- 1Geosciences Research Division, Scripps Institution of Oceanography, University of California San Diego, La Jolla, CA, USA
- 2Division of Environmental and Biomolecular Systems, Oregon Health and Science University, Beaverton, OR, USA
- 3Laboratoire de Génétique Moléculaire, Génomique et Microbiologie, Université de Strasbourg, Strasbourg, France
- 4Biomolecular Mass Spectrometry Facility, Department of Chemistry and Biochemistry, University of California San Diego, La Jolla, CA, USA
When iron-starved, the Mn(II)-oxidizing bacteria Pseudomonas putida strains GB-1 and MnB1 produce pyoverdines (PVDGB-1 and PVDMnB1), siderophores that both influence iron uptake and inhibit manganese(II) oxidation by these strains. To explore the properties and genetics of a PVD that can affect manganese oxidation, LC-MS/MS, and various siderotyping techniques were used to identify the peptides of PVDGB-1 and PVDMnB1 as being (for both PVDs): chromophore-Asp-Lys-OHAsp-Ser-Gly-aThr-Lys-cOHOrn, resembling a structure previously reported for P. putida CFML 90-51, which does not oxidize Mn. All three strains also produced an azotobactin and a sulfonated PVD, each with the peptide sequence above, but with unknown regulatory or metabolic effects. Bioinformatic analysis of the sequenced genome of P. putida GB-1 suggested that a particular non-ribosomal peptide synthetase (NRPS), coded by the operon PputGB1_4083-4086, could produce the peptide backbone of PVDGB-1. To verify this prediction, plasmid integration disruption of PputGB1_4083 was performed and the resulting mutant failed to produce detectable PVD. In silico analysis of the modules in PputGB1_4083-4086 predicted a peptide sequence of Asp-Lys-Asp-Ser-Ala-Thr-Lsy-Orn, which closely matches the peptide determined by MS/MS. To extend these studies to other organisms, various Mn(II)-oxidizing and non-oxidizing isolates of P. putida, P. fluorescens, P. marincola, P. fluorescens-syringae group, P. mendocina-resinovorans group, and P. stutzerii group were screened for PVD synthesis. The PVD producers (12 out of 16 tested strains) were siderotyped and placed into four sets of differing PVD structures, some corresponding to previously characterized PVDs and some to novel PVDs. These results combined with previous studies suggested that the presence of OHAsp or the flexibility of the pyoverdine polypeptide may enable efficient binding of Mn(III).
Introduction
The global manganese oxidation-reduction cycle, which depends on microbial activities that increase the manganese oxidation rate by up to 5 orders of magnitude (Hastings and Emerson, 1986; Tebo et al., 2004), strongly influences the cycling of organic compounds, pollutants, and many elements including carbon, arsenic, uranium, and chromium (Tebo et al., 2004). Among the most prevalent Mn(II)-oxidizing bacteria are various Pseudomonas species, which oxidize soluble Mn2+ to insoluble Mn(IV)oxides that accumulate in late logarithmic and early stationary growth phases (Toner et al., 2005). These Mn oxides coat the cells with dark brown precipitates of nanoparticulate MnO2, birnessite-type minerals that exhibit large surface areas and efficient adsorption of toxic metals and organics (Villalobos et al., 2003, 2006), contributing to the environmental importance of this process. Oxidation of Mn2+ by all tested pseudomonads is enzymatic and utilizes oxygen as an electron acceptor (Okazaki et al., 1997; Brouwers et al., 1999; Francis and Tebo, 2001).
The model Mn(II)-oxidizing Pseudomonas putida strains GB-1 and MnB1 are typical representatives of the widely-distributed and diverse group of several “fluorescent Pseudomonas” species, which synthesize fluorescent iron-chelating compounds (siderophores) called pyoverdines (PVDs) to scavenge iron in iron-starved conditions (Budzikiewicz, 1993; Albrecht-Gary et al., 1994; Schalk et al., 2002). However, PVDs also form strong complexes with Mn(III) and can inhibit the enzymatic formation of MnO2 by P. putida GB-1 and MnB1 (Parker et al., 2004, 2007), raising several interesting questions about the interplay of iron and manganese metabolisms in these organisms and in the environment.
The PVDs of various fluorescent pseudomonads share the same chromofluorophore [(1S)-5-amino-2,3-dihydro-8,9-dihydroxy-1H-pyrimido-[1,2-a]quinoline-1-carboxylic acid], but can differ in an attached peptide chain that is recognized by a strain-specific PVD uptake receptor on the cell surface (Fuchs et al., 2001; Clement et al., 2004; Schons et al., 2005, Shen et al., 2005). Most isolates synthesize and recognize a suite of PVDs, usually with the same peptide but with various modifications including the addition of acyl chains to the chromophore, sulfonation of the chromophore, or the formation of azotobactin, in which an extra 5-membered ring is added to the PVD chromophore (Fuchs et al., 2001). Both the peptide that comprises the backbone of a PVD and the chromophore are synthesized by non-ribosomal peptide synthetases (NRPSs) (Ravel and Cornelis, 2003). NRPSs are large enzymes containing multiple modules; the number and order of these modules generally correlate to the number and order of (modified or unmodified) amino acids in the peptides (Ravel and Cornelis, 2003). Among the domains found within each of the modules, the adenylation domains specifically recognize and activate corresponding amino acids. This property enables in silico predictions concerning the amino acid sequence corresponding to a particular NRPS (Rausch et al., 2005). The sequence of the peptide backbone of a PVD can also be indirectly achieved by siderotyping. Siderotyping compares unknown PVDs with standard PVDs in terms of isoelectric focusing, microbiological uptake studies with 59Fe-PVD standards of differing structures (which defines the specificity of a strain's PVD uptake receptor) and, if necessary, mass spectrometric (MS) techniques adapted to PVDs (Fuchs and Budzikiewicz, 2001; Fuchs et al., 2001; Meyer et al., 2008). Among other things, siderotyping rapidly screens whether a strain produces a previously-described or a novel PVD and can aid in determining the sequence of the peptide backbone, confirmed by MS/MS.
The present study aims: (1) to identify NRPSs responsible for synthesis of the peptide backbone of the PVD produced by P. putida GB-1, using genomic and genetic analyses; (2) to describe the structure of this PVD based on in silico predictions coupled with siderotyping and MS/MS determinations; and (3) assess whether there is any correlation between PVD structure or siderotype and the ability to oxidize Mn(II). Additionally, siderotyping of the PVDs synthesized by several other Mn(II)-oxidizing Pseudomonas species from diverse environments was also included to define a set of PVDs with varying peptide composition and differing uptake receptor specificity for use in future investigations of PVD effects on Mn(II) oxidation in pseudomonads.
Materials and Methods
In Silico Analysis
For phylogenetic analysis, a maximum likelihood tree was constructed with NRPS sequences of known capabilities. Sequences used include the PvdI, J and D proteins of P. aeruginosa PAO1 (PA2399-2402), Psyr_1957-1960 from P. syringae pv. syringae B728a (Ravel and Cornelis, 2003), and the SypC protein of P. syringae pv. syringae B301D (Scholz-Schroeder et al., 2003). The adenylation modules from each protein were identified by the NRPSpredictor (http://www-ab.informatik.uni-tuebingen.de/software/NRPSpredictor) (Rausch et al., 2005), which was also used to predict the identity of the amino acid incorporated into the peptide by each domain, for a total of 35 modules of ~150 amino acids, including the 8 modules from P. putida GB-1. Translated sequences were aligned using the MUSCLE multiple alignment program (Edgar, 2004) using the default parameters. A maximum likelihood phylogenetic tree was then calculated with the PROML program in the PHYLIP package (Felsenstein, 2005) using the Jones-Taylor-Thornton probability model (Jones et al., 1994). One hundred bootstrap replicates were calculated using these same conditions.
Mass Spectrometric Analysis
For mass spectrometric determinations, 72-h cultures (3–4 L) of P. putida CFML 90-51 (Sultana et al., 2000), P. putida MnB1(Caspi et al., 1998), and P. putida GB-1 (Corstjens et al., 1992), which had been grown at 20–25°C with shaking in low-iron casamino acids medium (Meyer et al., 1997), were centrifuged, filtered, and adjusted to pH 5.5 with HNO3. Each filtrate was adsorbed to a 35 cc SepPak C18 column, washed with 5 volumes of Milli-Q deionized water (18.2 MΩ), eluted with 50% methanol in deionized water, evaporated to dryness in a SpeedVac freeze-drier (Savant Corp.), and resuspended to 2–4 mM PVD in MilliQ water. The PVDs from strains CFML 90-51 and GB-1 were each diluted 10,000-fold with MilliQ water and mixed 50:50 with α-cyano-4-hydroxycinnamic acid (Agilent G2037A). Each mixture was spotted (1 μ L) on a stainless steel MALDI plate and analyzed on a 4800 MALDI-TOF/TOF mass spectrometer (Applied Biosystems) in reflector positive mode. Tandem mass spectrometry was acquired using 2 kV collision energy with collision-induced dissociation. The PVD of strain MnB1, which was analyzed at a different time, was diluted 1000-fold using 50% acetonitrile in 0.1% formic acid and analyzed by electrospray ionization on a QSTAR hybrid QqTOF mass spectrometer (Applied Biosystems) infused at 10 μL·min−1 in positive mode.
Plasmid Integration into PputGB1_4083
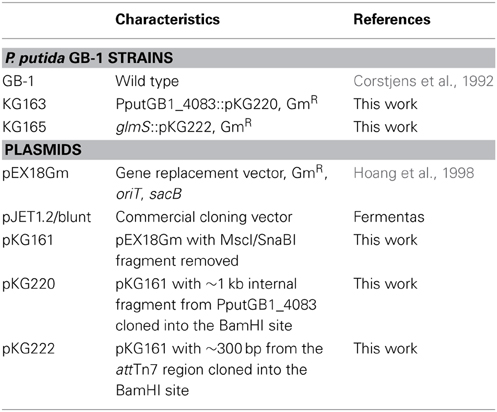
Strains and plasmids used are summarized in Table 1. To generate a plasmid integration disruption mutation of PputGB1_4083 (PputGB1_4083::pKG220), a homolog of pvdI, an ~1 kb region within the gene was amplified using primer fliF_2-R (ACGATGTCCAGGCGCACC). This primer was fortuitously discovered to anneal near the 3' end of PputGB1_4083 on opposite strands of the DNA, producing a 1 kb product. The PCR product was first cloned into the specialized PCR cloning plasmid pJET1.2/blunt (Fermentas) then subcloned into pKG161, a derivative of pEX18Gm (Table 1) from which the sacB gene had been deleted by digestion with MscI/SnaBI and self-ligation of the plasmid backbone. This plasmid (pKG220) was moved by conjugation into P. putida GB-1 (Geszvain and Tebo, 2010) and transconjugants were screened for Gm resistance. GmR colonies were screened for homologous recombination between the plasmid and the chromosome, resulting in integration of the plasmid into the chromosome, by isolating genomic DNA from candidate colonies and screening by PCR using the M13-F primer, which anneals within the plasmid, and the 4083_1-F primer (GGGCCGACCATCAGGTGAAAG), which anneals within PputGB1_4083 immediately upstream of the region present on pKG220. The ability to amplify an ~1 kb product with these primers indicated that the plasmid had integrated into the chromosome to generate PputGB1_4083::pKG220.
To address the concern that the presence of the plasmid backbone itself could affect the phenotype of the bacteria, we also generated a plasmid integration strain in which the plasmid was inserted downstream from PputGB1_5427 (glmS) in the attTn7 region of the chromosome. This region commonly tolerates insertions without affecting cell growth/behavior (Choi et al., 2005). The attTn7 region was amplified using primers glmS-F (GTTGGTTGTGTTCGCCGACG) and glmS-R (TTCAAGGCAGCGGAGGGG), and then cloned into pKG161 to generate plasmid pKG222 as described above. Plasmid integrants were generated as above and screened via PCR with the M13-F primer and the attTn7 region primer glmS_3-F (GCGCCGAACAACGAACTGC).
Test of NRPS Mutant
The wild-type equivalent, glmS::pKG222, and NRPS mutant, pvdI::pKG220, were grown in LB containing 50 μg ml−1 gentamicin and streaked out on LB plates supplemented with 36 μM FeSO4 with either 100 μM or 1 mM 2'-2' dipyridyl (Lehoux et al., 2000), CAS-CAA (Matthijs et al., 2004), or succinate medium (Meyer et al., 1997) to determine production of PVDs or siderophores.
Comparison of Mn-Oxidizing Strains
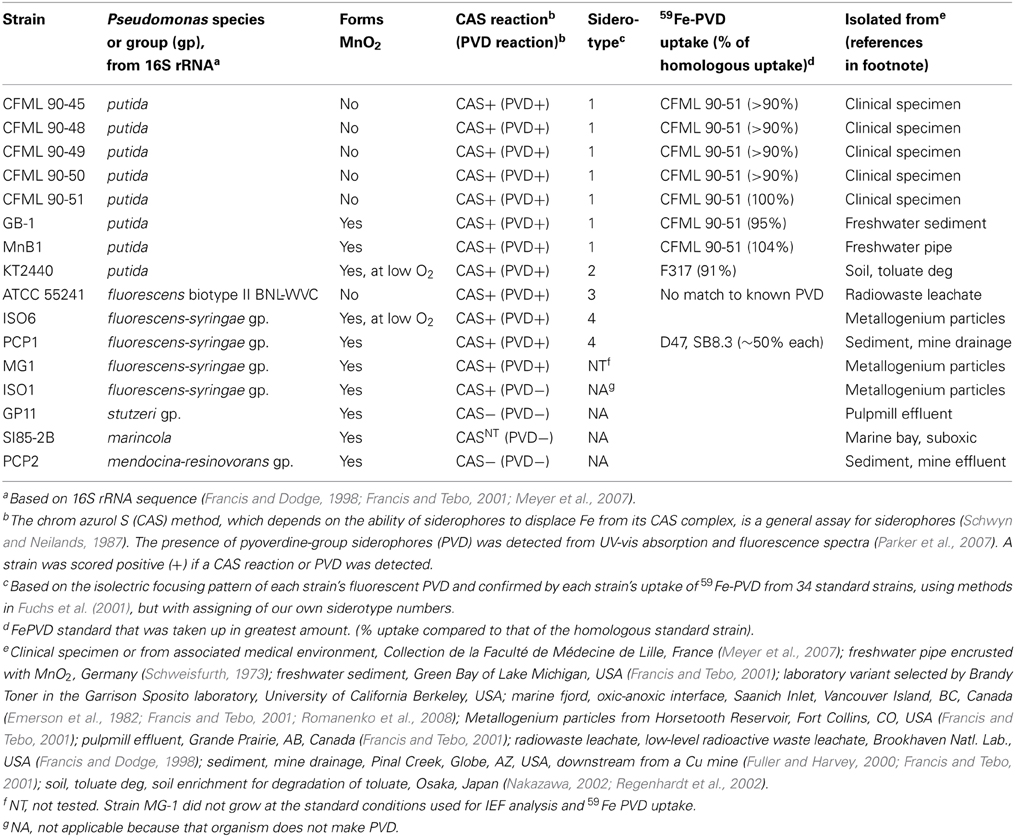
Table 2 lists the strains examined. For tests of siderophore production, each organism was serially transferred three times in each of two media: low-iron casamino acids medium (Meyer et al., 1997) and succinate minimal medium without added iron (Meyer et al., 1997). Chrom azurol S (CAS) tests of general siderophore presence in centrifuged and filtered culture supernatants were performed by the standard shuttle method (Schwyn and Neilands, 1987). For PVD detection, culture supernatants (ca. pH 7.7) were adjusted to pH 5, 6, or 8 with HCl or NaOH, transferred to quartz cuvettes (1 cm path length) and examined in a SpectraMax M2 scanning spectrophotometer-fluorimeter (Molecular Devices). PVD was identified by its known fluorescence (excitation at 405 nm; emission read at 470 and 535 nm, pH 8) and by its characteristic UV-vis absorbance properties as a function of pH (Albrecht-Gary et al., 1994; Parker et al., 2004, 2007), with screening for absorbance maxima at ca. 364 (pH 5), 380 (pH 6), and 400–405 nm (pH 7.5–8). Isoelectric focusing and biological uptake of 59Fe-PVD standards were as previously described (Fuchs et al., 2001; Meyer et al., 2007), with PVD standards from: Pseudomonas putida CFML 90-44, Pseudomonas sp. G76, Pseudomonas sp. G4, P. putida CFML 90-51, P. putida GS43, P. costantinii CFBP 5705T, P. fluorescens W, P. monteilii CFML 90-54, P. putida GS37, P. aeruginosa Pa6, Pseudomonas sp. 2908, P. putida WCS358, P. fluorescens PL7, Pseudomonas sp. B10, P. fluorescens 51W, P. fluorescens Pflii, Pseudomonas sp. CFML 96-188, P. fluorescens Pfl12, Pseudomonas sp. D47, P. thivervalensis ML45, P. fluorescens Pf0-1, P. putida AP3, Pseudomonas sp. G85, Pseudomonas sp. F317, and Pseudomonas sp. F360 (abbreviations: CFML, Collection de la Faculté de Médecine de Lille, France; CFBP, Collection Française de Bactéries Phytopathogènes, Angers, France).
Results
In Silico Identification of the Putative PVD Synthesis Operon in P. putida GB-1
The genome of P. putida GB-1 encodes an NRPS operon comprised of the genes PputGB1_4086 through PputGB1_4083 (Figure 1) (Markowitz et al., 2008). These four genes are annotated as encoding NRPSs and have homology to the P. aeruginosa PAO1 PVD synthesis genes pvdI/J and D. Furthermore, downstream of this operon is a putative TonB-dependent siderophore receptor gene (PputGB1_4082). A gene encoding a homolog of PvdO (PputGB1_4081) which appears to have some role in PVD formation (Yeterian et al., 2010) was also found to be present (Figure 1A). Upstream of the first gene in the putative operon—PputGB1_4086—is a sequence with a perfect match to the PvdS sigma recognition site (TAAAT-N16-CGT) (Ochsner et al., 2002) (Figure 1B). The alternative sigma factor PvdS is an iron-responsive extracytoplasmic function (ECF) sigma (Leoni et al., 2000), suggesting that expression of the genes PputGB1_4086-4083 is regulated by iron concentration as would be expected for a PVD synthesis operon. A PvdS recognition site is located upstream of the pvdI (PA2402) and pvdD (PA2399) genes of P. aeruginosa PAO1 as well (Ochsner et al., 2002). PputGB1_3810, a homolog of pvdS (PA2424), was found along with PputGB1_3809, a homolog of pvdL/psvA (PA2424), a putative NRPS for chromophore synthesis (Mossialos et al., 2002) (Figure 1A). Also found in this operon along with NRPSs for PVD peptide backbone synthesis is a homolog of a gene encoding SyrP (PputGB1_4087) which is an Asp hydroxylase required for synthesis of syringomycin (Singh et al., 2008).
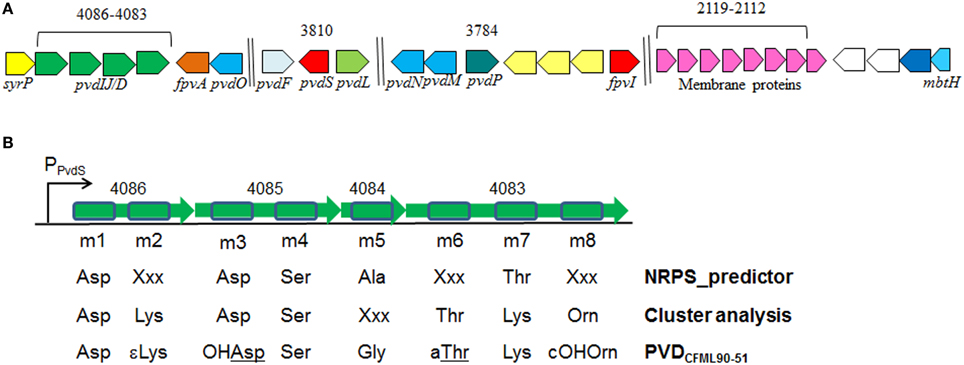
Figure 1. (A) Clusters of putative genes likely to be involved in PVD synthesis in P. putida GB-1. Identification is based on in silico comparison to known PVD-related genes, as described in the text. Genes are annotated according to Ravel and Cornelis (2003). Color schemes are based on Ravel and Cornelis (2003). Numbers on top are locus tags for P. putida GB-1 (PputGB1 numbers). Length is not to scale. (B) The 8 predicted adenylation domains found in the putative NRPSs (PputGB1_4083-4086) in P. putida GB-1, and the resulting peptides of each adenylation domain predicted using NRPSpredictor and cluster analysis in comparison with the reported peptide sequence of PVDCFML90-51. The NRPS genes are shown in green with the adenylation domains boxed in blue.
Generation of an NRPS Mutant
If the PputGB1_4086-4083 operon encodes the synthetic machinery of the PVD peptide in P. putida GB-1, disruption of this operon should lead to a loss of PVD synthesis, as has been shown to occur following mutation of the PVD peptide NRPS genes pvdI and pvdD in P. aeruginosa PAO1 (Merriman et al., 1995; Lehoux et al., 2000). Plasmid integration disruption was therefore performed on PputGB1_4083, which encodes a homolog to pvdI (PA2402 in P. aeruginosa PAO1). As would be expected, the plasmid integration mutant (KG163) did not fluoresce and lacked yellow-green pigments whereas a control strain in which the plasmid was integrated into the attTn7 site (KG165) did, suggesting the NRPS mutant KG163 did not produce PVDs (Figures 2A,B).
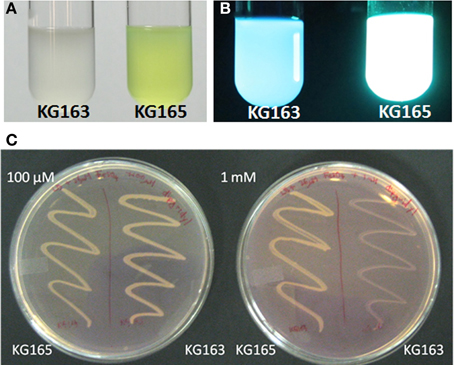
Figure 2. KG163 (PVD synthesis mutant of P. putida GB-1) and KG165 (WT equivalent) grown in succinate medium and visualized under normal light (A) and UV (B). KG163 and 165 grown on LB supplemented with 36 μM FeSO4 and different amounts of 2'-2' dipyridyl (C). Plate on the left has 100 μM and the right has 1 mM dipyridyl while the left side of each plate is the WT equivalent and on the right side of each plate is the NRPS mutant.
KG163 was also grown in increasing amounts of the iron-chelator dipyridyl to verify the decreased ability to synthesize PVD, which is needed to compete for iron under these conditions. As shown in Figure 2C, the growth of the NRPS mutant is inhibited by increasing amounts of dipyridyl while growth of KG165 was not. This result supports the conclusion that the inability to acquire iron and not toxicity of dipyridyl was responsible for the growth defect of the NRPS mutant, KG163. Based on these findings, we conclude that PputGB1_4083 is required for PVD synthesis.
In Silico Analyses of Adenylation Domains of NRPSs
To determine the peptide sequence of PVDGB−1, in silico analyses were performed using the NRPSpredictor website (http://www-ab.informatik.uni-tuebingen.de/software/NRPSpredictor) (Rausch et al., 2005). In silico analyses predicted 8 adenylation modules within the 4 genes of the putative PVD peptide synthesis operon (PputGB1_4086-4083) (Figure 1B). Also, it was possible to generate a preliminary prediction of the sequence of amino acids in the peptide: Asp1-Xxx2-Asp3-Ser4-Ala5-Xxx6-Thr7-Xxx8 (where Xxx indicates amino acids that could not be predicted using NRPSpredictor). To further investigate the nature of the amino acids recognized by each of the adenylation domains in the putative PVD peptide synthesis operon of P. putida GB-1, we compared the amino acid sequence of each domain to that of other pseudomonad PVD synthesis proteins that produce PVDs with known structures. In this analysis, most of the adenylation modules from P. putida GB-1 fell into distinct clusters with known adenylation modules in other organisms (Figure 3). The cluster analysis confirmed the conclusions of NRPS predictor for modules m1 (Asp), m3 (Asp), and m4 (Ser), but suggested a Lys at m7 instead of the Thr chosen by NRPS predictor (Figure 1B). For the three cases in which NRPS predictor failed to select an amino acid, the identifications by cluster analysis were: m2 (Lys), m6 (Thr), and m8 (Orn) (Figure 1B). More precisely, module m6 clusters with other Thr-incorporating modules from five different pseudomonads, and it appears to be distinct from the allo-Thr incorporating modules of SypC, although these belong to the more distantly related organism Pseudomonas syringae pv. syringae B301D. Module m8 falls into a cluster with two P. aeruginosa PAO1 modules that incorporate N5-formyl-N5-hydroxyornithine (hfOrn) but is located on a long branch and therefore might direct incorporation of a different amino acid. However, the P. putida GB-1 genome encodes a homolog to PvdA (PputGB1_2120), which is an enzyme responsible for the modification of ornithine to OHOrn (Visca et al., 2007) and thus strain GB-1 is predicted to have the biochemical potential to generate OHOrn. Module m5 appears to cluster somewhat near several Ala-incorporating modules from Pseudomonas syringae pv. syringae B301D, but lies on a long branch outside of that cluster, which may reflect a lack of sufficiently homologous sequences to accurately root this branch on the tree.
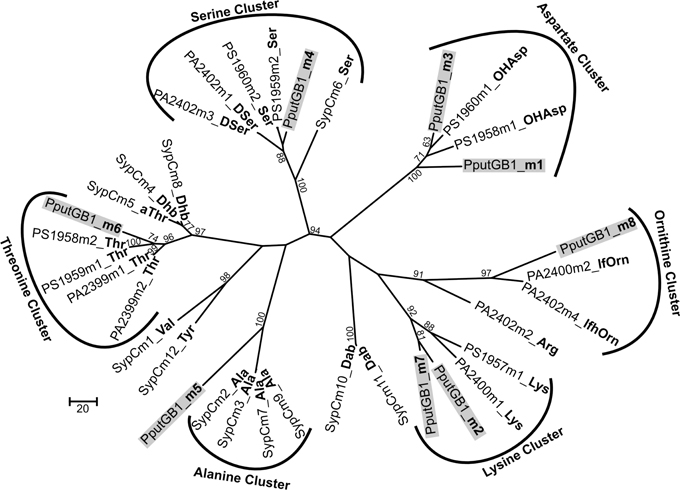
Figure 3. Phylogenetic cluster analysis using MUSCLE performed for selected adenylation domains among pseudomonads along with 8 adenylation domains found in the genome of P. putida GB-1 in PputGB1_4086-4083.
Chemical Composition and Siderotyping of PVDGB−1 and PVDMnB1
The PVDs of strains P. putida GB-1 and MnB1 were siderotyped in comparison to those of several isolates of known PVD structure (Table 2). Strains GB-1 and MnB1 were found to be in the same siderotype as P. putida CFML 90-51, which has been reported to have the following PVD peptide sequence: chromophore-Asp-Lys-OHAsp-Ser-Gly-aThr-Lys-cOHOrn (D-amino acids underlined) (Sultana et al., 2000). Since this sequence was strikingly similar to that predicted for PVDGB−1 by cluster analysis, it is included in Figure 1A for comparison. To determine whether this sequence also applied to strains GB-1 and MnB1, mass spectrophotometric (MS/MS) analysis was used to obtain molecular weights and also to identify the secondary MS/MS fragments of each PVD in partially purified preparations from the P. putida strains GB-1 and MnB1, in comparison to parallel preparations from P. putida CFML 90-51 (Table 3). The PVDs of all three organisms shared a major MS peak at the monoisotopic m/z of 1250–1251 Da (Table 3). This value, as well as the weights of secondary MS/MS fragmentation products, corresponded to the peptide sequence given above. Our data agreed with the previous report for PVDCFML90−51 (Sultana et al., 2000), except that the variable acyl side chain in our samples from strain GB-1 was malic acid amide (mala) instead of malic acid (mal) and the acyl side chain in our CFML 90-51 preparation was ~50% mala and ~50% mal, in contrast to the previous report in which only mal was found with CFML 90-51 (Sultana et al., 2000). Interestingly, MS/MS samples from all three strains, CFML 90-51, MnB1, and GB-1, also contained a major monoisotopic peak at m/z 1161 Da, which was identified to contain the same PVD peptide as in the 1250 Da peak, but with the PVD chromophore replaced by an azotobactin chromophore, a structure that is frequently co-produced with pyoverdine (Fuchs et al., 2001). Table 2 lists the MS apparent relative abundance of the peaks at m/z of 1250.4 and 1161.4 Da, expressed as a percentage of the total peak area of PVD-type siderophores produced by each strain. The samples also showed minor amounts of sulfonated PVD at 1333.5–1335.5 Da (2 and 6% of the total peak areas, respectively), and two minor peaks at 1232.5 and 1144 Da, which appear to represent water-loss reactions involving β elimination at various hydroxyl groups of the peptide, with the formation of double bonds conjugated to a carbonyl. Material from each peak at m/z 1250, 1161, and 1333 Da was further fragmented and analyzed by MS/MS. Since these secondary fragments from the three tested siderotype n° 1 strains were indistinguishable (data not shown), we conclude that these strains produced the same general set of PVD-type siderophores including sulfonated and non-sulfonated PVD and azotobactin.
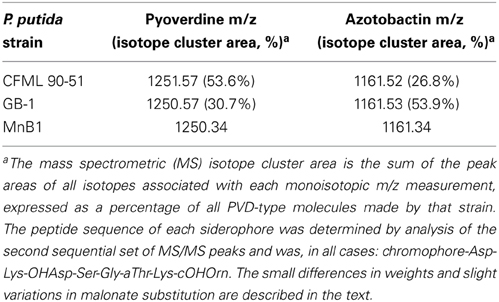
Table 3. Mass spectrometric (MS/MS) analysis of siderophores from several siderotype n° 1 strains of Pseudomonas putida.
Comparison of Mn-Oxidizing and Mn-Non-Oxidizing Strains
Pseudomonas isolates from diverse habitats and taxonomic groupings were characterized with respect to MnO2 formation and siderophore production (Table 2). The strains comprised six 16S rRNA groupings: P. putida, P. fluorescens, P. marincola, P. fluorescens-syringae group, P. mendocina-resinovorans group, and P. stutzeri group (Table 2). When iron starved, most strains were positive in the chrome azurol S (CAS) assay (Table 2), a standard method that detects the production of most types of siderophores (Schwyn and Neilands, 1987). The majority also formed PVD, as indicated by their release of green fluorescent compounds into the medium under iron-limiting, but not iron-replete, conditions (Table 2). The presence of PVD in culture fluids was confirmed by examination of fluorescence and absorbance spectra at pH 5, 6, and 8, both in the absence and presence of iron (Parker et al., 2004, 2007); in all positive cases the spectra were those expected for PVD (data not shown because they were so standard).
When the above strains were siderotyped based on the isoelectric focusing (IEF) patterns of their PVDs (Fuchs et al., 2001; Meyer, 2007), seven P. putida isolates showed identical IEF patterns (Table 2); these strains included two Mn(II) oxidizers (strains GB-1 and MnB1) and five isolates that did not detectably oxidize Mn(II) (the five CFML strains in Table 2). These seven strains also incorporated 59Fe-PVD from the reference organism CFML 90-51 as efficiently as their own 59Fe-PVD (Table 2), but did not internalize 24 other 59Fe-PVD standards of other siderotypes (see Methods). Based on our data and previous characterizations of the five CFML strains (Meyer et al., 2007), we conclude that these 7 isolates are of the same siderotype, here designated siderotype number 1 (n° 1), which is equivalent to n° 30 in Fuchs et al. (2001), siderovar n° 15 in Meyer et al. (2007), or n° 32 in Meyer et al. (2008).
The other tested strains were assigned to three new siderotypes (Table 2) that did not correspond to any grouping in Fuchs et al. (2001). P. putida KT2440 is here designated as siderotype n° 2, for which a PVD structure has been deduced from genome sequence (Ravel and Cornelis, 2003; Meyer et al., 2007). Previously, this siderotype was also assigned as siderovar n° 25 (Meyer et al., 2007) or as type 3 (Matthijs et al., 2009).
Siderotype n° 3, P. fluorescens ATCC 55241, did not take up any tested 59Fe-PVD standard and appears to have an as yet uncharacterized PVD (Table 2). The siderotype n° 4 strains were P fluorescens-syringae group Mn(II) oxidizers ISO6 and PCP1 which partially took up 59Fe-PVD of two reference strains, P. fluorescens SB8.3 and Pseudomonas sp. D47 (Table 2). PVD of Pseudomonas sp. D47 was previously classified as n° 29 and P. fluorescens sp. SB8.3 (=Ps4a) as n° 7 (Meyer et al., 2008). Although we have not determined the PVD structures of siderotypes n° 3 and n° 4, it is clear that they are not the same as those of siderotypes n° 1 or n° 2.
Discussion
P. putida strains GB-1 and MnB1 are Mn(II)-oxidizing bacteria that produce pyoverdines (PVDs), siderophores that affect iron uptake but may also substantially influence the metabolism of other metals, including Mn. To facilitate future studies of the potentially multi-faceted roles such as inhibiting Mn(II) oxidation played by PVD in the oxidation of Mn by pseudomonads, we have examined both the genetics of PVD production in strain GB-1 and the composition of the pyoverdines produced by P. putida GB-1, as well as several other Mn(II)-oxidizing strains.
Within the genome sequence of P. putida GB-1, a putative non-ribosomal peptide synthase (NRPS) operon capable of being involved in the synthesis of the PVDGB−1 peptide was identified: PputGB1_4086-4083. Disruption of PputGB1_4086-4083 resulted in a defect in PVD synthesis similar to that reported in other Pseudomonas strains with mutations in comparable NRPSs (Merriman et al., 1995; Lehoux et al., 2000). Furthermore, the peptide sequence based on in silico analyses of the adenylation domains of NRPS PputGB1_4086-4083 was consistent with the actual peptide sequence determined from MS/MS of PVDGB−1 samples. Therefore, it can be safely concluded that PputGB1_4086-4083 is the NRPS that governs PVD peptide synthesis in P. putida GB-1. In silico examination of the P. putida GB-1 genome also identified candidates (Figure 1) for many of the other genes needed for the synthesis, modification, and expression of PVDGB−1, including a NRPS for chromophore synthesis (PputGB1_3809), a TonB-dependent siderophore receptor gene (PputGB1_4082), an Asp hydroxylase (PputGB1_4087), an Orn hydroxylase (PputGB1_2120), and an iron-responsive ECF alternative sigma factor (PputGB1_3810) with recognition sites found in the promoters upstream of most of the above genes.
Based on siderotyping followed by MS/MS analysis (Tables 2, 3), we have determined the peptide sequences of PVDGB−1 and PVDMnB1 to be: chromophore-Asp-Lys-OHAsp-Ser-Gly-aThr-Lys-cOHOrn, identical to that reported (Sultana et al., 2000) and confirmed here (Table 3) for PVDCFML90−51. The OHAsp and the aThr in PVDCFML90−51 are known to be D isomers (Sultana et al., 2000). PVDGB−1 and PVDMnB1 probably also contain these two D amino acids because the degree of uptake of 59Fe-labeled PVDCFML90−51 by strains GB-1, MnB1, and CFML 90-51 was similar (Table 2), suggesting that the cellular uptake receptors of strains GB-1 and MnB1 did not detect a steric difference between PVDCFML90−51 and the endogenous PVD of each strain. However, the presence of D amino acids in PVDGB−1 and PVDMnB1 has not been directly tested.
The peptide sequence above suggests a metal-binding pocket formed by three moieties: (1) the catecholate of the chromophore, (2) the cyclic hydroxamate from cOHOrn, and (3) the α-OH-carboxylate from OHAsp. It is not yet clear how this structure, including the presence of a (OH)carboxylate donor group, leads to the higher thermodymamic stability constants for Mn(III), as compared to Fe(III), reported for these siderophores at physiological and alkaline pH (Parker et al., 2004; Harrington et al., 2012). However, an influence of OHcarboxylate on the preferential binding of Mn(III) has been recently suggested by an investigation showing that siderophores containing solely hydroxamates (e.g., desferrioxamine B, DFOB) or solely catecholates (e.g., protochelin) bind Fe(III) more strongly than Mn(III), whereas rhizoferrin, which complexes metals via a mixture of carboxylate and (OH)carboxylate groups, binds Mn(III) more strongly than Fe(III) (Harrington et al., 2012), as do several aminocarboxylate ligands (Hamm and Suwyn, 1967; Ahrland et al., 1990; Martell and Smith, 2003). Based on K-edge EXAFS comparisons of various Fe(III)- and Mn(III)-siderophore complexes in solution, Harrington et al. (2012) proposed that the greater flexibility of carboxylates vis-à-vis hydroxamates or catecholates allows the former to accommodate the Jahn-Teller-distorted coordination that is characteristic of Mn(III) but not Fe(III). This result suggests that (OH)carboxylates within siderophores, perhaps including the siderotype n° 1 PVDs studied here, may affect the preferential binding of Mn(III). However, the situation for pyoverdines probably also involves additional factors, because two mixed-moiety pyoverdines (PVDCFML90−51 and PVDPa1) both preferentially bound Mn(III) and both seemed to accommodate Jahn-Teller distortion, even though PVDCFML90−51 contains a (OH)carboxylate but PVDPa1 does not (Harrington et al., 2012). Perhaps these PVDs gain flexibility from their mixed donor groups, their polypeptide structure, or some other factor.
MS/MS analysis of the PVDs from P. putida strains GB-1 and MnB1 also indicated that each strain produced a set of three PVD-type siderophores sharing the same peptide tail but with differently modified chromophores: “classical” PVD, sulfonated PVD, and azotoactin (Table 3). Since all three are strongly fluorescent and since fluorescence was undetectable in the mutant KG163 (Figure 2B), it is likely that the peptide tail of all three PVD types in strain GB-1 is synthesized through the same NRPS operon, PputGB1_4083-4086, which makes sense since the peptides of all three PVD types showed identical MSMS fragmentation patterns. However, subsequent modifications could be subject to differing regulatory or catalytic pathways. It is currently unknown whether these three differing PVD types affect Mn metabolism or the complexation of Mn vis á vis Fe similarly or differently.
Azotobactin and PVD are both known to complex various metal cations (Braud et al., 2009; Wichard et al., 2009). However, azotobactin can also bind oxyanions such as molybdate and vanadate (Wichard et al., 2009). In contrast, the predominant PVD of P. aeruginosa PAO1 is not able to form complexes with vanadate, whereas the other main siderophore of strain PAO1, pyochelin, can (Baysse et al., 2000). This observation is consistent with other reports that PVDs do not play an important role with oxyanions (Wichard et al., 2009). Therefore, one function of azotobactin in P. putida GB-1 and related strains might be to complex oxyanions for uptake or detoxification, as was suggested for Azotobacter vinelandii (Wichard et al., 2009). Alternatively, sulfonated PVDs and azotobactin could be precursors or byproducts of PVD synthesis (Fuchs et al., 2001; Baysse et al., 2002). Further studies need to be performed to elucidate the respective roles of the multiple siderophores of these organisms, especially with regard to manganese oxidation.
Since the ability to oxidize Mn(II) occurs very commonly, but nonetheless sporadically, among a wide variety of Pseudomonas species (Francis and Tebo, 2001), it was no surprise that multiple PVD siderotypes were identified among the phylogenetically-diverse Mn(II)-oxidizing pseudomonads tested here (Table 2). Within P. putida, two siderotypes were identified (Table 2): n° 1 including strains CFML 90-51 and GB-1 and n° 2 consisting of strain KT2440. Although not included in this research, P. putida ATCC 12663 is also capable of oxidizing Mn(II), is closely related to P. putida GB-1 based on 16S data (Francis and Tebo, 2001), but produces a PVD that has been previously shown to be different from PVDCFML90−51 (Meyer et al., 2007). Since PVDCFML90−51 and PVDGB−1 are indistinguishable by MS/MS (Table 3) and siderotyping (Table 2), PVDGB−1 and PVDATCC12663 cannot be the same. Therefore, even among the Mn(II)-oxidizing P. putida at least three differing PVDs exist: PVDGB−1, PVDKT2440, and PVDATCC12663. This situation is in agreement with the conclusion of Meyer et al. (2007) that P. putida as currently defined is heterogeneous with respect to siderotype. It is also notable that the siderotype of a Mn oxidizer can be the same as that of a strain that does not oxidize Mn(II), as for P. putida GB-1 and CFML 90-51 (Table 2).
In summary, this study has combined in silico, genetic and chemical (siderotyping and MS/MS) approaches to explore the synthesis and nature of the suite of related PVDs (“classic” PVD, azotobactin, and sulfonated PVD) that were produced by the model Mn(II)-oxidizing organism Pseudomonas putida GB-1 at our growth conditions. In silico analysis indicated that position PputGB1_4083-4086 of the GB-1 genome contained NRPSs that could synthesize a peptide chain consistent with the PVDGB−1 peptide determined by MS/MS (chromophore-Asp-Lys-OHAsp-Ser-Gly-aThr-Lys-cOHOrn). Furthermore, mutation at PputGB1_4083 prevented PVD synthesis. A diverse selection of Mn-oxidizing Pseudomonas species were found to comprise at least three distinct PVD siderotypes, indicating differences in PVD structure and PVD uptake specificity that can be exploited in future studies concerning the ways that various PVDs can influence Mn metabolism, especially Mn(II) oxidation, in pseudomonads and other bacteria.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Katherine Barbeau, Elizabeth Komives, The Scripps Institution of Oceanography, and the U.C.S.D. Molecular Mass Spectrometry Facility for helpful discussions, technical assistance, and access to instruments. This publication was made possible by grants from the National Science Foundation (MCB-0630355 and OCE-1154307) and from the National Institute of Environmental Health Sciences (P42ES010337).
References
Ahrland, S., Dahlgren, Ã. S., and Persson, I. (1990). Stabilities and hydrolysis of some iron(III) and manganese(III) complexes with chelating ligands. Acta Agric. Scand. 40, 101–111. doi: 10.1080/00015129009438008
Albrecht-Gary, A. M., Blanc, S., Rochel, N., Ocaktan, A. Z., and Abdallah, M. A. (1994). Bacterial iron transport - coordination properties of pyoverdin Paa, a peptidic siderophore of Pseudomonas aeruginosa. Inorg. Chem. 33, 6391–6402. doi: 10.1021/ic00104a059
Baysse, C., Budzikiewicz, H., Uría Fernández, D., and Cornelis, P. (2002). Impaired maturation of the siderophore pyoverdine chromophore in Pseudomonas fluorescens ATCC 17400 deficient for the cytochrome c biogenesis protein CcmC. FEBS Lett. 523, 23–28. doi: 10.1016/S0014-5793(02)02915-0
Baysse, C., De Vos, D., Naudet, Y., Vandermonde, A., Ochsner, U., Meyer, J.-M., et al. (2000). Vanadium interferes with siderophore-mediated iron uptake in Pseudomonas aeruginosa. Microbiology 146, 2425–2434.
Braud, A., Hoegy, F., Jezequel, K., Lebeau, T., and Schalk, I. J. (2009). New insights into the metal specificity of the Pseudomonas aeruginosa pyoverdine-iron uptake pathway. Environ. Microbiol. 11, 1079–1091. doi: 10.1111/j.1462-2920.2008.01838.x
Brouwers, G. J., De Vrind, J. P., Corstjens, P. L., Cornelis, P., Baysse, C., and De Vrind-De Jong, E. W. (1999). cumA, a gene encoding a multicopper oxidase, is involved in Mn2+ oxidation in Pseudomonas putida GB-1. Appl. Environ. Microbiol. 65, 1762–1768.
Budzikiewicz, H. (1993). Secondary metabolites from fluorescent pseudomonads. FEMS Microbiol. Rev. 104, 209–228. doi: 10.1111/j.1574-6968.1993.tb05868.x
Caspi, R., Tebo, B. M., and Haygood, M. G. (1998). c-type cytochromes and manganese oxidation in Pseudomonas putida MnB1. Appl. Environ. Microbiol. 64, 3549–3555.
Choi, K.-H., Gaynor, J. B., White, K. G., Lopez, C., Bosio, C. M., Karkhoff-Schweizer, R. R., et al. (2005). A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2, 443–448. doi: 10.1038/nmeth765
Clement, E., Mesini, P. J., Pattus, F., and Schalk, I. J. (2004). The binding mechanism of pyoverdin with the outer membrane receptor FpvA in Pseudomonas aeruginosa is dependent on its iron-loaded status. Biochemistry 43, 7954–7965. doi: 10.1021/bi049768c
Corstjens, P. L., De Vrind, J. P., Westbroek, P., and De Vrind-De Jong, E. W. (1992). Enzymatic iron oxidation by Leptothrix discophora: identification of an iron-oxidizing protein. Appl. Environ. Microbiol. 58, 450–454.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Emerson, S., Kalhorn, S., Jacobs, L., Tebo, B. M., Nealson, K. H., and Rosson, R. A. (1982). Environmental oxidation rate of manganese(II): bacterial catalysis. Geochim. Cosmochim. Acta. 46, 1073–1079. doi: 10.1016/0016-7037(82)90060-6
Felsenstein, J. (2005). PHYLIP (Phylogeny Inference Package) Version 3.6. Distributed by the author. Seattle, WA: Department of Genome Sciences, University of Washington.
Francis, A. J., and Dodge, C. J. (1998). Remediation of soils and wastes contaminated with uranium and toxic metals. Environ. Sci. Technol. 32, 3993–3998. doi: 10.1021/es9803310
Francis, C. A., and Tebo, B. M. (2001). cumA multicopper oxidase genes from diverse Mn(II)-oxidizing and non-Mn(II)-oxidizing Pseudomonas strains. Appl. Environ. Microbiol. 67, 4272–4278. doi: 10.1128/AEM.67.9.4272-4278.2001
Fuchs, R., and Budzikiewicz, H. (2001). Structural sutides of pyoverdins by mass spectrometry. Curr. Org. Chem. 5, 265–288. doi: 10.2174/1385272013375562
Fuchs, R., Schafer, M., Geoffroy, V., and Meyer, J. M. (2001). Siderotyping a powerful tool for the characterization of pyoverdines. Curr. Top. Med. Chem. 1, 31–57. doi: 10.2174/1568026013395542
Fuller, C. C., and Harvey, J. W. (2000). Reactive uptake of trace metals in the hyporheic zone of a mining-contaminated stream, Pinal Creek, Arizona. Environ. Sci. Technol. 34, 1150–1155. doi: 10.1021/es990714d
Geszvain, K., and Tebo, B. M. (2010). Identification of a two-component regulatory pathway essential for Mn(II) oxidation in Pseudomonas putida GB-1. Appl. Environ. Microbiol. 76, 1224–1231. doi: 10.1128/AEM.02473-09
Hamm, R. E., and Suwyn, M. A. (1967). Preparation and characterization of some amino-polycarboxylate complexes of manganese(III). Inorg. Chem. 6, 139–142. doi: 10.1021/ic50047a032
Harrington, J. M., Parker, D. L., Bargar, J. R., Jarzecki, A. A., Tebo, B. M., Sposito, G., et al. (2012). Structural dependence of Mn complexation by siderophores: donor group dependence on complex stability and reactivity. Geochim. Cosmochim. Acta. 88, 106–119. doi: 10.1016/j.gca.2012.04.006
Hastings, D., and Emerson, S. (1986). Oxidation of manganese by spores of a marine Bacillus - kinetic and thermodynamic considerations. Geochim. Cosmochim. Acta. 50, 1819–1824. doi: 10.1016/0016-7037(86)90141-9
Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J., and Schweizer, H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. doi: 10.1016/S0378-1119(98)00130-9
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1994). A mutation data matrix for transmembrane proteins. FEBS Lett. 339, 269–275. doi: 10.1016/0014-5793(94)80429-X
Lehoux, D. E., Sanschagrin, F., and Levesque, R. C. (2000). Genomics of the 35-kb pvd locus and analysis of novel pvdIJK genes implicated in pyoverdine biosynthesis in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 190, 141–146. doi: 10.1111/j.1574-6968.2000.tb09276.x
Leoni, L., Orsi, N., De Lorenzo, V., and Visca, P. (2000). Functional analysis of PvdS, an iron starvation sigma factor of Pseudomonas aeruginosa. J. Bacteriol. 182, 1481–1491. doi: 10.1128/JB.182.6.1481-1491.2000
Markowitz, V. M., Chen, I. M. A., Palaniappan, K., Chu, K., Szeto, E., Grechkin, Y., et al. (2008). The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 38, D382–D390. doi: 10.1093/nar/gkp887
Martell, A. E., and Smith, R. M. (2003). Determination and Use of Stability Constants. Gaithersburg, MD: National Institute of Science and Technology (NIST).
Matthijs, S., Baysse, C., Koedam, N., Tehrani, K. A., Verheyden, L., Budzikiewicz, H., et al. (2004). The Pseudomonas siderophore quinolobactin is synthesized from xanthurenic acid, an intermediate of the kynurenine pathway. Mol. Microbiol. 52, 371–384. doi: 10.1111/j.1365-2958.2004.03999.x
Matthijs, S., Laus, G., Meyer, J.-M., Abbaspour-Tehrani, K., Schäfer, M., Budzikiewicz, H., et al. (2009). Siderophore-mediated iron acquisition in the entomopathogenic bacterium Pseudomonas entomophila L48 and its close relative Pseudomonas putida KT2440. Biometals 22, 951–964. doi: 10.1007/s10534-009-9247-y
Merriman, T. R., Merriman, M. E., and Lamont, I. L. (1995). Nucleotide sequence of pvdD, a pyoverdine biosynthetic gene from Pseudomonas aeruginosa: PvdD has similarity to peptide synthetases. J. Bacteriol. 177, 252–258.
Meyer, J.-M. (2007). “Siderotyping and bacterial taxonomy: a siderophore bank for a rapid identification at the species level of fluorescent and non-fluorescent pseudomonas,” in Soil Biology 12: Microbial Siderophores, eds A. Varma and S. Chincholkar (Berlin: Springer Verlag), 43–65.
Meyer, J.-M., Gruffaz, C., Raharinosy, V., Bezverbnaya, I., Schäfer, M., and Budzikiewicz, H. (2008). Siderotyping of fluorescent Pseudomonas: molecular mass determination by mass spectrometry as a powerful pyoverdine siderotyping method. Biometals 21, 259–271. doi: 10.1007/s10534-007-9115-6
Meyer, J.-M., Gruffaz, C., Tulkki, T., and Izard, D. (2007). Taxonomic heterogeneity, as shown by siderotyping, of strains primarily identified as Pseudomonas putida. Int. J. Syst. Evol. Microbiol. 57, 2543–2556. doi: 10.1099/ijs.0.65233-0
Meyer, J.-M., Stintzi, A., De Vos, D., Cornelis, P., Tappe, R., Taraz, K., et al. (1997). Use of siderophores to type pseudomonads: the three Pseudomonas aeruginosa pyoverdine systems. Microbiology 143, 35–43. doi: 10.1099/00221287-143-1-35
Mossialos, D., Ochsner, U., Baysse, C., Chablain, P., Pirnay, J.-P., Koedam, N., et al. (2002). Identification of new, conserved, non-ribosomal peptide synthetases from fluorescent pseudomonads involved in the biosynthesis of the siderophore pyoverdine. Mol. Microbiol. 45, 1673–1685. doi: 10.1046/j.1365-2958.2002.03120.x
Nakazawa, T. (2002). Travels of a Pseudomonas, from Japan around the world. Environ. Microbiol. 4, 782–786. doi: 10.1046/j.1462-2920.2002.00310.x
Ochsner, U. A., Wilderman, P. J., Vasil, A. I., and Vasil, M. L. (2002). GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45, 1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x
Okazaki, M., Sugita, T., Shimizu, M., Ohode, Y., Iwamoto, K., De Vrind-De Jong, E. W., et al. (1997). Partial purification and characterization of manganese-oxidizing factors of Pseudomonas fluorescens GB-1. Appl. Environ. Microbiol. 63, 4793–4799.
Parker, D. L., Morita, T., Mozafarzadeh, M. L., Verity, R., Mccarthy, J. K., and Tebo, B. M. (2007). Inter-relationships of MnO2 precipitation, siderophore-Mn-(III) complex formation, siderophore degradation, and iron limitation in Mn-(II)-oxidizing bacterial cultures. Geochim. Cosmochim. Acta. 71, 5672–5683. doi: 10.1016/j.gca.2007.03.042
Parker, D. L., Sposito, G., and Tebo, B. M. (2004). Manganese(III) binding to a pyoverdine siderophore produced by a manganese(II)-oxidizing bacterium. Geochim. Cosmochim. Acta. 68, 4809–4820. doi: 10.1016/j.gca.2004.05.038
Rausch, C., Weber, T., Kohlbacher, O., Wohlleben, W., and Huson, D. H. (2005). Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 33, 5799–5808. doi: 10.1093/nar/gki885
Ravel, J., and Cornelis, P. (2003). Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol. 11, 195–200. doi: 10.1016/S0966-842X(03)00076-3
Regenhardt, D., Heuer, H., Heim, S., Fernandez, D. U., Strompl, C., Moore, E. R. B., et al. (2002). Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ. Microbiol. 4, 912–915. doi: 10.1046/j.1462-2920.2002.00368.x
Romanenko, L. A., Uchino, M., Tebo, B. M., Tanaka, N., Frolova, G. M., and Mikhailov, V. V. (2008). Pseudomonas marincola sp. nov., isolated from marine environments. Int. J. Syst. Evol. Microbiol. 58, 706–710. doi: 10.1099/ijs.0.65406-0
Schalk, I. J., Abdallah, M. A., and Pattus, F. (2002). Recycling of pyoverdin on the FpvA receptor after ferric pyoverdin uptake and dissociation in Pseudomonas aeruginosa. Biochemistry 41, 1663–1671. doi: 10.1021/bi0157767
Scholz-Schroeder, B. K., Soule, J. D., and Gross, D. C. (2003). The sypA, sypB, and sypC synthetase genes encode twenty-two modules involved in the nonribosomal peptide synthesis of syringopeptin by Pseudomonas syringae pv. syringae B301D. Mol. Plant Microbe Interact. 16, 271–280. doi: 10.1094/MPMI.2003.16.4.271
Schons, V., Atkinson, R. A., Dugave, C., Graff, R., Mislin, G. L. A., Rochet, L., et al. (2005). The structure-activity relationship of ferric pyoverdine bound to its outer membrane transporter: implications for the mechanism of iron uptake. Biochemistry 44, 14069–14079. doi: 10.1021/bi051155s
Schweisfurth, R. (1973). Manganoxydierende Bakterien. I. Isolierung und Bestimmung einiger Stamme von Manganbacterien. Z. Allg. Mikrobiol. 13, 341–347. doi: 10.1002/jobm.3630130408
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–57. doi: 10.1016/0003-2697(87)90612-9
Shen, J.-S., Geoffroy, V., Neshat, S., Jia, Z., Meldrum, A., Meyer, J.-M., et al. (2005). FpvA-mediated ferric pyoverdine uptake in Pseudomonas aeruginosa: identification of aromatic residues in FpvA implicated in ferric pyoverdine binding and transport. J. Bacteriol. 187, 8511–8515. doi: 10.1128/JB.187.24.8511-8515.2005
Singh, G. M., Fortin, P. D., Koglin, A., and Walsh, C. T. (2008). β-Hydroxylation of the aspartyl residue in the phytotoxin syringomycin E: characterization of two candidate hydroxylases AspH and SyrP in Pseudomonas syringae. Biochemistry 47, 11310–11320. doi: 10.1021/bi801322z
Sultana, R., Siddiqui, B. S., Taraz, K., Budzikiewicz, H., and Meyer, J.-M. (2000). A pyoverdine from Pseudomonas putida CFML 90-51 with a Lys e-amino link in the peptide chain. Biometals 13, 147–152. doi: 10.1023/A:1009212729408
Tebo, B. M., Bargar, J. R., Clement, B. G., Dick, G. J., Murray, K. J., Parker, D., et al. (2004). Biogenic manganese oxides: properties and mechanisms of formation. Annu. Rev. Earth Planet. Sci. 32, 287–328. doi: 10.1146/annurev.earth.32.101802.120213
Toner, B., Fakra, S., Villalobos, M., Warwick, T., and Sposito, G. (2005). Spatially resolved characterization of biogenic manganese oxide production within a bacterial biofilm. Appl. Environ. Microbiol. 71, 1300–1310. doi: 10.1128/AEM.71.3.1300-1310.2005
Villalobos, M., Lanson, B., Manceau, A., Toner, B., and Sposito, G. (2006). Structural model for the biogenic Mn oxide produced by Pseudomonas putida. Am. Mineral. 91, 489–502. doi: 10.2138/am.2006.1925
Villalobos, M., Toner, B., Bargar, J., and Sposito, G. (2003). Characterization of the manganese oxide produced by Pseudomonas putida strain MnB1. Geochim. Cosmochim. Acta. 67, 2649–2662. doi: 10.1016/S0016-7037(03)00217-5
Visca, P., Imperi, F., and Lamont, I. (2007). “Pyoverdine synthesis and its regulation in fluorescent pseudomonads,” in Microbial Siderophores, eds A. Varma and S. B. Chincholkar (Berlin; Heidelberg: Springer-Verlag), 135–163.
Wichard, T., Bellenger, J.-P., Morel, F. O. M. M., and Kraepiel, A. M. L. (2009). Role of the siderophore azotobactin in the bacterial acquisition of nitrogenase metal cofactors. Environ. Sci. Technol. 43, 7218–7224. doi: 10.1021/es8037214
Keywords: siderophore, pyoverdine, azotobactin, manganese oxidation, iron
Citation: Parker DL, Lee S-W, Geszvain K, Davis RE, Gruffaz C, Meyer J-M, Torpey JW and Tebo BM (2014) Pyoverdine synthesis by the Mn(II)-oxidizing bacterium Pseudomonas putida GB-1. Front. Microbiol. 5:202. doi: 10.3389/fmicb.2014.00202
Received: 18 March 2014; Accepted: 16 April 2014;
Published online: 07 May 2014.
Edited by:
Partha Basu, Duquesne University, USACopyright © 2014 Parker, Lee, Geszvain, Davis, Gruffaz, Meyer, Torpey and Tebo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sung-Woo Lee, Division of Environmental and Biomolecular Systems, Oregon Health and Science University, 20000 NW Walker Rd., Beaverton, OR 97006, USA e-mail: sungwlz@gmail.com
 Dorothy L. Parker
Dorothy L. Parker Sung-Woo Lee
Sung-Woo Lee Kati Geszvain
Kati Geszvain Richard E. Davis2
Richard E. Davis2 Bradley M. Tebo
Bradley M. Tebo