- 1Plant Functional Biology and Climate Change Cluster, University of Technology, Sydney, Sydney, NSW, Australia
- 2The Mina and Everard Goodman Faculty of Life Sciences, Bar-Ilan University, Ramat Gan, Israel
The symbiotic dinoflagellates (genus Symbiodinium) inhabiting coral endodermal tissues are well known for their role as keystone symbiotic partners, providing corals with enormous amounts of energy acquired via photosynthesis and the absorption of dissolved nutrients. In the past few decades, corals reefs worldwide have been increasingly affected by coral bleaching (i.e., the breakdown of the symbiosis between corals and their dinoflagellate symbionts), which carries important socio-economic implications. Consequently, the number of studies focusing on the molecular and cellular processes underlying this biological phenomenon has grown rapidly, and symbiosis is now widely recognized as a major topic in coral biology. However, obtaining a clear image of the interplay between the environment and this mutualistic symbiosis remains challenging. Here, we review the potential of recent technological advances in molecular biology and approaches using stable isotopes to fill critical knowledge gaps regarding coral symbiotic function. Finally, we emphasize that the largest opportunity to achieve the full potential in this field arises from the integration of these technological advances.
Introduction
Reefs based on scleractinian corals are among the most productive and biologically diverse marine ecosystems on Earth (Moberg and Folke, 1999). At the heart of the success of corals is their symbiosis with dinoflagellate algae (zooxanthellae), which live within their tissues and provide each coral polyp with a wider metabolic repertoire (Anthony and Hoegh-Guldberg, 2003; Houlbreque and Ferrier-Pages, 2009). This fundamental symbiosis is known to enhance the ability of corals to synthesize a calcium carbonate skeleton (Gattuso et al., 1999), the structural basis of coral reef ecosystems, in an environment where nutrients are mostly limited.
Since 1979, populations of scleractinian corals have been reported as increasingly affected by mass coral bleaching, which involves the breakdown of the symbiosis between the cnidarian host and the dinoflagellate symbionts (Hoegh-Guldberg, 1999). Given that the economic value of coral reefs has been estimated around US $375 billion per year (Costanza et al., 1997) and that coral reefs support over 500 million people through the services and food that they provide, losing corals from reef systems would have substantial impacts on coastal populations worldwide. Consequently, many studies have focused on the causes and mechanisms of the disruption of this symbiosis in the past decades (more than 18,000 articles are available on Google scholar since 1990). However, despite the undoubted causal link between environmental stresses and bleaching, capturing the complexity of the interaction between coral-dinoflagellate symbiosis and its surrounding environment remains challenging, especially given that (i) the environment is complex and defined by a multitude of factors and (ii) the study of coral symbiosis is complex and hampered by the intertwined nature of coral-dinoflagellate symbiosis. Considering the complexity of coral symbiosis, here we identify specific knowledge gaps within the functioning of coral symbiosis, including immune defenses and metabolism, and argue that recent technological advances provide better tools to understand how the environment affects these functional processes.
Corals Grow in a Narrow Band of Environmental Conditions, Near Their Physiological Limits
Corals thrive in tropical waters, which happen to be warm, clear and generally oligotrophic (Muscatine and Porter, 1977). This narrow and consistent band of environmental conditions signifies that corals live and grow best near their physiological limits, especially with regards to three main factors: temperature, light and nutrients. Among these factors, the interaction between temperature and light has been intensively studied over the last decades because of a major interest in understanding thermal and light stress-related bleaching phenomena (Hoegh-Guldberg, 1999). One of the first sites of damage is the symbiont photosystem II apparatus (Lesser, 2006), a key component of photosynthetic pathways located within the chloroplast of Symbiodinium. This photosynthetic dysfunction results in the excessive production of reactive oxygen species (ROS) in the symbiont and promotes the degradation of host mitochondria, providing another potential site of harmful ROS production and oxidative stress (Dunn et al., 2012a). The excess ROS damages essential biological macromolecules and cellular structures, initiating a cascade of innate immune responses, which subsequently result in the release and/or degradation of the symbiotic dinoflagellates. Histological studies have revealed two possible mechanisms of symbiont degradation: (i) symbionts are degraded from the effects of ROS via programmed cell death (Strychar et al., 2004) or (ii) the coral host actively destroys the symbionts and ultimately expels them (Dunn et al., 2007b). The cellular mechanisms of coral bleaching have been the focus of many studies since the 1990’s, which are already well reviewed (for review, see: Weis, 2008). However, despite the clear involvement of cellular mechanisms such as exocytosis, host cell detachment, apoptosis, and necrosis, the cascade of immune responses and the modulation of cell death pathways leading to bleaching remain unsolved.
Technological Advances in Molecular Biology Progress Toward Unraveling Gene Function
By allowing researchers to simultaneously investigate genes and their level of expression, genomics and transcriptomics approaches have greatly improved our understanding of coral bleaching. In the late 2000s, the first microarrays studies had a tremendous impact on revealing the cellular foundation of thermal stress-induced coral bleaching (Desalvo et al., 2008; Rodriguez-Lanetty et al., 2009; Bellantuono et al., 2012). In the first medium–scale transcriptomics coral study, Desalvo et al. (2008) used a cDNA microarray containing more than 1300 genes of the coral Montastraea faveolata to measure gene expression changes associated with thermal stress. Their results suggested that oxidative stress in thermal-stressed corals causes a disruption of Ca2+ homeostasis and the initiation of cell death via apoptosis and necrosis. In a following study, Rodriguez-Lanetty et al. (2009), examined the effect of thermal stress on the early transcriptional response of aposymbiotic larvae of the reef-building coral Acropora millepora and show that elevated temperature compromise some critical components of the coral immune defenses including a mannose-binding C-type lectin. More recently, microarrays studies demonstrated that coral host transcriptomic states are correlated with different Symbiodinium genotypes (Desalvo et al., 2010) and that potential “early warning genes” and “severe heat-related genes” could be detected as a result of high heat stress (Maor-Landaw et al., 2014). The findings from Maor-Landaw et al. (2014) also suggest that during short-term heat stress, S. pistillata may divert cellular energy into mechanisms such as the ER-unfolded protein response (UPR) and ER-associated degradation (ERAD) at the expense of growth and biomineralization processes in an effort to survive and subsequently recover from the stress. The emergence of next-generation sequencing technologies has further increased the speed of coverage and decreased the cost through massively parallel sequencing methods. As a result, large–scale transcriptomics are well established as part of the coral biologist’s “toolbox” (Moya et al., 2012) and a total of approx. 600,000 sequences with more than 86,000 unique blast matches are available from various coral species through six assembled transcriptomes and one fully assembled genome (Shinzato et al., 2011) in databases such as systems biology of symbiosis1, compagen2, or cnidarian3. Genomic information on Symbiodinium are more scarce, due primarily to its very large genome size (approx. 2–4 Gb) and repetitive DNA (Leggat et al., 2011). However, since 2007, several expression sequence tags (ESTs) and subsequent gene expression studies were generated (Leggat et al., 2007; Rosic et al., 2010, 2011a,b), and more recently, the gene structure of the dinoflagellate has been further revealed by the draft assembly of the S. minutum nuclear genome (Shoguchi et al., 2013). Despite these enormous progresses, how the environment and genes ultimately interact to affect immune defenses and cell death in coral symbiosis remains poorly understood. Currently, the biggest challenges in this field are (i) to organize the growing body of molecular information into a clear mechanistic framework and testable hypotheses with a direct link to function and (ii) to direct hypothesis-driven research to aid in the examinations of gene function. While the first challenge is conceptual and beyond the scope of this paper, the latter is more technological and could be achieved using advanced functional approaches to silence/knockdown gene expression. In this respect, previous studies in the freshwater cnidarian Hydra magnipapillata have explored the use of the reverse-genetics technique RNA interference (RNAi), which consists of introducing a synthetic double strand of RNA into cells to selectively induce gene suppression. Unfortunately, this method has had limited success; the RNA delivery via electroporation often damages tissues and cells (Lohmann et al., 1999; Smith et al., 2000; Cardenas and Salgado, 2003). In a pioneering gene silencing study for a symbiotic cnidarian, Dunn et al. (2007a) reported the use of RNAi and chemical transfection delivery to suppress the expression of a gene coding for caspase, a proteolytic enzyme triggering cell death and bleaching in the symbiotic sea anemone Aiptasia pallida. Although their method was effective, the decrease obtained in caspase activity was not absolute (only 30%), most likely related to the instability or poor delivery of siRNA in vivo. Genetic modification is still in its infancy in symbiotic cnidarians, and efforts at using siRNA for gene silencing have often been hampered by the difficulty of effectively introducing it into cells of interest. As a result, no other attempts to use siRNA in corals have been published thus far.
Nanotechnology is a relatively new discipline, which in the last years is starting to be used for addressing questions related to biological systems. Thanks to nanotechnology, new materials can be developed that have new properties compared to existing properties. Nanomaterials have been shown to possess distinctive properties that contribute to promising applications in several fields. To give an example, fluorescent semiconductor nanoparticles (NPs) suffer less from photobleaching than conventional fluorophores. Thus, single-molecule based tracking of fluorescently labeled membrane-bound proteins received a great boost by moving from connectional fluorophores to fluorescent NPs (Bouzigues et al., 2007). The small size of the NPs gives them a high surface area-to-volume ratio and facilitates the interaction with several types of chemical species. NPs are also excellent candidates for drug delivery due to their capacity to interact with biomolecules and their possibility to be loaded with specific cargo (Bouzigues et al., 2007; Conde et al., 2012). Although NPs can be useful devices for delivering specific cargo in vitro and in vivo and their uptake is easily carried out, the specific control of the cargo release is still a challenge. Some years ago, a new concept was presented in which hollow particles, typically on the micrometer scale (so called polyelectrolyte capsules), were used as a carrier system. The cavity of the capsules can be loaded with a large variety and high quantity of cargo (De Koker et al., 2009; De Cock et al., 2010; Conde et al., 2012). If the walls of the capsules are modified with Au NPs, an optothermal opening can be made, providing the possibility to carry out a controlled release of the cargo (Skirtach et al., 2006; De Koker et al., 2009). This release is similar to the concept of caged Ca2+, in which Ca2+ ions are released from a chelator upon a flash of light. Capsules, however, allow for the release of a larger variety of cargo molecules, such as small drugs, proteins, or mRNA. Capsules can range in size from hundreds of nanometers to a few micrometers, depending on the size of the template that was used in synthesis (Skirtach et al., 2006; Wang et al., 2008). When less specific control of the release is required, biodegradable capsules are also a good alternative. It has been demonstrated that biodegradable capsules are degraded inside of the lysosome of cells, where their cargo is then released. Thus, nanotechnology can provide sophisticated carrier systems ranging from a few nanometers to a few micrometers, which allows for the controlled release of biologically active cargo. This next generation of carrier systems could be a game changer in the understanding of bleaching mechanisms, as they also include NPs with new biomaterials developed to fit the chemistry, biophysical structure, and biological function of siRNA. Many research groups are already reporting improved stability and delivery efficiency of siRNA (for review, see: Kozielski et al., 2013). However, in model organisms such as Nematods or Zebrafish, a reliable means of RNAi-mediated gene knockdown remains elusive (Kelly and Hurlstone, 2011). As such, a simpler alternative for gene knockdown, such as morpholino antisense oligos, is still the most effective method of gene suppression in Zebrafish. Morpholinos act by binding and blocking access to target mRNA (Nasevicius and Ekker, 2000). Since their first introduction in the early 2000’s, they been used in a range of model organisms, including sea urchin, ascidian, zebrafish, frog, chick, and mouse providing a relatively simple and rapid method to study gene function (Huang et al., 2012; for review see: Heasman, 2002). Although these better ways for gene suppression still require optimization and validation in coral symbiosis, their future development could be critical to address important hypotheses about gene function, such as the existence of symbiosis-specific genes.
Importance of Nutrients and Metabolic Function in Coral Symbiosis
Given the nutritional role of the symbionts, another intriguing question—and subsequent knowledge gap in coral symbiosis—arises as to whether bleaching may in part reflect a change in the nutritional status of the host–symbiont interaction. In this respect, recent studies have highlighted strong correlations between feeding, sustained photosynthetic activity and reduced bleaching (Ferrier-Pages et al., 2010; Beraud et al., 2013). Coral symbiosis requires the delicate balance of exchanged compounds between the symbiotic partners. The photosynthetically fixed carbon compounds translocated by the dinoflagellate symbiont to the host consist largely of non-nitrogenous compounds, such as glycerol, glucose, and succinate (Venn et al., 2008). These compounds are often referred to as “junk food,” as they directly support coral respiration and mucus production (Wild et al., 2004) but can only be used for coral growth when nitrogen and phosphorus are available from another source (Ferrier-Pages et al., 2010). Consequently, the ability to assimilate nitrogen and phosphorus by feeding on plankton (Houlbreque and Ferrier-Pages, 2009) or by absorbing nutrients dissolved in seawater, with a preference for phosphate, ammonium and nitrate (Grover et al., 2002), is a crucial attribute of the coral symbiosis.
Ecological stoichiometry is an increasingly broad field of research that evaluates how the relative quantity of specific chemical elements (C:N:P ratio) constrains or facilitates the movement of nutrients through an ecosystem (Sterner and Elser, 2002). Given that elemental ratios are unit less, using a stoichiometric approach allows for the tracking of the same response at scales where quantities differ greatly (for example, from single cells to community). Recent advances in this field of research have demonstrated an intimate link between biomass stoichiometry, environmental conditions and nutrient fluxes in a broad range of ecosystems (Taylor and Townsend, 2010). The same is certain to be the case for coral and coral reefs; the changes in biomass stoichiometry, driven by changes in macromolecular composition, reflect coral physiology (e.g., growth rate or accumulation of storage compounds) and in situ environmental conditions (e.g., temperature, nutrients, or light availability). Shifting the balance of supply versus demand for nitrogen and carbon, for example due to an environmental perturbation, may well result in the disruption of the coral–symbiont relationship, which is referred to as bleaching. The development of research investigating environmental impacts on the metabolic function of coral reefs is particularly necessary given that (i) reefs are rapidly deteriorating worldwide and that (ii) most coral reef research does not consider biomass stoichiometry and metabolic function when studying bleaching. Providing redress for such an omission may be critical in understanding the issue of coral reef degradation.
Techniques Based on Stable Isotopes to Quantify Metabolic Activity in Situ
The intertwined nature of the coral-dinoflagellate endosymbiosis has long hampered research on coral metabolic function, as studies often suffer from potential cross-contamination between coral host and dinoflagellate fractions (Yellowlees et al., 2008). In this context, the recent development of approaches combining incubations with stable isotopes and analysis of elements, DNA, RNA and other biomarkers has been a revolutionary step, allowing the detection of metabolically active microbes in their natural habitat (for a review on these techniques and their application in microbiology Musat et al., 2008; Orphan and House, 2009; Wagner, 2009; Murrell and Whiteley, 2011). Stable isotope probing (SIP) is a relatively recent method to track the metabolic fate of a compound “isotopically labeled.” In a seminal study, Radajewski et al. (2000), reported that 13C-DNA, produced during growth methylotrophic bacteria on a 13C-enriched carbon source could be resolved from 12C-DNA by density-gradient centrifugation, allowing both taxonomic and functional characterization by gene probing and sequence analysis (Radajewski et al., 2000). This method was described as DNA-SIP, and was soon after followed by RNA-SIP (Manefield et al., 2002), which provided another novel way to link the phylogeny of microorganisms to their function. Since then, SIP has been developed with the use of different stable isotopes including 15N and 18O (for review, see Murrell and Whiteley, 2011).
Stable isotope probing techniques are not the only way to track metabolic incorporation of stable isotopes. Metabolomics, which involves the quantitative analysis of all metabolites present in cells and tissues, can be used in combination with stable isotope and has the potential to play a key role in understanding coral symbiosis (for review, see: Gordon and Leggat, 2010). In a recent study, Dunn et al. (2012b) used stable isotopic incorporation from dissolved inorganic carbon (NaH13CO3) combined with HPLC-MS to investigate the lipogenesis in symbiotic cnidarian. Interestingly, their results indicated that fatty acids derived from photosynthetically fixed carbon were not used directly in host lipogenesis, suggesting that additional sources of carbon, such as host respiration and heterotrophy may be especially important for the lipogenesis of fatty acids in the cnidarian host. Another technique that over- comes the use of stable isotopes to detect metabolically active microbes in their natural environment is nano-scale secondary ion mass spectrometry (NanoSIMS). With NanoSIMS, secondary ions are extracted from the surface of a sample under the impact of a primary ions beam and subsequently analyzed using mass spectrometry providing imaging and quantification of up to seven isotopes of elements simultaneously (Wagner, 2009). When combined with stable isotopes incubation, NanoSIMS can be used to measure the relative metabolic contribution of different symbiotic partners (i.e., metabolic rates of individual host and symbiont cells) with single cell resolution (Pernice et al., 2012; Kopp et al., 2013). This technique can also be used in concert with in situ hybridizations to simultaneously identify individual cells and quantify their substrate uptake (Behrens et al., 2008; Musat et al., 2008). Although this combination of NanoSIMS and in situ hybridization has never been applied to coral, studies integrating these powerful methodologies can significantly improve our understanding of the functional diversity that exists at the very heart of reef-building corals (Pernice et al., 2014).
By allowing direct empirical evaluation of a proposed hypothesis, these functional approaches combining incubations with stable isotopes and biomarkers or elemental analysis can help addressing fundamental questions in coral symbiosis. Among others, an interesting hypothesis that remains poorly addressed so far concerning coral symbiosis, is whether photosynthetically fixed carbon may directly contribute to calcification of coral skeleton? Indeed, previous studies have demonstrated that photosynthesis and skeleton formation are tightly coupled in zooxanthellate scleractinian corals, calcification being, on average, three times higher in light than in darkness [for review, see Gattuso et al. (1999)]. However, the details of carbon supply to the calcification process are almost unknown. In this respect, elemental analyses combined with incubations using multiple stable isotopes focusing on skeletal formation [e.g., using 86Sr (Houlbreque et al., 2009)] and fixation of carbon via photosynthesis [e.g., using NaH13CO3 (Pernice et al., 2014)] have great potential to address this hypothesis and may be critical in understanding the link between photosynthesis and skeletal formation.
When integrated with recent advances in molecular biology, including “omics” approaches and next generation siRNA delivery systems, approaches based on incubation with stable isotopes could also provide important opportunities for system biology (Raes and Bork, 2008; Figure 1). Discovering the relationships between gene functions and the sum of all metabolic processes (i.e., nutrients and energy cycling) occurring within symbiotic corals could be an especially important next step to better understand the biology and functioning of this symbiosis.
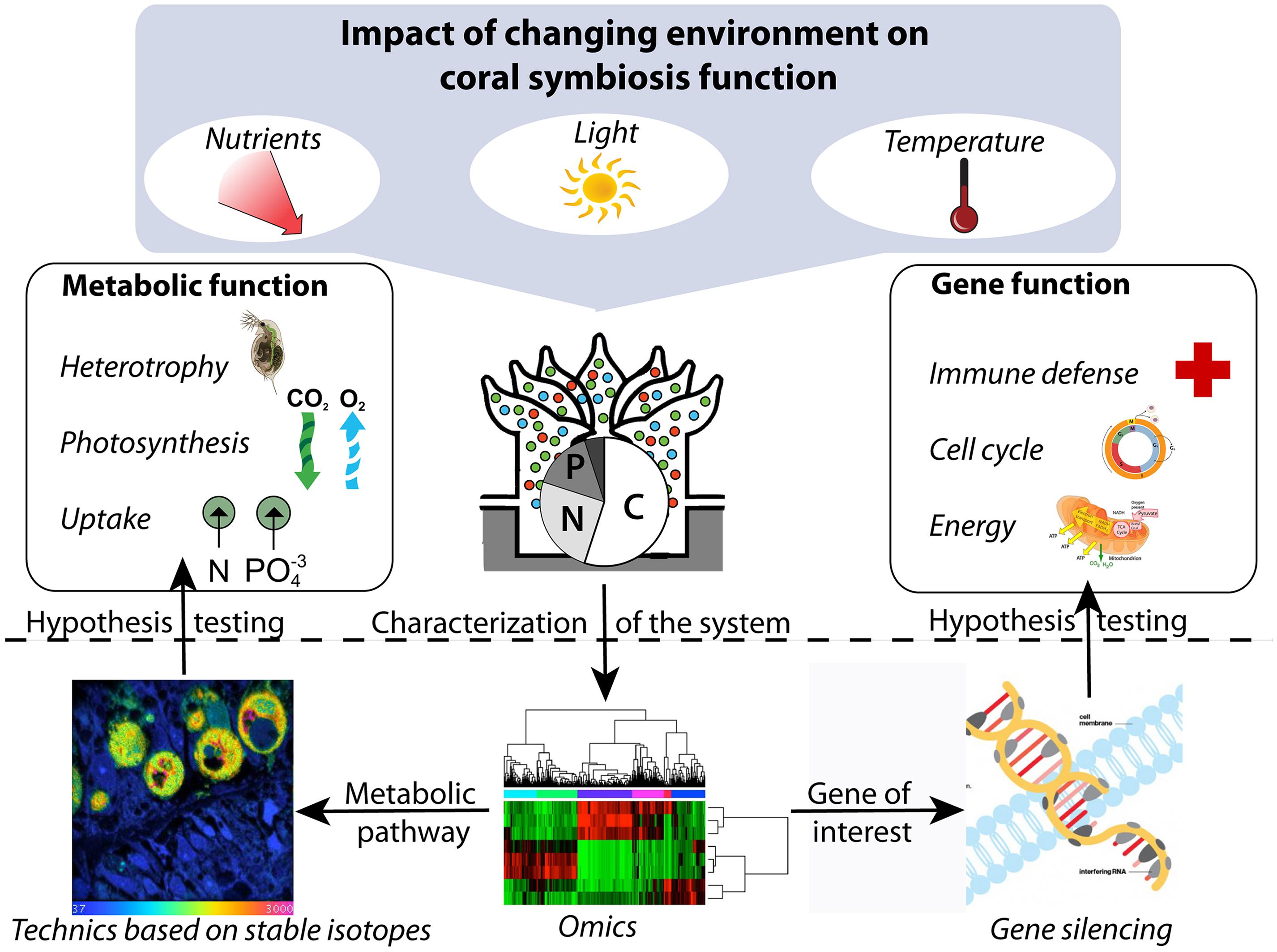
FIGURE 1. Conceptual framework integrating metabolic and gene function to study the impact of the environment on coral symbiosis. The “omics” approaches can be used to describe the system under different conditions and to identify genes and metabolic pathways of interest, which can subsequently be targeted by gene silencing and technics based on stable isotopes, respectively, to direct hypothesis-driven research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank two anonymous reviewers for their valuable and constructive comments on the first version of the manuscript, which helped improve the quality of this paper. Mathieu Pernice is supported by the Plant Functional Biology and Climate Change Cluster, University of Technology Sydney.
Footnotes
- ^ http://www.auburn.edu/~santosr/symbiosys.htm
- ^ http://www.compagen.org/links.html
- ^ http://data.centrescientifique.mc/blast/?option=home.php
References
Anthony, K. R. N., and Hoegh-Guldberg, O. (2003). Kinetics of photoacclimation in corals. Oecologia 134, 23–31. doi: 10.1007/s00442-002-1095-1
Behrens, S., Losekann, T., Pett-Ridge, J., Weber, P. K., Ng, W. O., Stevenson, B. S.,et al. (2008). Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl. Environ. Microbiol. 74, 3143–3150. doi: 10.1128/AEM.00191-08
Bellantuono, A., Hoegh-Guldberg, O., and Rodriguez-Lanetty, M. (2012). Resistance to thermal stress in corals without changes in symbiont composition. Proc. Roy. Soc. B 279, 1100–1107. doi: 10.1098/rspb.2011.1780
Beraud, E., Gevaert, F., Rottier, C., and Ferrier-Pages, C. (2013). The response of the scleractinian coral Turbinaria reniformis to thermal stress depends on the nitrogen status of the coral holobiont. J. Exp. Biol. 216, 2665–2674. doi: 10.1242/jeb.085183
Bouzigues, C., Morel, M., Triller, A., and Dahan, M. (2007). Asymmetric redistribution of GABA receptors during GABA gradient sensing by nerve growth cones analyzed by single quantum dot imaging. Proc. Natl. Acad. Sci. U.S.A. 104, 11251–11256. doi: 10.1073/pnas.0702536104
Cardenas, M. M., and Salgado, L. M. (2003). STK, the src homologue, is responsible for the initial commitment to develop head structures in Hydra. Dev. Biol. 264, 495–505. doi: 10.1016/j.ydbio.2003.08.022
Conde, J., Ambrosone, A., Sanz, V., Hernandez, Y., Marchesano, V., Tian, F.,et al. (2012). Design of multifunctional gold Nanoparticles for In Vitro and In Vivo gene silencing. ACS Nano 6, 8316–8324. doi: 10.1021/nn3030223
Costanza, R., dArge, R., deGroot, R., Farber, S., Grasso, M., Hannon, B.,et al. (1997). The value of the world’s ecosystem services and natural capital. Nature 387, 253–260. doi: 10.1038/387253a0
De Cock, L., De Koker, S., De Geest, B., Grooten, J., Vervaet, C., Remon, J.,et al. (2010). Polymeric multilayer capsules in drug delivery. Angew. Chem. Int. Ed. 49, 6954–6973. doi: 10.1002/anie.200906266
De Koker, S., De Geest, B., Singh, S., De Rycke, R., Naessens, T., Van Kooyk, Y.,et al. (2009). Polyelectrolyte microcapsules as antigen delivery vehicles to dendritic cells: uptake, processing, and cross-presentation of encapsulated antigens. Angew. Chem. Int. Ed. 48, 8485–8489. doi: 10.1002/anie.200903769
Desalvo, M. K., Rvoolstra, C., Sunagawa, S., Schwarz, J. A., Stillman, J. H., Coffroth, M. A.,et al. (2008). Differential gene expression during thermal stress and bleaching in the Caribbean coral Montastraea faveolata. Mol. Ecol. 17, 3952–3971. doi: 10.1111/j.1365-294X.2008.03879.x
Desalvo, M. K., Sunagawa, S., Fisher, P. L., Voolstra, C. R., Iglesias-Prieto, R., and Medina, M. (2010). Coral host transcriptomic states are correlated with Symbiodinium genotypes. Mol. Ecol. 19, 1174–1186. doi: 10.1111/j.1365-294X.2010.04534.x
Dunn, S. R., Pernice, M., Green, K., Hoegh-Guldberg, O., and Dove S. G. (2012a). Thermal stress promotes host mitochondrial degradation in symbiotic cnidarians: are the batteries of the reef going to run out? PLoS ONE 7:e39024. doi: 10.1371/journal.pone.0039024
Dunn, S. R., Thomas, M. C., Nette, G. W., and Dove, S. G. (2012b). A lipidomic approach to understanding free fatty acid lipogenesis derived from dissolved inorganic carbon within cnidarian-dinoflagellate symbiosis. PLoS ONE 7:e46801. doi: 10.1371/journal.pone.0046801
Dunn, S. R., Phillips, W. S., Green, D. R, and Weis, V. M. (2007a). Knockdown of actin and caspase gene expression by RNA interference in the symbiotic anemone Aiptasia pallida. Biol. Bull. 212, 250–258. doi: 10.2307/25066607
Dunn, S. R., Schnitzler, C. E., and Weis, V. M. (2007b). Apoptosis and autophagy as mechanisms of dinoflagellate symbiont release during cnidarian bleaching: every which way you lose. Proc. Biol. Sci. 274, 3079–3085. doi: 10.1098/rspb.2007.0711
Ferrier-Pages, C., Rottier, C., Beraud, E., and Levy, O. (2010). Experimental assessment of the feeding effort of three scleractinian coral species during a thermal stress: effect on the rates of photosynthesis. J. Exp. Mar. Biol. Ecol. 390, 118–124. doi: 10.1016/j.jembe.2010.05.007
Gattuso, J.-P., Allemand, D., and Frankignoulle, M. (1999). Photosynthesis and calcification at cellular, organismal and community levels in coral reefs: a review on interactions and control by carbonate chemistry. Am. Zool. 39, 160–183. doi: 10.1093/icb/39.1.160
Gordon, B. R., and Leggat, W. (2010). Symbiodinium-invertebrate symbioses and the role of metabolomics. Mar. Drugs 8, 2546–2568. doi: 10.3390/md8102546
Grover, R., Maguer, J.-F., Reynaud-Vaganay, S., and Ferrier-Pages, C. (2002). Uptake of ammonium by the scleractinian coral Stylophora pistillata: effect of feeding light, and ammonium concentrations. Limnol. Oceanogr. 47, 782–790. doi: 10.4319/lo.2002.47.3.0782
Heasman, J. (2002). Morpholino oligos: making sense of antisense? Dev. Biol. 243, 209–214. doi: 10.1006/dbio.2001.0565
Hoegh-Guldberg, O. (1999). Climate change, coral bleaching and the future of the world’s coral reefs. Mar. Freshw. Res. 50, 839–866. doi: 10.1071/MF99078
Houlbreque, F., and Ferrier-Pages, C. (2009). Heterotrophy in tropical scleractinian corals. Biol. Rev. Camb. Philos. Soc. 84, 1–17. doi: 10.1111/j.1469-185X.2008.00058.x
Houlbreque, F., Meibom, A., Cuif, J.-P., Stolarski, J., Marrocchi, Y., Ferrier-Pages, C.,et al. (2009). Strontium-86 labeling experiments show spatially heterogeneous skeletal formation in the scleractinian coral Porites porites. Geophys. Res. Lett. 36, L04604. doi: 10.1029/2008GL036782
Huang, P., Zhu, Z., Lin, S., and Zhang, B. (2012). Reverse genetic approaches in zebrafish. J. Genet. Genomics 39, 421–433. doi: 10.1016/j.jgg.2012.07.004
Kelly, A., and Hurlstone, A. F. (2011). The use of RNAi technologies for gene knockdown in zebrafish. Brief. Funct. Genomics 10, 189–196. doi: 10.1093/bfgp/elr014
Kopp, C., Pernice, M., Domart-Coulon, I., Djediat, C., Spangenberg, J. E., Alexander, D. T. L.,et al. (2013). Highly dynamic cellular-level response of symbiotic coral to a sudden increase in environmental nitrogen. mBio 4, 1–9. doi: 10.1128/mBio.00052-13
Kozielski, K. L., Tzeng, S. Y., and Green, J. J. (2013). Bioengineered nanoparticles for siRNA delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 5, 449–468. doi: 10.1002/wnan.1233
Leggat, W., Hoegh-Guldberg, O., Dove, S., and Yellowlees, D. (2007). Analysis of an EST library from the dinoflagellate (Symbiodinium sp.) symbiont of reef-building corals. J. Phycol. 43, 1010–1021. doi: 10.1111/j.1529-8817.2007.00387.x
Leggat, W., Yellowlees, D., and Medina, M. (2011). Recent progress in Symbiodinium transcriptomics. J. Exp. Mar. Biol. Ecol. 408, 120–125. doi: 10.1016/j.jembe.2011.07.032
Lesser, M. P. (2006). Oxidative stress in marine environments: biochemistry and physiological ecology. Annu. Rev. Physiol. 68, 253–278. doi: 10.1146/annurev.physiol.68.040104.110001
Lohmann, J. U., Endl, I., and Bosch, T. C. G. (1999). Silencing of developmental genes in Hydra. Dev. Biol. 214, 211–214. doi: 10.1006/dbio.1999.9407
Manefield, M., Whiteley, A., Griffiths, R., and Bailey, M. (2002). RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68, 5367–5373. doi: 10.1128/AEM.68.11.5367-5373.2002
Maor-Landaw, K., Karako-Lampert, S., Waldman, B.-A. H, Goffredo, S., Falini, G., Dubinsky, S.,et al. (2014). Gene expression profiles during short-term heat stress in the red sea coral Stylophora pistillata. Glob. Change Biol. doi: 10.1111/gcb.12592 [Epub ahead of print].
Moberg, F., and Folke, C. (1999). Ecological goods and services of coral reef ecosystems. Ecol. Econ. 29, 215–233. doi: 10.1016/S0921-8009(99)00009-9
Moya, A., Huisman, L., Ball, E. E., Hayward, D. C., Grasso, L. C., Chua, C. M.,et al. (2012). Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO2-driven acidification during the initiation of calcification. Mol. Ecol. 21, 2440–2454. doi: 10.1111/j.1365-294X.2012.05554.x
Murrell, J. C., and Whiteley, A. S. (2011). Stable Isotope Probing and Related Technologies. Washington, DC: American Society for Microbiology.
Musat, N., Halm, H., Winterholler, B., Hoppe, P., Peduzzi, S., Hillion, F.,et al. (2008). A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 17861–17866. doi: 10.3410/f.1127161.584886
Muscatine, L., and Porter, J. W. (1977). Reef corals: mutualistic symbioses adapted to nutrient-poor environments. BioScience 27, 454–460. doi: 10.2307/1297526
Nasevicius, A., and Ekker, S. C. (2000). Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26, 216–220. doi: 10.1038/79951
Orphan, V. J., and House, C. H. (2009). Geobiological investigations using secondary ion mass spectrometry: microanalysis of extant and paleo-microbial processes. Geobiology 7, 360–372. doi: 10.1111/j.1472-4669.2009.00201.x
Pernice, M., Dunn, S. R., Tonk, L., Dove, S., Domart-Coulon, I., Hoppe, P.,et al. (2014). A Nanoscale Secondary Ion Mass Spectrometry study of dinoflagellate functional diversity in reef-building corals. Environ. Microbiol. doi: 10.1111/1462-2920.12518 [Epub ahead of print].
Pernice, M., Meibom, A., Van Den Heuvel, A., Kopp, C., Domart-Coulon, I., Hoegh-Guldberg, O.,et al. (2012). A single-cell view of ammonium assimilation in coral-dinoflagellate symbiosis. ISME J. 6, 1314–1324. doi: 10.1038/ismej.2011.196
Radajewski, S., Ineson, P., Parekh, N. R., and Murrell, J. C. (2000). Stable-isotope probing as a tool in microbial ecology. Nature 403, 646–649. doi: 10.1038/35001054
Raes, J., and Bork, P. (2008). Molecular eco-systems biology: towards an understanding of community function. Nat. Rev. Microbiol. 6, 693–699. doi: 10.1038/nrmicro1935
Rodriguez-Lanetty, M., Harii, S., and Hoegh-Guldberg, O. (2009). Early molecular responses of coral larvae to hyperthermal stress. Mol. Ecol. 18, 5101–5114. doi: 10.1111/j.1365-294X.2009.04419.x
Rosic, N. N., Pernice, M., Dove, S., Dunn, S., and Hoegh-Guldberg, O. (2011a). Gene expression profiles of cytosolic heat shock proteins Hsp70 and Hsp90 from symbiotic dinoflagellates in response to thermal stress: possible implications for coral bleaching. Cell Stress Chaperones 16, 69–80. doi: 10.1007/s12192-010-0222-x
Rosic, N. N., Pernice, M., Rodriguez-Lanetty, M., and Hoegh-Guldberg, O. (2011b). Validation of housekeeping genes for gene expression studies in symbiodinium exposed to thermal and light stress. Mar. Biotechnol. 13, 355–365. doi: 10.1007/s10126-010-9308-9
Rosic, N. N., Pernice, M., Dunn, S., Dove, S., and Hoegh-Guldberg, O. (2010). Differential regulation by heat stress of novel cytochrome P450 genes from the dinoflagellate symbionts of reef-building corals. Appl. Environ. Microbiol. 76, 2823–2829. doi: 10.1128/aem.02984-09
Shinzato, C., Shoguchi, E., Kawashima, T., Hamada, M., Hisata, K., Tanaka, M.,et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–324. doi: 10.1038/nature10249
Shoguchi, E., Shinzato, C., Kawashima, T., Gyoja, F., Mungpakdee, S., Koyanagi, R.,et al. (2013). Draft assembly of the Symbiodinium minutum nuclear genome reveals Dinoflagellate gene structure. Curr. Biol. 23, 1399–1408. doi: 10.1016/j.cub.2013.05.062
Skirtach, A., Javier, A., Kreft, O., Köhler, K., Alberola, A., Möhwald, H.,et al. (2006). Laser-induced release of encapsulated materials inside living cells. Angew. Chem. Int. Ed. 45, 4612–4617. doi: 10.1002/anie.200504599
Smith, K. M., Gee, L., and Bode, H. R. (2000). HyAlx, an aristaless-related gene, is involved in tentacle formation in Hydra. Development 127, 4743–4752.
Sterner, R. W., and Elser, J. J. (2002). Ecological Stoichiometry: the Biology of Elements from Molecules to the Biosphere. Princeton, NJ: Princeton University Press.
Strychar, K. B., Coates, M., Sammarco, P. W., and Piva, T. J. (2004). Bleaching as a pathogenic response in scleractinian corals, evidenced by high concentrations of apoptotic and necrotic zooxanthellae. J. Exp. Mar. Biol. Ecol. 304, 99–121. doi: 10.1016/j.jembe.2003.11.023
Taylor, P. G., and Townsend, A. R. (2010). Stoichiometric control of organic carbon-nitrate relationships from soils to the sea. Nature 464, 1178–1181. doi: 10.1038/nature08985
Venn, A. A., Loram, J. E., and Douglas, A. E. (2008). Photosynthetic symbioses in animals. J. Exp. Bot. 59, 1069–1080. doi: 10.1093/jxb/erm328
Wagner, M. (2009). Single-cell ecophysiology of microbes as revealed by Raman microspectroscopy or secondary ion mass spectrometry imaging. Annu. Rev. Microbiol. 63, 411–429. doi: 10.1146/annurev.micro.091208.073233
Wang, Y., Angelatos, A., and Caruso, F. (2008). Template synthesis of nanostructured materials via layer-by-layer assembly. Chem. Mater. 20, 848–858. doi: 10.1021/cm7024813
Weis, V. M. (2008). Cellular mechanisms of Cnidarian bleaching: stress causes the collapse of symbiosis. J. Exp. Biol. 211, 3059–3066. doi: 10.1242/jeb.009597
Wild, C., Huettel, M., Klueter, A., Kremb, S. G., Rasheed, M. Y. M., and Jorgensen, B. B. (2004). Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 428, 66–70. doi: 10.1038/nature02344
Keywords: coral, symbiosis, Symbiodinium, genomics, stable isotopes
Citation: Pernice M and Levy O (2014) Novel tools integrating metabolic and gene function to study the impact of the environment on coral symbiosis. Front. Microbiol. 5:448. doi: 10.3389/fmicb.2014.00448
Received: 15 May 2014; Accepted: 05 August 2014;
Published online: 21 August 2014.
Edited by:
Monica Medina, Pennsylvania State University, USAReviewed by:
Mauricio Rodriguez-Lanetty, Florida International University, USAThomas Andrew Oliver, University of Hawaii at Manoa, USA
Copyright © 2014 Pernice and Levy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mathieu Pernice, Plant Functional Biology and Climate Change Cluster, University of Technology, Sydney, P. O. Box 123, Broadway, Sydney, NSW 2007, Australia e-mail: mat.pernice@gmail.com
 Mathieu Pernice
Mathieu Pernice Oren Levy
Oren Levy