- 1Physical Biosciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA, USA
- 2Biochemistry Division, University of Missouri, Columbia, MO, USA
- 3Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA, USA
Sulfate-reducing bacteria such as Desulfovibrio vulgaris Hildenborough are often found in environments with limiting growth nutrients. Using lactate as the electron donor and carbon source, and sulfate as the electron acceptor, wild type D. vulgaris shows motility on soft agar plates. We evaluated this phenotype with mutants resulting from insertional inactivation of genes potentially related to motility. Our study revealed that the cheA3 (DVU2072) kinase mutant was impaired in the ability to form motility halos. Insertions in two other cheA loci did not exhibit a loss in this phenotype. The cheA3 mutant was also non-motile in capillary assays. Complementation with a plasmid-borne copy of cheA3 restores wild type phenotypes. The cheA3 mutant displayed a flagellum as observed by electron microscopy, grew normally in liquid medium, and was motile in wet mounts. In the growth conditions used, the D. vulgaris ΔfliA mutant (DVU3229) for FliA, predicted to regulate flagella-related genes including cheA3, was defective both in flagellum formation and in forming the motility halos. In contrast, a deletion of the flp gene (DVU2116) encoding a pilin-related protein was similar to wild type. We conclude that wild type D. vulgaris forms motility halos on solid media that are mediated by flagella-related mechanisms via the CheA3 kinase. The conditions under which the CheA1 (DVU1594) and CheA2 (DVU1960) kinase function remain to be explored.
Introduction
Desulfovibrio vulgaris Hildenborough is an anaerobic model sulfate-reducing bacterium (SRB), representing the broad class of SRB that play an essential role in biogeochemical processes such as sulfur- and metal-cycling (Zhou et al., 2011). Motility, its relation to core physiology such as electron transfer (Tai et al., 2010), and the global nature of its regulation (Ueki et al., 2012) are key topics of research in both model and newly discovered anaerobic metal- and sulfate-reducing organisms (Takaki et al., 2010). The genomes of many organisms that occupy such ecological niches are now sequenced and reveal that some microbes have more than one putative chemotaxis-related gene cluster. Shewanella oneidensis MR1 encodes three chemotaxis gene clusters, one of which was shown to respond to electron acceptor concentrations (Bencharit and Ward, 2005; Li et al., 2007). Geobacter spp., are also anaerobic metal-reducing bacteria and encode six chemotaxis clusters, the functions of which are yet to be specifically elucidated (Tran et al., 2008).
D. vulgaris displays a single polar flagellum (Postgate and Campbell, 1966) and is documented to have motility on soft agar plates prepared with 0.7% (wt/vol) agarose and defined lactate/sulfate medium (Clark et al., 2007), with concentrations not considered limiting for either lactate or sulfate (Postgate, 1963; Mukhopadhyay et al., 2006). Aside from flagellar and pilin protein encoding genes, the genome of D. vulgaris encodes three separate chemotaxis clusters, each of which includes a putative cheA (Figures 1, S1). Here, we examine the observed motility in D. vulgaris as a function of lactate and sulfate in the medium and examine the role of several motility related genes in this phenotype.
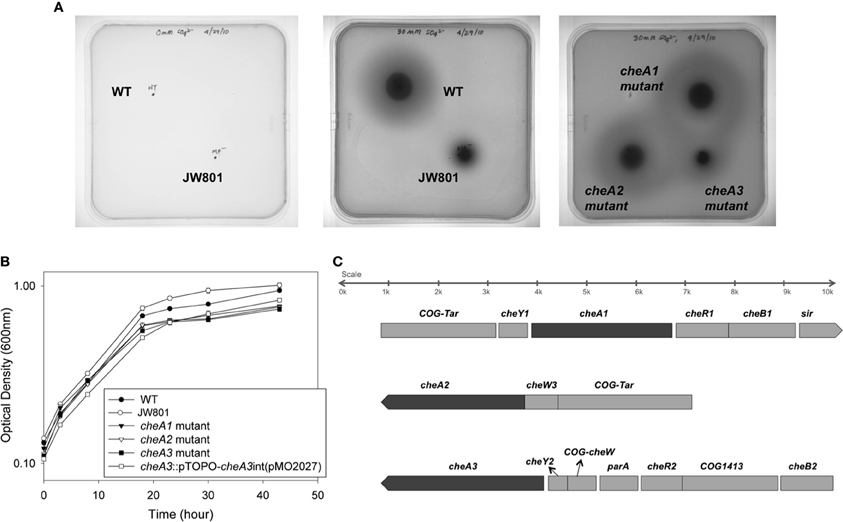
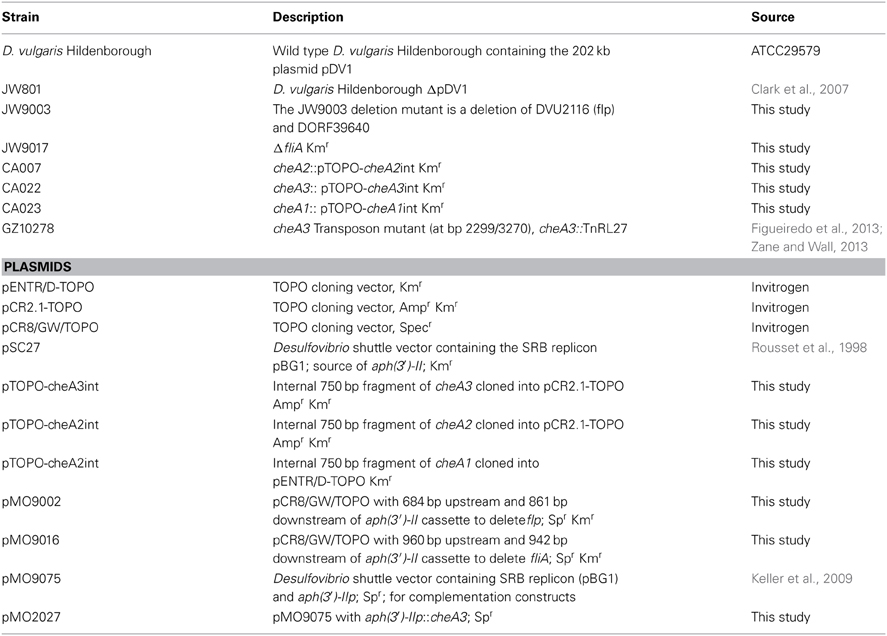
Figure 1. (A) Soft agar plate assays for D. vulgaris wild type, JW801, CA023 (cheA1 mutant), CA007 (cheA2 mutant), and CA022 (cheA3 mutant) strains in modified LS4D medium as described in methods with 0.4% (wt/vol) agar. Motility halos were imaged after 4 days of incubation in an anaerobic chamber at 30°C. No growth was observed for either the wild type or JW801 strain in the control plate that contains no sulfate (left panel). In LS4D plates, the wild type forms a motility halo, whereas, JW801 was impaired in forming a halo (center panel); the cheA3 mutant shows a defect in motility (right panel) relative to the cheA1 and cheA2 mutants and the wild type. (B) Growth assays of D. vulgaris wild type, JW801, CA023 (cheA1 mutant), CA007 (cheA2 mutant), CA022 (cheA3 mutant), and the cheA3 complemented strain, cheA3::pTOPO-cheA3int(pMO2027). Assays were done in LS4D medium at 30–32°C. Cultures were started at an approximate OD600 of 0.1 and grown until the late stationary phase. Data points are the averages of triplicate measurements. (C) Operons encoding the three cheA chemotaxis genes in D. vulgaris as predicted in http://www.microbesonline.org (Dehal et al., 2009). Top: cheA1; Middle: cheA2; and Bottom: cheA3. Arrowheads indicate the direction of transcription.
Materials and Methods
Bacterial Growth and Culture Maintenance
All strains and plasmids used in this study are listed in Table 1. D. vulgaris Hildenborough strain ATCC 29579 was obtained from the American Type Culture Collection (Manassas, VA, USA). Bacterial strains were grown and maintained as described previously (Mukhopadhyay et al., 2006). Unless noted otherwise, D. vulgaris was grown in defined LS4D medium with sodium lactate (60 mM) as the electron donor and sodium sulfate (30 mM) as the electron acceptor. Modified LS4D media, the MOYLS4 and MOY media reported previously (Zane et al., 2010), were used during construction of the cheA knock-out mutants. D. vulgaris strain JW801, lacking the native plasmid pDV1 (Clark et al., 2007), was used as a non-motile control and was grown similarly to the wild type. For growth of D. vulgaris mutant strains CA023, mutated in cheA1 (DVU1594); CA007, mutated in cheA2 (DVU1960); and CA022, mutated in cheA3 (DVU2072). The antibiotic G418 (Sigma Aldrich, St Louis, MO) was added to a final concentration of 400 μg/ml (Keller et al., 2009). For the complementation strain, cheA3::pTOPO-cheA3int(pMO2027), an additional antibiotic, spectinomycin (100 μg/ml), was added during growth. All D. vulgaris stocks were stored in 10% (vol/vol) glycerol at −80°C and were used as 10% (vol/vol) inocula into 10–30 ml of fresh medium and the cells were grown to mid-log phase (optical density at 600 nm (OD600) of 0.3 to 0.4).
Construction of CheA Insertional Mutants
Gene disruption mutants in the cheA genes were created by single crossover homologous recombination with suicide vectors containing 750-base pair internal gene regions. The internal gene fragments were produced by PCR amplification with primers listed in Table A1 and cloned into the pENTR/D-TOPO plasmid (Life Technologies, Grant Island, NY, USA). The suicide vectors were confirmed by sequencing and electroporated into wild-type D. vulgaris prepared as described previously (Keller et al., 2009). Transformants were recovered and colonies confirmed as described (Zane et al., 2010). Southern blot analysis was performed on all the mutant strains as described previously (Keller et al., 2009) to verify that the gene disruption occurred at the correct locus. The transposon mutant in cheA3 used for the Palleroni chamber assays (cheA3::TnRL27) was obtained from the D. vulgaris transposon mutant collection (Zane and Wall, 2013) cited in earlier reports (Fels et al., 2013; Figueiredo et al., 2013; Kazakov et al., 2013).
Complementation of CheA::pTOPO-CheA3int Mutant
The cheA3 gene was amplified by Herculase II (Agilent Technologies, Santa Clara, CA, USA) with primers listed in Table A1 and cloned into pMO9075 for expression from the aph(3')-II promoter. After selection of the recombinant plasmid and verification of the insert sequence, one isolate was named pMO2027. To obtain a complemented cheA3 mutant, cheA3 cells (CA022) were transformed with pMO2027 by electroporation as described (Keller et al., 2009), with the following exceptions: MOYLS4 (60/30, lactate/sulfate) medium was used throughout growth, electroporation, recovery and selective plating of the complemented mutant and the electroporation parameters were set at 1500 V, 250 Ω, and 25 μF. Following sequence verification of the plasmid recovered from the cheA3 mutant, one isolate was chosen as the complemented strain, cheA3::pTOPO-cheA3int (pMO2027), for comparison of phenotypes.
Construction of JW9017 (fliA) and JW9003 (flp) Deletion Mutants
The pMO9016 and pMO9002 plasmids for the marker-exchange deletion of fliA (DVU3229) and flp (DVU2116), respectively, were constructed by splicing by overlap extension (SOE) PCR (Horton et al., 1990) of three PCR amplimers as previously described (Zane et al., 2010). Transformation of the fliA and flp deletion plasmids into D. vulgaris was performed as previously described (Zane et al., 2010), with the exception that the G418-resistant transformants were selected from electroporated cells mixed into molten MOYLS4 medium with 400 μg G418/ml and poured into empty petri dishes for solidification.
Growth Assays
Cells were recovered overnight in 10 ml liquid MOYLS4 medium and used to inoculate 20–25 ml volume of fresh LS4D at a starting OD600 of 0.05–0.1. Growth assays were conducted in triplicate under anaerobic conditions at a temperature of 30–32°C. OD600 was monitored with a spectrophotometer (Agilent HP Diode Array Model 8452A, Agilent Technologies, Santa Clara, CA, USA) periodically as a function of time until the late stationary phase.
Soft Agar Plate Assays
Soft agar plate assays were used to study the motility as described in other reports (Li et al., 2007) with a few modifications. A modified formulation for LS4D medium was solidified with 0.4% (wt/vol) agar for motility assays. D. vulgaris cells were grown to an OD600 of 0.3–0.4 and 2 μl of cells were stabbed into the middle of the soft agar bed. For the sulfate disc assays, 0.4% (wt/vol) soft agar medium contained 12 mM sodium sulfate. A nylon membrane disc was pre-soaked in 30 mM sulfate or water and placed 0.5 cm from the center of inoculation immediately prior to inoculation. Plates were incubated at 30–32°C in the anaerobic chamber for 4–5 days to obtain a reasonable amount of motility. Photographs in Figures 1, 2 were taken under white light by a Biospectrum AC Imaging System (UVP, Upland, CA, USA) with the following constant instrumental parameters: exposure time: 634 μs; filter: SyBr Gold (485–655 nm); aperture: 1.2; zoom: 20%; focus: 80%; trans illumination: white. Figure 3A was imaged with a Nikon D5000 camera at the Veterinary Biomedical Communications at the University of Missouri-College of Veterinary Medicine. For the disc assays, images were taken with a white light (Figures 4A, S1) and 365 nm UV-light (Figure 4B) exposure using another UVP imaging system (UVP-chromato-Vue® C-75, UVP, Upland, CA, USA) mounted with a Canon G9 camera. After spraying 5 N sodium hydroxide over the agar bed, D. vulgaris cells fluoresce bright pink-orange under the 365 nm UV-light (Figure 4B), which is caused by the release of siroheme, the cofactor of bisulfite reductase desulfoviridin (Postgate, 1959).
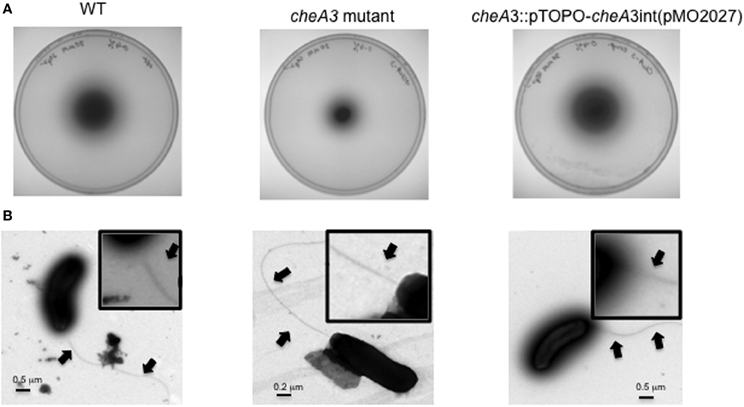
Figure 2. (A) Soft agar plate assays of D. vulgaris wild type, cheA3 mutant and cheA3 complement strain, cheA3::pTOPO-cheA3int(pMO2027) in LS4D medium with 0.4% (wt/vol) agar and 30 mM sulfate. (B) Transmission electron microscopic (TEM) images of the flagella of D. vulgaris wild type, cheA3 mutant and cheA3 complement strain, cheA3::pTOPO-cheA3int(pMO2027). In the main images and the enlarged inset views, arrows point to the flagella.
Figure 3. Soft agar plate assays (0.4% agar wt/vol) of D. vulgaris wild type, JW9017 (ΔfliA), and JW9003 (Δflp) in defined LS4D medium with 6 mM sulfate (A) or 30 mM sulfate (B). TEM images of JW9017 (C) and JW9003 (D) grown in defined LS4D medium show the presence of flagellum in the JW9003 strain but not in the JW9017 strain. Inset enlarged views are provided to indicate the flagellum clearly. Note: In rich media, a few cells in JW9017 show the presence of a shorter flagellum (Figure S4).
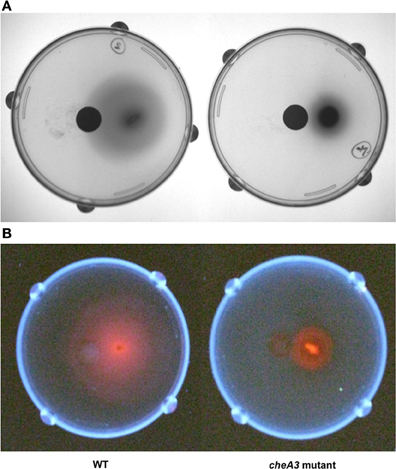
Figure 4. Soft agar plate disc assays of D. vulgaris wild type and cheA3 mutant strains with a nylon membrane disc soaked in 30 mM sulfate. Modified LS4D medium contained 0.4% (wt/vol) agar, 12 mM sodium sulfate, and 60 mM sodium lactate. Pictures were taken with white light (A) and UV-light (B). Sodium hydroxide solution (5 N) was sprayed over the surface of the agar bed before taking pictures under UV-light to enhance the fluorescence due to the presence of bisulfite reductase containing siroheme as a cofactor (Postgate, 1959).
Palleroni Chamber Assay
A capillary-based assay (Palleroni, 1976) was performed to provide quantitative measurement of the bacterial cell motility, as described previously (Sun et al., 2009). Briefly, 10 ml cultures grown to an OD of approximately 0.4–0.5 (mid-log) were spun down at ~5500 × g for 8 min at room temperature and resuspended in an equal volume of phosphate buffered saline (PBS). Each channel of the Palleroni chamber was filled with 550 μl of resuspended cells. The capillary (32 mm length, 1.1 mm inner diameter) was filled with one of the following solutions: 30 mM sulfate, 60 mM lactate or 1 × PBS (control) and placed horizontally into the Palleroni chamber. After the 15 min incubation period, contents from the capillary were dispensed into 135 μL of 1 × PBS. The micro-bicinchoninic acid (micro-BCA) assay (Pierce, Rockford, IL, USA) was used as per manufacturer's instruction to measure the protein from the cells, and served as a measure of the cell mass that entered the capillary during the assay. Absorbance was measured by the SpectraMax Pro microplate reader (Molecular Devices, Sunnyvale, CA). Dilutions of bovine serum albumin in 1 × PBS were used to prepare a standard curve.
Electron Microscopy
All electron microscopy samples were fixed in 2% (vol/vol) glutaraldehyde (EM grade, purchased from EMS, Hatfield, PA, USA) directly in the growth medium for several hours and then washed in phosphate buffered saline (PBS). For Transmission Electron Microscopy (TEM), a 5 μl sample was put onto a formvar and carbon coated copper grid (200 mesh, Ted Pella, Redding, CA, USA), which was freshly glow-discharged in order to make the carbon film hydrophilic. The sample was allowed to settle for 5 min and the liquid removed with filter paper. Immediately 5 μ l of a 2% (wt/vol) aqueous solution of uranyl acetate was put onto the grid and left for 1 min before also being dried with filter paper. Two quick washes (10 μl each) with distilled water followed. After drying, the grids were investigated with a Phillips Tecnai 12 electron microscope (FEI Company, Hillsboro, OR, USA) with a 120 kV accelerating voltage and magnifications typically between 2900 × and 9300 ×. A Gatan camera (Gatan, Pleasanton, CA, USA) was used for image acquisition.
Results and Discussion
Wild type D. vulgaris showed outward motility on soft agar plates over a period of four days relative to the JW801 strain, which lacked the native plasmid pDV1 (Figure 1A). JW801 is known to be non-motile (Clark et al., 2007), possibly due to a defect in flagellum formation, and served as a control. The levels of lactate and sulfate used in these assays were sufficient to permit robust growth of D. vulgaris in liquid medium (Postgate, 1963, 1979; Mukhopadhyay et al., 2006).
To investigate a potential role of cheA genes in this motility phenotype, gene disruption mutants in all three cheA loci were generated and examined on soft agar plates (Figure 1A). The cheA3 mutant showed a clear defect in this phenotype, whereas the remaining two cheA mutants were unaffected. All strains showed similar growth rates and maxima in liquid cultures of LS4D medium (Figure 1B). cheA3 is the terminal gene in an operon that encodes several chemotaxis genes and genes with other putative functions that have a role in motility (Figure 1C). For example, the parA homolog in Pseudomonas aeruginosa is known to affect motility, among other phenotypes (Lasocki et al., 2007). Though a polar mutation is unlikely, the cheA3 gene was complemented in the cheA3 mutant strain. The complemented mutant, cheA3::pTOPO-cheA3int(pMO2027), exhibited motility equivalent to the D. vulgaris wild type strain (Figure 2A), confirming the direct role of the CheA3 protein in this phenotype. Further, a visual examination of motility on a wet mount at 100 × magnification indicated all three strains to be motile (Supplementary video data). Consistent with this, high resolution TEM (Figure 2B) revealed that all three strains have flagella. Thus loss of motility in the cheA3 mutant in the soft agar plate is neither correlated with loss of motility in liquid medium nor with a defect in flagellum formation. Taken together, these observations suggest that the wild type motility observed in soft agar LS4D medium plates involve the sensor kinase CheA3 but not CheA1 or CheA2.
The ΔfliA mutant, but not a Δflp mutant, was found to be similarly defective in motility halo formation. FliA, a α28 RNA polymerase sigma factor, modulates the formation of the flagellar complex in the model Gram-negative bacterium Escherichia coli (Komeda, 1986). D. vulgaris also contains a fliA homolog (DVU3229), encoding a σ70 transcription factor, that is predicted to modulate 16 genes, including genes involved in the formation of the flagellum and cheA3 (Novichkov et al., 2010). The ΔfliA mutant (strain JW9017) was used to examine the role of the flagellum in the motility halo formation. In 0.4% (wt/vol) agar plates, this strain was severely impaired in halo formation (Figures 3A,B). TEM images of the FliA mutant confirmed it to be defective in flagellum formation (Figure 3C). Unlike the cheA3 mutant strain, the FliA mutant is non-motile as observed on wet mounts (data not shown). D. vulgaris also encodes genes for pilin formation, such as a putative flp gene (DVU2116) (Heidelberg et al., 2004). Flp pili are typically not known to mediate twitching motility and the D. vulgaris Δflp mutant (strain JW9003), when tested on 0.4% (wt/vol) soft agar plates showed no defect in the motility halo forming phenotype (Figures 3A,B). TEM images also show that the Δflp strain displays the polar flagellum (Figure 3D). While more characterization is required to confirm the motility mode leading to the halos in D. vulgaris, the evidence points toward a flagellum-based mechanism.
Upon using two different concentrations of sulfate in the soft agar plates, we observed larger motility halos for the lower concentration of sulfate (Figures 3A,B) in both the wild type and the Δflp mutant. In order to evaluate a possible correlation of D. vulgaris motility with sulfate, we performed two assays. First, we used a soft agar plate assay with a nylon membrane disc soaked in sulfate as described in the methods. An asymmetric motility of wild type D. vulgaris was observed toward the sulfate-soaked disc (Figure 4). Neither the halo nor the asymmetry was observed for the cheA3 mutant (Figure 4). The asymmetry was also not observed in the wild type D. vulgaris with either water-soaked (Figure S2) or lactate-soaked discs (Figure S3). Second, we conducted a Palleroni chamber-based assay, specifically used to test for swim-related phenotypes (Palleroni, 1976; Sun et al., 2009). For both the wild type and the cheA3 complemented strain, we observed a similar and significantly greater accumulation of cells in the capillary with lactate and sulfate, relative to the control (PBS) (Figure 5). The capillary assay results corroborate the ability of the wild type D. vulgaris to move toward sulfate and, unlike the plate-based assays, also toward lactate. Additional experiments will be required to examine the differences in D. vulgaris wild type motility toward lactate, between the soft agar plate assays and the capillary assays. Finally, consistent with the plate-based assays, the accumulation of cells for the cheA3 mutant was significantly lower in the conditions tested (Figure 5). Thus the cheA3 mutant may be generally impaired in directional motility.
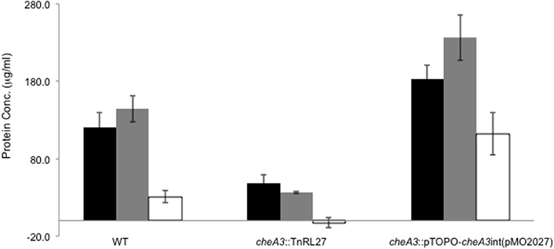
Figure 5. Palleroni chamber assay to examine the accumulation of cells in a capillary tube containing either 30 mM sulfate (black bar), 60 mM lactate (gray bar), or PBS (white bar) for the wild type, cheA3 mutant cheA3::TnRL27 and the cheA3 complement cheA3::pTOPO-cheA3int(pMO2027) strains. Assays were conducted in triplicate. Error bars are standard deviation of the means.
Possible causes for the observed outward motility on soft agar plates could be toward a nutrient as it gets depleted or away from an inhibitory compound that gets deposited during growth. The taxis observed toward nutrients in the capillary assay suggest the former to be the case. Terminal electron acceptors are known to be limiting in freshwater environments that SRB occupy (Hazen and Tabak, 2005). In Desulfovibrio spp., reports exist for aerotaxis (Eschemann et al., 1999), where some species have been shown to move toward low levels of oxygen (Fischer and Cypionka, 2006), and have even been postulated to use low levels of O2 as an electron acceptor (Cypionka, 2000). For D. vulgaris Hildenborough specifically, established electron acceptors supporting growth are sulfate (Postgate, 1963), thiosulfate and sulfite (Heidelberg et al., 2004). Even though D. vulgaris has been reported to reduce transition group metals such as iron, strontium, chromium and uranium (Lovley and Phillips, 1994; Payne et al., 2002; Park et al., 2008), sustained growth has not been reported during reduction of these metals (Payne et al., 2002; Park et al., 2008). As in Desulfovibrio spp., multiple chemotaxis modules are known to exist in many other bacteria, including Geobacter spp. (Tran et al., 2008), Vibrio cholerae (Gosink et al., 2002), P. aeruginosa (Kato et al., 1999), Rhodobacter sphaeroides (Gauden and Armitage, 1995; Martin et al., 2001), and M. xanthus (Yang et al., 1998). Where characterized, such as in S. oneidensis, only one CheA is responsible for movement toward electron acceptors (Bencharit and Ward, 2005; Li et al., 2007). Taken together, our results indicate that CheA3 may play this role in D. vulgaris Hildenborough. Homologs of the cheA3 in related bacteria (Figure S1), such as D. vulgaris Miyazaki and D. alaskensis G20, probably also perform the same function. As gene deletion mutant libraries become available in these bacteria, it will be possible to experimentally verify these predictions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Karen Clifford (University of Missouri, Columbia) for photographing the plates in Figures 3A,B. We thank Dr. Margie Romine (PNNL) for reviewing an earlier version of the manuscript. This work is part of ENIGMA, a Scientific Focus Area Program supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomics: GTL Foundational Science through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy. A portion of this work was supported by the U.S. Department of Energy Office of Science, Office of Biological and Environmental Research, Genomics Program:GTL BioHydrogen Production and BioEthanol contract DE-FG02-083464691.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00077/abstract
Figure S1 | The cheA3 histidine kinase operon is conserved between closely and distantly related Desulfovibrio species and few other organisms. Figure obtained from microbesonline.org (Dehal et al., 2009).
Figure S2 | Control assay for data presented in Figure 4. Soft agar plate disc assays of D. vulgaris wild type with a nylon membrane disc soaked in water. Modified LS4D medium in the agar contained 0.4% (wt/vol) agar, 12 mM sodium sulfate, and 60 mM sodium lactate.
Figure S3 | Soft agar plate disc assays of D. vulgaris wild type with a nylon membrane disc soaked in 60 mM lactate. Modified LS4D medium in the agar contained 0.4% (wt/vol) agar, 10 mM sodium lactate, and 30 mM sodium sulfate.
Figure S4 | TEM images of JW9017 mutant (lacking fliA) in rich media. Arrows are used to label possible truncated flagellum.
Supplementary video data | Video recording of D. vulgaris wild type, cheA3 mutant and cheA3 complement strain, cheA3::pTOPO-cheA3int(pMO2027) in wet mounts using a Samsung galaxy camera-phone held at the eye piece of a Leica DM4000 microscope at 100x magnification.
References
Bencharit, S., and Ward, M. J. (2005). Chemotactic responses to metals and anaerobic electron acceptors in Shewanella oneidensis MR-1. J. Bacteriol. 187, 5049–5053. doi: 10.1128/JB.187.14.5049-5053.2005
Clark, M. E., Edelmann, R. E., Duley, M. L., Wall, J. D., and Fields, M. W. (2007). Biofilm formation in Desulfovibrio vulgaris Hildenborough is dependent upon protein filaments. Environ. Microbiol. 9, 2844–2854. doi: 10.1111/j.1462-2920.2007.01398.x
Cypionka, H. (2000). Oxygen respiration by desulfovibrio species. Annu. Rev. Microbiol. 54, 827–848. doi: 10.1146/annurev.micro.54.1.827
Dehal, P. S., Joachimiak, M. P., Price, M. N., Bates, J. T., Baumohl, J. K., Chivian, D., et al. (2009). MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 38, D396–D400. doi: 10.1093/nar/gkp919
Eschemann, A., Kuhl, M., and Cypionka, H. (1999). Aerotaxis in Desulfovibrio. Environ. Microbiol. 1, 489–494. doi: 10.1046/j.1462-2920.1999.00057.x
Fels, S. R., Zane, G. M., Blake, S. M., and Wall, J. D. (2013). Rapid transposon liquid enrichment sequencing (TnLE-seq) for gene fitness evaluation in underdeveloped bacterial systems. Appl. Environ. Microbiol. 79, 7510–7517. doi: 10.1128/AEM.02051-13
Figueiredo, M. C., Lobo, S. A., Sousa, S. H., Pereira, F. P., Wall, J. D., Nobre, L. S., et al. (2013). Hybrid cluster proteins and flavodiiron proteins afford protection to Desulfovibrio vulgaris upon macrophage infection. J. Bacteriol. 195, 2684–2690. doi: 10.1128/JB.00074-13
Fischer, J. P., and Cypionka, H. (2006). Analysis of aerotactic band formation by Desulfovibrio desulfuricans in a stopped-flow diffusion chamber. FEMS Microbiol. Ecol. 55, 186–194. doi: 10.1111/j.1574-695X.2005.00024.x
Gauden, D. E., and Armitage, J. P. (1995). Electron transport-dependent taxis in Rhodobacter sphaeroides. J. Bacteriol. 177, 5853–5859.
Gosink, K. K., Kobayashi, R., Kawagishi, I., and Hase, C. C. (2002). Analyses of the roles of the three cheA homologs in chemotaxis of Vibrio cholerae. J. Bacteriol. 184, 1767–1771. doi: 10.1128/JB.184.6.1767-1771.2002
Hazen, T., and Tabak, H. (2005). Developments in bioremediation of soils and sediments polluted with metals and radionuclides: 2. field research on bioremediation of metals and radionuclides. Rev. Environ. Sci. Biotechnol. 4, 157–183. doi: 10.1007/s11157-005-2170-y
Heidelberg, J. F., Seshadri, R., Haveman, S. A., Hemme, C. L., Paulsen, I. T., Kolonay, J. F., et al. (2004). The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22, 554–559. doi: 10.1038/nbt959
Horton, R. M., Cai, Z. L., Ho, S. N., and Pease, L. R. (1990). Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8, 528–535.
Kato, J., Nakamura, T., Kuroda, A., and Ohtake, H. (1999). Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 63, 155–161. doi: 10.1271/bbb.63.155
Kazakov, A. E., Rajeev, L., Luning, E. G., Zane, G. M., Siddartha, K., Rodionov, D. A., et al. (2013). New family of tungstate-responsive transcriptional regulators in sulfate-reducing bacteria. J. Bacteriol. 195, 4466–4475. doi: 10.1128/JB.00679-13
Keller, K. L., Bender, K. S., and Wall, J. D. (2009). Development of a markerless genetic exchange system for Desulfovibrio vulgaris hildenborough and its use in generating a strain with increased transformation efficiency. Appl. Environ. Microbiol. 75, 7682–7691. doi: 10.1128/AEM.01839-09
Komeda, Y. (1986). Transcriptional control of flagellar genes in Escherichia coli K-12. J. Bacteriol. 168, 1315–1318.
Lasocki, K., Bartosik, A. A., Mierzejewska, J., Thomas, C. M., and Jagura-Burdzy, G. (2007). Deletion of the parA (soj) homologue in Pseudomonas aeruginosa causes ParB instability and affects growth rate, chromosome segregation, and motility. J. Bacteriol. 189, 5762–5772. doi: 10.1128/JB.00371-07
Li, J., Romine, M. F., and Ward, M. J. (2007). Identification and analysis of a highly conserved chemotaxis gene cluster in Shewanella species. FEMS Microbiol. Lett. 273, 180–186. doi: 10.1111/j.1574-6968.2007.00810.x
Lovley, D. R., and Phillips, E. J. (1994). Reduction of chromate by Desulfovibrio vulgaris and Its c(3) cytochrome. Appl. Environ. Microbiol. 60, 726–728.
Martin, A. C., Wadhams, G. H., and Armitage, J. P. (2001). The roles of the multiple CheW and CheA homologues in chemotaxis and in chemoreceptor localization in Rhodobacter sphaeroides. Mol. Microbiol. 40, 1261–1272. doi: 10.1046/j.1365-2958.2001.02468.x
Mukhopadhyay, A., He, Z., Alm, E. J., Arkin, A. P., Baidoo, E. E., Borglin, S. C., et al. (2006). Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J. Bacteriol. 188, 4068–4078. doi: 10.1128/JB.01921-05
Novichkov, P. S., Laikova, O. N., Novichkova, E. S., Gelfand, M. S., Arkin, A. P., Dubchak, I., et al. (2010). RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38, D111–D118. doi: 10.1093/nar/gkp894
Palleroni, N. J. (1976). Chamber for bacterial chemotaxis experiments. Appl. Environ. Microbiol. 32, 729–730.
Park, H., Lin, S., and Voordouw, G. (2008). Ferric iron reduction by Desulfovibrio vulgaris Hildenborough wild type and energy metabolism mutants. Antonie Van Leeuwenhoek 93, 79–85. doi: 10.1007/s10482-007-9181-3
Payne, R. B., Gentry, D. M., Rapp-Giles, B. J., Casalot, L., and Wall, J. D. (2002). Uranium reduction by Desulfovibrio desulfuricans strain G20 and a cytochrome c3 mutant. Appl. Environ. Microbiol. 68, 3129–3132. doi: 10.1128/AEM.68.6.3129-3132.2002
Postgate, J. (1959). A diagnostic reaction of Desulphovibrio desulphuricans. Nature 183, 481–482. doi: 10.1038/183481b0
Postgate, J. R. (1963). Versatile medium for the enumeration of sulfate-reducing bacteria. Appl. Microbiol. 11, 265–267.
Postgate, J. R., and Campbell, L. L. (1966). Classification of Desulfovibrio species, the nonsporulating sulfate-reducing bacteria. Bacteriol. Rev. 30, 732–738.
Rousset, M., Casalot, L., Rapp-Giles, B. J., Dermoun, Z., De Philip, P., Belaich, J. P., et al. (1998). New shuttle vectors for the introduction of cloned DNA in Desulfovibrio. Plasmid 39, 114–122.
Sun, Y., Gustavson, R. L., Ali, N., Weber, K. A., Westphal, L. L., and Coates, J. D. (2009). Behavioral response of dissimilatory perchlorate-reducing bacteria to different electron acceptors. Appl. Microbiol. Biotechnol. 84, 955–963. doi: 10.1007/s00253-009-2051-3
Tai, S. K., Wu, G., Yuan, S., and Li, K. C. (2010). Genome-wide expression links the electron transfer pathway of Shewanella oneidensis to chemotaxis. BMC Genomics 11:319. doi: 10.1186/1471-2164-11-319
Takaki, Y., Shimamura, S., Nakagawa, S., Fukuhara, Y., Horikawa, H., Ankai, A., et al. (2010). Bacterial lifestyle in a deep-sea hydrothermal vent chimney revealed by the genome sequence of the thermophilic bacterium Deferribacter desulfuricans SSM1. DNA Res. 17, 123–137. doi: 10.1093/dnares/dsq005
Tran, H. T., Krushkal, J., Antommattei, F. M., Lovley, D. R., and Weis, R. M. (2008). Comparative genomics of Geobacter chemotaxis genes reveals diverse signaling function. BMC Genomics 9:471. doi: 10.1186/1471-2164-9-471
Ueki, T., Leang, C., Inoue, K., and Lovley, D. R. (2012). Identification of multicomponent histidine-aspartate phosphorelay system controlling flagellar and motility gene expression in Geobacter species. J. Biol. Chem. 287, 10958–10966. doi: 10.1074/jbc.M112.345041
Yang, Z., Geng, Y., Xu, D., Kaplan, H. B., and Shi, W. (1998). A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol. Microbiol. 30, 1123–1130. doi: 10.1046/j.1365-2958.1998.01160.x
Zane, G. M., and Wall, J. (2013). Desulfovibrio vulgaris Hildenborough transposon mutant library: Available online at: http://desulfovibriomaps.biochem.missouri.edu/mutants/
Zane, G. M., Yen, H. C., and Wall, J. D. (2010). Effect of the deletion of qmoABC and the promoter-distal gene encoding a hypothetical protein on sulfate reduction in Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 76, 5500–5509. doi: 10.1128/AEM.00691-10
Zhou, J., He, Q., Hemme, C. L., Mukhopadhyay, A., Hillesland, K., Zhou, A., et al. (2011). How sulphate-reducing microorganisms cope with stress: lessons from systems biology. Nat. Rev. Microbiol. 9, 452–466. doi: 10.1038/nrmicro2575
Appendix
Keywords: sensor histidine kinase, cheA, soft agar plate assay, Palleroni chamber assay, electron acceptor, motility
Citation: Ray J, Keller KL, Catena M, Juba TR, Zemla M, Rajeev L, Knierim B, Zane GM, Robertson JJ, Auer M, Wall JD and Mukhopadhyay A (2014) Exploring the role of CheA3 in Desulfovibrio vulgaris Hildenborough motility. Front. Microbiol. 5:77. doi: 10.3389/fmicb.2014.00077
Received: 13 July 2013; Accepted: 12 February 2014;
Published online: 06 March 2014.
Edited by:
Biswarup Mukhopadhyay, Virginia Polytechnic Institute and State University, USAReviewed by:
Heribert Cypionka, University of Oldenburg, GermanyBirgit E. Scharf, Virginia Polytechnic Institute and State University, USA
Copyright © 2014 Ray, Keller, Catena, Juba, Zemla, Rajeev, Knierim, Zane, Robertson, Auer, Wall and Mukhopadhyay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aindrila Mukhopadhyay, Physical Biosciences Division, Lawrence Berkeley National Laboratory, 1 Cyclotron Rd., Berkeley, CA 94720, USA e-mail: amukhopadhyay@lbl.gov
†Present address: Kimberly L. Keller, Biology Department, William Woods University, Fulton, USA
 Jayashree Ray
Jayashree Ray Kimberly L. Keller
Kimberly L. Keller Michela Catena
Michela Catena Thomas R. Juba
Thomas R. Juba Marcin Zemla
Marcin Zemla Lara Rajeev
Lara Rajeev Bernhard Knierim
Bernhard Knierim Grant M. Zane
Grant M. Zane Jarrod J. Robertson2
Jarrod J. Robertson2 Manfred Auer
Manfred Auer Judy D. Wall
Judy D. Wall Aindrila Mukhopadhyay
Aindrila Mukhopadhyay