- 1Department of Food Technology, Doon P.G. College of Agriculture Science and Technology, Dehradun, India
- 2Department of Bioprocess and Food Engineering, Institute of Agriculture Sciences, Banaras Hindu University, Varanasi, India
- 3Department of Zoology and Biotechnology, H.N.B. Garhwal University, Srinagar, India
- 4Department of Microbiology, Sardar Bhagwan Singh Post Graduate Institute of Biomedical Sciences and Research, Dehradun, India
An actinomycetes strain designated as MN 2(6) was isolated from the solitary wasp mud nest. The isolate was identified using polyphasic taxonomy. It produced the extensive branched brown substrate and white aerial hyphae that changed into grayish black. The aerial mycelia produced the spiral spore chains with rugose spore surface. The growth was observed between temperature range of 27–37°C, pH 8–10 and below salt concentration of 6% (w/v). The comparative analysis of 16S rRNA gene sequence and phylogenetic relationship showed that strain MN 2(6) lies in clade with Streptomyces hygroscopicus subsp. hygroscopicus NRRL 2387T, Streptomyces sporocinereus NBRC 100766T and Streptomyces demainii NRRL B-1478T with which it shares a 16S rRNA gene sequence similarity of 99.3%. The strain MN 2(6) can be differentiated from type strains based on phenotypic characteristics. The strain MN 2(6) showed most promising activity against Gram-positive, Gram-negative bacteria, acid-fast bacilli and Candida species suggesting broad-spectrum characteristics of the active metabolite. Evaluation of anti-candidal activity of the metabolite of strain MN 2(6) by scanning electron microscopy (SEM) revealed changed external morphology of yeast. It kills the Candida cells due to the shrinkage and the cytosolic loss. However, further studies are required to elucidate the structure of the active metabolite produced by the isolate MN 2(6).
Introduction
In recent years searching new antibiotics has increased worldwide because of the serious problem of antibiotic resistance among the microbes. The need for new antibiotics has been met largely by the semisynthetic tailoring of natural product scaffolds discovered in the middle of the twentieth century (Clardy et al., 2006). The soil-derived microorganisms have been extensively screened for therapeutically important molecules. However, the frequency of discovering structurally new compounds is apparently decreasing these years and there is a need to seek unutilized microorganisms from unexplored sources (Brady et al., 2002). Moreover, the diversity of secondary metabolites depends more or less on the isolation source, namely, the habitat of the producers (Igarashi, 2004). The recent discovery of the novel primary and secondary metabolites from taxonomically unique population of actinomycetes suggest that these organisms could add a new dimension to microbial natural product research. In this context, new actinomycetes strains producing active compounds have been recently isolated from novel sources including saline, ocean, mangrove forests and niche habitats such as caves, beehives, pristine forests, lakes, rivers, shallow bird, solitary wasp mud nest, and other wetlands (Mukku et al., 2000; Mitra et al., 2008; Promnuan et al., 2009; Radhakrishnan et al., 2010; Poulsen et al., 2011; Kumar et al., 2012a). Assuming the above facts, the actinomycetes of the solitary wasp with regard to the occurrence of novel microbial flora have been studied previously (Kumar et al., 2012a,b). New species of the microorganisms have the potential to produce new metabolites, which justifies the isolation of new species at pharmaceutical research laboratories (Shomura et al., 1979). The present report highlights the taxonomy and antimicrobial activity of a new actinomycete strain isolated from the solitary wasp mud nest, a rare habitat.
Materials and Methods
Isolation, Identification, and Characterization
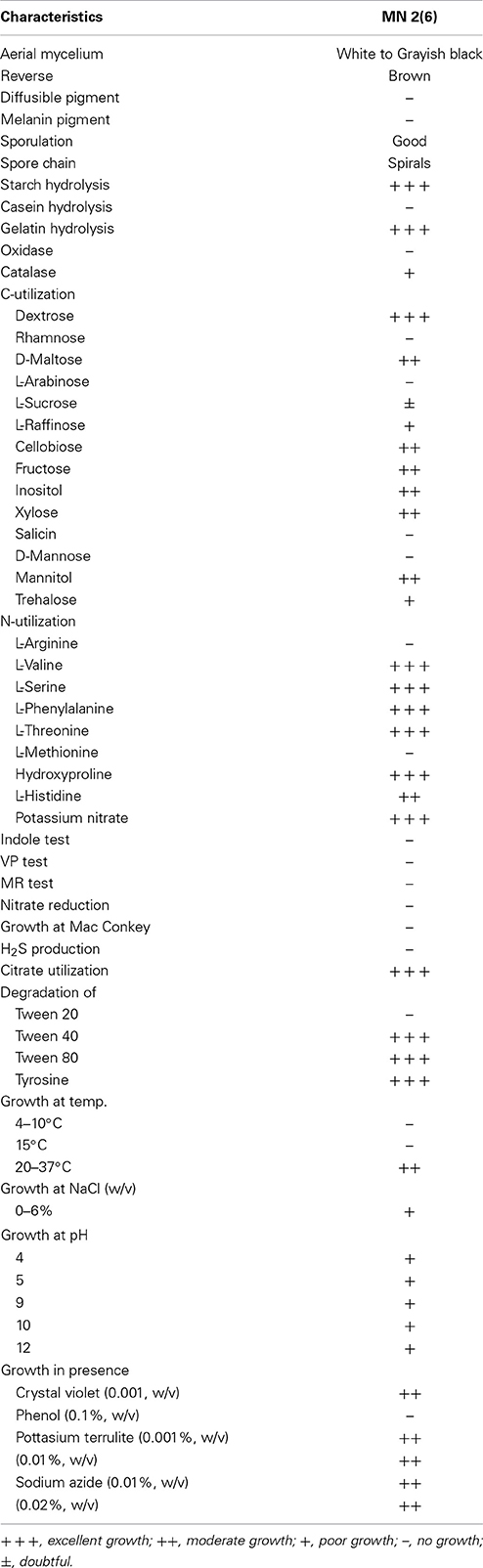
Strain MN 2(6) was isolated from the solitary wasp mud nest collected from Dehradun, India (Kumar et al., 2012a). Cultural characteristics of the strain MN 2(6) was examined every day grown on various International Streptomyces project (ISP) media (Shirling and Gottlieb, 1966). Micromorphology and sporulation were observed under light microscope by the inclined coverslip technique (Williams et al., 1989) on ISP-4 medium after incubating at 27°C for 7 days. The spore chain morphology and spore surface ornamentation were examined by scanning electron microscopy (SEM) (Zeiss EVO 40 EP) of 15-day old cultures grown on ISP-4 according to the method described previously (Kumar et al., 2011). Physiological characteristics were examined according to the methods described in the ISP (Shirling and Gottlieb, 1966) and Bergey's Manual of Systematic Bacteriology (Locci, 1989). Resistance to some antibiotics was detected by disc diffusion method. The isomeric forms of diaminopimelic acid (DAP) and the diagnostic sugar in the whole-cell hydrolysates were determined as described in MTCC Laboratory manual, IMTECH (1998), Chandigarh, India.
Molecular Identification
Chromosomal DNA of the strain MN 2(6) was prepared from cells grown in nutrient broth for 2–3 days incubation according to the method described earlier (Kumar et al., 2010). PCR amplification of the 16S rDNA of the strain MN 2(6) was done according to the methods described previously (Kumar et al., 2012a). The identification of phylogenetic neighbors was initially carried out by the BLASTN (Altschul et al., 1997) program against the database containing type strains with validly published prokaryotic names and representatives of uncultured phylotypes (Kim et al., 2012). The top 30 sequences with the highest scores were then selected for the calculation of pair wise sequence similarity using the global alignment algorithm (Myers and Miller, 1988), which was implemented at the EzTaxon-e server (http://eztaxon-e.ezbiocloud.net/; Kim et al., 2012). The isolate was identified using the EzTaxon-e server (http://eztaxon-e.ezbiocloud.net/; Kim et al., 2012) based on 16S rRNA sequence data. The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) and are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were a total of 1376 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007).
Production, Extraction, and Partial Purification of Metabolite
Production of metabolite was done in Glucose soybean meal medium (GS) as described in the previous study (Kumar et al., 2012c). The extraction and partial purification of antimicrobial metabolite from culture filtrate was done according to the method described previously (Kumar et al., 2012c).
Biological and Chemical Detection of Antimicrobial Compounds
Crude extract sample was subjected to thin-layer chromatography (TLC) and spotted onto silica gel plates (Merck Art 5735, Kieselgel 60F254), and then developed with chloroform: methanol: 25% ammonia (1:7:4, v/v) as the solvent mixture. The numbers of antibacterial active fractions were detected by bioautography (Odakura et al., 1984) on silica gel plates seeded with Micrococcus luteus and Candida albicans. Clear zone of inhibition indicated the position of antimicrobial compounds on the TLC plates, and the retention factor (Rf) value was calculated. The crude product was dissolved in methanol, and the absorption spectrum was recorded at 200–498.8 nm using UV–VIS spectrophotometer (Systronics double beam spectrophotometer). The partial chemical nature of antifungal metabolite was determined using in vitro assay. The polyene like activity of isolated antimicrobial metabolite was carried out by ergosterol agar plate method (Jain and Jain, 2006). The antimicrobial compound was partially characterized by spraying with chemical reagents such as 10% KOH ethanolic reagent, Millon's reagent, vanillin-HCl reagent, ninhydrin reagents, iodine vapors, 50% ethanolic H2SO4, and Dragendorf reagent.
Determination of Antimicrobial Activity (MIC Method)
Minimum Inhibitory Concentration was determined by the micro dilution method using a 96 well plate according to NCCLS (2008). Micro dilution methods involve the use of plastic microtiter plates (96 well). The two-fold serial dilution of the antibiotics was prepared from 512 to 0.004 μg/ml range (final concentration in Mueller Hinton broth). The MIC is the lowest concentration of the agent which completely inhibits the growth. The control containing no agent should be turbid (negative control) while control with standard antibiotic should be clear (positive control).
Scanning Electron Microscopy (SEM) for Study of Antifungal Action of Crude Extract
The anti-fungal action of extract was carried out by SEM. C. albicans was treated in vitro with half of the concentrations of MIC crude extract. The cells were harvested after 48 h of incubation at 35°C by centrifugation at 4°C for 5 min and were washed three times with phosphate buffer saline (PBS). The cells pellet was fixed in 3% (v/v) glutaraldehyde in PBS (pH 7.4) and dehydrated in increasing concentrations of ethanol (10%, v/v, increments, to 100%) (Lemar et al., 2005). Cells were mounted onto stubs. The upper surface of each stub was then coated, under vacuum, with a film of gold. The gold coating process was completed in 15–20 min. Once coated with gold, the specimens were ready for examination under scanning electron microscope (ZEISS EVO 40 EP). The gold coated metal stubs were viewed on the SEM at an accelerating voltage of 15 kV, a probe diameter of 102 Pa, to obtain secondary electron images. The field was scanned at low magnification until the line of growth was detected. Areas with clear and cells of yeasts were then selected for examination at higher magnification. Suitable fields in the preparation were photographed.
Results
The isolate MN 2(6) was isolated from the solitary wasp mud nest. It produced the extensive branched brown substrate and white aerial hyphae that changed in to grayish black. No diffusible pigments as well as melanin pigments were produced. The SEM revealed that the aerial mycelia produce spiral spore chains. The spore surface was rugose (Figure 1). The cultural characteristics of the Streptomyces sp. MN 2(6) are shown in Table 1. It showed growth on all the media except ISP-6 (Table 1). The growth was observed between the temperature range of 27–37°C and pH 8–10 and was found resistant against penicillin G (2 U), co-trimoxazole (25 μg), ciprofloxacin (5 μg), aztreonam (30 μg), cephradine (30 μg), erythromycin (10 μg), and cloxacillin (5 μg). At salt concentration above 6% (w/v), no growth was observed. The physiological characteristics are given in Table 2. Chemotaxonomic tests showed that whole-cell hydrolysates of isolate MN 2(6) were rich in the LL-diaminopimelic acid (LL-DAP), while no characteristic sugar indicated a chemotype I (Williams et al., 1989).

Figure 1. Scanning electron micrograph showing spore chain morphology of Streptomyces sp. MN 2(6); Bar, 2 μm.
An almost complete 16S rRNA gene sequence (1419 nt) for isolate MN 2(6) was determined in this study and submitted in Genbank under the accession number HM991286. The sequence was first analyzed by BLAST search and was then aligned with those of Streptomyces reference strains available in the Genbank database, which confirmed its identification as isolate MN 2(6) at genus level. Its position among the type strains of Streptomyces is shown in Figure 2. A phylogenetic tree was constructed based on 16S rRNA gene sequences to show the comparative relationship between strain MN 2(6) and other related Streptomyces species (Figure 2). The comparative analysis of 16S rRNA gene sequence and phylogenetic relationship showed that strain MN 2(6) lies in clade with S. hygroscopicus subsp. hygroscopicus NRRL 2387T, S. sporocinereus NBRC 100766T, and S. demainii NRRL B-1478T with which it shares a 16S rRNA gene sequence similarity of 99.3%.

Figure 2. Neighbor-joining tree showing the relationship between strain MN 2(6) and closely related members of the genus Streptomyces based on 16S rRNA gene sequences. Bootstrap percentages based on 1000 resamplings are listed at Nodes; only values above 70% are given. Bar 0.01 changes per nucleotide position. T indicates the type strains.
The antimicrobial activity of the strain MN 2(6) is given Table 3. The strain MN 2(6) inhibited Gram-positive bacteria, Gram-negative bacteria and yeast, suggesting broad-spectrum characteristics of the active metabolite. Lowest MIC value was recorded against M. smegmatis MTCC 6 (0.25 μg/mL), S. aureus MTCC 2940 (1.0 μg/mL), and Acinetobacter baumanii MTCC 1425 (1.0 μg/mL) followed by S. aureus MTCC 96 (2.0 μg/mL), S. aureus clinical-1 (β-lactamase) (2.0 μg/mL), E. faecalis MTCC 439 (2.0 μg/mL), Candida parapsilosis (2.0 μg/mL), C. albicans MTCC 1637 (2.0 μg/mL), and Candida tropicalis MTCC 2208 (2.0 μg/mL) while other pathogens showed higher value of MIC (Table 3). The partially purified product extracted with resin (Dianion™ HP-20) was checked for the number of bio-active compounds present in the methanol extract. One bio-active compound was found to have a Rf value of 0.78 (Figure S1). The chromogenic reactions were negative with 10% KOH ethanolic reagent, Millon's reagent, Dragendof reagent, and vanillin-HCl reagent, suggesting the absence of anthraquinones, phenol glycosides, heterocyclic compounds and myrrh constituents. However, it showed positive reactions with iodine vapors, 50% ethanolic H2SO4, and ninhydrin, indicating the presence of conjugated double bond, cardiac glycoside and free amine groups. The characteristics peak at 209 nm (Figure S2) indicates no chances of polyene class of antibiotics, which was further confirmed by ergosterol test. Evaluation of anti-candidal activity of the metabolite of strain MN 2(6) by SEM revealed control cells of C. albicans after 48 h of incubation showing normal oval shapes with smooth surfaces (Figure 3A). Control cells were lying apart, showing polar buds and bud scars, treatment of C. albicans at concentration of ½ MIC crude extract changed the external morphology of yeast (Figure 3B). The external morphology of the cells did not appear as smooth as that of untreated cells.
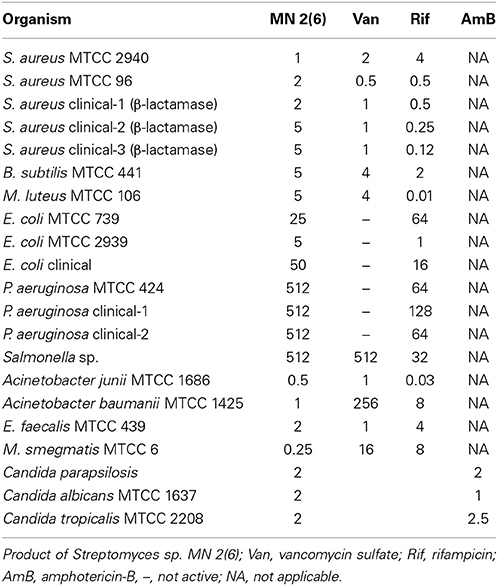
Table 3. In vitro MICs (μg/ml) of MN 2(6), Van, Rif, AmB against various bacterial pathogens and fungal pathogens by broth dilution method.
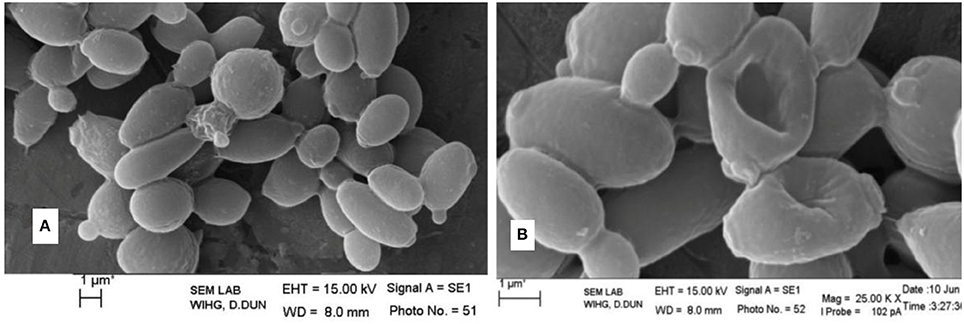
Figure 3. Effect of antifungal metabolite on Candida albicans cells; (A) without treatment; (B) treatment with active metabolite produced by MN 2(6).
Discussion
Microorganisms have served as a source for the majority of the drugs in use today (Demain and Snchez, 2009). Among the different microbes, actinomycetes have been and are the unparalleled source of bioactive metabolites. The metabolites from natural sources continue to play a key role in drug discovery and development by providing natural molecules with pharmaceutical potential and novel scaffolds for synthetic modification (Vincet et al., 2006). In this context, the isolate MN2 (6) of the previous study was taken for further studies. Based on cultural, physiochemical and chemo-taxonomical characteristics, it was found to belong the genus Streptomyces. The comparative analysis of 16S rRNA gene sequence and phylogenetic relationship showed that strain MN 2(6) lies in clade with S. hygroscopicus subsp. hygroscopicus NRRL 2387T, S. sporocinereus NBRC 100766T and S. demainii NRRL B-1478T with which it shares a 16S rRNA gene sequence similarity of 99.3%. The members of violaceusniger clade shared 16S rRNA gene similarities within the range 99.0–99.8%, values that correspond to between 3 and 11 nt differences at 1449 and 1446 locations, respectively. The highest 16S rRNA similarity found between the type strains belonging to this taxon a value equivalent to 7 nt differences at 1447 sites. Extensive DNA–DNA relatedness experiments indicated that members of the S. violaceusniger 16S rRNA gene clade that share similarities at or below 99.7%, that is, with four or more nucleotide differences, can be assigned to different genomic species (Goodfellow et al., 2007). As MN 2(6) shared 99.37% sequence similarity with type strains with nine nucleotide differences, hence may be novel species of Streptomyces belonging to violaceusniger clade. Although, strain MN 2(6) shares high 16S rDNA similarity value with S. demainii NRRL B-1478T, S. endus NRRL 2339T, S. sporocinereus NBRC 100766T, and S. hygroscopicus subsp. hugroscopicus NRRL 2387T but differs in various phenotypic characteristics. MN 2(6) produced white to whitish gray aerial and brown reverse mycelium while S. sporocinereus (Gause et al., 1983) produced sparse white aerial mycelium and green beige reverse mycelium. The spore surface of S. sporocinereus was smooth while it was rugose in case of MN 2(6). S. sporocinereus tolerate a salt concentration of 2.5% (w/v) whereas MN 2(6) can tolerate a salt concentration up to 6% (w/v). Xylose, inositol, raffinose and cellobiose are not utilized by of S. sporocinereus while the same was utilized by MN 2(6). Arginine was utilized by the type strains where as MN 2(6) does not utilize it. These characteristics differentiate MN 2(6) with type strain of S. sporocinereus. Similarly, MN 2(6) was differentiated from S. demainii (Goodfellow et al., 2007) in a number of characteristics. S. demainii produced gray aerial mycelium and grayish yellow reverse mycelium, reduced nitrate and hydrolysed casein whereas MN 2(6) did not give positive results for nitrate reduction and casein hydrolysis. Arabinose and salicin were utilized by S. demainii whereas as MN 2(6) did not utilize the same. Fructose and L-Histidine were not utilized by S. demainii while the same was utilize by MN 2(6). S. demainii grew at pH 4, 5, 9, and 10 only while MN 2(6) grew at a wide range of pH (4–10). MN 2(6) was also differentiated from S. hygroscopicus subsp. hugroscopicus (Goodfellow et al., 2007) in cultural characteristics, nitrate reduction and casein hydrolysis properties. The type strain of S. hygroscopicus subsp. hugroscopicus grew only at pH 5 whereas MN 2(6) grew at a pH range of 4–10. From the above discussion, clearly MN 2(6) is different from the type strains and may be novel species of Streptomyces belonging to violaceusniger clade.
The result of antimicrobial activity is comparable with the previous study (DeBoer et al., 1970; Furumai et al., 2003) and Streptomyces sp. MN2 (6) showed promising activity against Gram-positive, Gram-negative bacteria and Candida species. The result of bioautography indicates the presence of only one bioactive compound. The chemical characterization of this active compound suggests the absence of anthraquinones, phenol glycosides, heterocyclic compounds and myrrh constituents. However, it indicates the presence of conjugated double bond, cardiac glycoside and free amine groups (Kumar et al., 2012c). The characteristics peak at 209 nm indicates no chances of polyene class of antibiotics (Jain and Jain, 2006). However, the metabolite contained carboxy or peptide moiety in the compound as it showed maximum absorbance between 205 and 216 (Singh et al., 2009). Evaluation of anti-candidal activity of the metabolite of strain MN 2(6) by SEM may be helpful to understand the cell damage mechanism. The changes in morphology of yeast cells after treatment with Streptomyces sp. MN 2(6) are also consistent with findings of other researchers (Kitajima et al., 1976; Nurkanto and Julistiono, 2014). The shrinkage of cells was clearly observed in the electron micrograph which may be due to loss of cytosolic volume, which is mainly observed in case of polyene class of antibiotics (Kitajima et al., 1976). However, preliminary characterization of metabolites indicates that it is a non-polyene class of metabolite. These findings support that the stains MN 2(6) possess strong anti-candidal activity and killed pathogenic yeast due to considerable morphological changes.
From the results of the present study, it was concluded that the isolate may represent a novel species of Streptomyces belonging to violaceusniger clade and produce metabolite (s) which has a broad spectrum (active against Gram-positive, Gram-negative bacteria, acid-fast bacteria and yeast cells). It kills the Candida cells due to the shrinkage and cytosolic loss. However, further studies are required to elucidate the structure of the active metabolite produced by the isolate MN 2(6).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Gajraj S. Bisht and Vijay Kumar thank the Uttarakhand State Council of Science and Technology for grant received (UCS &lT/ R&D/LS/06-07/1158) to carry out this study and to the Management of S.B.S.P.G.I., Dehradun for providing necessary research facilities and Wadia Institute of Himalayan Geology, Dehradun, Uttarakhand, India for providing SEM facility.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00446/abstract
References
Altschul, S. F., Madden, T. L., Schaeffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Brady, S. F., Chao, C. J., and Clardy, J. (2002). New natural product families from an environmental DNA (eDNA) gene cluster. J. Am. Chem. Soc. 124, 9968–9969. doi: 10.1021/ja0268985
Clardy, J., Fischbach, M. A., and Walsh, C. T. (2006). New antibiotics from bacterial natural products. Nat. Biotechnol. 24, 1541–1550. doi: 10.1038/nbt1266
DeBoer, C., Meulman, P. A., Wnuk, R. J., and Peterson, D. H. (1970). Geldanamycin, a new antibiotic. J. Antibiot. (Tokyo) 23, 442–447. doi: 10.7164/antibiotics.23.442
Demain, A. L., and Snchez, S. (2009). Microbial drug discovery: 80 years of progress. J. Antibiot. 62, 5–16. doi: 10.1038/ja.2008.16
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Furumai, T., Yamakawa, T., Yoshida, R., and Igarashi, Y. (2003). Clethramycin, a new inhibitor of pollen tube growth with antifungal activity from Streptomyces hygroscopicus TP-A0623. I. Isolation, taxonomy, fermentation, isolation and biological properties. J. Antibiot. 56, 700–704. doi: 10.7164/antibiotics.56.700
Gause, G. F., Preobrazhenskaya, T. P., Sveshnikova, M. A., Terekhova, L. P., and Maximova, T. S. (1983). A Guide for the Determination of Actinomycetes. Genera Streptomyces, Streptoverticillium, and Chainia. Moscow: Nauka.
Goodfellow, M., Kumar, Y., Labeda, D. P., and Sembiring, L. (2007). The Streptomyces violaceusniger clade: a home for streptomycetes with rugose ornamented spores. Antonie Van Leeuwenhoek 92, 173–199. doi: 10.1007/s10482-007-9146-6
Igarashi, Y. (2004). Screening of novel bioactive compounds from plant-associated actinomycetes. Actinomycetologica 18, 63–66. doi: 10.3209/saj.18_63
IMTECH. (1998). Laboratory Manual for Identification of Actinomycetes. Chandigarh: Institute of Microbial Technology.
Jain, P. K., and Jain, P. C. (2006). Isolation characterization and antifungal activity of Streptomyces sampsonii GS 1322. Indian J. Exp. Biol. 45, 203–206.
Kim, O. S., Cho, Y. J., Lee, K., Yoon, S. H., Kim, M., Na, H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. doi: 10.1099/ijs.0.038075-0
Kitajima, Y., Sekiya, T., and Nozawa, Y. (1976). Freeze-fracture ultrastructural alterations induced by filipin, pimaricin, nystatin and amphotericin B in the plasmia membranes of Epidermophyton, Saccharomyces and red complex induced membrane lesions. Biochim. Biophys. Acta 455, 452–465. doi: 10.1016/0005-2736(76)90317-5
Kumar, K., Bharti, A., Gupta, V. K., Gusain, O. P., and Bisht, G. S. (2012a). Actinomycetes from solitary wasp mud nest and swallow bird mud nest: isolation and screening for their antibacterial activity. World J. Microbiol. Biotechnol. 28, 871–880. doi: 10.1007/s11274-011-0884-2
Kumar, V., Bharti, A., Gusain, O., and Bisht, G. S. (2011). Scanning electron microscopy of Streptomyces without use of any chemical fixatives. Scanning 33, 1–4. doi: 10.1002/sca.20261
Kumar, V., Bharti, A., Gusain, O. P., and Bisht, G. S. (2010). An improved method for isolation of genomic DNA from filamentous actinomycetes. J. Sci. Eng. Technol. Manag. 2, 10–13.
Kumar, V., Bharti, A., Negi, Y. K., Gusain, O. P., and Bisht, G. R. S. (2012b). Taxonomy and antimicrobial activity of a moderately salt tolerant and akaliphilic Streptomyces sp. MN 9(V) isolated from solitary wasp mud nest. Ann. Microbiol. 62, 979–985. doi: 10.1007/s13213-011-0337-z
Kumar, V., Gusain, O. P., Thakur, R. L., and Bisht, G. S. (2012c). Isolation, purification and partial characterization of an antibacterial agent produced by halotolerant alkaliphilic Streptomyces sp. EWC 7(2). Proc. Natl. Acad. Sci. India Sec. B Biol. Sci. 83, 199–206. doi: 10.1007/s40011-012-0117-y
Lemar, K. M., Ourania, P., Aon, M. A., Cortassa, S., Muller, C. T., Plummer, S., et al. (2005). Allyl alcohol and garlic (Allivum sativum) extract produce oxidative stress in Candida species. Microbiology 151, 3257–3265. doi: 10.1099/mic.0.28095-0
Locci, R. (1989). “Streptomyces and related genera,” in Bergey's Manual of Systematic Bacteriology, Vol. 4, eds S. T. Williams, M. E. Sharp, and J. G. Holt (Baltimore: Williams and Wilkins), 2451–2506.
Mitra, A., Santra, S. C., and Mukherjee, J. (2008). Distribution of actinomycetes, their antagonistic behaviour and the physico chemical characteristics of the world's largest tidal mangrove forest. Appl. Microbiol. Biotechnol. 80, 685–695. doi: 10.1007/s00253-008-1626-8
Mukku, V. J., Speitling, M., Laatsch, H., and Helmke, E. (2000). New butenolides from two marine streptomycetes. J. Nat. Prod. 63, 1570–1572. doi: 10.1021/np0001676
Myers, E. W., and Miller, W. (1988). Optimal alignments in linear space. Comput. Appl. Biosci. 4, 11–17.
NCCLS. (2008). Performance Standards for Antimicrobial Susceptibility Testing; Ninth Informational Supplement. NCCLS Document M100-S9. Wayne, PA: National Committee for Clinical Laboratory Standard, 120–126.
Nurkanto, A., and Julistiono, H. (2014). Screening and study of antifungal activity of leaf litter actinomycetes isolated from ternate island, Indonesia. Asian Pac. J. Trop. Biomed. 4, 886–891.
Odakura, Y., Kase, H., Itoh, S., Satoh, S., and Takasawa, S. (1984). Biosynthesis of asatromicin 421 and related antibiotics. Biosynthetic studies with blocked mutants of 422 Micromonospora olivasterospora. J. Antibiot. 12, 670–680.
Poulsen, M., Oh, D. C., Clardy, J., and Currie, C. R. (2011). Chemical analyses of wasp-associated Streptomyces bacteria reveal a prolific potential for natural products discovery. PLoS ONE 6:e16763. doi: 10.1371/journal.pone.0016763
Promnuan, Y., Kudo, T., and Chantawannakul, P. (2009). Actinomycetes isolated from beehives in Thailand. World J. Microbiol. Biotechnol. 25, 1685–1689. doi: 10.1007/s11274-009-0051-1
Radhakrishnan, M., Suganya, S., Balagurunathan, R., and Kumar, V. (2010). Preliminary screening for antibacterial and antimycobacterial activity of actinomycetes from less explored ecosystems. World J. Microbiol. Biotechnol. 26, 561–566. doi: 10.1007/s11274-009-0198-9
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Shirling, E. B., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Shomura, T., Yoshida, J., Amano, S., Kojima, M., Inouye, S., and Niida, T. (1979). Studies on Actinomycetales producing antibiotics only on agar culture. I. Screening, taxonomy and morphology productivity relationship of Streptomyces halstedii, strain SF 1993. J. Antibiot. 32, 425–427. doi: 10.7164/antibiotics.32.427
Singh, V., Praveen, V., Banga, J., and Tripathi, C. K. M. (2009). Antimicrobial activities of microbial strains from soil of stressed ecological niches of Eastern Uttar Pradesh, India. Indian J. Exp. Biol. 74, 298–303.
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 101, 11030–11035. doi: 10.1073/pnas.0404206101
Keywords: solitary wasp mud nest, Streptomyces sp., anti-candidal activity, violaceusniger clade, polyphasic taxonomy
Citation: Kumar V, Naik B, Gusain O and Bisht GS (2014) An actinomycete isolate from solitary wasp mud nest having strong antibacterial activity and kills the Candida cells due to the shrinkage and the cytosolic loss. Front. Microbiol. 5:446. doi: 10.3389/fmicb.2014.00446
Received: 04 June 2014; Accepted: 05 August 2014;
Published online: 21 August 2014.
Edited by:
Márcia Vanusa Da Silva, Universidade Federal de Pernambuco, BrazilReviewed by:
Dmitri Debabov, NovaBay Pharmaceuticals, USALuis Cláudio Nascimento Da Silva, University of Copenhagen, Denmark
Copyright © 2014 Kumar, Naik, Gusain and Bisht. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gajraj S. Bisht, Department of Microbiology, Sardar Bhagwan Singh Post Graduate Institute of Biomedical Sciences and Research, Balawala, 248161 Dehradun, India e-mail: grsbisht@gmail.com
 Vijay Kumar
Vijay Kumar Bindu Naik
Bindu Naik Omprakash Gusain
Omprakash Gusain Gajraj S. Bisht
Gajraj S. Bisht