- 1Division of Applied Life Sciences, Graduate School of Agriculture, Kyoto University, Kyoto, Japan
- 2Center for Fostering Young and Innovative Researchers, Nagoya Institute of Technology, Nagoya, Aichi, Japan
- 3Kobe University Research Center for Inland Seas, Awaji, Hyogo, Japan
- 4Japan Science and Technology Agency, CREST, Awaji, Hyogo, Japan
- 5Advanced Low Carbon Technology Research and Development Program, Japan Science and Technology Agency, Tokyo, Japan
This study investigated the potential local CH4 sink in various plant parts as a boundary environment of CH4 emission and consumption. By comparing CH4 consumption activities in cultures inoculated with parts from 39 plant species, we observed significantly higher consumption of CH4 associated with aquatic plants than other emergent plant parts such as woody plant leaves, macrophytic marine algae, and sea grass. In situ activity of CH4 consumption by methanotrophs associated with different species of aquatic plants was in the range of 3.7–37 μmol·h−1·g−1 dry weight, which was ca 5.7–370-fold higher than epiphytic CH4 consumption in submerged parts of emergent plants. The qPCR-estimated copy numbers of the particulate methane monooxygenase-encoding gene pmoA were variable among the aquatic plants and ranged in the order of 105–107 copies·g−1 dry weight, which correlated with the observed CH4 consumption activities. Phylogenetic identification of methanotrophs on aquatic plants based on the pmoA sequence analysis revealed a predominance of diverse gammaproteobacterial type-I methanotrophs, including a phylotype of a possible plant-associated methanotroph with the closest identity (86–89%) to Methylocaldum gracile.
Introduction
Methane (CH4) is the second most important greenhouse gas after CO2, and it accounts for 20–30% of the contribution of greenhouse gases to global warming (Solomon et al., 2007). Its concentration has rapidly increased by 2.5-fold since 1750 from approximately 700 to 1775 ppbv in 2005 (Solomon et al., 2007), although annual increases have varied. For long-term forecasting and control of atmospheric CH4, it is necessary to understand the CH4-flux, or the balance of sources and sinks in the environment. One of the most important processes of CH4 production is methanogenesis by Archaea. On the other hand, the largest CH4 sink is the dispersion (>80%) via reaction with hydroxyl-radicals in the troposphere, and the second sink is diffusion into the stratosphere. Consumption of CH4 by methanotrophs is the main biological CH4 sink, and is considered to contribute significantly to CH4 mitigation under both aerobic and anaerobic conditions (Conrad, 2009; Borrel et al., 2011). Active CH4 consumption is considered to occur at the oxic-anoxic interface. However, CH4 consumption in such environments is overlooked when the emission of CH4 is balanced against consumption. Local CH4 sinks have recently been investigated in various environments such as surface layers of wetlands, paddy fields, and sediments (Conrad, 2009). For example, more than 90% of the potentially emitted CH4 was consumed in the surface layer of lake sediment (Frenzel et al., 1990). Some important ecological CH4 sinks might be unaccounted for or not discovered.
Microbial consumption of CH4 is mainly conducted by methanotrophic bacteria. Methanotrophic bacteria belong to the phyla Proteobacteria (Hanson and Hanson, 1996), Verrucomicrobia including extremely acidophilic methanotrophs isolated from volcanic habitats (Op den Camp et al., 2009), and the candidate division NC10. NC10 includes a candidate oxygenic methanotroph that oxidizes CH4 by using O2 produced from nitrite (Ettwig et al., 2010). Among those methanotrophs, proteobacterial methanotrophs have been frequently detected as active CH4 utilizers in terrestrial environments, and grouped into the Gammaproteobacteria and Alphaproteobacteria. Gammaproteobacterial type I methanotrophs develop intracytoplasmic membranes (ICMs) as bundles of vesicular discs for CH4 oxidation and use the ribulose monophosphate pathway for formaldehyde fixation, while type II methanotrophs, which are members of the Alphaproteobacteria, possess ICMs composed of paired peripheral layers and utilize the serine pathway for formaldehyde fixation. The traditional type I and type II classification of methanotrophs were postulated based on these physiological characteristics. However, recent isolation of new methanotrophs revealed many exceptions to the traditional classification, giving more updated view on methanotrophs ecology, physiology, and phylogeny (Semrau et al., 2010; Borrel et al., 2011; Stein et al., 2012). Methane oxidization is catalyzed by two types of methane monooxygenases (MMO), soluble cytoplasmic MMO (sMMO), and membrane-bound particulate MMO (pMMO). While sMMO is found in a subset of methanotrophs, pMMO is present in all methanotrophs except Methylocella and Methyloferula (Dedysh et al., 2000; Dedysh, 2009; Vorobev et al., 2011). Therefore, the gene pmoA, encoding the beta subunit of pMMO, is often used as a biomarker to specifically detect aerobic methanotrophs. The sequences of pmoA are evolutionally conserved and reflect the 16S rRNA gene-based phylogeny of methanotrophs (McDonald et al., 2008).
Plant surfaces, not only in the rhizosphere, but also the emergent parts of plants, or phyllosphere, have recently attracted attention as sites for CH4 emission. Some plants have been reported to produce CH4 coupled with photosynthesis and emit CH4 from plant leaves (Keppler et al., 2006); such CH4 emissions account for 6% of the total CH4 emission on earth (Conrad, 2009). As is commonly observed in environments near CH4 emission-sources, a local CH4 sink in plants has also been reported for mosses (Raghoebarsing et al., 2005), rice roots, and emergent and aquatic plants (Heilman and Carlton, 2001; Sorrell et al., 2002).
CH4 is also expected to be consumed by plant-associated methanotrophs, which we have recently enriched and identified from various plant parts together with methanol-utilizing bacteria (Iguchi et al., 2012). The latter are well known as common inhabitants of the phyllosphere (Lidstrom, 2006; Knief et al., 2011; Vorholt, 2012). These findings suggested a possible contribution of methanotrophs to oxidation of CH4 in the phyllosphere. Therefore, studies on the interaction between plants and methanotrophs will provide useful information to understand and control the carbon cycle. However, such methanotroph-plant interactions have been investigated in very few plants, such as rice (Shrestha et al., 2008; Pfluger et al., 2011; Stein et al., 2012) and Sphagnum mosses (Raghoebarsing et al., 2005; Kip et al., 2011).
In this study, the local CH4 sink in plants was evaluated using plant parts sampled from various environments to determine the potential of methanotrophs to consume CH4 in different species of plants. After comparing the CH4-consuming activities of 39 different plant species, we discovered high CH4 consumption by methanotrophs on aquatic plants. Furthermore, methanotrophs in aquatic plants were quantified and identified based on pmoA sequence analysis.
Materials and Methods
CH4 Consumption in Cultures Inoculated with Plant Parts
As a first step to determine what kinds of plants have associations with CH4 consuming methanotrophs, CH4 consumption by plant samples was assayed by analyzing the changes of CH4 concentration in cultures inoculated with various plant parts listed in Supplemental Table 1. About one gram of plant material was placed into a glass-vial (60 mL capacity), which was filled with 20 ml of autoclaved mineral NMS medium (Whittenbury et al., 1970) supplemented with trace vitamins and trace elements (Yoshida et al., 2007). Submerged parts of emergent aquatic plants were used for incubations after washing by shaking in distilled water three times to remove soil. The incubation vials were closed with Teflon-coated butyl rubber caps. CH4 (5% v/v) was added to the vial by syringe and vials were incubated for 14 days at 28°C with shaking.
CH4 Consuming Activities of Methanotrophs in Aquatic Plants
Eight submerged aquatic plants and two aquatic plants with floating or aerial leaves (Figure 1) were sampled from a shallow eutrophic sub-basin (614 km2 of surface area and 3.5 m average depth) of Lake Biwa in Japan (35°4′ 32″N, 135°56′ 5″E), where the area covered with aquatic plants amounted to 52% of the total surface area in the basin in 2000 (Hamabata and Kobayashi, 2002). To analyze CH4 consumption activities by the aquatic plants, 5 g of plant material (wet weight) from each macrophyte was placed into 120 mL glass vials. Rice, Oryza sativa spp. japonica, cultivator Koshihikari, which had been growing for about 2 months and was 30 cm in height, was analyzed for comparison. The glass vials were supplemented with 1% (v/v) CH4 and closed with butyl-lubber caps. The vials were incubated for 30 h at 25°C in the dark and the CH4 in the headspace was analyzed every hour by using GC-FID as reported previously (Iguchi et al., 2011). Ten mL of lake water was used instead of aquatic plants to compare the CH4 consumption activities with those of aquatic plants. Aquatic plants and rice were washed by shaking three times in lake water and flood water from the rice field, respectively.
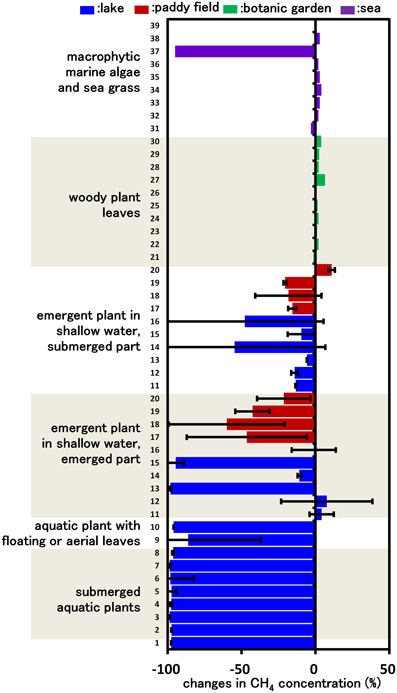
Figure 1. CH4 consumption in cultures inoculated with various plant parts. Columns indicate the changes in CH4 concentration after incubation for 14 days in vials containing mineral medium amended with 5% CH4 and plant parts. Each column indicates a single determination inoculated with each plant species (listed in Supplemental Table 1). Data for emergent and submerged aquatic plants represent the mean ± SD from three different glass bottles independently prepared in parallel, while data for tree leaves and sea weed are single determinations.
DNA Extraction from Water Plant Material
For analysis of methanotrophic microbial communities in the aquatic plants, approximately 0.5 g of plant material (wet weight) from each plant were used for DNA extraction. DNA was extracted from the aquatic plants by using an ISOIL DNA extraction kit (NIPPON GENE, Tokyo, Japan) according to manufacturer's instructions.
qPCR Targeting pmoA
qPCR targeting the pmoA gene using primers A189 and A682 (Holmes et al., 1995) was performed with a LightCycler FastStart DNA Master SYBR green I kit (Roche Molecular Biochemicals, Indianapolis, IN) and the LightCycler system (Roche Applied Science, Indianapolis, IN) as described previously (Yoshida et al., 2009). The PCR profile consisted of preheating at 95°C for 10 min and 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 10 s, and extension at 72°C for 20 s. The fluorescence signal was detected at 72°C during each cycle, and a melting curve was obtained by heating the product to 95°C and cooling it to 40°C. The calibration curves were graphed and the copy numbers of pmoA were calculated with LightCycler software version 3.5 (Roche Diagnostics) using serial dilutions of the pmoA amplicons from Methylococcus capsulatus Bath as a standard. The PCR efficiency was 90%, which was calculated from qPCR using serially diluted standard samples prepared from the pmoA amplicons of Methylococcus capsulatus Bath. Melting curve analyses demonstrated the absence of primer dimer formation and non-specific amplifications in all samples facilitating the use of 72°C as temperature of recording fluorescence.
Clone Library of pmoA Amplicons from Aquatic Plants
The extracted DNA samples were used as templates for amplification of pmoA by using two pmoA-specific primer sets, A189 and A682 (Holmes et al., 1995) and A189 and mb661 (Costello and Lidstrom, 1999). PCR using KOD Fx Neo (TOYOBO, Osaka, Japan) was performed by preheating the mixture to 94°C for 2 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 55°C for 30 s, and extension at 74°C for 30 s. The pmoA amplicons from aquatic plants were cloned by using a pCR®8/GW/TOPO® TA cloning kit (Invitrogen, Carlsbad, CA, USA) and sequenced using a BigDye Terminator v3.1 cycle sequencing kit and an ABI 3130 Genetic Analyzer (Applied Biosystems). The clones were chimera-checked by using chimera detection software, Bellerophon (Huber et al., 2004) and classified to operational taxonomic units (OTUs) having <0.07 of the evolutional distance (e.d.) in amino acid sequences by using Mothur (Schloss et al., 2009). The phylogenetic tree of the OTUs and relatives was drawn by the neighborhood joining method using MEGA5.1 (Tamura et al., 2011).
Enrichment and Isolation of Methanotrophs from Aquatic Plants
A piece (about 1 cm2) of aquatic plant material was inoculated into a glass-vial (30 mL capacity) containing 5 ml of NMS or AMS medium (Whittenbury et al., 1970) and 10% CH4 (v/v) as described above. The cultures were incubated at 28°C with shaking and subcultured in fresh medium when CH4 in the headspace was consumed. After 3–4 serial transfers, the cultures were serially diluted and spread onto 1.5% agar plates of NMS or AMS medium. The agar plates were incubated for 2–4 weeks at 28°C in a jar filled with a CH4/air mixture. About 100 colonies were picked and subcultured from each culture. The colonies were purified by repeating agar cultivation until a single colony morphotype was obtained. The isolates were phylogenetically identified by sequencing 16S rRNA and pmoA genes, which were amplified from cell lysates of the isolates (Yoshida et al., 2007).
Nucleotide Sequence Accession Numbers
The nucleotide sequence data reported in this paper have been deposited under DDBJ/EMBL/GenBank (http://www.ddbj.nig.ac.jp/Welcome-j.html) accession no. AB844797–AB845152, AB845287–AB845309, and AB845153–AB845175.
Results
CH4 Consumption in Cultures Inoculated with Various Parts of Plants
Our previous study indicated that methanotrophs inhabit emergent parts of various plant surfaces (Iguchi et al., 2012). In this work, we aimed to determine what types of plant could potentially serve as a niche for methanotrophs. Based on the assumption that CH4 consumption is only carried out by methanotrophic bacteria, we measured CH4 consumption in cultures inoculated with plant parts taken from various environments, thereby evaluating CH4 consumption activity by plant-associated methanotrophic consortia. After 14 days of incubation, none of the cultures inoculated with woody plant leaves showed detectable consumption of CH4. In contrast, cultures inoculated with several emergent plants, macrophytic marine algae and all aquatic plants consumed significant amounts of CH4 (Figure 1). In the emergent grass cultures, the cultures of emergent parts consumed higher amounts of CH4 than those of submerged parts. As a result of this screening, we found that aquatic plants showed a constant high CH4-consuming activity, and therefore the aquatic plants were subjected to further investigation.
CH4 Consumption by Aquatic Plants
Figure 2 shows the CH4 consumption by eight submerged aquatic plants and two aquatic plants with floating or aerial leaves. Within 30 h of incubation at 25°C, all 10 aquatic plants consumed significant amounts of CH4, in contrast to the vial containing lake water without aquatic plants, which did not consume CH4 within 30 h (data not shown). The CH4 consumption by aquatic plants was variable among the aquatic plants and sample preparations, exhibiting 3.7–37 μmol·h−1·g−1 dry weight of CH4 consumption in native submerged samples. In the control using rice as an emergent plant, the CH4 consumption was lower than that of aquatic plants with values of 0.018 ± 0.08, 0.29 ± 0.22, and 0.37 ± 0.03 μmol·h−1·g−1 dry weight for leaves, stems and washed roots, respectively. These results showed that aquatic plants had much higher CH4 consumption activity than the emergent plants.
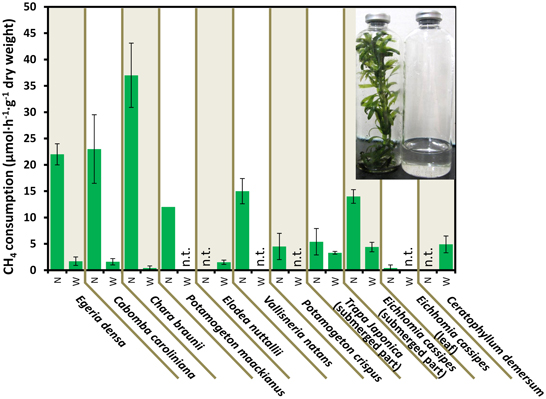
Figure 2. CH4 consumption in the aquatic plants. Data represent the mean ± SD from three different glass bottles independently prepared in parallel, except for the single determination with P. maackianus. The photo shows the incubated E. densa (left) and lake water (right) in bottles. N and W indicate unwashed native samples and samples washed by shaking three times in lake water, respectively. “n.t.” is “not determined.”
CH4 consumption by aquatic plants declined to 0.45–4.4 μmol·h−1·g−1 dry weight when bacterial biofilms were removed by washing. In Egeria densa, Cabomba caroliniana, and Chara braunii, 92–99% of the CH4 consumption was lost in the washed samples, whereas submerged parts of aquatic plants with floating or aerial leaves, Eichhomia cassipes and Trapa japonica retained 61 and 41% of the activity after washing, respectively. These results suggest that most methanotrophs were loosely associated with the surface of aquatic plants, but some associated tightly with or were within aquatic plants.
Quantitative Detection of Methanotrophs in Aquatic Plants
qPCR targeting a component of the pMMO-encoding gene, pmoA, produced amplicons of a single size from the extracted DNA of all CH4 consuming samples, i.e., E. densa, C. caroliniana, C. braunii, Potamogeton maackianus, Vallisneria natans, P. crispus, and submerged parts of T. japonica and E. cassipes. The qPCR-estimated copy numbers of pmoA from those samples were well correlated (r2 = 0.71) with the CH4 consumption activities (Figure 3), clearly indicating the involvement of methanotrophs in CH4 consumption. The populations of methanotrophs in aquatic plants were also variable. In the native sample, the copy numbers of pmoA were in the range from 4.6 ± 1.8 × 105 to 6.9 ± 0.77 × 107 copies·g−1 dry weight. The pmoA copy number declined to 990 ± 510 to 1.0 × 104 ± 890 copies·g−1 dry weight after washing, corresponding to 0.001–6.5% of the pmoA before washing. As observed with CH4 consumption activities, this result also supported a loose association of methanotrophs with the surface of aquatic plants. To date, populations of methanotrophs associated with emergent parts of plants have not been investigated in detail. Our present results indicate that methanotrophs associated with aquatic plants constitute a larger CH4 sink than those associated with the phyllosphere of terrestrial plants, and have demonstrated an unexpected contribution of aquatic plants to CH4 consumption.
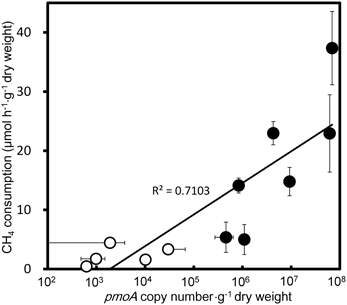
Figure 3. Correlation of qPCR estimated pmoA copy numbers and CH4 consumption activities in the aquatic plants. Filled circles and open circles represent the data from the native and slightly washed samples, respectively. Data represent the mean ± SD from three different samples independently prepared in parallel.
Phylogeny of Methanotrophs Associated with Aquatic Plants
A total of 358 cloned pmoA gene sequences were determined and classified into 21 OTUs (Figure 4 and Supplemental Table 2). These sequences were similar to those in members of the genus Methylosinus, which are type II methanotrophs in the Alphaproteobacteria, and the genera Methylosarcina, Methylomicrobium, Methylomonas, Methylovulum, Methylocaldum, and Methylococcus, which are type I methanotrophs in the Gammaproteobacteria. Three OTUs, OTU3, OTU19, and OTU21 were detected in all aquatic plants and represented 28, 20, and 11% of the total clones, respectively. The sequences of OTU3, the most abundant OTU, were most closely related (94–99% amino acid sequence identity) to pmoA sequences of Methylosarcina lacus LW14 (Kalyuzhnaya et al., 2005) among the available isolates, and were most closely related to pmoA-clones from rice roots (Shrestha et al., 2010), lake littoral wetlands (Siljanen et al., 2011), lake littoral sediments (Deutzmann et al., 2011), and rice field soil (Mayumi et al., 2010) (Figure 5). The sequences of OTU19, the second most abundant OTU, were 87–89% similar to the pmoA sequence from Methylocaldum gracile VKM-14L (Bodrossy et al., 1997) among isolated strains, and all of the clones were most closely related to pmoA clones from rice roots (Shrestha et al., 2008). The sequence of the third most abundant OTU (OTU21), which was 91–95% similar to the pmoA sequence of Methylococcaceae sp. strain OS501 (Iguchi et al., 2011) as the closest related isolate, was most closely related to a variety of environmental clones from littoral sediments of Lake Constance (Bussmann et al., 2004), CH4-consuming sludge (Osaka et al., 2008), and rice roots (Shrestha et al., 2010). In addition to those highly abundant OTUs, six OTUs had more than 5% frequency among the total clones and were detected in more than four aquatic plants as follows; the sequences of OTU1 were closely related to clones from lake sediment (Costello and Lidstrom, 1999; Pester et al., 2004), rice roots (Lüke et al., 2010), plant leaves (Iguchi et al., 2012) and onshore oil and gas field soil (Xu et al., 2013), the sequences of OTU2 were related to clones from landfill soils (Henneberger et al., 2012), the sequences of OTU4, OTU8, and OTU14 were related to clones from lake sediments (Costello and Lidstrom, 1999; Lin et al., 2004; Nercessian et al., 2005; Bussmann et al., 2006; Deutzmann et al., 2011; Siljanen et al., 2011), and the sequences of OTU15 were related to clones from rice roots (Shrestha et al., 2008; Lüke et al., 2010) and rice field soil (Ho et al., 2011).
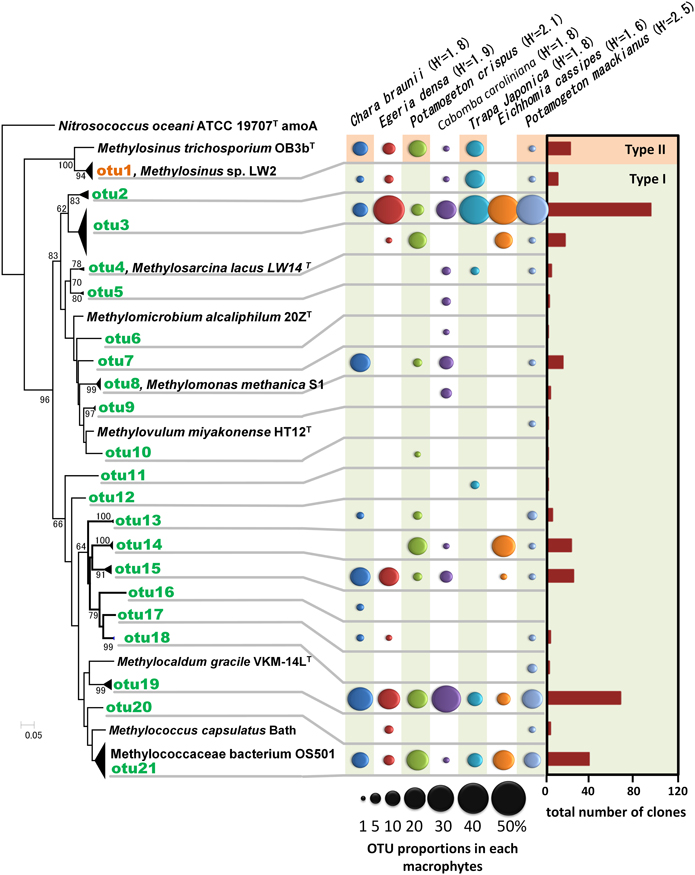
Figure 4. Neighbor-joining tree of methanotrophic phylotypes detected in the aquatic plants based on the deduced amino acid sequences of pmoA genes. Sequence of amoA of Nitrosococcus oceani was used as an outgroup to root the tree. H' values are Shannon diversity indices (Shannon and Weaver, 1948). Bootstrap values (>60%) obtained from 1000 resamplings are shown. Accession numbers for the sequences are AAC25091, ACN73467, ACN73466, AAA87220, AAF08211, AAB49821, AAC04380, BAL04120, ABD13901, YP 004915812, BAJ17641, AAA87218, AAG13081.
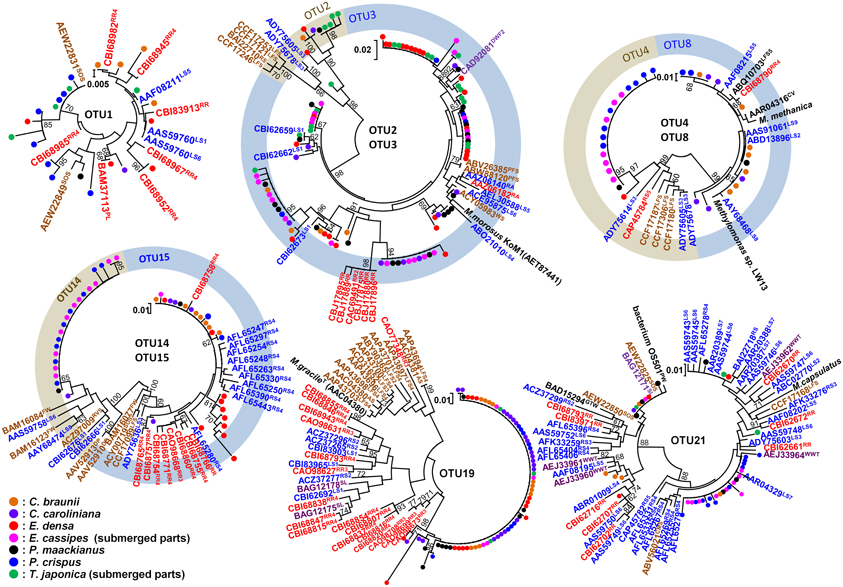
Figure 5. Neighbor-joining tree of individual clones in abundant OTUs and their related environmental clones based on deduced amino acid sequences of pmoA genes. The clones obtained in this study were indicated as closed circles. Bootstrap values (>60%) obtained from 1000 resamplings are shown. Related clones are indicated as accession numbers with superscript indicating their origins as follows; plants in red including RR, rice root and PL, plant leaf, saturated soil or sediment in blue including RS, rice field soil; LS, lake sediment; RA, river aquifer; TCE, TCE-contaminated aquifer; WS, wetland soil; and RVS, river sediment, unsaturated soil in brown including AS, agricultural soil; PFS, permafrost soil; LFS, landfill-cover soil; SOS, surface soils of onshore oil and gas fields; and CMS, coal mine soil, clone from water in purple including DWF, drinking water facility; SL, methane-consuming sludge; WWT, wastewater treatment plant; DP, drainage of peat land; GTW, subsurface geothermal water stream in a Japanese gold mine, and clones from biomat in black including FWI, freshwater iron-rich microbial mat; PW, pond water; and CV, biomat and water in cave.
Among the identified OTUs, all sequences of OTU19 as well as most sequences of OTU15 formed a cluster with the sequences of the clones obtained from rice roots, but not with other clusters. In particular, methanotrophs corresponding to OTU19 were widely and abundantly distributed in seven different aquatic plants and the sequences formed a cluster only with the sequences of the clones obtained from two different rice roots in different places. These results suggest that methanotrophs represented by OTU19 live in association with aquatic plants.
Enrichment and Isolation of Methanotrophs from Aquatic Plants
We attempted to enrich and isolate methanotrophs associated with C. demersum, E. densa, E. crassipes, T. japonica, P. crispus, and C. caroliniana. Since the use of NMS and AMS media could result in biased enrichment of particular methanotrophs (Hoefman et al., 2014), our results are not expected to directly reflect the population of methanotrophs on plant surfaces. Nevertheless, we wanted to isolate a variety of methanotrophs from plant surfaces, and therefore, NMS and AMS media were applied as the first trial. Indeed, our previous study also showed that a novel genus of type-I methanotroph, Methylovulum miyakonense (Iguchi et al., 2011) could be isolated by using NMS medium. Each culture that had been inoculated with a piece of an aquatic plant consumed all of the CH4 within a week, and maintained CH4 consumption activity after several serial transfers. For each sample, 1–9 colonies among 100 colonies grown on agar plates consumed CH4, and these colonies were further purified. The 100 picked colonies included 50 colonies from NMS medium and 50 colonies from AMS medium. Finally, 23 strains were isolated; these belonged to the genera Methylosinus, and Methylocystis based on sequencing of 16S rRNA and pmoA genes (Supplemental Figure 1). However, no type-I methanotrophs were isolated despite their abundance in aquatic plants. The nitrogen source used did not seem to result in isolation of specific methanotrophs, as similar strains were obtained using both NMS and AMS. Although there seems to have been a bias in the isolation of the dominant methanotrophs from aquatic plants due to the enrichment method, isolation of methanotrophic strains from aquatic plants would be the first step to reconstructing and establishing an ecological system of aquatic plants and methanotrophs having high CH4 consuming activity.
Discussion
We evaluated CH4 consuming activities of methanotrophs associated with different plant species sampled from various terrestrial and aquatic environments. Our study revealed that all aquatic plants from a eutrophic lake showed high CH4 consumption activity. The CH4 consumption activity (3.7–37 μmol·h−1·g−1 dry weight) was higher than that of rice roots (0.2–0.4 μmol·h−1·g−1 dry weight) (Eller and Frenzel, 2001) and roots and rhizomes of emergent aquatic plants (0.1–6.4 μmol·h−1·g−1 dry weight) (King, 1994) as soil-free epiphytic methanotrophic activities, as well as previous reports of Sphagnum moss (Raghoebarsing et al., 2005) and in aquatic plants (Sorrell et al., 2002). To evaluate and control the carbon cycle in aqueous environments, it is important to understand not only the atmospheric carbon cycling but also the local CH4 cycling and the derivative food chain in lakes.
Assuming the average CH4 consumption of native aquatic plants is common to all aquatic plants though the year, 160 t·y−1 of CH4 was potentially consumed by 2500 t of aquatic plants in the basin (calculated from 1995 data, Shiga Prefectural Fisheries Experiment Station 1998), which corresponds to 17% of the total CH4 emitted (770 t·y−1) from the lake surface of the basin (Kagotani et al., 1996). This large potential is suggested to represent substantial local CH4 cycling via methanotrophs associated with aquatic plants in this lake. The CH4 consumed by microbial communities associated with aquatic plants may have come from both dissolved CH4 in the water column and uptake from the sediment. In this lake sediment, 90% of the CH4 originating from organic matter in the deep sediment was consumed in the oxic surface (Murase et al., 2005). Therefore, the remaining 10% of the CH4 that passed though the oxic surface and was emitted into the water column represents CH4 that undergoes local cycling. Another route may be uptake from sediment via plant roots as observed in rice and other rooted aquatic plants. In this study, there were no big differences between unrooted samples, i.e., pieces of E. densa and C. demersum, and other rooted samples, suggesting that CH4 both from the sediment and the water column can be consumed by the aquatic plants. All aquatic plants assayed in this study, which were obtained from a eutrophic basin, consumed CH4, while aquatic plants sampled from oligotrophic environments did not (Sorrell et al., 2002). The Km for CH4 consumption in aquatic plants was reported to be in the range 3–6 μmol· L−1 (Sorrell et al., 2002), and this is not sufficient for uptake of atmospheric CH4. From this, we speculate that most of the CH4 utilized by methanotrophs in aquatic plants was produced in the sediment and taken up via the water column and roots.
In addition, methanotrophs corresponding to OTU19, which is related to Methylocaldum, may be selected through interaction with the plant surface. OTU19 was detected in all aquatic plants and the sequences clustered with clones from rice roots from two different sources (Shrestha et al., 2008; Lüke et al., 2010). The detection of OTU19 from different species of plants and the distinct clustering from other environmental clones suggests a particularly strong association of these methanotrophs with plant environments.
In conclusion, this study has shed light on the high potential of methanotrophs associated with aquatic plants as a local CH4 sink. We identified methanotrophs responsible for CH4 consumption as diverse uncultured type I methanotrophs in the gammaproteobacterial lineage, which were related to methanotrophs detected from rice roots, lake sediments, and rice fields.
Author Contributions
Naoko Yoshida performed all experiments and drafted the manuscript. Hiroyuki Iguchi and Akio Murakami participated in data interpretation and revising the draft. Hiroya Yurimoto and Yasuyoshi Sakai supervised the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported in part by Advanced Low Carbon Technology Research and Development Program (ALCA) of the Japan Science and Technology Agency, Grant-in-Aid for JSPS Fellows, and JSPS KAKENHI Grant Number: 08J03515.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00030/abstract
References
Bodrossy, L., Holmes, E. M., Holmes, A. J., Kovács, K. L., and Murrell, J. C. (1997). Analysis of 16S rRNA and methane monooxygenase gene sequences reveals a novel group of thermotolerant and thermophilic methanotrophs, Methylocaldum gen. nov. Arch. Microbiol. 168, 493–503. doi: 10.1007/s002030050527
Borrel, G., Jézéquel, D., Biderre-Petit, C., Morel-Desrosiers, N., Morel, J.-P., Peyret, P., et al. (2011). Production and consumption of methane in freshwater lake ecosystems. Res. Microbiol. 162, 832–847. doi: 10.1016/j.resmic.2011.06.004
Bussmann, I., Pester, M., Brune, A., and Schink, B. (2004). Preferential cultivation of type II methanotrophic bacteria from littoral sediments (Lake Constance). FEMS Microbiol. Ecol. 47, 179–189. doi: 10.1016/S0168-6496(03)00260-5
Bussmann, I., Rahalkar, M., and Schink, B. (2006). Cultivation of methanotrophic bacteria in opposing gradients of methane and oxygen. FEMS Microbiol. Ecol. 56, 331–344. doi: 10.1111/j.1574-6941.2006.00076.x
Conrad, R. (2009). The global methane cycle: recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 1, 285–292. doi: 10.1111/j.1758-2229.2009.00038.x
Costello, A. M., and Lidstrom, M. E. (1999). Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65, 5066–5074.
Dedysh, S. (2009). Exploring methanotroph diversity in acidic northern wetlands: molecular and cultivation-based studies. Microbiology 78, 655–669. doi: 10.1134/S0026261709060010
Dedysh, S. N., Liesack, W., Khmelenina, V. N., Suzina, N. E., Trotsenko, Y. A., Semrau, J. D., et al. (2000). Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50, 955–969. doi: 10.1099/00207713-50-3-955
Deutzmann, J. S., Wörner, S., and Schink, B. (2011). Activity and diversity of methanotrophic bacteria at methane seeps in eastern Lake Constance sediments. Appl. Environ. Microbiol. 77, 2573–2581. doi: 10.1128/AEM.02776-10
Eller, G., and Frenzel, P. (2001). Changes in activity and community structure of methane-oxidizing bacteria over the growth period of rice. Appl. Environ. Microbiol. 67, 2395–2403. doi: 10.1128/AEM.67.6.2395-2403.2001
Ettwig, K. F., Butler, M. K., Le Paslier, D., Pelletier, E., Mangenot, S., Kuypers, M. M., et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548. doi: 10.1038/nature08883
Frenzel, P., Thebrath, B., and Conrad, R. (1990). Oxidation of methane in the oxic surface layer of a deep lake sediment (Lake Constance). FEMS Microbiol. Lett. 73, 149–158. doi: 10.1111/j.1574-6968.1990.tb03935.x
Hamabata, E., and Kobayashi, Y. (2002). Present status of submerged macrophyte growth in Lake Biwa: recent recovery following a summer decline in the water level. Lakes Reserv. Res. Manag. 7, 331–338. doi: 10.1046/j.1440-1770.2002.00199.x
Heilman, M. A., and Carlton, R. G. (2001). Methane oxidation associated with submersed vascular macrophytes and its impact on plant diffusive methane flux. Biogeochemistry 52, 207–224. doi: 10.1023/A:1006427712846
Henneberger, R., Lüke, C., Mosberger, L., and Schroth, M. H. (2012). Structure and function of methanotrophic communities in a landfill cover soil. FEMS Microbiol. Ecol. 81, 52–65. doi: 10.1111/j.1574-6941.2011.01278.x
Ho, A., Lüke, C., Cao, Z., and Frenzel, P. (2011). Ageing well: methane oxidation and methane oxidizing bacteria along a chronosequence of 2000 years. Environ. Microbiol. Rep. 3, 738–743. doi: 10.1111/j.1758-2229.2011.00292.x
Hoefman, S., van der Ha, D., Boon, N., Vandamme, P., De Vos, P., and Heylen, K. (2014). Customized media based on miniaturized screening improve growth rate and cell yield of methane-oxidizing bacteria of the genus Methylomonas. Ant. Van Leeuwenhoek 105, 353–366. doi: 10.1007/s10482-013-0083-2
Holmes, A. J., Costello, A., Lidstrom, M. E., and Murrell, J. C. (1995). Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132, 203–208. doi: 10.1111/j.1574-6968.1995.tb07834.x
Huber, T., Faulkner, G., and Hugenholtz, P. (2004). Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20, 2317–2319. doi: 10.1093/bioinformatics/bth226
Iguchi, H., Sato, I., Sakakibara, M., Yurimoto, H., and Sakai, Y. (2012). Distribution of methanotrophs in the phyllosphere. Biosci. Biotechnol. Biochem. 76, 1580–1583. doi: 10.1271/bbb.120281
Iguchi, H., Yurimoto, H., and Sakai, Y. (2011). Stimulation of methanotrophic growth in cocultures by cobalamin excreted by Rhizobia. Appl. Environ. Microbiol. 77, 8509–8515. doi: 10.1128/AEM.05834-11
Kagotani, Y., Kanzaki, M., and Yoda, K. (1996). Methane budget determined at the ground and water surface level in various ecosystems in Shiga Prefecture, central Japan. Climate Res. 6, 79–88. doi: 10.3354/cr006079
Kalyuzhnaya, M. G., Stolyar, S. M., Auman, A. J., Lara, J. C., Lidstrom, M. E., and Chistoserdova, L. (2005). Methylosarcina lacus sp. nov., a methanotroph from Lake Washington, Seattle, USA, and emended description of the genus Methylosarcina. Int. J. Syst. Evol. Microbiol. 55, 2345–2350. doi: 10.1099/ijs.0.63405-0
Keppler, F., Hamilton, J. T. G., Brass, M., and Röckmann, T. (2006). Methane emissions from terrestrial plants under aerobic conditions. Nature 439, 187–191. doi: 10.1038/nature04420
King, G. M. (1994). Associations of methanotrophs with the roots and rhizomes of aquatic vegetation. Appl. Environ. Microbiol. 60, 3220–3227.
Kip, N., Dutilh, B. E., Pan, Y., Bodrossy, L., Neveling, K., Kwint, M. P., et al. (2011). Ultra deep pyrosequencing of pmoA amplicons confirms the prevalence of Methylomonas and Methylocystis in Sphagnum mosses from a Dutch peat bog. Environ. Microbiol. Rep. 3, 667–673. doi: 10.1111/j.1758-2229.2011.00260.x
Knief, C., Delmotte, N., Chaffron, S., Stark, M., Innerebner, G., Wassmann, R., et al. (2011). Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 6, 1378–1390. doi: 10.1038/ismej.2011.192
Lidstrom, M. E. (2006). “Aerobic methylotrophic bacteria,” in The Prokaryotes, eds S. Falkow, M. Dworkin, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (New York, NY: Springer), 618–634.
Lin, J. L., Radajewski, S., Eshinimaev, B. T., Trotsenko, Y. A., McDonald, I. R., and Murrell, J. C. (2004). Molecular diversity of methanotrophs in Transbaikal soda lake sediments and identification of potentially active populations by stable isotope probing. Environ. Microbiol. 6, 1049–1060. doi: 10.1111/j.1462-2920.2004.00635.x
Lüke, C., Krause, S., Cavigiolo, S., Greppi, D., Lupotto, E., and Frenzel, P. (2010). Biogeography of wetland rice methanotrophs. Environ. Microbiol. 12, 862–872. doi: 10.1111/j.1462-2920.2009.02131.x
Mayumi, D., Yoshimoto, T., Uchiyama, H., Nomura, N., and Nakajima-Kambe, T. (2010). Seasonal change in methanotrophic diversity and populations in a rice field soil assessed by DNA-stable isotope probing and quantitative real-time PCR. Microbes Environ. 25, 156–163. doi: 10.1264/jsme2.ME10120
McDonald, I. R., Bodrossy, L., Chen, Y., and Murrell, J. C. (2008). Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Environ. Microbiol. 74, 1305–1315. doi: 10.1128/AEM.02233-07
Murase, J., Sakai, Y., Kametani, A., and Sugimoto, A. (2005). Dynamics of methane in mesotrophic Lake Biwa, Japan. Ecol. Res. 20, 377–385. doi: 10.1007/s11284-005-0053-x
Nercessian, O., Noyes, E., Kalyuzhnaya, M. G., Lidstrom, M. E., and Chistoserdova, L. (2005). Bacterial populations active in metabolism of C1 compounds in the sediment of Lake Washington, a freshwater lake. Appl. Environ. Microbiol. 71, 6885–6899. doi: 10.1128/AEM.71.11.6885-6899.2005
Op den Camp, H. J. M., Islam, T., Stott, M. B., Harhangi, H. R., Hynes, A., Schouten, S., et al. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1, 293–306. doi: 10.1111/j.1758-2229.2009.00022.x
Osaka, T., Ebie, Y., Tsuneda, S., and Inamori, Y. (2008). Identification of the bacterial community involved in methane dependent denitrification in activated sludge using DNA stable isotope probing. FEMS Microbiol. Ecol. 64, 494–506. doi: 10.1111/j.1574-6941.2008.00473.x
Pester, M., Friedrich, M. W., Schink, B., and Brune, A. (2004). pmoA-based analysis of methanotrophs in a littoral lake sediment reveals a diverse and stable community in a dynamic environment. Appl. Environ. Microbiol. 70, 3138–3142. doi: 10.1128/AEM.70.5.3138-3142.2004
Pfluger, A. R., Wu, W.-M., Pieja, A. J., Wan, J., Rostkowski, K. H., and Criddle, C. S. (2011). Selection of Type I and Type II methanotrophic proteobacteria in a fluidized bed reactor under non-sterile conditions. Biores. Tech. 102, 9919–9926. doi: 10.1016/j.biortech.2011.08.054
Raghoebarsing, A. A., Smolders, A. J. P., Schmid, M. C., Rijpstra, W. I. C., Wolters-Arts, M., Derksen, J., et al. (2005). Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature 436, 1153–1156. doi: 10.1038/nature03802
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Semrau, J. D., DiSpirito, A. A., and Yoon, S. (2010). Methanotrophs and copper. FEMS Microbiol. Rev. 34, 496–531. doi: 10.1111/j.1574-6976.2010.00212.x
Shannon, C. E., and Weaver, W. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423, 623–656. doi: 10.1002/j.1538-7305.1948.tb01338.x
Shrestha, M., Abraham, W. R., Shrestha, P. M., Noll, M., and Conrad, R. (2008). Activity and composition of methanotrophic bacterial communities in planted rice soil studied by flux measurements, analyses of pmoA gene and stable isotope probing of phospholipid fatty acids. Environ. Microbiol. 10, 400–412. doi: 10.1111/j.1462-2920.2007.01462.x
Shrestha, M., Shrestha, P. M., Frenzel, P., and Conrad, R. (2010). Effect of nitrogen fertilization on methane oxidation, abundance, community structure, and gene expression of methanotrophs in the rice rhizosphere. ISME J. 4, 1545–1556. doi: 10.1038/ismej.2010.89
Siljanen, H. M., Saari, A., Krause, S., Lensu, A., Abell, G. C., Bodrossy, L., et al. (2011). Hydrology is reflected in the functioning and community composition of methanotrophs in the littoral wetland of a boreal lake. FEMS Microbiol. Ecol. 75, 430–445. doi: 10.1111/j.1574-6941.2010.01015.x
Solomon, S. D., Qin, M., Manning, Z., Chen, M., Marquis, K. B., Averyt, M., et al. (eds.). (2007). Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge; New York, NY: Cambridge University Press.
Sorrell, B. K., Downes, M. T., and Stanger, C. L. (2002). Methanotrophic bacteria and their activity on submerged aquatic macrophytes. Aquat. Bot. 72, 107–119. doi: 10.1016/S0304-3770(01)00215-7
Stein, L. Y., Roy, R., and Dunfield, P. F. (2012). “Aerobic methanotrophy and nitrification: processes and connections,” in eLS, (Chichester: John Wiley & Sons, Ltd). Available online at: http://onlinelibrary.wiley.com/doi/10.1002/9780470015902.a0022213/full
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840. doi: 10.1038/nrmicro2910
Vorobev, A. V., Baani, M., Doronina, N. V., Brady, A. L., Liesack, W., Dunfield, P. F., et al. (2011). Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int. J. Syst. Evol. Microbiol. 61, 2456–2463. doi: 10.1099/ijs.0.028118-0
Whittenbury, R., Phillips, K., and Wilkinson, J. (1970). Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 61, 205–218. doi: 10.1099/00221287-61-2-205
Xu, K., Tang, Y., Ren, C., Zhao, K., Wang, W., and Sun, Y. (2013). Activity, distribution, and abundance of methane-oxidizing bacteria in the near surface soils of onshore oil and gas fields. Appl. Microbiol. Biotechnol. 97, 7909–7918. doi: 10.1007/s00253-012-4500-7
Yoshida, N., Asahi, K., Sakakibara, Y., Miyake, K., and Katayama, A. (2007). Isolation and quantitative detection of tetrachloroethene (PCE)-dechlorinating bacteria in unsaturated subsurface soils contaminated with chloroethenes. J. Biosci. Bioeng. 104, 91–97. doi: 10.1263/jbb.104.91
Keywords: methanotroph, aquatic plants, pmoA, methane monooxygenase, methane sink
Citation: Yoshida N, Iguchi H, Yurimoto H, Murakami A and Sakai Y (2014) Aquatic plant surface as a niche for methanotrophs. Front. Microbiol. 5:30. doi: 10.3389/fmicb.2014.00030
Received: 15 November 2013; Accepted: 17 January 2014;
Published online: 03 February 2014.
Edited by:
Svetlana N. Dedysh, Russian Academy of Sciences, RussiaReviewed by:
Paul Bodelier, Netherlands Institute of Ecology (NIOO-KNAW), NetherlandsSvetlana N. Dedysh, Russian Academy of Sciences, Russia
Ingeborg Bussmann, Alfred-Wegener Institut, Germany
Copyright © 2014 Yoshida, Iguchi, Yurimoto, Murakami and Sakai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasuyoshi Sakai, Division of Applied Life Sciences, Graduate School of Agriculture, Kyoto University, Kitashirakawa-Oiwake, Sakyo-ku, Kyoto 606-8502, Japan e-mail: ysakai@kais.kyoto-u.ac.jp
 Naoko Yoshida
Naoko Yoshida Hiroyuki Iguchi
Hiroyuki Iguchi Hiroya Yurimoto
Hiroya Yurimoto Akio Murakami
Akio Murakami Yasuyoshi Sakai
Yasuyoshi Sakai