- 1Biotechnology Graduate Program, Biology Department and YJ-Science and Technology Research Center, American University in Cairo, New Cairo, Egypt
- 2Tourism Development Authority, Ministry of Tourism, Cairo, Egypt
The Red Sea possesses a unique geography, and its shores are rich in mangrove, macro-algal and coral reef ecosystems. Various sources of pollution affect Red Sea biota, including microbial life. We assessed the effects of industrialization on microbes along the Egyptian Red Sea coast at eight coastal sites and two lakes. The bacterial communities of sediment samples were analyzed using bacterial 16S rDNA pyrosequencing of V6-V4 hypervariable regions. The taxonomic assignment of 131,402 significant reads to major bacterial taxa revealed five main bacterial phyla dominating the sampled sites: Proteobacteria (68%), Firmicutes (13%), Fusobacteria (12%), Bacteriodetes (6%), and Spirochetes (0.03%). Further analysis revealed distinct bacterial consortia that primarily included (1) marine Vibrio spp.—suggesting a “marine Vibrio phenomenon”; (2) potential human pathogens; and (3) oil-degrading bacteria. We discuss two divergent microbial consortia that were sampled from Solar Lake West near Taba/Eilat and Saline Lake in Ras Muhammad; these consortia contained the highest abundance of human pathogens and no pathogens, respectively. Our results draw attention to the effects of industrialization on the Red Sea and suggest the need for further analysis to overcome the hazardous effects observed at the impacted sites.
Introduction
The Red Sea possesses a unique geography, as it is almost entirely locked by land, and its ecosystems are diverse, including mangrove, macro-algae and coral reefs (Alkershi and Menon, 2011). The Red Sea encompasses two gulfs, the Gulf of Suez and the Gulf of Aqaba, in addition to the Red Sea proper. The Gulf of Suez is entirely bordered by Egypt, while the borders of the Gulf of Aqaba are shared among four countries: Egypt, Israel, Jordan, and Saudi Arabia. The Red Sea proper is bordered by six countries: Egypt, Sudan, Eritrea, and Djibouti on the western shore and Saudi Arabia and Yemen on the eastern shore.
Severe pollution of the Red Sea has been reported multiple times from Egypt (Riegl and Velimirov, 1991; Ibrahim et al., 2011; El-Sorogy et al., 2013; El-Taher and Madkour, 2013), Israel (Loya and Rinkevich, 1980; Riegl and Velimirov, 1991; Loya, 2004; Abelson et al., 2005), Yemen (Alkershi and Menon, 2011), Saudi Arabia (Hanna and Muir, 1990; Badr et al., 2009; Montaser et al., 2010; Ali et al., 2011), Jordan (Al-Najjar et al., 2011), Sudan (Idrisa et al., 2007) and Eritrea (United Nations Development Programme, 2007). Pollution issues in the Red Sea are increasing in severity for several reasons: the Red Sea's small size (458,620 km2); the fact that it is bordered by eight countries; and its slow rate of water turnover (Medio et al., 2000).
The sources of pollution in the Red Sea include land-based sources (including urban development, industrial activities, dredging and filling, tourism and agriculture activities), oceanic sources (shipping, fishing, marine traffic and petroleum industries), and atmospheric sources (industries or port activities). Such severe pollution is likely to affect biological life and disturb the Red Sea's natural ecosystems (Regional Organization for the Conservation of the Environment of the Red Sea and Gulf of Aden, 2001). One of the major pollution threats studied is the health of the coral reefs and their ecosystem (Pandolfi et al., 2003; El-Sorogy et al., 2012). Reports on coral reef degradation and the impairment of coral growth and reproduction through algal overgrowth, increased sedimentation and coral disease, have been reported worldwide. Red Sea coral reefs constitute 3.8% of the worlds' coral reefs (PERSGA/GEF, 2004). In an assessment of 21 mangrove sites along the Gulf of Aqaba and the Egyptian Red Sea coastlines, covering ~550 hectares, mangrove degradation was reported from Egypt as a result of oil pollution, tourism, camel grazing and browsing and solid waste accumulation, primarily of plastics (PERSGA/GEF, 2004).
However, equally important is the effect of pollution on microbial life, a topic that has not been well studied in the Red Sea. Two studies have addressed human pathogens along the Red Sea coast (El-Shenawy and Farag, 2005; Ibrahim et al., 2011). In one study, the researchers measured the abundance of saprophytic (SB), salt-tolerant saprophytic (STSB), total coliforms (TC), Escherichia coli (EC) and fecal streptococci (FS) in 40 sites along the Egyptian Red Sea coastal waters and in the Aqaba and Suez Gulfs. Of these samples, 91.5% met “European and Egyptian current standards” (El-Shenawy and Farag, 2005). In 2011, TC, EC, and FS in water samples were used as indicators of microbial pollution. This study reported that the Suez Gulf is the most polluted among 194 sites in the Red Sea and Gulf of Aqaba (El-Shenawy and Farag, 2005; Ibrahim et al., 2011).
Conversely, the Red Sea's Egyptian coast has been better studied from perspectives other than the effect of pollution on the microbial life. For example, in 2005, the geochemistry of the sediments and seawater of four Red Sea lagoons were analyzed. The fauna of the Abu Ghoson lagoon was degraded due to the excessive shipping of phosphates, illmenites and feldspars through the port. Safaga Port encounters more than one pollution source causing heavy metal accumulation, including phosphate shipping, adjacent cement industry, land filling, navigation and construction activities, and shipyards (Abd El-Wahab et al., 2005; Mohamed, 2005). Safaga was shown to have the highest concentration of Fe, Pb, Mn, and Zn among the analyzed sites (Abd El-Wahab et al., 2005). The highest concentrations of “P” and “V” were detected in Qusseir Port, and phosphates were detected in high concentrations in its sediments (Mohamed, 2005). Similarly, Hamrawein Port sediments were reported to show distinctive brown coloration, which is characteristic of “P” presence. This is not surprising because Hamrawein Port is one of the oldest harbors for shipping phosphate (Mohamed et al., 2011). Sharm El-Maya is a shallow bay (~6 m deep) that is located in the southern suburb of Sharm El-Sheikh, and its southern reaches are connected to the Red Sea. This bay has been exposed to several oilspill accidents, including the accidental spill of 700 tons of fuel from a cargo ship in 1983 (Roberts and Sheppard, 1988; Khattab et al., 2006), the 1994 oil spill in Sharm El-Sheikh (Pilcher and Abou Zaid, 2000) and the 1999 oil spill accident in Sharm El-Maya (Morsy et al., 2010). All oil spills represent a major threat to the bay ecosystem, including sea grass and coral patches. Additionally, the bay acts as a nursery for commercially valuable fish (Morsy et al., 2010). The combination of the oil spills with the structure and ecosystem of this bay resulted in the entrapment of sediments and oil particles, which caused deleterious effects on coral reef reproduction and the photosynthetic cycle (Loya and Rinkevich, 1980; Al-Halasah and Ammary, 2007).
In addition to the coastal sites of the Red Sea, lakes near the sea can be affected by industrialization. In this study, we analyzed two lakes: Solar Lake at the Gulf of Aqaba (Solar Lake-W) and Saline Lake inside the Ras Muhammed National Park protected area (Saline Lake-RM). Solar Lake-W was selected for this study because it is the only Egyptian Red Sea site where the microbial community has been thoroughly studied. Saline Lake-RM is a petroleum-impacted site that has limited impact from tourism, which allowed us to assess one pollution impact in isolation.
Solar Lake was discovered in 1967 by the workers of the Eilat Nature Reserve (Por, 1968; Eckstein, 1970). They reported the presence of hot water at the bottom of the lake, which was interpreted as hot brines. Later, it was attributed to solar radiation (Eckstein, 1970). When the lake was discovered, it was reported to have the dimensions 80 × 40 m and to be 30 m away from the Red Sea coast, and it was characterized as a meromictic lake (Por, 1968).
Solar Lake is rich in H2S and was thought to release high concentrations of H2S compared with the production reported from non-polluted waters (Cohen et al., 1975). This has been attributed to the activity of cyanobacterial mats (Cohen et al., 1975). Several strains of cyanobacteria have been isolated from Solar Lake. In 1975, Oscillatoria limnetica was isolated from the H2S-rich layer of the lake (Cohen et al., 1975). Cyanobacterial laminites was detected in different layers of the lake and aided in tracking the age and history of the lake (Cohen et al., 1977). Cyanobacteria control the diurnal cycle of the lake through photosynthesis and O2–H2S regulation (Jorgensen et al., 1979b). Cyanobacteria anoxygenic photosynthesis is a major contributor to elemental sulfur production and sulfur cycling in the lake (Jorgensen et al., 1979a). Dactylococcopsis salina, a gas vacuolated cyanobacterium, was also isolated from the Solar Lake (Walsby et al., 1983). Additionally, a novel species of Desulfovibrio oxyclinae was isolated from the Solar Lake cyanobacterial mat. This species was demonstrated to adapt to wide variation in oxygen and sulfide concentrations (Krekeler et al., 1997). Archaeal 16S rDNA analysis demonstrated that halobacteria dominate the archaeal community of the lake and halophilic methanogens were identified in the sulfide- and methane-rich layer (Cytryn et al., 2000).
Few studies have addressed microbial life in Saline Lake-RM. A Gram-negative, haloalkaliphilic and facultative anaerobic bacteria, Salinivibrio sharmensis, and Halomonos sinaiensis were isolated from Saline Lake (Romano et al., 2007, 2011).
Much work in the Red Sea has focused on visible marine pollution, its various sources and its effects on coral reefs, mangroves and fisheries. However, neglected topics include research on microbial life in Red Sea sediments or water and how pollution affects the distribution and abundance of the microbial communities. Here, we taxonomically identify microbial communities cultured from sediment samples collected at sites that have been impacted by petroleum, industry and tourism.
Materials and Methods
Site Description and Sample Collection
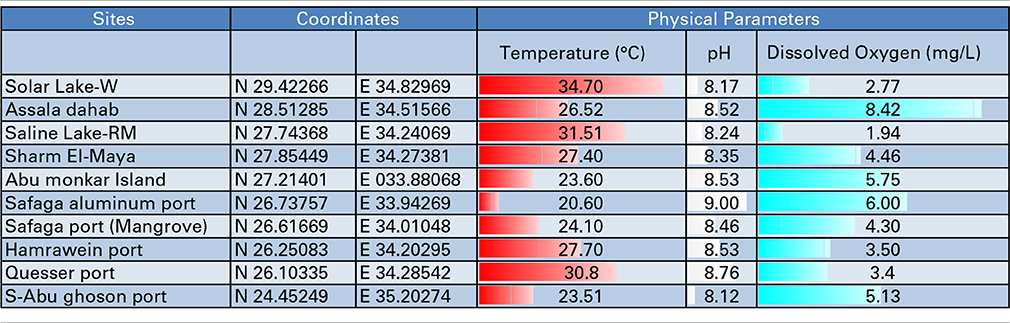
Eight coastal sites and two lakes on the Red Sea Egyptian coast were selected for sampling based on the industrial impacts affecting each (Table 1, Figure 1). Six of the ten sites lie on the Red Sea proper, two are on the Sinai Peninsula and two are on the Gulf of Aqaba. The ten analyzed sites included four ports for shipping aluminum (Safaga Aluminum Port), ilmenite (S-Abu Ghoson Port) and phosphate (Qusseir Port and Hamrawein Port), a site previously reported to have suffered oil spills (Sharm El-Maya) and a tourism impacted site (Assala Dahab). Two sites were considered to be non-impacted sites: a protected site (Abu Monkar Island) and a protected mangrove area (Safaga Port-mangrove), which showed oil deposits. The two lakes (Saline Lake-RM and Solar Lake-W) showed different impacts. Saline Lake, which is inside Ras Muhammed, was thought to be a protected area; however we found extensive oil deposits evident in the soft sediments of its dense salt marches. Solar Lake-W is the west shore of the Lake and lies in the direction of the mountains, not toward the rift. The water on the west side of the lake was characterized by green coloration and H2S odor, and salt deposits surrounded the lake. Core sampling on the western side of the lake released black fumes (data not shown). Surface sediment samples were collected from the eight sites along the Egypt Red Sea coast in addition to the two lakes in Sinai Peninsula (Table 1, Figure 1). The samples were collected using a basic homemade stainless steel core (5 cm diameter/0.5 m length) and an AMS Multi-Stage Sludge and Sediment Sampler (using one 12″ plastic liner; cat. no. 403.31). The samples were collected near the shore at depths ranging from 0.5 to 1 meter from the sea surface. The middle part of the core (~0.25 m depth) was taken for further analysis to minimize the contamination from the seawater or the shore sand during the on-site handling process.
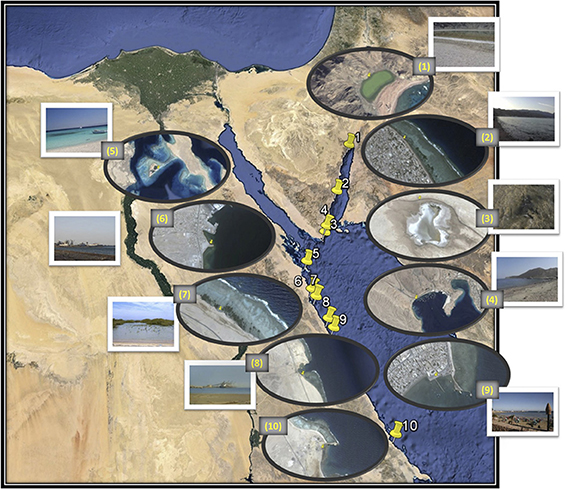
Figure 1. Map of the locations of the eight coastal sites (numbered 2, 4, and 5–10) and the two lakes (numbered 1 and 3). Image generated using Google, Image Landsat Data SIOA, NOAA, U.S. Navy, NGA, GEBCO. The adjacent zoomed image was generated by Image © 2014 DigitalGlobe (sample sites 1, 5, and 6), Image © 2014 CNES/Astrium (sample sites 2 and 4), Data SIOA, NOAA, U.S. Navy, NGA, GEBCO, Image © 2014 CNES/Astrium (sample site 3) and Image © 2014 CNES/Astrium, Data SIOA, NOAA, U.S. Navy, NGA, GEBCO, Image © 2014 TerraMetrics (sample sites 7, 8, 9, and 10). Sample sites: 1. Solar Lake-W, 2. Assala-Dahab, 3. Saline Lake-RM, 4. Sharm El-Maya, 5. Abu-MonkarIsland, 6. Safaga Port (Aluminum), 7. Safaga Port-mangrove, 8. Hamrawein Port, 9. Qusseir port and 10.S-Abu Ghoson port.
Bacterial Culturing and DNA Preparation
A few grams (~5 g) of the collected sediments was inoculated, on site, directly in 20-mL of freshly prepared Difco™ Marine Broth 2216, using 50 ml falcon tubes to allow for aeration. The cultures were incubated with random mixing for 3 days at room temperature. After delivering the cultures to the lab, the cultures were inverted several times, and 1 ml was taken from each culture for bacterial DNA extraction. The DNA was extracted using QIAamp® DNA Blood Mini Kit (cat no. 51106) following the Protocol for Bacteria in the kit's mini-handbook. The prepared DNA was kept at −80°C for sequencing.
PCR Amplification, 454 Pyrotag Sequencing and 16S rDNA Analysis
For bacterial taxonomic assignment, prepared genomic bacterial DNA was used to amplify the bacterial 16S rDNA hyper-variable regions V6 and V4as previously described (Sogin et al., 2006). The bacterial primers utilized in this study have been described (Siam et al., 2012). The amplicons recovered were subjected to pyrosequencing by 454 GS FLX Titanium technology (454 Life Sciences). V6-V4 reads were deposited in NCBI SRA under the accessions SRR1437688-SRR1437697.
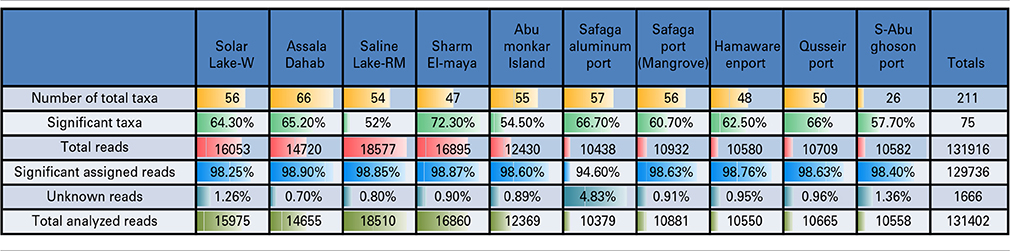
The resources on the Visualization and Analysis of Microbial Population Structures (VAMPS) website, hosted by the Josephine Bay Paul Center, MBL, Woods Hole (http://vamps.mbl.edu/resources/databases.php) were used for the phylogenetic analysis and taxonomic assignment of the reads to major bacterial taxa. Fisher's exact test was used to determine the species/genera that differed significantly in abundance across the different sites (p < 0.05, Bonferroni-corrected). The total number of raw reads (no significance test applied) and the number of assigned taxa are presented in Table 2. The significant reads are the reads that passed the cut-off value of the former test. We selected known taxa from these significant reads (significant taxa).
Results
Samples and Physical Parameters
We attempted to characterize the microbial community along the Egyptian Red Sea coast and two lakes, as illustrated in Figure 1. The coordinates, temperature gradients, pH and dissolved oxygen of the sampling sites are illustrated in Table 1. We started in Solar Lake-W, which is situated near the border of Egypt and Israel on the Gulf of Aqaba (northeastern Egypt), and ended with S-Abu Ghoson Port at the south coast of the Red Sea (southern Egypt). We observed the highest temperatures in the two lakes investigated, Solar Lake-W and Saline Lake-RM, measuring 34.7 and 31.7°C, respectively. The lowest temperature (20.6°C) and highest pH (9.00) were detected in SafagaPort (Aluminum). The remaining sites showed pH ranges of 8.5 ± 0.085. Solar Lake-W and Saline Lake-RM had higher salinity than did the coastal sites (Edwards and Head, 1987; Thompson et al., 2013); the lakes' measured 107.9 and 149.8 ppt, respectively. More variation in the dissolved oxygen was observed in our samples, with Assala-Dahab showing the highest dissolved oxygen saturation (8.42 mg/L), followed by Safaga Port (Aluminum) and Abu-Monkar Island, measuring 6.00 and 5.75 mg/L, respectively (Table 1). Aside from Assala-Dahab, in which the oxygen saturation is considered to be greater than the saturation level of the Red Sea, the dissolved oxygen saturation level in the remaining sites was within the previously reported range (4.8–6.5 ml/L; Institute of Marine Research, 2012).
Pyrotag 16S rDNA Diversity and Taxonomic Assignments
A total of 131,916 reads were generated from the cultured Red Sea coastal sediments and the two lakes using pyrotag sequencing. The taxonomic assignment of the reads to major bacterial genera detected 211 different genera. Following significance testing, we concluded that only 75 genera were significantly detected; these were represented by a total of 131,402 reads. Only 1.3% of these were unassigned reads, including both unassigned bacteria and unassigned organisms. The label “unassigned organism” indicates a taxon of unknown bacteria, archaea or eukarya (Table 2). We compared the diversity of bacterial phyla reads across the sites. Using the media and conditions described in the materials and methods section, five major bacterial phyla were cultured from the 10 sites sampled, predominantly Proteobacteria (68%), followed by Firmicutes (13%), Fusobacteria (12%), Bacteriodetes (6%) and Spirochetes (0.03%) (Figure 2). Members of Proteobacteria included γ-proteobacteria (92%), followed by δ–proteobacteria (7%) and ε-proteobacteria (1%) (Figure 2, Table 3). Very low proportions of unassigned organisms were detected across sites (0.9% collectively).
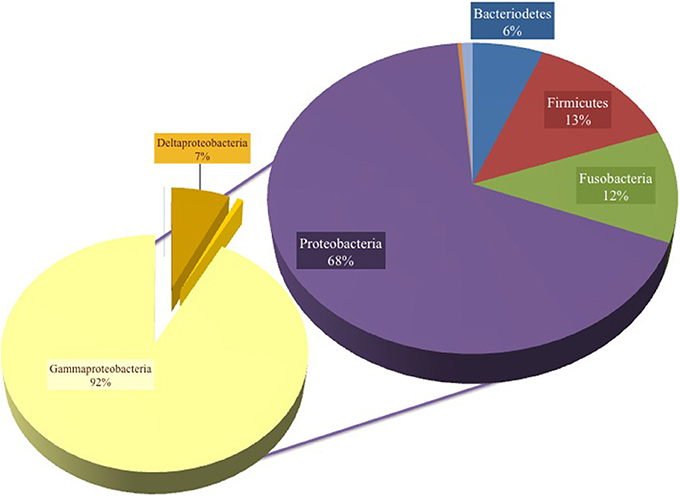
Figure 2. Pie chart representation of the total cultured phyla in all samples, showing 68% Proteobacteria, 13% Firmicutes, 12% Fusobacteria, 6% Bacteroidetes and unknown bacteria and organism phyla (percent abundance not shown). The composition was 92% γ-proteobacteria, 7% δ-proteobacteria and the remaining Proteobacteria were assigned as ε-proteobacteria.
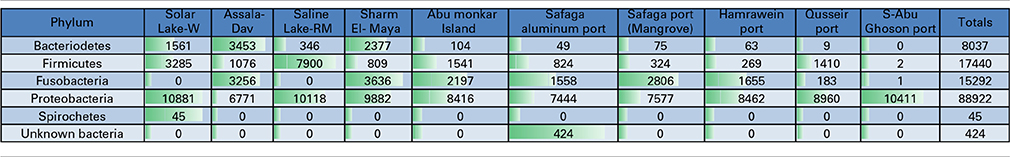
Table 3. Total number of reads assigned at the phylum level in Red Sea coastal samples and two lakes.
Proteobacteria-assigned reads predominated at all of the sampled sites. No other phyla were significantly detected in S-Abu Ghoson Port. Fusobacteria were not detected in the two lakes. Firmicutes represent the second most abundant phylum in the two lakes and Qusseir Port. Bacteriodetes, Firmicutes and Fusobacteria were detected in the remaining coastal samples, but in varying abundances. For example, Assala-Dahab showed the highest incidence of Bacteriodetes (43/23.6% total Bacteriodetes/total culture), followed by Sharm El-Maya (30/14.1% total Bacteriodetes/total culture). Sharm-El-Maya showed the highest incidence of Fusobacteria (23.7%), followed by Assala-Dahab (21.3%). Fusobacteria was the second most abundant phylum in all of the sites, except Qusseir Port, S-Abu Ghoson Port and the two lakes. Unassigned organisms were the second most abundant phylum in S-Abu Ghoson Port. Interestingly, all reads for Spirochetes were detected in Solar Lake-W. A significant number of reads (424) representing 0.3% of the total analyzed reads were assigned as unknown bacterial phylum; these were only detected in Safaga Port (Aluminum) (Table 3).
Bacterial Consortia Along the Red Sea Coast
An average of 34 ± 1.6 reads were assigned to the remaining seven Red Sea Coast samples, with rare reads constituting 28 ± 1.5. The S-Abu Ghoson Port reads were unique from the other coastal sites: 15 bacterial reads were identified, of which 11 were considered rare. Unknown species of Photobacterium (50%) and Photobacterium halotolerans (24%) predominated among the bacterial-assigned reads in S-Abu Ghoson Port. Unassigned Vibrio spp. predominated among the reads in the remaining Red Sea Coast samples (53.4%), followed by Propionigenium maris (15.4%). Qusseir Port and S-Abu Ghoson Port were exceptions (Table S1).
Table 4A shows a preliminary taxonomic assignment of V6-V4 reads to the previously reported pathogenic bacteria (strictly infecting humans). Note that the V6-V4 reads are relatively short for assigning taxa at the species level. However, we observed that Clostridium botulinum and three assigned species of Vibrio represented the pathogenic bacteria detected in the cultures of these Red Sea sites. In total, Vibrio parahaemolyticus was the most abundant pathogenic bacterium (48.5/0.67% of the total pathogenic bacteria/total reads), followed by Clostridium botulinum (35.8/0.5% of the total pathogenic bacteria/total reads). The pattern of the pathogenic bacteria (distribution and abundance) in the Solar Lake-W was different from the remaining sites, as Clostridium botulinum was detected only at this site. As expected, the taxonomic assignment of Saline Lake-RM reads did not match any known pathogenic bacteria, followed by Abu-Monkar Island and Assala-Dahab, which showed the lowest abundance of reads to pathogenic bacteria (0.8/0.1 and 2.2/0.3% of the total detected pathogenic bacteria/the site's total cultured bacteria, respectively).
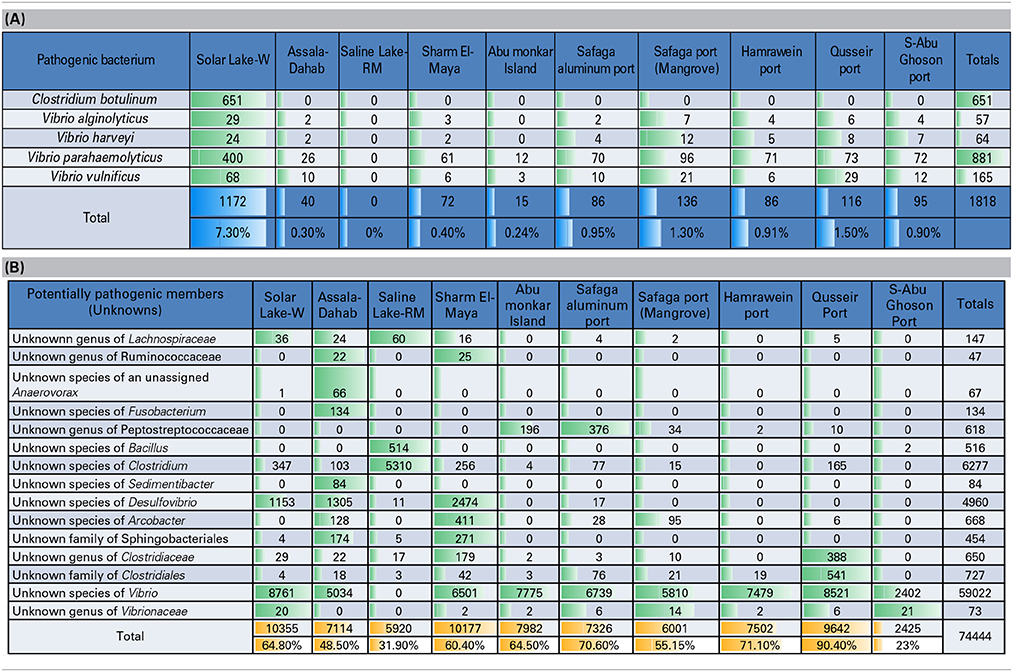
Table 4. (A) Preliminary assignment of total number of reads to known human pathogens, (B) Total number of reads assigned to potential pathogens (previously reported genera/species included human pathogens and non-pathogens).
We also detected reads for which previously identified genera/species are reported to include both pathogenic and non-pathogenic members. We refer to these as potential pathogens (Table 4B). Our culture approach has detected these potentially pathogenic bacteria, including five unknown families of Lachnospiraceae, Ruminococcaceae, Peptostreptococcaceae, Clostridiaceae and Vibrionaceae. Additionally, eight unknown species of unassigned Anaerovorax, Fusobacterium, Bacillus, Clostridium, Sedimentibacter, Desulfovibrio, Arcobacter, and Vibrio. Additionally, two unknown orders of Sphingobacteriales and Clostridiales were detected, and we categorized them as potentially pathogenic bacteria.
Solar Lake-W and Saline Lake-RM Bacterial Consortia
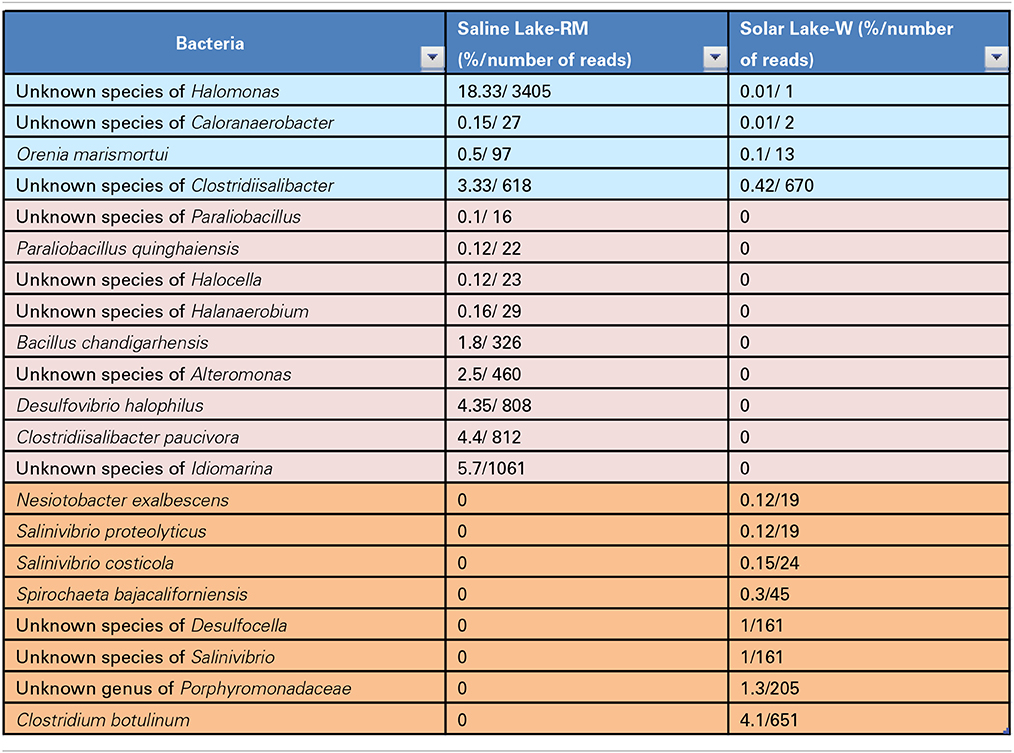
In total, 36 and 28 bacterial reads were cultured and amplified from the Solar Lake-W and Saline Lake-RM, respectively. Four bacterial reads were unique to these two lakes, including reads assigned to Orenia marismortui and unknown species of Caloranaerobacter, Clostridiisalibacter and Halomonas. Eight and nine reads were unique to Solar Lake-W and Saline Lake-RM, respectively (Table 5). Of the 36 bacterial reads cultured from Solar Lake-W, 21 are considered rare bacterial reads (less than 1%). Unknown species of Vibrio dominated the cultured community (55%). The remaining reads constituted six species of Vibrio (Vibrio parahaemolyticus-2.5%; Figure 3). Surprisingly, 9% of the culture was assigned to the genus Clostridium, with 4.1% assigned as Clostridium botulinum. Seven percent of the culture was assigned to Desulfovibrio and 4% to Clostridibacter. Conversely, an unknown genus of Marinlabiacae and Geosporobacter genus represented 7% and 5% of the total bacterial reads of this lake, respectively (Figure 3).
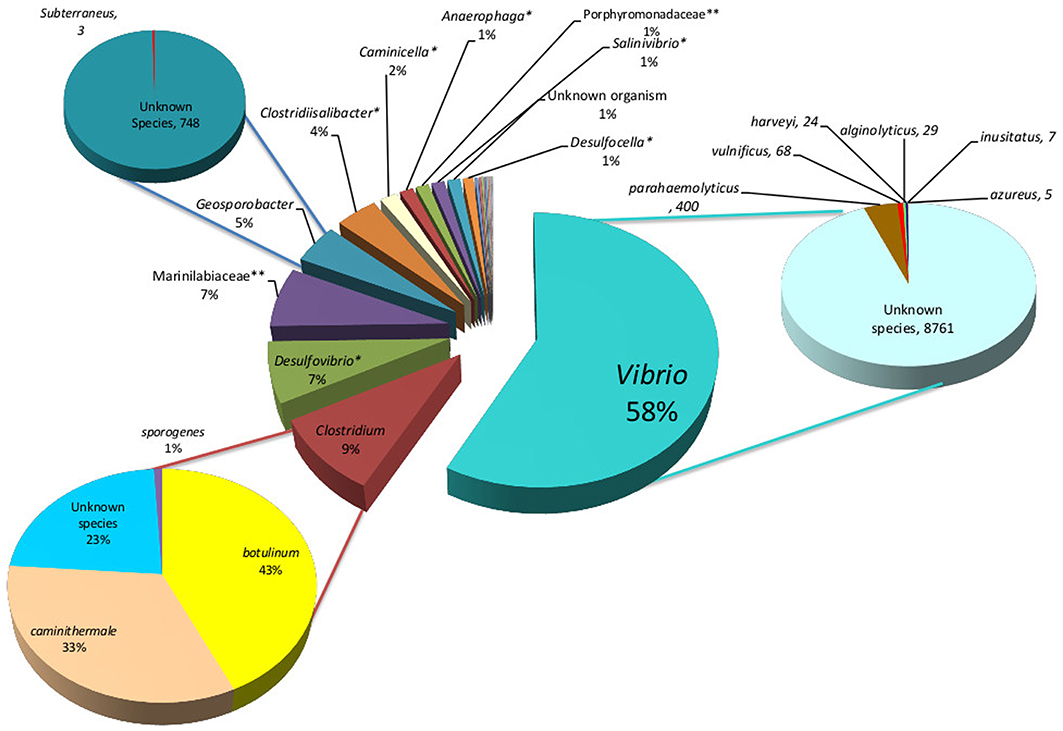
Figure 3. Solar Lake West genera are presented in the middle pie chart, predominated by Vibrio (58%), Clostridium (9%), unknown species of Desulfovibrio (7%) and Geosporobacter (5%). The Vibrio, Desulfovibrio and Geosporobacter are predominantly unknown species. The predominant Clostridium species is C. botulinum (43%). *unknown species;**unknown genus.
Of the 28 bacterial reads in the Saline Lake-RM, 17 were considered to be rare bacterial taxa (less than 1%). Saline Lake-RM was dominated by an unknown species of Clostridium (29%), followed by an unknown species of Marinobacter (24%), an unknown species of Halomonas (18%), and an unknown species of Idiomarina (5.7%). Only six bacterial reads were assigned at the species level: Clostridiisalibacter paucivorans (4.3%), Desulfovibrio halophilus (4.3%), Bacillus chandigarhensis (1.8%), Orenia marismortui (0.5%), Paraliobacillus quinghaiensis (0.12%), and Clostridium caminithermale (0.01%).
Four more unknown species were significantly detected: unknown species of Clostridiisalibacter, Bacillus, Alteromonas and Anaerophaga account for 3.3, 2.8, 2.5, and 1.8% of assigned reads, respectively. The rare bacterial taxa (below 1%) include two species, nine genera, two families and two orders (Figure 4).
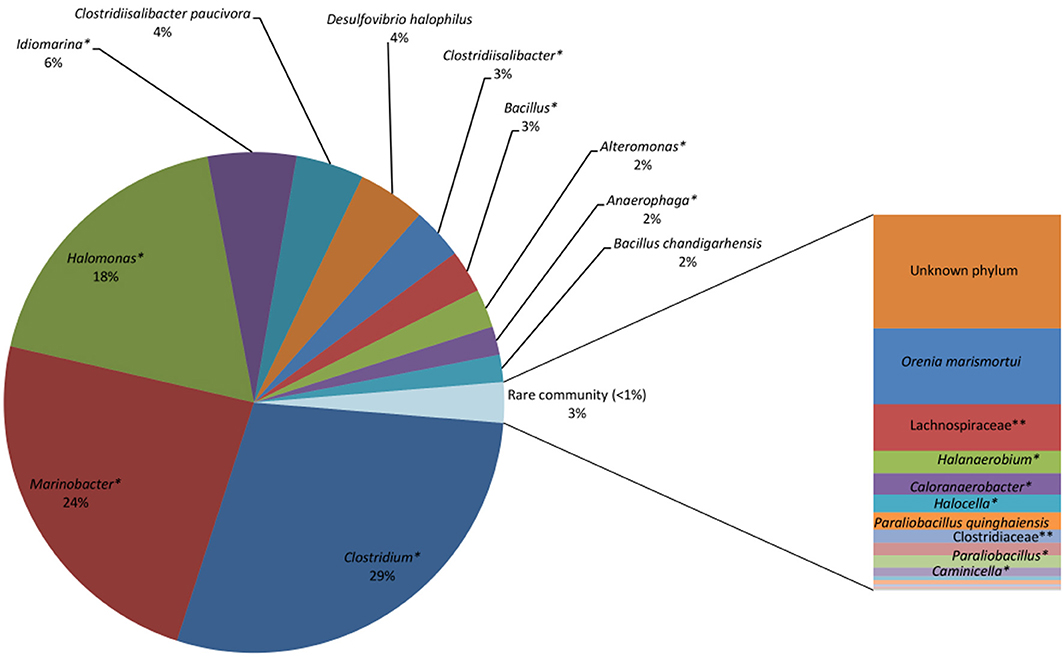
Figure 4. Saline Lake bacterial consortia showing predominantly unknown species, including unknown species (*) of Clostridium (29%), Marinobacter (24%), Halomonas (18%), Idiomarina (6%), Clostridiisalibacter (3%), Bacillus (3%), and Alteromonas (2%). Known species identified include Clostridiisalibacterpaucivora (4%), Desulfovibrio halophiles (4%) and Bacillus chandigarhensis (2%). *Unknown species
Discussion
We analyzed the microbial community in sites that have been impacted by, land-based, oceanic and atmospheric pollution sources along the Red Sea coast. Additionally, we assessed the microbial community in two lakes on the Sinai Peninsula: Solar Lake-W and the Saline Lake-RM, which are believed to have seeps from the Red Sea (Aharon et al., 1977). Following sediment cultures, we used the V6-V4 hypervariable region and amplified a significant number of pyrotags; 131,916 16S rDNA reads were obtained, of which 211 were assigned to major bacterial taxa. We analyzed the 75 significant taxa detected (131,402 reads) and grouped them into two major categories: human pathogens and oil-degrading bacteria.
We used a culture media that allows the cultivation of heterotrophic marine bacteria. Therefore, this study examines a portion of the bacterial community in these environments. Our cultured marine surface sediment samples had several taxa in common. For example, γ-proteobacteria dominated all of the cultured bacteria in the10 sampled sites. Several previous culture-independent approaches showed a significant dominance of γ-proteobacteria in marine sites (Liao et al., 2009), followed by Firmicutes, Fusobacteria and Bacteriodetes. A previous study identified Firmicutes and Bacteriodetes from RasMuhammed sponges using a culturing approach (Radwan et al., 2010; Aboul-Ela et al., 2012). Fusobacteria have previously been detected in the Red Sea; however, they were detected in deep sediments of a brine pool (Siam et al., 2012). Most species of Fusobacteria and Bacteriodetes are anaerobes. Fusobacteria have been cultured from surface sediments samples of the Wadden Sea, Germany, under strict anaerobic conditions (Köpke et al., 2005). Note that strict anaerobic conditions were not implemented in our culture conditions; however, anaerobic bacteria such as Fusobacteria, Bacteriodetes and Clostridium were detected. Because our culture conditions are most likely enriched for aerobes, it is likely that such anaerobes are more abundant in situ. This finding suggests that we may have cultured the predominant phyla in our studied sites; however, rare taxa are as important in the microbial community. This imposes a limitation on our study, in that it strictly focuses on the numerically dominant bacterial taxa. A culture-independent approach would allow the study of the entire bacterial community.
Several human pathogens were detected in our Red Sea samples, including known Vibrio and Clostridium species. Vibrio species are naturally detected in marine environments (Johnson et al., 2010). Vibrio spp. have been found to dominate “plastisphere” (i.e., plastic marine debris; Zettler et al., 2013). In this culture-independent study, 10 different Vibrio species were detected and were dominated by an unknown species of Vibrio. Surprisingly, our culture-dependent approach detected 28 Vibrio species; of these, 12 were significantly detected in our cultures, which were dominated by an unknown species of Vibrio. This “marine Vibrio phenomenon” was detected at all of our sites except Saline Lake-RM. In addition to the striking dominance of this unknown species of Vibrio, three of the remaining 11 known Vibrio species were human pathogens. Several Vibrio species have been reported to cause gastrointestinal, skin and other infections (Thompson et al., 2004), including V. parahemolyticus, V. vulnificus and V. alginolyticus. Note that V. vulnificus, V. splendinus and V. sinaloensis were not detected in the “plastisphere” Vibrio community (Zettler et al., 2013). V. shilonii, which was detected in our cultures, is known to cause coral bleaching (Banin et al., 2000; Thompson et al., 2004). V. fortis and V. harveyi may contribute to coral bleaching (Thompson et al., 2004). Solar Lake-W was most dominated by pathogenic bacteria, which constituted 7.3% of the Solar Lake-W bacterial culture; these were primarily Vibrio parahaemolyticus. One limitation in our study is the short read length of the V6-V4 region, which does not provide optimal resolution at the species level. We therefore cannot assign taxa to the species level based solely on V6-V4 16S rRNA. However our results imply that the “marine Vibrio phenomenon” may pose a pathogenicity risk for human and/or marine life.
Similarly, other human pathogens were detected in our sediment samples. Most importantly, Clostridium botulinum was uniquely and significantly detected in Solar Lake-W. Clostridial members are known for their ability to survive under harsh conditions through spore formation. Clostridium botulinum outbreak detection includes the detection of spores from the contaminated environment, which may include soil and aquatic environments (e.g., marine sediments; Neimanis and Speck, 2012). This may explain the detection of strict anaerobes under our aerobic culture conditions: we could have isolated bacterial DNA from bacterial spores. Additionally, it is not uncommon to detect Clostridium in marine sediments, as this is one of its natural habitats(Neimanis and Speck, 2012). However, C. botulinum is a serious human pathogen (Neimanis and Speck, 2012). Botulism was first reported in 1991 from a traditional Egyptian salted raw fish, known as “faseikh” (Weber et al., 1993). Since then, no botulism cases have been reported in Egypt (Horowitz, 2010). In a 2011 study C. botulinum was isolated from food samples in Assiut, Egypt, but no botulism cases were reported (Ahmed et al., 2011).
In contrast to all of the other sites, Saline Lake-RM had no cultured/detected known pathogenic Vibrio or Clostridium. The bacterial community in Saline Lake-RM is predominated by a different and unknown bacterial community that is more likely to play a role in hydrocarbon metabolism. Other unknown bacterial families, genera and species were detected in different samples along the Red Sea coast (Table 4B). Related members of these bacterial groups were shown to be human pathogens. Due to the lack of assignments of bacterial reads to known families, genera or species, we may consider them potential pathogens. Further analysis on these groups should be performed to identify their pathogenic potential.
It was not surprising to detect oil-degrading bacteria along the Red Sea, particularly at industrialized sites. However, the oil-degrading bacterial consortia detected in the Red Sea coastal samples were distinct from those detected in the two lakes. Propionigenium maris, Psychrilyobacter, Tepidibacter and Photobacterium were mainly detected in the Red Sea coastal samples. Propionigenium maris is a marine debrominating bacteria (Watson et al., 2000). This species was one of the most abundant species in most of the analyzed sites. Psychrilyobacter produces H2 and acetate and has the ability to degrade hexahydro-1,3,5-trinitro-1,3,5-triazine and octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine, two nitramine explosives (Zhao et al., 2009). Tepidibacter was isolated from an oil field in China (Tan et al., 2012). Photobacteriumspecies show mercuric resistance (Reyes et al., 1999), are stable in biodiesel production at high methanol concentrations (Yang et al., 2009) and have high oil emulsification activity (Ryu et al., 2006). A bacterial consortium constituting 21 taxa was unique to the two lakes (Table 5). Additionally, three unknown species of Marinobacter and Bacillus were detected in Saline Lake-W only, and Anaerophaga was detected in both lakes. The unknown species of Marinobacter was the second most abundant taxa in Saline Lake-RM (24%). Previous studies have identified several strains of Marinobacter as oil-degrading bacteria that can degrade aliphatic hydrocarbons under oxic conditions (Cohen, 2002; Duran, 2010). An unknown species of Idiomarina was also uniquely identified in Saline Lake-W (5.7%). A strain of crude oil-degrading bacteria Idiomarina xiamenensis was isolated from surface water enriched in crude oil (Wang et al., 2010, 2011). Interestingly, the sulfate-reducing bacterium Desulfovibrio halophilus was isolated for the first time from the Solar Lake (Caumette et al., 1991). However, in our study, we have only detected it in Saline Lake-RM. This species was also detected in brine stratal water of an oil field (Welsh et al., 1996). This bacterium is known to accumulate organic solutes under high salt conditions (Welsh et al., 1996), such as those in Saline Lake-RM. We identified an oil-degrading bacterial consortium in Saline Lake-RM. Anaerophaga (detected in the two lakes only) was present in blackish-oily sedimentary residues in an oil separation tank (Denger et al., 2002; Schink, 2010). It is worth noting that all of the previous studies on Solar Lake isolated specific cyanobacteria and analyzed the cyanobacterial mats (Krumbein et al., 1977; Jorgensen et al., 1979b; Teske et al., 1998; Wieland and Kühl, 2000). Strikingly, our approach did not detect or assign any of the reads to cyanobateria in the Solar Lake culture. Note that cyanobacteria were not detected in uncultured sediments (data not shown). Taken together with our results, the studies conducted on Solar Lake West near Taba/Eilat between its discovery in 1967 and the last reported study in 1983 suggest that the microbial community in this lake has varied greatly during the past 20 years. This finding draws attention to the importance of microbial studies in monitoring and conserving marine environments.
This study molecularly characterized cultured microbial consortia along Egypt's Red Sea coast, with a focus on industrialized sites. Our results demonstrate the dominance of Vibrio spp. (human pathogens, coral pathogens and predominantly unknown species), common marine bacteria, hydrocarbon-degrading bacteria and other human pathogenic bacteria. The oil-degrading bacterial consortia were distinctly unique in the Red Sea coast compared with the two lakes sampled, suggesting different hydrocarbon exposures in these two ecosystem types. Additionally, the human pathogen consortia were dominated by Vibrio spp., which is different from the Saline Lake-W, where no known bacterial pathogens were detected. This study provides preliminary evidence for the use of bacterial consortia to assess the impact of industrialization on marine environments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is supported by the YJ-Science and Technology Research Center grant to Rania Siam. We thank Amged Ouf for technical assistance with the sample collection.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00363/abstract
Table S1 | Bacterial communities comprisingmore than 0.1% in each site are represented.
References
Abd El-Wahab, M., Dar, M. A., and Mohammad, T. A. (2005). Sediments, coral reefs and seawater interactions in some coastal lagoons, Red Sea, Egypt. Egypt. J. Aquat. Res. 31, 69–85.
Abelson, A., Olinky, R., and Gaines, S. (2005). Coral recruitment to the reefs of Eilat, Red Sea: temporal and spatial variation, and possible effects of anthropogenic disturbances. Mar. Pollut. Bull. 50, 576–582. doi: 10.1016/j.marpolbul.2005.02.021
Aboul-Ela, H. M., Shreadah, M. A., Abdel-Monem, N. M., Yakout, G. A., and Van Soest, R. W. (2012). Isolation, cytotoxic activity and phylogenetic analysis of Bacillus sp. bacteria associated with the red sea sponge Amphimedon ochracea. Adv. Biosci. Biotechnol. 3, 815–823. doi: 10.4236/abb.2012.37101
Aharon, P., Kolodny, Y., and Sass, E. (1977). Recent hot brine dolomitization in the “Solar Lake,” Gulf of Elat, isotopici, chemical, and mineralogical study. J. Geol. 85, 27–48. doi: 10.1086/628267
Ahmed, S. H., Badary, M. S., Mohamed, W. A., and Elkhawaga, A. A. (2011). Multiplex PCR for Detection and Genotyping of C. botulinum Types A, B, E and F Neurotoxin Genes in Some Egyptian Food Products. J. Am. Sci. 7, 176–190. doi: 10.1128/AEM.67.12.5694-5699.2001
Al-Halasah, N., and Ammary, B. (2007). “Potential risk and control of contamination in the Gulf of Aqaba-Jordan,” in Managing Critical Infrastructure Risks, eds I. Linkov, R. J. Wenning, and G. A. Kiker (Amman: Springer), 435–444. doi: 10.1007/978-1-4020-6385-5_25
Al-Najjar, T., Rasheed, M., Ababneh, Z., Ababneh, A., and Al-Omarey, H. (2011). Heavy metals pollution in sediment cores from the Gulf of Aqaba, Red Sea. Nat. Sci. 3, 775–782. doi: 10.4236/ns.2011.39102
Ali, A. A., Elazein, E. M., and Alian, M. A. (2011). Investigation of heavy metals pollution in water, sediment and fish at Red Sea- Jeddah Coast- KSA at two different locations. J. Appl. Environ. Biol. Sci. 1, 630–637.
Alkershi, A., and Menon, N. (2011). Phytoplankton in polluted waters of the Red Sea coast of Yemen. J. Mar. Biol. Ass. India 53, 1–6. doi: 10.6024/jmbai.2011.53.2.01630-01
Badr, N. B., El-Fiky, A. A., Mostafa, A. R., and Al-Mur, B. A. (2009). Metal pollution records in core sediments of some Red Sea coastal areas, Kingdom of Saudi Arabia. Environ. Monit. Assess. 155, 509–526. doi: 10.1007/s10661-008-0452-x
Banin, E., Israel, Y., Kushmaro, T., Loya, Y., Orr, E., and Rosenberg, E. (2000). Penetration of the coral-bleaching bacterium Vibrio shiloi into Oculina patagonica. Appl. Environ. Microbiol. 66, 3031–3036. doi: 10.1128/AEM.66.7.3031-3036.2000
Caumette, P., Cohen, Y., and Matheron, R. (1991). Isolation and Characterization of Desulfovibrio halophilus sp. nov., a Halophilic Sulfate-reducing Bacterium Isolated from Solar Lake (Sinai). System. Appl. Microbiol. 14, 33–38. doi: 10.1016/S0723-2020(11)80358-9
Cohen, Y. (2002). Bioremediation of oil by marine microbial mats. Int. Microbiol. 5, 189–193. doi: 10.1007/s10123-002-0089-5
Cohen, Y., Krumbcin, W. E., Goldberg, M., and Shilo, M. (1977). Solar Lake (Sinai). I. Physical and chemical limnology. Limnol. Oceanogr. 22, 597–609. doi: 10.4319/lo.1977.22.4.0597
Cohen, Y., Padan, E., and Shilo, M. (1975). Facultative anoxygenic photosynthesis in the cyanobacterium Oscillatoria limnetica. J. Bacteriol. 123, 855–861.
Cytryn, E., Minz, D., Oremland, R. S., and Cohen, Y. (2000). Distribution and diversity of archaea corresponding to the Limnological cycle of a hypersaline stratified Lake (Solar Lake, Sinai, Egypt). Appl. Environ. Microbiol. 66, 3269–3276. doi: 10.1128/AEM.66.8.3269-3276.2000
Denger, K., Warthmann, R., Ludwig, W., and Schink, B. (2002). Anaerophaga thermohalophila gen. nov., sp. nov., a moderately thermohalophilic, strictly anaerobic fermentative bacterium. Int. J. Syst. Evol. Microbiol. 52, 173–178.
Duran, R. (2010). “Part 1 The Microbes; 5- Marinobacter,” in Handbook of Hydrocarbon and Lipid Microbiology, ed K. N. Timmis (Springer), 1725–1735. doi: 10.1007/978-3-540-77587-4_122
Eckstein, Y. (1970). Physicochemical limnology and geology of a meromictic pond on the Red Sea shore. Limnol. Oceanogr. 15, 363–372. doi: 10.4319/lo.1970.15.3.0363
Edwards, A., and Head, S. (1987). Red Sea, Key Environments. International Union for Conservation of Nature and Natural Resources. Oxford; New York: Pergamon Press.
El-Shenawy, M. A., and Farag, A. E. (2005). Spatial and temporal variability of saprophytic and water quality bacteria along the coast of the Aqaba and Suez Gulfs and Red Sea, Egypt. Microbial. Ecol. Health Dis. 17, 94–102. doi: 10.1080/08910600510037217
El-Sorogy, A., El-Kammar, A., Ziko, A., Aly, M., and Nour, H. (2013). Gastropod shells as pollution indicators, Red Sea coast, Egypt. J. Afr. Earth Sci. 87, 93–99. doi: 10.1016/j.jafrearsci.2013.08.004
El-Sorogy, A. S., Mohamed, M. A., and Nour, H. E. (2012). Heavy metals contamination of the Quaternary coral reefs, Red Sea coast, Egypt. Environ. Earth Sci. 67, 777–785. doi: 10.1007/s12665-012-1535-0
El-Taher, A., and Madkour, H. A. (2013). Environmental and radio-ecological studies on shallow marine sediments from harbour areas along the Red Sea coast of Egypt for identification of anthropogenic impacts. Isotopes Environ. Health Stud. 50, 120–133. doi: 10.1080/10256016.2013.826211
Hanna, R. G., and Muir, G. L. (1990). Red sea corals as biomonitors of trace metal pollution. Environ. Monit. Assess. 14, 211–222. doi: 10.1007/BF00677917
Horowitz, B. Z. (2010). Type E botulism. Clin. Toxicol. 48, 880–895. doi: 10.3109/15563650.2010.526943
Ibrahim, H. A., Farag, A. M., Beltagy, E. A., and El-Shenawy, M. A. (2011). Microbial pollution indicators along the Egyptian coastal waters of Suez and Aqaba Gulfs and Red Sea. J. Egypt Public Health Assoc. 86, 111–118. doi: 10.1097/01.EPX.0000407209.05371.82
Idrisa, A. M., Eltayebb, M. A. H., Potgieter-Vermaakc, S. S., Van Griekenc, R., and Potgieterc, J. H. (2007). Assessment of heavy metals pollution in Sudanese harbors along the Red Sea Coast. Microchem. J. 87, 104–112. doi: 10.1016/j.microc.2007.06.004
Institute of Marine Research. (2012). The Dissolved Oxygen. [Online]. Red Sea University. Available online at: www.imr.sd/index.php/component/content/article/38-slide/73-the-dissolved-oxygen-.html (Accessed March 3, 2014).
Johnson, C. N., Flowers, A. R., Noriea, N. F., Zimmerman, A. M., Bowers, J. C., Depaola, A., et al. (2010). Relationships between Environmental Factors and Pathogenic Vibrios in the Northern Gulf of Mexico. Appl. Environ. Microbiol. 26, 7076–7084. doi: 10.1128/AEM.00697-10
Jorgensen, B. B., Kuenen, J. G., and Cohen, Y. (1979a). Microbial transformations of sulfur compounds in a stratified lake (Solar Lake, Sinai). Limnol. Oceanogr. 24, 799–822. doi: 10.4319/lo.1979.24.5.0799
Jorgensen, B. B., Revsbech, N. P., Blackburn, T. H., and Cohen, Y. (1979b). Diurnal cycle of oxygen and sulfide microgradients and microbial photosynthesis in a cyanobacterial mat sediment. Appl. Environ. Microbiol. 38, 46–58.
Khattab, R. M., Temraz, T. A., Kotb, M. M., and Hanafy, M. H. (2006). Assessment of oil pollution in Sharm El-Maiya Bay, Sharm El-Sheikh, South Sinai, Egypt. Catrina 1, 33–40.
Köpke, B., Wilms, R., Engelen, B., Cypionka, H., and Sass, H. (2005). Microbial diversity in coastal subsurface sediments: a cultivation approach using various electron acceptors and substrate gradients. Appl. Environ. Microbiol. 71:7819. doi: 10.1128/AEM.71.12.7819-7830.2005
Krekeler, D., Sigalevich, P., Teske, A., Cypionka, H., and Cohen, Y. (1997). A sulfate-reducing bacterium from the oxic layer of a microbial mat from Solar Lake (Sinai), Desulfovibrio oxyclinae sp. nov. Arch. Microbiol. 167, 369–375. doi: 10.1007/s002030050457
Krumbein, W. E., Cohen, Y., and Shilo, M. (1977). Solar Lake (Sinai). 4. Stromatolitic Cyanobacterial Mats. Limnol. Oceanogr. 22, 635–656. doi: 10.4319/lo.1977.22.4.0635
Liao, L., Xu, X. W., Wang, C.-S., Zhang, D.-S., and Wu, M. (2009). Bacterial and archaeal communities in the surface sediment from the northern slope of the South China Sea. Zhejiang Univ. Sci. B 10, 890–901. doi: 10.1631/jzus.B0920181
Loya, Y. (2004). “The coral reefs of eilat – past,present and future: three decades of coral community structure studies,” in Coral Health and Disease, eds E. Roseneberg and Y. Loya (Berlin; Heidelberg: Springer), 1–34.
Loya, Y., and Rinkevich, B. (1980). Effects of oil pollution on coral reef communities. Mar. Ecol. Prog. Ser. 3, 167–180. doi: 10.3354/meps003167
Medio, D., Sheppard, C., and Gascoigne, J. (2000). “Section II: country profiles,” in Coral Reefs of the Indian Ocean: Their Ecology and Conservation: Their Ecology and Conservation, ed T. R. Mcclanahan (Cary, NC: Oxford University Press), 81–471.
Mohamed, A. W. (2005). Geochemistry and Sedimentology of Core Sediments and the Influence of Human Activities; Qusier, Safaga And Hurghada Harbors, Red Sea Coast, Egypt. Egypt. J. Aquat. Res. 31, 93–103.
Mohamed, M. A. E., Madkour, H. A., and El-Saman, M. I. (2011). Impact of anthropogenic activities and natural inputs on oceanographic characteristics of water and geochemistry of surface sediments in different sites along the Egyptian Red Sea Coast. Afr. J. Environ. Sci. Technol. 5, 494–511. doi: 10.5897/AJEST10.282
Montaser, M., Mahfouz, M. E., El-Shazly, S. A., Abdel-Rahman, G. H., and Bakry, S. (2010). Toxicity of heavy metals on fish at Jeddah coast KSA: Metallothionein expression as a biomarker and Histopathological study on liver and gills. World J. Fish Mar. Sci. 2, 174–185.
Morsy, M. A., Soliman, F. A., Khattab, R. M., Rashed, M. A., and El-Masry, N. N. (2010). Implications of environmental monitoring of oil pollution in Sharm El-Maya Bay, Sharm El-Sheikh, Egypt. CATRINA 5, 97–103.
Neimanis, A., and Speck, S. (2012). “Clostridium species and Botulism,” in Infectious Diseases of Wild Mammals and Birds in Europe, eds D. Gavier-Widen, A. Meredith, and J. P. Duff (Somerset, NJ: Wiley), 570.
Pandolfi, J. M., Bradbury, R. H., Sala, E., Hughes, T. P., Bjorndal, K. A., Cooke, R. G., et al. (2003). Global trajectories of the long-term decline of coral reef ecosystems. Science 301, 955–958. doi: 10.1126/science.1085706
PERSGA/GEF. (2004). Status of Mangroves in the Red Sea and Gulf of Aden. PERSGA Technical Series No. 11. PERSGA. Jeddah.
Pilcher, N., and Abou Zaid, M. (2000). The Status of Coral Reefs in Egypt-2000. Cairo: PERSGA (Regional organization for the Protection of the Environment of the Red Sea and Gulf of Aden).
Radwan, M., Hanora, A., Zan, J., Mohamed, N. M., Abo-Elmatty, D. M., Abou-El-Ela, S. H., et al. (2010). Bacterial community analyses of two Red Sea Sponges. Mar. Biotechnol. 12, 350–360. doi: 10.1007/s10126-009-9239-5
Regional Organization for the Conservation of the Environment of the Red Sea and Gulf of Aden. (2001). Strategic Action Programme for the Red Sea and Gulf of Aden: Country Reports. Washington, DC: World Bank. Available online at: https://openknowledge.worldbank.org/handle/10986/13870 License: CC BY 3.0 IGO
Reyes, N. S., Frischer, M. E., and Sobecky, P. A. (1999). Characterization of mercury resistance mechanisms in marine sediment microbial communities. FEMS Microbiol. Ecol. 30, 273–284. doi: 10.1111/j.1574-6941.1999.tb00655.x
Riegl, B., and Velimirov, B. (1991). “How many damaged corals in Red Sea reef systems? A quantitative survey,” in Coelenterate Biology: Recent Research on Cnidaria and Ctenophora, eds R. B. Williams, P. F. S. Cornelius, R. G. Hughes, and E. A. Robso (Belgium: Springer), 249–256.
Roberts, C., and Sheppard, C. (1988). Oil spill at Sharm-el-Sheikh. Mar. Pollut. Bull. 19, 92–93. doi: 10.1016/0025-326X(88)90697-2
Romano, I., Lama, L., Orlando, P., Nicolaus, B., Giordano, A., and Gambacorta, A. (2007). Halomonas sinaiensis sp. nov., a novel halophilic bacterium isolated from a salt lake inside Ras Muhammad Park, Egypt. Extremophiles 11, 789–796. doi: 10.1007/s00792-007-0100-3
Romano, I., Orlando, P., Gambacorta, A., Nicolaus, B., Dipasquale, L., Pascual, J., et al. (2011). Salinivibrio sharmensis sp. nov., a novel haloalkaliphilic bacterium from a saline lake in Ras Mohammed Park (Egypt). Extremophiles 15, 213–220. doi: 10.1007/s00792-010-0349-9
Ryu, H. S., Kim, H. K., Choi, W. C., Kim, M. H., Park, S. Y., Han, N. S., et al. (2006). New cold-adapted lipase from Photobacterium lipolyticum sp. nov. that is closely related to filamentous fungal lipases. Appl. Microbiol. Biotechnol. 70, 321–326. doi: 10.1007/s00253-005-0058-y
Schink, B. (2010). “Bergey's manual of systematic bacteriology,” in The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes, Vol. 4, eds N. R. Krieg, W. Ludwig, W. B. Whitman, B. P. Hedlund, B. J. Paster, J. T. Staley, et al. (New York, NY: Williams and Wilkins), 51–54.
Siam, R., Mustafa, G. A., Sharaf, H., Moustafa, A., Ramadan, A. R., Antunes, A., et al. (2012). Unique prokaryotic consortia in geochemically distinct sediments from Red Sea Atlantis II and discovery deep brine pools. PLoS ONE 7:e42872. doi: 10.1371/journal.pone.0042872
Sogin, M. L., Morrison, H. G., Huber, J. A., Mark Welch, D., Huse, S. M., Neal, P. R., et al. (2006). Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U.S.A. 103, 12115–12120. doi: 10.1073/pnas.0605127103
Tan, H. Q., Wu, X. Y., Zhang, X. Q., Wu, M., and Zhu, X. F. (2012). Tepidibacter mesophilus sp. nov., a mesophilic fermentative anaerobe isolated from soil polluted by crude oil, and emended description of the genus Tepidibacter. Int. J. Syst. Evol. Microbiol. 62, 66–70. doi: 10.1099/ijs.0.027409-0
Teske, A., Ramsing, N. B., Habicht, K., Fukui, M., Kuver, J., Jørgensen, B. B., et al. (1998). Sulfate-reducing bacteria and their activities in cyanobacterial mats of solar lake (Sinai, Egypt). Appl. Environ. Microbiol. 64, 2943–2951.
Thompson, F. L., Iida, T., and Swings, J. (2004). Biodiversity of Vibrios. Microbiol. Mol. Biol. Rev. 68, 403–431. doi: 10.1128/MMBR.68.3.403-431.2004
Thompson, L. R., Field, C., Romanuk, T., Ngugi, D. K., Siam, R., Dorry, H. E., et al. (2013). Patterns of ecological specialization among microbial populations in the Red Sea and diverse oligotrophic marine environments. Ecol. Evol. 3, 1780–1797. doi: 10.1002/ece3.593
United Nations Development Programme. (2007). Eritrea's Coastal Marine and Island Biodiversity Conservation Project. eds Alain Jeudy de Grissac and Kaleab Negussie. Available online at: www.er.undp.org/energy&environment/ECMIB.html
Walsby, A. E., Van Rijn, J., and Cohen, Y. (1983). The biology of a new gas-vacuolate cyanobacterium, Dactylococcopsis salina sp. nov., in Solar Lake. Proc. R. Soc. Lond. B 217, 417–447. doi: 10.1098/rspb.1983.0019
Wang, L., Lai, Q., Fu, Y., Chen, H., Wang, W., Wang, J., et al. (2011). Idiomarina xiamenensis sp. nov., isolated from surface seawater, and proposal to transfer Pseudidiomarina aestuarii to the genus Idiomarina as Idiomarina aestuarii comb. nov. Int. J. Syst. Evol. Microbiol. 61, 969–973. doi: 10.1099/ijs.0.022970-0
Wang, L., Wang, W., Lai, Q., and Shao, Z. (2010). Gene diversity of CYP153A and AlkB alkane hydroxylases in oil-degrading bacteria isolated from the Atlantic Ocean. Environ. Microbiol. 12, 1230–1242. doi: 10.1111/j.1462-2920.2010.02165.x
Watson, J., Matsui, G. Y., Leaphart, A., Wiegel, J., Rainey, F. A., and Lovell, C. R. (2000). Reductively debrominating strains of Propionigenium maris from burrows of bromophenol-producing marine infauna. Int. J. Syst. Evol. Microbiol. 50, 1035–1042. doi: 10.1099/00207713-50-3-1035
Weber, J. T., Hibbs, R. G., Darwish, A., Mishu, B., Corwin, A. L., Rakha, M., et al. (1993). A massive outbreak of type E botulism associated with traditional salted fish in Cairo. J. Infect. Dis. 167, 451–454. doi: 10.1093/infdis/167.2.451
Welsh, D. T., Lindsay, Y. E., Caumette, P., Herbert, R. A., and Hannan, J. (1996). Identification of trehalose and glycine betaine as compatible solutes in the moderately halophilic sulfate reducing bacterium, Desulfovibrio halophilus. FEMS Microbiol. Lett. 140, 203–207. doi: 10.1111/j.1574-6968.1996.tb08337.x
Wieland, A., and Kühl, M. (2000). Irradiance and temperature regulation of oxygenic photosynthesis and O2 consumption in a hypersaline cyanobacterial mat (Solar Lake, Egypt). Mar. Biol. 137, 71–85. doi: 10.1007/s002270000331
Yang, K. S., Sohn, J.-H., and Kim, H. K. (2009). Catalytic properties of a lipase from Photobacterium lipolyticum for biodiesel production containing a high methanol concentration. J. Biosci. Bioeng. 107, 599–604. doi: 10.1016/j.jbiosc.2009.01.009
Zettler, E. R., Mincer, T. J., and Amaral-Zettler, L. A. (2013). Life in the “plastisphere”: microbial communities on plastic marine debris. Environ. Sci. Technol. 47, 7137-7146. doi: 10.1021/es401288x
Keywords: Red Sea, Taba/Eilat, 16S rRNA, mangrove, oil/hydrocarbons, Vibrios, Clostridium botulinum, Solar lake
Citation: Mustafa GA, Abd-Elgawad A, Abdel-Haleem AM and Siam R (2014) Egypt's Red Sea coast: phylogenetic analysis of cultured microbial consortia in industrialized sites. Front. Microbiol. 5:363. doi: 10.3389/fmicb.2014.00363
Received: 31 March 2014; Accepted: 27 June 2014;
Published online: 11 August 2014.
Edited by:
Karla B. Heidelberg, University of Southern California, USAReviewed by:
Xiang Xiao, Shanghai JiaoTong University, ChinaGuang Gao, Chinese Academy of Sciences, China
Zhenfeng Liu, University of Southern California, USA
Copyright © 2014 Mustafa, Abd-Elgawad, Abdel-Haleem and Siam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rania Siam, Biology Department, American University in Cairo, New Cairo Campus, AUC Avenue, PO Box 74, New Cairo 11835, Egypt e-mail: rsiam@aucegypt.edu
 Ghada A. Mustafa
Ghada A. Mustafa Amr Abd-Elgawad
Amr Abd-Elgawad Alyaa M. Abdel-Haleem
Alyaa M. Abdel-Haleem Rania Siam
Rania Siam