2011). Soon after, this equatorial ring is duplicated and moves apart due to nascent peptidoglycan insertion between the rings. This “peripheral” elongation continues until new internal hemispheres are formed (Massidda et al., 2013). Presumably, the presence of the equatorial rings (analogous to the “piecrust” features seen in S. aureus; Wheeler et al., 2011) in the daughter cells alone (Figure 2E) can serve as a marker for the cell equator and hence the site for future cell division (Zapun et al., 2008) through an as yet-unidentified mechanism.
Neisseria gonorrhoeae
Neisseria gonorrhoeae (Ng) is a Gram-negative coccus that divides in two alternating perpendicular planes (Westling-Häggström et al., 1977) resulting in a tetrad of daughter cells. N. gonorrhoeae lacks a Noc or SlmA homolog but encodes MinC, MinD, and MinE. In contrast to E. coli, deletion of minCNg (Ramirez-Arcos et al., 2001) as well as minDNg (Szeto et al., 2001) led to abnormal cell division, lysis and reduced viability of gonococcal cells highlighting their importance for cell division in this organism. Furthermore, heterologous production of a GFP-tagged MinDNg in round E. coli rodA mutants showed GFP-MinDNg oscillating along the axis parallel to the septa (Ramirez-Arcos et al., 2002). This oscillation pattern along the long axis of the cell could generate a MinCD inhibitory concentration gradient which is lowest in the plane that is perpendicular to a previous division event thereby allowing septum formation within the next plane (Figure 2F; Ramirez-Arcos et al., 2002; Pinho et al., 2013).
Conclusion and Perspectives
While there are some recurring themes for the spatial regulation of cell division in bacteria, such as the use of the ParA/MinD family of ATPases in several organisms (Lutkenhaus, 2012; Treuner-Lange et al., 2013), many bacteria do it differently, with the proteins and mechanisms being less conserved than was once thought. The ultimate purpose of division site regulation is to ensure that cytokinesis produces viable daughter cells. The diversity of mechanisms by which bacteria can achieve this is further exemplified by a recent study of Mycobacterium spp., which showed that Z ring positioning is essentially random in these organisms and that cell division can occur over nucleoids. This appears to be compensated by post-septal DNA transport rather than strict control of division site placement (Singh et al., 2013).
There are several factors that might have enabled bacteria to evolve different positioning mechanisms. These include cell shape (e.g., rod versus coccus), mode of peptidoglycan synthesis (elongation/division modes), lifestyle (division occurs asymmetrically in C. crescentus for example, and is only essential during sporulation in Streptomyces), as well as the repertoire of FtsZ-binding proteins present in the organism. A striking recent example of niche-specific division site selection comes from the study of a rod-shaped γ-proteobacterium that adheres via one of its poles to the nematode Laxus oneistus. To ensure that both daughter cells remain attached to the host, this bacterium grows by increasing in width and forms its septum parallel rather than perpendicular to the length axis (Leisch et al., 2012). We are likely to find more variations on the division-positioning mechanism as more bacterial species are examined. Establishing the mechanistic details of these will be important, and will provide a more complete and universal understanding of how division site positioning is integrated with the cell cycle and physiology of the bacterial cell.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adams, D. W., and Errington, J. (2009). Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7, 642–653. doi: 10.1038/nrmicro2198
Atilano, M. L., Pereira, P. M., Yates, J., Reed, P., Veiga, H., Pinho, M. G., et al. (2010). Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 107, 18991–18996. doi: 10.1073/pnas.1004304107
Barak, I., and Wilkinson, A. J. (2007). Division site recognition in Escherichia coli and Bacillus subtilis. FEMS Microbiol. Rev. 31, 311–326. doi: 10.1111/j.1574-6976.2007.00067.x
Bates, D., and Kleckner, N. (2005). Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121, 899–911. doi: 10.1016/j.cell.2005.04.013
Bernard, R., Marquis, K. A., and Rudner, D. Z. (2010). Nucleoid occlusion prevents cell division during replication fork arrest in Bacillus subtilis. Mol. Microbiol. 78, 866–882. doi: 10.1111/j.1365-2958.2010.07369.x
Bernhardt, T. G., and de Boer, P. A. (2005). SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18, 555–564. doi: 10.1016/j.molcel.2005.04.012
Bramkamp, M., Emmins, R., Weston, L., Donovan, C., Daniel, R. A., and Errington, J. (2008). A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol. Microbiol. 70, 1556–1569. doi: 10.1111/j.1365-2958.2008.06501.x
Bramkamp, M., and van Baarle, S. (2009). Division site selection in rod-shaped bacteria. Curr. Opin. Microbiol. 12, 683–688. doi: 10.1016/j.mib.2009.10.002
Cambridge, J., Blinkova, A., Magnan, D., Bates, D., and Walker, J. R. (2013). A replication-inhibited un-segregated nucleoid at mid-cell blocks Z-ring formation and cell division independently of SOS and the SlmA nucleoid occlusion protein in Escherichia coli. J. Bacteriol. 196, 36–49. doi: 10.1128/JB.01230-12
Cho, H., Mcmanus, H. R., Dove, S. L., and Bernhardt, T. G. (2011). Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc. Natl. Acad. Sci. U.S.A. 108, 3773–3778. doi: 10.1073/pnas.1018674108
Dajkovic, A., Lan, G., Sun, S. X., Wirtz, D., and Lutkenhaus, J. (2008). MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 18, 235–244. doi: 10.1016/j.cub.2008.01.042
De Boer, P. A. (2010). Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13, 730–737. doi: 10.1016/j.mib.2010.09.015
Gilbo, C. M., Beaton, C. D., and Coles, N. W. (1967). Electron microscopy of the lysis of Staphylococcus aureus cell walls by aeromonas lytic factor. J. Bacteriol. 93, 1972–1975.
Harry, E., Monahan, L., and Thompson, L. (2006). Bacterial cell division: the mechanism and its precision. Int. Rev. Cytol. 253, 27–94. doi: 10.1016/S0074-7696(06)53002-5
Harry, E. J., Rodwell, J., and Wake, R. G. (1999). Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Mol. Microbiol. 33, 33–40. doi: 10.1046/j.1365-2958.1999.01439.x
Higgins, M. L., and Shockman, G. D. (1970). Model for cell wall growth of Streptococcus faecalis. J. Bacteriol. 101, 643–648.
Hu, Z. L., Mukherjee, A., Pichoff, S., and Lutkenhaus, J. (1999). The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. U.S.A. 96, 14819–14824. doi: 10.1073/pnas.96.26.14819
Kiekebusch, D., Michie, K. A., Essen, L. O., Lowe, J., and Thanbichler, M. (2012). Localized dimerization and nucleoid binding drive gradient formation by the bacterial cell division inhibitor MipZ. Mol. Cell 46, 245–259. doi: 10.1016/j.molcel.2012.03.004
Land, A. D., Tsui, H.-C. T., Kocaoglu, O., Vella, S. A., Shaw, S. L., Keen, S. K., et al. (2013). Requirement of essential Pbp2x and GpsB for septal ring closure in Streptococcus pneumoniae D39. Mol. Microbiol. 90, 939–955. doi: 10.1111/mmi.12408
Leisch, N., Verheul, J., Heindl, N. R., Gruber-Vodicka, H. R., Pende, N., Den Blaauwen, T., et al. (2012). Growth in width and FtsZ ring longitudinal positioning in a gammaproteobacterial symbiont. Curr. Biol. 22, R831–R832. doi: 10.1016/j.cub.2012.08.033
Lenz, P., and Sogaard-Andersen, L. (2011). Temporal and spatial oscillations in bacteria. Nat. Rev. Microbiol. 9, 565–577. doi: 10.1038/nrmicro2612
Lutkenhaus, J. (2007). Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76, 14.11–14.24.
Lutkenhaus, J. (2012). The ParA/MinD family puts things in their place. Trends Microbiol. 20, 411–418. doi: 10.1016/j.tim.2012.05.002
Lutkenhaus, J., Pichoff, S., and Du, S. (2012). Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton (Hoboken) 69, 778–790. doi: 10.1002/cm.21054
Margolin, W. (2000). Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24, 531–548. doi: 10.1111/j.1574-6976.2000.tb00554.x
Margolin, W. (2005). FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6, 862–871. doi: 10.1038/nrm1745
Massidda, O., Nováková, L., and Vollmer, W. (2013). From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division? Environ. Microbiol. 15, 3133–3157. doi: 10.1111/1462-2920.12189
Matias, V. R., and Beveridge, T. J. (2007). Cryo-electron microscopy of cell division in Staphylococcus aureus reveals a mid-zone between nascent cross walls. Mol. Microbiol. 64, 195–206. doi: 10.1111/j.1365-2958.2007.05634.x
Migocki, M. D., Freeman, M. K., Wake, R. G., and Harry, E. J. (2002). The Min system is not required for precise placement of the midcell Z ring in Bacillus subtilis. EMBO Rep. 3, 1163–1167. doi: 10.1093/embo-reports/kvf233
Monahan, L. G., and Harry, E. J. (2013). Identifying how bacterial cells find their middle: a new perspective. Mol. Microbiol. 87, 231–234. doi: 10.1111/mmi.12114
Moriya, S., Rashid, R. A., Rodrigues, C. D., and Harry, E. J. (2010). Influence of the nucleoid and the early stages of DNA replication on positioning the division site in Bacillus subtilis. Mol. Microbiol. 76, 634–647. doi: 10.1111/j.1365-2958.2010.07102.x
Nevo-Dinur, K., Govindarajan, S., and Amster-Choder, O. (2012). Subcellular localization of RNA and proteins in prokaryotes. Trends Genet. 28, 314–322. doi: 10.1016/j.tig.2012.03.008
Patrick, J. E., and Kearns, D. B. (2008). MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol. Microbiol. 70, 1166–1179. doi: 10.1111/j.1365-2958.2008.06469.x
Pinho, M. G., Kjos, M., and Veening, J.-W. (2013). How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat. Rev. Microbiol. 11, 601–614. doi: 10.1038/nrmicro3088
Ramirez-Arcos, S., Szeto, J., Beveridge, T., Victor, C., Francis, F., and Dillon, J. (2001). Deletion of the cell-division inhibitor MinC results in lysis of Neisseria gonorrhoeae. Microbiology 147, 225–237.
Ramirez-Arcos, S., Szeto, J., Dillon, J. A., and Margolin, W. (2002). Conservation of dynamic localization among MinD and MinE orthologues: oscillation of Neisseria gonorrhoeae proteins in Escherichia coli. Mol. Microbiol. 46, 493–504. doi: 10.1046/j.1365-2958.2002.03168.x
Regamey, A., Harry, E. J., and Wake, R. G. (2000). Mid-cell Z ring assembly in the absence of entry into the elongation phase of the round of replication in bacteria: co-ordinating chromosome replication with cell division. Mol. Microbiol. 38, 423–434. doi: 10.1046/j.1365-2958.2000.02130.x
Rodrigues, C. D., and Harry, E. J. (2012). The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization. PLoS Genet. 8:e1002561. doi: 10.1371/journal.pgen.1002561
Rothfield, L., Taghbalout, A., and Shih, Y. L. (2005). Spatial control of bacterial division-site placement. Nat. Rev. Microbiol. 3, 959–968. doi: 10.1038/nrmicro1290
Rudner, D. Z., and Losick, R. (2010). Protein subcellular localization in bacteria. Cold Spring Harb. Perspect. Biol. 2, a000307. doi: 10.1101/cshperspect.a000307
Schlag, M., Biswas, R., Krismer, B., Kohler, T., Zoll, S., Yu, W., et al. (2010). Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol. Microbiol. 75, 864–873. doi: 10.1111/j.1365-2958.2009.07007.x
Shapiro, L., Mcadams, H. H., and Losick, R. (2009). Why and how bacteria localize proteins. Science 326, 1225–1228. doi: 10.1126/science.1175685
Singh, B., Nitharwal, R. G., Ramesh, M., Pettersson, B. M., Kirsebom, L. A., and Dasgupta, S. (2013). Asymmetric growth and division in Mycobacterium spp.: compensatory mechanisms for non-medial septa. Mol. Microbiol. 88, 64–76. doi: 10.1111/mmi.12169
Smits, W. K., Goranov, A. I., and Grossman, A. D. (2010). Ordered association of helicase loader proteins with the Bacillus subtilis origin of replication in vivo. Mol. Microbiol. 75, 452–461. doi: 10.1111/j.1365-2958.2009.06999.x
Szeto, J., Ramirez-Arcos, S., Raymond, C., Hicks, L. D., Kay, C. M., and Dillon, J.-A. R. (2001). Gonococcal MinD affects cell division in Neisseria gonorrhoeae and Escherichia coli and exhibits a novel self-interaction. J. Bacteriol. 183, 6253–6264. doi: 10.1128/JB.183.21.6253-6264.2001
Thanbichler, M. (2009). Spatial regulation in Caulobacter crescentus. Curr. Opin. Microbiol. 12, 715–721. doi: 10.1016/j.mib.2009.09.013
Thanbichler, M., and Shapiro, L. (2006). MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126, 147–162. doi: 10.1016/j.cell.2006.05.038
Tonthat, N. K., Arold, S. T., Pickering, B. F., Van Dyke, M. W., Liang, S., Lu, Y., et al. (2011). Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 30, 154–164. doi: 10.1038/emboj.2010.288
Tonthat, N. K., Milam, S. L., Chinnam, N., Whitfill, T., Margolin, W., and Schumacher, M. A. (2013). SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid. Proc. Natl. Acad. Sci. U.S.A. 110, 10586–10591. doi: 10.1073/pnas.1221036110
Touhami, A., Jericho, M. H., and Beveridge, T. J. (2004). Atomic force microscopy of cell growth and division in Staphylococcus aureus. J. Bacteriol. 186, 3286–3295. doi: 10.1128/JB.186.11.3286-3295.2004
Traag, B. A., and van Wezel, G. P. (2008). The SsgA-like proteins in actinomycetes: small proteins up to a big task. Antonie Van Leeuwenhoek 94, 85–97. doi: 10.1007/s10482-008-9225-3
Treuner-Lange, A., Aguiluz, K., Van Der Does, C., Gomez-Santos, N., Harms, A., Schumacher, D., et al. (2013). PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus. Mol. Microbiol. 87, 235–253. doi: 10.1111/mmi.12094
Turner, R. D., Ratcliffe, E. C., Wheeler, R., Golestanian, R., Hobbs, J. K., and Foster, S. J. (2010). Peptidoglycan architecture can specify division planes in Staphylococcus aureus. Nat. Commun. 1, 26. doi: 10.1038/ncomms1025
Tzagoloff, H., and Novick, R. (1977). Geometry of cell division in Staphylococcus aureus. J. Bacteriol. 129, 343–350.
Veiga, H., Jorge, A. M., and Pinho, M. G. (2011). Absence of nucleoid occlusion effector Noc impairs formation of orthogonal FtsZ rings during Staphylococcus aureus cell division. Mol. Microbiol. 80, 1366–1380. doi: 10.1111/j.1365-2958.2011.07651.x
Westling-Häggström, B., Elmros, T., Normark, S., and Winblad, B. (1977). Growth pattern and cell division in Neisseria gonorrhoeae. J. Bacteriol. 129, 333–342.
Wheeler, R., Mesnage, S., Boneca, I. G., Hobbs, J. K., and Foster, S. J. (2011). Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol. Microbiol. 82, 1096–1109. doi: 10.1111/j.1365-2958.2011.07871.x
Willemse, J., Borst, J. W., De Waal, E., Bisseling, T., and Van Wezel, G. P. (2011). Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 25, 89–99. doi: 10.1101/gad.600211
Wu, L. J., and Errington, J. (2004). Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117, 915–925. doi: 10.1016/j.cell.2004.06.002
Wu, L. J., and Errington, J. (2012). Nucleoid occlusion and bacterial cell division. Nat. Rev. Microbiol. 10, 8–12. doi: 10.1038/nrmicro2671
Wu, L. J., Franks, A. H., and Wake, R. G. (1995). Replication through the terminus region of the Bacillus subtilis chromosome is not essential for the formation of a division septum that partitions the DNA. J. Bacteriol. 177, 5711–5715.
Wu, L. J., Ishikawa, S., Kawai, Y., Oshima, T., Ogasawara, N., and Errington, J. (2009). Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 28, 1940–1952. doi: 10.1038/emboj.2009.144
Yu, W., Herbert, S., Graumann, P. L., and Götz, F. (2010). Contribution of SMC (structural maintenance of chromosomes) and SpoIIIE to chromosome segregation in staphylococci. J. Bacteriol. 192, 4067–4073. doi: 10.1128/JB.00010-10
Yu, X. C., and Margolin, W. (1999). FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol. 32, 315–326. doi: 10.1046/j.1365-2958.1999.01351.x
Keywords: cell division, ftsZ, min system, nucleoid occlusion, Z ring, division regulation, bacterial cell division
Citation: Monahan LG, Liew ATF, Bottomley AL and Harry EJ (2014) Division site positioning in bacteria: one size does not fit all. Front. Microbiol. 5:19. doi: 10.3389/fmicb.2014.00019
Received: 28 November 2013; Accepted: 13 January 2014;
Published online: 03 February 2014.
Edited by:
Marc Bramkamp, Ludwig Maximilians University Munich, GermanyReviewed by:
Daniel B. Kearns, Indiana University, USAJan-Willem Veening, University of Groningen, Netherlands
Copyright © 2014 Monahan, Liew, Bottomley and Harry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elizabeth J. Harry, The ithree Institute, University of Technology, P.O. Box 123, Ultimo, Sydney, NSW 2007, Australia e-mail: liz.harry@uts.edu.au
 Leigh G. Monahan
Leigh G. Monahan Andrew T. F. Liew
Andrew T. F. Liew Amy L. Bottomley
Amy L. Bottomley Elizabeth J. Harry
Elizabeth J. Harry