- School of Dentistry, University of California-Los Angeles, Los Angeles, CA, USA
The oral cavity contains more than 700 microbial species that are engaged in extensive cell–cell interactions. These interactions contribute to the formation of highly structured multispecies communities, allow them to perform physiological functions, and induce synergistic pathogenesis. Co-adhesion between oral microbial species influences their colonization of oral cavity and effectuates, to a large extent, the temporal and spatial formation of highly organized polymicrobial community architecture. Individual species also compete and collaborate with other neighboring species through metabolic interactions, which not only modify the local microenvironment such as pH and the amount of oxygen, making it more suitable for the growth of other species, but also provide a metabolic framework for the participating microorganisms by maximizing their potential to extract energy from limited substrates. Direct physical contact of bacterial species with its neighboring co-habitants within microbial community could initiate signaling cascade and achieve modulation of gene expression in accordance with different species it is in contact with. In addition to communication through cell–cell contact, quorum sensing (QS) mediated by small signaling molecules such as competence-stimulating peptides (CSPs) and autoinducer-2 (AI-2), plays essential roles in bacterial physiology and ecology. This review will summarize the evidence that oral microbes participate in intercellular communications with co-inhabitants through cell contact-dependent physical interactions, metabolic interdependencies, as well as coordinative signaling systems to establish and maintain balanced microbial communities.
Introduction
With the respect to microbial flora, the human oral cavity is one of the most densely populated sites of the human body, consisting of as many as 600–800 bacterial species (Paster et al., 2006; Dewhirst et al., 2010). Extensive clinical studies have indicated that the oral microbial flora is responsible for two major human diseases: dental caries and periodontitis (Marsh, 1994). The environmental diversity of the oral cavity promotes the establishment of distinct microbial communities. For example, the supragingival plaque is known to be dominated by Gram-positive streptococci and the subgingival plaque is populated with mainly Gram-negative anaerobic bacteria (Marsh, 1994). These microbial inhabitants have co-evolved not only with their host, but also with each other, leading to extensive intercellular communications across species. The ecological equilibrium of a well-organized multispecies oral microbial community is maintained through competitive and cooperative interactions between microorganisms of the same or different species at the cellular and molecular levels.
Intercellular communications are essential for the development of temporal and spatial organization of oral microbial communities, and are involved in processes such as provision of attachment sites, modification of local microenvironment, cooperative nutrient utilization, as well as synergistic and competitive interactions (Kuramitsu et al., 2007). In addition, intercellular signaling initiated by cell–cell contact, quorum sensing, and other diffusible signaling molecules allow oral microbial species to coordinate their physiological behaviors and pathogenesis (Jenkinson and Lamont, 2005; Kolenbrander et al., 2006). All of these intercellular interactions between oral microbes enable them to select co-residents, promote the establishment of a highly structured and diverse microbial community, and play an essential role in microbial pathogenesis.
Physical Interactions
The ability of species to adhere to salivary pellicles or to bacteria that are already attached to the surface is among the key factors that determine whether a species can perpetuate in the oral cavity. Initial adhesion of the early colonizers of oral microbial communities invariably involves binding to saliva components that are adsorbed to solid surfaces such as teeth or to desquamating surfaces such as epithelial tissue. Oral streptococci are believed to be among the earliest inhabitants on tooth surfaces due to their capability to adhere directly to salivary pellicle and comprise about 80% of the early colonizers (Avila et al., 2009). Meanwhile, Actinomyces has also been identified in the inner portion of the dental biofilms, suggesting their early colonizer nature (Palmer et al., 2003; Dige et al., 2009). Other early colonizers are comprised of Veillonella and Neisseria (Li et al., 2004). Several studies have identified adhesins and receptors for early colonizing microorganisms to adhere specifically to salivary pellicle and even determine the unique domains on these molecules involved in binding. For example, it has been shown that Hsa mediated binding of Streptococcus gordonii to sialic acid-containing receptors in salivary pellicle (Ding et al., 2010) and the region containing lysine in the salivary components may have binding activity to S. gordonii and Streptococcus sanguinis. Meanwhile, the scavenger receptor cysteine-rich domain peptide 2 (SRCRP 2) region may also function as a receptor for the binding of streptococci (Hamada et al., 2004).
Biofilm matrix also plays a role in maintaining microbial colonization, therefore contributing to oral biofilm development. Streptococcus mutans, a predominantly prevalent caries-associated species in humans (Loesche, 1986), utilizes extracellular polysaccharides synthesized by glucosyltransferases (Gtfs) to facilitate its firm attachment to teeth and promote tight cell clustering (Koo et al., 2010; Xiao et al., 2012). The Gtfs exoenzymes of S. mutans were shown to adhere strongly not only to the salivary pellicle that covers the surface of teeth but also to bacterial surfaces, and could produce glucans in the adsorbed state (Vacca-Smith and Bowen, 1998; Bowen and Koo, 2011). In view of the cohesive attribute (Loesche, 1986; Tamesada et al., 2004), glucans synthesized on the pellicle provides additional bacterial binding sites, while the polymers on the cell surfaces of same species or other residents increase cell–cell cohesion (Figure 1). For example, GtfB, one of Gtfs, binds effectively to both yeast and hyphae cell forms of Candida albicans in an enzymatically active form and the glucans formed in situ not only enhances the binding of S. mutans cells to C. albicans cells but also promotes the colonization of C. albicans on the tooth surface (Gregoire et al., 2011). Among the numerous types, the rigid alpha 1, 3 linked glucans are particularly suited for cohesion (Rölla et al., 1985). Due to its important role in promoting microbial adherence and clustering, many oral bacterial species modulate their behavior within biofilm by interfering with matrix structure. Actinobacillus actinomycetemcomitans, the predominant pathogen in aggressive periodontitis was reported to be able to produce a matrix-degrading enzyme dispersin B for degrading poly-N-acetylglucosamine (PNAG), a major polysaccharide component of biofilm matrix, and thus, caused detachment and dispersion of A. actinomycetemcomitans biofilm cells (Kaplan et al., 2003). Streptococcus salivarius produces a fructosyltransferase (FTF) and an exo-beta-D-fructosidase (FruA) to hinder further polymicrobial community development with other oral bacteria such as S. mutans (Ogawa et al., 2011).
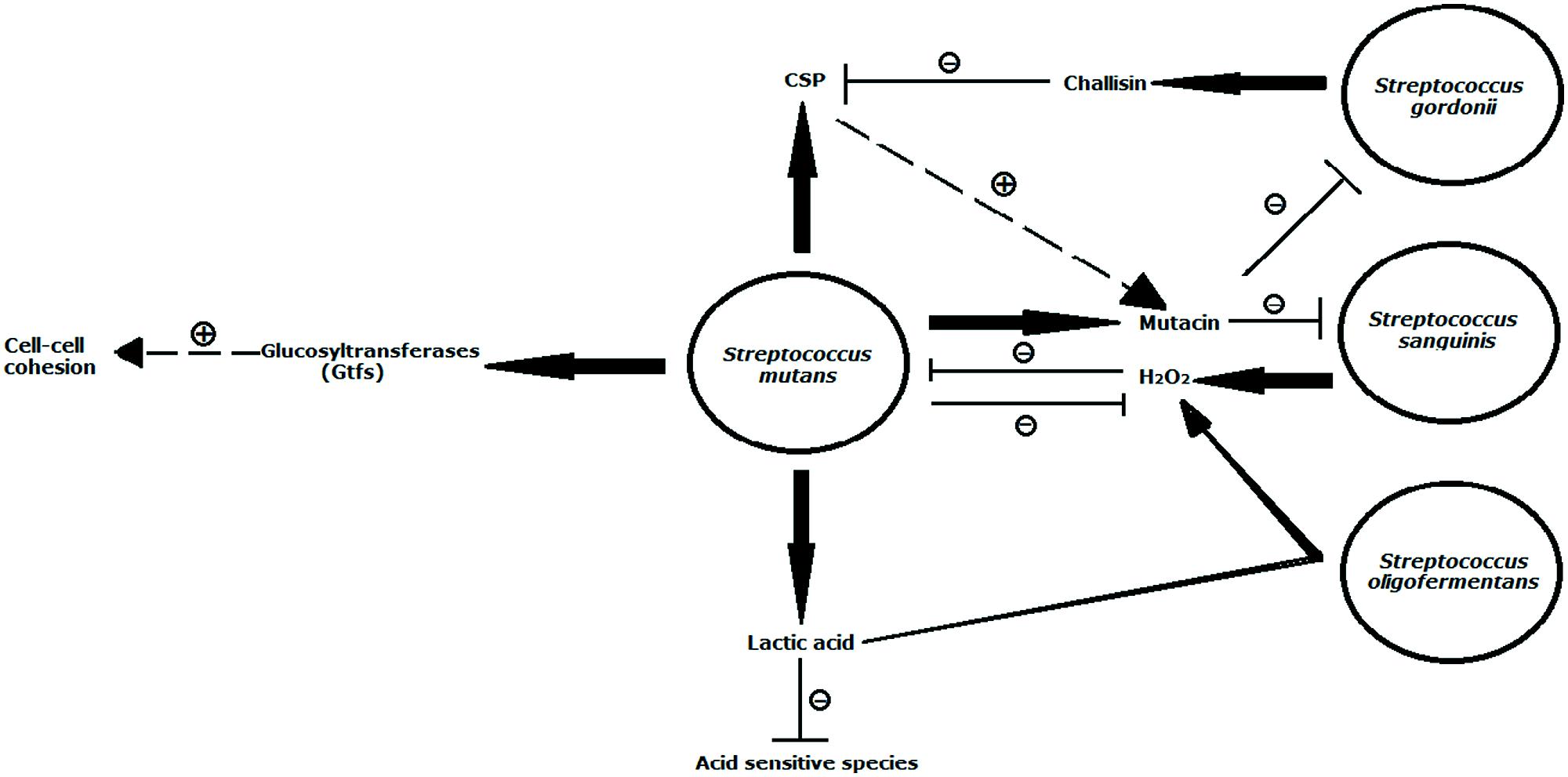
FIGURE 1. Illustration of interspecies interactions between Streptococcus mutans and selected oral streptococci that have been studied on a molecular level. Depicted is the production of lactic acid, mutacin, glucosyltransferases (Gtfs), and CSP, which also enhances mutacin production by S. mutans. Lactic acid has been found to inhibit proliferation of a variety of acid-sensitive anaerobe species, but it was recently discovered to serve as a substrate for S. oligofermentans for H2O2 production. This compound is also generated by S. sanguinis and was previously shown to inhibit growth of S. mutans. In order to counteract this antagonistic molecules, S. mutans produces some oxidative stress factors such as Dps-like protein (Dpr) and superoxide dismutase (Sod) in modulating its competitive fitness by alleviating H2O2-mediated interspecies competition exerted by other streptococcal co-residents. S. sanguinis and S. gordoniiare are sensitive to the mutacins produced by S. mutans. The Gtfs exoenzymes of S. mutans adhere to the cell surfaces of same species or other residents in an enzymatically active form and the glucans formed in situ increase cell–cell cohesion. S. gordonii utilizes challisin to reduce mutacin production by reducing the levels of the stimulating factor CSP. The dashed arrow in combination with + indicates stimulation, solid arrows symbolize production, and ⊣ in combination with – designates inhibition. Model is modified from Kuramitsu et al. (2007).
After the initial attachment of early colonizers to salivary receptors, the early colonizers could transform teeth or oral soft tissue surface to a bacterial surface. The adhesins on cell surface of early colonizers enable the sequential addition of the partner species. Oral streptococci express a diversity of cell surface molecules functioning as adhesins that recognize diverse bacterial receptors, such as pili, surface fibrils and Antigen I/II family protein, indicating that streptococci have a broad pairwise match with succeeding colonizers. It has been documented that oral streptococci recruit strains of Actinomyces by a highly selective cell surface interactions between specific streptococcal cell wall polysaccharides and Actinomyces sp. type 2 fimbriae (Cisar et al., 1979). Additionally, an inducible high-affinity Mn2+ transporter of S. gordonii (Kolenbrander et al., 1998a) was found to mediate its coadhesion with Actinomyces naeslundii (Kolenbrander et al., 1994). The close spatial association between streptococci and Actinomyces, as well as between streptococci and Veillonella was visualized in early stage of dental biofilms in vivo (Palmer et al., 2003; Chalmers et al., 2007).
The bacteria that can recruit subsequent colonizing species through binding to early and late colonizers constitute middle colonizers. Fusobacterium nucleatum is considered a “bridge-organism” that links early commensal colonizers and late colonizing species including periodontal pathogens and is important in the succession of genera in oral multispecies communities. Multivalent adhesins on the cell surface of F. nucleatum, with their distinct selectivity to early and late colonizers, contribute greatly to the development of polymicrobial communities within the oral cavity. It has been shown that the distinctive corncob structure visible in mature plaque is formed by the coadhesion between F. nucleatum and streptococci (Lancy et al., 1983). In addition to streptococci, other early colonizers such as A. naeslundii and Veillonella parvula also bind to F. nucleatum (Biyikoǧlu et al., 2012). Among the multiple adhesins of F. nucleatum, an arginine-inhibitable adhesin (RadD) is responsible for its adherence to streptococci; while some serotype-specific adhesins are involved in binding to periodontopathogens including Porphyromonas gingivalis, A. actinomycetemcomitans and Treponema denticola (Rosen and Sela, 2006; Rosen et al., 2008; Rupani et al., 2008; Kaplan et al., 2009). The coadhesion between F. nucleatum and these late colonizing Gram-negative species are usually mediated by lectin–carbohydrate interactions. For instance, the recognition between a galactose-specific lectin-like adhesin expressed by F. nucleatum and the sugar moiety present in the capsule and lipopolysaccharide of P. gingivalis contributes to coadhesion between the two species (Rosen and Sela, 2006). Similar galactose-containing receptors are also found on cell surface of A. actinomycetemcomitans (Rupani et al., 2008) and T. denticola (Rosen et al., 2008). As for Tannerella forsythia which is associated with severe and chronic periodontitis, its cell surface glycoproteins in S-layer act as adhesins in coadhesion with F. nucleatum (Shimotahira et al., 2013). Apart from protein–carbohydrate interactions, some researchers have reported that leucine-rich repeat in proteins from T. denticola and T. forsythia are involved in protein–protein interactions with each other and with F. nucleatum (Ikegami et al., 2004; Sharma et al., 2005). Even though T. forsythia possesses an S-layer at the outermost cell surface, a portion of the protein is exposed on the cell surface, one of them being OmpA-like protein, which was proposed to play a role in intercellular adhesion (Abe et al., 2011). Through selective recognition of adhesins on cell surface of F. nucleatum, these periodontitis associated species are integrated into microbial communities. For example, Filifactor alocis, an emerging important periodontal pathogen was found to preferentially accumulate at sites rich in F. nucleatum (Wang et al., 2013).
Besides F. nucleatum, P. gingivalis also exhibits a broad range of pairwise interactions with other microbial community members. P. gingivalis was shown to be able to assemble into a heterotypic microbial complex with S. gordonii and F. nucleatum (Lamont et al., 2002; Periasamy and Kolenbrander, 2009b). The specific molecular interactions between P. gingivalis and S. gordonii have been well characterized and occur through Mfa-Ssp and fimbrial adhesin (FimA)-GAPDH adhesin–receptor pairs (Maeda et al., 2004; Kuboniwa and Lamont, 2010). P. gingivalis’ short fimbriae (Mfa) bind to S. gordonii AgI/II family proteins SspA and SspB (Lamont et al., 2002; Park et al., 2005), whereas P. gingivalis long fimbriae (FimA) mediate its attachment to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of Streptococcus cristatus (Maeda et al., 2004). Further researches revealed that the region defined by residues 1167–1250 of S. gordonii SspB protein was essential motif for P. gingivalis binding (Brooks et al., 1997), and both NITVK motif and the nuclear receptor box of SspB contributed to the Mfa–SspB interaction (Demuth et al., 2001; Daep et al., 2011). The binding domain of GAPDH present on the streptococcal surface specific for P. gingivalis long fimbriae exists within the region encompassing amino acid residues 166–183 of GAPDH (Nagata et al., 2009). Although Mfa and FimA participate in the coadhesion of P. gingivalis to streptococci, another P. gingivalis surface molecule InlJ retards the development of P. gingivalis–S. gordonii community (Capestany et al., 2006), which allows P. gingivalis to fine-tune the extent of buildup of microbial communities. Besides, Kuboniwa et al. (2009) reported that P. gingivalis had a decreased ability to partner with S. gordonii and F. nucleatum when being defective in HmuR, a major hemin uptake protein. Consistent with the interspecies interaction described above, biofilm visualization by confocal microscopy confirmed that P. gingivalis colonized in vitro biofilms mainly within regions where S. gordonii accumulated (Kuboniwa et al., 2006).
Late colonizers are species with weak colonizing ability that require partner species to integrate into microbial communities and are comprised primarily of anaerobic, Gram-negative bacteria (Kolenbrander et al., 2006; Zijnge et al., 2010). As a late colonizer, T. denticola is unable to adhere to oral surfaces such as teeth or epithelial tissue; however, in the presence of P. gingivalis, it could colonize oral biofilms. In sub-gingival dental biofilms, T. denticola is always found more external relative to P. gingivalis (Ishihara et al., 2004). The chymotrypsin-like proteinase (CTLP), found within a high-molecular-mass complex on the cell surface of T. denticola, mediates its adherence to other potential periodontal pathogens including P. gingivalis, F. nucleatum, Prevotella intermedia and Parvimonas micra, and thus is crucial for T. denticola to integrate into multispecies oral microbial communities (Cogoni et al., 2012).
Physiological Interactions
Metabolites Mediated Cooperation
The high cell density of the oral microbial community impels individual resident to compete and collaborate with its neighboring species in order to survive in a hostile environment. An important factor in determining the microbial colonization in multispecies communities is the availability of nutrients. Metabolic communications among oral microbes may occur through the excretion of a metabolite by one bacterium that can be utilized as a nutrient by another species (Takahashi et al., 2010; Liu et al., 2012), or through syntrophic biochemical enzymes to cooperatively metabolize a substrate (Kolenbrander et al., 2002).
In the oral cavity, the short-chain acids such as lactate and acetate produced by early colonizers via sugar metabolism serve as carbon and energy sources for succeeding colonizers. This cross-feeding ensures sequential microbial colonization (Kolenbrander, 2011). Oral streptococci are well known by its ability to generate lactic acid as a by-product of sugar fermentation, whereas some neighboring species, such as Veillonellae sp. (Kumar et al., 2005; Chalmers et al., 2008) are unable to ferment sugars, but use lactic acid as a preferred source of carbon and energy. The ability of A. actinomycetemcomitans to utilize L-lactate, a carbohydrate metabolic by-product of S. gordonii as nutrient source allows it to integrate and become much more competitive within oral polymicrobial communities (Brown and Whiteley, 2007; Ramsey et al., 2011; Figure 2). The consumption of lactic acid by these lactic acid-utilizing bacteria minimizes lactic acid accumulation, which otherwise would cause streptococci to repress amylase synthesis and reduce the utilization efficiency of fermentable sugars by both partners (Egland et al., 2004). P. gingivalis and T. denticola are also metabolically connected. Succinate produced by T. denticola supports the growth of P. gingivalis, which, in turn, enhances the growth of T. denticola by providing isobutyric acid and proteinaceous substrates (Grenier, 1992; Nilius et al., 1993). Besides short-chain acids, Porphyromonas sp. can acquire vitamin K-like growth factors from Veillonella (Hojo et al., 2009).
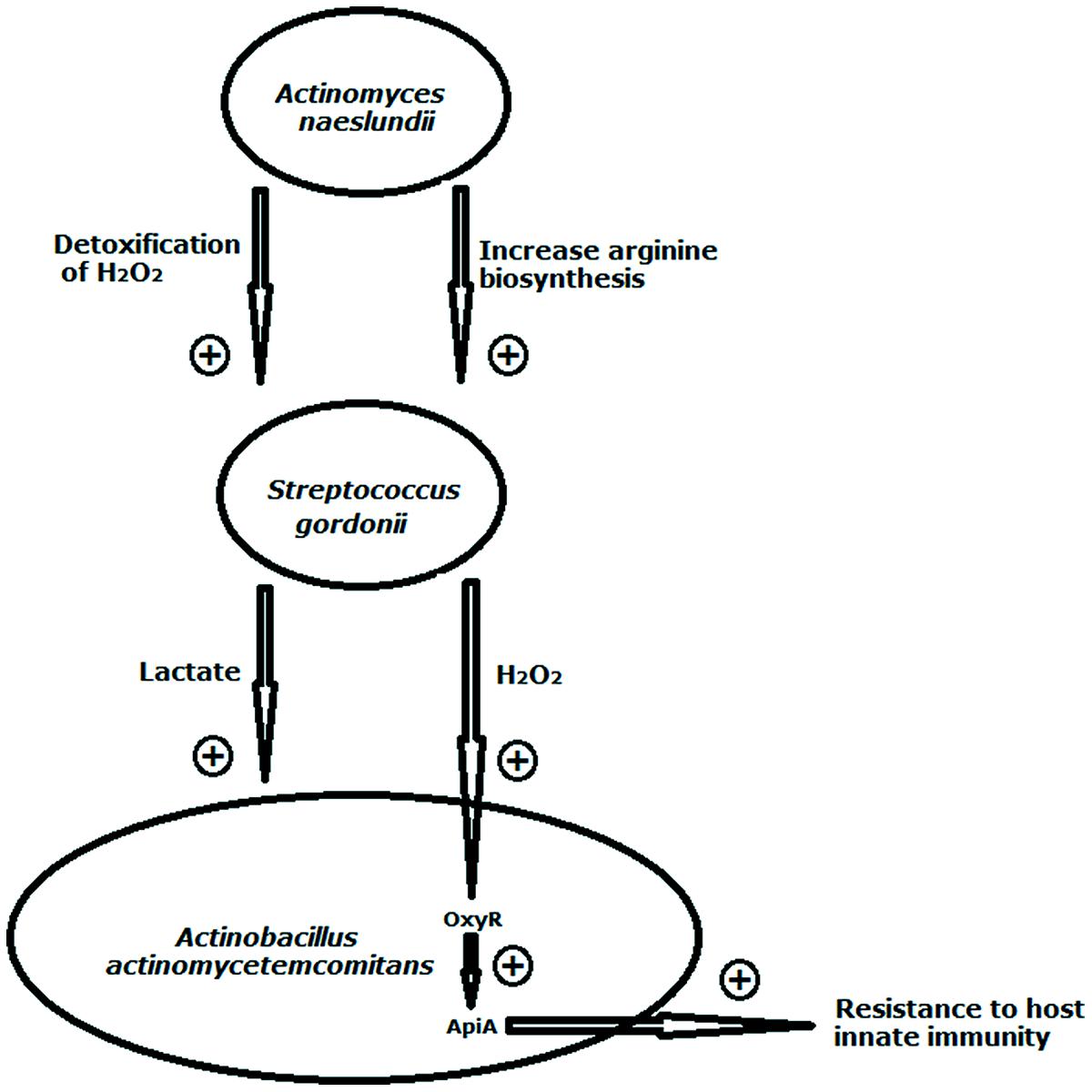
FIGURE 2. Illustration of interspecies interactions between Streptococcus gordonii and its co-residents Actinomyces naeslundii and Actinobacillus actinomycetemcomitans. S. gordonii benefits from its coaggregation with A. naeslundii both in tolerating protein oxidation by H2O2 and in upregulating arginine biosynthesis. A. actinomycetemcomitans utilizes L-lactate, a carbohydrate metabolic by-product of S. gordonii as nutrient source. The consumption of lactic acid by the lactic acid-utilizing bacteria minimizes lactic acid accumulation, which otherwise would inhibit streptococcal growth. S. gordonii-generated H2O2 can stimulate activation of the A. actinomycetemcomitans OxyR regulator to increase transcription of apiA that leads to enhanced resistance to host innate immunity. The arrow in combination with + indicates stimulation. Model compiled from Brown and Whiteley (2007), Jakubovics et al. (2008a, b), and Ramsey and Whiteley (2009).
Another type of metabolic interaction involves the complementary substrate utilization. Individual species in the oral cavity usually do not contain complete metabolic pathways to degrade complex salivary components. However, the breakdown of these compounds can be achieved through cooperation among multiple species. The synergistic and concerted action of Streptococcus mitis, S. gordonii, Streptococcus crostatus, and A. naeslundii with overlapping patterns of enzyme activity in salivary mucin catabolism has been reported (Wickström et al., 2009). Similarly, co-culture of S. sanguinis and S. oralis obtains significantly higher degradation efficiency of mucin than in monospecies culture (van der Hoeven and Camp, 1991). Therefore, this type of metabolic cooperation can benefit the participating species through maximizing their potential to extract energy from limited substrates.
Metabolic synergistic partnership usually occurs among community residents that often co-localize within the same niche and are metabolically compatible. In a three-species flow cell, P. gingivalis could not grow together with Streptococcus oralis–A. actinomycetemcomitans and S. oralis–F. nucleatum two-species community but displayed mutualistic growth when paired with S. gordonii–S. oralis, or V. parvula–A. actinomycetemcomitans, as well as with V. parvula–F. nucleatum two-species community (Periasamy and Kolenbrander, 2009a, b). The results indicated that P. gingivalis and S. oralis are incompatible, whereas S. gordonii could assist P. gingivalis to overcome this incompatibility, which also indicated that S. oralis was much more restricted in its successful partnership compared to its relative S. gordonii. Furthermore, in the presence of V. parvula, S. oralis displays positive partnership either with P. gingivalis or F. nucleatum. These findings illustrate the high species selectivity during multispecies community development.
Metabolites Mediated Competition
Interspecies antagonism reflects the competition among bacterial species for limited space and nutritional resources, and is the fundamental driving force in defining the structure and activity of polymicrobial oral communities. H2O2 secreted by viridans streptococci, including S. sanguinis, S. oralis, S. mitis, S. gordonii, Streptococcus parasanguinis, and a few S. mutans strains, plays an important role in interspecies competitive interaction (Figure 1). An inverse correlation between Streptococcus oligofermentans and S. mutans has been reported and thus suggested a possible antagonistic interaction between the two species (Tong et al., 2003). Tong et al. (2007) showed that S. oligofermentans possesses the capacity to convert its competitor’s main “weapon” (lactic acid) into an inhibitory chemical (H2O2) against S. mutans in order to gain a competitive growth advantage. This fascinating ability may be an example of a counteroffensive strategy used during chemical warfare within the oral microbial community. Furthermore, the inhibitory nature of H2O2 helps to structure the microbial composition of multispecies communities and contributes to an overall ecological homeostasis. It has been known that high levels of cariogenic S. mutans in the oral cavity correlate with low levels of S. sanguinis (Caufield et al., 2000). An epidemiological survey demonstrated the presence of putative periodontopathogens such as P. gingivalis, A. actinomycetemcomitans, and Eikenella corrodens in a destructive periodontitis site was correlated with the absence of certain viridans streptococci including S. sanguinis and Streptococcus intermedius, and vice versa (Hillman et al., 1985).
In addition to H2O2, bacteriocin or bacteriocin-like activities in multispecies communities play an essential role in modulating the ecological balance of oral microbial communities (Figure 1). Some oral commensal streptococci, such as S. sanguinis, S. gordonii, and S. oligofermentans, can suppress the overgrowth of cariogenic species S. mutans through the production of bacteriocins (Kreth et al., 2005a; Heng et al., 2007; Tong et al., 2008). S. mutans, in turn, gains competitive edge by producing lactic acid and mutacin to antagonize the growth of other oral commensal streptococci (Loesche, 1986; Qi et al., 2001). Lactobacillus paracasei can also produce bacteriocin to inhibit the growth of P. gingivalis, P. intermedia, T. forsythia, S. salivarius, and S. sanguinis (Pangsomboon et al., 2006, 2009). Prevotella nigrescens (P. nigrescens) ATCC 25261 was reported to produce a novel bacteriocin Nigrescin against P. gingivalis, P. intermedia, T. forsythia, and Actinomyces sp. (Kaewsrichan et al., 2004). More recently, it has been suggested that P. gingivalis gingipains secreted in the subgingival environment are the main inhibitor in promoting the detachment of antecedent A. actinomycetemcomitans cells from the surface (Takasaki et al., 2013).
Some species have evolved specific mechanisms to counteract the antagonistic molecules such as H2O2 and bacteriocin (Figure 1). For example, S. mutans produces Dps-like protein (Dpr), superoxide dismutase (Sod) and glutathione synthetase (GshAB) in modulating its competitive fitness by alleviating H2O2-mediated interspecies competition exerted by other streptococcal co-residents, thus promoting its survival and persistence within the oral microbial communities (Fujishima et al., 2013; Zheng et al., 2013). Meanwhile, S. mutans expresses a cell-envelop associated eukaryotic serine/threonine protein kinase (STPK) to resist peroxide (Zhu and Kreth, 2010). Another example is that S. gordonii secrets challisin to inhibit bacteriocin production through interfering with competence-stimulating peptide (CSP)-dependent quorum sensing system in S. mutans (Wang and Kuramitsu, 2005).
pH/Oxygen Mediated Interactions
The co-inhabitants within the oral microbial community usually favor mutual growth in harsh conditions by modifying surrounding pH or oxygen tension. For example, as pH drops caused by acid-producing bacteria such as streptococci and actinomyces following sugar fermentation, acid-tolerant bacteria, including S. mutans and lactobacilli, will out-compete acid-sensitive bacteria and have great impact on the microbial composition within community (Figure 1). Meanwhile, the lactic acid-utilizing bacteria such as Veillonella atypica and A. actinomycetemcomitans could prevent the acid buildup in dental biofilms, thus restoring microenvironment pH toward neutrality even in the presence of lactic acid bacteria and fermentable carbohydrates. As a result, the acid-sensitive species such as P. gingivalis are protected from acid attack (Takahashi, 2003). Acid-neutralizing activity could also be derived from the decrease in acidity during the fermentation of amino acid into organic acids and ammonia by certain periodontopathogens (Takahashi, 2003). For instance, it has been shown that F. nucleatum can grow at a wide pH range of 5.0–7.0, while P. gingivalis is susceptible to pH below 6.5. A study by Takahashi (2003) demonstrated that F. nucleatum can generate ammonia from glutamic and aspartic acid, subsequently elevating the pH to favor P. gingivalis growth. Besides adjusting local pH, F. nucleatum is also aerotolerant and its metabolic activity can reduce the concentration of oxygen to levels that can be tolerated by obligate anaerobes such as P. gingivalis, P. Nigrescens, and T. denticola (Bradshaw and Marsh, 1998; Bradshaw et al., 1998; Kolenbrander et al., 1998b). Without F. nucleatum, the number of obligate anaerobes would decrease sharply in oxygenated environments (Bradshaw et al., 1998). Meanwhile, F. nucleatum can support P. gingivalis growth by providing a capnophilic microenvironment when growing in a CO2 depleted environment (Diaz et al., 2002).
DNA Mediated Interactions
Cell lysis resulting from H2O2 and bacteriocin has been conjectured to provide genomic DNAs to the microbial communities (Zhu and Kreth, 2012). Extracellular DNA can be trapped within microbial biofilm matrix (Nadell et al., 2009) and might help to stabilize the biofilm structure (Barnes et al., 2012). Perry et al. (2009) found that treating S. mutans biofilms with DNase I reduced the surface-associated biomass by approximately 20%.
Genetic exchange occurs within oral microbial communities because residents are in close proximity. Horizontal gene transfer is the main driving force in bacterial evolution of antibiotic resistance and pathogenicity (Roberts and Mullany, 2010). Genome sequencing of several oral bacteria has revealed the presence of “pathogenicity islands” on the chromosomes of species such as P. gingivalis (Curtis et al., 1999). Conjugation, transformation, and transduction have been commonly observed in a variety of oral bacterial species (LeBlanc et al., 1978; Kuramitsu and Trapa, 1984; Yeung, 1999; Li et al., 2001b). Many genera of oral bacteria, including Actinomyces, Bifidobacterium, Fusobacterium, Haemophilus, Peptostreptococcus, Streptococcus, and Veillonella contain conjugative transposons that facilitate intercellular DNA transfer through conjugation (Rice, 1998). It is well known that oral streptococci are naturally competent, and it is possible that the DNA in the extracellular matrix is transmitted among them (Warburton et al., 2007). A study reported by Kreth et al. (2005b) proposed that S. mutans might utilize competence-induced bacteriocin to kill and lyse the neighboring species colonizing the same ecological niche and take up their DNA. Besides streptococci, P. gingivalis is able to exchange fimbrial allele types I and IV via natural competence as an adaptive process to modify behavior (Kerr et al., 2014). A. actinomycetemcomitans isolates from periodontal pockets have also been found to contain bacteriophages (Sandmeier et al., 1995).
Microbial Intercellular Signaling
Within dense and constricted oral microbial community, intercellular signaling allows residents to regulate their gene expressions in accordance with their environments, and coordinate their behaviors in virulence factors regulation, metabolic adaptation, competence initiation and polymicrobial biofilm development.
Contact-Dependent Signaling
In response to physical association with host tissues or bacterial cell surface, bacteria could initiate signal transduction cascades to regulate the expression of genes involved in adherence (Zainal-Abidin et al., 2012; Sarkar et al., 2014). For example, attachment of S. gordonii to salivary pellicle on the tooth surface triggers upregulated expression of AgI/II family protein Ssp (Dû and Kolenbrander, 2000), which are adhesins to salivary proteins (Demuth et al., 1996), as well as Actinomyces and P. gingivlais (Lamont et al., 2002). Further study revealed that this transcriptional regulation was mediated by a two-component system BrfAB (Zhang et al., 2004). Thus, the direct contact with saliva enhances S. gordonii’s adherence to tooth surface and the recruitment of late colonizing species such as Actinomyces, which will contribute to the spatiotemporal development of multispecies communities. Meanwhile, signaling can also be triggered when different bacterial species are in direct physical contact. P. gingivalis was reported to acquire increased adhesive capacities on various substrata through up-regulated expression of gingipain upon contact with T. denticola (Meuric et al., 2013). The transcriptional responses as a result of cell–cell contact often are not limited to a few genes. A study showed that thirty-three genes were differentially regulated during accretion of P. gingivalis in heterotypic biofilms with S. gordonii (Simionato et al., 2006). The functions of the regulated genes were predominantly related to metabolism and energy production. Another study on P. gingivalis assembled into a mixed community with F. nucleatum and S. gordonii revealed that over 400 genes were differentially regulated in response to cell–cell contact (Kuboniwa et al., 2009). Such signaling is important in the integration of P. gingivalis into early biofilms dominated by Gram-positive bacteria. In addition to facilitating bacteria colonization of microbial community, contact-inducible gene expression could also enhance microbial growth. S. gordonii was shown to benefit from its coaggregation with A. naeslundii in arginine-restricted conditions through three fold changes in the expression of the genes involved in arginine biosynthesis (Jakubovics et al., 2008a; Figure 2). This signaling was not observed in co-cultures where there was no coaggregation between the two species, suggesting that there is a specific transcriptional response after cell–cell contact. Contact-inducible transcriptional change could prepare bacteria to better cope with the environmental stress, such as the exposure to H2O2 within the multispecies communities. An example comes from our observation of S. sanguinis and F. nucleatum (He et al., 2012). We found that upon contact with S. sanguinis, F. nucleatum acquired increased resistance to H2O2 and also significantly inhibited H2O2 production by S. sanguinis. This event was not observed in a F. nucleatum mutant deficient in radD, which encodes an outer membrane protein adhesin responsible for coadhesion with streptococci. This defense strategy of F. nucleatum prevents antagonism by other oral bacteria and allows integration into the developing oral microbial community. Similarly, coaggregation with A. naeslundii protects S. gordonii from oxidative damage by H2O2 (Jakubovics et al., 2008b; Figure 2).
On the other hand, cell contact-dependent signaling has also been shown to reduce adhesion. Park et al. (2006) observed that following contact with S. gordonii, the short fimbriae adhesin Mfa of P. gingivalis was down-regulated. Moreover, a cascade of tyrosine phosphorylation/dephosphorylation events in P. gingivalis initiated by contact with S. gordonii constrains the heterotypic community development between the two species (Maeda et al., 2008; Chawla et al., 2010). Similarly, cell–cell contact between T. forsythia and F. nucleatum or T. forsythia and P. gingivalis also initiates a signal transduction cascade that causes down-regulated expression of BspA leucine-rich repeat protein adhesin in T. forsythia (Inagaki et al., 2005), resulting in reduced adherence ability. There is a signaling down-regulation in microbial biosynthesis pathway as well. A study showed S. gordonii and F. nucleatum reduced the abundance of P. gingivalis proteins responsible for several vitamin synthesis and pyrimidine biosynthesis in a contact-dependent manner (Kuboniwa et al., 2009).
Some degree of specificity exists in response to contact-dependent signaling. The study of Hendrickson et al. (2012) demonstrated that, when in contact with P. gingivalis, S. gordonii up-regulates the expression of the glycolysis pathway while when in contact with F. nucleatum the pentose phosphate pathway is up-regulated.
Quorum Sensing
In addition to contact-dependent signaling, microbial intercellular signaling can also be mediated by secreted diffusible molecules. Quorum sensing mediated by autoinducing diffusible molecules (autoinducer) is a system of stimulus and response correlated to population density, which allows bacteria to coordinate gene expression on a population-wide scale. A significant portion of a bacterial genome (4–10%) and proteome (20% or more) can be influenced by quorum sensing signaling (Dunman et al., 2001; Arevalo-Ferro et al., 2003; Schuster et al., 2003; Wagner et al., 2003). Common classes of autoinducer are N-Acyl Homoserine Lactones (N-AHL) found in Gram-negative bacteria, autoinducing peptides (AIPs) produced by Gram-positive bacteria, and a family of autoinducers known as autoinducer-2 (AI-2) which is widespread among both Gram-negative and Gram-positive bacteria (Miller and Bassler, 2001) and is known as the most conserved quorum sensing signal molecule.
In Gram-negative bacteria, N-AHL signal is mainly generated by an N-AHL synthase of the LuxI family of proteins, and is recognized by an N-AHL receptor protein belonging to the LuxR family of transcriptional regulators (Ryan and Dow, 2008). Most Gram-negative bacteria use analogous LuxI/LuxR-type circuits for intraspecies signaling in which only members of the same species are able to recognize and respond to the signal. Meanwhile, there are reports suggesting their involvement in interspecies signaling (Huber et al., 2001; Riedel et al., 2001). The N-AHL intra- and interspecies signaling role of oral microbes needs to be further investigated. In this review we mainly focus on cell–cell signaling via AIPs and AI-2.
AIPs
In Gram-positive bacteria, recognition of and response to the AIPs occur not by direct binding to a cognate receptor but through a two-component signal transduction system, in which AIPs binds to a membrane-bound histidine kinase sensor and the binding leads to phosphorylation of response regulator proteins that ultimately bind to the promoter of target genes to regulate gene expression (Senadheera and Cvitkovitch, 2008).
Competence-stimulating peptide-mediated quorum sensing has been identified in various oral streptococcal species including S. mutans, S. gordonii, and S. intermedius (Håvarstein et al., 1997; Cvitkovitch, 2001; Petersen et al., 2004). The proposed role of CSP-mediated quorum sensing is to alter genes transcription and proteins synthesis involved in biofilm formation, competence development, bacteriocin synthesis, stress resistance, and autolysis (Petersen et al., 2004; Senadheera and Cvitkovitch, 2008; Perry et al., 2009; Senadheera et al., 2009).
Although AIPs are generally species-specific and sometimes strain-specific, they can be detected by and induce response in other species. A study showed that S. mutans CSP could be sensed by C. albicans and resulted in induction of its germ tube formation (Jarosz et al., 2009). In Enterococcus faecalis (E. faecalis), Fsr quorum-sensing system exerts inhibitive effect on C. albicans hyphal morphogenesis (Cruz et al., 2013). These two observations demonstrate the potential signaling role of AIPs in interkingdom communication. In order to perpetuate in complex oral multispecies community, microbial species can contend with its competitor by decreasing the concentration of the extracellular AIPs secreted by the competitor. P. gingivalis was found to be able to inactivate S. mutans CSP and abolish CSP-induced natural transformation (Wang et al., 2011a). Wang and Kuramitsu (2005) reported that S. gordonii, S. sanguinis, S. mitis, and S. oralis inhibited S. mutans’ mutacin production by degrading its CSP. S. gordonii can secrete serine protease challisin to inactivate S. mutans CSP, subsequently impairing S. mutans biofilm formation (Wang et al., 2011b; Figure 1). S. salivarius was also reported to strongly inhibit S. mutans biofilm formation via inactivating CSP produced by S. mutans, which provides a competitive advantage to S. salivarius against its competing bacteria (Tamura et al., 2009).
AI-2
AI-2 is a group of molecules derived from spontaneous rearrangement of S-4,5-dihydroxy-2,3-pentanedione (DPD), whose production is the catabolism of S-adenosylhomocysteine by conserved LuxS (Schauder et al., 2001b). It is widely distributed in Gram-positive and Gram-negative bacteria and plays a widespread role in bacterial communication across species boundaries (Hardie and Heurlier, 2008; Jayaraman and Wood, 2008). AI-2 has been well-characterized as a universal interspecies signal molecule in regulating formation of multispecies biofilms (Bassler et al., 1997; Schauder and Bassler, 2001a; Whitmore and Lamont, 2011). For example, mixed biofilm formation by A. naeslundii and S. oralis in flowing saliva was shown to be dependent on the production of AI-2 by S. oralis (Rickard et al., 2006). A luxS mutant of S. oralis, which cannot produce AI-2, does not form dual-species biofilms with A. naeslundii. This defect can be restored with either genetic complementation or supplementation with synthetic AI-2. Streptococcus-derived AI-2 can also regulate gene expression in P. gingivalis (Chung et al., 2001). Meanwhile, S. gordonii and F. nucleatum can sense P. gingivalis AI-2 as well and translate the signal into a biofilm phenotype (McNab et al., 2003; Saito et al., 2008). In addition to streptococcal AI-2, P. gingivlais can response to AI-2 signal secreted by A. actinomycetemcomitans (Fong et al., 2001). AI-2 secreted by F. nucleatum is able to elicit changes in the expression of the representative adhesins of the so-called “red-complex” (P. gingivalis, T. denticola, and T. forsythia), and thus, increase the colonization of these periodontopathogens in oral multispecies biofilms (Jang et al., 2013a).
The concentration of AI-2 is critical for interspecies mutualism. It has been reported that above and below optimal AI-2 concentration dual-species biofilm formation of S. oralis and A. naeslundii was suppressed (Rickard et al., 2006). F. nucleatum, a middle colonizer of oral microbial communities, applies differential regulation of AI-2 on biofilm growth of two oral streptococci by exerting a stimulatory effect on S. gordonii and an inhibitory effect on S. oralis (Jang et al., 2013b). Cuadra-Saenz et al. (2012) found that S. gordonii produce more AI-2 than S. oralis. They suggested that AI-2 could alter the structure and composition of pioneering oral streptococcal population, thereafter influencing the subsequent succession of other species into oral microbial communities (Cuadra-Saenz et al., 2012). Commensal bacteria such as S. oralis and A. naeslundii have been shown to respond to AI-2 levels that are below those produced by species associated with oral disease, for example F. nucleatum (Frias et al., 2001). Commensal species produce and respond to AI-2 signals at picomolar concentrations, which are much lower than that of periodontopathogens (Kolenbrander et al., 2006). Fusobacteria are usually detectable in oral microbial biofilms after 8 h of biofilm development (Li et al., 2004). As entry of Fusobacteria increases on the tooth surface and local AI-2 concentration rises, Fusobacteria would initiate signal-dependent changes in gene expression regulated by AI-2. By contrast, commensal bacteria are inhibited by the increasing concentration of AI-2. Accordingly, high levels of AI-2 increase the accumulation of periodontopathogens and reduce the growth of commensal bacteria. Considering the different AI-2 concentrations, some researchers propose AI-2 as a modulator for multispecies microbial communities in transition from a beneficial community to a pathogenic community (Kolenbrander et al., 2010).
In addition to interspecies interaction, AI-2 has been reported to play a role in interkingdom signaling. C. albicans can sense AI-2 produced by S. gordonii to induce numerous transcriptional changes, including increased production of hyphae and the activation or repression of three mitogen-activated protein kinases involved in morphogenetic switching (Bamford et al., 2009).
During the development of highly organized microbial communities, individual species must be able to overcome antagonistic effects from other community co-inhabitants in order to perpetuate in the multispecies community (Vollaard and Clasener, 1994; He et al., 2012). Xavier et al. (2007) found that some species of enteric bacteria can degrade AI-2 signaling and interfere with other competing species’ ability to assess and respond correctly to changes in cell population density. No such interactions have yet been observed for oral microbial community members.
Recent studies also have demonstrated an emerging role for AI-2 as an intraspecies signal molecule besides its well-characterized function in interspecies signaling. In A. actinomycetemcomitans and S. mutans, AI-2 is required in modulating their biofilm development and virulence (Merritt et al., 2003; Shao et al., 2007; Torres-Escobar et al., 2013). While in P. gingivalis, AI-2 regulates iron (hemin) acquisition (James et al., 2006) and oxidative stress-related genes (Yuan et al., 2005).
Non-Autoinducing Molecules Dependent Signaling
In addition to autoinducers, oral microbial species can sense and respond to extracellular non-autoinducing diffusible molecules to achieve gene regulation. It has been shown that a diffusible molecule dependent signaling exists between S. gordonii and V. atypica besides their cross-feeding through lactic acid. S. gordonii responds to maltose or a related sugar from the lipopolysaccharide released by V. atypica by up-regulating the expression of streptococcal transcription factor CcpA, which is required for amylase synthesis (Egland et al., 2004; Johnson et al., 2009). Increased amylase activity on a starch substrate causes S. gordonii to generate more lactate, the primary energy source for V. atypica. Thus, one or both species involved in this event benefit from this metabolic requirements-driven interspecies interaction. Furthermore, this type of signaling occurred only in S. gordonii cells located within a few micrometers of V. atypica, emphasizing that diffusible signals between species are designed to function over very short distances, on the order of 1 μm. In addition to coordinative effect, metabolites can induce antagonistic signaling as well. For example, extracellular arginine deiminase of S. cristatus and S. intermedius downregulates expression of FimA of P. gingivalis, and consequently P. gingivalis cannot integrate into communities together with these organisms (Xie et al., 2007; Christopher et al., 2010). In contrast, lack of the similar signaling pathway in S. gordonii makes it more compatible with P. gingivalis compared to S. cristatus and S. intermedius.
Acidic pH and H2O2 could play a role in interspecies signaling, which may affect gene expression patterns of bacteria cells growing within oral microbial communities (Li and Burne, 2001a; Deng et al., 2007). Under low microenvironmental pH, 14% of the S. mutans genome displays altered gene expression including quorum sensing systems (Gong et al., 2009). In response to H2O2-induced oxidative stress, there were three major protein systems of F. nucleatum changed (Steeves et al., 2011). S. gordonii-generated H2O2 can stimulate activation of the A. actinomycetemcomitans OxyR regulator to increase transcription of apiA that leads to enhanced resistance to host innate immunity (Ramsey and Whiteley, 2009; Figure 2).
Other than growth inhibition, antibiotics at sub-inhibitory concentration were reported to be potent global regulators of transcription for bacteria within microbial communities (Yim et al., 2006). In S. mutans GS5, low concentrations of antibiotics could up-regulate expression of bacteriocin immunity protein gene, which affected its sensitivity to a variety of antimicrobial agents (Matsumoto-Nakano and Kuramitsu, 2006).
Conclusion
Recently, there is an increasing appreciation of the essential roles of inter-cellular communications in polymicrobial oral biofilm development, environmental adaptation, and virulent factors regulation. Adhesins and receptors-mediated binding benefits microbial community residents by providing a broader habitat range and influences the temporal and spatial formation of highly organized polymicrobial community architecture. Furthermore, the close physical association allows residents to obtain metabolic support from their neighboring species. Cooperative metabolic interactions either via cross-feeding or through cooperatively metabolizing substrate maximize co-residents’ potential to extract energy from limited substrates. In addition to synergistic interactions, oral bacterial species are also engaged in intense competition for limited space and nutritional resources using compounds such as bacteriocin and H2O2, which plays a crucial role in defining the structure and activity of oral microbial communities. Intercellular signaling within the same or between different bacterial species can be achieved by contact dependent mechanisms or mediated by diffusible signal molecules. These signaling events play significant roles in coordinating gene expression involved in microbial physiology and pathobiology. Among the secreted signaling molecules, AIPs and AI-2 appear to be involved in both intra-species quorum sensing and a variety of interspecies interactions across oral species. More comprehensive investigations of microbial intercellular interactions will shed light on the complexity of multispecies oral microbial communities and may provide novel approaches for controlling microbial community-based pathogenesis.
Conflict of Interest Statement
Dr. Shi is a part time chief science officer of C3 Jian Inc., which has licensed technologies from UC regents.
Acknowledgments
This work was supported by a NIH grant DE020102 and DE021108.
References
Abe, T., Murakami, Y., Nagano, K., Hasegawa, Y., Moriguchi, K., Ohno, N.,et al. (2011). OmpA-like.protein influences cell shape and adhesive activity of Tannerella forsythia. Mol. Oral Microbiol. 26, 374–387. doi: 10.1111/j.2041-1014.2011.00625.x
Arevalo-Ferro, C., Hentzer, M., Reil, G., Görg, A., Kjelleberg, S., Givskov, M.,et al. (2003). Identification of quorumsensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 5, 1350–1369. doi: 10.1046/j.1462-2920.2003.00532.x
Avila, M., Ojcius, D. M., and Yilmaz, Ö. (2009). The oral microbiota: living with a permanent guest. DNA Cell Biol. 28, 405–411. doi: 10.1089/dna.2009.0874
Bamford, C. V., d’Mello, A., Nobbs, A. H., Dutton, L. C., Vickerman, M. M., and Jenkinson, H. F. (2009). Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect. Immun. 77, 3696–3704. doi: 10.1128/IAI.00438-09
Barnes, A. M., Ballering, K. S., Leibman, R. S., Wells, C. L., and Dunny, G. M. (2012). Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. MBio 3, e00193–e00212. doi: 10.1128/mBio.00193-12
Bassler, B. L., Greenberg, E. P., and Stevens, A. M. (1997). Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179, 4043–4045.
Biyikoǧlu, B., Ricker, A., and Diaz, P. I. (2012). Strain-specific colonization patterns and serum modulation of multi-species oral biofilm development. Anaerobe 18, 459–470. doi: 10.1016/j.anaerobe.2012.06.003
Bowen, W. H., and Koo, H. (2011). Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45, 69–86. doi: 10.1159/000324598
Bradshaw, D. J., and Marsh, P. D. (1998). Analysis of pH-driven disruption of oral microbial communities in vitro. Caries Res. 32, 456–462. doi: 10.1159/000016487
Bradshaw, D. J., Marsh, P. D., Watson, G. K., and Allison, C. (1998). Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect. Immun. 66, 4729–4732.
Brooks, W., Demuth, D. R., Gil, S., and Lamont, R. J. (1997). Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect. Immun. 65, 3753–3758.
Brown, S. A., and Whiteley, M. (2007). A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J. Bacteriol. 189, 6407–6414. doi: 10.1128/JB.00554-07
Capestany, C. A., Kuboniwa, M., Jung, I. Y., Park, Y., Tribble, G. D., and Lamont, R. J. (2006). Role of the Porphyromonas gingivalis InlJ protein in homotypic and heterotypic biofilm development. Infect. Immun. 74, 3002–3005. doi: 10.1128/IAI.74.5.3002-3005.2006
Caufield, P. W., Dasanayake, A. P., Li, Y., Pan, Y., Hsu, J., and Hardin, J. M. (2000). Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect. Immun. 68, 4018–4023. doi: 10.1128/IAI.68.7.4018-4023.2000
Chalmers, N. I., Palmer, R. J., Cisar, J. O., and Kolenbrander, P. E. (2008). Characterization of a Streptococcus sp.–Veillonella sp. community micromanipulated from dental plaque. J. Bacteriol. 190, 8145–8154. doi: 10.1128/JB.00983-08
Chalmers, N. I., Palmer, R. J. Jr., Du-Thumm, L., Sullivan, R., Shi, W., and Kolenbrander, P. E. (2007). Use of quantum dot luminescent probes to achieve single-cell resolution of human oral bacteria in biofilms. Appl. Environ. Microbiol. 73, 630–636. doi: 10.1128/AEM.02164-06
Chawla, A., Hirano, T., Bainbridge, B. W., Demuth, D. R., Xie, H., and Lamont, R. J. (2010). Community signaling between Streptococcus gordonii and Porphyromonas gingivalis is controlled by the transcriptional regulator CdhR. Mol. Microbiol. 78, 1510–1522. doi: 10.1111/j.1365-2958.2010.07420.x
Christopher, A. B., Arndt, A., Cugini, C., and Davey, M. E. (2010). A streptococcal effector protein that inhibits Porphyromonas gingivalis biofilm development. Microbiology 156, 3469–3477. doi: 10.1099/mic.0.042671-0
Chung, W. O., Park, Y., Lamont, R. J., McNab, R., Barbieri, B., and Demuth, D. R. (2001). Signaling system in Porphyromonas gingivalis based on a LuxS protein. J. Bacteriol. 183, 3903–3909. doi: 10.1128/JB.183.13.3903-3909.2001
Cisar, J. O., Kolenbrander, P. E., and McIntire, F. C. (1979). Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect. Immun. 24, 742–752.
Cogoni, V., Morgan-Smith, A., Fenno, J. C., Jenkinson, H. F., and Dymock, D. (2012). Treponema denticola chymotrypsin-like proteinase (CTLP) integrates spirochaetes within oral microbial communities. Microbiology 158, 759–770. doi: 10.1099/mic.0.055939-0
Cruz, M. R., Graham, C. E., Gagliano, B. C., Lorenz, M. C., and Garsin, D. A. (2013). Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect. Immun. 81, 189–200. doi: 10.1128/IAI.00914-12
Cuadra-Saenz, G., Rao, D. L., Underwood, A. J., Belapure, S. A., Campagna, S. R., Sun, Z.,et al. (2012). Autoinducer-2 influences interactions amongst pioneer colonizing streptococci in oral biofilms. Microbiology 158, 1783–1795. doi: 10.1099/mic.0.057182-0
Curtis, M. A., Hawley, S. A., and Aduse-Opoku, J. (1999). The rag locus of Porphyromonas gingivalis: a novel pathogenicity island. J. Periodontol. Res. 34, 400–440. doi: 10.1111/j.1600-0765.1999.tb02273.x
Cvitkovitch, D. G. (2001). Genetic competence and transformation in oral streptococci. Crit. Rev. Oral Biol. Med. 12, 217–243. doi: 10.1177/10454411010120030201
Daep, C. A., Novak, E. A., Lamont, R. J., and Demuth, D. R. (2011). Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect. Immun. 79, 67–74. doi: 10.1128/IAI.00361-10
Demuth, D. R., Duan, Y., Brooks, W., Holmes, A. R., McNab, R., and Jenkinson, H. F. (1996). Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol. Microbiol. 20, 403–413. doi: 10.1111/j.1365-2958.1996.tb02627.x
Demuth, D. R., Irvine, D. C., Costerton, J. W., Cook, G. S., and Lamont, R. J. (2001). Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect. Immun. 69, 5736–5741. doi: 10.1128/IAI.69.9.5736-5741.2001
Deng, D. M., Liu, M. J., ten Cate, J. M., and Crielaard, W. (2007). The VicRK system of Streptococcus mutans responds to oxidative stress. J. Dent. Res. 86, 606–610. doi: 10.1177/154405910708600705
Dewhirst, F. E., Chen, T., Izard, J., Paster, B. J., Tanner, A. C. R., Yu, W. H.,et al. (2010). The human oral microbiome. J. Bacteriol. 192, 5002–5017. doi: 10.1128/JB.00542-10
Diaz, P. I., Zilm, P. S., and Rogers, A. H. (2002). Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbondioxide-depleted environments. Microbiology 148, 467–472.
Dige, I., Raarup, M. K., Nyengaard, J. R., Kilian, M., and Nyvad, B. (2009). Actinomyces naeslundii in initial dental biofilm formation. Microbiology 155, 2116–2126. doi: 10.1099/mic.0.027706-0
Ding, A. M., Palmer, R. J. Jr., Cisar, J. O., and Kolenbrander, P. E. (2010). Shear-enhanced oral microbial adhesion. Appl. Environ. Microbiol. 76, 1294–1297. doi: 10.1128/AEM.02083-09
Dû, L. D., and Kolenbrander, P. E. (2000). Identification of saliva-regulated genes of Streptococcus gordonii DL1 by differential display using random arbitrarily primed PCR. Infect. Immun. 68, 4834–4837. doi: 10.1128/IAI.68.8.4834-4837.2000
Dunman, P. M., Murphy, E., Haney, S., Palacios, D., Tucker-Kellogg, G., Wu, S.,et al. (2001). Transcription profiling–based identification of Staphylococcus aureus genes regulated by the agr and/ or sarA loci. J. Bacteriol. 183, 7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001
Egland, P. G., Palmer, R. J., and Kolenbrander, P. E. (2004). Interspecies communication in Streptococcus gordonii–Veillonella atypicabiofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. U.S.A. 101, 16917–16922. doi: 10.1073/pnas.0407457101
Fong, K. P., Chung, W. O., Lamont, R. J., and Demuth, D. R. (2001). Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69, 7625–7634. doi: 10.1128/IAI.69.12.7625-7634.2001
Frias, J., Olle, E., and Alsina, M. (2001). Periodontal pathogens produce quorum sensing signal molecules. Infect. Immun. 69, 3431–3434. doi: 10.1128/IAI.69.5.3431-3434.2001
Fujishima, K., Kawada-Matsuo, M., Oogai, Y., Tokuda, M., Torii, M., and Komatsuzawa, H. (2013). dpr and sod in Streptococcus mutans are involved in coexistence with S. sanguinis, and PerR is associated with resistance to H2O2. Appl. Environ. Microbiol. 79, 1436–1443. doi: 10.1128/AEM.03306-12
Gong, Y., Tian, X. L., Sutherland, T., Sisson, G., Mai, J., Ling, J.,et al. (2009). Global transcriptional analysis of acid-inducible genes in Streptococcus mutans: multiple two-component systems involved in acid adaptation. Microbiology 155, 3322–3332. doi: 10.1099/mic.0.031591-0
Gregoire, S., Xiao, J., Silva, B. B., Gonzalez, I., Agidi, P. S., Klein, M. I.,et al. (2011). Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl. Environ. Microbiol. 77, 6357–6367. doi: 10.1128/AEM.05203-11
Grenier, D. (1992). Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infect. Immun. 60, 5298–5301.
Hamada, T., Kawashima, M., Watanabe, H., Tagami, J., and Senpuku, H. (2004). Molecular interactions of surface protein peptides of Streptococcus gordonii with human salivary components. Infect. Immun. 72, 4819–4826. doi: 10.1128/IAI.72.8.4819-4826.2004
Hardie, K. R., and Heurlier, K. (2008). Establishing bacterial communities by “word of mouth”: LuxS and autoinducer 2 in biofilm development. Nat. Rev. Microbiol. 6, 635–643. doi: 10.1038/nrmicro1916
Håvarstein, L. S., Hakenbeck, R., and Gaustad, P. (1997). Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179, 6589–6594.
He, X., Hu, W., Kaplan, C. W., Guo, L., Shi, W., and Lux, R. (2012). Adherence to streptococci facilitates Fusobacterium nucleatum integration into an oral microbial community. Microb. Ecol. 63, 532–542. doi: 10.1007/s00248-011-9989-2
Hendrickson, E. L., Wang, T., Dickinson, B. C., Whitmore, S. E., Wright, C. J., Lamont, R. J.,et al. (2012). Proteomics of Streptococcus gordonii within a model developing oral microbial community. BMC Microbiol. 12:211. doi: 10.1186/1471-2180-12-211
Heng, N. C. K., Tagg, J. R., and Tompkins, G. R. (2007). Competence-dependent bacteriocin production by Streptococcus gordonii DL1 (Challis). J. Bacteriol. 189, 1468–1472. doi: 10.1128/JB.01174-06
Hillman, J. D., Socransky, S. S., and Shivers, M. (1985). The relationships between streptococcal species and periodontopathic bacteria in human dental plaque. Arch. Oral Biol. 30, 791–795. doi: 10.1016/0003-9969(85)90133-5
Hojo, K., Nagaoka, S., Ohshima, T., and Maeda, N. (2009). Bacterial interactions in dental biofilm development. J. Dent. Res. 88, 982–990. doi: 10.1177/0022034509346811
Huber, B., Riedel, K., Hentzer, M., Heydorn, A., Gotschlich, A., Givskov, M.,et al. (2001). The cep quorumsensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147, 2517–2528.
Ikegami, A., Honma, K., Sharma, A., and Kuramitsu, H. K. (2004). Multiple functions of the leucine-rich repeat protein LrrA of Treponema denticola. Infect. Immun. 72, 4619–4627. doi: 10.1128/IAI.72.8.4619-4627.2004
Inagaki, S., Kuramitsu, H. K., and Sharma, A. (2005). Contact-dependent regulation of a Tannerella forsythia virulence factor, BspA, in biofilms. FEMS Microbiol. Lett. 249, 291–296. doi: 10.1016/j.femsle.2005.06.032
Ishihara, K., Nabuchi, A., Ito, R., Miyachi, K., Kuramitsu, H. K., and Okuda, K. (2004). Correlation between detection rates of periodontopathic bacterial DNA in carotid coronary stenotic artery plaque and in dental plaque samples. J. Clin. Microbiol. 42, 1313–1315. doi: 10.1128/JCM.42.3.1313-1315.2004
Jakubovics, N. S., Gill, S. R., Iobst, S. E., Vickerman, M. M., and Kolenbrander, P. E. (2008a). Regulation of gene expression in a mixed-genus community: stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii. J. Bacteriol. 190, 3646–3657. doi: 10.1128/JB.00088-08
Jakubovics, N. S., Gill, S. R., Vickerman, M. M., and Kolenbrander, P. E. (2008b). Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol. Ecol. 66, 637–644. doi: 10.1111/j.1574-6941.2008.00585.x
James, C. E., Hasegawa, Y., Park, Y., Yeung, V., Tribble, G. D., Kuboniwa, M.,et al. (2006). LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect. Immun. 74, 3834–3844. doi: 10.1128/IAI.01768-05
Jang, Y. J., Choi, Y. J., Lee, S. H., Jun, H. K., and Choi, B. K. (2013a). Autoinducer 2 of Fusobacterium nucleatum as a target molecule to inhibit biofilm formation of periodontopathogens. Arch. Oral Biol. 58, 17–27. doi: 10.1016/j.archoralbio.2012.04.016
Jang, Y. J., Sim, J., Jun, H. K., and Choi, B. K. (2013b). Differential effect of autoinducer 2 of Fusobacterium nucleatum on oral streptococci. Arch. Oral Biol. 58, 1594–1602. doi: 10.1016/j.archoralbio.2013.08.006
Jarosz, L. M., Deng, D. M., van der Mei, H. C., Crielaard, W., and Krom, B. P. (2009). Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot. Cell 8, 1658–1664. doi: 10.1128/EC.00070-09
Jayaraman, A., and Wood, T. K. (2008). Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu. Rev. Biomed. Eng. 10, 145–167. doi: 10.1146/annurev.bioeng.10.061807.160536
Jenkinson, H. F., and Lamont, R. J. (2005). Oral microbial communities in sickness and in health. Trends Microbiol. 13, 589–595. doi: 10.1016/j.tim.2005.09.006
Johnson, B. P., Jensen, B. J., Ransom, E. M., Heinemann, K. A., Vannatta, K. M., Egland, K. A.,et al. (2009). Interspecies signaling between Veillonella atypica and Streptococcus gordonii requires the transcription factor CcpA. J. Bacteriol. 191, 5563–5565. doi: 10.1128/JB.01226-08
Kaewsrichan, J., Douglas, C. W. I., Nissen-Meyer, J., Fimland, G., and Teanpaisan, R. (2004). Characterization of a bacteriocin produced by Prevotella nigrescens ATCC 25261. Lett. Appl. Microbiol. 39, 451–458. doi: 10.1111/j.1472-765X.2004.01608.x
Kaplan, C. W., Lux, R., Haake, S. K., and Shi, W. (2009). The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol. Microbiol. 71, 35–47. doi: 10.1111/j.1365-2958.2008.06503.x
Kaplan, J. B., Ragunath, C., Ramasubbu, N., and Fine, D. H. (2003). Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J. Bacteriol. 185, 4693–4698. doi: 10.1128/JB.185.16.4693-4698.2003
Kerr, J. E., Abramian, J. R., Dao, D. H., Rigney, T. W., Fritz, J., Pham, T.,et al. (2014). Genetic exchange of fimbrial alleles exemplifies the adaptive virulence strategy of Porphyromonas gingivalis. PLoS ONE 13:e91696. doi: 10.1371/journal.pone.0091696
Kolenbrander, P. E. (2011). Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source. Int. J. Oral 3, 49–54. doi: 10.4248/IJOS11025
Kolenbrander, P. E., Andersen, R. N., Baker, R. A., Jenkinson, H. F. (1998a). The adhesion-associated sca operon in Streptococcus gordonii encodes an inducible high-affinity ABC transporter for Mn2+ uptake. J. Bacteriol. 180, 290–295.
Kolenbrander, P. E., Andersen, R. N., and Moore, L. V. (1998b). Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect. Immun. 57, 3194–3203.
Kolenbrander, P. E., Andersen, R. N., Blehert, D. S., Egland, P. G., Foster, J. S., and Palmer, R. J. Jr. (2002). Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66, 486–505. doi: 10.1128/MMBR.66.3.486-505.2002
Kolenbrander, P. E., Andersen, R. N., and Ganeshkumar, N. (1994). Nucleotide sequence of the Streptococcus gordonii PK488 coaggregation adhesin gene, scaA, and ATP-binding cassette. Infect. Immun. 62, 4469–4480.
Kolenbrander, P. E., Palmer, R. J. Jr., Periasamy, S., and Jakubovics, N. S. (2010). Oral multispecies biofilm development and the key role of cell–cell distance. Nat. Rev. Microbiol. 8, 471–480. doi: 10.1038/nrmicro2381
Kolenbrander, P. E. Jr., Palmer, R. J., Rickard, A. H., Jakubovics, N. S., Chalmers, N. I., and Diaz, P. I. (2006). Bacterial interactions and successions during plaque development. Periodontol. 2000 42, 47–79. doi: 10.1111/j.1600-0757.2006.00187.x
Koo, H., Xiao, J., Klein, M. I., and Jeon, J. G. (2010). Exopolysaccharides (EPS) produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J. Bacteriol. 192, 3024–3032. doi: 10.1128/JB.01649-09
Kreth, J., Merritt, J., Shi, W., and Qi, F. (2005a). Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 187, 7193–203. doi: 10.1128/JB.187.21.7193-7203.2005
Kreth, J., Merritt, J., Shi, W., and Qi, F. (2005b). Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol. Microbiol. 57, 392–404. doi: 10.1111/j.1365-2958.2005.04695.x
Kuboniwa, M., Hendrickson, E. L., Xia, Q., Wang, T., Xie, H., Hackett, M.,et al. (2009). Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 9:98. doi: 10.1186/1471-2180-9-98
Kuboniwa, M., and Lamont, R. J. (2010). Subgingival biofilm formation. Periodontol. 2000 52, 38–52. doi: 10.1111/j.1600-0757.2009.00311.x
Kuboniwa, M., Tribble, G. D., Lamer, C. E., Killic, A. O., Tao, L., Herzberg, M.,et al. (2006). Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol. Microbiol. 60, 121–139. doi: 10.1111/j.1365-2958.2006.05099.x
Kumar, P. S., Griffen, A. L., Moeschberger, M. L., and Leys, E. J. (2005). Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 43, 3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005
Kuramitsu, H. K., He, X., Lux, R., Anderson, M. H., and Shi, W. (2007). Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71, 653–670. doi: 10.1128/MMBR.00024-07
Kuramitsu, H. K., and Trapa, V. (1984). Genetic exchange between oral streptococci during mixed growth. J. Gen. Microbiol. 130, 2497–2500.
Lamont, R. J., EI-Sabaeny, A., Park, Y., Cook, G. S., Costerton, J. W., and Demuth, D. R. (2002). Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gigivalis biofiloms on streptococcal substrates. Microbiology 148, 1627–1636.
Lancy, P. Jr., Dirienzo, J. M., Appelbaum, B., Rosan, B., and Holt, S. C. (1983). Corncob formation between Fusobacterium nucleatum and Streptococcus sanguis. Infect. Immun. 40, 303–309.
LeBlanc, D. J., Hawley, R. J., Lee, L. N., and Martin, E. J. St. (1978). “Conjugal” transfer of plasmid DNA among oral streptococci. Proc. Natl. Acad. Sci. U.S.A. 75, 3484–3487. doi: 10.1073/pnas.75.7.3484
Li, J., Helmerhorst, E. J., Leone, C. W., Troxler, R. F., Yaskell, T., Haffajee, A. D.,et al. (2004). Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 97, 1311–1318. doi: 10.1111/j.1365-2672.2004.02420.x
Li, Y., and Burne, R. A. (2001a). Regulation of the gtfBC and ftf genes of Streptococcus mutans in biofims in response to pH and carbohydrate. Microbiology 147, 2841–2848.
Li, Y. H., Lau, P. C. Y., Lee, J. H., Ellen, R. P., and Cvitkovitch, D. G. (2001b). Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183, 897–908. doi: 10.1128/JB.183.3.897-908.2001
Liu, B., Faller, L. L., Klitgord, N., Mazumdar, V., Ghodsi, M., Sommer, D. D.,et al. (2012). Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS ONE 7:e37919. doi: 10.1371/journal.pone.0037919
Loesche, W. J. (1986). Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50, 353–380.
Maeda, K., Nagata, H., Nonaka, A., Kataoka, K., Tanaka, M., and Shizukuishi, S. (2004). Oral streptococcal glyceraldehyde-3-phosphate dehydrogenase mediates interaction with Porphyromonas gingivalis fimbriae. Microbes Infect. 6, 1163–1170. doi: 10.1016/j.micinf.2004.06.005
Maeda, K., Tribble, G. D., Tucker, C. M., Anaya, C., Shizukuishi, S., Lewis, J. P.,et al. (2008). A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol. Microbiol. 69, 1153–1164. doi: 10.1111/j.1365-2958.2008.06338.x
Marsh, P. D. (1994). Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 8, 263–271.
Matsumoto-Nakano, M., and Kuramitsu, H. K. (2006). Role of bacteriocin immunity proteins in the antimicrobial sensitivity of Streptococcus mutans. J. Bacteriol. 188, 8095–8102. doi: 10.1128/JB.00908-06
McNab, R., Ford, S. K., El-Sabaeny, A., Barbieri, B., Cook, G. S., and Lamont, R. J. (2003). LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185, 274–284. doi: 10.1128/JB.185.1.274-284.2003
Merritt, J., Qi, F., Goodman, S. D., Anderson, M. H., and Shi, W. (2003). Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71, 1972–1979. doi: 10.1128/IAI.71.4.1972-1979.2003
Meuric, V., Martin, B., Guyodo, H., Rouillon, A., Tamanai-Shacoori, Z., Barloy-Hubler, F.,et al. (2013). Treponema denticola improves adhesive capacities of Porphyromonas gingivalis. Mol. Oral Microbiol. 28, 40–53. doi: 10.1111/omi.12004
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Nadell, C. D., Xavier, J. B., and Foster, K. R. (2009). The sociobiology of bio-films. FEMS Microbiol. Rev. 33, 206–224. doi: 10.1111/j.1574-6976.2008.00150.x
Nagata, H., Iwasaki, M., Maeda, K., Kuboniwa, M., Hashino, E., Toe, M.,et al. (2009). Identification of the binding domain of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase for Porphyromonas gingivalis major fimbriae. Infect. Immun. 77, 5130–5138. doi: 10.1128/IAI.00439-09
Nilius, A. M., Spencer, S. C., and Simonson, L. G. (1993). Stimulation of in vitro growth of Treponema denticola by extracellular growth factors produced by Porphyromonas gingivalis. J. Dent. Res. 72, 1027–1031. doi: 10.1177/00220345930720060601
Ogawa, A., Furukawa, S., Fujita, S., Mitobe, J., Kawarai, T., Narisawa, N.,et al. (2011). Inhibition of Streptococcus mutans biofilm formation by Streptococcus salivarius FruA. Appl. Environ. Microbiol. 77, 1572–1580. doi: 10.1128/AEM.02066-10
Palmer, R. J. Jr., Gordon, S. M., Cisar, J. O., and Kolenbrander, P. E. (2003). Co-aggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 185, 3400–3409. doi: 10.1128/JB.185.11.3400-3409.2003
Pangsomboon, K., Bansal, S., Martin, G. P., Suntinanalert, P., Kaewnopparat, S., and Srichana, T. (2009). Further characterization of a bacteriocin produced by Lactobacillus paracasei HL32. J. Appl. Microbiol. 106, 1928–1940. doi: 10.1111/j.1365-2672.2009.04146.x
Pangsomboon, K., Kaewnopparat, S., Pitakpornpreecha, T., and Srichana, T. (2006). Antibacterial activity of a bacteriocin from Lactobacillus paracasei HL32 against Porphyromonas gingivalis. Arch. Oral Biol. 51, 784–793. doi: 10.1016/j.archoralbio.2006.03.008
Park, Y., James, C. E., Yoshimura, F., and Lamont, R. J. (2006). Expression of the short fimbriae of Porphyromonas gingivalis is regulated in oral bacterial consortia. FEMS Microbiol. Lett. 262, 65–71. doi: 10.1111/j.1574-6968.2006.00357.x
Park, Y., Simionato, M. R., Sekiya, K., Murakami, Y., James, D., Chen, W.,et al. (2005). Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect. Immun. 73, 3983–3989. doi: 10.1128/IAI.73.7.3983-3989.2005
Paster, B. J., Olsen, I., Aas, J. A., and Dewhirst, F. E. (2006). The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000 2, 80–87. doi: 10.1111/j.1600-0757.2006.00174.x
Periasamy, S., and Kolenbrander, P. E. (2009a). Aggregatibacter actinomycetemcomitans builds mutualistic biofilm communities with Fusobacterium nucleaum and Veillonella species in saliva. Infect. Immun. 77, 3542–3551. doi: 10.1128/IAI.00345-09
Periasamy, S., and Kolenbrander, P. E. (2009b). Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J. Bacteriol. 191, 6804–6811. doi: 10.1128/JB.01006-09
Perry, J. A., Cvitkovitch, D. G., and Levesque, C. M. (2009). Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol. Lett. 299, 261–266. doi: 10.1111/j.1574-6968.2009.01758.x
Petersen, F. C., Pecharki, D., and Scheie, A. A. (2004). Biofilm mode of growth of Streptococcus intermedius favored by a competence-stimulating signaling peptide. J. Bacteriol. 186, 6327–6331. doi: 10.1128/JB.186.18.6327-6331.2004
Qi, F., Chen, P., and Caufield, P. W. (2001). The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 67, 15–21. doi: 10.1128/AEM.67.1.15-21.2001
Ramsey, M. M., Rumbaugh, K. P., and Whiteley, M. (2011). Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 7:e1002012. doi: 10.1371/journal.ppat.1002012
Ramsey, M. M., and Whiteley, M. (2009). Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc. Natl. Acad. Sci. U.S.A. 106, 1578–1583. doi: 10.1073/pnas.0809533106
Rice, L. B. (1998). Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42, 1871–1877.
Rickard, A. H., Palmer, R. J., Blehert, D. S., Campagna, S. R., Semmelhack, M. F., Egland, P. G.,et al. (2006). Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 60, 1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x
Riedel, K., Hentzer, M., Geisenberger, O., Huber, B., Steidle, A., Wu, H.,et al. (2001). N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147, 3249–3262.
Roberts, A. P., and Mullany, P. (2010). Oral biofilms: a reservoir of transferable, bacterial, antimicrobial resistance. Expert Rev. Anti Infect. Ther. 8, 1441–1450. doi: 10.1586/eri.10.106
Rölla, G., Scheie, A. A., and Ciardi, J. E. (1985). Role of sucrose in plaque formation. Scand. J. Dent. Res. 93, 105–111.
Rosen, G., Genzler, T., and Sela, M. N. (2008). Coaggregation of Treponema denticola with Porphyromonas gingivalis and Fusobacterium nucleatum is mediated by the major outer sheath protein of Treponema denticola. FEMS Microbiol. Lett. 289, 59–66. doi: 10.1111/j.1574-6968.2008.01373.x
Rosen, G., and Sela, M. N. (2006). Coaggregation of Porphyromonas gingivalis and Fusobacterium nucleatum PK 1594 is mediated by capsular polysaccharide and lipopolysaccharide. FEMS Microbiol. Lett. 256, 304–310. doi: 10.1111/j.1574-6968.2006.00131.x
Rupani, D., Izano, E. A., Schreiner, H. C., Fine, D. H., and Kaplan, J. B. (2008). Aggregatibacter actinomycetemcomitansserotype f O-polysaccharide mediates coaggregation with Fusobacterium nucleatum. Oral Microbiol. Immunol. 23, 127–130. doi: 10.1111/j.1399-302X.2007.00399.x
Ryan, R. P., and Dow, J. M. (2008). Diffusible signals and interspecies communication in bacteria. Microbiology 154, 1845–1858. doi: 10.1099/mic.0.2008/017871-0
Saito, Y., Fujii, R., Nakagawa, K. I., Kuramitsu, H. K., Okuda, K., and Ishihara, K. (2008). Stimulation of Fusobacterium nucleatum biofilm formation by Porphyromonas gingivalis. Oral Microbiol. Immunol. 23, 1–6. doi: 10.1111/j.1399-302X.2007.00380.x
Sandmeier, H., van Winkelhoff, A. J., Bär, K., Ankli, E., Maeder, M., and Meyer, J. (1995). Temperate bacteriophages are common among Actinobacillus actinomycetemcomitansisolates from periodontal pockets. J. Periodontol. Res. 30, 418–425. doi: 10.1111/j.1600-0765.1995.tb01296.x
Sarkar, J., McHardy, I. H., Simanian, E. J., Shi, W., and Lux, R. (2014). Transcriptional responses of Treponema denticola to other oral bacterial species. PLoS ONE 9:e88361. doi: 10.1371/journal.pone.0088361
Schauder, S., and Bassler, B. L. (2001a). The languages of bacteria. Genes Dev. 15, 1468–1480. doi: 10.1101/gad.899601
Schauder, S., Shokat, K., Surette, M. G., and Bassler, B. B. (2001b). The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41, 463–476. doi: 10.1046/j.1365-2958.2001.02532.x
Schuster, M., Lostroh, C. P., Ogi, T., and Greenberg, E. P. (2003). Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185, 2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003
Senadheera, D., and Cvitkovitch, D. G. (2008). Quorum sensing and biofilm formation by Streptococcus mutans. Adv. Exp. Med. Biol. 631, 178–188. doi: 10.1007/978-0-387-78885-2_12
Senadheera, D., Krastel, K., Mair, R., Persadmehr, A., Abranches, J., Burne, R. A.,et al. (2009). Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J. Bacteriol. 191, 6415–6424. doi: 10.1128/JB.00793-09
Shao, H., Lamont, R. J., and Demuth, D. R. (2007). Autoinducer 2 is required for biofilm growth of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect. Immun. 75, 4211–4218. doi: 10.1128/IAI.00402-07
Sharma, A., Inagaki, S., Sigurdson, W., and Kuramitsu, H. K. (2005). Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiol. Immunol. 20, 39–42. doi: 10.1111/j.1399-302X.2004.00175.x
Shimotahira, N., Oogai, Y., Kawada-Matsuo, M., Yamada, S., Fukutsuji, K., Nagano, K.,et al. (2013). The surface layer of Tannerella forsythia contributes to serum resistance and oral bacterial coaggregation. Infect. Immun. 81, 1198–1206. doi: 10.1128/IAI.00983-12
Simionato, M. R., Tucker, C. M., Kuboniwa, M., Lamont, G., Demuth, D. R., Tribble, G. D.,et al. (2006). Porphyromonas gingivalis genes involved in community development with Streptococcus gordonii. Infect. Immun. 74, 6419–6428. doi: 10.1128/IAI.00639-06
Steeves, C. H., Potrykus, J., Barnett, D. A., and Bearne, S. L. (2011). Oxidative stress response in the opportunistic oral pathogen Fusobacterium nucleatum. Proteomics 11, 2027–2037. doi: 10.1002/pmic.201000631
Takahashi, N. (2003). Acid-neutralizing activity during amino acid fermentation by Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol. Immunol. 18, 109–113. doi: 10.1034/j.1399-302X.2003.00054.x
Takasaki, K., Fujise, O., Miura, M., Hamachi, T., and Maeda, K. (2013). Porphyromonas gingivalis displays a competitive advantage over Aggregatibacter actinomycetemcomitans in co-cultured biofilm. J. Periodontol. Res. 48, 286–292. doi: 10.1111/jre.12006
Takahashi, N., Washio, J., and Mayanagi, G. (2010). Metabolomics of supragingival plaque and oral bacteria. J. Dent. Res. 89, 1383–1388. doi: 10.1177/0022034510377792
Tamesada, M., Kawabata, S., Fujiwara, T., and Hamada, S. (2004). Synergistic effects of streptococcal glucosyltransferases on adhesive biofilm formation. J. Dent. Res. 83, 874–879. doi: 10.1177/154405910408301110
Tamura, S., Yonezawa, H., Motegi, M., Nakao, R., Yoneda, S., Watanabe, H.,et al. (2009). Inhibiting effects of Streptococcus salivarius on competence-stimulating peptide-dependent biofilm formation by Streptococcus mutans. Oral Microbiol. Immunol. 24, 152–161. doi: 10.1111/j.1399-302X.2008.00489.x
Tong, H., Chen, W., Merritt, J., Qi, F., Shi, W., and Dong, X. (2007). Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol. Microbiol. 63, 872–880. doi: 10.1111/j.1365-2958.2006.05546.x
Tong, H., Chen, W., Shi, W., Qi, F., and Dong, X. (2008). SO-LAAO, a novel L-amino acid oxidase that enables Streptococcus oligofermentans to outcompete Streptococcus mutans by generating H2O2 from peptone. J. Bacteriol. 190, 4716–4721. doi: 10.1128/JB.00363-08
Tong, H., Gao, X., and Dong, X. (2003). Streptococcus oligofermentans sp. nov., a novel oral isolate from caries-free humans. Int. J. Syst. Evol. Microbiol. 53, 1101–1104. doi: 10.1099/ijs.0.02493-0
Torres-Escobar, A., Juárez-Rodríguez, M. D., Lamont, R. J., and Demuth, D. R. (2013). Transcriptional regulation of Aggregatibacter actinomycetemcomitans lsrACDBFG and lsrRK operons and their role in biofilm formation. J. Bacteriol. 195, 56–65. doi: 10.1128/JB.01476-12
Vacca-Smith, A. M., and Bowen, W. H. (1998). Binding properties of streptococcal glucosyltransferases for hydroxyapatite, saliva-coated hydroxyapatite, and bacterial surfaces. Arch. Oral Biol. 43, 103–110. doi: 10.1016/S0003-9969(97)00111-8
van der Hoeven, J. S., and Camp, P. J. (1991). Synergistic degradation of mucin by Streptococcus oralis and Streptococcus sanguis in mixed chemostat cultures. J. Dent. Res. 70, 1041–1044. doi: 10.1177/00220345910700070401
Vollaard, E., and Clasener, H. (1994). Colonization resistance. Antimicrob. Agents Chemother. 38, 409–414. doi: 10.1128/AAC.38.3.409
Wagner, V. E., Bushnell, D., Passador, L., Brooks, A. I., and Iglewski, B. H. (2003). Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185, 2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003
Wang, B. Y., Alvarez, P., Hong, J., and Kuramitsu, H. K. (2011a). Periodontal pathogens interfere with quorum-sensing-dependent virulence properties in Streptococcus mutans. J. Periodontol. Res. 46, 105–110. doi: 10.1111/j.1600-0765.2010.01319.x
Wang, B. Y., Deutch, A., Hong, J., and Kuramitsu, H. K. (2011b). Proteases of an early colonizer can hinder Streptococcus mutans colonization in vitro. J. Dent. Res. 90, 501–505. doi: 10.1177/0022034510388808
Wang, B. Y., and Kuramitsu, H. K. (2005). Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl. Environ. Microbiol. 71, 354–362. doi: 10.1128/AEM.71.1.354-362.2005
Wang, Q., Wright, C. J., Dingming, H., Uriarte, S. M., Lamont, R. J. (2013). Oral community interactions of Filifactor alocis in vitro. PLoS ONE 8:e76271. doi: 10.1371/journal.pone.0076271
Warburton, P. J., Palmer, R. M., Munson, M. A., and Wade, W. G. (2007). Demonstration of in vivo transfer of doxycycline resistance mediated by a novel transposon. J. Antimicrob. Chemother. 60, 973–980. doi: 10.1093/jac/dkm331
Whitmore, S. E., and Lamont, R. J. (2011). The pathogenic persona of community-associated oral streptococci. Mol. Microbiol. 81, 305–314. doi: 10.1111/j.1365-2958.2011.07707.x
Wickström, C., Herzberg, M. C., Beighton, D., and Svensäter, G. (2009). Proteolytic degradation of human salivary MUC5B by dental biofilms. Microbiology 155, 2866–2872. doi: 10.1099/mic.0.030536-0
Xavier, K. B., Bassler, B. L., Xavier, K. B., Miller, S. T., Lu, W., Kim, J. H.,et al. (2007). Phosphorylation and processing of the quorum-sensing molecule autoinducer-2 in enteric bacteria. ACS Chem. Biol. 2, 128–136. doi: 10.1021/cb600444h
Xiao, J., Klein, M. I., Falsetta, M. L., Lu, B., Delahunty, C. M., and Yates, J. R. III,et al. (2012). The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 8:e1002623. doi: 10.1371/journal.ppat.1002623
Xie, H., Lin, X., Wang, B. Y., Wu, J., and Lamont, R. J. (2007). Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology 153, 3228–3234. doi: 10.1099/mic.0.2007/009050-0
Yeung, M. K. (1999). Molecular and genetic analysis of Actinomyces species. Oral Biol. Med. 10, 120–138. doi: 10.1177/10454411990100020101
Yim, G., Wang, H. H., and Davies, J. (2006). The truth about antibiotics. Int. J. Med. Microbial. 296, 163–170. doi: 10.1016/j.ijmm.2006.01.039
Yuan, L., Hillman, J. D., and Progulske-Fox, A. (2005). Microarray analysis of quorum sensing regulated genes in Porphyromonas gingivalis. Infect. Immun. 73, 4146–4154. doi: 10.1128/IAI.73.7.4146-4154.2005
Zainal-Abidin, Z., Veith, P. D., Dashper, S. G., Zhu, Y., Catmull, D. V., Chen, Y. Y.,et al. (2012). Differential proteomic analysis of a polymicrobial biofilm. J. Proteome Res. 11, 4449–4464. doi: 10.1021/pr300201c
Zhang, Y., Lei, Y., Khammanivong, A., and Herzberg, M. C. (2004). Identification of a novel two-component system in Streptococcus gordonii V288 involved in biofilm formation. Infect. Immun. 72, 3489–3494. doi: 10.1128/IAI.72.6.3489-3494.2004
Zheng, X., Zhang, K., Zhou, X., Liu, C., Li, M., Li, Y.,et al. (2013). Involvement of gshAB in the interspecies competition within oral biofilm. J. Dent. Res. 92, 819–824. doi: 10.1177/0022034513498598
Zhu, L., and Kreth, J. (2010). Role of Streptococcus mutans eukaryotic-type serine/threonine protein kinase in interspecies interactions with Streptococcus sanguinis. Arch. Oral Biol. 55, 385–390. doi: 10.1016/j.archoralbio.2010.03.012
Zhu, L., and Kreth, J. (2012). The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid. Med. Cell Longev. 2012:717843. doi: 10.1155/2012/717843
Keywords: oral microbial community, coadhesion, signaling transduction, metabolic interactions, cell-cell communication
Citation: Guo L, He X and Shi W (2014) Intercellular communications in multispecies oral microbial communities. Front. Microbiol. 5:328. doi: 10.3389/fmicb.2014.00328
Received: 29 March 2014; Paper pending published: 16 May 2014;
Accepted: 14 June 2014; Published online: 01 July 2014.
Edited by:
Manfred Auer, Lawrence Berkeley Lab, USAReviewed by:
Hui Wu, University of Alabama at Birmingham, USARichard Lamont, University of Louisville, USA
Copyright © 2014 Guo, He and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenyuan Shi, School of Dentistry, University of California-Los Angeles, 10833 Le Conte Avenue, Los Angeles, CA 90095, USA e-mail: wenyuan@ucla.edu
 Lihong Guo
Lihong Guo Xuesong He
Xuesong He Wenyuan Shi
Wenyuan Shi