- 1Sustainable Agro-Ecosystems and Bioresources Department, Research and Innovation Centre, Fondazione Edmund Mach, San Michele all'Adige, Italy
- 2Department of Environmental Sciences, Faculty of Biological Sciences, Quaid-i-Azam University, Islamabad, Pakistan
- 3Dipartimento di Scienze Agrarie e Ambientali - Produzione, Territorio, Agroenergia, Università degli Studi di Milano, Milano, Italy
Generally, plants are not considered as hosts for human and animal pathogens (HAP). The recent produce-associated outbreaks of food-borne diseases have drawn attention toward significant deficiencies in our understanding of the ecology of HAP, and their potential for interkingdom transfer. To examine the association of microorganisms classified as HAP with plants, we surveyed the presence and distribution of HAP bacterial taxa (henceforth HAPT, for brevity's sake) in the endosphere of grapevine (Vitis vinifera L.) both in the plant stems and leaves. An enrichment protocol was used on leaves to detect taxa with very low abundance in undisturbed tissues. We used pyrosequencing and phylogenetic analyses of the 16S rDNA gene. We identified several HAPT, and focused on four genera (Propionibacterium, Staphylococcus, Clostridium, and Burkholderia). The majority of the bacterial sequences in the genus Propionibacterium, from grapevine leaf and stem, were identified as P. acnes. Clostridia were detected in leaves and stems, but their number was much higher in leaves after enrichment. HAPT were indentified both in leaves and wood of grapevines. This depicts the ability of these taxa to be internalized within plant tissues and maintain their population levels in a variety of environments. Our analysis highlighted the presence of HAPT in the grapevine endosphere and unexpected occurrence of these bacterial taxa in this atypical environment.
Introduction
Endophytes (non-pathogenic microorganisms living inside plant tissues and cells) are common inhabitants of interior plant parts (the endosphere) universally present and found in all the species of plants studied up till now (Schulz and Boyle, 2006). Some endophytes are able to promote plant growth and to suppress plant diseases (Compant et al., 2005; Lugtenberg and Kamilova, 2009).
Intriguingly, among these endosphere-dwellers, we can occasionally find some human and animal pathogens (HAP) (Holden et al., 2009; Kirzinger et al., 2011). HAP not only contaminate plant surfaces, but also actively interact with plants and can colonize them as alternative hosts (Holden et al., 2009). Some human bacterial pathogens are capable to colonize inner plant tissues (Tyler and Triplett, 2008), a phenomenon that in most cases can be considered as an opportunistic exploitation of a short-term habitat (Campisano et al., 2014). Generally, pathogens are studied solely for their harmful impact on human and animal health, causing disease and epidemics. On the other hand, the regular interaction of human and animal carriers with their environment puts these pathogens in contact with alternative niches, including additional hosts (Enders et al., 1993; Lenz et al., 2003). It is then unsurprising that previous works found out well known and potential HAP undergoing an endophytitc stage in their lifestyle (Kirzinger et al., 2011). Scientific literature often reports that members of the family Enterobactericeae, including pathogenic Salmonella and Shigella genus strains, Vibrio cholerae strains, and the human opportunistic pathogen Pseudomonas aeruginosa were found on plants or inside plants (Akhtyamova, 2013). Salmonella enterica strains have been isolated as endophytic colonizers of barley roots, spreading to the rhizodermis layers (Kutter et al., 2006).
Enteric bacterial pathogens, usually transmitted through foods, are well adapted to vertebrate hosts and generally colonize the gut (Wagenaar, 2008). Some have humans as their principal host, while many others are persistent in animal populations, adapted to a particular reservoir or environment, and affect humans only incidentally (Lynch et al., 2009). HAP on plants are generally thought of as having a reservoir in the intestines of a vertebrate host and, once discarded in manure, coming into direct or indirect contact with epigeous or hypogeous plant tissues in a variety of ways. Traditionally, they were considered to be fleeting on plant surfaces, persisting inertly in cracks, wounds, and stomatal openings. They were considered unable to aggressively modify or to communicate with the plant. However, it is now apparent that enteric bacteria do not just land on and reside in plants. These pathogens can stick tightly to produce, multiply, and enter into the tissues of leaves or fruits, in some cases even moving into inner plant parts (Berger et al., 2010; Erickson, 2012). Whether endophytic growth may actually be part of HAP life cycle is under debate. Findings such as those mentioned before, reporting an endophytic stage for HAP, would explain why existing surface decontamination procedures may be inefficient in removing contaminants from plant produce (Rosenblueth and Martinez-Romero, 2006; Teplitski et al., 2009; Saldaña et al., 2011; Barak and Schroeder, 2012; Olaimat and Holley, 2012). The diffusion of HAP in ecosystems are of scrupulous importance from the perspectives of both biology and evolution of cross-kingdom pathogenesis and/or adaptation (Lenz et al., 2003; Kirzinger et al., 2011). While the majority of published research has focused on describing the enteric HAP, there is no doubt that other HAP can also interact with plants as part of their life-cycle. Here, we surveyed the presence and distribution of HAP taxa (HAPT) in the endosphere of grapevine (Vitis vinifera L.). We used pyrosequencing of the bacterial 16S rDNA gene to identify sequences belonging to genera where HAP are abundant. We present the first (to the authors' best knowledge) report of the occurrence of bacteria in taxa potentially pathogenic to human and animals in the grapevine endosphere.
Materials and Methods
Sample Collection and DNA Extraction
Grapevines samples were taken in Northern Italy. For stem endophytes analysis, 12 plants (7 plants cv Chardonnay and 5 plants cv Merlot) were sampled from vineyards in Trentino, Italy with farmers permission (sites are mapped here: http://goo.gl/maps/7AI7j) during the fall of 2010, from October 27th to November 11th. One lateral shoot per plant was removed and stored briefly at 5–10°C. Plant material was pre-processed as described previously (Pancher et al., 2012) and DNA was extracted from surface-disinfected and aseptically peeled grapevine stems. Briefly, plant material was pulverized in sterile steel jars using liquid nitrogen and a mixer-mill. DNA was extracted from each sample using FastDNA spin kit for soil and a FastPrep-24 mixer (MP Biomedical, USA) according to standard manufacturer protocols. Plants used for leaf endophytes analysis (cv Barbera) were sampled from a vineyard in Lombardia, Italy (site is mapped here: http://goo.gl/5Nfh9R) on October 15th, 2007. Plant leaves were surface-sterilized as previously described (Bulgari et al., 2009) and aseptically prepared for DNA extraction. In a subset of leaf samples DNA isolation was preceded by a microbe enrichment strategy as described in previous studies (Jiao et al., 2006; Bulgari et al., 2009, 2011). Briefly, plant tissues were sterilized, grounded in liquid nitrogen, and aseptically incubated at 28°C for 12 h in gentle agitation in an enzymatic solution (0.1% macerozyme, 1% cellulase, 0.7M mannitol, 5 mM N-morpholinoethanesulfonic acid, 9 mM CaCl2, and 65 μM KH2PO4). In another subsample, DNA isolation was performed directly after surface sterilization. DNA was extracted for each sample according to the protocol described by Prince et al. (1993) modified by the addition of lysozyme (3 mg/ml), L-lysine (0.15 mol/l), EGTA (6 mmol/l, pH 8.0), and by the incubation at 37°C for 30 min, before the lysis step.
Pyrosequencing of Endophytic Communities
To obtain amplicons for pyrosequencing, we amplified the 16S rDNA gene from each sample using High Fidelity FastStart DNA polymerase (Roche, USA) and the universal primers 799f/1520r with 454 adaptors and a sample-specific 10-mer barcode (designed following the instructions for Roche 454 technology1). These primers allow selective amplification of bacterial DNA, targeting 16S rDNA hypervariable regions v5-v9 (Chelius and Triplett, 2001) and minimize the chance of amplification of plastid DNA (Ghyselinck et al., 2013). The PCR product was separated on 1% agarose gel and gel-purified using Invitrogen PureLink (Invitrogen, USA). DNA was quantified via quantitative PCR using the Library quantification kit—Roche 454 titanium (KAPA Biosystems, USA) and pooled in a final amplicon library. The 454 pyrosequencing was carried out at the Sequencing Platform facility in Fondazione Edmund Mach, on the GS FLX+ system using the new XL+ chemistry dedicated to long reads of up to 800 bp, following the manufacturer's recommendations. The new XL+ chemistry coupled with the unidirectional sequencing strategy led to the sequencing of multiple variable regions on a single read and to overcome the bottleneck associated with a short read approach.
Data Analysis
We generated 16S rDNA gene sequences relative to endophytes from three batches of samples: leaf endophytes with bacterial enrichment, leaf endophytes without enrichment, and stem endophytes. We used Roche 454 GS FLX+ sequencing as described above and analyzed the sequencing output using a standard Qiime pipeline (Caporaso et al., 2010). As a first step we demultiplexed and filtered sequences on the basis of quality score and read length (only sequences with a minimum average score of 20 and length between 250 and 1000 bp were retained), in order to remove short and low quality reads. Moreover, in this step we removed sequences with more than 6 ambiguous bases or with homopolymers longer than 6 bases. We performed chimera identification and filtering using usearch61 (Edgar, 2010). After removing chimeras, we used uclust (Edgar, 2010) for clustering all the sequences into Operational Taxonomic Units (OTUs), by applying a similarity threshold of 97%. This is commonly used to represent species level similarity (although this threshold does not necessarily match with what is regarded as species for many microorganisms) (Crawford et al., 2009). For each OTU we picked a representative sequence using Qiime and we used these sequences to assign a taxonomical identity to each OTU using the RDP classifier (Wang et al., 2007). In this step we used a confidence threshold of 0.8 and e-value ≤ 0.001 against the Greengenes 97% reference data set. Based on taxonomical identity, we manually selected OTUs corresponding to HAPT from all three datasets. To understand the distance between these OTUs and database reference strains, we downloaded 16S rDNA sequences of HAP and non-pathogenic microorganisms from NCBI (www.ncbi.nlm.nih.gov) (Altschul et al., 1997) and aligned them to the representative sequences assigned to the same taxon in our datasets. For the alignment we used MUSCLE (Edgar, 2004; Caporaso et al., 2010) and filtered the alignment using Qiime. Then, we built the phylogenetic trees shown in Figures 1–4 from these alignments using DNAML (Maximum Likelihood) in BioEdit (Hall, 1999) and rendered them using Itol (Letunic and Bork, 2006, 2011). Information on OTU type and abundance was co-displayed with the generated trees. OTUs were colored according to the origin of samples (types: stem, leaf enriched, leaf non-enriched, HAP reference strain, non-pathogenic reference strain). Abundance was displayed as circles for every OTU (abundance is proportional to the circle radius).
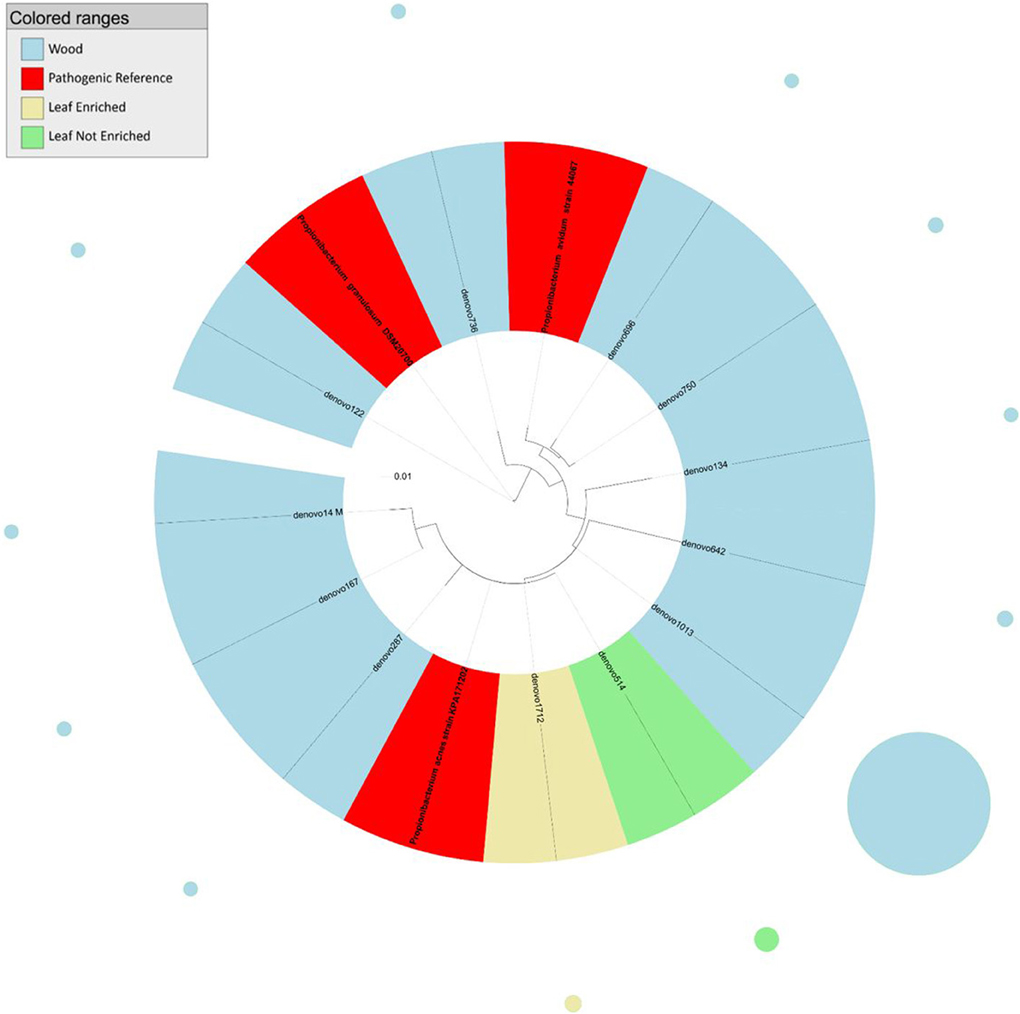
Figure 1. Phylogenetic relationships based on partial 16S rDNA gene sequences obtained from pyrosequencing (this work) and closely related Propionibacterium sequences, retrieved from GenBank (Pathogenic and non-pathogenic Reference). The tree was built using a maximum likelihood method and rendered using iTOL. The relative abundance of each OTU is reported as circles and it is proportional to circle radius. Sequences tag and relative abundance circles were colored according to the source dataset.
The 16S rDNA gene nucleotide sequences, representative of the OTUs identified in this study, are available with NCBI GenBank accession numbers KJ851800-KJ851922 (Supplementary Table 1).
Results
We investigated the composition of bacterial endophytic communities in the grapevine endosphere (both in the leaf and the stem) by pyrosequencing the bacterial 16S rDNA gene. Sequence analysis revealed the presence of several OTUs (70 from enriched leaves, 13 from non-enriched leaves, and 40 from stems, see Table 1) classified as HAPT or non-pathogenic taxa closely related to HAPT. We focused our subsequent analysis on four genera containing potential pathogens: Propionibacterium, Staphylococcus, Clostridium, and Burkholderia.
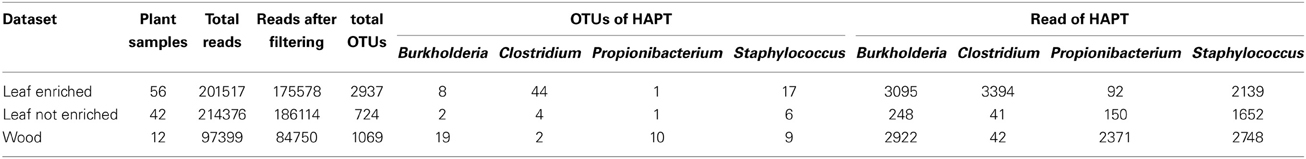
Table 1. Specifics for each datasets: number of samples collected, number of reads retained after quality filtering, OTUs retained after singleton removal, OTUs assigned to HAPTs and number of reads clustered in these OTUs.
We inferred phylogenetic trees from the alignment of 16S rDNA nucleotide sequences. OTUs assigned to these taxa were identified, with varying abundance, in enriched and non-enriched leaves and in stems of grapevine. Most OTUs from grapevine leaves were assigned to genera Clostridium (44 and 4 OTUs from enriched- and not-enriched leaves, respectively) and Staphylococcus (17 and 6 OTUs from enriched- and not-enriched leaves, respectively), albeit the most abundant OTU from enriched leaf samples (denovo115) was associated with non-pathogenic species of the genus Burkholderia (Figure 2). The most abundant OTU from non-enriched leaves (denovo703) was assigned to a cluster including both pathogenic and non-pathogenic species of the genus Staphylococcus. On the other hand, most OTUs from endophytic bacteria in grapevine stems were assigned to genera Burkholderia (19 OTUs) and Propionibacterium (10 OTUs), although the most abundant OTU (denovo979) was taxonomically closer to pathogenic species Staphylococcus epidermidis (Figure 3).
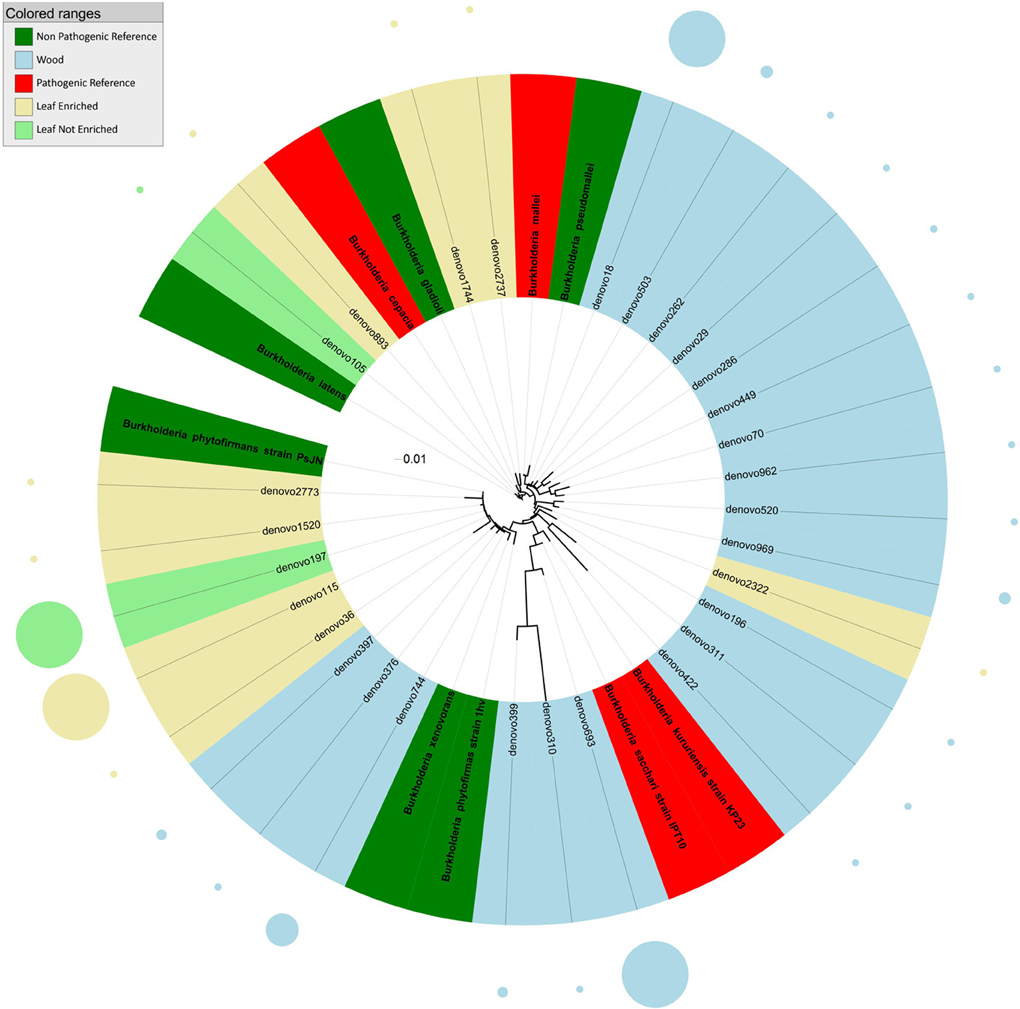
Figure 2. Phylogenetic relationships based on partial 16S rDNA gene sequences obtained from pyrosequencing (this work) and closely related Burkholderia sequences, retrieved from GenBank (Pathogenic and non-pathogenic Reference). The tree was built using a maximum likelihood method and rendered using iTOL. The relative abundance of each OTU is reported as circles and it is proportional to circle radius. Sequences tag and relative abundance circles were colored according to the source dataset.
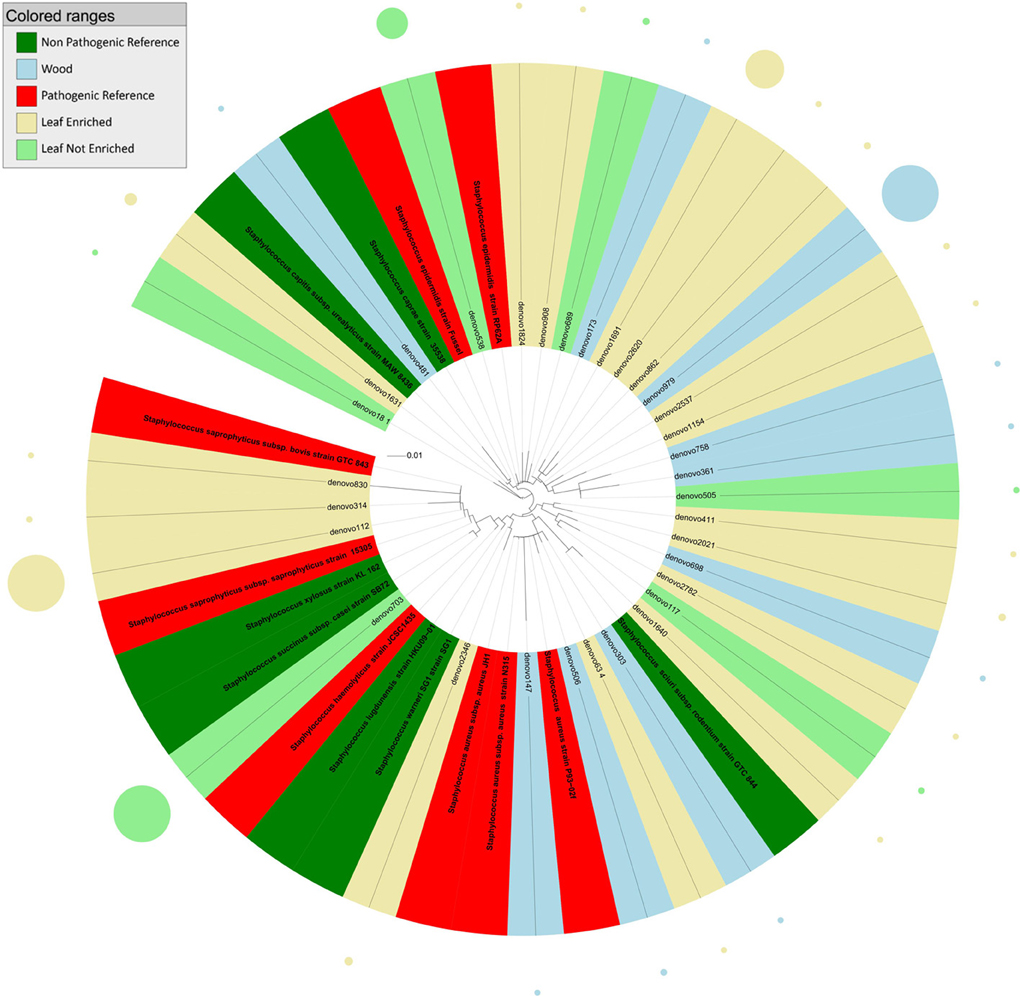
Figure 3. Phylogenetic relationships based on partial 16S rDNA gene sequences obtained from pyrosequencing (this work) and closely related Staphylococcus sequences, retrieved from GenBank (Pathogenic and non-pathogenic Reference). The tree was built using a maximum likelihood method and rendered using iTOL. The relative abundance of each OTU is reported as circles and it is proportional to circle radius. Sequences tag and relative abundance circles were colored according to the source dataset.
The analysis of the phylogenetic tree of propionibacteria showed three main clades within the Propionibacterium genus (Figure 1). In detail, all sequences from grapevine leaves and most sequences from the stem clustered (cluster 1) with Propionibacterium acnes (which we previously demonstrated to be endocelluarly associated with grapevine stems, Campisano et al., 2014), while other sequences from the stem clustered with the species P. granulosum (cluster 2) and P. avidum (cluster 3).
Phylogenetic analysis indicated that sequences from grapevine endosphere assigned to the genus Burkholderia formed four distinct clusters (Figure 2). The first cluster included endophytic bacteria from grapevine leaves grouping with HAP and plant pathogens such as B. latens, B. cepacia, and B. gladioli. The second cluster included bacteria, identified exclusively in stems, grouping with Burkholderia mallei (HAP) and B. pseudomallei (non-pathogenic endophyte). The third cluster included endophytic bacteria from grapevine leaves and stem, grouping with environmental and endophytic bacteria such as Burkholderia phytofirmans and B. xenovorans. The forth cluster included HAP species of the genus Burkholderia (B. kururiensis and B. sacchari), and consisted of endophytic bacteria identified exclusively in grapevine wood tissues.
The tree obtained by analysis of 16S rDNA sequences assigned to the genus Staphylococcus (Figure 3) revealed that endophytes were distributed in six main clusters. Cluster 1 included sequences from grapevine leaf endophytes strictly related to the HAP species S. saprophyticus and S. haemolyticus, and to the non-pathogenic species S. succinus, S. xylosus, S. lugdunensis, and S. warneri. Cluster 2 included sequences from grapevine leaves and stems grouping with the pathogenic S. aureus, and one OTU from leaf strictly related to non-pathogenic species S. sciuri. The majority of OTUs from grapevine leaf and wood endophytic bacteria clustered together with the pathogenic species S. epidermidis (cluster 3). Cluster 4 included one OTU representing sequences from plant stems grouping closely to non-pathogenic species S. caprae and S. urealyticus. The remaining grapevine sequences, from both leaf and stem, grouped in two unassigned clusters (these OTUs shared sequence identity <97% in comparison with 16S rDNA gene nucleotide sequences previously deposited in GenBank). Blastn (best-hit) of these sequences indicated that the cluster 5 (represented by denovo411 and denovo2021) is related to S. warneri, while the cluster 6 (represented by denovo698, denovo2782, and denovo117) to S. auricularis.
Sequences identified as genus Clostridium (Figure 4) clustered into three groups relative to the tree inferred from the 16S rDNA sequence alignment. Most endophyte sequences in this genus were obtained from grapevine leaf. In detail, the first cluster included endophytic bacteria grouping with HAP such as Clostridium difficile, C. tetani, and with non-pathogenic environmental species (C. vincentii and C. bijerinckii). The second cluster included grapevine endophytic bacteria closely related to HAP Clostridium botulinum and C. perfrigerans, and non-pathogenic bacteria C. drakei and C. ghonii. The third cluster contained grapevine endophytic bacteria of the genus Clostridium related to unassigned species.
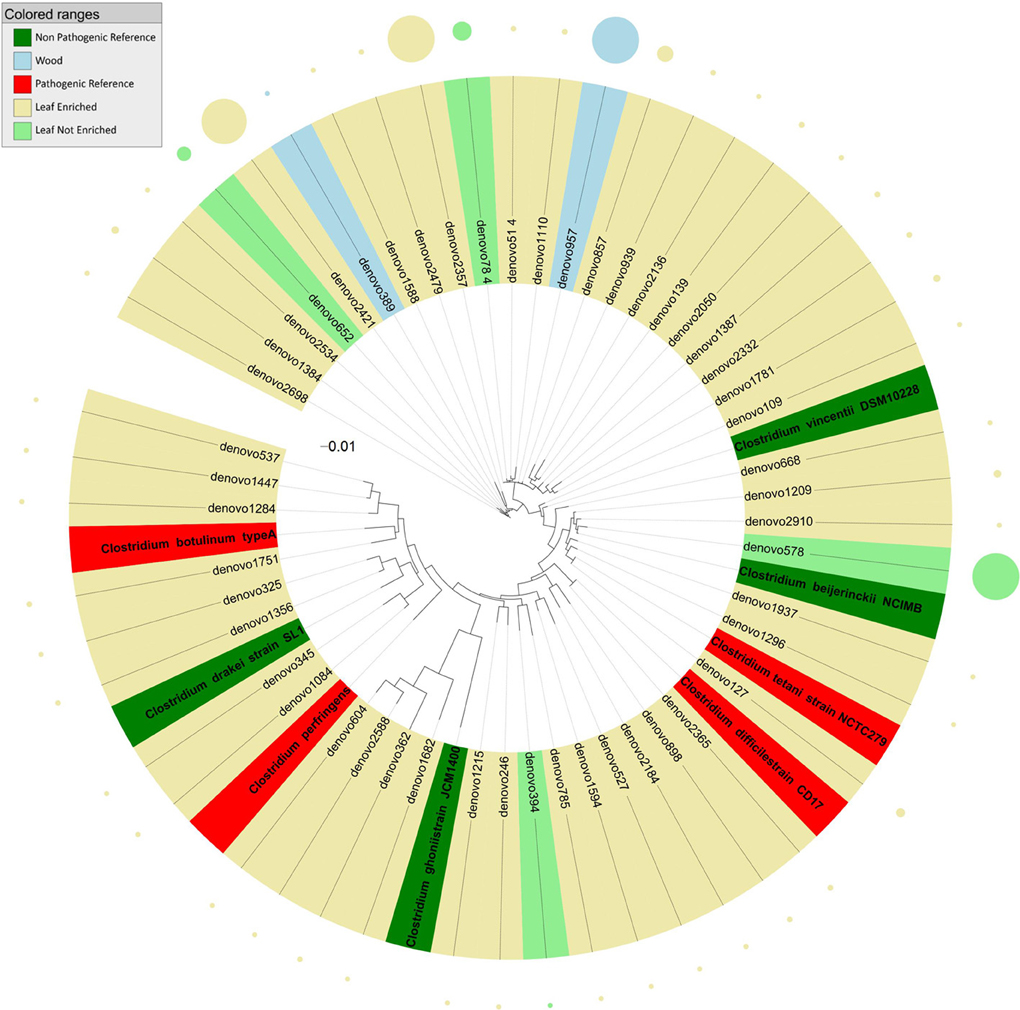
Figure 4. Phylogenetic relationships based on partial 16S rDNA gene sequences obtained from pyrosequencing (this work) and closely related Clostridium sequences, retrieved from GenBank (Pathogenic and non-pathogenic Reference). The tree was built using a maximum likelihood method and rendered using iTOL. The relative abundance of each OTU is reported as circles and it is proportional to circle radius. Sequences tag and relative abundance circles were colored according to the source dataset.
Discussion
In the present study, we investigated microbial communities in the endosphere of grapevine leaves and stems by pyrosequencing and phylogenetic analyses of the 16S rDNA gene. Several endophytic OTUs belonging to four genera (Propionibacterium, Staphylococcus, Clostridium, and Burkholderia), known to include some species recognized as human- and animal-pathogens (HAP), were identified within the characterized microbial communities. Such OTUs were identified in enriched and not-enriched leaves and in grapevine stems. As reported in literature, the specific treatment of leaf samples with cell hydrolytic enzymes (microbe enrichment strategy) releases all microbes living in association with plant tissues (Jiao et al., 2006; Bulgari et al., 2009, 2011), improving the bacterial display and identification. The majority of the bacterial sequences in the genus Propionibacterium, from grapevine leaf and stem, were identified as P. acnes. This species is among the causing agents of acnes and its members are generally associated with human skin, where they feed on fatty acids secreted by sebaceous glands (Webster et al., 1981; Zouboulis, 2004). It also colonizes the human gut (Perry and Lambert, 2011), and is reported as an opportunistic pathogen in post-surgical infections (Nisbet et al., 2007). We previously identified genomic changes in P. acnes type Zappae, tightly associated with grapevines (Campisano et al., 2014). The wide diversity in sequences assigned to genus Propionibacterium shows that these endophytes are mostly present in the stem woody tissues (where the majority of sequences is amplified) and only marginally present in leaves, reinforcing the notion of a tight symbiosis of these bacteria with the plant.
The genus Burkholderia includes more than 60 species, some of which are known plant-dwellers (endophytes and epiphytes, Compant et al., 2008). Some species of Burkholderia, such as B. mallei and B. cepacia (Burkholderia cepacia complex, BCC), are recognized as HAP and plant pathogens (Govan et al., 1996; LiPuma, 1998; Coenye and Vandamme, 2003). Recently, Burkholderia species had gained considerable importance owing to their pathogenicity, but two findings had a strong impact on their ecological perception: (i) the identification of nitrogen fixation in Burkholderia species other than B. vietnamiensis (which belongs to the BCC), such as B. brasilensis M130 and B. kururiensis, and (ii) the description of legume-nodulating Burkholderia and their subsequent characterization as genuine endosymbionts (Suarez-Moreno et al., 2012). We found sequences clustering with such pathogenic Burkholderia in grapevine leaves. Stem-associated Burkholderia OTUs either grouped with non-pathogenic (or plant beneficial) species such as B. phytofirmans (Sessitsch et al., 2005) and B. xenovorans (Caballero-Mellado et al., 2007), or formed a separate phylogenetic group very similar to B. kururiensis and B. sacchari, by blastn analysis of best hit.
We can speculate that the presence of potentially pathogenic Burkholderia associated sequences in leaf green tissues but not in the woody stems represents a difference in the ecology of the plant-beneficial burkholderias from the HAP ones. All tissues in the leaf are in close proximity of the plant surface, while stem-associated tissues are possibly more difficult to colonize by transient colonizers. This may explain why plant-beneficial burkholderias were found mainly in the plant woody stem, while HAP ones only survive in leaves. We also noted that highly represented Burkholderia OTUs belong to the cluster including plant-associated species B. phytofirmans and B. xenovorans or to the clusters including B. kururiensis and B. sacchari, while the potentially pathogenic ones are in much lower numbers. B. phytofirmans is one of most studied endophytes. This strain was visualized colonizing the grapevine root surface, entering the endorhiza and spreading to grape inflorescence stalks, pedicels and then to immature berries through xylem vessels (Compant et al., 2008). We also reported in this work as one of the most abundant burkholderias in grapevine.
OTUs from grapevine leaves were mostly identified as genera Clostridium and Staphylococcus. These taxa include endophytic OTUs closely related to the pathogenic species Staphylococcus saprophyticus, S. aureus, and S. epidermidis. Quite interestingly, most Staphylococcus sequences appeared to associate taxonomically with S. epidermidis (Figure 3). S. epidermidis colonizes the epithelial surfaces of every human being. Furthermore, it is one of the most common causes of nosocomial infections. In addition to the abundant prevalence of S. epidermidis on the human skin, this high incidence is mainly due to the exceptional capacity of S. epidermidis to stick to the surfaces of indwelling medical devices during device insertion (Otto, 2008, 2009). At our knowledge this is the first report of pathogenic Staphylococcus associated sequences in plants.
Almost all the endophytic sequences assigned to the genus Clostridium were amplified from grapevine leaf DNA. This group included sequences grouping with HAP such as Clostridium difficile, C. perfrigens, C. botulinum, and C. tetani. A major factor important to the colonization of plants is how long bacteria persist in the soil before dying. Clostridium spores persisted in soil for 16 months and were found on the leaves of parlsey grown in the contaminated soil (Girardin et al., 2005). We can speculate that the presence of Clostridium, human and animal pathogenic taxa (HAPT) in grapevine leaves could be related to long–lasting spore contamination. Honey sometimes contains spores of C. botulinum, which may cause infant botulism in humans 1 year old and younger. The toxin eventually paralyzes the infant's breathing muscles (Tanzi and Gabay, 2002). C. difficile can flourish when other bacteria in the gut are killed during antibiotic therapy, leading to pseudomembranous colitis (a cause of antibiotic-associated diarrhea).
HAPT were indentified both in leaves and wood of grapevines. This depicts the ability of these pathogens to be internalized within plant tissues. Isolation of human pathogenic enterobacteria from within the tissue of fresh and minimally prepared produce has been reported (Eblen et al., 2004; Shi et al., 2007; Soto et al., 2007; Holden et al., 2009). Furthermore, it appears that once a plant has been colonized by bacteria there is the potential for vertical transmission to successive generations, as demonstrated for S. typhimurium on tomatoes (Guo et al., 2001). The exploration of endophytic communities, using metagenome-based community analyses (Bulgarelli et al., 2012; Lundberg et al., 2012) coupled with the exploration of the pathogenic potential of pathogens, are beginning to reveal that many HAP are capable of exploiting plant hosts. This means the apparent pathogens may have adapted to the plants and have become plant symbionts, for at least one stage of their life cycle. This also shows the ability and potential of HAP to persist on multiple hosts, with plants serving as intermediate hosts or reservoirs for them. The ability of these pathogens to maintain their population levels in a variety of environments likely increases their pan genome and evolutionary potential (Campisano et al., 2014). Although we focused on four genera (Propionibacterium, Staphylococcus, Clostridium, and Burkholderia) only, we identified several other taxa known for harboring HAP bacteria. Our analysis highlighted the presence of potential HAPT in the grapevine endosphere and, to the authors' knowledge, represents the first report of the unexpected occurrence of these bacterial taxa in this atypical (yet crucially important for agriculture) environment.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by Provincia Autonoma di Trento, progetto PAT—Call 2 Team 2009—Incoming–Mecagrafic. We thank COST Action FA1103 for supporting this work.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00327/abstract
Footnotes
1. ^http://www.liv.ac.uk/media/livacuk/centreforgenomicresearch/The_GS_FLX_Titanium_Chemistry_Extended_MID_Set.pdf
References
Akhtyamova, N. (2013). Human pathogens on or within the plant and useful endophytes. Cell Dev. Biol. 2:e119. doi: 10.4172/2168-9296.1000e119
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Barak, J. D., and Schroeder, B. K. (2012). Interrelationships of food safety and plant pathology: the life cycle of human pathogens on plants. Annu. Rev. Phytopathol. 50, 241–266. doi: 10.1146/annurev-phyto-081211-172936
Berger, C. N., Sodha, S. V., Shaw, R. K., Griffin, P. M., Pink, D., Hand, P., et al. (2010). Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 12, 2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Bulgari, D., Casati, P., Brusetti, L., Quaglino, F., Brasca, M., Daffonchio, D., et al. (2009). Endophytic bacterial diversity in grapevine (Vitis vinifera L.) leaves described by 16SrRNA gene sequence analysis and length heterogeneity-PCR. J. Microbiol. 47, 393–401. doi: 10.1007/s12275-009-0082-1
Bulgari, D., Casati, P., Crepaldi, P., Daffonchio, D., Quaglino, F., Brusetti, L., et al. (2011). Restructuring of endophytic bacterial communities in grapevine yellows-diseased and recovered Vitis vinifera L. plants. Appl. Environ. Microbiol. 77, 5018–5022. doi: 10.1128/AEM.00051-11
Caballero-Mellado, J., Onofre-Lemus, J., Estrada-de los Santos, P., and Martınez-Aguilar, L. (2007). The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl. Environ. Microbiol. 73, 5308–5319. doi: 10.1128/AEM.00324-07
Campisano, A., Ometto, L., Compant, S., Pancher, M., Antonielli, L., Yousaf, S., et al. (2014). Interkingdom transfer of the acne causing agent, Propionibacterium acnes, from human to grapevine. Mol. Biol. Evol. 31, 1059–1065. doi: 10.1093/molbev/msu075
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chelius, M. K., and Triplett, E. W. (2001). The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 41, 252–263. doi: 10.1007/s002480000087
Coenye, T., and Vandamme, P. (2003). Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5, 719–729. doi: 10.1046/j.1462-2920.2003.00471.x
Compant, S., Duffy, B., Nowak, J., Clement, C., and Barka, E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
Compant, S., Nowak, J., Coenye, T., Clement, C., and Ait Barka, E. (2008). Diversit and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 32, 607–626. doi: 10.1111/j.1574-6976.2008.00113.x
Crawford, P. A., Crowley, J. R., Sambandam, N., Muegge, B. D., Costello, E. K., Hamady, M., et al. (2009). Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc. Natl. Acad. Sci. U.S.A. 106, 11276–11281. doi: 10.1073/pnas.0902366106
Eblen, B. S., Walderhaug, M. O., Edelson-Mammel, S., Chirtel, S. J., De Jesus, A., Merker, R. I., et al. (2004). Potential for internalization, growth, and survival of Salmonella and Escherichia coli O157:H7 in oranges. J. Food Protect. 67, 1578–1584.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Enders, U., Karch, H., Toyka, K. V., Michels, M., Zielasek, J., Pette, M., et al. (1993). The spectrum of immune responses to Campylobacter jejuniand glycoconjugates in Guillain-Barre syndrome and in other neuroimmunological disorders. Ann. Neurol. 34, 136–144. doi: 10.1002/ana.410340208
Erickson, M. (2012). Internalization of fresh produce by foodborne pathogens. Annu. Rev. Food Sci. Technol. 3, 283–310. doi: 10.1146/annurev-food-022811-101211
Ghyselinck, J., Pfeiffer, S., Heylen, K., Sessitsch, A., and De Vos, P. (2013). The effect of primer choice and short read sequences on the outcome of 16S rRNAgene based diversity studies. PLoS ONE 8:e71360. doi: 10.1371/journal.pone.0071360
Girardin, H., Morris, C. E., Albagnac, C., Dreux, N., Glaux, C., and Nguyen-The, C. (2005). Behaviour of the pathogen surrogates Listeria innocua and Clostridium sporogenes during production of parsley in fields fertilized with contaminated amendments. FEMS Microbiol. Ecol. 54, 287–295. doi: 10.1016/j.femsec.2005.04.003
Govan, J. R., Hughes, J. E., and Vandamme, P. (1996). Burkholderiacepacia: medical, taxonomic and ecological issues. J. Med. Microbiol. 45, 395–407. doi: 10.1099/00222615-45-6-395
Guo, X., Chen, J., Brackett, R. E., and Beuchat, L. R. (2001). Survival of Salmonellae on and in tomato plants from the time of inoculation at flowering and early stages of fruit development through fruit ripening. Appl. Environ. Microbiol. 67, 4760–4764. doi: 10.1128/AEM.67.10.4760-4764.2001
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Holden, N., Pritchard, L., and Toth, I. (2009). Colonization outwith the colon: plants as an alternative environmental reservoir for human pathogenic enterobacteria. FEMS Microbiol. Rev. 33, 689–703. doi: 10.1111/j.1574-6976.2008.00153.x
Jiao, J. Y., Wang, H. X., Zeng, Y., and Shen, Y. M. (2006). Enrichmentfor microbes living in association with plant tissues. J. Appl. Microbiol. 100, 830–837. doi: 10.1111/j.1365-2672.2006.02830.x
Kirzinger, M. W. B., Nadarasah, G., and Stavrinides, J. (2011). Insights into cross-kingdom plant pathogenic bacteria. Genes 2, 980–997. doi: 10.3390/genes2040980
Kutter, S., Hartmann, A., and Schmid, M. (2006). Colonization of barley (Hordeum vulgare) with Salmonella enterica and Listeria spp. FEMS Microbiol. Ecol. 56, 262–271. doi: 10.1111/j.1574-6941.2005.00053.x
Lenz, L. L., Mohammadi, S., Geissler, A., and Portnoy, D. A. (2003). SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 12432–12437. doi: 10.1073/pnas.2133653100
Letunic, I., and Bork, P. (2006). Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Letunic, I., and Bork, P. (2011). Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478. doi: 10.1093/nar/gkr201
LiPuma, J. J. (1998). Burkholderia cepacia. Management issues and new insights. Clin. Chest Med. 19, 473–486. doi: 10.1016/S0272-5231(05)70094-0
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsisthaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Lynch, M. F., Tauxe, R. V., and Hedberg, C. W. (2009). The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol. Infect. 137, 307–315. doi: 10.1017/S0950268808001969
Nisbet, M., Briggs, S., Ellis-Pegler, R., Thomas, M., and Holland, D. (2007). Propionibacterium acnes: an under-appreciated cause of post-neurosurgical infection. J. Antimicrob. Chemother. 60, 1097–1103. doi: 10.1093/jac/dkm351
Olaimat, A. N., and Holley, R. A. (2012). Factors influencing the microbial safety of fresh produce: a review. Food Microbiol. 32, 1–19. doi: 10.1016/j.fm.2012.04.016
Otto, M. (2008). Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 322, 207–228. doi: 10.1007/978-3-540-75418-3_10
Otto, M. (2009). Staphylococcus epidermidis - the “accidental” pathogen. Nat. Rev. Microbiol. 7, 555–567. doi: 10.1038/nrmicro2182
Pancher, M., Ceol, M., Corneo, P. E., Longa, C. M., Yousaf, S., Pertot, I., et al. (2012). Fungal endophytic communities in grapevines (Vitisvinifera L.) respond to crop management. Appl. Environ. Microbiol. 78, 4308–4317. doi: 10.1128/AEM.07655-11
Perry, A., and Lambert, P. (2011). Propionibacterium acnes: infection beyond the skin. Expert. Rev. Anti. Infect. Ther. 9, 1149–1156. doi: 10.1586/eri.11.137
Prince, J. P., Davis, R. E., Wolf, T. K., Lee, I. M., Mogen, B. D., Dally, E. L., et al. (1993). Moleculardetection of diverse mycoplasma like organisms (MLOs) associatedwith grapevine yellows and their classification with asteryellows MLOs. Phytopathology 83, 1130–1137. doi: 10.1094/Phyto-83-1130
Rosenblueth, M., and Martinez-Romero, E. (2006). Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 19, 827–837. doi: 10.1094/MPMI-19-0827
Saldaña, Z., Sánchez, E., Xicohtencatl-Cortes, J., Puente, J. L., and Giron, J. A. (2011). Surface structures involved in plant stomata and leaf colonization by Shiga-toxigenic Escherichia coli O157:H7. Front. Microbiol. 2:119. doi: 10.3389/fmicb.2011.00119
Sessitsch, A., Coenye, T., Sturz, A. V., Vandamme, P., Ait Barka, E., Faure, D., et al. (2005). Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant beneficial properties. Int. J. Syst. Evol. Microbiol. 55, 1187–1192. doi: 10.1099/ijs.0.63149-0
Shi, X., Namvar, A., Kostrzynska, M., Hora, R., and Warriner, K. (2007). Persistence and growth of different Salmonellaserovars on preand postharvest tomatoes. J. Food Protect. 70, 2725–2731.
Soto, M., Chavez, G., Baez, M., Martinez, C., and Chaidez, C. (2007). Internalization of Salmonella typhimurium into mango pulp and prevention of fruit pulp contamination by chlorine and copper ions. Int. J. Environ. Health Res. 17, 453–459. doi: 10.1080/09603120701695593
Suarez-Moreno, Z. R., Caballero-Mellado, J., Coutinho, B. G., Mendonça-Previato, L., James, E. K., and Venturi, V. (2012). Common features of environmental and potentially beneficial plant-associated burkholderia. Microb. Ecol. 63, 249–256. doi: 10.1007/s00248-011-9929-1
Tanzi, M. G., and Gabay, M. P. (2002). Association between honey consumption and infant botulism. Pharmacotherapy 22, 1479–1483. doi: 10.1592/phco.22.16.1479.33696
Teplitski, M., Barak, J. D., and Schneider, K. R. (2009). Human enteric pathogens in produce: un-answered ecological questions with direct implications for food safety. Curr. Opin. Biotechnol. 20, 166–171. doi: 10.1016/j.copbio.2009.03.002
Tyler, H. L., and Triplett, E. W. (2008). Plants as a habitat for beneficial and/or human pathogenic bacteria. Annu. Rev. Phytopathol. 46, 53–73. doi: 10.1146/annurev.phyto.011708.103102
Wagenaar, J. (2008). “Poultry colonization with Campylobacter and its control at the primary production level, Chapter 37,” in Campylobacter, eds C. S. I. N. Nachamkin, C. M. Scymanski, and M. Blaser (Washington, DC: ASM Press), 667–678.
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Webster, G. F., Ruggieri, M. R., and McGinley, K. J. (1981). Correlation of Propionibacterium acnes populations with the presence of triglycerides on nonhuman skin. Appl. Environ. Microbiol. 41, 1269–1270.
Keywords: endosphere, grapevine, pathogens, bacteria, pyrosequencing
Citation: Yousaf S, Bulgari D, Bergna A, Pancher M, Quaglino F, Casati P and Campisano A (2014) Pyrosequencing detects human and animal pathogenic taxa in the grapevine endosphere. Front. Microbiol. 5:327. doi: 10.3389/fmicb.2014.00327
Received: 01 April 2014; Accepted: 14 June 2014;
Published online: 08 July 2014.
Edited by:
Nicola Holden, The James Hutton Institute, UKReviewed by:
Murali Gopal, Central Plantation Crops Research Institute, IndiaAna Catarina Gomes, Biocant, Portugal
Vittoria Catara, University of Catania, Italy
Copyright © 2014 Yousaf, Bulgari, Bergna, Pancher, Quaglino, Casati and Campisano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Campisano, Fondazione Edmund Mach, via E. Mach 1, 38010 San Michele all'Adige, Italy e-mail: andrea.campisano@fmach.it
†These authors have contributed equally to this work.
 Sohail Yousaf1,2†
Sohail Yousaf1,2† Daniela Bulgari
Daniela Bulgari Alessandro Bergna
Alessandro Bergna Michael Pancher
Michael Pancher Fabio Quaglino
Fabio Quaglino Andrea Campisano
Andrea Campisano